Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
-
Suping Zhang
Abstract
Dental pulp stem cells hold significant prospects for tooth regeneration and repair. However, a comprehensive understanding of the molecular differences between dental pulp stem cells (DPSC, from permanent teeth) and stem cells from human exfoliated deciduous teeth (SHED, from deciduous teeth) remains elusive, which is crucial for optimizing their therapeutic potential. To address this gap, we employed a novel data-independent acquisition (DIA) proteomics approach to compare the protein expression profiles of DPSC and SHED. Based on nano-LC-MS/MS DIA proteomics, we identified over 7,000 proteins in both cell types. By comparing their expression levels, 209 differentially expressed proteins were identified. Subsequent Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses, along with protein–protein interaction network construction, revealed significant metabolic differences and key regulatory nodes. DPSC exhibited significantly higher expression of proteins belonging to the NDUFB family, SMARC family, RPTOR and TLR3. These proteins are known to be involved in critical cellular processes such as mitochondrial energy metabolism, mTOR-related autophagy pathway, and innate immune response. Conversely, SHED displayed elevated expression of AKR1B family, which participated in glycerolipid metabolism and adipogenic differentiation, PRKG1, MGLL and UQCRB proteins associated with thermogenesis. These findings highlight the specific proteomic landscape of DPSC and SHED, suggesting their distinct biological roles and potential applications.
1 Introduction
Dental pulp stem cells (DPSC) and stem cells from human exfoliated deciduous teeth (SHED) represent the two primary subtypes of tooth-derived pulp stem cells [1,2]. Both cell types originate from the dental pulp, the soft tissue within the tooth, distinguishing them from periodontal stem cells such as periodontal ligament stem cells (PDLSC), gingival mesenchymal stem cells (GMSC), and stem cells from apical papilla (SCAP), which are isolated from the surrounding periodontal tissues [3,4]. Since their discovery and characterization, these dental-derived mesenchymal stem cells have attracted significant research interest due to their inherent mesenchymal stem cell properties, including self-renewal, multi-differentiation potential, and low immunogenicity [5,6]. These properties make them highly promising candidates for applications in regenerative medicine and tissue engineering [7,8].
However, due to the distinct origin in terms of host ages and tooth types (permanent and deciduous), DPSC and SHED exhibit significant differences in their biological characteristics and clinical applications. Research indicated that SHED possessed a higher concentration of anti-aging signals, suggesting a potential for enhanced rejuvenation of aging cells most due to their origin from youth tissue [9,10]. Comparative studies have revealed disparities in the proliferation rate, differentiation capability, and cytokine expression profiles between DPSC and SHED. It showed that SHED demonstrated a higher proliferation rate and differentiation capacity in vitro, along with superior mineralization potential compared to DPSC [11]. Moreover, SHED exhibited a more pronounced expression of cytokines associated with immunomodulation, odontogenesis, and osteogenesis, while DPSC demonstrated elevated expression of angiogenesis-related cytokines [12] and exhibited significant potential in bone tissue engineering and nerve tissue repairing [13,14].
Despite these intriguing findings, a comprehensive understanding of the protein molecular basis underlying the functional differences between DPSC and SHED remains elusive. Proteins, as the ultimate products of gene expression, are fundamental to cellular function. They are integral to a wide range of essential cellular processes, including metabolic catalysis, signal transduction, substance transport, immune defense, and cell division. Understanding the proteomic landscape of these two stem cell types is critical for optimizing their therapeutic applications. While DPSC and SHED share similar biological characteristics, their differences at the protein level may impact their efficacy and safety in specific applications.
In this study, we aim to conduct a comparative proteomic analysis of DPSC and SHED utilizing the innovative data-independent acquisition (DIA) proteomics technology [15]. By dissecting the protein profiles of these two stem cell populations, we aim to uncover the protein molecular basis for their distinct biological properties and guide future research into their therapeutic applications in tooth regenerative and tissue engineering. This investigation will shed light on the protein molecular basis of their functional differences and provide valuable insights for optimizing their clinical applications.
2 Materials and methods
2.1 Subjects and cell culture
This study involved the isolation and in vitro culture of DPSC and SHED. DPSC was harvested from impacted third molars or other extracted teeth of healthy human subjects aged 16–29 years. SHED was obtained from exfoliated deciduous teeth of children aged 5–8 years. Informed consent was obtained from all participants or their guardians prior to the collection of any samples. Teeth were collected from Suzhou Stomatological Hospital. All experiments were conducted at Soochow University, adhering to all relevant national regulations, institutional policies, and the tenets of the Helsinki Declaration. This research involving human subjects received ethical approval from the ethics committees of Suzhou Stomatological Hospital and Soochow University (Approval Number: SUDA20230727H08).
The tooth samples were placed in PBS buffer after extraction and sent to the laboratory for processing within 12 h. Following the extraction of pulp tissue, primary dental stem cells were cultured using either an adherent culture method or enzymatic digestion with collagenase I (3 mg/mL)/dispase (4 mg/mL) (Sigma, USA) as described previously [16]. Once the cells had grown into fibroblasts, they were subcultured in α-modification of Eagle’s medium (α-MEM, HyClone, USA) supplemented with 15% fetal bovine serum (Gibco, USA), 100 U/mL penicillin‒streptomycin (Beyotime, China), 100 mM l-ascorbic acid phosphate (Sigma, USA), and 2 mM l-glutamine (Gibco, USA). Cultures were maintained in a humidified incubator at 37°C with 5% CO2.
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance with the tenets of the Helsinki Declaration, and has been approved by the Ethics Committees of Suzhou Stomatological Hospital and Soochow University (Approval Number: SUDA20230727H08).
2.2 Cell proliferation and identification
Cell proliferation was assessed using a cell counting method. Briefly, 2 × 104 cells were seeded into individual wells of six-well plates and counted at Days 1, 2, 3, and 4. Cell numbers were recorded at each time point. The cell proliferation rate was calculated based on the cell number of the seeded time. Cell proliferation rate = (No1–4 − 20,000)/20,000 × 100%.
Dental stem cell identification was performed via flow cytometry (BD Calibur, USA) based on surface marker expression. Cells were collected and stained with antibodies targeting CD29, CD44, CD73, CD90, CD105, CD146, and CD166 (BioLegend, USA). The positive expression rate was analyzed using flow cytometry. Additionally, CD34 and CD45 staining was conducted to exclude potential hematopoietic cell. Data were presented as mean ± SD and statistically analyzed using GraphPad Prism 8.0.
2.3 Protein sample preparation and DIA proteome detection
This study employed next-generation label-free DIA quantitative proteomics technology. Six cell samples, including three DPSCs and three SHEDs from different individuals, were selected for DIA proteomic analysis. Cells were collected at passage 3, with a minimum of 5 × 106 cells required for protein extraction. For protein sample preparation, the cell pellets were suspended in cell lysis buffer containing sodium dodecyl sulfate (SDS) and 1× cocktail (with EDTA), followed by ultrasonic dissociation. The lysate was then centrifuged at 25,000g for 15 min at 4°C. The supernatant was incubated with 10 mM dithiothreitol at 37°C for 30 min, followed by the addition of 55 mM iodoacetamide and incubation in the dark for 45 min. Five volumes of pre-cooled acetone were added to the lysate and incubated at −20°C for 2 h. After centrifugation at 25,000g for 15 min at 4°C, the supernatant was discarded. The protein precipitate was resuspended in an appropriate amount of protein lysate without SDS and sonicated. The final protein sample was obtained after centrifugation at 25,000g for 15 min at 4°C.
Following tryptic digestion and high pH reversed-phase separation, the peptide samples were analyzed by nano-liquid chromatography coupled with tandem mass spectrometry (nano-LC‒MS/MS) for data-dependent acquisition (DDA) library construction and DIA quantitative detection. This process was outsourced and completed at BGI, China. For DDA library construction, a pooled sample containing an equal amount of all six samples was subjected to LC–MS/MS for DDA library construction. For DIA analysis, each individual sample was separated using a 120 min high-performance liquid chromatography gradient prior to MS analysis. The gradient was as follows: 0–5 min, 5% mobile phase B (98% acetonitrile, 0.1% formic acid); 5–90 min, linear increase of mobile phase B from 5 to 25%; 90–100 min, increase of mobile phase B from 25 to 35%; 100–105 min, increase of mobile phase B from 35 to 80%; 105–115 min, 80% mobile phase B; 115–120 min, 5% mobile phase B. The LC-separated peptides were ionized by nano-electrospray ionization and analyzed using a Q-Exactive HF tandem mass spectrometer (Thermo Fisher Scientific, San Jose, CA) in DIA mode. The main MS settings were ion source voltage 1.9 kV, MS scan range 400–1,250 m/z, MS resolution 120,000, and maximum injection time (MIT) 50 ms. The MS scan range was divided into 45 continuous windows for DIA–MS/MS analysis. MS/MS fragmentation was performed using higher-energy collision dissociation with automatic MIT. Fragment ions were detected in the Orbitrap with a resolution of 30,000. The collision energy was set to a distributed mode of 22.5, 25, and 27.5, with an automatic gain control target of 1E6.
2.4 Data analysis and differential protein identification
The data analysis consisted of three primary stages: spectral library construction from DDA data, large-scale DIA data acquisition, and subsequent data analysis. Spectral library construction was achieved using DDA data identified by the Andromeda search engine within MaxQuant. These identified results served as the foundation for constructing a comprehensive spectral library. For large-scale DIA data, mProphet algorithm was applied for analytical quality control, ensuring the reliability of the vast quantitative data generated.
MSstats version 2.0 software package was used for quantitative and differential protein analysis [17]. The selection of the protein database is crucial for accurate protein identification. In this study, we chose the non-redundant proteome database, RefSeq, obtained from the National Center for Biotechnology Information. The DIA data were further refined through retention time calibration using iRT peptides. To mitigate false positives, a target-decoy model, commonly used in SWATH-MS analysis, was implemented, achieving a 1% false discovery rate (FDR). This stringent filtering resulted in highly significant quantitative findings. These data were then preprocessed based on the defined comparison groups (SHED vs DPSC). Subsequent statistical significance testing, conducted using a specific model, identified differentially expressed proteins (DEPs) between DPSC and SHED. A stringent filtering process was applied, utilizing a fold change threshold of greater than 2 and a p-value less than 0.05, to obtain statistically significant DEPs.
Hierarchical clustering was employed to group these DEPs, providing a visual representation of DEP expression at the sample level. Additionally, enrichment analysis was conducted on the identified DEPs to further explore their biological significance. Figure S1 displays the process of DIA proteomics.
2.5 Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
To delve into the functional implications of the identified DEPs, we conducted GO and KEGG pathway enrichment analyses. The GO analysis, facilitated by the GO plot package within the MSstats software, aimed to elucidate the enrichment of GO terms related to cellular components (CC), biological processes, and molecular functions associated with the differential protein expression.
Furthermore, KEGG pathway enrichment analysis was performed to identify significantly over-represented pathways in the dataset. The classification and bubble chart visualization of KEGG pathways provided a comprehensive overview of pathway enrichment. The Phyper function within R software was utilized for enrichment analysis, calculating the p-value for each pathway. Subsequently, the p-value was corrected for multiple comparisons using the FDR to obtain the Q-value. A Q-value threshold of 0.05 was employed to define statistically significant enrichment.
2.6 Protein–protein interaction (PPI) network analysis
Proteins often interact with each other to form functional complexes and execute cellular processes. To investigate these interactions, we conducted PPI network analysis by comparing our dataset with the STRING database. This analysis resulted in a network interaction diagram depicting the relationships between DEPs, focusing on the top 100 interactions with the highest confidence levels. This combined approach, encompassing GO and KEGG pathway enrichment analysis alongside PPI network analysis, provided a multifaceted understanding of the functional consequences of differential protein expression between SHED and DPSC groups.
3 Results
3.1 Primary stem cell culture and identification
Primary stem cells were isolated using a combined approach of enzymatic digestion and tissue adherence. Cells migrated out of the tissue explants within 7–10 days of culture and were subsequently sub-cultured into 60 mm dishes after 14–20 days. Figure 1a demonstrates the morphology of the cultured cells. Once cells reached confluence, they exhibited a relatively uniform, spindle-shaped, fibroblast-like morphology and displayed characteristic colony formation. Cell proliferation analysis revealed no significant differences in growth rate between DPSC and SHED (Figure 1b), which might be related to the small sample size.

Cell morphology, proliferation, and identification. (a) Microscopic morphology of the cells. (b) Cell proliferation of the cells in different time points. (c) Representative FCM chart illustrating surface marker expression on the cells. (d) Statistical histogram of the expression of markers in DPSC and SHED.
To confirm the stem cell nature of the cultured cells, surface marker expression was assessed by flow cytometry. Both DPSC and SHED exhibited high expression of mesenchymal stem cell markers, including CD29, CD44, CD73, CD90, CD105, CD146, and CD166, while lacking expression of hematopoietic markers CD34 and CD45 (Figure 1c and d). These findings indicate that the cultured cells were indeed dental-derived mesenchymal stem cells.
3.2 Differential protein analysis and GO annotation reveals the subcellular protein localization
Following successful culture of primary dental pulp tissue into stable cell lines, a comparative proteomics study using DIA was performed on six individual DPSC and SHED cell lines (three replicates per group). A total of 8,509 proteins were identified across all cells, with 7,662 proteins exhibiting comparable expression levels between DPSC and SHED (data uploaded in public database iProX: IPX0009667000). Database and software analysis revealed an average of 7,114 ± 184 proteins in DPSC and 7,136 ± 138 proteins in SHED. Figure S2 shows the CV value and PCA analysis of DIA quality control between DPSC and SHED. For clinical human samples, due to the individual differences between samples, the experiment is considered stable and reliable when the median CV value is below 30%. In this study, the median CV values of DPSC and SHED groups were between 15 and 25%, indicating that the experiment was stable and reliable.
Differential expression analysis identified 107 proteins upregulated and 102 proteins downregulated in SHED compared to DPSC, with 7,453 proteins showing no significant changes (Supplementary File 1 with 209 DEPs). The volcano plot in Figure 2a visually represents the DEPs between DPSC and SHED. Figure 2b depicts the clustering analysis of these DEPs.
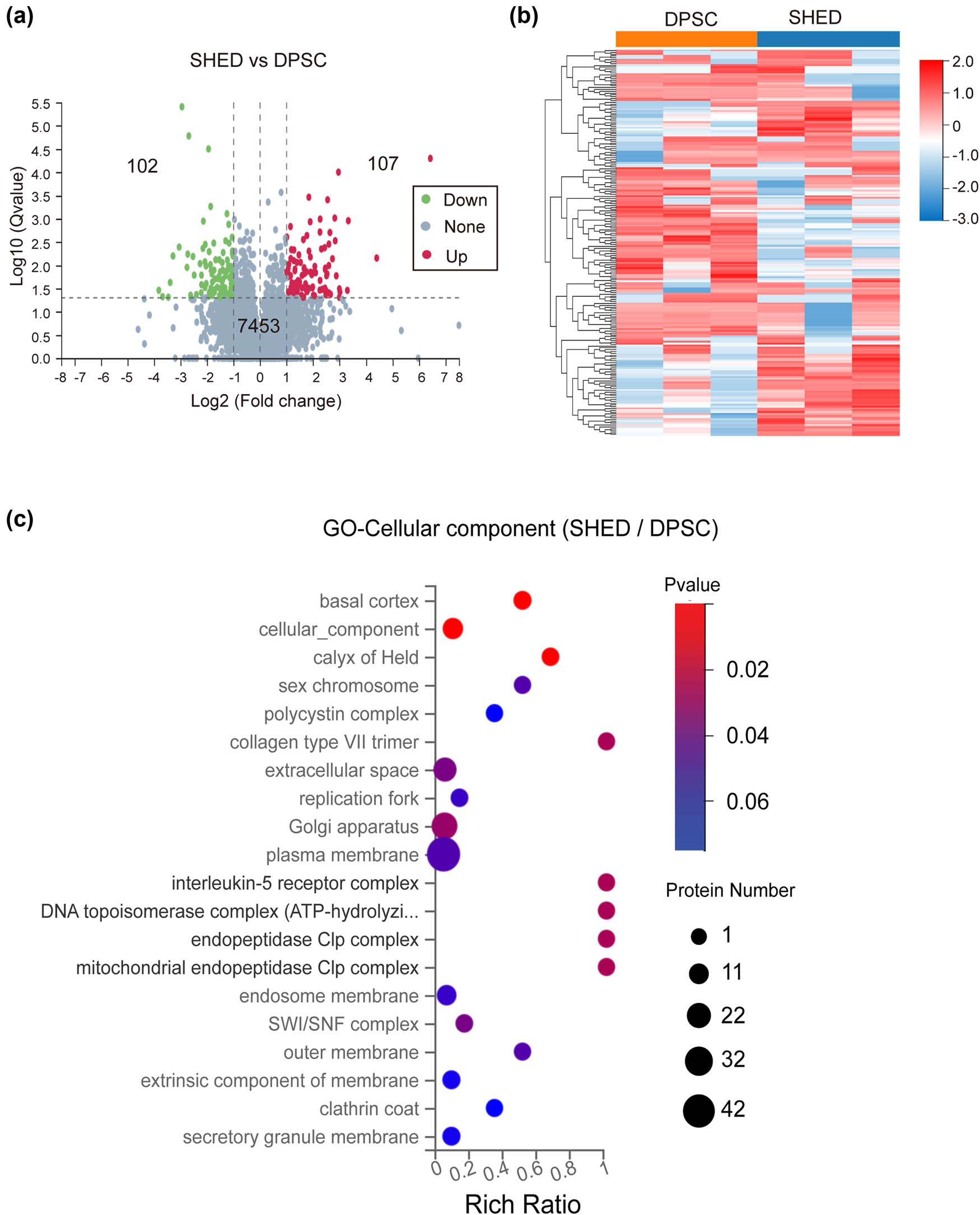
DEP analysis. (a) Volcano plot of the up- and down-regulated proteins between SHED and DPSC. (b) Clustering heatmap of the DEPs. (c) GO analysis of the CC for the differential proteins.
GO analysis of the DEPs revealed that a significant portion (42 proteins) were localized to the plasma membrane, with a notable number also found in the Golgi apparatus (32 proteins) or the extracellular space (22 proteins). The remaining DEPs were localized to other cellular compartments (Figure 2c).
3.3 KEGG classification and pathway enrichment highlights metabolic differences
The DEPs were subjected to KEGG pathway classification, revealing significant enrichment across diverse functional categories (Figure 3). Notably, a substantial number of proteins (57) were associated with human diseases, indicating potential implications for cellular dysfunction and disease pathogenesis. Further investigation revealed a prominent role of metabolism, with 28 proteins enriched in metabolic pathways.

KEGG pathway classification of the differential proteins between SHED and DPSC. The X-axis represents the numbers of proteins associated with each pathway, while the Y-axis displays the enriched pathway names.
Subsequent pathway enrichment analysis (Figure 4a) revealed distinct metabolic pathways that characterize the differences of DPSC and SHED. The bubble chart visualization highlighted the most significantly enriched pathways, with thermogenesis, glycerolipid metabolism, folate biosynthesis, and pentose/glucuronate interconversions emerging as key contributors to the observed metabolic divergence between these cell types.
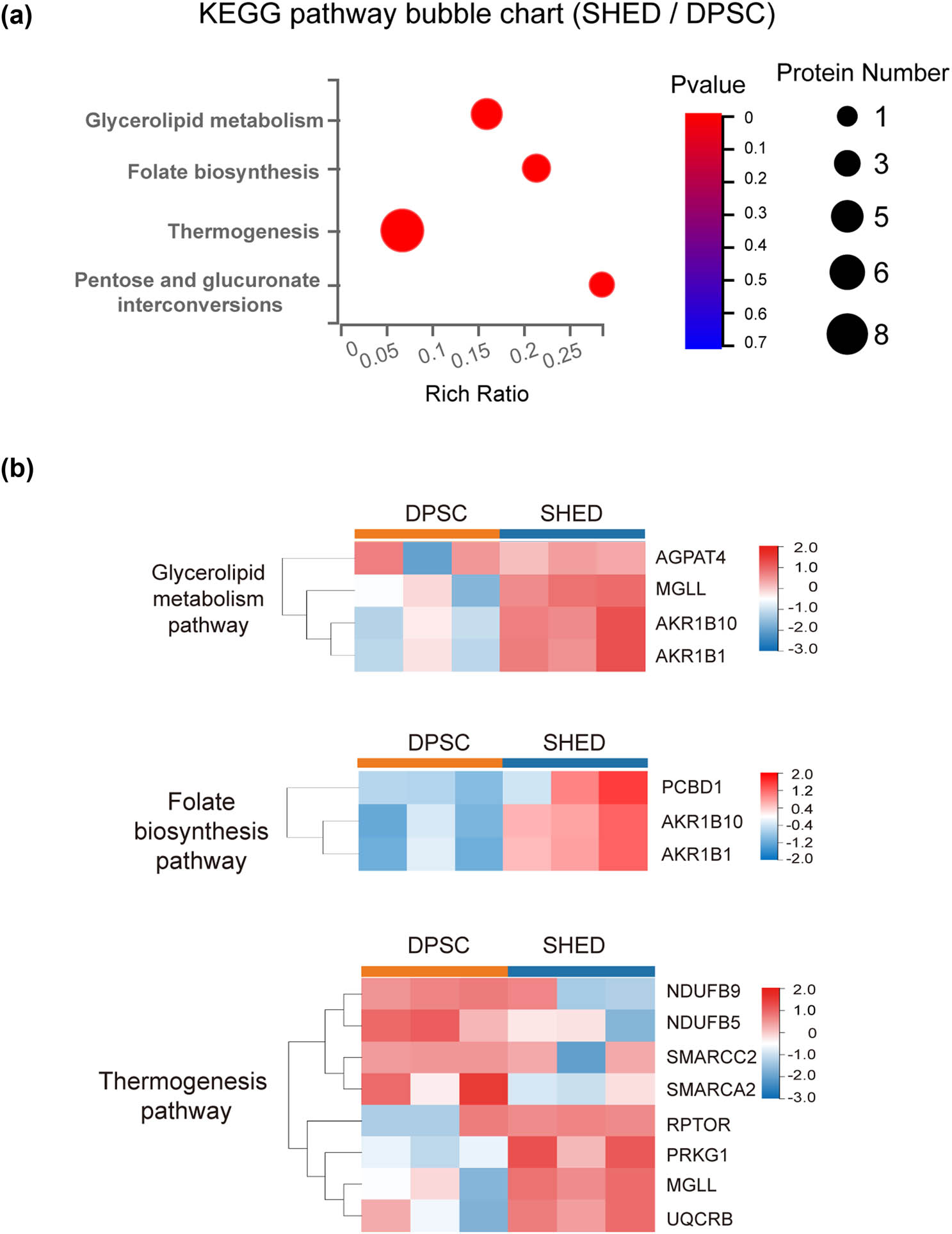
Bubble chart of KEGG pathway enrichment and clustering of representative pathways. (a) Bubble chart illustrating the significant KEGG pathway enrichment. (b) Clustering of the three most significantly enriched pathways.
Protein-pathway associations were as follows: eight proteins enriched in thermogenesis pathway, including NDUFB9, NDUFB5, SMARCC2, SMARCA2, RPTOR, PRKG1, MGLL, and UQCRB. Four proteins, AGPAT4, MGLL, AKR1B10, and AKR1B1, exhibited enrichment in glycerolipid metabolism pathway. Folate biosynthesis pathway encompassed three proteins: AKR1B10, AKR1B1, and PCBD1. AKR1B10 and AKR1B1 were also involved in pentose/glucuronate interconversions (Figure 4b).
Table 1 provides a detailed description of these proteins, highlighting differential expression patterns. Eight proteins (NDUFB family, SMARC, RPTOR, etc.) showed elevated expression in DPSC, while six proteins (AKR1B family, PRKG1, MGLL, etc.) were upregulated in SHED.
List of the protein names and descriptions of the KEGG differential proteins
| Protein name | Description | Ratio (SHED/DPSC) |
|---|---|---|
| NDUFB9 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 | 0.300 |
| NDUFB5 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5 | 0.474 |
| SMARCC2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 | 0.371 |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily a member 2 | 0.494 |
| RPTOR | Regulatory-associated protein of mTOR | 0.350 |
| AGPAT4 | 1-Acylglycerol-3-phosphate acyltransferase 4 | 0.162 |
| TLR3 | Toll-like receptor 3 | 0.304 |
| SRRM2 | Serine/arginine repetitive matrix protein 2 | 0.12 |
| PRKG1 | Protein kinase cGMP-dependent 1 | 2.662 |
| MGLL | Monoglyceride lipase | 2.183 |
| UQCRB | Ubiquinol-cytochrome c reductase binding protein | 2.088 |
| AKR1B10 | Aldo-keto reductase family 1 member B10 | 5.790 |
| AKR1B1 | Aldo-keto reductase family 1 member B1 | 6.653 |
| PCBD1 | Pterin-4-alpha-carbinolamine dehydratase 1 | 2.271 |
3.4 PPI network analysis unveils key regulatory nodes
The PPI network analysis (Figure 5) revealed a complex interplay of protein interactions, highlighting four distinct modules enriched in specific pathways and proteins. These modules converged around four central proteins – RPTOR, MGLL, SRRM2, and TLR3 – acting as pivotal regulatory nodes within the network. RPTOR, with its extensive connections to seven proteins or pathways, emerges as a key player in stem cell functions. Its involvement in the mTOR-related autophagy pathway warrants particular attention. MGLL, linked to four proteins or pathways, participates in crucial metabolic processes, including thermogenesis and glycerolipid metabolism. Further investigation into the roles and interactions of these proteins within the PPI network is essential to comprehensively understand their contributions to the observed metabolic differences between DPSC and SHED.

PPI network of the differential proteins. (a) PPI network of the differential proteins between DPSC and SHED. Red nodes represent upregulated proteins, while blue nodes represent downregulated proteins. The size of each nodes reflects the degree of relationship density. (b) Clustering heatmap depicting the four most significant protein interaction networks.
4 Discussion
Proteomics, a fundamental technique for protein screening and functional analysis, has attained significant prominence within the scientific community. DIA mass spectrometry offers a distinct advantage over DDA by systematically fragmenting all detectable ions within a wide m/z range, regardless of their intensity. This comprehensive approach results in a wider dynamic range, enhanced reproducibility, improved sensitivity, greater accuracy in quantification, and potentially increased proteome coverage [18]. DIA has proven to be a widely adopted methodology in quantitative proteomic studies, particularly in plasma/serum proteomics, facilitating the identification of potential biomarkers across a diverse spectrum of diseases such as in 3P medicine (predictive, preventive, and personalized), cancer, etc. [19,20].
This study presents the first application of DIA proteomic technology to elucidate the proteomic landscape of DPSC and SHED. The comprehensive analysis identified over 7,000 proteins and 209 DEPs between DPSC and SHED, revealing distinct metabolic profiles between these two stem cell types. KEGG pathway and PPI network analysis revealed significant enrichment of proteins involved in metabolic pathways, particularly thermogenesis, glycerolipid metabolism, and folate biosynthesis. Notably, we identified the NDUFB protein family, SMARC family, RPTOR, and TLR3 proteins as prominent proteins in DPSC, while the AKR1B protein family, MGLL, and UQCRB were significantly enriched in SHED. Furthermore, RPTOR and MGLL emerged as central nodes in the PPI network, indicating their critical roles in cellular function. Importantly, these proteins were not previously reported in other dental stem cell proteomic studies.
Previous research on dental-related stem cells, such as DPSC, PDLSC, and SCAP, primarily relied on traditional two-dimensional gel electrophoresis proteomics. These studies primarily identified high-abundance proteins such as cytoskeletal proteins and cell adhesion molecules [21,22]. While subsequent gel-free high-throughput proteomic methods, like iTRAQ, tandem mass tags (TMT), and other shotgun approaches, have significantly expanded the repertoire of identified proteins and their associated functions and pathways, these studies mainly focused on comparisons between DPSC and other stem cells, such as PDLSC, SCAP, etc. [23–25], leaving the differential proteomic landscape between DPSC and SHED largely unexplored. A study investigating odontogenic differentiation from human DPSC using TMT-based proteomic analysis [23] identified 223 DEPs involved in cellular processes, metabolic processes, and biological regulation, and the protein expression levels of FBN1 and TGF-β2 validated by WB were consistent with the proteomic analysis. Lei et al. [24,25] employed TMT proteomics to compare the protein profiles of DPSC with those of PDLSC/SCAP. They identified high-scoring sub-networks in DPSC enriched with neural-related molecules, encoding cell vesicle transport and mitochondrial energy transfer to regulate cell proliferation and secretion factors. Additionally, a significant number of cell adhesion molecules were identified among the highly expressed molecules in PDLSC, indicating their potential role in providing cell attachment functions. While SCAP was characterized by a high expression of Niemann-Pick C1, a protein implicated in immune stability of SCAP.
Regarding the study between DPSC and SHED, Zainol Abidin et al. conducted a label-free quantitative proteomic analysis to investigate the differential expression of osteoblast-related proteins during osteogenic differentiation in DPSC and SHED. Their findings revealed significant differences in the expression of three proteins (PSMD11/RPN11, PLS3, and CLIC1) in stem cells from SHED, while a single protein (SYNCRIP) exhibited differential expression in DPSC [26]. However, a comprehensive proteomic analysis of undifferentiated DPSC and SHED has remained elusive until now. This research addressed this significant gap in the current area.
In the DEPs, DPSC exhibited significantly higher expression of the NDUFB (NADH dehydrogenase) family proteins, essential components of the mitochondrial electron transport chain responsible for energy production. This observation aligns with previous studies indicating the up-regulation of NDUF-related proteins during the differentiation of human GMSCs into neuron-like cells, suggesting a potential role for NDUFB in cell differentiation and metabolic reprogramming [27]. Furthermore, DPSC demonstrated elevated expression of RPTOR, a crucial component of the rapamycin target complex 1 (mTORC1), which plays a central role in the mTOR signaling pathway and governs essential cellular processes including body development, differentiation, and cell homeostasis [28]. Studies in mice have implicated RPTOR in maintaining the spermatogonial stem cell pool [29] and its deletion in hematopoietic stem/progenitor cells has been shown to lead to self-destructive innate immunity [30]. The increased RPTOR expression in DPSC suggests a higher autophagy capacity and enhanced regulation of cellular processes through the mTOR pathway.
Conversely, SHED displayed significantly higher expression of the AKR1B protein family, known for catalyzing the reduction of various aldehydes, including the aldehyde form of glucose [31]. This role in glucose metabolism is particularly relevant in diabetic complications, where AKR1B reduces glucose to sorbitol. Studies have shown that AKR1B1 expression increases during the differentiation of human multipotent adipose-derived stem cells, paralleling PGF2α release, while PGF2α receptor levels decline in early differentiation [32]. The elevated expression of AKR1B in SHED suggests a potential association with enhanced adipogenic differentiation ability. Other prominent proteins up-regulated in SHED include MGLL and UQCRB. MGLL, an enzyme responsible for hydrolyzing the endocannabinoid 2-arachidonoylglycerol, plays a crucial role in various physiological processes, including pain and nociperception [33]. UQCRB, a component of the mitochondrial respiratory chain, has been shown to play a pivotal role in hypoxic signaling-induced epithelial–mesenchymal transition (EMT), potentially serving as a therapeutic target for reversing EMT by inhibiting mitochondrial ROS production [34]. Additionally, N-acetyltransferase 10 has been demonstrated to repress Uqcr11 and Uqcrb, promoting heart regeneration by regulating their expression in mouse and human cardiomyocytes [35], further emphasizing the importance of UQCRB in metabolic reprogramming.
In conclusion, this study successfully established a DIA proteomics approach to analyze dental stem cells derived from permanent and deciduous teeth, leading to the identification of DEPs and enrichment of KEGG pathways. The findings revealed distinct metabolic profiles and key regulatory nodes between DPSC and SHED. Specifically, DPSC displayed higher expression of proteins belonging to the NDUFB family, SMARC family, RPTOR, and TLR3, implicated in pathways related to mitochondrial energy metabolism, mTOR-related autophagy, and immune signaling. In contrast, SHED exhibited higher expression of the AKR1B family, MGLL, and UQCRB, suggesting a stronger capacity for adipogenic differentiation and involvement in glycerolipid metabolism and thermogenesis pathways.
The identification of key regulatory proteins such as NDUFB, RPTOR, AKR1B, and MGLL provides valuable insights into the underlying mechanisms governing the differences between DPSC and SHED, and opens avenues for further investigation into their roles in stem cell function and potential therapeutic applications. However, it is important to acknowledge the limitations inherent in the use of a limited sample size and the absence of validation. Further research with larger sample sizes is necessary to validate these findings and confirm the observed differences.
-
Funding information: This research was supported by the Science and Technology Development Plan Project of Suzhou, China (SYSD2019067), the Jiangsu Province Preventive Medicine Research Project (Ym2023016), and the Construction Plan of Scientific Innovation Team of Suzhou Vocational Health College (SZWZYTD202202).
-
Author contributions: All authors have contributed to this manuscript and have agreed to its submission to the journal. They have reviewed all results and approved the final version. S.Z., Y.L., J.W., and F.M. designed the study; S.Z. and Y.L. conducted all experiments; J.D., Y.G., and D.Y. collected the tooth samples and assisted with cell culture experiments; M.J., L.C., and N.Y. analyzed the data; S.Z. and F.M. drafted and revised the manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625–30.10.1073/pnas.240309797Search in Google Scholar PubMed PubMed Central
[2] Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100:5807–12.10.1073/pnas.0937635100Search in Google Scholar PubMed PubMed Central
[3] Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364(9429):149–55.10.1016/S0140-6736(04)16627-0Search in Google Scholar PubMed
[4] Ge S, Mrozik KM, Menicanin D, Gronthos S, Bartold PM. Isolation and characterization of mesenchymal stem cell-like cells from healthy and inflamed gingival tissue: potential use for clinical therapy. Regen Med. 2012;7(6):819–32.10.2217/rme.12.61Search in Google Scholar PubMed
[5] Sui B, Wu D, Xiang L, Fu Y, Kou X, Shi S. Dental pulp stem cells: from discovery to clinical application. J Endod. 2020;46:S46–55.10.1016/j.joen.2020.06.027Search in Google Scholar PubMed
[6] Liu P, Zhang Y, Ma Y, Tan S, Ren B, Liu S, et al. Application of dental pulp stem cells in oral maxillofacial tissue engineering. Int J Med Sci. 2022;19:310–20.10.7150/ijms.68494Search in Google Scholar PubMed PubMed Central
[7] Xie Z, Shen Z, Zhan P, Yang J, Huang Q, Huang S, et al. Functional dental pulp regeneration: basic research and clinical translation. Int J Mol Sci. 2021;22:8991.10.3390/ijms22168991Search in Google Scholar PubMed PubMed Central
[8] Zhang W, Yelick PC. Tooth repair and regeneration: potential of dental stem cells. Trends Mol Med. 2021;27:501–11.10.1016/j.molmed.2021.02.005Search in Google Scholar PubMed PubMed Central
[9] Jin S, Wang Y, Wu X, Li Z, Zhu L, Niu Y, et al. Young exosome bio-nanoparticles restore aging-impaired tendon stem/progenitor cell function and reparative capacity. Adv Mater. 2023;35(18):e2211602.10.1002/adma.202211602Search in Google Scholar PubMed
[10] Xing C, Hang Z, Guo W, Li Y, Shah R, Zhao Y, et al. Stem cells from human exfoliated deciduous teeth rejuvenate the liver in naturally aged mice by improving ribosomal and mitochondrial proteins. Cytotherapy. 2023;25(12):1285–92.10.1016/j.jcyt.2023.08.015Search in Google Scholar PubMed
[11] Wang X, Sha XJ, Li GH, Yang FS, Ji K, Wen LY, et al. Comparative characterization of stem cells from human exfoliated deciduous teeth and dental pulp stem cells. Arch Oral Biol. 2012;57:1231–40.10.1016/j.archoralbio.2012.02.014Search in Google Scholar PubMed
[12] Kang CM, Shin MK, Jeon M, Lee YH, Song JS, Lee JH. Distinctive cytokine profiles of stem cells from human exfoliated deciduous teeth and dental pulp stem cells. J Dent Sci. 2022;17:276–83.10.1016/j.jds.2021.03.019Search in Google Scholar PubMed PubMed Central
[13] Bai X, Cao R, Wu D, Zhang H, Yang F, Wang L. Dental pulp stem cells for bone tissue engineering: a literature review. Stem Cell Int. 2023;2023:7357179.10.1155/2023/7357179Search in Google Scholar PubMed PubMed Central
[14] Gonmanee T, Thonabulsombat C, Vongsavan K, Sritanaudomchai H. Differentiation of stem cells from human deciduous and permanent teeth into spiral ganglion neuron-like cells. Arch Oral Biol. 2018;88:34–41.10.1016/j.archoralbio.2018.01.011Search in Google Scholar PubMed
[15] Krasny L, Huang PH. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Mol Omics. 2021;17:29–42.10.1039/D0MO00072HSearch in Google Scholar PubMed
[16] Mrozik K, Gronthos S, Shi S, Bartold PM. A method to isolate, purify, and characterize human periodontal ligament stem cells. Methods Mol Biol. 2017;1537:413–27.10.1007/978-1-4939-6685-1_24Search in Google Scholar PubMed
[17] Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–6.10.1093/bioinformatics/btu305Search in Google Scholar PubMed
[18] Li J, Smith LS, Zhu HJ. Data-independent acquisition (DIA): an emerging proteomics technology for analysis of drug-metabolizing enzymes and transporters. Drug Discov Today Technol. 2021;39:49–56.10.1016/j.ddtec.2021.06.006Search in Google Scholar PubMed PubMed Central
[19] Ge Z, Feng P, Zhang Z, Liang Z, Chen R, Li J. Identification of novel serum protein biomarkers in the context of 3P medicine for intravenous leiomyomatosis: a data-independent acquisition mass spectrometry-based proteomics study. EPMA J. 2023;14:613–29.10.1007/s13167-023-00338-0Search in Google Scholar PubMed PubMed Central
[20] Rao J, Wan X, Tou F, He Q, Xiong A, Chen X, et al. Molecular characterization of advanced colorectal cancer using serum proteomics and metabolomics. Front Mol Biosci. 2021;8:687229.10.3389/fmolb.2021.687229Search in Google Scholar PubMed PubMed Central
[21] Eleuterio E, Trubiani O, Sulpizio M, Di Giuseppe F, Pierdomenico L, Marchisio M, et al. Proteome of human stem cells from periodontal ligament and dental pulp. PLoS One. 2013;8(8):e71101.10.1371/journal.pone.0071101Search in Google Scholar PubMed PubMed Central
[22] Pivoriuūnas A, Surovas A, Borutinskaite V, Matuzeviccius D, Treigyte G, Savickiene J, et al. Proteomic analysis of stromal cells derived from the dental pulp of human exfoliated deciduous teeth. Stem Cell Dev. 2010;19(7):1081–93.10.1089/scd.2009.0315Search in Google Scholar PubMed
[23] Xiao X, Xin C, Zhang Y, Yan J, Chen Z, Xu H, et al. Characterization of odontogenic differentiation from human dental pulp stem cells using TMT-based proteomic analysis. Biomed Res Int. 2020;2020:3871496.10.1155/2020/3871496Search in Google Scholar PubMed PubMed Central
[24] Lei T, Wang J, Liu Y, Chen P, Zhang Z, Zhang X, et al. Proteomic profile of human stem cells from dental pulp and periodontal ligament. J Proteom. 2021;245:104280.10.1016/j.jprot.2021.104280Search in Google Scholar PubMed
[25] Lei T, Zhang X, Chen P, Li Q, Du H. Proteomic profile of human dental follicle stem cells and apical papilla stem cells. J Proteom. 2021;231:103928.10.1016/j.jprot.2020.103928Search in Google Scholar PubMed
[26] Zainol Abidin IZ, Manogaran T, Abdul Wahab RM, Karsani SA, Yazid MD, Yazid F, et al. Label-free quantitative proteomic analysis of ascorbic acid-induced differentially expressed osteoblast-related proteins in dental pulp stem cells from deciduous and permanent teeth. Curr Stem Cell Res Ther. 2023;18:417–28.10.2174/1574888X17666220627145424Search in Google Scholar PubMed
[27] Cai S, Lei T, Bi W, Sun S, Deng S, Zhang X, et al. Chitosan hydrogel supplemented with metformin promotes neuron-like cell differentiation of gingival mesenchymal stem cells. Int J Mol Sci. 2022;23:3276.10.3390/ijms23063276Search in Google Scholar PubMed PubMed Central
[28] Liu M, Zhang J, Pinder BD, Liu Q, Wang D, Yao H, et al. WAVE2 suppresses mTOR activation to maintain T cell homeostasis and prevent autoimmunity. Science. 2021;371:eaaz4544.10.1126/science.aaz4544Search in Google Scholar PubMed
[29] Serra N, Velte EK, Niedenberger BA, Kirsanov O, Geyer CB. The mTORC1 component RPTOR is required for maintenance of the foundational spermatogonial stem cell pool in mice. Biol Reprod. 2019;100:429–39.10.1093/biolre/ioy198Search in Google Scholar PubMed PubMed Central
[30] Yin Y, Lei S, Li L, Yang X, Yin Q, Xu T, et al. RPTOR methylation in the peripheral blood and breast cancer in the Chinese population. Genes Genomics. 2022;44:435–43.10.1007/s13258-021-01182-0Search in Google Scholar PubMed
[31] Banerjee S. Aldo keto reductases AKR1B1 and AKR1B10 in cancer: molecular mechanisms and signaling networks. Adv Exp Med Biol. 2021;1347:65–82.10.1007/5584_2021_634Search in Google Scholar PubMed
[32] Pastel E, Pointud JC, Loubeau G, Dani C, Slim K, Martin G, et al. Aldose reductases influence prostaglandin F2α levels and adipocyte differentiation in male mouse and human species. Endocrinology. 2015;156:1671–84.10.1210/en.2014-1750Search in Google Scholar PubMed
[33] Clemente JC, Nulton E, Nelen M, Todd MJ, Maguire D, Schalk-Hihi C, et al. Screening and characterization of human monoglyceride lipase active site inhibitors using orthogonal binding and functional assays. J Biomol Screen. 2012;17:629–40.10.1177/1087057112441012Search in Google Scholar PubMed
[34] Monti E, Mancini A, Marras E, Gariboldi MB. Targeting mitochondrial ROS production to reverse the epithelial–mesenchymal transition in breast cancer cells. Curr Issues Mol Biol. 2022;44:5277–93.10.3390/cimb44110359Search in Google Scholar PubMed PubMed Central
[35] Ma W, Tian Y, Shi L, Liang J, Ouyang Q, Li J, et al. N-acetyltransferase 10 represses Uqcr11 and Uqcrb independently of ac4C modification to promote heart regeneration. Nat Commun. 2024;15:2137.10.1038/s41467-024-46458-7Search in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Genetic variants in VWF exon 26 and their implications for type 1 Von Willebrand disease in a Saudi Arabian population
- Lipoxin A4 improves myocardial ischemia/reperfusion injury through the Notch1-Nrf2 signaling pathway
- High levels of EPHB2 expression predict a poor prognosis and promote tumor progression in endometrial cancer
- Knockdown of SHP-2 delays renal tubular epithelial cell injury in diabetic nephropathy by inhibiting NLRP3 inflammasome-mediated pyroptosis
- Exploring the toxicity mechanisms and detoxification methods of Rhizoma Paridis
- Concomitant gastric carcinoma and primary hepatic angiosarcoma in a patient: A case report
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
- Retraction
- Retraction of “Down-regulation of miR-539 indicates poor prognosis in patients with pancreatic cancer”
Articles in the same Issue
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Genetic variants in VWF exon 26 and their implications for type 1 Von Willebrand disease in a Saudi Arabian population
- Lipoxin A4 improves myocardial ischemia/reperfusion injury through the Notch1-Nrf2 signaling pathway
- High levels of EPHB2 expression predict a poor prognosis and promote tumor progression in endometrial cancer
- Knockdown of SHP-2 delays renal tubular epithelial cell injury in diabetic nephropathy by inhibiting NLRP3 inflammasome-mediated pyroptosis
- Exploring the toxicity mechanisms and detoxification methods of Rhizoma Paridis
- Concomitant gastric carcinoma and primary hepatic angiosarcoma in a patient: A case report
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
- Retraction
- Retraction of “Down-regulation of miR-539 indicates poor prognosis in patients with pancreatic cancer”

