Abstract
Lung epithelial cells and fibroblasts poorly express angiotensin-converting enzyme 2, and the study aimed to investigate the role of the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on inflammation and epithelial–mesenchymal transition (EMT) in two lung cell lines and to understand the potential mechanism. Lung epithelial cells (BEAS-2B) and fibroblasts (MRC-5) were treated with the spike protein, then inflammatory and EMT phenotypes were detected by enzyme-linked immunosorbent assay, Transwell, and western blot assays. RNA-sequence and bioinformatic analyses were performed to identify dysregulated genes. The roles of the candidate genes were further investigated. The results showed that treatment with 1,000 ng/mL of spike protein in two lung cell lines caused increased levels of IL-6, TNF-α, CXCL1, and CXCL3, and the occurrence of EMT. RNA-sequence identified 4,238 dysregulated genes in the spike group, and 18 candidate genes were involved in both inflammation- and EMT-related processes. GADD45A had the highest verified fold change (abs), and overexpression of GADD45A promoted the secretion of cytokines and EMT in the two lung cell lines. In conclusion, the spike protein induces inflammation and EMT in lung epithelial cells and fibroblasts by upregulating GADD45A, providing a new target to inhibit inflammation and EMT.
1 Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a pandemic of acute respiratory disease, named “coronavirus disease 2019” (COVID-19), with more than 240 million confirmed cases and 4.8 million deaths as of October 19, 2021 [1]. It has been reported that 81% of patients have mild disease with common symptoms of fever and cough, and 14% have severe disease requiring ventilation in an intensive care unit [2,3]. At the end of 2021, Molnupiravir and Nirmatrelvir-Ritonavir, two oral antiviral drugs, were approved for the treatment of patients with mild-to-moderate COVID-19 [4]. Before the approval and application of the drugs, vaccination and necessary personal preventive behaviors were still the most effective ways to prevent the transmission of SARS-CoV-2 [3].
Although most cases show mild symptoms when SARS-CoV-2 replicates in the lung epithelium in the early stages, some cases progress to complex symptoms, including respiratory dysfunction and systemic hyperinflammation [5]. Furthermore, multiple pieces of evidence indicate that prolonged COVID-19 can lead to lung fibrosis [6]. SARS-CoV-2 is a positive-sense single-stranded RNA virus that encodes four structural proteins: the envelope (E), nucleocapsid (N), spike (S), and membrane (M). SARS-CoV-2 has 79.5% similarity with SARS-CoV-1 at the genomic level [7]. One of the major differences is that 27 mutations are involved in the coding of the S protein [8], which is responsible for attachment to host cells in the initiation stage. This could explain the much higher affinity between angiotensin-converting enzyme 2 (ACE2) and S proteins [9,10] and the stronger infectivity [11] of SARS-CoV-2 than SARS-CoV-1. However, only a small subset of alveolar type II (AT2) cells express ACE2 [12], which is not expressed in other epithelial cells [13]. Therefore, inflammation and further fibrosis induced by SARS-CoV-2 or the S protein may be affected by non-AT2 cells independent of ACE2.
Notably, recent clues point out that the S protein is involved in the production of chemokines in non-AT2 cells in the initiation stage, which generally occurs in the amplification and consummation stages [14]. For instance, various inflammatory cytokines and chemokines have been detected in human lung epithelial cells stimulated with extracellular S protein [15]. Other studies also proved that this inflammatory reaction could be triggered by the S protein, independent of active viral infection and replication. Additionally, the effector cells include bronchial epithelial, microvascular endothelial, and airway epithelial cells [16,17]. Another study indicated that only the S protein, but not the E, N, or M proteins, can induce epithelial–mesenchymal transition (EMT) marker changes in breast cancer cells [18]. This could increase the risk of metastasis in cancer patients once they are infected. EMT is a complex process in which cells are transformed into mesenchymal cells through specific procedures, accompanied by a series of cytoskeletal and morphological changes, resulting in cell migration and invasion. This phenotypic alteration of cells is involved in cancer metastasis [19], chronic inflammation [19], chronic obstructive pulmonary disease, and lung fibrosis [20]. The effect of the S protein on the EMT of lung epithelial cells has not yet been investigated.
Lung fibroblasts are known to play pivotal roles in chronic respiratory disease and are involved in inflammation and lung fibrogenesis [21,22]. On the one hand, they are able to produce inflammatory cytokines [23]; on the other, they are regarded as effector cells through interactions with the injured alveolar epithelium in lung fibrogenesis [22]. Whether the inflammatory or migratory phenotype of lung fibroblasts can be directly induced or altered by the S protein is also unknown. These changes may be involved in the obvious symptoms or disease progression induced by SARS-CoV-2, including lung inflammation and fibrosis.
In the present study, we investigated the role of the S protein in inflammatory cytokine production and EMT in two non-AT2 cell lines – lung epithelial cells (BEAS-2B) and lung fibroblasts (MRC-5) – which poorly express ACE2 [13,24]. RNA-sequencing (RNA-seq) was performed to identify abnormally expressed genes, and bioinformatics analysis was used for the annotation of biological roles and screening of genes of interest. Finally, the functions of the candidate gene, growth arrest, and DNA damage-inducible gene alpha (GADD45A) in lung cells were investigated. Our study provides a new mechanism for understanding the role of S protein in lung inflammation and fibrosis induced by SARS-CoV-2.
2 Materials and methods
2.1 Cells and cell culture
Human lung epithelial cells (BEAS-2B) and human embryonic lung fibroblasts (MRC-5) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). BEAS-2B cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) supplemented with 10% FBS and 1% penicillin/streptomycin. MRC-5 cells were cultured in minimum essential medium (MEM, Gibco) supplemented with the same supplements. All the cells were incubated at 37℃ with 5% CO2.
2.2 Cytotoxicity
The cells were seeded in 96-well plates at a density of 3,000 cells/well. Different concentrations (0, 10, 100, 500, 1,000, and 2,000 ng/mL) of recombinant S protein, purchased from ABclonal Technology Co., Ltd (Wuhan, China), were co-cultured in the wells for 3 days. The 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to detect the viability of the cells daily at the same time points. Briefly, the supernatant of wells was removed and 10 µL of 5 mg/mL MTT (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) mixed with the 90 µL of separate complete medium of each cell was added into each well. After incubation for 3 h, the unreacted MTT solution was removed carefully and 100 µL dimethyl sulfoxide was added to resolve the formazan, followed by incubation for another 30 min. The absorbance of each well was measured at 490 nm using a microplate reader. The relative cellular viability was used to determine cytotoxicity.
2.3 Enzyme linked immunosorbent assay (ELISA)
After the cells were treated with S protein (100 ng/mL) for 48 h, the supernatant was collected for ELISA. The production of inflammation cytokines (IL-6, TNF-α) and chemokines (CXCL1, CXCL3) were detected using the ELISA kits following the manufacturer’s instructions. The cytokine production levels were calculated using separate standard curves. Human IL-6 and TNF-α ELISA kits were purchased from ABclonal Technology Co., Ltd (Wuhan, China), and human CXCL1 and CXCL3 ELISA kits were provided by MULTISCIENCES (LIANKE) Biotech, Co., Ltd (Hangzhou, China).
2.4 Transwell assay
The cells were collected and resuspended in a separate medium without FBS supplementation at a density of 4 × 105 cells/mL. Subsequently, 100 µL of the cell suspension was added to the center of the upper chamber of the Transwell insert (8 µm pore size; EMD Millipore, Billerica, MA, USA), and 600 µL of complete medium was added to the lower chamber. After 24 h of incubation, the upper chambers were collected and washed twice with PBS. Subsequently, the cells retained on the upper surface were removed, and those that migrated to the bottom surface of the chamber were fixed with methanol for 20 min and stained with 1% crystal violet for another 20 min. Finally, the migrated cells were photographed under a light microscope and five fields were randomly selected for cell counting.
2.5 Western blot
Total protein was extracted using a protein extraction kit (KeyGen, Jiangsu, China) and quantified using a BCA protein concentration detection kit (Beyotime, Shanghai, China) following the manufacturer’s instructions. Ten micrograms of protein were added to 12% SDS-PAGE gels for electrophoresis, and the separated proteins were transferred to polyvinylidene fluoride membranes. Next, 5% non-fat milk was added, and the membranes were incubated for 1 h. After washing thrice with tris buffered saline with tween-20, the membranes were cut and incubated with the diluted antibodies overnight, followed by incubation with the secondary antibody for 2 h. Subsequently, the signal of the desired protein was enhanced using an ECL kit (ABclonal) and captured on a gel imaging analysis system (Tanon, Shanghai, China). Primary antibodies against E-cadherin (1:1,000), Vimentin (1:1,000), N-cadherin (1:1,000), and GAPDH (1:1,000) were purchased from ABclonal. GAPDH was used as a loading control. The gray values of the bands were analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.6 RNA-seq
After cells were treated with S protein for 48 h, total RNA from each cell sample was collected using TRIzol reagent (Ambion, Thermo Fisher Scientific) and extracted using the RNeasy mini kit (Qiagen, Germany). Next, the RNA was sent to Sinotech Genomics Co., Ltd (Shanghai, China) for cDNA library construction. Briefly, mRNA molecules were first purified using poly T oligo-attached magnetic beads and then fragmented into small pieces for the synthesis of first- and second-strand cDNA. After an end repair, cDNA fragments was added with a single “A” base and ligated to the adapters. Finally, the cDNA was enriched by PCR to create a cDNA library and then sequenced on an Illumina NovaSeq 6000 (Illumina, USA). Dysregulated genes were screened using the following filtering criteria: fold-change (absolute) [FC(abs)] >2.
2.7 Construction of transcription factor (TF) regulatory network
TRANSFAC (http://genexplain.com/transfac) was used to predict the potential TFs of the differentially expressed genes (DEGs), and only TFs included in the identified DEGs were retained to explore their transcriptional regulatory roles. Finally, the TF regulatory network was visualized using Cytoscape software.
2.8 Bioinformatical analyses
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway analyses were performed to analyze the biological functions and potentially relevant signaling pathways of dysregulated genes. Gene numbers >2 and a p < 0.05 were used as thresholds to screen relevant GO terms and KEGG pathways. The terms and pathways were ranked in descending order according to the enrichment factor, and the top 30 terms and pathways were selected and presented in a separate bubble chart. The Venn diagram analysis was performed by Sinotech Genomics Co., Ltd (Shanghai, China).
2.9 qPCR
Total RNA was extracted using a universal RNA purification kit (TIANGEN, Beijing, China) following the manufacturer’s instructions and quantified using a microspectrophotometer (Allsheng, Hangzhou, China). RNA (500 ng) was reverse transcribed into cDNA using the PrimeScript RT Master Mix kit (TAKARA) according to the manufacturer’s protocol. Subsequently, 1 µL of cDNA was premixed with 0.5 µL of each primer, 10 µL of SYBR Green (TAKARA), and 8 µL of deionized water; the reaction system was transferred to a Real-time Thermal Cycler X960 (Heal-force, Shanghai, China) for amplification. The reaction was started at 95℃ for 5 min, followed by 40 cycles of 95℃ for 10 s and 60℃ for 60 s. Data were analyzed using the 2−ΔΔCq method, and GAPDH was used as a normalization control. Primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China), and the sequences are listed in Table 1.
Primer sequences used for qPCR
| Gene name | sequences (5′–3′) | |
|---|---|---|
| Forward | Reverse | |
| FGF18 | GATGGGGACAAGTATGCCCAG | GCCTTTGCGGTTCATGCAC |
| CHRM1 | CTCTATACCACGTACCTGCTCA | CCGAGTCACGGAGAAGTAGC |
| MAPK15 | GGGCCTATGGCATTGTGTG | TCTCTGGGCATCTGTCTTATCC |
| FGFR2 | AGCACCATACTGGACCAACAC | GGCAGCGAAACTTGACAGTG |
| NODAL | CAGTACAACGCCTATCGCTGT | TGCATGGTTGGTCGGATGAAA |
| SERPINE1 | ACCGCAACGTGGTTTTCTCA | TTGAATCCCATAGCTGCTTGAAT |
| ANGPT2 | AACTTTCGGAAGAGCATGGAC | CGAGTCATCGTATTCGAGCGG |
| PTPRR | ACCTATCGCCCATCACATTACA | GCGGTGGTAGCTTTGATCTCA |
| IL24 | TTGCCTGGGTTTTACCCTGC | AAGGCTTCCCACAGTTTCTGG |
| GADD45A | GAGAGCAGAAGACCGAAAGGA | CACAACACCACGTTATCGGG |
| GAPDH | CTGGGCTACACTGAGCACC | AAGTGGTCGTTGAGGGCAATG |
2.10 Cell transfection
Cells were seeded in a six-well plate at a density of 3 × 105 cells/mL, and the vectors containing full-length GADD45A were transfected into the cells using Lipofectamine ®2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer’s instructions. Two days after transfection, the cells were collected for qPCR validation. The overexpression vector of GADD45A was synthetized by Sangon Biotech Co., Ltd (Shanghai, China), and an empty vector was used as a negative control (NC).
2.11 Statistical analysis
Data were presented as mean ± standard deviation, and each experiment was performed at least three times. All analyses were performed using SPSS software (version 16.0; SPSS, Inc.). Student’s t-test was used to evaluate the statistical significance of the differences between two groups. Statistical significance was set at p < 0.05.
3 Results
3.1 The S protein induced inflammatory molecule secretion and EMT of lung cells
First, the cytotoxicity of the S protein was detected using the MTT assay. The results showed that the viability of the two cell lines treated with S protein was similar to that of the control group at a dose of 10–500 ng/mL at all treatment time points, and a small reduction in viability was observed in MRC-5 cells treated with 1,000 and 2,000 ng/mL for 48 or 72 h, respectively (Figure 1a). This indicated a slight cytotoxicity of the S protein, and the highest safe dose and treatment time (1,000 ng/mL and 48 h) were selected for the following assays. The production of inflammatory cytokines (IL-6, TNF-α) were increased by ∼1-fold, and the chemokines (CXCL1, CXCL3) were upregulated by 2–3-fold after cells were treated with S protein for 48 h through ELISA detection (Figure 1b). Transwell assay results showed that the migration abilities of both cell types were significantly enhanced compared to those of the control group (Figure 1c). EMT of cells also occurred with higher levels of vimentin and N-cadherin and lower levels of E-cadherin (Figure 1d).
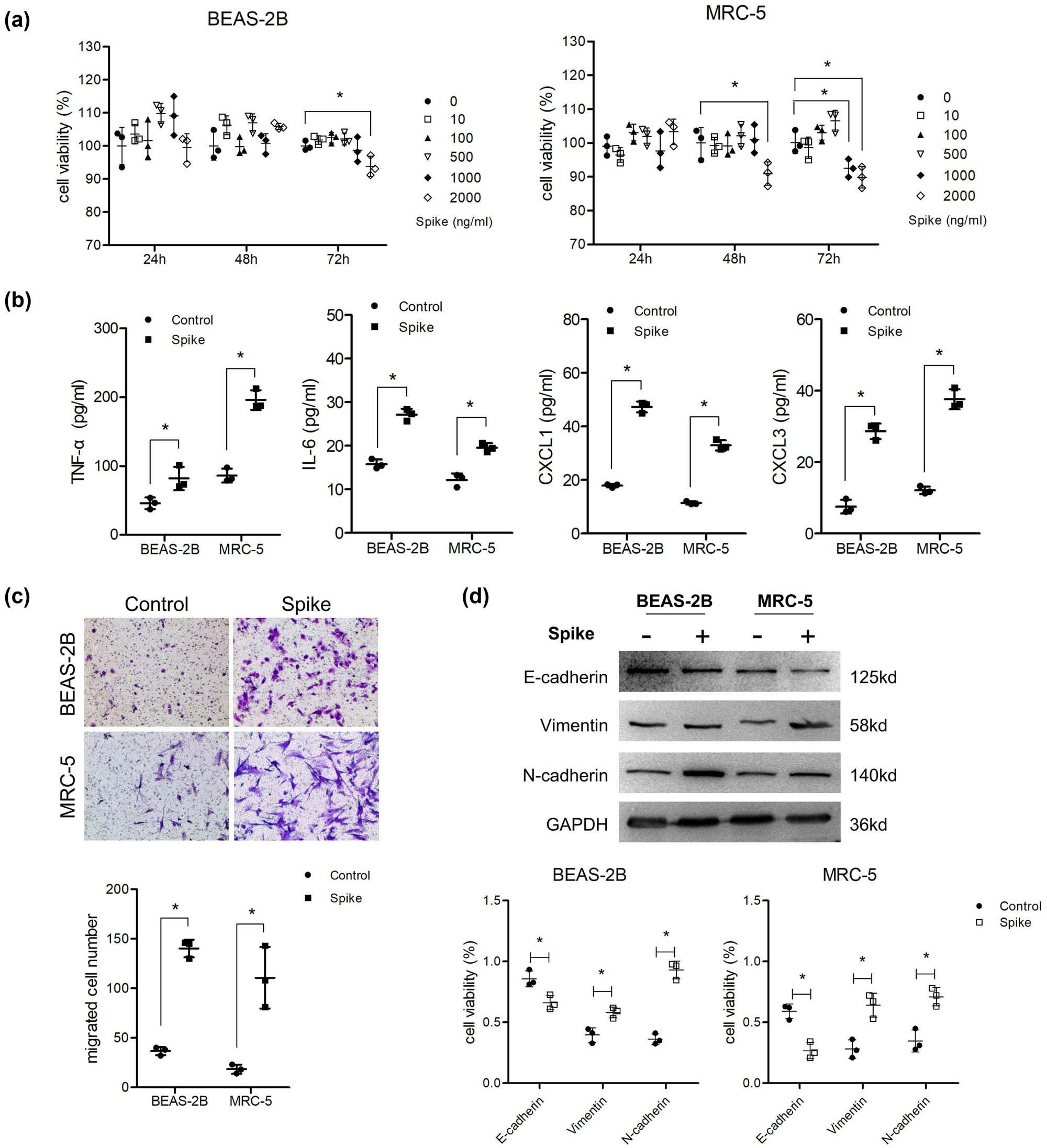
Spike protein induced inflammatory molecule secretion and EMT of lung cells. (a) Cells were co-cultured with different concentrations of S protein for 24, 48, and 72 h. MTT was used to detect cell viability (n = 3). After cells were treated with 1,000 ng/mL S protein for 48 h, (b) ELISA was used to analyze the supernatant levels of TNF-α, IL-6, CXCL1, and CXCL2 (n = 3). (c) Transwell assay was performed to detect migration ability (n = 3) and (d) western blotting was used to detect the protein levels of E-cadherin, vimentin, and N-cadherin (n = 3).
3.2 Dysregulated genes identified by RNA-seq after S protein treatment
After the cells were treated with S protein for 48 h, RNA-seq was used to identify abnormally expressed genes. As shown in Figure 2a, 4,238 dysregulated genes were screened (including 2,144 upregulated and 2,094 downregulated genes) under the threshold of [FC(abs)] >2. The heat map in Figure 2b indicates that the expression profiles of the four samples varied significantly, and several clusters of gene expression trends between the two different samples in the spike group were consistent. Table 2 lists the alterations in the expression of the top ten genes with the highest FC (abs) values, which varied from approximately 40 to 400. Furthermore, a TF regulatory network of all dysregulated genes was constructed to uncover the regulatory relationships between genes. Only a small number of genes were identified, including five TFs (POU5F1B, FOXO6, FOXD2, FOSL1, and NFIX) and 24 target genes (Figure 2c). Two TFs (FOXO6 and FOXD2) shared 11 target genes.

Dysregulated genes identified by RNA-seq after S protein treatment. RNA-seq was performed to identify the dysregulated genes after the cells were treated with 1,000 ng/mL S protein for 48 h (n = 1). (a) Scatter plot of dysregulated genes. Sp, spike group; Con: Control group. (b) Heat map of dysregulated genes. Each line indicates a gene and each column indicates a cell sample. (c) TF regulatory network constructed from dysregulated genes. Purple rounded rectangles, genes; yellow rounded rectangles, TF.
Top ten dysregulated genes after Spike protein treatment
| Gene id | Gene name | log2FC | log2FC abs | FC abs | Up/down | |
|---|---|---|---|---|---|---|
| 1 | ENSG00000283515 | AC020915.4 | −8.57 | 8.57 | 379.60 | Down |
| 2 | ENSG00000274049 | INO80B-WBP1 | 7.74 | 7.74 | 213.32 | Up |
| 3 | ENSG00000068781 | STON1-GTF2A1L | 6.58 | 6.58 | 95.48 | Up |
| 4 | ENSG00000285238 | AC006064.6 | −6.55 | 6.55 | 93.69 | Down |
| 5 | ENSG00000244563 | RPS26P19 | −6.23 | 6.23 | 74.81 | Down |
| 6 | ENSG00000275121 | AC211486.4 | 6.15 | 6.15 | 70.78 | Up |
| 7 | ENSG00000248405 | PRR5-ARHGAP8 | −6.11 | 6.11 | 68.90 | Down |
| 8 | ENSG00000242419 | PCDHGC4 | −5.70 | 5.70 | 51.83 | Down |
| 9 | ENSG00000127954 | STEAP4 | −5.29 | 5.29 | 39.06 | Down |
| 10 | ENSG00000103426 | CORO7-PAM16 | −5.24 | 5.24 | 37.78 | Down |
3.3 Screen of candidate genes regulating inflammation and EMT following GO and KEGG analyses
GO analysis indicated that the dysregulated genes were mainly enriched in several terms, including G protein-coupled acetylcholine receptor activity, water transmembrane transporter activity, and the complement receptor-mediated signaling pathway (Table S1). KEGG pathway enrichment analysis showed that the genes were mainly involved in rheumatoid arthritis, inflammatory bowel disease, and p53, IL-17, and PPAR signaling pathways (Figure 3).
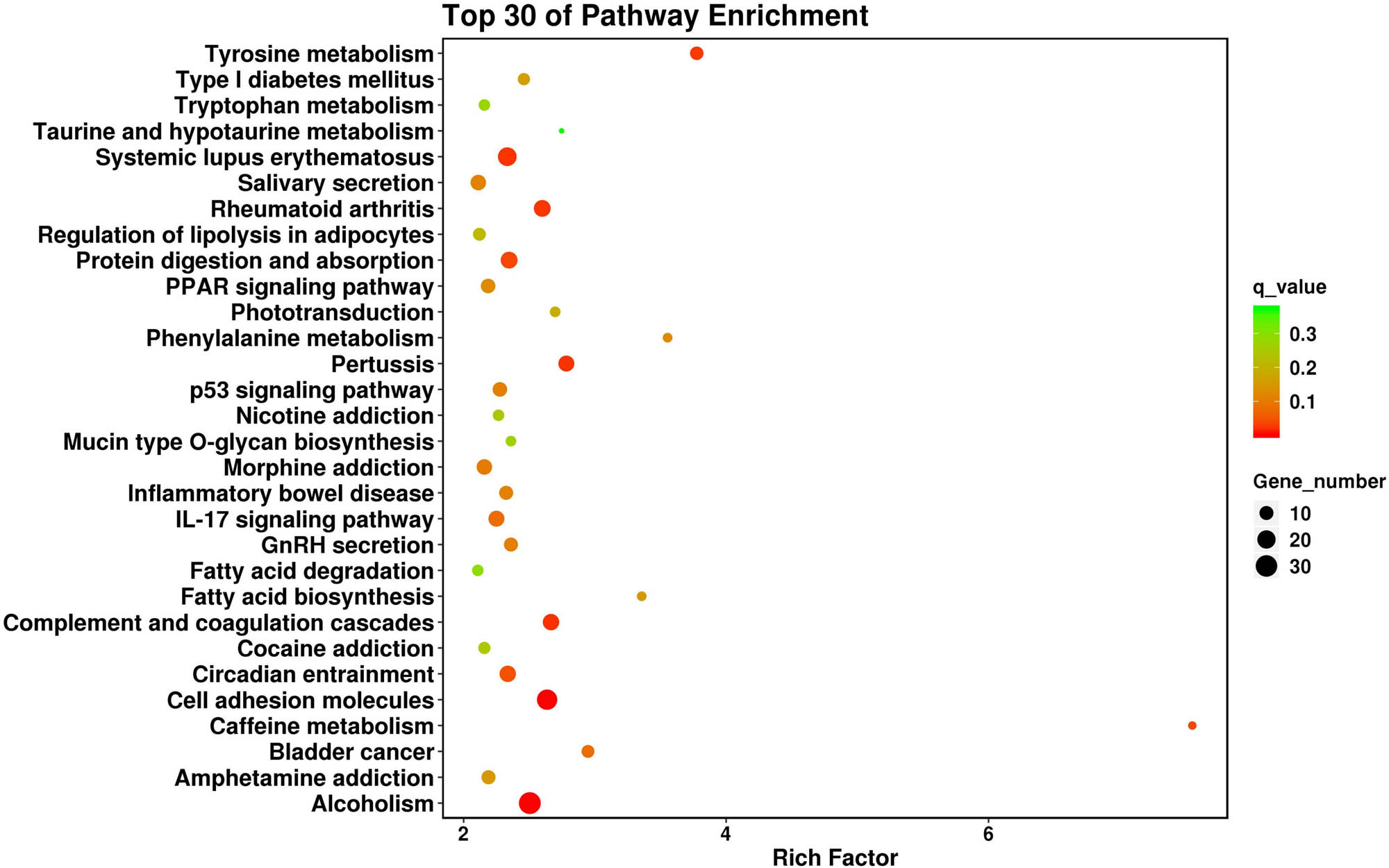
Top 30 of KEGG pathway enrichment analysis of the dysregulated genes.
It was relatively difficult to screen candidates for the altered phenotype of cells following analysis of the top 30 terms or pathways. Therefore, several keywords (“cytokine,” “motility,” and “actin cytoskeleton”) were used to screen relevant terms and pathways in Tables S1 and S2. As a result, two terms (GO:0004896 and GO:0005125) and two pathways (hsa04060 and hsa04061) were screened out under the threshold of p < 0.05 (Table 3). Many inflammatory cytokines (IL24, IL20, and IL1A) and chemokines (CXCL1, CXCL6, CXCL3, and CXCL5) were upregulated, which is consistent with our ELISA results. In addition, 61 genes were enriched in the negative regulation of cell motility and regulation of the actin cytoskeleton; these genes may play important roles in the regulation of migration ability and EMT. Venn diagram analysis indicated that three genes (IL24, NODAL, and HMGB1) were involved in both collection 1 (cytokines) and collection 2 (motility/actin cytoskeleton) (Figure 4a). Next, eight pathways involved in the signal transduction of SARS-CoV-2 infection were selected from Table S2, and a total of 18 overlapping genes were obtained by Venn diagram analysis of collections 2 and 3 (pathway). The screened genes are listed in Table 4 according to the FC(abs) values.
Genes enriched in cytokine- and migration-related biological processes and pathways
| Keywords (collection) | GO/pathway_ID | GO term/pathway_DES | Up_ genes | Down_ genes | Gene_up_list | Gene_down_list |
|---|---|---|---|---|---|---|
| Cytokine (collection 1) | hsa04060 | Cytokine-cytokine receptor interaction | 28 | 12 | EDA2R, IL13RA2, CXCL1, BMP2, GDF15, IL24, FAS, IL23R, IL1A, NODAL, INHBB, IL20, IL12RB2, CXCL6, TNFSF15, CCR1, TNFRSF10C, TNFRSF9, MSTN, CSF2RB, CXCL8, CXCL3, TNFSF13B, IL9R, CXCL5, IL1RL1, IL31RA, GDF1 | CNTFR, IL1R2, TNFRSF8, TNFSF10, EDAR, BMP8B, IL13, BMP6, IL34, INHBE, CCR10, CXCL16 |
| hsa04061 | Viral protein interaction with cytokine and cytokine receptor | 9 | 3 | CXCL1, IL24, IL20, CXCL6, CCR1, TNFRSF10C, CXCL8, CXCL3, CXCL5 | TNFSF10, IL34, CCR10 | |
| GO:0004896 | Cytokine receptor activity | 10 | 5 | IL1RL1, IL12RB2, IL13RA2, IL31RA, CSF2RB, IL17RD, IL9R, IL23R, CCRL2, CCR1 | CNTFR, GFRA3, GPR17, IL1R2, CCR10 | |
| GO:0005125 | Cytokine activity | 20 | 10 | CXCL8, CMTM2, TNFSF15, CXCL5, BMP2, GDF15, CXCL6, CXCL3, WNT5B, MSTN, NODAL, NDP, IL20, IL24, SPP1, IL1A, GDF1, INHBB, CXCL1, TNFSF13B | INHBE, HMGB1, BMP6, IL34, AREG, CXCL16, BMP8B, IL13, TNFSF10, WNT8B | |
| Motility/actin cytoskeleton (collection 2) | GO:2000146 | Negative regulation of cell motility | 18 | 23 | TIE1, MIR503, MIR221, MIR10A, STC1, MIR424, NODAL, NAV3, SLIT2, SERPINE1, DDT, IL24, TMIGD3, PTPRR, GADD45A, ARHGDIB, DNAI3, HAS1 | CDH1, MEOX2, HMGB1,GPR18, CHRD, LDLRAD4, MIR149, TMEFF2, ANGPT2,CLDN5, SERPINF1, MARVELD3, KRT16, IFITM1, MIR199A1, MYOCD, PADI2, SPOCK3, SVBP, MAPK15, MIR29B2, TACSTD2, C5AR2 |
| hsa04810 | Regulation of actin cytoskeleton | 8 | 12 | SCIN, CYFIP2, PIP5K1B, ITGAX, EGF, CHRM5, PAK6, PIK3R1 | ACTR3C, FGFR3, FGF18, PDGFD, FGF22, PPP1R12B, CHRM4, IQGAP2, CHRM1, FGF7, FGFR2, LPAR5 | |
| Pathway (collection 3) | hsa03320 | PPAR signaling pathway | 4 | 7 | MMP1, SLC27A6, ACSL4, ACSBG, | LPL, FABP7, FABP3, ACSBG2, FABP4, PLIN1, AQP7 |
| hsa04115 | p53 signaling pathway | 11 | 10 | CCND1, SESN2, SERPINE1, FAS, MDM2, SERPINB5, GADD45A, CCND2, ZMAT3, TP53I3 | RRM2 | |
| hsa04657 | IL-17 signaling pathway | 14 | 10 | CXCL1, MMP3, CXCL6, FOSL1, MMP1, FOSB, PTGS2, CXCL8, CXCL3, CXCL5 | MUC5B, IL13, JUN, MAPK15 | |
| hsa04010 | MAPK signaling pathway | 11 | 22 | DUSP2, HSPA2, DUSP4, FAS, IL1A, TGFA, GADD45A, FLT4, PTPRR, EGF, DUSP16 | FGFR3, FGF18, FLT1, RASGRP2, PDGFD, FGF22, ANGPT2, CACNA1I, RASGRP3, EFNA3, DUSP9, NFATC1, CACNG6, EFNA2, AREG, IGF2, EFNA1, CACNG7, JUN, FGF7, CACNA2D3, FGFR2 | |
| hsa04151 | PI3K-Akt signaling pathway | 16 | 22 | TLR4, CCND1, VWF, LAMC2, MDM2, TGFA, GNG13, GNG2, FLT4, EGF, CCND2, SPP1, PIK3R1, LAMB3, C8orf44-SGK3, GNG8 | FGFR3, DDIT4, FGF18, FLT1, LPAR6, TNXB, PDGFD, FGF22, ANGPT2, GNG7, GNG4, EFNA3, COL4A3, EFNA2, AREG, IGF2, EFNA1, CHRM1, FGF7, FGFR2, LPAR5, VTN | |
| hsa04668 | TNF signaling pathway | 8 | 3 | CXCL1, FAS, MMP3, CXCL6, PTGS2, PIK3R1, CXCL3, CXCL5 | RIPK3, SOCS3, JUN | |
| hsa04064 | NF-κB signaling pathway | 9 | 1 | TLR4, EDA2R, CXCL1, GADD45A, PTGS2, CXCL8, CXCL3, TNFSF13B, LBP | EDAR | |
| hsa04630 | JAK-STAT signaling pathway | 12 | 3 | IL13RA2, CCND1, IL24, IL23R, IL20, IL12RB2, EGF, SOCS7, CCND2, CSF2RB, PIK3R1, ENSIL9R | CNTFR, IL13, SOCS3 |

Screen and validation of candidate genes. (a) Venn diagram analysis of overlapped genes screened between collection 2 and collection 1 or 3. (b) qPCR validation of the fold changes of the top ten overlapped genes in two lung cells treated with 1,000 ng/mL S protein for 48 h (n = 3).
Eighteen overlapped genes screened between collection 2 and collection 1 or 3
| Gene id | Gene name | log2FC | log2FC abs | FC abs | Up/down | |
|---|---|---|---|---|---|---|
| 1 | ENSG00000156427 | FGF18 | −3.17 | 3.17 | 9.01 | Down |
| 2 | ENSG00000168539 | CHRM1 | −2.81 | 2.81 | 7.01 | Down |
| 3 | ENSG00000181085 | MAPK15 | −1.95 | 1.95 | 3.86 | Down |
| 4 | ENSG00000066468 | FGFR2 | −1.85 | 1.85 | 3.62 | Down |
| 5 | ENSG00000156574 | NODAL | 1.81 | 1.81 | 3.49 | Up |
| 6 | ENSG00000106366 | SERPINE1 | 1.76 | 1.76 | 3.39 | Up |
| 7 | ENSG00000091879 | ANGPT2 | −1.65 | 1.65 | 3.13 | Down |
| 8 | ENSG00000184574 | LPAR5 | −1.50 | 1.50 | 2.84 | Down |
| 9 | ENSG00000153233 | PTPRR | 1.46 | 1.46 | 2.75 | Up |
| 10 | ENSG00000162892 | IL24 | 1.38 | 1.38 | 2.61 | Up |
| 11 | ENSG00000116717 | GADD45A | 1.31 | 1.31 | 2.49 | Up |
| 12 | ENSG00000138798 | EGF | 1.25 | 1.25 | 2.38 | Up |
| 13 | ENSG00000170962 | PDGFD | −1.25 | 1.25 | 2.37 | Down |
| 14 | ENSG00000070388 | FGF22 | −1.22 | 1.22 | 2.34 | Down |
| 15 | ENSG00000145675 | PIK3R1 | 1.18 | 1.18 | 2.27 | Up |
| 16 | ENSG00000068078 | FGFR3 | −1.17 | 1.17 | 2.25 | Down |
| 17 | ENSG00000189403 | HMGB1 | −1.15 | 1.15 | 2.23 | Down |
| 18 | ENSG00000140285 | FGF7 | −1.14 | 1.14 | 2.20 | Down |
3.4 Overexpression of GADD45A promoted inflammatory molecule secretion and EMT of lung cells
The top five upregulated and five downregulated candidates were selected for qPCR validation. The results in Figure 4b show that the log2FC values of some genes (GFG18, CHRM1, and MAPK15) in MRC-5 were very low (nearly 0), and that the values of FGFR2 and ANGPT2 were not consistent between the two cell lines; however, the remaining five genes increased. Among the five upregulated genes, GADD45A had the highest validated FC (abs) in both cell lines (Figure 4b). Next, the overexpression vector of GADD45A was transfected into lung cells and confirmed by qPCR (Figure 5a). ELISA results indicated that lung cells overexpressing GADD45A secreted higher levels of inflammatory molecules than those in the NC group (Figure 5b). Furthermore, the migration ability of the two lung cell lines was enhanced after the overexpression of GADD45A (Figure 5c). Consistently, GADD45A induced significant EMT in lung cells (Figure 5d).

Overexpression of GADD45A promoted inflammatory molecule secretion and EMT of lung cells. (a) Cells were transfected with GADD45A overexpression vector, and qPCR was performed to detect the transfection effect (n = 3). After cells overexpressed with GADD45A, (b) ELISA was used to analyze the supernatant levels of TNF-α, IL-6, CXCL1, and CXCL2 (n = 3). (c) Transwell assay was performed to detect migration ability (n = 3) and (d) western blotting was used to detect the protein levels of E-cadherin, vimentin, and N-cadherin (n = 3).
4 Discussion
Lung inflammation is the most common symptom of SARS-CoV-2, and fibrosis occurs in some COVID-19 cases [6]. Mutation of the S protein of SARS-CoV-2 [8] and the inflammation and EMT phenotype triggered by the S protein (but not other structural proteins) [15,18] indicate the potential complex roles of the S protein in the progression of relevant symptoms and diseases after infection. Because the expression level of ACE2 in AT2 cells is very limited, let alone in other lung cells [12,13], focusing on non-AT2 cells is helpful in understanding and determining the changes that occur in the early stages of infection. To date, inflammation triggered by a single S protein has been confirmed in macrophages and epithelial cells [16,25,26]. On the other hand, lung fibroblasts – the major cells involved in chronic respiratory disease and lung fibrogenesis – are often neglected. Additionally, the EMT of lung epithelial cells or fibroblasts induced by the S protein remains unknown. Therefore, in the present study, the role of the S protein in fibroblasts (MRC-5) was also investigated to uncover the potential shared mechanism of inflammation and EMT induced by the S protein in both lung epithelial cells and fibroblasts.
In contrast to macrophages, the production of inflammatory molecules stimulated by S protein in epithelial cells is much slower, often requiring 12 or 24 h [15]. In the present study, 48 h was selected as the stimulation time for the two cell types for both phenotype and RNA-seq analyses. Our result indicated that S protein treatment significantly increased proinflammatory molecule expression (IL-6, TNF-α, CXCL1, CXCL3) from the two cells. In addition, EMT of the two lung cell lines was detected after S protein stimulation. EMT could be triggered by multiple pathways including Wnt, nuclear factor (NF)-kappa B (κB), and transforming growth factor β pathways. Additionally, NF-κB activated by inflammatory cytokines are thought to further accentuate EMT in the lung [20]. Moreover, cells undergoing partial EMT exhibit enhanced inflammation [27]. This suggests that the S protein triggers EMT and inflammation in lung cells, resulting in consistently increased levels of EMT and inflammation. Once proinflammatory cytokines and chemokines enter the bloodstream, a number of immune cells are recruited to initiate a defense mechanism, which may promote a cytokine storm [28].
To investigate the potential mechanism, RNA-seq was performed to screen for abnormally expressed genes in lung cells. As expected, the gene expression profiles of the two cell types differed significantly. Fortunately, several clusters of gene expression trends were consistent after S protein treatment in the absence of a p-value. Although the top ten genes showed a significant fold change, clues supporting their roles in inflammation or EMT were less definite, according to the bioinformatics analysis results. Therefore, GO terms and KEGG pathways related to inflammation and EMT were directly screened and several candidates were obtained through overlapping analysis. Recent studies have indicated that multiple pathways are involved in SARS-CoV-2 infection. The NF-κB pathway is a classical pathway regulating inflammation, and the activation of NF-κB has been confirmed in various cell types from COVID-19 patients [29] or post-stimulation with S protein [30] and its subunit S1 [31]. Activation of the p38 mitogen-activated protein kinase (MAPK) signaling pathway is considered one of the major pathways involved in many viral infections, and treatment with p38 or MEK inhibitors effectively inhibits the infectivity of SARS-CoV-2 and its S protein [32,33]. Furthermore, the PPAR [34], PI3K/AKT [16], and p53 [35] signaling pathways were all activated in SARS-CoV-2 and S protein-infected samples. Therefore, an overlapping analysis of genes enriched in these pathways and EMT-related biological processes is important for screening candidate genes.
GADD45A belongs to the GADD45 family and is transcriptionally activated by various stress stimuli. The protein encoded by GADD45A is small (18 kDa) and plays an important role in regulating DNA repair, cell proliferation, survival, differentiation, and immune responses by interacting with several intracellular signaling molecules [36,37]. Recent studies have indicated the complex roles of GADD45A in inflammation. Mathew et al. found that lung GADD45A was significantly enhanced after a single-dose thoracic radiation, and mice deficient in GADD45A showed increased susceptibility to radiation-induced lung injury and higher levels of inflammatory cytokines [38]. This indicates that GADD45A is an important modulator of lung inflammatory responses. GADD45b can also be induced in T cells by the pro-inflammatory cytokines IL-12 and IL-18 [39]. Additionally, studies have uncovered the important role of GADD45A in regulating migration by functioning as a target of miRNAs [40,41]. These findings reflect the dual roles of GADD45A in regulating inflammation and EMT, which are consistent with the results of the overlapping analysis and phenotype verification assay.
In the present study, GADD45A was upregulated after lung cells were treated with S protein, which can be sorted as an environmental stimulus. The p38/MAPK pathway is considered the major activated pathway when GADD45A responds to stimuli [36], and it could be the upstream signal transduction pathway for increased GADD45A. We also noted that GADD45A is one of the target genes of the TF POU class 5 homeobox 1 B (POU5F1B), which plays important roles in carcinogenesis [42,43]. Whether abnormal expression of this TF contributes to the upregulation of GADD45A requires further investigation. A previous study indicated that proinflammatory cytokines induce the expression of GADD45A [39]; hence, we proposed that autocrine inflammatory molecules (induced by the S protein) could further promote the expression of GADD45A.
Taken together, our study indicates that the S protein is able to induce inflammation and EMT in lung epithelial cells and fibroblasts through the upregulation of GADD45A, which provides a new target to inhibit inflammation and relevant fibrosis. More importantly, our study showed that lung fibroblasts are another group of effector cells of the S protein, providing new target cells to combat the virus. The gene expression profiles of lung cells in response to the S protein also provided a number of candidate genes for a better understanding of the mechanism of the S protein.
-
Funding information: This study was funded by Shanghai Municipal Health Commission (Grant number: 202040099).
-
Author contributions: M.Z., J.C., and W.M. conceived and designed the experiments. J.C., W.M., and H.C. performed the experiments, J.C., Z.W., and J.L. acquired the data, J.C. and X.W. analyzed and interpreted the data. J.C., W.M., X.W., and H.C. wrote the article. Z.J. and B.C. reviewed and revised the article.
-
Conflict of interest: The authors declare no conflict of interest.
-
Data availability statement: The datasets generated during and/or analysed during the current study are available in supporting information files, and from the corresponding author on reasonable request.
References
[1] World Health Organization website. https://covid19.who.int/. obtained on 19 Oct, 2021.Suche in Google Scholar
[2] Song C, Li Z, Li C, Huang M, Liu J, Fang Q, et al. SARS-CoV-2: the monster causes COVID-19. Front Cell Infect Microbiol. 2022;12:835750. 10.3389/fcimb.2022.835750.Suche in Google Scholar PubMed PubMed Central
[3] Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–54. 10.1038/s41579-020-00459-7.Suche in Google Scholar PubMed PubMed Central
[4] Saravolatz LD, Depcinski S, Sharma M. Molnupiravir and Nirmatrelvir-Ritonavir: oral coronavirus disease 2019 antiviral drugs. Clin Infect Dis. 2023;76(1):165–71. 10.1093/cid/ciac180.Suche in Google Scholar PubMed PubMed Central
[5] Romagnoli S, Peris A, De Gaudio AR, Geppetti P. SARS-CoV-2 and COVID-19: from the bench to the bedside. Physiol Rev. 2020;100(4):1455–66. 10.1152/physrev.00020.2020.Suche in Google Scholar PubMed PubMed Central
[6] Mylvaganam RJ, Bailey JI, Sznajder JI, Sala MA, Northwestern Comprehensive CCC. Recovering from a pandemic: pulmonary fibrosis after SARS-CoV-2 infection. Eur Respir Rev. 2021;30(162):210194. 10.1183/16000617.0194-2021.Suche in Google Scholar PubMed PubMed Central
[7] Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. COVID-19, SARS and MERS: are they closely related. Clin Microbiol Infect. 2020;26(6):729–34. 10.1016/j.cmi.2020.03.026.Suche in Google Scholar PubMed PubMed Central
[8] Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–8. 10.1016/j.chom.2020.02.001.Suche in Google Scholar PubMed PubMed Central
[9] Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–60. 10.1007/s11427-020-1637-5.Suche in Google Scholar PubMed PubMed Central
[10] Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–20. 10.1038/s41586-020-2180-5.Suche in Google Scholar PubMed
[11] Yang TJ, Yu PY, Chang YC, Hsu SD. D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation. J Biol Chem. 2021;297(4):101238. 10.1016/j.jbc.2021.101238.Suche in Google Scholar PubMed PubMed Central
[12] Ortiz ME, Thurman A, Pezzulo AA, Leidinger MR, Klesney-Tait JA, Karp PH, et al. Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMedicine. 2020;60:102976. 10.1016/j.ebiom.2020.102976.Suche in Google Scholar PubMed PubMed Central
[13] McAlinden KD, Lu W, Ferdowsi PV, Myers S, Markos J, Larby J, et al. Electronic cigarette aerosol is cytotoxic and increases ACE2 expression on human airway epithelial cells: implications for SARS-CoV-2 (COVID-19). J Clin Med. 2021;10(5):1028. 10.3390/jcm10051028.Suche in Google Scholar PubMed PubMed Central
[14] Masoomikarimi M, Garmabi B, Alizadeh J, Kazemi E, Azari Jafari A, Mirmoeeni S, et al. Advances in immunotherapy for COVID-19: a comprehensive review. Int Immunopharmacol. 2021;93:107409. 10.1016/j.intimp.2021.107409.Suche in Google Scholar PubMed PubMed Central
[15] Khan S, Shafiei MS, Longoria C, Schoggins J, Savani RC, Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. bioRxiv. 2021. 10.1101/2021.03.16.435700.Suche in Google Scholar PubMed PubMed Central
[16] Li F, Li J, Wang PH, Yang N, Huang J, Ou J, et al. SARS-CoV-2 spike promotes inflammation and apoptosis through autophagy by ROS-suppressed PI3K/AKT/mTOR signaling. Biochim Biophys Acta Mol Basis Dis. 2021;1867(12):166260. 10.1016/j.bbadis.2021.166260.Suche in Google Scholar PubMed PubMed Central
[17] Anand G, Perry AM, Cummings CL, St Raymond E, Clemens RA, Steed AL. Surface proteins of SARS-CoV-2 drive airway epithelial cells to induce IFN-dependent inflammation. J Immunol. 2021;206(12):3000–9. 10.4049/jimmunol.2001407.Suche in Google Scholar PubMed PubMed Central
[18] Lai YJ, Chao CH, Liao CC, Lee TA, Hsu JM, Chou WC, et al. Epithelial–mesenchymal transition induced by SARS-CoV-2 required transcriptional upregulation of snail. Am J Cancer Res. 2021;11(5):2278–90.Suche in Google Scholar
[19] Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. 10.1016/j.cell.2016.06.028.Suche in Google Scholar PubMed
[20] Bartis D, Mise N, Mahida RY, Eickelberg O, Thickett DR. Epithelial–mesenchymal transition in lung development and disease: does it exist and is it important? Thorax. 2014;69(8):760–5. 10.1136/thoraxjnl-2013-204608.Suche in Google Scholar PubMed
[21] Hackett TL, Vriesde N, Al-Fouadi M, Mostaco-Guidolin L, Maftoun D, Hsieh A, et al. The role of the dynamic lung extracellular matrix environment on fibroblast morphology and inflammation. Cells. 2022;11(2):185. 10.3390/cells11020185.Suche in Google Scholar PubMed PubMed Central
[22] John AE, Joseph C, Jenkins G, Tatler AL. COVID-19 and pulmonary fibrosis: a potential role for lung epithelial cells and fibroblasts. Immunol Rev. 2021;302(1):228–40. 10.1111/imr.12977.Suche in Google Scholar PubMed PubMed Central
[23] Flavell SJ, Hou TZ, Lax S, Filer AD, Salmon M, Buckley CD. Fibroblasts as novel therapeutic targets in chronic inflammation. Br J Pharmacol. 2008;153(Suppl 1):S241–6. 10.1038/sj.bjp.0707487.Suche in Google Scholar PubMed PubMed Central
[24] Wu Q, Kumar N, Lafuse WP, Ahumada OS, Saljoughian N, Whetstone E, et al. Influenza A virus modulates ACE2 expression and SARS-CoV-2 infectivity in human cardiomyocytes. iScience. 2022;25(12):105701. 10.1016/j.isci.2022.105701.Suche in Google Scholar PubMed PubMed Central
[25] Shirato K, Takanari J, Kizaki T. Standardized extract of asparagus officinalis stem attenuates SARS-CoV-2 spike protein-induced IL-6 and IL-1beta production by suppressing p44/42 MAPK and Akt phosphorylation in murine primary macrophages. Molecules. 2021;26(20):6189. 10.3390/molecules26206189.Suche in Google Scholar PubMed PubMed Central
[26] Barhoumi T, Alghanem B, Shaibah H, Mansour FA, Alamri HS, Akiel MA, et al. SARS-CoV-2 coronavirus spike protein-induced apoptosis, inflammatory, and oxidative stress responses in THP-1-like-macrophages: potential role of angiotensin-converting enzyme inhibitor (Perindopril). Front Immunol. 2021;12:728896. 10.3389/fimmu.2021.728896.Suche in Google Scholar PubMed PubMed Central
[27] Grande MT, Sanchez-Laorden B, Lopez-Blau C, De Frutos CA, Boutet A, Arevalo M, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med. 2015;21(9):989–97. 10.1038/nm.3901.Suche in Google Scholar PubMed
[28] Yang L, Xie X, Tu Z, Fu J, Xu D, Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct Target Ther. 2021;6(1):255. 10.1038/s41392-021-00679-0.Suche in Google Scholar PubMed PubMed Central
[29] Hariharan A, Hakeem AR, Radhakrishnan S, Reddy MS, Rela M. The role and therapeutic potential of NF-kappa-B pathway in severe COVID-19 patients. Inflammopharmacology. 2021;29(1):91–100. 10.1007/s10787-020-00773-9.Suche in Google Scholar PubMed PubMed Central
[30] Patra T, Meyer K, Geerling L, Isbell TS, Hoft DF, Brien J, et al. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16(12):e1009128. 10.1371/journal.ppat.1009128.Suche in Google Scholar PubMed PubMed Central
[31] Shirato K, Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7(2):e06187. 10.1016/j.heliyon.2021.e06187.Suche in Google Scholar PubMed PubMed Central
[32] Cheng Y, Sun F, Wang L, Gao M, Xie Y, Sun Y, et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics. 2020;10(26):12223–40. 10.7150/thno.50992.Suche in Google Scholar PubMed PubMed Central
[33] Zhou L, Huntington K, Zhang S, Carlsen L, So EY, Parker C, et al. Natural killer cell activation, reduced ACE2, TMPRSS2, cytokines G-CSF, M-CSF and SARS-CoV-2-S pseudovirus infectivity by MEK inhibitor treatment of human cells. bioRxiv. 2020. 10.1101/2020.08.02.230839.Suche in Google Scholar PubMed PubMed Central
[34] Cao X, Tian Y, Nguyen V, Zhang Y, Gao C, Yin R, et al. Spike protein of SARS-CoV-2 activates macrophages and contributes to induction of acute lung inflammation in male mice. FASEB J. 2021;35(9):e21801. 10.1096/fj.202002742RR.Suche in Google Scholar PubMed PubMed Central
[35] Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–70. 10.1080/22221751.2020.1747363.Suche in Google Scholar PubMed PubMed Central
[36] Hollander MC, Fornace AJ Jr. Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene. 2002;21(40):6228–33. 10.1038/sj.onc.1205774.Suche in Google Scholar PubMed
[37] Schmitz I. Gadd45 proteins in immunity. Adv Exp Med Biol. 2013;793:51–68. 10.1007/978-1-4614-8289-5_4.Suche in Google Scholar PubMed
[38] Mathew B, Takekoshi D, Sammani S, Epshtein Y, Sharma R, Smith BD, et al. Role of GADD45a in murine models of radiation- and bleomycin-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2015;309(12):L1420–9. 10.1152/ajplung.00146.2014.Suche in Google Scholar PubMed PubMed Central
[39] Yang J, Zhu H, Murphy TL, Ouyang W, Murphy KM. IL-18-stimulated GADD45 beta required in cytokine-induced, but not TCR-induced, IFN-gamma production. Nat Immunol. 2001;2(2):157–64. 10.1038/84264.Suche in Google Scholar PubMed
[40] Yang Y, Shang H. Silencing lncRNA-DGCR5 increased trophoblast cell migration, invasion and tube formation, and inhibited cell apoptosis via targeting miR-454-3p/GADD45A axis. Mol Cell Biochem. 2021;476(9):3407–21. 10.1007/s11010-021-04161-x.Suche in Google Scholar PubMed
[41] Tan Y, Xiao D, Xu Y, Wang C. Long non-coding RNA DLX6-AS1 is upregulated in preeclampsia and modulates migration and invasion of trophoblasts through the miR-376c/GADD45A axis. Exp Cell Res. 2018;370(2):718–24. 10.1016/j.yexcr.2018.07.039.Suche in Google Scholar PubMed
[42] Yu J, Zhang J, Zhou L, Li H, Deng ZQ, Meng B. The octamer-binding transcription factor 4 (OCT4) Pseudogene, POU domain class 5 transcription factor 1B (POU5F1B), is upregulated in cervical cancer and down-regulation inhibits cell proliferation and migration and induces apoptosis in cervical cancer cell lines. Med Sci Monit. 2019;25:1204–13. 10.12659/MSM.912109.Suche in Google Scholar PubMed PubMed Central
[43] Pan Y, Zhan L, Chen L, Zhang H, Sun C, Xing C. POU5F1B promotes hepatocellular carcinoma proliferation by activating AKT. Biomed Pharmacother. 2018;100:374–80. 10.1016/j.biopha.2018.02.023.Suche in Google Scholar PubMed
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Exosomes derived from mesenchymal stem cells overexpressing miR-210 inhibits neuronal inflammation and contribute to neurite outgrowth through modulating microglia polarization
- Current situation of acute ST-segment elevation myocardial infarction in a county hospital chest pain center during an epidemic of novel coronavirus pneumonia
- circ-IARS depletion inhibits the progression of non-small-cell lung cancer by circ-IARS/miR-1252-5p/HDGF ceRNA pathway
- circRNA ITGA7 restrains growth and enhances radiosensitivity by up-regulating SMAD4 in colorectal carcinoma
- WDR79 promotes aerobic glycolysis of pancreatic ductal adenocarcinoma (PDAC) by the suppression of SIRT4
- Up-regulation of collagen type V alpha 2 (COL5A2) promotes malignant phenotypes in gastric cancer cell via inducing epithelial–mesenchymal transition (EMT)
- Inhibition of TERC inhibits neural apoptosis and inflammation in spinal cord injury through Akt activation and p-38 inhibition via the miR-34a-5p/XBP-1 axis
- 3D-printed polyether-ether-ketone/n-TiO2 composite enhances the cytocompatibility and osteogenic differentiation of MC3T3-E1 cells by downregulating miR-154-5p
- Propofol-mediated circ_0000735 downregulation restrains tumor growth by decreasing integrin-β1 expression in non-small cell lung cancer
- PVT1/miR-16/CCND1 axis regulates gastric cancer progression
- Silencing of circ_002136 sensitizes gastric cancer to paclitaxel by targeting the miR-16-5p/HMGA1 axis
- Short-term outcomes after simultaneous gastrectomy plus cholecystectomy in gastric cancer: A pooling up analysis
- SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways
- Molecular mechanism by which the Notch signaling pathway regulates autophagy in a rat model of pulmonary fibrosis in pigeon breeder’s lung
- lncRNA TPT1-AS1 promotes cell migration and invasion in esophageal squamous-cell carcinomas by regulating the miR-26a/HMGA1 axis
- SIRT1/APE1 promotes the viability of gastric cancer cells by inhibiting p53 to suppress ferroptosis
- Glycoprotein non-metastatic melanoma B interacts with epidermal growth factor receptor to regulate neural stem cell survival and differentiation
- Treatments for brain metastases from EGFR/ALK-negative/unselected NSCLC: A network meta-analysis
- Association of osteoporosis and skeletal muscle loss with serum type I collagen carboxyl-terminal peptide β glypeptide: A cross-sectional study in elder Chinese population
- circ_0000376 knockdown suppresses non-small cell lung cancer cell tumor properties by the miR-545-3p/PDPK1 pathway
- Delivery in a vertical birth chair supported by freedom of movement during labor: A randomized control trial
- UBE2J1 knockdown promotes cell apoptosis in endometrial cancer via regulating PI3K/AKT and MDM2/p53 signaling
- Metabolic resuscitation therapy in critically ill patients with sepsis and septic shock: A pilot prospective randomized controlled trial
- Lycopene ameliorates locomotor activity and urinary frequency induced by pelvic venous congestion in rats
- UHRF1-induced connexin26 methylation is involved in hearing damage triggered by intermittent hypoxia in neonatal rats
- LINC00511 promotes melanoma progression by targeting miR-610/NUCB2
- Ultra-high-performance liquid chromatography-tandem mass spectrometry analysis of serum metabolomic characteristics in people with different vitamin D levels
- Role of Jumonji domain-containing protein D3 and its inhibitor GSK-J4 in Hashimoto’s thyroiditis
- circ_0014736 induces GPR4 to regulate the biological behaviors of human placental trophoblast cells through miR-942-5p in preeclampsia
- Monitoring of sirolimus in the whole blood samples from pediatric patients with lymphatic anomalies
- Effects of osteogenic growth peptide C-terminal pentapeptide and its analogue on bone remodeling in an osteoporosis rat model
- A novel autophagy-related long non-coding RNAs signature predicting progression-free interval and I-131 therapy benefits in papillary thyroid carcinoma
- WGCNA-based identification of potential targets and pathways in response to treatment in locally advanced breast cancer patients
- Radiomics model using preoperative computed tomography angiography images to differentiate new from old emboli of acute lower limb arterial embolism
- Dysregulated lncRNAs are involved in the progress of myocardial infarction by constructing regulatory networks
- Single-arm trial to evaluate the efficacy and safety of baclofen in treatment of intractable hiccup caused by malignant tumor chemotherapy
- Genetic polymorphisms of MRPS30-DT and NINJ2 may influence lung cancer risk
- Efficacy of immune checkpoint inhibitors in patients with KRAS-mutant advanced non-small cell lung cancer: A retrospective analysis
- Pyroptosis-based risk score predicts prognosis and drug sensitivity in lung adenocarcinoma
- Upregulation of lncRNA LANCL1-AS1 inhibits the progression of non-small-cell lung cancer via the miR-3680-3p/GMFG axis
- CircRANBP17 modulated KDM1A to regulate neuroblastoma progression by sponging miR-27b-3p
- Exosomal miR-93-5p regulated the progression of osteoarthritis by targeting ADAMTS9
- Downregulation of RBM17 enhances cisplatin sensitivity and inhibits cell invasion in human hypopharyngeal cancer cells
- HDAC5-mediated PRAME regulates the proliferation, migration, invasion, and EMT of laryngeal squamous cell carcinoma via the PI3K/AKT/mTOR signaling pathway
- The association between sleep duration, quality, and nonalcoholic fatty liver disease: A cross-sectional study
- Myostatin silencing inhibits podocyte apoptosis in membranous nephropathy through Smad3/PKA/NOX4 signaling pathway
- A novel long noncoding RNA AC125257.1 facilitates colorectal cancer progression by targeting miR-133a-3p/CASC5 axis
- Impact of omicron wave and associated control measures in Shanghai on health management and psychosocial well-being of patients with chronic conditions
- Clinicopathological characteristics and prognosis of young patients aged ≤45 years old with non-small cell lung cancer
- TMT-based comprehensive proteomic profiling identifies serum prognostic signatures of acute myeloid leukemia
- The dose limits of teeth protection for patients with nasopharyngeal carcinoma undergoing radiotherapy based on the early oral health-related quality of life
- miR-30b-5p targeting GRIN2A inhibits hippocampal damage in epilepsy
- Long non-coding RNA AL137789.1 promoted malignant biological behaviors and immune escape of pancreatic carcinoma cells
- IRF6 and FGF1 polymorphisms in non-syndromic cleft lip with or without cleft palate in the Polish population
- Comprehensive analysis of the role of SFXN family in breast cancer
- Efficacy of bronchoscopic intratumoral injection of endostar and cisplatin in lung squamous cell carcinoma patients underwent conventional chemoradiotherapy
- Silencing of long noncoding RNA MIAT inhibits the viability and proliferation of breast cancer cells by promoting miR-378a-5p expression
- AG1024, an IGF-1 receptor inhibitor, ameliorates renal injury in rats with diabetic nephropathy via the SOCS/JAK2/STAT pathway
- Downregulation of KIAA1199 alleviated the activation, proliferation, and migration of hepatic stellate cells by the inhibition of epithelial–mesenchymal transition
- Exendin-4 regulates the MAPK and WNT signaling pathways to alleviate the osteogenic inhibition of periodontal ligament stem cells in a high glucose environment
- Inhibition of glycolysis represses the growth and alleviates the endoplasmic reticulum stress of breast cancer cells by regulating TMTC3
- The function of lncRNA EMX2OS/miR-653-5p and its regulatory mechanism in lung adenocarcinoma
- Tectorigenin alleviates the apoptosis and inflammation in spinal cord injury cell model through inhibiting insulin-like growth factor-binding protein 6
- Ultrasound examination supporting CT or MRI in the evaluation of cervical lymphadenopathy in patients with irradiation-treated head and neck cancer
- F-box and WD repeat domain containing 7 inhibits the activation of hepatic stellate cells by degrading delta-like ligand 1 to block Notch signaling pathway
- Knockdown of circ_0005615 enhances the radiosensitivity of colorectal cancer by regulating the miR-665/NOTCH1 axis
- Long noncoding RNA Mhrt alleviates angiotensin II-induced cardiac hypertrophy phenotypes by mediating the miR-765/Wnt family member 7B pathway
- Effect of miR-499-5p/SOX6 axis on atrial fibrosis in rats with atrial fibrillation
- Cholesterol induces inflammation and reduces glucose utilization
- circ_0004904 regulates the trophoblast cell in preeclampsia via miR-19b-3p/ARRDC3 axis
- NECAB3 promotes the migration and invasion of liver cancer cells through HIF-1α/RIT1 signaling pathway
- The poor performance of cardiovascular risk scores in identifying patients with idiopathic inflammatory myopathies at high cardiovascular risk
- miR-2053 inhibits the growth of ovarian cancer cells by downregulating SOX4
- Nucleophosmin 1 associating with engulfment and cell motility protein 1 regulates hepatocellular carcinoma cell chemotaxis and metastasis
- α-Hederin regulates macrophage polarization to relieve sepsis-induced lung and liver injuries in mice
- Changes of microbiota level in urinary tract infections: A meta-analysis
- Identification of key enzalutamide-resistance-related genes in castration-resistant prostate cancer and verification of RAD51 functions
- Falls during oxaliplatin-based chemotherapy for gastrointestinal malignancies – (lessons learned from) a prospective study
- Outcomes of low-risk birth care during the Covid-19 pandemic: A cohort study from a tertiary care center in Lithuania
- Vitamin D protects intestines from liver cirrhosis-induced inflammation and oxidative stress by inhibiting the TLR4/MyD88/NF-κB signaling pathway
- Integrated transcriptome analysis identifies APPL1/RPS6KB2/GALK1 as immune-related metastasis factors in breast cancer
- Genomic analysis of immunogenic cell death-related subtypes for predicting prognosis and immunotherapy outcomes in glioblastoma multiforme
- Circular RNA Circ_0038467 promotes the maturation of miRNA-203 to increase lipopolysaccharide-induced apoptosis of chondrocytes
- An economic evaluation of fine-needle cytology as the primary diagnostic tool in the diagnosis of lymphadenopathy
- Midazolam impedes lung carcinoma cell proliferation and migration via EGFR/MEK/ERK signaling pathway
- Network pharmacology combined with molecular docking and experimental validation to reveal the pharmacological mechanism of naringin against renal fibrosis
- PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma
- miR-141-3p accelerates ovarian cancer progression and promotes M2-like macrophage polarization by targeting the Keap1-Nrf2 pathway
- lncRNA OIP5-AS1 attenuates the osteoarthritis progression in IL-1β-stimulated chondrocytes
- Overexpression of LINC00607 inhibits cell growth and aggressiveness by regulating the miR-1289/EFNA5 axis in non-small-cell lung cancer
- Subjective well-being in informal caregivers during the COVID-19 pandemic
- Nrf2 protects against myocardial ischemia-reperfusion injury in diabetic rats by inhibiting Drp1-mediated mitochondrial fission
- Unfolded protein response inhibits KAT2B/MLKL-mediated necroptosis of hepatocytes by promoting BMI1 level to ubiquitinate KAT2B
- Bladder cancer screening: The new selection and prediction model
- circNFATC3 facilitated the progression of oral squamous cell carcinoma via the miR-520h/LDHA axis
- Prone position effect in intensive care patients with SARS-COV-2 pneumonia
- Clinical observation on the efficacy of Tongdu Tuina manipulation in the treatment of primary enuresis in children
- Dihydroartemisinin ameliorates cerebral I/R injury in rats via regulating VWF and autophagy-mediated SIRT1/FOXO1 pathway
- Knockdown of circ_0113656 assuages oxidized low-density lipoprotein-induced vascular smooth muscle cell injury through the miR-188-3p/IGF2 pathway
- Low Ang-(1–7) and high des-Arg9 bradykinin serum levels are correlated with cardiovascular risk factors in patients with COVID-19
- Effect of maternal age and body mass index on induction of labor with oral misoprostol for premature rupture of membrane at term: A retrospective cross-sectional study
- Potential protective effects of Huanglian Jiedu Decoction against COVID-19-associated acute kidney injury: A network-based pharmacological and molecular docking study
- Clinical significance of serum MBD3 detection in girls with central precocious puberty
- Clinical features of varicella-zoster virus caused neurological diseases detected by metagenomic next-generation sequencing
- Collagen treatment of complex anorectal fistula: 3 years follow-up
- LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through down-regulating SP-A by sponging to miR-424
- Efficacy analysis of empirical bismuth quadruple therapy, high-dose dual therapy, and resistance gene-based triple therapy as a first-line Helicobacter pylori eradication regimen – An open-label, randomized trial
- SMOC2 plays a role in heart failure via regulating TGF-β1/Smad3 pathway-mediated autophagy
- A prospective cohort study of the impact of chronic disease on fall injuries in middle-aged and older adults
- circRNA THBS1 silencing inhibits the malignant biological behavior of cervical cancer cells via the regulation of miR-543/HMGB2 axis
- hsa_circ_0000285 sponging miR-582-3p promotes neuroblastoma progression by regulating the Wnt/β-catenin signaling pathway
- Long non-coding RNA GNAS-AS1 knockdown inhibits proliferation and epithelial–mesenchymal transition of lung adenocarcinoma cells via the microRNA-433-3p/Rab3A axis
- lncRNA UCA1 regulates miR-132/Lrrfip1 axis to promote vascular smooth muscle cell proliferation
- Twenty-four-color full spectrum flow cytometry panel for minimal residual disease detection in acute myeloid leukemia
- Hsa-miR-223-3p participates in the process of anthracycline-induced cardiomyocyte damage by regulating NFIA gene
- Anti-inflammatory effect of ApoE23 on Salmonella typhimurium-induced sepsis in mice
- Analysis of somatic mutations and key driving factors of cervical cancer progression
- Hsa_circ_0028007 regulates the progression of nasopharyngeal carcinoma through the miR-1179/SQLE axis
- Variations in sexual function after laparoendoscopic single-site hysterectomy in women with benign gynecologic diseases
- Effects of pharmacological delay with roxadustat on multi-territory perforator flap survival in rats
- Analysis of heroin effects on calcium channels in rat cardiomyocytes based on transcriptomics and metabolomics
- Risk factors of recurrent bacterial vaginosis among women of reproductive age: A cross-sectional study
- Alkbh5 plays indispensable roles in maintaining self-renewal of hematopoietic stem cells
- Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients
- Correlation between microvessel maturity and ISUP grades assessed using contrast-enhanced transrectal ultrasonography in prostate cancer
- The protective effect of caffeic acid phenethyl ester in the nephrotoxicity induced by α-cypermethrin
- Norepinephrine alleviates cyclosporin A-induced nephrotoxicity by enhancing the expression of SFRP1
- Effect of RUNX1/FOXP3 axis on apoptosis of T and B lymphocytes and immunosuppression in sepsis
- The function of Foxp1 represses β-adrenergic receptor transcription in the occurrence and development of bladder cancer through STAT3 activity
- Risk model and validation of carbapenem-resistant Klebsiella pneumoniae infection in patients with cerebrovascular disease in the ICU
- Calycosin protects against chronic prostatitis in rats via inhibition of the p38MAPK/NF-κB pathway
- Pan-cancer analysis of the PDE4DIP gene with potential prognostic and immunotherapeutic values in multiple cancers including acute myeloid leukemia
- The safety and immunogenicity to inactivated COVID-19 vaccine in patients with hyperlipemia
- Circ-UBR4 regulates the proliferation, migration, inflammation, and apoptosis in ox-LDL-induced vascular smooth muscle cells via miR-515-5p/IGF2 axis
- Clinical characteristics of current COVID-19 rehabilitation outpatients in China
- Luteolin alleviates ulcerative colitis in rats via regulating immune response, oxidative stress, and metabolic profiling
- miR-199a-5p inhibits aortic valve calcification by targeting ATF6 and GRP78 in valve interstitial cells
- The application of iliac fascia space block combined with esketamine intravenous general anesthesia in PFNA surgery of the elderly: A prospective, single-center, controlled trial
- Elevated blood acetoacetate levels reduce major adverse cardiac and cerebrovascular events risk in acute myocardial infarction
- The effects of progesterone on the healing of obstetric anal sphincter damage in female rats
- Identification of cuproptosis-related genes for predicting the development of prostate cancer
- Lumican silencing ameliorates β-glycerophosphate-mediated vascular smooth muscle cell calcification by attenuating the inhibition of APOB on KIF2C activity
- Targeting PTBP1 blocks glutamine metabolism to improve the cisplatin sensitivity of hepatocarcinoma cells through modulating the mRNA stability of glutaminase
- A single center prospective study: Influences of different hip flexion angles on the measurement of lumbar spine bone mineral density by dual energy X-ray absorptiometry
- Clinical analysis of AN69ST membrane continuous venous hemofiltration in the treatment of severe sepsis
- Antibiotics therapy combined with probiotics administered intravaginally for the treatment of bacterial vaginosis: A systematic review and meta-analysis
- Construction of a ceRNA network to reveal a vascular invasion associated prognostic model in hepatocellular carcinoma
- A pan-cancer analysis of STAT3 expression and genetic alterations in human tumors
- A prognostic signature based on seven T-cell-related cell clustering genes in bladder urothelial carcinoma
- Pepsin concentration in oral lavage fluid of rabbit reflux model constructed by dilating the lower esophageal sphincter
- The antihypertensive felodipine shows synergistic activity with immune checkpoint blockade and inhibits tumor growth via NFAT1 in LUSC
- Tanshinone IIA attenuates valvular interstitial cells’ calcification induced by oxidized low density lipoprotein via reducing endoplasmic reticulum stress
- AS-IV enhances the antitumor effects of propofol in NSCLC cells by inhibiting autophagy
- Establishment of two oxaliplatin-resistant gallbladder cancer cell lines and comprehensive analysis of dysregulated genes
- Trial protocol: Feasibility of neuromodulation with connectivity-guided intermittent theta-burst stimulation for improving cognition in multiple sclerosis
- LncRNA LINC00592 mediates the promoter methylation of WIF1 to promote the development of bladder cancer
- Factors associated with gastrointestinal dysmotility in critically ill patients
- Mechanisms by which spinal cord stimulation intervenes in atrial fibrillation: The involvement of the endothelin-1 and nerve growth factor/p75NTR pathways
- Analysis of two-gene signatures and related drugs in small-cell lung cancer by bioinformatics
- Silencing USP19 alleviates cigarette smoke extract-induced mitochondrial dysfunction in BEAS-2B cells by targeting FUNDC1
- Menstrual irregularities associated with COVID-19 vaccines among women in Saudi Arabia: A survey during 2022
- Ferroptosis involves in Schwann cell death in diabetic peripheral neuropathy
- The effect of AQP4 on tau protein aggregation in neurodegeneration and persistent neuroinflammation after cerebral microinfarcts
- Activation of UBEC2 by transcription factor MYBL2 affects DNA damage and promotes gastric cancer progression and cisplatin resistance
- Analysis of clinical characteristics in proximal and distal reflux monitoring among patients with gastroesophageal reflux disease
- Exosomal circ-0020887 and circ-0009590 as novel biomarkers for the diagnosis and prediction of short-term adverse cardiovascular outcomes in STEMI patients
- Upregulated microRNA-429 confers endometrial stromal cell dysfunction by targeting HIF1AN and regulating the HIF1A/VEGF pathway
- Bibliometrics and knowledge map analysis of ultrasound-guided regional anesthesia
- Knockdown of NUPR1 inhibits angiogenesis in lung cancer through IRE1/XBP1 and PERK/eIF2α/ATF4 signaling pathways
- D-dimer trends predict COVID-19 patient’s prognosis: A retrospective chart review study
- WTAP affects intracranial aneurysm progression by regulating m6A methylation modification
- Using of endoscopic polypectomy in patients with diagnosed malignant colorectal polyp – The cross-sectional clinical study
- Anti-S100A4 antibody administration alleviates bronchial epithelial–mesenchymal transition in asthmatic mice
- Prognostic evaluation of system immune-inflammatory index and prognostic nutritional index in double expressor diffuse large B-cell lymphoma
- Prevalence and antibiogram of bacteria causing urinary tract infection among patients with chronic kidney disease
- Reactive oxygen species within the vaginal space: An additional promoter of cervical intraepithelial neoplasia and uterine cervical cancer development?
- Identification of disulfidptosis-related genes and immune infiltration in lower-grade glioma
- A new technique for uterine-preserving pelvic organ prolapse surgery: Laparoscopic rectus abdominis hysteropexy for uterine prolapse by comparing with traditional techniques
- Self-isolation of an Italian long-term care facility during COVID-19 pandemic: A comparison study on care-related infectious episodes
- A comparative study on the overlapping effects of clinically applicable therapeutic interventions in patients with central nervous system damage
- Low intensity extracorporeal shockwave therapy for chronic pelvic pain syndrome: Long-term follow-up
- The diagnostic accuracy of touch imprint cytology for sentinel lymph node metastases of breast cancer: An up-to-date meta-analysis of 4,073 patients
- Mortality associated with Sjögren’s syndrome in the United States in the 1999–2020 period: A multiple cause-of-death study
- CircMMP11 as a prognostic biomarker mediates miR-361-3p/HMGB1 axis to accelerate malignant progression of hepatocellular carcinoma
- Analysis of the clinical characteristics and prognosis of adult de novo acute myeloid leukemia (none APL) with PTPN11 mutations
- KMT2A maintains stemness of gastric cancer cells through regulating Wnt/β-catenin signaling-activated transcriptional factor KLF11
- Evaluation of placental oxygenation by near-infrared spectroscopy in relation to ultrasound maturation grade in physiological term pregnancies
- The role of ultrasonographic findings for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative breast cancer
- Construction of immunogenic cell death-related molecular subtypes and prognostic signature in colorectal cancer
- Long-term prognostic value of high-sensitivity cardiac troponin-I in patients with idiopathic dilated cardiomyopathy
- Establishing a novel Fanconi anemia signaling pathway-associated prognostic model and tumor clustering for pediatric acute myeloid leukemia patients
- Integrative bioinformatics analysis reveals STAT2 as a novel biomarker of inflammation-related cardiac dysfunction in atrial fibrillation
- Adipose-derived stem cells repair radiation-induced chronic lung injury via inhibiting TGF-β1/Smad 3 signaling pathway
- Real-world practice of idiopathic pulmonary fibrosis: Results from a 2000–2016 cohort
- lncRNA LENGA sponges miR-378 to promote myocardial fibrosis in atrial fibrillation
- Diagnostic value of urinary Tamm-Horsfall protein and 24 h urine osmolality for recurrent calcium oxalate stones of the upper urinary tract: Cross-sectional study
- The value of color Doppler ultrasonography combined with serum tumor markers in differential diagnosis of gastric stromal tumor and gastric cancer
- The spike protein of SARS-CoV-2 induces inflammation and EMT of lung epithelial cells and fibroblasts through the upregulation of GADD45A
- Mycophenolate mofetil versus cyclophosphamide plus in patients with connective tissue disease-associated interstitial lung disease: Efficacy and safety analysis
- MiR-1278 targets CALD1 and suppresses the progression of gastric cancer via the MAPK pathway
- Metabolomic analysis of serum short-chain fatty acid concentrations in a mouse of MPTP-induced Parkinson’s disease after dietary supplementation with branched-chain amino acids
- Cimifugin inhibits adipogenesis and TNF-α-induced insulin resistance in 3T3-L1 cells
- Predictors of gastrointestinal complaints in patients on metformin therapy
- Prescribing patterns in patients with chronic obstructive pulmonary disease and atrial fibrillation
- A retrospective analysis of the effect of latent tuberculosis infection on clinical pregnancy outcomes of in vitro fertilization–fresh embryo transferred in infertile women
- Appropriateness and clinical outcomes of short sustained low-efficiency dialysis: A national experience
- miR-29 regulates metabolism by inhibiting JNK-1 expression in non-obese patients with type 2 diabetes mellitus and NAFLD
- Clinical features and management of lymphoepithelial cyst
- Serum VEGF, high-sensitivity CRP, and cystatin-C assist in the diagnosis of type 2 diabetic retinopathy complicated with hyperuricemia
- ENPP1 ameliorates vascular calcification via inhibiting the osteogenic transformation of VSMCs and generating PPi
- Significance of monitoring the levels of thyroid hormone antibodies and glucose and lipid metabolism antibodies in patients suffer from type 2 diabetes
- The causal relationship between immune cells and different kidney diseases: A Mendelian randomization study
- Interleukin 33, soluble suppression of tumorigenicity 2, interleukin 27, and galectin 3 as predictors for outcome in patients admitted to intensive care units
- Identification of diagnostic immune-related gene biomarkers for predicting heart failure after acute myocardial infarction
- Long-term administration of probiotics prevents gastrointestinal mucosal barrier dysfunction in septic mice partly by upregulating the 5-HT degradation pathway
- miR-192 inhibits the activation of hepatic stellate cells by targeting Rictor
- Diagnostic and prognostic value of MR-pro ADM, procalcitonin, and copeptin in sepsis
- Review Articles
- Prenatal diagnosis of fetal defects and its implications on the delivery mode
- Electromagnetic fields exposure on fetal and childhood abnormalities: Systematic review and meta-analysis
- Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae
- Saddle pulmonary embolism in the setting of COVID-19 infection: A systematic review of case reports and case series
- Vitamin C and epigenetics: A short physiological overview
- Ebselen: A promising therapy protecting cardiomyocytes from excess iron in iron-overloaded thalassemia patients
- Aspirin versus LMWH for VTE prophylaxis after orthopedic surgery
- Mechanism of rhubarb in the treatment of hyperlipidemia: A recent review
- Surgical management and outcomes of traumatic global brachial plexus injury: A concise review and our center approach
- The progress of autoimmune hepatitis research and future challenges
- METTL16 in human diseases: What should we do next?
- New insights into the prevention of ureteral stents encrustation
- VISTA as a prospective immune checkpoint in gynecological malignant tumors: A review of the literature
- Case Reports
- Mycobacterium xenopi infection of the kidney and lymph nodes: A case report
- Genetic mutation of SLC6A20 (c.1072T > C) in a family with nephrolithiasis: A case report
- Chronic hepatitis B complicated with secondary hemochromatosis was cured clinically: A case report
- Liver abscess complicated with multiple organ invasive infection caused by hematogenous disseminated hypervirulent Klebsiella pneumoniae: A case report
- Urokinase-based lock solutions for catheter salvage: A case of an upcoming kidney transplant recipient
- Two case reports of maturity-onset diabetes of the young type 3 caused by the hepatocyte nuclear factor 1α gene mutation
- Immune checkpoint inhibitor-related pancreatitis: What is known and what is not
- Does total hip arthroplasty result in intercostal nerve injury? A case report and literature review
- Clinicopathological characteristics and diagnosis of hepatic sinusoidal obstruction syndrome caused by Tusanqi – Case report and literature review
- Synchronous triple primary gastrointestinal malignant tumors treated with laparoscopic surgery: A case report
- CT-guided percutaneous microwave ablation combined with bone cement injection for the treatment of transverse metastases: A case report
- Malignant hyperthermia: Report on a successful rescue of a case with the highest temperature of 44.2°C
- Anesthetic management of fetal pulmonary valvuloplasty: A case report
- Rapid Communication
- Impact of COVID-19 lockdown on glycemic levels during pregnancy: A retrospective analysis
- Erratum
- Erratum to “Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway”
- Erratum to: “Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p”
- Retraction
- Retraction of “Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients”
- Retraction of “circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis”
- Retraction of “miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells”
- Retraction of “SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis”
- Retraction of “circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury”
- Retraction of “lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells”
- Special issue Linking Pathobiological Mechanisms to Clinical Application for cardiovascular diseases
- Effect of cardiac rehabilitation therapy on depressed patients with cardiac insufficiency after cardiac surgery
- Special issue The evolving saga of RNAs from bench to bedside - Part I
- FBLIM1 mRNA is a novel prognostic biomarker and is associated with immune infiltrates in glioma
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part III
- Development of a machine learning-based signature utilizing inflammatory response genes for predicting prognosis and immune microenvironment in ovarian cancer
Artikel in diesem Heft
- Research Articles
- Exosomes derived from mesenchymal stem cells overexpressing miR-210 inhibits neuronal inflammation and contribute to neurite outgrowth through modulating microglia polarization
- Current situation of acute ST-segment elevation myocardial infarction in a county hospital chest pain center during an epidemic of novel coronavirus pneumonia
- circ-IARS depletion inhibits the progression of non-small-cell lung cancer by circ-IARS/miR-1252-5p/HDGF ceRNA pathway
- circRNA ITGA7 restrains growth and enhances radiosensitivity by up-regulating SMAD4 in colorectal carcinoma
- WDR79 promotes aerobic glycolysis of pancreatic ductal adenocarcinoma (PDAC) by the suppression of SIRT4
- Up-regulation of collagen type V alpha 2 (COL5A2) promotes malignant phenotypes in gastric cancer cell via inducing epithelial–mesenchymal transition (EMT)
- Inhibition of TERC inhibits neural apoptosis and inflammation in spinal cord injury through Akt activation and p-38 inhibition via the miR-34a-5p/XBP-1 axis
- 3D-printed polyether-ether-ketone/n-TiO2 composite enhances the cytocompatibility and osteogenic differentiation of MC3T3-E1 cells by downregulating miR-154-5p
- Propofol-mediated circ_0000735 downregulation restrains tumor growth by decreasing integrin-β1 expression in non-small cell lung cancer
- PVT1/miR-16/CCND1 axis regulates gastric cancer progression
- Silencing of circ_002136 sensitizes gastric cancer to paclitaxel by targeting the miR-16-5p/HMGA1 axis
- Short-term outcomes after simultaneous gastrectomy plus cholecystectomy in gastric cancer: A pooling up analysis
- SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways
- Molecular mechanism by which the Notch signaling pathway regulates autophagy in a rat model of pulmonary fibrosis in pigeon breeder’s lung
- lncRNA TPT1-AS1 promotes cell migration and invasion in esophageal squamous-cell carcinomas by regulating the miR-26a/HMGA1 axis
- SIRT1/APE1 promotes the viability of gastric cancer cells by inhibiting p53 to suppress ferroptosis
- Glycoprotein non-metastatic melanoma B interacts with epidermal growth factor receptor to regulate neural stem cell survival and differentiation
- Treatments for brain metastases from EGFR/ALK-negative/unselected NSCLC: A network meta-analysis
- Association of osteoporosis and skeletal muscle loss with serum type I collagen carboxyl-terminal peptide β glypeptide: A cross-sectional study in elder Chinese population
- circ_0000376 knockdown suppresses non-small cell lung cancer cell tumor properties by the miR-545-3p/PDPK1 pathway
- Delivery in a vertical birth chair supported by freedom of movement during labor: A randomized control trial
- UBE2J1 knockdown promotes cell apoptosis in endometrial cancer via regulating PI3K/AKT and MDM2/p53 signaling
- Metabolic resuscitation therapy in critically ill patients with sepsis and septic shock: A pilot prospective randomized controlled trial
- Lycopene ameliorates locomotor activity and urinary frequency induced by pelvic venous congestion in rats
- UHRF1-induced connexin26 methylation is involved in hearing damage triggered by intermittent hypoxia in neonatal rats
- LINC00511 promotes melanoma progression by targeting miR-610/NUCB2
- Ultra-high-performance liquid chromatography-tandem mass spectrometry analysis of serum metabolomic characteristics in people with different vitamin D levels
- Role of Jumonji domain-containing protein D3 and its inhibitor GSK-J4 in Hashimoto’s thyroiditis
- circ_0014736 induces GPR4 to regulate the biological behaviors of human placental trophoblast cells through miR-942-5p in preeclampsia
- Monitoring of sirolimus in the whole blood samples from pediatric patients with lymphatic anomalies
- Effects of osteogenic growth peptide C-terminal pentapeptide and its analogue on bone remodeling in an osteoporosis rat model
- A novel autophagy-related long non-coding RNAs signature predicting progression-free interval and I-131 therapy benefits in papillary thyroid carcinoma
- WGCNA-based identification of potential targets and pathways in response to treatment in locally advanced breast cancer patients
- Radiomics model using preoperative computed tomography angiography images to differentiate new from old emboli of acute lower limb arterial embolism
- Dysregulated lncRNAs are involved in the progress of myocardial infarction by constructing regulatory networks
- Single-arm trial to evaluate the efficacy and safety of baclofen in treatment of intractable hiccup caused by malignant tumor chemotherapy
- Genetic polymorphisms of MRPS30-DT and NINJ2 may influence lung cancer risk
- Efficacy of immune checkpoint inhibitors in patients with KRAS-mutant advanced non-small cell lung cancer: A retrospective analysis
- Pyroptosis-based risk score predicts prognosis and drug sensitivity in lung adenocarcinoma
- Upregulation of lncRNA LANCL1-AS1 inhibits the progression of non-small-cell lung cancer via the miR-3680-3p/GMFG axis
- CircRANBP17 modulated KDM1A to regulate neuroblastoma progression by sponging miR-27b-3p
- Exosomal miR-93-5p regulated the progression of osteoarthritis by targeting ADAMTS9
- Downregulation of RBM17 enhances cisplatin sensitivity and inhibits cell invasion in human hypopharyngeal cancer cells
- HDAC5-mediated PRAME regulates the proliferation, migration, invasion, and EMT of laryngeal squamous cell carcinoma via the PI3K/AKT/mTOR signaling pathway
- The association between sleep duration, quality, and nonalcoholic fatty liver disease: A cross-sectional study
- Myostatin silencing inhibits podocyte apoptosis in membranous nephropathy through Smad3/PKA/NOX4 signaling pathway
- A novel long noncoding RNA AC125257.1 facilitates colorectal cancer progression by targeting miR-133a-3p/CASC5 axis
- Impact of omicron wave and associated control measures in Shanghai on health management and psychosocial well-being of patients with chronic conditions
- Clinicopathological characteristics and prognosis of young patients aged ≤45 years old with non-small cell lung cancer
- TMT-based comprehensive proteomic profiling identifies serum prognostic signatures of acute myeloid leukemia
- The dose limits of teeth protection for patients with nasopharyngeal carcinoma undergoing radiotherapy based on the early oral health-related quality of life
- miR-30b-5p targeting GRIN2A inhibits hippocampal damage in epilepsy
- Long non-coding RNA AL137789.1 promoted malignant biological behaviors and immune escape of pancreatic carcinoma cells
- IRF6 and FGF1 polymorphisms in non-syndromic cleft lip with or without cleft palate in the Polish population
- Comprehensive analysis of the role of SFXN family in breast cancer
- Efficacy of bronchoscopic intratumoral injection of endostar and cisplatin in lung squamous cell carcinoma patients underwent conventional chemoradiotherapy
- Silencing of long noncoding RNA MIAT inhibits the viability and proliferation of breast cancer cells by promoting miR-378a-5p expression
- AG1024, an IGF-1 receptor inhibitor, ameliorates renal injury in rats with diabetic nephropathy via the SOCS/JAK2/STAT pathway
- Downregulation of KIAA1199 alleviated the activation, proliferation, and migration of hepatic stellate cells by the inhibition of epithelial–mesenchymal transition
- Exendin-4 regulates the MAPK and WNT signaling pathways to alleviate the osteogenic inhibition of periodontal ligament stem cells in a high glucose environment
- Inhibition of glycolysis represses the growth and alleviates the endoplasmic reticulum stress of breast cancer cells by regulating TMTC3
- The function of lncRNA EMX2OS/miR-653-5p and its regulatory mechanism in lung adenocarcinoma
- Tectorigenin alleviates the apoptosis and inflammation in spinal cord injury cell model through inhibiting insulin-like growth factor-binding protein 6
- Ultrasound examination supporting CT or MRI in the evaluation of cervical lymphadenopathy in patients with irradiation-treated head and neck cancer
- F-box and WD repeat domain containing 7 inhibits the activation of hepatic stellate cells by degrading delta-like ligand 1 to block Notch signaling pathway
- Knockdown of circ_0005615 enhances the radiosensitivity of colorectal cancer by regulating the miR-665/NOTCH1 axis
- Long noncoding RNA Mhrt alleviates angiotensin II-induced cardiac hypertrophy phenotypes by mediating the miR-765/Wnt family member 7B pathway
- Effect of miR-499-5p/SOX6 axis on atrial fibrosis in rats with atrial fibrillation
- Cholesterol induces inflammation and reduces glucose utilization
- circ_0004904 regulates the trophoblast cell in preeclampsia via miR-19b-3p/ARRDC3 axis
- NECAB3 promotes the migration and invasion of liver cancer cells through HIF-1α/RIT1 signaling pathway
- The poor performance of cardiovascular risk scores in identifying patients with idiopathic inflammatory myopathies at high cardiovascular risk
- miR-2053 inhibits the growth of ovarian cancer cells by downregulating SOX4
- Nucleophosmin 1 associating with engulfment and cell motility protein 1 regulates hepatocellular carcinoma cell chemotaxis and metastasis
- α-Hederin regulates macrophage polarization to relieve sepsis-induced lung and liver injuries in mice
- Changes of microbiota level in urinary tract infections: A meta-analysis
- Identification of key enzalutamide-resistance-related genes in castration-resistant prostate cancer and verification of RAD51 functions
- Falls during oxaliplatin-based chemotherapy for gastrointestinal malignancies – (lessons learned from) a prospective study
- Outcomes of low-risk birth care during the Covid-19 pandemic: A cohort study from a tertiary care center in Lithuania
- Vitamin D protects intestines from liver cirrhosis-induced inflammation and oxidative stress by inhibiting the TLR4/MyD88/NF-κB signaling pathway
- Integrated transcriptome analysis identifies APPL1/RPS6KB2/GALK1 as immune-related metastasis factors in breast cancer
- Genomic analysis of immunogenic cell death-related subtypes for predicting prognosis and immunotherapy outcomes in glioblastoma multiforme
- Circular RNA Circ_0038467 promotes the maturation of miRNA-203 to increase lipopolysaccharide-induced apoptosis of chondrocytes
- An economic evaluation of fine-needle cytology as the primary diagnostic tool in the diagnosis of lymphadenopathy
- Midazolam impedes lung carcinoma cell proliferation and migration via EGFR/MEK/ERK signaling pathway
- Network pharmacology combined with molecular docking and experimental validation to reveal the pharmacological mechanism of naringin against renal fibrosis
- PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma
- miR-141-3p accelerates ovarian cancer progression and promotes M2-like macrophage polarization by targeting the Keap1-Nrf2 pathway
- lncRNA OIP5-AS1 attenuates the osteoarthritis progression in IL-1β-stimulated chondrocytes
- Overexpression of LINC00607 inhibits cell growth and aggressiveness by regulating the miR-1289/EFNA5 axis in non-small-cell lung cancer
- Subjective well-being in informal caregivers during the COVID-19 pandemic
- Nrf2 protects against myocardial ischemia-reperfusion injury in diabetic rats by inhibiting Drp1-mediated mitochondrial fission
- Unfolded protein response inhibits KAT2B/MLKL-mediated necroptosis of hepatocytes by promoting BMI1 level to ubiquitinate KAT2B
- Bladder cancer screening: The new selection and prediction model
- circNFATC3 facilitated the progression of oral squamous cell carcinoma via the miR-520h/LDHA axis
- Prone position effect in intensive care patients with SARS-COV-2 pneumonia
- Clinical observation on the efficacy of Tongdu Tuina manipulation in the treatment of primary enuresis in children
- Dihydroartemisinin ameliorates cerebral I/R injury in rats via regulating VWF and autophagy-mediated SIRT1/FOXO1 pathway
- Knockdown of circ_0113656 assuages oxidized low-density lipoprotein-induced vascular smooth muscle cell injury through the miR-188-3p/IGF2 pathway
- Low Ang-(1–7) and high des-Arg9 bradykinin serum levels are correlated with cardiovascular risk factors in patients with COVID-19
- Effect of maternal age and body mass index on induction of labor with oral misoprostol for premature rupture of membrane at term: A retrospective cross-sectional study
- Potential protective effects of Huanglian Jiedu Decoction against COVID-19-associated acute kidney injury: A network-based pharmacological and molecular docking study
- Clinical significance of serum MBD3 detection in girls with central precocious puberty
- Clinical features of varicella-zoster virus caused neurological diseases detected by metagenomic next-generation sequencing
- Collagen treatment of complex anorectal fistula: 3 years follow-up
- LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through down-regulating SP-A by sponging to miR-424
- Efficacy analysis of empirical bismuth quadruple therapy, high-dose dual therapy, and resistance gene-based triple therapy as a first-line Helicobacter pylori eradication regimen – An open-label, randomized trial
- SMOC2 plays a role in heart failure via regulating TGF-β1/Smad3 pathway-mediated autophagy
- A prospective cohort study of the impact of chronic disease on fall injuries in middle-aged and older adults
- circRNA THBS1 silencing inhibits the malignant biological behavior of cervical cancer cells via the regulation of miR-543/HMGB2 axis
- hsa_circ_0000285 sponging miR-582-3p promotes neuroblastoma progression by regulating the Wnt/β-catenin signaling pathway
- Long non-coding RNA GNAS-AS1 knockdown inhibits proliferation and epithelial–mesenchymal transition of lung adenocarcinoma cells via the microRNA-433-3p/Rab3A axis
- lncRNA UCA1 regulates miR-132/Lrrfip1 axis to promote vascular smooth muscle cell proliferation
- Twenty-four-color full spectrum flow cytometry panel for minimal residual disease detection in acute myeloid leukemia
- Hsa-miR-223-3p participates in the process of anthracycline-induced cardiomyocyte damage by regulating NFIA gene
- Anti-inflammatory effect of ApoE23 on Salmonella typhimurium-induced sepsis in mice
- Analysis of somatic mutations and key driving factors of cervical cancer progression
- Hsa_circ_0028007 regulates the progression of nasopharyngeal carcinoma through the miR-1179/SQLE axis
- Variations in sexual function after laparoendoscopic single-site hysterectomy in women with benign gynecologic diseases
- Effects of pharmacological delay with roxadustat on multi-territory perforator flap survival in rats
- Analysis of heroin effects on calcium channels in rat cardiomyocytes based on transcriptomics and metabolomics
- Risk factors of recurrent bacterial vaginosis among women of reproductive age: A cross-sectional study
- Alkbh5 plays indispensable roles in maintaining self-renewal of hematopoietic stem cells
- Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients
- Correlation between microvessel maturity and ISUP grades assessed using contrast-enhanced transrectal ultrasonography in prostate cancer
- The protective effect of caffeic acid phenethyl ester in the nephrotoxicity induced by α-cypermethrin
- Norepinephrine alleviates cyclosporin A-induced nephrotoxicity by enhancing the expression of SFRP1
- Effect of RUNX1/FOXP3 axis on apoptosis of T and B lymphocytes and immunosuppression in sepsis
- The function of Foxp1 represses β-adrenergic receptor transcription in the occurrence and development of bladder cancer through STAT3 activity
- Risk model and validation of carbapenem-resistant Klebsiella pneumoniae infection in patients with cerebrovascular disease in the ICU
- Calycosin protects against chronic prostatitis in rats via inhibition of the p38MAPK/NF-κB pathway
- Pan-cancer analysis of the PDE4DIP gene with potential prognostic and immunotherapeutic values in multiple cancers including acute myeloid leukemia
- The safety and immunogenicity to inactivated COVID-19 vaccine in patients with hyperlipemia
- Circ-UBR4 regulates the proliferation, migration, inflammation, and apoptosis in ox-LDL-induced vascular smooth muscle cells via miR-515-5p/IGF2 axis
- Clinical characteristics of current COVID-19 rehabilitation outpatients in China
- Luteolin alleviates ulcerative colitis in rats via regulating immune response, oxidative stress, and metabolic profiling
- miR-199a-5p inhibits aortic valve calcification by targeting ATF6 and GRP78 in valve interstitial cells
- The application of iliac fascia space block combined with esketamine intravenous general anesthesia in PFNA surgery of the elderly: A prospective, single-center, controlled trial
- Elevated blood acetoacetate levels reduce major adverse cardiac and cerebrovascular events risk in acute myocardial infarction
- The effects of progesterone on the healing of obstetric anal sphincter damage in female rats
- Identification of cuproptosis-related genes for predicting the development of prostate cancer
- Lumican silencing ameliorates β-glycerophosphate-mediated vascular smooth muscle cell calcification by attenuating the inhibition of APOB on KIF2C activity
- Targeting PTBP1 blocks glutamine metabolism to improve the cisplatin sensitivity of hepatocarcinoma cells through modulating the mRNA stability of glutaminase
- A single center prospective study: Influences of different hip flexion angles on the measurement of lumbar spine bone mineral density by dual energy X-ray absorptiometry
- Clinical analysis of AN69ST membrane continuous venous hemofiltration in the treatment of severe sepsis
- Antibiotics therapy combined with probiotics administered intravaginally for the treatment of bacterial vaginosis: A systematic review and meta-analysis
- Construction of a ceRNA network to reveal a vascular invasion associated prognostic model in hepatocellular carcinoma
- A pan-cancer analysis of STAT3 expression and genetic alterations in human tumors
- A prognostic signature based on seven T-cell-related cell clustering genes in bladder urothelial carcinoma
- Pepsin concentration in oral lavage fluid of rabbit reflux model constructed by dilating the lower esophageal sphincter
- The antihypertensive felodipine shows synergistic activity with immune checkpoint blockade and inhibits tumor growth via NFAT1 in LUSC
- Tanshinone IIA attenuates valvular interstitial cells’ calcification induced by oxidized low density lipoprotein via reducing endoplasmic reticulum stress
- AS-IV enhances the antitumor effects of propofol in NSCLC cells by inhibiting autophagy
- Establishment of two oxaliplatin-resistant gallbladder cancer cell lines and comprehensive analysis of dysregulated genes
- Trial protocol: Feasibility of neuromodulation with connectivity-guided intermittent theta-burst stimulation for improving cognition in multiple sclerosis
- LncRNA LINC00592 mediates the promoter methylation of WIF1 to promote the development of bladder cancer
- Factors associated with gastrointestinal dysmotility in critically ill patients
- Mechanisms by which spinal cord stimulation intervenes in atrial fibrillation: The involvement of the endothelin-1 and nerve growth factor/p75NTR pathways
- Analysis of two-gene signatures and related drugs in small-cell lung cancer by bioinformatics
- Silencing USP19 alleviates cigarette smoke extract-induced mitochondrial dysfunction in BEAS-2B cells by targeting FUNDC1
- Menstrual irregularities associated with COVID-19 vaccines among women in Saudi Arabia: A survey during 2022
- Ferroptosis involves in Schwann cell death in diabetic peripheral neuropathy
- The effect of AQP4 on tau protein aggregation in neurodegeneration and persistent neuroinflammation after cerebral microinfarcts
- Activation of UBEC2 by transcription factor MYBL2 affects DNA damage and promotes gastric cancer progression and cisplatin resistance
- Analysis of clinical characteristics in proximal and distal reflux monitoring among patients with gastroesophageal reflux disease
- Exosomal circ-0020887 and circ-0009590 as novel biomarkers for the diagnosis and prediction of short-term adverse cardiovascular outcomes in STEMI patients
- Upregulated microRNA-429 confers endometrial stromal cell dysfunction by targeting HIF1AN and regulating the HIF1A/VEGF pathway
- Bibliometrics and knowledge map analysis of ultrasound-guided regional anesthesia
- Knockdown of NUPR1 inhibits angiogenesis in lung cancer through IRE1/XBP1 and PERK/eIF2α/ATF4 signaling pathways
- D-dimer trends predict COVID-19 patient’s prognosis: A retrospective chart review study
- WTAP affects intracranial aneurysm progression by regulating m6A methylation modification
- Using of endoscopic polypectomy in patients with diagnosed malignant colorectal polyp – The cross-sectional clinical study
- Anti-S100A4 antibody administration alleviates bronchial epithelial–mesenchymal transition in asthmatic mice
- Prognostic evaluation of system immune-inflammatory index and prognostic nutritional index in double expressor diffuse large B-cell lymphoma
- Prevalence and antibiogram of bacteria causing urinary tract infection among patients with chronic kidney disease
- Reactive oxygen species within the vaginal space: An additional promoter of cervical intraepithelial neoplasia and uterine cervical cancer development?
- Identification of disulfidptosis-related genes and immune infiltration in lower-grade glioma
- A new technique for uterine-preserving pelvic organ prolapse surgery: Laparoscopic rectus abdominis hysteropexy for uterine prolapse by comparing with traditional techniques
- Self-isolation of an Italian long-term care facility during COVID-19 pandemic: A comparison study on care-related infectious episodes
- A comparative study on the overlapping effects of clinically applicable therapeutic interventions in patients with central nervous system damage
- Low intensity extracorporeal shockwave therapy for chronic pelvic pain syndrome: Long-term follow-up
- The diagnostic accuracy of touch imprint cytology for sentinel lymph node metastases of breast cancer: An up-to-date meta-analysis of 4,073 patients
- Mortality associated with Sjögren’s syndrome in the United States in the 1999–2020 period: A multiple cause-of-death study
- CircMMP11 as a prognostic biomarker mediates miR-361-3p/HMGB1 axis to accelerate malignant progression of hepatocellular carcinoma
- Analysis of the clinical characteristics and prognosis of adult de novo acute myeloid leukemia (none APL) with PTPN11 mutations
- KMT2A maintains stemness of gastric cancer cells through regulating Wnt/β-catenin signaling-activated transcriptional factor KLF11
- Evaluation of placental oxygenation by near-infrared spectroscopy in relation to ultrasound maturation grade in physiological term pregnancies
- The role of ultrasonographic findings for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative breast cancer
- Construction of immunogenic cell death-related molecular subtypes and prognostic signature in colorectal cancer
- Long-term prognostic value of high-sensitivity cardiac troponin-I in patients with idiopathic dilated cardiomyopathy
- Establishing a novel Fanconi anemia signaling pathway-associated prognostic model and tumor clustering for pediatric acute myeloid leukemia patients
- Integrative bioinformatics analysis reveals STAT2 as a novel biomarker of inflammation-related cardiac dysfunction in atrial fibrillation
- Adipose-derived stem cells repair radiation-induced chronic lung injury via inhibiting TGF-β1/Smad 3 signaling pathway
- Real-world practice of idiopathic pulmonary fibrosis: Results from a 2000–2016 cohort
- lncRNA LENGA sponges miR-378 to promote myocardial fibrosis in atrial fibrillation
- Diagnostic value of urinary Tamm-Horsfall protein and 24 h urine osmolality for recurrent calcium oxalate stones of the upper urinary tract: Cross-sectional study
- The value of color Doppler ultrasonography combined with serum tumor markers in differential diagnosis of gastric stromal tumor and gastric cancer
- The spike protein of SARS-CoV-2 induces inflammation and EMT of lung epithelial cells and fibroblasts through the upregulation of GADD45A
- Mycophenolate mofetil versus cyclophosphamide plus in patients with connective tissue disease-associated interstitial lung disease: Efficacy and safety analysis
- MiR-1278 targets CALD1 and suppresses the progression of gastric cancer via the MAPK pathway
- Metabolomic analysis of serum short-chain fatty acid concentrations in a mouse of MPTP-induced Parkinson’s disease after dietary supplementation with branched-chain amino acids
- Cimifugin inhibits adipogenesis and TNF-α-induced insulin resistance in 3T3-L1 cells
- Predictors of gastrointestinal complaints in patients on metformin therapy
- Prescribing patterns in patients with chronic obstructive pulmonary disease and atrial fibrillation
- A retrospective analysis of the effect of latent tuberculosis infection on clinical pregnancy outcomes of in vitro fertilization–fresh embryo transferred in infertile women
- Appropriateness and clinical outcomes of short sustained low-efficiency dialysis: A national experience
- miR-29 regulates metabolism by inhibiting JNK-1 expression in non-obese patients with type 2 diabetes mellitus and NAFLD
- Clinical features and management of lymphoepithelial cyst
- Serum VEGF, high-sensitivity CRP, and cystatin-C assist in the diagnosis of type 2 diabetic retinopathy complicated with hyperuricemia
- ENPP1 ameliorates vascular calcification via inhibiting the osteogenic transformation of VSMCs and generating PPi
- Significance of monitoring the levels of thyroid hormone antibodies and glucose and lipid metabolism antibodies in patients suffer from type 2 diabetes
- The causal relationship between immune cells and different kidney diseases: A Mendelian randomization study
- Interleukin 33, soluble suppression of tumorigenicity 2, interleukin 27, and galectin 3 as predictors for outcome in patients admitted to intensive care units
- Identification of diagnostic immune-related gene biomarkers for predicting heart failure after acute myocardial infarction
- Long-term administration of probiotics prevents gastrointestinal mucosal barrier dysfunction in septic mice partly by upregulating the 5-HT degradation pathway
- miR-192 inhibits the activation of hepatic stellate cells by targeting Rictor
- Diagnostic and prognostic value of MR-pro ADM, procalcitonin, and copeptin in sepsis
- Review Articles
- Prenatal diagnosis of fetal defects and its implications on the delivery mode
- Electromagnetic fields exposure on fetal and childhood abnormalities: Systematic review and meta-analysis
- Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae
- Saddle pulmonary embolism in the setting of COVID-19 infection: A systematic review of case reports and case series
- Vitamin C and epigenetics: A short physiological overview
- Ebselen: A promising therapy protecting cardiomyocytes from excess iron in iron-overloaded thalassemia patients
- Aspirin versus LMWH for VTE prophylaxis after orthopedic surgery
- Mechanism of rhubarb in the treatment of hyperlipidemia: A recent review
- Surgical management and outcomes of traumatic global brachial plexus injury: A concise review and our center approach
- The progress of autoimmune hepatitis research and future challenges
- METTL16 in human diseases: What should we do next?
- New insights into the prevention of ureteral stents encrustation
- VISTA as a prospective immune checkpoint in gynecological malignant tumors: A review of the literature
- Case Reports
- Mycobacterium xenopi infection of the kidney and lymph nodes: A case report
- Genetic mutation of SLC6A20 (c.1072T > C) in a family with nephrolithiasis: A case report
- Chronic hepatitis B complicated with secondary hemochromatosis was cured clinically: A case report
- Liver abscess complicated with multiple organ invasive infection caused by hematogenous disseminated hypervirulent Klebsiella pneumoniae: A case report
- Urokinase-based lock solutions for catheter salvage: A case of an upcoming kidney transplant recipient
- Two case reports of maturity-onset diabetes of the young type 3 caused by the hepatocyte nuclear factor 1α gene mutation
- Immune checkpoint inhibitor-related pancreatitis: What is known and what is not
- Does total hip arthroplasty result in intercostal nerve injury? A case report and literature review
- Clinicopathological characteristics and diagnosis of hepatic sinusoidal obstruction syndrome caused by Tusanqi – Case report and literature review
- Synchronous triple primary gastrointestinal malignant tumors treated with laparoscopic surgery: A case report
- CT-guided percutaneous microwave ablation combined with bone cement injection for the treatment of transverse metastases: A case report
- Malignant hyperthermia: Report on a successful rescue of a case with the highest temperature of 44.2°C
- Anesthetic management of fetal pulmonary valvuloplasty: A case report
- Rapid Communication
- Impact of COVID-19 lockdown on glycemic levels during pregnancy: A retrospective analysis
- Erratum
- Erratum to “Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway”
- Erratum to: “Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p”
- Retraction
- Retraction of “Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients”
- Retraction of “circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis”
- Retraction of “miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells”
- Retraction of “SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis”
- Retraction of “circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury”
- Retraction of “lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells”
- Special issue Linking Pathobiological Mechanisms to Clinical Application for cardiovascular diseases
- Effect of cardiac rehabilitation therapy on depressed patients with cardiac insufficiency after cardiac surgery
- Special issue The evolving saga of RNAs from bench to bedside - Part I
- FBLIM1 mRNA is a novel prognostic biomarker and is associated with immune infiltrates in glioma
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part III
- Development of a machine learning-based signature utilizing inflammatory response genes for predicting prognosis and immune microenvironment in ovarian cancer

