Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
-
Junyue Xing
, Hao Tang
and Lin Liu
Abstract
Pulmonary atresia (PA) is a severe cyanotic congenital heart disease. Although some genetic mutations have been described to be associated with PA, the knowledge of pathogenesis is insufficient. The aim of this research was to use whole-exome sequencing (WES) to determine novel rare genetic variants in PA patients. We performed WES in 33 patients (27 patient–parent trios and 6 single probands) and 300 healthy control individuals. By applying an enhanced analytical framework to incorporate de novo and case–control rare variation, we identified 176 risk genes (100 de novo variants and 87 rare variants). Protein‒protein interaction (PPI) analysis and Genotype-Tissue Expression analysis revealed that 35 putative candidate genes had PPIs with known PA genes with high expression in the human heart. Expression quantitative trait loci analysis revealed that 27 genes that were identified as novel PA genes that could be affected by the surrounding single nucleotide polymorphism were screened. Furthermore, we screened rare damaging variants with a threshold of minor allele frequency at 0.5% in the ExAC_EAS and GnomAD_exome_EAS databases, and the deleteriousness was predicted by bioinformatics tools. For the first time, 18 rare variants in 11 new candidate genes have been identified that may play a role in the pathogenesis of PA. Our research provides new insights into the pathogenesis of PA and helps to identify the critical genes for PA.
1 Introduction
Pulmonary atresia (PA) is a severe cyanotic congenital heart disease that accounts for 1.3–3.4% of congenital heart malformations [1,2]. Nonetheless, the incidence of PA in different populations varies [3]. The main feature of PA is the lack of connection between the ventricular artery and the pulmonary artery. Without surgical intervention, the early mortality rate is as high as 80%, and very few children can live to adulthood [4]. According to whether there is a ventricular septal defect, PA is divided into two types: PA with ventricular septal defect (PA-VSD) and PA with the intact ventricular septum (PA-IVS) [5,6,7]. The occurrence times of the two types of PA are distinct. Compared with PA-VSD, the damage from PA-IVS occurs in later pregnancy. PA-VSD damage occurs before the complete formation of the ventricular septum, while PA-IVS damage occurs after the completion of the ventricular septum formation [8]. PA infants usually have heart failure and cyanosis in the clinic. During the foetal period, prenatal ultrasonography has a high detection rate of PA-VSD, which is beneficial for the choice of surgical method. However, not all potential children can be detected by ultrasonography, especially those with main pulmonary collateral arteries [9]. At present, the main treatment for PA is still surgical treatment [10]. Although surgical techniques have been improved, the prognosis of PA is still poor [4].
Abnormal cardiac embryonic development is the main cause of congenital heart disease, so genetic research on congenital heart disease has always concentrated on the process of cardiac development. PA is a type of conic arterial trunk malformation, and its mechanism may be related to developmental abnormalities of the conic arterial trunk. Although the 22q11 microdeletion was found in approximately 8–17% of patients with conic arterial malformation associated with other systemic abnormalities, it rarely occurred in patients with isolated conic arterial malformation [11,12]. Through animal models, it was found that Nkx2.5, Isl1, Foxh1, Foxc1, Fgf8, Tbx1, and other genes played a role in the development, proliferation, and migration of cardiac neural crest cells and secondary heart field cells [13,14,15,16,17]. If these genes are deleted or mutated, conic arterial trunk malformation might occur [18]. The removal of the second heart area in HH14 chicken embryos led to PA, and the removal of the second heart area in HH18 chicken embryos led to coronary artery abnormalities [19]. Blocking the FGF signalling pathway in HH14 chicken embryos can also lead to PA. Nonetheless, the aforementioned studies are rarely verified in human heart development. Genetic evaluation of PA-IVS cases in identical twins demonstrated that WFDC8 and WFDC9 had a 55 kb deletion, but this was not found to be associated with genetic diseases, and the clinical significance was unclear [20]. Some PA candidate genes have been screened by whole-exome sequencing (WES). Fifty-six rare variations of seven new candidate genes (DNAH10, DST, FAT1, HMCN1, HNRNPC, TEP1, and TYK2) were screened in PA/VSD (n = 60) and PA/IVS (n = 20), and it was found that candidate genes of PA/VSD and PA/IVS were different. Three new rare copy number variations (CNVs), 16p11.2 del (PPP4C), 5q35.3 del (FLT4), and 5p13.1 del (RICTOR), were found in PA-VSD [21]. However, the function of these candidate genes still needs to be validated by further research [22].
The aforementioned PA studies suggest that many PA-related genes have not been found. With the development of next-generation sequencing technology, WES is increasingly being used in disease research, and it has gradually become a common screening method for molecular genetics research of diseases. In this study, to screen new PA pathogenic genes, WES was performed on peripheral blood samples of both 27 familial cases and 6 sporadic cases of PA. Bioinformatics screening was performed to determine the candidate genes to lay the foundation for exploring the molecular genetic pathogenesis of PA.
2 Materials and methods
2.1 Study population
Our cohort included 33 unrelated PA cases (27 familial and 6 sporadic cases) and 300 healthy control individuals (healthy children’s parents from the Chigene Chinese Gene-Phenotype Database) diagnosed by echocardiography, cardiac catheterization, or surgery. Enhanced association analysis was conducted by using two approaches: family-based de novo variant analysis and nonfamily rare variant association analysis (RVAS). The TADA-de novo model of TADA software was used to analyse the pathogenicity of de novo variants, and the significant top 100 genes are listed. EPACTS software was used for RVAS analysis of 33 cases and control samples from the Chigene Database. skat, b.callapse, b.madsen, and b.wcnt are gene-wise or group-wise tests of epacts software for gene-based effect analysis.
2.2 Sample collection
For the DNA extraction and whole-exome library construction, umbilical cord blood or foetal tissue genomic DNA was extracted using the Blood Genome Column Medium Extraction Kit (Kangweishiji, China) according to the manufacturer’s instructions. The extracted DNA samples were subjected to quality control using a Qubit 2.0 fluorimeter and electrophoresis with a 0.8% agarose gel for further protocols. Protein-coding exome enrichment was performed using xGenExome Research Panel v1.0 (IDT, Iowa, USA), which consists of 429,826 individually synthesized and quality-controlled probes. This targets a 39 Mb protein-coding region (19,396 genes) of the human genome and covers 51 Mb of end-to-end tiled probe space.
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations, institutional policies, and in accordance with the tenets of the Helsinki Declaration, and has been approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University (2020-KY-142).
2.3 Sequencing
High-throughput sequencing was performed by an Illumina NovaSeq 6000 series sequencer (PE150), and not less than 99% of the target sequences were sequenced. The sequencing process was performed by Chigene.
2.4 Bioinformatics analysis
For quality control, raw data were processed by fastp for adapter removal and low-quality read filtering.
For SNV and indel screening, paired-end reads were aligned to the Ensemble GRCh37/hg19 reference genome. The results were sorted with SAMtools contrast, and the repeat sequence was marked with Picard. According to the analysis process of GATK, the mutation sites were identified to obtain the single nucleotide polymorphism (SNP) and the insertion or deletion (Indel) information of small fragments in each sample. All variation loci were compared with 1000Genome, ExAC, GnomAD, and other databases, and low-frequency variants and de novo variants were screened out.
For CNV screening, adaptor sequences and reads of low quality were removed. Reads were mapped to reference genome hg19 using the BWA software. CNVs of 100 KB and greater in length were detected using Chigene independently developed software packages for CNV detection. Intervals of CNV were detected and screened according to public CNV databases. The CNV databases Decipher, ClinVar, OMIM, DGV, and ClinGen were used as references to annotate the pathogenic classification of each screened CNV.
For variant annotation and pathogenicity prediction, the online system independently developed by Chigene was used to annotate database-based minor allele frequencies (MAFs) and ACMG practice guideline-based pathogenicity of every yielded gene variant, and the system also provided serial software packages for conservative analysis and protein product structure prediction. The databases for MAF annotation, including the dbSNP, ExAC_EAS, GnomAD_exome_EAS, Sift, Polypen2_hdiv, and MutationTaster software packages, were used to predict protein product structure variations. As a prioritized pathogenicity annotation to ACMG guidelines, the OMIM, HGMD, and ClinVar databases were used to confirm the pathogenicity of every variant. Since clinical sample data were used in this study, there was a hospital enrichment effect leading to selection bias in the results. In general, the size of the hospital enrichment effect is approximately 5–10 times. In this study, we selected a fivefold enrichment threshold for screening. The calculation method of the hospital enrichment effect divides the observed prevalence rate by the estimated prevalence rate ([case_sample_number/33]/[control_sample_number/300] > 5). To predict functional changes in variants at splicing sites, the MaxEntScan, dbscSNV, and GTAG software packages were used instead of the product structure prediction software. The protein sequences were downloaded from the UniProt website. The amino acid sequences of humans were compared with those of other species by MEGA 7 sequence alignment software. The change value of free energy (ΔΔG) was quantitatively calculated by I-Mutant to predict the effect of amino acid mutation on protein stability online. I-Mutant is free software for analysing the stability of the protein. ΔΔG is an energy prediction parameter that is used for quantitative calculation of the protein stability free energy difference caused by a single amino acid mutation. This reflects the effect of thermal denaturation of amino acid mutations on protein stability. The effect of a single mutation on the overall stability of a protein can be analysed based on the PDB ID or sequence primary structure information. If the protein tends to be stable due to mutation, then ΔΔG > 0; otherwise, the protein stability decreases after the mutation. The SWISS-MODEL online website was used to predict the protein structures of the wild type and mutant type.
2.5 Protein‒protein interaction (PPI) analysis
PPI data were extracted from the STRING database. The statistically significant candidate genes from de novo and RVAS and the known PA genes in the HPO (Human Phenotype Ontology) database and Phenolyzer database were combined with the STRING service platform, and the species was limited to Homo sapiens to construct the protein interaction network. The key PPI network was obtained with a confidence score ≥0.4 as the screening condition.
2.6 Genotype-tissue expression (GTEx)–expression quantitative trait loci (eQTL) analysis
Using the open-source tissue expression data provided by GTEx v8, the differential expression of candidate genes obtained by PPI analysis in related normal tissues (left ventricle and atrial appendage) was analysed, and genes with high expression in the heart were selected. eQTL analysis was executed by FastQTL software to correlate genotype and gene expression. Genes whose expression levels can be affected by surrounding SNPs were screened.
3 Results
3.1 Screening process of candidate genes
Pedigree-based de novo analysis and case‒control-based RVAS research strategies were adopted to evaluate the novel genetic risk genes of PA patients (Figure 1). As shown in Figure 1, through the analysis of WES data from 33 PA cases and 300 healthy control individuals, 345,770 highly reliable variations were successfully obtained. The TADA-De novo model was used for association analysis, and the top 100 significant genes were listed according to P < 0.05 and at least one de novo missense variation of each gene (Table S1). Four models were used in RVAS research: b.collapse.epacts, b.madsen.epacts, b.wcnt.epacts, and skat.epacts. Eighty-seven rare variations of genes were screened out using MAF < 5% and P ≤ 5 × 10−6 by combining the genes analysed by four models (Table S2). Gene interaction network and expression analysis of GTEx indicated that 35 candidate genes highly expressed in the human heart had PPIs with known PA genes. There were 27 genes that could be affected by SNPs in the eQTL analysis. According to the ACMG standard guidelines, 18 rare pathogenic variations in 11 genes were screened out in the ExAC_EAS and GnomAD_exome_EAS databases with MAF < 0.5%.
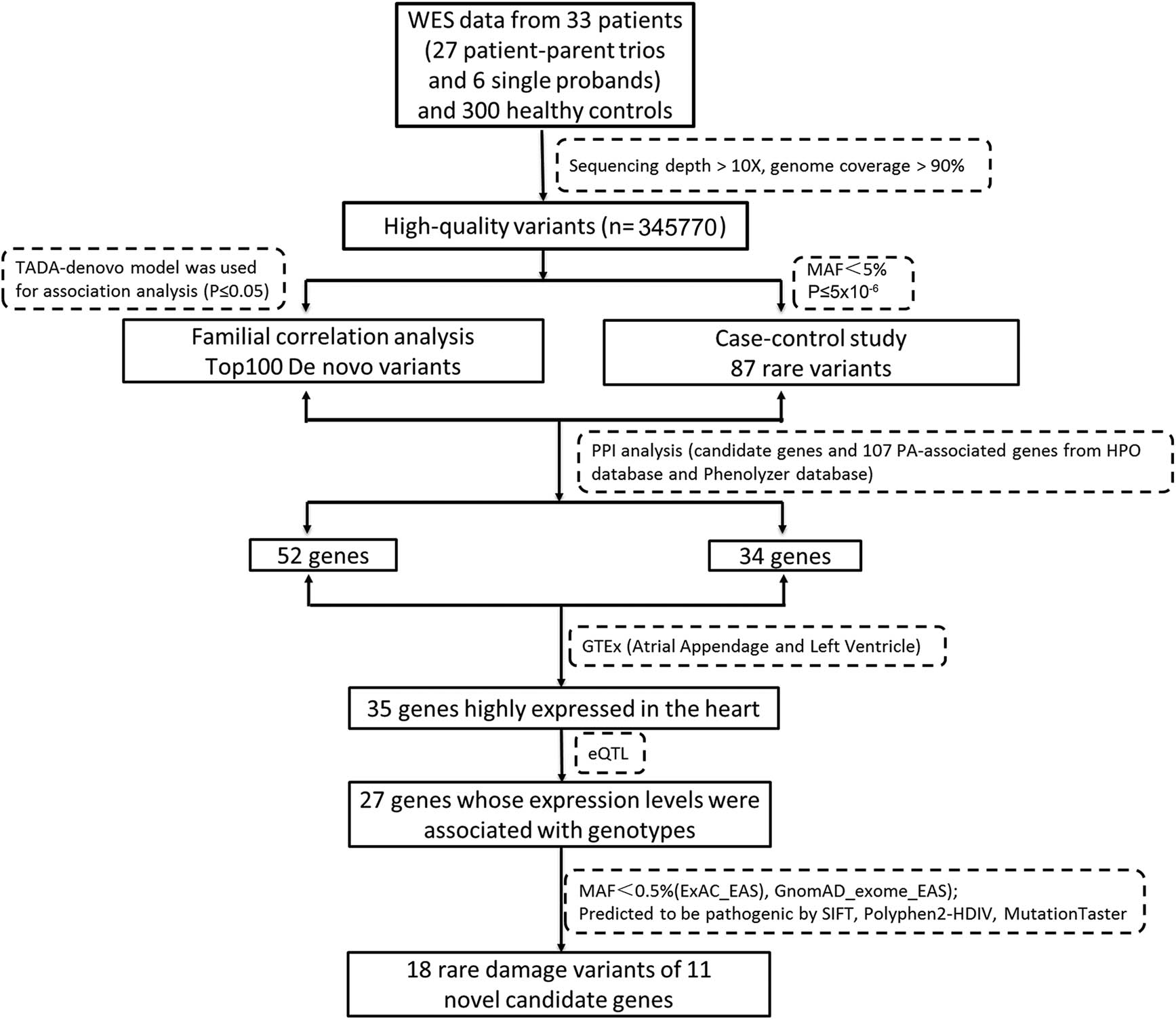
The workflow chart of the study. The workflow shows the analytical strategies for the different steps taken in WES analysis. After mutation invocation and annotation, candidate genes were collected by network analysis and gene expression analysis.
3.2 Overall comparison of variants between the case and control samples
The variation classification, variation type, and variation number in the SNV class were statistically analysed (Figure 2). In the 59,376 nucleotide sites of the case samples, 42,680 transitional and 16,696 transversional sites with 5,828 indels were found, and the ratio between transition and transversion was 2.56. In the 70,485 nucleotide sites of the control samples, 49,999 transitional and 20,486 transversional sites with 8,111 indels were found, and the ratio between transition and transversion was 2.44. The ratio of transversional sites to transitional sites is greater than 2, indicating that base substitution is not affected by the saturation effect.

Comparisons of rare damaging variants between the case and control groups. The number of variants in each variant classification, SNV class, and variant type is presented in (a–c), respectively.
3.3 Gene networks
To determine the potential contribution of these pedigree-based and case–control-based candidate genes to PA, 107 known PA-related genes reported in the HPO and Phenolyzer databases were analysed (Table S3). The STRING tool was employed to obtain PPI relationships for the known PA genes. Using a confidence score of ≥0.4, the network of PPI relationships exhibited 78 key genes, including 52 de novo variants (Figure 3a) and 34 rare variants (Figure 3b) (Table S4). Except for the VWF gene, the other screened genes were identified as novel PA genes.

PPI network between known PA-associated genes (yellow nodes) and del candidate genes (blue nodes). Network interaction of proteins (confidence score ≥0.4) was performed by using Cytoscape, bioinformatics software with the STRING database. Fifty-two de novo variants (a) and 34 rare variants (b) interacting with known PA genes were screened out.
3.4 Correlation analysis between variants and expression levels
GTEx is a database with tissue-specific gene expression and regulation. Samples from 54 different parts of the human body were simultaneously subjected to transcriptome sequencing and genotyping analysis [23]. The process by which DNA mutations affect mRNA expression is called eQTL analysis. The expression levels of all samples were analysed by the GTEx database, and 35 highly expressed candidate genes in the heart (atrial appendage and left ventricle) were screened (Table S5, Figure S1). The eQTL analysis was performed by FastQTL software to correlate genotype with gene expression. Twenty-seven genes that were identified as novel PA genes that could be affected by the surrounding SNP were screened (Table S5).
3.5 Hospital enrichment effect filtration and pathogenicity analysis of rare variants
Rare variants were screened in the ExAC and gnomAD databases of the East Asian population with a threshold of MAF <0.5%. We selected fivefold for filtering hospital enrichment effect filtration. The SIFT, Polyphen2, and MutationTaster databases were used to analyse the harmfulness of rare mutations. Finally, we screened 18 harmful rare variants in 11 genes (Table S6). These mutation sites have not been reported in the literature and are not included in the dbSNP database.
3.6 Protein structure prediction analysis
Most mutations have ΔΔG < 0, which are unstable mutations. Mutant proteins with ΔΔG < 1 showed significant instability. The ΔΔG of the BCLAF1 p.R271S, PABPC1 p.L562S, and CTBP2 p.G117S gene mutation sites were all less than 1, indicating that the protein stability was significantly decreased (Table S7). The amino acid mutations of succinate dehydrogenase-flavin (SDHA), BCLAF1, PABPC1, CTBP2, and CNN2 are located in highly conserved regions (Figure S2). Usually, mutations in highly conserved regions are pathogenic.
The number of hydrogen bonds in PABPC1 and CTBP2 changed (Figure 4). PABPC1 p.L562S was found in three patients, and mutations of this site were not found in the healthy group. The amino acid changes from leucine to serine, and the mutant has an additional hydrogen bond. CTBP2 p.G117S was found in eight patients, and this site did not change in the healthy group. The mutant had an additional four hydrogen bonds. CTBP2 p. K8X stopped translation, resulting in a complete loss of the functional domain. This mutation was found in 17 patients, but not in the healthy control group. The physicochemical properties of side-chain radical of BCLAF1 p.R271S, BCLAF1 p.Q165K, PABPC1 p.L562S, CTBP2 p.T182M, CTBP2 p.G117S, and CNN2 p.R266Q changed.

The structure prediction of the wild-type proteins and variants of PABPC1 and CTBP2. (a) Crystal structure of wild-type PABPC1. Crystal structure data were taken from RSCB PDB, I.D.: 1G9L. (b) Putative structure of the PABPC1 p.L562S mutant. A hydrogen bond was added between Ser562 and Gln558 (red arrow). (c) Crystal structure of wild-type CTBP2. Crystal structure data were taken from RSCB PDB, I.D.: 2OME. (d) Putative structure of the CTBP2 p.G117S mutant. Four hydrogen bonds have been added between Ser117, Ile113, Lys114, Gly120, and Asn358 (red arrows). (e) Putative structure of the CTBP2 p.K8X mutant.
4 Discussion
To explore the potential risk genes for PA, WES was performed for 33 patients and 300 healthy control individuals. For the first time, 18 rare damage variations in 11 new candidate genes were found to play a role in the pathogenesis of PA. This study expanded the range of PA pathogenic genes and provided new evidence for molecular genetics research on PA.
In mammalian embryonic development, the heart is the first organ to complete development. Abnormal embryonic development is the main cause of congenital heart disease. Studies have discovered that at the early stage of embryonic development, the heart needs to activate a large number of mRNAs for rapid division and differentiation. The expression of genes related to early embryonic development in mammals is primarily regulated at the post-transcriptional level [24]. The development of the heart valve starts with the endothelial-to-mesenchymal transition (EndoMT). First, some endothelial cells in the atrioventricular canal and outflow tract migrate to the cardiac glial area through the EndoMT to form mesenchymal cells. Subsequently, these mesenchymal cells highly proliferate and fuse to form a dense endocardial cushion with a cellular structure. Finally, the endocardial cushion is reshaped to produce an extracellular collagen matrix and form a mature valve [25,26,27]. Some transcription factors specifically highly expressed in the heart may be involved in the process of endothelial mesenchymal cell transformation, such as the GATA family (zinc finger protein transcription factor). GATA4 and GATA6 are highly expressed during endocardial cushion formation [28]. A mouse model was used to show that the expression of GATA4 is necessary for the transformation of endothelial–mesenchymal cells in the atrioventricular endocardial cushion of mice [29]. SDHA, BCLAF1, PABPC1, CTBP2, and CNN2 were found to be closely related to heart development.
PABPC1 p.L562 is located in the poly(A)-binding protein (PABP) functional domain. PABP is a class of highly conserved RNA-binding proteins that are ubiquitous in eukaryotes and can specifically recognize and bind to polyadenosine sequences. Eukaryotic proteins play an essential role in gene expression by binding to the 3′-poly(A) tail of mRNA. Although they lack catalytic activity, they can provide scaffolds in cells that bind to poly(A) tails and interact with other proteins to mediate transcription and translation and transport, and regulate mRNA degradation. PABPC1 is largely involved in a variety of post-transcriptional regulations to regulate gene expression, such as the regulation of mRNA translation initiation and microRNA-mediated expression. This post-transcriptional regulation of gene expression can regulate protein synthesis without changing transcriptional activity, allowing cells to respond quickly to external signals. Dynamic changes in poly(A) tail length can regulate the translation of specific mRNAs [30]. When the mRNA leaves the nucleus with a poly(A) tail of 250–300 A bases, the length gradually shortens and begins to extend when the mRNA is reactivated [31].
Increased expression of PABPC1 in cardiomyocytes can induce physiological cardiac hypertrophy [32]. In the hearts of adults and mice, when the length of the poly(A) tail was shortened, the expression level of PABPC1 decreased. When heart disease occurred, the poly(A) tail became longer and the expression level of PABPC1 increased. This indicates that PABPC1 regulates its own mRNA translation through the poly(A) tail [32]. The expression level of PABPC1 is low during early egg formation in Xenopus laevis. The expression level of PABPC1 increased rapidly after blastocyst transformation to early embryonic development, and PABPC1 was still found in the heart, brain, and other organs of adult Xenopus laevis [24]. Studies using model biological organisms, such as Drosophila, Caenorhabditis elegans, Saccharomyces cerevisiae, and Xenopus laevis, have identified that mutations of PABPC1 can lead to multiple organ malformations, including cardiac malformations [33]. Previous studies have demonstrated that PABPC1 plays an important role in cardiac differentiation and development.
CTBP2 p.G117 is located in the d-isomer-specific 2-hydroxydehydrogenase catalytic domain (2-Hacid_dh) of the CTBP2 protein. The NAD binding domain is the largest component of the 2-hydroxydehydrogenase catalytic domain of the CTBP2 protein. CTBP2 p.K8X terminated the translation at the eighth amino acid, and the protein functional domain was completely lost. C-terminal binding protein (CTBP) was initially identified as a protein that interacts with C-terminal elements and is highly conserved in eukaryotes [34]. CTBP family proteins are mainly used as transcriptional corepressors. dCTBP (the drosophila homologue) was shown to function as a transcriptional corepressor. CTBP can also activate the transcription process [35,36]. A CTBP2-knockout mouse experiment showed that the expression of the Wnt3A target gene Brachyury in E10.5 embryos was significantly lower than that in E9.5 embryos. This indicates that CTBP2 may be a Wnt-mediated transcriptional activator of Brachyury [37]. CTBP2 plays a unique regulatory role in the development of mice. The lack of CTBP leads to the inhibition of the activity of a large number of transcription factors, which leads to defects in cardiac morphology in mice. Cardiac embryos show pericardial dilation, and the development of the heart tube is blocked [37].
The p.R271S and p.Q165K variants of BCLAF1 are located in the THRAP3-BCLAF1 functional domain. BCLAF1 (Bcl-2-related transcription factor) is a multifunctional protein with a DNA binding domain and RS functional domain rich in arginine-serine. BCLAF1 is involved in the regulation of transcription and post-transcriptional processing. It can be used as a transcription factor and plays an important role in mRNA precursor splicing [38,39,40]. BCLAF1 plays an important role in the growth and development of mice. BCLAF1-deficient mice did not show embryonic death [41] but exhibited arrested lung development, resulting in death shortly after birth [42]. BCLAF1 is a harmful molecule in cardiac I/R injury. Cardiomyocyte-specific overexpression of BCLAF1 aggravated cardiac I/R injury, while partial knockout of BCLAF1 alleviated cardiac I/R injury. This is the first evidence of the underlying mechanism of BCLAF1 in cardiac I/R injury and may indicate its role as a potential therapeutic target for ischaemic heart disease [43].
The p.V446A and p.A449V residues of SDHA are located in the FAD binding domain. Succinate dehydrogenase (SDH), also known as succinate ubiquinone oxidoreductase or mitochondrial complex II, is the only multisubunit enzyme that integrates into the inner membrane of mitochondria in the tricarboxylic acid (TCA) cycle. SDH plays an important role in TCA and the aerobic respiratory chain [44]. Mutations in SDH cause the disruption and overflow of electron transfer from mitochondrial complex II [45] and even reverse the transfer process of succinic acid to fumaric acid in TCA, which may be the basis of multisystem lesions [46]. It has been reported that when SDH mutations occur, the respiration ability of mitochondria completely disappears, and a large number of reactive oxygen species (ROS) are produced, leading to the death of cardiac myocytes [47]. SDHA is the only active group in the SDH structure and is the key factor in the TCA cycle and aerobic respiration chain. SDHB, SDHC, and SDHD are inactive groups that only play a role in maintaining structural stability [48]. As a key enzyme in mitochondria, SDHA is involved in energy metabolism. At normal oxygen levels, hypoxia-inducing factor (HIF-1α) is a major regulator of cell metabolism. The SDHA gene has a strong ability to balance selection recognition, and SDHA mutants regulate cellular oxygen balance by influencing it. SDHA gene mutations decrease SDH activity, resulting in succinic acid accumulation. HIF-1α hydroxylation is accompanied by the conversion of α-ketoglutaric acid to succinic acid. Additionally, the accumulation of succinic acid can inhibit the hydroxylation and degradation of HIF-1α, resulting in an increase in HIF-1α protein and its target gene expression. Increasing succinic acid in the mitochondria by succinic dehydrogenase drives more ROS production. The SDHA c.1351C > T mutation has been reported in cases of mitochondrial metabolic disorders accompanied by cardiac damage [49]. Cardiomyopathy associated with SDHA defects has been reported in neonates. Rare cases of autosomal recessive neonatal isolated dilated cardiomyopathy are associated with the SDHA G555E mutation [50].
There are four noncovalent forces (hydrogen bonds, hydrophobic forces, electrostatic forces, and van der Waals forces) and two covalent forces (sulphide bonds and metal ion binding forces) that maintain protein structure. The amino acid composition of a protein is related to its thermal stability. Hydrogen bonds are a strong force existing in the peptide chain and generally exist in proteins. Aliphatic amino acids (such as alanine, isoleucine, leucine, and valine) are mostly embedded in the protein, and the internal conformation stability of the protein is improved through hydrophobic interactions within the protein, thereby improving its temperature tolerance. Ionizable residues (such as arginine, aspartate, histidine, glutamic acid, and lysine) mainly maintain the stability of the external structure of the protein by forming a charge force, thereby promoting the overall stability of the protein. The interaction of aromatic amino acids (such as phenylalanine, tryptophan, and tyrosine) includes the interaction between aromatic groups and cation-π interactions, which plays an important role in stabilizing proteins.
The number of hydrogen bonds between PABPC1 and CTBP2 changed. PABPC1 p.L562 S changed from leucine (hydrophobic amino acid) to serine (polar noncharged amino acid), and one hydrogen bond was added. CTBP2 p.G117S added four hydrogen bonds. There were no hydrogen bond changes at other mutant sites, but the polarity of side chain groups at almost all mutant sites of BCLAF1, PABPC1, CTBP2, and CNN2 changed. Mutated at sites 446 and 449 of SDHA, alanine and valine residues were replaced with each other. The side chain of valine (isopropyl) is larger than that of alanine (methyl), and both are hydrophobic amino acids, which are generally distributed in protein molecules. Specifically, if the mutation is distributed in the active site, it can change the structure of the active site and may cause the loss of protein activity. The protein stabilities of BCLAF1, PABPC1, and CTBP2 were significantly decreased. It can be inferred that the functional domains of PABPC1 and CTBP2 play an important role in the occurrence of PA.
5 Conclusion
In this study, 18 novel mutations in 11 genes were discovered by WES and bioinformatics prediction analysis. These mutations have not been reported in the literature, and the dbSNP database is not included. Along with the structure of proteins, the relationship between the functional domains of new mutation sites and PA was analysed and discussed. The potential role of these novel mutation sites in the transcriptional regulation of cardiac embryonic development is worth exploring. This research study expanded the PA pathogenic gene spectrum and laid a foundation for the translation of basic medicine concepts for clinical genetic counselling, prenatal diagnosis, and gene therapy.
Acknowledgement
We would like to thank our patients for agreeing to donate their personal data for our study and to have these data published.
-
Funding information: The work was supported by the National Natural Science Foundation of China (82071950, 82100294), Natural Science Foundation of Henan Province for Excellent Young Scientists (202300410364), Medical Science and Technology Project of Henan Province (SB201901099, SBGJ202003015, and SBGJ202003001). Science and Technology Research Project of Henan Province (212102310046), and Opening Foundation of National Health Commission Key Laboratory of Birth Defects Prevention (ZD202006).
-
Author contributions: Conceived and designed the experiment: L. Liu, H. Tang, and C. Z. Wang; clinical support: T. B. Fan, C. Y. Cui, and Y. N. Li; collected the samples and conducted the experiments: J. Y. Xing and H. D. Wang; analysed exome data: Y. Y. Xie, S. Wang, and W. Y. Gu; wrote the manuscript: Y. Y. Xie, J. Y. Xing, C. Z. Wang, H. Tang, and L. Liu. All the authors have reviewed the manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analysed during the current study are available from the corresponding author on a reasonable request. This study used the hg19/GRCh37 version of the human genome as the reference genome. Genes can be retrieved on the USCS genome Browser website (https://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/hg19.fa.gz).
References
[1] Mackie AS, Gauvreau K, Perry SB, del Nido PJ, Geva T. Echocardiographic predictors of aortopulmonary collaterals in infants with tetralogy of fallot and pulmonary atresia. J Am Coll Cardiol. 2003;41(5):852–7.10.1016/S0735-1097(02)02960-1Search in Google Scholar
[2] Honjo O, Al-Radi OO, MacDonald C, Tran KC, Sapra P, Davey LD, et al. The functional intraoperative pulmonary blood flow study is a more sensitive predictor than preoperative anatomy for right ventricular pressure and physiologic tolerance of ventricular septal defect closure after complete unifocalization in patients with pulmonary atresia, ventricular septal defect, and major aortopulmonary collaterals. Circulation. 2009;120(11 Suppl):S46–52.10.1161/CIRCULATIONAHA.108.844084Search in Google Scholar PubMed
[3] Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112(1 Pt 1):101–7.10.1542/peds.112.1.101Search in Google Scholar PubMed
[4] Leonard H, Derrick G, O’Sullivan J, Wren C. Natural and unnatural history of pulmonary atresia. Heart. 2000;84(5):499–503.10.1136/heart.84.5.499Search in Google Scholar PubMed PubMed Central
[5] van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–7.10.1016/j.jacc.2011.08.025Search in Google Scholar PubMed
[6] Gao M, He X, Zheng J. Advances in molecular genetics for pulmonary atresia. Cardiol Young. 2017;27(2):207–16.10.1017/S1047951116001487Search in Google Scholar PubMed
[7] Digilio MC, Marino B, Grazioli S, Agostino D, Giannotti A, Dallapiccola B. Comparison of occurrence of genetic syndromes in ventricular septal defect with pulmonic stenosis (classic tetralogy of Fallot) versus ventricular septal defect with pulmonic atresia. Am J Cardiol. 1996;77(15):1375–6.10.1016/S0002-9149(96)00212-3Search in Google Scholar
[8] Kutsche LM, Van Mierop LH. Pulmonary atresia with and without ventricular septal defect: a different etiology and pathogenesis for the atresia in the 2 types? Am J Cardiol. 1983;51(6):932–5.10.1016/S0002-9149(83)80168-4Search in Google Scholar PubMed
[9] Yang SH, Luo PH, Tian XX. Prenatal diagnosis of pulmonary atresia with ventricular septal defect. J Med Ultrason. 2018;45(2):341–44.10.1007/s10396-017-0809-2Search in Google Scholar PubMed
[10] Tomov ML, Perez L, Ning L, Chen H, Jing B, Mingee A, et al. A 3D Bioprinted in vitro model of pulmonary artery atresia to evaluate endothelial cell response to microenvironment. Adv Healthc Mater. 2021;10(20):e2100968.10.1002/adhm.202100968Search in Google Scholar PubMed PubMed Central
[11] Devriendt K, Eyskens B, Swillen A, Dumoulin M, Gewillig M, Fryns JP. The incidence of a deletion in chromosome 22Q11 in sporadic and familial conotruncal heart disease. Eur J Pediatr. 1996;155(8):721.10.1007/BF01957162Search in Google Scholar PubMed
[12] Gelb BD. Molecular genetics of congenital heart disease. Curr Opin Cardiol. 1997;12(3):321–8.10.1097/00001573-199705000-00014Search in Google Scholar PubMed
[13] Seo S, Kume T. Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol. 2006;296(2):421–36.10.1016/j.ydbio.2006.06.012Search in Google Scholar PubMed
[14] Brade T, Gessert S, Kühl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311(2):297–310.10.1016/j.ydbio.2007.08.004Search in Google Scholar PubMed
[15] Washington Smoak I, Byrd NA, Abu-Issa R, Goddeeris MM, Anderson R, Morris J, et al. Sonic hedgehog is required for cardiac outflow tract and neural crest cell development. Dev Biol. 2005;283(2):357–72.10.1016/j.ydbio.2005.04.029Search in Google Scholar PubMed
[16] Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, et al. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17(2):269–81.10.1101/gad.1048903Search in Google Scholar PubMed PubMed Central
[17] Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127(17):3865–76.10.1242/dev.127.17.3865Search in Google Scholar PubMed
[18] Ward C, Stadt H, Hutson M, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol. 2005;284(1):72–83.10.1016/j.ydbio.2005.05.003Search in Google Scholar PubMed
[19] Hutson MR, Zhang P, Stadt HA, Sato AK, Li YX, Burch J, et al. Cardiac arterial pole alignment is sensitive to FGF8 signaling in the pharynx. Dev Biol. 2006;295(2):486–97.10.1016/j.ydbio.2006.02.052Search in Google Scholar PubMed
[20] De Stefano D, Li P, Xiang B, Hui P, Zambrano E. Pulmonary atresia with intact ventricular septum (PA-IVS) in monozygotic twins. Am J Med Genet A. 2008;146a(4):525–8.10.1002/ajmg.a.32160Search in Google Scholar PubMed
[21] Shi X, Zhang L, Bai K, Xie H, Shi T, Zhang R, et al. Identification of rare variants in novel candidate genes in pulmonary atresia patients by next generation sequencing. Comput Struct Biotechnol J. 2020;18:381–92.10.1016/j.csbj.2020.01.011Search in Google Scholar PubMed PubMed Central
[22] Xie H, Hong N, Zhang E, Li F, Sun K, Yu Y. Identification of rare copy number variants associated with pulmonary atresia with ventricular septal defect. Front Genet. 2019;10:15.10.3389/fgene.2019.00015Search in Google Scholar PubMed PubMed Central
[23] Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET, et al. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60.10.1126/science.1262110Search in Google Scholar PubMed PubMed Central
[24] Cosson B, Couturier A, Le Guellec R, Moreau J, Chabelskaya S, Zhouravleva G, et al. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol Cell. 2002;94(4–5):217–31.10.1016/S0248-4900(02)01195-4Search in Google Scholar PubMed
[25] Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335.10.1016/S0074-7696(05)43005-3Search in Google Scholar PubMed
[26] Schroeder JA, Jackson LF, Lee DC, Camenisch TD. Form and function of developing heart valves: coordination by extracellular matrix and growth factor signaling. J Mol Med (Berl). 2003;81(7):392–403.10.1007/s00109-003-0456-5Search in Google Scholar PubMed
[27] Yutzey KE, Colbert M, Robbins J. Ras-related signaling pathways in valve development: ebb and flow. Physiol (Bethesda). 2005;20:390–7.10.1152/physiol.00035.2005Search in Google Scholar PubMed
[28] Nemer G, Nemer M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev Biol. 2003;254(1):131–48.10.1016/S0012-1606(02)00026-XSearch in Google Scholar PubMed
[29] Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133(18):3607–18.10.1242/dev.02519Search in Google Scholar PubMed PubMed Central
[30] Bag J, Bhattacharjee RB. Multiple levels of post-transcriptional control of expression of the poly (A)-binding protein. RNA Biol. 2010;7(1):5–12.10.4161/rna.7.1.10256Search in Google Scholar PubMed
[31] Burgess HM, Gray NK. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans. 2010;38(6):1517–22.10.1042/BST0381517Search in Google Scholar PubMed
[32] Chorghade S, Seimetz J, Emmons R, Yang J, Bresson SM, Lisio M, et al. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. Elife. 2017;6:e24139.10.7554/eLife.24139Search in Google Scholar PubMed PubMed Central
[33] Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, et al. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci U S A. 2011;108(19):7844–9.10.1073/pnas.1017664108Search in Google Scholar PubMed PubMed Central
[34] Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12(2):469–78.10.1002/j.1460-2075.1993.tb05679.xSearch in Google Scholar PubMed PubMed Central
[35] Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280(5360):101–4.10.1126/science.280.5360.101Search in Google Scholar PubMed
[36] Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17(7):2067–78.10.1093/emboj/17.7.2067Search in Google Scholar PubMed PubMed Central
[37] Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22(15):5296–307.10.1128/MCB.22.15.5296-5307.2002Search in Google Scholar PubMed PubMed Central
[38] Kasof GM, Goyal L, White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol. 1999;19(6):4390–404.10.1128/MCB.19.6.4390Search in Google Scholar PubMed PubMed Central
[39] Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25(8):381–8.10.1016/S0968-0004(00)01604-2Search in Google Scholar PubMed
[40] Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13(3):302–9.10.1016/S0955-0674(00)00212-XSearch in Google Scholar PubMed
[41] Sarras H, Alizadeh Azami S, McPherson JP. In search of a function for BCLAF1. ScientificWorldJournal. 2010;10:1450–61.10.1100/tsw.2010.132Search in Google Scholar PubMed PubMed Central
[42] McPherson JP, Sarras H, Lemmers B, Tamblyn L, Migon E, Matysiak-Zablocki E, et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009;16(2):331–9.10.1038/cdd.2008.167Search in Google Scholar PubMed
[43] Zhang Y, Zhang X. The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia-reperfusion injury. Nat Commun. 2021;12(1):522.10.1038/s41467-020-20844-3Search in Google Scholar PubMed PubMed Central
[44] Huang LS, Borders TM, Shen JT, Wang CJ, Berry EA. Crystallization of mitochondrial respiratory complex II from chicken heart: a membrane-protein complex diffracting to 2.0 A. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):380–7.10.1107/S0907444905000181Search in Google Scholar PubMed PubMed Central
[45] Chouchani ET, Pell VR, Gaude E, Aksentijević D, Sundier SY, Robb EL, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–35.10.1038/nature13909Search in Google Scholar PubMed PubMed Central
[46] Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496(7444):238–42.10.1038/nature11986Search in Google Scholar PubMed PubMed Central
[47] Grimm S. Respiratory chain complex II as general sensor for apoptosis. Biochim Biophys Acta. 2013;1827(5):565–72.10.1016/j.bbabio.2012.09.009Search in Google Scholar PubMed
[48] Dhingra R, Kirshenbaum LA. Succinate dehydrogenase/complex II activity obligatorily links mitochondrial reserve respiratory capacity to cell survival in cardiac myocytes. Cell Death Dis. 2015;6(10):e1956.10.1038/cddis.2015.310Search in Google Scholar PubMed PubMed Central
[49] Courage C, Jackson CB, Hahn D, Euro L, Nuoffer JM, Gallati S, et al. SDHA mutation with dominant transmission results in complex II deficiency with ocular, cardiac, and neurologic involvement. Am J Med Genet A. 2017;173(1):225–30.10.1002/ajmg.a.37986Search in Google Scholar PubMed
[50] Levitas A, Muhammad E, Harel G, Saada A, Caspi VC, Manor E, et al. Familial neonatal isolated cardiomyopathy caused by a mutation in the flavoprotein subunit of succinate dehydrogenase. Eur J Hum Genet. 2010;18(10):1160–5.10.1038/ejhg.2010.83Search in Google Scholar PubMed PubMed Central
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”

