Abstract
In this study, genetic diversity and germplasm identification of 28 alfalfa germplasm cultivars materials were evaluated by analyzing their internal transcribed spacer 2 (ITS2), trnL-F, and psbA-trnH sequences to provide the innovative reference of alfalfa varieties genetic diversity and identify research. The results showed that the fragment average length of ITS2, trnL-F, and psbA-trnH sorting sequences were 455.7 bp, 230.3 bp, and 345.6 bp, respectively. The ITS2 sequence was too conservative to reflect the individual differences between intercultivars and intracultivars in the preliminary experiment. Furthermore, trnL-F and psbA-trnH sequence differences were relatively small between intercultivars but significant between intracultivars. Alfalfa cultivars were divided into four groups by sequence similarity clustering. Alfalfa cultivars trnL-F and psbA-trnH sequences have apparent differences, showing that chloroplast conservative sequences were independent evolution. Compared with trnL-F and psbA-trnH sequences of alfalfa cultivars, psbA-trnH sequence has abundant variation sites and can better reflect the differences between cultivars than the trnL-F sequence. Therefore, the psbA-trnH sequence can identify different alfalfa cultivars and establish the DNA sequence fingerprint.
1 Introduction
Alfalfa (Medicago sativa L.) is originated in Asia Minor, the Caucasus, Iran, Turkmenistan, and Central Asia [1] and is one of the most important leguminous forage in the world due to its high yield, high quality, and strong adaptability. M. sativa L. is the main research object of breeding, and the resources of alfalfa varieties are the most important basic production materials for animal husbandry and ecological improvement. By 2020, 113 national-level alfalfa varieties have been approved and registered in China [2]. Alfalfa and hybrid alfalfa are the main varieties formed by breeding. Compared with the developed countries in animal husbandry, such as the United States, Canada, New Zealand, and Australia, the number of varieties formed is small, and the breeding efficiency is low. Genetic research and identification of alfalfa germplasm resources have important theoretical and practical significance for alfalfa breeding and the development of the alfalfa industry.
Genetic diversity is the basis of species diversity and ecosystem diversity. It studies the degree of difference between species at the molecular level and reflects the genetic variation within species and between different populations [3]. Research on alfalfa genetics and genetic diversity in China mainly focuses on population morphology, agronomic traits, resistance and physiological characteristics, molecular markers, and DNA sequence analysis. Genetic diversity and identification of cultivar groups based on conserved sequences, i.e., chloroplast and ribosomal genome sequences, have been reported in alfalfa. The chloroplast genome is a closed double-stranded circular molecular structure, generally 115–210 Kb [4]. Chloroplast genes have relatively conservative characteristics, a small genome, a large amount of DNA information, a large copy number, and a high mutation rate of noncoding regions. The psbA-trnH gene sequence is the gene sequence of the noncoding region of chloroplast RNA, and the noncoding region is the region that cannot be transcribed, which is messenger RNA. The sequence is easy to be amplified and sequenced. It contains a large amount of information and has stable maternal inheritance, which can be efficiently applied to compare differences between different groups within a species. The trnL-trnF (trnL-F) gene sequence is the chloroplast RNA coding gene and the noncoding region gene sequence [5]. Sequence fragments have the advantages of simple sequence, convenient amplification, high evolution rate, little influence on the external environment, and easy mutation and variation of bases between different populations. It is widely used in interspecific and subspecific level genetic evolution phylogeny research. The chloroplast gene trnL-F sequence, psbA-trnH sequence, and nuclear gene internal transcribed spacer (ITS) sequence marker systems are mainly used in plant phylogeny research, species genetic diversity, and medicinal use in China. There are few reports on identifying plant Chinese medicinal materials in the herbage field. For example, datureae plants analysis, Bupleurum marginatum var. analysis based on internal transcribed spacer 2 (ITS2) barcode [6,7], mainly concentrates on forage and the phylogenetic research of Poaceae materials. In relevant studies, phylogenetic analysis has been conducted on the materials of the genus Alkali, the genera Astragali radix, the Medicago, and their relatives [8,9,10,11,12,13]. Ribosomal DNA ITS sequences are a family of genes encoding ribosomal RNA in the nucleus of plant cells and are nuclear gene fragments [6]. The sequence of the ribosome coding region is generally highly conserved, and there are few reports on the study of the conserved ribosome sequence in alfalfa in China. In this study, 28 alfalfa varieties and germplasm materials were taken as the research objects, and their ITS2, trnL-F, and psbA-trnH sequences were analyzed for genetic and germplasm identification. The genetic structure characteristics and genetic diversity of alfalfa variety germplasm resources in different populations were analyzed. The research has a theoretical reference for screening excellent alfalfa germplasm, establishing an alfalfa variety identification system, and exploring and utilizing alfalfa variety germplasm resources. Furthermore, it fills the gap of research on the genetic and variety identification characteristics of alfalfa in China combined with trnL-F, psbA-trnH, and other conserved sequences.
2 Materials and methods
2.1 Experiment material
The test materials are 28 alfalfa varieties (materials) from different countries and regions (Table 1). The seeds of the experimental germplasm resources were obtained from the National Medium-term Forage Germplasm Resource Bank of the Grassland Research Institute, Chinese Academy of Agricultural Sciences. The test material was planted in the Agricultural and Animal Husbandry Interlaced Area Experimental Demonstration Base of the Grassland Research Institute of the Chinese Academy of Agricultural Sciences in 2012 and was planted as a single plant. In 2018, alfalfa was sampled at the branching stage, and 10 fresh and young leaf samples were collected from each material, placed in an ice box, and brought back. Then, samples were stored in a −80°C ultra-low temperature refrigerator in the laboratory for future use.
Alfalfa varieties (materials)
| Code | Name | Species | Origin | Analysis method |
|---|---|---|---|---|
| 1 | Aohan | Medicago sativa L. | China | ITS2 |
| 2 | Junggar | Medicago sativa L. | China | trnL-F, psbA-trnH |
| 3 | Zhaodong | Medicago sativa L. | China | trnL-F, psbA-trnH |
| 4 | Tianshui | Medicago sativa L. | China | trnL-F |
| 5 | Longdong | Medicago sativa L. | China | psbA-trnH |
| 6 | Xinjiang Daye | Medicago sativa L. | China | trnL-F, psbA-trnH |
| 7 | Gannong No. 3 | Medicago sativa L. | China | trnL-F, psbA-trnH |
| 8 | Zhongmu No. 2 | Medicago sativa L. | China | trnL-F, psbA-trnH |
| 9 | Tumu No. 1 | Medicago varia Martin. | China | trnL-F, psbA-trnH |
| 10 | Xinmu No. 1 | Medicago varia Martin. | China | trnL-F, psbA-trnH |
| 11 | Caoyuan No. 1 | Medicago varia Martin. | China | trnL-F, psbA-trnH |
| 12 | Caoyuan No. 2 | Medicago varia Martin. | China | ITS2, trnL-F, psbA-trnH |
| 13 | Gannong No. 1 | Medicago varia Martin. | China | trnL-F, psbA-trnH |
| 14 | Gannong No. 2 | Medicago varia Martin. | China | trnL-F, psbA-trnH |
| 15 | Japan 90 | Medicago sativa L. | Japan | trnL-F |
| 16 | Bear No. 1 | Medicago sativa L. | US | psbA-trnH |
| 17 | Czech 26-1 | Medicago sativa L. | Czech | trnL-F, psbA-trnH |
| 18 | Golden Queen | Medicago sativa L. | US | psbA-trnH |
| 19 | Soviet Union 1209 | Medicago sativa L. | Russia | trnL-F, psbA-trnH |
| 20 | Soviet Union 6220 | Medicago sativa L. | Russia | trnL-F, psbA-trnH |
| 21 | UK | Medicago sativa L. | UK | psbA-trnH |
| 22 | AmeriGraze 401Z | Medicago sativa L. | US | trnL-F |
| 23 | Apollo supreme | Medicago sativa L. | US | trnL-F, psbA-trnH |
| 24 | Rumble | Medicago sativa L. | Canada | trnL-F, psbA-trnH |
| 25 | WL-320 | Medicago sativa L. | US | trnL-F |
| 26 | WL-323 | Medicago sativa L. | US | trnL-F, psbA-trnH |
| 27 | Canada | Medicago sativa L. | Canada | trnL-F, psbA-trnH |
| 28 | Medicago falcata | Medicago falcata L. | China | trnL-F, psbA-trnH |
There were differences in the number of materials selected for the alfalfa varieties tested by different sequences, and two materials were selected for ITS2 (Table 1). Among them, the ITS2 sequence research was conducted on Medicago sativa L.cv.Aohan and Medicago varia Martin.cv.Caoyuan No. 2, and Aohan and Caoyuan No. 2 are two species, and there should be big differences in gene sequences, but two copies were found in the preliminary identification test. The DNA sequences of the materials showed little difference (Figure 1). Therefore, other corresponding material experiments were not carried out in the follow-up. The ITS2 sequence was unsuitable for analyzing genetic diversity among alfalfa varieties or species. According to the presence or absence of materials in the research process, 24 alfalfa materials were selected for the psbA-trnH sequence, and 23 materials were selected for the trnL-F sequence.
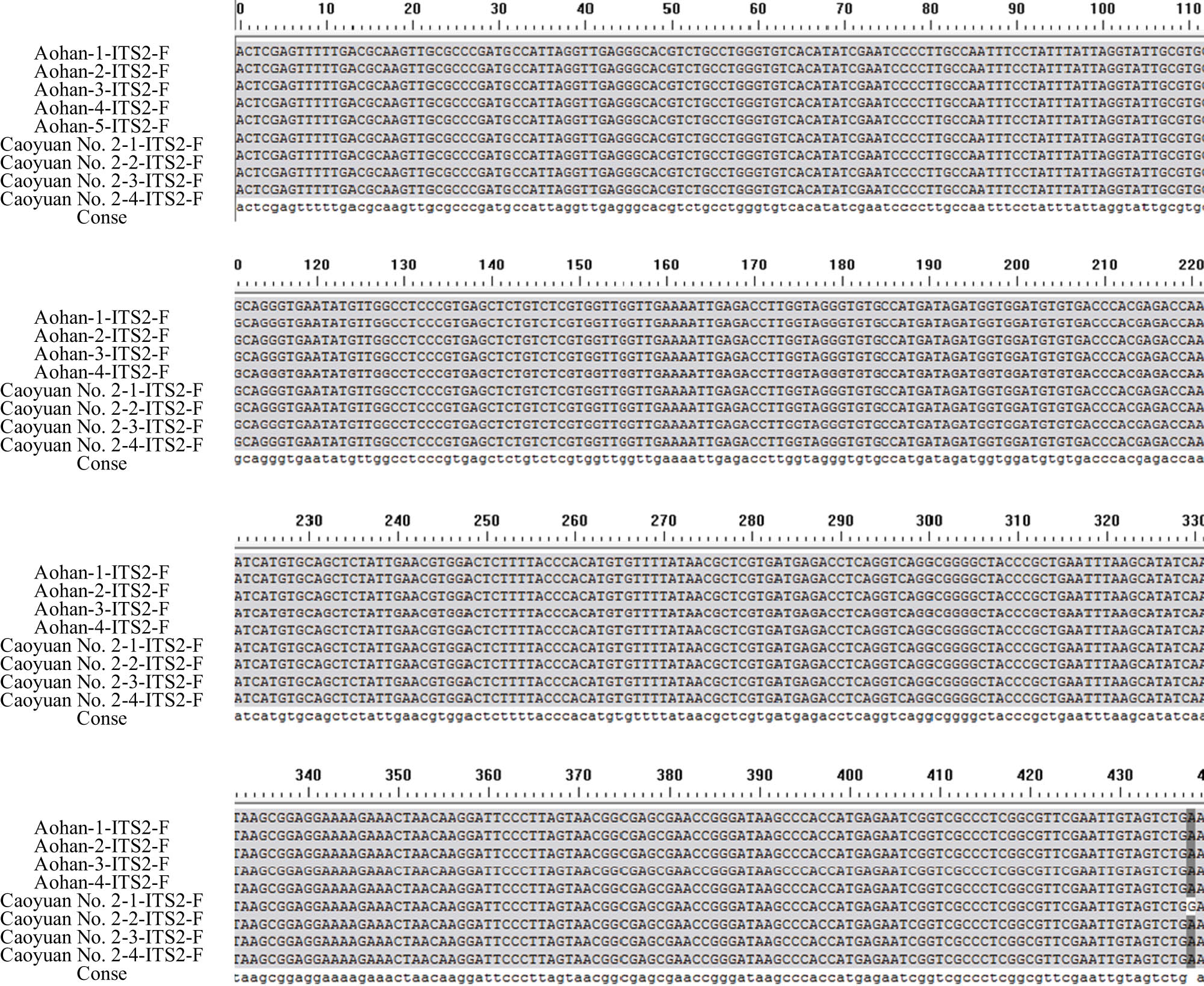
ITS2 sequences of different alfalfa varieties (materials).
2.2 DNA extraction
The DNA extraction method of single leaf material was extracted by DNA extraction kit (CTAB Plant genome extraction kit, BLKW, Beijing). DNA concentration and purity were detected by the 1.0% agarose detection method and trace UV/Vis spectrophotometer, respectively.
2.3 Primer and polymerase chain reaction (PCR) amplification procedure
The primers used in the experiment are shown in Table 2. The research results of NCBI number are presented in Table 2 (list first number only). ITS2 sequence PCR amplification system 30 μL includes 10–20 ng template 1 μL, 10 mol L−1 primers 1 μL each, PCR buffer 3 μL, dNTP 1 μL, Mg2+ 2 μL, Taq DNase 1 μL, and ddH2O 20 μL. The amplification program was an annealing temperature of 56°C, pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1.5 min, 40 cycles, and extension at 72°C for 10 min. trnL-F, psbA-trnH gene interval sequence amplification system 30 μL includes 10–20 ng template 1.5 μL, PCR Mix 15 μL, ddH2O 10.5 μL, primer F 1.5 μL, and primer R 1.5 μL. Program: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 49°C for 30 s, extension at 72°C for 90 s, 30 cycles, and extension at 72°C for 7 min. All PCR amplification products were stored in a refrigerator at 4°C for future use.
Primer message
| Sequence | Primer (5′ to 3′) | Temperature (°C) | Amplicon size (bp) |
| ITS2, NCBI:MT610943.1… | F: ATGCGATACTTGGTGTGAAT | 56 | 20 |
| R: GACGCTTCTCCAGACTACAAT | 21 | ||
| trnL-F, NCBI:MW271002.1… | F: GTTATGCATGAACGTAATGCTC | 49 | 22 |
| R: CGCGCATGGTGGATTCACAATCC | 23 | ||
| psbA-trnH, NCBI:KP174827.1… | F: GGTTCAAGTCCCTCTATCCC | 49 | 20 |
| R: ATTTGAACTGGTGACACGAG | 21 |
trnL-F/psbA-trnH sequence PCR 30 μL reaction system includes 1.5 μL diluted DNA solution, 15 μL PCR Mix, 10.5 μL ddH2O, 1.5 μL primer F, and 1.5 μL primer R. They were mixed to form the reaction system and centrifuged for use. The amplification program was optimized for reaction conditions. The amplification is performed as follows: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 49°C for 30 s, and extension at 72°C for 30 cycles for 90 s, and then extended at 72°C for 7 min and stored at 4°C for future use.
The amplified products were subjected to 1.0% agarose gel electrophoresis, stained with nucleic acid dyes, and observed and photographed with a gel imaging system.
2.4 PCR product sequencing
The PCR amplification products were purified and used for the sequencing reaction. The ITS2, trnL-F, and psbA-trnH sequences of all product samples were determined by Shenzhen Huada Gene Technology Co., Ltd. Sequencing was performed by direct sequencing of PCR products, and each sample was sequenced in forward and reverse directions to ensure the sequencing accuracy.
2.5 Data analysis
The DNA sequence fragments obtained by sequencing were used for sequencing quality evaluation. Forward and reverse sequence sequencing results were analyzed by Codoncode Aligner, seqMan, and DNAMan. Furthermore, CLUSTALX 2.0 software was used for sequence alignment. The genetic distances of the aligned sequences were calculated by MEGA 7.0 software, and the molecular phylogenetic tree was established by the kimura method. The confidence of each branch of the phylogenetic tree was tested by bootstrap (1,000 repetitions), and gaps were always treated as missing.
3 Results and analysis
3.1 ITS2 sequence polymorphism analysis of alfalfa
Aohan and Caoyuan No. 2 were selected as research materials, and 18 forward and reverse ITS2 sequences were obtained by PCR amplification, recovery, and sequencing. The sequencing results of the nine genotypes ITS2 with unidirectional sequences are shown in Figure 1. The results showed that the effective sequence was 454–457 bp in full length, with an average of 455.7 bp. The nucleic acid bases of the two alfalfa populations differed very little. The contents of T, C, A, and G were 26.0, 22.3, 24.2, and 27.5%, with variations ranging from 25.9∼26.2, 22.1∼22.7, 23.8∼24.5, and 27.2∼27.9%, respectively. The average content of A and T is 50.2%, which is slightly larger than that of G and C. It can be seen from the unidirectional sequence of ITS2 that the corresponding sequences of Aohan and Caoyan No. 2 have basically no sequence differences between populations and local populations after removing the primer sequences and irregular sequences at the initial stage of sequencing. This result indicated that ITS2 was too conservative for alfalfa resources. In addition, no insertion and deletion variation was found between the corresponding sequences, and the corresponding ITS2 sequences were not suitable for application as genetic diversity markers. Aohan and Caoyuan No. 2 are M. sativa L. and Medicago varia Martin, which are different alfalfa species. The ITS2 sequences show identical sequences among species, i.e., no difference between species. In the selected five Aohan and four Caoyuan No. 2, the intraspecific sequences are also the same, i.e., no intraspecific difference. There is no polymorphism in applying the ITS2 sequence in alfalfa to study interspecific and intraspecific genetic diversity.
3.2 trnL-F sequence polymorphism analysis of alfalfa
A total of 115 forward and reverse trnL-F sequences were obtained by PCR amplification, recovery, and sequencing of 23 alfalfa germplasm materials. After removing primer sequences, hybrid and repetitive sequence fragments, 23 alfalfa materials (genotypes), and trnL-F unidirectional sequences were sorted out. The results are shown in Figure 2. The effective sequence length is about 240 bp, averaging 230.3 bp. Nucleic acid bases of alfalfa materials are different. The average content of G + C is 32.4%, and the variation is within 30.6–34.6%. The average ambiguous site rate in the spliced sequences is 1.59%. The polymorphism analysis of trnL-F sequences of 23 alfalfa germplasm materials showed that the total number of sequence loci is 242, the haplotype diversity (Hd) is 0.478, and the nucleotide polymorphism (Pi) is 0.322, with an average number of nucleotide differences (k) of 1.974.

TrnL-F sequences of different alfalfa varieties (materials).
3.3 psbA-trnH sequence polymorphism analysis of alfalfa
A total of 120 forward and reverse psbA-trnH sequences were obtained by PCR amplification, recovery, and sequencing of psbA-trnH sequences of 24 alfalfa germplasm materials. After removing primer sequences, hybrid and repetitive fragments, 24 unidirectional sequences of psbA-trnH sequences of materials (genotypes) were sorted out. The results are shown in Figure 3. The effective sequence length is about 345 bp, with an average of 345.6 bp. The average content of G + C in the alfalfa psbA-trnH sequence is 18.27%, and the variation is within 21.4–17.6%. The average ambiguous site rate in the spliced sequences is 8.96%. The polymorphism analysis of psbA-trnH sequences of 24 alfalfa germplasm materials showed that the total number of loci is 352 bp, the haplotype diversity (Hd) is 0.582, and the nucleotide polymorphism (Pi) is 0.457, with an average number of nucleotide differences (k) of 2.682. The total number of variation sites (Vs) in the psbA-trnH sequence of each material is 86, accounting for 25.13% of the total number of sites. There are 44 single nucleotide variation sites (Ss) and 13 parsimony informative sites (Ps). Among the 84 single-nucleotide variation sites, 17 are two-base variations. The type of sequence base variation is mainly substitution variation, including T-C, G-A transition, T-A, G-T, C-A transversion, and 2–4 bp insertion and deletion variation.

PsbA-trnH sequences of different alfalfa varieties (materials).
3.4 Cluster analysis of trnL-F and psbA-trnH sequence in alfalfa
The similarity of trnL-F sequences of 23 alfalfa germplasms was clustered (Figure 4). Analysis showed that the trnL-F sequence homology similarity between Japan 90 and Xinjiang Daye as well as Zhaodong and Zhongmu No. 2 is the largest, both of which are 100%. Tumu No. 1 and other alfalfa varieties have the lowest sequence homology similarity of trnL-F, with a similarity of 94%. The trnL-F sequence homology of M. falcata and most materials is low, ranging from 94 to 95%.

Alfalfa similarity clustering of trnL-F sequences.
The test materials were clustered using the trnL-F sequence homology similarity of 96% as the classification standard. Among them, the materials (varieties) such as Junggar, M. falcata, Tumu No. 1, Gannong No. 3, and Soviet Union 6220 were grouped into one category, and other varieties were grouped into one category.
The clustering results of trnL-F sequence homology similarity between M. falcata and other test materials did not reach the outgroup (homogeneity) level with the highest sequence similarity. This result reflects that the trnL-F sequence is relatively conserved in alfalfa species such as M. falcata L. and M. sativa L. Comparison of M. falcata trnL-F sequence with other trnL-F sequences showed that ATTT at the 100 bp site and AT at the 116–117 bp site are specific bases and can be used as the basis for the identification of M. falcata L. and M. sativa L. resources.
Figure 5 shows that the similarity of psbA-trnH sequences of 24 alfalfa germplasms was clustered. Analysis suggested that the homology among the alfalfa varieties Bear No. 1, Caoyuan No. 1, Caoyuan No. 2, Gannong No. 1, Gannong No. 2, M. falcata, Longdong, Rumble, Soviet Union 1209, Tumu No. 1, and WL-323, Apollo supreme, Canadian alfalfa and Zhaodong alfalfa, Gannong No. 3, and Soviet Union 6220 alfalfa groups are the highest, all of which are 100%. The psbA-trnH sequence homology similarity between Junggar alfalfa and other alfalfa is 95%. With the psbA-trnH sequence homology similarity of 96% as the classification criterion, the test materials can be grouped into one category except for Junggar alfalfa. The psbA-trnH sequence homology of alfalfa and most alfalfa materials can reach 100%. The average length of the psbA-trnH sequence is 345.6 bp. The sequence is rich in polymorphisms such as single-nucleotide variation sites, parsimony information sites, and insertion and deletion fragments. The psbA-trnH sequencing results can better identify alfalfa variety resources and can be applied to alfalfa variety identification.

Alfalfa similarity clustering of psbA-trnH sequences.
3.5 Sequence comparison of trnL-F and psbA-trnH in alfalfa and DNA barcode formation analysis
The trnL-F sequences of 24 alfalfa varieties (materials) and the psbA-trnH sequences of 23 alfalfa varieties (materials) show sequence differences. The corresponding sequences can form DNA sequence barcodes to identify alfalfa varieties. According to Figures 2 and 3, the psbA-trnH sequence variation sites are abundant for all sequenced alfalfa materials. The trnL-F sequence forms 67 variation sites and 84 psbA-trnH sequences, according to statistics. Table 3 shows that the sequencing fragments are sorted and aligned within a single variety, and the psbA-trnH variation sites of the materials within varieties range from 0 to 5, with an average of 0.79. The trnL-F variant sites range from 0 to 14, averaging 5.09. Among the variation rates of trnL-F and psbA-trnH sequences within varieties, the average content of G + C fragments of trnL-F sequences in each variety is 32.4%. The highest single nucleotide variation rates among varieties are shown in Zhongmu No. 2 and Canadian alfalfa, both with 1.46%. The average G + C content of the psbA-trnH sequence fragment is 18.3%. The highest intravariety variation rate is Czech 26-1 alfalfa, with a variation rate of 4.78%. Based on the comparison results of alfalfa DNA sequences between and within varieties, the DNA identification barcodes of alfalfa psbA-trnH sequences are more abundant in polymorphisms between varieties. In addition, the variation rate within varieties is relatively low, and the identification of alfalfa varieties has formed a clearer DNA barcode sequence.
Basic information of alfalfa varieties psbA-trnH and trnL-F sequences
| Name | psbA-trnH | trnL-F | ||||||
|---|---|---|---|---|---|---|---|---|
| Fragment | G + C | Mutation | Mutation rate | Fragment | G + C | Mutation | Mutation rate | |
| Golden Queen | 341 | 16.72 | 0 | 0 | — | — | — | — |
| Caoyuan No. 1 | 341 | 17.01 | 0 | 0 | 233 | 32.62 | 10 | 4.29 |
| Caoyuan No. 2 | 341 | 17.01 | 0 | 0 | 233 | 32.62 | 10 | 4.29 |
| Apollo supreme | 341 | 19.06 | 0 | 0 | 232 | 30.6 | 8 | 3.45 |
| Gannong No. 1 | 341 | 17.01 | 1 | 0.29 | 231 | 32.47 | 4 | 1.73 |
| Gannong No. 2 | 341 | 17.01 | 2 | 0.59 | 231 | 33.33 | 2 | 0.87 |
| Gannong No. 3 | 341 | 19.06 | 0 | 0 | 230 | 33.04 | 2 | 0.87 |
| Medicago falcata | 349 | 16.91 | 5 | 1.43 | 231 | 33.33 | 1 | 0.43 |
| Canada alfalfa | 341 | 17.6 | 5 | 1.46 | 230 | 33.04 | 5 | 2.17 |
| Czech 26-1 | 345 | 18.26 | 0 | 0 | 230 | 31.74 | 11 | 4.78 |
| Rumble | 341 | 21.11 | 0 | 0 | 230 | 32.17 | 1 | 0.43 |
| Soviet Union 1209 | 341 | 21.41 | 0 | 0 | 229 | 33.19 | 3 | 1.31 |
| Soviet Union 6220 | 341 | 19.35 | 0 | 0 | 230 | 33.04 | 3 | 1.3 |
| Tumu No. 1 | 341 | 21.41 | 0 | 0 | 231 | 31.6 | 4 | 1.73 |
| Xinjiang Daye | 345 | 19.71 | 2 | 0.58 | 229 | 32.31 | 5 | 2.18 |
| Xinmu No. 1 | 343 | 20.99 | 0 | 0 | 231 | 32.03 | 9 | 3.9 |
| Zhaodong | 341 | 18.77 | 0 | 0 | 229 | 34.06 | 3 | 1.31 |
| Zhongmu No. 2 | 341 | 20.53 | 4 | 1.46 | 230 | 32.17 | 4 | 1.74 |
| Junggar | 345 | 18.84 | 0 | 0 | 233 | 33.05 | 0 | 0 |
| AmeriGraze | 341 | 21.41 | 0 | 0 | 231 | 32.03 | 1 | 0.43 |
| BEAR | 342 | 21.05 | 0 | 0 | — | — | — | — |
| WL323 | 341 | 17.3 | 0 | 0 | 232 | 32.76 | 4 | 1.72 |
| Longdong | 341 | 19.94 | 0 | 0 | — | — | — | — |
| UK | 344 | 21.22 | 0 | 0 | — | — | — | — |
| WL320 | — | — | — | — | 222 | 32.88 | 9 | 4.05 |
| Japan 90 | — | — | — | — | 228 | 30.7 | 14 | 6.14 |
| Tianshui | — | — | — | — | 229 | 32.31 | 4 | 1.75 |
4 Discussion
With the innovation and progress of identification technology, fingerprints and DNA barcoding have become the core research fields of identification technology. They are widely used in biological research and are considered the most efficient and accurate technical system for identifying biogenetic characteristics from root genes and biological functional tissues. The quality of alfalfa cultivar resources determines the yield potential and resistance to a large extent. Research on the development of an accurate, reliable, rapid, and simple variety identification method is important for identifying the authenticity of alfalfa varieties, strengthening the construction of the seed quality standard system, and improving the quality of alfalfa seeds. Currently, the identification of alfalfa varieties mainly relies on conventional identification methods. The corresponding fingerprint technology is affected by factors such as the detection object, technology, and equipment, and few standardized detection procedures have been established. Standardized test procedures need to be strengthened. Fingerprints and DNA barcoding have the advantages of rapidity, accuracy, and little impact on environmental pressures in identifying the genetic variation and genetic composition of alfalfa varieties.
Although many scientists have done a lot of research on DNA fingerprints, the identification of alfalfa varieties is still a difficult problem. This article mainly explored the use of ITS2, trnL-F, and psbA-trnH markers to identify alfalfa varieties. We use only 28 genotypes, but the technology we have established can group these alfalfa varieties well. The preliminary verification of these technologies shows that this is of great value for the identification of alfalfa varieties and is worth promoting and exploring.
Ribosomal DNA ITS is a family of genes encoding ribosomal RNA in the nucleus of plant cells [14]. Ribosomal coding region sequences are generally highly conserved, similar to chloroplast-conserved sequences. The corresponding spacer sequence is characterized by a large amount of mutation information, rapid mutation, simple sequences, convenient amplification, high evolution rates, less influence by the external environment, and no functional restrictions. Currently, it is widely used in the study of genetic evolutionary phylogeny at the interspecific and subspecific levels. In reference [15], six Leguminosae forages were tested, and ITS cannot be used as DNA barcoding candidate sequences due to low amplification efficiency. In reference [16], 156 alfalfa populations were tested. The ITS fragments of other species, except for a few relatives, also showed a high ability of species to define. The results of the former are preferred. Comparing the results of different studies revealed that the selected primer fragments differ, showing different results. The screening and identification of primers are critical. Comprehensive analysis showed that the ITS2 primer used in this research needs to be further optimized, and screening should be performed in many species, i.e., to distinguish the DNA barcoding potential of different alfalfa varieties.
Chloroplast gene trnL-F and psbA-trnH sequences are chloroplast spacer gene fragments with high amplification success rate and short purpose fragment length [17]. The chloroplast gene trnL-F and psbA-trnH sequence and nuclear gene ITS sequence marker systems are mainly used in plant phylogeny research, species genetic diversity, and identification of medicinal plants in china. Among them, the systematic classification of medicinal plants and the identification of Chinese medicinal materials are the most widely used, and the corresponding technology tends to be mature. Currently, Rosaceae, Rutaceae, Honeysuckle, Ginseng, Cistanche, Euphorbia, Shegan, Chonglou, and other families, genera, and species have initially established DNA barcodes and fingerprints based on conserved chloroplast sequences [18,19,20,21,22,23,24]. The genetic diversity and molecular phylogenetic classification of medicinal plant germplasm resources are mainly carried out in the families, genera, and species of sage, Schisandra, Trillium, Ophiopogon japonicus, Crow garlic, tulip, and onion [25,26,27,28]. The chloroplast gene trnL-F, psbA-trnH sequence, and the nuclear gene ITS sequence marker systems are rarely reported in forages in China, mainly in Sichuan Agricultural University and Lanzhou University, with more phylogenetic studies in gramineae materials. In reference [7], ITS sequences and psbA-trnH sequences were used for phylogenetic analysis on 37 materials of 6 genera, including Elymus. In reference [8], the psbA-trnH sequence was used to study the genetic diversity of 87 populations of M. japonica. In reference [9], ndhF, psbA-trnH, and trnL-F gene sequences were adopted to study the developmental phylogeny of 67 materials of the genus Gossypium and its relatives. In reference [6], ITS sequences were used to conduct phylogenetic analysis on 41 materials of the genus Laysporium and its relatives. The application of legume and alfalfa mainly includes the classification of pathogen populations, the legume phylogenetic relationships, and species delimitation [29,30,31]. In reference [16], samples representing 21 naturally distributed species in China were collected, and the chloroplast genomes of 75 individuals representing 20 species were assembled. The results showed that 18 species are well delimited except for Medicago sativa, Medicago falcata, and Melilotus officinalis. The primers used in this study verified the feasibility of identifying alfalfa species as DNA barcoding and formed alfalfa variety identification barcoding using psbA-trnH and trnL-F. Compared with the 23 materials used in the study, psbA-trnH sequence identification was clearer and more accurate, and the effect of distinguishing different alfalfa varieties was better. The studies on genetic diversity and variety identification of alfalfa have a one-sided analysis regarding morphological characteristics, agronomic traits, functional proteins, isozymes, and molecular markers. Conservative gene sequences can effectively complement their research shortcomings. To study the new and effective methods of alfalfa genetic diversity, further in-depth related research can be conducted to make the research methods and technical means more suitable for the research of alfalfa genetic diversity and variety identification.
5 Conclusion
In this study, the widely used sequences of ribosomal ITS2 and chloroplast trnL-F and psbA-trnH were used to analyze the genetic diversity and variety identification of alfalfa varieties. The alfalfa ITS2, trnL-F, and psbA-trnH sequences were sequenced to obtain the sequenced fragments with average lengths of 455.7 bp, 230.3 bp, and 345.6 bp, respectively. Among them, the alternative fragments of the ITS2 sequence showed high conservation both within and among varieties. The trnL-F and psbA-trnH sequences differed little within varieties but had obvious differences among varieties. According to the alfalfa similarity clustering of trnL-F and psbA-trnH sequences, the tested alfalfa varieties were clustered into four categories. The similarities of trnL-F and psbA-trnH sequences were different among alfalfa varieties, and the differences were large, indicating that the conserved chloroplast sequences in each variety population were independently evolved. The comparison of the trnL-F and psbA-trnH sequences of alfalfa varieties showed that the psbA-trnH sequence had more variant sites and richer sequence polymorphisms. Thus, the sequence is more suitable for forming clear DNA barcoding for alfalfa variety identification. During alfalfa variety identification and genetic diversity analysis, the ITS2 sequence was too conservative. The initial sequencing results in the previous experiment could not reflect the differences between varieties and individuals within varieties. The corresponding polymorphisms of the chloroplast gene sequences are abundant, which can better reflect the differences between varieties and are more suitable for identifying different alfalfa varieties.
-
Funding information: This work was supported by the Inner Mongolia autonomous region seed industry demonstration projects of major science and technology innovation (high quality alfalfa varieties breeding and industrial demonstration 2022JBGS0020).
-
Author contributions: Wang Y. and Wang M.J. conceived and designed the research. Xu C.B. and Tong L.G. conducted experiments. Zhang X.M. analyzed the data. Wang Y. wrote the paper. All authors read and approve of the manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Ahlgren GH. Forage crops. New York: McGraw-Hill; 1949.Search in Google Scholar
[2] Wang XL. Identification of cold tolerance and screening of cold tolerant germplasm of alfalfa [Doctoral dissertation]. Huhoot: Inner Mongolia Agricultural University; 2021. Chinese.Search in Google Scholar
[3] Xue DY. Status and protection of biological genetic resources in China. Beijing: China Environmental Science Press; 2005. Chinese.Search in Google Scholar
[4] Zhang YJ, Li DZ. Advances in phylogenomics based on complete chloroplast genomes. Plant Diversity Resour. 2011;33(4):365–75. Chinese.Search in Google Scholar
[5] Taberlet P, Gielly L, Pautou G. Universal primers for amplification of three non-codingregions of chloroplast DNA. Plant Mol Biol. 1991;17(5):1105–9.10.1007/BF00037152Search in Google Scholar PubMed
[6] Ding XX, Su HL, Bao S, Tu JJ, Qiu XH, Huang ZH. Molecular identification of datureae plants based on DNA barcoding. Mol Plant Breed 1–14. [2022-12-16]. https://kns.cnki.net/kcms/detail/46.1068.S.20221104.1617.011.html. Chinese.Search in Google Scholar
[7] Xia ZD, LiuX, Feng ML, Xi XH, Tong LG. Chinese traditional and herbal drugs. 2020;51(2):6062–69. Chinese.Search in Google Scholar
[8] Sha LN. Studies on morphology, cytology and molecular phylogenetics in the genus of Leymus [Doctoral dissertation]. Yaan: Sichuan Agricultural University; 2008. Chinese.Search in Google Scholar
[9] Gao G. Phylogenetic and evolution analysis of Elymus in China [Doctoral dissertation]. Yaan: Sichuan Agricultural University; 2015. Chinese.Search in Google Scholar
[10] Xin YR. Genetic diversity analysis of Elymus sibiricus. L based on chloroplast trnL-F and psbA-trnH sequences [Master dissertation]. Yaan: Sichuan Agricultural University; 2018. Chinese.Search in Google Scholar
[11] Lei YX. Phylogenetic and evolution analysis of Roegheria and its related genera (Triticeae; Poaceae) [Master dissertation]. Yaan: Sichuan Agricultural University; 2018. Chinese.Search in Google Scholar
[12] Geng YP, Ma YZ, Wang F, Xie XD, Zhang PF, Liu YL. Identification of Astragali Radix and its adulterants based on psbA-trnH sequence. J Shanxi Agric Sci. 2020;48(5):664–8. Chinese.Search in Google Scholar
[13] Mu ZJ, Zhang YP, Du XL, Huang CS, Yu LL, Wang XY. Identification of leguminous ethnic medicine and its mixed falsify based on the ITS2 sequence. J Chin Med Mater. 2022;45(5):1093–8. Chinese.Search in Google Scholar
[14] Hamby RK, Zimmer EA. Ribosomal RNA as a phylogenetic tool in plant systematics. Molecular systematics of plants. New York: Chapman & Hall; 1992. p. 50–9110.1007/978-1-4615-3276-7_4Search in Google Scholar
[15] Li YQ. Screening of universal DNA barcodes for common forages and establishment of database [Master dissertation]. Lanzhou: Gansu Agricultural University; 2017. Chinese.Search in Google Scholar
[16] Chen JY. Phylogenetic analysis and taxonomic study of Medicago sensu lato in China [Master dissertation]. Lanzhou: Lanzhou University; 2021. Chinese.Search in Google Scholar
[17] Wang CZ, Li P, Ding JY, Peng X, Yuan CS. Simultaneous identification of Bulbus Fritillariae cirrhosae using PCR-RFLP analysis. Phytomedicine. 2007;14:628–32.10.1016/j.phymed.2006.09.008Search in Google Scholar PubMed
[18] Wang CZ, Li P, Ding JY, Yuan CS. Identification of Fritillaria pallidiflora using diagnostic PCR and PCR-RFLP based on nuclear ribosomal DNA internal transcribed spacer sequences. Planta Med. 2005;71(4):384–6.10.1055/s-2005-864112Search in Google Scholar PubMed
[19] Gao T, Yao H, Song JY, Zhu YJ, Liu C, Chen SL. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evolut Biol. 2010;10:324.10.1186/1471-2148-10-324Search in Google Scholar PubMed PubMed Central
[20] Qin MJ, Huang Y, Yang G. RbcL sequence analysis of Belamcahda chihehsis and related medicinal plants of Iris. Acta Pharm Sin. 2003;38(2):147–52. Chinese.Search in Google Scholar
[21] Zhu YJ, Chen SL, Yao H. DNA barcoding the medicinal plants of the genus paris. Acta Pharm Sin. 2010;45(3):376–82. Chinese.Search in Google Scholar
[22] Luo K, Chen SL, Chen KL. Study on plant universal DNA barcode based on Rutaceae. Sci Sin Vitae. 2010;40(4):342–51. Chinese.Search in Google Scholar
[23] Pang XH, Song JY, Zhu YJ. Applying plant DNA barcodes for Rosaceae species identification. Cladistics. 2011;27:165–70.10.1111/j.1096-0031.2010.00328.xSearch in Google Scholar PubMed
[24] Gao T, Yao H, Song JY, Liu C, Zhu YJ, Ma XY, et al. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. Ethnopharmacol. 2010;130:116–21.10.1016/j.jep.2010.04.026Search in Google Scholar PubMed
[25] Wang Y, Li DH, Zhang YT. Analys is of ITS sequences of some med icinal plants and their related species in Salvia. Acta Pharm Sin. 2007;12:1309–13. Chinese.Search in Google Scholar
[26] Wang YH, Zhang SZ, Gao JP. Phylogeny of schisandraceae based on the cpDNA rbcL sequences. J Fudan Univ (Nat Sci). 2003;42(4):550–4. Chinese.Search in Google Scholar
[27] He TH, Rao GY, You RL, Ge S, Zhang DM. Genetic structure and heterozygosity variation between generations of ophiopogon xylorrhizus (Liliaceae s.l.), an endemic species in Yunnan, Southwest China. Biochem Genet. 2001;39(3–4):93–8.Search in Google Scholar
[28] Nguyen NH, Driscoll HE, Specht CD. A molecular phylogeny of the wild onions (Allium; Alliaceae) with a focus on the western North American center of diversity. Mol Phylogenet Evol. 2008;47:1157–72.10.1016/j.ympev.2007.12.006Search in Google Scholar PubMed
[29] Wang XJ, Dong WP, Zhou SL. Analysis alfalfa plant evolution path based on the chloroplast genome in china. Acta Ecol Sin. 2022;42(15):6125–36. Chinese.Search in Google Scholar
[30] Igbari AD, Ogundipe OT. Phylogenetic patterns in the tribe Acacieae (Caesalpinioideae: Fabaceae) based on rbcL, matK, trnL-F and ITS sequence data. Asia Pac J Mol Biol Biotechnol. 2019;27(2):103–15. 10.35118/apjmbb.2019.027.2.13.Search in Google Scholar
[31] Tao XL. The study on the complete chloroplast genome of Medicago sativa and Vicia sativa [Master dissertation]. Lanzhou: Lanzhou University; 2017. Chinese.Search in Google Scholar
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”
Articles in the same Issue
- Biomedical Sciences
- Systemic investigation of inetetamab in combination with small molecules to treat HER2-overexpressing breast and gastric cancers
- Immunosuppressive treatment for idiopathic membranous nephropathy: An updated network meta-analysis
- Identifying two pathogenic variants in a patient with pigmented paravenous retinochoroidal atrophy
- Effects of phytoestrogens combined with cold stress on sperm parameters and testicular proteomics in rats
- A case of pulmonary embolism with bad warfarin anticoagulant effects caused by E. coli infection
- Neutrophilia with subclinical Cushing’s disease: A case report and literature review
- Isoimperatorin alleviates lipopolysaccharide-induced periodontitis by downregulating ERK1/2 and NF-κB pathways
- Immunoregulation of synovial macrophages for the treatment of osteoarthritis
- Novel CPLANE1 c.8948dupT (p.P2984Tfs*7) variant in a child patient with Joubert syndrome
- Antiphospholipid antibodies and the risk of thrombosis in myeloproliferative neoplasms
- Immunological responses of septic rats to combination therapy with thymosin α1 and vitamin C
- High glucose and high lipid induced mitochondrial dysfunction in JEG-3 cells through oxidative stress
- Pharmacological inhibition of the ubiquitin-specific protease 8 effectively suppresses glioblastoma cell growth
- Levocarnitine regulates the growth of angiotensin II-induced myocardial fibrosis cells via TIMP-1
- Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study
- Single-cell transcription analysis reveals the tumor origin and heterogeneity of human bilateral renal clear cell carcinoma
- Identification of iron metabolism-related genes as diagnostic signatures in sepsis by blood transcriptomic analysis
- Long noncoding RNA ACART knockdown decreases 3T3-L1 preadipocyte proliferation and differentiation
- Surgery, adjuvant immunotherapy plus chemotherapy and radiotherapy for primary malignant melanoma of the parotid gland (PGMM): A case report
- Dosimetry comparison with helical tomotherapy, volumetric modulated arc therapy, and intensity-modulated radiotherapy for grade II gliomas: A single‑institution case series
- Soy isoflavone reduces LPS-induced acute lung injury via increasing aquaporin 1 and aquaporin 5 in rats
- Refractory hypokalemia with sexual dysplasia and infertility caused by 17α-hydroxylase deficiency and triple X syndrome: A case report
- Meta-analysis of cancer risk among end stage renal disease undergoing maintenance dialysis
- 6-Phosphogluconate dehydrogenase inhibition arrests growth and induces apoptosis in gastric cancer via AMPK activation and oxidative stress
- Experimental study on the optimization of ANM33 release in foam cells
- Primary retroperitoneal angiosarcoma: A case report
- Metabolomic analysis-identified 2-hydroxybutyric acid might be a key metabolite of severe preeclampsia
- Malignant pleural effusion diagnosis and therapy
- Effect of spaceflight on the phenotype and proteome of Escherichia coli
- Comparison of immunotherapy combined with stereotactic radiotherapy and targeted therapy for patients with brain metastases: A systemic review and meta-analysis
- Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation
- Association between the VEGFR-2 -604T/C polymorphism (rs2071559) and type 2 diabetic retinopathy
- The role of IL-31 and IL-34 in the diagnosis and treatment of chronic periodontitis
- Triple-negative mouse breast cancer initiating cells show high expression of beta1 integrin and increased malignant features
- mNGS facilitates the accurate diagnosis and antibiotic treatment of suspicious critical CNS infection in real practice: A retrospective study
- The apatinib and pemetrexed combination has antitumor and antiangiogenic effects against NSCLC
- Radiotherapy for primary thyroid adenoid cystic carcinoma
- Design and functional preliminary investigation of recombinant antigen EgG1Y162–EgG1Y162 against Echinococcus granulosus
- Effects of losartan in patients with NAFLD: A meta-analysis of randomized controlled trial
- Bibliometric analysis of METTL3: Current perspectives, highlights, and trending topics
- Performance comparison of three scaling algorithms in NMR-based metabolomics analysis
- PI3K/AKT/mTOR pathway and its related molecules participate in PROK1 silence-induced anti-tumor effects on pancreatic cancer
- The altered expression of cytoskeletal and synaptic remodeling proteins during epilepsy
- Effects of pegylated recombinant human granulocyte colony-stimulating factor on lymphocytes and white blood cells of patients with malignant tumor
- Prostatitis as initial manifestation of Chlamydia psittaci pneumonia diagnosed by metagenome next-generation sequencing: A case report
- NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2
- Association of interleukin-10 rs1800896, rs1800872, and interleukin-6 rs1800795 polymorphisms with squamous cell carcinoma risk: A meta-analysis
- Exosomal HBV-DNA for diagnosis and treatment monitoring of chronic hepatitis B
- Shear stress leads to the dysfunction of endothelial cells through the Cav-1-mediated KLF2/eNOS/ERK signaling pathway under physiological conditions
- Interaction between the PI3K/AKT pathway and mitochondrial autophagy in macrophages and the leukocyte count in rats with LPS-induced pulmonary infection
- Meta-analysis of the rs231775 locus polymorphism in the CTLA-4 gene and the susceptibility to Graves’ disease in children
- Cloning, subcellular localization and expression of phosphate transporter gene HvPT6 of hulless barley
- Coptisine mitigates diabetic nephropathy via repressing the NRLP3 inflammasome
- Significant elevated CXCL14 and decreased IL-39 levels in patients with tuberculosis
- Whole-exome sequencing applications in prenatal diagnosis of fetal bowel dilatation
- Gemella morbillorum infective endocarditis: A case report and literature review
- An unusual ectopic thymoma clonal evolution analysis: A case report
- Severe cumulative skin toxicity during toripalimab combined with vemurafenib following toripalimab alone
- Detection of V. vulnificus septic shock with ARDS using mNGS
- Novel rare genetic variants of familial and sporadic pulmonary atresia identified by whole-exome sequencing
- The influence and mechanistic action of sperm DNA fragmentation index on the outcomes of assisted reproduction technology
- Novel compound heterozygous mutations in TELO2 in an infant with You-Hoover-Fong syndrome: A case report and literature review
- ctDNA as a prognostic biomarker in resectable CLM: Systematic review and meta-analysis
- Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: A case report
- Phylogenetic analysis of promoter regions of human Dolichol kinase (DOLK) and orthologous genes using bioinformatics tools
- Collagen changes in rabbit conjunctiva after conjunctival crosslinking
- Effects of NM23 transfection of human gastric carcinoma cells in mice
- Oral nifedipine and phytosterol, intravenous nicardipine, and oral nifedipine only: Three-arm, retrospective, cohort study for management of severe preeclampsia
- Case report of hepatic retiform hemangioendothelioma: A rare tumor treated with ultrasound-guided microwave ablation
- Curcumin induces apoptosis in human hepatocellular carcinoma cells by decreasing the expression of STAT3/VEGF/HIF-1α signaling
- Rare presentation of double-clonal Waldenström macroglobulinemia with pulmonary embolism: A case report
- Giant duplication of the transverse colon in an adult: A case report and literature review
- Ectopic thyroid tissue in the breast: A case report
- SDR16C5 promotes proliferation and migration and inhibits apoptosis in pancreatic cancer
- Vaginal metastasis from breast cancer: A case report
- Screening of the best time window for MSC transplantation to treat acute myocardial infarction with SDF-1α antibody-loaded targeted ultrasonic microbubbles: An in vivo study in miniswine
- Inhibition of TAZ impairs the migration ability of melanoma cells
- Molecular complexity analysis of the diagnosis of Gitelman syndrome in China
- Effects of maternal calcium and protein intake on the development and bone metabolism of offspring mice
- Identification of winter wheat pests and diseases based on improved convolutional neural network
- Ultra-multiplex PCR technique to guide treatment of Aspergillus-infected aortic valve prostheses
- Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- Immune checkpoint inhibitors in cancer patients with COVID-19
- Utility of methylene blue mixed with autologous blood in preoperative localization of pulmonary nodules and masses
- Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma
- Berberine suppressed sarcopenia insulin resistance through SIRT1-mediated mitophagy
- DUSP2 inhibits the progression of lupus nephritis in mice by regulating the STAT3 pathway
- Lung abscess by Fusobacterium nucleatum and Streptococcus spp. co-infection by mNGS: A case series
- Genetic alterations of KRAS and TP53 in intrahepatic cholangiocarcinoma associated with poor prognosis
- Granulomatous polyangiitis involving the fourth ventricle: Report of a rare case and a literature review
- Studying infant mortality: A demographic analysis based on data mining models
- Metaplastic breast carcinoma with osseous differentiation: A report of a rare case and literature review
- Protein Z modulates the metastasis of lung adenocarcinoma cells
- Inhibition of pyroptosis and apoptosis by capsaicin protects against LPS-induced acute kidney injury through TRPV1/UCP2 axis in vitro
- TAK-242, a toll-like receptor 4 antagonist, against brain injury by alleviates autophagy and inflammation in rats
- Primary mediastinum Ewing’s sarcoma with pleural effusion: A case report and literature review
- Association of ADRB2 gene polymorphisms and intestinal microbiota in Chinese Han adolescents
- Tanshinone IIA alleviates chondrocyte apoptosis and extracellular matrix degeneration by inhibiting ferroptosis
- Study on the cytokines related to SARS-Cov-2 in testicular cells and the interaction network between cells based on scRNA-seq data
- Effect of periostin on bone metabolic and autophagy factors during tooth eruption in mice
- HP1 induces ferroptosis of renal tubular epithelial cells through NRF2 pathway in diabetic nephropathy
- Intravaginal estrogen management in postmenopausal patients with vaginal squamous intraepithelial lesions along with CO2 laser ablation: A retrospective study
- Hepatocellular carcinoma cell differentiation trajectory predicts immunotherapy, potential therapeutic drugs, and prognosis of patients
- Effects of physical exercise on biomarkers of oxidative stress in healthy subjects: A meta-analysis of randomized controlled trials
- Identification of lysosome-related genes in connection with prognosis and immune cell infiltration for drug candidates in head and neck cancer
- Development of an instrument-free and low-cost ELISA dot-blot test to detect antibodies against SARS-CoV-2
- Research progress on gas signal molecular therapy for Parkinson’s disease
- Adiponectin inhibits TGF-β1-induced skin fibroblast proliferation and phenotype transformation via the p38 MAPK signaling pathway
- The G protein-coupled receptor-related gene signatures for predicting prognosis and immunotherapy response in bladder urothelial carcinoma
- α-Fetoprotein contributes to the malignant biological properties of AFP-producing gastric cancer
- CXCL12/CXCR4/CXCR7 axis in placenta tissues of patients with placenta previa
- Association between thyroid stimulating hormone levels and papillary thyroid cancer risk: A meta-analysis
- Significance of sTREM-1 and sST2 combined diagnosis for sepsis detection and prognosis prediction
- Diagnostic value of serum neuroactive substances in the acute exacerbation of chronic obstructive pulmonary disease complicated with depression
- Research progress of AMP-activated protein kinase and cardiac aging
- TRIM29 knockdown prevented the colon cancer progression through decreasing the ubiquitination levels of KRT5
- Cross-talk between gut microbiota and liver steatosis: Complications and therapeutic target
- Metastasis from small cell lung cancer to ovary: A case report
- The early diagnosis and pathogenic mechanisms of sepsis-related acute kidney injury
- The effect of NK cell therapy on sepsis secondary to lung cancer: A case report
- Erianin alleviates collagen-induced arthritis in mice by inhibiting Th17 cell differentiation
- Loss of ACOX1 in clear cell renal cell carcinoma and its correlation with clinical features
- Signalling pathways in the osteogenic differentiation of periodontal ligament stem cells
- Crosstalk between lactic acid and immune regulation and its value in the diagnosis and treatment of liver failure
- Clinicopathological features and differential diagnosis of gastric pleomorphic giant cell carcinoma
- Traumatic brain injury and rTMS-ERPs: Case report and literature review
- Extracellular fibrin promotes non-small cell lung cancer progression through integrin β1/PTEN/AKT signaling
- Knockdown of DLK4 inhibits non-small cell lung cancer tumor growth by downregulating CKS2
- The co-expression pattern of VEGFR-2 with indicators related to proliferation, apoptosis, and differentiation of anagen hair follicles
- Inflammation-related signaling pathways in tendinopathy
- CD4+ T cell count in HIV/TB co-infection and co-occurrence with HL: Case report and literature review
- Clinical analysis of severe Chlamydia psittaci pneumonia: Case series study
- Bioinformatics analysis to identify potential biomarkers for the pulmonary artery hypertension associated with the basement membrane
- Influence of MTHFR polymorphism, alone or in combination with smoking and alcohol consumption, on cancer susceptibility
- Catharanthus roseus (L.) G. Don counteracts the ampicillin resistance in multiple antibiotic-resistant Staphylococcus aureus by downregulation of PBP2a synthesis
- Combination of a bronchogenic cyst in the thoracic spinal canal with chronic myelocytic leukemia
- Bacterial lipoprotein plays an important role in the macrophage autophagy and apoptosis induced by Salmonella typhimurium and Staphylococcus aureus
- TCL1A+ B cells predict prognosis in triple-negative breast cancer through integrative analysis of single-cell and bulk transcriptomic data
- Ezrin promotes esophageal squamous cell carcinoma progression via the Hippo signaling pathway
- Ferroptosis: A potential target of macrophages in plaque vulnerability
- Predicting pediatric Crohn's disease based on six mRNA-constructed risk signature using comprehensive bioinformatic approaches
- Applications of genetic code expansion and photosensitive UAAs in studying membrane proteins
- HK2 contributes to the proliferation, migration, and invasion of diffuse large B-cell lymphoma cells by enhancing the ERK1/2 signaling pathway
- IL-17 in osteoarthritis: A narrative review
- Circadian cycle and neuroinflammation
- Probiotic management and inflammatory factors as a novel treatment in cirrhosis: A systematic review and meta-analysis
- Hemorrhagic meningioma with pulmonary metastasis: Case report and literature review
- SPOP regulates the expression profiles and alternative splicing events in human hepatocytes
- Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway
- PTX3 promotes IVIG resistance-induced endothelial injury in Kawasaki disease by regulating the NF-κB pathway
- Pancreatic ectopic thyroid tissue: A case report and analysis of literature
- The prognostic impact of body mass index on female breast cancer patients in underdeveloped regions of northern China differs by menopause status and tumor molecular subtype
- Report on a case of liver-originating malignant melanoma of unknown primary
- Case report: Herbal treatment of neutropenic enterocolitis after chemotherapy for breast cancer
- The fibroblast growth factor–Klotho axis at molecular level
- Characterization of amiodarone action on currents in hERG-T618 gain-of-function mutations
- A case report of diagnosis and dynamic monitoring of Listeria monocytogenes meningitis with NGS
- Effect of autologous platelet-rich plasma on new bone formation and viability of a Marburg bone graft
- Small breast epithelial mucin as a useful prognostic marker for breast cancer patients
- Continuous non-adherent culture promotes transdifferentiation of human adipose-derived stem cells into retinal lineage
- Nrf3 alleviates oxidative stress and promotes the survival of colon cancer cells by activating AKT/BCL-2 signal pathway
- Favorable response to surufatinib in a patient with necrolytic migratory erythema: A case report
- Case report of atypical undernutrition of hypoproteinemia type
- Down-regulation of COL1A1 inhibits tumor-associated fibroblast activation and mediates matrix remodeling in the tumor microenvironment of breast cancer
- Sarcoma protein kinase inhibition alleviates liver fibrosis by promoting hepatic stellate cells ferroptosis
- Research progress of serum eosinophil in chronic obstructive pulmonary disease and asthma
- Clinicopathological characteristics of co-existing or mixed colorectal cancer and neuroendocrine tumor: Report of five cases
- Role of menopausal hormone therapy in the prevention of postmenopausal osteoporosis
- Precisional detection of lymph node metastasis using tFCM in colorectal cancer
- Advances in diagnosis and treatment of perimenopausal syndrome
- A study of forensic genetics: ITO index distribution and kinship judgment between two individuals
- Acute lupus pneumonitis resembling miliary tuberculosis: A case-based review
- Plasma levels of CD36 and glutathione as biomarkers for ruptured intracranial aneurysm
- Fractalkine modulates pulmonary angiogenesis and tube formation by modulating CX3CR1 and growth factors in PVECs
- Novel risk prediction models for deep vein thrombosis after thoracotomy and thoracoscopic lung cancer resections, involving coagulation and immune function
- Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms
- Evaluation of effects of small-incision approach treatment on proximal tibia fracture by deep learning algorithm-based magnetic resonance imaging
- An online diagnosis method for cancer lesions based on intelligent imaging analysis
- Medical imaging in rheumatoid arthritis: A review on deep learning approach
- Predictive analytics in smart healthcare for child mortality prediction using a machine learning approach
- Utility of neutrophil–lymphocyte ratio and platelet–lymphocyte ratio in predicting acute-on-chronic liver failure survival
- A biomedical decision support system for meta-analysis of bilateral upper-limb training in stroke patients with hemiplegia
- TNF-α and IL-8 levels are positively correlated with hypobaric hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats
- Stochastic gradient descent optimisation for convolutional neural network for medical image segmentation
- Comparison of the prognostic value of four different critical illness scores in patients with sepsis-induced coagulopathy
- Application and teaching of computer molecular simulation embedded technology and artificial intelligence in drug research and development
- Hepatobiliary surgery based on intelligent image segmentation technology
- Value of brain injury-related indicators based on neural network in the diagnosis of neonatal hypoxic-ischemic encephalopathy
- Analysis of early diagnosis methods for asymmetric dementia in brain MR images based on genetic medical technology
- Early diagnosis for the onset of peri-implantitis based on artificial neural network
- Clinical significance of the detection of serum IgG4 and IgG4/IgG ratio in patients with thyroid-associated ophthalmopathy
- Forecast of pain degree of lumbar disc herniation based on back propagation neural network
- SPA-UNet: A liver tumor segmentation network based on fused multi-scale features
- Systematic evaluation of clinical efficacy of CYP1B1 gene polymorphism in EGFR mutant non-small cell lung cancer observed by medical image
- Rehabilitation effect of intelligent rehabilitation training system on hemiplegic limb spasms after stroke
- A novel approach for minimising anti-aliasing effects in EEG data acquisition
- ErbB4 promotes M2 activation of macrophages in idiopathic pulmonary fibrosis
- Clinical role of CYP1B1 gene polymorphism in prediction of postoperative chemotherapy efficacy in NSCLC based on individualized health model
- Lung nodule segmentation via semi-residual multi-resolution neural networks
- Evaluation of brain nerve function in ICU patients with Delirium by deep learning algorithm-based resting state MRI
- A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis
- Markov model combined with MR diffusion tensor imaging for predicting the onset of Alzheimer’s disease
- Effectiveness of the treatment of depression associated with cancer and neuroimaging changes in depression-related brain regions in patients treated with the mediator-deuterium acupuncture method
- Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis
- Monitoring and evaluation of anesthesia depth status data based on neuroscience
- Exploring the conformational dynamics and thermodynamics of EGFR S768I and G719X + S768I mutations in non-small cell lung cancer: An in silico approaches
- Optimised feature selection-driven convolutional neural network using gray level co-occurrence matrix for detection of cervical cancer
- Incidence of different pressure patterns of spinal cerebellar ataxia and analysis of imaging and genetic diagnosis
- Pathogenic bacteria and treatment resistance in older cardiovascular disease patients with lung infection and risk prediction model
- Adoption value of support vector machine algorithm-based computed tomography imaging in the diagnosis of secondary pulmonary fungal infections in patients with malignant hematological disorders
- From slides to insights: Harnessing deep learning for prognostic survival prediction in human colorectal cancer histology
- Ecology and Environmental Science
- Monitoring of hourly carbon dioxide concentration under different land use types in arid ecosystem
- Comparing the differences of prokaryotic microbial community between pit walls and bottom from Chinese liquor revealed by 16S rRNA gene sequencing
- Effects of cadmium stress on fruits germination and growth of two herbage species
- Bamboo charcoal affects soil properties and bacterial community in tea plantations
- Optimization of biogas potential using kinetic models, response surface methodology, and instrumental evidence for biodegradation of tannery fleshings during anaerobic digestion
- Understory vegetation diversity patterns of Platycladus orientalis and Pinus elliottii communities in Central and Southern China
- Studies on macrofungi diversity and discovery of new species of Abortiporus from Baotianman World Biosphere Reserve
- Food Science
- Effect of berrycactus fruit (Myrtillocactus geometrizans) on glutamate, glutamine, and GABA levels in the frontal cortex of rats fed with a high-fat diet
- Guesstimate of thymoquinone diversity in Nigella sativa L. genotypes and elite varieties collected from Indian states using HPTLC technique
- Analysis of bacterial community structure of Fuzhuan tea with different processing techniques
- Untargeted metabolomics reveals sour jujube kernel benefiting the nutritional value and flavor of Morchella esculenta
- Mycobiota in Slovak wine grapes: A case study from the small Carpathians wine region
- Elemental analysis of Fadogia ancylantha leaves used as a nutraceutical in Mashonaland West Province, Zimbabwe
- Microbiological transglutaminase: Biotechnological application in the food industry
- Influence of solvent-free extraction of fish oil from catfish (Clarias magur) heads using a Taguchi orthogonal array design: A qualitative and quantitative approach
- Chromatographic analysis of the chemical composition and anticancer activities of Curcuma longa extract cultivated in Palestine
- The potential for the use of leghemoglobin and plant ferritin as sources of iron
- Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM
- Bioengineering and Biotechnology
- Biocompatibility and osteointegration capability of β-TCP manufactured by stereolithography 3D printing: In vitro study
- Clinical characteristics and the prognosis of diabetic foot in Tibet: A single center, retrospective study
- Agriculture
- Biofertilizer and NPSB fertilizer application effects on nodulation and productivity of common bean (Phaseolus vulgaris L.) at Sodo Zuria, Southern Ethiopia
- On correlation between canopy vegetation and growth indexes of maize varieties with different nitrogen efficiencies
- Exopolysaccharides from Pseudomonas tolaasii inhibit the growth of Pleurotus ostreatus mycelia
- A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- Melatonin enhances salt tolerance in sorghum by modulating photosynthetic performance, osmoregulation, antioxidant defense, and ion homeostasis
- Effects of plant density on alfalfa (Medicago sativa L.) seed yield in western Heilongjiang areas
- Identification of rice leaf diseases and deficiency disorders using a novel DeepBatch technique
- Artificial intelligence and internet of things oriented sustainable precision farming: Towards modern agriculture
- Animal Sciences
- Effect of ketogenic diet on exercise tolerance and transcriptome of gastrocnemius in mice
- Combined analysis of mRNA–miRNA from testis tissue in Tibetan sheep with different FecB genotypes
- Isolation, identification, and drug resistance of a partially isolated bacterium from the gill of Siniperca chuatsi
- Tracking behavioral changes of confined sows from the first mating to the third parity
- The sequencing of the key genes and end products in the TLR4 signaling pathway from the kidney of Rana dybowskii exposed to Aeromonas hydrophila
- Development of a new candidate vaccine against piglet diarrhea caused by Escherichia coli
- Plant Sciences
- Crown and diameter structure of pure Pinus massoniana Lamb. forest in Hunan province, China
- Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources
- Tissue culture and rapid propagation technology for Gentiana rhodantha
- Effects of cadmium on the synthesis of active ingredients in Salvia miltiorrhiza
- Cloning and expression analysis of VrNAC13 gene in mung bean
- Chlorate-induced molecular floral transition revealed by transcriptomes
- Effects of warming and drought on growth and development of soybean in Hailun region
- Effects of different light conditions on transient expression and biomass in Nicotiana benthamiana leaves
- Comparative analysis of the rhizosphere microbiome and medicinally active ingredients of Atractylodes lancea from different geographical origins
- Distinguish Dianthus species or varieties based on chloroplast genomes
- Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury
- Study on fresh processing key technology and quality influence of Cut Ophiopogonis Radix based on multi-index evaluation
- An advanced approach for fig leaf disease detection and classification: Leveraging image processing and enhanced support vector machine methodology
- Erratum
- Erratum to “Protein Z modulates the metastasis of lung adenocarcinoma cells”
- Erratum to “BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells”
- Retraction
- Retraction to “Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats”

