Abstract
Testis size is important for identifying breeding animals with adequate sperm production. The aim of this study was to survey the expression profile of mRNA and miRNA in testis tissue from rams carrying different FecB genotypes, including the wild-type and heterozygous genotypes in Tibetan sheep. Comparative transcriptome profiles for ovine testes were established for wild-type and heterozygote Tibetan sheep by next-generation sequencing. RNA-seq results identified 3,910 (2,034 up- and 1,876 downregulated) differentially expressed (DE) genes and 243 (158 up- and 85 downregulated) DE microRNAs (miRNAs) in wild-type vs heterozygote sheep, respectively. Combined analysis of mRNA-seq and miRNA-seq revealed that 20 miRNAs interacted with 48 true DE target genes in wild-type testes compared to heterozygous genotype testes. These results provide evidence for a functional series of genes operating in Tibetan sheep testis. In addition, quantitative real-time PCR analysis showed that the expression trends of randomly selected DE genes in testis tissues from different genotypes were consistent with high-throughput sequencing results.
1 Introduction
In mammals, testes are male reproductive organs that produce sperm, secrete androgenic hormones, and maintain male characteristics [1]. In production, we usually use a scrotal circumference as a proxy for testicular size, which is significantly and positively correlated with testicular size, thus indirectly indicating that the trait is easy to measure and has high heritability [2]. Studies have found positive correlations between testicular size and sperm motility, production, concentration, and ejaculate volume and a negative association with abnormal sperm percentage in livestock animals, including cattle, sheep, goat, and pig [3,4,5,6]. Testis size affects the age at puberty and the postpartum interval of offspring in females [7,8]. Therefore, the selection of elite sires based on testicular size is an effective method to improve reproductive performances. Thus, the testes are one of the important indicators for predicting male reproductive ability.
Testicles descend before puberty is reached and grow in size after puberty, increasing in volume by more than 500% [9]. This growth is due to lytic function, sperm production, and interstitial fluid and Sertoli cell fluid production. Sertoli cells and germ cells rapidly proliferate at birth and undergo a radical change in morphology and function at the onset of puberty [9]. This is due to a complex biological process, involving genetic and microRNA modulation, which is essential for testis development and sperm function in adult males. A gene called INHBA has been shown to affect testis morphogenesis, testicular cell proliferation, and testis weight in mice, and has a potential role in regulating testis size [10,11,12]. Among the regulators of spermatogenesis, small RNAs, including microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs), have recently attracted great attention [13]. miRNAs are small (∼22 nucleotides) endogenous RNAs that negatively regulate gene expression by targeting the 3′-untranslated region (3′-UTR) [14] or coding region [15] of mRNAs. Studies have shown that miRNAs play an important role in testicular development and spermatogenesis in mammals. New generation sequencing tools on mouse testis tissues at 8, 16, and 24 d found key miRNAs such as miR-34c, miR-221, miR-222, miR-20, and miR-106a [16]. Research has shown that miR-34c may inhibit the proliferation of male reproductive stem cells in dairy goats, and differentially expressed (DE) mRNAs and miRNAs have been identified in pig testis tissues at different developmental stages [17]. Expression profile and biological function studies on testicular tissues of yak, cattle, and cattle-yak have found that miRNAs are involved in apoptosis and are associated with the blockade of cattle-yak spermatogenesis [18]. Another well-known gene that controls reproductive traits in sheep, BMPR-IB (FecB), has been studied by many researchers, focusing on ewes [19,20,21]. Ewes carrying the heterozygous genotype tend to produce more lambs, and, conversely, individuals carrying the wild type produce fewer lambs [22]. The relationship between ram fertility and FecB has not been elucidated, and we propose that a molecular mechanism exists for the higher fertility of rams carrying the heterozygotes compared to wild-type individuals. Although these studies describe mRNA and miRNA expression profiles of testicular tissues from different livestock animals, the genetic mechanisms of the testis tissue from different genotypes on FecB locus have not been investigated. Studies on the expression profiles of mRNA and miRNA in testis tissue of rams carrying wild type and heterozygotes in Tibetan sheep are lacking.
An indigenous sheep breed (Tibetan sheep) was selected for this study. This sheep is widely distributed as livestock in the Qinghai-Tibet Plateau (QTP) and adjacent areas in China. Tibetan sheep are well adapted to the harsh natural environment and are irreplaceable livestock in the high-altitude areas of QTP. They are the main source of meat in Tibet and have been found to have a low rate of double birth. Qiao et al. found FecB mutations in the ewes of Tibetan sheep [23,24]. Using a molecular marker of the FecB gene, the core population of multiparous Tibetan sheep was established, and the technical problems of low fecundity in ewe were solved. However, ram fertility is poorly researched. Mounting evidence indicates a series of genes that are critical for testis development, especially testis size and volume. AKT was shown to act as an anti-apoptosis gene in animal growth development and plays an important role in mouse testicular spermatogenesis [25]. Moreover, PLK1 was shown to be critical for testis development, and EHD4 is required for the survival of germ cells and attainment of normal prepubertal testicular size in mice [26]. EPAS1 is required for spermatogenesis in postnatal mouse testes. Postnatal EPAS1 ablation leads to male infertility, with reduced testis size and weight [27]. KITl is crucial for the testicular environment postnatally, and KIT is important for the proliferation and survival of spermatogonia [28].
Tibetan sheep reach sexual maturity at 8 months. In our preliminary research, we made a breakthrough in identifying the presence of FecB in Tibetan sheep and using this molecular marker for selection to expand the core population of Tibetan sheep. Because the relationship between ram fertility and FecB has not been elucidated in Tibetan sheep, we propose a potential molecular mechanism for the higher fertility of rams carrying the heterozygotes compared to the wild type. FecB has been reported more frequently in ewes and rarely in rams. This study is the first to study the relationship between FecB and testicular transcriptome in rams. In this study, we used Solexa deep sequencing to investigate mRNA, miRNA profiles, and construct mRNA–miRNA networks for testes from Tibetan sheep with different genotypes. This work provides a theoretical basis for the study of ram fertility and provides insights into testicular development in ruminants.
2 Materials and methods
2.1 Animals and sample collection
Blood from 40 individual rams was randomly tested from the Tibetan sheep population for genotyping at the FecB gene locus (A746G) according to molecular detection methods for the major gene FecB in sheep [29]. In general, there are three genotypes at the FecB mutation locus: AA (also known as ++ or wild type), AG (heterozygous), and GG (homozygous). Based on the genotyping results of the FecB locus and the preciousness of heterozygote rams, which can be irreversibly damaged by slaughter, we selected six rams, including three wild types and three heterozygotes, for high-throughput sequencing. To ensure that the environmental conditions were similar throughout the experiment, all the lambs were housed in a well-ventilated room with controlled temperature and humidity. Testicular tissues were collected as described by Xu et al. [30]. All the animals and samples used in this study were collected according to the protocol approved by the institutional animal care and use committee of Qinghai University.
-
Ethical approval: The research related to animal use has been complied with all the relevant national regulations and institutional policies for the care and use of animals, and has been approved by the institutional animal care and use committee of Qinghai University.
2.2 RNA isolation and quantification
A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. The total RNA and small RNA were isolated using TRIzol® reagent (TransGen Biotech, Beijing, China). RNA purity was checked using a NanoPhotometer spectrophotometer (IMPLEN, CA, USA). RNA degradation and contamination were monitored on 1% agarose gel. RNA concentration and integrity were measured using Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA) and RNA Nano 6000Assy Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA), respectively. The experimental protocols were performed according to the manufacturer’s technical instructions.
2.3 mRNA sequencing and analysis
cDNA libraries were prepared by first isolating total RNA from testis tissue. Total RNA was then reverse transcribed into cDNA, which was then amplified to create cDNA molecules. cDNA libraries were used for transcriptome sequencing. In this study, six RNA libraries were constructed and named TWT1, TWT2, TWT3, THT1, THT2, and THT3. Sequencing libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations. After cluster generation, the libraries were sequenced on an Illumina HiSeq Xten platform and 150 bp-paired end reads were generated. Clean reads were obtained by filtering out adapter sequences and removing low-quality reads from raw data. Simultaneously, Q30 and GC content of clean data were calculated. RNA-seq reads were aligned to the sheep reference genome (Oar_v3.1) using Tophat 2.0.10 [31] with default parameters. The gene expression level was calculated by reads per kilo-base per million reads after the read numbers mapped to each gene were counted by HTSeq v0.6.1 [32]. DE genes (DEGs) were examined using the DEGseq R package (1.12.0) [33]. A corrected P-value of 0.05 and an absolute value of log2 (fold change) of 1.5 were set as the thresholds for significant differential expression. Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the GOseq R packages based on Wallenius non-central hyper-geometric distribution [34].
2.4 miRNA sequencing and analysis
miRNA libraries were prepared by first isolating total RNA from testis tissue. Total RNA was processed with a miRNA-specific reverse transcription step to create a miRNA library. miRNA libraries were used for miRNA expression profiling. Six small RNA libraries were constructed: SRWT1, SRWT2, SRWT3, SRHT1, SRHT2, and SRHT3. Sequencing libraries were generated using NEBNext® Multiplex small RNA Library Prep Set for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Small RNAs ligated with 50 and 30 adapters were reverse transcribed and amplified. Library preparations were sequenced on the Illumina HiSeq Xten platform according to the manufacturer’s instructions. After removing adapter sequences, reads containing poly-N, and low-quality reads, the clean reads were mapped to the Repeatmasker and Rfam database to remove tags originating from repeated sequences, rRNA, tRNA, snRNA, and snoRNA. The miRNA expression levels were estimated by transcript per million values. Differential expression analysis of miRNA was performed using the DEGseq R package [33]. P-value was adjusted using q-value, and q-value < 0.01 and |log2 (fold change)| > 1 were set as the threshold for significantly differential expression by default.
2.5 Construction of an mRNA–miRNA regulatory network in testicular tissue of different genotypes
miRNAs can be incompletely complementary base-paired to the target mRNA to inhibit its expression. Spermatogenesis is subject to various factors: several miRNAs and their target genes are involved in the regulation of this complex process. True DE target genes (TDETGs) came as a result of the overlap of the target genes of DE miRNAs with the DEGs. The methods to construct mRNA–miRNA networks consist of three key points. First, data collection was performed by collecting DE miRNA and mRNA data from both the wild-type and heterozygote groups. Second, data analysis was performed using RNAhybrid tools [35] to compare the data and identify miRNA and mRNA sequences that are DE in the same tissues. Third, network construction was performed using Cytoscape software [36] to create a network diagram with DE miRNA and mRNA nodes.
2.6 Validation of mRNA by qRT-PCR
Total RNA was extracted using TRIzol® reagent (TransGen Biotech, Beijing, China) as described by the manufacturer. After acquiring high-quality total RNA, mRNAs were reverse transcribed using the RevertAid™ First Strand cDNA Synthesis Kit (TransGen Biotech, Beijing, China). Quantitative real-time PCR (qRT-PCR) analyses on the mRNAs were performed using SYBR Green PCR Master Mix (ABI, USA, 4304437) in a Bio-rad CFX96 real-time system. Beta actin was used as an internal control for each mRNA. Primer sequences of all mRNAs were designed using primer 5 (Table 1). For mRNA quantification, the reaction conditions were: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 59°C for 50 s. Relative mRNA expression was evaluated using the 2−△△CT method [37]. Differences were considered significant when P < 0.05 using one-way analysis of variance. All statistical analyses were performed using general linear models in R. Visualization of graphs was done with the ggplot package [38] in R.
mRNA primers for reverse-transcriptase quantitative PCR for different genotype testis tissue
| Gene name | Accession numbers | Primer sequence (5″ → 3″) | Product size (bp) | Annealing temperature (°C) | |
|---|---|---|---|---|---|
| AKT | NM_001161857 | Forward | CGCCGATGATCGATAACCCT | 156 | 60 |
| Reverse | CAGCGGTCTGCTACTTCCTT | ||||
| EHD4 | XM_004010473 | Forward | TGGCAGTTTTGGTGGTATGC | 219 | 60 |
| Reverse | CCTTTATTCCTCCTGGAGCCC | ||||
| TGFBR3 | XM_042250942 | Forward | TGTGGTTGGTTCGATGCAGA | 224 | 60 |
| Reverse | CTCCTCCCCGTGTTTAGCAG | ||||
| ODF3 | XM_004019747 | Forward | TTTCTGCCCGAACCAAGACC | 176 | 60 |
| Reverse | CACGTCAGTCTGGTGGTAGG | ||||
| Reverse | CCATGTCTCGGTAGCACTGG | ||||
| INHA | NM_001308579 | Forward | AGAGCCGCCCTCAATATCTC | 146 | 60 |
| Reverse | GGTTGGGCACCATCTCATACT | ||||
| VIM | XM_004014247 | Forward | GGGACAAGATAGCTACCCGC | 260 | 60 |
| Reverse | GTCGAAGGCCCGTGTAAAGA | ||||
| COL1A1 | XM_027974705 | Forward | GGTGTCGGCGGGTTGTC | 289 | 60 |
| Reverse | TTGTACGCCCCCAGAATTG | ||||
| COL1A2 | XM_004007726 | Forward | TGGACCAATGGGGTTGATGG | 272 | 60 |
| Reverse | CCCTAATGCCCTTGAAGCCA | ||||
| ACTB | NM_001009784 | Forward | CCTGCGGCATTCACGAA | 134 | 60 |
| Reverse | GCGGATGTCGACGTCACA | ||||
3 Results
3.1 mRNA expression analysis from testes in different genotypes
To obtain differential comparative transcriptome profiles for testes of different genotypes, six libraries, namely TWT1, TWT2, TWT3, THT1, THT2, and THT3, were constructed and sequenced by Illumina HiSeq Xten platform. The major characteristics of the six libraries are summarized in Table 2. The Q30 values of the six libraries were >88.47%, and GC content was ∼52.49%. Thus, sequencing quality was feasible. Approximately 65.68–74.72% of clean reads were mapped to the sheep reference genome (Oar_v3.1), of which 63.27–73.03% were uniquely mapped, whereas 1.70–2.62% showed multiple matches. Finally, 2,034 up- and 1,876 downregulated genes were identified in the wild-type group compared to the heterozygote group. These genes may be regulators of testicular growth and spermatogenesis.
Basic characteristics of mRNA-seq
| Sample | TWT1 | TWT2 | TWT3 | THT 1 | THT 2 | THT 3 |
|---|---|---|---|---|---|---|
| Group | Wide type | Wide type | Wide type | Heterozygote | Heterozygote | Heterozygote |
| Raw reads | 20,588,268 | 22,046,944 | 22,051,621 | 21,648,924 | 21,719,049 | 28,164,621 |
| Clean reads | 20,577,992 | 22,035,926 | 22,040,601 | 21,638,105 | 21,708,195 | 28,024,500 |
| Q30 | 88.47 | 89.50 | 88.66 | 89.24 | 89.36 | 88.49 |
| GC content (%) | 54.51 | 55.03 | 55.81 | 52.15 | 53.00 | 52.75 |
| Total reads | 41,155,984 | 44,071,852 | 44,081,202 | 43,276,210 | 43,416,390 | 56,049,000 |
| Mapped reads | 27,032,277 (65.68%) | 29,254,170 (66.38%) | 28,955,373 (65.69%) | 32,513,302 (75.13%) | 32,261,454 (74.31%) | 41,110,731 (73.35%) |
| Unique mapped reads | 26,040,780 (63.27%) | 28,099,343 (63.76%) | 27,866,465 (63.22%) | 31,764,931 (73.40%) | 31,549,418 (72.67%) | 40,123,163 (71.59%) |
| Multiple map reads | 991,497 (2.41%) | 1,154,827 (2.62%) | 1,088,908 (2.47%) | 748,371 (1.73%) | 712,036 (1.64%) | 987,568 (1.76%) |
| Reads map to ‘+’ | 13,233,281 (32.15%) | 14,285,546 (32.41%) | 14,166,568 (32.14%) | 16,123,649 (37.26%) | 16,011,892 (36.88%) | 20,356,053 (36.32%) |
| Reads map to ‘−’ | 13,260,067 (32.22%) | 14,297,503 (32.44%) | 14,166,223 (32.14%) | 16,093,216 (37.19%) | 15,979,783 (36.81%) | 20,336,161 (36.28%) |
Clean reads = read numbers of single-ended sequencing (150 bp). Total reads = read numbers of double-ended sequencing. Total mapped% = total mapped/clean reads; total mapped: clean reads mapped on the sheep genome. Unique mapped% = unique mapped/total mapped; unique mapped: reads mapped only one region to the reference genome. Multiple mapped% = multiple mapped/total mapped; multiple mapped: reads mapped more than one region to the reference genome.
Sequencing analysis revealed that AKT, EHD4, TGFBR3, ODF3, INHA, VIM, COLIA1, and COLIA2 were DE in the testes of different genotypes. An extensive literature research revealed that these genes are closely related to testicular development, sperm morphology, and spermatogenesis. Therefore, we selected these genes for qPCR validation. To confirm the gene expression pattern identified by RNA sequencing, eight genes including four up- (AKT, EHD4, TGFBR3, ODF3) and four downregulated (INHA, VIM, COLIA1, COLIA2) genes were selected for real-time RT-PCR using testis samples from different genotypes. The results showed that the expression patterns of eight genes were consistent with those of the RNA sequencing results (Figure 1), implying that our RNA-seq results and analysis methods were highly reliable. GO enrichment analyses showed that the DEGs were significantly enriched for GO terms involving cell morphogenesis, tissue development, and spermatogenesis (Figure 2).
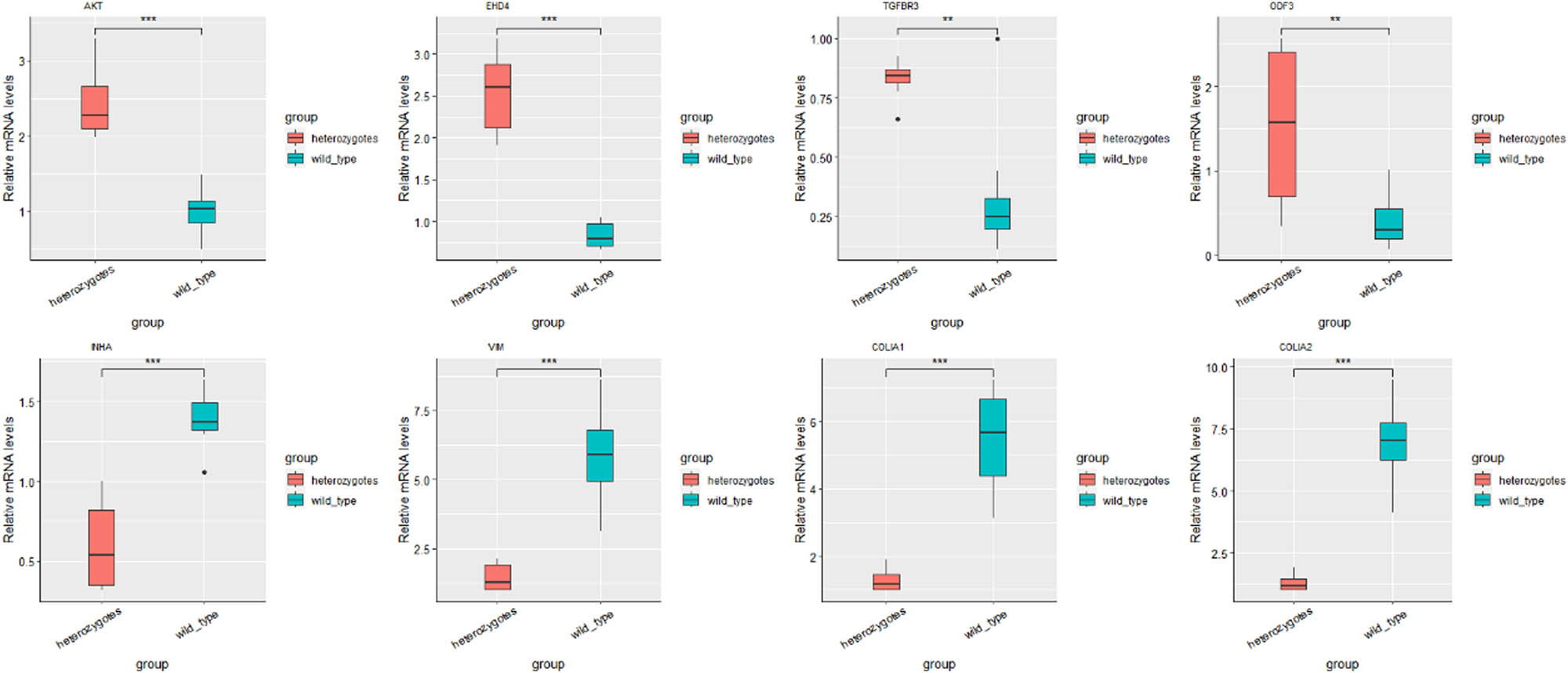
Differentially expressed mRNAs were examined in different genotype testis tissue. One asterisk (P value <0.05) indicates a significant difference between two groups, two asterisks (P value <0.01) indicate a possible highly significant difference between two groups, and three asterisks (P value <0.001) indicate a highly significant difference between two groups.

GO enrichment analysis of DEGs in wild type vs heterozygous group. BP: biological process, MF: molecular function, and CC: cellular component.
3.2 miRNA expression analysis from testes of different genotypes
The Q30 values of the wild type and heterozygotes were more than 97.17%. The read length demonstrated a bimodal with 21–23 and 26–32 nt in six libraries (Figure 3), which was consistent with the classical fragments of miRNAs. These results showed that the sequencing quality was feasible. After obtaining clean reads, the clean reads were categorized using some database. Subsequently, unannotated clean reads of sRNA were mapped to the sheep reference genome (Ovis_aries3.1). Approximately 58.74–65.55% of clean reads were mapped to the sheep reference genome (Ovis_aries3.1) (Table 3). The results indicated that the diversity and distribution of miRNAs in testes samples from different genotypes, including wild type and heterozygous, are relatively abundant. These results suggest that miRNAs are involved in testicular development. Finally, 243 DE miRNAs, including 158 up- and 85 downregulated miRNAs, were found in the wild type compared to the heterozygote group.

Length distribution and abundance of sequences in each library in miRNA analysis.
Proportion of sRNA unannotated clean reads mapped to reference genome
| Sample | Group | Total_reads | Mapped_reads | Percentage | Mapped_reads (+) | Percentage | Mapped_reads (-) | Percentage |
|---|---|---|---|---|---|---|---|---|
| SRWT1 | Wide type | 22,349,308 | 13,127,773 | 58.74 | 9,489,962 | 42.46 | 3,637,811 | 16.28 |
| SRWT2 | Wide type | 38,961,364 | 25,407,601 | 65.21 | 19,440,026 | 49.90 | 5,967,575 | 15.32 |
| SRWT3 | Wide type | 9,818,354 | 6,435,474 | 65.55 | 4,801,936 | 48.91 | 1,633,538 | 16.64 |
| SRHT1 | Heterozygote | 26,304,263 | 15,124,368 | 57.50 | 7,559,945 | 28.74 | 7,564,423 | 28.76 |
| SRHT2 | Heterozygote | 22,876,684 | 14,065,416 | 61.48 | 7,931,266 | 34.67 | 6,134,150 | 26.81 |
| SRHT3 | Heterozygote | 21,153,623 | 12,620,989 | 59.66 | 6,866,318 | 32.46 | 5,754,671 | 27.20 |
3.3 Integrated function analysis of genes and miRNA expression
Compared to the heterozygote group, 48 TDETGs were observed in the wild type, including 16 upregulated TDETGs corresponding to 8 downregulated miRNAs and 32 downregulated TDETGs corresponding to 12 upregulated miRNAs (Table 4). miRNAs and protein-coding genes are involved in regulatory networks to regulate spermatogenesis and testes development. In terms of miRNAs, which can positively and negatively regulate target genes, both types of networks were constructed. Type I belongs to the regulatory network of upregulated mRNAs interacting with downregulated miRNAs in wild type compared to heterozygote group. Type II belongs to the regulatory network of downregulated mRNAs interacting with upregulated miRNAs in the wild type compared to the heterozygote group (Figure 4). A total of three miRNAs and ten genes were present in the type I network, whereas two miRNAs and ten genes were present in the type II network. We found that oar-miR-27a, miR-125b, miR-127, EHD4, TGFBR3, and PDGF were all active in the regulatory network.
Typical differential expression miRNAs and target genes involving testicular development, including positive and negative regulation in wild type compared to heterozygote group
| High expression of miRNA | Low expression of target genes | Lower expression of miRNA | Higher expression of target genes |
|---|---|---|---|
| oar-miR-27a | EHD4;PDGFRB; AKT;GSG2;NOS3;PLD4;TGFBR3 | oar-miR-99a | ODF3;CFL1;CLGN;INHA |
| oar-miR-127 | PKD2;DDX3X;CCDC87;TGFBR3;AKT | novel-64 | NSUN7;ANKRD31;MARCH8;GSG2 |
| novel-55 | SPN;MAP3K8 | oar-let-7b | MARCH8;EPAS1 |
| novel-28 | SH3PXD2B;HR;RBCK1;STARD13;BCL9L;IFI35;PDGFRB | oar-miR-125a | VIM;COLAAI1;COLAI2;GNAS;INHA |
| novel-18 | NBL1;LOC101109421 | novel-40 | ENSOARG00000001264 |
| novel-30 | IGFN1;NFKB;IL4R;VWF;DOCK1 | novel-62 | ESPL1;CCDC87 |
| novel-45 | KDELC2;PTPN21 | novel-45 | PPP2R4;EHD4 |
| novel-89 | SARDH | novel-68 | KIAA1328;TGFBR3 |
| novel-56 | WDR45 | ||
| novel-28 | CD248;FBLN2 | ||
| novel-19 | NBL1; PDGFRB | ||
| novel-59 | STARD13 |

Regulatory network mRNA interaction miRNA; (a) represents Type I, where up-regulated mRNA interacts with down-regulated miRNA in wild type compared to the heterozygous group, (b) represents Type II, where down-regulated mRNA interacts with up-regulated miRNA in wild type compared to the heterozygous group.Blue labels represent miRNA and green labels represent mRNA.
4 Discussion
Xizang sheep, also known as Tibetan sheep, are widespread in QTP and adjacent areas. Using molecular markers of the FecB gene, the core population of multiparous Tibetan sheep was established and the technical problems of low fecundity in ewe were solved. However, ram fertility is poorly researched. Therefore, we investigated testicular genetics in wild-type rams and heterozygote rams to find effective molecular markers for measuring ram fertility.
Sequencing results revealed a big difference in the wild type compared to the heterozygote group. In general, clean reads were assigned to three major regions, including exonic, intergenic, and intronic. Approximately 65.68–74.72% of clean reads were mapped to the sheep reference genome, of which 63.27–73.03% were uniquely mapped. The efficiency of mapping to the reference genome was low and we give several possible reasons for this. First, the Tibetan sheep selected are breed specific and may differ from the reference genome. Second, the sheep reference genome does not contain the Y chromosome, and we sequenced testicular tissue. A total of 2,034 up- and 1,876 downregulated genes were identified in the wild type compared to the heterozygote group. These genes may be important for testicular growth and spermatogenesis.
The length distribution of small RNAs was concentrated in the two peaks of 19–24 and 26–32 nt. Notably, the clean reads of the wild-type group were mainly miRNA based. Interestingly, the miRNA level gradually decreased, and the level of piRNA gradually increased in the heterozygote testes; these data were coherent with previous descriptions. These results indicate that the regulation of spermatogenesis is complex. Consistently, Hirano et al. [39] performed small RNA-seq analysis in the marmoset and found that piRNA is dominant in adult testes. Li et al. used high-throughput sequencing to compare miRNA expression in immature and mature pigs and reported that the proportion of miRNAs was higher before sexual maturity in large white pigs, but not in Luchuan pigs [40]. Lian et al. found a significant difference in the proportion of small RNAs before and after sexual maturation [41]. Similar to the results of this experiment, the proportion of miRNAs in the wild-type group was significantly higher than that in the heterozygote group. These results suggest that both miRNA and piRNA may be involved in testicular development during sexual maturity.
TDETGs were used in this sequencing study to identify potential regulatory factors that influence testicular development. A total of 20 potential miRNAs were identified, including 8 down- and 12 upregulated miRNAs. These miRNAs include five known miRNAs (oar-miR-27a, oar-let-7b, oar-miR-125b, oar-miR-99a, and oar-miR-127), and newly discovered miRNAs. The functions of some known miRNAs have been reported. miR-27a is a differentiation marker for spermatogonial stem cells and is highly expressed in dairy goat testis cells [42]. In the mRNA–miRNA regulatory network constructed in this study, we screened miR-27a, suggesting that miR-27a may have potential regulatory effects on testicular development and differentiation of spermatogonial stem cells in sheep. Thus, these studies show that miRNA function is conserved in the vast majority of animals. In addition, studies in pigs have shown that miR-27a can be used as an internal reference gene for assessing the quality of cryopreserved sperm [43]. Studies have shown that miR-125b plays a crucial role in regulating zygote genome activation in oocytes and embryos [44]. miR-125b can target the degradation of the PAP gene, leading to an increase in testosterone concentration, a decrease in sperm count, and an increase in abnormal sperm ratio in male mice [45]. Related studies have shown that miR-125b is highly expressed in germ cells and directly regulates the secretion of testosterone by targeting PAP, increasing the mitochondrial DNA copy number in sperm cells, and ultimately affecting semen quality [45]. This indicates that miR-125b has a positive effect on reproductive performance and could be used as a drug and diagnostic target for male sterility. In the mRNA–miRNA regulatory network constructed in this study, we screened miR-125b, suggesting that miR-125b plays a key role in the development of sheep testes, especially hormone secretion and energy metabolism. miR-125b was found to play an important role in sheep reproduction. Although other miRNAs, such as miR-99 and miR-299, have been identified, their roles in reproduction are unknown [46,47]. Studies have shown that they can regulate stem cell self-renewal, ovarian cancer cells, and glioma, suggesting that their roles in reproduction should not be neglected.
Furthermore, we also found that the miRNA target genes were involved in reproduction. For example, the target gene EHD4 of miR-27a, which was originally described as a protein expressed in the extracellular matrix, has been shown to be a cytosolic and endosomal vesicle-associated protein [48]. George et al. [26] found that although Ehd4 −/− mice were fertile male, their 31 days postpartum testicular weight decreased by 50%, and apoptosis gradually increased. Other defects, including reduced seminiferous tubule diameter, abnormal regulation of spermatogenic epithelium, and abnormal head of slender sperm cells, are also present in Ehd4 −/− mice, which eventually result in lower sperm counts and lower fertility. In adult testes, EHD4 is highly expressed in primary spermatocytes and EHD4 deletion alters the levels of other EHD proteins in an age-dependent manner. The results suggest that EHD4 plays a role in the normal development of mitosis and late germ cells, and that EHD protein-mediated endocytic cycling is important for germ cell development and testicular function [26]. The target gene TGFBR3 is regulated by miR-127 and miR-27a. Beta glycans (TGFBR3) belong to one of the ligands of the transforming growth factor beta (TGFβ) superfamily. Sarraj et al. [49] found that Tgfbr3 −/− mice developed severe fine cords at 12.5–13.5 days after conception and their fetal stromal cell function was affected; their experimental data show that TGFBR3 is necessary for the normal formation of the spermatic cord, the development of fetal stromal cells, and the establishment of endocrine function. TGFBR3 was also screened in the network constructed in this study, suggesting that this gene is important for fine testicular formation, interstitial cell development, and endocrine function in sheep. The findings confirmed that miR-27a and miR-127 potentially control testis size. Testicular development is controlled by complex levels of gene regulatory proteins, growth factors, cell adhesion molecules, signaling molecules, and hormones that interact with each other through controlled relationships, usually in a short period of time. Platelet-derived growth factor (PDGF) is a key regulator of connective tissue cells during embryogenesis and pathogenesis, including PDGFA, PDGFB, PDGFRA, and PDGFRB. The results of this study suggested that PDGFRB is a target of miR-27a. Mice with mutations in PDGF had normal testicular development before birth, normal fetal cytoplasmic cell numbers, and normal masculine functions. However, after birth, the gene mutation caused testicular shrinkage, loss of mesenchymal cells, decreased testosterone, and impaired spermatogenesis [50]. Another interesting finding is regulation of the target genes COLIA1 and COLIA2 by miR-125. There is evidence that COLIA1 and COLIA2 are involved in type A spermatogonia and play a role in mediating the isolation and migration of germ cells during spermatogenesis [51]. The aforementioned evidence revealed that these genes may play a key role in regulating testicular development.
5 Conclusions
In this study, 20 potential miRNAs, including 8 down- and 12 upregulated miRNAs were identified by constructing mRNA–miRNA regulatory networks. The miRNA target genes, namely, TGFBR3, PDGFRB, Col1a2, and EHD4, are related to reproduction. Therefore, these miRNA molecules can be used as potential candidate small molecules to regulate the testicular size and provide a theoretical reference for the genetic regulation of testicular development in sheep.
-
Funding information: This work was supported by the Youth Fund of Qinghai Provincial Science and Technology Department (2021-ZJ-978Q), the Key Project of the Youth Foundation of Qinghai University (2020-QNY-1).
-
Author contributions: Wu Sun designed the experiments and drafted the manuscript. Sequencing data processing and visualization were done by Wu Sun and Xiayang Jin. Shike Ma and Yuhong Ma carried out quantitative experiments.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Coulter GH, Foote RH. Bovine testicular measurements as indicators of reproductive performance and their relationship to productive traits in cattle: A review. Theriogenology. 1979;11(4):297–311.10.1016/0093-691X(79)90072-4Search in Google Scholar PubMed
[2] Abba Y, Igbokwe IO. Testicular and related size evaluations in nigerian sahel goats with optimal cauda epididymal sperm reserve. Vet Med Int. 2015;2015:357519.10.1155/2015/357519Search in Google Scholar PubMed PubMed Central
[3] Condorelli R, Calogero AE, La Vignera S. Relationship between testicular volume and conventional or nonconventional sperm parameters. Int J Endocrinol. 2013;2013:145792.10.1155/2013/145792Search in Google Scholar PubMed PubMed Central
[4] Yassen AM, Mahmoud MN. Relationship between body weight and testicular size in buffalo bulls. J Agric Sci. 1972;78(3):367–70.10.1017/S0021859600026265Search in Google Scholar
[5] Hagiya K, Hanamure T, Hayakawa H, Abe H, Baba T, Muranishi Y, et al. Genetic correlations between yield traits or days open measured in cows and semen production traits measured in bulls. Animal. 2018;12(10):2027–31.10.1017/S1751731117003470Search in Google Scholar PubMed
[6] Jacyno E, Kawęcka M, Pietruszka A, Sosnowska A. Phenotypic correlations of testes size with semen traits and the productive traits of young boars. Reprod Domest Anim. 2015;50(6):926–30.10.1111/rda.12610Search in Google Scholar PubMed
[7] Kastelic JP. Understanding and evaluating bovine testes. Theriogenology. 2014;81(1):18–23.10.1016/j.theriogenology.2013.09.001Search in Google Scholar PubMed
[8] Van Melis MH, Eler JP, Rosa GJ, Ferraz JB, Figueiredo LG, Mattos EC, et al. Additive genetic relationships between scrotal circumference, heifer pregnancy, and stayability in Nellore cattle. J Anim Sci. 2010;88(12):3809–13.10.2527/jas.2009-2127Search in Google Scholar PubMed
[9] Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction. 2003;125(6):769–84.10.1530/rep.0.1250769Search in Google Scholar PubMed
[10] Mendis SH, Meachem SJ, Sarraj MA, Loveland KL. Activin A balances Sertoli and germ cell proliferation in the fetal mouse testis. Biol Reprod. 2011;84(2):379–91.10.1095/biolreprod.110.086231Search in Google Scholar PubMed
[11] Mithraprabhu S, Mendis S, Meachem SJ, Tubino L, Matzuk MM, Brown CW, et al. Activin bioactivity affects germ cell differentiation in the postnatal mouse testis in vivo. Biol Reprod. 2010;82(5):980–90.10.1095/biolreprod.109.079855Search in Google Scholar PubMed PubMed Central
[12] Tomaszewski J, Joseph A, Archambeault D, Yao HH. Essential roles of inhibin beta A in mouse epididymal coiling. Proc Natl Acad Sci U S A. 2007;104(27):11322–7.10.1073/pnas.0703445104Search in Google Scholar PubMed PubMed Central
[13] Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–7.10.1126/science.1130164Search in Google Scholar PubMed
[14] Krützfeldt J, Stoffel M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab. 2006;4(1):9–12.10.1016/j.cmet.2006.05.009Search in Google Scholar PubMed
[15] Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23(4):604–15.10.1101/gr.139758.112Search in Google Scholar PubMed PubMed Central
[16] Sree S, Radhakrishnan K, Indu S, Kumar PG. Dramatic changes in 67 miRNAs during initiation of first wave of spermatogenesis in Mus musculus testis: global regulatory insights generated by miRNA-mRNA network analysis. Biol Reprod. 2014;91(3):69.10.1095/biolreprod.114.119305Search in Google Scholar PubMed
[17] Li M, Yu M, Liu C, Zhu H, Hua J. Expression of miR-34c in response to overexpression of Boule and Stra8 in dairy goat male germ line stem cells (mGSCs). Cell Biochem Funct. 2013;31(4):281–8.10.1002/cbf.2970Search in Google Scholar PubMed
[18] Xu C, Wu S, Zhao W, Mipam T, Liu J, Liu W, et al. Differentially expressed microRNAs between cattleyak and yak testis. Sci Rep. 2018;8(1):592.10.1038/s41598-017-18607-0Search in Google Scholar PubMed PubMed Central
[19] Liu SF, Jiang YL, Du LX. Studies of BMPR-IB and BMP15 as candidate genes for fecundity in little tailed han sheep. Acta Genet. Sin. 2003;30(8):755–60.Search in Google Scholar
[20] Wen YL, Guo XF, Ma L, Zhang XS, Chu MX. The expression and mutation of BMPR1B and its association with litter size in small-tail Han sheep (Ovis aries). Arch Anim Breed. 2021;64(1):211–21.10.5194/aab-64-211-2021Search in Google Scholar PubMed PubMed Central
[21] Yang Z, Yang X, Liu G, Deng M, Sun B, Guo Y, et al. Polymorphisms in BMPR-IB gene and their association with litter size trait in Chinese Hu sheep. Anim Biotechnol. 2020;10(1):1–10.Search in Google Scholar
[22] Yue C, Bai WL, Zheng YY, Hui TY, Sun JM, Guo D, et al. Correlation analysis of candidate gene SNP for high-yield in Liaoning cashmere goats with litter size and cashmere performance. Anim Biotechnol. 2021;32(1):43–50.10.1080/10495398.2019.1652188Search in Google Scholar PubMed
[23] Qiao HS, Yang BH, Yue YJ, Ma SK, Qiao GY, Zha XT. Detection and analysis of BMPR-IB gene polymorphism in plateau Tibetan sheep ram population. Heilongjiang Anim Sci Vet. Med. 2018;16:56–8.Search in Google Scholar
[24] Qiao GY, Qiao HS, Ma SK, Zha XT. Multiparous reproductive performance analysis of plateau Tibetan sheep. Chinese J Anim Husb Vet Med. 2017;10:17–8.Search in Google Scholar
[25] Santos-Ahmed J, Brown C, Smith SD, Weston P, Rasoulpour T, Gilbert ME, et al. Akt1 protects against germ cell apoptosis in the postnatal mouse testis following lactational exposure to 6-N-propylthiouracil. Reprod Toxicol. 2011;31(1):17–25.10.1016/j.reprotox.2010.09.012Search in Google Scholar PubMed PubMed Central
[26] George M, Rainey MA, Naramura M, Ying G, Harms DW, Vitaterna MH, et al. Ehd4 is required to attain normal prepubertal testis size but dispensable for fertility in male mice. Genesis. 2010;48(5):328–42.10.1002/dvg.20620Search in Google Scholar PubMed PubMed Central
[27] Gruber M, Mathew LK, Runge AC, Garcia JA, Simon MC. EPAS1 is required for spermatogenesis in the postnatal mouse testis. Biol Reprod. 2010;82(6):1227–36.10.1095/biolreprod.109.079202Search in Google Scholar PubMed PubMed Central
[28] Deshpande S, Agosti V, Manova K, Moore MA, Hardy MP, Besmer P. Kit ligand cytoplasmic domain is essential for basolateral sorting in vivo and has roles in spermatogenesis and hematopoiesis. Dev Biol. 2010;337(2):199–210.10.1016/j.ydbio.2009.10.022Search in Google Scholar PubMed PubMed Central
[29] Chu MX, Di R, Ye SC, Fang L, Liu ZH, Peng ZL, et al. Establishment of molecular detection methods for high prolificacy major gene FecB in sheep and its application. J Agric Biotechnol. 2009;17(1):52–8.Search in Google Scholar
[30] Xu H, Sun W, Pei S, Li W, Li F, Yue X. Identification of key genes related to postnatal testicular development based on transcriptomic data of testis in Hu sheep. Front Genet. 2022;12:773695.10.3389/fgene.2021.773695Search in Google Scholar PubMed PubMed Central
[31] Ghosh S, Chan CK. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol Biol. 2016;1374:339–61.10.1007/978-1-4939-3167-5_18Search in Google Scholar PubMed
[32] Simon A, Theodor PP, Wolfgang H. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;2:166–9.10.1093/bioinformatics/btu638Search in Google Scholar PubMed PubMed Central
[33] Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–38.10.1093/bioinformatics/btp612Search in Google Scholar PubMed
[34] Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14.10.1186/gb-2010-11-2-r14Search in Google Scholar PubMed PubMed Central
[35] Tafer H, Hofacker IL. RNAplex: A fast tool for RNA–RNA interaction search. Bioinformatics. 2008;24(22):2657–63.10.1093/bioinformatics/btn193Search in Google Scholar PubMed
[36] Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.10.1101/gr.1239303Search in Google Scholar PubMed PubMed Central
[37] Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8.10.1006/meth.2001.1262Search in Google Scholar PubMed
[38] Gustavsson EK, Zhang D. ggtranscript: an R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics. 2022;38(15):3844–6.10.1093/bioinformatics/btac409Search in Google Scholar PubMed PubMed Central
[39] Hirano T, Iwasaki YW, Lin ZY, Imamura M, Seki NM, Sasaki E, et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA. 2014;20(8):1223–37.10.1261/rna.045310.114Search in Google Scholar PubMed PubMed Central
[40] Li Y, Li J, Fang C, Shi L, Tan J, Xiong Y, et al. Genome-wide differential expression of genes and small RNAs in testis of two different porcine breeds and at two different ages. Sci Rep. 2016;6:26852.10.1038/srep26852Search in Google Scholar PubMed PubMed Central
[41] Lian C, Sun B, Niu S, Yang R, Liu B, Lu C, et al. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J. 2012;279(6):964–75.10.1111/j.1742-4658.2012.08480.xSearch in Google Scholar PubMed
[42] Wu J, Liao M, Zhu H, Kang K, Mu H, Song W, et al. CD49f-positive testicular cells in Saanen dairy goat were identified as spermatogonia-like cells by miRNA profiling analysis. J Cell Biochem. 2014;115(10):1712–23.10.1002/jcb.24835Search in Google Scholar PubMed
[43] Zhang Y, Zeng CJ, He L, Ding L, Tang KY, Peng WP. Selection of endogenous reference microRNA genes for quantitative reverse transcription polymerase chain reaction studies of boar spermatozoa cryopreservation. Theriogenology. 2015;83(4):634–41.10.1016/j.theriogenology.2014.10.027Search in Google Scholar PubMed
[44] Kim KH, Seo YM, Kim EY, Lee SY, Kwon J, Ko JJ, et al. The miR-125 family is an important regulator of the expression and maintenance of maternal effect genes during preimplantational embryo development. Open Biol. 2016;6(11):160181.10.1098/rsob.160181Search in Google Scholar PubMed PubMed Central
[45] Li L, Zhu Y, Chen T, Sun J, Luo J, Shu G, et al. miR-125b-2 knockout in testis is associated with targeting to the PAP Gene, mitochondrial copy number, and impaired sperm quality. Int J Mol Sci. 2019;20(1):148–56.10.3390/ijms20010148Search in Google Scholar PubMed PubMed Central
[46] Khalaj M, Woolthuis CM, Hu W, Durham BH, Chu SH, Qamar S, et al. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med. 2017;214(8):2453–70.10.1084/jem.20161595Search in Google Scholar PubMed PubMed Central
[47] Zhao R, Liu Q, Lou C. MicroRNA-299-3p regulates proliferation, migration and invasion of human ovarian cancer cells by modulating the expression of OCT4. Arch Biochem Biophys. 2018;651:21–7.10.1016/j.abb.2018.05.007Search in Google Scholar PubMed
[48] Kuo HJ, Tran NT, Clary SA, Morris NP, Glanville RW. Characterization of EHD4, an EH domain-containing protein expressed in the extracellular matrix. J Biol Chem. 2001;276(46):43103–10.10.1074/jbc.M106128200Search in Google Scholar PubMed
[49] Sarraj MA, Escalona RM, Umbers A, Chua HK, Small C, Griswold M, et al. Fetal testis dysgenesis and compromised Leydig cell function in Tgfbr3 (beta glycan) knockout mice. Biol Reprod. 2010;82(1):153–62.10.1095/biolreprod.109.078766Search in Google Scholar PubMed
[50] Mariani S, Basciani S, Arizzi M, Spera G, Gnessi L. PDGF and the testis. Trends Endocrinol Metab. 2002;13(1):11–7.10.1016/S1043-2760(01)00518-5Search in Google Scholar PubMed
[51] He Z, Feng L, Zhang X, Geng Y, Parodi DA, Suarez-Quian C, et al. Expression of Col1a1, Col1a2 and procollagen I in germ cells of immature and adult mouse testis. Reproduction. 2005;130(3):333–41.10.1530/rep.1.00694Search in Google Scholar PubMed
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”

