Abstract
Triple-negative breast cancer (TNBC) is an aggressive subtype with limited treatment options and high mortality rates. It remains a prevailing clinical need to distinguish whether the patient can benefit from therapy, such as chemotherapy. By integrating single-cell and global transcriptome data, we have for the first time identified TCL1A+ B cell functions that are prognostically relevant in TNBC. This finding broadens the perspective of traditional tumor-infiltrating lymphocytes in predicting survival, especially the potential value of B cells in TNBC. Single-cell RNA-seq data from five TNBC patients were collected to identify the association between immune cell populations and clinical outcomes. Functional analysis was according to gene set enrichment analysis using pathways from MsigDB. Subsequently, the gene signature of TCL1A+ B cells based on differential expression genes of TCL1A+ B cells versus other immune cells was used to explore the correlation with tumor microenvironment (TME) and construct a prognostic signature using a non-parametric and unsupervised method. We identified TCL1A+ B cells as a cluster of B cells associated with clinical outcomes in TNBC. Functional analysis demonstrated its function in B cell activation and regulation of immune response. The highly enriched TCL1A+ B cell population was found to be associated with a thermal TME with anti-tumor effects. A high abundance of TCL1A+ B cell population is positively correlated with a favorable therapeutic outcome, as indicated by longer overall survival. The present study suggests that TCL1A+ B cells play a key role in the treatment and prognostic prediction of TNBC, although further studies are needed to validate our findings. Moreover, the integration of transcriptome data at various resolutions provides a viable approach for the discovery of novel prognostic markers.
1 Introduction
Triple-negative breast cancer (TNBC) is defined as a kind of breast cancer with the absence of estrogen receptor (ER), progesterone receptor, and human epidermal growth factor receptor 2 (HER-2). Because of the lack of molecular targets of therapeutic agents, therapeutic options are limited for TNBC patients [1,2]. Chemotherapy is currently the primary established treatment option for patients with TNBC that significantly improve the prognosis of TNBC patients [3]. Many patients will receive neoadjuvant chemotherapy before surgery so as to reduce the size of the primary lesion and lower the tumor stage to meet the goal of surgical applicability and improve the rate of surgical breast conservation. Some studies have found that 90% of patients undergoing neoadjuvant chemotherapy experience a reduction in tumor size, and 30% of patients even achieve complete remission [4,5]. EGFR overexpression is common in TNBC, which is directly related to the poor prognosis of patients. Treatment with cetuximab can improve the survival rate of patients, which has a significant effect. For women with TNBC who have a BRCA mutation and whose cancer no longer responds to standard breast cancer chemotherapeutic agents, platinum-based chemotherapeutic drug agents such as cisplatin or carboplatin chemotherapy agents or targeted agents known as poly-ADP-ribose polymerase inhibitors such as Lynparza or Talzenna may be considered. In addition, PD-1/PD-L1 inhibitors have also shown sound therapeutic effects in immunotherapy. However, not all patients can obtain clinical benefits from the chemotherapy; approximately 50% of patients had no pathological complete response [4,6–8]. A novel, better treatment option, and personality clinical intervention strategy are still urgently needed.
With the development of single-cell RNA-seq (scRNA-seq) technology, individual transcriptomic gene expression data can be obtained. It provides new chances to explore the tumor microenvironment (TME) heterogeneity of breast cancer and TNBC and perform the association between immune cell populations and clinical outcomes [9–12]. However, the cohort size and the access to clinical outcomes generally become the limitation of related studies.
The prognosis prediction capability of tumor infiltration lymphocytes, particularly T cells, has been described across multiple indications [13–15]. Recent studies have also demonstrated that higher infiltration of B cells might show a favorable prognosis in non-small cell lung cancer, soft tissue sarcoma, colorectal cancer, and renal cell carcinoma [16–19]. As such, it is unclear whether B cells or B cell subpopulations are beneficial in correlation with clinical outcomes or can be used as a prediction factor in TNBC.
This study integrated the scRNA-seq and bulk RNA-seq data with a bioinformatic method to explore the association between immune cell populations and clinical benefits. We identified TCL1A+ B cells, similar to follicular B cells, which positively correlated to favorable clinical outcomes. Furthermore, we constructed a gene signature to demonstrate the correlation between the TME and the B cell subpopulation and uncover its value in predicting prognosis.
2 Materials and methods
2.1 Public data collection
The scRNA-seq data of five TNBC patients with primary tumors were collected from the Gene Expression Omnibus database. GSE169246 [9] (PD-L1 blockade-induced temporal single cell dynamics in TNBC), which utilized single-cell RNA- and ATAC-sequencing to examine the dynamics of immune cells in metastatic TNBC patients treated with paclitaxel or its combination with atezolizumab. The investigator evaluated the clinical responses based on radiologic assessments of tumor sizes every 8 weeks according to the Response Evaluation Criteria in Solid Tumors (RECIST, version 1.1) [20]. Notably, after the eighth week of treatment initiation, the final response was defined according to the reaction of patients, including PR (responder, complete response/partial response samples) and SD stable disease, progression disease samples. All five patients received chemotherapy with paclitaxel monotherapy.
Two bulk RNA-seq cohorts were utilized in our study. The first one of 116 TNBC patients and their clinical information were downloaded and extracted from the TCGA-BRCA dataset based on ER, progesterone receptor, and HER2 status. Another bulk RNA-seq data of 319 TNBC patients and corresponding clinical information were collected from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) project in the cBioportal database [21]. Similar to the TCGA dataset, ER, progesterone receptor, and HER2 gene information was applied to select TNBC patients.
2.2 scRNA-seq data processing
The unique molecular identifier (UMI) matrix, barcode list, and features information of five patients were used as input data into Seurat (v4.1) [22]. We utilized the library size correction method to normalize the raw expression matrix with the NormalizeData function. FindVariableFeatures was applied to find out the highly variable genes (HGVs). Normalized expression data were further scaled with the top 2,000 HGVs and regressed with the percentage of mitochondrial gene counts, UMI counts, and patient and cell cycle scores. To reduce noise and remove the batch effect, we performed principal component analysis with 50 principal components (PCs) and selected the top 30 PCs for the RunHarmony function of the harmony package [23]. “Patient” was the variable to remove during the integration. We next constructed a k nearest neighbor graph based on the harmony space with the FindNeighbor function. The Louvain algorithm was applied to cluster the cells using 0.5 as resolution. Uniform manifold approximation and projection (UMAP) was used with the same harmony spaces for visualization, and the expression level of genes was shown by the FeaturePlot function.
We detected differential expression genes (DEGs) using the FindAllMarkers function to annotate the immune cell clusters. Marker genes annotated the significant cell populations. To further identify the specific group, marker genes between cluster 12 and other clusters and cluster 12 versus cluster 9 were detected by FindMakers function with different idents.
2.3 Definition of gene signature
From the results of DEGs between cluster 12 and others, genes with log2FC ≥1 and adjusted p-value <0.05 were considered as signature genes of TCL1A+ B cells. To further confirm the uniqueness of the gene signature, we utilized the AddModuleScore function to evaluate the abundance of the above gene signature in each cell and displayed it using the RidgePlot function.
2.4 Pathway and immune cell populations analysis
To investigate the function of TCL1A+ B cells in scRNA-seq data, DEGs with log2FC ≥1 and adjusted p-value <0.05 were considered significantly differential genes to downstream analysis. The gene symbols were converted into Entrez id using the org.Hs.eg.db package. We analyzed biological processes using the enrichGO function from the clusterProfiler package according to the significant DEGs [24]. The top 30 pathways were displayed by dot plot. In addition, we utilized an R package called fast gene set enrichment analysis (GSEA) (fuse) to calculate the empirical enrichment score null distributions simultaneously for gene sets from C2-CP canonical pathways of MsigDB.
To evaluate the association between TCL1A+ B cells and TME in bulk RNA-seq, we performed the abundance score using the given method with gene signatures of TCL1A+ B cells [25]. The patients from TCGA-TNBC were further classified into two groups based on the median value of abundance score. An abundance score of four immune cell populations, including activated CD4+ T cells, activated CD8+ T cells, effector memory CD4+ T cells, and effector memory CD8+ T cells, and four hot-tumor gene sets were conducted by the given method as well.
Raw counts data of TCGA-TNBC were downloaded using the TCGAbiolinks package, and group information was inherited from the above analysis. We analyzed gene rank by a negative binomial generalized linear model to estimate dispersion and logarithmic fold changes incorporating data-driven prior distributions using DESeq2. GSEA was used to evaluate the normalized enrichment scores of pathways from the Reactome pathway database [26–28].
2.5 Survival analysis
The TCGA-TNBC and METABRIC-TNBC described above were used to evaluate the prognostic performance of TCL1A+ B cells. We utilized the GSVA method for gene signature to obtain the abundance score of TCL1A+ B cells. The patients from each cohort were classified into high and low groups according to the median value of the abundance score of the given signature, respectively. The Cox proportional hazards regression model was used to evaluate hazard ratios (HRs). The median overall survival (OS) and survival curves were performed using the Kaplan–Meier method.
2.6 Statistical analysis
Statistical analysis was performed as described in the figure legends. Correlation coefficients were calculated by Pearson correlation. Survival was measured by the Kaplan–Meier method. Statistical significance was determined by Cox regression, Wilcoxon test, and Benjamini–Hochberg method.
3 Results
3.1 Association of immune cell populations with clinical outcome
The scRNA-seq data of five TNBC patients were collected from a published study. Two patients were defined as having a partial response, and the others are stable diseases based on RECIST. We utilized the harmony package to integrate all samples and generated 21 cell clusters from 26,384 single cells with Seurat package (Figure 1a). Based on the expression of marker genes, the 14 cell clusters were classified into eight significant immune cell populations (Figure 1b), including CD8+ T cells (CD3D+CD8A+CD4−, clusters 1, 6, 8), CD4+ T cells (CD3D+CD4+CD8A−, cluster 2), regulatory T cells (Tregs) (FOXP3+, cluster 3), follicular helper T cells (Tfh) (CD200+, cluster 7), B cells (CD19+MS4A1+, cluster 9, 12), plasma B cells (MZB1+, cluster 4), macrophage (LYZ+C1QC +, cluster 5), and pDC (LILRA4+, cluster 14).
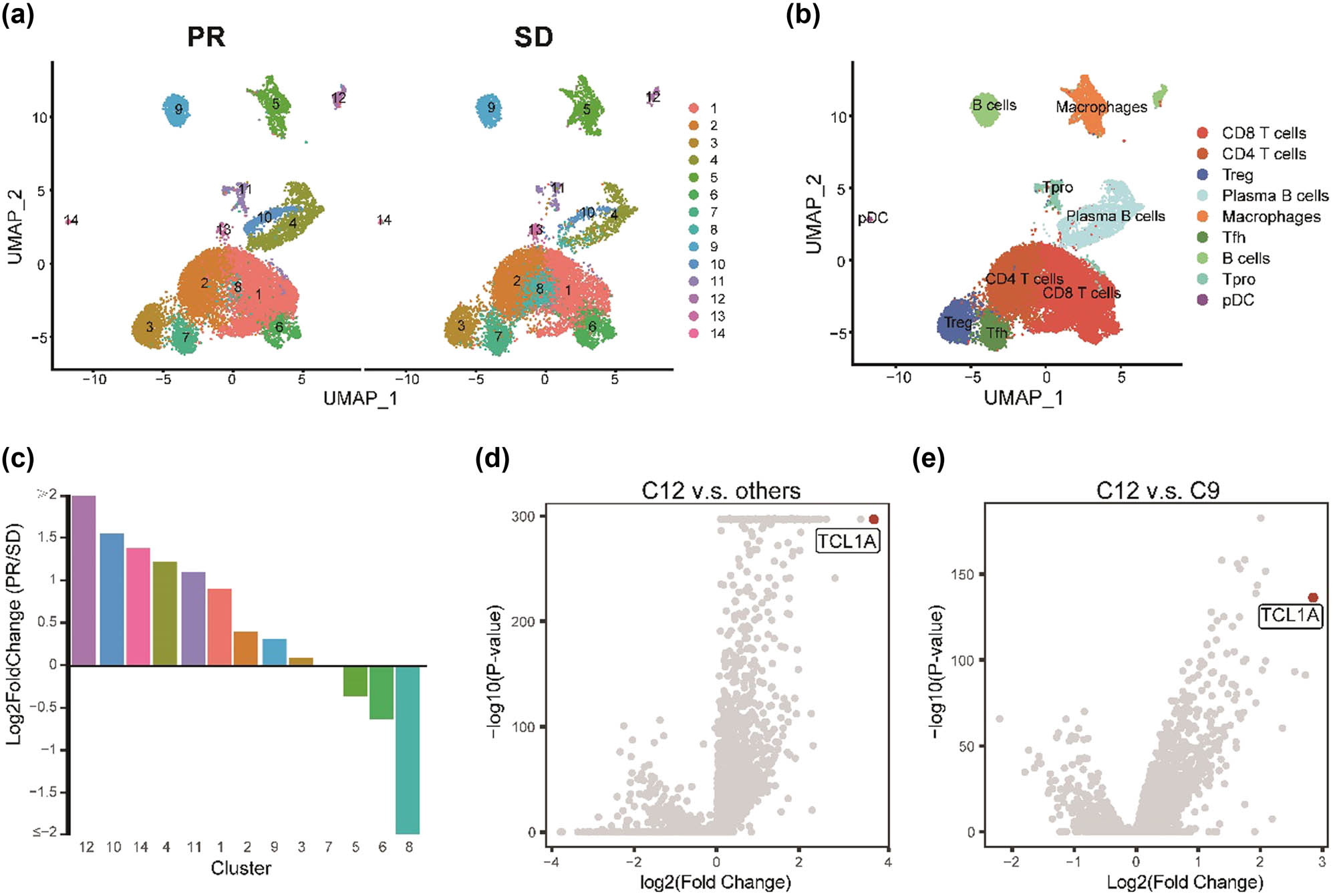
Identification of immune cell populations by scRNA-seq. (a) UMAP plot of immune cell clusters between PR and SD from five TNBC patients. (b) UMAP plot of nine significant immune cell populations. (c) The fold change of the percentages of each of the 14 clusters comparing the PR and SD. (d) Scatter plot of DEGs between cluster 12 and other clusters. (e) Scatter plot of DEGs between cluster 12 and cluster 9. Red point of (d) and (e) represents the top 1 DEG ranked by log2 fold change.
To investigate the association of immune cells with clinical outcome, we utilized integrative analysis by combining cells from all patients (Figure 1c). Usingtwo-fold differences as a biologically significant threshold, we identified two clusters, cluster 8 and cluster 12, which exceed this criterion. Furthermore, cluster 12 was 2.04-fold higher in the PR group versus the SD group, whereas cluster 8 was 3.74-fold higher in the SD group than in the PR group. We also tested the distribution of cluster 12 at the patient level (Figure S1); the findings provide support for the integrated analysis, indicating that patients in the PR group exhibited a significantly higher proportion of cluster 12, which is associated with a pro-tumoral environment, compared to almost all patients in the SD group. Unfortunately, the statistical analysis cannot be done due to the small size of the samples.
To further annotate cluster 12, we intersected the DEGs between cluster 12 versus other groups and cluster 12 versus another B cell cluster (cluster 9) (Figure 1d and e, Table S1). TCL1A (TCL1 family AKT coactivator A), as the highest expression differential gene in both groups, was used to annotate cluster 12. Cluster 12 is thus named TCL1A+ B cells.
Overall, we identified two clusters; a subset of CD8+ T cells and a subset of B cells were highly related to clinical outcomes in TNBC. In contrast, only the B cell subset, TCL1A+ B cells, positively correlate with PR patients.
3.2 Functional exploration of TCL1A+ B cells
We investigated the function of TCL1A+ B cells in the above scRNA-seq data. By comparing transcriptomic differences of TCL1A+ B cells versus other immune cell populations, we found that TCL1A+ B cells significantly enriched in B cell-related immune response and regulation of T cell activation biological processes (Figure 2a), such as B cell activation, activation of the immune response, and positive regulation of T cell activation, indicating the association with anti-tumor immune response. Moreover, compared to another B cell population (cluster 9), the proliferation, differentiation, and activation function was more robust in TCL1A+ B cells than in another (Figure 2b).

Enrichment biological processes of TCL1A+ B cells. (a) Enrichment biological processes between TCL1A+ B cells and other immune cell populations. (b) Enrichment biological processes between TCL1A+ B cells and another B cell populations.
To explore the potential functional differences of TCL1A+ B cells between the PR and SD groups, we utilized GSEA with pathways from five pathway databases based on gene rank from the two groups. The results showed that TCL1A+ B cells from SD patients are more likely to be enriched in hypoxia, NFAT, and TNF-alpha pathways, correlated with tumor invasion and metastasis, indicating a pro-tumoral environment (Table S3).
3.3 Higher TCL1A+ B cells are associated with an anti-TME
Despite TCL1A being selected to represent cluster 12, it was expressed by a few cells of another B cell population as well (Figure 3a). Based on previous studies [29,30], the high expression level of BCL6, NEIL1, and MEF2B proposed that our TCL1A+ B cells are more similar to follicular B cells. To perform the abundance of TCL1A+ B cells in bulk transcriptomic data, we first constructed a signature with 165 genes (Table S2). Examining the signature in scRNA-seq data showed that the signature could further distinguish the TCL1A+ B cells from other immune cell populations, especially other B cells (Figure 3b).
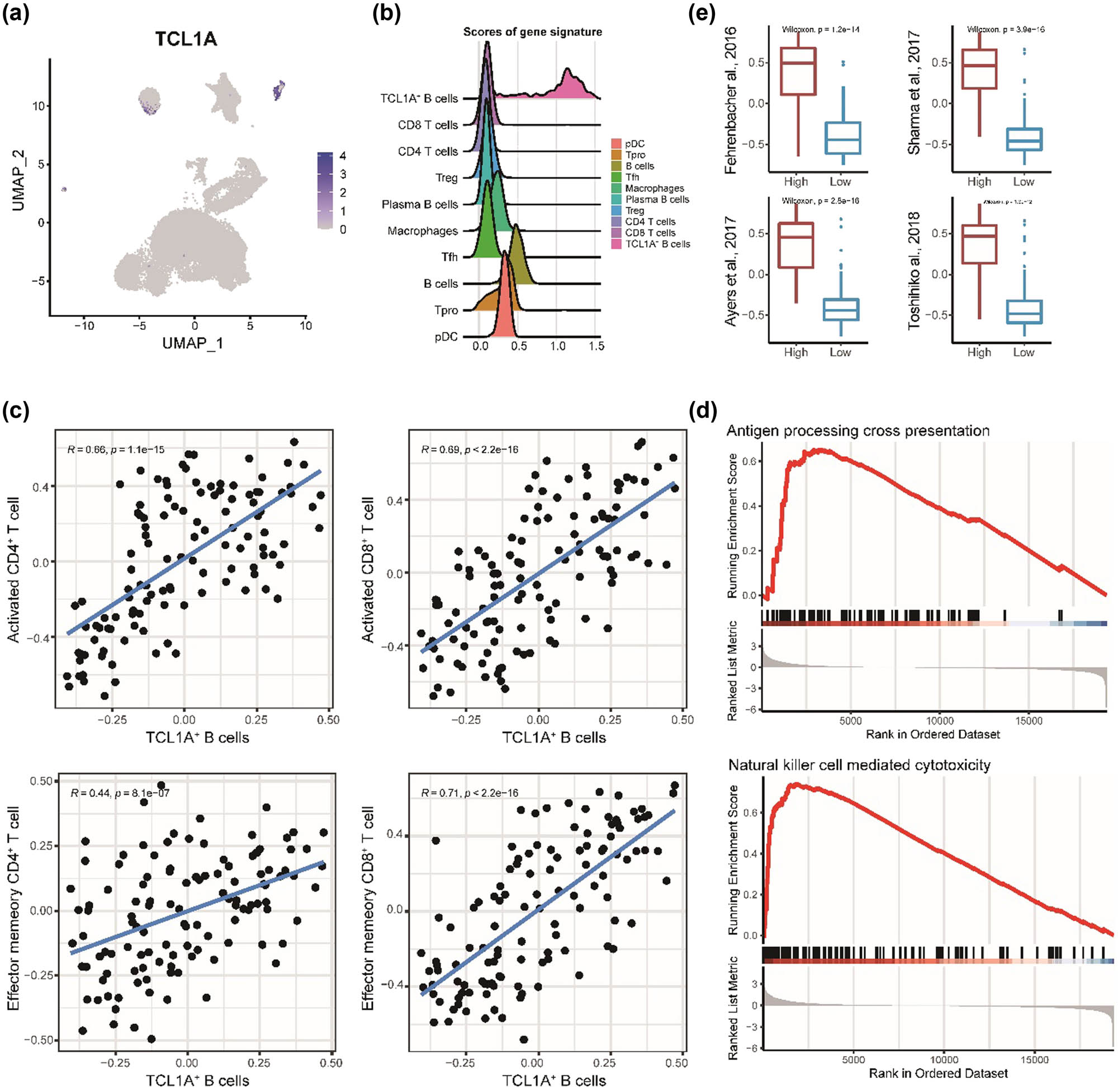
Association between TCL1A+ B cells and TME. (a) Scatter plot of TCL1A expression, color gradation represents the expression level. (b) Ridge plot of the density of the mean expression value of TCL1A+ B cell gene signature in significant cell populations. (c) Scatter plot of correlation between TCL1A+ B cells and activated CD4 + T cells, activated CD8 + T cells, effector memory CD4 + T cells, and effector memory CD8 + T cells. (d) GSEA results of two pathways between two groups. (e) Box plots of four hot-tumor scores in two groups. Two groups represent high abundance of TCL1A+ B cells and low abundance of TCL1A+ B cells with median abundance score as a cut-off.
To investigate the association between TCL1A+ B cells and tumor environment in the TCGA-TNBC cohort, which is a subset of the TCGA-BRCA cohort, we estimated the abundance of TCL1A+ B cells in each patient through the GSVA score of the gene signature. By correlation analysis, we found that TCL1A+ B cells are strongly positively correlated with activated CD4+ and CD8+ T cells and effector memory CD4+ and CD8+ T cells (Figure 3c), which have played crucial roles in the adaptive immune response. Interestingly, GSEA results showed that antigen processing cross-presentation and natural killer cell-mediated cytotoxicity were enriched in a high group of TCL1A+ B cells (Figure 3d), proposing that innate immune response was promoted, even though antigen processing and presentation was involved in adaptive immune as well. To characterize the TME, we collected four gene sets to represent hot-tumor. The hot-tumor scores were significantly increased in the TCL1A+ B cells high group with all gene sets (Figure 3e). Taken together, our results described that the TME of the high abundance TCL1A+ B cells’ patients is related to hot-tumor with an anti-tumor response.
3.4 Higher TCL1A+ B cells are correlated with a favorable prognosis
Next, we utilized the same analysis approach that classified TCGA-TNBC patients into two groups based on the TCL1A+ B cells to evaluate the correlation with OS. The survival analysis showed that OS time in the high abundance group is significantly longer than in the low group (HR: 0.365, 95% CI: 0.128–1.036, p-value: 0.048, Figure 4a). However, the number of TNBC patients in TCGA is small (n = 116); to further confirm the results, we collected another cohort with 319 TNBC patients. Concordant survival result of METABRIC cohort was observed in the high and low TCL1A+ B cells group (HR: 0.606, 95% CI: 0.444–0.827, p-value: 0.0014, Figure 4b). Notably, the median OS time is 2.43-fold longer in the high group (mOS: 20.44 years) than in the low group (mOS: 8.42 years), indicating that the patients who had a higher abundance of TCL1A+ B cells preferred to have a better clinical outcome and longer OS time.

Survival analysis of TCL1A+ B cells: (a) Kaplan–Meier curves of OS to the abundance of TCL1A+ B cells in TCGA-TNBC and (b) METABRIC-TNBC. High represents the high abundance of TCL1A+ B cells, and low means the low abundance of TCL1A+ B cells, with the median abundance score of each cohort as a cut-off.
4 Discussion
TNBC is an aggressive subtype of breast cancer with limited therapeutic options and high rates of mortality. Identifying patients more likely to obtain clinical benefit from therapy is essential. The improvement of sequencing technology allows for exploring the tumor environment in a single-cell solution. With the clinical information, we can construct an insight between immune cells and outcomes to detect which resistant cell population is related to a favorable clinical outcome. The innovation of this study is to integrate single-cell and global transcriptome data to discover new survival predictive markers, and to reveal the mechanism by which TCL1A+ B cells regulate the TME and affect the survival of TNBC. This provides new ideas for exploring individualized treatment strategies based on TCL1A-B cells in the future.
Our study collected scRNA-seq data from five TNBC patients to construct an immune cell atlas for primary tumors with eight significant resistant cell populations. We further identified a cluster of B cells, annotated based on the common DEG, TCL1A, between the group and other immune cells and the group and another B cell cluster. TCL1A, as a protein-coding gene, is also known as T cell leukemia/lymphoma protein 1A, which has been reported to enhance the phosphorylation and activation of AKT1, AKT2, and AKT3, improve cell proliferation, stabilize mitochondrial membrane potential, and promote cell survival [31–34]. Recently, integrated transcriptomic and epigenetic analysis suggests it also acts as a transcription factor and coregulator of NF-κB p65 in lymphoblastoid cell lines [35]. In Raji Burkitt lymphoma, TCL1A has been connected with TP63 [36]. These findings indicate that TCL1A is essential in multiple biological processes, even in the tumor. Moreover, TCL1A has been proposed as a potential therapeutic target in B cell lymphoma necessary for malignant B cell survival, indicating its association with B cells [37].
Studies have shown the association between B cells and clinical outcomes in solid tumors, such as head and neck squamous cell carcinoma and lung cancer [38,39]. A previous study of the total RNA-seq of flow-sorted samples reported that B cells with TCL1A expression correlated with improved survival time in cervical tumors, suggesting its prediction potential [40]. However, the study of B cells, particularly TCL1A+ B cells, and clinical benefit in TNBC is limited. Based on fold change analysis, we found that TCL1A+ B cells are more than two-fold in PR patients than in SD patients in TNBC. The sample-based analysis revealed that one PR patient had a higher fraction of TCL1A+ B cells than all other SD patients (Figure S1). The results propose that TCL1A+ B cells are associated with favorable clinical outcomes.
A recent review, which discusses various B cell signatures in normal and diseased tissues, has shown that TCL1A can be used as a marker for both naïve B cells and follicular B cells in different tissues [41]. However, our analysis proved that it is more similar to follicular B cells, which generally existed in the germinal center, rather than naïve B cells based on the gene marker of BCL6, NEIL1, and MEF2B [29,30]. The functional analysis of the TCL1A+ B cells showed that it was mainly involved in B cell activation, immune response, and regulation of T cell activation, partially differing from the function of naïve B cells. One major limitation of this study is that the cohort size of scRNA-seq data is limited due to our strict restriction of the sample site, so sample-based analysis does not have significant results. We are also collecting data and planning to reappear the findings with a large cohort in the future.
To further investigate TCL1A+ B cells in bulk RNA-seq, we constructed a gene signature to evaluate the abundance of such cells. With multiple analysis strategies, we found that TCL1A+ B cells are positively correlated with several T cells, for example, activated CD8 T cells, which have been proven to support anti-tumor immune response. The results proposed that innate immune response has been enhanced as well. Interestingly, examination by four hot-tumor gene signatures showed that tumors with a high abundance of TCL1A+ B cells showed hot-tumor characterization, indicating that an immune checkpoint inhibitors-based therapy strategy might be helpful in such patients [42]. Finally, survival analysis in two cohorts demonstrated longer survival time in higher TCL1A+ B cells patients, suggesting its potential to predict prognosis in TNBC.
5 Conclusion
According to our study, TCL1A+ B cells as a B cell subset are associated with better clinical outcomes in TNBC patients treated with chemotherapy. By integrating single-cell and global transcriptome data, we found that higher abundance of TCL1A+ B cells was associated with a stronger anti-midstream immune microenvironment and longer OS. These results suggest that TCLIA + B cells play an important role in the prediction of TNBC treatment and prognosis, but further studies are needed to verify our findings. By combining transcriptome data with different segmentation rates, our study reveals a feasible pathway to find novel biomarkers for predicting prognosis.
In our study, we identified an association between TCL1A+ B cells and the clinical outcome of TNEC. Functional analysis showed that TCL1A+ B cells were related to the activation of B cells and immune regulation [2]. In addition, we constructed a gene signature based on differentially expressed genes between TCL1A+ B cells and other immune cells to demonstrate the relationship with the TME, and constructed a prognostic signature using non-parametric and unsupervised methods.
We also found that the OS time of the TCL1A+ B cell high abundance group was significantly longer than that of the low abundance group in TCGA-TNBC and METABRIC-TNBC patients. These data suggest that patients with higher abundance of TCL1A+ B cells tend to achieve better clinical treatment outcomes and longer survival [4]. When studies considered drug efficacy, TCL1A+ B cells were found to also predict clinical response, but this needs further confirmation.
In conclusion, our study fully confirmed the potential therapeutic predictive and prognostic relevance of TCL1A+ B cells in TNBC patients. If validated, this would provide new targets for fine-tuning treatment strategies to improve treatment outcomes.
-
Funding information: The study was supported by Natural Science Foundation of Fujian Province (2017J01201); Medical Innovation Project of Fujian Provincial Health Commission (2015-CXB-15).
-
Author contributions: P.H. supervised the project, designed experiments, and drafted the manuscript. P.H. and Y.L. collected the data and performed bioinformatic analysis. P.H. and N.Z.W. revised the manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016 Nov;13(11):674–90.10.1038/nrclinonc.2016.66Search in Google Scholar PubMed PubMed Central
[2] Mo H, Xu B. Progress in systemic therapy for triple-negative breast cancer. Front Med. 2021 Feb;15(1):1–10.10.1007/s11684-020-0741-5Search in Google Scholar PubMed
[3] Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020 Jun;22(1):15–25.10.1186/s13058-020-01296-5Search in Google Scholar PubMed PubMed Central
[4] Sikov W, Berry D, Perou CM, Saharan B, Cirrincione CT, Tolaney S, et al. Abstract S2-05: Event-free and overall survival following neoadjuvant weekly paclitaxel and dose-dense AC+/− carboplatin and/or bevacizumab in triple-negative breast cancer: outcomes from CALGB 40603 (Alliance). Cancer Res. 2016;76:S2-05.10.1158/1538-7445.SABCS15-S2-05Search in Google Scholar
[5] Hu X-C, Zhang J, Xu B-H, Cai L, Ragaz J, Wang Z-H, et al. Cisplatin plus gemcitabine versus paclitaxel plus gemcitabine as first-line therapy for metastatic triple-negative breast cancer (CBCSG006): a randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2015;16(4):436–46.10.1016/S1470-2045(15)70064-1Search in Google Scholar PubMed
[6] von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol. 2014 Jun;15(7):747–56.10.1016/S1470-2045(14)70160-3Search in Google Scholar PubMed
[7] Bardia A, Hurvitz SA, Tolaney SM, Loirat D, Punie K, Oliveira M, et al. Sacituzumab govitecan in metastatic triple-negative breast cancer. N Engl J Med. 2021 Apr;384(16):1529–41.10.1056/NEJMoa2028485Search in Google Scholar PubMed
[8] Martín M, Chan A, Dirix L, O’Shaughnessy J, Hegg R, Manikhas A, et al. A randomized adaptive phase II/III study of buparlisib, a pan-class I PI3K inhibitor, combined with paclitaxel for the treatment of HER2- advanced breast cancer (BELLE-4). Ann Oncol. 2017 Feb;28(2):313–20.10.1093/annonc/mdw562Search in Google Scholar PubMed
[9] Zhang YY, Chen HY, Mo HN, Hu XD, Gao RR, Zhao YH, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell. 2021 Dec;39(12):1578.10.1016/j.ccell.2021.09.010Search in Google Scholar PubMed
[10] Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021 Sep;53(9):1334.10.1038/s41588-021-00911-1Search in Google Scholar PubMed PubMed Central
[11] Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021 May;27(5):820.10.1038/s41591-021-01323-8Search in Google Scholar PubMed
[12] Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, et al. The single-cell pathology landscape of breast cancer. Nature. 2020 Feb;578(7796):615.10.1038/s41586-019-1876-xSearch in Google Scholar PubMed
[13] Liu BL, Zhang YY, Wang DF, Hu XD, Zhang ZM. Single-cell meta-analyses reveal responses of tumor-reactive CXCL13(+) T cells to immune-checkpoint blockade. Nat Cancer. 2022 Sep;3(9):1123.10.1038/s43018-022-00433-7Search in Google Scholar PubMed
[14] Yost KE, Satpathy AT, Wells DK, Qi YY, Wang CL, Kageyama R, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019 Aug;25(8):1251.10.1038/s41591-019-0522-3Search in Google Scholar PubMed PubMed Central
[15] Liu BL, Hu XD, Feng KC, Gao RR, Xue ZQ, Zhang SJ, et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat Cancer. 2022 Jan;3(1):108.10.1038/s43018-021-00292-8Search in Google Scholar PubMed
[16] Cabrita R, Lauss M, Sanna A, Donia M, Larsen MS, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma (vol 577, pg 561, 2020). Nature. 2020 Apr;580(7801):E1–E.10.1038/s41586-020-2155-6Search in Google Scholar PubMed
[17] Helmink BA, Reddy SM, Gao JJ, Zhang SJ, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020 Jan;577(7791):549.10.1038/s41586-019-1922-8Search in Google Scholar PubMed PubMed Central
[18] Petitprez F, de Reynies A, Keung EZ, Chen TWW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020 Jan;577(7791):556.10.1038/s41586-019-1906-8Search in Google Scholar PubMed
[19] Xia J, Xie ZJ, Niu GM, Lu Z, Wang ZQ, Xing Y, et al. Single-cell landscape and clinical outcomes of infiltrating B cells in colorectal cancer. Immunology. 2023 Jan;168(1):135–51.10.1111/imm.13568Search in Google Scholar PubMed
[20] Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009 Jan;45(2):228–47.10.1016/j.ejca.2008.10.026Search in Google Scholar PubMed
[21] Pereira B, Chin SF, Rueda OM, Vollan HKM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.10.1038/ncomms11479Search in Google Scholar PubMed PubMed Central
[22] Hao YH, Hao S, Andersen-Nissen E, Mauck WM, Zheng SW, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021 Jun;184(13):3573.10.1016/j.cell.2021.04.048Search in Google Scholar PubMed PubMed Central
[23] Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019 Dec;16(12):1289.10.1038/s41592-019-0619-0Search in Google Scholar PubMed PubMed Central
[24] Wu TZ, Hu EQ, Xu SB, Chen MJ, Guo PF, Dai ZH, et al. ClusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation-Amsterdam. 2021 Aug;2(3):100141.10.1016/j.xinn.2021.100141Search in Google Scholar PubMed PubMed Central
[25] Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013 Jan 16;14:7.10.1186/1471-2105-14-7Search in Google Scholar PubMed PubMed Central
[26] Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003 Jul;34(3):267–73.10.1038/ng1180Search in Google Scholar PubMed
[27] Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005 Oct;102(43):15545–50.10.1073/pnas.0506580102Search in Google Scholar PubMed PubMed Central
[28] Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022 Jan;50(D1):D687–92.10.1093/nar/gkab1028Search in Google Scholar PubMed PubMed Central
[29] Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012 May;247:172–83.10.1111/j.1600-065X.2012.01112.xSearch in Google Scholar PubMed
[30] Brescia P, Schneider C, Holmes AB, Shen Q, Hussein S, Pasqualucci L, et al. MEF2B instructs germinal center development and acts as an oncogene in B cell lymphomagenesis. Cancer Cell. 2018 Sep;34(3):453.10.1016/j.ccell.2018.08.006Search in Google Scholar PubMed PubMed Central
[31] Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S, et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci USA. 2000 Mar 28;97(7):3028–33.10.1073/pnas.97.7.3028Search in Google Scholar PubMed PubMed Central
[32] Laine J, Kunstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000 Aug;6(2):395–407.10.1016/S1097-2765(00)00039-3Search in Google Scholar PubMed
[33] Laine J, Kunstle G, Obata T, Noguchi M. Differential regulation of Akt kinase isoforms by the members of TCL1 oncogene family. J Biol Chem. 2002 Feb;277(5):3743–51.10.1074/jbc.M107069200Search in Google Scholar PubMed
[34] Kunstle G, Laine J, Pierron G, Kagami S, Nakajima H, Hoh F, et al. Identification of Akt association and oligomerization domains of the Akt kinase coactivator TCL1. Mol Cell Biol. 2002 Mar;22(5):1513–25.10.1128/MCB.22.5.1513-1525.2002Search in Google Scholar PubMed PubMed Central
[35] Ho MF, da Rocha EL, Zhang C, Ingle JN, Goss PE, Shepherd LE, et al. TCL1A, a novel transcription factor and a coregulator of nuclear factor kB p65: single nucleotide polymorphism and estrogen dependence. J Pharmacol Exp Ther. 2018 Jun;365(3):700–10.10.1124/jpet.118.247718Search in Google Scholar PubMed PubMed Central
[36] Gaudio E, Paduano F, Pinton S, D’Agostino S, Rocca R, Costa G, et al. TCL1A interacts with TP63 and enhances the survival of Raji Burkitt lymphoma cell line. Brit J Haematol. 2018 Nov;183(3):509–12.10.1111/bjh.14989Search in Google Scholar PubMed
[37] Aggarwal M, Villuendas R, Gomez G, Rodriguez-Pinilla SM, Sanchez-Beato M, Alvarez D, et al. TCL1A expression delineates biological and clinical variability in B-cell lymphoma. Modern Pathol. 2009 Feb;22(2):206–15.10.1038/modpathol.2008.148Search in Google Scholar PubMed
[38] Kim SS, Shen S, Miyauchi S, Sanders PD, Franiak-Pietryga I, Mell L, et al. B cells improve overall survival in HPV-associated squamous cell carcinomas and are activated by radiation and PD-1 blockade. Clin Cancer Res. 2020 Jul;26(13):3345–59.10.1158/1078-0432.CCR-19-3211Search in Google Scholar PubMed PubMed Central
[39] Wang SS, Liu W, Ly D, Xu H, Qu LM, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol. 2019 Jan;16(1):6–18.10.1038/s41423-018-0027-xSearch in Google Scholar PubMed PubMed Central
[40] Punt S, Corver WE, van der Zeeuw SAJ, Kielbasa SM, Osse EM, Buermans HPJ, et al. Whole-transcriptome analysis of flow-sorted cervical cancer samples reveals that B cell expressed TCL1A is correlated with improved survival. Oncotarget. 2015 Nov;6(36):38681–94.10.18632/oncotarget.4526Search in Google Scholar PubMed PubMed Central
[41] Morgan D, Tergaonkar V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 2022 Mar;43(3):210–29.10.1016/j.it.2022.01.003Search in Google Scholar PubMed
[42] Gerard CL, Delyon J, Wicky A, Homicsko K, Cuendet MA, Michielin O. Turning tumors from cold to inflamed to improve immunotherapy response. Cancer Treat Rev. 2021 Dec;101:102227.10.1016/j.ctrv.2021.102227Search in Google Scholar PubMed
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”

