Abstract
Hereditary hearing loss, an auditory neuropathy disorder, is characterized by its high prevalence and significant impact on the quality of life of those affected. In Chinese populations, the most prevalent gap junction beta-2 (GJB2) mutation hotspot is c.235delC. Currently available genetic tests require expensive instruments and specialized technicians or have long testing cycles and high costs, and therefore cannot meet point-of-care testing (POCT) requirements. The objective of this study was to evaluate the viability of a POCT kit. In only 42 min, we successfully identified the GJB2 mutation site c.235delC by integrating CRISPR-Cas nucleic acid detection with recombinase-aided amplification (RAA) and a lateral flow dipstick (LFD) method. This method has the capacity to detect low-abundance nucleic acids (as low as 102 copies/μL) and low mutation frequency (20%), in addition to accurately distinguishing wild-type, homozygous, and heterozygous mutation. This approach was utilized to assess blood samples from a total of 31 deaf patients and 5 healthy volunteers. All results were subsequently confirmed through the implementation of Sanger sequencing. Our detection results were consistent with Sanger sequencing results. The diagnostic sensitivity and specificity were 100%. The combination of CRISPR-Cas13a and LFD may be a promising method for POCT of deafness genes.
Graphical abstract
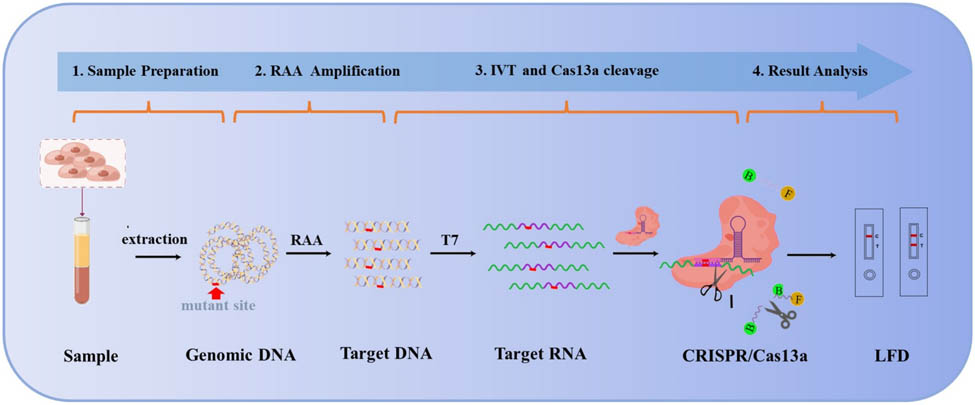
1 Introduction
The gap junction beta-2 (GJB2) is located on chromosome 13q11-q12 and encodes the connexin 26 (Cx26) [1]. GJB2 mutation has been identified as a significant genetic factor contributing to non-syndromic hearing impairment, accounting for a range of 13–42% of cases [2,3]. In healthy populations, the carrier rate of GJB2 mutation is estimated to be approximately 2.86% [4]. The most common mutation sites in GJB2 include c.235delC, c.35delG, c.176_191del16, c.299_300delAT, etc. [5]. In Chinese populations, the most common GJB2 mutation site is c.235delC, accounting for 77% of GJB2 mutations [6].
As the most prevalent variant type for GJB2, missense variants have the capacity to disrupt the function of Cx26. Cx26 molecules have the ability to form gap junction (GJ) channels through extracellular docking. GJs facilitate intercellular communication through the diffusion of ions, metabolites, and small signaling molecules, which are imperative for the maintenance of normal function and fluid environment homeostasis of the inner ear. The primary pathogenic mechanisms of recessive pathogenic variants include trafficking defects, abnormal hemichannel activity, and abnormal GJ channel function [7]. These may result in potassium recycling disruption, adenosine-triphosphate-calcium signaling propagation disruption, and energy supply dysfunction, which lead to Cx26-related hearing loss [8]. The majority of patients with this type of deafness are congenital and have severe to profound hearing loss [9,10].
Early diagnosis and treatment are the most effective measures for controlling hereditary deafness. Current methods used to screen hereditary deafness genes include gene chip microarray [11], relevant fluorescence polymerase chain reaction (PCR) method [12], time-of-flight mass spectrometry [13], and next-generation sequencing [14]. However, currently available genetic tests require expensive instruments and specialized technicians or have long testing cycles and high costs and therefore cannot meet point-of-care testing (POCT) requirements. There is an urgent need for a rapid, accurate, and equipment-free detection method for deafness genes to better support clinical diagnosis and management of hereditary deafness.
The clustered regularly interspaced short palindromic repeat-CRISPR-associated protein (CRISPR-Cas) system has been demonstrated to not only stimulate a surge in gene editing [15] but also to result in advancements in molecular diagnostics. It has developed as a rapid, accurate, and portable nucleic acid detection technology [16,17]. The CRISPR-Cas system can also be used to detect gene mutations and single-nucleotide polymorphisms (SNPs) due to its ability to specifically recognize target nucleic acids [18]. Jin et al. [19] designed a fast and sensitive detection system for SLC26A4 pathogenic mutations using CRISPR/Cas12a, but the results were fluorescence dependent, requiring a microplate reader for the detection, and the binding of the Cas12a-crRNA complex to the target DNA was limited by protospacer adjacent motif (PAM) sequences. However, the Cas13a protein does not require PAM sequences to recognize its targets, and the CRISPR-Cas13a system has significantly low off-target effects. Furthermore, the CRISPR-Cas13a system exhibits potential for SNP detection following the tuning of crRNAs [20].
The present study demonstrated the capacity to rapidly and accurately detect the GJB2 mutation site c.235delC by combining CRISPR-Cas13a with RAA and lateral flow dipstick (LFD). This provides a novel and feasible strategy for the use of POCT for the detection of deafness genes.
2 Materials and methods
2.1 Materials
Oligonucleotides (primers), ssRNA-FB reporters, crRNAs, and PUC57 plasmids with customized gene sequences were synthesized by Sangon Biotech Co., Ltd (Shanghai, China) or purchased from the aforementioned company. NTP Mix, RNase inhibitor murine, and T7 RNA polymerase were purchased from New England Biolabs. The RAA Nucleic Acid Amplification Kit (Basic) was purchased from Hangzhou ZC Bio-Sci&Tech Co., Ltd (Hangzhou, China). The LwaCas13a protein was purchased from Nanjing Kingsley Biotechnology Co., Ltd (Nanjing, China). RNase Alert v2 was purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). Nanogold (AuNPs) coated with rabbit anti-FAM antibody (IgG) was purchased from Xi’an Qiyue Biotechnology Co., Ltd (Xi’an, China).
2.2 RAA amplification of the target gene region
The design and synthesis of 30–32 bp RAA primers were conducted, with the ligation of the T7 promoter sequence for the T7 RNA polymerase (5′-AATTCTAATACGACTCACTATAG-3′) to the 5′ end of the forward primer (Table S1). The c.235delC of the GJB2 mutation target was approximately 170–200 bp. RAA amplification was then performed using commercial RAA kits that had been fine-tuned according to the instructions. Specifically, 25 μL of A buffer, 2.5 μL of forward primer (10 μM), 2.5 μL of reverse primer (10 μM), 7.5 μL of RNase-free water, 10 μL of DNA template, and 2.5 μL of B buffer were added to each dry powder tube. After mixing, the reaction was incubated at 37°C for 20 min. The products of the RAA reaction were then removed for the cleavage assay and verified by electrophoresis on a 2% agarose gel.
2.3 Cas13a fluorescent cleavage assay
The CRISPR fluorescence reaction system was optimized with modifications to the concentrations of each reactant, as outlined in Kellner et al. [17]. The reaction system consisted of a total of 25 μL, including 1 μL rNTP mix (final concentration of 1 mM), 0.5 μL RNAase inhibitor (final concentration of 2 IU/μL), 0.5 μL LwaCas13a protein (final concentration of 20 nM), 0.25 μL T7 RNA polymerase (final concentration of 1 IU/μL), 0.25 μL MgCl2 (final concentration of 10 mM), 0.5 μL HEPES buffer (final concentration of 20 mM), 1.5 μL fluorescent reporter RNA (final concentration of 120 nM), 1.5 μL crRNA (final concentration of 120 nM), 5 μL RAA amplification product, and 14.25 μL RNase-free water. The above reaction solution was thoroughly mixed and placed in a fluorescence quantitative PCR instrument (excitation light/emission light wavelengths of 490 nm/520 nm, reaction temperature set at 37°C, fluorescence was collected every 2 min for a total of 15 times), and fluorescence intensity changes were recorded.
2.4 Preparation of lateral flow test strips
The lateral flow test strips comprised a sample pad, a gold standard pad, an absorbent pad, a PVC backing, a nitrocellulose membrane (NC membrane), a test line (T line), and a control line (C line). Initially, the gold standard pad was prepared by means of the supersaturated impregnation method. In a subsequent step, nanogolds (AuNPs) encapsulated with rabbit anti-FAM antibody (IgG) were resuspended in 1.5 mL of the embedding solution (0.25% TritonX-100, 8% sucrose, 20 mM Na3PO4, 5% bovine serum albumin). Thereafter, the AuNP solution was dripped onto the polyester membrane, which was then allowed to dry naturally, to prepare the gold standard pad. The NC membrane was then attached to the PCV base plate, containing T and C lines, which were sprayed with streptavidin (SA) (1 mg/mL) and mouse anti-rabbit IgG antibody (1 mg/mL), respectively. It should be noted that the T and C lines were 6 mm apart. The NC membrane was subjected to a drying process at a temperature of 37°C for a duration of 1 h. Subsequently, the sample pads, gold standard pads, and absorbent pads were affixed to the base plate, ensuring a 2 mm overlap, and then stored at 4°C in a dry environment.
2.5 Cas13a cleavage assay on lateral flow test strips
The CRISPR reaction system for test strip assays was adapted from the fluorescent CRISPR reaction system in the appropriate proportions. The reaction system thus consists of a total of 60 μL, including 2.4 μL rNTP mix (final concentration of 1 mM), 1.2 μL RNAase inhibitor (final concentration of 2 IU/μL), 1.2 μL LwaCas13a protein (final concentration of 20 nM), 0.6 μL T7 RNA polymerase (final concentration of 1 IU/μL), 0.6 μL MgCl2 (final concentration of 10 mM), 1.2 μL HEPES buffer (final concentration of 20 mM), 3 μL the reporter RNA for test strip assays (final concentration of 100 nM), 3.6 μL crRNA (final concentration of 120 nM), 5 μL RAA amplification product, and 41.2 μL RNase-free water. The above reaction system was thoroughly mixed and placed in a metal bath at 37°C for 20 min. Thereafter, all of the reaction products were then added to the sample wells of the CRISPR test strips. The results were then read after a period of 2 min had elapsed. The resultant data from the test strips were interpreted as follows: a negative result is indicated by the visibility of the T line (test line) and the C line (control line); conversely, a positive result is indicated by the disappearance of the T line and the visibility of the C line.
2.6 Clinical sample collection and DNA extraction
Blood samples were collected from 31 deaf patients and 5 healthy volunteers at the Third Hospital of Changsha. Genomic DNA was extracted from the blood samples using the Rapid Blood Genomic DNA Isolation Kit. The total yield of DNA extracted from the blood samples ranged between 40 and 250 ng/μL.
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations and institutional policies and in accordance with the tenets of the Helsinki Declaration and has been approved by the Institutional Ethics Committee of the Third Hospital of Changsha (KY-EC-2023-003).
2.7 Statistical analyses
Statistical analyses were conducted utilizing GraphPad Prism 9.5 software. The measurement data were expressed as the mean ± standard deviation. All experiments in this study were performed in triplicates. The diagnostic sensitivity and specificity were used to assess the performance of CRISPR-Cas13a combined with LFD against Sanger sequencing.
3 Results
3.1 The principle of the detection
In this study, to distinguish the genotype of the GJB2 c.235delC mutation site, dual crRNAs were introduced, i.e., wild-type crRNAs and mutant-type crRNAs. The workflow of the CRISPR-Cas13a molecular detection technology is illustrated in Figure 1a. This technology comprises target gene amplification, in vitro transcription of amplification products, Cas13a trans-cleavage, and result analysis. Specifically, the target gene was used to generate a large number of double-stranded DNA fragments containing the sequence of the locus to be detected using RAA isothermal amplification. The target RNA was then generated by in vitro transcription using T7 polymerase and was bound to the Cas13a-crRNA complex, which activated the trans-cleavage activity of the Cas13a protein on the ssRNA-FQ and ssRNA-FB reporters. The cleavage of these reporters, either ssRNA-FQ or ssRNA-FB, resulted in the production of fluorescent signals or lateral flow chromatographic changes.
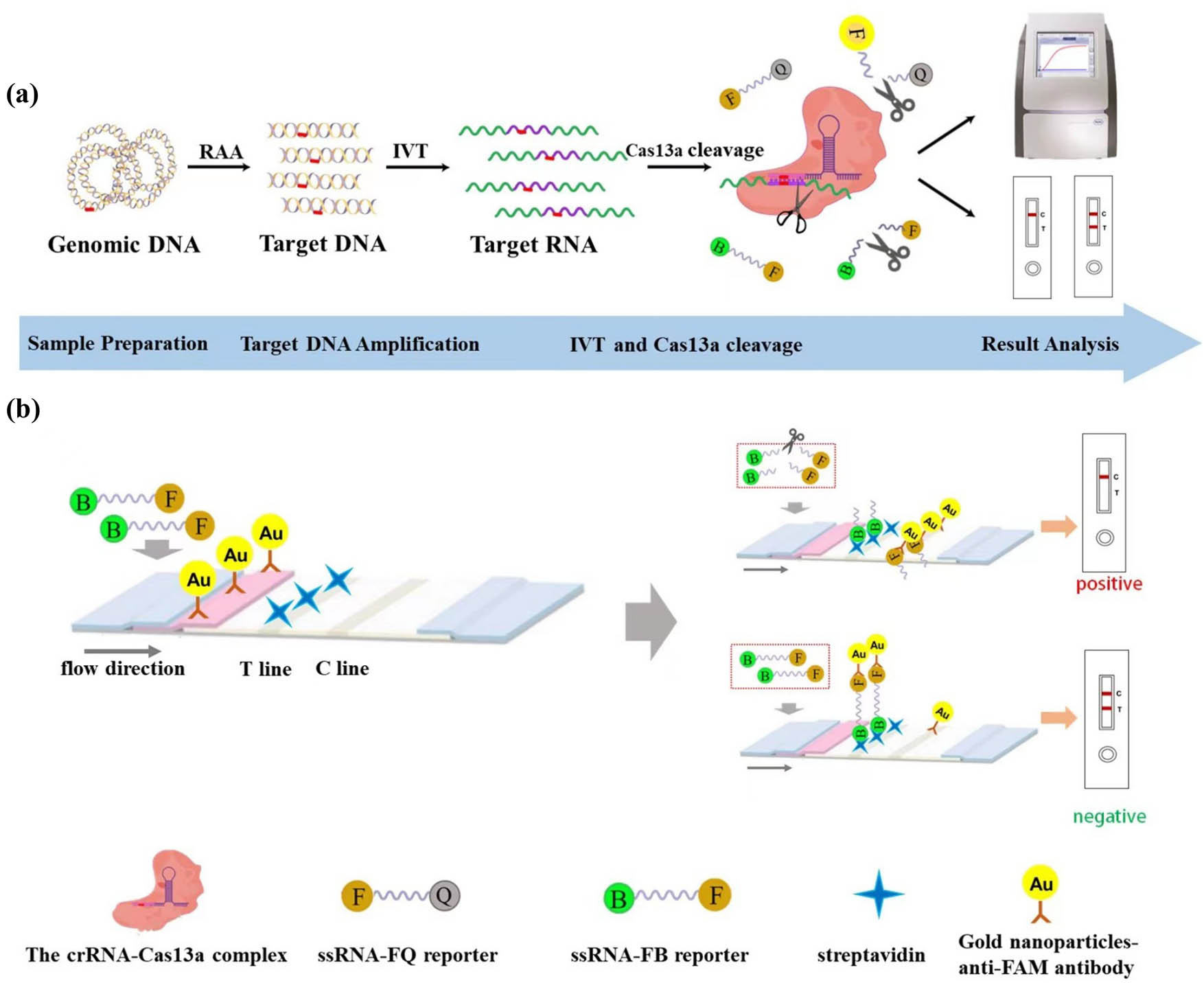
Illustration of the detection principle, which is based on the CRISPR/Cas13a system when combined with an LFD. (a) Schematic representation of a rapid GJB2 detection method employing CRISPR/Cas13a. The workflow consists of sample preparation, target DNA amplification, TIV and Cas13a cleavage, and the result analysis. The ssRNA-FQ reporter and the ssRNA-FB reporter are cleaved when the crRNA is fully recognized by the target RNA, resulting in a fluorescent or chromatographic signal. RAA, recombinase aided amplification; IVT, in vitro transcription; F, fluorophore; Q, quencher; B, biotin. (b) The working principle of lateral flow test strips. The lateral flow test strip contains the test line (T line) and the control line (C line). The result of the test is considered positive if the T line disappears and the C line appears or negative if both the T and C lines are visible. F, fluorophore; B, biotin; Au, gold nanoparticles.
The lateral flow strips based on CRISPR-Cas13a were colloidal gold test strips, the working principle of which is shown in Figure 1b. The test strips exhibited the following characteristics. The gold-labeled pad contained colloidal gold particles encapsulated with rabbit anti-FAM antibody; the test line (T line) was sprayed with SA; the control line (C line) was sprayed with mouse-anti-rabbit IgG antibody; and the ssRNA-FB reporter was a single-stranded RNA of approximately 22 nt in length, with the 5′ end modified with biotin and the 3′ end modified with FAM (Table S1). The trans-cleavage activity of the Cas13a protein was not activated when the crRNA failed to recognize the target RNA. The FAM group located at the 3′ end of the ssRNA-FB reporter gene has been demonstrated to bind to the rabbit anti-FAM antibody on the colloidal gold particles, thus forming a “biotin-FAM-anti-FAM immune complex” polymer. During the lateral flow chromatography process, a portion of colloidal gold bound to SA on the T line of the test strip with the “Biotin-FAM-anti-FAM antibody-colloidal gold” polymer formed a visible T line and another portion of colloidal gold that was not bound to the ssRNA-FB reporter on the test strip bound to the rabbit anti-FAM antibody on the colloidal gold particles to form a visible C line. In the presence of target RNA in the reaction system, Cas13a trans-cleavage activity was activated, and the ssRNA-FB reporter gene was cleaved, thus preventing the formation of the “biotin-FAM-anti-FAM immune complex” polymer cannot be formed. In the lateral flow chromatography, the result of the “T line disappears and C line appears” was due to the combination of all colloidal gold particles with mouse anti-rabbit IgG antibody in the C line through rabbit anti-FAM antibody.
3.2 Design and screening of RAA primers
As demonstrated in Figure 2a, two forward primers and three reverse primers were designed near the c.235 site of GJB2, respectively. Thereafter, one of the forward primers and one of the reverse primers were combined to form six pairs of primers. To evaluate the amplification performance of these six pairs of primers, wild-type DNA templates (Table S1) were amplified using the RAA Nucleic Acid Amplification Kit (Basic) and then analyzed by agarose gel electrophoresis. The experimental results are shown in Figure 2b, using primer combinations F1/R1, F1/R2, F1/R3, F2/R1, F2/R2, and F2/R3. The F1/R1 experimental group was found to be more effective. To ensure the reliability of the results, we also analyzed the gray scale values of each target band using ImageJ software, thus excluding the possibility of subjective visual errors. The outcomes are presented in Figure 2c, which illustrates the mean grey scale values of the amplification target bands for primer combinations F1/R1, F1/R2, F1/R3, F2/R1, F2/R2, and F2/R3 relative to the positive control. The mean gray scale values were 2.37, 1.67, 1.80, 1.74, 1.79, and 1.69, respectively, which further validates the hypothesis that the amplification efficiency of primer combination F1/R1 was optimal. (It should be noted that the influence of the length of the amplification products was negligible due to the almost similar size of the amplification products.)
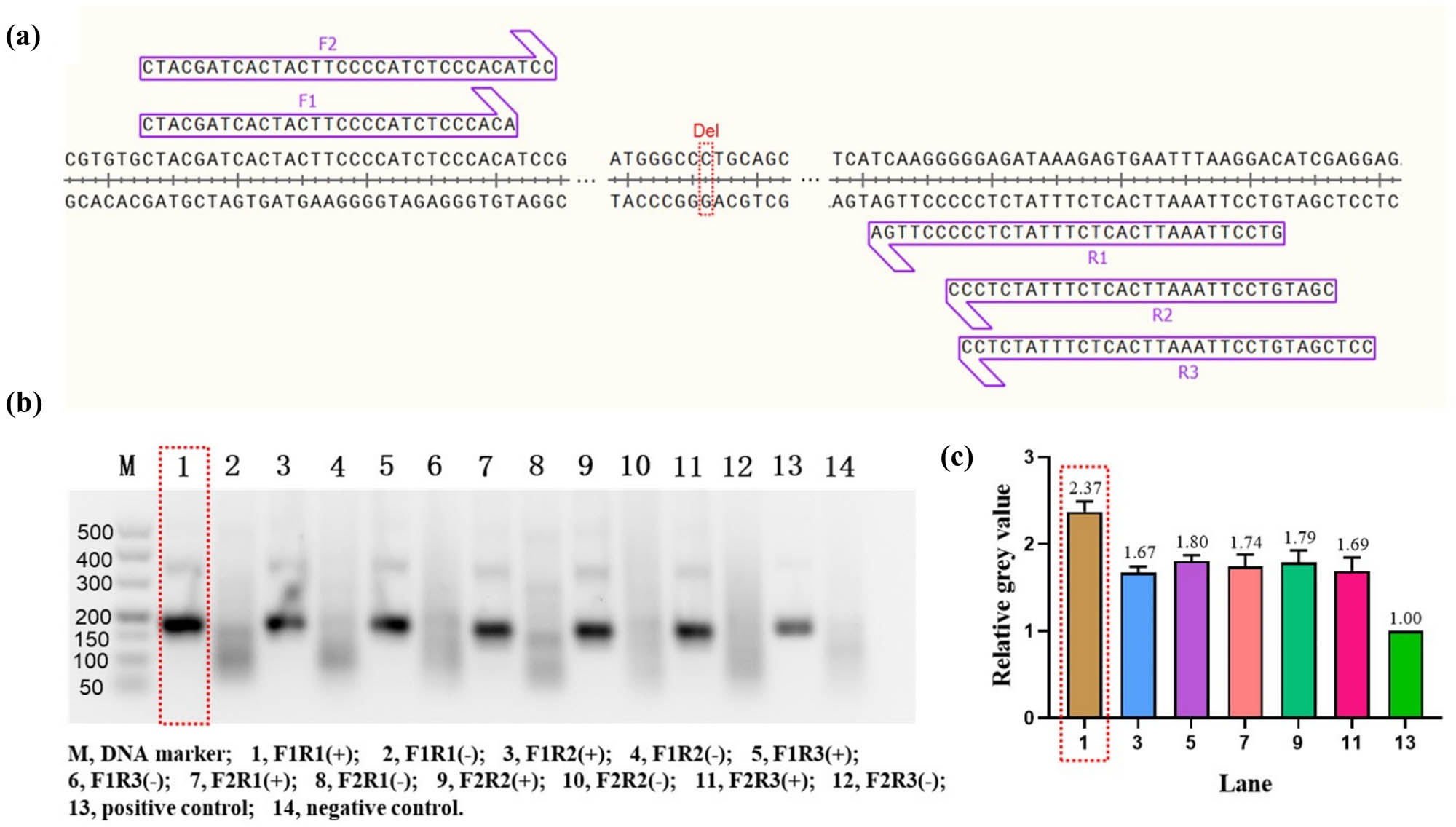
Design and screening of RAA primers. (a) Schematic representation of the RAA primers design. The red dashed rectangular box marks the c.235 locus of GJB2 and “Del” is an abbreviation for deletion. F, forward primer; R, reverse primer. (b) Agarose gel electrophoresis analysis of RAA amplification products. The best results are marked by the red dashed rectangular box. “+” indicates that DNA template was added during RAA amplification. “–” indicates that RNase-free water was substituted during RAA amplification. (c) Gray value analysis of agarose gel electrophoresis bands. All bands were compared to the positive experimental group to form relative gray values. The best results are marked by the red dashed rectangular box. Error bars in Figure 2c present the standard deviation (SD) based on three independent gray value analyses.
3.3 Design and screening of crRNAs
The crRNA sequence of CRISPR-Cas13 system consists of a backbone region with a hairpin structure at the 5′ end and a spacer region that determines its specificity. In addition, it has been shown that there is a difference in the error tolerance of different base sites in the spacer sequence, which may affect the ability of CRISPR-Cas to detect SNP [21]. As shown in Figure 3a, we designed three crRNAs at the c.235 loci of GJB2 for each wild-type DNA template and mutant DNA template and then compared the efficacy of crRNAs using the RAA-CRISPR fluorescence assay. The results are shown in Figure 3b. A strong fluorescence signal appeared when wild-type DNA templates were detected using wild-type crRNA-1, while there was no obvious fluorescence signal when mutant DNA templates were detected, which was comparable to the negative control. Meanwhile, a strong fluorescence signal appeared when mutant DNA templates were detected using mutant crRNA-2, while no obvious fluorescence signal appeared when wild-type DNA templates were detected, which was comparable to the negative control. All other crRNAs tested to detect wild-type or mutant DNA templates showed cross-reactivity or failed to achieve efficient detection performance, so none of them was an ideal choice. Therefore, we finally selected wild-type crRNA-1 and mutant crRNA-2 to form a dual-crRNA system for subsequent experiments because of their better specificity and efficient detection performance.
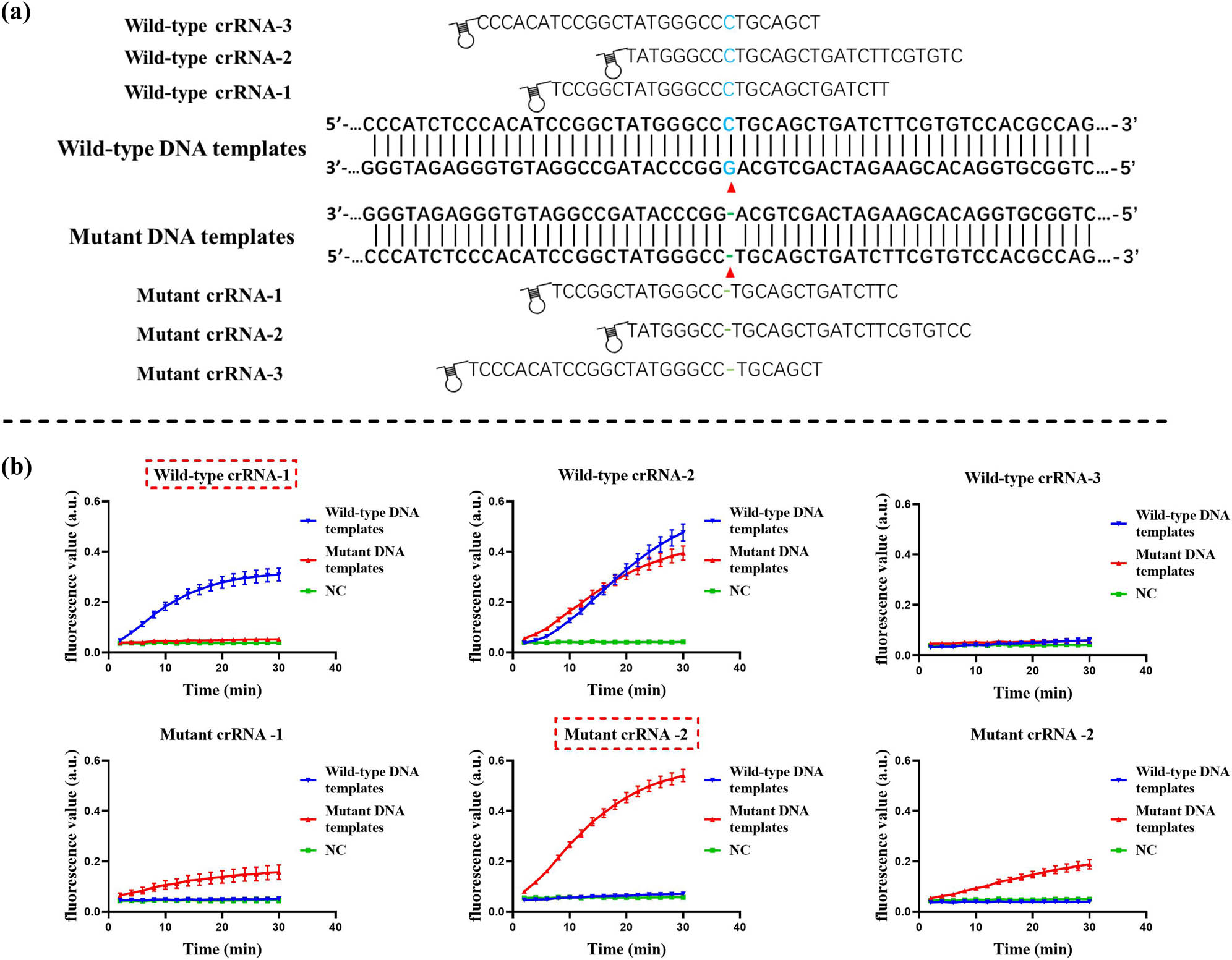
Design and screening of crRNAs. (a) Schematic representation of the crRNA design. The red triangles indicate the c.235delC mutation site of GJB2. (b) Screening of crRNAs by the CRISPR fluorescence-based assay. The best results are marked by the red dashed rectangular box. Error bars represent standard deviation (SD) from three independent tests.
3.4 The detection analytical sensitivity
After optimizing crRNAs in the dual-crRNA detection system, the analytical sensitivity of the CRISPR fluorescence-based detection system and the CRISPR chromatography test strip-based detection system was tested using a plasmid DNA concentration gradient method. We diluted the wild-type and mutant plasmids to 104, 103, 102, 101, 100, and 10−1 copies/µL, respectively, and used these two plasmids as templates for the CRISPR fluorescence-based assay and the CRISPR chromatography test strip-based assay to evaluate their detection sensitivity. As shown in Figure 4a and b, when the concentration of wild-type DNA template or mutant DNA template was greater than or equal to 101 copies/μL, a strong fluorescence signal appeared using the dual crRNA detection system based on the CRISPR fluorescence assay, whereas no obvious fluorescence signal appeared when the concentration was less than 101 copies/μL. Similarly, when the concentration of the wild-type DNA template or the mutant DNA template was greater than or equal to 102 copies/μL, positive results appeared, and even when the concentration of wild-type DNA template was 101 copies/μL, positive results were obtained in two out of three tests (Figure 4c and d). This high detection sensitivity is sufficient for the clinical detection of blood samples.
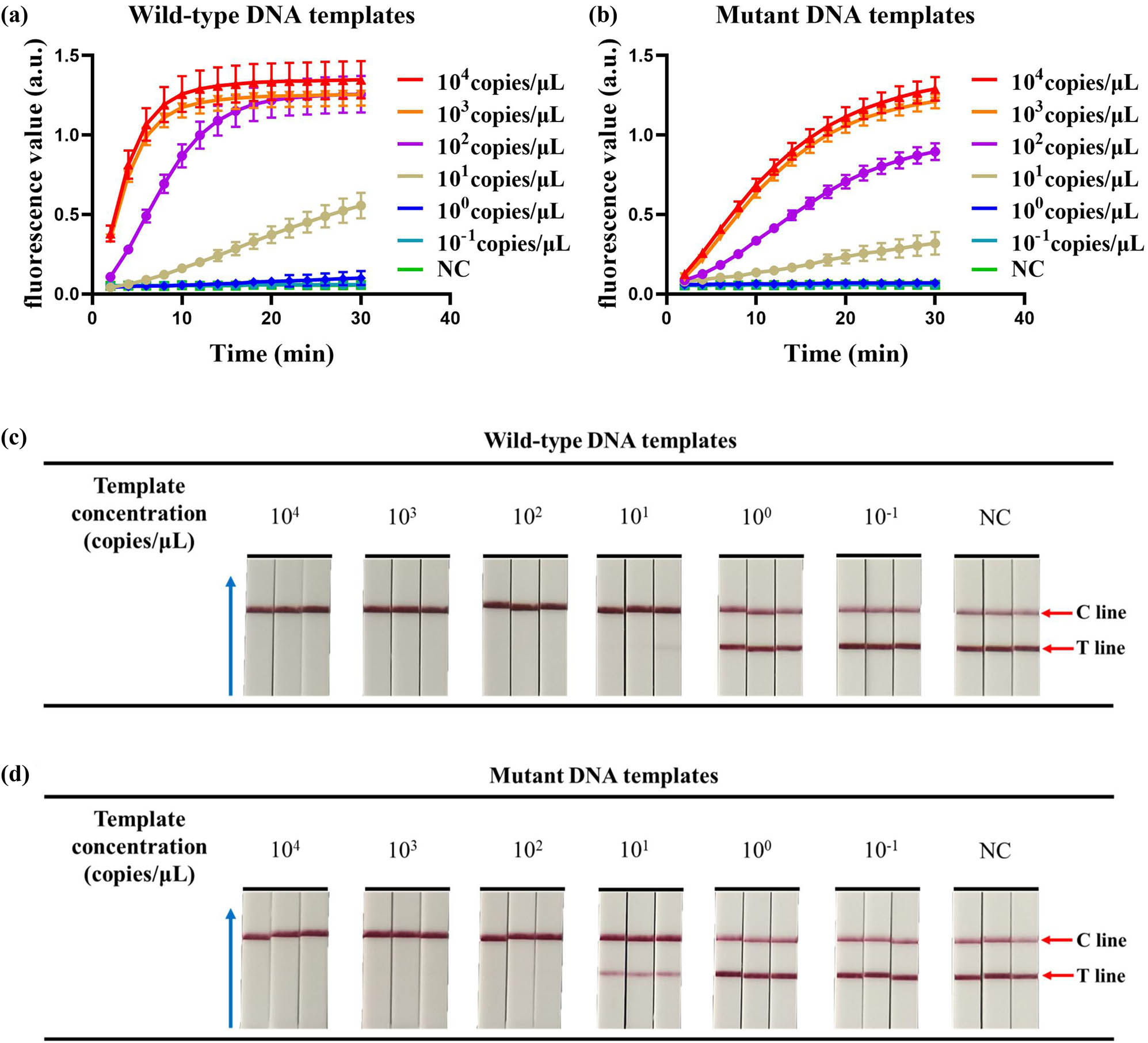
Detection of analytical sensitivity. (a) The detection sensitivity of wild-type DNA templates by the CRISPR fluorescence-based assay. (b) The detection sensitivity of mutant DNA templates by the CRISPR fluorescence-based assay. (c) The detection sensitivity of wild-type DNA templates by the CRISPR combined with LFD assay. (d) The detection sensitivity of mutant DNA templates by the CRISPR combined with LFD assay. The blue arrow points to the direction of flow of the test strip. NC, negative control. The error bars represent the standard deviation (SD) from three independent tests.
3.5 The ability to detect mutation frequencies
To evaluate the ability of this dual-crRNA detection system to identify mutant genotypes, we mixed wild-type DNA templates and mutant DNA templates at a concentration of 104 copies/μL at 100, 50, 40, 30, 20, 10, and 0% mutation rates and then assayed them with CRISPR fluorescence-based and CRISPR chromatography strip-based assays, respectively. As shown in Figure 5a and b, when the mutation frequency was greater than or equal to 10%, the mutant crRNA detection system using CRISPR-based fluorescence detection showed a strong fluorescence signal, whereas when the mutation frequency was 100%, the wild-type crRNA detection system using CRISPR-based fluorescence detection did not show a significant fluorescence signal. Similarly, when the mutation frequency was higher than or equal to 20%, the mutant crRNA detection system using CRISPR chromatography strip-based assay showed a positive result, whereas when the mutation frequency was 100%, the wild-type crRNA detection system using CRISPR chromatography strip-based assay showed a negative result (Figure 5c and d). The above results indicate that the CRISPR detection system utilizing this dual-crRNA system can effectively distinguish mutant genotypes.
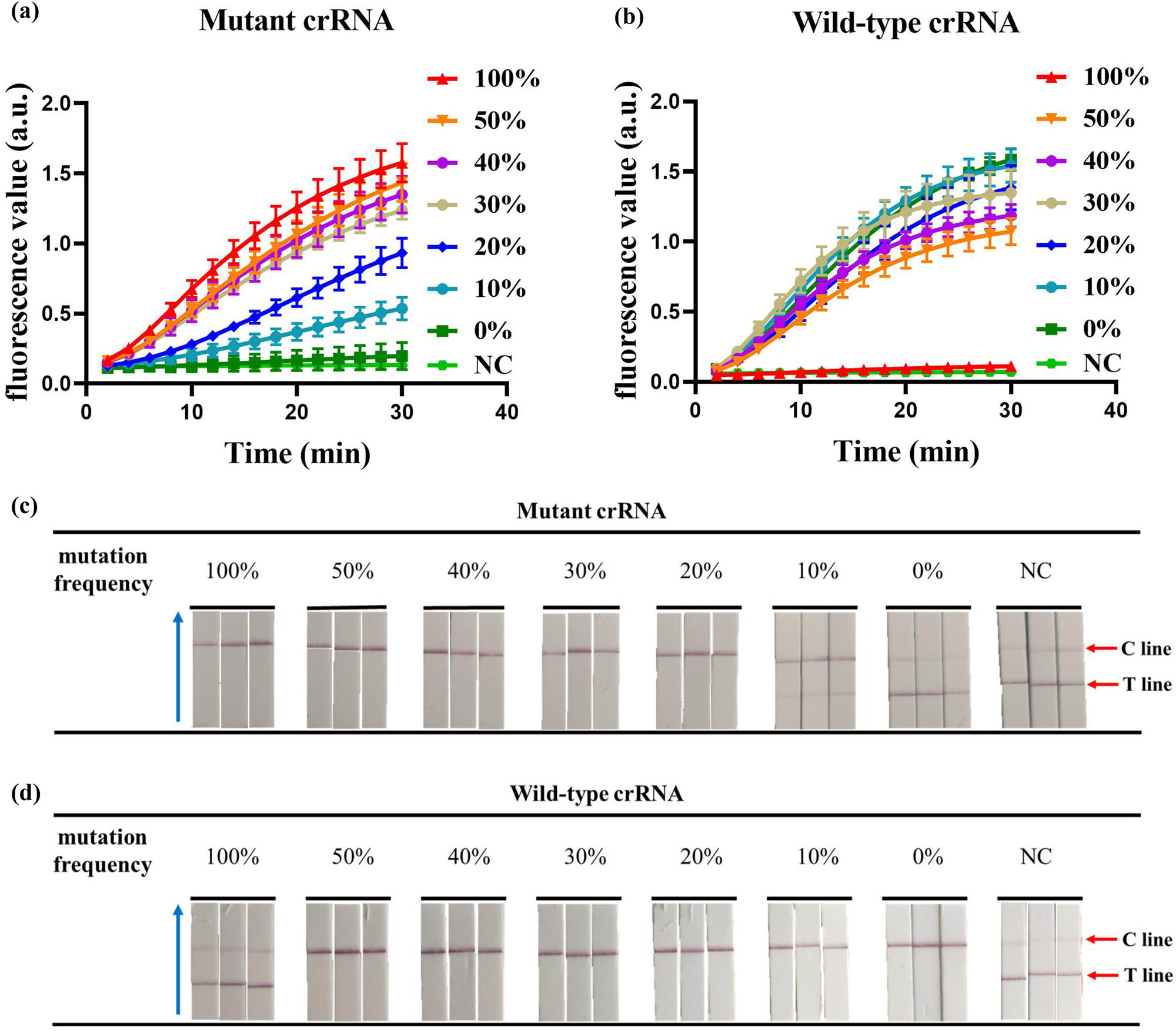
Detection of mutation frequency. (a) Detection of mutation frequency by the CRISPR fluorescence-based assay using mutant crRNA. (b) Detection of mutation frequency by the CRISPR fluorescence-based assay using wild-type crRNA. (c) Detection of mutation frequency by the CRISPR combined with LFD assay using mutant crRNA. (d) Detection of mutation frequency by the CRISPR combined with LFD assay using wild-type crRNA. The blue arrow points to the direction of flow of the test strip. NC, negative control. Error bars represent the standard deviation (SD) from three independent tests.
3.6 Clinical samples mutation detection
The c.235delC site was analyzed in blood samples from 31 deaf patients and 5 healthy volunteers using a CRISPR-Cas13a-based assay in combination with LFD and Sanger sequencing. Positive LFD result using wild-type crRNA and negative LFD result using mutant crRNA indicated that the genotype was wild type. Positive LFD result using wild-type crRNA and mutant crRNA indicated that the genotype was heterozygous mutation; Negative LFD result using wild-type crRNA and positive LFD result using mutant crRNA indicated that the genotype was homozygous mutation (Figure 6a). Blood samples from 6 deaf patients were tested, and the CRISPR chromatographic strip-based assay showed both heterozygous mutations (Figure 6b). CRISPR chromatography strip-based assay showed wild types in other samples including 25 deaf patients and 5 healthy volunteers (Figure 6b). We only showed representative results, and complete results were detailed in the supplementary material (Figure S1). The homozygous mutation was not available in our results due to a lack of sufficient clinical samples. At the same time, all samples were genotyped using Sanger sequencing. The results of the CRISPR-Cas13a combined with the LFD assay were 100% (36/36) consistent with the results of the Sanger sequencing assay (Figure 6c). Compared with the Sanger sequencing method, the diagnostic sensitivity and specificity of CRISPR-Cas13a combined with LFD were found to be 100% (6/[6 + 0]) and 100% (30/[0 + 30]), respectively (Table 1).
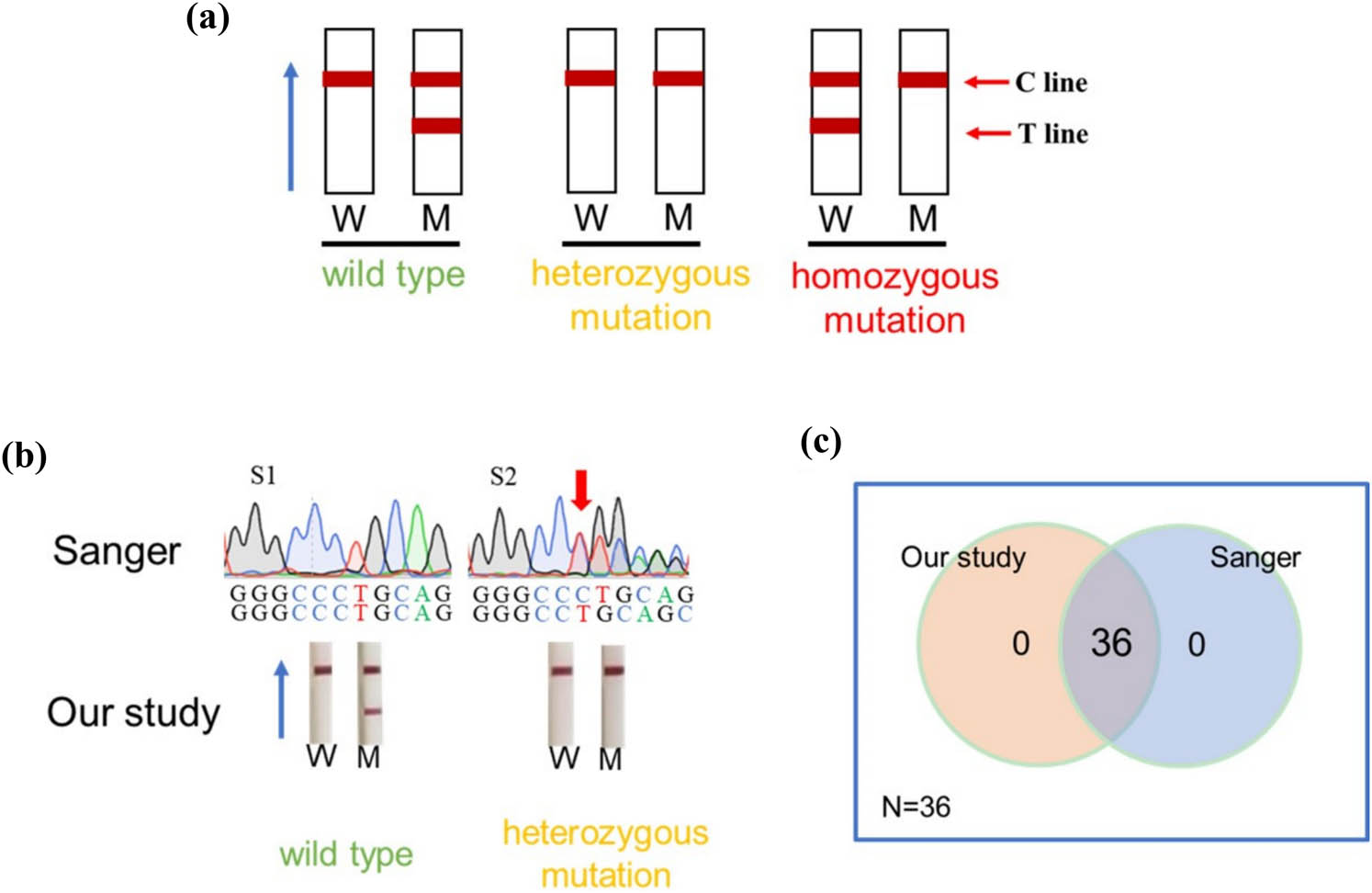
Detection of clinical samples. (a) The schematic diagram of allele discrimination by CRISPR-Cas13a combined with LFD. W indicates the LFD using wild-type crRNA; M indicates the LFD using mutant crRNA. (b) Representative results of clinical sample analyses using CRISPR-Cas13a combined with LFD and Sanger sequencing. Red arrows mark the c.235delC mutation site. (c) The Venn diagram shows the number of samples with consistent results using the CRISPR-Cas13a combined with LFD and the Sanger sequencing assay.
Comparison of results between Sanger sequencing and CRISPR-Cas13a combined with LFD
| CRISPR-Cas13a combined with LFD | Sanger sequencing | |
|---|---|---|
| Heterozygous mutation | Wild type | |
| Heterozygous mutation | 6 | 0 |
| Wild type | 0 | 30 |
4 Discussion
The World Report on Hearing estimated that more than 1.5 billion people worldwide are affected by deafness. It is noteworthy that more than half of the observed cases of hearing impairment in newborns are attributable to genetic factors [22]. The implementation of widespread genetic screening and diagnosis of deafness in specific populations has the potential to reduce the birth of deaf children and is identified as a key factor in the early detection, diagnosis, and treatment of hereditary deafness. Currently, genetic testing for hereditary deafness is limited to specific genetic laboratories or experimental centers that are unable to meet POCT requirements of deafness genes and is generally expensive.
SNPs are major genetic targets for hearing loss screening. The most prevalent variant is GJB2 c.235delC in the Chinese newborn population and East Asia [6]. CRISPR/Cas-mediated gene editing has received increasing attention as a prospective approach for modeling and treating hereditary deafness, although there are very few applications in the genetic diagnosis of deafness. The CRISPR-Cas13 system, which is characterized by its ability to undergo cis-cleavage and trans-cleavage, has been extensively and effectively utilized for the rapid and sensitive detection of nucleic acids. Feng Zhang’s team [16] developed a platform termed SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) that utilized isothermal amplification and Cas13 to detect RNA or DNA mutations through lateral flow. The technology demonstrates its potential as a platform for rapid, portable, visual nucleic acid detection and instrument-free analysis. The ability to detect SNPs allows SHERLOCK to perform rapid and accurate allele typing. By combining CRISPR-Cas12a with isothermal amplification, Doudna et al. [23] developed a method called DNA endonuclease-targeted CRISPR trans reporter (DETECTR), which facilitates the rapid and specific detection of DNA. Nalefski et al. [24] examined the nucleic acid detection properties of Cas13 and Cas12a enzymes. Their findings revealed that the activation of Cas12a trans-activity was rate-limited by steps occurring after target binding. Conversely, the Cas13a RNase was rapidly activated following target binding. Moreover, CRISPR-Cas13a binding to target RNA does not require PAM sequences. In our study, a rapid and accurate assay for the c.235delC mutation site of GJB2 was established by utilizing the powerful amplification performance of RAA, the efficient and specific discriminatory ability of the CRISPR-Cas13a system, and the simplicity and portability of the LFD. We introduced a dual-crRNA CRISPR system that significantly improved the sensitivity of the SNP genotyping including wild-type, homozygous and heterozygous mutation. The method is highly sensitive and can stably detect 102 copies/μL of DNA plasmid template, which is sufficient for the detection of clinical samples. The entire detection process, including RAA isothermal amplification (39°C, 20 min), CRISPR cleavage (37°C, 20 min), and result presentation via LFD (room temperature, 2 min), took only 42 min in total and did not require costly instrumentation beyond a simple temperature control device. The results could be judged by naked eyes. We tested blood samples from 31 deaf patients and 5 healthy volunteers using this approach, including 30 wild types and 6 heterozygous mutations of c.235delC. The detection results were consistent with Sanger sequencing results. These results showed that it is feasible to detect deafness genes using CRISPR-Cas13a combined with RAA and LFD. It has been indicated that the SHERLOCK platform has the advantage of low cost compared to other platforms such as TaqMan qPCR. A typical single-plex reaction is estimated to cost as little as $0.60 [17,21].
Generally, detecting multiple target molecules in a single reaction is necessary for many applications. SHERLOCK exploited differences in the recognition of reporter probe sequences by different Cas13 homolog proteins and Cas12 to screen multiple orthogonal diagnostic systems for the detection of multiple targets [16]. Using real-time PCR and antibody-based lateral flow systems, Xu’s team devised a fast visual diagnostic system for the detection of multiple carbapenemase genes [25]. Cas13’s well-characterized preference to cleave polynucleotides renders it particularly well-suited for multiplexed assays [26]. The present study will focus on developing a multiplex assay that combines the most common deafness gene variants, which has considerable potential. The rapid development of CRISPR-based diagnostics (CRISPR-dx) technology has made it feasible to create multiplex lateral-flow assays with several test lines for distinct targets.
However, this method also has some limitations: first, this technique requires specific crRNA, which limits its ability to detect only known mutant genes, not unknown mutant genes. Furthermore, RAA isothermal amplification and CRISPR cleavage require two steps, along with many reaction components, which complicates the process. The overarching objective of this project is to formulate a method that can be executed in various healthcare settings with only a small thermostatic water bath and pipettes, and with simple training for operators. At the grassroots level, the lack of supplies and relevant knowledge among medical staff requires that all testing methods must be simple and low-cost. Due to the aforementioned limitations, our testing methods are currently incapable of meeting the grassroots community’s needs.
CRISPR-based diagnostics (CRISPR-Dx) is a promising technology for detecting nucleic acids while concomitantly reducing equipment requirements. We are optimistic about research into the rapid pretreatment of clinical samples, based on a large number of studies on cellular nucleic acid-free extraction from blood and saliva samples [27]. The assay, designated SHINE (streamlined highlighting of infections to navigate epidemics), has been shown to simplify the workflow by ambient sample lysis procedure and the lyophilization of test reagents, thereby enabling isothermal amplification and CRISPR-Cas13 detection in a single step [28]. We will focus on sample processing, nucleic acid amplification-free, multiple tests, and integrated simple detection devices, which will significantly improve the accessibility of diagnostic tests.
5 Conclusions
Our study demonstrated that this method is not only capable of detecting low abundance nucleic acids (as low as 102 copies/μL) and low mutation frequency (20%) but also accurately distinguishes between wild-type, homozygous and heterozygous mutation without expensive instrumentation. This CRISPR-based lateral flow chromatography test strip provides a new viable solution for POCT of deafness gene mutation locus, which could be widely used in the future at the grassroots level, where there is a lack of technicians and equipment.
Acknowledgments
The authors are grateful for the reviewers’ valuable comments that improved the manuscript.
-
Funding information: This study was supported by the Changsha Municipal Natural Science Foundation (No. kq2208457).
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and consented to its submission to the journal, reviewed all the results, and approved the final version of the manuscript. YD: conceptualization, methodology, project administration, writing – original draft, writing – review and editing. JX: conceptualization, methodology, writing – original draft. MY: conceptualization, methodology, writing – original draft. YH: methodology, writing – review and editing. YY: conceptualization, investigation, supervision, writing – review and editing.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Mikstiene V, Jakaitiene A, Byckova J, Gradauskiene E, Preiksaitiene E, Burnyte B, et al. The high frequency of GJB2 gene mutation c.313_326del14 suggests its possible origin in ancestors of Lithuanian population. BMC Genet. 2016;17:45.10.1186/s12863-016-0354-9Search in Google Scholar PubMed PubMed Central
[2] Xiong YK, Chen MH, Wang HW, Chen LJ, Huang HL, Xu LP. Mutation analysis of GJB2, SLC26A4, GJB3 and mtDNA12SrRNA genes in 251 non-syndromic hearing loss patients in Fujian, China. Int J Pediatr Otorhinolaryngol. 2024;176:111777.10.1016/j.ijporl.2023.111777Search in Google Scholar PubMed
[3] Friis IJ, Aaberg K, Edholm B. Causes of hearing loss and implantation age in a cohort of Danish pediatric cochlear implant recipients. Int J Pediatr Otorhinolaryngol. 2023;171:111640.10.1016/j.ijporl.2023.111640Search in Google Scholar PubMed
[4] Liu YQ, Wang L, Yuan LL, Li YQ, Chen ZS, Yang BC, et al. Hereditary deafness carrier screening in 9,993 Chinese individuals. Front Genet. 2024;14:1327258.10.3389/fgene.2023.1327258Search in Google Scholar PubMed PubMed Central
[5] Zhang J, Wang HY, Yan CB, Guan J, Yin LW, Lan L, et al. The frequency of common deafness-associated variants among 3,555,336 newborns in China and 141,456 individuals across seven populations worldwide. Ear Hearing. 2022;44:232–41.10.1097/AUD.0000000000001274Search in Google Scholar PubMed
[6] Feng J, Zeng ZR, Luo SJ, Liu XX, Luo Q, Yang K, et al. Carrier frequencies, trends, and geographical distribution of hearing loss variants in China: The pooled analysis of 2,161,984 newborns. Heliyon. 2024;10:e24850.10.1016/j.heliyon.2024.e24850Search in Google Scholar PubMed PubMed Central
[7] Mao L, Wang YQ, An L, Zeng BP, Wang YY, Frishman D, et al. Molecular mechanisms and clinical phenotypes of GJB2 missense variants. Biology (Basel). 2023;12:505.10.3390/biology12040505Search in Google Scholar PubMed PubMed Central
[8] Chen PH, Wu WJ, Zhang JF, Chen JM, Li Y, Sun LH, et al. Pathological mechanisms of connexin26-related hearing loss: Potassium recycling, ATP-calcium signaling, or energy supply? Front Mol Neurosci. 2022;15:976388.10.3389/fnmol.2022.976388Search in Google Scholar PubMed PubMed Central
[9] Wang HY, Gao Y, Guan J, Lan L, Yang J, Xiong WP, et al. Phenotypic heterogeneity of post-lingual and/or milder hearing loss for the patients with the GJB2 c.235delC homozygous mutation. Front Cell Dev Biol. 2021;9:647240.10.3389/fcell.2021.647240Search in Google Scholar PubMed PubMed Central
[10] Lin YF, Lin HC, Tsai CL, Hsu YC. GJB2 mutation spectrum in the Taiwanese population and genotype-phenotype comparisons in patients with hearing loss carrying GJB2 c.109G > A and c.235delC mutations. Hear Res. 2022;413:108135.10.1016/j.heares.2020.108135Search in Google Scholar PubMed
[11] Gardner P, Oitmaa E, Messner A, Hoefsloot L, Metspalu A, Schrijver I. Simultaneous multigene mutation detection in patients with sensorineural hearing loss through a novel diagnostic microarray: a new approach for newborn screening follow-up. Pediatrics. 2006;118:985–94.10.1542/peds.2005-2519Search in Google Scholar PubMed
[12] Yang HY, Luo HY, Zhang GW, Zhang JQ, Peng ZY, Xiang JL. A multiplex PCR amplicon sequencing assay to screen genetic hearing loss variants in newborns. BMC Med Genomics. 2021;14:61.10.1186/s12920-021-00906-1Search in Google Scholar PubMed PubMed Central
[13] Wang Y, Chen WC, Liu ZZ, Xing W, Zhang HY. Comparison of the mutation spectrum of common deafness-causing genes in 509 patients with nonsyndromic hearing loss in 4 different areas of china by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Int Adv Otol. 2021;17:492–9.10.5152/iao.2021.21086Search in Google Scholar PubMed PubMed Central
[14] Reda Del Barrio S, García Fernández A, Quesada-Espinosa JF, Sánchez-Calvín MT, Gómez-Manjón I, Sierra-Tomillo O, et al. Genetic diagnosis of childhood sensorineural hearing loss. Acta Otorrinolaringol Esp. 2024;75:83–93.10.1016/j.otoeng.2023.07.002Search in Google Scholar PubMed
[15] Zhang HM, Qin CH, An CM, Zheng XW, Wen SX, Chen WJ, et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer. 2021;20:126.10.1186/s12943-021-01431-6Search in Google Scholar PubMed PubMed Central
[16] Patchsung M, Homchan A, Aphicho K, Suraritdechachai S, Wanitchanon T, Pattama A, et al. A multiplexed Cas13-based assay with point-of-care attributes for simultaneous COVID-19 diagnosis and variant surveillance. CRISPR J. 2023;6:99–115.10.1089/crispr.2022.0048Search in Google Scholar PubMed PubMed Central
[17] Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012.10.1038/s41596-019-0210-2Search in Google Scholar PubMed PubMed Central
[18] Blanluet C, Huyke DA, Ramachandran A, Avaro AS, Santiago JG. Detection and discrimination of single nucleotide polymorphisms by quantification of CRISPR-cas catalytic efficiency. Anal Chem. 2022;94:15117–23.10.1021/acs.analchem.2c03338Search in Google Scholar PubMed
[19] Jin XH, Zhang L, Wang XJ, An LS, Huang SS, Dai P, et al. Novel CRISPR/Cas12a-based genetic diagnostic approach for SLC26A4 mutation-related hereditary hearing loss. Eur J Med Genet. 2022;65:104406.10.1016/j.ejmg.2021.104406Search in Google Scholar PubMed
[20] Bai XP, Gao PQ, Qian KL, Yang JD, Deng HJ, Fu TW, et al. A highly sensitive and specific detection method for Mycobacterium tuberculosis fluoroquinolone resistance mutations utilizing the CRISPR-Cas13a system. Front Microbiol. 2022;13:847373.10.3389/fmicb.2022.847373Search in Google Scholar PubMed PubMed Central
[21] Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–42.10.1126/science.aam9321Search in Google Scholar PubMed PubMed Central
[22] WHO. World report on hearing. Geneva: World Health Organization; 2021. Available from: World report on hearing (who.int).Search in Google Scholar
[23] Chen JS, Ma E, Harrington LB, Costa MD, Tian XR, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–9.10.1126/science.aar6245Search in Google Scholar PubMed PubMed Central
[24] Nalefski EA, Patel N, Leung PJK, Islam Z, Kooistra RM, Parikh I, et al. Kinetic analysis of Cas12a and Cas13a RNA-Guided nucleases for development of improved CRISPR-Based diagnostics. iScience. 2021;24:102996.10.1016/j.isci.2021.102996Search in Google Scholar PubMed PubMed Central
[25] Xu YJ, Luo J, Lai W, Da JJ, Yang B, Luo XR, et al. Multiplex lateral flow test strip for detection of carbapenemase genes using barcoded tetrahedral DNA capture probe-based biosensing interface. Mikrochim Acta. 2023;190:360.10.1007/s00604-023-05903-ySearch in Google Scholar PubMed
[26] Kongkaew R, Uttamapinant C, Patchsung M. Point-of-care CRISPR-based diagnostics with premixed and freeze-dried reagents. J Vis Exp. 2024;210:e66703.10.3791/66703Search in Google Scholar PubMed
[27] Adedokun G, Alipanah M, Fan ZH. Sample preparation and detection methods in point-of-care devices towards future at-home testing. Lab Chip. 2024;24:3626–50.10.1039/D3LC00943BSearch in Google Scholar PubMed PubMed Central
[28] Arizti-Sanz J, Bradley A, Zhang YB, Boehm CK, Freije CA, Grunberg ME, et al. Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants. Nat Biomed Eng. 2022;6:932–43.10.1038/s41551-022-00889-zSearch in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Safety assessment and modulation of hepatic CYP3A4 and UGT enzymes by Glycyrrhiza glabra aqueous extract in female Sprague–Dawley rats
- Adult-onset Still’s disease with hemophagocytic lymphohistiocytosis and minimal change disease
- Role of DZ2002 in reducing corneal graft rejection in rats by influencing Th17 activation via inhibition of the PI3K/AKT pathway and downregulation of TRAF1
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Genetic variants in VWF exon 26 and their implications for type 1 Von Willebrand disease in a Saudi Arabian population
- Lipoxin A4 improves myocardial ischemia/reperfusion injury through the Notch1-Nrf2 signaling pathway
- High levels of EPHB2 expression predict a poor prognosis and promote tumor progression in endometrial cancer
- Knockdown of SHP-2 delays renal tubular epithelial cell injury in diabetic nephropathy by inhibiting NLRP3 inflammasome-mediated pyroptosis
- Exploring the toxicity mechanisms and detoxification methods of Rhizoma Paridis
- Concomitant gastric carcinoma and primary hepatic angiosarcoma in a patient: A case report
- YAP1 inhibition protects retinal vascular endothelial cells under high glucose by inhibiting autophagy
- Identification of secretory protein related biomarkers for primary biliary cholangitis based on machine learning and experimental validation
- Integrated genomic and clinical modeling for prognostic assessment of radiotherapy response in rectal neoplasms
- Stem cell-based approaches for glaucoma treatment: a mini review
- Bacteriophage titering by optical density means: KOTE assays
- Neutrophil-related signature characterizes immune landscape and predicts prognosis of esophageal squamous cell carcinoma
- Integrated bioinformatic analysis and machine learning strategies to identify new potential immune biomarkers for Alzheimer’s disease and their targeting prediction with geniposide
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- Histone modification and non-coding RNAs in skin aging: emerging therapeutic avenues
- A multiplicative behavioral model of DNA replication initiation in cells
- Biogenic gold nanoparticles synthesized from Pergularia daemia leaves: a novel approach for nasopharyngeal carcinoma therapy
- Creutzfeldt-Jakob disease mimicking Hashimoto’s encephalopathy: steroid response followed by decline
- Impact of semaphorin, Sema3F, on the gene transcription and protein expression of CREB and its binding protein CREBBP in primary hippocampal neurons of rats
- Iron overloaded M0 macrophages regulate hematopoietic stem cell proliferation and senescence via the Nrf2/Keap1/HO-1 pathway
- Revisiting the link between NADPH oxidase p22phox C242T polymorphism and ischemic stroke risk: an updated meta-analysis
- Exercise training preferentially modulates α1D-adrenergic receptor expression in peripheral arteries of hypertensive rats
- Overexpression of HE4/WFDC2 gene in mice leads to keratitis and corneal opacity
- Tumoral calcinosis complicating CKD-MBD in hemodialysis: a case report
- Mechanism of KLF4 Inhibition of epithelial-mesenchymal transition in gastric cancer cells
- Dissecting the molecular mechanisms of T cell infiltration in psoriatic lesions via cell-cell communication and regulatory network analysis
- Circadian rhythm-based prognostic features predict immune infiltration and tumor microenvironment in molecular subtypes of hepatocellular carcinoma
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Molecular identification and control studies on Coridius sp. (Hemiptera: Dinidoridae) in Al-Khamra, south of Jeddah, Saudi Arabia
- 10.1515/biol-2025-1218
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Response of hybrid grapes (Vitis spp.) to two biotic stress factors and their seedlessness status
- Metabolomic profiling reveals systemic metabolic reprogramming in Alternaria alternata under salt stress
- Effects of mixed salinity and alkali stress on photosynthetic characteristics and PEPC gene expression of vegetable soybean seedlings
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Causal effects of trace elements on congenital foot deformities and their subtypes: a Mendelian randomization study with gut microbiota mediation
- Honey meets acidity: a novel biopreservative approach against foodborne pathogens
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Fluorescent detection of sialic acid–binding lectins using functionalized quantum dots in ELISA format
- Smart tectorigenin-loaded ZnO hydrogel nanocomposites for targeted wound healing: synthesis, characterization, and biological evaluation
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
- Retraction
- Retraction of “Down-regulation of miR-539 indicates poor prognosis in patients with pancreatic cancer”
Articles in the same Issue
- Safety assessment and modulation of hepatic CYP3A4 and UGT enzymes by Glycyrrhiza glabra aqueous extract in female Sprague–Dawley rats
- Adult-onset Still’s disease with hemophagocytic lymphohistiocytosis and minimal change disease
- Role of DZ2002 in reducing corneal graft rejection in rats by influencing Th17 activation via inhibition of the PI3K/AKT pathway and downregulation of TRAF1
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Genetic variants in VWF exon 26 and their implications for type 1 Von Willebrand disease in a Saudi Arabian population
- Lipoxin A4 improves myocardial ischemia/reperfusion injury through the Notch1-Nrf2 signaling pathway
- High levels of EPHB2 expression predict a poor prognosis and promote tumor progression in endometrial cancer
- Knockdown of SHP-2 delays renal tubular epithelial cell injury in diabetic nephropathy by inhibiting NLRP3 inflammasome-mediated pyroptosis
- Exploring the toxicity mechanisms and detoxification methods of Rhizoma Paridis
- Concomitant gastric carcinoma and primary hepatic angiosarcoma in a patient: A case report
- YAP1 inhibition protects retinal vascular endothelial cells under high glucose by inhibiting autophagy
- Identification of secretory protein related biomarkers for primary biliary cholangitis based on machine learning and experimental validation
- Integrated genomic and clinical modeling for prognostic assessment of radiotherapy response in rectal neoplasms
- Stem cell-based approaches for glaucoma treatment: a mini review
- Bacteriophage titering by optical density means: KOTE assays
- Neutrophil-related signature characterizes immune landscape and predicts prognosis of esophageal squamous cell carcinoma
- Integrated bioinformatic analysis and machine learning strategies to identify new potential immune biomarkers for Alzheimer’s disease and their targeting prediction with geniposide
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- Histone modification and non-coding RNAs in skin aging: emerging therapeutic avenues
- A multiplicative behavioral model of DNA replication initiation in cells
- Biogenic gold nanoparticles synthesized from Pergularia daemia leaves: a novel approach for nasopharyngeal carcinoma therapy
- Creutzfeldt-Jakob disease mimicking Hashimoto’s encephalopathy: steroid response followed by decline
- Impact of semaphorin, Sema3F, on the gene transcription and protein expression of CREB and its binding protein CREBBP in primary hippocampal neurons of rats
- Iron overloaded M0 macrophages regulate hematopoietic stem cell proliferation and senescence via the Nrf2/Keap1/HO-1 pathway
- Revisiting the link between NADPH oxidase p22phox C242T polymorphism and ischemic stroke risk: an updated meta-analysis
- Exercise training preferentially modulates α1D-adrenergic receptor expression in peripheral arteries of hypertensive rats
- Overexpression of HE4/WFDC2 gene in mice leads to keratitis and corneal opacity
- Tumoral calcinosis complicating CKD-MBD in hemodialysis: a case report
- Mechanism of KLF4 Inhibition of epithelial-mesenchymal transition in gastric cancer cells
- Dissecting the molecular mechanisms of T cell infiltration in psoriatic lesions via cell-cell communication and regulatory network analysis
- Circadian rhythm-based prognostic features predict immune infiltration and tumor microenvironment in molecular subtypes of hepatocellular carcinoma
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Molecular identification and control studies on Coridius sp. (Hemiptera: Dinidoridae) in Al-Khamra, south of Jeddah, Saudi Arabia
- 10.1515/biol-2025-1218
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Response of hybrid grapes (Vitis spp.) to two biotic stress factors and their seedlessness status
- Metabolomic profiling reveals systemic metabolic reprogramming in Alternaria alternata under salt stress
- Effects of mixed salinity and alkali stress on photosynthetic characteristics and PEPC gene expression of vegetable soybean seedlings
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Causal effects of trace elements on congenital foot deformities and their subtypes: a Mendelian randomization study with gut microbiota mediation
- Honey meets acidity: a novel biopreservative approach against foodborne pathogens
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Fluorescent detection of sialic acid–binding lectins using functionalized quantum dots in ELISA format
- Smart tectorigenin-loaded ZnO hydrogel nanocomposites for targeted wound healing: synthesis, characterization, and biological evaluation
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
- Retraction
- Retraction of “Down-regulation of miR-539 indicates poor prognosis in patients with pancreatic cancer”

