Abstract
Background
The objective of this research was to investigate the involvement of RAB39B in acute myeloid leukemia (AML) using bioinformatics analysis and in vitro experiments for validation.
Methods
In this article, RNA sequencing data from The Cancer Genome Atlas and genotype-tissue expression were utilized to analyze the expression of RAB39BA and identify differentially expressed genes.
Results
AML exhibited elevated expression of RAB39B in diverse tumor types. In laboratory experiments, it has been demonstrated that RAB39B exhibits a significant expression level in AML cell lines when compared to normal peripheral blood monocytes. Moreover, RAB39B is closely linked to the growth and programmed cell death of AML cells.
Conclusion
In conclusion, RAB39B shows potential as a biomarker for the identification and prediction of AML, contributing to the growth and cell death processes in AML.
1 Introduction
AML, also known as acute myeloid leukemia, is a cancerous tumor characterized by its significant diversity, unpredictable outlook, and elevated fatality rate. At present, the primary factors that determine risk stratification and treatment options primarily consist of cytogenetic and molecular abnormalities [1,2]. Gene mutations occurring during the processes of cell proliferation, differentiation, and apoptosis serve as the foundation for the development of AML. Nevertheless, the development of AML is highly intricate and has yet to be precisely understood. The current targeted drugs or conventional combination chemotherapy are still not effective enough for treating AML, with the overall survival (OS) rate for AML patients still hovering at a low level, below 30% at the 5-year survival rate [3]. A significant contributor to this poor prognosis is the profound heterogeneity exhibited by AML in terms of its biological characteristics and clinical manifestations [4].
Over the past decade, extensive research has unraveled a lot of genetic mutations, chromosomal abnormalities, and gene expression patterns in AML [5]. These studies have highlighted the profound impact of specific genetic alterations on patient outcomes. Mutations in NPM1 or CEBPA are frequently observed in AML and are generally indicative of a favorable prognosis, potentially slowing disease progression by modulating processes such as cellular differentiation and apoptosis [6,7]. However, it is important to note that not all AML patients harbor these mutations. Moreover, even among those who do, there can be significant variability in clinical courses and treatment outcomes. Conversely, mutations in genes such as FLT3, IDH1/IDH2, RUNX1, and TP53 are not only associated with distinct AML subtypes but also correlate with adverse prognosis and influence patient responsiveness to therapy [7,8,9,10]. Despite the remarkable advancements in AML biomarker research, current cytogenetic and molecular markers still fall short in fully elucidating the complex biological underpinnings and clinical heterogeneity of this disease. Hence, the discovery and recognition of novel biomarkers can enhance our comprehension of the molecular processes involved in AML development and facilitate the creation of personalized diagnostic and therapeutic strategies. This, in turn, could significantly contribute to the management, prognostic categorization, and advancement of targeted medications for AML [11,12].
Rab proteins, small GTPases, regulate the movement of vesicles in the cells of eukaryotic organisms. Rab39B belongs to the Rab protein family. The expression of Rab39B can be observed in different tissues of humans. It is situated on the Xq28 chromosome and is composed of two exons that cover a length of 3764 base pairs in the genomic DNA of humans [13]. The primary emphasis of previous research on Rab39B has been on its involvement in neurodevelopmental abnormalities, such as Parkinson’s disease, cognitive impairment, and autism spectrum disorder [14,15]. According to reports, cancer researchers have found a high expression of Rab39B in germ cell tumors, gastric stromal tumors, and diffuse large B-cell lymphomas [16,17,18]. Additional research has indicated that Rab39B is linked to the tumor microenvironment and the outlook of pancreatic adenocarcinoma [19]. Furthermore, RAB39B is believed to be involved in regulating the PI3K/Akt/mTOR signaling pathway to affect cell behavior [20]. Apoptosis and autophagy pathways have been linked to Rab39B in recent research [21,22]. Emerging evidence also highlights Rab39B’s potential role in intracellular vesicle trafficking and signaling pathways related to cancer cell proliferation, migration, and invasion, suggesting it may represent a novel therapeutic target [16,23,24]. Inhibiting Rab39B’s function or expression could disrupt these crucial processes, offering a new thought for AML treatment [16]. Nevertheless, the precise molecular mechanisms linked to Rab39B remain undisclosed, particularly in relation to mechanisms related to tumors. Until now, there has been a lack of research on the molecular mechanism, biological role, and future outlook of Rab39B in AML.
Through the examination of information obtained from accessible databases, we assessed the variation in RAB39B expression in both pan-cancer and AML. To comprehend the potential roles of RAB39B in AML, we employed protein‒protein interactions (PPIs), coexpressed genes, immune infiltration, m6A-related genes, cuproptosis-related genes, and ceRNA networks. Furthermore, we employed multidimensional analysis to assess the autonomous predictive significance of RAB39B in AML. Additionally, we performed in vitro assays to confirm the distinct manifestation of RAB39B and assess its impact on the growth and programmed cell death of AML cells.
So far, research on the expression, prognosis, and mechanism of RAB39B in AML is still blank. Our study provides the possibility for RAB39B as a new biomarker for AML and establishes sufficient scientific evidence for clinical decision-making and risk management.
2 Materials and methods
2.1 Analysis of biological information and sources of data
The Cancer Genome Atlas (TCGA) and genotype-tissue expression (GTEx) pan-cancer RNA-seq data were uniformly processed from UCSC XENA (https://xenabrowser.net/datapages/). The data of AML samples from HTSeq FPKM and HTSeq Count were acquired from TCGA platform, whereas the RNA-seq data from GTEx, encompassing normal tissue samples, were utilized to facilitate a comparative analysis of RAB39B expression levels between AML and normal samples for subsequent analysis [25,26].
2.2 LinkedOmics analysis
The LinkedOmics database, accessible at http://www.linkedomics.org/login.php, houses a comprehensive collection of multiomics and clinical data derived from 32 different cancer types as part of TCGA project [27]. Using the Link Finder module of Linked Omics, we rigorously conducted a search for differentially expressed genes (DEGs) that are associated with RAB39B in AML (n = 150). The Pearson correlation coefficient was utilized to perform statistical analysis on genes coexpressed with RAB39B, and the corresponding outcomes were presented using volcano plots and heatmaps. The LinkInterpreter module was utilized to perform Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis to annotate genes that were coexpressed with RAB39B.
2.3 Search Tool for the Retrieval of Interacting Genes (STRING) analysis
The purpose of STRING (https://cn.string-db.org/) was to combine all identified and anticipated connections among proteins, encompassing both physical interactions and functional associations [28]. Using the STRING database, we performed an analysis and built a PPI network for RAB39B in this research.
2.4 Construction of the ceRNA network
starBase is a freely available platform that systematically investigates RNA and protein RNA interaction networks through the analysis of 37 CLIP-seq datasets, including PAR CLIP, HITS CLIP, iCLIP, and CLASH [29]. starBase was utilized to predict the prospective miRNA of RAB39B. Screen for shared miRNAs predicted by PITA, miRmap, and TargetScan. Furthermore, we examined the association between the expression of these potential medications and RAB39B to identify miRNAs that are better suited for ceRNA circumstances.
MiRNet2.0 (www.mirnet.ca/miRNet/home.xhtml) is a web-based tool that provides a comprehensive and interactive analysis platform for exploring miRNA‒target interactions. Visit the homepage of ca/miRNet.xhtml) is a network visualization analysis platform centered on miRNA [30]. Using miRNet2.0, we built a correlation network to predict the target lncRNAs for miR-152-3p and miR-582-5p. We also screened lncRNAs that had a more significant correlation with miRNAs. Ultimately, the ceRNA network for DLBCL was constructed by utilizing the negative correlation between the expression levels of miRNA mRNA and miRNA lncRNA, with the key lncRNA miRNA mRNA (RAB39B) being included.
2.5 Analysis of immune infiltration
Conduct immune infiltration analysis utilizing single sample gene set enrichment analysis (ssGSEA) for single sample gene set enrichment analysis. The immune infiltration status of the corresponding cloud data was calculated using the markers of 24 immune cells provided in the Immunity article [31], utilizing the ssGSEA algorithm from the GSVA package [1.46.0] of R (4.2.1). Investigation of genes associated with cuproptosis and genes related to m6A methylation. The RNAseq data (level 3) and corresponding clinical information of AML tumors were acquired from TCGA dataset available at https//portal.gdc.com. Subsequently, the R software package pheatmap was utilized to construct the multigene correlation map. We examined the correlation between RAB39B and m6A-associated genes, along with genes related to cuproptosis [32,33]. Spearman’s correlation analysis was employed to depict the association among numerical variables lacking a normal distribution, and P values below 0.05 were deemed statistically significant.
2.6 Prognostic correlation analysis
The pROC package was utilized for receiver operating characteristic (ROC) analysis of the data, and the diagnostic ROC curve was then visualized using ggplot2. To investigate the prognostic significance of RAB39B in AML, Kaplan‒Meier curves were generated using GEPIA2 (http//gepia2.cancer-pku.cn/) [34]. Proportional hazards hypothesis testing and Cox regression analysis were conducted using the survival package. Additionally, the rms package was utilized to create nomogram-related models, visualize them, and generate nomograms and calibration plots. A calibration curve can be evaluated graphically by mapping the predicted probability of the nomogram to the observed rate, with the 45° line indicating the best value predicted.
2.7 Cell culture
OCI-AML-3, KG-1, MOLM-13, and MV4-11, which are human AML cell lines, were acquired from Cell Biotechnology (Shanghai) Co., Ltd. and cultured following the suggested procedure. According to previous research [35], AML cells were transfected with siRNA (sense 3′-UCAUUCUUCAGAAGAGGUUTT-5′; antisense 3′-AACCUCUUCUGAAGAAUGATT-3000′).
2.8 Quantitative polymerase chain reaction performed in real time
Quantitative PCR (qPCR) was used to analyze the expression of the AML cell lines OCI-AML-3, KG-1, MOLM-13, and MV4-11. The RAB39B primers used were 5′-CTGGGATACAGCGGGTCAAG-3′ (forward primer) and 5′-GAAGGACCTGCGGTTGGTAA-3′ (reverse primer) [35].
2.9 Western blot
The collected cells were mixed with lysis buffer and protein inhibitor, followed by lysis on ice for 30 min. Subsequently, centrifugation at a speed of 12,000 rpm was performed for 15 min. The liquid above the sediment was collected, and a specific quantity of 5 times concentrated sample buffer was introduced. The specimen was subjected to a temperature of 100°C for a duration of 10 min. The protein concentration was measured utilizing a Denovix ultra micro ultraviolet‒visible spectrophotometer. The protein gel was transferred onto a PVDF membrane, followed by blocking with 5% skim milk for a duration of 2 h. Subsequently, the membrane was incubated overnight with the primary antibody (Catalog Number.12162-1-AP, Proteintech) at 4°C. The membrane was exposed to the secondary antibody (ZB2301, Beijing Zhongshan) at ambient temperature for a duration of 60 min. Photographs were taken using a Tanon fully automatic chemiluminescence imaging system, and the grayscale values were read using the system’s analysis software. The trial was conducted three times.
2.10 Cell proliferation detection
Cells were placed in 96-well plates and incubated for the necessary duration to conduct the cell proliferation assay. The experimental group was treated with CCK-8 reagent (Beijing Suo Laibao Technology Co., Ltd.) by adding it to each well. The treated wells were then placed in a 37°C, 5% CO2 incubator and incubated for a duration of 2 h. A microplate reader was used to measure the OD value of the solution at a wavelength of 450 nm.
2.11 Flow cytometry
To analyze apoptosis, we utilized a kit from Hangzhou Lianke Biotechnology Co., Ltd., to detect cell apoptosis.
The cells were rinsed with PBS. Then, trypsin (without EDTA) was introduced for cell digestion. Next, the culture medium was added to halt the reaction and prevent excessive digestion. After that, centrifugation was performed to eliminate the supernatant. The cells were washed with cold PBS, and the cell concentration was adjusted to 1 × 106/mL. Centrifuge at 1,500 rpm for 5 min and discard the supernatant. Next, 500 µL of 1× binding buffer working solution was added. Gently resuspend the cells and introduce 5 µL of ANNEXIN V-FITC and 10 µL of PI. Prepare separate staining tubes and a blank tube for the cells. The blank tube should not contain any staining agent, while the two single staining tubes should have 5 µL of ANNEXIN V-FITC and 10 µL of PI. Following a gentle swirling motion, it was allowed to sit in a dimly lit area at ambient temperature for a duration of 5 min.
The Beckman FC500 flow cytometry instrument was utilized to load the device, calibrate the voltage of the empty tube, adjust the compensation of the tube with single staining, configure the parameters, and upload the sample tube for data analysis.
2.12 Statistical analysis
R (4.2.1) was used for the analysis and visualization of all statistical data and graphics. The chi-square test, Fisher’s exact test, Kruskal‒Wallis test, and Wilcoxon signed-rank test were utilized for the analysis of clinical information. Statistically significant results were defined as P values <0.05 in all analyses.
3 Results
3.1 Expression of RAB39B in both pan-cancer and AML
The information obtained from UCSC XENA undergoes a standardized Tour process, where it is transformed into RNAseq data in the TPM format of TCGA and GTEx. A notable variation in RAB39 expression was observed among 26 cancer types (Figure 1a), including AML (LAML), when comparing the expression of RAB39B in normal and tumor samples from TCGA and GTEx databases (Figure 1b).
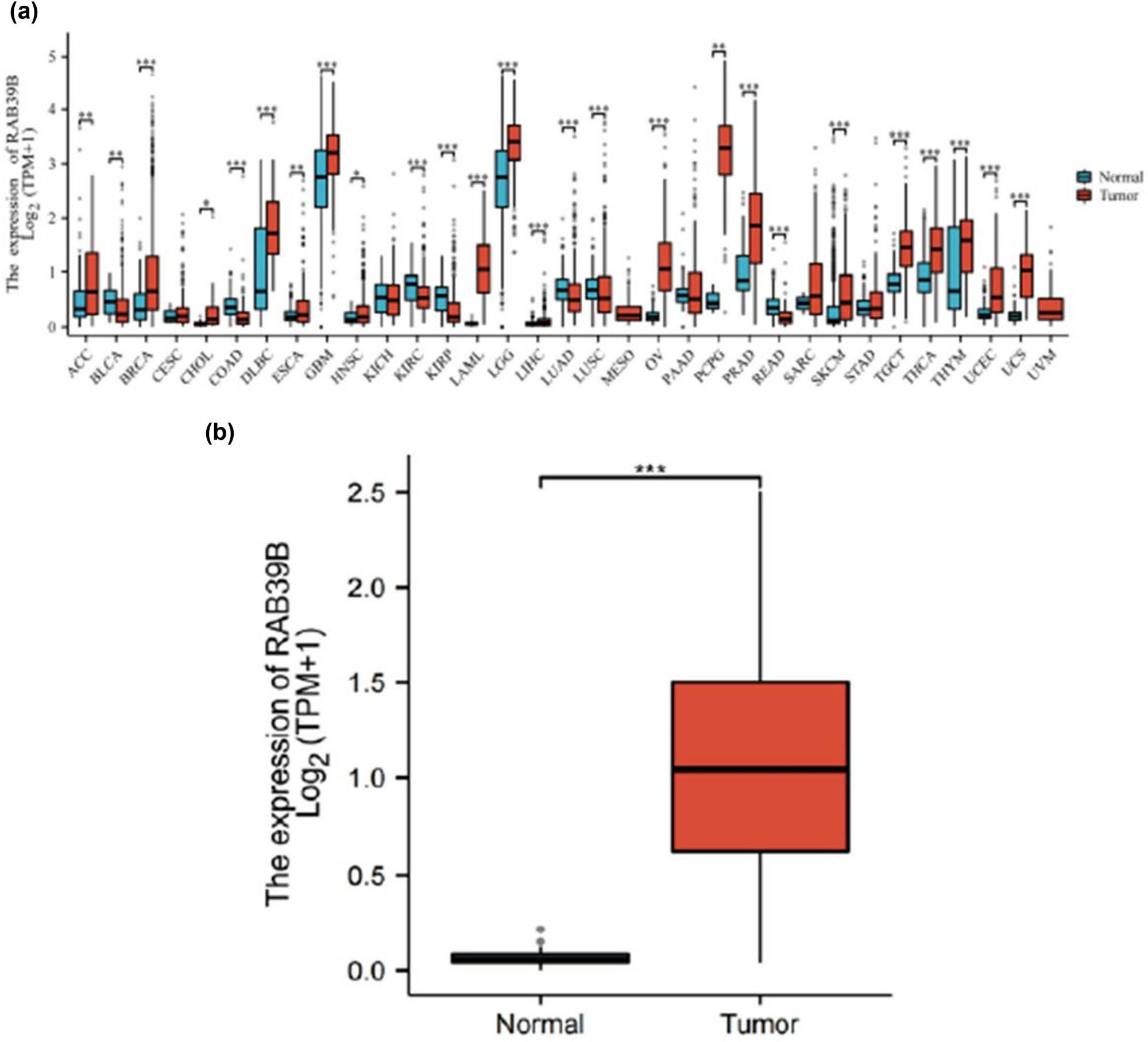
AML samples exhibited greater expression of RAB39B than normal samples. (a) Comparison of the expression levels of RAB39B in both normal and pan-cancer samples. (b) Comparison of RAB39B expression levels in normal and AML samples. A comparison of the two groups was conducted using the Wilcoxon rank sum test. NS: P = 0.05 or higher; *P < 0.05; **P < 0.01; ***P < 0.001.
3.2 RAB39B PPI network and co-expression analysis in AML
By utilizing the STRING database, we set a medium confidence score threshold of >0.400 to construct a PPI network for RAB39B (Figure 2a), resulting in the identification of a total of 10 genes associated with RAB39B. The analysis indicated that RAB39B was linked to WDR41, a complex that possesses GEF activity and controls autophagy. This complex includes SMCR8, C9orf72, RAB8A, RABGGTA, GDI1, GDI2, SQSTM1, VPS35, and DNAJC6. RAB8A belongs to the Ras oncogene family among these correlated proteins, whereas SMCR8, C9orf72, WDR41, and SQSTM1 are linked to autophagy.
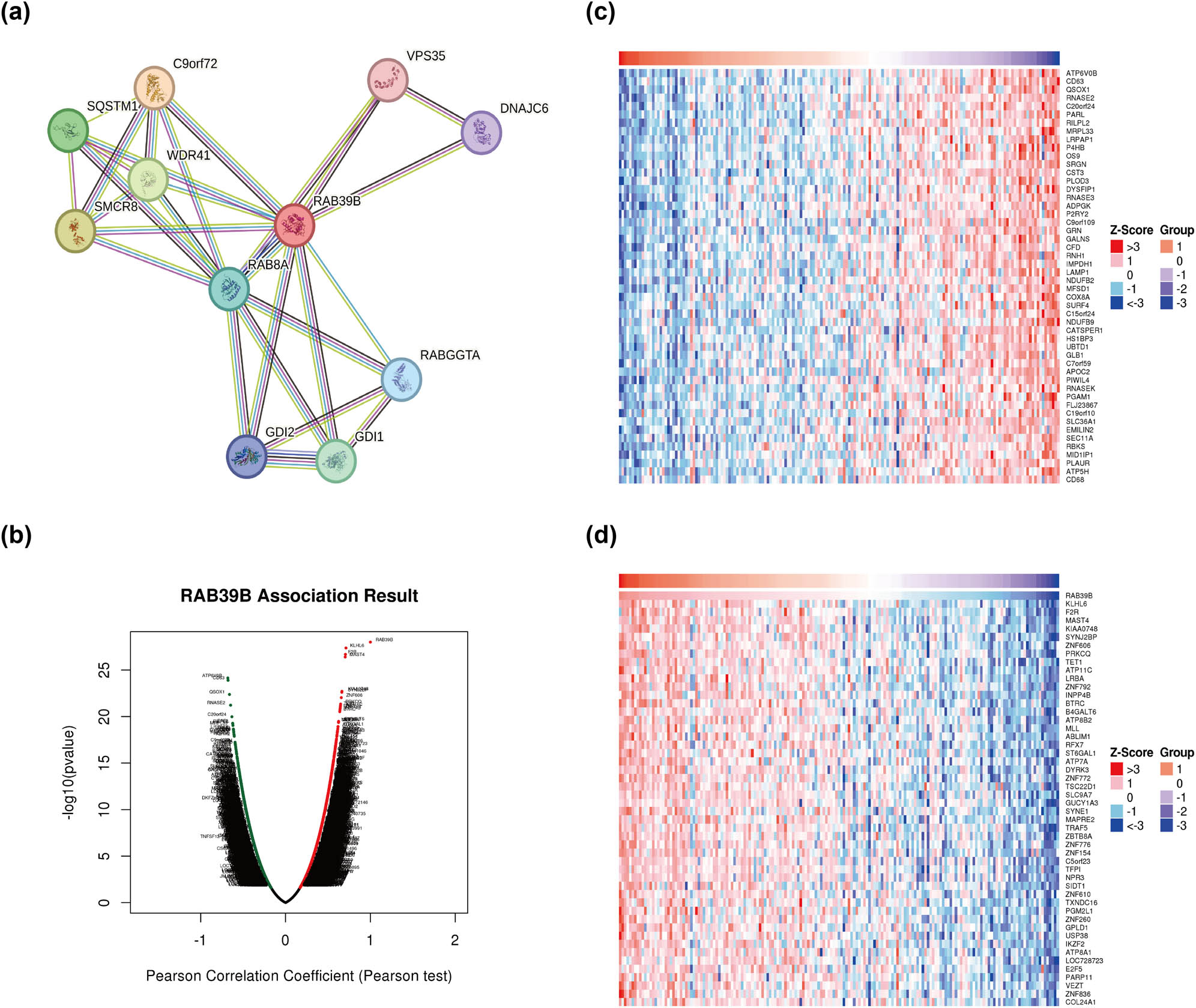
Analysis of genes associated with RAB39B and their enrichment. (a) RAB39B’s PPI network. (b) AML coexpressed genes were analyzed by LinkedOmics and visualized by Volcano plots. (c) The top 50 genes that have a positive correlation with RAB39B in AML. (d) The 50 most negatively correlated genes with RAB39B in AML.
To gain a deeper understanding of the biological role of RAB39B, we examined the coexpression of RAB39B in AML through the utilization of the LinkedOmics database. The analysis reveals that a total of 5,710 genes (dark red dots) exhibit a positive correlation with RAB39B, whereas 3,712 genes (dark green dots) demonstrate a negative correlation with it (Figure 2b). The heatmap displayed a favorable association (Figure 2c) and a connection with RAB39B (Figure 2d). Furthermore, the LinkInterpreter module was used to perform GO and KEGG enrichment analyses on coexpressed genes of RAB39B. According to the GO functional annotation, the coexpressed gene RAB39B primarily participates in the replication, transportation, and synthesis of DNA and proteins (Figure 3a–c). According to the KEGG functional annotation, RAB39B and its associated genes primarily participate in the Wnt signaling pathway, Fanconi anemia pathway, T-cell receptor signaling pathway, and phosphatidylinositol signaling pathway. Additionally, they play a role in the synthesis and regulation of nucleotides and proteins. Furthermore, these genes also play a role in controlling the metabolic pathways of AML, including amino acid metabolism, carbon metabolism, nitrogen metabolism, phospholipid metabolism, and other biochemical processes (Figure 3d). These findings indicate that RAB39B may play a significant biological function in the development of AML.
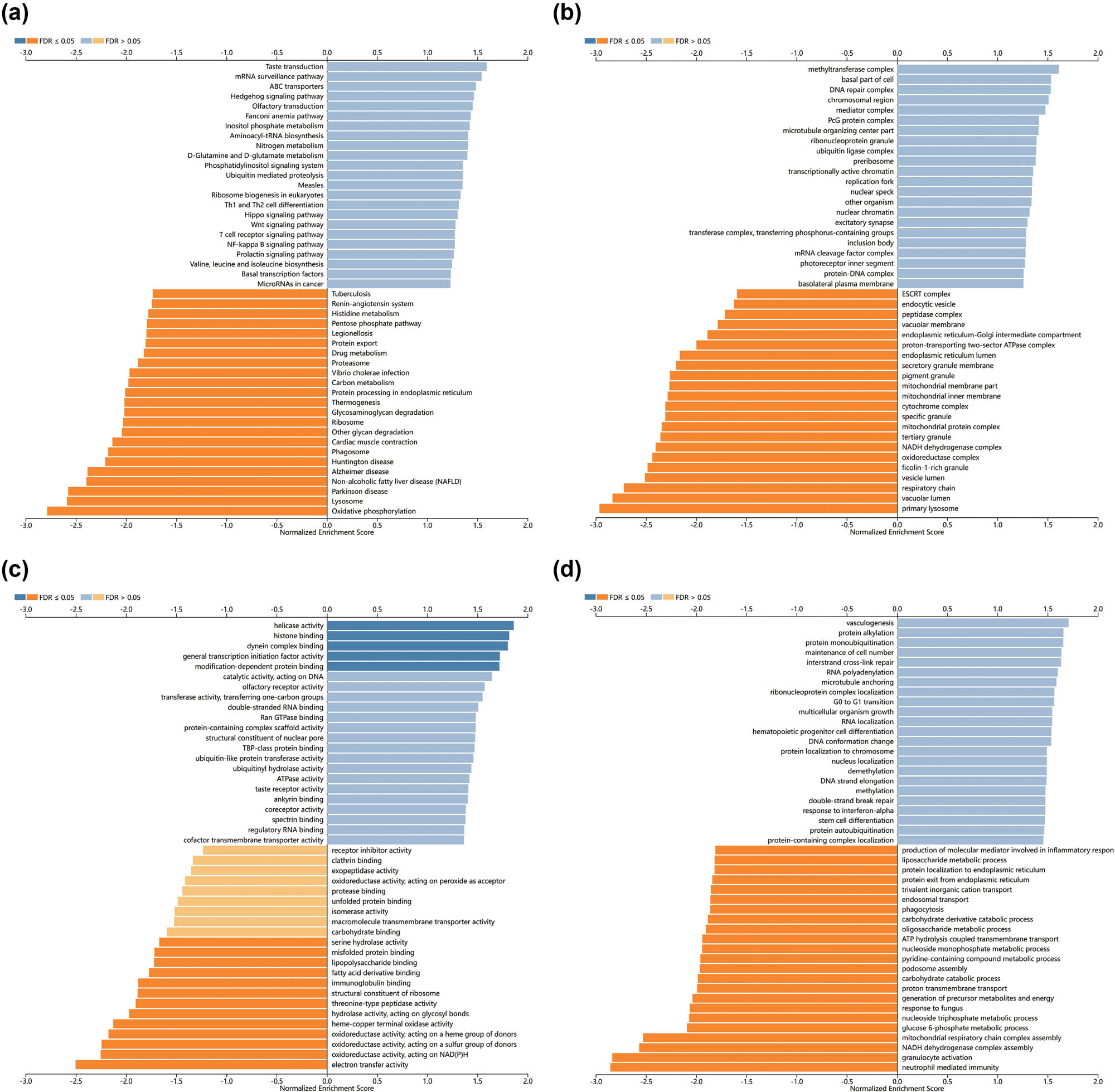
Enrichment analysis of RAB39B coexpressed genes in AML. GO biological process (a), cellular component (b), and molecular function (c) analyses were used to annotate coexpressed genes of RAB39B. (d) KEGG pathway analysis of RAB39B coexpressed genes.
3.3 Analysis of RAB39B in AML using immunofluorescence
For the correlation analysis of the association between AML and 24 distinct immune cells, we utilized data extracted from TCGA database and employed Spearman’s method to assess the connection with RAB39B expression. According to Figure 4a, RAB39B exhibited a strong positive correlation with T helper cells (r = 0.614), CD8 T cells (r = 0.440), B cells (r = 0.366), and T cells (r = 0.321), but showed a negative correlation with dendritic cells (DCs, r = −0.288). Figure 4b displays a notable distinction in the scatter plot depicting the relationship between the enrichment score of specific immune cells and the expression level of RAB39B. The surface morphology of T helper cells, T cells, CD8 T cells, and B cells showed a notable variation between low and high expression of RAB39B, as depicted in Figure 4c.
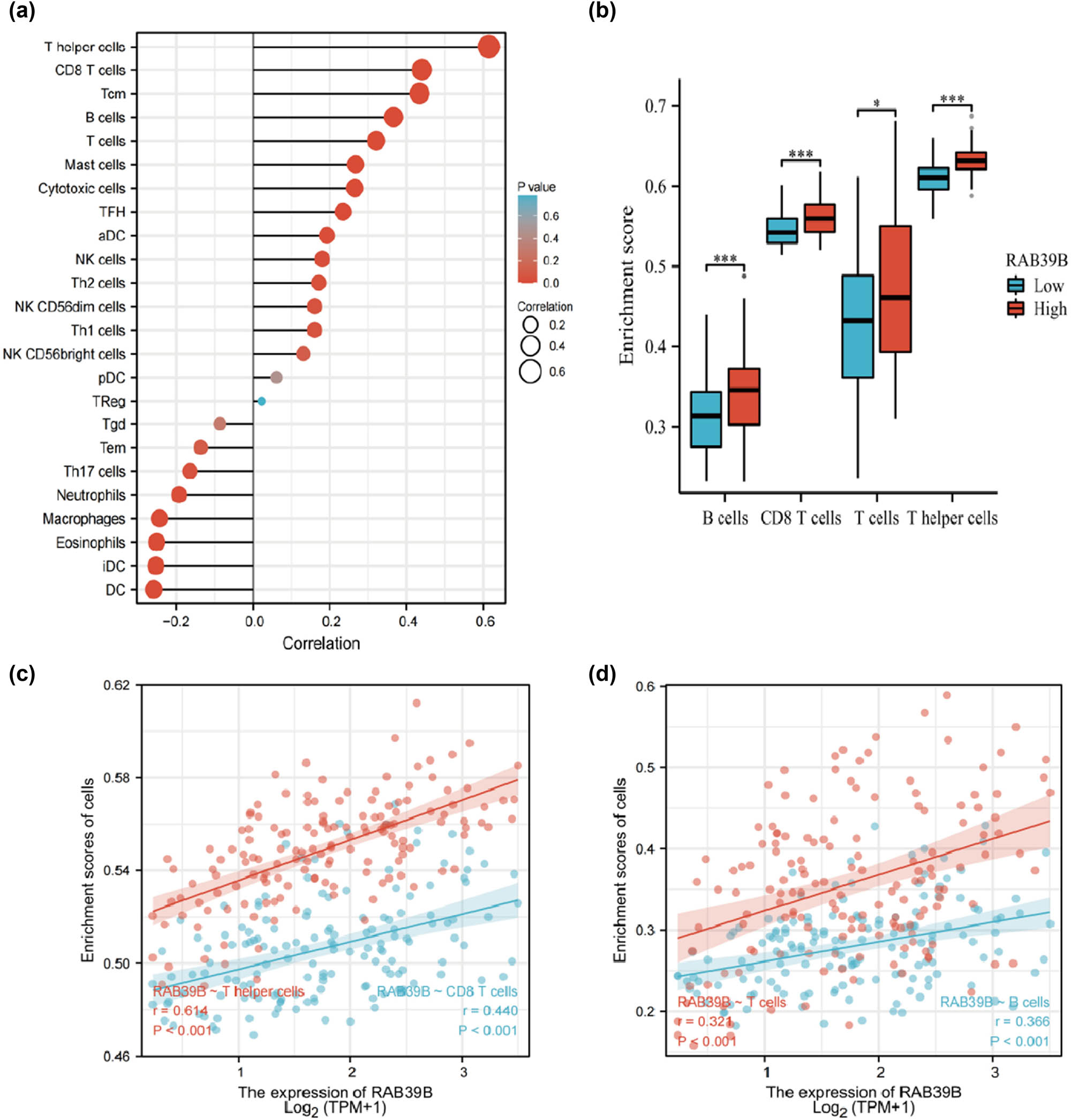
The correlation between RAB39B and immune markers in AML. (a) Correlation between the expression of RAB39B and 24 different types of immune cells. (b) Scatter plots depicting the correlation between RAB39B expression and the level of immune infiltration by T cells, B cells, T helper cells, and CD8 T cells. (c, d) The presence of T helper cells, T cells, CD8 T cells, and B cells is observed in cases with both low and high expression levels of RAB39B.
3.4 Building ceRNA networks associated with RAB39B in AML
The starBase database was utilized to screen the target miRANA of RAB39B in PITA, miRmap, and TargetScan. Thirty-six, 22, and 18 target miRNAs were obtained, respectively. From these three databases (Figure 5a), a sum of nine miRNAs that are commonly found was acquired. To construct ceRNAs, we screened miRNAs that were negatively correlated with RAB39B due to the regulation of target genes by miRNAs. In AML (Figure 5b), a noteworthy inverse association was discovered between hsa-miR-152-3p (r = −0.199, p-value = 7.12 × 10−2) and hsa-miR-582-5p (r = −0.180, p-value = 1.04 × 10−1), as well as the expression of RAB39B.
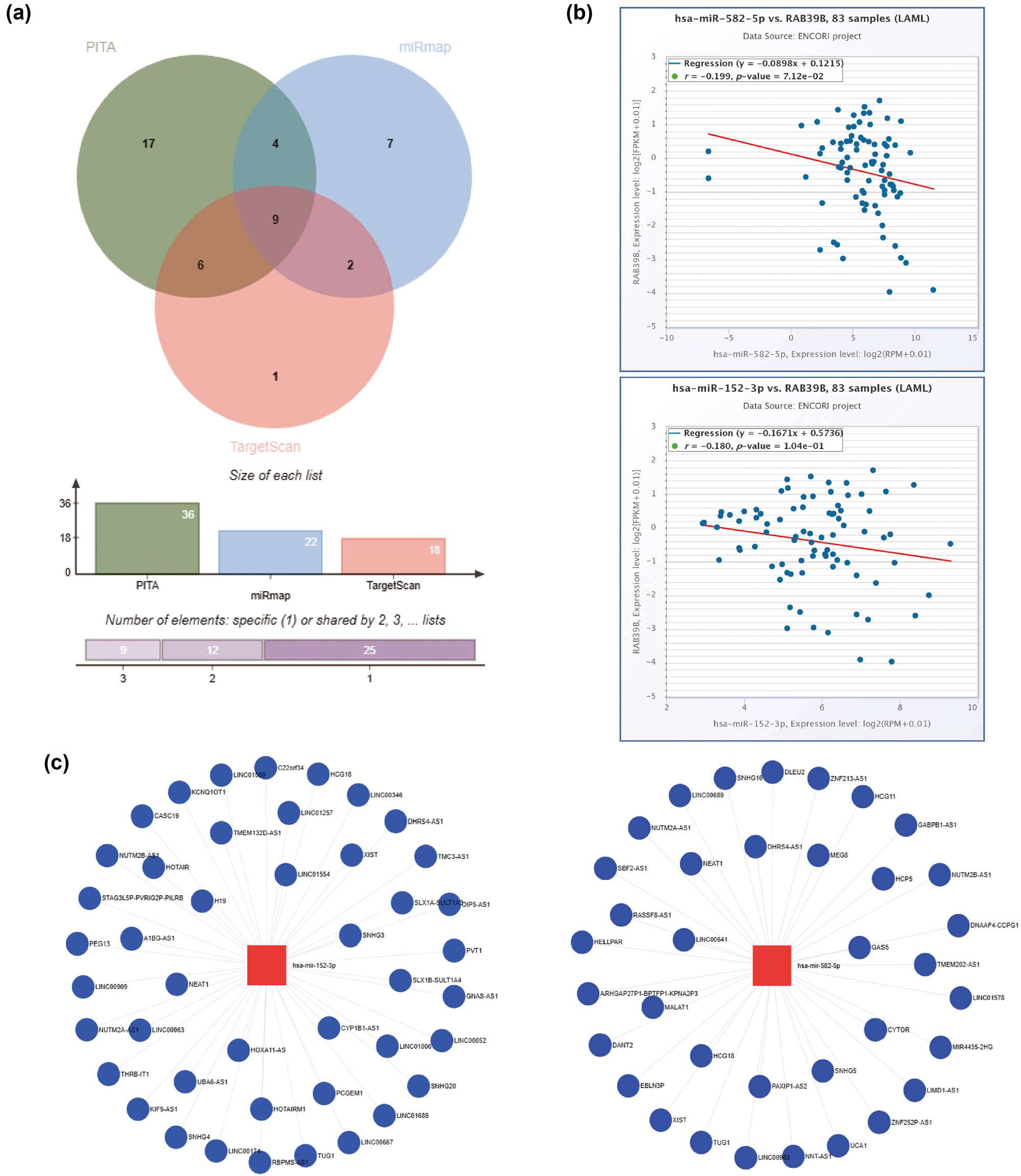
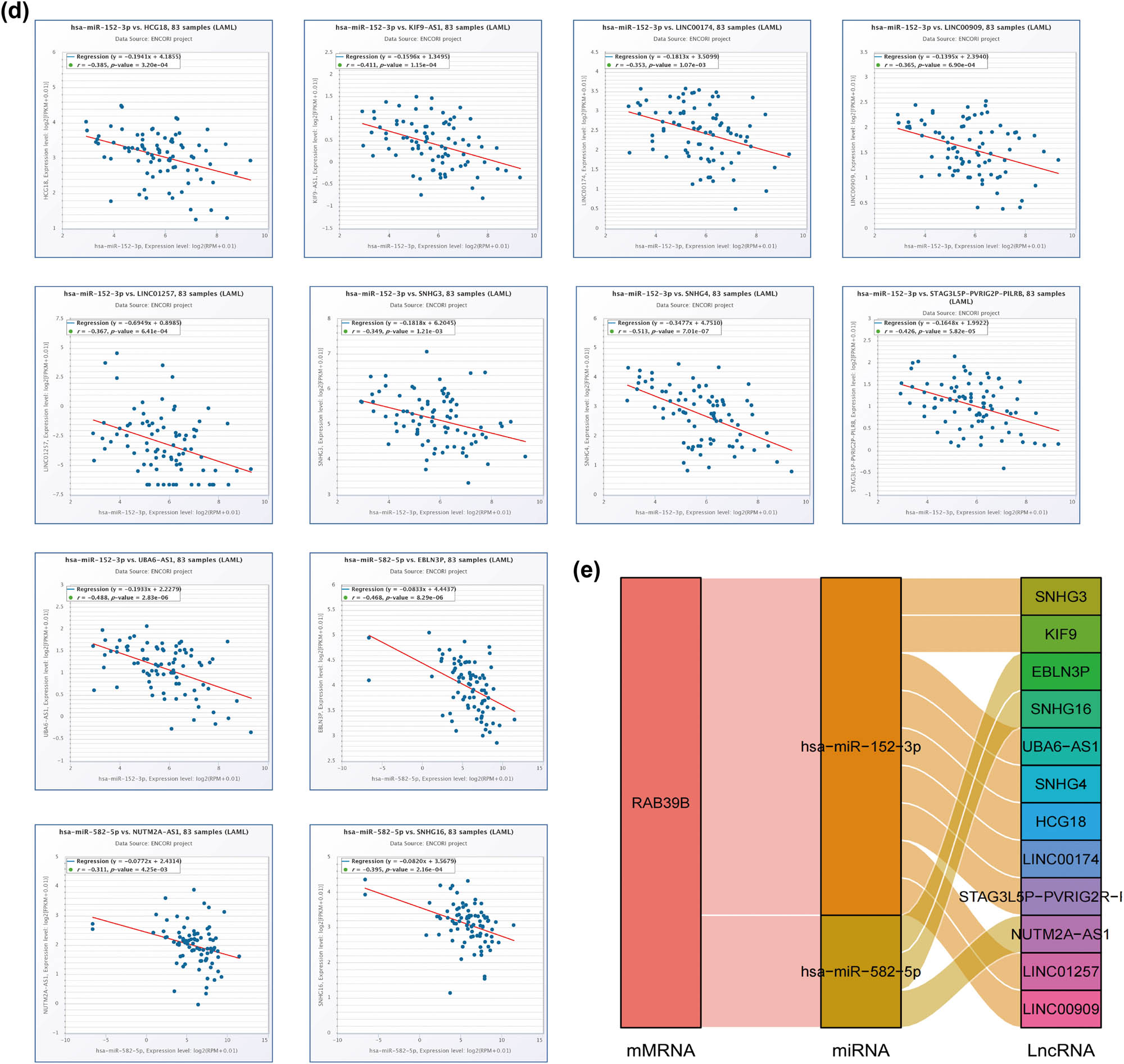
Construction of a ceRNA network associated with RAB39B in AML. (a) A Venn diagram illustrating the potential targets of RAB39B as predicted by PITA, miRmap, and TargetScan. (b) Scatter plots illustrating the correlation between RAB39B and the miRNAs of interest. (c) miRNet provides predicted target lncRNAs for hsa-miR-152-3p and hsa-miR-582-5p. (d) Scatter plots depict the correlation of hsa-miR-152-3p or hsa-miR-582-5p with the target lncRNAs. (e) The ceRNA hypothesis is supported by a sandwich figure illustrating the regulatory network of lncRNA miRNA mRNA (RAB39B).
According to the ceRNA network hypothesis, a negative correlation was observed between lncRNAs and miRNAs (Figure 5c). A correlation analysis was performed, considering lncRNAs that exhibited a correlation coefficient lower than –0.30 with miRNA. Figure 5d shows that EBLN3P, NUTM2A-AS1, and SNHG16 exhibited a negative correlation with hsa-miR-582-5p, while SNHG3, KIF9, UBA6-AS1, SNHG4, HCG18, LINC00174, STAG3L5P-PVRIG2R-PILRB, LINC01257, and LINC00909 displayed a negative correlation with hsa-miR-152-3p. Using the Sangi Figure (Figure 5e), we created a total of 12 ceRNA networks.
3.5 The interaction between RAB39B and disconfidptosis in AML
To examine the correlation between RAB39B and m6A methylation, we explored the association between RAB39B and 20 m6A-associated genes in AML. A strong connection was discovered between RAB39B and 15 m6A-associated genes, namely, YTHDC1, YTHDC2, YTHDF1, YTHDF2, ZC3H13, WTAP, VIRMA, RBMX, RBM15B, RBM15, METTL3, METTL14, IGF2BP3, FTO, and HNRNPC (as shown in Figure 6a). Figure 6b presents scatter plots demonstrating the correlations between RAB39B and the top six most strongly correlated m6A-associated genes (YTHDC1, YTHDC2, YTHDF1, YTHDF2, ZC3H13, and WTAP), confirming the association of RAB39B with m6A methylation in AML.
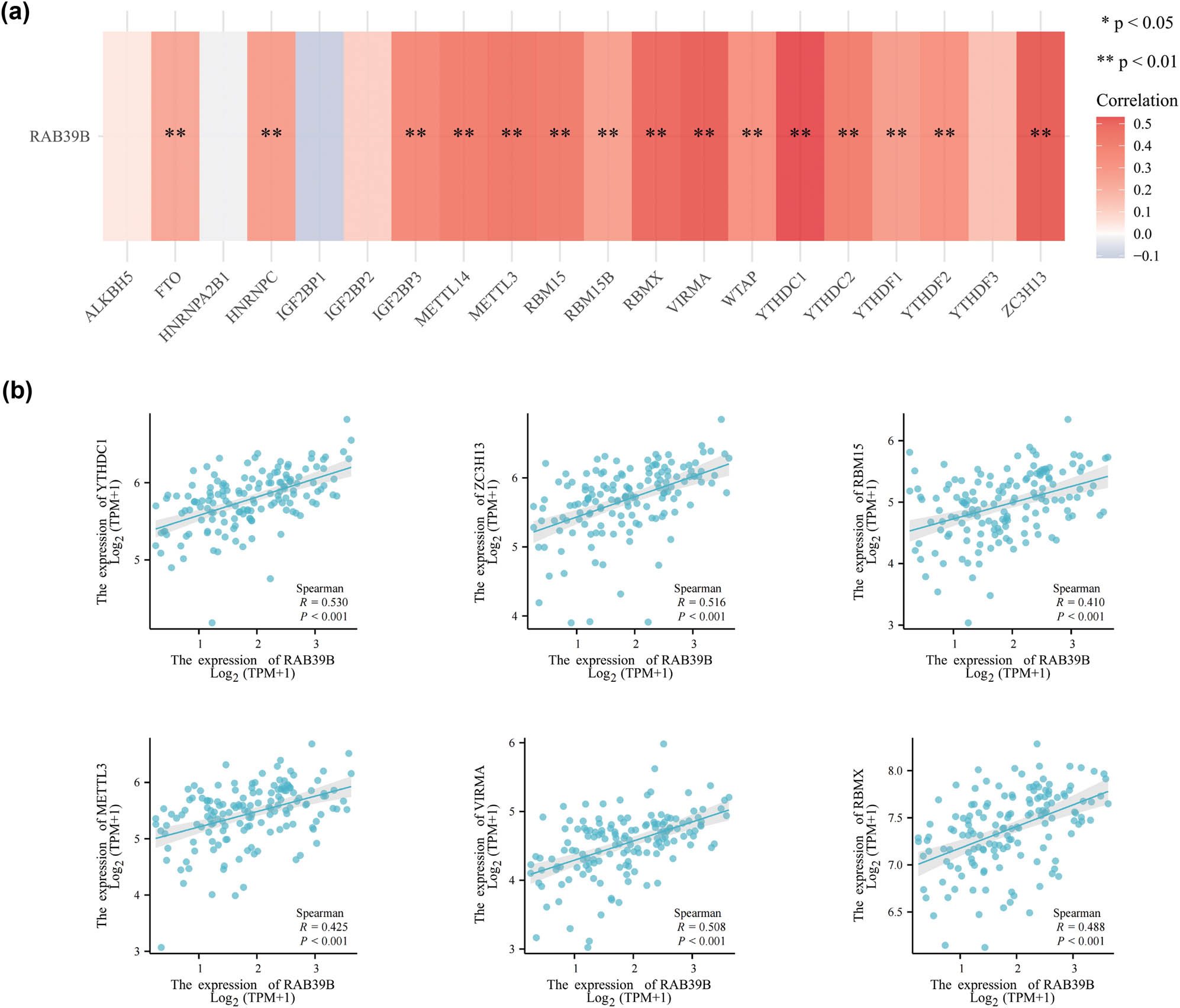
The association between RAB39B and m6A-associated genes in AML. (a) Examination of the association between RAB39B expression and the expression of genes related to m6A in TCGA AML cohort. (b) Scatter plots illustrating the relationship between RAB39B and genes associated with m6A.
3.6 Interaction between RAB39B and necroptosis in AML
Furthermore, we also investigated the correlation between RAB39B and cuprotosis. Out of the 15 genes associated with cuprotosis, 9 genes exhibited a notable and favorable relationship with the expression of RAB39B. These genes include ATP7A, ATP7B, DBT, DLAT, DLST, GCSH, GLS, LIAS, and LIPI (as shown in Figure 7a). The scatter plot of the top 6 correlations is depicted in Figure 7b. It suggests that RAB39B may play an important role in regulating cuprotosis-related processes.
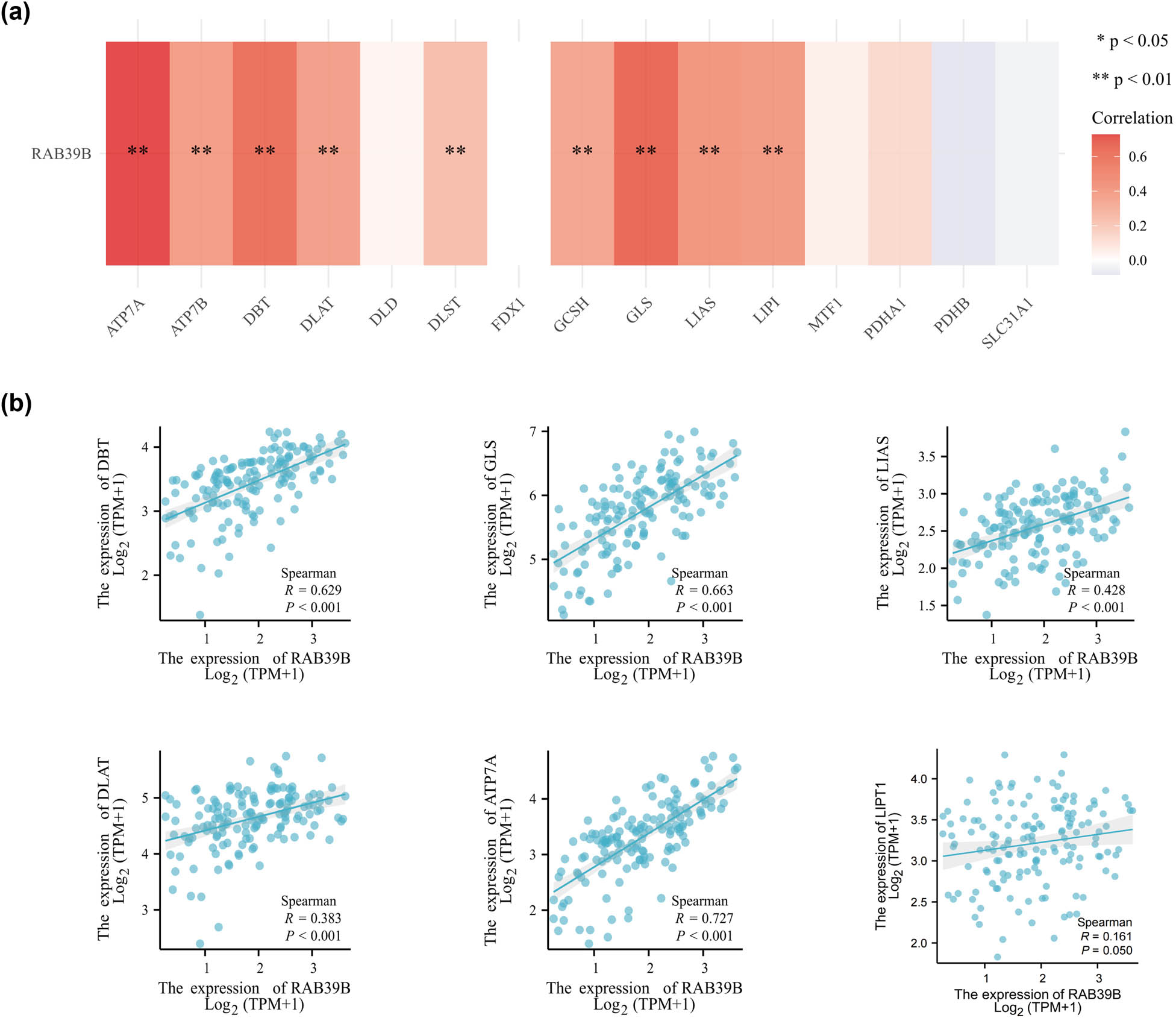
The relationship between RAB39B and genes associated with cuproptosis in AML. (a) Examining the association between the expression of RAB39B and genes related to cuproptosis in TCGA AML cohort. (b) Scatter plots illustrating the relationship between RAB39B and genes associated with cuproptosis.
3.7 Correlation between the expression of RAB39B and clinical characteristics and cytogenetic risk
The clinical characteristics of 150 AML patients (Table 1) retrieved from TCGA database demonstrate that RAB39B expression levels are significantly correlated with cytogenetic risk, FAB classifications (excluding the M3 type), bone marrow (BM) blast counts, and specific genetic mutations in FLT3 and NPM1. To be more specific, AML patients with high expression of RAB39B exhibited a significant association with adverse cytogenetic risk (P < 0.001) and specific FAB classifications (P < 0.001), excluding the M3 type due to its distinct treatment regimen involving ATRA. Additionally, high RAB39B expression was correlated with higher BM blast counts (P = 0.012) and showed a trend toward correlation with higher peripheral blood (PB) blast counts (P = 0.072) and white blood cell (WBC) counts (P = 0.060). Notably, patients with FLT3 mutations had lower expression of RAB39B (P = 0.026), while those with NPM1 mutations also tended to have lower RAB39B expression (P = 0.012). However, age, gender, race, WBC counts, PB blast counts, and other mutations (IDH1 R132, IDH1 R140, IDH1 R172, and RAS) did not show significant associations with RAB39B expression.
Correlation between the expression of RAB39B and clinicopathological characteristics in AML samples retrieved from TCGA database
| Characteristics | Low expression of RAB39B | High expression of RAB39B | P value |
|---|---|---|---|
| n | 75 | 75 | |
| Gender, n (%) | 0.412 | ||
| Female | 36 (24%) | 31 (20.7%) | |
| Male | 39 (26%) | 44 (29.3%) | |
| Race, n (%) | 0.593 | ||
| Asian and Black or African American | 8 (5.4%) | 6 (4%) | |
| White | 67 (45%) | 68 (45.6%) | |
| Age, n (%) | 0.869 | ||
| ≤60 | 43 (28.7%) | 44 (29.3%) | |
| >60 | 32 (21.3%) | 31 (20.7%) | |
| IDH1 R132 mutation, n (%) | 0.412 | ||
| Negative | 68 (45.9%) | 67 (45.3%) | |
| Positive | 5 (3.4%) | 8 (5.4%) | |
| IDH1 R140 mutation, n (%) | 0.961 | ||
| Negative | 69 (46.6%) | 67 (45.3%) | |
| Positive | 6 (4.1%) | 6 (4.1%) | |
| IDH1 R172 mutation, n (%) | 1.000 | ||
| Negative | 74 (50%) | 72 (48.6%) | |
| Positive | 1 (0.7%) | 1 (0.7%) | |
| FLT3 mutation, n (%) | 0.026 | ||
| Negative | 45 (30.8%) | 56 (38.4%) | |
| Positive | 29 (19.9%) | 16 (11%) | |
| RAS mutation, n (%) | 0.731 | ||
| Negative | 70 (47%) | 71 (47.7%) | |
| Positive | 5 (3.4%) | 3 (2%) | |
| NPM1 mutation, n (%) | 0.012 | ||
| Negative | 52 (34.9%) | 64 (43%) | |
| Positive | 23 (15.4%) | 10 (6.7%) | |
| BM blasts (%), n (%) | 0.012 | ||
| ≤20 | 37 (24.7%) | 22 (14.7%) | |
| >20 | 38 (25.3%) | 53 (35.3%) | |
| PB blasts (%), n (%) | 0.072 | ||
| ≤70 | 30 (20%) | 41 (27.3%) | |
| >70 | 45 (30%) | 34 (22.7%) | |
| WBC count (× 10 9/L ), n (%) | 0.060 | ||
| ≤20 | 32 (21.5%) | 44 (29.5%) | |
| >20 | 42 (28.2%) | 31 (20.8%) | |
| Cytogenetic risk, n (%) | <0.001 | ||
| Favorable | 25 (16.9%) | 5 (3.4%) | |
| Intermediate/normal | 38 (25.7%) | 44 (29.7%) | |
| Poor | 11 (7.4%) | 25 (16.9%) | |
| FAB classifications, n (%) | <0.001 | ||
| M0 | 1 (0.7%) | 14 (10.4%) | |
| M1 | 12 (8.9%) | 23 (17.1%) | |
| M2 | 13 (9.6%) | 25 (18.5%) | |
| M4 | 21 (15.6%) | 8 (5.9%) | |
| M5 | 14 (10.4%) | 1 (0.7%) | |
| M6 | 0 (0%) | 2 (1.5%) | |
| M7 | 0 (0%) | 1 (0.7%) |
To further confirm the connection between AML clinical factors and RAB39B (Table 2), logistic regression analysis was employed. RAB39B expression showed a positive correlation with cytogenetic risk (P = 0.009) and BM blasts (P = 0.013) but exhibited a negative correlation with FLT3 mutation (P = 0.028) and NPM1 mutation (P = 0.014).
The relationship between the clinicopathological factors of AML and RAB39B expression by using logistic regression analysis
| Characteristics | Total (N) | OR (95% CI) | P value |
|---|---|---|---|
| Age (>60 vs ≤60) | 150 | 0.947 (0.298–1.595) | 0.869 |
| WBC count (×109/L) (>20 vs ≤20) | 149 | 0.537 (–0.113–1.187) | 0.061 |
| Cytogenetic risk (poor vs favorable and intermediate/normal) | 148 | 2.922 (2.121–3.724) | 0.009 |
| BM blasts (%) (>20 vs ≤20) | 150 | 2.346 (1.673–3.018) | 0.013 |
| PB blasts (%) (>70 vs ≤70) | 150 | 0.553 (–0.095–1.201) | 0.073 |
| FLT3 mutation (positive vs negative) | 146 | 0.443 (–0.282–1.169) | 0.028 |
| IDH1 R132 mutation (positive vs negative) | 148 | 1.624 (0.457–2.791) | 0.416 |
| IDH1 R140 mutation (positive vs negative) | 148 | 1.030 (–0.151–2.210) | 0.961 |
| IDH1 R172 mutation (positive vs negative) | 148 | 1.028 (–1.763–3.819) | 0.985 |
| NPM1 mutation (positive vs negative) | 149 | 0.353 (–0.474–1.181) | 0.014 |
The OS of AML patients was influenced by RAB39B (high vs low, P = 0.028), cytogenetic risk (favorable and intermediate/normal vs poor, P < 0.019), and age (≤60 vs >60, P < 0.001) in the univariate Cox model (Table 3). Age (≤60 vs >60, P < 0.001) emerged as the sole independent variable linked to OS in patients with AML, as per the multivariate Cox model.
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 139 | ||||
| ≤60 | 78 | Reference | Reference | ||
| >60 | 61 | 3.321 (2.156–5.116) | <0.001 | 3.193 (2.042–4.992) | <0.001 |
| Cytogenetic risk | 137 | ||||
| Favorable and intermediate/normal | 106 | Reference | Reference | ||
| Poor | 31 | 1.803 (1.102–2.951) | 0.019 | 1.356 (0.820–2.243) | 0.235 |
| Gender | 139 | ||||
| Female | 62 | Reference | |||
| Male | 77 | 1.024 (0.671–1.564) | 0.912 | ||
| Race | 138 | ||||
| Asian and Black or African American | 11 | Reference | |||
| White | 127 | 1.200 (0.485–2.966) | 0.693 | ||
| WBC count (×109/L) | 138 | ||||
| ≤20 | 74 | Reference | |||
| >20 | 64 | 1.156 (0.757–1.764) | 0.503 | ||
| BM blasts (%) | 139 | ||||
| ≤20 | 58 | Reference | |||
| >20 | 81 | 1.159 (0.754–1.780) | 0.502 | ||
| PB blasts (%) | 139 | ||||
| ≤70 | 65 | Reference | |||
| >70 | 74 | 1.224 (0.802–1.869) | 0.349 | ||
| FLT3 mutation | 135 | ||||
| Negative | 96 | Reference | |||
| Positive | 39 | 1.266 (0.798–2.009) | 0.316 | ||
| IDH1 R132 mutation | 137 | ||||
| Negative | 125 | Reference | |||
| Positive | 12 | 0.586 (0.237–1.448) | 0.247 | ||
| IDH1 R140 mutation | 137 | ||||
| Negative | 126 | Reference | |||
| Positive | 11 | 1.129 (0.564–2.258) | 0.733 | ||
| IDH1 R172 mutation | 137 | ||||
| Negative | 135 | Reference | |||
| Positive | 2 | 0.608 (0.085–4.376) | 0.621 | ||
| NPM1 mutation | 138 | ||||
| Negative | 105 | Reference | |||
| Positive | 33 | 1.134 (0.704–1.826) | 0.606 | ||
| RAS mutation | 138 | ||||
| Negative | 130 | Reference | |||
| Positive | 8 | 0.642 (0.235–1.756) | 0.388 | ||
| RAB39B | 71 | ||||
| Low | 38 | Reference | Reference | ||
| High | 33 | 1.980 (1.076–3.641) | 0.028 | 1.520 (0.776–3.041) | 0.274 |
3.8 Prognostic correlation analysis of RAB39B in AML
Through ROC curve analysis of data from UCSC XENA, we investigated the ability of RAB39B to distinguish between healthy cells and AML cells. The result (Figure 8a) revealed an area under the curve of 0.991, highlighting its high sensitivity and specificity in identifying AML. Furthermore, utilizing the GEPIA2 database to examine the Kaplan‒Meier survival plot, it was observed that an elevated level of RAB39B expression was linked to an unfavorable prognosis (HR = 2.3, P < 0.05) (Figure 8b).
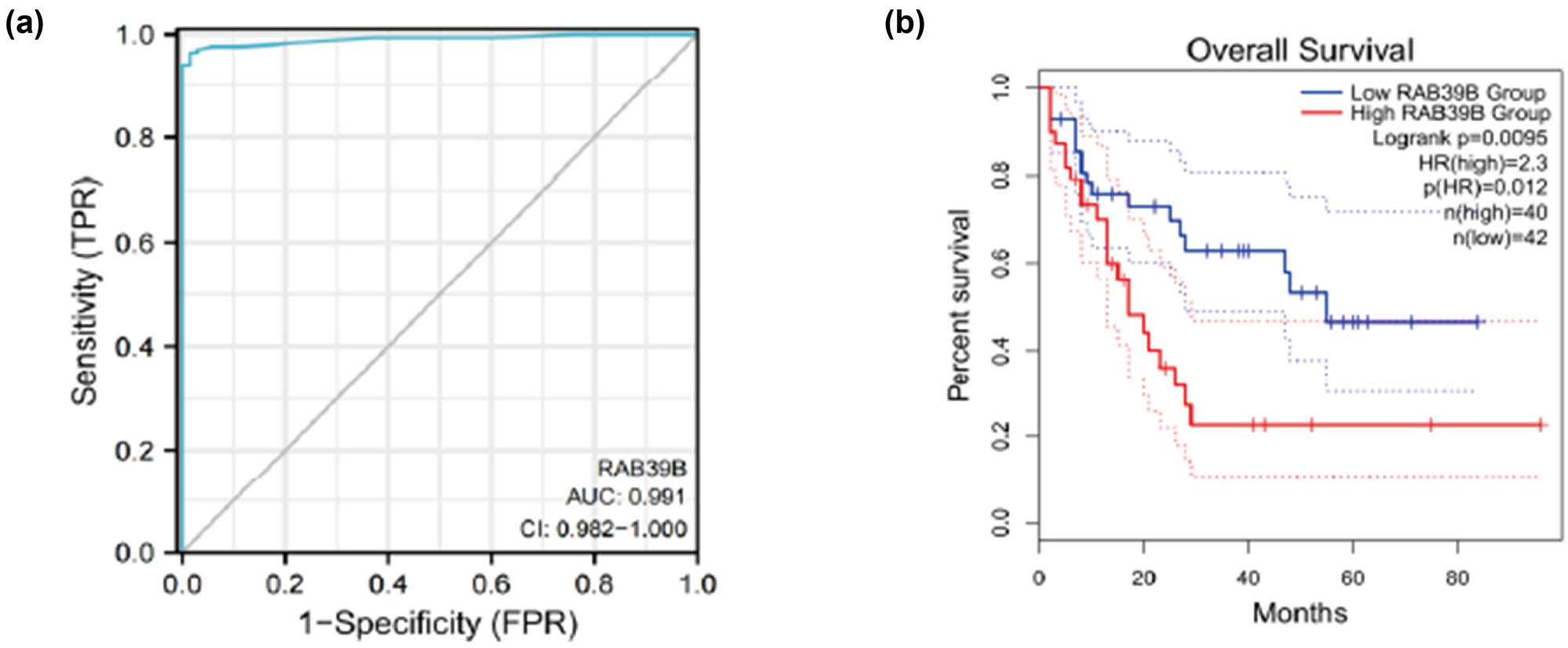
(a) Draw ROC curve chart based on prognostic analysis. (b) Draw a Kaplan Meier curve based on prognostic analysis.
3.9 The prognostic model of RAB39B in AML
Based on the results of the previous multivariate Cox regression analysis, we identified three independent prognostic factors: age, cytogenetic risk, and RAB39B expression. Utilizing these factors, we constructed an AML prognostic prediction nomogram model (Figure 9a) to estimate the 1-, 2-, and 3-year survival rates of AML patients. In Figure 9b, the calibration curves for 1-, 2-, and 3-year survival rates all closely approximate the 45° line, indicating that this prognostic prediction model exhibits high accuracy in predicting the survival rates of AML patients.
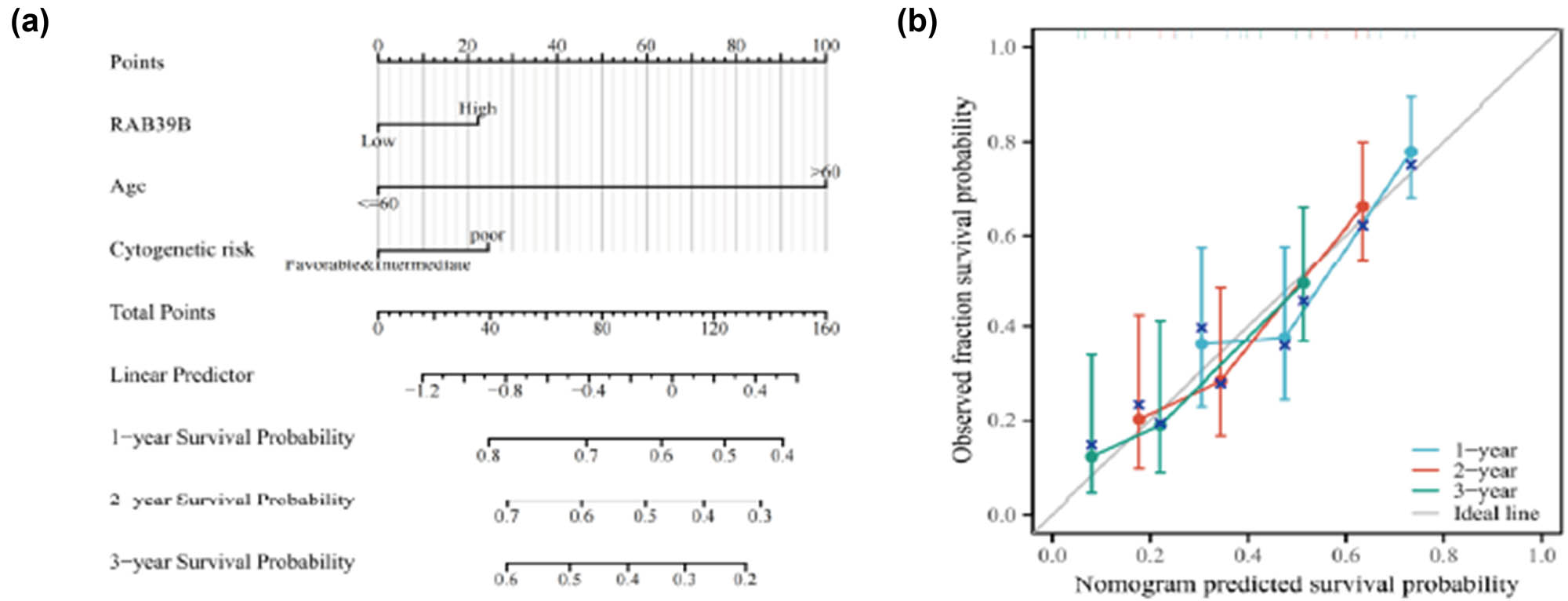
An AML prognostic predictive model for RAB39B. (a) A nomogram is available to estimate the likelihood of surviving for 1, 2, and 3 years for AML. (b) A calibration plot of the nomogram for predicting OS at 1, 2, and 3 years.
3.10 In vitro validation of RAB39B’s function in AML
Initially, Western blot analysis was employed to identify the contrasting expression of RAB39B in various cellular lineages (OCI-AML-3, KG-1, MOLM-13, and MV4-11 cell lines). The expression of RAB39B was higher in all four of these AML cell lines than in regular peripheral blood monocytes (Figure 10a). Given that the MOML-13 and MV4-11 cell lines exhibited relatively significant differences in RAB39B expression in the Western blot analysis, we therefore focused our subsequent research on these two representative cell lines. Through siRNA transfection (Figure 10b), the expression of RAB39B was inhibited in the MOML-13 and MV4-11 cell lines. In the CCK-8 study, it was observed that the growth of AML cell lines declined following the suppression of RAB39B (as depicted in Figure 10c). Conversely, the suppression of RAB39B also resulted in an elevation of AML cell apoptosis (as illustrated in Figure 10d).
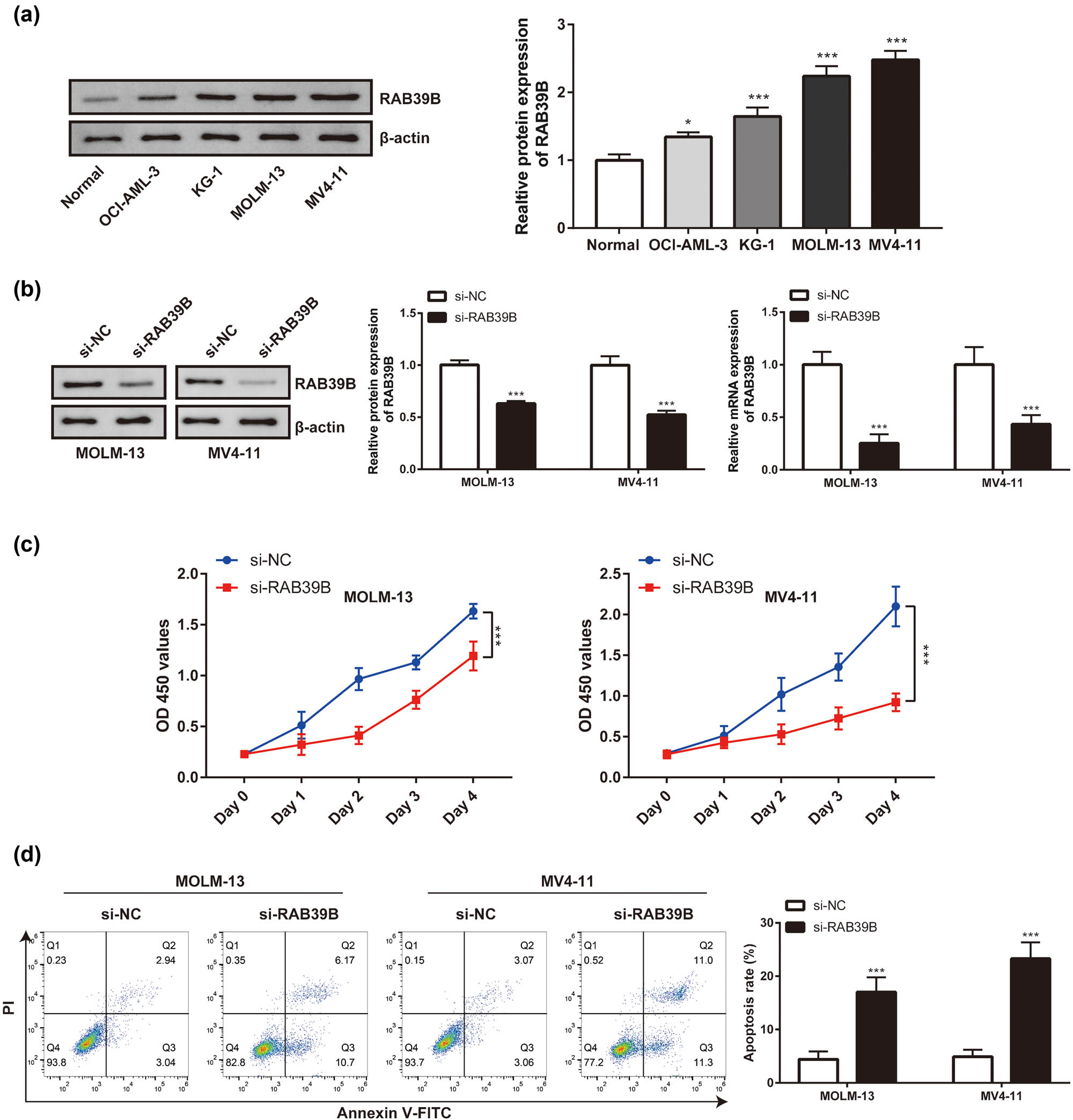
AML is linked to the expression and proliferation of RAB39B in cells. (a) The expression of RAB39B in various cell lines was detected using Western blotting and qPCR. (b) qPCR and Western blotting techniques were used to measure the transfection effectiveness of RAB39B siRNA in MOML-13 and MV4-11 cells. (c) The CCK-8 proliferation experiment revealed that suppressing RAB39B can result in reduced AML cell proliferation. (d) Knocking down RAB39B leads to increased apoptosis in AML cells.
4 Discussion
AML is the prevailing form of acute leukemia among adults. Despite thorough investigation to discover predictive biomarkers, AML continues to be a condition characterized by notable prognostic variations and elevated fatality rates. Rab proteins, which are small GTPases, play a role in controlling the movement of vesicles within eukaryotic cells. Rab39B is a member of the Rab protein family and can be found on the Xq28 region of the human chromosome. Rab proteins, which play a significant role in controlling vesicle trafficking, have emerged as crucial regulators of various cellular processes, including cell growth, differentiation, survival, autophagy, and apoptosis [14,15,20,21,38]. Rab proteins impact membrane trafficking and trafficking complexes, thereby regulating cell metabolism, apoptosis, and autophagy. They also have a significant role in tumors, neurological diseases, and other genetic diseases, ultimately influencing multiple signaling pathways.
Several RAB genes have been discovered to have a connection with tumors. Various RAB genes have been linked to tumors, including RAB27A [39], which serves as a predictive indicator for pancreatic ductal adenocarcinoma; RAB13 and RAB3D, which are elevated in pan-cancer [40,41]; and RAB1A and RAB6A, which have significant functions in rectal cancer and bile duct cancer [42,43], respectively. Numerous atypical Rab proteins are believed to be linked to malignant tumors. Research indicates that RAB39B exhibits increased expression in various types of tumors, such as germ cell tumors, gastric stromal tumors, and diffuse large B-cell lymphoma [16,17,18]. Moreover, the removal and excessive expression of RAB39B impact cellular autophagic flow [35,44]. Nevertheless, the current lack of comprehensive research on the mechanism and prognostic significance of RAB39B in AML remains evident.
Through analysis of TCGA data, we found that RAB39B has significant expression differences in most cancers and AMLs of pan-cancer. In vitro experiments confirmed this as well. By conducting PPI and coexpression gene enrichment analysis, we discovered that RAB39B is probably engaged in substance conveyance, DNA replication and conveyance, substance metabolism, and more within AML cells. Consequently, it governs the functions of apoptosis and autophagy in AML. Furthermore, numerous pathways associated with tumors were identified, including the Wnt signaling pathway, T-cell receptor signaling pathway, and phosphatidylinositol signaling pathway. Notably, the Fanconi anemia pathway, which plays a crucial role in DNA damage repair, was found to interact with multiple pathways and contribute to the progression of AML [45]. By analyzing these data, we can gain insight into the possible roles of RAB39B in the onset and progression of AML.
The presence of immune cells in the tumor microenvironment is a crucial factor, as studies have indicated that it plays a significant role in both tumor progression and response to treatment. The cellular foundation of antitumor immunity is primarily composed of tumor-infiltrating immune cells, particularly T cells. Additionally, studies have demonstrated the significance of various immune cells, such as DCs, B lymphocytes, and natural killer cells. B lymphocytes and dendritic cells play a multifaceted role in the growth of tumors and the surrounding environment, serving as both catalysts for tumor advancement and, conversely, impediments to it [46]. Viewing AML from a different angle may shed light on the immune evasion mechanism, leading to the development of novel treatment approaches and a deeper comprehension of immune cell infiltration within the AML tumor microenvironment. This, in turn, can enhance our understanding of the role of RAB39B in AML. Analysis of immune infiltration revealed a strong positive correlation between RAB39B and T helper cells, T cells, CD8 T cells, and B cells but a negative correlation with DCs. The findings indicate that RAB39B might contribute to unfavorable outcomes in AML through its impact on immune infiltration. In addition, in the past few years, new targeted T-cell therapy methods for AML have been developing, including the construction of T-cell receptors, reactivation of endogenous T-cell responses through immune checkpoint inhibitors, and so on. At present, these immunotherapies are not yet complete and require new biomarkers and immune checkpoints to better regulate the tumor microenvironment and cell therapy. From this, we can foresee the potential of RAB39B in tumor immunotherapy [47].
Using miRNA shas-miR-152-3p and hsa-miR-582-5p, along with lncRNAs EBLN3P, NUTM2A-AS1, SNHG16, SNHG3, KIF9, UBA6-AS1, SNHG4, HCG18, LINC00174, STAG3L5P-PVRIG2R-PILRB, LINC01257, and LINC00909, we established a network consisting of lncRNA‒miRNA‒mRNA (RAB39B). Among them, EBLN3P, SNHG proteins and HCG18 [48,49,50,51] have been identified as driving factors for various tumors. At present, we have only constructed a credible network model, and further experiments are needed to verify this finding.
The study on m6A methylation and AML revealed that YTHDF1, METTL14, YTHDC2, ZC3H13, RBM15, WTAP, METTL3, METTL14, FTO, IGF2BP3, and ALKBH5 are among the m6A genes implicated in the development of AML. The heatmap indicates that all the aforementioned genes, excluding ALKBH5, exhibit a strong correlation with RAB39B in AML. This finding enhances our comprehension of the mechanism underlying RAB39B in AML [52,53,54,55,56,57,58,59]. The literature states that AML has three cuproptosis genes, namely, GCSH, LIPT1, and DLAT [60,61], which possess individual and noteworthy prognostic significance. All three genetic factors in our research exhibited a strong correlation with RAB39B. The presence of these occurrences indicates a connection between m6A methylation and cuproptosis in AML, suggesting that RAB39B could exert a significant influence on these vital biological mechanisms. We should conduct further experiments to continue to explore this issue.
To validate the correlation between RAB39B and the survival rate of AML patients, we employed ROC curves and Kaplan‒Meier survival analysis. The ROC curve shows that RAB39B can distinguish between tumor cells and normal cells, and the expression of RAB39B is significantly correlated with the survival rate of AML patients. Through logistic, univariate, and multivariate Cox analyses, we additionally assessed the expression of RAB39B and other clinical-pathological factors associated with OS in AML. We can observe that the expression of RAB39B is associated with NPM1 and FLT3 mutations. Studies have found that AML associated with these two mutations generally has a good prognosis, and this relationship is worth further exploration to seek new treatment options through rab protein on the basis of specific mutations [62]. RAB39B, cytogenetic risk, and age can all be considered separate predictors in the univariate Cox analysis. Incorporating RAB39B, cytogenetic risk, and age into the construction of prognostic column charts and calibration charts, the calibration charts showed consistent results with RAB39B column charts for 1, 2, and 3 years. The findings suggest that an elevated level of RAB39B is linked to an unfavorable prognosis, making it a reliable marker for prognosticating AML outcomes.
Ultimately, we performed an in vitro confirmation of the initial analysis, and the findings indicated a variation in RAB39B expression between AML cell lines and typical human peripheral blood monocytes. By suppressing the expression of RAB39B, we observed a reduction in AML cell growth and an elevation in cell death. The preliminary stage of the in vitro experiment ensured the functional analysis of RAB39B in AML.
5 Conclusions
The objective of our study was to investigate the function and predictive significance of RAB39B in AML. We discovered an upsurge in RAB39B expression in AML for the first time. Additionally, we determined the potential pathways and mechanisms through which RAB39B contributes to the development of AML. Furthermore, we conducted a novel analysis of the correlation between RAB39B expression and AML immune infiltration, m6A modification, copper-induced cell death, ceRNA network, and prognosis. We also conducted concise validation using in vitro experiments. Currently, we believe that RAB39B has the potential to function as both a diagnostic and prognostic indicator for AML.
Abbreviations
- AML
-
Acute myeloid leukemia
- TCGA
-
The Cancer Genome Atlas
- GTEx
-
Genotype-tissue expression
- DEGs
-
Differentially expressed genes
- GSEA
-
Gene set enrichment analysis
- GO
-
Gene Ontology
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- STRING
-
Search Tool for the Retrieval of Interacting Genes
- PPI
-
Protein–protein interaction
- ROC
-
Receiver operating characteristic
Acknowledgements
We acknowledge Yuxin Sun, Yuning Sun, and Yilun Liu for helping with the technical elements of the experiments and the data analysis.
-
Funding information: None.
-
Author contributions: CS and MXL designed and implemented the study. CS wrote the manuscript and performed analytical calculations and statistical analysis. JSW conducted a general review. All authors read and approved the final manuscript. This study has no funding support.
-
Conflict of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
-
Data availability statement: The datasets used in the current study are publicly available data from UCSC Xena platform (https://xena.ucsc.edu/), The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/), The LinkedOmics database (http://www.linkedomics.org/login.php), STRING (https://cn.string-db.org/), starBase (https://starbase.sysu.edu.cn/), and miRNet2.0 (www.mirnet.ca/miRNet/home.xhtml). Further inquiries can be obtained directly from the corresponding author in a reasonable request.
References
[1] Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–52.10.1056/NEJMra1406184Suche in Google Scholar PubMed
[2] Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–89.10.1056/NEJMoa1112304Suche in Google Scholar PubMed PubMed Central
[3] Hemminki K, Zitricky F, Forsti A, Kontro M, Gjertsen BT, Severinsen MT, et al. Age-specific survival in acute myeloid leukemia in the Nordic countries through a half century. Blood Cancer J. 2024;14(1):44.10.1038/s41408-024-01033-7Suche in Google Scholar PubMed PubMed Central
[4] Wysota M, Konopleva M, Mitchell S. Novel therapeutic targets in acute myeloid leukemia (AML). Curr Oncol Rep. 2024;26(4):409–20.10.1007/s11912-024-01503-ySuche in Google Scholar PubMed PubMed Central
[5] Jayavelu AK, Wolf S, Buettner F, Alexe G, Haupl B, Comoglio F, et al. The proteogenomic subtypes of acute myeloid leukemia. Cancer Cell. 2022;40(3):301–17 e12.10.1016/j.ccell.2022.02.006Suche in Google Scholar PubMed
[6] Zhao M, Liao M, Gale RP, Zhang M, Wu L, Yan N, et al. Acute myeloid leukemia with normal cytogenetics and NPM1-mutation: Impact of mutation topography on outcomes. Biomedicines. 2024;12(12):2921.10.3390/biomedicines12122921Suche in Google Scholar PubMed PubMed Central
[7] Han X, Liu X, Wan K, Yan H, Zhang M, Liu H, et al. The clinical features and outcomes of elderly patients with acute myeloid leukemia: a real word research. Clin Exp Med. 2025;25(1):27.10.1007/s10238-024-01536-4Suche in Google Scholar PubMed PubMed Central
[8] Wei Q, Hu S, Xu J, Loghavi S, Daver N, Toruner GA, et al. Detection of KMT2A partial tandem duplication by optical genome mapping in myeloid neoplasms: Associated cytogenetics, gene mutations, treatment responses, and patient outcomes. Cancers (Basel). 2024;16(24):4193.10.3390/cancers16244193Suche in Google Scholar PubMed PubMed Central
[9] Di J, Sheng T, Arora R, Stocks-Candelaria J, Wei S, Lutz C, et al. The validation of digital PCR-based minimal residual disease detection for the common mutations in IDH1 and IDH2 genes in patients with acute myeloid leukemia. J Mol Diagn. 2025;27:100–810.1016/j.jmoldx.2024.11.002Suche in Google Scholar PubMed PubMed Central
[10] Gerritsen M, In ‘t Hout FEM, Knops R, Mandos BLR, Decker M, Ripperger T, et al. Acute myeloid leukemia associated RUNX1 variants induce aberrant expression of transcription factor TCF4. Leukemia. 2025;39:520–3.10.1038/s41375-024-02470-wSuche in Google Scholar PubMed
[11] Dohner H, Wei AH, Lowenberg B. Towards precision medicine for AML. Nat Rev Clin Oncol. 2021;18(9):577–90.10.1038/s41571-021-00509-wSuche in Google Scholar PubMed
[12] Yang X, Wang J. Precision therapy for acute myeloid leukemia. J Hematol Oncol. 2018;11(1):3.10.1186/s13045-018-0603-7Suche in Google Scholar PubMed PubMed Central
[13] Cheng H, Ma Y, Ni X, Jiang M, Guo L, Ying K, et al. Isolation and characterization of a human novel RAB (RAB39B) gene. Cytogenet Genome Res. 2002;97(1–2):72–5.10.1159/000064047Suche in Google Scholar PubMed
[14] Giannandrea M, Bianchi V, Mignogna ML, Sirri A, Carrabino S, D’Elia E, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86(2):185–95.10.1016/j.ajhg.2010.01.011Suche in Google Scholar PubMed PubMed Central
[15] Tang BL. RAB39B’s role in membrane traffic, autophagy, and associated neuropathology. J Cell Physiol. 2021;236(3):1579–92.10.1002/jcp.29962Suche in Google Scholar PubMed
[16] Xu C, Liang T, Liu J, Fu Y. RAB39B as a chemosensitivity-related biomarker for diffuse large B-cell lymphoma. Front Pharmacol. 2022;13:931501.10.3389/fphar.2022.931501Suche in Google Scholar PubMed PubMed Central
[17] Kou Y, Zhao Y, Bao C, Wang Q. Comparison of gene expression profile between tumor tissue and adjacent non-tumor tissue in patients with gastric gastrointestinal stromal tumor (GIST). Cell Biochem Biophys. 2015;72(2):571–8.10.1007/s12013-014-0504-5Suche in Google Scholar PubMed
[18] Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Guetgemann I, et al. Gene expression profiling identifies new biological markers of neoplastic germ cells. Anticancer Res. 2007;27(5A):3091–100.Suche in Google Scholar
[19] He QL, Jiang HX, Zhang XL, Qin SY. Relationship between a 7-mRNA signature of the pancreatic adenocarcinoma microenvironment and patient prognosis (a STROBE-compliant article). Medicine (Baltimore). 2020;99(29):e21287.10.1097/MD.0000000000021287Suche in Google Scholar PubMed PubMed Central
[20] Zhang W, Ma L, Yang M, Shao Q, Xu J, Lu Z, et al. Cerebral organoid and mouse models reveal a RAB39b-PI3K-mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 2020;34(7–8):580–97.10.1101/gad.332494.119Suche in Google Scholar PubMed PubMed Central
[21] Chiu CC, Weng YH, Yeh TH, Lu JC, Chen WS, Li AH, et al. Deficiency of RAB39B activates ER stress-induced pro-apoptotic pathway and causes mitochondrial dysfunction and oxidative stress in dopaminergic neurons by impairing autophagy and upregulating alpha-synuclein. Mol Neurobiol. 2023;60(5):2706–28.10.1007/s12035-023-03238-6Suche in Google Scholar PubMed
[22] Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016;2(9):e1601167.10.1126/sciadv.1601167Suche in Google Scholar PubMed PubMed Central
[23] Gopal Krishnan PD, Golden E, Woodward EA, Pavlos NJ, Blancafort P. Rab GTPases: Emerging oncogenes and tumor suppressive regulators for the editing of survival pathways in cancer. Cancers (Basel). 2020;12(2):259.10.3390/cancers12020259Suche in Google Scholar PubMed PubMed Central
[24] Jin H, Tang Y, Yang L, Peng X, Li B, Fan Q, et al. Rab GTPases: Central coordinators of membrane trafficking in cancer. Front Cell Dev Biol. 2021;9:648384.10.3389/fcell.2021.648384Suche in Google Scholar PubMed PubMed Central
[25] Li K, Chen L, Zhang H, Wang L, Sha K, Du X, et al. High expression of COMMD7 is an adverse prognostic factor in acute myeloid leukemia. Aging (Albany NY). 2021;13(8):11988–2006.10.18632/aging.202901Suche in Google Scholar PubMed PubMed Central
[26] Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–8.10.1038/s41587-020-0546-8Suche in Google Scholar PubMed PubMed Central
[27] Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63.10.1093/nar/gkx1090Suche in Google Scholar PubMed PubMed Central
[28] Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–12.10.1093/nar/gkaa1074Suche in Google Scholar PubMed PubMed Central
[29] Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7.10.1093/nar/gkt1248Suche in Google Scholar PubMed PubMed Central
[30] Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–51.10.1093/nar/gkaa467Suche in Google Scholar PubMed PubMed Central
[31] Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95.10.1016/j.immuni.2013.10.003Suche in Google Scholar PubMed
[32] Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–61.10.1126/science.abf0529Suche in Google Scholar PubMed PubMed Central
[33] Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X, et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18(1):137.10.1186/s12943-019-1066-3Suche in Google Scholar PubMed PubMed Central
[34] Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60.10.1093/nar/gkz430Suche in Google Scholar PubMed PubMed Central
[35] Niu M, Zheng N, Wang Z, Gao Y, Luo X, Chen Z, et al. RAB39B deficiency impairs learning and memory partially through compromising autophagy. Front Cell Dev Biol. 2020;8:598622.10.3389/fcell.2020.598622Suche in Google Scholar PubMed PubMed Central
[36] Wang B, Wang W, Li Q, Guo T, Yang S, Shi J, et al. High expression of microtubule-associated protein TBCB predicts adverse outcome and immunosuppression in acute myeloid leukemia. J Cancer. 2023;14(10):1707–24.10.7150/jca.84215Suche in Google Scholar PubMed PubMed Central
[37] Lu X, Li Y, Yang Y, Zhuang W, Chai X, Gong C. OLFML2A overexpression predicts an unfavorable prognosis in patients with AML. J Oncol. 2023;2023:6017852.10.1155/2023/6017852Suche in Google Scholar PubMed PubMed Central
[38] Koss DJ, Campesan S, Giorgini F, Outeiro TF. Dysfunction of RAB39B-mediated vesicular trafficking in lewy body diseases. Mov Disord. 2021;36(8):1744–58.10.1002/mds.28605Suche in Google Scholar PubMed
[39] Wang Q, Ni Q, Wang X, Zhu H, Wang Z, Huang J. High expression of RAB27A and TP53 in pancreatic cancer predicts poor survival. Med Oncol. 2015;32(1):372.10.1007/s12032-014-0372-2Suche in Google Scholar PubMed
[40] Zhang XD, Liu ZY, Luo K, Wang XK, Wang MS, Huang S, et al. Clinical implications of RAB13 expression in pan-cancer based on multi-databases integrative analysis. Sci Rep. 2023;13(1):16859.10.1038/s41598-023-43699-2Suche in Google Scholar PubMed PubMed Central
[41] Yang J, Liu W, Lu X, Fu Y, Li L, Luo Y. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget. 2015;6(13):11125–38.10.18632/oncotarget.3575Suche in Google Scholar PubMed PubMed Central
[42] Yang L, Zhu Z, Zheng Y, Yang J, Liu Y, Shen T, et al. RAB6A functions as a critical modulator of the stem-like subsets in cholangiocarcinoma. Mol Carcinog. 2023;62(10):1460–73.10.1002/mc.23589Suche in Google Scholar PubMed
[43] Thomas JD, Zhang YJ, Wei YH, Cho JH, Morris LE, Wang HY, et al. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 2014;26(5):754–69.10.1016/j.ccell.2014.09.008Suche in Google Scholar PubMed PubMed Central
[44] Wang Z, Niu M, Zheng N, Meng J, Jiang Y, Yang D, et al. Increased level of RAB39B leads to neuronal dysfunction and behavioural changes in mice. J Cell Mol Med. 2023;27(9):1214–26.10.1111/jcmm.17704Suche in Google Scholar PubMed PubMed Central
[45] Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25(3):331–43.10.1016/j.molcel.2007.01.003Suche in Google Scholar PubMed
[46] Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17(8):807–21.10.1038/s41423-020-0488-6Suche in Google Scholar PubMed PubMed Central
[47] Daver N, Alotaibi AS, Bucklein V, Subklewe M. T-cell-based immunotherapy of acute myeloid leukemia: current concepts and future developments. Leukemia. 2021;35(7):1843–63.10.1038/s41375-021-01253-xSuche in Google Scholar PubMed PubMed Central
[48] Zeng H, Zhou S, Cai W, Kang M, Zhang P. LncRNA SNHG1: Role in tumorigenesis of multiple human cancers. Cancer Cell Int. 2023;23(1):198.10.1186/s12935-023-03018-1Suche in Google Scholar PubMed PubMed Central
[49] Xu Y, Luan G, Li Z, Liu Z, Qin G, Chu Y. Tumour-derived exosomal lncRNA SNHG16 induces telocytes to promote metastasis of hepatocellular carcinoma via the miR-942-3p/MMP9 axis. Cell Oncol (Dordr). 2023;46(2):251–64.10.1007/s13402-023-00782-0Suche in Google Scholar PubMed
[50] Yang J, Yang Y. Long noncoding RNA endogenous bornavirus-like nucleoprotein acts as an oncogene by regulating microRNA-655-3p expression in T-cell acute lymphoblastic leukemia. Bioengineered. 2022;13(3):6409–19.10.1080/21655979.2022.2044249Suche in Google Scholar PubMed PubMed Central
[51] Chen YP, Zhang DX, Cao Q, He CK. LncRNA HCG18 promotes osteosarcoma cells proliferation, migration, and invasion in by regulating miR-34a/RUNX2 pathway. Biochem Genet. 2023;61(3):1035–49.10.1007/s10528-022-10294-5Suche in Google Scholar PubMed
[52] Hong YG, Yang Z, Chen Y, Liu T, Zheng Y, Zhou C, et al. The RNA m6A reader YTHDF1 is required for acute myeloid leukemia progression. Cancer Res. 2023;83(6):845–60.10.1158/0008-5472.CAN-21-4249Suche in Google Scholar PubMed
[53] Liao X, Chen L, Liu J, Hu H, Hou D, You R, et al. m(6)A RNA methylation regulators predict prognosis and indicate characteristics of tumour microenvironment infiltration in acute myeloid leukaemia. Epigenetics. 2023;18(1):2160134.10.1080/15592294.2022.2160134Suche in Google Scholar PubMed PubMed Central
[54] Zhang N, Shen Y, Li H, Chen Y, Zhang P, Lou S, et al. The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. Exp Mol Med. 2022;54(2):194–205.10.1038/s12276-022-00735-xSuche in Google Scholar PubMed PubMed Central
[55] Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601.10.1038/s41586-021-03536-wSuche in Google Scholar PubMed PubMed Central
[56] Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677–91 e10.10.1016/j.ccell.2019.03.006Suche in Google Scholar PubMed PubMed Central
[57] Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205 e9.10.1016/j.stem.2017.11.016Suche in Google Scholar PubMed PubMed Central
[58] Cheng Y, Gao Z, Zhang T, Wang Y, Xie X, Han G, et al. Decoding m(6)A RNA methylome identifies PRMT6-regulated lipid transport promoting AML stem cell maintenance. Cell Stem Cell. 2023;30(1):69–85 e7.10.1016/j.stem.2022.12.003Suche in Google Scholar PubMed
[59] Wang J, Li Y, Wang P, Han G, Zhang T, Chang J, et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling axis. Cell Stem Cell. 2020;27(1):81–97 e8.10.1016/j.stem.2020.04.001Suche in Google Scholar PubMed
[60] Li Y, Kan X. Cuproptosis-related genes MTF1 and LIPT1 as novel prognostic biomarker in acute myeloid leukemia. Biochem Genet. 2024;62(2):1136–59.10.1007/s10528-023-10473-ySuche in Google Scholar PubMed
[61] Li P, Li J, Wen F, Cao Y, Luo Z, Zuo J, et al. A novel cuproptosis-related LncRNA signature: Prognostic and therapeutic value for acute myeloid leukemia. Front Oncol. 2022;12:966920.10.3389/fonc.2022.966920Suche in Google Scholar PubMed PubMed Central
[62] Pratcorona M, Brunet S, Nomdedeu J, Ribera JM, Tormo M, Duarte R, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013;121(14):2734–8.10.1182/blood-2012-06-431122Suche in Google Scholar PubMed
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Network pharmacological analysis and in vitro testing of the rutin effects on triple-negative breast cancer
- Impact of diabetes on long-term survival in elderly liver cancer patients: A retrospective study
- Knockdown of CCNB1 alleviates high glucose-triggered trophoblast dysfunction during gestational diabetes via Wnt/β-catenin signaling pathway
- Risk factors for severe adverse drug reactions in hospitalized patients
- Analysis of the effect of ALA-PDT on macrophages in footpad model of mice infected with Fonsecaea monophora based on single-cell sequencing
- Development and validation of headspace gas chromatography with a flame ionization detector method for the determination of ethanol in the vitreous humor
- CMSP exerts anti-tumor effects on small cell lung cancer cells by inducing mitochondrial dysfunction and ferroptosis
- Predictive value of plasma sB7-H3 and YKL-40 in pediatric refractory Mycoplasma pneumoniae pneumonia
- Antiangiogenic potential of Elaeagnus umbellata extracts and molecular docking study by targeting VEGFR-2 pathway
- Comparison of the effectiveness of nurse-led preoperative counseling and postoperative follow-up care vs standard care for patients with gastric cancer
- Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis
- Adhered macrophages as an additional marker of cardiomyocyte injury in biopsies of patients with dilated cardiomyopathy
- Association between statin administration and outcome in patients with sepsis: A retrospective study
- Exploration of the association between estimated glucose disposal rate and osteoarthritis in middle-aged and older adults: An analysis of NHANES data from 2011 to 2018
- A comparative analysis of the binary and multiclass classified chest X-ray images of pneumonia and COVID-19 with ML and DL models
- Lysophosphatidic acid 2 alleviates deep vein thrombosis via protective endothelial barrier function
- Transcription factor A, mitochondrial promotes lymph node metastasis and lymphangiogenesis in epithelial ovarian carcinoma
- Serum PM20D1 levels are associated with nutritional status and inflammatory factors in gastric cancer patients undergoing early enteral nutrition
- Hydromorphone reduced the incidence of emergence agitation after adenotonsillectomy in children with obstructive sleep apnea: A randomized, double-blind study
- Vitamin D replacement therapy may regulate sleep habits in patients with restless leg syndrome
- The first-line antihypertensive nitrendipine potentiated the therapeutic effect of oxaliplatin by downregulating CACNA1D in colorectal cancer
- Health literacy and health-related quality of life: The mediating role of irrational happiness
- Modulatory effects of Lycium barbarum polysaccharide on bone cell dynamics in osteoporosis
- Mechanism research on inhibition of gastric cancer in vitro by the extract of Pinellia ternata based on network pharmacology and cellular metabolomics
- Examination of the causal role of immune cells in non-alcoholic fatty liver disease by a bidirectional Mendelian randomization study
- Clinical analysis of ten cases of HIV infection combined with acute leukemia
- Investigating the cardioprotective potential of quercetin against tacrolimus-induced cardiotoxicity in Wistar rats: A mechanistic insights
- Clinical observation of probiotics combined with mesalazine and Yiyi Baitouweng Decoction retention enema in treating mild-to-moderate ulcerative colitis
- Diagnostic value of ratio of blood inflammation to coagulation markers in periprosthetic joint infection
- Sex-specific associations of sex hormone binding globulin and risk of bladder cancer
- Core muscle strength and stability-oriented breathing training reduces inter-recti distance in postpartum women
- The ERAS nursing care strategy for patients undergoing transsphenoidal endoscopic pituitary tumor resection: A randomized blinded controlled trial
- The serum IL-17A levels in patients with traumatic bowel rupture post-surgery and its predictive value for patient prognosis
- Impact of Kolb’s experiential learning theory-based nursing on caregiver burden and psychological state of caregivers of dementia patients
- Analysis of serum NLR combined with intraoperative margin condition to predict the prognosis of cervical HSIL patients undergoing LEEP surgery
- Commiphora gileadensis ameliorate infertility and erectile dysfunction in diabetic male mice
- The correlation between epithelial–mesenchymal transition classification and MMP2 expression of circulating tumor cells and prognosis of advanced or metastatic nasopharyngeal carcinoma
- Tetrahydropalmatine improves mitochondrial function in vascular smooth muscle cells of atherosclerosis in vitro by inhibiting Ras homolog gene family A/Rho-associated protein kinase-1 signaling pathway
- A cross-sectional study: Relationship between serum oxidative stress levels and arteriovenous fistula maturation in maintenance dialysis patients
- A comparative analysis of the impact of repeated administration of flavan 3-ol on brown, subcutaneous, and visceral adipose tissue
- Identifying early screening factors for depression in middle-aged and older adults: A cohort study
- Perform tumor-specific survival analysis for Merkel cell carcinoma patients undergoing surgical resection based on the SEER database by constructing a nomogram chart
- Unveiling the role of CXCL10 in pancreatic cancer progression: A novel prognostic indicator
- High-dose preoperative intraperitoneal erythropoietin and intravenous methylprednisolone in acute traumatic spinal cord injuries following decompression surgeries
- RAB39B: A novel biomarker for acute myeloid leukemia identified via multi-omics and functional validation
- Impact of peripheral conditioning on reperfusion injury following primary percutaneous coronary intervention in diabetic and non-diabetic STEMI patients
- Clinical efficacy of azacitidine in the treatment of middle- and high-risk myelodysplastic syndrome in middle-aged and elderly patients: A retrospective study
- The effect of ambulatory blood pressure load on mitral regurgitation in continuous ambulatory peritoneal dialysis patients
- Expression and clinical significance of ITGA3 in breast cancer
- Single-nucleus RNA sequencing reveals ARHGAP28 expression of podocytes as a biomarker in human diabetic nephropathy
- rSIG combined with NLR in the prognostic assessment of patients with multiple injuries
- Toxic metals and metalloids in collagen supplements of fish and jellyfish origin: Risk assessment for daily intake
- Exploring causal relationship between 41 inflammatory cytokines and marginal zone lymphoma: A bidirectional Mendelian randomization study
- Gender beliefs and legitimization of dating violence in adolescents
- Effect of serum IL-6, CRP, and MMP-9 levels on the efficacy of modified preperitoneal Kugel repair in patients with inguinal hernia
- Effect of smoking and smoking cessation on hematological parameters in polycythemic patients
- Pathogen surveillance and risk factors for pulmonary infection in patients with lung cancer: A retrospective single-center study
- Necroptosis of hippocampal neurons in paclitaxel chemotherapy-induced cognitive impairment mediates microglial activation via TLR4/MyD88 signaling pathway
- Celastrol suppresses neovascularization in rat aortic vascular endothelial cells stimulated by inflammatory tenocytes via modulating the NLRP3 pathway
- Cord-lamina angle and foraminal diameter as key predictors of C5 palsy after anterior cervical decompression and fusion surgery
- GATA1: A key biomarker for predicting the prognosis of patients with diffuse large B-cell lymphoma
- Influencing factors of false lumen thrombosis in type B aortic dissection: A single-center retrospective study
- MZB1 regulates the immune microenvironment and inhibits ovarian cancer cell migration
- Integrating experimental and network pharmacology to explore the pharmacological mechanisms of Dioscin against glioblastoma
- Trends in research on preterm birth in twin pregnancy based on bibliometrics
- Four-week IgE/baseline IgE ratio combined with tryptase predicts clinical outcome in omalizumab-treated children with moderate-to-severe asthma
- Single-cell transcriptomic analysis identifies a stress response Schwann cell subtype
- Acute pancreatitis risk in the diagnosis and management of inflammatory bowel disease: A critical focus
- Effect of subclinical esketamine on NLRP3 and cognitive dysfunction in elderly ischemic stroke patients
- Interleukin-37 mediates the anti-oral tumor activity in oral cancer through STAT3
- CA199 and CEA expression levels, and minimally invasive postoperative prognosis analysis in esophageal squamous carcinoma patients
- Efficacy of a novel drainage catheter in the treatment of CSF leak after posterior spine surgery: A retrospective cohort study
- Comprehensive biomedicine assessment of Apteranthes tuberculata extracts: Phytochemical analysis and multifaceted pharmacological evaluation in animal models
- Relation of time in range to severity of coronary artery disease in patients with type 2 diabetes: A cross-sectional study
- Dopamine attenuates ethanol-induced neuronal apoptosis by stimulating electrical activity in the developing rat retina
- Correlation between albumin levels during the third trimester and the risk of postpartum levator ani muscle rupture
- Factors associated with maternal attention and distraction during breastfeeding and childcare: A cross-sectional study in the west of Iran
- Mechanisms of hesperetin in treating metabolic dysfunction-associated steatosis liver disease via network pharmacology and in vitro experiments
- The law on oncological oblivion in the Italian and European context: How to best uphold the cancer patients’ rights to privacy and self-determination?
- The prognostic value of the neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and prognostic nutritional index for survival in patients with colorectal cancer
- Factors affecting the measurements of peripheral oxygen saturation values in healthy young adults
- Comparison and correlations between findings of hysteroscopy and vaginal color Doppler ultrasonography for detection of uterine abnormalities in patients with recurrent implantation failure
- The effects of different types of RAGT on balance function in stroke patients with low levels of independent walking in a convalescent rehabilitation hospital
- Causal relationship between asthma and ankylosing spondylitis: A bidirectional two-sample univariable and multivariable Mendelian randomization study
- Correlations of health literacy with individuals’ understanding and use of medications in Southern Taiwan
- Correlation of serum calprotectin with outcome of acute cerebral infarction
- Comparison of computed tomography and guided bronchoscopy in the diagnosis of pulmonary nodules: A systematic review and meta-analysis
- Curdione protects vascular endothelial cells and atherosclerosis via the regulation of DNMT1-mediated ERBB4 promoter methylation
- The identification of novel missense variant in ChAT gene in a patient with gestational diabetes denotes plausible genetic association
- Molecular genotyping of multi-system rare blood types in foreign blood donors based on DNA sequencing and its clinical significance
- Exploring the role of succinyl carnitine in the association between CD39⁺ CD4⁺ T cell and ulcerative colitis: A Mendelian randomization study
- Dexmedetomidine suppresses microglial activation in postoperative cognitive dysfunction via the mmu-miRNA-125/TRAF6 signaling axis
- Analysis of serum metabolomics in patients with different types of chronic heart failure
- Diagnostic value of hematological parameters in the early diagnosis of acute cholecystitis
- Pachymaran alleviates fat accumulation, hepatocyte degeneration, and injury in mice with nonalcoholic fatty liver disease
- Decrease in CD4 and CD8 lymphocytes are predictors of severe clinical picture and unfavorable outcome of the disease in patients with COVID-19
- METTL3 blocked the progression of diabetic retinopathy through m6A-modified SOX2
- The predictive significance of anti-RO-52 antibody in patients with interstitial pneumonia after treatment of malignant tumors
- Exploring cerebrospinal fluid metabolites, cognitive function, and brain atrophy: Insights from Mendelian randomization
- Development and validation of potential molecular subtypes and signatures of ocular sarcoidosis based on autophagy-related gene analysis
- Widespread venous thrombosis: Unveiling a complex case of Behçet’s disease with a literature perspective
- Uterine fibroid embolization: An analysis of clinical outcomes and impact on patients’ quality of life
- Discovery of lipid metabolism-related diagnostic biomarkers and construction of diagnostic model in steroid-induced osteonecrosis of femoral head
- Serum-derived exomiR-188-3p is a promising novel biomarker for early-stage ovarian cancer
- Enhancing chronic back pain management: A comparative study of ultrasound–MRI fusion guidance for paravertebral nerve block
- Peptide CCAT1-70aa promotes hepatocellular carcinoma proliferation and invasion via the MAPK/ERK pathway
- Electroacupuncture-induced reduction of myocardial ischemia–reperfusion injury via FTO-dependent m6A methylation modulation
- Hemorrhoids and cardiovascular disease: A bidirectional Mendelian randomization study
- Cell-free adipose extract inhibits hypertrophic scar formation through collagen remodeling and antiangiogenesis
- HALP score in Demodex blepharitis: A case–control study
- Assessment of SOX2 performance as a marker for circulating cancer stem-like cells (CCSCs) identification in advanced breast cancer patients using CytoTrack system
- Risk and prognosis for brain metastasis in primary metastatic cervical cancer patients: A population-based study
- Comparison of the two intestinal anastomosis methods in pediatric patients
- Factors influencing hematological toxicity and adverse effects of perioperative hyperthermic intraperitoneal vs intraperitoneal chemotherapy in gastrointestinal cancer
- Endotoxin tolerance inhibits NLRP3 inflammasome activation in macrophages of septic mice by restoring autophagic flux through TRIM26
- Lateral transperitoneal laparoscopic adrenalectomy: A single-centre experience of 21 procedures
- Petunidin attenuates lipopolysaccharide-induced retinal microglia inflammatory response in diabetic retinopathy by targeting OGT/NF-κB/LCN2 axis
- Procalcitonin and C-reactive protein as biomarkers for diagnosing and assessing the severity of acute cholecystitis
- Factors determining the number of sessions in successful extracorporeal shock wave lithotripsy patients
- Development of a nomogram for predicting cancer-specific survival in patients with renal pelvic cancer following surgery
- Inhibition of ATG7 promotes orthodontic tooth movement by regulating the RANKL/OPG ratio under compression force
- A machine learning-based prognostic model integrating mRNA stemness index, hypoxia, and glycolysis‑related biomarkers for colorectal cancer
- Glutathione attenuates sepsis-associated encephalopathy via dual modulation of NF-κB and PKA/CREB pathways
- FAHD1 prevents neuronal ferroptosis by modulating R-loop and the cGAS–STING pathway
- Association of placenta weight and morphology with term low birth weight: A case–control study
- Investigation of the pathogenic variants induced Sjogren’s syndrome in Turkish population
- Nucleotide metabolic abnormalities in post-COVID-19 condition and type 2 diabetes mellitus patients and their association with endocrine dysfunction
- TGF-β–Smad2/3 signaling in high-altitude pulmonary hypertension in rats: Role and mechanisms via macrophage M2 polarization
- Ultrasound-guided unilateral versus bilateral erector spinae plane block for postoperative analgesia of patients undergoing laparoscopic cholecystectomy
- Profiling gut microbiome dynamics in subacute thyroiditis: Implications for pathogenesis, diagnosis, and treatment
- Delta neutrophil index, CRP/albumin ratio, procalcitonin, immature granulocytes, and HALP score in acute appendicitis: Best performing biomarker?
- Anticancer activity mechanism of novelly synthesized and characterized benzofuran ring-linked 3-nitrophenyl chalcone derivative on colon cancer cells
- H2valdien3 arrests the cell cycle and induces apoptosis of gastric cancer
- Prognostic relevance of PRSS2 and its immune correlates in papillary thyroid carcinoma
- Association of SGLT2 inhibition with psychiatric disorders: A Mendelian randomization study
- Motivational interviewing for alcohol use reduction in Thai patients
- Luteolin alleviates oxygen-glucose deprivation/reoxygenation-induced neuron injury by regulating NLRP3/IL-1β signaling
- Polyphyllin II inhibits thyroid cancer cell growth by simultaneously inhibiting glycolysis and oxidative phosphorylation
- Relationship between the expression of copper death promoting factor SLC31A1 in papillary thyroid carcinoma and clinicopathological indicators and prognosis
- CSF2 polarized neutrophils and invaded renal cancer cells in vitro influence
- Proton pump inhibitors-induced thrombocytopenia: A systematic literature analysis of case reports
- The current status and influence factors of research ability among community nurses: A sequential qualitative–quantitative study
- OKAIN: A comprehensive oncology knowledge base for the interpretation of clinically actionable alterations
- The relationship between serum CA50, CA242, and SAA levels and clinical pathological characteristics and prognosis in patients with pancreatic cancer
- Identification and external validation of a prognostic signature based on hypoxia–glycolysis-related genes for kidney renal clear cell carcinoma
- Engineered RBC-derived nanovesicles functionalized with tumor-targeting ligands: A comparative study on breast cancer targeting efficiency and biocompatibility
- Relationship of resting echocardiography combined with serum micronutrients to the severity of low-gradient severe aortic stenosis
- Effect of vibration on pain during subcutaneous heparin injection: A randomized, single-blind, placebo-controlled trial
- The diagnostic performance of machine learning-based FFRCT for coronary artery disease: A meta-analysis
- Comparing biofeedback device vs diaphragmatic breathing for bloating relief: A randomized controlled trial
- Serum uric acid to albumin ratio and C-reactive protein as predictive biomarkers for chronic total occlusion and coronary collateral circulation quality
- Multiple organ scoring systems for predicting in-hospital mortality of sepsis patients in the intensive care unit
- Single-cell RNA sequencing data analysis of the inner ear in gentamicin-treated mice via intraperitoneal injection
- Suppression of cathepsin B attenuates myocardial injury via limiting cardiomyocyte apoptosis
- Influence of sevoflurane combined with propofol anesthesia on the anesthesia effect and adverse reactions in children with acute appendicitis
- Review Articles
- The effects of enhanced external counter-pulsation on post-acute sequelae of COVID-19: A narrative review
- Diabetes-related cognitive impairment: Mechanisms, symptoms, and treatments
- Microscopic changes and gross morphology of placenta in women affected by gestational diabetes mellitus in dietary treatment: A systematic review
- Review of mechanisms and frontier applications in IL-17A-induced hypertension
- Research progress on the correlation between islet amyloid peptides and type 2 diabetes mellitus
- The safety and efficacy of BCG combined with mitomycin C compared with BCG monotherapy in patients with non-muscle-invasive bladder cancer: A systematic review and meta-analysis
- The application of augmented reality in robotic general surgery: A mini-review
- The effect of Greek mountain tea extract and wheat germ extract on peripheral blood flow and eicosanoid metabolism in mammals
- Neurogasobiology of migraine: Carbon monoxide, hydrogen sulfide, and nitric oxide as emerging pathophysiological trinacrium relevant to nociception regulation
- Plant polyphenols, terpenes, and terpenoids in oral health
- Laboratory medicine between technological innovation, rights safeguarding, and patient safety: A bioethical perspective
- End-of-life in cancer patients: Medicolegal implications and ethical challenges in Europe
- The maternal factors during pregnancy for intrauterine growth retardation: An umbrella review
- Intra-abdominal hypertension/abdominal compartment syndrome of pediatric patients in critical care settings
- PI3K/Akt pathway and neuroinflammation in sepsis-associated encephalopathy
- Screening of Group B Streptococcus in pregnancy: A systematic review for the laboratory detection
- Giant borderline ovarian tumours – review of the literature
- Leveraging artificial intelligence for collaborative care planning: Innovations and impacts in shared decision-making – A systematic review
- Cholera epidemiology analysis through the experience of the 1973 Naples epidemic
- Risk factors of frailty/sarcopenia in community older adults: Meta-analysis
- Supplement strategies for infertility in overweight women: Evidence and legal insights
- Scurvy, a not obsolete disorder: Clinical report in eight young children and literature review
- A meta-analysis of the effects of DBS on cognitive function in patients with advanced PD
- Protective role of selenium in sepsis: Mechanisms and potential therapeutic strategies
- Strategies for hyperkalemia management in dialysis patients: A systematic review
- C-reactive protein-to-albumin ratio in peripheral artery disease
- Case Reports
- Delayed graft function after renal transplantation
- Semaglutide treatment for type 2 diabetes in a patient with chronic myeloid leukemia: A case report and review of the literature
- Diverse electrophysiological demyelinating features in a late-onset glycogen storage disease type IIIa case
- Giant right atrial hemangioma presenting with ascites: A case report
- Laser excision of a large granular cell tumor of the vocal cord with subglottic extension: A case report
- EsoFLIP-assisted dilation for dysphagia in systemic sclerosis: Highlighting the role of multimodal esophageal evaluation
- Molecular hydrogen-rhodiola as an adjuvant therapy for ischemic stroke in internal carotid artery occlusion: A case report
- Coronary artery anomalies: A case of the “malignant” left coronary artery and its surgical management
- Rapid Communication
- Biological properties of valve materials using RGD and EC
-
A single oral administration of flavanols enhances short
-term memory in mice along with increased brain-derived neurotrophic factor - Letter to the Editor
- Role of enhanced external counterpulsation in long COVID
- Expression of Concern
- Expression of concern “A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma”
- Expression of concern “Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway”
- Expression of concern “circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8”
- Corrigendum
- Corrigendum to “Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism”
- Corrigendum to “Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis”
- Corrigendum to “The progress of autoimmune hepatitis research and future challenges”
- Retraction
- Retraction of “miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway”
- Retraction of: “LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through downregulating SP-A by sponging to miR-424”
- Retraction of: “SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways”
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part II
- Unveiling novel biomarkers for platinum chemoresistance in ovarian cancer
- Lathyrol affects the expression of AR and PSA and inhibits the malignant behavior of RCC cells
- The era of increasing cancer survivorship: Trends in fertility preservation, medico-legal implications, and ethical challenges
- Bone scintigraphy and positron emission tomography in the early diagnosis of MRONJ
- Meta-analysis of clinical efficacy and safety of immunotherapy combined with chemotherapy in non-small cell lung cancer
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part IV
- Exploration of mRNA-modifying METTL3 oncogene as momentous prognostic biomarker responsible for colorectal cancer development
- Special Issue The evolving saga of RNAs from bench to bedside - Part III
- Interaction and verification of ferroptosis-related RNAs Rela and Stat3 in promoting sepsis-associated acute kidney injury
- The mRNA MOXD1: Link to oxidative stress and prognostic significance in gastric cancer
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part II
- Dynamic changes in lactate-related genes in microglia and their role in immune cell interactions after ischemic stroke
- A prognostic model correlated with fatty acid metabolism in Ewing’s sarcoma based on bioinformatics analysis
- Red cell distribution width predicts early kidney injury: A NHANES cross-sectional study
- Special Issue Diabetes mellitus: pathophysiology, complications & treatment
- Nutritional risk assessment and nutritional support in children with congenital diabetes during surgery
- Correlation of the differential expressions of RANK, RANKL, and OPG with obesity in the elderly population in Xinjiang
- A discussion on the application of fluorescence micro-optical sectioning tomography in the research of cognitive dysfunction in diabetes
- A review of brain research on T2DM-related cognitive dysfunction
- Metformin and estrogen modulation in LABC with T2DM: A 36-month randomized trial
- Special Issue Innovative Biomarker Discovery and Precision Medicine in Cancer Diagnostics
- CircASH1L-mediated tumor progression in triple-negative breast cancer: PI3K/AKT pathway mechanisms
Artikel in diesem Heft
- Research Articles
- Network pharmacological analysis and in vitro testing of the rutin effects on triple-negative breast cancer
- Impact of diabetes on long-term survival in elderly liver cancer patients: A retrospective study
- Knockdown of CCNB1 alleviates high glucose-triggered trophoblast dysfunction during gestational diabetes via Wnt/β-catenin signaling pathway
- Risk factors for severe adverse drug reactions in hospitalized patients
- Analysis of the effect of ALA-PDT on macrophages in footpad model of mice infected with Fonsecaea monophora based on single-cell sequencing
- Development and validation of headspace gas chromatography with a flame ionization detector method for the determination of ethanol in the vitreous humor
- CMSP exerts anti-tumor effects on small cell lung cancer cells by inducing mitochondrial dysfunction and ferroptosis
- Predictive value of plasma sB7-H3 and YKL-40 in pediatric refractory Mycoplasma pneumoniae pneumonia
- Antiangiogenic potential of Elaeagnus umbellata extracts and molecular docking study by targeting VEGFR-2 pathway
- Comparison of the effectiveness of nurse-led preoperative counseling and postoperative follow-up care vs standard care for patients with gastric cancer
- Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis
- Adhered macrophages as an additional marker of cardiomyocyte injury in biopsies of patients with dilated cardiomyopathy
- Association between statin administration and outcome in patients with sepsis: A retrospective study
- Exploration of the association between estimated glucose disposal rate and osteoarthritis in middle-aged and older adults: An analysis of NHANES data from 2011 to 2018
- A comparative analysis of the binary and multiclass classified chest X-ray images of pneumonia and COVID-19 with ML and DL models
- Lysophosphatidic acid 2 alleviates deep vein thrombosis via protective endothelial barrier function
- Transcription factor A, mitochondrial promotes lymph node metastasis and lymphangiogenesis in epithelial ovarian carcinoma
- Serum PM20D1 levels are associated with nutritional status and inflammatory factors in gastric cancer patients undergoing early enteral nutrition
- Hydromorphone reduced the incidence of emergence agitation after adenotonsillectomy in children with obstructive sleep apnea: A randomized, double-blind study
- Vitamin D replacement therapy may regulate sleep habits in patients with restless leg syndrome
- The first-line antihypertensive nitrendipine potentiated the therapeutic effect of oxaliplatin by downregulating CACNA1D in colorectal cancer
- Health literacy and health-related quality of life: The mediating role of irrational happiness
- Modulatory effects of Lycium barbarum polysaccharide on bone cell dynamics in osteoporosis
- Mechanism research on inhibition of gastric cancer in vitro by the extract of Pinellia ternata based on network pharmacology and cellular metabolomics
- Examination of the causal role of immune cells in non-alcoholic fatty liver disease by a bidirectional Mendelian randomization study
- Clinical analysis of ten cases of HIV infection combined with acute leukemia
- Investigating the cardioprotective potential of quercetin against tacrolimus-induced cardiotoxicity in Wistar rats: A mechanistic insights
- Clinical observation of probiotics combined with mesalazine and Yiyi Baitouweng Decoction retention enema in treating mild-to-moderate ulcerative colitis
- Diagnostic value of ratio of blood inflammation to coagulation markers in periprosthetic joint infection
- Sex-specific associations of sex hormone binding globulin and risk of bladder cancer
- Core muscle strength and stability-oriented breathing training reduces inter-recti distance in postpartum women
- The ERAS nursing care strategy for patients undergoing transsphenoidal endoscopic pituitary tumor resection: A randomized blinded controlled trial
- The serum IL-17A levels in patients with traumatic bowel rupture post-surgery and its predictive value for patient prognosis
- Impact of Kolb’s experiential learning theory-based nursing on caregiver burden and psychological state of caregivers of dementia patients
- Analysis of serum NLR combined with intraoperative margin condition to predict the prognosis of cervical HSIL patients undergoing LEEP surgery
- Commiphora gileadensis ameliorate infertility and erectile dysfunction in diabetic male mice
- The correlation between epithelial–mesenchymal transition classification and MMP2 expression of circulating tumor cells and prognosis of advanced or metastatic nasopharyngeal carcinoma
- Tetrahydropalmatine improves mitochondrial function in vascular smooth muscle cells of atherosclerosis in vitro by inhibiting Ras homolog gene family A/Rho-associated protein kinase-1 signaling pathway
- A cross-sectional study: Relationship between serum oxidative stress levels and arteriovenous fistula maturation in maintenance dialysis patients
- A comparative analysis of the impact of repeated administration of flavan 3-ol on brown, subcutaneous, and visceral adipose tissue
- Identifying early screening factors for depression in middle-aged and older adults: A cohort study
- Perform tumor-specific survival analysis for Merkel cell carcinoma patients undergoing surgical resection based on the SEER database by constructing a nomogram chart
- Unveiling the role of CXCL10 in pancreatic cancer progression: A novel prognostic indicator
- High-dose preoperative intraperitoneal erythropoietin and intravenous methylprednisolone in acute traumatic spinal cord injuries following decompression surgeries
- RAB39B: A novel biomarker for acute myeloid leukemia identified via multi-omics and functional validation
- Impact of peripheral conditioning on reperfusion injury following primary percutaneous coronary intervention in diabetic and non-diabetic STEMI patients
- Clinical efficacy of azacitidine in the treatment of middle- and high-risk myelodysplastic syndrome in middle-aged and elderly patients: A retrospective study
- The effect of ambulatory blood pressure load on mitral regurgitation in continuous ambulatory peritoneal dialysis patients
- Expression and clinical significance of ITGA3 in breast cancer
- Single-nucleus RNA sequencing reveals ARHGAP28 expression of podocytes as a biomarker in human diabetic nephropathy
- rSIG combined with NLR in the prognostic assessment of patients with multiple injuries
- Toxic metals and metalloids in collagen supplements of fish and jellyfish origin: Risk assessment for daily intake
- Exploring causal relationship between 41 inflammatory cytokines and marginal zone lymphoma: A bidirectional Mendelian randomization study
- Gender beliefs and legitimization of dating violence in adolescents
- Effect of serum IL-6, CRP, and MMP-9 levels on the efficacy of modified preperitoneal Kugel repair in patients with inguinal hernia
- Effect of smoking and smoking cessation on hematological parameters in polycythemic patients
- Pathogen surveillance and risk factors for pulmonary infection in patients with lung cancer: A retrospective single-center study
- Necroptosis of hippocampal neurons in paclitaxel chemotherapy-induced cognitive impairment mediates microglial activation via TLR4/MyD88 signaling pathway
- Celastrol suppresses neovascularization in rat aortic vascular endothelial cells stimulated by inflammatory tenocytes via modulating the NLRP3 pathway
- Cord-lamina angle and foraminal diameter as key predictors of C5 palsy after anterior cervical decompression and fusion surgery
- GATA1: A key biomarker for predicting the prognosis of patients with diffuse large B-cell lymphoma
- Influencing factors of false lumen thrombosis in type B aortic dissection: A single-center retrospective study
- MZB1 regulates the immune microenvironment and inhibits ovarian cancer cell migration
- Integrating experimental and network pharmacology to explore the pharmacological mechanisms of Dioscin against glioblastoma
- Trends in research on preterm birth in twin pregnancy based on bibliometrics
- Four-week IgE/baseline IgE ratio combined with tryptase predicts clinical outcome in omalizumab-treated children with moderate-to-severe asthma
- Single-cell transcriptomic analysis identifies a stress response Schwann cell subtype
- Acute pancreatitis risk in the diagnosis and management of inflammatory bowel disease: A critical focus
- Effect of subclinical esketamine on NLRP3 and cognitive dysfunction in elderly ischemic stroke patients
- Interleukin-37 mediates the anti-oral tumor activity in oral cancer through STAT3
- CA199 and CEA expression levels, and minimally invasive postoperative prognosis analysis in esophageal squamous carcinoma patients
- Efficacy of a novel drainage catheter in the treatment of CSF leak after posterior spine surgery: A retrospective cohort study
- Comprehensive biomedicine assessment of Apteranthes tuberculata extracts: Phytochemical analysis and multifaceted pharmacological evaluation in animal models
- Relation of time in range to severity of coronary artery disease in patients with type 2 diabetes: A cross-sectional study
- Dopamine attenuates ethanol-induced neuronal apoptosis by stimulating electrical activity in the developing rat retina
- Correlation between albumin levels during the third trimester and the risk of postpartum levator ani muscle rupture
- Factors associated with maternal attention and distraction during breastfeeding and childcare: A cross-sectional study in the west of Iran
- Mechanisms of hesperetin in treating metabolic dysfunction-associated steatosis liver disease via network pharmacology and in vitro experiments
- The law on oncological oblivion in the Italian and European context: How to best uphold the cancer patients’ rights to privacy and self-determination?
- The prognostic value of the neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and prognostic nutritional index for survival in patients with colorectal cancer
- Factors affecting the measurements of peripheral oxygen saturation values in healthy young adults
- Comparison and correlations between findings of hysteroscopy and vaginal color Doppler ultrasonography for detection of uterine abnormalities in patients with recurrent implantation failure
- The effects of different types of RAGT on balance function in stroke patients with low levels of independent walking in a convalescent rehabilitation hospital
- Causal relationship between asthma and ankylosing spondylitis: A bidirectional two-sample univariable and multivariable Mendelian randomization study
- Correlations of health literacy with individuals’ understanding and use of medications in Southern Taiwan
- Correlation of serum calprotectin with outcome of acute cerebral infarction
- Comparison of computed tomography and guided bronchoscopy in the diagnosis of pulmonary nodules: A systematic review and meta-analysis
- Curdione protects vascular endothelial cells and atherosclerosis via the regulation of DNMT1-mediated ERBB4 promoter methylation
- The identification of novel missense variant in ChAT gene in a patient with gestational diabetes denotes plausible genetic association
- Molecular genotyping of multi-system rare blood types in foreign blood donors based on DNA sequencing and its clinical significance
- Exploring the role of succinyl carnitine in the association between CD39⁺ CD4⁺ T cell and ulcerative colitis: A Mendelian randomization study
- Dexmedetomidine suppresses microglial activation in postoperative cognitive dysfunction via the mmu-miRNA-125/TRAF6 signaling axis
- Analysis of serum metabolomics in patients with different types of chronic heart failure
- Diagnostic value of hematological parameters in the early diagnosis of acute cholecystitis
- Pachymaran alleviates fat accumulation, hepatocyte degeneration, and injury in mice with nonalcoholic fatty liver disease
- Decrease in CD4 and CD8 lymphocytes are predictors of severe clinical picture and unfavorable outcome of the disease in patients with COVID-19
- METTL3 blocked the progression of diabetic retinopathy through m6A-modified SOX2
- The predictive significance of anti-RO-52 antibody in patients with interstitial pneumonia after treatment of malignant tumors
- Exploring cerebrospinal fluid metabolites, cognitive function, and brain atrophy: Insights from Mendelian randomization
- Development and validation of potential molecular subtypes and signatures of ocular sarcoidosis based on autophagy-related gene analysis
- Widespread venous thrombosis: Unveiling a complex case of Behçet’s disease with a literature perspective
- Uterine fibroid embolization: An analysis of clinical outcomes and impact on patients’ quality of life
- Discovery of lipid metabolism-related diagnostic biomarkers and construction of diagnostic model in steroid-induced osteonecrosis of femoral head
- Serum-derived exomiR-188-3p is a promising novel biomarker for early-stage ovarian cancer
- Enhancing chronic back pain management: A comparative study of ultrasound–MRI fusion guidance for paravertebral nerve block
- Peptide CCAT1-70aa promotes hepatocellular carcinoma proliferation and invasion via the MAPK/ERK pathway
- Electroacupuncture-induced reduction of myocardial ischemia–reperfusion injury via FTO-dependent m6A methylation modulation
- Hemorrhoids and cardiovascular disease: A bidirectional Mendelian randomization study
- Cell-free adipose extract inhibits hypertrophic scar formation through collagen remodeling and antiangiogenesis
- HALP score in Demodex blepharitis: A case–control study
- Assessment of SOX2 performance as a marker for circulating cancer stem-like cells (CCSCs) identification in advanced breast cancer patients using CytoTrack system
- Risk and prognosis for brain metastasis in primary metastatic cervical cancer patients: A population-based study
- Comparison of the two intestinal anastomosis methods in pediatric patients
- Factors influencing hematological toxicity and adverse effects of perioperative hyperthermic intraperitoneal vs intraperitoneal chemotherapy in gastrointestinal cancer
- Endotoxin tolerance inhibits NLRP3 inflammasome activation in macrophages of septic mice by restoring autophagic flux through TRIM26
- Lateral transperitoneal laparoscopic adrenalectomy: A single-centre experience of 21 procedures
- Petunidin attenuates lipopolysaccharide-induced retinal microglia inflammatory response in diabetic retinopathy by targeting OGT/NF-κB/LCN2 axis
- Procalcitonin and C-reactive protein as biomarkers for diagnosing and assessing the severity of acute cholecystitis
- Factors determining the number of sessions in successful extracorporeal shock wave lithotripsy patients
- Development of a nomogram for predicting cancer-specific survival in patients with renal pelvic cancer following surgery
- Inhibition of ATG7 promotes orthodontic tooth movement by regulating the RANKL/OPG ratio under compression force
- A machine learning-based prognostic model integrating mRNA stemness index, hypoxia, and glycolysis‑related biomarkers for colorectal cancer
- Glutathione attenuates sepsis-associated encephalopathy via dual modulation of NF-κB and PKA/CREB pathways
- FAHD1 prevents neuronal ferroptosis by modulating R-loop and the cGAS–STING pathway
- Association of placenta weight and morphology with term low birth weight: A case–control study
- Investigation of the pathogenic variants induced Sjogren’s syndrome in Turkish population
- Nucleotide metabolic abnormalities in post-COVID-19 condition and type 2 diabetes mellitus patients and their association with endocrine dysfunction
- TGF-β–Smad2/3 signaling in high-altitude pulmonary hypertension in rats: Role and mechanisms via macrophage M2 polarization
- Ultrasound-guided unilateral versus bilateral erector spinae plane block for postoperative analgesia of patients undergoing laparoscopic cholecystectomy
- Profiling gut microbiome dynamics in subacute thyroiditis: Implications for pathogenesis, diagnosis, and treatment
- Delta neutrophil index, CRP/albumin ratio, procalcitonin, immature granulocytes, and HALP score in acute appendicitis: Best performing biomarker?
- Anticancer activity mechanism of novelly synthesized and characterized benzofuran ring-linked 3-nitrophenyl chalcone derivative on colon cancer cells
- H2valdien3 arrests the cell cycle and induces apoptosis of gastric cancer
- Prognostic relevance of PRSS2 and its immune correlates in papillary thyroid carcinoma
- Association of SGLT2 inhibition with psychiatric disorders: A Mendelian randomization study
- Motivational interviewing for alcohol use reduction in Thai patients
- Luteolin alleviates oxygen-glucose deprivation/reoxygenation-induced neuron injury by regulating NLRP3/IL-1β signaling
- Polyphyllin II inhibits thyroid cancer cell growth by simultaneously inhibiting glycolysis and oxidative phosphorylation
- Relationship between the expression of copper death promoting factor SLC31A1 in papillary thyroid carcinoma and clinicopathological indicators and prognosis
- CSF2 polarized neutrophils and invaded renal cancer cells in vitro influence
- Proton pump inhibitors-induced thrombocytopenia: A systematic literature analysis of case reports
- The current status and influence factors of research ability among community nurses: A sequential qualitative–quantitative study
- OKAIN: A comprehensive oncology knowledge base for the interpretation of clinically actionable alterations
- The relationship between serum CA50, CA242, and SAA levels and clinical pathological characteristics and prognosis in patients with pancreatic cancer
- Identification and external validation of a prognostic signature based on hypoxia–glycolysis-related genes for kidney renal clear cell carcinoma
- Engineered RBC-derived nanovesicles functionalized with tumor-targeting ligands: A comparative study on breast cancer targeting efficiency and biocompatibility
- Relationship of resting echocardiography combined with serum micronutrients to the severity of low-gradient severe aortic stenosis
- Effect of vibration on pain during subcutaneous heparin injection: A randomized, single-blind, placebo-controlled trial
- The diagnostic performance of machine learning-based FFRCT for coronary artery disease: A meta-analysis
- Comparing biofeedback device vs diaphragmatic breathing for bloating relief: A randomized controlled trial
- Serum uric acid to albumin ratio and C-reactive protein as predictive biomarkers for chronic total occlusion and coronary collateral circulation quality
- Multiple organ scoring systems for predicting in-hospital mortality of sepsis patients in the intensive care unit
- Single-cell RNA sequencing data analysis of the inner ear in gentamicin-treated mice via intraperitoneal injection
- Suppression of cathepsin B attenuates myocardial injury via limiting cardiomyocyte apoptosis
- Influence of sevoflurane combined with propofol anesthesia on the anesthesia effect and adverse reactions in children with acute appendicitis
- Review Articles
- The effects of enhanced external counter-pulsation on post-acute sequelae of COVID-19: A narrative review
- Diabetes-related cognitive impairment: Mechanisms, symptoms, and treatments
- Microscopic changes and gross morphology of placenta in women affected by gestational diabetes mellitus in dietary treatment: A systematic review
- Review of mechanisms and frontier applications in IL-17A-induced hypertension
- Research progress on the correlation between islet amyloid peptides and type 2 diabetes mellitus
- The safety and efficacy of BCG combined with mitomycin C compared with BCG monotherapy in patients with non-muscle-invasive bladder cancer: A systematic review and meta-analysis
- The application of augmented reality in robotic general surgery: A mini-review
- The effect of Greek mountain tea extract and wheat germ extract on peripheral blood flow and eicosanoid metabolism in mammals
- Neurogasobiology of migraine: Carbon monoxide, hydrogen sulfide, and nitric oxide as emerging pathophysiological trinacrium relevant to nociception regulation
- Plant polyphenols, terpenes, and terpenoids in oral health
- Laboratory medicine between technological innovation, rights safeguarding, and patient safety: A bioethical perspective
- End-of-life in cancer patients: Medicolegal implications and ethical challenges in Europe
- The maternal factors during pregnancy for intrauterine growth retardation: An umbrella review
- Intra-abdominal hypertension/abdominal compartment syndrome of pediatric patients in critical care settings
- PI3K/Akt pathway and neuroinflammation in sepsis-associated encephalopathy
- Screening of Group B Streptococcus in pregnancy: A systematic review for the laboratory detection
- Giant borderline ovarian tumours – review of the literature
- Leveraging artificial intelligence for collaborative care planning: Innovations and impacts in shared decision-making – A systematic review
- Cholera epidemiology analysis through the experience of the 1973 Naples epidemic
- Risk factors of frailty/sarcopenia in community older adults: Meta-analysis
- Supplement strategies for infertility in overweight women: Evidence and legal insights
- Scurvy, a not obsolete disorder: Clinical report in eight young children and literature review
- A meta-analysis of the effects of DBS on cognitive function in patients with advanced PD
- Protective role of selenium in sepsis: Mechanisms and potential therapeutic strategies
- Strategies for hyperkalemia management in dialysis patients: A systematic review
- C-reactive protein-to-albumin ratio in peripheral artery disease
- Case Reports
- Delayed graft function after renal transplantation
- Semaglutide treatment for type 2 diabetes in a patient with chronic myeloid leukemia: A case report and review of the literature
- Diverse electrophysiological demyelinating features in a late-onset glycogen storage disease type IIIa case
- Giant right atrial hemangioma presenting with ascites: A case report
- Laser excision of a large granular cell tumor of the vocal cord with subglottic extension: A case report
- EsoFLIP-assisted dilation for dysphagia in systemic sclerosis: Highlighting the role of multimodal esophageal evaluation
- Molecular hydrogen-rhodiola as an adjuvant therapy for ischemic stroke in internal carotid artery occlusion: A case report
- Coronary artery anomalies: A case of the “malignant” left coronary artery and its surgical management
- Rapid Communication
- Biological properties of valve materials using RGD and EC
-
A single oral administration of flavanols enhances short
-term memory in mice along with increased brain-derived neurotrophic factor - Letter to the Editor
- Role of enhanced external counterpulsation in long COVID
- Expression of Concern
- Expression of concern “A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma”
- Expression of concern “Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway”
- Expression of concern “circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8”
- Corrigendum
- Corrigendum to “Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism”
- Corrigendum to “Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis”
- Corrigendum to “The progress of autoimmune hepatitis research and future challenges”
- Retraction
- Retraction of “miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway”
- Retraction of: “LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through downregulating SP-A by sponging to miR-424”
- Retraction of: “SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways”
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part II
- Unveiling novel biomarkers for platinum chemoresistance in ovarian cancer
- Lathyrol affects the expression of AR and PSA and inhibits the malignant behavior of RCC cells
- The era of increasing cancer survivorship: Trends in fertility preservation, medico-legal implications, and ethical challenges
- Bone scintigraphy and positron emission tomography in the early diagnosis of MRONJ
- Meta-analysis of clinical efficacy and safety of immunotherapy combined with chemotherapy in non-small cell lung cancer
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part IV
- Exploration of mRNA-modifying METTL3 oncogene as momentous prognostic biomarker responsible for colorectal cancer development
- Special Issue The evolving saga of RNAs from bench to bedside - Part III
- Interaction and verification of ferroptosis-related RNAs Rela and Stat3 in promoting sepsis-associated acute kidney injury
- The mRNA MOXD1: Link to oxidative stress and prognostic significance in gastric cancer
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part II
- Dynamic changes in lactate-related genes in microglia and their role in immune cell interactions after ischemic stroke
- A prognostic model correlated with fatty acid metabolism in Ewing’s sarcoma based on bioinformatics analysis
- Red cell distribution width predicts early kidney injury: A NHANES cross-sectional study
- Special Issue Diabetes mellitus: pathophysiology, complications & treatment
- Nutritional risk assessment and nutritional support in children with congenital diabetes during surgery
- Correlation of the differential expressions of RANK, RANKL, and OPG with obesity in the elderly population in Xinjiang
- A discussion on the application of fluorescence micro-optical sectioning tomography in the research of cognitive dysfunction in diabetes
- A review of brain research on T2DM-related cognitive dysfunction
- Metformin and estrogen modulation in LABC with T2DM: A 36-month randomized trial
- Special Issue Innovative Biomarker Discovery and Precision Medicine in Cancer Diagnostics
- CircASH1L-mediated tumor progression in triple-negative breast cancer: PI3K/AKT pathway mechanisms


