Abstract
Excessive intracellular copper accumulation triggers cuproptosis, a novel regulated cell death process with therapeutic potential. Analyzing 566 The Cancer Genome Atlas samples alongside lung adenocarcinoma (LUAD)-specific microarray and single-cell sequencing data, we identified 109 cuproptosis-associated genes, of which C1QBP and PFKP emerged as key prognostic markers. Four-gene risk model stratified patients into high- and low-risk groups with distinct survival outcomes, where high-risk scores correlated with advanced TNM stages. Clinical validation confirmed that elevated C1QBP/PFKP expression in LUAD tissues predicted shorter progression-free survival. Functional assays demonstrated that silencing C1QBP or PFKP increased intracellular copper concentration, suppressed proliferation, and inhibited invasion, mechanistically linking these genes to cuproptosis dysregulation. Our findings nominate C1QBP/PFKP as actionable targets for LUAD therapy, offering both prognostic biomarkers and copper-metabolism-directed treatment strategies.
1 Introduction
Lung cancer is the most prevalent cancer globally and a major cause of cancer-related deaths around the world. Non-small-cell lung cancer makes up more than 80% of all lung cancer cases, with lung adenocarcinoma (LUAD) being the most frequently occurring subtype [1]. The northern region is characterized by a cold climate and heavy industry, and studies have shown that air pollution and cold weather are associated with a higher incidence of lung cancer [2,3]. This combination of environmental and industrial factors creates a unique risk profile for lung cancer in the northern region. Despite significant improvements in treatment strategies for LUAD in recent years, the prognosis remains poor [4].
Cuproptosis is a novel regulatory mechanism of apoptotic cell death that occurs due to the excessive accumulation of copper ions in cells, leading to mitochondrial lipid peroxidation and alterations in iron-sulfur clusters [5,6,7]. Unlike apoptosis (programmed cell death) and ferroptosis (iron-dependent lipid peroxidation), cuproptosis is driven by irreversible mitochondrial copper accumulation, leading to distinct aggregation of lipoylated tricarboxylic acid (TCA) cycle proteins and catastrophic energy collapse PMID: 38801962.
Numerous studies have reported that intracellular copper can participate in the development of various tumors through multiple mechanisms, suggesting it could be a potential therapeutic target [6]. However, the mechanisms regulating cuproptosis in tumor cells in LUAD remain unclear, necessitating urgent research into therapeutic targets targeting this regulatory mechanism. Beyond cancer, cuproptosis plays established roles in neurodegenerative diseases – notably Alzheimer’s, where copper chelation therapy is under clinical evaluation (PMID: 35691251). In cardiovascular systems, copper-dependent cardiomyocyte death exacerbates heart failure progression. These findings underscore the broad pathophysiological relevance of copper homeostasis (PMID: 36774340).
This study identified 109 prognostically significant cuproptosis-related genes using single-factor Cox regression analysis. After excluding conflicting genes, we selected 11 key genes for further analysis. Finally, we validated the tissue expression and in vitro functions of two genes, C1QBP and PFKP, which were found to be involved in copper ion metabolism in LUAC and affect tumor cell proliferation and invasion capabilities.
2 Materials and methods
2.1 Data collection
We acquired RNA-seq data in TPM format along with clinical information for LUAD from The Cancer Genome Atlas (TCGA) database, selecting 566 samples, which include 59 normal and 507 LUAD tissues. In addition to the TCGA data, we included LUAD-related microarray and single-cell sequencing data specifically collected from northern region patients, which were downloaded from the NCBI GEO database (GSE13213, GSE31210, GSE37745, GSE68465, and GSE72094). Clinical details for these datasets are summarized in Table S1.
2.2 Screening of cuproptosis-related genes
We studied 347 cuproptosis-related genes with false discovery rate < 0.05, as detailed in Table S2. Expression data for these genes were extracted from the TCGA dataset and integrated with follow-up information. Univariate Cox regression analysis, performed with the R package “survival,” identified prognostic genes related to cuproptosis. The risk and favorable genes were compared using the R package “VennDiagram” [8].
2.3 Development and validation of the risk prediction model
Univariate Cox regression analysis identified prognostic genes in the training group. Additionally, multivariate Cox regression was conducted, with variable selection guided by the Akaike information criterion (AIC), using the R package “survival. Risk scores were calculated using the “predict” function from the R “stats” package, and patients were divided into high-risk and low-risk groups according to the median risk score. Prognostic differences were assessed using the survfit function and the log-rank test. The R package “timeROC” was used to calculate the area under the receiver operating characteristic (ROC) curve (AUC) for 1, 2, and 3 years. The model was validated using internal test groups, external datasets (GSE13213, GSE31210, GSE68465, GSE72094), and the TCGA dataset to ensure accuracy and robustness.
2.4 Immune infiltration and immunotherapy analysis
The levels of immune cell infiltration were assessed using the CIBERSORT algorithm [9]. The R package “estimate” assessed stromal, immune, and ESTIMATE scores to evaluate the tumor microenvironment. Immune phenotype scores (IPS) predicted patient responses to immunotherapy and provided insights into tumor immune characteristics [10].
2.5 Clinical sample collection
Clinical samples were collected from LUAD patients who had been confirmed by histopathology and were scheduled for surgery at the hospital. None of these patients had undergone prior systemic anti-tumor therapy. Tumor and adjacent normal tissues, each approximately 1 cm³, were excised during surgery, immediately fixed in formalin for immunohistochemistry, and stored in liquid nitrogen for quantitative PCR (qPCR).
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations and institutional policies and in accordance with the tenets of the Helsinki Declaration and has been approved by the authors’ institutional review board or equivalent committee.
2.6 Immunohistochemistry
Immunohistochemical analysis of clinical LUAD and adjacent normal tissues was performed to detect C1QBP, PFKP, and cuproptosis markers (FDX1, LIAS, HSP70) using the DAB staining method. The expression levels of these proteins were quantified and analyzed for their association with cuproptosis markers.
2.7 qPCR
qPCR was conducted on LUAD tissues, adjacent normal tissues, and A549 cell lines before and after siRNA interference. This analysis quantified mRNA levels of C1QBP, PFKP, and cuproptosis markers (FDX1, LIAS, HSP70) to validate their role in cuproptosis regulation in LUAD. The primer sequences (5′−3′) were as follows:
FDX1-F CTGGCTTGTTCAACCTGTCACC
FDX1-R GATTTGGCAGCCCAACCGTGAT
LIAS-F GCCAAGAAGGTTCAGCCTGATG
LIAS-R GTCTACATCTGCCTCACGAAGTG
HSP70-F GACCTGCCAATCGAATCAGC
HSP70-R CTGCGTTCTTAGCATCATTCCGC
PFKP-F CCAGTCCAGAGATGTGCCG
PFKP-R TGGCCGAAGATGAAGAGCG
C1QBP-F GCTGCTTATGGAGATGGACAA
C1QBP-R CCAGGACACAGAGGCAACA
Gene expression was calculated using the 2−ΔΔCt method with GAPDH as endogenous control. Reactions were performed in triplicate under the following conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s using QuantStudio 6 Pro system (Applied Biosystems).
2.8 Cell culture and treatment
The A549 LUAD cell line was purchased from the National Center for Model Organisms/Chinese Academy of Sciences Cell Bank. Cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin under 5% CO2 at 37°C. Gene silencing of C1QBP and PFKP was achieved using RNA interference (RNAi). siRNA sequences (5′−3′) were as follows:
PFKP: GCAGAACUCUUCAAUGAUATT; UAUCAUUG AAGAGUUCUGCTT
C1QBP: GGAAGAUGCCUCUGAUUAUTT; AUAAU AGAGGCAUCUUCCTT
Cells were transfected with 50 nM siRNA using Lipofectamine 3000 (Invitrogen) for 48 h. Scramble siRNA (5′-UUCUCCGAACGUGUCACGU-3′) served as a negative control. Knockdown efficiency was verified at 24, 48, and 72 h post-transfection by Western blotting.
2.9 Intracellular copper ion concentration detection
Intracellular copper ion concentrations in A549 cells, both before and after C1QBP and PFKP interference, were measured using a complexometric and colorimetric method. The copper assay kit (Elabscience) was used for the experiment. The copper ion concentration (μmol/g protein) was calculated using the following formula (1):
where ΔA580 is the OD value of the standard well − OD value of the blank well (OD value when the standard concentration is 0), a = slope of the standard curve, b is the intercept of the standard curve, f is the dilution factor of the sample before adding to the detection system, and Cpr is the protein concentration of the sample before adding to the detection system (g protein/L)
The standard curve is fitted by the following formula:
where y is the OD value of the standard well − OD value of the blank well (OD value when the standard concentration is zero) and x is the concentration of the standard.
2.10 Cell function assays
Cell viability was evaluated using the cell counting kit-8 (CCK8) assay to investigate the effects of C1QBP and PFKP on A549 cells. Proliferation was measured with the BeyoClick™ EdU-488 Cell Proliferation Detection Kit. Clonogenic ability was evaluated through a plate cloning assay, and cell migration was assessed using the Transwell invasion assay.
2.11 Image analysis
Images were acquired using an Olympus BX53 microscope (400× magnification). Scale bars were calibrated with ImageJ (v1.53) and inserted using Adobe Illustrator. Arrows annotate regions discussed in the text.
3 Results
3.1 Identification of cuproptosis-related prognostic genes in LUAD
We performed a univariate Cox regression analysis to identify genes linked to prognosis in LUAD patients, resulting in 109 cuproptosis-related genes. Among these, 106 were identified as risk factors and 3 as favorable factors (Table S3). After excluding conflicting genes REXO2 and COQ7, we categorized the remaining 345 genes into two groups: negative (n = 31) and positive (n = 314) target genes.
We then intersected the risk factors with the negative target genes, identifying eight genes: PRKDC, PGP, SLC16A1, C1QBP, CLUH, HNRNPU, PFKP, and PCBP2. Intersecting favorable factors with positive target genes yielded three genes: PET100, SGF29, and MPC1 (Figure 1a). Thus, we identified 11 cuproptosis-related genes for further exploration of their roles in LUAD progression.
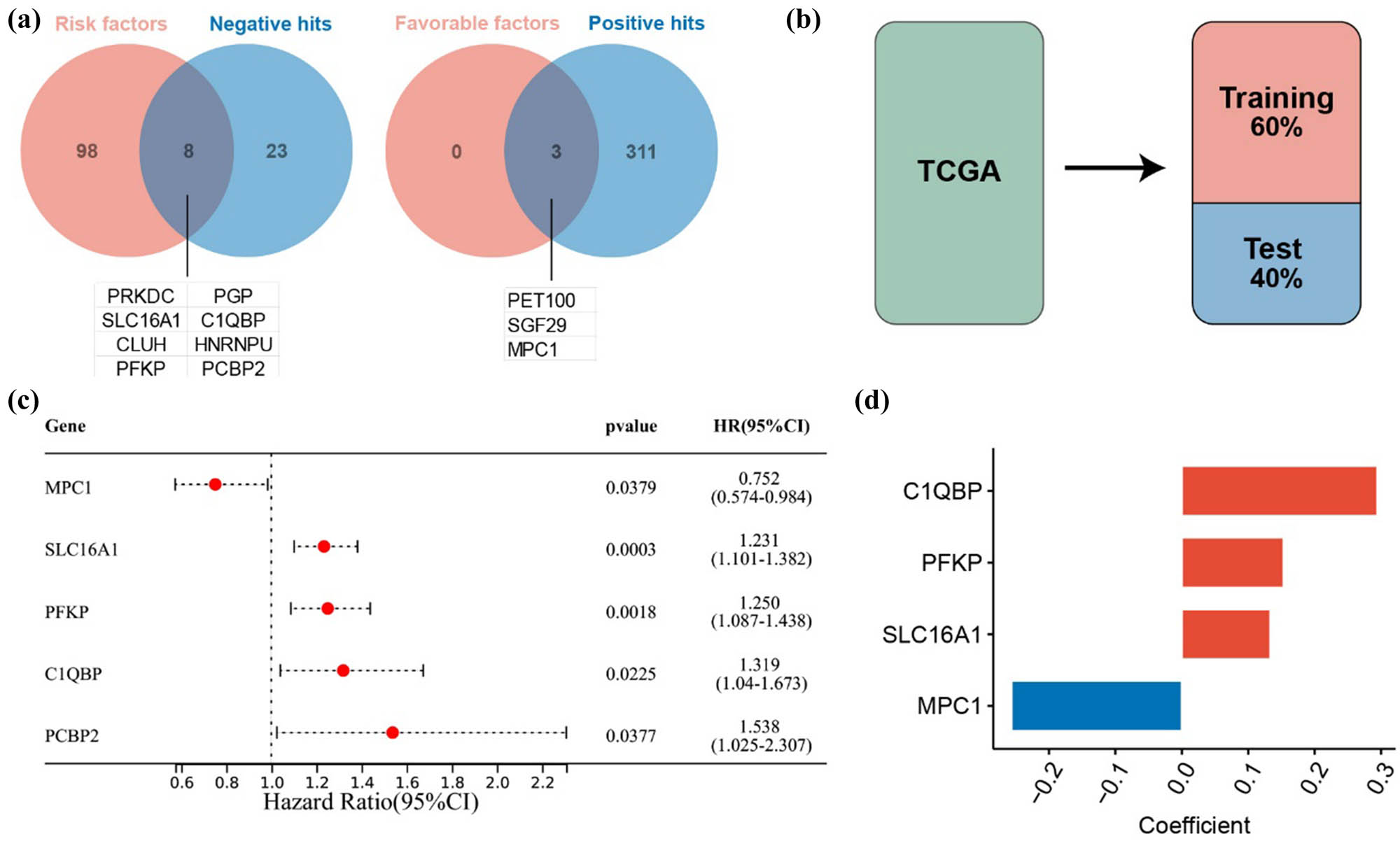
Establishment of the prognostic model for cuproptosis-related genes. (a) Venn diagram showing the intersection of prognostic genes and cuproptosis-related genes. (b) Division of the TCGA cohort into training and testing groups. (c) Univariate Cox regression analysis based on the training group. (d) Regression coefficients of the model genes.
3.2 Development of a prognostic model using cuproptosis-related genes
The TCGA dataset was randomly divided into two groups: 60% for training and 40% for testing, in order to develop a prognostic model (Figure 1b). Univariate Cox regression in the training group identified five candidate genes associated with overall survival (OS) (Figure 1c). Multivariate Cox regression with AIC selection finalized the model genes as C1QBP, PFKP, SLC16A1, and MPC1 (Figure 1d). Patients were divided into high-risk and low-risk groups according to the median risk score. The model demonstrated significant prognostic differentiation between these groups (Figure 2a), achieving AUC values of 0.653, 0.673, and 0.655 for 1-, 2-, and 3-year assessments, respectively.

Prognostic analysis of the model. The predictive ability of the model was validated using Kaplan–Meier survival analysis and ROC curves in (a) the training group, (b) the internal testing group, (c) the GSE13213 dataset, (d) the GSE31210 dataset, (e) the GSE68465 dataset, and (f) the GSE72094 dataset.
3.3 Internal testing and external validation of the prognostic model
Validation within the internal testing group revealed that high-risk patients experienced significantly lower OS (P = 0.0024). The 1-, 2-, and 3-year AUC values were 0.766, 0.716, and 0.733, respectively (Figure 2b). The model’s robustness was confirmed across external datasets, including GSE13213, GSE31210, GSE68465, and GSE72094, where high-risk patients also exhibited poorer survival rates (Figure 2c–f). These validations affirm the model’s stability and predictive accuracy across diverse datasets.
3.4 Analysis of risk scores and clinical characteristics
In both training and testing groups of the TCGA dataset, high-risk scores correlated with significantly lower OS (P < 0.001), with 1-, 2-, and 3-year AUC values of 0.671, 0.684, and 0.673 (Figure 3a and b). High-risk scores were associated with male gender (P < 0.001), advanced disease stages (P < 0.001), and lymph node involvement (P < 0.001) (Figure 3c). High-risk patients were significantly more prevalent among males (P < 0.001), those with advanced disease (P = 0.007), and those with lymph node involvement (P = 0.002) (Figure 3d). Risk scores were confirmed as a significant prognostic factor (hazard ratio [HR] = 2.13, P < 0.001) and as an independent prognostic indicator (HR = 2.13, P < 0.001) (Figure 3e and f).

Analysis of risk score and clinical characteristics. (a) Kaplan–Meier curves of patients in high and low-risk groups in the TCGA dataset. (b) ROC curves of the model in the TCGA dataset. (c) Differential analysis of RiskScore across clinical characteristic groups (including age, gender, stage, T stage, N stage, M stage). (d) Distribution of clinical characteristic groups, including age, gender, stage, T stage, M stage, and N stage. (e) Univariate Cox regression analysis. (f) Multivariate Cox regression analysis.
3.5 Immune microenvironment and immunotherapy
The CIBERSORT algorithm showed increased infiltration of CD8+ T cells, activated memory CD4+ T cells, and M0 and M1 macrophages in high-risk patients. In contrast, there was a decrease in resting memory CD4+ T cells, monocytes, resting mast cells, and dendritic cells (Figure 4a). The ESTIMATE algorithm indicated that the high-risk group had lower stromal, immune, and ESTIMATE scores (Figure 4b). The expression of immune checkpoint genes was generally higher in the low-risk group, except for CD276, TNFSF4, and CD274 (Figure 4c). IPS suggested that patients in the low-risk category might benefit more from immunotherapy (Figure 4d).

Immune infiltration and immunotherapy analysis. (a) Analysis of 22 immune cells using CIBERSORT. (b) Analysis of immune infiltration using ESTIMATE. (c) Differential expression of immune checkpoint genes between high and low-risk groups. (d) IPS prediction of immunotherapy efficacy. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
3.6 Upregulation of C1QBP and PFKP in LUAD
The expression of C1QBP was significantly upregulated in LUAD tissues compared to adjacent non-cancerous tissues (P < 0.001) and associated with worse prognosis (P = 0.018) (Figure 5a–c). This trend was consistent across six GEO datasets (Figure 5d–i). Analysis of the GSE149655 single-cell sequencing dataset in the TISCH online database (http://tisch.comp-genomics.org/home/) revealed C1QBP expression predominantly in alveolar cells, monocytes/macrophages, and endothelial cells (Figure 5j).

Upregulation of C1QBP in LUAD tissues. (a) Differential expression of C1QBP in the TCGA dataset. (b) Differential expression of C1QBP in paired samples from the TCGA dataset. (c) KM curve of C1QBP in the TCGA dataset. (d) Expression of C1QBP in the GSE10072 dataset, (e) GSE32863 dataset, (f) GSE40791 dataset, (g) GSE43458 dataset, (h) GSE63459 dataset, and (i) GSE75037 dataset. (j) Analysis of the GSE149655 dataset using the TISCH database. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
PFKP was also significantly upregulated in LUAD tissues compared to adjacent tissues (P < 0.001) and associated with poor prognosis (P = 0.018) (Figure 6a–c). This upregulation was confirmed across six GEO datasets (Figure 6d–i). Single-cell analysis showed PFKP primarily in CD8+ T cells and endothelial cells (Figure 6j).

Upregulation of PFKP in LUAD tissues. (a) Differential expression of PFKP in the TCGA dataset. (b) Differential expression of PFKP in paired samples from the TCGA dataset. (c) KM curve of PFKP in the TCGA dataset. (d) Expression of PFKP in the GSE10072 dataset, (e) GSE32863 dataset, (f) GSE40791 dataset, (g) GSE43458 dataset, (h) GSE63459 dataset, and (i) GSE75037 dataset. (j) Analysis of the GSE149655 dataset using the TISCH database. ***P ≤ 0.001, ****P ≤ 0.0001.
3.7 Differential expression of cuproptosis-related genes C1QBP and PFKP in LUAD tissues
Molecular biology experiments on tumor tissues from 5 LUAD patients revealed higher expression of C1QBP and PFKP in tumors compared to adjacent tissues (Figure 7a and b). PFKP was highly expressed in the extracellular matrix of tumors, while C1QBP was more cytoplasmic in tumors. Cuproptosis markers FDX1 and LIAS were less expressed in tumors but prominent in adjacent tissues. qPCR validated the higher mRNA expression levels of C1QBP and PFKP in tumor tissues (Figure 7c).

The cuproptosis related genes C1QBP and PFKP are differentially expressed in LUAD tissues. (a) Representative IHC images of C1QBP in LUAD (scale bar: 100μm, arrows indicate positively stained tumor cells). (b) Quantification of staining intensity (mean ± SEM, n = 5, *P < 0.05 by Student’s t-test). (c) Relative mRNA expression levels of target genes C1QBP and PFKP and cuproptosis marker genes FDX1 and LIAS. Adenocarcinoma of the lung tumor tissue (LUAD), control paracancerous tissue (CTRL). N = 3, t-test, *P ≤ 0.05. (d) Relative mRNA expression before and after RNA interference. N = 3, t-test, *P ≤ 0.05. (e) Cell copper ion concentration. One-way analysis of variance test, A549 N = 5, A549-siC1QBP N = 3, A549-siPFKP N = 3, *P ≤ 0.05. **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
3.8 Regulation of copper ion concentration by C1QBP and PFKP
Previous research has shown that copper-induced cell death results from the accumulation of copper ions, which disrupts iron-sulfur cluster proteins and induces stress responses related to protein toxicity [6]. Key measurable indicators include intracellular copper ion concentration, pyruvate, α-ketoglutarate, FDX1, DLAT, LIAS, and HSP70 [5]. In this study, we used FDX1 and LIAS as detection targets for copper-induced cell death to investigate the role of C1QBP and PFKP in LUAD in vitro.
RNAi in A549 cells confirmed successful silencing of C1QBP and PFKP (Figure 7d). Intracellular copper ion concentrations were significantly higher in A549-siC1QBP and A549-siPFKP cells compared to original A549 cells, indicating that downregulation of these proteins increases copper ion levels (Figure 7e). This suggests that C1QBP and PFKP might influence LUAD progression by modulating cuproptosis signaling. Using the copper colorimetric assay kit, we detected the copper ion concentrations as shown in Tables S4 and S5.
3.9 Regulation of cuproptosis markers by C1QBP and PFKP
To further investigate the potential functions of C1QBP and PFKP in LUAD, we conducted additional experiments using the A549 LUAD cell line. Western blot analysis showed elevated FDX1 protein levels in A549-siC1QBP and A549-siPFKP cells compared to original A549 cells (Figure 8a and b), indicating increased cuproptosis levels following downregulation of C1QBP and PFKP.

C1QBP and PFKP can regulate cuproptosis. (a) Western blot of FDX1 in A549, A549-siC1QBP and A549-siPFKP. (b) Relative expression of cuproptosis maker genes mRNA in A549, A549-siC1QBP and A549-siPFKP. One-way analysis of variance (ANOVA) test, A549 N = 3, A549-siC1QBP N = 3, A549-siPFKP N = 3, *P ≤ 0.05. (c) Cell viability of A549, A549-siC1QBP and A549-siPFKP. (d) EdU-positive cell percentage. One-way ANOVA test, N = 3, *P ≤ 0.05. (e) EdU fluorescence staining of A549, A549-siC1QBP and A549-siPFKP. **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
3.10 Effect of C1QBP and PFKP on cell proliferation and invasion
CCK8 assays revealed significantly lower cell viability in A549-siC1QBP and A549-siPFKP cells (Table S6, Figure 8c). EdU assays showed reduced DNA replication activity in these cells (Figure 8d and e). Transwell assays demonstrated decreased invasion ability in C1QBP and PFKP interference groups (Figure 9a and b), and colony formation assays showed fewer colonies (Figure 9c and d). These results suggest that C1QBP and PFKP regulate LUAD cell proliferation and invasion.

C1QBP and PFKP can regulate the in vitro proliferation and invasion of LUAD cell. (a) Images of cells in the inner chamber of the Transwell. (b) Cell counts per field by Transwell. One-way analysis of variance (ANOVA) test, N = 3, ***P ≤ 0.0001. (c) Images of cell clone formation. (d) Cell clone formation. One-way ANOVA test, N = 3, **P ≤ 0.01.
4 Discussion
Tumor progression is characterized by the uncontrolled growth of malignant cells, which often exhibit heightened metabolic demands compared to normal cells. This increased demand for nutrients and oxygen leads to severe deficiencies in the tumor microenvironment, which in turn impacts tumor survival and growth [11]. Copper, a crucial cofactor for numerous enzymes, plays a vital role in maintaining cellular functions [7,12]. Under normal conditions, copper concentrations are tightly regulated to avoid toxic accumulation, thereby preserving cellular balance [13].
Recent studies have introduced a novel concept in copper metabolism: copper-dependent cell death [14]. This form of cell death differs from traditional programmed cell deaths like ferroptosis and apoptosis. Copper directly interacts with lipid components in the TCA cycle, causing acylated proteins to aggregate and resulting in the loss of iron-sulfur cluster proteins. This process leads to protein toxicity and ultimately cell death [14]. This discovery highlights the importance of copper homeostasis and its impact on immune cell infiltration.
The notion of “copper-dependent cell proliferation” has emerged, describing how copper regulates tumor growth through both enzymatic and non-enzymatic mechanisms [15]. However, when copper levels exceed a critical threshold, it becomes toxic, inducing a type of cell death referred to as “cuproptosis.” This process involves copper-induced disruptions in mitochondrial pathways, suggesting a potential therapeutic strategy for cancer [6].
Our study utilized RNA-seq data and clinical information from TCGA and the NCBI GEO database, focusing on 347 cuproptosis-related genes. We identified 109 genes with prognostic significance through univariate Cox regression analysis, from which 11 key genes were selected after excluding contradictory ones.
We developed a prognostic model using TCGA data, dividing the dataset into training and testing groups. Univariate and multivariate Cox regression analyses identified four significant genes (C1QBP, PFKP, SLC16A1, and MPC1). The model effectively categorized patients into high-risk and low-risk groups based on median risk scores, with high-risk patients showing significantly poorer OS in both the training and validation groups. The model’s robustness was confirmed across external datasets (GSE13213, GSE31210, GSE68465, GSE72094).
The risk score correlated with clinical features, revealing higher scores in male patients, those with advanced stages, and those with lymph node metastasis. Cox regression analysis revealed that the risk score is an independent predictor of prognosis. Immune infiltration analysis showed increased CD8+ T-cell and M0/M1 macrophage infiltration in high-risk patients, while low-risk patients exhibited higher stromal and immune scores [PMID: 38773982] [PMID: 39664584]. Immune phenotype scoring suggested that patients classified as low-risk may have a better response to immunotherapy. Our findings revealed that C1QBP and PFKP were upregulated in LUAD tissues and linked to a poorer prognosis. Single-cell analysis confirmed their differential expression across various cell types within LUAD tissues. PFKP’s role in tumors involves regulating energy metabolism and influencing glycolytic pathways, which are crucial under hypoxic conditions. High PFKP expression enhances glycolysis, supporting rapid tumor cell proliferation and invasion [16,17]. C1QBP, on the other hand, regulates immune responses, oxidative stress, and cell cycle control and is associated with intracellular redox balance and oxidative stress responses [18,19,20,21].
Experimental validation of C1QBP and PFKP revealed their higher expression in LUAD tissues than in adjacent non-cancerous tissues. Silencing these genes increased the expression of cuproptosis markers (FDX1, LIAS), suggesting their involvement in cuproptosis regulation. Additionally, reduced cell proliferation and invasion were observed in cells with silenced C1QBP and PFKP, indicating their potential role as oncogenes.
Combining bioinformatics, histological, and in vitro data, we hypothesize that C1QBP and PFKP regulate LUAD progression through cellular cuproptosis mechanisms. These findings provide important insights into potential therapeutic strategies that focus on copper metabolism for cancer treatment. Further research is needed to explore how manipulating intracellular copper levels can affect tumor biology and provide new avenues for LUAD therapy. Therapeutic inhibition of C1QBP/PFKP may be achievable using existing strategies: PFKP is targeted by metabolic inhibitors like 3PO in preclinical models [PMID: 38473963], while C1QBP interacts with chemosensitizers such as obatoclax [citation]. Copper chelators could synergize with these approaches to induce cuproptosis, though combinatorial efficacy requires testing [PMID: 28107702] [PMID: 39664584].
This study has the following limitations. First, while the model shows predictive value (AUC 0.65–0.73), we acknowledge this modest performance may limit standalone clinical application. However, our risk score could complement existing biomarkers through multi-modal integration, similar to how inflammatory scores enhance prognostic systems like TNM staging. Future studies should validate combinatorial approaches. Second, while our data demonstrate copper-dependent C1QBP/PFKP associations – supported by FDX1/LIAS correlations – we recognize these are indirect measures of cuproptosis. Direct validation of copper-induced mitochondrial dysfunction remains an important goal for future studies. Third, while immunohistochemistry validated the overexpression of C1QBP/PFKP in LUAD tissues (n = 5), the small sample size restricts generalizability. Larger-scale pathological studies are needed to confirm the spatial distribution of these proteins across tumor stages and subtypes.
In summary, while research on cuproptosis-related genes in LUAD is still evolving, C1QBP and PFKP appear to play significant roles in tumor progression. Understanding the concentration-specific effects of copper on normal versus tumor cells could reveal new therapeutic strategies for LUAD. The insights gained from this study pave the way for future research into targeted cuproptosis therapies in cancer treatment.
5 Conclusion
C1QBP and PFKP are critical cuproptosis-related genes in LUAD, with their high expression linked to poor prognosis and increased tumor progression; targeting these genes could offer potential therapeutic strategies for managing LUAD.
-
Funding information: This study was supported by Beijing Kangmeng Charity Foundation Medical Research Development Fund Project, No. km227009.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and consented to its submission to the journal, reviewed all the results, and approved the final version of the manuscript. Y.J.L. contributed to writing – original draft, and formal analysis. X.Z.D. contributed to the investigation. X.L.Y. contributed to writing – review and editing.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Global Burden of Disease 2019 Cancer Collaboration, Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: A Systematic analysis for the global burden of disease study 2019. JAMA Oncol. 2022;8(3):420–44.Suche in Google Scholar
[2] Wang H, Zeng H, Miao H, Shu C, Guo Y, Ji JS. Climate factors associated with cancer incidence: An ecological study covering 33 cancers from population-based registries in 37 countries. PLOS Clim. 2024;3(3):e0000362.10.1371/journal.pclm.0000362Suche in Google Scholar
[3] Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 Cancer Groups, 1990 to 2016: A systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4(11):1553–68.10.1200/JCO.2018.36.15_suppl.1568Suche in Google Scholar
[4] Succony L, Rassl DM, Barker AP, McCaughan FM, Rintoul RC. Adenocarcinoma spectrum lesions of the lung: Detection, pathology and treatment strategies. Cancer Treat Rev. 2021;99:102237.10.1016/j.ctrv.2021.102237Suche in Google Scholar PubMed
[5] Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–61.10.1126/science.abf0529Suche in Google Scholar PubMed PubMed Central
[6] Xie J, Yang Y, Gao Y, He J. Cuproptosis: mechanisms and links with cancers. Mol Cancer. 2023;22(1):46.10.1186/s12943-023-01732-ySuche in Google Scholar PubMed PubMed Central
[7] Chen L, Min J, Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct Target Ther. 2022;7(1):378.10.1038/s41392-022-01229-ySuche in Google Scholar PubMed PubMed Central
[8] Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 2011;12:35.10.1186/1471-2105-12-35Suche in Google Scholar PubMed PubMed Central
[9] Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7.10.1038/nmeth.3337Suche in Google Scholar PubMed PubMed Central
[10] Scire J, Huisman JS, Grosu A, Angst DC, Lison A, Li J, et al. estimateR: an R package to estimate and monitor the effective reproductive number. BMC Bioinf. 2023;24(1):310.10.1186/s12859-023-05428-4Suche in Google Scholar PubMed PubMed Central
[11] Nia HT, Munn LL, Jain RK. Physical traits of cancer. Science. 2020;370(6516):eaaz0868.10.1126/science.aaz0868Suche in Google Scholar PubMed PubMed Central
[12] Scheiber I, Dringen R, Mercer JF. Copper: effects of deficiency and overload. Met Ions Life Sci. 2013;13:359–87.10.1007/978-94-007-7500-8_11Suche in Google Scholar PubMed
[13] Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4(3):176–85.10.1038/nchembio.72Suche in Google Scholar PubMed
[14] Kahlson MA, Dixon SJ. Copper-induced cell death. Science. 2022;375(6586):1231–2.10.1126/science.abo3959Suche in Google Scholar PubMed
[15] Ge EJ, Bush AI, Casini A, Cobine PA, Cross JR, DeNicola GM, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer. 2022;22(2):102–13.10.1038/s41568-021-00417-2Suche in Google Scholar PubMed PubMed Central
[16] Wang H, Penaloza T, Manea AJ, Gao X. PFKP: More than phosphofructokinase. Adv Cancer Res. 2023;160:1–15.10.1016/bs.acr.2023.03.001Suche in Google Scholar PubMed PubMed Central
[17] Chen J, Zou L, Lu G, Grinchuk O, Fang L, Ong DST, et al. PFKP alleviates glucose starvation-induced metabolic stress in lung cancer cells via AMPK-ACC2 dependent fatty acid oxidation. Cell Discov. 2022;8(1):52.10.1038/s41421-022-00406-1Suche in Google Scholar PubMed PubMed Central
[18] Wang Q, Chai D, Sobhani N, Sun N, Neeli P, Zheng J, et al. C1QBP regulates mitochondrial plasticity to impact tumor progression and antitumor immune response. Front Physiol. 2022;13:1012112.10.3389/fphys.2022.1012112Suche in Google Scholar PubMed PubMed Central
[19] Wang J, Huang CL, Zhang Y. Complement C1q Binding Protein (C1QBP): physiological functions, mutation-associated mitochondrial cardiomyopathy and current disease models. Front Cardiovasc Med. 2022;9:843853.10.3389/fcvm.2022.843853Suche in Google Scholar PubMed PubMed Central
[20] Tian H, Chai D, Wang G, Wang Q, Sun N, Jiang G, et al. Mitochondrial C1QBP is essential for T cell antitumor function by maintaining mitochondrial plasticity and metabolic fitness. Cancer Immunol Immunother. 2023;72(7):2151–68.10.1007/s00262-023-03407-5Suche in Google Scholar PubMed PubMed Central
[21] Zhai X, Liu K, Fang H, Zhang Q, Gao X, Liu F, et al. Mitochondrial C1qbp promotes differentiation of effector CD8(+) T cells via metabolic-epigenetic reprogramming. Sci Adv. 2021;7(49):eabk0490.10.1126/sciadv.abk0490Suche in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
Artikel in diesem Heft
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Hyperosmolar hyperglycemic state with severe hypernatremia coexisting with central diabetes insipidus: A case report and literature review
- Efficacy and mechanism of escin in improving the tissue microenvironment of blood vessel walls via anti-inflammatory and anticoagulant effects: Implications for clinical practice
- Merkel cell carcinoma: Clinicopathological analysis of three patients and literature review
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”


