Abstract
In order to study the sequence characteristics, gene order, and codon usage of the mitochondrial genome of Haemaphysalis hystricis, and to explore its phylogenetic relationship, a total of 36 H. hystricis isolated from dogs were used as sample in this study. The mitochondrial genome of a H. hystricis was amplified with several pairs of specific primers by PCR, and was sequenced by first generation sequencing. The mitochondrial genome of H. hystricis was 14,719 bp in size, and it contained 37 genes including 13 protein coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and AT-rich region. Each PCG sequence had different lengths, the sequence longest and shortest gene were nad5 (1,652 bp) and atp8 (155 bp), respectively, among the 13 PCGs. All PCGs used ATN as their initiation codon, 10 of 13 PCGs used TAN as their termination codon, and 3 of which had incomplete termination codon (TA/T). Most of the 22 tRNAs with different sizes could form the classical cloverleaf structures expect for tRNA-Ala, tRNA-Ser1, tRNA-Ser2, and tRNA-Glu, and there were base mismatch (U-U and U-G) in all the 22 tRNAs sequences. Two rRNAs, namely rrnL and rrnS, had different lengths, rrnL located between tRNA-Leu1 and tRNA-Val, and rrnS located between tRNA-Val and tRNA-Ile, respectively. Two AT (D-loop) control areas with different lengths were in the mitochondrial genome, the NCRL was located between tRNA-Leu2 and tRNA-Cys, and the NCRS was located between rrnS and tRNA-Ile. The complete mitochondrial genome sequence of H. hystricis was AT preferences, and the gene order is the same as that of other Haemaphysalis family ticks. However, phylogenetic analysis showed that H. hystricis was most closely related to Haemaphysalis longicornis among the selected ticks. The mitochondrial genome not only enriches the genome database, provides more novel genetic markers for identifying tick species, and studying its molecular epidemiology, population genetics, systematics, but also have implications for the diagnosis, prevention, and control of ticks and tick-borne diseases in animals and humans.
1 Introduction
Ticks, as an obligatory blood-feeding parasites [1,2], could parasitize humans and various animals including birds, mammalians, and crawls [3,4], and can also transmit numerous pathogenic microorganisms such as bacteria, fungi, viruses, and protozoa [5,6], which can lead to disease in invaded animals [7,8], and sometimes even cause death [9]. At present, ticks are classified into three families, namely the Argasidae, the Ixodidae, and the Nuttalliellidae, which encompass 867 species of tick that are recorded in the world [10]. The Ixodidae family, comprising 682 species, is the largest among the three families, which encompass 5 subfamilies and 13 genus [11]. Haemaphysalis hystricis (H. hystricis) belongs to Acarina, Ixodidae, and Haemaphysalis [12], which usually parasitizes on dogs and other hosts including humans at times [13,14]. As a common and important vector, H. hystricis carries and transmits a number of pathogens, including Babesia canis, Ehrlichia canis, and Rickettsia conorii [15,16], posing a huge threat to the health of both humans and animals [17]. Moreover, H. hystricis is widely distributed in China and others countries [12], and has become a dominant tick species in some areas of China. However, current knowledge about H. hystricis is only limited to its morphology and biology, and limited molecular information of this tick is reported, especially in genetics and molecular epidemiology owing to the lack of genome information and suitable genetic markers. Thus, the study on mitochondrial genome of H. hystricis is a key step for finding the genetic marker applying to its molecular epidemiology, population genetics, and systematics study, and have implications for studying the diagnosis, prevention, and control of ticks and tick-borne diseases in animals and humans in future [11,18].
Generally, for identification and classification of ticks, the traditional taxonomic method according to morphological structure of parasites is still a main method [19,20]. It can clearly display the morphological characteristics of various parts of the insect body but not study the insects’ gene variation, genetic structure, and phylogenetic relationship at the molecular level and the traditional taxonomic method is highly dependent on the integrity of the sample and the experience of the appraiser. However, when insects are at different developmental stages or when they have extremely similar morphological structure, the traditional taxonomic method cannot effectively identify the parasite species [21]. Therefore, it is a crucial step in parasitology research to apply others methods to accurately identify the parasite species. With the rapid development of molecular biotechnology, especially the popularization of PCR technology, the parasitology study is also developing toward the molecular level. Currently, as more and more genome information of parasite is deposited to GenBank, including mt genome, ribosomal genome, and RNAs, various gene markers used in parasite species identification are found, including cox1, nad1, nad4, 18S rRNA, ITS-1, and ITS-2 [22,23]. However, some researchers believe that the complete mt genome sequence supplied information is more than that of fragment gene sequence [24], hence the mt genome sequence is more suitable for analyzing parasitic gene variation, genetic structure, and phylogenetic study [25–27]. Mt genome of metazoan (expect for lice and flea) is a double stranded circular DNA molecule [28–30], which has rapid evolutionary rate, matrilineal inheritance, multiple copies, and no genetic recombination characteristics [28,31]. Currently, mt genome data have become a valuable source for studying parasitology, which provide numerous molecular genetic markers. These genetic markers from mt genome sequence have been widely applied for the genetics and molecular identification of many zoonotic ectoparasites including ticks and t insects that suck animal blood [32–34]. Therefore, based on the study status of mt genome of H. hystricis, we sequenced the complete mt genome of a tick representative H. hystricis collected from a hound dog in Huaihua district, Hunan province, China, and analyzed its sequence characteristics, gene order, and codons usage. The phylogenetic relationship of H. hystricis and other ticks from Haemaphysalis family was performed by the concatenated amino acid sequence of 13 protein-coding genes (PCGs) in this study. Thereby, the aims of this study were (i) to determine the complete mt genome of H. hystricis; (ii) to analyze its sequence characteristic, gene order, codon usage, and the secondary structure of tRNAs and rRNAs; (iii) to reveal the phylogenetic relationship of H. hystricis and other ticks; and (iv) to provide novel mitochondrial resources and genetic markers to this ectoparasite.
2 Materials and methods
2.1 Parasites and DNA extraction
A total of 36 adult ticks of H. hystricis were obtained from a hound dog in Huaihua district, Hunan province, China. After being removed from the surface of the dog’s body, ticks were washed three times with physiological saline in a glass culture dish, and were identified morphologically for species under an optical microscope according to the existing keys and descriptions [12]. The genomic DNA was extracted from an individual sample using the blood and tissue DNA extraction kit (TIANGEN) and sodium SDS/proteinase K according to the manufacturer’s recommendations, was then purified by the DNA purification kit (TIANGEN), and was finally eluted into 50 µL using ddH2O. The rest of the ticks were fixed in 70% (v/v) ethanol at room temperature, and the extracted DNA was stored at −20°C until use.
-
Ethical approval: The research related to animal use has been complied with all the relevant national regulations and institutional policies for the care and use of animals and has been approved by the Animal Ethics Committee of HuaiHua Vocational and Technical College.
2.2 PCR amplification and sequencing
The complete mt genome of H. hystricis was divided into five long fragments to amplify by long PCR with ten pairs of primers as shown in Table 1. The primers to amplify target genes were designed based on that of conserved gene sequences on mt genome of other ticks that have a closer phylogenetic relationship, and were then biosynthesized by BGI-Shenzhen (Shenzhen, China). The long gene fragments of mt genome of H. hystricis were amplified with specific primers by long PCR. PCR amplifications were performed in a volume of 50 μL containing 2 mM of MgCl2, 0.2 mM each of deoxyribonucleoside triphosphate, 2.5 μL of 10× rTaq buffer, 2.5 μM of each primer, 1.25 U rTaq polymerase (Takara), 1 μL of DNA sample, and 22 μL of ddH2O. The cycling conditions consisted of predenaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 53–56°C for 45 s and extension at 72°C for 1.5 min, followed by a final extension at 72°C for 5 min. After amplification, the PCR products (5 μL) were examined on a 1.5% agarose gel and visualized under a UV transilluminator. The PCR products were bidirectionally sequenced with a primer walking strategy by Shenggong Biotechnology (Shanghai, China).
Primers used to amplify the complete mt genome of H. hystricis
| Amplified region | Sequence (5′–3′) |
|---|---|
| cox1 | F: TTTAGTTGAAAGAGGAGCCG |
| R: TGATTCCTGTTAGTCCTCCAAC | |
| nad1 | F: TTCTTCAATAGCTTAAATAATTCC |
| R: GTTTTATTAANAGNAGCTTTTTTCACTC | |
| rrnS | F: GCACTTTCCAGTACTTTAACTTTG |
| R: TCTCTAGTTAATTTNGTGCCAGC | |
| cytb | F: GTTATTCCAACTTTTAAATTCAATG |
| R: TTGGTGTAATAATAAAAAATAAGAAG | |
| cox1-nad1 | F: TGATTCTTTGGACACCCAGA |
| R: CATTCAAAGTATTCTCTTATTGGATC | |
| nad1- rrnS | F: CCTGCCTTATTTGGTCCTTT |
| R: GGCGGTATTTTAAGCTTTTC | |
| rrnS-cytb | F: GCACTTTCCAGTACTTTAACTTTG |
| R: TTGGTGTAATAATAAAAAATAAGAAG | |
| cytb-cox1 | F: GTTATTCCAACTTTTAAATTCAATG |
| R: TGATTCCTGTTAGTCCTCCAAC |
2.3 Sequence analyses
Each obtained sequence was corrected by Chromas software. Sequences were assembled manually and aligned by comparing with the whole mt genome sequences of Haemaphysalis longicornis and Haemaphysalis flava available from the GenBank. The gene boundaries in this mt genome were identified by the computer program MAFFT 7.122. Based on the comparison with that of ticks reported previously, the translation initiation and translation termination codons were identified using the computer program Expasy (https://web.expasy.org/translate) [35]. For analyzing sequence characteristics, the four bases A, T, G, and C contents were calculated by the computer program DNAMAN, and the A + T and G + C contents were also calculated, respectively, based on A, T, G, and C contents. Most of the putative secondary structures of tRNAs were identified by the computer program tRNAscan-SE [36], and those of the tRNAs’ secondary structure unidentified by tRNAscan-SE was recognized by eye [28]. Meanwhile, all anticodon sequences of tRNA genes were recognized manually. However, two rRNA genes with different sizes in this mt genome were predicted by comparing with the mt genome sequences of H. longicornis and H. flava reported previously.
2.4 Phylogenetic analyses
Thirteen PCG sequences and two rRNAs gene sequences were concatenated into a single amino acid sequence according to the gene order in mt genome of H. hystricis. However, 18 formerly sequenced ticks’ mt genomes downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank), including H. longicornis (MG721210), Ixodes persulcatus (NC004370), Ixodes hexagonus (AF081828), Ixodes holocyclus (AB075955), Ixodes pavlovskyi (NC023831), Haemaphysalis formosensis (NC020334), Haemaphysalis concinna (NC034785), Haemaphysalis inermis (NC020335), Dermacentor nuttalli (NC028528), Dermacentor silvarum (NC026552), Dermacentor nitens (NC023349), Amblyomma triguttatum (AB113317), Amblyomma cajennense (OP901707), Rhipicephalus australis (KC503255), Rhipicephalus simus (KJ739594), Rhipicephalus sanguineus (AF081829), Rhipicephalus microplus (KP143546), and H. flava (MG604958) were used as inner group. Later, the mt genome sequence of Ornithodoros savignyi (KJ133599) and Argas persicus (OM368320) downloaded from GenBank were also used as out-group. The computer software MAFFT 7.122 was used to align all amino acid sequences, and the Gblocks online server (http://molevol.cmima.csic.es/castresana/Gblocksserver.html) was used to align and exclude the ambiguous regions in the concatenated sequence using less stringent selection options under setting default parameters [37]. These concatenated sequences were subjected to phylogenetic analysis using maximum likelihood (ML) and Bayesian Inference (BI; MrBayes), respectively. The computer software ProtTest 2.4 was used to select the most suitable model of evolution, and the GTR + G + R model was selected based on the Akaike information criterion [38]. The computer software PhyML 3.0 was used to conduct the ML and BI analysis using the subtree pruning and regrafting method, and the ML and BI trees were constructed by FigTreev.1.4 with the default parameters [39]. In addition, Bootstrap frequency was calculated using 100 bootstrap replicates.
3 Results and analysis
3.1 Mitogenome organization and composition
The complete mt genome of H. hystricis was 14,719 bp in size (Figure 1), and the mt genome sequence has been deposited in GenBank under the accession number PP396414. The mt genome of H. hystricis is a typical double stranded circular DNA molecule, which totally contains 37 genes including 13 PCGs, 22 tRNAs, 2 rRNAs, and 2 non-coding regions (Figure 1; Table 2). The gene order on the mt genome is identical to that of the ticks from Haemaphysalis family, but significantly different from that of soft ticks. All genes are transcribed in the same direction. The nucleotide composition of the entire mt genome of H. hystricis is A = 5,649 (38.4%), T = 5,719 (38.9%), G = 1,450 (9.9%), and C = 1,901 (12.9%), and its A + T content was 77.3%, G + C content was 22.8%, showing obvious base AT preference, in accordance with mt genomes of metazoan animals sequenced to date.
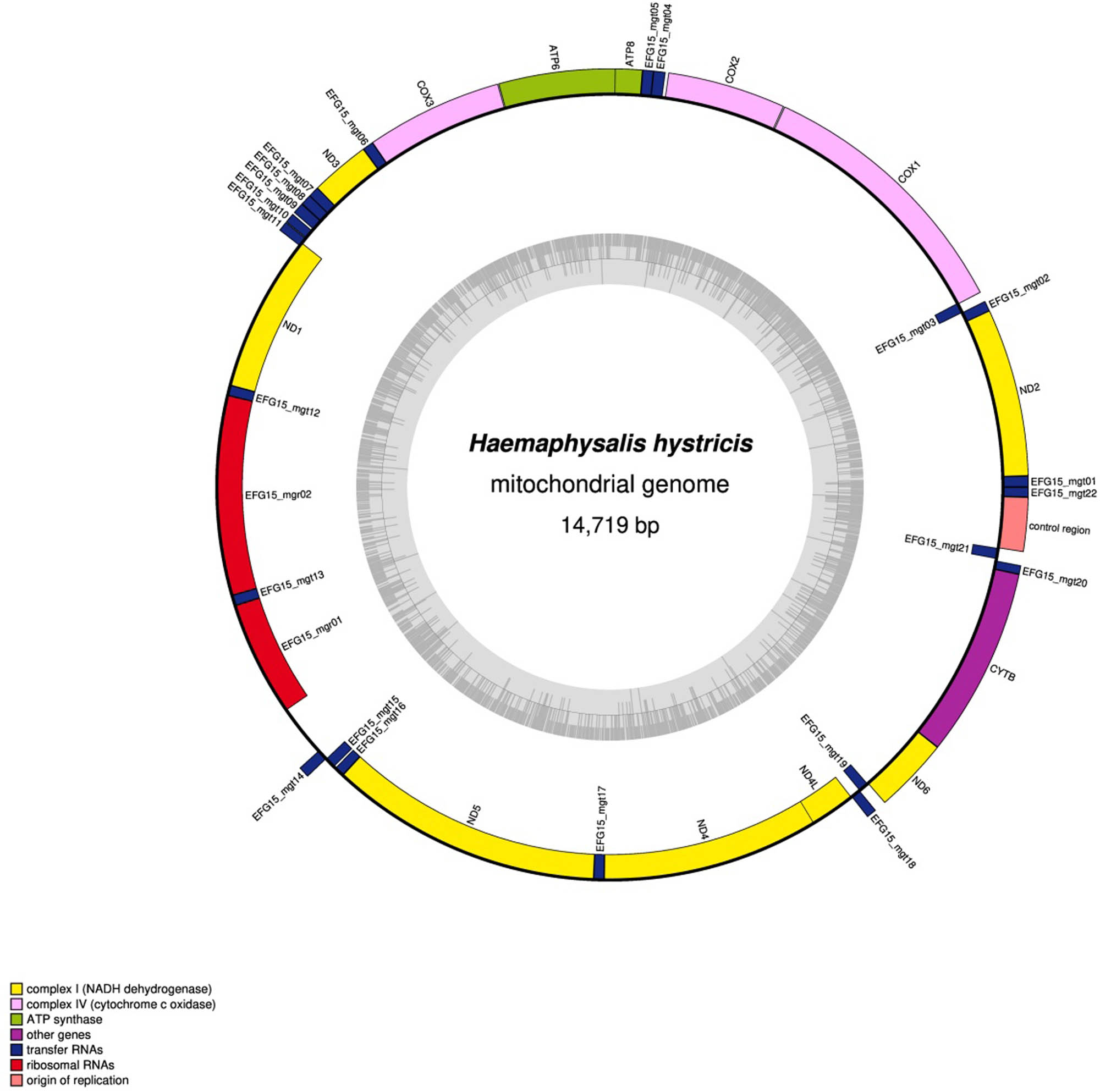
Complete mt genome of H. hystricis.
Organization of H. hystricis mitochondrial genome
| Gene | Position | Length (bp) | Start codons | Stop codons | Anti-codons |
|---|---|---|---|---|---|
| tRNA-Met | 1–62 | 62 | CAT | ||
| nad2 | 63–1,019 | 957 | ATT | TAA | |
| tRNA-Trp | 1,018–1,078 | 61 | TCA | ||
| tRNA-Tyr | 1,077–1,140 | 64 | GTA | ||
| cox1 | 1,133–2,671 | 1,539 | ATT | TAA | |
| cox2 | 2,676–3,350 | 675 | ATG | TAA | |
| tRNA-Lys | 3,367–3,434 | 68 | CTT | ||
| tRNA-Asp | 3,434–3,498 | 65 | GTC | ||
| atp8 | 3,499–3,654 | 156 | ATT | TAA | |
| atp6 | 3,647–4,314 | 668 | ATG | TAA | |
| cox3 | 4,319–5,098 | 780 | ATG | TAA | |
| tRNA-Gly | 5,098–5,159 | 62 | TCC | ||
| nad3 | 5,160–5,498 | 339 | ATT | TAG | |
| tRNA-Ala | 5,497–5,549 | 53 | TGC | ||
| tRNA-Arg | 5,558–5,621 | 64 | TCG | ||
| tRNA-Asn | 5,622–5,687 | 66 | GTT | ||
| tRNA-Ser1 | 5,698–5,755 | 58 | TCT | ||
| tRNA-Glu | 5,762–5,815 | 54 | TTC | ||
| nad1 | 5,793–6,749 | 957 | ATT | TAA | |
| tRNA-Leu1 | 6,750–6,811 | 62 | TAA | ||
| rrnL | 6,812–8,017 | 1,206 | |||
| tRNA-Val | 8,018–8,080 | 63 | TAC | ||
| rrnS | 8,082–8,775 | 694 | |||
| NCRS | 8,776–9,083 | 308 | |||
| tRNA-Ile | 9,084–9,150 | 67 | GAT | ||
| tRNA-Gln | 9,151–9,227 | 77 | TTG | ||
| tRNA-Phe | 9,228–9,288 | 61 | GAA | ||
| nad5 | 9,291–10,942 | 1,652 | ATT | TA | |
| tRNA-His | 10,943–11,005 | 63 | GTG | ||
| nad4 | 11,006–12,320 | 1,315 | ATG | T | |
| nad4L | 12,314–12,589 | 276 | ATG | TAA | |
| tRNA-Thr | 12,592–12,651 | 60 | TGT | ||
| tRNA-Pro | 12,652–12,714 | 63 | TGG | ||
| nad6 | 12,717–13,146 | 430 | ATC | T | |
| cytb | 13,147–14,226 | 1,080 | ATG | TAA | |
| tRNA-Ser2 | 14,227–14,277 | 51 | TGA | ||
| tRNA-Leu2 | 14,289–14,353 | 65 | TAG | ||
| NCRL | 14,354–14,662 | 309 | |||
| tRNA-Cys | 14,663–14,718 | 56 | GCA |
3.2 PCGs and codon usage
The mt genome of H. hystricis totally contains 13 PCGs including nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad1, nad5, nad4, nad4L, nad6, and cytb, of which the longest sequence is nad5 gene of 1,652 bp, and the shortest sequence is atp8 gene that has 155 bp in size. Moreover, in this mt genome, all PCGs used ATN as their initiation codon. For example, the nad2, cox1, atp8, nad3, nad1, and nad5 genes start with ATT, the cox2, atp6, cox3, nad4, nad4L, and cytb genes start with ATG, and the nad6 use ATC, respectively (Table 2). In the same way, 10 of 13 PCGs, namely nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad1, nad4L, and cytb, have complete termination codon (TAN), of which the cox1, atp8, atp6, nad1, nad5, nad4L, nad6, and cytb use TAA as termination codon, the nad3 uses TAG as termination codon. Three of 13 PCGs including nad5, nad4, and nad5 have incomplete termination codon (TA/T), of which nad4 and nad6 genes use T as termination codon, and the nad5 gene uses TA as termination codon. Furthermore, all PCG sequences display base AT preference, which means that their AT content is significantly higher than the GC content.
3.3 Intergenic spacers and overlapping sequences
There are 13 gene gaps in the mt genome of H. hystricis, which varies from 1 to 16 bp. The longest gene gap is located between cox2 and tRNA-Lys. Similarly, eight gene overlap regions with different lengths are found in this mt genome, which varies from 1 to 23 bp. The longest gene overlap region is located s between tRNA-Glu and nad1. The length of all gene gaps is 386 bp accounting for 2.62%.
3.4 Transfer RNA genes (tRNA)
The mt genome of H. hystricis totally contains 22 tRNA genes that range from 51 (tRNA-Ser2) to 77 bp (tRNA-Gln) in size. These tRNAs gene orders were identified in other ticks from Haemaphysalis family mt genome. All 22 tRNA genes could form typical clover structures expect for tRNA-Ala, tRNA-Ser1, tRNA-Ser2, and tRNA-Glu. Their predicted secondary structures are shown in Figure 2, which are like that of other hard ticks. According to Figure 2, we found that the majority of tRNAs display base mismatch phenomenon such as U-U and G-U based on base complementary pairing principle. However, all tRNAs lack components of the secondary structure. For instance, tRNA-Gly, tRNA-Arg, tRNA-Ile, tRNA-Leu2, and tRNA-Lys lack a TψC loop, the tRNA-Met, tRNA-Trp, tRNA-Asn, tRNA-Leu1, tRNA-His, tRNA-Asp, and tRNA-Pro lack a TψC arm, the tRNA-Ala and tRNA-Ser2 lack an amino acid acceptor arm, and all tRNAs of H. hystricis lack a variable loop.
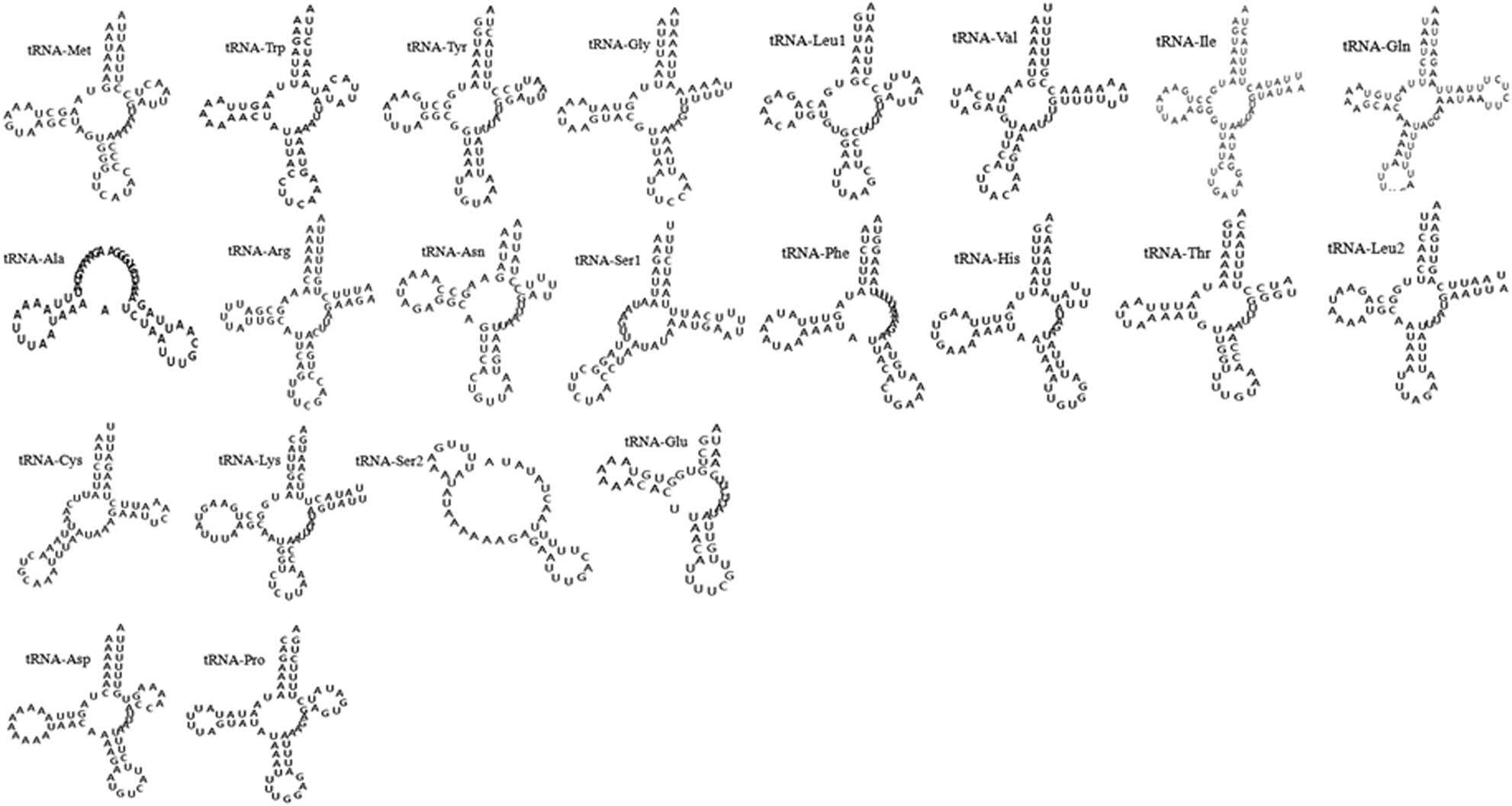
Putative secondary structures of the 22 tRNA genes of H. hystricis.
3.5 Ribosomal RNA genes
The mt genome of H. hystricis contains two rRNAs with different sizes, namely rrnL and rrnS. The rrnL gene of H. hystricis is located between tRNA-Leu1 and tRNA-Val, and rrnS gene is located between tRNA-Val and tRNA-Ile. The rrnL gene sequence of 1,206 bp is longer than the rrnS gene (694 bp) in size. The A + T contents of the rrnL and rrnS genes are 83.2 and 79.7%, respectively, which is similar to some of the other hard ticks.
3.6 A + T-rich region
The mt genome of H. hystricis contains two non-coding regions (A + T-rich region), namely the long non-coding region and the short non-coding region. The long non-coding region was named NCRL, and the short non-coding region was named NCRS in this study. For H. hystricis, the NCRL is located between the tRNA-Leu2 and tRNA-Cys, and the NCRS is located between the rrnS and tRNA-Ile. The lengths of NCRL and NCRS are 309 and 308 bp, respectively. However, the A + T content of the NCRL and NCRS are 65.0 and 68.1%, respectively, which means that their sequences show base AT preference.
3.7 Phylogenetic analyses
Phylogenetic analyses of H. hystricis with selected 18 species of ticks from five genera were performed by ML and BI based on concatenated sequences of 13 PCGs and 2 rRNAs. The phylogenetic trees were reconstructed by computer programs Clustal X, PhyML 3.0, and FigTree v1.3.1, as shown in Figures 3 and 4. According to Figures 3 and 4, we observed that the phylogenetic trees were both divided into five major clades (Clade I–V). Within the two trees, I. persulcatus, I. hexagonus, I. holocyclus, and I. pavlovskyi together clustered into a branch, and the node values were 90 and 57 in the ML and BI trees, respectively; D. nuttalli, D. silvarum, and D. nitens also formed a branch; A. cajennense and A. triguttatum together formed a branch; R. australis, R. simus, R. sanguineus, and R. microplus together clustered into a branch; H. hystricis, H. longicornis, H. formosensis, H. concinna, H. inermis, and H. flava together formed a branch, but this branch was divided into three small branches again, H. concinna, H. formosensis, and H. flava together formed the first small branch, H. longicornis and H. hystricis together formed the second small branch, H. inermis individually formed the third small branch, respectively. Based on the phylogenetic analyses results, we found that H. hystricis was more closely related to H. longicornis than the other selected species of ticks, which is consistent with that of the study on ticks classification [40].
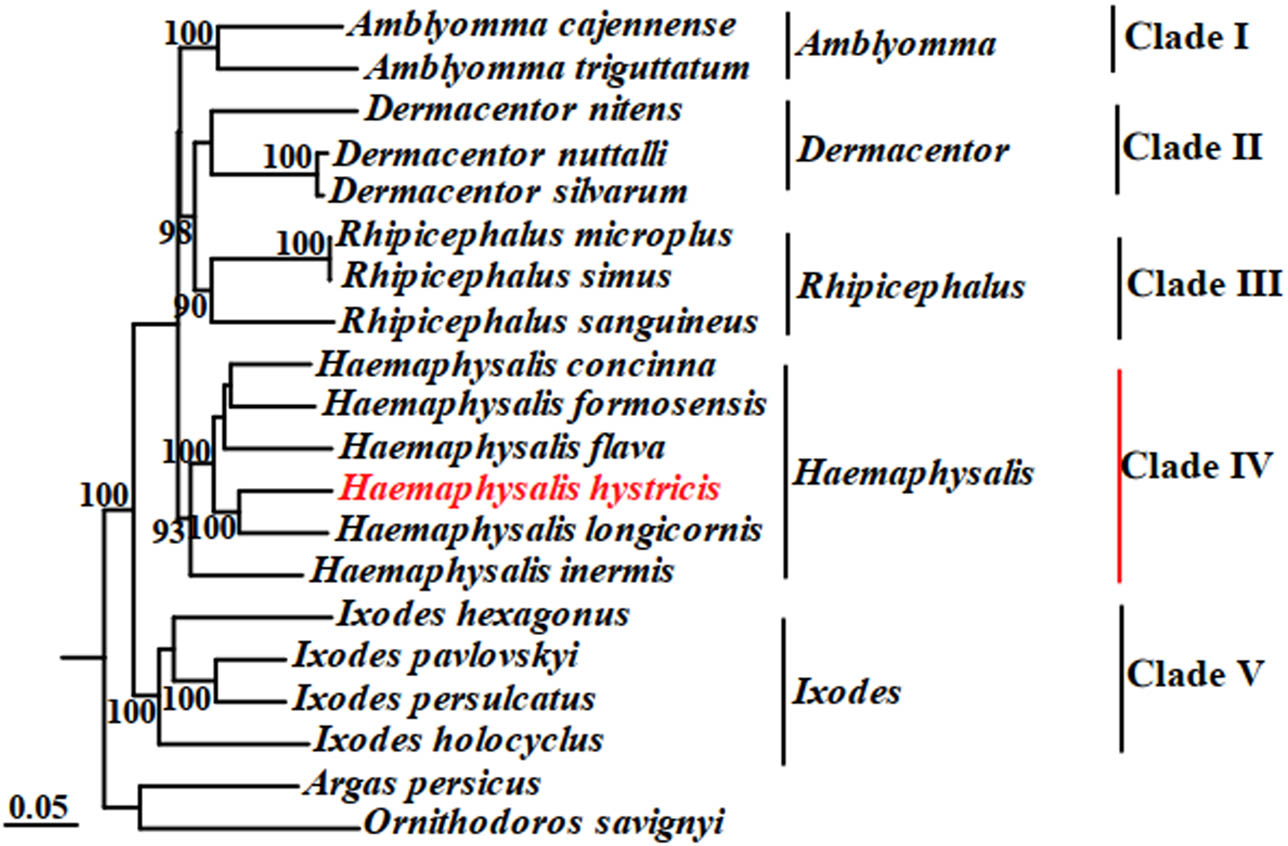
Phylogenetic tree (ML) of H. hystricis by concatenated sequences of 13 PCGs and 2 rRNAs.
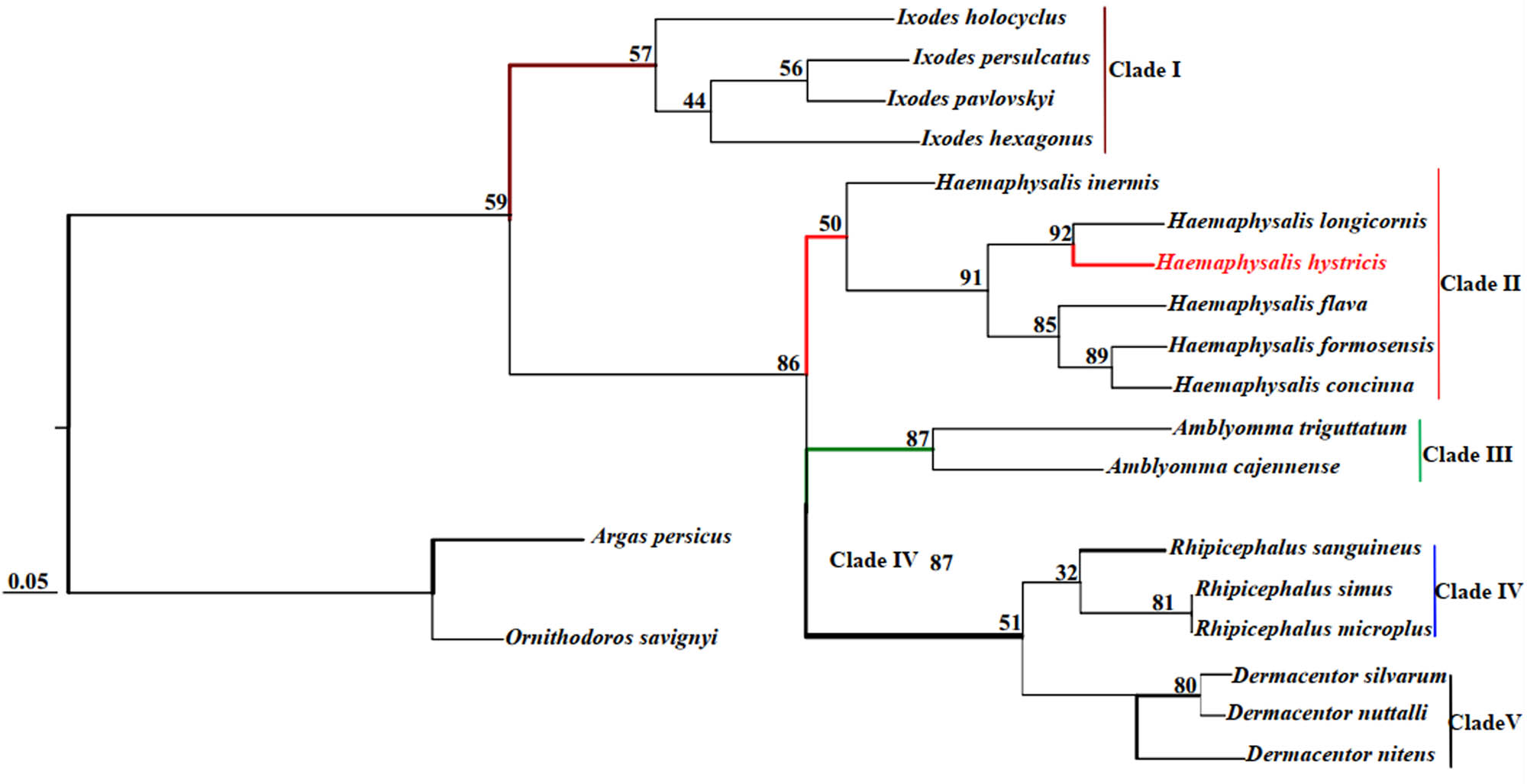
Phylogenetic tree (BI) of H. hystricis by concatenated sequences of 13 PCGs and 2 rRNAs.
4 Discussion
In the recent years, as the study on the classification and identification of parasites has developed toward the molecular level, many molecular biology techniques were applied widely to parasitology research, especially PCR. Compared to traditional morphological methods, not only the molecular biology technology is completely unaffected by individual differences, sample integrity, appraiser experience, developmental stages of the insect, and its environment, but also the classification and identification results obtained have a high degree of accuracy [41]. Currently, molecular biology technology has become an important auxiliary means for classification and identification of parasites [42–44]. The most crucial step in using molecular biotechnology to identify and classify parasites is how to find and select appropriate genetic markers. According to the massive research literature, mt genome of metazoan animals and some of its gene sequence all have characteristics such as rapid genetic evolution, lack of gene recombination, and small intra-species differences but large inter-species differences [28,31]. Due to the information provided by the entire mt genome is far more than that of fragmented gene sequences, so some researchers think that the mt genome is more valuable as genetic marker for studying species identification, genetic structure, gene variation, haplotype polymorphism, and phylogenetic relationship of parasites [24,45]. Thus, we sequenced the mt genome of H. hystricis by PCR, and analyzed its sequence characteristics, gene composition, arrangement order, and codons usage; meanwhile, we explored phylogenetic relationship between H. hystricis and other ticks by 13 PCG sequences.
Based on the sequencing, annotation, gene composition, and arrangement order results, the entire mt genome of H. hystricis contains 37 genes (13 PCGs, 22 tRNAs, 2 rRNAs), which is in accordance with other metazoan animals’ mt genome reported previously [46–48]. Compared with other ticks from Haemaphysalis family, we found that the mt genome of H. hystricis is far longer than that of H. longicornis, H. formosensis, H. concinna, and H. flava, but is significantly shorter than that of H. inermis, which may be closely related to the interval region, overlapping sequence, and D-loop region contained in their mt genome. Generally, most of the scholars believed that the number and length of D-loop region contained in the mt genome directly affect the length of the mt genome [49,50]. However, some scholars opined that the D-loop region sequence in mt genome lose bases, which affects the length of the mt genome [51,52], but Zhang and Hewitt thought that the number of concatenated duplicate copies in D-loop region directly affect the length of the mt genome [53]. However, the mt genome of H. hystricis and its 13 PCG sequences both show base AT preference clearly, which is consistent with that of the results of the sequence characteristics of mt genome of metazoan animals [54,55]. Some researchers pointed out that the mt genome of metazoan animals has the characteristics of multiple copy numbers and fast evolution rate [28,31], which may be related to its base AT preference. To the best of our knowledge, a nucleotide sequence has high AT content meaning that it has a low melting point temperature causing DNA double stranded to dissociate into single strand, which greatly increases the probability of base mutation and improves the speed of evolution.
According to the results of the codons usage, we found that all PCGs used a standard ATN start codon in mt genome of H. hystricis. Six PCGs (nad2, cox1, atp8, nad3, nad5, and nad6) started with ATT (Ile), five PCGs including cox2, atp6, cox3, nad4, and nad4L started with ATG (Met), and nad1 started with ATA (Met), which is consistent with other ticks [28,56,57]. All initiation codons of this mt gene also appear in other mt gene of other arthropods including most Acari species, which suggests that the pattern of initiation codon usage is relatively conservative in arthropod mitochondrial genomes [58]. However, those unusual start codons such as TTG and GTG that are predicted in D. pteronyssinus and D. farinae [57] are not found in H. hystricis, suggesting that the mt genome of H. hystricis is no alternative start codons. A total of eight PCGs applied the complete termination codon (TAA and TAG), while four PCGs (nad2, cox3, nad3, and nad4) ended at a single T residue, and the cytb ended at TA residue, which is also consistent with some ticks and arthropod [28,57]. The mitochondrial genome of metazoans with incomplete stop codon is common, and some studies believed that these incomplete stop codons are the results of gene polyadenylation after transcription [28,57,59]. In H. hystricis, the PCGs that use T as termination codon may end with a TGA codon; however, in this gene, as the GA nucleotide in the termination codon overlaps with the 5′end of downstream genes, they are annotated as ending with the T to minimize overlapping [57]. Usually, the termination codon of the cytb gene in mt genome of metazoans is considered to be complete by cleaving the transcripts of the polycistron, and form the stop codon by mRNA polyadenylation [60].
According to the results of the tRNAs analysis, we found that most of tRNAs (except for tRNA-Ala, tRNA-Ser1, tRNA-Ser2, and tRNA-Glu) have typical secondary structures, but five tRNAs lack a TψC loop, and all tRNAs lack the variable loop. Generally, the tRNA losing the D-arm is common, especially for the tRNA-Ser1, tRNA-Ser2, tRNA-Leu1, and tRNA-Leu2. Some studies pointed out that the trnS1 lost D-arm has been considered as a typical feature in mt genome of all chelicerate [61]. The tRNA-Ser1 in the mt genome of H. hystricis does not lose the D-arm, which agrees with the viewpoint that this structure in other tRNAs of arthropods is less common as proposed by Sun et al. [57]. Besides, all tRNAs of H. hystricis do not lose the D-arm, which is contrary to those study results that tRNAs lacking D-arm were found in mt genome of the sea spiders, the scorpion, Ixodida, Mesostigmata, and Acariformes. Furthermore, we found that the majority of tRNAs in H. hystricis exhibits base mismatch phenomenon (U-U and G-U), which is consistent with the results reported previously [31,56]. Some researchers opined that the base mismatch may be closely related to the rare base in tRNAs [62].
According to the results of the rRNAs analysis, we found that the rrnL is longer than rrnS in size, and both are located in close proximity to each other on the N-strand and next to the largest non-coding regions, which is consistent with many mt genome of arthropods including H. longicornis, H. formosensis, H. concinna, H. inermis, and H. flava reported previously [53], but is significantly different from the mt genome of D. pteronyssinus, D. farinae, and P. persimilis. The rrnL and rrnS genes in the mt genome of D. pteronyssinus, D. farinae, and P. persimilis are both located on the J- strand, very close to each other, and distant from the largest non-coding regions [63,64]. These results suggested that the transcription mechanisms of rRNA gene of metazoans may be different, and it requires further investigation. Besides, the sequences of rrnS and rrnL both show AT preferences.
According to the results of the non-coding regions analysis, we found that there are gene gaps, overlapping regions, and two non-coding regions in H. hystricis mitogenome. However, the number of gene gap is far more than that of overlapping region, which suggests that mt genome of H. hystricis is not very tight. Two non-coding regions, also known as D-loop region, in the mt genome of H. hystricis have different lengths, and both display AT preferences. Some researchers point out that the non-coding region of arthropods is rich in tandem AT repeat sequences, its A + T content ranges from 80 to 98% [65], and these AT repeat sequences directly affect the entire mitochondrial genome in size. Moreover, the non-coding region contains a large number of replication starting points, transcription starting points, and control sequences [66], which forms a secondary structure of the stem ring through hydrogen bonding, and there are highly conserved sequences related to the replication, transcription, and translation processes of the mitochondrial genomes such as TATA and GAAT on both sides of the stem ring structure.
According to the results of the phylogenetic analysis, we found that there is a close phylogenetic relationship between H. hystricis and H. longicornis among the selected ticks from Haemaphysalis family, followed by H. flava, H. formosensis, H. concinna, and H. inermis. Compared to other genus ticks, the phylogenetic relationship of Haemaphysalis and Amblyomma ticks is the closest, followed by Dermacentor and Rhipicephalus genera, and the Ixodes genus is the latest. It suggests that the mt genome of H. hystricis is an effective tool for studying its molecular epidemiology, population genetics, and systematics. Sequencing mt genome of H. hystricis, not only enriches the genome database, but also has implications for the diagnosis, prevention, and control of ticks and tick-borne diseases in animals and humans.
5 Conclusion
In the present study, we determined the complete mtDNA data of H. hystricis and compared with that of other ticks from Haemaphysalis and other genera. According to the results, we found that the mt gene arrangement for H. hystricis is the same as that of the selected ticks from Haemaphysalis family, and its sequence shows AT preference clearly. Most of the tRNA genes of H. hystricis are able to form typical clover structures expect for tRNA-Ala, tRNA-Ser1, tRNA-Ser2, and tRNA-Glu, and there are base mismatch phenomenon in the structures of tRNAs. The mt genome of H. hystricis has two rRNAs and non-coding regions of distinct AT preference. Phylogenetic analyses also indicate that H. hystricis is more closely related to H. longicornis than to other ticks. However, the mt genome has implications for further studying the molecular identification, population genetics, systematics of ticks, as well as the diagnosis, prevention, and control of tick-borne diseases in animals and humans.
Acknowledgements
This work was supported in part by Scientific Research Fund of Hunan Provincial Education Department (No. 23B1072), the Natural Science Fund of Hunan Provincial (No. 2022JJ50319), the Project support was provided by the Research Foundation of Engineering Research Center for the Prevention and Control of Animal Original Zoonosis, Fujian Province University (2022K004), and the Key Laboratory Project Fund of Huaihua (Nos. 2023R2208, 2020R2206).
-
Funding information: Authors state no funding involved.
-
Author contributions: All authors have contributed to this manuscript and have agreed to its submission to the journal. They have reviewed all results and approved the final version. LI.Z.B, and H.C.Q designed the study; X.M., H.H., and S.M. conducted all experiments; LI.Z.B and Y.T. analyzed the data; LI.Z.B and H.C.Q drafted and revised the manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Xu XL, Cheng TY, Yang H. Enolase, a plasminogen receptor isolated from salivary gland transcriptome of the Ixodid tick Haemaphysalis flava. Parasitol Res. 2016;115(5):1955–64.10.1007/s00436-016-4938-0Search in Google Scholar PubMed
[2] Liu L, Cheng TY, Yan F. Expression pattern of subA in different tissues and bloodfeeding status in Haemaphysalis flava. Exp Appl Acarol. 2016;70(4):511–22.10.1007/s10493-016-0088-4Search in Google Scholar PubMed
[3] Sormunen JJ, Klemola T, Vesterinen EJ. Ticks (Acari: Ixodidae) parasitizing migrating and local breeding birds in Finland. Exp Appl Acarol. 2022;86(1):145–56.10.1007/s10493-021-00679-3Search in Google Scholar PubMed PubMed Central
[4] Suzin A, da Silva MX, Tognolli MH, Vogliotti A, Adami SF, Moraes MFD, et al. Ticks on humans in an Atlantic rainforest preserved ecosystem in Brazil: species, life stages, attachment sites, and temporal pattern of infestation. Ticks Tick Borne Dis. 2022;13(1):101862.10.1016/j.ttbdis.2021.101862Search in Google Scholar PubMed
[5] Greay TL, Oskam CL, Gofton AW, Rees RL, Ryan UM, Irwin Peter J. A survey of ticks (Acari: Ixodidae) of companion animals in Australia. Parasit Vectors. 2016;9(1):207.10.1186/s13071-016-1480-ySearch in Google Scholar PubMed PubMed Central
[6] Silaghi C, Beck R, Oteo JA, Pfeffer M, Sprong H. Neoehrlichiosis: an emergingtick-borne zoonosis caused by Candidatus Neoehrlichia mikurensis. Exp Appl Acarol. 2016;68:279–97.10.1007/s10493-015-9935-ySearch in Google Scholar PubMed
[7] Zhao GP, Wang YX, Fan ZW, Ji Y, Liu MJ, Zhang WH, et al. Mapping ticks and tick-borne pathogens in China. Nat Commun. 2021;12(1):1075.10.1038/s41467-021-21375-1Search in Google Scholar PubMed PubMed Central
[8] Madison-Antenucci S, Kramer LD, Gebhardt LL, Kauffman E. Emerging tick-borne diseases. Clin Microbiol Rev. 2020;33(2):e00083-18.10.1128/CMR.00083-18Search in Google Scholar PubMed PubMed Central
[9] Cave GL, Richardson EA, Chen K, Watson DW, Roe RM. Acaricidal biominerals and mode-of-action studies against adult blacklegged ticks, Ixodes scapularis. Microorganisms. 2023;11(8):1906.10.3390/microorganisms11081906Search in Google Scholar PubMed PubMed Central
[10] Burger TD, Shao R, Beati L, Miller H, Barker SC. Phylogenetic analysis of ticks (Acari: Ixodida) using mitochondrial genomes and nuclear rRNA genes indicates that the genus Amblyomma is polyphyletic. Mol Phylogenet Evol. 2012;64:45–55.10.1016/j.ympev.2012.03.004Search in Google Scholar PubMed
[11] Li ZB, Liu GH, Cheng TY. Molecular characterization of hard tick Haemaphysalis longicornis from China by sequences of the internal transcribed spacers of ribosomal DNA. Exp Appl Acarol. 2018;74(2):171–6.10.1007/s10493-018-0210-xSearch in Google Scholar PubMed PubMed Central
[12] Deng GF. Economic insect fauna of China. Vol. 15, Beijing: Science Press; 1978.Search in Google Scholar
[13] Jongejan F, Su BL, Yang HJ, Berger L, Bevers J, Liu PC, et al. Molecular evidence for the transovarial passage of Babesia gibsoni in Haemaphysalis hystricis (Acari: Ixodidae) ticks from Taiwan: a novel vector for canine babesiosis. Parasit Vectors. 2018;11(1):134.10.1186/s13071-018-2722-ySearch in Google Scholar PubMed PubMed Central
[14] Su BL, Liu PC, Fang JC, Jongejan F. Correlation between Babesia species affecting dogs in Taiwan and the local distribution of the vector ticks. Vet Sci. 2023;10(3):227.10.3390/vetsci10030227Search in Google Scholar PubMed PubMed Central
[15] Li ZB, Fu YT, Cheng TY, Yao GM, Hou QH, Li F, et al. Mitochondrial gene heterogeneity and population genetics of Haemaphysalis longicornis (Acari: Ixodidae) in China. Acta Parasitol. 2019;64(2):360–6.10.2478/s11686-019-00053-9Search in Google Scholar PubMed
[16] Yuasa Y, Tsai YL, Chang CC, Hsu TH, Chou CC. The prevalence of Anaplasma platys and a potential novel Anaplasma species exceed that of Ehrlichia canis in asymptomatic dogs and Rhipicephalus sanguineus in Taiwan. J Vet Med Sci. 2017;79(9):1494–502.10.1292/jvms.17-0224Search in Google Scholar PubMed PubMed Central
[17] Khoo JJ, Lim FS, Tan KK, Chen FS, Phoon WH, Khor CS, et al. Detection in malaysia of a Borrelia sp. from Haemaphysalis hystricis (Ixodida: Ixodidae). J Med Entomol. 2017;54(5):1444–8.10.1093/jme/tjx131Search in Google Scholar PubMed
[18] Li J, Liu X, Mu J, Yu X, Fei Y, Chang J, et al. Emergence of a novel Ehrlichia minasensis strain, harboring the major immunogenic glycoprotein trp36 with unique tandem repeat and C-terminal region sequences, in Haemaphysalis hystricis ticks removed from free-ranging sheep in Hainan province, China. Microorganisms. 2019;7(9):369.10.3390/microorganisms7090369Search in Google Scholar PubMed PubMed Central
[19] Testé E, Hernández-Rodríguez M, Veltjen E, Bécquer ER, Rodríguez-Meno A, Palmarola A, et al. Integrating morphological and genetic limits in the taxonomic delimitation of the cuban taxa of Magnoliasubsect, Talauma (Magnoliaceae). PhytoKeys. 2022;213:35–66.10.3897/phytokeys.213.82627.figure7Search in Google Scholar
[20] Nava S, Estrada-Peña A, Petney T, Beati L, Labruna MB, Szabó MP, et al. The taxonomic status of Rhipicephalus sanguineus (Latreille, 1806). Vet Parasitol. 2015;208:2–8.10.1016/j.vetpar.2014.12.021Search in Google Scholar PubMed
[21] Barker SC, Walker AR, Campelo D. A list of the 70 species of Australian ticks; diagnostic guides to and species accounts of Ixodes holocyclus (paralysis tick), Ixodes cornuatus (southern paralysis tick) and Rhipicephalus australis (Australian cattle tick); and consideration of the place of Australia in the evolution of ticks with comments on four controversial ideas. Int J Parasitol. 2014;44:941–53.10.1016/j.ijpara.2014.08.008Search in Google Scholar PubMed
[22] Song S, Shao R, Atwell R, Barker S, Vankan D. Phylogenetic and phylogeographic relationships in Ixodes holocyclus and Ixodes cornuatus (Acari: Ixodidae) inferred from cox1 and ITS2 sequences. Int J Parasitol. 2011;41:871–80.10.1016/j.ijpara.2011.03.008Search in Google Scholar PubMed
[23] Kovalev SY, Golovljova IV, Mukhacheva TA. Natural hybridization between Ixodes ricinus and Ixodes persulcatus ticks evidenced by molecular genetics methods. Ticks TickBorne Dis. 2016;7:113–8.10.1016/j.ttbdis.2015.09.005Search in Google Scholar PubMed
[24] Deng YP, Yi JN, Fu YT, Nie Y, Zhang Y, Liu GH. Comparative analyses of the mitochondrial genomes of the cattle tick Rhipicephalus microplus clades A and B from China. Parasitol Res. 2022;121(6):1789–97.10.1007/s00436-022-07501-ySearch in Google Scholar PubMed
[25] Palomares-Rius JE, Tsai IJ, Karim N, Akiba M, Kato T, Maruyama H, et al. Genome-wide variation in the pinewood nematode Bursaphelenchus xylophilus and its relationship with pathogenic traits. BMC Genomics. 2015;16:845.10.1186/s12864-015-2085-0Search in Google Scholar PubMed PubMed Central
[26] Anderson DL, Morgan MJ. Genetic and morphological variation of bee-parasitic Tropilaelaps mites (Acari: Laelapidae): new and re-defined species. Exp Appl Acarol. 2007;43(1):1–24.10.1007/s10493-007-9103-0Search in Google Scholar PubMed
[27] Zhang Y, Nie Y, Deng YP, Liu GH, Fu YT. The complete mitochondrial genome sequences of the cat flea Ctenocephalides felis felis (Siphonaptera: Pulicidae) support the hypothesis that C. felis isolates from China and USA were the same C. f. felis subspecies. Acta Trop. 2021;217:105880.10.1016/j.actatropica.2021.105880Search in Google Scholar PubMed
[28] Liu GH, Li S, Zou FC, Wang CR, Zhu XQ. The complete mitochondrial genome of rabbit pinworm Passalurus ambiguus: genome characterization and phylogenetic analysis. Parasitol Res. 2016;115(1):423–9.10.1007/s00436-015-4778-3Search in Google Scholar PubMed
[29] Lu XY, Zhang QF, Jiang DD, Du CH, Xu R, Guo XG, et al. Characterization of the complete mitochondrial genome of Ixodes granulatus (Ixodidae) and its phylogenetic implications. Parasitol Res. 2022;121(8):2347–58.10.1007/s00436-022-07561-0Search in Google Scholar PubMed
[30] Wang C, Wang L, Liu Y, Deng L, Wei M, Wu K, et al. The mitochondrial genome of the giant panda tick Haemaphysalis flava (Acari, Ixodidae) from Southwest China. Mitochondrial DNA B Resour. 2020;5(2):1188–90.10.1080/23802359.2020.1731350Search in Google Scholar PubMed PubMed Central
[31] Liu GH, Chen F, Chen YZ, Song HQ, Lin RQ, Zhou DH, et al. Complete mitochondrial genome sequence data provides genetic evidence that the brown dog tick Rhipicephalus sanguineus (Acari: Ixodidae) represents a species complex. Int J Biol Sci. 2013;9(4):361–9.10.7150/ijbs.6081Search in Google Scholar PubMed PubMed Central
[32] Li LY, Deng YP, Zhang Y, Wu Y, Fu YT, Liu GH, et al. Characterization of the complete mitochondrial genome of Culex vishnui (Diptera: Culicidae), one of the major vectors of Japanese encephalitis virus. Parasitol Res. 2023;122(6):1403–14.10.1007/s00436-023-07840-4Search in Google Scholar PubMed
[33] Kneubehl AR, Muñoz-Leal S, Filatov S, de Klerk DG, Pienaar R, Lohmeyer KH, et al. Amplification and sequencing of entire tick mitochondrial genomes for a phylogenomic analysis. Sci Rep. 2022;12(1):19310.10.1038/s41598-022-23393-5Search in Google Scholar PubMed PubMed Central
[34] Zou Y, Chen M, Wu T, Tian T, Wen Z. Identification and analysis of the complete mitochondrial genome of Acheilognathus omeiensis (Cypriniformes, Cyprinidae). Mitochondrial DNA B Resour. 2018;3(1):270–1.10.1080/23802359.2018.1443032Search in Google Scholar PubMed PubMed Central
[35] Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–52.10.1385/1-59259-584-7:531Search in Google Scholar PubMed
[36] Lowe TM, Eddy SR. TRNAscan – SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64.10.1093/nar/25.5.955Search in Google Scholar PubMed PubMed Central
[37] Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77.10.1080/10635150701472164Search in Google Scholar PubMed
[38] Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5.10.1093/bioinformatics/bti263Search in Google Scholar PubMed
[39] Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704.10.1080/10635150390235520Search in Google Scholar PubMed
[40] Liu Y, Wang L, Wang L, Deng L, Wei M, Wu K, et al. Characterization of the complete mitogenome sequence of the giant panda tick Haemaphysalis hystricis. Mitochondrial DNA B Resour. 2020;5(2):1191–3.10.1080/23802359.2020.1731352Search in Google Scholar PubMed PubMed Central
[41] Coimbra-Dores MJ, Maia-Silva M, Marques W, Oliveira AC, Rosa F, Dias D. Phylogenetic insight on Mediterranean and Afrotropical Rhipicephalus species (Acari: Ixodida) based on mitochondrial DNA. Exp Appl Acarol. 2018;75:107–28.10.1007/s10493-018-0254-ySearch in Google Scholar PubMed
[42] Yi JN, Jin YC, Liu JH, Li F, Li ZB, Cheng TY. Genetic variation in three mitochondrial genes among cattle tick Rhipicephalus microplus originating from four provinces of China. Trop Biomed. 2019;36(1):297–303.Search in Google Scholar
[43] Kulakova NV, Khasnatinov MA, Sidorova EA, Adel’shin RV, Belikov SI. Molecular identification and phylogeny of Dermacentor nuttalli (Acari: Ixodidae). Parasitol Res. 2014;113:1787–93.10.1007/s00436-014-3824-xSearch in Google Scholar PubMed
[44] Kanduma EG, Mwacharo JM, Githaka NW, Kinyanjui PW, Njuguna JN, Kamau LM, et al. Analyses of mitochondrial genes reveal two sympatric but genetically divergent lineages of Rhipicephalus appendiculatus in Kenya. Parasit Vectors. 2016;9:353.10.1186/s13071-016-1631-1Search in Google Scholar PubMed PubMed Central
[45] Chen Z, Xuan Y, Liang G, Yang X, Yu Z, Barker SC, et al. Precise annotation of tick mitochondrial genomes reveals multiple copy number variation of short tandem repeats and one transposon-like element. BMC Genomics. 2020;21(1):488.10.1186/s12864-020-06906-2Search in Google Scholar PubMed PubMed Central
[46] Hyeon JY, McGinnis H, Sims M, Helal ZH, Kim J, Chung DH, et al. Complete mitochondrial genome of Asian longhorned tick, Haemaphysalis longicornis, Neumann, 1901 (Acari: Ixodida: Ixodidae) identified in the United States. Mitochondrial DNA B Resour. 2021;6(8):2402–5.10.1080/23802359.2021.1922100Search in Google Scholar PubMed PubMed Central
[47] Sui S, Yang Y, Fang Z, Wang J, Wang J, Fu Y, et al. Complete mitochondrial genome and phylogenetic analysis of Ixodes persulcatus (taiga tick). Mitochondrial DNA B Resour. 2016;2(1):3–4.10.1080/23802359.2016.1202737Search in Google Scholar PubMed PubMed Central
[48] Almeida C, Simões R, Coimbra-Dores MJ, Rosa F, Dias D. Mitochondrial DNA analysis of Rhipicephalus sanguineus s.l. from the western Iberian peninsula. Med Vet Entomol. 2017;31(2):167–77.10.1111/mve.12222Search in Google Scholar PubMed
[49] Lu X, Jiang D, Du C, Rao C, Yin J, Fang Y, et al. Complete mitochondrial genome and phylogenetic analysis of Ixodes acutitarsus (Acari: Ixodidae). Mitochondrial DNA B Resour. 2022;7(6):1134–6.10.1080/23802359.2022.2089066Search in Google Scholar PubMed PubMed Central
[50] Chen B, Liu YF, Lu XY, Jiang DD, Wang X, Zhang QF, et al. Complete mitochondrial genome of Ctenophthalmus quadratus and Stenischia humilis in China provides insights into fleas phylogeny. Front Vet Sci. 2023;10:1255017.10.3389/fvets.2023.1255017Search in Google Scholar PubMed PubMed Central
[51] Zhang J, Peng C, Xu S, Zhao Y, Zhang X, Zhang S, et al. Mitochondrial displacement loop region single nucleotide polymorphisms and mitochondrial DNA copy number associated with risk of ankylosing spondylitis. Int J Rheum Dis. 2023;26(11):2157–62.10.1111/1756-185X.14876Search in Google Scholar PubMed
[52] Toh YL, Wong E, Chae JW, Yap NY, Yeo AHL, Shwe M, et al. Association of mitochondrial DNA content and displacement loop region sequence variations with cancer-related fatigue in breast cancer survivors receiving chemotherapy. Mitochondrion. 2020;54:65–71.10.1016/j.mito.2020.07.004Search in Google Scholar PubMed
[53] Zhang DX, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997;25(2):99–120.10.1016/S0305-1978(96)00042-7Search in Google Scholar
[54] Amzati GS, Pelle R, Muhigwa JB, Kanduma EG, Djikeng A, Madder M, et al. Mitochondrial phylogeography and population structure of the cattle tick Rhipicephalus appendiculatus in the African Great Lakes region. Parasit Vectors. 2018;11(1):329.10.1186/s13071-018-2904-7Search in Google Scholar PubMed PubMed Central
[55] Guo DH, Zhang Y, Fu X, Gao Y, Liu YT, Qiu JH, et al. Complete mitochondrial genomes of Dermacentor silvarum and comparative analyses with another hard tick Dermacentor nitens. Exp Parasitol. 2016;169:22–7.10.1016/j.exppara.2016.07.004Search in Google Scholar PubMed
[56] Wang Y, Zhu FC, He LS, Danchin A. Unique tRNA gene profile suggests paucity of nucleotide modifications in anticodons of a deep-sea symbiotic Spiroplasma. Nucleic Acids Res. 2018;46(5):2197–203.10.1093/nar/gky045Search in Google Scholar PubMed PubMed Central
[57] Sun ET, Li CP, Nie LW, Jiang YX. The complete mitochondrial genome of the brown leg mite, Aleuroglyphus ovatus (Acari: Sarcoptiformes): evaluation of largest non-coding region and unique tRNAs. Exp Appl Acarol. 2014;64(2):141–57.10.1007/s10493-014-9816-9Search in Google Scholar PubMed
[58] Dermauw W, Van Leeuwen T, Vanholme B, Tirry L. The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods. BMC Genom. 2009;10:107.10.1186/1471-2164-10-107Search in Google Scholar PubMed PubMed Central
[59] Tuli MD, Li H, Pan X, Li S, Zhai J, Wu Y, et al. Heteroplasmic mitochondrial genomes of a Raillietina tapeworm in wild Pangolin. Parasit Vectors. 2022;15(1):204.10.1186/s13071-022-05301-ySearch in Google Scholar PubMed PubMed Central
[60] Cattaneo R. Different types of messenger RNA editing. Annu Rev Genet. 1991;25:71–88.10.1146/annurev.genet.25.1.71Search in Google Scholar
[61] Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216.10.1016/S0074-7696(08)62066-5Search in Google Scholar
[62] Murata A, Nakamori M, Nakatani K. Modulating RNA secondary and tertiary structures by mismatch binding ligands. Methods. 2019;167:78–91.10.1016/j.ymeth.2019.05.006Search in Google Scholar PubMed
[63] Dermauw W, Vanholme B, Tirry L, Van Leeuwen T. Mitochondrial genome analysis of the predatory mite Phytoseiulus persimilis and a revisit of the Metaseiulus occidentalis mitochondrial genome. Genome. 2010;53:285–301.10.1139/G10-004Search in Google Scholar PubMed
[64] Klimov PB, OConnor BM. Improved tRNA prediction in the American house dust mite reveals widespread occurrence of extremely short minimal tRNAs in acariform mites. BMC Genom. 2009;10:598.10.1186/1471-2164-10-598Search in Google Scholar PubMed PubMed Central
[65] Shao R, Campbell NJ, Barker SC. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera). Mol Biol Evol. 2001;18(5):858–65.10.1093/oxfordjournals.molbev.a003867Search in Google Scholar PubMed
[66] Saito S, Tamura K, Aotsuka T. Replication origin of mitochondrial DNA in insects. Genetics. 2005;171(4):1695–705.10.1534/genetics.105.046243Search in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”
Articles in the same Issue
- Biomedical Sciences
- Mechanism of triptolide regulating proliferation and apoptosis of hepatoma cells by inhibiting JAK/STAT pathway
- Maslinic acid improves mitochondrial function and inhibits oxidative stress and autophagy in human gastric smooth muscle cells
- Comparative analysis of inflammatory biomarkers for the diagnosis of neonatal sepsis: IL-6, IL-8, SAA, CRP, and PCT
- Post-pandemic insights on COVID-19 and premature ovarian insufficiency
- Proteome differences of dental stem cells between permanent and deciduous teeth by data-independent acquisition proteomics
- Optimizing a modified cetyltrimethylammonium bromide protocol for fungal DNA extraction: Insights from multilocus gene amplification
- Preliminary analysis of the role of small hepatitis B surface proteins mutations in the pathogenesis of occult hepatitis B infection via the endoplasmic reticulum stress-induced UPR-ERAD pathway
- Efficacy of alginate-coated gold nanoparticles against antibiotics-resistant Staphylococcus and Streptococcus pathogens of acne origins
- Battling COVID-19 leveraging nanobiotechnology: Gold and silver nanoparticle–B-escin conjugates as SARS-CoV-2 inhibitors
- Neurodegenerative diseases and neuroinflammation-induced apoptosis
- Impact of fracture fixation surgery on cognitive function and the gut microbiota in mice with a history of stroke
- COLEC10: A potential tumor suppressor and prognostic biomarker in hepatocellular carcinoma through modulation of EMT and PI3K-AKT pathways
- High-temperature requirement serine protease A2 inhibitor UCF-101 ameliorates damaged neurons in traumatic brain-injured rats by the AMPK/NF-κB pathway
- SIK1 inhibits IL-1β-stimulated cartilage apoptosis and inflammation in vitro through the CRTC2/CREB1 signaling
- Rutin–chitooligosaccharide complex: Comprehensive evaluation of its anti-inflammatory and analgesic properties in vitro and in vivo
- Knockdown of Aurora kinase B alleviates high glucose-triggered trophoblast cells damage and inflammation during gestational diabetes
- Calcium-sensing receptors promoted Homer1 expression and osteogenic differentiation in bone marrow mesenchymal stem cells
- ABI3BP can inhibit the proliferation, invasion, and epithelial–mesenchymal transition of non-small-cell lung cancer cells
- Changes in blood glucose and metabolism in hyperuricemia mice
- Rapid detection of the GJB2 c.235delC mutation based on CRISPR-Cas13a combined with lateral flow dipstick
- IL-11 promotes Ang II-induced autophagy inhibition and mitochondrial dysfunction in atrial fibroblasts
- Short-chain fatty acid attenuates intestinal inflammation by regulation of gut microbial composition in antibiotic-associated diarrhea
- Application of metagenomic next-generation sequencing in the diagnosis of pathogens in patients with diabetes complicated by community-acquired pneumonia
- NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation
- Phytol-mixed micelles alleviate dexamethasone-induced osteoporosis in zebrafish: Activation of the MMP3–OPN–MAPK pathway-mediating bone remodeling
- Association between TGF-β1 and β-catenin expression in the vaginal wall of patients with pelvic organ prolapse
- Primary pleomorphic liposarcoma involving bilateral ovaries: Case report and literature review
- Effects of de novo donor-specific Class I and II antibodies on graft outcomes after liver transplantation: A pilot cohort study
- Sleep architecture in Alzheimer’s disease continuum: The deep sleep question
- Ephedra fragilis plant extract: A groundbreaking corrosion inhibitor for mild steel in acidic environments – electrochemical, EDX, DFT, and Monte Carlo studies
- Langerhans cell histiocytosis in an adult patient with upper jaw and pulmonary involvement: A case report
- Inhibition of mast cell activation by Jaranol-targeted Pirin ameliorates allergic responses in mouse allergic rhinitis
- Aeromonas veronii-induced septic arthritis of the hip in a child with acute lymphoblastic leukemia
- Clusterin activates the heat shock response via the PI3K/Akt pathway to protect cardiomyocytes from high-temperature-induced apoptosis
- Research progress on fecal microbiota transplantation in tumor prevention and treatment
- Low-pressure exposure influences the development of HAPE
- Stigmasterol alleviates endplate chondrocyte degeneration through inducing mitophagy by enhancing PINK1 mRNA acetylation via the ESR1/NAT10 axis
- AKAP12, mediated by transcription factor 21, inhibits cell proliferation, metastasis, and glycolysis in lung squamous cell carcinoma
- Association between PAX9 or MSX1 gene polymorphism and tooth agenesis risk: A meta-analysis
- A case of bloodstream infection caused by Neisseria gonorrhoeae
- Case of nasopharyngeal tuberculosis complicated with cervical lymph node and pulmonary tuberculosis
- p-Cymene inhibits pro-fibrotic and inflammatory mediators to prevent hepatic dysfunction
- GFPT2 promotes paclitaxel resistance in epithelial ovarian cancer cells via activating NF-κB signaling pathway
- Transfer RNA-derived fragment tRF-36 modulates varicose vein progression via human vascular smooth muscle cell Notch signaling
- RTA-408 attenuates the hepatic ischemia reperfusion injury in mice possibly by activating the Nrf2/HO-1 signaling pathway
- Decreased serum TIMP4 levels in patients with rheumatoid arthritis
- Sirt1 protects lupus nephritis by inhibiting the NLRP3 signaling pathway in human glomerular mesangial cells
- Sodium butyrate aids brain injury repair in neonatal rats
- Interaction of MTHFR polymorphism with PAX1 methylation in cervical cancer
- Convallatoxin inhibits proliferation and angiogenesis of glioma cells via regulating JAK/STAT3 pathway
- The effect of the PKR inhibitor, 2-aminopurine, on the replication of influenza A virus, and segment 8 mRNA splicing
- Effects of Ire1 gene on virulence and pathogenicity of Candida albicans
- Small cell lung cancer with small intestinal metastasis: Case report and literature review
- GRB14: A prognostic biomarker driving tumor progression in gastric cancer through the PI3K/AKT signaling pathway by interacting with COBLL1
- 15-Lipoxygenase-2 deficiency induces foam cell formation that can be restored by salidroside through the inhibition of arachidonic acid effects
- FTO alleviated the diabetic nephropathy progression by regulating the N6-methyladenosine levels of DACT1
- Clinical relevance of inflammatory markers in the evaluation of severity of ulcerative colitis: A retrospective study
- Zinc valproic acid complex promotes osteoblast differentiation and exhibits anti-osteoporotic potential
- Primary pulmonary synovial sarcoma in the bronchial cavity: A case report
- Metagenomic next-generation sequencing of alveolar lavage fluid improves the detection of pulmonary infection
- Uterine tumor resembling ovarian sex cord tumor with extensive rhabdoid differentiation: A case report
- Genomic analysis of a novel ST11(PR34365) Clostridioides difficile strain isolated from the human fecal of a CDI patient in Guizhou, China
- Effects of tiered cardiac rehabilitation on CRP, TNF-α, and physical endurance in older adults with coronary heart disease
- Changes in T-lymphocyte subpopulations in patients with colorectal cancer before and after acupoint catgut embedding acupuncture observation
- Modulating the tumor microenvironment: The role of traditional Chinese medicine in improving lung cancer treatment
- Alterations of metabolites related to microbiota–gut–brain axis in plasma of colon cancer, esophageal cancer, stomach cancer, and lung cancer patients
- Research on individualized drug sensitivity detection technology based on bio-3D printing technology for precision treatment of gastrointestinal stromal tumors
- CEBPB promotes ulcerative colitis-associated colorectal cancer by stimulating tumor growth and activating the NF-κB/STAT3 signaling pathway
- Oncolytic bacteria: A revolutionary approach to cancer therapy
- A de novo meningioma with rapid growth: A possible malignancy imposter?
- Diagnosis of secondary tuberculosis infection in an asymptomatic elderly with cancer using next-generation sequencing: Case report
- Hesperidin and its zinc(ii) complex enhance osteoblast differentiation and bone formation: In vitro and in vivo evaluations
- Research progress on the regulation of autophagy in cardiovascular diseases by chemokines
- Anti-arthritic, immunomodulatory, and inflammatory regulation by the benzimidazole derivative BMZ-AD: Insights from an FCA-induced rat model
- Immunoassay for pyruvate kinase M1/2 as an Alzheimer’s biomarker in CSF
- The role of HDAC11 in age-related hearing loss: Mechanisms and therapeutic implications
- Evaluation and application analysis of animal models of PIPNP based on data mining
- Therapeutic approaches for liver fibrosis/cirrhosis by targeting pyroptosis
- Fabrication of zinc oxide nanoparticles using Ruellia tuberosa leaf extract induces apoptosis through P53 and STAT3 signalling pathways in prostate cancer cells
- Haplo-hematopoietic stem cell transplantation and immunoradiotherapy for severe aplastic anemia complicated with nasopharyngeal carcinoma: A case report
- Modulation of the KEAP1-NRF2 pathway by Erianin: A novel approach to reduce psoriasiform inflammation and inflammatory signaling
- The expression of epidermal growth factor receptor 2 and its relationship with tumor-infiltrating lymphocytes and clinical pathological features in breast cancer patients
- Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights
- BAP1 complexes with YY1 and RBBP7 and its downstream targets in ccRCC cells
- Hypereosinophilic syndrome with elevated IgG4 and T-cell clonality: A report of two cases
- Electroacupuncture alleviates sciatic nerve injury in sciatica rats by regulating BDNF and NGF levels, myelin sheath degradation, and autophagy
- Polydatin prevents cholesterol gallstone formation by regulating cholesterol metabolism via PPAR-γ signaling
- RNF144A and RNF144B: Important molecules for health
- Analysis of the detection rate and related factors of thyroid nodules in the healthy population
- Artesunate inhibits hepatocellular carcinoma cell migration and invasion through OGA-mediated O-GlcNAcylation of ZEB1
- Endovascular management of post-pancreatectomy hemorrhage caused by a hepatic artery pseudoaneurysm: Case report and review of the literature
- Efficacy and safety of anti-PD-1/PD-L1 antibodies in patients with relapsed refractory diffuse large B-cell lymphoma: A meta-analysis
- SATB2 promotes humeral fracture healing in rats by activating the PI3K/AKT pathway
- Overexpression of the ferroptosis-related gene, NFS1, corresponds to gastric cancer growth and tumor immune infiltration
- Understanding risk factors and prognosis in diabetic foot ulcers
- Atractylenolide I alleviates the experimental allergic response in mice by suppressing TLR4/NF-kB/NLRP3 signalling
- FBXO31 inhibits the stemness characteristics of CD147 (+) melanoma stem cells
- Immune molecule diagnostics in colorectal cancer: CCL2 and CXCL11
- Inhibiting CXCR6 promotes senescence of activated hepatic stellate cells with limited proinflammatory SASP to attenuate hepatic fibrosis
- Cadmium toxicity, health risk and its remediation using low-cost biochar adsorbents
- Pulmonary cryptococcosis with headache as the first presentation: A case report
- Solitary pulmonary metastasis with cystic airspaces in colon cancer: A rare case report
- RUNX1 promotes denervation-induced muscle atrophy by activating the JUNB/NF-κB pathway and driving M1 macrophage polarization
- Morphometric analysis and immunobiological investigation of Indigofera oblongifolia on the infected lung with Plasmodium chabaudi
- The NuA4/TIP60 histone-modifying complex and Hr78 modulate the Lobe2 mutant eye phenotype
- Experimental study on salmon demineralized bone matrix loaded with recombinant human bone morphogenetic protein-2: In vitro and in vivo study
- A case of IgA nephropathy treated with a combination of telitacicept and half-dose glucocorticoids
- Analgesic and toxicological evaluation of cannabidiol-rich Moroccan Cannabis sativa L. (Khardala variety) extract: Evidence from an in vivo and in silico study
- Wound healing and signaling pathways
- Combination of immunotherapy and whole-brain radiotherapy on prognosis of patients with multiple brain metastases: A retrospective cohort study
- To explore the relationship between endometrial hyperemia and polycystic ovary syndrome
- Research progress on the impact of curcumin on immune responses in breast cancer
- Biogenic Cu/Ni nanotherapeutics from Descurainia sophia (L.) Webb ex Prantl seeds for the treatment of lung cancer
- Dapagliflozin attenuates atrial fibrosis via the HMGB1/RAGE pathway in atrial fibrillation rats
- Glycitein alleviates inflammation and apoptosis in keratinocytes via ROS-associated PI3K–Akt signalling pathway
- ADH5 inhibits proliferation but promotes EMT in non-small cell lung cancer cell through activating Smad2/Smad3
- Apoptotic efficacies of AgNPs formulated by Syzygium aromaticum leaf extract on 32D-FLT3-ITD human leukemia cell line with PI3K/AKT/mTOR signaling pathway
- Novel cuproptosis-related genes C1QBP and PFKP identified as prognostic and therapeutic targets in lung adenocarcinoma
- Bee venom promotes exosome secretion and alters miRNA cargo in T cells
- Treatment of pure red cell aplasia in a chronic kidney disease patient with roxadustat: A case report
- Comparative bioinformatics analysis of the Wnt pathway in breast cancer: Selection of novel biomarker panels associated with ER status
- Kynurenine facilitates renal cell carcinoma progression by suppressing M2 macrophage pyroptosis through inhibition of CASP1 cleavage
- RFX5 promotes the growth, motility, and inhibits apoptosis of gastric adenocarcinoma cells through the SIRT1/AMPK axis
- ALKBH5 exacerbates early cardiac damage after radiotherapy for breast cancer via m6A demethylation of TLR4
- Phytochemicals of Roman chamomile: Antioxidant, anti-aging, and whitening activities of distillation residues
- Circadian gene Cry1 inhibits the tumorigenicity of hepatocellular carcinoma by the BAX/BCL2-mediated apoptosis pathway
- The TNFR-RIPK1/RIPK3 signalling pathway mediates the effect of lanthanum on necroptosis of nerve cells
- Longitudinal monitoring of autoantibody dynamics in patients with early-stage non-small-cell lung cancer undergoing surgery
- The potential role of rutin, a flavonoid, in the management of cancer through modulation of cell signaling pathways
- Construction of pectinase gene engineering microbe and its application in tobacco sheets
- Construction of a microbial abundance prognostic scoring model based on intratumoral microbial data for predicting the prognosis of lung squamous cell carcinoma
- Sepsis complicated by haemophagocytic lymphohistiocytosis triggered by methicillin-resistant Staphylococcus aureus and human herpesvirus 8 in an immunocompromised elderly patient: A case report
- Sarcopenia in liver transplantation: A comprehensive bibliometric study of current research trends and future directions
- Advances in cancer immunotherapy and future directions in personalized medicine
- Can coronavirus disease 2019 affect male fertility or cause spontaneous abortion? A two-sample Mendelian randomization analysis
- Heat stroke associated with novel leukaemia inhibitory factor receptor gene variant in a Chinese infant
- PSME2 exacerbates ulcerative colitis by disrupting intestinal barrier function and promoting autophagy-dependent inflammation
- Ecology and Environmental Science
- Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity
- The complete mitochondrial genome analysis of Haemaphysalis hystricis Supino, 1897 (Ixodida: Ixodidae) and its phylogenetic implications
- Epidemiological characteristics and risk factors analysis of multidrug-resistant tuberculosis among tuberculosis population in Huzhou City, Eastern China
- Indices of human impacts on landscapes: How do they reflect the proportions of natural habitats?
- Genetic analysis of the Siberian flying squirrel population in the northern Changbai Mountains, Northeast China: Insights into population status and conservation
- Diversity and environmental drivers of Suillus communities in Pinus sylvestris var. mongolica forests of Inner Mongolia
- Global assessment of the fate of nitrogen deposition in forest ecosystems: Insights from 15N tracer studies
- Fungal and bacterial pathogenic co-infections mainly lead to the assembly of microbial community in tobacco stems
- Influencing of coal industry related airborne particulate matter on ocular surface tear film injury and inflammatory factor expression in Sprague-Dawley rats
- Temperature-dependent development, predation, and life table of Sphaerophoria macrogaster (Thomson) (Diptera: Syrphidae) feeding on Myzus persicae (Sulzer) (Homoptera: Aphididae)
- Eleonora’s falcon trophic interactions with insects within its breeding range: A systematic review
- Agriculture
- Integrated analysis of transcriptome, sRNAome, and degradome involved in the drought-response of maize Zhengdan958
- Variation in flower frost tolerance among seven apple cultivars and transcriptome response patterns in two contrastingly frost-tolerant selected cultivars
- Heritability of durable resistance to stripe rust in bread wheat (Triticum aestivum L.)
- Molecular mechanism of follicular development in laying hens based on the regulation of water metabolism
- Animal Science
- Effect of sex ratio on the life history traits of an important invasive species, Spodoptera frugiperda
- Plant Sciences
- Hairpin in a haystack: In silico identification and characterization of plant-conserved microRNA in Rafflesiaceae
- Widely targeted metabolomics of different tissues in Rubus corchorifolius
- The complete chloroplast genome of Gerbera piloselloides (L.) Cass., 1820 (Carduoideae, Asteraceae) and its phylogenetic analysis
- Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants
- Correlation analysis between semen routine parameters and sperm DNA fragmentation index in patients with semen non-liquefaction: A retrospective study
- Plasticity of the anatomical traits of Rhododendron L. (Ericaceae) leaves and its implications in adaptation to the plateau environment
- Effects of Piriformospora indica and arbuscular mycorrhizal fungus on growth and physiology of Moringa oleifera under low-temperature stress
- Effects of different sources of potassium fertiliser on yield, fruit quality and nutrient absorption in “Harward” kiwifruit (Actinidia deliciosa)
- Comparative efficiency and residue levels of spraying programs against powdery mildew in grape varieties
- The DREB7 transcription factor enhances salt tolerance in soybean plants under salt stress
- Using plant electrical signals of water hyacinth (Eichhornia crassipes) for water pollution monitoring
- Food Science
- Phytochemical analysis of Stachys iva: Discovering the optimal extract conditions and its bioactive compounds
- Review on role of honey in disease prevention and treatment through modulation of biological activities
- Computational analysis of polymorphic residues in maltose and maltotriose transporters of a wild Saccharomyces cerevisiae strain
- Optimization of phenolic compound extraction from Tunisian squash by-products: A sustainable approach for antioxidant and antibacterial applications
- Liupao tea aqueous extract alleviates dextran sulfate sodium-induced ulcerative colitis in rats by modulating the gut microbiota
- Toxicological qualities and detoxification trends of fruit by-products for valorization: A review
- Polyphenolic spectrum of cornelian cherry fruits and their health-promoting effect
- Optimizing the encapsulation of the refined extract of squash peels for functional food applications: A sustainable approach to reduce food waste
- Advancements in curcuminoid formulations: An update on bioavailability enhancement strategies curcuminoid bioavailability and formulations
- Impact of saline sprouting on antioxidant properties and bioactive compounds in chia seeds
- The dilemma of food genetics and improvement
- Bioengineering and Biotechnology
- Impact of hyaluronic acid-modified hafnium metalorganic frameworks containing rhynchophylline on Alzheimer’s disease
- Emerging patterns in nanoparticle-based therapeutic approaches for rheumatoid arthritis: A comprehensive bibliometric and visual analysis spanning two decades
- Application of CRISPR/Cas gene editing for infectious disease control in poultry
- Preparation of hafnium nitride-coated titanium implants by magnetron sputtering technology and evaluation of their antibacterial properties and biocompatibility
- Preparation and characterization of lemongrass oil nanoemulsion: Antimicrobial, antibiofilm, antioxidant, and anticancer activities
- Corrigendum
- Corrigendum to “Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells”
- Corrigendum to “Effects of Ire1 gene on virulence and pathogenicity of Candida albicans”

