Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
-
Yanxia Zhou
Abstract
In the early stage, our research group cloned Echinococcus granulosus-specific antigen, EgG1Y162, from protoscolex and adult worms of E. granulosus. In order to enhance the immunogenicity of the vaccine, we prepared a recombinant vaccine by tandemly linking EgG1Y162, splicing the protein and linker at the gene level. This approach is expected to improve the immunogenicity of the vaccine by enhancing the molecular weight of the protein and increasing the antigenic epitopes. Bioinformatics was used to predict the physicochemical properties, transmembrane domain, protein structure, and T-/B-cell antigenic epitope of different recombinant proteins, EgG1Y162-linker-EgG1Y162. Finally, the linker sequence, “GGGGSGGG,” which had the least influence on the migration of recombinant protein T/B epitope and can fold normally in series with EgG1Y162, was selected to design the recombinant vaccine. The plasmid was produced using genetic engineering techniques, and the recombinant protein, EGG1Y162-GGGGSGGG-EgG1Y162, was induced to be expressed and purified. EgG1Y162-GGGGSGGG-EgG1Y162 was identified to be correctly expressed with 100% specificity. Compared with EgG1Y162, EgG1Y162-GGGGSGGG-EgG1Y162 was more likely to promote dendritic cell maturation. EgG1Y162-GGGGSGGG-EgG1Y162 was speculated to have the potential to improve antigen immunogenicity by increasing the molecular weight and antigenic epitope.
1 Introduction
Echinococcosis, also known as hydatid disease, is a zoonotic parasitic disease caused by the infection of an intermediate host with Echinococcosis granulosus [1]. It has two subtypes, cystic echinococcosis and vesicular echinococcosis. Cystic echinococcosis is more serious and causes high mortality. Cystic echinococcosis is distributed worldwide and common in countries and regions with developed animal husbandry. The current treatment for echinococcosis is still not ideal and has limitations. Therefore, the development of a vaccine is of great interest and is an ideal way to prevent echinococcosis [2,3,4,5]. The selection of specific antigenic targets is the primary task of preparing recombinant vaccines [6,7]. In the study of E. granulosus [8], Cao et al. discovered a new gene, EgG1Y162, which was sent to the gene bank. In addition, a series of experiments confirmed that the recombinant protein, EgG1Y162, could react with the serum of dogs infected with E. granulosus, which indicates that the antigen has high specificity and sensitivity in the immune response of the body. Relevant studies found that specific antibodies can be increased in vivo after mice were immunized with EgG1Y162 antigen, which promotes the proliferation of lymphocytes and participates in the cellular and humoral immunity of the body [9]. The prediction and analysis of EgG1Y162 epitope by bioinformatics revealed that this antigen has abundant epitope information, which can enhance the immune response of an organism and induce immune protection [10]. Zhang et al. showed that the recombinant protein EgG1Y162 had good antigenicity [11]. In the present study, recombinant vaccines were prepared by tandemly linking two EgG1Y162 proteins with linker sequences to increase the immunogenicity and immunoreactivity of the vaccine. Three linker sequences, namely, GSGGSG, GGGGSGGG, and GSGGSGGGSGGSGGG, were used to design the recombinant vaccines. The resultant vaccines were compared in terms of physicochemical properties, structure, and antigenic epitopes by bioinformatics method [10], and the most suitable linker sequence was selected. The recombinant protein EgG1Y162-linker-EgG1Y162 was successfully prepared and identified, and the immune effect and mechanism of the recombinant protein were preliminarily explored.
2 Materials and methods
2.1 Materials
2.1.1 Sequences of EgG1Y162 and EgG1Y162–linker–EgG1Y162 proteins
The amino acid sequence of the EgG1Y162 protein, which has a total length of 120 aa, was stored in GenBank with accession number AB458259. The recombinant proteins, EgG1Y162-GSGGSG-EgG1Y162, EgG1Y162-GGGGSGGG-EgG1Y162, and EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162, had total lengths of 246, 248, and 255 aa, respectively.
EgG1Y162:
VDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGF
EgG1Y162-GSGGSG-EgG1Y162:
VDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGFGSGVDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGF
EgG1Y162-GGGGSGGG-EgG1Y162:
VDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGFGGGGSGGGVDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGF
EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162:
VDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGFGSGGSGGSGGSGGGVDPELMAKLTKELKTTLPEHFRWIHVGSRSLELGWNATGLANLHADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLKPSTFYEVVQAFKGGSQVFKYTGFIRTLAPGEDGADRASGF
2.1.2 Bioinformatics software
The following softwares were used in the bioinformatics analysis: ProtParam (http://web.expasy.org/protparam/), Transmembrane Helices Hidden Markov Model (TMHMM) server (http:/www.cbs.dtu.dk/services/TMHMM-2.0/) [5], Self-Optimized Prediction Method with Alignment (SOPMA, https://npsa-prabi.ibcp.fr/cgibin/npsa_automat.pl? page = npsa_sopma.html), BepiPred1.0 server (http://www.cbs.dtu.dk/services/BepiPred-1.0/), Immune Epitope Database (IEDB, http://tools.iedb.org/main/bcell/, http://tools.iedb.org/mhcii/), SVMTriP (http://sysbio.unl.edu/SVMTriP/prediction.php), Propred (http://imed.med.ucm.es/Tools/rankpep.html), and SYFPEITHI (http://www.syfpeithi.de/bin/mhcserver.dll/epitopeprediction).
2.1.3 Plasmids and reagents
The glycerol expression bacteriophage EgG1Y162 was kept in our laboratory. Plasmid EgG1Y162-GGGGGSGGG-EgG1Y162 was constructed and synthesized by Shanghai Biotechnology Company, China. Escherichia coli BL21 (DE3) receptor cells were obtained from Shanghai Weidi Biotechnology Co., Ltd (China). Lysogeny broth (LB) liquid and solid media were purchased from Shanghai Biotechnology Co. (China). Agarose was purchased from Invitrogen, USA. Plasmid extraction kit was purchased from Beijing Tiangen Co. (China). Polymerase chain reaction (PCR) kits, Q cutting enzymes (EcoRI and SalI), and DNA Maker2000 were purchased from Dalian TaKaRa Co. (Kyoto, Japan). Pre-stained Protein Maker was purchased from Thermo Fisher Scientific (Massachusetts, US). Isopropyl β-D-thiogalactoside (IPTG) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel preparation kit were purchased from Solarbio (Beijing, China). Polyvinylidene difluoride membrane (PVDF) was purchased from GE Healthcore (Little Chalfont, UK). His-Tag Mouse antibody was purchased from Cell Signaling Technology (BD biosciences, Franklin Lakes, US). Goat anti-mouse IgG-horseradish peroxidase (HRP) was purchased from Absin Biotechnology Co. (Shanghai, China). Goat anti-human IgG-HRP was purchased from Beijing Bioss Biotechnology Co. (China). High-sensitivity enhanced chemiluminescence (ECL) test kit was purchased from Comwin Biotech Co., Ltd (Beijing, China). HisTrap purification columns were purchased from General Electric, USA. RPMI 1640 medium, fetal bovine serum (BI), and double antibodies were purchased from Hyclone. Erythrocyte lysate was purchased from Beijing Solarbio Technology Co. (China). RmIL-4 and rmGM-CSF were purchased from PeproTech (New Jersey, US). Fluorescein isothiocyanate (FITC)-coupled anti-HIS antibody was purchased from Beijing Bioss Biotechnology Co. (China). Recombinant cytokine lysis dilution kit was purchased from Hangzhou Unitech Biotechnology Co. (China).
2.1.4 Experimental animals
Specific pathogen-free C57 mice (6–8 weeks, 20 ± 2 g) were purchased from the Experimental Animal Center of Xinjiang Medical University (License No:SCXK(Xin)2016-0003).
-
Ethical approval: The research related to animal use has been complied with all the relevant national regulationsand institutional policies for the care and use of animals.
2.2 Methods
2.2.1 Bioinformatics prediction method
ProtParam was initially used to analyze the physicochemical properties of each recombinant protein, including protein molecular mass, theoretical isoelectric point, extinction coefficient, and other theoretical properties. Then, the TMHMM server was used to predict and analyze the transmembrane region of each recombinant protein, and SOPMA was used in the secondary structure prediction of each recombinant protein with the transmembrane region to improve the reliability of the pretreatment. The 3D model of recombinant proteins was established using the online software, I-TASSER. BepiPred1.0 server and SVMTriP were used to analyze the possible dominant linear epitopes of each recombinant protein on B cells. Propred, IEDB, and SYFPEITHI were used to predict the T-cell antigenic epitopes of each recombinant protein. HLA-DRB1*0701 was selected as the parameter. The epitope with the highest SYFPEITHI score (the higher the score, the higher the probability that the sequence will be the dominant epitope) and lowest IEDB percentage rank (the lower the percent rank, the higher the probability of the sequence being the dominant epitope) was determined. Multiple results were compared, and the prediction results of secondary structures were referred to exclude the structures that could not easily form epitopes. Based on the predicted results of B-cell and T-cell epitopes, the length and location of each predicted epitope were evaluated. The sequences that were too short to form epitopes were excluded, and peptides with high multiple prediction repetition rates were selected as the possible dominant T/B combined epitopes of recombinant proteins.
2.2.2 Identification of prokaryotic expression plasmid and induction expression, purification, and identification of recombinant protein
The synthesized plasmid, pET30a-EgG1Y162-GGGGSGGG-EgG1Y162, was identified by double digestion with EcoRI and SalI. The correctly identified plasmid was transformed into the host bacterium, E. coli BL21 (DE3). The single clone was selected and inoculated in 20 mL of LB liquid medium containing 30 μg/mL kanamycin and incubated overnight on a shaking table at 220 rpm at 37°C. The next day, the overnight culture was inoculated at a ratio of 1:50 in LB liquid medium containing 30 μg/mL kanamycin to expand the culture. The bacterial culture solution was cultured on a shaking table at 220 rpm at 37°C until the optical density of the bacterial solution was 0.6–0.8 and induced for 6 h with a final IPTG concentration of 0.5 mmol/L at 28°C. The target protein in the supernatant was obtained and purified, and the expression of recombinant protein was identified by Western blot. His-Tag antibodies (Cell Signaling Techonology, Beverly, MA, US, Cat. N.#9991) were used for Western blot detection.
2.2.3 Western blot analysis of recombinant protein expression
The purified proteins were electrotransferred to the PVDF membrane after SDS–PAGE. Then, the membrane was closed with 5% skimmed milk powder at room temperature for 1 h and repeatedly washed three times with 1× TBST for 15 min each time. Finally, the primary antibody was added and incubated overnight at 4°C on a shaking table. Cysticercosis mouse serum and normal mouse serum were used as primary antibodies which were diluted at a concentration of 1:150 and incubated overnight. The antibodies were discarded the next day, and the PVDF membrane was washed three times with 1× TBST for 15 min each time. Goat anti-mouse IgG-HRP (Absin, Shanghai, China, Cat. N.abs20039)(1:3000 dilution) was added, incubated for 2 h at room temperature, and washed three times with 1× TBST for 15 min each time. ECL was added for color development, and the results were observed. Specificity was calculated using the formula:
2.2.4 Maturation of mouse bone marrow-derived dendritic cells (DCs) after 24 h stimulation with recombinant protein
The mice were sacrificed by cervical dislocation. The tibia and femur were removed under aseptic conditions, and the muscle tissue was stripped. Appropriate PBS was absorbed with a syringe and inserted into the epiphysis to clean the bone cavity and obtain bone marrow cells. The cells were filtered through a filter and centrifuged, and the supernatant was discarded. The cells were added with an appropriate amount of erythrocyte lysate, mixed, and placed in a refrigerator at 4°C for 10 min, and centrifuged for 5 min at room temperature, and the supernatant was discarded. The bone marrow cells were suspended in a complete culture medium, inoculated into six-well plates with rmGM-CSF and rmIL-4, incubated in a CO2 incubator with half volume of fluid change every day, and added with the corresponding cytokines until day 7 to obtain immature dendritic cells (imDCs). EgG1Y162 (final concentration 500 ng/mL) and EgG1Y162-GGGGSGGG-EgG1Y162 (final concentration 500 ng/mL) were co-cultured with imDC for 24 h, and the cell suspension was collected. Then, flow cytometric identification was performed. The cells (1 × 106) were placed in a flow tube, added with PBS (2 mL), and centrifuged at 1,000 rpm for 5 min. The supernatant was discarded, and 50 μL 1× PBS containing 1 μL CD86 + antibody, 1 μL CD11c + antibody, 1 μL CD45 + antibody, and 1 μL I-Ab antibody was added and incubated at 4°C for 30 min in the dark. The cells were washed by adding 1 mL 1× PBS and centrifuged at 1,000 rpm and 4°C for 5 min. The cells were repeatedly cleaned with PBS and then added with 300 μL PBS to resuscitate the cells. The percentage of mature DC was detected within 4 h.
2.3 Statistics
All data were shown by Mean Value ± Standard Deviation
3 Results
3.1 Bioinformatics analysis showed that “GGGGSGGG” was connected to the EgG1Y162 protein as the linker sequence and none of the epitopes shifted
3.1.1 Comparison of the physicochemical properties of recombinant proteins and prediction of transmembrane regions
The physicochemical properties of the recombinant proteins were analyzed by using ProtParam (http://web.expasy.org/protparam/). The result showed that the EgG1Y162 protein is composed of 120 aa and has a molecular mass of 13515.49 Ka, a theoretical isoelectric point (pI) value of 9.22; a chemical molecular formula of C619H960N164O174S1, an extinction coefficient of 18,450, an instability index of 29.98 (>40 means unstable protein, and < 40 is stable protein), and a grand average of hydropathicity index (GRAVY) of −0.263 (the overall GRAVY range is between −2 and 2; negative values indicate hydrophilic proteins). The EgG1Y162-GSGGSG-EgG1Y162 protein is composed of 246 aa and has a molecular mass of 27415.32 Da, theoretical pI value of 9.34, a chemical formula of C1252H1940N334O355S2, an extinction coefficient of 36,900, an instability index of 30.04, and a GRAVY of −0.270. The EgG1Y162-GGGGSGGG-EgG1Y162 protein is composed of 248 aa and has a protein molecular mass of 27499.40 Da, a theoretical pI value of 9.34, a chemical formula of C1255H1944N336O356S2, an extinction coefficient of 36,900, an instability index of 31.87, and a GRAVY of −0.269. The EgG1Y162-GSGGSG-EgG1Y162 protein is composed of 246 aa and has a molecular mass of 27415.32 Da, a theoretical pI value of 9.34, a chemical formula of C1252H1940N334O355S2, an extinction coefficient of 36,900, an instability index of 30.04, and a GRAVY of −0.270. The EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162 protein is composed of 255 aa and has a molecular mass of 27988.84 Da, a theoretical pI value of 9.34, chemical formula of C1272H1971N343O366S2, an extinction coefficient of 36,900, an instability index of 31.75, and a GRAVY of −0.278. The TMHMM server was used to analyze the transmembrane region of each recombinant protein. The transmembrane protein regions were greater than 1, which indicates that each recombinant protein is an extracellular protein that can be fully contacted by antigen-presenting cells and initiate a strong immune response from T and B cells. The results are shown in Figure 1.

Transmembrane domain of each recombinant protein. (a) EgG1Y162; (b) EGG1Y162-GSGGSG-EGG1Y162; (c) EGG1Y162-GGGGSGGG-EGG1Y162; (d) EGG1y162-GSGGSGGSGGG-EGG1Y162.
3.1.2 Prediction of the secondary structures of each recombinant protein
The secondary structures of each recombinant protein were predicted and analyzed by SOPMA online software. The predicted results demonstrate the secondary structures of each recombinant protein and the proportions of various structural domains present in the protein, including alpha helix, extended strand, beta turn, and random coil, as shown in Figure 2.

SOMPA analysis of the secondary structures of recombinant proteins: (a) EgG1Y162; (b) EgG1Y162-GSGGSG-EgG1Y162; (c) EgG1Y162-GGGGSGGG-EgG1Y162; (d) EgG1Y162-GSGGSGGGSGSGGG-EgG1Y162.
3.1.3 Analysis of the T-/B-cell antigenic epitopes of recombinant proteins
BepiPred1.0 Server [12], SVMTriP, IEBD, and SYFPEITHI software were used to predict the possible dominant antigenic epitopes of EgG1Y162 and the EgG1Y162-linker-EgG1Y162 proteins on T/B cells. The predicted results showed that EgG1Y162 has three B-cell antigen epitopes, which are located at 19–28, 60–75, and 108–117 aa (Table 1), and three dominant T-cell antigen epitopes, which are located at 7–21, 57–69, and 77–111 aa (Table 2). Recombinant protein EgG1Y162-GSGGSG-EgG1Y162 has six B-cell antigen epitopes, which are located at 19–26, 60–68, 108–130, 147–154, 186–205, and 235–242 aa (Table 3), and six dominant T-cell antigen epitopes, which are located at 7–37, 45–69, 95–111, 133–160, 171–195, and 221–237 aa (Table 4). Recombinant protein EgG1Y162-GGGGSGGG-EgG1Y162 has six B-cell antigen epitopes, which are located at 19–28, 60–79, 108–132, 149–166, 188–207, and 236–244 aa (Table 5), and six dominant T-cell epitopes, which are located at 10–37, 45–69, 95–111, 135–165, 173–198, and 220–237 aa (Table 6). Recombinant protein EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162 has six B-cell antigen epitopes, which are located at 19–26, 57–70, 108–139, 157–178, 195–205, and 243–252 aa (Table 7), and six dominant T-cell epitopes, which are located at 8–21, 45–69, 95–111, 155–172, 180–207, and 230–246 aa (Table 8). The T-/B-cell antigenic epitopes of EgG1Y162 and EgG1Y162-linker-EgG1Y162 recombinant proteins are shown in Table 9. EgG1Y162-GGSGGG-EgG1Y162 had less epitope migration compared with EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162 and EgG1Y162-GSGGSG-EgG1Y162. Therefore, the recombinant protein EgG1Y162-linker-EgG1Y162 whose linker sequence was “GGGGSGGG” was selected for verification.
Prediction results of the B-cell epitope of recombinant protein EgG1Y162
| Bepi Pred1.0 | SVMTriP | IEDB | |||
|---|---|---|---|---|---|
| Position | Sequence | Position | Sequence | Position | Sequence |
| 108–120 | APGEDGADRASGF | 19–38 | EHFRWIHVGSRSLELGWNAT | 14–28 | KTTLPEHFRWIHVGS |
| 60–79 | TFKYRNVPIERQKLTLEGLK | 55–75 | YTTYVTFKYRNVPIERQKLTL | ||
| 105–117 | RTLAPGEDGADRA | ||||
Prediction results of the T-cell epitope of recombinant protein EgG1Y162
| SYFPEITHI | IEDB | ||||
|---|---|---|---|---|---|
| Position | Sequence | Score | Position | Sequence | Rank |
| 32–46 | ELGWNATGLANLHAD | 26 | 22–36 | RWIHVGSRSLELGWN | 0.90 |
| 2–16 | DPELMAKLTKELKTT | 24 | 23–37 | WIHVGSRSLELGWNA | 1.70 |
| 95–109 | SQVFKYTGFIRTLAP | 18 | 42–56 | NLHADHIKLTANLYT | 3.60 |
| 18–32 | PEHFRWIHVGSRSLE | 16 | 57–71 | TYVTFKYRNVPIERQ | 11.00 |
| 80–94 | PSTFYEVVVQAFKGG | 16 | 58–72 | YVTFKYRNVPIERQK | 11.00 |
| 4–18 | ELMAKLTKELKTTLP | 14 | 7–21 | AKLTKELKTTLPEHF | 12.00 |
| 47–61 | HIKLTANLYTTYVTF | 14 | 77–91 | GLKPSTFYEVVVQAF | 12.00 |
| 62–76 | KYRNVPIERQKLTLE | 14 | 92–106 | KGGSQVFKYTGFIRT | 12.00 |
| 77–91 | GLKPSTFYEVVVQAF | 14 | 97–111 | VFKYTGFIRTLAPGE | 12.00 |
| 104–118 | IRTLAPGEDGADRAS | 14 | |||
Prediction results of the B-cell epitopes of recombinant protein EgG1Y162-GSGGSG-EgG1Y162
| Bepi Pred1.0 | SVMTriP | IEDB | |||
|---|---|---|---|---|---|
| Position | Sequence | Position | Sequence | Position | Sequence |
| 108–130 | APGEDGADRASGFGSGGSGVDPE | 19–38 | EHFRWIHVGSRSLELGWNAT | 15–26 | TTLPEHFRWIHV |
| 234–246 | APGEDGADRASGF | 145–164 | EHFRWIHVGSRSLELGWNAT | 57–68 | TYVTFKYRNVPI |
| 60–79 | TFKYRNVPIERQKLTLEGLK | 107–145 | LAPGEDGADRASGFGSGGSGVDPELMAKLTKELKTTLPE | ||
| 186–205 | TFKYRNVPIERQKLTLEGLK | 147–154 | FRWIHVGS | ||
| 181–206 | YTTYVTFKYRNVPIERQKLTLEGLKP | ||||
| 235–242 | PGEDGADR | ||||
Prediction results of the T-cell epitope of recombinant protein EgG1Y162-GSGGSG-EgG1Y162
| Propred | SYFPEITHI | IEDB | ||||||
|---|---|---|---|---|---|---|---|---|
| Position | Sequence | Score | Position | Sequence | Score | Position | Sequence | Score |
| 173–181 | HIKLTANLY | 13.661 | 32–46 | ELGWNATGLANLHAD | 26 | 23–37 | WIHVGSRSLELGWNA | 1.70 |
| 47–55 | HIKLTANLY | 13.661 | 146–160 | HFRWIHVGSRSLELG | 26 | 149–163 | WIHVGSRSLELGWNA | 1.70 |
| 170–178 | HADHIKLTA | 12.667 | 158–172 | ELGWNATGLANLHAD | 26 | 42–56 | NLHADHIKLTANLYT | 3.60 |
| 44–52 | HADHIKLTA | 12.667 | 2–16 | DPELMAKLTKELKTT | 26 | 168–182 | NLHADHIKLTANLYT | 3.60 |
| 128–142 | DPELMAKLTKELKTT | 24 | 10–24 | TKELKTTLPEHFRWI | 8.30 | |||
| 10–24 | TKELKTTLPEHFRWI | 24 | 136–150 | TKELKTTLPEHFRWI | 8.30 | |||
| 22–36 | RWIHVGSRSLELGWN | 24 | 57–71 | TYVTFKYRNVPIERQ | 11.00 | |||
| 45–59 | ADHIKLTANLYTTYV | 22 | 183–197 | TYVTFKYRNVPIERQ | 11.00 | |||
| 75–89 | LEGLKPSTFYEVVVQ | 22 | 58–72 | YVTFKYRNVPIERQK | 11.00 | |||
| 171–185 | ADHIKLTANLYTTYV | 22 | 184–198 | YVTFKYRNVPIERQK | 11.00 | |||
| 201–215 | LEGLKPSTFYEVVVQ | 22 | 7–21 | AKLTKELKTTLPEHF | 12.00 | |||
| 55–69 | YTTYVTFKYRNVPIE | 18 | 133–147 | AKLTKELKTTLPEHF | 12.00 | |||
| 95–109 | SQVFKYTGFIRTLAP | 18 | 92–106 | KGGSQVFKYTGFIRT | 12.00 | |||
| 100–114 | YTGFIRTLAPGEDGA | 18 | 77–91 | GLKPSTFYEVVVQAF | 12.00 | |||
| 117–131 | ASGFGSGGSGVDPEL | 18 | 203–217 | GLKPSTFYEVVVQAF | 12.00 | |||
| 181–195 | YTTYVTFKYRNVPIE | 18 | 218–232 | KGGSQVFKYTGFIRT | 12.00 | |||
| 221–235 | SQVFKYTGFIRTLAP | 18 | 97–111 | VFKYTGFIRTLAPGE | 12.00 | |||
| 226–240 | YTGFIRTLAPGEDGA | 18 | 223–237 | VFKYTGFIRTLAPGE | 12.00 | |||
Prediction results of the B-cell epitope of recombinant protein EgG1Y162-GGGGSGGG-EgG1Y162
| Bepi Pred1.0 | SVMTriP | IEDB | |||
|---|---|---|---|---|---|
| Position | Sequence | Position | Sequence | Position | Sequence |
| 108–132 | APGEDGADRASGFGGGGSGGGVDPE | 19–38 | EHFRWIHVGSRSLELGWNAT | 14–28 | KTTLPEHFRWIHVGS |
| 236–248 | APGEDGADRASGF | 147–166 | EHFRWIHVGSRSLELGWNAT | 38–43 | TGLANL |
| 60–79 | TFKYRNVPIERQKLTLEGLK | 56–80 | TTYVTFKYRNVPIERQKLTLEGLKP | ||
| 188–207 | TFKYRNVPIERQKLTLEGLK | 108–147 | APGEDGADRASGFGGGGSGGGVDPELMAKLTKELKTTLPE | ||
| 149–154 | FRWIHV | ||||
| 166–171 | TGLANL | ||||
| 183–208 | YTTYVTFKYRNVPIERQKLTLEGLKP | ||||
| 221–230 | GGSQVFKYTG | ||||
| 236–244 | APGEDGADR | ||||
Prediction results of the T-cell epitope of recombinant protein EgG1Y162-GGGGSGGG-EgG1Y162
| Propred | SYFPEITHI | IEDB | ||||||
|---|---|---|---|---|---|---|---|---|
| Position | Sequence | Score | Position | Sequence | Score | Position | Sequence | Score |
| 175–183 | HIKLTANLY | 13.661 | 32–46 | ELGWNATGLANLHAD | 26 | 21–35 | FRWIHVGSRSLELGW | 0.70 |
| 47–55 | HIKLTANLY | 13.661 | 148–162 | HFRWIHVGSRSLELG | 26 | 150–164 | RWIHVGSRSLELGWN | 0.90 |
| 172–180 | HADHIKLTA | 12.667 | 160–174 | ELGWNATGLANLHAD | 26 | 23–37 | WIHVGSRSLELGWNA | 1.70 |
| 44–52 | HADHIKLTA | 12.667 | 2–16 | DPELMAKLTKELKTT | 24 | 151–165 | WIHVGSRSLELGWNA | 1.70 |
| 130–144 | DPELMAKLTKELKTT | 24 | 42–56 | NLHADHIKLTANLYT | 3.60 | |||
| 10–24 | TKELKTTLPEHFRWI | 22 | 170–184 | NLHADHIKLTANLYT | 3.60 | |||
| 22–36 | RWIHVGSRSLELGWN | 22 | 56–70 | TTYVTFKYRNVPIER | 11.00 | |||
| 45–59 | ADHIKLTANLYTTYV | 22 | 184–198 | TTYVTFKYRNVPIER | 11.00 | |||
| 75–89 | LEGLKPSTFYEVVVQ | 22 | 186–200 | YVTFKYRNVPIERQK | 11.00 | |||
| 138–152 | TKELKTTLPEHFRWI | 22 | 7–21 | AKLTKELKTTLPEHF | 12.00 | |||
| 173–187 | ADHIKLTANLYTTYV | 22 | 135–149 | AKLTKELKTTLPEHF | 12.00 | |||
| 203–217 | LEGLKPSTFYEVVVQ | 22 | 77–91 | GLKPSTFYEVVVQAF | 12.00 | |||
| 55–69 | YTTYVTFKYRNVPIE | 18 | 205–219 | GLKPSTFYEVVVQAF | 12.00 | |||
| 95–109 | SQVFKYTGFIRTLAP | 18 | 92–106 | KGGSQVFKYTGFIRT | 12.00 | |||
| 100–114 | YTGFIRTLAPGEDGA | 18 | 220–234 | KGGSQVFKYTGFIRT | 12.00 | |||
| 117–131 | ASGFGGGGSGGGVDP | 18 | 97–111 | VFKYTGFIRTLAPGE | 12.00 | |||
| 183–197 | YTTYVTFKYRNVPIE | 18 | 225–235 | VFKYTGFIRTLAPGE | 12.00 | |||
| 223–237 | SQVFKYTGFIRTLAP | 18 | ||||||
| 228–242 | YTGFIRTLAPGEDGA | 18 | ||||||
Prediction results of the B-cell epitope of recombinant protein EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162
| Bepi Pred1.0 | KSVMTriP | IEDB | |||
|---|---|---|---|---|---|
| Position | Sequence | Position | Sequence | position | Sequence |
| 108–139 | APGEDGADRASGFGSGGSGGGSGGSGGGVDPE | 19–38 | EHFRWIHVGSRSLELGWNAT | 10–26 | TKELKTTLPEHFRWIHV |
| 243–255 | APGEDGADRASGF | 154–173 | EHFRWIHVGSRSLELGWNAT | 38–43 | TGLANL |
| 60–79 | TFKYRNVPIERQKLTLEGLK | 57–70 | TYVTFKYRNVPIER | ||
| 195–214 | TFKYRNVPIERQKLTLEGLK | 107–154 | LAPGEDGADRASGFGSGGSGGGSGGSGGGVDPELMAKLTKELKTTLPE | ||
| 157–161 | RWIHV | ||||
| 173–178 | TGLANL | ||||
| 192–205 | TYVTFKYRNVPIER | ||||
| 241–252 | TLAPGEDGADRA | ||||
Prediction results of the T-cell epitope of recombinant protein EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162
| Propred | SYFPEITHI | IEDB | ||||||
|---|---|---|---|---|---|---|---|---|
| Position | Sequence | Score | Position | Sequence | Score | Position | Sequence | Score |
| 182–190 | HIKLTANLY | 13.661 | 32–46 | ELGWNATGLANLHAD | 26 | 154–168 | EHFRWIHVGSRSLEL | 0.64 |
| 47–55 | HIKLTANLY | 13.661 | 155–169 | HFRWIHVGSRSLELG | 26 | 22–36 | RWIHVGSRSLELGWN | 0.90 |
| 179–187 | HADHIKLTA | 12.667 | 2–16 | DPELMAKLTKELKTT | 24 | 23–37 | WIHVGSRSLELGWNA | 1.70 |
| 44–52 | HADHIKLTA | 12.667 | 137–151 | DPELMAKLTKELKTT | 24 | 158–172 | WIHVGSRSLELGWNA | 1.70 |
| 10–24 | TKELKTTLPEHFRWI | 22 | 42–56 | NLHADHIKLTANLYT | 3.60 | |||
| 45–59 | ADHIKLTANLYTTYV | 22 | 177–191 | NLHADHIKLTANLYT | 3.60 | |||
| 75–89 | ADHIKLTANLYTTYV | 22 | 8–22 | KLTKELKTTLPEHFR | 11.00 | |||
| 180–194 | ADHIKLTANLYTTYV | 22 | 56–70 | TTYVTFKYRNVPIER | 11.00 | |||
| 210–224 | LEGLKPSTFYEVVVQ | 22 | 191–205 | TTYVTFKYRNVPIER | 11.00 | |||
| 165–179 | SLELGWNATGLANLH | 20 | 58–72 | YVTFKYRNVPIERQK | 11.00 | |||
| 55–69 | YTTYVTFKYRNVPIE | 18 | 193–207 | YVTFKYRNVPIERQK | 11.00 | |||
| 95–109 | SQVFKYTGFIRTLAP | 18 | 7–21 | AKLTKELKTTLPEHF | 12.00 | |||
| 100–114 | YTGFIRTLAPGEDGA | 18 | 142–156 | AKLTKELKTTLPEHF | 12.00 | |||
| 117–131 | ASGFGSGGSGGGSGG | 18 | 77–91 | GLKPSTFYEVVVQAF | 12.00 | |||
| 190–204 | YTTYVTFKYRNVPIE | 18 | 212–226 | GLKPSTFYEVVVQAF | 12.00 | |||
| 230–244 | SQVFKYTGFIRTLAP | 18 | 92–106 | KGGSQVFKYTGFIRT | 12.00 | |||
| 235–249 | YTGFIRTLAPGEDGA | 18 | 227–241 | KGGSQVFKYTGFIRT | 12.00 | |||
| 230–244 | SQVFKYTGFIRTLAP | 12.00 | ||||||
| 97–111 | VFKYTGFIRTLAPGE | 12.00 | ||||||
| 232–246 | VFKYTGFIRTLAPGE | 12.00 | ||||||
Prediction of T-/B-cell antigen co-epitopes of each recombinant protein
| Recombinant protein | Position | T/B combined epitope | |
|---|---|---|---|
| B-Cell antigen epitopes | T-Cell antigen epitopes | ||
| EgG1Y162 | 19–28 | 7–18 | AKLTKELKTTLPEHFRWI HVGS |
| 60–75 | 47–71 | HIKLTANLYTTYVTFKYRNVPIERQ KLTL | |
| 108–117 | 95–118 | SQVFKYTGFIRTLAPGEDGADRAS | |
| EgG1Y162-GSGGSG-EgG1Y162 | 19–26 | 7–37 | AKLTKELKTTLPEHFRWIHVGSRSLELGWNA |
| 60–68 | 45–69 | ADHIKLTANLYTTYVTFKYRNVPIE | |
| 108–130 | 95–111 | SQVFKYTGFIRTLAPGE DGADRASGFGSGGSGVDPE | |
| 147–154 | 133–160 | AKLTKELKTTLPEHFRWIHVGSRSLELG | |
| 186–205 | 171–195 | ADHIKLTANLYTTYVTFKYRNVPIE RQKLTLEGLK | |
| 235–242 | 221–237 | SQVFKYTGFIRTLAPGE DGADR | |
| EgG1Y162-GGGGSGGG-EgG1Y162 | 19–28 | 10–37 | TKELKTTLPEHFRWIHVGSRSLELGWNA |
| 60–79 | 45–69 | ADHIKLTANLYTTYVTFKYRNVPIE RQKLTLEGLK | |
| 108–132 | 95–111 | SQVFKYT GFIRTLAPGEDGADRASGFGGG | |
| 149–166 | 135–165 | AKLTKELKTTLPEHFRWIHVGSRSLELGWNA T | |
| 188–207 | 173–198 | ADHIKLTANLYTTYVTFKYRNVPIER QKLTLEGLK | |
| 236–244 | 220–237 | KGGSQVFKYTGFIRTEAP GEDGADR | |
| EgG1Y162-GSGGSGGGSGGSGGG-EgG1Y162 | 19–26 | 8–21 | KLTKELKTTLPEHF RWIHV |
| 57–70 | 45–69 | ADHIKLTANLYTTYVTFKYRNVPIE R | |
| 108–139 | 95–111 | SQVFKYTGFIRTLAPGE DGADRASGFGSG | |
| 157–178 | 155–172 | HFRWIHVGSRSLELGWNA TGLANL | |
| 195–205 | 180–207 | ADHIKLTANLYTTYVTFKYRNVPIERQK | |
| 243–252 | 230–246 | SQVFKYTGFIRTLAPGE DGADRA | |
Note: T-cell epitopes are in italics, and B-cell epitopes are underlined.
3.1.4 Tertiary structure prediction
I-TASSER [13,14] was used to predict the tertiary structures of EgG1Y162 and EgG1Y162-GGGGSGGG-EgG1Y162. In the prediction of the tertiary structure of EgG1Y162 (Figure 3a), the C-score was −2.25 (C-score ranges from −5 to 2, and a higher score shows a model with higher confidence), the template modeling (TM) score was 0.45 ± 0.14 (TM > 0.5 shows a correct topological model, and TM < 0.17 suggests a randomly similar model), and the root mean square deviation (RMSD) was 10.9 ± 4.6 Å. In the prediction of the tertiary structure of EgG1Y162-GGGGSGGG-EgG1Y162 (Figure 3b), the C-score was −1.55; the TM score was 0.52 ± 0.15, and the RMSD was 7.6 ± 4.3 Å. The results showed that in EgG1Y162-GGGGSGGG-EgG1Y162, the two EgG1Y162 in series can be expressed normally.
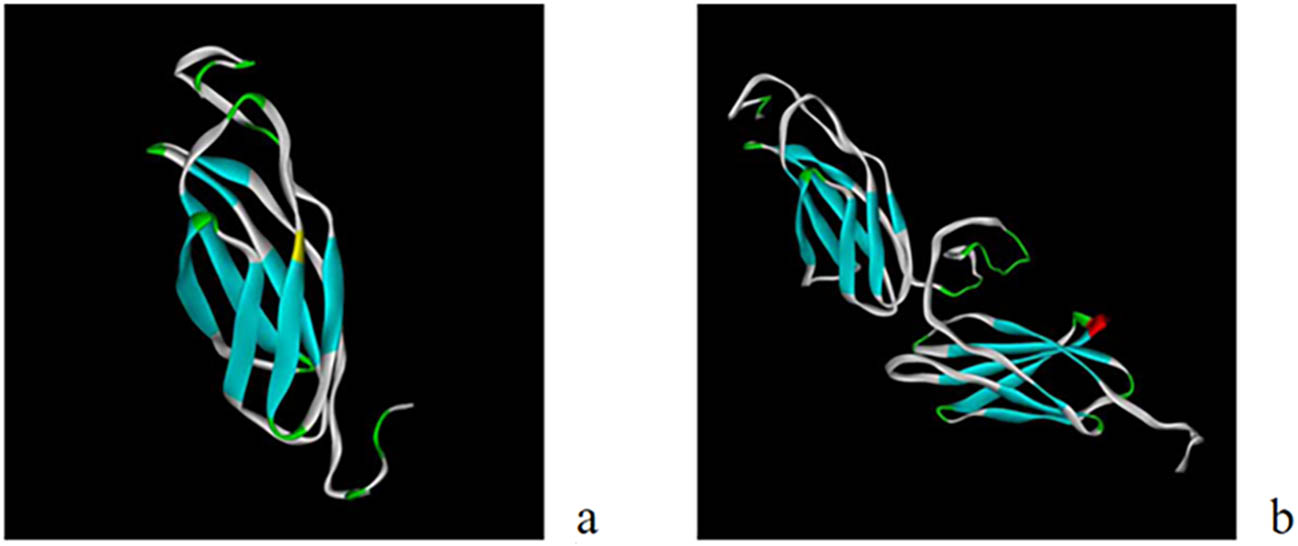
Prediction of the tertiary structure of recombinant protein: (a) EgG1Y162; (b) EgG1Y162-GGGGSGGG-EgG1Y162.
3.2 EgG1Y162-GGGGSGGG-EgG1Y162 promoted DC maturation by increasing antigenic epitopes, presumably with possible strong immunogenicity
3.2.1 The recombinant plasmid was correctly constructed and the prokaryotic expression of recombinant protein was correctly induced
The recombinant plasmid, pET30a-EgG1Y162-GGGGSGGG-EgG1Y162, was double digested by EcoRI and SalI pairs, and the target fragments with sizes of about 5,400 and 756 bp, which were consistent with the expected size, were obtained by 1% agarose gel electrophoresis (Figure 4a). The induced expression of purified recombinant proteins, HIS-EgG1Y162 and HIS-EgG1Y162-GGGGGSGGG-EgG1Y162, showed distinct bands at 20.5 (Figure 4b) and 35 kDa (Figure 4c), respectively, which were in accordance with the expected results.

Identification of recombinant plasmid pET30a-EgG1Y162-GGGGSGGG-EgG1Y162 digestion and protein purification: (a) identification of recombinant plasmid pET30a-EgG1Y162-GGGGSGGG-EgG1Y162 digestion. M: DL5000; 1 and 2: Recombinant plasmids digested by EcoRI and SalI, respectively. (b) Western blot analysis of purified EgG1Y162. M: protein molecular quality standard; 1–3: EgG1Y162. (c) Western blot analysis of purified HIS-EgG1Y162-GGGGSGGG-EgG1Y162.
3.2.2 Specificity of EgG1Y162-GGGGGSGGG-EgG1Y162
The serum of 10 normal mice was analyzed by Western blot. None of the serum of the normal mice showed obvious reaction bands at about 35 kDa, whereas obvious reaction bands were found in the serum of 8 mice infected with E. granulosus at about 35 kDa of the target band. The calculated specificity was 100% as shown in Figure 5.

Western blot identification of mouse sera. M: protein molecular quality standard; (a) 1–8: sera of cystic hydatid mouse; (b) 1–10: normal mouse sera.
3.2.3 EgG1Y162-GGGGSGGG-EgG1Y162 promoted the maturation of the mouse bone morrow-derived DCs
After the mouse bone marrow-derived DCs were stimulated with recombinant protein for 24 h, the percentage of mature DCs in the EgG1Y162-GGGGSGGG-EgG1Y162-stimulated group (21.533 ± 0.777%) was significantly higher than that of the EgG1Y162-stimulated group (9.37 ± 0.800%) as shown in Figure 6 (t = 18.883, P < 0.01).

Flow cytometry analysis of the percentage of mature DCs after EgG1Y162 and EgG1Y162-GGGGSGGG-EgG1Y162 stimulation for 24 h.
4 Discussion
Echinococcosis is an endemic and natural zoonotic disease caused by E. granulosus infection in intermediate livestock, especially in the pastoral areas of Northwest China [15]. It has a severe impact on human health, and the extent of the disease gradually increases, which will seriously restrict economic development [16,17]. The current treatment method for hydatid disease is still relatively limited, and the effect of surgery or medical treatment is not ideal [18,19]. Therefore, vaccination is an effective way to prevent the infection of hydatid disease, and the development of vaccine has become a hot spot of current research [20]. Our research group previously demonstrated that the antigen EgG1Y162 of E. granulosus has good immunogenicity and can induce the body to produce an effective immune response for immune protection [21]. The immunogenicity of the antigen has a great relationship with its state and time of existence in the body. An antigen with a larger molecular weight and a more stable protein state is stronger, exists in the organism for a longer period of time, has a stronger immunogenicity, and can produce a stronger immune response; thus, it can enable the organism to obtain better protection. In the present study, we tried to cascade antigen EgG1Y162 by selecting an appropriate linker sequence and increasing the molecular weight of the recombinant protein to achieve a better immune response and improve the immunogenicity of the antigen.
With the development of computer bioinformatics technology, bioinformatics has been increasingly used to predict protein structures [22]. Therefore, in this study, the structures of EgG1Y162-linker-EgG1Y162 recombinant proteins with different linker sequences were predicted by bioinformatics methods. The results showed that the physicochemical properties, hydrophilicity, and stability of EgG1Y162 did not change after the incorporation of a series of long, medium, and short linker sequences. We further examined whether the T-/B-cell antigen epitope shifted in the three EgG1Y162-linker-EgG1Y162 recombinant proteins to select the best linker sequence. Therefore, SYFPEITHI [23], IEDB [24], BepiPred [25], and SVMTriP [26] were used to predict the epitopes of the recombinant proteins. HLA-DRB1*0701 was selected as the parameter. Ten results were compared to determine which epitope had the highest SYFPEITHI score (the higher the score, the more likely the sequence is to be the dominant epitope) and the lowest IEDB percentile rank (the lower the percentile rank, the more likely the sequence is to be the dominant epitope). Based on the results of secondary structures, the structures that could not form an epitope were excluded. In EgG1Y162-GSGGSG-EgG1Y162, the T- and B-cell antigen epitopes did not change or shift. We found through data integration that the T-/B-cell antigen epitopes of EgG1Y162-GGGGSGGG-EgG1Y162 are located at 10–37 (equivalent to 10–37 aa on EgG1Y162, “TKELKTTLPEHFRWIHVGSRSLELGWNA”), 45–79 (equivalent to 45–79 aa on EgG1Y162, “ADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLK”), 95–132 (equivalent to 95–120 aa on EgG1Y162, “SQVFKYTGFIRTLAPGEDGADRASGFGGG”), 135–166 (equivalent to 7–38 aa on EgG1Y162, “AKLTKELKTTLPEHFRWIHVGSRSLELGWNAT”), and 173–207 (equivalent to 45–79 aa on EgG1Y162, “ADHIKLTANLYTTYVTFKYRNVPIERQKLTLEGLK”), and 220–244 aa (equivalent to 92–116 aa on EgG1Y162, “KGGSQVFKYTGFIRTEAPGEDGADR”). The analysis showed that among the three recombinant proteins, the epitope of EgG1Y162-GGGGSGGG-EgG1Y162 almost had no offset. This result also suggests that EgG1Y162 could be connected in series by linker sequence “GGGGSGGG” to increase the molecular weight of the protein without affecting the original epitope.
Tertiary structure prediction was performed on EgG1Y162 and EgG1Y162-GGGGSGGG-EgG1Y162 to determine whether linker sequence “GGGGSGGG” affects the expression of proteins on both sides. Tertiary structure prediction is performed through three methods: homology modeling, line string method, and ab initio prediction method. I-TASSER [14,27] online prediction server uses two sets of algorithms, namely, homology modeling and line string method, to model and predict the protein’s tertiary structure; its accuracy and reliability are much higher than those of Swiss Model and PHYRE, and its 3D model is based on the multi-line LOMETS and iterative TASSER. Predictive models were derived to match the database of BioLiP protein functions, and the first one provided is the most reliable template parameter [28]. In the tertiary structure prediction of EgG1Y162, the C-score was −1.55, the TM score was 0.52 ± 0.15, and the RMSD was 7.6 ± 4.3 Å. In the tertiary structure prediction of EgG1Y162-GGGGSGGG-EgG1Y162, the C-score was −2.25, the TM score was 0.45 ± 0.14, and the RMSD was 10.9 ± 4.6 Å. The prediction results of the 3D structures of the two proteins showed that the contiguous proteins can be correctly folded with the addition of the linker sequence “GGGGSGGG,” which further confirmed the feasibility of selecting the linker sequence “GGGGSGGG.” The recombinant antigen is linked to the same antigen to increase the specific epitope, so as to cause a strong immune response. There are relevant studies linking different antigens to prepare a multi-epitope vaccine, which also has significant immune effects and good safety, providing a new idea for the follow-up experiments of this study [29,30,31].
A large number of recombinant proteins need to be induced to conduct animal immunization experiments. The research group explored the induced expression conditions of the recombinant protein and found that the IPTG concentration of recombinant plasmid pET30a-EgG1Y162 was 0.2 mmol/L at 28°C, and the induced expression of recombinant protein EgG1Y162 was the highest in the supernatant after 6 h of induction. The protein expression level of recombinant plasmid pET30a-EgG1Y162-GGGGSGGG-EgG1Y162 was the highest in the supernatant at 28°C, the final IPTG concentration was 0.5 mmol/L, and the induction lasted for 6 h. In the Western blot method, we applied the His-Tag tag antibody for the initial identification of the proteins. The clear bands at approximately 20.5 and 35 kDa indicated that the EgG1Y162 and EgG1Y162-GGGGSGGG-EgG1Y162 were successfully induced. In further serological validation, the recombinant protein EgG1Y162-GGGGSGGG-EgG1Y162 antigenicity was analyzed in the sera of diseased mice and normal mice, and the specificity of the protein was found to be 100% in both cases.
The antigen presentation process of vaccine entry into the organism starts with dedicated APCs [24,25]. APCs are the first gate to trigger the immune response after vaccine entry into the organism [32]. DCs are the most functional APCs in the body. Under normal conditions, most DCs in the body are in the immature stage, and imDCs have a strong capacity for antigen endocytosis and processing [33]. After a series of processes, such as antigen uptake and inflammatory factor activation, DCs change from immature to mature, and mature DCs highly express antigen-presenting molecules, MHC class II molecules, and co-stimulatory molecules, such as CD54, CD40, CD80, and CD86, which initiate MHC-II class-restricted CD4 + Th2 responses, promote the antibody production of B cells and cell-mediated immunity, and play a good immunoprotective effect [34,35,36]. Purified HIS-EgG1Y162 and HIS-EgG1Y162-GGGGSGGG-EgG1Y162 were separately co-cultured with the DCs obtained by in vitro induction. Compared with HIS-EgG1Y162, DCs were more likely to mature under HIS-EgG1Y162-GGGGSGGG-EgG1Y162 stimulation. This finding suggests that EgG1Y162-GGGGSGGG-EgG1Y162 stimulation promoted the maturation of mouse bone marrow-derived DCs and thus enhanced the immune response. We preliminarily speculated that the recombinant vaccine, EgG1Y162-GGGGSGGG-EgG1Y162, had improved immunogenicity because of the increased molecular weight and repeating T-/B-cell epitope.
In summary, we predicted and screened the best linker sequence by bioinformatics prediction and successfully induced the expression of the recombinant vaccine, EgG1Y162-GGGGSGGG-EgG1Y162. The results of serological identification in mice showed that the recombinant vaccine had good specificity. The antigen epitopes contained in the recombinant vaccine were increased by increasing the molecular weight of the recombinant vaccine to promote the maturation of DC and improve the immunogenicity of the vaccine. The design and preparation of recombinant protein EgG1Y162-GGGGGSGGG-EgG1Y162 provide a new idea for the optimization and improvement of the vaccine against encapsulated diseases. The development of a safe and effective hydatid vaccine is only the first step toward eliminating hydatid, and we need to anticipate implementation strategies and acceptance. Therefore, a comprehensive evaluation of the reliability, safety, and benefits of a candidate vaccine needs to be carried out before it can be put into clinical trials.
-
Funding information: This project is financially supported by the National Natural Science Foundation of China (81760656), the National Natural Science Foundation of China (32260192) and the Natural Science Fund of Xinjiang Uygur Autonomous Region (2018D01C157).
-
Author contributions: Xiaotao Zhou, Jianbing Ding, and Chunbao Cao designed and conceived the research; bioinformatic analysis was performed by Yanxia Zhou, Shangqi Zhao, and Yanmin Li; the experiment was performed by Yanxia Zhou and Shangqi Zhao; Mingkai Yu, Jia Zheng, and Qiaoqiao Gong analyzed the data; the manuscript was drafted by Yanxia Zhou and revised by Xiaotao Zhou; all authors read and approved the final manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Otero-Abad B, Torgerson PR. A systematic review of the epidemiology of echinococcosis in domestic and wild animals. PLoS Negl Trop Dis. 2013;7:6e2249.10.1371/journal.pntd.0002249Suche in Google Scholar PubMed PubMed Central
[2] Zhang F, Pang N, Zhu Y, Zhou D, Zhao H, Hu J, et al. CCR7(lo)PD-1(hi) CXCR5( +) CD4( +) T cells are positively correlated with levels of IL-21 in active and transitional cystic echinococcosis patients. BMC Infect Dis. 2015;15:457.10.1186/s12879-015-1156-9Suche in Google Scholar PubMed PubMed Central
[3] Zhang W, Zhang Z, Shi B, Li J, You H, Tulson G, et al. Vaccination of dogs against Echinococcus granulosus, the cause of cystic hydatid disease in humans. J Infect Dis. 2006;194:7966–74.10.1086/506622Suche in Google Scholar PubMed
[4] Pandey RK, Bhatt TK, Prajapati VK. Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Sci Rep. 2018;8:11125.10.1038/s41598-018-19456-1Suche in Google Scholar PubMed PubMed Central
[5] Jazouli M, Lightowlers MW, Bamouh Z, Gauci CG, Tadlaoui K, Ennaji MM, et al. Immunological responses and potency of the EG95NC(-) recombinant sheep vaccine against cystic echinococcosis. Parasitol Int. 2020;78:102149.10.1016/j.parint.2020.102149Suche in Google Scholar PubMed
[6] Safavi A, Kefayat A, Ghahremani F, Mahdevar E, Moshtaghian J. Immunization using male germ cells and gametes as rich sources of cancer/testis antigens for inhibition of 4T1 breast tumors’ growth and metastasis in BALB/c mice. Int Immunopharmacol. 2019;74:105719.10.1016/j.intimp.2019.105719Suche in Google Scholar PubMed
[7] Shakibapour M, Kefayat A, Reza Mofid M, Shojaie B, Mohamadi F, Maryam Sharafi S, et al. Anti-cancer immunoprotective effects of immunization with hydatid cyst wall antigens in a non-immunogenic and metastatic triple-negative murine mammary carcinoma model. Int Immunopharmacol. 2021;99:107955.10.1016/j.intimp.2021.107955Suche in Google Scholar PubMed
[8] Cao CB, Ma XM, Ding JB, Jia HY, Mamuty W, Ma HM, et al. Cloning and sequence analysis of the egG1Y162 gene of Echinococcus granulosus. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2009;27:2177–9.Suche in Google Scholar
[9] Ma X, Zhao H, Zhang F, Zhu Y, Peng S, Ma H, et al. Activity in mice of recombinant BCG-EgG1Y162 vaccine for Echinococcus granulosus infection. Hum Vaccin Immunother. 2016;12:1170–5.10.1080/21645515.2015.1064564Suche in Google Scholar PubMed PubMed Central
[10] Zhao X, Zhang F, Li Z, Wang H, An M, Li Y, et al. Bioinformatics analysis of EgA31 and EgG1Y162 proteins for designing a multi-epitope vaccine against Echinococcus granulosus. Infect Genet Evol. 2019;73:98–108.10.1016/j.meegid.2019.04.017Suche in Google Scholar PubMed
[11] Zhang F, Ma X, Zhu Y, Wang H, Liu X, Zhu M, et al. Identification, expression and phylogenetic analysis of EgG1Y162 from Echinococcus granulosus. Int J Clin Exp Pathol. 2014;7:5655–64.Suche in Google Scholar
[12] Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2.10.1186/1745-7580-2-2Suche in Google Scholar PubMed PubMed Central
[13] Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:4725–38.10.1038/nprot.2010.5Suche in Google Scholar PubMed PubMed Central
[14] Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinforma. 2008;9:40.10.1186/1471-2105-9-40Suche in Google Scholar PubMed PubMed Central
[15] Ozturk G, Aydinli B, Yildirgan MI, Basoglu M, Atamanalp SS, Polat KY, et al. Posttraumatic free intraperitoneal rupture of liver cystic echinococcosis: a case series and review of literature. Am J Surg. 2007;194:3313–6.10.1016/j.amjsurg.2006.11.014Suche in Google Scholar PubMed
[16] Wen H, Vuitton L, Tuxun T, Li J, Vuitton DA, Zhang W, et al. Echinococcosis: Advances in the 21st century. Clin Microbiol Rev. 2019;32:2.10.1128/CMR.00075-18Suche in Google Scholar PubMed PubMed Central
[17] Messaoudi H, Zayene B, Ben Ismail I, Lajmi M, Lahdhili H, Hachicha S, et al. Bilateral pulmonary hydatidosis associated with uncommon muscular localization. Int J Surg Case Rep. 2020;76:130–3.10.1016/j.ijscr.2020.09.070Suche in Google Scholar PubMed PubMed Central
[18] Gonder N, Demir IH, Kilincoglu V. The effectiveness of combined surgery and chemotherapy in primary hydatid cyst of thigh muscles, a rare localization and its management. J Infect Chemother. 2021;27:3533–536.10.1016/j.jiac.2020.10.027Suche in Google Scholar PubMed
[19] Zhang W, McManus DP. Recent advances in the immunology and diagnosis of echinococcosis. FEMS Immunol Med Microbiol. 2006;47:124–41.10.1111/j.1574-695X.2006.00060.xSuche in Google Scholar PubMed
[20] Milhau N, Almouazen E, Bouteille S, Hellel-Bourtal I, Azzouz-Maache S, Benavides U, et al. In vitro evaluations on canine monocyte-derived dendritic cells of a nanoparticles delivery system for vaccine antigen against Echinococcus granulosus. PLoS One. 2020;15:2e0229121.10.1371/journal.pone.0229121Suche in Google Scholar PubMed PubMed Central
[21] Zhang F, Li S, Zhu Y, Zhang C, Li Y, Ma H, et al. Immunization of mice with egG1Y162-1/2 provides protection against Echinococcus granulosus infection in BALB/c mice. Mol Immunol. 2018;94:183–9.10.1016/j.molimm.2018.01.002Suche in Google Scholar PubMed
[22] Sha T, Li Z, Zhang C, Zhao X, Chen Z, Zhang F, et al. Bioinformatics analysis of candidate proteins Omp2b, P39 and BLS for Brucella multivalent epitope vaccines. Microb Pathog. 2020;147:104318.10.1016/j.micpath.2020.104318Suche in Google Scholar PubMed
[23] Schuler MM, Nastke MD, Stevanovikc S. SYFPEITHI: Database for searching and T-cell epitope prediction. Methods Mol Biol. 2007;409:75–93.10.1007/978-1-60327-118-9_5Suche in Google Scholar PubMed
[24] Gonzalez-Diaz H, Perez-Montoto LG, Ubeira FM. Model for vaccine design by prediction of B-epitopes of IEDB given perturbations in peptide sequence, in vivo process, experimental techniques, and source or host organisms. J Immunol Res. 2014;2014:768515.10.1155/2014/768515Suche in Google Scholar PubMed PubMed Central
[25] Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W1W24–9.10.1093/nar/gkx346Suche in Google Scholar PubMed PubMed Central
[26] Yao B, Zheng D, Liang S, Zhang C. SVMTriP: A method to predict B-Cell linear antigenic epitopes. Methods Mol Biol. 2020;2131:299–307.10.1007/978-1-0716-0389-5_17Suche in Google Scholar PubMed
[27] Zheng W, Zhang C, Bell EW, Zhang Y. I-TASSER gateway: A protein structure and function prediction server powered by XSEDE. Future Gener Comput Syst. 2019;99:73–85.10.1016/j.future.2019.04.011Suche in Google Scholar PubMed PubMed Central
[28] Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:4702–10.10.1002/prot.20264Suche in Google Scholar PubMed
[29] Safavi A, Kefayat A, Sotoodehnejadnematalahi F, Salehi M, Modarressi MH. Production, purification, and in vivo evaluation of a novel multiepitope peptide vaccine consisted of immunodominant epitopes of SYCP1 and ACRBP antigens as a prophylactic melanoma vaccine. Int Immunopharmacol. 2019;76:105872.10.1016/j.intimp.2019.105872Suche in Google Scholar PubMed
[30] Safavi A, Kefayat A, Mahdevar E, Ghahremani F, Nezafat N, Modarressi MH. Efficacy of co-immunization with the DNA and peptide vaccines containing SYCP1 and ACRBP epitopes in a murine triple-negative breast cancer model. Hum Vaccin Immunother. 2021;17:122–34.10.1080/21645515.2020.1763693Suche in Google Scholar PubMed PubMed Central
[31] Mahdevar E, Kefayat A, Safavi A, Behnia A, Hejazi SH, Javid A, et al. Immunoprotective effect of an in silico designed multiepitope cancer vaccine with BORIS cancer-testis antigen target in a murine mammary carcinoma model. Sci Rep. 2021;11:123121.10.1038/s41598-021-01770-wSuche in Google Scholar PubMed PubMed Central
[32] Jiraviriyakul A, Songjang W, Kaewthet P, Tanawatkitichai P, Bayan P, Pongcharoen S. Honokiol-enhanced cytotoxic T lymphocyte activity against cholangiocarcinoma cells mediated by dendritic cells pulsed with damage-associated molecular patterns. World J Gastroenterol. 2019;25:293941–55.10.3748/wjg.v25.i29.3941Suche in Google Scholar PubMed PubMed Central
[33] Karimi K, Boudreau JE, Fraser K, Liu H, Delanghe J, Gauldie J, et al. Enhanced antitumor immunity elicited by dendritic cell vaccines is a result of their ability to engage both CTL and IFNgamma-producing NK cells. Mol Ther. 2008;16:2411–8.10.1038/sj.mt.6300347Suche in Google Scholar PubMed
[34] Qian C, Cao X. Dendritic cells in the regulation of immunity and inflammation. Semin Immunol. 2018;35:3–11.10.1016/j.smim.2017.12.002Suche in Google Scholar PubMed
[35] Zheng H, Liu L, Zhang H, Kan F, Wang S, Li Y, et al. Correction: Dendritic cells pulsed with placental gp96 promote tumor-reactive immune responses. PLoS One. 2019;14:6e0218362.10.1371/journal.pone.0218362Suche in Google Scholar PubMed PubMed Central
[36] Xu Y, Tang X, Yang M, Zhang S, Li S, Chen Y, et al. Interleukin 10 gene-modified bone marrow-derived dendritic cells attenuate liver fibrosis in mice by inducing regulatory T cells and inhibiting the TGF-beta/smad signaling pathway. Mediators Inflamm. 2019;2019:4652596.10.1155/2019/4652596Suche in Google Scholar PubMed PubMed Central
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”
Artikel in diesem Heft
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”

