Abstract
The poultry industry faces multifaceted challenges, including escalating demand for poultry products, climate change impacting feed availability, emergence of novel avian pathogens, and antimicrobial resistance. Traditional disease control measures are costly and not always effective, prompting the need for complementary methods. Gene editing (GE, also called genome editing) technologies, particularly CRISPR/Cas9, offer promising solutions. This article summarizes recent advancements in utilizing CRISPR/Cas GE to enhance infectious disease control in poultry. It begins with an overview of modern GE techniques, highlighting CRISPR/Cas9’s advantages over other methods. The potential applications of CRISPR/Cas in poultry infectious disease prevention and control are explored, including the engineering of innovative vaccines, the generation of disease-resilient birds, and in vivo pathogen targeting. Additionally, insights are provided regarding regulatory frameworks and future perspectives in this rapidly evolving field.
1 Introduction
The term “poultry” encompasses a wide variety of domesticated avian species raised for eggs, meat, and feathers. These include chickens, Muscovy ducks, mallard ducks, turkeys, guinea fowl, geese, quail, pigeons, ostriches, and pheasants. Chickens dominate globally, comprising 94% of the world’s poultry population in 2020 and contributing 90 and 93% to poultry meat and egg production, respectively. Other species play regionally significant roles; for example, ducks are prevalent in Asia, turkeys are concentrated in North America and Europe, and guinea fowl and geese are primarily found in Africa and Asia [1].
Poultry are raised globally under systems ranging from simple shelters in rural areas to fully automated, large-scale operations. In developing countries, indigenous poultry often rely on foraging and minimal management, making intensive rearing economically unviable due to low productivity. In areas with limited consumption growth, such as parts of Africa, family-level production remains significant, often led by women [2]. Commercial production systems dominate globally, producing most poultry meat and eggs. These systems utilize selected breeds requiring optimal nutrition, disease prevention, and confinement. Their efficiency is driven by poultry’s high feed conversion, rapid reproduction, and short production cycles, enabling quick responses to demand and advancements in genetics, health, and feeding practices. Sophisticated housing is generally limited to large-scale operations due to cost [3].
Poultry serve as the predominant source of animal proteins [3,4], with global hen egg production reaching 86 million tonnes in 2021 [5], and over 140 million tonnes of poultry meat produced in 2023, representing 40% of the world’s total meat production [6]. However, the poultry industry grapples with multifaceted challenges, from the escalating demand for poultry products due to population growth to the dwindling availability of feed resulting from climate change and the growing allocation of arable lands for human food cultivation. Additionally, the emergence of novel avian pathogens, characterized by heightened virulence and adaptability, poses a significant threat to flock health and production [7–9]. Compounding these challenges is the alarming rise of antimicrobial resistance, presenting a field-wide problem with bacterial diseases in poultry becoming increasingly challenging to treat due to this resistance [10]. This issue is further exacerbated by the stringent regulations and evolving restrictions on the use of antimicrobials in poultry production [11–13], which have traditionally been pivotal in disease management strategies.
Traditional strategies for controlling infectious diseases in poultry have relied on a multifaceted approach encompassing biosecurity protocols [14], stringent sanitation practices, widespread vaccination initiatives [15], routine testing, and sanitary culling efforts [16]. This comprehensive framework has demonstrated varying degrees of success in managing numerous significant poultry ailments. Notably, certain diseases, such as pullorum disease and fowl typhoid, have been effectively eradicated from commercial poultry populations in several developed regions owing to the diligent implementation of these control measures [17].
Despite this effectiveness, these approaches have some drawbacks and limitations. One significant concern is their substantial cost. The expenditure can vary significantly depending on several factors, such as the scale of the operation, the specific disease being targeted, and geographic location. The financial burden can be particularly high during disease outbreaks, where rapid response efforts, increased surveillance, and mass culling may be required to prevent further spread. For instance, during outbreaks, the culling of large numbers of birds is often necessary, resulting in significant economic losses. Furthermore, this practice can also impact food security. A prime example is the High Pathogenic Avian Influenza (HPAI) epidemic in Europe, where approximately 50 million birds were culled within the span of a year, from October 2021 to September 2022, in affected farms [18].
The ongoing evolution of pathogens and the emergence of novel strains have severely undermined the efficacy of conventional methods, especially vaccination programs [19]. Consequently, there have been substantial economic losses attributed to decreased productivity, increased mortality rates, and the requirement for costly disease management measures. Compounding this issue is the lack of protective vaccines for specific diseases, further exacerbating the challenge. Lymphoid leukosis, a neoplastic disease, stands out as a prime example, as no treatments or vaccines are currently available to mitigate its impact [20].
Additionally, some poultry-related microbes pose a potential threat to human health through food safety issues, exposing the public to contaminated meat and eggs; this is the case, for example, for non-typhoidal Salmonella [21,22] and Campylobacter [21]. Additionally, other poultry pathogens, such as some avian influenza viruses, can be transmitted to humans, posing also a public health concern [23].
As a result, the poultry industry faces an urgent need for complementary control methods to bolster its defenses against infectious diseases. In this regard, genome editing (GE) techniques offer a promising avenue for innovation [24]. The Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system represents one of the gene-editing technologies that have the potential to revolutionize disease control strategies in poultry [25]. One of its primary advantages lies in its capacity to develop disease-resilient poultry by precisely modifying the host’s genome [26]. Additionally, it facilitates the engineering of innovative vaccines [27], while its ability to target pathogens in vivo holds considerable therapeutic promise, opening new avenues for combating poultry diseases with unprecedented precision and effectiveness [28]. Thus far, significant advancements in utilizing CRISPR/Cas technology have been achieved primarily in two poultry species, namely chicken and quail, with chicken leading the progress.
This article provides a comprehensive and up-to-date examination of CRISPR/Cas gene editing in poultry infectious disease control. It synthesizes recent research with a focus on practical applications, including the precise engineering of disease-resistant poultry, the development of next-generation vaccines, and in vivo pathogen targeting. CRISPR/Cas9 is highlighted as a transformative tool for advancing poultry health and improving disease management strategies.
While much of the existing work primarily addresses viral infections, this article takes a more integrative approach by extending the discussion to bacterial and protozoal diseases. By bridging multiple facets of CRISPR research in poultry, it offers a broader and more comprehensive perspective on its potential for disease control across different poultry species.
Additionally, it examines the challenges of translating this technology into commercial practice, addressing regulatory and ethical considerations, feasibility constraints, and the evolving legal landscape.
2 Modern gene-editing techniques
2.1 Overview
GE technology enables precise modifications to the genetic material, allowing for the addition, removal, or alteration of DNA or RNA within the genome [29–31].
Compared to conventional transgenic techniques, where exogenous DNA, typically recombinant DNA sequences, is randomly inserted into the genome, GE techniques offer distinct advantages. They enable the introduction of site-specific mutations without introducing additional genetic mutations into the genome, potentially yielding modifications that are indistinguishable from naturally occurring variants.
Modern GE techniques harness programmable DNA nucleases, known as genome editors, capable of inducing precise double-strand breaks (DSBs) at targeted locations within the genome, earning them the title of site-directed nucleases [32]. This capability allows researchers to precisely modify specific DNA sites, enabling a wide range of applications [33,34]. The DSBs triggered in the genome activate endogenous DNA repair mechanisms, resulting in specific genetic alterations through two main pathways: non-homologous end joining (NHEJ) [35,36] or homology-directed repair (HDR) [37]. NHEJ, the most common cellular repair mechanism, is also highly error-prone and often leads to insertions and deletions (indels) at the repair site resulting in frameshifts and functional knockouts (KO). This process can be utilized to create null mutation alleles for multiple purposes such as studying gene function. In contrast, HDR is a less frequent mechanism but it enables knock-in (KI) strategies. Depending on the objective-whether it is to insert a gene or inhibit gene function-HDR or NHEJ is preferred, respectively [38].
Site-directed nucleases encompass zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9 nucleases. Their emergence has revolutionized the realm of genetic modification in both plants and animals including poultry, primarily due to the high efficiencies achieved in targeted mutagenesis.
ZFNs, created in 1996, combine zinc finger modules with the DNA cleavage domain of the restriction enzyme FokI [39]. This fusion allows ZFNs to induce DSBs efficiently, leading to widespread use in GE across various organisms since 2001 [40–48].
The advent of TALENs in 2010 represented another pivotal advancement in the realm of designer nucleases [49,50]. Unlike ZFNs, TALENs utilized DNA-binding modules derived from TALE proteins, which offered greater flexibility and ease of generation [51].
Finally, CRISPR/Cas9 emerges as the most prevalent and advanced technique for GE. The construction of the CRISPR/Cas9 system is less expensive and intricate process compared to the ZFNs and TALENs systems, requiring mainly the synthesis of a short specific RNA sequence molecule for GE at a specific locus. Moreover, this system is more effective due to its broader accessibility to target sites and higher target specificity. This is attributed to the availability of computational tools for designing guide RNAs (sgRNAs) in the CRISPR/Cas9 system which enhances the predictability of guide specificity and contributes to minimizing off-target effects [52,53].
2.2 The rise and evolution of CRISPR/Cas technology
CRISPR/Cas technology has transformed the field of GE, recognized as the “Breakthrough of the Year” in 2015 by the journal Science [54]. The widespread adoption of the CRISPR/Cas system for GE has been propelled by its simplicity, specificity, efficiency, precision, and capability for multiplex targeting. This rapid adoption is further fueled by the open accessibility of the technology, granting researchers quick and affordable access to cutting-edge tools for their projects [55].
2.2.1 Discovery and development
The CRISPR system was first identified in Escherichia coli in 1987 [56], with further research revealing its presence in other bacteria and archaea [57]. By 2007, it was understood as a bacterial defense mechanism against phages [58], acting similarly to an adaptive immune system [59–61]. In 2012, the components necessary for GE were identified, including CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), which guide the Cas9 protein to specific DNA targets to introduce double-stranded breaks [62,63]. This breakthrough led to the development of the single guide RNA (sgRNA) system, simplifying the application of CRISPR/Cas9 in GE [64]. This fundamental discovery paved the way for extensive applications across various organisms, including poultry.
The discovery of CRISPR/Cas9 sparked a competitive drive to apply the system in eukaryotic cells. By January 2013, multiple research teams had demonstrated successful GE in human cells [65–68], and the first applications on germline cells soon followed [69]. That same year, the dCas9 protein, a version of Cas9 lacking nuclease activity, enabled gene regulation techniques like CRISPR activation (CRISPRa) [70] and interference (CRISPRi) [71] by fusing dCas9 with transcription regulators. In 2015, the discovery of SaCas9, a compact Cas9 variant suitable for delivery in adeno-associated viruses, expanded the system’s versatility [72]. While the Cas12a (Cpf1) protein broadened the range of targetable sites [73,74]. By 2016, base editing emerged [75] to allow precise DNA modifications without double-strand breaks, reducing off-target effects [76]. In 2017, the CRISPR/Cas13 system was identified [77], allowing RNA editing [78,79] and modifying gene function through mRNA degradation [80,81]. Further improvements led to Cas9 variants with expanded targeting flexibility and higher fidelity [82–84]. Prime editing, introduced in 2019 [85], is built upon base editing, enabling precise edits – including substitutions, insertions, and deletions – without double-strand breaks or donor templates.
This timeline of discoveries and advancements highlights the rapid evolution of CRISPR technology. A summary of the key milestones is provided in Figure 1.

Discovery and development of the CRISPR/Cas systems.
2.2.2 Classification of the CRISPR/Cas systems
In recent years, there has been a significant rise in the number and diversity of identified CRISPR/Cas systems. Based on the Cas gene signature and the specific targeting mechanism, the new classification categorizes CRISPR/Cas systems into 2 classes, 6 types and 33 subtypes [86], compared with 5 types and 16 subtypes in 2015 [87].
CRISPR/Cas9 system is the most widely used and extensively studied system for GE. This system gained prominence due to its simplicity, efficiency, and versatility in targeted GE [62].
2.2.3 Limitations of the CRISPR/Cas systems
One major concern with using CRISPR/Cas9 GE technology is the potential for off-target effects, where unintended genetic modifications arise due to the system acting on sites similar but not identical to the target sequence [88]. These off-target effects occur when the Cas9 protein binds to protospacer adjacent motif (PAM)-like sequences and/or when the guide RNA (gRNA) binds to sequences that share similarity with the target site, especially when only a few base pair mismatches are present [89]. Such unintended alterations can result in small insertions and deletions (indels) or even large structural variations, such as chromosomal rearrangements. While some mutations may be biologically silent, others can have unpredictable effects, some of which may be harmful to the host, including the disruption of essential genes, immune responses, or oncogene activation [89–91].
To mitigate these risks, multiple strategies have been implemented. One approach involves utilizing bioinformatics tools to predict and assess potential off-target sites, allowing for the design of gRNAs with improved specificity [52,92]. In parallel, researchers have explored the use of Cas9 orthologs such as SaCas9 [93], St1Cas9 [94], and St3Cas9 [95], which are derived from bacterial species other than Streptococcus pyogenes (the source of the commonly used wild type [WT] SpCas9). These orthologs recognize more complex PAM sequences, thereby reducing the likelihood of unintended DNA cleavage. Moreover, some Cas9 orthologs exhibit inherent differences in nuclease activity and guide RNA interactions, further enhancing precision. Another refinement involves engineering high-fidelity SpCas9 variants such as eSpCas9 [83], SpCas9-HF1 [96], and HypaCas9 [97], which have been specifically designed to reduce off-target cleavage while preserving on-target efficiency. Additionally, the use of Cas9 nickases – mutant variants that introduce single-strand rather than double-strand breaks – enhances specificity. By requiring two gRNAs to target opposite strands near the desired editing site, this strategy significantly reduces off-target effects compared to conventional Cas9 [98,99].
Optimizing delivery methods also plays a crucial role in improving specificity. One effective approach is the use of Cas9-sgRNA ribonucleoproteins (RNPs), which offer rapid cellular clearance compared to plasmid-based delivery systems. Because RNPs degrade quickly after GE, they minimize the window for unintended edits, thereby enhancing specificity [100]. Similarly, the direct injection of an adenoviral CRISPR/Cas9 vector into quail blastoderm allowed precise GE as no mutations were detected in off-target regions, and vector integration was avoided [101].
Employing anti-CRISPR proteins temporarily inhibits Cas9 activity after the desired edit is made, thereby reducing the duration of potential off-target effects [102]. Moreover, given that off-target effects may vary depending on the cell cycle stage, synchronizing cells to a specific phase during CRISPR editing has been proposed as a means to further minimize errors [103].
Base editing [75,104] and prime editing [105] techniques offer alternative CRISPR methods with potentially fewer off-target effects compared to conventional Cas9-mediated DSBs.
Finally, validating CRISPR/Cas9-mediated gene editing through sequencing-based approaches [106] is critical for assessing both the intended and unintended genetic modifications. However, no single molecular assay can fully capture the genetic landscape of edited organisms or address all possible allele variations. A comprehensive molecular characterization is necessary to gain an in-depth understanding of the genetic changes induced. If undesired mutations are detected, iterative optimization of the CRISPR system can be performed to enhance precision and further reduce off-target effects [107].
3 Application of CRISPR/Cas gene-editing techniques in poultry
The last 15 years have witnessed the rapid development of gene-editing technology. ZFN-mediated gene editing has yet to be reported in poultry. In contrast, TALEN-mediated gene targeting allowed the successful generation of ovalbumin (OVA) knockout chickens in 2014. Cultured primordial germ cells (PGCs) were transfected with plasmids encoding OVA-TALENs. This resulted in deletions in 33% of PGC cultures. The modified PGCs were transplanted into recipient embryos, producing chimeric roosters that, upon reaching sexual maturity, generated OVA heterozygous knockout chicks with a 10% efficiency [108]. In a similar approach, a 2017 study utilized TALENs combined with HDR to generate sterile hens. Cultured PGCs were transfected with TALEN-encoding plasmids, achieving an 8.1% editing efficiency. Heterozygous male PGCs were then transplanted into recipient embryos, and one of the resulting founder roosters successfully produced genetically modified offspring with a 6% efficiency [109]. These findings highlight the potential of TALENs for precise gene editing in poultry, though with relatively low efficiency.
On the contrary, in 2016, a study in chickens demonstrated successful germline gene editing through CRISPR-mediated homologous recombination in PGCs. An additional loxP site was inserted into the immunoglobulin heavy chain (IgH) locus via HDR, resulting in variable germline transmission rates across different PGC lines, with some reaching up to 90% [110].
Also, in 2016, Oishi et al. [111] achieved over 90% mutation efficiency in cultured chicken PGCs by targeting the OVA and ovomucoid genes using CRISPR/Cas9. Transplantation of CRISPR-modified ovomucoid PGCs into recipient embryos resulted in germline chimeric roosters, which transmitted the mutation to offspring at a rate of approximately 50%. Similarly, using CRISPR/Cas9, Koslová et al. [112] achieved a remarkably high efficiency of homologous recombination, with 88% of PGC clones successfully acquiring the precise deletion of the three nucleotides encoding the tryptophan residue at position 38 (W38) in both chNHE1 alleles.
Building on these studies and numerous other investigations [113,114], compelling evidence has demonstrated the superior efficiency of CRISPR/Cas9 over TALENs for gene editing in poultry. Beyond its higher mutation and transmission rates, CRISPR/Cas9 has surpassed TALENs as the preferred genome-editing tool due to its simplicity, speed, affordability, and greater target specificity. These advantages have driven its rapid adoption as the primary choice for precise genetic modifications across various organisms, including poultry.
CRISPR/Cas GE holds promise for addressing global food security challenges by enhancing production performances [98,115,116] and disease control measures [117–120]. It also offers opportunities to enhance animal welfare [121], create specific disease models [122], and develop poultry bioreactors [114].
The technique, which is capable of precisely targeting nearly any genomic location, has the potential to enhance traditional methods of disease prevention, control, or elimination. A primary benefit is the ability to develop disease-resilient poultry through genetic modifications in the host genome [123]. CRISPR/Cas genome editors can be applied to explore pathogen–host interactions, facilitate the engineering of vaccines, and in vivo pathogen targeting (Figure 2).
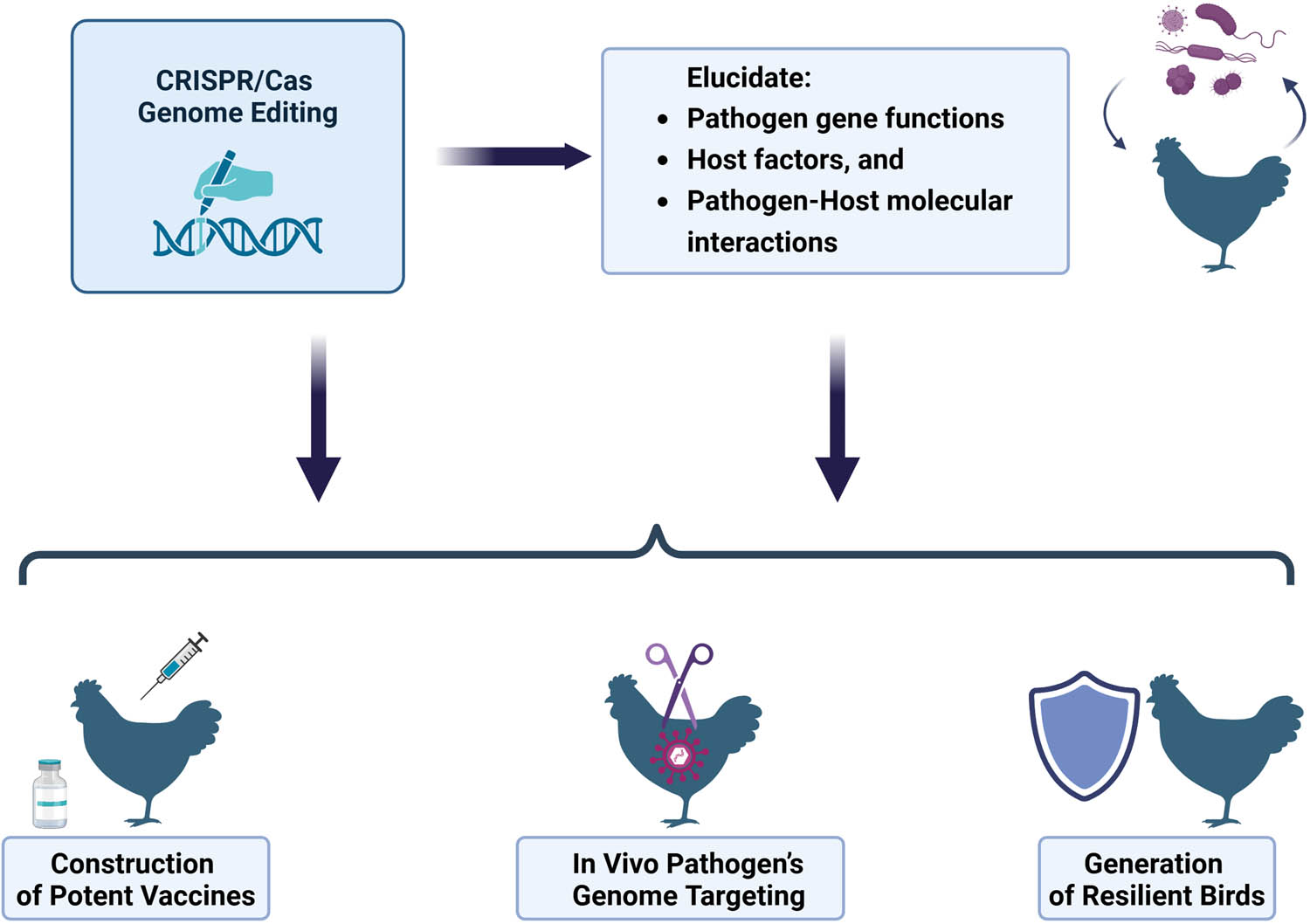
Leveraging CRISPR/Cas to control poultry infectious diseases.
While the application of CRISPR/Cas in avian species is still emerging, notable progress has been achieved, primarily in two poultry species, namely chickens and quails, with chickens leading the way.
Beyond GE, certain CRISPR/Cas systems have found valuable applications in molecular diagnostics. Diagnostic tools based on CRISPR/Cas12a [124–128] and CRISPR/Cas13a [129–135] exhibit high efficacy, sensitivity, and speed in detecting poultry pathogens, with promising potential for point-of-care use.
By linking with transcriptional regulators or domains, catalytically inactive Cas9 (dCas9) can be employed to induce either the activation or repression of RNA transcription [136]. In this context, Williams et al. developed a novel genome and epigenome engineering toolkit enabling the manipulation of endogenous gene expression and enhancer activity in chicken embryos [137].
3.1 Use of CRISPR/Cas to elucidate pathogen’s gene functions, host factors, and pathogens–host molecular interactions
Advancements in genomics, particularly genome sequencing of both host and pathogens, empower researchers to identify candidate genes involved in infection and defense mechanisms.
CRISPR genome screening involves using CRISPR/Cas9 technology to systematically target and modify genes within a genome. The objective of this approach is to identify and study the function of specific genes by observing the resulting changes in cellular or organismal behavior [138,139]. This powerful tool offers new avenues for identifying relevant genes in hosts and pathogens during infection, as well as for investigating pathogen–host interactions [28,139–141]. These findings can be leveraged for identifying advantageous alleles for selective breeding within a poultry population, vaccine development [142] and to generate disease-resistant birds [143]. The successful editing process can be achieved with high efficiency by combining the suitable delivery strategy of Cas9 and sgRNA [144] with the right cells and CRISPR database tools.
Several studies have demonstrated the utility of CRISPR/Cas9 for functional gene analysis in poultry pathogens and host cells (Table 1).
Studies using CRISPR/Cas to elucidate pathogen’s gene functions, host factors, and pathogen–host molecular interactions
| Reference | Crispr/Cas9*-based gene edition | Key findings/study contribution |
|---|---|---|
| [145] | Deletion of the Meq and pp38 genes from the CVI988 vaccine strain of MDV | The first successful utilization of the CRISPR/Cas9 GE system for MDV-1 viral gene knockout |
| [146] | Deletion of the viral gene pp38 in MDV-transformed LCLs | The study contradicts prior reports and showcases that pp38 gene is dispensable for the transformed state of MDV-transformed LCLs |
| [147] | Deletion of MDV-miR-M4 from the MDV-induced lymphoma-derived lymphoblastoid cell line MDCC-HP8 |
|
| [148] | Deletions of the Meq- or the mid-clustered miRNAs from vvMDV strain RB-1B virus | Establishing a new platform for mutagenesis of viral miRNAs encoded by the MDV-1 |
| [149] | Deletion of the MDV-2 glycoprotein B (gB) in LCLs | Highlighting the potential of targeted GE as an antiviral strategy against pathogenic MDV-1 and other viruses affecting chickens |
| [150] | Deletion of MDV-1 pp38 creating a mutant (GX0101∆pp38) for the rapid screening and identification of pp38-specific monoclonal antibodies |
|
| [151] | Generation of Meq-deleted MDV-1 viruses |
|
| [152] | Remove the exon encoding the 33 amino acid insertion from chicken ANP32A (lacking in mammals) or knockout the entire protein in chicken cells | Influenza A viruses do not replicate in chicken cells that lack ANP32A |
| [153] | Deletion of the entire 99 nucleotide (nt) fifth exon (A99) of cANP32A | Chicken ANP32A only, not ANP32B and ANP32E, plays a pivotal role in supporting vPol activity of AIV |
| [154] | Deletion of candidate virulent ssaU gene encoding type III secretion system from S. Gallinarum genome |
|
| [155] | Deletion of the SpvB gene from a large virulent plasmid of S. Gallinarum strain (SG18) |
|
| [156] |
|
|
| [157] | *: In this study, Cas12a was used instead of Cas9 | Demonstration that FnCas12a could trigger GE in E. tenella |
| Knocking-in a coding cassette for an enhanced yellow fluorescent protein (EYFP) and dihydrofolate reductase–thymidylate synthase gene (DHFR) | ||
| [158] | Disruption of ETH2_0411800 | ETH2_0411800 is non-essential for E. tenella’s growth and development |
| [159] | Disruption of the chicken TBK1 (chTBK1) gene in chicken embryonic fibroblast DF-1 | chTBK1 was revealed to be indispensable in STING-mediated IFN-β activation in chicken cells |
| [160] | Recombination activating gene 1 (RAG1) knockout aiming the generation of an immunodeficient chicken model | Highlighting the pivotal role of RAG1 in chicken immature B cell development, Ig gene conversion during embryonic stages, and demonstrates the dose-dependent regulatory role of RAG1 during immune cell development |
| [161] | Deletion of chicken protein arginine methyltransferase 5 (prmt5) gene from DF-1 cell line | Uncovering a functional link of chPRMT5 in suppression of IFN-b production and interferon-stimulated gene expression |
| [162] | IRF7 knockout in DF-1 cells | IRF7 has a role in host antiviral response against the AIV in chickens |
In 2018, Zhang et al. used CRISPR/Cas9 to knock out the Meq and pp38 genes in serotype-1 Marek’s disease virus (MDV-1), revealing the potential of CRISPR for studying viral gene functions [145]. In 2019, the team extended this work to MDV-transformed cell lines, finding that pp38 deletion enhanced cell proliferation, suggesting it is non-essential for these cells’ transformation [146]. They also showed that MDV-miR-M4, a microRNA previously linked to tumor formation, is not necessary for maintaining the transformed phenotype [147]. By utilizing the CRISPR/Cas9 system with a double-guide RNA transfection/virus infection strategy, the team successfully established a new platform for mutagenesis of viral miRNAs encoded by the MDV-1 [148]. Further, in 2022, they successfully deleted MDV-2 glycoprotein B (gB) from MDV-transformed cells, showcasing GE’s potential as an antiviral strategy [149].
For MDV-specific antibody production, Teng et al. used CRISPR/Cas9 to create a pp38-deleted mutant for screening monoclonal antibodies (mAbs), identifying mAb 31G7 with high specificity [150]. Later, they developed Meq-specific mAbs using a hydrophilic polypeptide of Meq protein and hybridoma technology, proposing this technique for efficient mAb generation against viral proteins [151].
Avian influenza viruses (AIV) depend on specific host factors for replication, with variations in these factors affecting the virus’s ability to replicate in avian versus mammalian hosts [152]. In chickens, acidic nuclear phosphoprotein 32 family member A (ANP32A) has been identified as a critical host restriction factor for AIV. Park et al. used CRISPR/Cas9 to investigate the roles of ANP32 family members, finding that deleting a segment of chicken ANP32A, which includes an extra 33 amino acids not found in mammals, led to a significant reduction in viral polymerase activity. This result underscores the essential role of chicken ANP32A in supporting the AIV replication [153] and aligns with the observations of Long et al., who demonstrated that deleting ANP32B in chicken cells did not affect AIV polymerase activity. This indicates that ANP32A is the essential factor for AIV replication in chickens, while ANP32B and ANP32E are not involved in this process [152].
Salmonella Gallinarum, responsible for fowl typhoid, presents a significant economic threat to the poultry industry, particularly in developing countries. In 2022, researchers from the University of the Punjab, Pakistan, conducted gene deletion studies on S. Gallinarum [154,155]. Tahir et al. used CRISPR/Cas9 to delete the ssaU gene, which encodes the type III secretion system (TTSS), revealing its essential role in the pathogen’s virulence. The resulting mutant strain was non-virulent and unable to colonize poultry organs, suggesting potential use in live vaccine development [154]. Similarly, Basit et al. knocked out the SpvB gene, finding that infected chickens showed no signs of disease, supporting the use of CRISPR/Cas9 to develop attenuated vaccine strains [155].
Hu et al. applied CRISPR/Cas9 in Eimeria tenella, enabling single-gene and whole-gene family functional analysis. They identified 23 essential genes from the ApiAp2 transcription factor family, advancing the understanding of parasite development [156].
Cheng et al. optimized a transfection protocol utilizing FnCas12a protein for editing E. tenella, thereby enhancing opportunities for dissecting gene function and advancing the development of anticoccidial drugs and vaccines for Eimeria species [157]. CRISPR/Cas9-mediated disruption of ETH2_0411800 suggests that this gene is non-essential for E. tenella’s growth and development [158].
For poultry immune function studies, CRISPR/Cas9 facilitated gene knockouts in chicken DF-1 cells. One study demonstrated that the TBK1 gene is vital for STING-mediated IFN-β activation in chicken cells [159]. Lee et al. used CRISPR/Cas9 to generate an immunodeficient chicken model by knocking out the RAG1 gene [160].
Zeng et al. investigated the role of protein arginine methyltransferase 5 (PRMT5) in chicken cells. They generated a prmt5 gene-deficient DF-1 cell line using CRISPR/Cas9, which displayed increased IFN-b production compared to wild-type cells. This suggests a functional link of chPRMT5 in the suppression of IFN-b production and interferon-stimulated gene expression in chicken cells [161].
Additionally, research on IRF7-deficient chicken cells revealed increased viral replication of low pathogenic avian influenza virus, highlighting IRF7’s role in antiviral responses [162].
3.2 Construction of vaccines
Vaccines, alongside robust biosecurity measures, are central to disease prevention in the poultry industry. With the rise of emerging infectious diseases, enhancing vaccine strategies is critical. Recombinant multivalent vectored vaccines, which protect against multiple pathogens, are particularly valuable [163]. They can reduce selection pressure on field strains, streamline vaccination processes, lower production costs, and improve poultry welfare by reducing the need for multiple injections [164]. Vectored vaccines also elicit both cellular and humoral immune responses, and they facilitate DIVA (Differentiation Between Infected And Vaccinated Animals) strategies [165].
Over the past three decades, recombinant vectored vaccines, especially those using fowl pox virus and turkey herpesvirus (HVT), have become essential for controlling major viral diseases in poultry [166]. Advances in genetic engineering now enable the insertion of multiple foreign genes into vectors, allowing for broader disease protection [164]. CRISPR/Cas9 has further revolutionized this field by enabling the rapid development of multivalent vaccines that can simultaneously protect against several avian diseases [167,168] (Table 2).
Studies using CRISPR/Cas to develop poultry vaccines
| Vector | Antigen encoded by the inserted expression cassette | Disease(s) targeted by the candidate vaccine1 | Reference |
|---|---|---|---|
| HVT | AIV H7N9 HA | AI (H7HA subtypes) | [176] |
| IBDV VP2 | IBD | [177] | |
| ILTV gD-gI and the AIV H9N2 HA (insertion into the previously developed HVT-IBDV VP2 viral genome) |
|
[168] | |
| F (Fusion) gene of NDV | ND | [178] | |
| OmpH gene from P. multocida | Fowl cholera | [180] | |
| IBDV (G2d strain) VP2 gene | IBD (especially due to the IBDV G2d strain) | [181] | |
| HA of AIV H9N2 (Y280 strain) | AI (H9N2 Y280 strain) | [182] | |
|
IBDV (G2d) + AIV (H9N2/Y280) | [183] | |
| Insertion of mCherry cassette aiming the identification of new potential sites for the insertion of foreign genes. | A novel intergenic site HVT-005/006 was identified. | [175]2 | |
| HA of the H9N2 was inserted in this new site | Confirmation of the suitability of HVT-005/006 site for inserting foreign genes | ||
| DEV | HPAIV H5N1 HA |
|
[187] |
| Pre-membrane proteins (PrM) and envelope glycoprotein (E) of DTMUV | |||
| H5N8-HA | Avian influenza (H5Nx subtypes) | [186] | |
| GFP was used as a tag and removed later by Cre-Lox | |||
| Outer membrane protein H (ompH) | Fowl cholera | [167,189] | |
| FAdV-4 | Fusion protein of RFP and FAdV-4 Fiber-1 | Hepatitis hydropericardium syndrome (HHS) | [191] |
| Fusion protein of EGFP and FAdV-4 Fiber-2 | [192] | ||
| Fiber-2 without N-terminal 7–40aa | [193] | ||
| EGFP replacing Fiber-2 | [194] | ||
| Fiber of FAdV-8b | HHS and IBH | [195] | |
| HA of the H9N2 AIV |
|
[196] | |
| Eimeria acervulina |
|
Coccidiosis (E. acervulina) | [197] |
| ILTV | NDV Fusion (F) protein (in addition to the deletion of vector’s thymidine kinase [TK] and unique short 4 [US4] genes) | ILT and ND | [198] |
1In addition to Marek’s disease for chicken when the vector is HVT and in addition to Duck Viral Enteritis in susceptible species when the vector is DEV. 2In this study, the objective was to identify novel insertion sites rather than the construction of a vaccine targeting a specific disease. GFP: green fluorescence protein, RFP: red fluorescence protein, EGFP: enhanced green fluorescence protein, EYFP: enhanced yellow fluorescent protein.
3.2.1 Hvt-based candidate vaccines constructed using CRISPR/Cas9-mediated gene-editing technique
HVT has been used for decades as a vaccine against Marek’s disease (MD) [169], a highly contagious poultry disease characterized by the development of T-cell lymphomas and nerve enlargement. Vaccination, coupled with sanitation and selective breeding, is the primary control strategy for MD. HVT, with its large double-stranded DNA genome [170], can accommodate foreign genes encoding immunogenic proteins. Its non-pathogenic nature and ability to induce long-lasting immunity made it an early choice for expressing foreign antigens, enabling HVT to simultaneously provide immunity against MD and other viral diseases [171–173] avoiding interference between individual vaccines [174].
CRISPR/Cas9 has further advanced HVT vaccine development by streamlining the creation of multivalent vaccines, increasing the potential for HVT to serve as a versatile vector in poultry vaccination strategies.
Zai et al. identified a new insertion site within HVT, enabling stable expression of foreign genes, demonstrated by the successful integration of H9N2 hemagglutinin [175]. They recommended screening the entire HVT genome to discover additional sites for foreign gene insertion.
Chang et al. developed a bivalent HVT vaccine by inserting the H7N9 hemagglutinin gene into a specific intergenic region of HVT [176]. Similarly, Tang et al. [168] created a triple-insert recombinant vaccine by incorporating the infectious laryngotracheitis virus (ILTV) gD-gI gene and AIV H9N2 hemagglutinin into an HVT strain already expressing the VP2 protein from infectious bursal disease virus (IBDV) [177]. This innovative vaccine offers protection against three major avian diseases alongside MD.
The rHVT-F vaccine, expressing the fusion (F) protein of genotype XII Newcastle disease virus (NDV), provided full protection in chickens assessed five days post-challenge [178]. In a follow-up study, the F gene of genotype XII NDV was inserted into two different sites within the HVT genome. A single dose of the resulting vaccines provided sustained protection for at least 52 weeks post-vaccination [179].
Fowl cholera, caused by the highly transmissible bacterium Pasteurella multocida, is a significant avian ailment with global implications. Apinda et al. engineered rHVT-OmpH, carrying an outer membrane protein gene from P. multocida. This recombinant vaccine induced strong immunity and protected ducks from the pathogen, showing HVT’s potential for non-chicken hosts [180].
To counter the IBDV (G2d) variant, researchers developed rHVT-VP2, achieving full protection against this challenging strain [181]. More recently, the rHVT/Y280 vaccine was engineered by inserting the hemagglutinin (HA) gene of H9N2/Y280 into the HVT genome, conferring protection against H9N2/Y280 [182]. Subsequently, the VP2 gene of IBDV (G2d) was added to this recombinant virus, creating the dual-insert rHVT-VP2-HA vaccine for broader immunization coverage [183].
3.2.2 Duck enteritis virus (DEV)-based candidate vaccines constructed using CRISPR/Cas9-mediated gene-editing technique
DEV, a highly fatal alpha-herpesvirus affecting ducks, geese, and swans [184], has been repurposed as a vector for recombinant multivalent vaccines due to its large genome and restricted host range [185–188]. Zou et al. developed a novel recombinant DEV (rDEV) encoding genes for HPAIV H5N1 and duck tembusu virus (DTMUV), creating a trivalent vaccine (C-KCE-HA/PrM-E). Ducks vaccinated with this candidate showed strong immune responses and were protected against all three pathogens [187].
Using an NHEJ-CRISPR/Cas9 and Cre-Lox system, another rDEV was engineered to express influenza antigens. This system allowed for green fluorescence protein (GFP) tagging, followed by its removal [186]. Apinda et al. applied a similar method to develop rDEV vaccines expressing the P. multocida OmpH gene at two genomic sites [167,189]. These recombinant viruses matched the growth characteristics of wild-type DEV [189] and successfully protected ducklings against both DEV and P. multocida, without inducing any clinical symptoms or vaccine-related pathology [167].
3.2.3 Fowl adenovirus-based candidate vaccines constructed using CRISPR/Cas9-mediated gene-editing technique
Hepatitis-hydropericardium syndrome (HHS), caused by highly virulent Fowl Adenovirus (FAdV) serotypes, especially FAdV-4, poses a significant economic threat to the poultry industry [190]. Researchers from Hangzhou University in China have developed several live attenuated recombinant FAdV-4 vaccine candidates using CRISPR/Cas9 technology, publishing their findings in six articles between 2021 and 2023. These candidates include FAdV4-RFP_F1, which expresses the fusion protein of red fluorescence protein (RFP) and Fiber-1 [191]; and FA4-EGFP, which expresses the enhanced green fluorescence protein EGFP-Fiber-2 fusion protein [192]. Another candidate, FAV-4_Del, involves a deletion within Fiber-2 [193], while FAdV4-EGFP-rF2 replaces Fiber-2 entirely with EGFP [194]. These vaccines showed significant attenuation and provided complete protection against FAdV-4 in chicken trials [191–194]. The efficacy of FAdV4-EGFP-rF2 as a recombinant vaccine candidate, despite the knockout of the entire fiber-2 gene, illustrates its dispensability for both FAdV-4 virus replication and effective protection [194]. To broaden protection, the team created FA4-F8b, a recombinant virus expressing FAdV-8b fiber, aimed at preventing both HHS and inclusion body hepatitis (IBH). FA4-F8b was inactivated due to its high pathogenicity in 2-week-old SPF chicks, but it still provided effective protection against both FAdV-4 and FAdV-8b after inactivation [195].
In their latest study, the researchers used a double-fluorescence system to further modify FadV-4 [192], producing FAdV4-HA(H9), which expresses the HA gene from H9N2 AIV. This candidate vaccine was attenuated, induced early immune responses, and reduced H9N2 replication in chickens [196].
3.2.4 Candidate recombinant vaccines based on other vectors
3.2.4.1 Eimeria acervulina
Zhang et al. achieved stable transfection of E. acervulina, with confirmed expression of the AIV H9N2 M2 (M2e) protein in the cytoplasm of sporozoites. The fecundity of the modified parasite (EaM2e) matched that of the wild type [197]. Subsequent investigations are required to determine whether EaM2e can serve as a live vaccine vector.
3.2.4.2 Infectious laryngotracheitis virus
Atasoy et al. used a CRISPR/Cas9 system combined with the Cre-Lox system to simultaneously delete virulence factors and insert foreign genes into the ILTV genome. They successfully removed the thymidine kinase (TK) and unique short 4 (US4) genes while adding the NDV fusion (F) gene. This method did not impair ILTV replication or F protein expression, providing a promising tool for creating attenuated and multivalent vaccine vectors [198].
3.3 In vivo pathogen’s genome targeting
CRISPR/Cas could be used to precisely cut pathogen’s genomes in a targeted sequence offering the potential to prevent or treat infections [199].
Li et al. efficiently edited the long terminal repeats of reticuloendotheliosis virus (REV) using CRISPR/Cas9, resulting in the inhibition of viral protein expression and the disruption of the proviral genome in chicken cells. Furthermore, they successfully delivered the CRISPR/Cas9 system into REV-infected chickens using an attenuated MDV vaccine strain as a vector. This led to a reduction in REV viral load and alleviation of associated symptoms [200]. This marks the first instance of using herpesvirus-delivered CRISPR/Cas9 to confer resistance against avian retroviruses in chickens, providing a novel strategy against viral infections.
Challagula et al. explore the use of CRISPR/Cas13a to selectively disrupt RNA in chicken fibroblast DF1 cells, particularly focusing on its potential as an antiviral strategy against the influenza A virus (IAV). The team designed multiple CRISPR RNAs (crRNAs) against IAV genes, demonstrating reduced viral titers in cells transfected with these crRNAs. The study suggests that Cas13a’s precision and lack of off-target effects make it a promising tool for functional studies and antiviral strategies in chickens, with the potential to combat HPAIV strains [201].
The same year, Challagula et al. reported the development of transgenic chickens expressing Cas9 and guide RNAs (gRNAs) targeting the ICP4 gene of MDV. These chickens showed significantly reduced MDV replication when challenged with the virus. The designed gRNAs specifically interfered with MDV replication in transgenic chicken cells but not with HVT, suggesting that CRISPR/Cas9 can be used as an antiviral approach to control MDV infection in chickens without impeding the use of HVT as a vector for recombinant vaccines [202].
Mohsin et al. applied CRISPR/Cas9 to disrupt E. tenella genes, showing a remarkable reduction in lesion and oocyst scores, supporting its use against parasitic infections [203].
Liu et al. assessed the effectiveness of using MDV as a delivery system for the CRISPR/Cas9 gene-editing tool to target and disrupt the reverse-transcribed products of the avian leukosis virus subgroup J (ALV-J) RNA genome during its infection cycle in vitro and in vivo. They showed that the engineered MDV, expressing ALV-J-targeting CRISPR/Cas9, successfully resisted ALV-J challenges in host cells. This outcome demonstrates the CRISPR/Cas9 system’s effectiveness as a treatment against ALV-J infection and suggests the potential of MDV as an efficient delivery system for CRISPR/Cas9 in chickens [204].
Recent advancements in CRISPR/Cas9 GE highlight its transformative potential in targeting integrated viral genomes. A recent study demonstrated the safety and efficacy of CRISPR/Cas9 for in vivo editing of proviral DNA in simian immunodeficiency virus (SIV)-infected rhesus macaques. Targeting multiple regions of the SIV genome, the study achieved functional biodistribution to SIV reservoirs without off-target effects or abnormal pathology. Notably, macaques receiving higher doses showed improved lymphocyte counts, underscoring the therapeutic promise of this approach [205]. Like ALV-J, SIV is a retrovirus, and the ability to excise its DNA from host genomes provides a compelling proof of concept for using CRISPR to tackle integrated viral genomes. This breakthrough opens possibilities for eradicating persistent retroviral pathogens in poultry, such as Marek’s disease virus or lymphoid leukosis virus. By demonstrating the feasibility of excising integrated viral DNA in vivo, these findings set a precedent for developing CRISPR-based therapies for persistent poultry pathogens.
Several species of Mycoplasma cause substantial economic losses in livestock. The challenge in studying and addressing these bacteria lies in the lack of efficient recombination and genome engineering tools, hindering the production of mutant strains for identifying virulence factors and developing improved vaccine strains for many Mycoplasma species. Ipoutcha et al. developed an effective CRISPR-derived genetic tool for introducing targeted mutations in three major pathogenic species among them, the avian species Mycoplasma gallisepticum (Mgal). The team employed an inducible dCas9-cytidine deaminase system to disrupt several major virulence factors in these pathogens. Individual mutants of potential virulence genes were isolated [206].
3.4 Generation of disease-resilient poultry
In the context of production animals, disease resilience refers to an animal’s ability to maintain productive performance in the face of infection [207,208], and it encompasses two key components: disease resistance and disease tolerance. Disease resistance is defined as the individual’s ability to inhibit or limit pathogen replication within the host [209], while disease tolerance refers to the infected host’s capacity to reduce the impact of infection on health and performance, enabling it to sustain high levels of health or production despite a given pathogen load within the host [120].
Traditional breeding methods, which involve techniques such as cross-breeding and selective breeding, have been relied upon to enhance desirable traits in livestock, including disease resilience. However, they are limited by the natural genetic variation within populations and can be time-consuming [210,211]. While conventional breeding programs have led to gradual improvements, they may encounter challenges in obtaining disease resilience if many genes are involved [120].
The use of precise gene editing (PGE) in poultry is considered a transformative technology, offering the potential to revolutionize the breeding of desired traits in livestock [118]. PGE allows for the swift incorporation of new or existing beneficial mutations within a species or closely related ones that do not typically interbreed into elite breeding animals while avoiding the introduction of unwanted traits typical of traditional selective breeding [212].
In certain instances, although similar genetic improvements could theoretically be achieved through traditional breeding methods, GE expedites the process by bypassing the need for multiple generations of selective crossings or the identification of rare animals carrying desired genetic variants [119]. While GE is not intended to replace traditional breeding, it complements it by providing breeders with increased genetic variation to select from [213,214].
With recent advances in precision genome targeting, the generation of genetically modified poultry is now more attainable than ever [215]. The introduction of resilience alleles into a poultry population could be obtained by editing host factors crucial for pathogens’ entry or replication. Viruses, for example, rely on specific host cell receptor molecules to enter target cells [216]. Through the precise targeting and removal or modification of these receptors, CRISPR/Cas9-mediated GE has the potential to effectively hinder viral infections [217]. However, regulatory frameworks and public acceptance are still evolving, and further research is needed to fully realize the potential of GE.
The development of porcine reproductive and respiratory syndrome (PRRS)-resistant pigs using CRISPR/Cas9 GE highlights the transformative potential of this technology in combating economically devastating livestock diseases. By precisely targeting the porcine CD163 gene, which encodes a receptor essential for viral entry, researchers achieved complete resistance to the PRRS virus [218–221]. This groundbreaking solution addresses one of the most economically damaging illnesses for swine producers [222,223]. A scaled gene-editing program successfully introduced this resistance trait into four genetically diverse and elite porcine lines, ensuring its relevance and applicability for commercial breeding populations [224]. This achievement exemplifies how precise GE can target critical host factors to confer disease resistance and serves as a model for similar innovations in poultry.
3.4.1 In vitro testing for resistance to pathogens
Preliminary in vitro trials using specific cell cultures are essential for evaluating the efficacy and safety of genetic modifications aimed at developing disease-resistant poultry.
Lymphoid leukosis is caused by avian leukosis viruses (ALVs), which are categorized into subgroups A, B, C, D, and J [225]. Control measures focus on eradicating the virus from breeding flocks [20]. This strategy has substantially reduced the frequency of the disease in commercial flocks. Given the susceptibility of all studied chicken lines to ALV infection [226], there is considerable interest in developing resilient chicken lines.
To induce resistance to infections by ALV subgroups B [227], J [228], and A [229], Lee et al. utilized CRISPR/Cas9-based GE to modify viral receptor genes in DF-1 chicken fibroblasts. The tumor virus locus B (tvb) gene, encoding the TVB receptor, which is essential for ALV subgroup B entry into host cells, was efficiently modified, conferring resistance to ALV subgroup B [227]. For ALV-J, they altered the ALV-J receptor: the chicken Na+/H+ exchange 1, (chNHE1) by targeting the tryptophan residue at position 38 (Trp38) [228] previously characterized as involved in viral attachment and entry [230]. The targeted mutation resulted in a complete resistance to viral infection. Similarly, disruption of exon 2 within the tva gene in DF-1 fibroblasts conferred resistance to ALV subgroup A. Using a sequential approach, they modified all three receptor genes to block ALV subgroups A, B, and J, demonstrating the potential for generating cells resistant to various viral pathogens by targeting distinct receptors for cellular entry [229].
Koslová et al. similarly used CRISPR/Cas9 to introduce frame-shifting mutations in chicken cell line DF-1 at the tvc, tva, and tvj loci, conferring resistance to ALV subgroups C, A, and J [231]. These findings paved the way for creating ALV-resistant chickens using CRISPR/Cas9 [112].
The limited pathogenicity of AIV in waterfowl, such as ducks, is attributed to the presence of the retinoic acid-inducible gene I (RIG-I) [232], in contrast to chickens, which lack this gene and exhibit severe disease when infected with HPAIV. To confer resistance to AIV in chickens, RIG-I was successfully knocked into chicken DF-1 cells, establishing a RIG-I-dependent immune response without overexpression of RIG-I or disruption of host genes [233].
Another study involved replacing the C-terminal domain (CTD) of chicken melanoma differentiation-associated protein 5 (cMDA5) with that of RIG-I. The engineered cMDA5 gene was then expressed in cMDA5 knockout DF-1 cells. This modification resulted in an enhanced interferon-mediated immune response and a notable reduction in the titer of IAV [234].
3.4.2 Advances in methods for in vivo gene editing utilizing the CRISPR/Cas9 system in poultry
3.4.2.1 Cultured PGCs-mediated gene editing in poultry
PGCs, the embryonic precursors of sperm and egg cells, can be isolated from various stages of embryonic development [235–237] and cultured while maintaining their germline competency [238]. When cultured PGCs are introduced into the bloodstream of recipient embryos, they migrate to the gonads and integrate into the germline, resulting in the creation of a germline chimera [239,240]. Cultured PGCs allowed the generation of the first knockout chickens in 2013 [241]. The in vitro genetic editing of chicken PGCs using CRISPR/Cas9 system has become a standard practice in many chicken research laboratories, opening up numerous potential applications for genetically edited chickens [110,113,242].
When injected into a host chick embryo, edited PGCs integrate into the host embryo’s gonads alongside the native PGCs, diminishing the likelihood of offspring deriving from the donor PGCs in subsequent mating [243]. To address this, methods to reduce or eliminate native PGCs have been explored, but they often fail to eradicate all germ cells and pose significant toxicity risks to the host embryo. Recently, Ballantyne et al. developed a surrogate host chicken line allowing for conditional ablation of both male and female germlines. By using CRISPR/Cas9-mediated HDR to target the DAZL gene, which is exclusively expressed in the germ cell lineage, they induced apoptosis in the host’s germ cells upon activation of caspase-9 protein by a chemical compound. This enables efficient colonization of the host’s gonads by edited PGCs. Direct mating of these surrogates facilitates the production of pure-breed homozygous edited offspring, reducing generation time and increasing the number of homozygous genome-edited offspring [244].
While cultured PGCs serve as effective tools for GE in poultry, there are limitations to this method. Notably, only chicken PGCs have been reliably cultured long-term in vitro. This limitation makes it difficult to isolate and amplify genome-edited PGCs in other species. Moreover, PGC-based techniques are time-intensive, involving the selection of edited PGCs, microinjection, and raising G0 germline chimeras until sexual maturity to obtain edited offspring.
3.4.2.2 In vivo transfection of PGCs
The direct injection of genome engineering tools into the circulatory system of the developing embryo just before the PGCs migrate to the gonads enabled the transformation of circulating PGCs and the generation of transgenic chickens [215,245]. This method could be applied to species that lack the long-term PGC culture method.
3.4.2.3 Sperm Transfection–Assisted Gene Editing (STAGE)
STAGE entails directly transfecting spermatozoa with Cas9 mRNA and sgRNA. Using these modified sperm for adult hen insemination allows for the direct production of edited progeny. Although GE has been achieved successfully in chicken embryos using STAGE, the efficiency of producing edited offspring remains relatively low, indicating the need for further enhancements [246].
3.4.2.4 Viral infection
The direct injection of the adenoviral CRISPR/Cas9 vector into the avian blastoderms was successfully applied to generate genome-edited quail [101] and was later applied to generate edited chicken and duck [247].
3.4.2.5 Other methods
Intracytoplasmic sperm injection (ICSI)-mediated GE technology is a rapid method to generate targeted gene knockout in poultry [248,249]. Nevertheless, implementing this technique demands substantial technical expertise and effort, along with specialized equipment. The current hatching success rate remains modest, indicating the need for additional research.
3.4.3 Disease-resilient chickens developed using CRISPR/Cas9 systems
ALV-J replication relies on the functional cellular receptor chNHE1, where a crucial amino acid for virus entry is the tryptophan residue number 38 (W38) located in its extracellular segment [228,230]. Building on these findings, Koslová et al. deleted W38 in chicken PGCs. Edited PGCs underwent an orthotopic transplantation and successfully developed, with significantly elevated efficacy, an inbred chicken line CB resistant to ALV-J infection with no observable side effects in edited birds [112]. Also, by precisely deleting W38, Hellmich et al. successfully conferred ALV-J resistance in a commercial egg-type chicken line [250]. Edited birds challenged by a highly pathogenic ALV-J displayed no pathological clinical signs or lesions [251].
Although earlier studies reported promising outcomes, recent in vitro and in vivo findings by Matoušková et al. reveal that minor modifications to the ALV-J receptor NHE1, specifically the deletion of a single amino acid (W38), initially block ALV-J effectively but may ultimately be circumvented by viral adaptations in the envelope protein. These findings suggest that more substantial receptor alterations may be required to achieve durable resistance [252].
In order to generate a chicken line resistant to ALV A and K, Koslová et al. edited chicken PGCS (CPGCs) by introducing a frame-shifting deletion into the chicken tva gene coding the Tva cell surface protein serving as the entry receptor for ALV A and K. Edited cells are then transplanted into the testes of sterilized recipient roosters. The resulting tva−/− chickens demonstrated complete resistance to ALV A and K in both in vitro and in vivo assessments, contrasting with their susceptible tva+/+ and tva+/− siblings. The tva knockout chickens exhibited a specific disorder in cobalamin/vitamin B12 metabolism, aligning with the recognized role of Tva as a receptor for cobalamin [253]. To address this concern, the authors suggest a more targeted modification of Tva by changing a specific amino acid residue crucial for virus binding, such as the C40W substitution found in the tva allele.
Globally, the poultry production industry faces significant challenges from AIV H9N2 infections and HPAI outbreaks, resulting in heavy economic losses. Moreover, several AIV serotypes are zoonotic with the risk of the emergence of strains with pandemic potential [254].
Controlling AI through poultry vaccination faces challenges due to rapid and continuous antigenic drift in field viruses and global limitations in vaccine production and supply [255]. GE has emerged as a promising solution to develop AIV-resistant poultry [256].
In chicken cells, the AI viral RNA polymerase depends on chicken ANP32A proteins for replication [257], while ANP32B is inactive [153,258]. Researchers used CRISPR/Cas9 to induce N129I and D130N substitutions into the ANP32A gene [259] to impede AIV infection and transmission in chickens. The residues were altered in CPGCs, and gene-edited chickens were subsequently derived from these modified cells [143]. Gene-edited chickens showed resistance to AIV (H9N2-UDL) infection through natural transmission routes when exposed to infected birds without displaying health issues. However, very high inoculation dose led to breakthrough infections with various amino acid substitutions detected in the viral polymerase genes, enabling the enzyme to utilize the edited ANP32A protein and suboptimal ANP32 family members [143]. This unintended outcome underscores the importance of a robust GE strategy and subsequent evaluation, including challenges with various AI phenotypes at non-physiological exposure levels to eliminate the possibility of adapted viruses eruption.
Ultimately, editing all three members of the ANP32 family in chicken cells, resulting in no virus polymerase activity, illustrated a proof of principle for combining multiple edits in host genes to confer sterile resistance. However, the potential deleterious effects on animal health highlight the need for careful consideration [143]. For a successful strategy against AI, the authors suggest multiple edits targeting the proviral potential of ANP32A, B, and E in order to eliminate the risk of escape mutants [143].
This study marks a significant milestone in genetic research as GE has successfully generated viable chickens partially resistant to influenza virus A infection for the first time.
4 Conclusion and perspectives
Poultry, serving as a significant protein source, is facing various challenges, including infectious diseases, resulting in considerable economic losses and public health concerns.
Over the past decade, CRISPR/Cas9-mediated GE technology has undergone rapid advancement. Due to its precision, efficiency, versatility, and simplicity, the system has revolutionized genetic modification, offering the potential to enhance the prevention and control of poultry infectious diseases.
Through targeted modifications at specific loci, this technology has significantly advanced our understanding of host–pathogen interactions. The insights gleaned have contributed to the swift creation of novel candidate poultry vaccines and have facilitated the development of disease-resistant birds. Moreover, this technology enables in vivo targeting of pathogens, marking a pivotal stride forward in bolstering infectious disease prevention and control efforts.
Leveraging CRISPR/Cas9-based strategies, innovative multivalent vectored vaccines have been engineered, offering the potential for simultaneous protection against up to four major poultry diseases [168]. The efficacy of the developed candidate vaccines has been remarkable, coupled with a notable absence of adverse reactions. This suggests a promising trajectory toward the commercial availability of CRISPR-engineered poultry vaccines in the market.
Recent studies have showcased the efficacy of CRISPR/Cas technology in targeting a multitude of poultry pathogens. CRISPR/Cas13a has been effectively utilized to disrupt AIV RNA in chicken cells, demonstrating its potential as an antiviral strategy. Additionally, CRISPR/Cas9 has been successfully employed to target specific DNA loci in the genomes of viruses, bacteria, and parasites, resulting in the inhibition of their replication within the host.
Producing permanent disease resilience, which can be passed down through generations, is a key objective in poultry production. It allows for the reduction of culling, vaccination costs, and surveillance programs. By integrating GE with poultry breeding programs, it becomes possible to develop poultry lines with enhanced disease resilience.
Targeted deletion of the W38 residue in the ALV-J receptor NHE1 confers initial resistance in chickens [112,250]; however, recent findings have demonstrated the eruption of resistant viral strains, suggesting a need for more extensive receptor modifications [252].
Target edition to the ANP32A gene resulted in resistance to AIV H9N2-UDL without adverse effects on health or productivity [143]. However, challenges remain, as evidenced by breakthrough infections observed at higher viral doses, highlighting the importance of robust editing strategies and continued evaluation to mitigate potential risks of viral adaptation.
Despite the tremendous potential of CRISPR/Cas systems, several challenges need to be addressed. Off-target effects, delivery methods, ethical concerns, public acceptance, and regulatory discrepancies across countries remain key barriers to widespread application.
A major concern is whether gene-edited poultry will be accepted by the public, as past experiences with genetically modified organisms (GMOs) suggest that public perception plays a crucial role in the adoption of new biotechnologies [260]. GMOs produced using earlier technologies faced strong opposition, often driven by concerns over food safety, environmental impact, and corporate control over food production [261]. Given this historical context, genome-edited poultry, despite not containing foreign transgenes like traditional GMOs, may still encounter similar challenges in public acceptance [262]. Misconceptions about gene editing and distrust in regulatory institutions could contribute to resistance unless proactive efforts are made to communicate the distinctions between CRISPR-based GE and conventional genetic modification. One of the central factors influencing public acceptance is the purpose of GE. Reports suggest that people are more likely to support gene editing when it is used to enhance animal health and welfare, such as preventing infectious diseases, rather than for productivity gains that primarily benefit producers [263]. Public attitudes toward genome-edited poultry are also shaped by the perceived risks associated with off-target effects and genetic stability. Ethical concerns related to animal welfare emphasize the importance of minimizing unintended genetic modifications, as unforeseen mutations could lead to physiological or behavioral changes with negative implications for livestock well-being [264].
Ensuring that GE aligns with ethical considerations, including minimizing animal suffering and maintaining genetic diversity, could help build broader acceptance [262].
Transparent communication, rigorous safety assessments, and strong governance frameworks are necessary to foster trust in genome-editing applications. Engaging the public in discussions on the ethical and societal implications of this technology – while ensuring that regulatory policies align with broader societal values – will be crucial in determining the long-term acceptance of gene-edited poultry. Ultimately, fostering trust in the institutions responsible for gene editing and demonstrating tangible benefits for both animals and consumers will be key to integrating CRISPR-based innovations into poultry production responsibly and sustainably.
Regulatory hurdles remain a significant challenge in the commercialization of genome-edited poultry, including those produced using CRISPR/Cas9. Current frameworks in many countries fail to distinguish genome-edited animals from transgenic organisms, subjecting them to lengthy and costly approval processes. For example, in the United States, the FDA regulates genetically modified animals as “New Animal Drugs,” leading to approval timelines exceeding 15 years, as seen with genetically engineered pigs [265] and fish [266]. Similarly, in the European Union, rigid policies do not differentiate between transgenic and genome-edited animals, further complicating approvals [267]. In contrast, countries like Brazil [268] and Argentina [269] have streamlined their regulations, recognizing genome-edited animals (without foreign DNA insertions) as equivalent to traditionally bred animals, avoiding unnecessary regulatory barriers. China, a leading country in CRISPR/Cas9 research [270,271], actively supports GE advancements but still regulates genome-edited animals under GMO laws, with no commercial approvals to date [272]. However, regulatory landscapes are evolving, reflecting a global trend toward balancing innovation and regulation [273]. The European Parliament’s recent vote on New Genomic Techniques (NGTs), which exempts gene-edited plants deemed indistinguishable from conventionally bred ones from GMO legislation, signals a shift toward more adaptable policies [274].
Amid current challenges, the optimism surrounding CRISPR/Cas systems remains well-founded. With the ongoing advancements in next-generation sequencing and artificial intelligence, CRISPR/Cas applications are expected to broaden, encompassing additional poultry species such as turkeys, geese, ducks, and guinea fowl. Researchers are hopeful that continued advancements and collaborative efforts will address current limitations, ultimately enabling the full potential of GE technologies. This optimistic trajectory points toward a future where CRISPR/Cas can be harnessed to achieve sustainable and resilient poultry production on a global scale, transforming the industry and providing significant benefits for disease resistance, environmental sustainability, and animal welfare.
Acknowledgments
The author is grateful for the reviewer’s valuable comments that improved the manuscript. Open Access funding provided by the Qatar National Library.
-
Funding information: Open Access funding has been provided by the Qatar National Library.
-
Author contribution: The author confirms the sole responsibility for the conception, comprehensive review of the literature, and manuscript preparation.
-
Conflict of interest: Author states no conflict of interest.
-
Data availability statement: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
[1] FAO. Poultry species | Gateway to poultry production and products | [Internet]. [cited 2025 Jan 4]. https://www.fao.org/poultry-production-products/production/poultry-species/en/.Search in Google Scholar
[2] FAO. Production systems | Gateway to poultry production and products | [Internet]. [cited 2025 Jan 4]. https://www.fao.org/poultry-production-products/production/production-systems/en/.Search in Google Scholar
[3] Thorp B. The poultry industry. In: Barrow P, Nair V, Baigent S, Atterbury R, Clark M, editors. Poultry health: A guide for professionals. Wallingford Boston, MA: CABI; 2021. p. 25–8. (CABI Books).Search in Google Scholar
[4] Vaillancourt JP. Poultry production in the world. In: Brugère-Picoux J, Vaillancourt JP, editors. Manual of poultry diseases. Paris: AFAS; 2015. p. 2–8.Search in Google Scholar
[5] FAO. World food and agriculture – Statistical yearbook 2023 [Internet]. Rome, Italy: FAO; 2023. [cited 2024 Feb 14] http://www.fao.org/documents/card/en/c/cc8166en.Search in Google Scholar
[6] FAO. Meat market review, emerging trends and outlook 2023. Rome: FAO; 2023.Search in Google Scholar
[7] Rajkhowa TK, Zodinpuii D, Bhutia LD, Islam SJ, Gogoi A, Hauhnar L, et al. Emergence of a novel genotype of class II New Castle Disease virus in North Eastern States of India. Gene. 2023 May 15;864:147315. [cited 2024 Mar 16]. https://www.sciencedirect.com/science/article/pii/S0378111923001567.10.1016/j.gene.2023.147315Search in Google Scholar PubMed
[8] Saif YM, Swayne DE, Pantin-Jackwood MJ, Spackman E, Johnson TJ, Day JM, et al. Emerging diseases and diseases of complex or unknown etiology. In: Swayne DE, Boulianne M, Logue CM, McDougald LR, McDougald LR, Suarez DL, editors. Diseases of poultry. 14th edn. Hoboken, New Jersey, USA: Wiley-Blackwell; 2020. p. 1383–410.10.1002/9781119371199.ch33Search in Google Scholar
[9] Abozeid HH. Global emergence of infectious bronchitis virus variants: Evolution, immunity, and vaccination challenges. Transbound Emerg Dis. 2023;2023(1):1144924. [cited 2024 Oct 25] https://onlinelibrary.wiley.com/doi/abs/10.1155/2023/1144924.10.1155/2023/1144924Search in Google Scholar PubMed PubMed Central
[10] Abreu R, Semedo-Lemsaddek T, Cunha E, Tavares L, Oliveira M. Antimicrobial drug resistance in poultry production: Current status and innovative strategies for bacterial control. Microorg. 2023 Apr 6;11(4):953. [cited 2024 Mar 16] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10141167/.10.3390/microorganisms11040953Search in Google Scholar PubMed PubMed Central
[11] Ndahi MD, Hendriksen R, Helwigh B, Card RM, Fagbamila IO, Abiodun-Adewusi OO, et al. Determination of antimicrobial use in commercial poultry farms in Plateau and Oyo States, Nigeria. Antimicrob Resist Infect Control. 2023 Apr 10;12(1):30. [cited 2024 Mar 16] https://aricjournal.biomedcentral.com/articles/10.1186/s13756-023-01235-x.10.1186/s13756-023-01235-xSearch in Google Scholar PubMed PubMed Central
[12] Carrique-Mas JJ, Hue LT, Dung LT, Thuy NT, Padungtod P. Restrictions on antimicrobial use in animals producing meat, milk and eggs, Viet Nam. Bull World Health Organ. 2023;101(3):223–5. [cited 2025 Apr 15] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9948497/.10.2471/BLT.22.289187Search in Google Scholar PubMed PubMed Central
[13] Wallinga D, Smit LAM, Davis MF, Casey JA, Nachman KE. A review of the effectiveness of current US policies on antimicrobial use in meat and poultry production. Curr Env Health Rep. 2022 Jun 1;9(2):339–54. [cited 2024 Oct 25] 10.1007/s40572-022-00351-x.Search in Google Scholar PubMed PubMed Central
[14] Souillard R, Allain V, Dufay-Lefort AC, Rousset N, Amalraj A, Spaans A, et al. Biosecurity implementation on large-scale poultry farms in Europe: A qualitative interview study with farmers. Prev Vet Med. 2024 Mar 1;224:106119. [cited 2024 Mar 16] https://www.sciencedirect.com/science/article/pii/S0167587724000059.10.1016/j.prevetmed.2024.106119Search in Google Scholar PubMed
[15] Ravikumar R, Chan J, Prabakaran M. Vaccines against major poultry viral diseases: Strategies to improve the breadth and protective efficacy. Viruses. 2022 Jun;14(6):1195. [cited 2023 Nov 17] https://www.mdpi.com/1999-4915/14/6/1195.10.3390/v14061195Search in Google Scholar PubMed PubMed Central
[16] Simancas-Racines A, Cadena-Ullauri S, Guevara-Ramírez P, Zambrano AK, Simancas-Racines D. Avian influenza: Strategies to manage an outbreak. Pathogens. 2023 Apr;12(4):610. [cited 2024 Mar 16] https://www.mdpi.com/2076-0817/12/4/610.10.3390/pathogens12040610Search in Google Scholar PubMed PubMed Central
[17] World Organisation for Animal Health (WOAH). Fowl typhoid and pullorum disease. In: Manual of diagnostic tests and vaccines for terrestrial animals. Paris, France: WOAH; 2018. [cited 2025 Apr 15] https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.03.07_FOWL_TYPHOID.pdf.Search in Google Scholar
[18] EFSA (European Food Safety Authority). Avian influenza cases in poultry and water birds on the rise | EFSA [Internet]. 2022 [cited 2024 Feb 5]. https://www.efsa.europa.eu/en/news/avian-influenza-cases-poultry-and-water-birds-rise.Search in Google Scholar
[19] Houta MH, Hassan KE, El-Sawah AA, Elkady MF, Kilany WH, Ali A, et al. The emergence, evolution and spread of infectious bronchitis virus genotype GI-23. Arch Virol. 2021 Jan;166(1):9–26. [cited 2024 Mar 16] http://link.springer.com/10.1007/s00705-020-04920-z.10.1007/s00705-020-04920-zSearch in Google Scholar PubMed PubMed Central
[20] Fandiño S, Gomez-Lucia E, Benítez L, Doménech A. Avian leukosis: Will we be able to get rid of it? Anim Open Access J MDPI. 2023 Jul 19;13(14):2358. [cited 2024 Mar 16]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10376345/.10.3390/ani13142358Search in Google Scholar PubMed PubMed Central
[21] Thames HT, Theradiyil Sukumaran A. A review of salmonella and campylobacter in broiler meat: Emerging challenges and food safety measures. Foods. 2020 Jun 11;9(6):776. [cited 2024 Mar 16] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7353592/.10.3390/foods9060776Search in Google Scholar PubMed PubMed Central
[22] Solís D, Cordero N, Quezada-Reyes M, Escobar-Astete C, Toro M, Navarrete P, et al. Prevalence of salmonella in eggs from conventional and cage-free egg production systems and the role of consumers in reducing household contamination. Foods. 2023 Nov 28;12(23):4300. [cited 2024 Mar 16] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10706720/.10.3390/foods12234300Search in Google Scholar PubMed PubMed Central
[23] Kang M, Wang LF, Sun BW, Wan WB, Ji X, Baele G, et al. Zoonotic infections by avian influenza virus: changing global epidemiology, investigation, and control. Lancet Infect Dis. 2024 Aug;24(8):e522–31.10.1016/S1473-3099(24)00234-2Search in Google Scholar PubMed
[24] Doran T, Challagulla A, Cooper C, Tizard M, Jenkins K. Genome editing in poultry - opportunities and impacts. Natl Inst Biosci J. 2017;1. [cited 2025 Apr 15] https://www.nibjournal.ed.ac.uk/nibjournal/article/view/1742/pdf.10.2218/natlinstbiosci.1.2016.1742Search in Google Scholar
[25] Khwatenge CN, Nahashon SN. Recent advances in the application of CRISPR/Cas9 gene editing system in poultry species. Front Genet. 2021;12:627714. [cited 2023 Nov 15] https://www.frontiersin.org/articles/10.3389/fgene.2021.627714.10.3389/fgene.2021.627714Search in Google Scholar PubMed PubMed Central
[26] Proudfoot C, Lillico S, Tait-Burkard C. Genome editing for disease resistance in pigs and chickens. Anim Front. 2019 Jun 25;9(3):6–12. [cited 2023 Nov 27]. 10.1093/af/vfz013.Search in Google Scholar PubMed PubMed Central
[27] Vilela J, Rohaim MA, Munir M. Application of CRISPR/Cas9 in understanding avian viruses and developing poultry vaccines. Front Cell Infect Microbiol. 2020 Nov 24;10:581504. [cited 2023 Nov 12] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7732654/.10.3389/fcimb.2020.581504Search in Google Scholar PubMed PubMed Central
[28] Söllner JH, Mettenleiter TC, Petersen B. Genome editing strategies to protect livestock from viral infections. Viruses. 2021 Oct;13(10):1996. [cited 2023 Nov 17] https://www.mdpi.com/1999-4915/13/10/1996.10.3390/v13101996Search in Google Scholar PubMed PubMed Central
[29] WHO. Human genome editing [Internet]. [cited 2023 Dec 27]. https://www.who.int/health-topics/human-genome-editing Search in Google Scholar
[30] European Parliament. Directorate General for Parliamentary Research Services. Genome editing in humans: A survey of law, regulation and governance principles. LU: Publications Office; 2022. [cited 2023 Dec 27] https://data.europa.eu/doi/10.2861/07058.Search in Google Scholar
[31] International Organization for Standardization. ISO 5058-1:2021(en), Biotechnology – Genome editing – Part 1: Vocabulary [Internet]. 2021 [cited 2023 Dec 27] https://www.iso.org/obp/ui/#iso:std:iso:5058:-1:ed-1:v1:en.Search in Google Scholar
[32] Badr A, El-Shazly HH. Genome editing by site-directed nucleases and its applications in producing climate change resilient crop plants. In: Khan Z, Shahwar D, Heikal Y, editors. Genome editing and global food security. 1st edn. London, United Kingdom: Routledge; 2023. p. 17–41.10.4324/9781003382102-2Search in Google Scholar
[33] Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014 May;15(5):321–34. [cited 2024 Mar 1] https://www.nature.com/articles/nrg3686.10.1038/nrg3686Search in Google Scholar PubMed
[34] Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014 Nov 28;346(6213):1258096. [cited 2024 Jan 4] https://www.science.org/doi/10.1126/science.1258096.10.1126/science.1258096Search in Google Scholar PubMed
[35] Stinson BM, Loparo JJ. Repair of DNA double-strand breaks by the non-homologous end joining pathway. Annu Rev Biochem. 2021 Jun 20;90:137–64. [cited 2024 Mar 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8899865/.10.1146/annurev-biochem-080320-110356Search in Google Scholar PubMed PubMed Central
[36] Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end joining pathway. Annu Rev Biochem. 2010;79:181–211. [cited 2024 Mar 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3079308/.10.1146/annurev.biochem.052308.093131Search in Google Scholar PubMed PubMed Central
[37] Cannan WJ, Pederson DS. Mechanisms and consequences of double-strand DNA break formation in chromatin. J Cell Physiol. 2016 Jan;231(1):3–14. [cited 2024 Mar 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4994891/.10.1002/jcp.25048Search in Google Scholar PubMed PubMed Central
[38] Barkova OY, Larkina TA, Krutikova AA, Polteva EA, Shcherbakov YS, Peglivanyan GK, et al. Innovative approaches to genome editing in chickens. Cytol Genet. 2022 Apr 1;56(2):196–207. [cited 2024 Jan 10] 10.3103/S0095452722020037.Search in Google Scholar
[39] Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996 Feb 6;93(3):1156–60. [cited 2023 Dec 25] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC40048/.10.1073/pnas.93.3.1156Search in Google Scholar PubMed PubMed Central
[40] Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001 Jan;21(1):289–97. [cited 2023 Dec 25] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC88802/.10.1128/MCB.21.1.289-297.2001Search in Google Scholar PubMed PubMed Central
[41] Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003 May 2;300(5620):764–4. [cited 2023 Dec 25] https://www.science.org/doi/10.1126/science.1079512.10.1126/science.1079512Search in Google Scholar PubMed
[42] Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002 Jul;161(3):1169–75. [cited 2024 Feb 26]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462166/.10.1093/genetics/161.3.1169Search in Google Scholar PubMed PubMed Central
[43] Meng X, Noyes MB, Zhu L(Julie), Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc finger nucleases. Nat Biotechnol. 2008 Jun;26(6):695–701. [cited 2024 Feb 26] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2502069/.10.1038/nbt1398Search in Google Scholar PubMed PubMed Central
[44] Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, et al. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2011 Apr 26;108(17):7052–7. [cited 2024 Feb 26] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3084115/.10.1073/pnas.1102030108Search in Google Scholar PubMed PubMed Central
[45] Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009 Jul 24;325(5939):433–3. [cited 2023 Dec 28] https://www.sciencemag.org/lookup/doi/10.1126/science.1172447.10.1126/science.1172447Search in Google Scholar PubMed PubMed Central
[46] Meyer M, de Angelis MH, Wurst W, Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15022–6. [cited 2023 Dec 28] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2930558/.10.1073/pnas.1009424107Search in Google Scholar PubMed PubMed Central
[47] Flisikowska T, Thorey IS, Offner S, Ros F, Lifke V, Zeitler B, et al. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS One. 2011 Jun 13;6(6):e21045. [cited 2023 Dec 28] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3113902/.10.1371/journal.pone.0021045Search in Google Scholar PubMed PubMed Central
[48] Yu S, Luo J, Song Z, Ding F, Dai Y, Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011 Nov;21(11):1638–40. [cited 2023 Dec 28] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3364726/.10.1038/cr.2011.153Search in Google Scholar PubMed PubMed Central
[49] Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011 Feb;29(2):143–8. [cited 2023 Dec 29] https://www.nature.com/articles/nbt.1755.10.1038/nbt.1755Search in Google Scholar PubMed
[50] Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010 Oct;186(2):757–61. [cited 2023 Dec 29] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2942870/.10.1534/genetics.110.120717Search in Google Scholar PubMed PubMed Central
[51] Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011 Sep 30;333(6051):1843–6. [cited 2023 Dec 29] https://www.science.org/doi/10.1126/science.1204094.10.1126/science.1204094Search in Google Scholar PubMed
[52] Li C, Chu W, Gill RA, Sang S, Shi Y, Hu X, et al. Computational tools and resources for CRISPR/cas genome editing. Genomics Proteom Bioinforma. 2023 Feb 1;21(1):108–26. 10.1016/j.gpb.2022.02.006. [cited 2023 Dec 20].Search in Google Scholar PubMed PubMed Central
[53] Liu G, Zhang Y, Zhang T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput Struct Biotechnol J. 2019 Nov 29;18:35–44. [cited 2024 Mar 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6921152/.10.1016/j.csbj.2019.11.006Search in Google Scholar PubMed PubMed Central
[54] Travis J. Making the cut CRISPR genome-editing technology shows its power. Science. 2015 Dec 18;350(6267):1456–7. [cited 2023 Dec 18] https://www.science.org/doi/10.1126/science.350.6267.1456.10.1126/science.350.6267.1456Search in Google Scholar PubMed
[55] LaManna CM, Barrangou R. Enabling the rise of a CRISPR world. CRISPR J. 2018 Jun 1;1(3):205–8. [cited 2023 Dec 17] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6636870/.10.1089/crispr.2018.0022Search in Google Scholar PubMed PubMed Central
[56] Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987 Dec;169(12):5429–33. [cited 2023 Dec 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC213968/.10.1128/jb.169.12.5429-5433.1987Search in Google Scholar PubMed PubMed Central
[57] Mojica FJM, Juez G, Rodriguez‐Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified Pst I sites. Mol Microbiol. , 1993 Aug;9(3):613–21. [cited 2023 Dec 20] https://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.1993.tb01721.x.10.1111/j.1365-2958.1993.tb01721.xSearch in Google Scholar PubMed
[58] Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007 Mar 23;315(5819):1709–12. [cited 2023 Dec 20] https://www.science.org/doi/10.1126/science.1138140.10.1126/science.1138140Search in Google Scholar PubMed
[59] Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011 Jun;14(3):321–7. [cited 2023 Dec 21] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3119747/.10.1016/j.mib.2011.03.005Search in Google Scholar PubMed PubMed Central
[60] Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011 Dec 15;45(1):273–97. [cited 2023 Dec 21] https://www.annualreviews.org/doi/10.1146/annurev-genet-110410-132430.10.1146/annurev-genet-110410-132430Search in Google Scholar PubMed
[61] Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012 Feb;482(7385):331–8. [cited 2023 Dec 21] https://www.nature.com/articles/nature10886.10.1038/nature10886Search in Google Scholar PubMed
[62] Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816–21. [cited 2023 Dec 22] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6286148/.10.1126/science.1225829Search in Google Scholar PubMed PubMed Central
[63] Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci. 2012 Sep 25;109(39):E2579–86. [cited 2023 Dec 17] https://www.pnas.org/doi/full/10.1073/pnas.1208507109.10.1073/pnas.1208507109Search in Google Scholar PubMed PubMed Central
[64] Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Nov;8(11):2281–308. [cited 2023 Dec 29] https://www.nature.com/articles/nprot.2013.143.10.1038/nprot.2013.143Search in Google Scholar PubMed PubMed Central
[65] Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339(6121):823–6. [cited 2023 Dec 3] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3712628/.10.1126/science.1232033Search in Google Scholar PubMed PubMed Central
[66] Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013 Feb 2;339(6121):819. [cited 2023 Dec 17] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3795411/.10.1126/science.1231143Search in Google Scholar PubMed PubMed Central
[67] Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013 Jan 29;2:e00471. [cited 2023 Dec 22] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3557905/.10.7554/eLife.00471Search in Google Scholar PubMed PubMed Central
[68] Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013 Mar;31(3):230–2. [cited 2023 Dec 22] https://www.nature.com/articles/nbt.2507.10.1038/nbt.2507Search in Google Scholar PubMed
[69] Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient in vivo genome editing using RNA-guided nucleases. Nat Biotechnol. 2013 Mar;31(3):227–9. [cited 2023 Dec 22] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3686313/.10.1038/nbt.2501Search in Google Scholar PubMed PubMed Central
[70] Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013 Oct;10(10):977–9. [cited 2023 Dec 22] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3794058/.10.1038/nmeth.2598Search in Google Scholar PubMed PubMed Central
[71] Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013 Jul 18;154(2):442–51. [cited 2023 Dec 22] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3770145/.10.1016/j.cell.2013.06.044Search in Google Scholar PubMed PubMed Central
[72] Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015 Apr 9;520(7546):186–91. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4393360/.10.1038/nature14299Search in Google Scholar PubMed PubMed Central
[73] Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell. 2015 Oct 22;163(3):759–71. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4638220/.10.1016/j.cell.2015.09.038Search in Google Scholar PubMed PubMed Central
[74] Kim Y, Cheong SA, Lee JG, Lee SW, Lee MS, Baek IJ, et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016 Aug;34(8):808–10. [cited 2023 Dec 31] https://www.nature.com/articles/nbt.3614.10.1038/nbt.3614Search in Google Scholar PubMed
[75] Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016 Apr 20;533(7603):420–4. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4873371/.10.1038/nature17946Search in Google Scholar PubMed PubMed Central
[76] Xu T, Zhong J, Huang Z, Yu L, Zheng J, Xie L, et al. Optimization of the base editor BE4max in chicken somatic cells. Poult Sci. 2022 Sep 13;101(12):102174. [cited 2024 Feb 12] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9573927/.10.1016/j.psj.2022.102174Search in Google Scholar PubMed PubMed Central
[77] Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPR-Cas13a. Nature. 2017 Oct 12;550(7675):280–4. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5706658/.10.1038/nature24049Search in Google Scholar PubMed PubMed Central
[78] Adler BA, Hessler T, Cress BF, Lahiri A, Mutalik VK, Barrangou R, et al. Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing. Nat Microbiol. 2022 Dec;7(12):1967–79. [cited 2023 Dec 31] https://www.nature.com/articles/s41564-022-01258-x.10.1038/s41564-022-01258-xSearch in Google Scholar PubMed PubMed Central
[79] Ogasawara S, Ebashi S. RNA overwriting of cellular mRNA by Cas13b-directed RNA-dependent RNA polymerase of influenza A virus. Int J Mol Sci. 2023 Jun 11;24(12):10000. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10298375/.10.3390/ijms241210000Search in Google Scholar PubMed PubMed Central
[80] Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting Type VI-D CRISPR effectors. Cell. 2018 Apr 19;173(3):665–76.e14. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5910255/.10.1016/j.cell.2018.02.033Search in Google Scholar PubMed PubMed Central
[81] Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, et al. RNA editing with CRISPR-Cas13. Science. 2017 Nov 24;358(6366):1019–27. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5793859/.10.1126/science.aaq0180Search in Google Scholar PubMed PubMed Central
[82] Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino A, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013 Sep 12;154(6):1380–9. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3856256/.10.1016/j.cell.2013.08.021Search in Google Scholar PubMed PubMed Central
[83] Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016 Jan 1;351(6268):84–8. [cited 2025 Feb 11] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4714946/.10.1126/science.aad5227Search in Google Scholar PubMed PubMed Central
[84] Schmid-Burgk JL, Gao L, Li D, Gardner Z, Strecker J, Lash B, et al. Highly parallel profiling of Cas9 variant specificity. Mol Cell. 2020 May 21;78(4):794–800.e8. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7370240/.10.1016/j.molcel.2020.02.023Search in Google Scholar PubMed PubMed Central
[85] Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019 Dec;576(7785):149–57. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6907074/.10.1038/s41586-019-1711-4Search in Google Scholar PubMed PubMed Central
[86] Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020 Feb;18(2):67–83. [cited 2023 Dec 21] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8905525/.10.1038/s41579-019-0299-xSearch in Google Scholar PubMed PubMed Central
[87] Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015 Nov;13(11):722–36. [cited 2023 Dec 21] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5426118/.10.1038/nrmicro3569Search in Google Scholar PubMed PubMed Central
[88] Guo C, Ma X, Gao F, Guo Y. Off-target effects in CRISPR/Cas9 gene editing. Front Bioeng Biotechnol. 2023 Mar 9;11:1143157. [cited 2024 Mar 14] https://www.frontiersin.org/articles/10.3389/fbioe.2023.1143157.10.3389/fbioe.2023.1143157Search in Google Scholar PubMed PubMed Central
[89] Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated Genome engineering. Mol Ther Nucleic Acids. 2015 Jan 1;4:e264. [cited 2025 Feb 11] https://www.cell.com/molecular-therapy-family/nucleic-acids/abstract/S2162-2531(16)30049-X.10.1038/mtna.2015.37Search in Google Scholar PubMed PubMed Central
[90] Anderson KR, Haeussler M, Watanabe C, Janakiraman V, Lund J, Modrusan Z, et al. CRISPR off-target analysis in genetically engineered rats and mice. Nat Methods. 2018 Jul;15(7):512–4. [cited 2025 Feb 5] https://www.nature.com/articles/s41592-018-0011-5.10.1038/s41592-018-0011-5Search in Google Scholar PubMed PubMed Central
[91] Mengstie MA, Azezew MT, Dejenie TA, Teshome AA, Admasu FT, Teklemariam AB, et al. Recent advancements in reducing the Off-target effect of CRISPR-Cas9 genome editing. Biol Targets Ther. 2024 Jan 18;18:21–8. [cited 2025 Feb 11] https://www.dovepress.com/recent-advancements-in-reducing-the-off-target-effect-of-crispr-cas9-g-peer-reviewed-fulltext-article-BTT.10.2147/BTT.S429411Search in Google Scholar PubMed PubMed Central
[92] Naeem M, Alkhnbashi OS. Current bioinformatics tools to optimize CRISPR/Cas9 experiments to reduce off-target effects. Int J Mol Sci. 2023 Mar 27;24(7):6261. [cited 2024 Mar 14] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10094584/.10.3390/ijms24076261Search in Google Scholar PubMed PubMed Central
[93] Yang ZX, Fu YW, Zhao JJ, Zhang F, Li SA, Zhao M, et al. Superior fidelity and distinct editing outcomes of SaCas9 compared with SpCas9 in genome editing. Genomics Proteom Bioinforma. 2023 Dec 1;21(6):1206–20. [cited 2025 Feb 11] https://www.sciencedirect.com/science/article/pii/S1672022922001681.10.1016/j.gpb.2022.12.003Search in Google Scholar PubMed PubMed Central
[94] Agudelo D, Carter S, Velimirovic M, Duringer A, Rivest JF, Levesque S, et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020 Jan;30(1):107–17. [cited 2025 Feb 11] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6961573/.10.1101/gr.255414.119Search in Google Scholar PubMed PubMed Central
[95] Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, et al. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther. 2016 Jan 12;24(3):636. [cited 2025 Feb 11] https://pmc.ncbi.nlm.nih.gov/articles/PMC4786917/.10.1038/mt.2015.218Search in Google Scholar PubMed PubMed Central
[96] Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen N, Zheng Z, et al. High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature. 2016 Jan 28;529(7587):490–5. [cited 2025 Feb 11] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4851738/.10.1038/nature16526Search in Google Scholar PubMed PubMed Central
[97] Ikeda A, Fujii W, Sugiura K, Naito K. High-fidelity endonuclease variant HypaCas9 facilitates accurate allele-specific gene modification in mouse zygotes. Commun Biol. 2019 Oct 10;2(1):1–7. [cited 2025 Feb 11] https://www.nature.com/articles/s42003-019-0627-8.10.1038/s42003-019-0627-8Search in Google Scholar PubMed PubMed Central
[98] Kim GD, Lee JH, Song S, Kim SW, Han JS, Shin SP, et al. Generation of myostatin-knockout chickens mediated by D10A-Cas9 nickase. FASEB J. 2020;34(4):5688–96. [cited 2023 Dec 8] https://onlinelibrary.wiley.com/doi/abs/10.1096/fj.201903035R.10.1096/fj.201903035RSearch in Google Scholar PubMed
[99] Klermund J, Rhiel M, Kocher T, Chmielewski KO, Bischof J, Andrieux G, et al. On- and off-target effects of paired CRISPR-Cas nickase in primary human cells. Mol Ther. 2024 May 1;32(5):1298–310. [cited 2025 Apr 16]. 10.1016/j.ymthe.2024.03.006.Search in Google Scholar PubMed PubMed Central
[100] Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014 Jun;24(6):1012–9. [cited 2025 Feb 7] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4032847/.10.1101/gr.171322.113Search in Google Scholar PubMed PubMed Central
[101] Lee J, Ma J, Lee K. Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail. Proc Natl Acad Sci. 2019 Jul 2;116(27):13288–92. [cited 2023 Dec 6] https://www.pnas.org/doi/full/10.1073/pnas.1903230116.10.1073/pnas.1903230116Search in Google Scholar PubMed PubMed Central
[102] Choudhary N, Tandi D, Verma RK, Yadav VK, Dhingra N, Ghosh T, et al. A comprehensive appraisal of mechanism of anti-CRISPR proteins: an advanced genome editor to amend the CRISPR gene editing. Front Plant Sci. 2023 Jun 23;14:1164461. [cited 2024 Mar 14] https://www.frontiersin.org/articles/10.3389/fpls.2023.1164461/full.10.3389/fpls.2023.1164461Search in Google Scholar PubMed PubMed Central
[103] Matsumoto D, Tamamura H, Nomura W. A cell cycle-dependent CRISPR-Cas9 activation system based on an anti-CRISPR protein shows improved genome editing accuracy. Commun Biol. 2020 Oct 23;3(1):1–10. [cited 2024 Mar 14] https://www.nature.com/articles/s42003-020-01340-2.10.1038/s42003-020-01340-2Search in Google Scholar PubMed PubMed Central
[104] Zhou S, Cai B, He C, Wang Y, Ding Q, Liu J, et al. Programmable base editing of the sheep genome revealed no genome-wide off-target mutations. Front Genet. 2019 Mar 15;10:215. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6428697/.10.3389/fgene.2019.00215Search in Google Scholar PubMed PubMed Central
[105] Liu Y, Li X, He S, Huang S, Li C, Chen Y, et al. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020 Apr 28;6:27. [cited 2023 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7186222/.10.1038/s41421-020-0165-zSearch in Google Scholar PubMed PubMed Central
[106] Atkins A, Chung CH, Allen AG, Dampier W, Gurrola TE, Sariyer IK, et al. Off-target analysis in gene editing and applications for clinical translation of CRISPR/Cas9 in HIV-1 therapy. Front Genome Ed. 2021;3:673022.10.3389/fgeed.2021.673022Search in Google Scholar PubMed PubMed Central
[107] Bunton-Stasyshyn RK, Codner GF, Teboul L. Screening and validation of genome-edited animals. Lab Anim; 2021 Jul 1;56(1):69–82. [cited 2025 Feb 10] https://journals.sagepub.com/doi/full/10.1177/00236772211016922.10.1177/00236772211016922Search in Google Scholar PubMed PubMed Central
[108] Park TS, Lee HJ, Kim KH, Kim JS, Han JY. Targeted gene knockout in chickens mediated by TALENs. Proc Natl Acad Sci. 2014 Sep 2;111(35):12716–21. [cited 2025 Feb 10] https://www.pnas.org/doi/abs/10.1073/pnas.1410555111.10.1073/pnas.1410555111Search in Google Scholar PubMed PubMed Central
[109] Taylor L, Carlson DF, Nandi S, Sherman A, Fahrenkrug SC, McGrew MJ. Efficient TALEN-mediated gene targeting of chicken primordial germ cells. Development. 2017 Jan 1;144(5):928–34. [cited 2025 Feb 11] https://journals.biologists.com/dev/article/doi/10.1242/dev.145367/264331/Efficient-TALEN-mediated-gene-targeting-of-chicken.10.1242/dev.145367Search in Google Scholar PubMed PubMed Central
[110] Dimitrov L, Pedersen D, Ching KH, Yi H, Collarini EJ, Izquierdo S, et al. Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. Kormann MSD, editor. PLoS One. 2016 Apr 21;11(4):e0154303. [cited 2023 Dec 31] https://dx.plos.org/10.1371/journal.pone.0154303.10.1371/journal.pone.0154303Search in Google Scholar PubMed PubMed Central
[111] Oishi I, Yoshii K, Miyahara D, Kagami H, Tagami T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci Rep. 2016 Apr 6;6(1):23980. [cited 2023 Dec 6] https://www.nature.com/articles/srep23980.10.1038/srep23980Search in Google Scholar PubMed PubMed Central
[112] Koslová A, Trefil P, Mucksová J, Reinišová M, Plachý J, Kalina J, et al. Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus. Proc Natl Acad Sci. 2020 Jan 28;117(4):2108–12. [cited 2023 Nov 11] https://www.pnas.org/doi/10.1073/pnas.1913827117.10.1073/pnas.1913827117Search in Google Scholar PubMed PubMed Central
[113] Idoko-Akoh A, Taylor L, Sang HM, McGrew MJ. High fidelity CRISPR/Cas9 increases precise monoallelic and biallelic editing events in primordial germ cells. Sci Rep. 2018 Oct 11;8(1):15126. [cited 2023 Dec 31] https://www.nature.com/articles/s41598-018-33244-x.10.1038/s41598-018-33244-xSearch in Google Scholar PubMed PubMed Central
[114] Oishi I, Yoshii K, Miyahara D, Tagami T. Efficient production of human interferon beta in the white of eggs from ovalbumin gene–targeted hens. Sci Rep. 2018 Jul 5;8(1):10203. [cited 2023 Dec 6] https://www.nature.com/articles/s41598-018-28438-2.10.1038/s41598-018-28438-2Search in Google Scholar PubMed PubMed Central
[115] Park TS, Park J, Lee JH, Park JW, Park BC. Disruption of G0/G1 switch gene 2 (G0S2) reduced abdominal fat deposition and altered fatty acid composition in chicken. FASEB J. 2019 Jan;33(1):1188–98. [cited 2025 Apr 16] https://faseb.onlinelibrary.wiley.com/doi/10.1096/fj.201800784R.10.1096/fj.201800784RSearch in Google Scholar PubMed
[116] Mishu MA, Nath SK, Sohidullah M, Hossain MT. Advancement of animal and poultry nutrition: Harnessing the power of CRISPR-Cas genome editing technology. J Adv Vet Anim Res. 2024 Jun 21;11(2):483. [cited 2024 Oct 24]. https://pmc.ncbi.nlm.nih.gov/articles/PMC11296187/.10.5455/javar.2024.k798Search in Google Scholar PubMed PubMed Central
[117] Wang S, Qu Z, Huang Q, Zhang J, Lin S, Yang Y, et al. Application of gene editing technology in resistance breeding of livestock. Life. 2022 Jul 18;12(7):1070. [cited 2024 Mar 20]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9325061/.10.3390/life12071070Search in Google Scholar PubMed PubMed Central
[118] Gao F, Li P, Yin Y, Du X, Cao G, Wu S, et al. Molecular breeding of livestock for disease resistance. Virology. 2023 Oct 1;587:109862. [cited 2024 Mar 20] https://www.sciencedirect.com/science/article/pii/S0042682223001757.10.1016/j.virol.2023.109862Search in Google Scholar PubMed
[119] Bhat SA, Malik AA, Ahmad SM, Shah RA, Ganai NA, Shafi SS, et al. Advances in genome editing for improved animal breeding: A review. Vet World. 2017 Nov;10(11):1361–6. [cited 2024 Mar 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5732344/.10.14202/vetworld.2017.1361-1366Search in Google Scholar PubMed PubMed Central
[120] Knap PW, Doeschl-Wilson A. Why breed disease-resilient livestock, and how? Genet Sel Evol. 2020 Oct 14;52(1):60. [cited 2023 Dec 10] 10.1186/s12711-020-00580-4.Search in Google Scholar PubMed PubMed Central
[121] Liu Z, Wu T, Xiang G, Wang H, Wang B, Feng Z, et al. Enhancing animal disease resistance, production efficiency, and welfare through precise genome editing. Int J Mol Sci. 2022 Jan;23(13):7331. [cited 2024 Mar 20] https://www.mdpi.com/1422-0067/23/13/7331.10.3390/ijms23137331Search in Google Scholar PubMed PubMed Central
[122] Chojnacka-Puchta L, Sawicka D. CRISPR/Cas9 gene editing in a chicken model: current approaches and applications. J Appl Genet. 2020 May 1;61(2):221–9. [cited 2025 Feb 11] 10.1007/s13353-020-00537-9.Search in Google Scholar PubMed PubMed Central
[123] Gul H, Habib G, Khan IM, Rahman SU, Khan NM, Wang H, et al. Genetic resilience in chickens against bacterial, viral and protozoal pathogens. Front Vet Sci. 2022 Nov 10;9:1032983. [cited 2024 Apr 16] https://www.frontiersin.org/articles/10.3389/fvets.2022.1032983/full.10.3389/fvets.2022.1032983Search in Google Scholar PubMed PubMed Central
[124] Gong J, Jiang Y, Zhang D, Li T, Fu L, Dou X. One-tube detection of Salmonella Typhimurium using LAMP and CRISPR-Cas12b. Microbiol Spectr. 2024 Aug 27;12(10):e01271. [cited 2024 Oct 24] https://pmc.ncbi.nlm.nih.gov/articles/PMC11448145/.10.1128/spectrum.01271-24Search in Google Scholar PubMed PubMed Central
[125] Yu Z, Shao Y, Shi D, Dong Y, Zhang Y, Cheng F, et al. A rapid, ultrasensitive, and highly specific method for detecting fowl adenovirus serotype 4 based on the LAMP-CRISPR/Cas12a system. Poult Sci. 2024 Jun 28;103(9):104048. [cited 2024 Oct 24] https://pmc.ncbi.nlm.nih.gov/articles/PMC11315145/.10.1016/j.psj.2024.104048Search in Google Scholar PubMed PubMed Central
[126] Zhou X, Wang S, Ma Y, Li Y, Deng G, Shi J, et al. Rapid detection of avian influenza virus based on CRISPR-Cas12a. Virol J. 2023 Nov 13;20(1):261.10.1186/s12985-023-02232-7Search in Google Scholar PubMed PubMed Central
[127] Zhou X, Wang S, Ma Y, Jiang Y, Li Y, Shi J, et al. On-site and visual detection of the H5 subtype avian influenza virus based on RT-RPA and CRISPR/Cas12a. Viruses. 2024 May 10;16(5):753.10.3390/v16050753Search in Google Scholar PubMed PubMed Central
[128] Zhang Q, Yu G, Ding X, Zhang K, Sun W, Li Q, et al. A rapid simultaneous detection of duck hepatitis A virus 3 and novel duck reovirus based on RPA CRISPR Cas12a/Cas13a. Int J Biol Macromol. 2024 Aug 1;274:133246. [cited 2024 Oct 25] https://www.sciencedirect.com/science/article/abs/pii/S0141813024040510.10.1016/j.ijbiomac.2024.133246Search in Google Scholar PubMed
[129] Zhang Z, Wang C, Chen X, Zhang Z, Shi G, Zhai X, et al. Based on CRISPR-Cas13a system, to establish a rapid visual detection method for avian influenza viruses. Front Vet Sci. 2024 Jan 8;10:1272612. [cited 2024 Mar 14] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10800881/.10.3389/fvets.2023.1272612Search in Google Scholar PubMed PubMed Central
[130] Yin D, Yin L, Wang J, Shen X, Pan X, Hou H, et al. Visual detection of duck tembusu virus with CRISPR/Cas13: A sensitive and specific point-of-care detection. Front Cell Infect Microbiol. 2022;12:848365. [cited 2023 Nov 17] https://www.frontiersin.org/articles/10.3389/fcimb.2022.848365.10.3389/fcimb.2022.848365Search in Google Scholar PubMed PubMed Central
[131] Wu Y, Li Y, Wen X. CRISPR-Cas13a-based detection method for avian influenza virus. Front Microbiol. 2023 Oct 11;14:1288951. [cited 2024 Mar 13] https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2023.1288951/full.10.3389/fmicb.2023.1288951Search in Google Scholar PubMed PubMed Central
[132] Li J, Zhang Z, Zhang Z, Chen X, Wang C, Zhai X, et al. Rapid detection of avian leukemia virus using CRISPR/Cas13a based lateral flow dipstick. Front Vet Sci. 2024 Aug 16;11:1424238. [cited 2024 Oct 24] https://pmc.ncbi.nlm.nih.gov/articles/PMC11362082/.10.3389/fvets.2024.1424238Search in Google Scholar PubMed PubMed Central
[133] Chen SS, Yang YL, Wang HY, Guo TK, Azeem RM, Shi CW, et al. CRISPR/Cas13a-based genome editing for establishing the detection method of H9N2 subtype avian influenza virus. Poult Sci. 2024 Oct;103(10):104068.10.1016/j.psj.2024.104068Search in Google Scholar PubMed PubMed Central
[134] Chen S, Li Y, Liao R, Liu C, Zhou X, Wang H, et al. Detection of avian leukosis virus subgroup J (ALV-J) using RAA and CRISPR-Cas13a combined with fluorescence and lateral flow assay. Int J Mol Sci. 2024 Oct;25(19):10780.10.3390/ijms251910780Search in Google Scholar PubMed PubMed Central
[135] Xu Q, Zhang Y, Sadigh Y, Tang N, Chai J, Cheng Z, et al. Specific and sensitive visual proviral DNA detection of major pathogenic avian leukosis virus subgroups using CRISPR-associated nuclease Cas13a. Viruses. 2024 Jul 20;16(7):1168. [cited 2024 Oct 25] https://pmc.ncbi.nlm.nih.gov/articles/PMC11281634/.10.3390/v16071168Search in Google Scholar PubMed PubMed Central
[136] Cai R, Lv R, Shi X, Yang G, Jin J. CRISPR/dCas9 tools: Epigenetic mechanism and application in gene transcriptional regulation. Int J Mol Sci. 2023 Jan;24(19):14865. [cited 2024 Mar 22] https://www.mdpi.com/1422-0067/24/19/14865.10.3390/ijms241914865Search in Google Scholar PubMed PubMed Central
[137] Williams RM, Senanayake U, Artibani M, Taylor G, Wells D, Ahmed AA, et al. Genome and epigenome engineering CRISPR toolkit for in vivo modulation of cis-regulatory interactions and gene expression in the chicken embryo. Dev Camb Engl. 2018 Feb 15;145(4):dev160333. [cited 2024 Feb 17] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5869010/.10.1242/dev.160333Search in Google Scholar PubMed PubMed Central
[138] Bock C, Datlinger P, Chardon F, Coelho MA, Dong MB, Lawson KA, et al. High-content CRISPR screening. Nat Rev Methods Primer. 2022 Feb 10;2(1):1–23. [cited 2023 Dec 3] https://www.nature.com/articles/s43586-021-00093-4.10.1038/s43586-021-00093-4Search in Google Scholar
[139] Srivastava K, Pandit B. Genome-wide CRISPR screens and their applications in infectious disease. Front Genome Ed. 2023 Sep 19;5:1243731. [cited 2024 Mar 11] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10546192/.10.3389/fgeed.2023.1243731Search in Google Scholar PubMed PubMed Central
[140] Doerflinger M, Forsyth W, Ebert G, Pellegrini M, Herold MJ. CRISPR/Cas9 – The ultimate weapon to battle infectious diseases? Cell Microbiol. , 2017;19(2):e12693. [cited 2024 Feb 18] https://onlinelibrary.wiley.com/doi/abs/10.1111/cmi.12693.10.1111/cmi.12693Search in Google Scholar PubMed
[141] Chulanov V, Kostyusheva A, Brezgin S, Ponomareva N, Gegechkori V, Volchkova E, et al. CRISPR screening: Molecular tools for studying virus–host interactions. Viruses. 2021 Nov;13(11):2258. [cited 2024 Mar 11] https://www.mdpi.com/1999-4915/13/11/2258.10.3390/v13112258Search in Google Scholar PubMed PubMed Central
[142] Zahedipour F, Zahedipour F, Zamani P, Jaafari MR, Sahebkar A. Harnessing CRISPR technology for viral therapeutics and vaccines: from preclinical studies to clinical applications. Virus Res. 2024 Mar;341:199314.10.1016/j.virusres.2024.199314Search in Google Scholar PubMed PubMed Central
[143] Idoko-Akoh A, Goldhill DH, Sheppard CM, Bialy D, Quantrill JL, Sukhova K, et al. Creating resistance to avian influenza infection through genome editing of the ANP32 gene family. Nat Commun. 2023 Oct 10;14(1):6136. [cited 2023 Nov 9] https://www.nature.com/articles/s41467-023-41476-3.10.1038/s41467-023-41476-3Search in Google Scholar PubMed PubMed Central
[144] Ahn J, Lee J, Park JY, Oh KB, Hwang S, Lee CW, et al. Targeted genome editing in a quail cell line using a customized CRISPR/Cas9 system. Poult Sci. 2017 May 1;96(5):1445–50. [cited 2024 Feb 8] https://www.sciencedirect.com/science/article/pii/S0032579119313185.10.3382/ps/pew435Search in Google Scholar PubMed
[145] Zhang Y, Tang N, Sadigh Y, Baigent S, Shen Z, Nair V, et al. Application of CRISPR/Cas9 gene editing system on MDV-1 genome for the study of gene function. Viruses. 2018 May 24;10(6):279. [cited 2023 Dec 14] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6024840/.10.3390/v10060279Search in Google Scholar PubMed PubMed Central
[146] Zhang Y, Luo J, Tang N, Teng M, Reddy VRAP, Moffat K, et al. Targeted editing of the pp38 gene in Marek’s disease virus-transformed cell lines using CRISPR/Cas9 system. Viruses. 2019 May;11(5):391. [cited 2023 Nov 10] https://www.mdpi.com/1999-4915/11/5/391.10.3390/v11050391Search in Google Scholar PubMed PubMed Central
[147] Zhang Y, Tang N, Luo J, Teng M, Moffat K, Shen Z, et al. Marek’s disease virus-encoded microRNA 155 ortholog critical for the induction of lymphomas is not essential for the proliferation of transformed cell lines. J Virol. 2019 Aug 13;93(17):e00713–19. [cited 2024 Jan 9] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6694823/.10.1128/JVI.00713-19Search in Google Scholar PubMed PubMed Central
[148] Luo J, Teng M, Zai X, Tang N, Zhang Y, Mandviwala A, et al. Efficient mutagenesis of Marek’s disease virus-encoded microRNAs using a CRISPR/Cas9-based gene editing system. Viruses. 2020 Apr 20;12(4):466. [cited 2024 Jan 9] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7232411/.10.3390/v12040466Search in Google Scholar PubMed PubMed Central
[149] Zhang Y, Li W, Tang N, Moffat K, Nair V, Yao Y. Targeted deletion of glycoprotein B gene by CRISPR/Cas9 nuclease inhibits gallid herpesvirus type 3 in dually infected Marek’s disease virus-transformed lymphoblastoid cell line MSB-1. J Virol. , 2022 Mar 23;96(6):e02027–21. [cited 2024 Jan 24]. https://journals.asm.org/doi/10.1128/jvi.02027-21.10.1128/jvi.02027-21Search in Google Scholar PubMed PubMed Central
[150] Teng M, Zhou ZY, Yao Y, Nair V, Zhang GP, Luo J. A new strategy for efficient screening and identification of monoclonal antibodies against oncogenic avian herpesvirus utilizing CRISPR/Cas9-based gene-editing technology. Viruses. 2022 Sep;14(9):2045. [cited 2023 Dec 13] https://www.mdpi.com/1999-4915/14/9/2045.10.3390/v14092045Search in Google Scholar PubMed PubMed Central
[151] Teng M, Liu JL, Luo Q, Zheng LP, Yao Y, Nair V, et al. Efficient cross-screening and characterization of monoclonal antibodies against Marek’s disease specific meq oncoprotein using CRISPR/Cas9-gene-edited viruses. Viruses. 2023 Apr;15(4):817. [cited 2023 Nov 17] https://www.mdpi.com/1999-4915/15/4/817.10.3390/v15040817Search in Google Scholar PubMed PubMed Central
[152] Long JS, Idoko-Akoh A, Mistry B, Goldhill DH, Staller E, Schreyer J, et al. Avian ANP32B does not support influenza A virus polymerase and influenza A virus relies exclusively on ANP32A in chicken cells. bioRxiv. 2019;512012. [cited 2024 Oct 26] https://www.biorxiv.org/content/10.1101/512012v1.Search in Google Scholar
[153] Park YH, Chungu K, Lee SB, Woo SJ, Cho HY, Lee HJ, et al. Host-specific restriction of avian influenza virus caused by differential dynamics of ANP32 family members. J Infect Dis. 2020 Jan 1;221(1):71–80. [cited 2023 Nov 10] 10.1093/infdis/jiz506.Search in Google Scholar PubMed
[154] Tahir H, Basit A, Tariq H, Haider Z, Ullah A, Hayat Z, et al. Coupling CRISPR/Cas9 and lambda red recombineering system for genome editing of salmonella gallinarum and the effect of ssaU knock-out mutant on the virulence of bacteria. Biomedicines. 2022 Dec;10(12):3028. [cited 2023 Nov 17] https://www.mdpi.com/2227-9059/10/12/3028.10.3390/biomedicines10123028Search in Google Scholar PubMed PubMed Central
[155] Basit A, Tahir H, Haider Z, Tariq H, Ullah A, Rehman SU. CRISPR/Cas9-based deletion of SpvB gene from Salmonella gallinarum leads to loss of virulence in chicken. Front Bioeng Biotechnol. 2022 Jun 13;10:885227. [cited 2024 Apr 16]. https://www.frontiersin.org/articles/10.3389/fbioe.2022.885227/full.10.3389/fbioe.2022.885227Search in Google Scholar PubMed PubMed Central
[156] Hu D, Tang X, Ben Mamoun C, Wang C, Wang S, Gu X, et al. Efficient single-gene and gene family editing in the apicomplexan parasite Eimeria tenella using CRISPR-Cas9. Front Bioeng Biotechnol. 2020 Feb 25;8:128. [cited 2024 Apr 16]. https://www.frontiersin.org/article/10.3389/fbioe.2020.00128/full.10.3389/fbioe.2020.00128Search in Google Scholar PubMed PubMed Central
[157] Cheng P, Zhang Z, Yang F, Cai S, Wang L, Wang C, et al. FnCas12a/crRNA-mediated genome editing in Eimeria tenella. Front Genet. 2021 Sep 22;12:738746. [cited 2024 Apr 16]. https://www.frontiersin.org/articles/10.3389/fgene.2021.738746/full.10.3389/fgene.2021.738746Search in Google Scholar PubMed PubMed Central
[158] Chen L, Tang X, Sun P, Hu D, Zhang Y, Wang C, et al. Comparative transcriptome profiling of Eimeria tenella in various developmental stages and functional analysis of an ApiAP2 transcription factor exclusively expressed during sporogony. Parasit Vectors. 2023 Jul 19;16:241. [cited 2024 Oct 25]. https://pmc.ncbi.nlm.nih.gov/articles/PMC10354945/.10.1186/s13071-023-05828-8Search in Google Scholar PubMed PubMed Central
[159] Cheng Y, Lun M, Liu Y, Wang H, Yan Y, Sun J. CRISPR/Cas9-mediated chicken TBK1 gene knockout and its essential role in STING-mediated IFN-β induction in chicken cells. Front Immunol. 2019 Jan 4;9:3010. [cited 2024 Apr 16]. https://www.frontiersin.org/article/10.3389/fimmu.2018.03010/full.10.3389/fimmu.2018.03010Search in Google Scholar PubMed PubMed Central
[160] Lee KY, Choi HJ, Park KJ, Woo SJ, Kim YM, Han JY. Development and characterization of a CRISPR/Cas9-mediated RAG1 knockout chicken model lacking mature B and T cells. Front Immunol. 2022 Aug 11;13:892476. [cited 2024 Feb 16]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9403712/.10.3389/fimmu.2022.892476Search in Google Scholar PubMed PubMed Central
[161] Zeng Q, Cao J, Xie F, Zhu L, Wu X, Hu X, et al. CRISPR-Cas9-mediated chicken prmt5 gene knockout and its critical role in interferon regulation. Poult Sci. 2024 Mar;103(3):103344. [cited 2024 Apr 16]. https://linkinghub.elsevier.com/retrieve/pii/S0032579123008647.10.1016/j.psj.2023.103344Search in Google Scholar PubMed PubMed Central
[162] Kim TH, Kern C, Zhou H. Knockout of IRF7 highlights its modulator function of host response against avian influenza virus and the involvement of MAPK and TOR signaling pathways in chicken. Genes. 2020 Apr;11(4):385. [cited 2024 Apr 22]. https://www.mdpi.com/2073-4425/11/4/385.10.3390/genes11040385Search in Google Scholar PubMed PubMed Central
[163] Criado MF, Kassa A, Bertran K, Kwon JH, Sá e Silva M, Killmaster L, et al. Efficacy of multivalent recombinant herpesvirus of turkey vaccines against high pathogenicity avian influenza, infectious bursal disease, and Newcastle disease viruses. Vaccine. 2023 May 2;41(18):2893–904. [cited 2024 Mar 18] https://www.sciencedirect.com/science/article/pii/S0264410X23003493.10.1016/j.vaccine.2023.03.055Search in Google Scholar PubMed
[164] Abdelaziz K, Helmy YA, Yitbarek A, Hodgins DC, Sharafeldin TA, Selim MSH. Advances in poultry vaccines: leveraging biotechnology for improving vaccine development, stability, and delivery. Vaccines. 2024 Feb;12(2):134. [cited 2024 Oct 24] https://www.mdpi.com/2076-393X/12/2/134.10.3390/vaccines12020134Search in Google Scholar PubMed PubMed Central
[165] Suarez DL. DIVA vaccination strategies for avian influenza virus. Avian Dis. 2012;56(4):836–44, http://www.jstor.org/stable/23322209.10.1637/10207-041512-Review.1Search in Google Scholar PubMed
[166] Hein R, Koopman R, García M, Armour N, Dunn JR, Barbosa T, et al. Review of poultry recombinant vector vaccines. Avian Dis. 2021 Aug 26;65(3):438–52. [cited 2023 Nov 17] 10.1637/0005-2086-65.3.438.Search in Google Scholar PubMed
[167] Apinda N, Muenthaisong A, Chomjit P, Sangkakam K, Nambooppha B, Rittipornlertrak A, et al. Simultaneous protective immune responses of ducks against duck plague and fowl cholera by recombinant duck enteritis virus vector expressing Pasteurella multocida OmpH gene. Vaccines. 2022 Aug;10(8):1358. [cited 2023 Nov 17] https://www.mdpi.com/2076-393X/10/8/1358.10.3390/vaccines10081358Search in Google Scholar PubMed PubMed Central
[168] Tang N, Zhang Y, Sadigh Y, Moffat K, Shen Z, Nair V, et al. Generation of a triple insert live avian herpesvirus vectored vaccine using CRISPR/Cas9-based gene editing. Vaccines. 2020 Mar;8(1):97. [cited 2023 Nov 13] https://www.mdpi.com/2076-393X/8/1/97.10.3390/vaccines8010097Search in Google Scholar PubMed PubMed Central
[169] Okazaki W, Purchase HG, Burmester BR. Protection against Marek’s disease by vaccination with a herpesvirus of Turkeys. Avian Dis. 1970;14(2):413–29. [cited 2024 Jan 13] https://www.jstor.org/stable/1588488.10.2307/1588488Search in Google Scholar
[170] Afonso CL, Tulman ER, Lu Z, Zsak L, Rock DL, Kutish GF. The genome of Turkey herpesvirus. J Virol. 2001 Jan;75(2):971–8. [cited 2024 Jan 14] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC113993/.10.1128/JVI.75.2.971-978.2001Search in Google Scholar PubMed PubMed Central
[171] Sondermeijer PJA, Claessens JAJ, Jenniskens PE, Adrian Mockett AP, Thijssen RAJ, Willemse MJ, et al. Avian herpesvirus as a live viral vector for the expression of heterologous antigens. Vaccine. 1993 Jan;11(3):349–58. [cited 2024 Jan 13] https://linkinghub.elsevier.com/retrieve/pii/0264410X93901987.10.1016/0264-410X(93)90198-7Search in Google Scholar PubMed
[172] Palya V, Tatár-Kis T, Arafa ASA, Felföldi B, Mató T, Setta A. Efficacy of a Turkey herpesvirus vectored newcastle disease vaccine against genotype VII.1.1 virus: Challenge route affects shedding pattern. Vaccines. 2021 Jan 11;9(1):37. [cited 2024 Mar 18] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7826937/.10.3390/vaccines9010037Search in Google Scholar PubMed PubMed Central
[173] Gimeno IM, Cortes AL, Faiz N, Villalobos T, Badillo H, Barbosa T. Efficacy of various HVT vaccines (conventional and recombinant) against Marek’s disease in broiler chickens: Effect of dose and age of vaccination. Avian Dis. 2016;60(3):662–8. [cited 2024 Mar 18] https://www.jstor.org/stable/26431727.10.1637/11415-040116-Reg.1Search in Google Scholar PubMed
[174] Dunn JR, Dimitrov KM, Miller PJ, Garcia M, Turner-Alston K, Brown A, et al. Evaluation of protective efficacy when combining Turkey herpesvirus–vector vaccines. Avian Dis. 2018 Dec 17;63(1):75. [cited 2024 Jan 14] https://bioone.org/journals/avian-diseases/volume-63/issue-1/11979-092818-Reg.1/Evaluation-of-Protective-Efficacy-When-Combining-Turkey-HerpesvirusVector-Vaccines/10.1637/11979-092818-Reg.1.full.10.1637/11979-092818-Reg.1Search in Google Scholar PubMed
[175] Zai X, Shi B, Shao H, Qian K, Ye J, Yao Y, et al. Identification of a novel insertion site HVT-005/006 for the generation of recombinant Turkey herpesvirus vector. Front Microbiol. 2022 May 25;13:886873. [cited 2024 Mar 9] https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2022.886873/full.10.3389/fmicb.2022.886873Search in Google Scholar PubMed PubMed Central
[176] Chang P, Ameen F, Sealy JE, Sadeyen JR, Bhat S, Li Y, et al. Application of HDR-CRISPR/Cas9 and erythrocyte binding for rapid generation of recombinant Turkey herpesvirus-vectored avian influenza virus vaccines. Vaccines. 2019 Dec;7(4):192. [cited 2023 Nov 13] https://www.mdpi.com/2076-393X/7/4/192.10.3390/vaccines7040192Search in Google Scholar PubMed PubMed Central
[177] Tang N, Zhang Y, Pedrera M, Chang P, Baigent S, Moffat K, et al. A simple and rapid approach to develop recombinant avian herpesvirus vectored vaccines using CRISPR/Cas9 system. Vaccine. 2018 Jan 29;36(5):716–22. [cited 2023 Dec 4] https://www.sciencedirect.com/science/article/pii/S0264410X17317784.10.1016/j.vaccine.2017.12.025Search in Google Scholar PubMed PubMed Central
[178] Calderón K, Rojas-Neyra A, Carbajal-Lévano B, Luján-Valenzuela L, Ticona J, Isasi-Rivas G, et al. A recombinant Turkey herpesvirus expressing the F protein of newcastle disease virus genotype XII generated by NHEJ-CRISPR/Cas9 and Cre-LoxP systems confers protection against genotype XII challenge in chickens. Viruses. 2022 Apr 11;14(4):793. [cited 2024 Jan 10] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9030537/.10.3390/v14040793Search in Google Scholar PubMed PubMed Central
[179] Shi B, Yang G, Xiao Y, Qian K, Shao H, Xu M, et al. Long-term protection against virulent newcastle disease virus (NDV) in chickens immunized with a single dose of recombinant Turkey herpesvirus expressing NDV F protein. Vaccines. 2024 Jun;12(6):604. [cited 2024 Aug 28] https://www.mdpi.com/2076-393X/12/6/604.10.3390/vaccines12060604Search in Google Scholar PubMed PubMed Central
[180] Apinda N, Yao Y, Zhang Y, Muenthaisong A, Sangkakam K, Nambooppha B, et al. Efficiency of NHEJ-CRISPR/Cas9 and Cre-LoxP engineered recombinant Turkey herpesvirus expressing Pasteurella multocida OmpH protein for fowl cholera prevention in ducks. Vaccines. 2023 Sep;11(9):1498. [cited 2023 Nov 12] https://www.mdpi.com/2076-393X/11/9/1498.10.3390/vaccines11091498Search in Google Scholar PubMed PubMed Central
[181] Zhang JF, Park JY, Kim SW, Choi YR, Cha SY, Jang HK, et al. Development of a highly efficient CRISPR/Cas9-mediated herpesvirus of Turkey-based vaccine against novel variant infectious bursal disease virus. Vaccines. 2024 Mar;12(3):226. [cited 2024 Mar 20] https://www.mdpi.com/2076-393X/12/3/226.10.3390/vaccines12030226Search in Google Scholar PubMed PubMed Central
[182] Zhang JF, Kim SW, Shang K, Park JY, Choi YR, Jang HK, et al. Protection of chickens against H9N2 avian influenza isolates with a live vector vaccine expressing influenza hemagglutinin gene derived from Y280 avian influenza virus. Animals. 2024 Jan;14(6):872. [cited 2024 Apr 17] https://www.mdpi.com/2076-2615/14/6/872.10.3390/ani14060872Search in Google Scholar PubMed PubMed Central
[183] Zhang JF, Shang K, Kim SW, Park JY, Wei B, Jang HK, et al. Simultaneous construction strategy using two types of fluorescent markers for HVT vector vaccine against infectious bursal disease and H9N2 avian influenza virus by NHEJ-CRISPR/Cas9. Front Vet Sci. 2024 May 13;11:1385958. [cited 2024 Oct 25] https://pmc.ncbi.nlm.nih.gov/articles/PMC11135205/.10.3389/fvets.2024.1385958Search in Google Scholar PubMed PubMed Central
[184] Dhama K, Kumar N, Saminathan M, Tiwari R, Karthik K, Kumar MA, et al. Duck virus enteritis (duck plague) – a comprehensive update. Vet Q. 2017 Jan 1;37(1):57–80. [cited 2024 Jan 20] 10.1080/01652176.2017.1298885.Search in Google Scholar PubMed
[185] Liu J, Chen P, Jiang Y, Wu L, Zeng X, Tian G, et al. A duck enteritis virus-vectored bivalent live vaccine provides fast and complete protection against H5N1 avian influenza virus infection in ducks. J Virol. , 2011 Nov;85(21):10989–98. [cited 2024 Jan 20] https://journals.asm.org/doi/10.1128/jvi.05420-11.10.1128/JVI.05420-11Search in Google Scholar PubMed PubMed Central
[186] Chang P, Yao Y, Tang N, Sadeyen JR, Sealy J, Clements A, et al. The application of NHEJ-CRISPR/Cas9 and Cre-Lox system in the generation of bivalent duck enteritis virus vaccine against avian influenza virus. Viruses. 2018 Feb 13;10(2):81. [cited 2023 Nov 23] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5850388/.10.3390/v10020081Search in Google Scholar PubMed PubMed Central
[187] Zou Z, Huang K, Wei Y, Chen H, Liu Z, Jin M. Construction of a highly efficient CRISPR/Cas9-mediated duck enteritis virus-based vaccine against H5N1 avian influenza virus and duck Tembusu virus infection. Sci Rep. 2017 May 3;7:1478. [cited 2023 Nov 13] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5431151/.10.1038/s41598-017-01554-1Search in Google Scholar PubMed PubMed Central
[188] Li H, Wang Y, Han Z, Wang Y, Liang S, Jiang L, et al. Recombinant duck enteritis viruses expressing major structural proteins of the infectious bronchitis virus provide protection against infectious bronchitis in chickens. Antivir Res. 2016 Jun 1;130:19–26. [cited 2024 Jan 20] https://www.sciencedirect.com/science/article/pii/S0166354216301243.10.1016/j.antiviral.2016.03.003Search in Google Scholar PubMed PubMed Central
[189] Apinda N, Yao Y, Zhang Y, Reddy VRAP, Chang P, Nair V, et al. CRISPR/Cas9 editing of duck enteritis virus genome for the construction of a recombinant vaccine vector expressing ompH gene of pasteurella multocida in two novel insertion sites. Vaccines. 2022 May;10(5):686. [cited 2024 Apr 16] https://www.mdpi.com/2076-393X/10/5/686.10.3390/vaccines10050686Search in Google Scholar PubMed PubMed Central
[190] Ye J, Liang G, Zhang J, Wang W, Song N, Wang P, et al. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg Microbes Infect. 2016 Jan 1;5(1):1–12. [cited 2024 Jan 23] 10.1038/emi.2016.50.Search in Google Scholar PubMed PubMed Central
[191] Mu Y, Xie Q, Wang W, Lu H, Lian M, Gao W, et al. A novel fiber-1-edited and highly attenuated recombinant serotype 4 fowl adenovirus confers efficient protection against lethal challenge. Front Vet Sci. 2021 Nov 22;8:759418. [cited 2024 Apr 16] https://www.frontiersin.org/articles/10.3389/fvets.2021.759418/full.10.3389/fvets.2021.759418Search in Google Scholar PubMed PubMed Central
[192] Xie Q, Cao S, Zhang W, Wang W, Li L, Kan Q, et al. A novel fiber-2-edited live attenuated vaccine candidate against the highly pathogenic serotype 4 fowl adenovirus. Vet Res. 2021 Feb 27;52(1):35. [cited 2023 Nov 17] 10.1186/s13567-021-00907-z.Search in Google Scholar PubMed PubMed Central
[193] Xie Q, Wang W, Li L, Kan Q, Fu H, Geng T, et al. Domain in Fiber-2 interacted with KPNA3/4 significantly affects the replication and pathogenicity of the highly pathogenic FAdV-4. Virulence. 2021 Dec 31;12(1):754–65. [cited 2023 Nov 17] 10.1080/21505594.2021.1888458.Search in Google Scholar PubMed PubMed Central
[194] Xie Q, Wang W, Kan Q, Mu Y, Zhang W, Chen J, et al. FAdV-4 without fiber-2 is a highly attenuated and protective vaccine candidate. Microbiol Spectr. 2022 Feb 2;10(1):e01436–21. [cited 2024 Jan 23] https://journals.asm.org/doi/10.1128/spectrum.01436-21.10.1128/spectrum.01436-21Search in Google Scholar PubMed PubMed Central
[195] Lu H, Xie Q, Zhang W, Zhang J, Wang W, Lian M, et al. A novel recombinant FAdV-4 virus with fiber of FAdV-8b provides efficient protection against both FAdV-4 and FAdV-8b. Viruses. 2022 Feb;14(2):376. [cited 2024 Jan 23] https://www.mdpi.com/1999-4915/14/2/376.10.3390/v14020376Search in Google Scholar PubMed PubMed Central
[196] Guo Y, Xu Z, Chao Y, Cao X, Jiang H, Li H, et al. An efficient double-fluorescence approach for generating fiber-2-edited recombinant serotype 4 fowl adenovirus expressing foreign gene. Front Microbiol. 2023 Mar 31;14:1160031. [cited 2024 Apr 16] https://www.frontiersin.org/articles/10.3389/fmicb.2023.1160031/full.10.3389/fmicb.2023.1160031Search in Google Scholar PubMed PubMed Central
[197] Zhang S, Tang X, Wang S, Shi F, Duan C, Bi F, et al. Establishment of recombinant eimeria acervulina expressing multi-copies M2e derived from avian influenza virus H9N2. Vaccines. 2021 Jul;9(7):791. [cited 2024 Feb 17] https://www.mdpi.com/2076-393X/9/7/791.10.3390/vaccines9070791Search in Google Scholar PubMed PubMed Central
[198] Atasoy MO, Rohaim MA, Munir M. Simultaneous deletion of virulence factors and insertion of antigens into the infectious laryngotracheitis virus using NHEJ-CRISPR/Cas9 and Cre–Lox system for construction of a stable vaccine vector. Vaccines. 2019 Dec 5;7(4):207. [cited 2023 Nov 13] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6963826/.10.3390/vaccines7040207Search in Google Scholar PubMed PubMed Central
[199] Najafi S, Tan SC, Aghamiri S, Raee P, Ebrahimi Z, Jahromi ZK, et al. Therapeutic potentials of CRISPR-Cas genome editing technology in human viral infections. Biomed Pharmacother. 2022 Apr;148:112743. [cited 2024 Mar 13]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8872819/.10.1016/j.biopha.2022.112743Search in Google Scholar PubMed PubMed Central
[200] Li K, Liu Y, Xu Z, Zhang Y, Yao Y, Nair V, et al. Prevention of avian retrovirus infection in chickens using CRISPR-Cas9 delivered by Marek’s disease virus. Mol Ther - Nucleic Acids. 2020 Sep 4;21:343–53. [cited 2023 Dec 4] https://www.cell.com/molecular-therapy-family/nucleic-acids/abstract/S2162-2531(20)30170-0.10.1016/j.omtn.2020.06.009Search in Google Scholar PubMed PubMed Central
[201] Challagulla A, Schat KA, Doran TJ. In vitro inhibition of influenza virus using CRISPR/Cas13a in chicken cells. Methods Protoc. 2021 Jun;4(2):40. [cited 2023 Nov 17] https://www.mdpi.com/2409-9279/4/2/40.10.3390/mps4020040Search in Google Scholar PubMed PubMed Central
[202] Challagulla A, Jenkins KA, O’Neil TE, Shi S, Morris KR, Wise TG, et al. In vivo inhibition of Marek’s disease virus in transgenic chickens expressing Cas9 and gRNA against ICP4. Microorganisms. 2021 Jan;9(1):164. [cited 2023 Dec 3] https://www.mdpi.com/2076-2607/9/1/164.10.3390/microorganisms9010164Search in Google Scholar PubMed PubMed Central
[203] Mohsin M, Li Y, Zhang X, Wang Y, Huang Z, Yin G, et al. Development of CRISPR-CAS9 based RNA drugs against Eimeria tenella infection. Genomics. 2021 Nov 1;113(6):4126–35. [cited 2024 Feb 17] https://www.sciencedirect.com/science/article/pii/S0888754321003827.10.1016/j.ygeno.2021.10.019Search in Google Scholar PubMed
[204] Liu Y, Xu Z, Zhang Y, Yu M, Wang S, Gao Y, et al. Marek’s disease virus as a CRISPR/Cas9 delivery system to defend against avian leukosis virus infection in chickens. Vet Microbiol. 2020 Mar 1;242:108589. [cited 2023 Nov 12] https://www.sciencedirect.com/science/article/pii/S0378113519312891.10.1016/j.vetmic.2020.108589Search in Google Scholar PubMed
[205] Burdo TH, Chen C, Kaminski R, Sariyer IK, Mancuso P, Donadoni M, et al. Preclinical safety and biodistribution of CRISPR targeting SIV in non-human primates. Gene Ther. 2024 May;31(5):224–33. [cited 2025 Jan 3] https://www.nature.com/articles/s41434-023-00410-4.10.1038/s41434-023-00410-4Search in Google Scholar PubMed PubMed Central
[206] Ipoutcha T, Rideau F, Gourgues G, Arfi Y, Lartigue C, Blanchard A, et al. Genome editing of veterinary relevant mycoplasmas using a CRISPR-Cas base editor system. Dozois CM, editor. Appl Environ Microbiol. 2022 Sep 13;88(17):e00996–22. [cited 2024 Apr 16] https://journals.asm.org/doi/10.1128/aem.00996-22.10.1128/aem.00996-22Search in Google Scholar PubMed PubMed Central
[207] Bai X, Plastow GS. Breeding for disease resilience: Opportunities to manage polymicrobial challenge and improve commercial performance in the pig industry. Cabi Agric Biosci. 2022;3(1):6. [cited 2024 Apr 20] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8761052/.10.1186/s43170-022-00073-ySearch in Google Scholar PubMed PubMed Central
[208] Albers GA, Gray GD, Piper LR, Barker JS, Le Jambre LF, Barger IA. The genetics of resistance and resilience to Haemonchus contortus infection in young merino sheep. Int J Parasitol. 1987 Oct;17(7):1355–63.10.1016/0020-7519(87)90103-2Search in Google Scholar PubMed
[209] Råberg L, Graham AL, Read AF. Decomposing health: tolerance and resistance to parasites in animals. Philos Trans R Soc Lond B Biol Sci. 2009 Jan 12;364(1513):37–49.10.1098/rstb.2008.0184Search in Google Scholar PubMed PubMed Central
[210] Saxena VK, Kolluri G. Selection methods in poultry breeding: From genetics to genomics. In: Liu X, editor. Application of genetics and genomics in poultry science. London, United Kingdom: InTech; 2018. [cited 2024 Apr 16]. http://www.intechopen.com/books/application-of-genetics-and-genomics-in-poultry-science/selection-methods-in-poultry-breeding-from-genetics-to-genomics.10.5772/intechopen.77966Search in Google Scholar
[211] Clark J, Whitelaw B. A future for transgenic livestock. Nat Rev Genet. 2003 Oct;4(10):825–33. [cited 2024 Mar 5]. https://www.nature.com/articles/nrg1183.10.1038/nrg1183Search in Google Scholar PubMed PubMed Central
[212] Bishop TF, Van Eenennaam AL, Dickinson MH, Vosshall LB, Dow JAT. Genome editing approaches to augment livestock breeding programs. J Exp Biol. 2020 Feb 1;223(Suppl_1):jeb207159. [cited 2025 Apr 16] https://journals.biologists.com/jeb/article/223/Suppl_1/jeb207159/224598/Genome-editing-approaches-to-augment-livestock.10.1242/jeb.207159Search in Google Scholar PubMed
[213] Ballantyne M, Doddamani D J, McGrew M. The use of genome editing in poultry breeding. In Advances in poultry genetics and genomics. Cambridge, United Kingdom: Burleigh Dodds Science Publishing; 2020. p. 523–40.10.19103/AS.2020.0065.29Search in Google Scholar
[214] Hickey Jm, Bruce C, Whitelaw A, Gorjanc G. Promotion of alleles by genome editing in livestock breeding programmes. J Anim Breed Genet. 2016;133(2):83–4. [cited 2024 Mar 5] https://onlinelibrary.wiley.com/doi/abs/10.1111/jbg.12206.10.1111/jbg.12206Search in Google Scholar PubMed
[215] Tyack SG, Jenkins KA, O’Neil TE, Wise TG, Morris KR, Bruce MP, et al. A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells. Transgenic Res. 2013 Dec;22(6):1257–64. [cited 2024 Jan 1] http://link.springer.com/10.1007/s11248-013-9727-2.10.1007/s11248-013-9727-2Search in Google Scholar PubMed
[216] Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005 Jul 18;170(2):317–25. [cited 2024 Feb 13] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2171413/.10.1083/jcb.200503059Search in Google Scholar PubMed PubMed Central
[217] Schusser B, Doran T. Advances in genetic engineering of the avian genome. In Avian immunology. 3rd edn. London: Academic Press; 2022. p. 559–72.10.1016/B978-0-12-818708-1.00022-1Search in Google Scholar
[218] Wells KD, Bardot R, Whitworth KM, Trible BR, Fang Y, Mileham A, et al. Replacement of porcine CD163 scavenger receptor cysteine-rich domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome virus. J Virol. 2017 Jan 3;91(2):e01521–16. [cited 2025 Jan 4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5215333/.10.1128/JVI.01521-16Search in Google Scholar PubMed PubMed Central
[219] Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, et al. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017 Feb 23;13(2):e1006206. [cited 2025 Jan 4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5322883/.10.1371/journal.ppat.1006206Search in Google Scholar PubMed PubMed Central
[220] Yang H, Zhang J, Zhang X, Shi J, Pan Y, Zhou R, et al. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antivir Res. 2018 Mar 1;151:63–70. [cited 2025 Jan 4] https://www.sciencedirect.com/science/article/abs/pii/S0166354217307337.10.1016/j.antiviral.2018.01.004Search in Google Scholar PubMed
[221] Guo C, Wang M, Zhu Z, He S, Liu H, Liu X, et al. Highly efficient generation of pigs harboring a partial deletion of the CD163 SRCR5 domain, which are fully resistant to porcine reproductive and respiratory syndrome virus 2 infection. Front Immunol. 2019 Aug 8;10:1846. [cited 2025 Jan 4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6694839/.10.3389/fimmu.2019.01846Search in Google Scholar PubMed PubMed Central
[222] Renken C, Nathues C, Swam H, Fiebig K, Weiss C, Eddicks M, et al. Application of an economic calculator to determine the cost of porcine reproductive and respiratory syndrome at farm-level in 21 pig herds in Germany. Porc Health Manag. 2021 Jan 4;7(1):3. [cited 2025 Jan 4] 10.1186/s40813-020-00183-x.Search in Google Scholar PubMed PubMed Central
[223] Meléndez-Arce R, Vargas-Leitón B, Steeneveld W, Van Nes A, Stegeman JA, Romero- Zuñiga JJ. Stochastic model to assess bioeconomic impact of PRRS on pig farms in Costa Rica. Prev Vet Med. 2023 Nov;220:106032. [cited 2025 Jan 4] https://linkinghub.elsevier.com/retrieve/pii/S0167587723001964.10.1016/j.prevetmed.2023.106032Search in Google Scholar PubMed
[224] Burger BT, Beaton BP, Campbell MA, Brett BT, Rohrer MS, Plummer S, et al. Generation of a commercial-scale founder population of porcine reproductive and respiratory syndrome virus resistant pigs using CRISPR-Cas. CRISPR J. 2024 Feb;7(1):12–28. [cited 2025 Jan 4] https://www.liebertpub.com/doi/10.1089/crispr.2023.0061.10.1089/crispr.2023.0061.correxSearch in Google Scholar PubMed
[225] Nair V. Neoplastic diseases. In: Swayne DE, Boulianne M, Logue CM, McDougald LR, Nair V, Suarez DL, editors. Diseases of poultry. 14th edn. Hoboken, New Jersey, USA: Wiley-Blackwell; 2020. p. 587–625.Search in Google Scholar
[226] Payne LN, Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012 Feb 1;41(1):11–9. [cited 2024 Feb 6] 10.1080/03079457.2011.646237.Search in Google Scholar PubMed
[227] Lee HJ, Lee KY, Park YH, Choi HJ, Yao Y, Nair V, et al. Acquisition of resistance to avian leukosis virus subgroup B through mutations on tvb cysteine-rich domains in DF-1 chicken fibroblasts. Vet Res. 2017 Sep 13;48(1):48. [cited 2023 Nov 29] 10.1186/s13567-017-0454-1.Search in Google Scholar PubMed PubMed Central
[228] Lee HJ, Lee KY, Jung KM, Park KJ, Lee KO, Suh JY, et al. Precise gene editing of chicken Na+/H+ exchange type 1 (chNHE1) confers resistance to avian leukosis virus subgroup J (ALV-J). Dev Comp Immunol. 2017 Dec 1;77:340–9. [cited 2023 Nov 29] https://www.sciencedirect.com/science/article/pii/S0145305X17304329.10.1016/j.dci.2017.09.006Search in Google Scholar PubMed
[229] Lee HJ, Park KJ, Lee KY, Yao Y, Nair V, Han JY. Sequential disruption of ALV host receptor genes reveals no sharing of receptors between ALV subgroups A, B, and J. J Anim Sci Biotechnol. 2019 Apr 2;10(1):23. [cited 2023 Nov 29] 10.1186/s40104-019-0333-x.Search in Google Scholar PubMed PubMed Central
[230] Kučerová D, Plachý J, Reinišová M, Šenigl F, Trejbalová K, Geryk J, et al. Nonconserved tryptophan 38 of the cell surface receptor for subgroup J avian leukosis virus discriminates sensitive from resistant avian species. J Virol. 2013 Aug;87(15):8399–407. [cited 2023 Dec 7] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3719790/.10.1128/JVI.03180-12Search in Google Scholar PubMed PubMed Central
[231] Koslová A, Kučerová D, Reinišová M, Geryk J, Trefil P, Hejnar J. Genetic resistance to avian leukosis viruses induced by CRISPR/Cas9 editing of specific receptor genes in chicken cells. Viruses. 2018 Nov;10(11):605. [cited 2023 Dec 6] https://www.mdpi.com/1999-4915/10/11/605.10.3390/v10110605Search in Google Scholar PubMed PubMed Central
[232] Barber MRW, Aldridge JR, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):5913–8. [cited 2024 Jan 4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2851864/.10.1073/pnas.1001755107Search in Google Scholar PubMed PubMed Central
[233] Ichikawa K, Motoe Y, Ezaki R, Matsuzaki M, Horiuchi H. Knock-in of the duck retinoic acid-inducible gene I (RIG-I) into the Mx gene in DF-1 cells enables both stable and immune response-dependent RIG-I expression. Biochem Biophys Rep. 2021 Sep 1;27:101084. [cited 2024 Feb 18] https://www.sciencedirect.com/science/article/pii/S2405580821001783.10.1016/j.bbrep.2021.101084Search in Google Scholar PubMed PubMed Central
[234] Woo SJ, Choi HJ, Park YH, Rengaraj D, Kim JK, Han JY. Amplification of immunity by engineering chicken MDA5 combined with the C terminal domain (CTD) of RIG-I. Appl Microbiol Biotechnol. 2022 Feb 1;106(4):1599–613. [cited 2024 Feb 18] 10.1007/s00253-022-11806-4.Search in Google Scholar PubMed
[235] Kang KS, Lee HC, Kim HJ, Lee HG, Kim YM, Lee HJ, et al. Spatial and temporal action of chicken primordial germ cells during initial migration. Reproduction. 2015 Feb 1;149(2):179–87. [cited 2024 Feb 13] https://rep.bioscientifica.com/view/journals/rep/149/2/179.xml.10.1530/REP-14-0433Search in Google Scholar PubMed
[236] Hu T, Taylor L, Sherman A, Keambou Tiambo C, Kemp SJ, Whitelaw B, et al. A low-tech, cost-effective and efficient method for safeguarding genetic diversity by direct cryopreservation of poultry embryonic reproductive cells. eLife. 2022;11:e74036. [cited 2024 Feb 14] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8789256/.10.7554/eLife.74036Search in Google Scholar PubMed PubMed Central
[237] Li BC, Chen GH, Qin J, Wu XS, Wu SL, Cai ZT. Suitable stages for isolation and culture PGCs from chicken embryos. Int J Poult Sci. 2005 Oct 15;4(11):885–90. [cited 2024 Jan 1] https://www.scialert.net/abstract/?doi=ijps.2005.885.890.10.3923/ijps.2005.885.890Search in Google Scholar
[238] Whyte J, Glover JD, Woodcock M, Brzeszczynska J, Taylor L, Sherman A, et al. FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Rep. 2015 Nov 19;5(6):1171–82. [cited 2024 Jan 1] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4682126/.10.1016/j.stemcr.2015.10.008Search in Google Scholar PubMed PubMed Central
[239] Song Y, Duraisamy S, Ali J, Kizhakkayil J, Jacob VD, Mohammed MA, et al. Characteristics of long-term cultures of avian primordial germ cells and gonocytes1. Biol Reprod. 2014 Jan 1;90(1):15. 1–8. [cited 2024 Feb 13] 10.1095/biolreprod.113.113381.Search in Google Scholar PubMed
[240] Ezaki R, Ichikawa K, Matsuzaki M, Horiuchi H. Targeted knock-in of a fluorescent protein gene into the chicken vasa homolog locus of chicken primordial germ cells using CRIS-PITCh method. J Poult Sci. 2022 Apr 25;59(2):182–90. [cited 2024 Jan 1] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9039151/.10.2141/jpsa.0210067Search in Google Scholar PubMed PubMed Central
[241] Schusser B, Collarini EJ, Yi H, Izquierdo SM, Fesler J, Pedersen D, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc Natl Acad Sci. 2013 Dec 10;110(50):20170–5. [cited 2023 Dec 8] https://www.pnas.org/doi/full/10.1073/pnas.1317106110.10.1073/pnas.1317106110Search in Google Scholar PubMed PubMed Central
[242] Sid H, Schusser B. Applications of gene editing in chickens: A new era is on the horizon. Front Genet. 2018;9:456. [cited 2023 Nov 15] https://www.frontiersin.org/articles/10.3389/fgene.2018.00456.10.3389/fgene.2018.00456Search in Google Scholar PubMed PubMed Central
[243] Han JY, Lee HJ. Genome editing mediated by primordial germ cell in chicken. In: Hatada I, editor. Genome editing in animals: Methods and protocols. New York, NY: Humana Press; 2017. p. 301–12. (Methods in Molecular Biology).10.1007/978-1-0716-3016-7_23Search in Google Scholar PubMed
[244] Ballantyne M, Woodcock M, Doddamani D, Hu T, Taylor L, Hawken RJ, et al. Direct allele introgression into pure chicken breeds using Sire Dam Surrogate (SDS) mating. Nat Commun. 2021 Jan 28;12(1):659. [cited 2024 Feb 17] https://www.nature.com/articles/s41467-020-20812-x.10.1038/s41467-020-20812-xSearch in Google Scholar PubMed PubMed Central
[245] Challagulla A, Jenkins KA, O’Neil TE, Morris KR, Wise TG, Tizard ML, et al. Germline engineering of the chicken genome using CRISPR/Cas9 by in vivo transfection of PGCs. Anim Biotechnol. 2020;34(4):775–84. [cited 2024 Feb 13] https://www.tandfonline.com/doi/full/10.1080/10495398.2020.1789869.10.1080/10495398.2020.1789869Search in Google Scholar PubMed
[246] Cooper CA, Challagulla A, Jenkins KA, Wise TG, O’Neil TE, Morris KR, et al. Generation of gene edited birds in one generation using sperm transfection assisted gene editing (STAGE). Transgenic Res. 2017 Jun 1;26(3):331–47. [cited 2024 Feb 3] 10.1007/s11248-016-0003-0.Search in Google Scholar PubMed
[247] Lee J, Kim DH, Karolak MC, Shin S, Lee K. Generation of genome-edited chicken and duck lines by adenovirus-mediated in vivo genome editing. Proc Natl Acad Sci. 2022 Nov 8;119(45):e2214344119. [cited 2023 Dec 6] https://www.pnas.org/doi/full/10.1073/pnas.2214344119.10.1073/pnas.2214344119Search in Google Scholar PubMed PubMed Central
[248] Mizushima S, Sasanami T, Ono T, Kuroiwa A. Current approaches to and the application of intracytoplasmic sperm injection (ICSI) for avian genome editing. Genes. 2023 Mar 20;14(3):757. [cited 2024 Mar 5] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10048369/.10.3390/genes14030757Search in Google Scholar PubMed PubMed Central
[249] Mizushima S, Sasanami T, Ono T, Matsuzaki M, Kansaku N, Kuroiwa A. Cyclin D1 gene expression is essential for cell cycle progression from the maternal-to-zygotic transition during blastoderm development in Japanese quail. Dev Biol. 2021 Aug 1;476:249–58. [cited 2024 Mar 8] https://www.sciencedirect.com/science/article/pii/S0012160621001007.10.1016/j.ydbio.2021.04.005Search in Google Scholar PubMed
[250] Hellmich R, Sid H, Lengyel K, Flisikowski K, Schlickenrieder A, Bartsch D, et al. Acquiring resistance against a retroviral infection via CRISPR/Cas9 targeted genome editing in a commercial chicken line. Front Genome Ed. 2020 May 28;2:3. [cited 2024 Apr 16] https://www.frontiersin.org/article/10.3389/fgeed.2020.00003/full.10.3389/fgeed.2020.00003Search in Google Scholar PubMed PubMed Central
[251] Kheimar A, Klinger R, Bertzbach LD, Sid H, Yu Y, Conradie AM, et al. A genetically engineered commercial chicken line is resistant to highly pathogenic avian leukosis virus subgroup J. Microorganisms. 2021 May;9(5):1066. [cited 2023 Nov 17] https://www.mdpi.com/2076-2607/9/5/1066.10.3390/microorganisms9051066Search in Google Scholar PubMed PubMed Central
[252] Matoušková M, Plachý J, Kučerová D, Pecnová Ľ, Reinišová M, Geryk J, et al. Rapid adaptive evolution of avian leukosis virus subgroup J in response to biotechnologically induced host resistance. PLOS Pathog. 2024 Aug 15;20(8):e1012468. [cited 2024 Oct 25] https://pmc.ncbi.nlm.nih.gov/articles/PMC11349186/.10.1371/journal.ppat.1012468Search in Google Scholar PubMed PubMed Central
[253] Koslová A, Trefil P, Mucksová J, Krchlíková V, Plachý J, Krijt J, et al. Knock-out of retrovirus receptor gene tva in the chicken confers resistance to avian leukosis virus subgroups A and K and affects cobalamin (Vitamin B12)-dependent level of methylmalonic acid. Viruses. 2021 Dec;13(12):2504. [cited 2023 Nov 17] https://www.mdpi.com/1999-4915/13/12/2504.10.3390/v13122504Search in Google Scholar PubMed PubMed Central
[254] Richard M, de Graaf M, Herfst S. Avian influenza A viruses: From zoonosis to pandemic. Future Virol. 2014 May 1;9(5):513–24. [cited 2024 Mar 16] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4157675/.10.2217/fvl.14.30Search in Google Scholar PubMed PubMed Central
[255] Nielsen SS, Alvarez J, Bicout DJ, Calistri P, Canali E, Drewe JA, et al. Vaccination of poultry against highly pathogenic avian influenza – part 1. Available vaccines and vaccination strategies. EFSA J. 2023 Oct 10;21(10):e08271. [cited 2024 Jan 4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10563699/.10.2903/j.efsa.2023.p211001Search in Google Scholar PubMed PubMed Central
[256] Looi FY, Baker ML, Townson T, Richard M, Novak B, Doran TJ, et al. Creating disease resistant chickens: A viable solution to avian influenza? Viruses. 2018 Oct;10(10):561. [cited 2023 Nov 17] https://www.mdpi.com/1999-4915/10/10/561.10.3390/v10100561Search in Google Scholar PubMed PubMed Central
[257] Wandzik JM, Kouba T, Cusack S. Structure and function of influenza polymerase. Cold Spring Harb Perspect Med. 2021 Sep;11(9):a038372. [cited 2024 Jan 18] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8415296/.10.1101/cshperspect.a038372Search in Google Scholar PubMed PubMed Central
[258] Long JS, Idoko-Akoh A, Mistry B, Goldhill DH, Staller E, Schreyer J, et al. Avian ANP32B does not support influenza A virus polymerase and influenza A virus relies exclusively on ANP32A in chicken cells. Microbiology. 2019 Jan. [cited 2024 Jan 4] http://biorxiv.org/lookup/doi/10.1101/512012.10.1101/512012Search in Google Scholar
[259] Zhang H, Zhang Z, Wang Y, Wang M, Wang X, Zhang X, et al. Fundamental contribution and host range determination of ANP32A and ANP32B in influenza A virus polymerase activity. J Virol. 2019 Jun 14;93(13):e00174–19. [cited 2023 Nov 9] 10.1128/jvi.00174-19. https://journals.asm.org/doi/full/10.1128/jvi.00174-19.Search in Google Scholar
[260] Paull J. The failures of genetically modified organisms (GMOS): Resistance, regulation, and rejection. Agrofor. 2019 Oct 10;4(3):139–52. [cited 2025 Feb 11] http://doisrpska.nub.rs/index.php/AGR/article/view/7635.10.7251/AGRENG1903139PSearch in Google Scholar
[261] Marris C. Public views on GMOs: Deconstructing the myths. EMBO Rep. 2001 Jul 7;2(7):545–8. [cited 2025 Feb 11] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1083956/.10.1093/embo-reports/kve142Search in Google Scholar PubMed PubMed Central
[262] Ishii T. Genome-edited livestock: Ethics and social acceptance. Anim Front. 2017 Apr 1;7(2):24–32. [cited 2025 Feb 11] 10.2527/af.2017.0115.Search in Google Scholar
[263] Nuffield Council on Bioethics. Genome editing and farmed animal breeding: Social and ethical issues. London, United Kingdom: Nuffield Council on Bioethics; 2021 Dec. https://cdn.nuffieldbioethics.org/wp-content/uploads/Genome-editing-and-farmed-animal-breeding-FINAL-WEB-PDF.pdf.Search in Google Scholar
[264] Nuffield Council on Bioethics. Public dialogue on genome editing in farmed animals. London, United Kingdom: NCOB and BBSRC; 2022. p. 102. https://cdn.nuffieldbioethics.org/wp-content/uploads/NCOB-BBSRC-Sciencewise-Genome-editing-and-farmed-animals-dialogue-Oct-2022.pdf.Search in Google Scholar
[265] U.S. Food and Drug Administration. FDA. FDA; 2020 [cited 2025 Feb 11]. FDA approves first-of-its-kind intentional genomic alteration in line of domestic pigs for both human food, potential therapeutic uses. https://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-intentional-genomic-alteration-line-domestic-pigs-both-human-food. Search in Google Scholar
[266] U.S. Food and Drug Administration. FDA. FDA; 2024 [cited 2025 Feb 11]. AquAdvantage Salmon. https://www.fda.gov/animal-veterinary/intentional-genomic-alterations-igas-animals/aquadvantage-salmon. Search in Google Scholar
[267] Genetic Literacy Project K. European Union: Animals [Internet]. Global gene editing regulation tracker. 2024 [cited 2024 Mar 24]. https://crispr-gene-editing-regs-tracker.geneticliteracyproject.org/european-union-animals/. Search in Google Scholar
[268] Genetic Literacy Project. Brazil: Animals [Internet]. Global gene editing regulation tracker. 2024 [cited 2025 Feb 11]. https://crispr-gene-editing-regs-tracker.geneticliteracyproject.org/brazil-animals/. Search in Google Scholar
[269] Genetic Literacy Project. Argentina: Animals [Internet]. Global gene editing regulation tracker. 2024 [cited 2025 Feb 11]. https://crispr-gene-editing-regs-tracker.geneticliteracyproject.org/argentina-animals/. Search in Google Scholar
[270] Halmi MFA, Zulkifli MAF, Zaman KHK. CRISPR-Cas9 genome editing: A brief scientometric insight on scientific production and knowledge structure. J Scientometr Res. 2024 Dec 26;12(2):395–403 [cited 2024 Dec 31] https://jscires.org/full-text/6568/.10.5530/jscires.12.2.035Search in Google Scholar
[271] Liu H, Lv Z, Zhang G, Wang X, Wang Y, Wang K. Knowledge mapping and current trends of global research on CRISPR in the field of cancer. Front Cell Dev Biol. 2023 May 2;11:1178221. [cited 2024 Dec 31] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10185797/.10.3389/fcell.2023.1178221Search in Google Scholar PubMed PubMed Central
[272] Genetic Literacy Project K. China: Animals [Internet]. Global gene editing regulation tracker. 2024 [cited 2024 Mar 24]. https://crispr-gene-editing-regs-tracker.geneticliteracyproject.org/china-animals/.Search in Google Scholar
[273] Lim D, Choi I. Global trends in regulatory frameworks for animal genome editing in agriculture. J Anim Reprod Biotechnol. 2023 Dec 31;38(4):247–53. [cited 2024 Mar 24] https://www.e-jarb.org/journal/view.html?doi=10.12750/JARB.38.4.247.10.12750/JARB.38.4.247Search in Google Scholar
[274] European Parliament. Directorate General for Parliamentary Research Services. European Parliament. 2024 [cited 2024 Mar 25]. New Genomic Techniques: MEPs back rules to support green transition of farmers. https://www.europarl.europa.eu/news/en/press-room/20240202IPR17320/new-genomic-techniques-meps-back-rules-to-support-green-transition-of-farmers.Search in Google Scholar
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Safety assessment and modulation of hepatic CYP3A4 and UGT enzymes by Glycyrrhiza glabra aqueous extract in female Sprague–Dawley rats
- Adult-onset Still’s disease with hemophagocytic lymphohistiocytosis and minimal change disease
- Role of DZ2002 in reducing corneal graft rejection in rats by influencing Th17 activation via inhibition of the PI3K/AKT pathway and downregulation of TRAF1
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Genetic variants in VWF exon 26 and their implications for type 1 Von Willebrand disease in a Saudi Arabian population
- Lipoxin A4 improves myocardial ischemia/reperfusion injury through the Notch1-Nrf2 signaling pathway
- High levels of EPHB2 expression predict a poor prognosis and promote tumor progression in endometrial cancer
- Knockdown of SHP-2 delays renal tubular epithelial cell injury in diabetic nephropathy by inhibiting NLRP3 inflammasome-mediated pyroptosis
- Exploring the toxicity mechanisms and detoxification methods of Rhizoma Paridis
- Concomitant gastric carcinoma and primary hepatic angiosarcoma in a patient: A case report
- YAP1 inhibition protects retinal vascular endothelial cells under high glucose by inhibiting autophagy
- Identification of secretory protein related biomarkers for primary biliary cholangitis based on machine learning and experimental validation
- Integrated genomic and clinical modeling for prognostic assessment of radiotherapy response in rectal neoplasms
- Stem cell-based approaches for glaucoma treatment: a mini review
- Bacteriophage titering by optical density means: KOTE assays
- Neutrophil-related signature characterizes immune landscape and predicts prognosis of esophageal squamous cell carcinoma
- Integrated bioinformatic analysis and machine learning strategies to identify new potential immune biomarkers for Alzheimer’s disease and their targeting prediction with geniposide
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- Histone modification and non-coding RNAs in skin aging: emerging therapeutic avenues
- A multiplicative behavioral model of DNA replication initiation in cells
- Biogenic gold nanoparticles synthesized from Pergularia daemia leaves: a novel approach for nasopharyngeal carcinoma therapy
- Creutzfeldt-Jakob disease mimicking Hashimoto’s encephalopathy: steroid response followed by decline
- Impact of semaphorin, Sema3F, on the gene transcription and protein expression of CREB and its binding protein CREBBP in primary hippocampal neurons of rats
- Iron overloaded M0 macrophages regulate hematopoietic stem cell proliferation and senescence via the Nrf2/Keap1/HO-1 pathway
- Revisiting the link between NADPH oxidase p22phox C242T polymorphism and ischemic stroke risk: an updated meta-analysis
- Exercise training preferentially modulates α1D-adrenergic receptor expression in peripheral arteries of hypertensive rats
- Overexpression of HE4/WFDC2 gene in mice leads to keratitis and corneal opacity
- Tumoral calcinosis complicating CKD-MBD in hemodialysis: a case report
- Mechanism of KLF4 Inhibition of epithelial-mesenchymal transition in gastric cancer cells
- Dissecting the molecular mechanisms of T cell infiltration in psoriatic lesions via cell-cell communication and regulatory network analysis
- Circadian rhythm-based prognostic features predict immune infiltration and tumor microenvironment in molecular subtypes of hepatocellular carcinoma
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Molecular identification and control studies on Coridius sp. (Hemiptera: Dinidoridae) in Al-Khamra, south of Jeddah, Saudi Arabia
- 10.1515/biol-2025-1218
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Response of hybrid grapes (Vitis spp.) to two biotic stress factors and their seedlessness status
- Metabolomic profiling reveals systemic metabolic reprogramming in Alternaria alternata under salt stress
- Effects of mixed salinity and alkali stress on photosynthetic characteristics and PEPC gene expression of vegetable soybean seedlings
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Causal effects of trace elements on congenital foot deformities and their subtypes: a Mendelian randomization study with gut microbiota mediation
- Honey meets acidity: a novel biopreservative approach against foodborne pathogens
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Fluorescent detection of sialic acid–binding lectins using functionalized quantum dots in ELISA format
- Smart tectorigenin-loaded ZnO hydrogel nanocomposites for targeted wound healing: synthesis, characterization, and biological evaluation
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
- Retraction
- Retraction of “Down-regulation of miR-539 indicates poor prognosis in patients with pancreatic cancer”
Articles in the same Issue
- Safety assessment and modulation of hepatic CYP3A4 and UGT enzymes by Glycyrrhiza glabra aqueous extract in female Sprague–Dawley rats
- Adult-onset Still’s disease with hemophagocytic lymphohistiocytosis and minimal change disease
- Role of DZ2002 in reducing corneal graft rejection in rats by influencing Th17 activation via inhibition of the PI3K/AKT pathway and downregulation of TRAF1
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Genetic variants in VWF exon 26 and their implications for type 1 Von Willebrand disease in a Saudi Arabian population
- Lipoxin A4 improves myocardial ischemia/reperfusion injury through the Notch1-Nrf2 signaling pathway
- High levels of EPHB2 expression predict a poor prognosis and promote tumor progression in endometrial cancer
- Knockdown of SHP-2 delays renal tubular epithelial cell injury in diabetic nephropathy by inhibiting NLRP3 inflammasome-mediated pyroptosis
- Exploring the toxicity mechanisms and detoxification methods of Rhizoma Paridis
- Concomitant gastric carcinoma and primary hepatic angiosarcoma in a patient: A case report
- YAP1 inhibition protects retinal vascular endothelial cells under high glucose by inhibiting autophagy
- Identification of secretory protein related biomarkers for primary biliary cholangitis based on machine learning and experimental validation
- Integrated genomic and clinical modeling for prognostic assessment of radiotherapy response in rectal neoplasms
- Stem cell-based approaches for glaucoma treatment: a mini review
- Bacteriophage titering by optical density means: KOTE assays
- Neutrophil-related signature characterizes immune landscape and predicts prognosis of esophageal squamous cell carcinoma
- Integrated bioinformatic analysis and machine learning strategies to identify new potential immune biomarkers for Alzheimer’s disease and their targeting prediction with geniposide
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- TRIM21 accelerates ferroptosis in intervertebral disc degeneration by promoting SLC7A11 ubiquitination and degradation
- Histone modification and non-coding RNAs in skin aging: emerging therapeutic avenues
- A multiplicative behavioral model of DNA replication initiation in cells
- Biogenic gold nanoparticles synthesized from Pergularia daemia leaves: a novel approach for nasopharyngeal carcinoma therapy
- Creutzfeldt-Jakob disease mimicking Hashimoto’s encephalopathy: steroid response followed by decline
- Impact of semaphorin, Sema3F, on the gene transcription and protein expression of CREB and its binding protein CREBBP in primary hippocampal neurons of rats
- Iron overloaded M0 macrophages regulate hematopoietic stem cell proliferation and senescence via the Nrf2/Keap1/HO-1 pathway
- Revisiting the link between NADPH oxidase p22phox C242T polymorphism and ischemic stroke risk: an updated meta-analysis
- Exercise training preferentially modulates α1D-adrenergic receptor expression in peripheral arteries of hypertensive rats
- Overexpression of HE4/WFDC2 gene in mice leads to keratitis and corneal opacity
- Tumoral calcinosis complicating CKD-MBD in hemodialysis: a case report
- Mechanism of KLF4 Inhibition of epithelial-mesenchymal transition in gastric cancer cells
- Dissecting the molecular mechanisms of T cell infiltration in psoriatic lesions via cell-cell communication and regulatory network analysis
- Circadian rhythm-based prognostic features predict immune infiltration and tumor microenvironment in molecular subtypes of hepatocellular carcinoma
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Molecular identification and control studies on Coridius sp. (Hemiptera: Dinidoridae) in Al-Khamra, south of Jeddah, Saudi Arabia
- 10.1515/biol-2025-1218
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Response of hybrid grapes (Vitis spp.) to two biotic stress factors and their seedlessness status
- Metabolomic profiling reveals systemic metabolic reprogramming in Alternaria alternata under salt stress
- Effects of mixed salinity and alkali stress on photosynthetic characteristics and PEPC gene expression of vegetable soybean seedlings
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Causal effects of trace elements on congenital foot deformities and their subtypes: a Mendelian randomization study with gut microbiota mediation
- Honey meets acidity: a novel biopreservative approach against foodborne pathogens
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Fluorescent detection of sialic acid–binding lectins using functionalized quantum dots in ELISA format
- Smart tectorigenin-loaded ZnO hydrogel nanocomposites for targeted wound healing: synthesis, characterization, and biological evaluation
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
- Retraction
- Retraction of “Down-regulation of miR-539 indicates poor prognosis in patients with pancreatic cancer”

