Abstract
Hepatocellular carcinoma (HCC) is a frequently diagnosed malignancy with a high mortality rate. Cisplatin (CDDP) is a widely applied anti-cancer drug. However, a large population of liver cancer patients developed CDDP resistance. The polypyrimidine tract binding protein (PTBP1) is an RNA-binding protein involving in progressions of diverse cancers. Here we report PTBP1 was significantly upregulated in liver tumors and cell lines. Silencing PTBP1 effectively sensitized HCC cells to CDDP. From the established CDDP-resistant HCC cell line (HepG2 CDDP Res), we observed that CDDP-resistant cells were more sensitive to CDDP under low glutamine supply compared with that in HCC parental cells. CDDP-resistant HCC cells displayed elevated glutamine metabolism rate. Consistently, PTBP1 promotes glutamine uptake and the glutamine metabolism key enzyme, glutaminase (GLS) expression. Bioinformatics analysis predicted that the 3′-UTR of GLS mRNA contained PTBP1 binding motifs which were further validated by RNA immunoprecipitation and RNA pull-down assays. PTBP1 associated with GLS 3′-UTR to stabilize GLS mRNA in HCC cells. Finally, we demonstrated that the PTBP1-promoted CDDP resistance of HCC cells was through modulating the GLS–glutamine metabolism axis. Summarily, our findings uncovered a PTBP1-mediated CDDP resistance pathway in HCC, suggesting that PTBP1 is a promisingly therapeutic target to overcome chemoresistance of HCC.
1 Introduction
Liver cancer, which is a common malignancy with a high mortality rate and poor prognosis, is one of the most leading causes of cancer-associated death [1,2]. Hepatocellular carcinoma (HCC) is the major (70–85%) subtype of liver cancer [3]. Currently, surgical removal is the primarily therapeutic approach against HCC [4]. However, early symptoms of primary HCC are difficult to characterize [4]. Thus, chemo- and radio-therapies have been applied to HCC patients in middle or advanced stage when significant symptoms were observed [5]. Cisplatin (CDDP) is a platinum-based chemotherapeutic drug, acting through interacting with purine bases to induce DNA lesions, leading to cancer cell death [5,6]. Although CDDP has been widely used for cancer treatment in clinical practice, side effects and development of CDDP resistance has resulted in substantial barriers to limit its wide applications [7]. Therefore, it is imperative to identify the molecular pathways and mechanisms of CDDP-resistant HCC.
Tumor cells exhibit metabolic profiles that they demand higher metabolic rates such as glucose and glutamine metabolism [8]. Accumulating studies uncovered critical roles of glutamine metabolism in tumorigenesis and development using in vitro and in vivo models [9,10]. Moreover, the dysregulated cellular metabolism rate of cancer cells was tightly correlated to chemoresistance [11] suggesting that blocking glutamine metabolism of HCC cells could potentially enhance the therapeutic outcomes of CDDP.
Polypyrimidine tract binding protein (PTBP1) is an RNA-binding protein which plays important biological roles in diverse processes of cancer cells [12]. PTBP1 was known to act as a tumor-promotive regulator in various cancers, including gastric cancer [13], breast cancer [14], colon cancer [15], and lung cancer [16]. Furthermore, studies have proven that PTBP1 was positively associated with glucose metabolism of cancer cells through modulating the pyruvate kinase M2 isoform [17]. However, the precise roles and molecular targets of PTBP1 in regulating glutamine metabolism and CDDP resistance of liver cancer cells have not been elucidated.
In this study, the biological roles of PTBP1 in CDDP-resistant liver cancer were investigated. Bioinformatics analysis predicted the 3′-UTR of glutaminase (GLS) contained binding motifs of PTBP1. Molecular mechanisms of the PTBP1–GLS association were explored. This study proposes that targeting the PTBP1–GLS–glutamine metabolism axis could be a potential strategy to overcome chemoresistance of liver cancer.
2 Materials and methods
2.1 HCC specimen collections
This study was approved by the Ethics Committee of Tianjin Second People’s Hospital. Forty HCC tumor specimens and matched normal liver tissues were collected from liver cancer patients from July 2018 to April 2020 in the Liver Diseases Branch, Tianjin Second People’s Hospital, Tianjin, China. No chemo- or radio-therapy was applied before biopsy. After surgery, tissues were immediately stored in liquid nitrogen. All patients signed the informed consent. The required minimum sample number (n = 35) was estimated by power analysis.
2.2 Cell culture and reagents
HCC cell lines HepG2, Huh7, CA3, SNU-878, and SNU-182 as well as human normal liver cell line, L02 were purchased from the Shanghai Institute for Biological Science (China). Cells were cultured with RPMI 1640 medium with 10% fetal bovine serum plus 100 U/mL penicillin and 100 U/mL streptomycin under 5% CO2 at 37°C. Establishment of CDDP-resistant HepG2 cell line was performed according to previous descriptions [18]. Monoclonal anti-PTBP1 (rabbit, #8776), anti-GLS (rabbit, #56750), anti-MDR1/ABCB1 (rabbit, #13342), and anti-β-actin (rabbit, #4970) were purchased from Cellsignaling Tech (Danvers, MA, USA). CDDP was purchased from Sigma-Aldrich (Shanghai, China).
2.3 Bioinformatics analysis
The association between PTBP1 and GLS and the binding motifs of PTBP1 on 3′-UTR of GLS were predicted from starBase of ENCORI http://starbase.sysu.edu.cn/. Kaplan–Meier plotter survival rate for liver cancer patients with low or high PTBP1 level was analyzed from https://kmplot.com. Expressions of PTBP1 and GLS from normal or tumorous liver tissues were analyzed from TCGA database by https://ualcan.path.uab.edu. The expression correlation between PTBP1 and GLS in liver cancer was analyzed by Pearson correlation coefficient analysis from starBase of ENCORI http://starbase.sysu.edu.cn/.
2.4 Transfection with siRNA or plasmid DNA
Silencing of GLS or PTBP1 gene expression was conducted by transfection of specific siRNA. HepG2 cells were incubated in six-well plates at a density of 5 × 105 cells/well for overnight. Cells were transfected with siRNA targeting human GLS or PTBP1 using the Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. PTBP1 overexpression plasmid and control plasmid were obtained from Origene.com. The control siRNA, siGLS, and siPTBP1 were synthesized by Gene-Pharma Co. (Shanghai, China). siRNA knockdown efficiency was validated by western blot in HCC cells. Plasmid DNA was transfected at 1 µg/mL and siRNAs were transfected at 25 nM for 48 h. Transfection was performed in triplicate.
2.5 Total RNA isolation and qRT-PCR
Total RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, cultured cells were lysed with lysis buffer and proteinase K. DNase I solution (Thermo Fisher Scientific) was added to digest the genomic DNA. The quality and quantity of RNA samples were measured by a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Rockford, IL, USA). The first-strand cDNA was synthesized from RNA sample (1 µg) using iScript™ RT Supermix for RT-qPCR® (Bio-Rad, Hercules, CA, USA). The qRT-PCR reactions were conducted using the SYBR Green qPCR SuperMix reagents (Thermo Fisher Scientific Inc., Waltham, MA, USA) with typical amplification parameters of 95°C for 30 s, followed by 40 cycles at 98°C for 10 s and 60°C for 30 s. Primers for qRT-PCR were: PTBP1: Forward: 5′-GCATCGACTTTTCCAAGCTC-3′, Reverse: 5′-GGAAACCAGCTCCTGCATAC-3′; GLS: Forward: 5′-CAGAAGGCACAGACATGGTTGG-3′, Reverse: 5′-GGCAGAAACCACCATTAGCCAG-3′; β-actin: Forward: 5′-CTGAGAGGGAAATCGTGCGT-3′, Reverse: 5′-CCACAGGATTCCATACCCAAGA-3′. β-Actin was used as an internal control. The relative expressions were calculated using the 2−ΔΔCT method. The experiments were performed in triplicate and repeated three times.
2.6 Cell viability analysis
HCC cells were seeded in 96-well plates containing 0.2 mL medium for 24 h. After treatment with CDDP for 48 h, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added into cell culture medium, followed by incubation at 37°C for 4 h. Dimethyl sulfoxide (150 μL per well) was added and incubated for 1 h. The absorbance was measured at 570 nm. The experiments were performed in triplicate and repeated three times.
2.7 Cell apoptosis analysis
Apoptosis rate of HCC cells in response to CDDP treatment was examined using Annexin V-fluorescein isothiocyanate (FITC) and PI (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions. Briefly, after treatments, cells (5 × 105 cells/well in six-well plate) were harvested, washed, suspended in 100 µL binding buffer, and then stained with Annexin V-FITC and PI for 15 min in the dark at room temperature. Apoptotic cells were measured using the Cytoflex (Beckman Coulter, Inc.). FlowJo software was used to analyze the cytometric data. The experiments were performed in triplicate and repeated three times.
2.8 Clonogenic assay
After treatment with CDDP, parental or CDDP-resistant liver cancer cells were seeded onto six-well plates at a density of 5 × 104 cells/well with normal or low glutamine medium. Cells were cultured for 2 weeks, and the medium was re-freshed every 3 days. The colonies were fixed by 4% paraformaldehyde and stained with 0.1% crystal violet for 5 min. Plates were washed extensively with phosphate-buffered saline (PBS). Colonies with >50 cells/colony were counted and recorded under a bright field microscopy. The experiments were repeated three times.
2.9 RNA immunoprecipitation (RIP)
Total RNA was isolated from HCC cells using an Rneasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. RIP assay was performed using a Magna RIPTM RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA). Briefly, cells were lysed in RIP lysis buffer. Anti-IgG (control) or anti-PTBP1 (dilution 1:50) antibody with A/G immunomagnetic beads were added into cell lysates to immunoprecipitate PTBP1–RNA complexes. Purified RNA samples were subjected to qPCR analysis using GLS specific primers to determine the enrichment of GLS mRNA. The experiments were repeated three times.
2.10 RNA pull-down assay
The GLS mRNA-associated PTBP1 protein was examined by RNA pull-down assay. Briefly, negative control RNA (antisense RNA of binding motif) and the predicted binding motif of GLS were labeled using a biotin RNA labeling mix (Roche, Shanghai, China). The biotin labeled RNA was treated with RNase-free DNase I and purified by an RNeasy mini kit (Qiagen, Shanghai, China), followed by incubating with proteins from HCC cell. The streptavidin agarose beads (Invitrogen, Carlsbad, CA, USA) were then added into the mixture. After washing, the amount of PTBP1 protein in the RNA–protein complex was determined by western blot. The experiments were repeated three times.
2.11 RNA stability
The effect of PTBP1 on the stability of GLS mRNA was evaluated by detecting the half-life of GLS mRNA. Cells were treated with 5 μg/mL actinomycin D for 0, 2, 4, 6, and 8 h. Total RNA was collected by TRIzol reagent. The expression level of GLS mRNA was measured by qRT-PCR. The experiments were performed in triplicate and repeated three times.
2.12 Western blot
Liver cancer cells were harvested and washed at 4°C with PBS. Cells were lysed on ice with RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 1× protease inhibitors cocktail (Thermo Fisher Scientific) for 20 min, followed by centrifugation at 12,000×g for 20 min at 4°C. Equal amount of protein sample (40 µL) from each group was separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes. Membranes were blocked with 4% bovine serum albumin in phosphate-buffered saline with Tween 20 (PBST) for 1 h at room temperature, followed by incubation with primary antibodies at 1:1,000 dilution at 4°C for overnight. After complete washing by PBST, membranes were incubated with secondary antibodies at room temperature for 1 h. β-Actin was loading control. Membranes were detected by the enhanced chemiluminescence (Applygen Technologies Inc., Beijing, China) and the ChemiDoc Imaging Systems (Bio-Rad, Hercules, CA, USA). The experiments were repeated three times.
2.13 Statistical analysis
Statistical analysis was performed using the Prism software version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data were presented as mean ± standard deviation. Two-tailed student’s t-test was applied to analyze the statistical significance between two groups. Significance of difference among three or more groups was analyzed by one-way ANOVA. p-Value <0.05 was considered statistically significant.
3 Results
3.1 PTBP1 is upregulated in liver cancer and associated with CDDP resistance
Bioinformatics analysis showed that PTBP1 was markedly overexpressed in various cancers, including liver cancer (Figure A1, Figure 1a). Moreover, the expression levels of PTBP1 were positively associated with the grades (Figure A2a) and metastasis status (Figure A2b) of liver cancer, suggesting a potentially oncogenic role of PTBP1 in liver cancer. To verify this, PTBP1 expressions were examined in human HCC specimens and their adjacent normal liver tissues from 40 HCC patients. qRT-PCR and immunohistochemistry showed that PTBP1 was remarkably highly expressed in cancerous tissues compared with normal tissues (Figure 1b and c). In addition, Kaplan–Meier plotter survival analysis illustrated that HCC patients with higher PTBP1 were significantly associated with lower survival rates (Figure 1d). Furthermore, both mRNA and protein levels of PTBP1 were significantly elevated in HCC cell lines, including HepG2, SNU-878, Huh7, C3A, and SNU-182 cells compared with normal hepatocytes cell line, L02 (Figure 1e and f). To explore the biological functions of PTBP1 in chemosensitivity, specific PTBP1 siRNA was transfected into HepG2 cells to knockdown PTBP1 expression (Figure 1g). Subsequently, HepG2 cells without or with PTBP1 silencing were exposed to increased concentrations of CDDP. Consistent results from cell viability assay and cell apoptosis assay demonstrated that silencing PTBP1 effectively enhanced the cytotoxicity of CDDP on HCC cells (Figure 1h and i). Taken together, these results suggest that PTBP1 is associated with poor response to CDDP treatment and exhibits an oncogenic role in HCC.

PTBP1 is positively correlated with liver cancer and CDDP resistance. (a) Expressions of PTBP1 in liver cancer tissues and normal liver tissues analyzed from TCGA database. (b) Expressions of PTBP1 in liver cancer tissues (n = 40) and normal liver tissues (n = 40) examined by qRT-PCR. (c) Representative IHC staining of PTBP1 in normal and tumorous liver tissues. (d) Kaplan–Meier plotter survival analysis shows the correlation between PTBP1 expression levels and survival rates of liver cancer cells. (e) mRNA and (f) protein expressions of PTBP1 are shown in one normal hepatocyte cell line and five HCC cell lines. (g) HepG2 cells were transfected with control siRNA or PTBP1 siRNA, expressions of PTBP1 were detected by western blot. (h) The above transfected cells were treated with CDDP at the indicated concentrations for 48 h. Cell response to CDDP was determined by cell viability assay and (i) Annexin V apoptosis assay. *p < 0.05; **p < 0.01; ***p < 0.001.
3.2 CDDP-resistant HCC cells display glutamine addictive phenotypes
We then investigated the underlying cellular mechanisms of the PTBP1-promoted CDDP resistance. Accumulating evidence unveiled that the dysregulated cancer metabolism was tightly associated with tumorigenesis, progressions as well as chemoresistance [9,10]. To assess the roles of glutamine metabolism in CDDP sensitivity, we established a CDDP-resistant liver cancer cell line (HepG2 CDDP Res) via exposing cells to elevated concentrations of CDDP. HepG2 parental cells were maintained in cell culture medium and treated with CDDP (4–16 μg/mL) for at least 4 weeks to select the survival (CDDP Res) cells. As shown in Figure 2a, HepG2 CDDP Res cells could tolerate higher concentrations of CDDP than HepG2 parental cells (Figure 2a and b). The CDDP IC50 of HepG2 CDDP Res was 35.24 µg/mL, which is significantly higher than that of HepG2 parental cells (8.24 µg/mL) (Figure 2a). Consistent results from clonogenic assay demonstrated that CDDP treatment at 8 µg/mL only slightly inhibited cell survival of HepG2 CDDP-resistant cells. However, apparently cell death was observed in HepG2 parental cells under the same concentration of CDDP treatment (Figure 2b). Moreover, the ABCB1 protein expression was significantly upregulated in CDDP-resistant HepG2 cells (Figure A3). Expectedly, PTBP1 was significantly upregulated in HepG2 CDDP-resistant cells compared to that in HepG2 parental cells (Figure 2c). We then evaluated the glutamine metabolism characteristics in CDDP-resistant liver cancer cells. The glutamine uptake (Figure 2d) and GLS activity (Figure 2e), two glutamine metabolism readouts were significantly elevated in HepG2 CDDP-resistant cells. Furthermore, under low glutamine supply, CDDP-resistant cells were more inhibited by CDDP treatment, while the HepG2 parental cells were less affected under the same CDDP treatment (Figure 2f). Subsequently, MTT assay (Figure 2g) and cell apoptosis assay (Figure 2h) demonstrated that low glutamine treatment effectively re-sensitized CDDP-resistant cells to CDDP treatment. Summarily, these results revealed that CDDP-resistant HCC cells were more dependent on glutamine metabolism, suggesting that targeting glutamine metabolism is a potentially therapeutic approach for treatment of chemoresistant liver cancer.
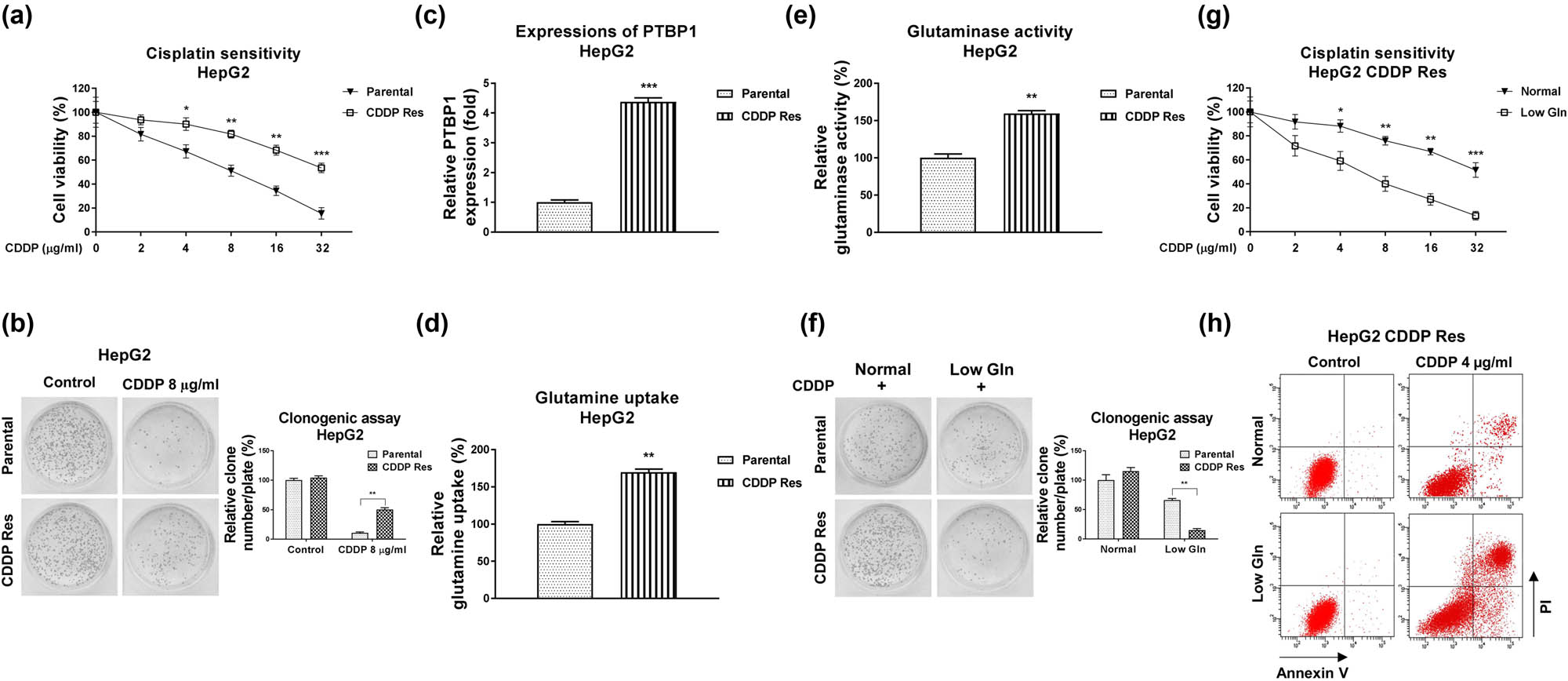
CDDP-resistant liver cancer cells show glutamine addictive phenotypes. (a) HepG2 parental cells and CDDP-resistant cells were treated with CDDP at the indicated concentrations for 48 h. Cell response to CDDP was determined by cell viability assay and (b) clonogenic assay. (c) Expressions of PTBP1 in HepG2 parental cells and CDDP-resistant cells were examined by Western blot. (d) Glutamine uptake and (e) GLS activity were examined in HepG2 parental and CDDP-resistant cells. (f) HepG2 parental cells and CDDP-resistant cells were cultured with normal or low glutamine condition, cells were treated with CDDP at the indicated concentrations. The cell survival rates were examined by clonogenic assay, (g) cell viability assay, and (h) Annexin V apoptosis assay. *p < 0.05; **p < 0.01; ***p < 0.001.
3.3 PTBP1 promotes glutamine metabolism of HCC cells
Given the above results revealed a PTBP1-promoted CDDP resistance and a positive correlation between glutamine metabolism and CDDP resistance in HCC cells, we then evaluated whether PTBP1 directly regulated glutamine metabolism of HCC cells. As shown in Figure 3a, knocking down of PTBP1 significantly blocked the protein expression of GLS, which catalyzes the speed-limiting reaction of glutamine metabolism in HCC cells and was significantly upregulated in liver cancer (Figure A4). Consistently, silencing PTBP1 effectively suppressed glutamine uptake (Figure 3b) and GLS activity of HCC cells (Figure 3c). We then assessed the effects of PTBP1 knockdown on glutamine metabolism of CDDP-resistant HCC cells. Expected results demonstrated that knocking down of PTBP1 significantly downregulated the GLS expression (Figure 3d) and suppressed glutamine metabolism (Figure 3e and f) of HepG2 CDDP Res cells. In summary, these results indicated that PTBP1 positively regulates glutamine metabolism of liver cancer cells.

PTBP1 promotes glutamine metabolism of HCC cells. (a) HepG2 cells were transfected with control siRNA or siPTBP1. Expressions of GLS were examined by western blot. (b) Glutamine uptake and (c) GLS activity from the above transfected cells were examined. (d) HepG2 CDDP Res cells were transfected with control siRNA or siPTBP1. Expressions of GLS were examined by western blot. (e) Glutamine uptake and (f) GLS activity from the above transfected cells were examined. *p < 0.05; **p < 0.01.
3.4 PTBP1 binds to 3′-UTR of GLS to upregulate GLS expressions via preventing its RNA degradation
Recent studies revealed that PTBP1 acted as a typical RNA-binding protein [12]. Thus, the post-transcriptional regulation of target genes such as regulating RNA stability might be a potential molecular mechanism of the PTBP1-promoted glutamine metabolism. We then analyzed the downstream RNA targets of PTBP1. Analysis from TCGA database indicated that GLS was significantly upregulated in liver cancer (Figure A4) and positively associated with the grades (Figure A5A) and metastasis status (Figure A5b) of liver cancer. Previous studies indicated that PTBP1 preferentially bond to polypyrimidine-rich stretches of RNAs [12,19]. We then assessed whether GLS mRNA contains PTBP1 binding elements. Bioinformatics analysis illustrated a PTBP1–RNA interaction between PTBP1 and GLS 3′-UTR which contains multiple PTBP1 binding motifs (Figure 4a). Among them, one motif showed the strongest binding capacity with PTBP1 (Figure 4a). Expectedly, a significantly positive correlation between PTBP1 and GLS was observed in HCC patients (Figure 4b), suggesting that PTBP1 positively regulates GLS mRNA through binding to its 3′-UTR region. We then hypothesized PTBP1-enhanced mRNA stability of GLS to upregulate its expression. To test that, RIP assay was performed using PTBP1-specific antibody in HepG2 cells. qRT-PCR results demonstrated that GLS 3′-UTR region was enriched in PTBP1-precipitated RNA fragments (Figure 4c). In addition, RNA pull-down assay was performed using biotin-labeled GLS 3′-UTR to pull down specific binding proteins. Results in Figure 4d showed that significant amount of PTBP1 was pulled down by GLS 3′-UTR. To examine whether the PTBP1-upregulated GLS expressions was through binding to 3′-UTR of GLS, PTBP1 was silenced in HepG2 cells and the PTBP1–GLS interaction was analyzed by RIP. Expectedly, HepG2 cells with lower PTBP1 bond less amount of mRNA fragments (Figure 4e). We then evaluated whether PTBP1 affects mRNA stability of GLS. RNA stability assays were performed to compare the half-life of GLS mRNAs in control and PTBP1-silenced HCC cells. Results showed that the half-life of GLS mRNA was significantly suppressed in PTBP1-silencing cells compared to that in control cells (Figure 4f). Taken together, these results validated that PTBP1 upregulated GLS expression through binding to GLS 3′-UTR, resulting in GLS mRNA stabilization in HCC cells.

PTBP1 binding to 3′-UTR of GLS mRNA to regulate its mRNA stability. (a) Predicted binding motifs of PTBP1 on GLS 3′-UTR. (b) Expression correlation analysis between GLS and PTBP1 in liver cancers. (c) RIP was performed in HepG2 cells using IgG control or anti-PTBP1 antibody. GLS mRNA amounts in the PTBP1-immunoprecipitated fraction were measured by qRT-PCR. (d) RNA pull-down assay was performed in HepG2 cells. The PTBP1 protein which was associated with GLS mRNA was assayed by western blotting. (e) HepG2 cells were transfected with control siRNA or PTBP1 siRNA for 48 h, RIP experiments were performed and the GLS mRNA abundance in immunoprecipitated fraction was determined by qRT-PCR. (f) HepG2 cells were treated with 5 μg/mL actinomycin D for 0, 2, 4, 6, and 8 h, the relative half-life of GLS mRNA levels were measured by qRT-PCR. *p < 0.05; ***p < 0.001.
3.5 Blocking PTBP1 re-sensitizes CDDP-resistant HCC cells through suppressing glutamine metabolism
Finally, we assessed whether the PTBP1-promoted CDDP resistance was through upregulating the GLS-mediated glutamine metabolism. Silencing GLS (Figure 5a) effectively blocked glutamine uptake (Figure 5b), GLS activity (Figure 5c), and the CDDP resistance of HepG2 (Figure 5d). Moreover, HepG2 CDDP Res cells with PTBP1 overexpression (Figure 5e) displayed significantly elevated glutamine uptake (Figure 5f), GLS activity (Figure 5g), and the CDDP resistance (Figure 5h). Consequently, these phenotypes were further overridden by treatment of GLS inhibitor, BPTES, which is a noncompetitive inhibitor of GLS to specifically block the GLS enzymatic activity but not affect the protein expression (Figure 5e–g). Taken together, the above results demonstrated a PTBP1–GLS–glutamine metabolism axis in regulating CDDP resistance of liver cancer cell.

Roles of the PTBP1–GLS–glutamine metabolism pathway in CDDP-resistant HCC cells. (a) HepG2 CDDP-resistant cells were transfected with control siRNA or si GLS, expressions of GLS were determined by western blot. (b) Glutamine uptake, (c) GLS activity, and (d) CDDP sensitivity were examined in the above transfected cells. (e) HepG2 CDDP-resistant cells were transfected with control plasmid or GLS overexpression plasmid. Cells were treated with control or GLS inhibitor, BPTES. Expressions of GLS were determined by western blot. (f) Glutamine uptake, (g) GLS activity, and (h) CDDP sensitivity were examined in the above transfected cells. *p < 0.05; **p < 0.01; ***p < 0.001.
4 Discussion
Liver cancer is a prevalent malignancy with a high mortality rate and poor prognosis throughout the world. HCC is the most common liver cancer subtype [1–3]. Currently, the platinum-based anti-cancer drugs have been widely applied to improve the clinical outcomes of liver cancer patients who were diagnosed at middle or advanced stage [5,6]. However, development of drug resistance impaired the clinical applications of CDDP. This study unveiled a PTBP1-promoted CDDP resistance in HCC cells through modulating glutamine metabolism. PTBP1 was significantly upregulated in liver tumors and cell lines. Moreover, PTBP1 was positively associated with CDDP resistance, indicating that PTBP1 is a potentially diagnostic biomarker and therapeutic target against chemoresistant liver cancer.
Cancer cells display a new hallmark that they reprogram glucose metabolism by utilization of glucose toward aerobic glycolysis instead of mitochondrial oxidative phosphorylation, a phenomenon called “Warburg effect” [20]. Not surprisingly, cancer cells demand a variety of nutrients such as glucose, glutamine, and lipids as carbon sources and energy for hyper-proliferation [20]. Since oncogenic shift of cellular metabolism rendered cancer cells addicted to glutamine, molecular targets and signaling pathway involved in glutamine metabolism could be potentially therapeutic approaches. Therefore, in light of the glutamine metabolism in the CDDP-resistant HCC cells, investigating new therapeutic targets and underlying molecular mechanisms are urgent tasks to overcome CDDP resistance. From the established CDDP-resistant liver cancer cell line, we observed that the glutamine metabolism was remarkably elevated. Moreover, under low glutamine supply, CDDP-resistant HepG2 cells exhibited higher CDDP sensitivity than that from HepG2 parental cells. We further demonstrated that PTBP1 stimulated glutamine metabolism of HCC cells. These results consistently revealed that PTBP1 promoted CDDP resistance of HCC cells through modulating the glutamine metabolism.
As an RNA binding protein, PTBP1 preferentially binds to polypyrimidine-rich stretches of RNA to regulate various RNA processions such as RNA metabolism, alternative splicing, translation, stability, translocation, and pre-mRNA processing [12]. In addition, PTBP1 was an important regulator in multiple cancers [13–16]. However, the roles of PTBP1 in glutamine metabolism as well as the direct mechanisms of GLS regulation have not been investigated. Here, we highlighted a PTBP1 binding motif in 3′-UTR of GLS. The binding of PTBP1 on GLS 3′-UTR was validated by RIP assay and RNA pull-down assay. Previous studies reported that PTBP1 bond to 3′-UTR of downstream mRNAs to stabilize them by preventing the degradation protein UPF1 from binding to 3′-UTRs [21]. Results from RNA stability assay consistently showed that PTBP1 bond to GLS 3′-UTR to stabilize its mRNA. These results consolidated that PTBP1 upregulated GLS expressions through direct binding to 3′-UTR of GLS, leading to the stabilization of GLS mRNA. However, this study still has limitations that the majority of discovery was from in vitro assay. An in vivo xenograft mouse model will be analyzed in our further works.
In summary, this study unveiled a PTBP1-promoted CDDP resistance in HCC cells through promoting glutamine metabolism by stabilizing the GLS mRNA. Our discovery highlights a novel molecular axis for overcoming chemoresistant liver cancer.
Acknowledgement
None.
-
Funding information: This study was supported by an internal research funding resource from Tianjin Second People’s Hospital.
-
Author contributions: R.M.T. and Y.L. designed the experiments. R.M.T., Y.F.L., X.J.Sh., and Y.L. carried out the experiments and analyzed the data. All authors participated into the manuscript preparation.
-
Conflict of interest: The authors declare no potential conflicts of interest.
-
Data availability statement: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Appendix

Expression of PTBP1 across TCGA cancers (normal vs tumor).

Expression of PTBP1 in HCC tissues based on (a) Tumor grad and (b) nodal metastasis status.

Expressions of ABCB1 in parental and CDDP resistant HepG2 cells.

Expression of GLS in normal liver tissue and HCC tissues from TCGA database.

Expression of GLS in HCC tissues based on (a) Tumor grad and (b) nodal metastasis status.
References
[1] Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019 Apr;380(15):1450–62.10.1056/NEJMra1713263Search in Google Scholar PubMed
[2] El Jabbour T, Lagana SM, Lee H. Update on hepatocellular carcinoma: pathologists’ review. World J Gastroenterol. 2019 Apr;25(14):1653–65.10.3748/wjg.v25.i14.1653Search in Google Scholar PubMed PubMed Central
[3] Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020 Jan;1873(1):188314.10.1016/j.bbcan.2019.188314Search in Google Scholar PubMed PubMed Central
[4] Demir T, Lee SS, Kaseb AO. Systemic therapy of liver cancer. Adv Cancer Res. 2021;149:257–94.10.1016/bs.acr.2020.12.001Search in Google Scholar PubMed
[5] Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017 Mar;34(2):153–9.10.1053/j.semdp.2016.12.011Search in Google Scholar PubMed
[6] Czarnomysy R, Radomska D, Szewczyk OK, Roszczenko P, Bielawski K. Platinum and palladium complexes as promising sources for antitumor treatments. Int J Mol Sci. 2021 Jul;22(15):8271.10.3390/ijms22158271Search in Google Scholar PubMed PubMed Central
[7] Makovec T. Cisplatin and beyond: molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiol Oncol. 2019 Mar;53(2):148–58.10.2478/raon-2019-0018Search in Google Scholar PubMed PubMed Central
[8] Li T, Le A. Glutamine metabolism in cancer. Adv Exp Med Biol. 2018;1063:13–32.10.1007/978-3-319-77736-8_2Search in Google Scholar PubMed
[9] Kodama M, Oshikawa K, Shimizu H, Yoshioka S, Takahashi M, Izumi Y, et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat Commun. 2020 Mar;11(1):1320.10.1038/s41467-020-15136-9Search in Google Scholar PubMed PubMed Central
[10] Matés JM, Campos-Sandoval JA, Santos-Jiménez JL, Márquez J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett. 2019 Dec;467:29–39.10.1016/j.canlet.2019.09.011Search in Google Scholar PubMed
[11] Cocetta V, Ragazzi E, Montopoli M. Links between cancer metabolism and cisplatin resistance. Int Rev Cell Mol Biol. 2020;354:107–64.10.1016/bs.ircmb.2020.01.005Search in Google Scholar PubMed
[12] Zhu W, Zhou BL, Rong LJ, Ye L, Xu HJ, Zhou Y, et al. Roles of PTBP1 in alternative splicing, glycolysis, and oncogenesis. J Zhejiang Univ Sci B. 2020 Feb;21(2):122–36.10.1631/jzus.B1900422Search in Google Scholar PubMed PubMed Central
[13] Ren ZH, Shang GP, Wu K, Hu CY, Ji T. WGCNA Co-expression network analysis reveals ILF3-AS1 functions as a CeRNA to regulate PTBP1 expression by sponging miR-29a in gastric cancer. Front Genet. 2020 Feb;11:39.10.3389/fgene.2020.00039Search in Google Scholar PubMed PubMed Central
[14] Wang X, Li Y, Fan Y, Yu X, Mao X, Jin F. PTBP1 promotes the growth of breast cancer cells through the PTEN/Akt pathway and autophagy. J Cell Physiol. 2018 Nov;233(11):8930–9.10.1002/jcp.26823Search in Google Scholar PubMed PubMed Central
[15] Cheng C, Xie Z, Li Y, Wang J, Qin C, Zhang Y. PTBP1 knockdown overcomes the resistance to vincristine and oxaliplatin in drug-resistant colon cancer cells through regulation of glycolysis. Biomed Pharmacother. 2018 Dec;108:194–200.10.1016/j.biopha.2018.09.031Search in Google Scholar PubMed
[16] Cho CY, Chung SY, Lin S, Huang JS, Chen YL, Jiang SS, et al. PTBP1-mediated regulation of AXL mRNA stability plays a role in lung tumorigenesis. Sci Rep. 2019 Nov;9(1):16922.10.1038/s41598-019-53097-2Search in Google Scholar PubMed PubMed Central
[17] Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016 Apr;35(16):2031–9.10.1038/onc.2015.270Search in Google Scholar PubMed PubMed Central
[18] Wu S, Zhang T, Du J. Ursolic acid sensitizes cisplatin-resistant HepG2/DDP cells to cisplatin via inhibiting Nrf2/ARE pathway. Drug Des Dev Ther. 2016 Oct;10:3471–81.10.2147/DDDT.S110505Search in Google Scholar PubMed PubMed Central
[19] Fritz SE, Ranganathan S, Wang CD, Hogg JR. The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay. J Biol Chem. 2020 Aug;295(33):11613–25.10.1074/jbc.RA120.013824Search in Google Scholar PubMed PubMed Central
[20] Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020 Oct;9(10):2308.10.3390/cells9102308Search in Google Scholar PubMed PubMed Central
[21] Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife. 2016 Jan;5:e11155.10.7554/eLife.11155Search in Google Scholar PubMed PubMed Central
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Exosomes derived from mesenchymal stem cells overexpressing miR-210 inhibits neuronal inflammation and contribute to neurite outgrowth through modulating microglia polarization
- Current situation of acute ST-segment elevation myocardial infarction in a county hospital chest pain center during an epidemic of novel coronavirus pneumonia
- circ-IARS depletion inhibits the progression of non-small-cell lung cancer by circ-IARS/miR-1252-5p/HDGF ceRNA pathway
- circRNA ITGA7 restrains growth and enhances radiosensitivity by up-regulating SMAD4 in colorectal carcinoma
- WDR79 promotes aerobic glycolysis of pancreatic ductal adenocarcinoma (PDAC) by the suppression of SIRT4
- Up-regulation of collagen type V alpha 2 (COL5A2) promotes malignant phenotypes in gastric cancer cell via inducing epithelial–mesenchymal transition (EMT)
- Inhibition of TERC inhibits neural apoptosis and inflammation in spinal cord injury through Akt activation and p-38 inhibition via the miR-34a-5p/XBP-1 axis
- 3D-printed polyether-ether-ketone/n-TiO2 composite enhances the cytocompatibility and osteogenic differentiation of MC3T3-E1 cells by downregulating miR-154-5p
- Propofol-mediated circ_0000735 downregulation restrains tumor growth by decreasing integrin-β1 expression in non-small cell lung cancer
- PVT1/miR-16/CCND1 axis regulates gastric cancer progression
- Silencing of circ_002136 sensitizes gastric cancer to paclitaxel by targeting the miR-16-5p/HMGA1 axis
- Short-term outcomes after simultaneous gastrectomy plus cholecystectomy in gastric cancer: A pooling up analysis
- SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways
- Molecular mechanism by which the Notch signaling pathway regulates autophagy in a rat model of pulmonary fibrosis in pigeon breeder’s lung
- lncRNA TPT1-AS1 promotes cell migration and invasion in esophageal squamous-cell carcinomas by regulating the miR-26a/HMGA1 axis
- SIRT1/APE1 promotes the viability of gastric cancer cells by inhibiting p53 to suppress ferroptosis
- Glycoprotein non-metastatic melanoma B interacts with epidermal growth factor receptor to regulate neural stem cell survival and differentiation
- Treatments for brain metastases from EGFR/ALK-negative/unselected NSCLC: A network meta-analysis
- Association of osteoporosis and skeletal muscle loss with serum type I collagen carboxyl-terminal peptide β glypeptide: A cross-sectional study in elder Chinese population
- circ_0000376 knockdown suppresses non-small cell lung cancer cell tumor properties by the miR-545-3p/PDPK1 pathway
- Delivery in a vertical birth chair supported by freedom of movement during labor: A randomized control trial
- UBE2J1 knockdown promotes cell apoptosis in endometrial cancer via regulating PI3K/AKT and MDM2/p53 signaling
- Metabolic resuscitation therapy in critically ill patients with sepsis and septic shock: A pilot prospective randomized controlled trial
- Lycopene ameliorates locomotor activity and urinary frequency induced by pelvic venous congestion in rats
- UHRF1-induced connexin26 methylation is involved in hearing damage triggered by intermittent hypoxia in neonatal rats
- LINC00511 promotes melanoma progression by targeting miR-610/NUCB2
- Ultra-high-performance liquid chromatography-tandem mass spectrometry analysis of serum metabolomic characteristics in people with different vitamin D levels
- Role of Jumonji domain-containing protein D3 and its inhibitor GSK-J4 in Hashimoto’s thyroiditis
- circ_0014736 induces GPR4 to regulate the biological behaviors of human placental trophoblast cells through miR-942-5p in preeclampsia
- Monitoring of sirolimus in the whole blood samples from pediatric patients with lymphatic anomalies
- Effects of osteogenic growth peptide C-terminal pentapeptide and its analogue on bone remodeling in an osteoporosis rat model
- A novel autophagy-related long non-coding RNAs signature predicting progression-free interval and I-131 therapy benefits in papillary thyroid carcinoma
- WGCNA-based identification of potential targets and pathways in response to treatment in locally advanced breast cancer patients
- Radiomics model using preoperative computed tomography angiography images to differentiate new from old emboli of acute lower limb arterial embolism
- Dysregulated lncRNAs are involved in the progress of myocardial infarction by constructing regulatory networks
- Single-arm trial to evaluate the efficacy and safety of baclofen in treatment of intractable hiccup caused by malignant tumor chemotherapy
- Genetic polymorphisms of MRPS30-DT and NINJ2 may influence lung cancer risk
- Efficacy of immune checkpoint inhibitors in patients with KRAS-mutant advanced non-small cell lung cancer: A retrospective analysis
- Pyroptosis-based risk score predicts prognosis and drug sensitivity in lung adenocarcinoma
- Upregulation of lncRNA LANCL1-AS1 inhibits the progression of non-small-cell lung cancer via the miR-3680-3p/GMFG axis
- CircRANBP17 modulated KDM1A to regulate neuroblastoma progression by sponging miR-27b-3p
- Exosomal miR-93-5p regulated the progression of osteoarthritis by targeting ADAMTS9
- Downregulation of RBM17 enhances cisplatin sensitivity and inhibits cell invasion in human hypopharyngeal cancer cells
- HDAC5-mediated PRAME regulates the proliferation, migration, invasion, and EMT of laryngeal squamous cell carcinoma via the PI3K/AKT/mTOR signaling pathway
- The association between sleep duration, quality, and nonalcoholic fatty liver disease: A cross-sectional study
- Myostatin silencing inhibits podocyte apoptosis in membranous nephropathy through Smad3/PKA/NOX4 signaling pathway
- A novel long noncoding RNA AC125257.1 facilitates colorectal cancer progression by targeting miR-133a-3p/CASC5 axis
- Impact of omicron wave and associated control measures in Shanghai on health management and psychosocial well-being of patients with chronic conditions
- Clinicopathological characteristics and prognosis of young patients aged ≤45 years old with non-small cell lung cancer
- TMT-based comprehensive proteomic profiling identifies serum prognostic signatures of acute myeloid leukemia
- The dose limits of teeth protection for patients with nasopharyngeal carcinoma undergoing radiotherapy based on the early oral health-related quality of life
- miR-30b-5p targeting GRIN2A inhibits hippocampal damage in epilepsy
- Long non-coding RNA AL137789.1 promoted malignant biological behaviors and immune escape of pancreatic carcinoma cells
- IRF6 and FGF1 polymorphisms in non-syndromic cleft lip with or without cleft palate in the Polish population
- Comprehensive analysis of the role of SFXN family in breast cancer
- Efficacy of bronchoscopic intratumoral injection of endostar and cisplatin in lung squamous cell carcinoma patients underwent conventional chemoradiotherapy
- Silencing of long noncoding RNA MIAT inhibits the viability and proliferation of breast cancer cells by promoting miR-378a-5p expression
- AG1024, an IGF-1 receptor inhibitor, ameliorates renal injury in rats with diabetic nephropathy via the SOCS/JAK2/STAT pathway
- Downregulation of KIAA1199 alleviated the activation, proliferation, and migration of hepatic stellate cells by the inhibition of epithelial–mesenchymal transition
- Exendin-4 regulates the MAPK and WNT signaling pathways to alleviate the osteogenic inhibition of periodontal ligament stem cells in a high glucose environment
- Inhibition of glycolysis represses the growth and alleviates the endoplasmic reticulum stress of breast cancer cells by regulating TMTC3
- The function of lncRNA EMX2OS/miR-653-5p and its regulatory mechanism in lung adenocarcinoma
- Tectorigenin alleviates the apoptosis and inflammation in spinal cord injury cell model through inhibiting insulin-like growth factor-binding protein 6
- Ultrasound examination supporting CT or MRI in the evaluation of cervical lymphadenopathy in patients with irradiation-treated head and neck cancer
- F-box and WD repeat domain containing 7 inhibits the activation of hepatic stellate cells by degrading delta-like ligand 1 to block Notch signaling pathway
- Knockdown of circ_0005615 enhances the radiosensitivity of colorectal cancer by regulating the miR-665/NOTCH1 axis
- Long noncoding RNA Mhrt alleviates angiotensin II-induced cardiac hypertrophy phenotypes by mediating the miR-765/Wnt family member 7B pathway
- Effect of miR-499-5p/SOX6 axis on atrial fibrosis in rats with atrial fibrillation
- Cholesterol induces inflammation and reduces glucose utilization
- circ_0004904 regulates the trophoblast cell in preeclampsia via miR-19b-3p/ARRDC3 axis
- NECAB3 promotes the migration and invasion of liver cancer cells through HIF-1α/RIT1 signaling pathway
- The poor performance of cardiovascular risk scores in identifying patients with idiopathic inflammatory myopathies at high cardiovascular risk
- miR-2053 inhibits the growth of ovarian cancer cells by downregulating SOX4
- Nucleophosmin 1 associating with engulfment and cell motility protein 1 regulates hepatocellular carcinoma cell chemotaxis and metastasis
- α-Hederin regulates macrophage polarization to relieve sepsis-induced lung and liver injuries in mice
- Changes of microbiota level in urinary tract infections: A meta-analysis
- Identification of key enzalutamide-resistance-related genes in castration-resistant prostate cancer and verification of RAD51 functions
- Falls during oxaliplatin-based chemotherapy for gastrointestinal malignancies – (lessons learned from) a prospective study
- Outcomes of low-risk birth care during the Covid-19 pandemic: A cohort study from a tertiary care center in Lithuania
- Vitamin D protects intestines from liver cirrhosis-induced inflammation and oxidative stress by inhibiting the TLR4/MyD88/NF-κB signaling pathway
- Integrated transcriptome analysis identifies APPL1/RPS6KB2/GALK1 as immune-related metastasis factors in breast cancer
- Genomic analysis of immunogenic cell death-related subtypes for predicting prognosis and immunotherapy outcomes in glioblastoma multiforme
- Circular RNA Circ_0038467 promotes the maturation of miRNA-203 to increase lipopolysaccharide-induced apoptosis of chondrocytes
- An economic evaluation of fine-needle cytology as the primary diagnostic tool in the diagnosis of lymphadenopathy
- Midazolam impedes lung carcinoma cell proliferation and migration via EGFR/MEK/ERK signaling pathway
- Network pharmacology combined with molecular docking and experimental validation to reveal the pharmacological mechanism of naringin against renal fibrosis
- PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma
- miR-141-3p accelerates ovarian cancer progression and promotes M2-like macrophage polarization by targeting the Keap1-Nrf2 pathway
- lncRNA OIP5-AS1 attenuates the osteoarthritis progression in IL-1β-stimulated chondrocytes
- Overexpression of LINC00607 inhibits cell growth and aggressiveness by regulating the miR-1289/EFNA5 axis in non-small-cell lung cancer
- Subjective well-being in informal caregivers during the COVID-19 pandemic
- Nrf2 protects against myocardial ischemia-reperfusion injury in diabetic rats by inhibiting Drp1-mediated mitochondrial fission
- Unfolded protein response inhibits KAT2B/MLKL-mediated necroptosis of hepatocytes by promoting BMI1 level to ubiquitinate KAT2B
- Bladder cancer screening: The new selection and prediction model
- circNFATC3 facilitated the progression of oral squamous cell carcinoma via the miR-520h/LDHA axis
- Prone position effect in intensive care patients with SARS-COV-2 pneumonia
- Clinical observation on the efficacy of Tongdu Tuina manipulation in the treatment of primary enuresis in children
- Dihydroartemisinin ameliorates cerebral I/R injury in rats via regulating VWF and autophagy-mediated SIRT1/FOXO1 pathway
- Knockdown of circ_0113656 assuages oxidized low-density lipoprotein-induced vascular smooth muscle cell injury through the miR-188-3p/IGF2 pathway
- Low Ang-(1–7) and high des-Arg9 bradykinin serum levels are correlated with cardiovascular risk factors in patients with COVID-19
- Effect of maternal age and body mass index on induction of labor with oral misoprostol for premature rupture of membrane at term: A retrospective cross-sectional study
- Potential protective effects of Huanglian Jiedu Decoction against COVID-19-associated acute kidney injury: A network-based pharmacological and molecular docking study
- Clinical significance of serum MBD3 detection in girls with central precocious puberty
- Clinical features of varicella-zoster virus caused neurological diseases detected by metagenomic next-generation sequencing
- Collagen treatment of complex anorectal fistula: 3 years follow-up
- LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through down-regulating SP-A by sponging to miR-424
- Efficacy analysis of empirical bismuth quadruple therapy, high-dose dual therapy, and resistance gene-based triple therapy as a first-line Helicobacter pylori eradication regimen – An open-label, randomized trial
- SMOC2 plays a role in heart failure via regulating TGF-β1/Smad3 pathway-mediated autophagy
- A prospective cohort study of the impact of chronic disease on fall injuries in middle-aged and older adults
- circRNA THBS1 silencing inhibits the malignant biological behavior of cervical cancer cells via the regulation of miR-543/HMGB2 axis
- hsa_circ_0000285 sponging miR-582-3p promotes neuroblastoma progression by regulating the Wnt/β-catenin signaling pathway
- Long non-coding RNA GNAS-AS1 knockdown inhibits proliferation and epithelial–mesenchymal transition of lung adenocarcinoma cells via the microRNA-433-3p/Rab3A axis
- lncRNA UCA1 regulates miR-132/Lrrfip1 axis to promote vascular smooth muscle cell proliferation
- Twenty-four-color full spectrum flow cytometry panel for minimal residual disease detection in acute myeloid leukemia
- Hsa-miR-223-3p participates in the process of anthracycline-induced cardiomyocyte damage by regulating NFIA gene
- Anti-inflammatory effect of ApoE23 on Salmonella typhimurium-induced sepsis in mice
- Analysis of somatic mutations and key driving factors of cervical cancer progression
- Hsa_circ_0028007 regulates the progression of nasopharyngeal carcinoma through the miR-1179/SQLE axis
- Variations in sexual function after laparoendoscopic single-site hysterectomy in women with benign gynecologic diseases
- Effects of pharmacological delay with roxadustat on multi-territory perforator flap survival in rats
- Analysis of heroin effects on calcium channels in rat cardiomyocytes based on transcriptomics and metabolomics
- Risk factors of recurrent bacterial vaginosis among women of reproductive age: A cross-sectional study
- Alkbh5 plays indispensable roles in maintaining self-renewal of hematopoietic stem cells
- Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients
- Correlation between microvessel maturity and ISUP grades assessed using contrast-enhanced transrectal ultrasonography in prostate cancer
- The protective effect of caffeic acid phenethyl ester in the nephrotoxicity induced by α-cypermethrin
- Norepinephrine alleviates cyclosporin A-induced nephrotoxicity by enhancing the expression of SFRP1
- Effect of RUNX1/FOXP3 axis on apoptosis of T and B lymphocytes and immunosuppression in sepsis
- The function of Foxp1 represses β-adrenergic receptor transcription in the occurrence and development of bladder cancer through STAT3 activity
- Risk model and validation of carbapenem-resistant Klebsiella pneumoniae infection in patients with cerebrovascular disease in the ICU
- Calycosin protects against chronic prostatitis in rats via inhibition of the p38MAPK/NF-κB pathway
- Pan-cancer analysis of the PDE4DIP gene with potential prognostic and immunotherapeutic values in multiple cancers including acute myeloid leukemia
- The safety and immunogenicity to inactivated COVID-19 vaccine in patients with hyperlipemia
- Circ-UBR4 regulates the proliferation, migration, inflammation, and apoptosis in ox-LDL-induced vascular smooth muscle cells via miR-515-5p/IGF2 axis
- Clinical characteristics of current COVID-19 rehabilitation outpatients in China
- Luteolin alleviates ulcerative colitis in rats via regulating immune response, oxidative stress, and metabolic profiling
- miR-199a-5p inhibits aortic valve calcification by targeting ATF6 and GRP78 in valve interstitial cells
- The application of iliac fascia space block combined with esketamine intravenous general anesthesia in PFNA surgery of the elderly: A prospective, single-center, controlled trial
- Elevated blood acetoacetate levels reduce major adverse cardiac and cerebrovascular events risk in acute myocardial infarction
- The effects of progesterone on the healing of obstetric anal sphincter damage in female rats
- Identification of cuproptosis-related genes for predicting the development of prostate cancer
- Lumican silencing ameliorates β-glycerophosphate-mediated vascular smooth muscle cell calcification by attenuating the inhibition of APOB on KIF2C activity
- Targeting PTBP1 blocks glutamine metabolism to improve the cisplatin sensitivity of hepatocarcinoma cells through modulating the mRNA stability of glutaminase
- A single center prospective study: Influences of different hip flexion angles on the measurement of lumbar spine bone mineral density by dual energy X-ray absorptiometry
- Clinical analysis of AN69ST membrane continuous venous hemofiltration in the treatment of severe sepsis
- Antibiotics therapy combined with probiotics administered intravaginally for the treatment of bacterial vaginosis: A systematic review and meta-analysis
- Construction of a ceRNA network to reveal a vascular invasion associated prognostic model in hepatocellular carcinoma
- A pan-cancer analysis of STAT3 expression and genetic alterations in human tumors
- A prognostic signature based on seven T-cell-related cell clustering genes in bladder urothelial carcinoma
- Pepsin concentration in oral lavage fluid of rabbit reflux model constructed by dilating the lower esophageal sphincter
- The antihypertensive felodipine shows synergistic activity with immune checkpoint blockade and inhibits tumor growth via NFAT1 in LUSC
- Tanshinone IIA attenuates valvular interstitial cells’ calcification induced by oxidized low density lipoprotein via reducing endoplasmic reticulum stress
- AS-IV enhances the antitumor effects of propofol in NSCLC cells by inhibiting autophagy
- Establishment of two oxaliplatin-resistant gallbladder cancer cell lines and comprehensive analysis of dysregulated genes
- Trial protocol: Feasibility of neuromodulation with connectivity-guided intermittent theta-burst stimulation for improving cognition in multiple sclerosis
- LncRNA LINC00592 mediates the promoter methylation of WIF1 to promote the development of bladder cancer
- Factors associated with gastrointestinal dysmotility in critically ill patients
- Mechanisms by which spinal cord stimulation intervenes in atrial fibrillation: The involvement of the endothelin-1 and nerve growth factor/p75NTR pathways
- Analysis of two-gene signatures and related drugs in small-cell lung cancer by bioinformatics
- Silencing USP19 alleviates cigarette smoke extract-induced mitochondrial dysfunction in BEAS-2B cells by targeting FUNDC1
- Menstrual irregularities associated with COVID-19 vaccines among women in Saudi Arabia: A survey during 2022
- Ferroptosis involves in Schwann cell death in diabetic peripheral neuropathy
- The effect of AQP4 on tau protein aggregation in neurodegeneration and persistent neuroinflammation after cerebral microinfarcts
- Activation of UBEC2 by transcription factor MYBL2 affects DNA damage and promotes gastric cancer progression and cisplatin resistance
- Analysis of clinical characteristics in proximal and distal reflux monitoring among patients with gastroesophageal reflux disease
- Exosomal circ-0020887 and circ-0009590 as novel biomarkers for the diagnosis and prediction of short-term adverse cardiovascular outcomes in STEMI patients
- Upregulated microRNA-429 confers endometrial stromal cell dysfunction by targeting HIF1AN and regulating the HIF1A/VEGF pathway
- Bibliometrics and knowledge map analysis of ultrasound-guided regional anesthesia
- Knockdown of NUPR1 inhibits angiogenesis in lung cancer through IRE1/XBP1 and PERK/eIF2α/ATF4 signaling pathways
- D-dimer trends predict COVID-19 patient’s prognosis: A retrospective chart review study
- WTAP affects intracranial aneurysm progression by regulating m6A methylation modification
- Using of endoscopic polypectomy in patients with diagnosed malignant colorectal polyp – The cross-sectional clinical study
- Anti-S100A4 antibody administration alleviates bronchial epithelial–mesenchymal transition in asthmatic mice
- Prognostic evaluation of system immune-inflammatory index and prognostic nutritional index in double expressor diffuse large B-cell lymphoma
- Prevalence and antibiogram of bacteria causing urinary tract infection among patients with chronic kidney disease
- Reactive oxygen species within the vaginal space: An additional promoter of cervical intraepithelial neoplasia and uterine cervical cancer development?
- Identification of disulfidptosis-related genes and immune infiltration in lower-grade glioma
- A new technique for uterine-preserving pelvic organ prolapse surgery: Laparoscopic rectus abdominis hysteropexy for uterine prolapse by comparing with traditional techniques
- Self-isolation of an Italian long-term care facility during COVID-19 pandemic: A comparison study on care-related infectious episodes
- A comparative study on the overlapping effects of clinically applicable therapeutic interventions in patients with central nervous system damage
- Low intensity extracorporeal shockwave therapy for chronic pelvic pain syndrome: Long-term follow-up
- The diagnostic accuracy of touch imprint cytology for sentinel lymph node metastases of breast cancer: An up-to-date meta-analysis of 4,073 patients
- Mortality associated with Sjögren’s syndrome in the United States in the 1999–2020 period: A multiple cause-of-death study
- CircMMP11 as a prognostic biomarker mediates miR-361-3p/HMGB1 axis to accelerate malignant progression of hepatocellular carcinoma
- Analysis of the clinical characteristics and prognosis of adult de novo acute myeloid leukemia (none APL) with PTPN11 mutations
- KMT2A maintains stemness of gastric cancer cells through regulating Wnt/β-catenin signaling-activated transcriptional factor KLF11
- Evaluation of placental oxygenation by near-infrared spectroscopy in relation to ultrasound maturation grade in physiological term pregnancies
- The role of ultrasonographic findings for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative breast cancer
- Construction of immunogenic cell death-related molecular subtypes and prognostic signature in colorectal cancer
- Long-term prognostic value of high-sensitivity cardiac troponin-I in patients with idiopathic dilated cardiomyopathy
- Establishing a novel Fanconi anemia signaling pathway-associated prognostic model and tumor clustering for pediatric acute myeloid leukemia patients
- Integrative bioinformatics analysis reveals STAT2 as a novel biomarker of inflammation-related cardiac dysfunction in atrial fibrillation
- Adipose-derived stem cells repair radiation-induced chronic lung injury via inhibiting TGF-β1/Smad 3 signaling pathway
- Real-world practice of idiopathic pulmonary fibrosis: Results from a 2000–2016 cohort
- lncRNA LENGA sponges miR-378 to promote myocardial fibrosis in atrial fibrillation
- Diagnostic value of urinary Tamm-Horsfall protein and 24 h urine osmolality for recurrent calcium oxalate stones of the upper urinary tract: Cross-sectional study
- The value of color Doppler ultrasonography combined with serum tumor markers in differential diagnosis of gastric stromal tumor and gastric cancer
- The spike protein of SARS-CoV-2 induces inflammation and EMT of lung epithelial cells and fibroblasts through the upregulation of GADD45A
- Mycophenolate mofetil versus cyclophosphamide plus in patients with connective tissue disease-associated interstitial lung disease: Efficacy and safety analysis
- MiR-1278 targets CALD1 and suppresses the progression of gastric cancer via the MAPK pathway
- Metabolomic analysis of serum short-chain fatty acid concentrations in a mouse of MPTP-induced Parkinson’s disease after dietary supplementation with branched-chain amino acids
- Cimifugin inhibits adipogenesis and TNF-α-induced insulin resistance in 3T3-L1 cells
- Predictors of gastrointestinal complaints in patients on metformin therapy
- Prescribing patterns in patients with chronic obstructive pulmonary disease and atrial fibrillation
- A retrospective analysis of the effect of latent tuberculosis infection on clinical pregnancy outcomes of in vitro fertilization–fresh embryo transferred in infertile women
- Appropriateness and clinical outcomes of short sustained low-efficiency dialysis: A national experience
- miR-29 regulates metabolism by inhibiting JNK-1 expression in non-obese patients with type 2 diabetes mellitus and NAFLD
- Clinical features and management of lymphoepithelial cyst
- Serum VEGF, high-sensitivity CRP, and cystatin-C assist in the diagnosis of type 2 diabetic retinopathy complicated with hyperuricemia
- ENPP1 ameliorates vascular calcification via inhibiting the osteogenic transformation of VSMCs and generating PPi
- Significance of monitoring the levels of thyroid hormone antibodies and glucose and lipid metabolism antibodies in patients suffer from type 2 diabetes
- The causal relationship between immune cells and different kidney diseases: A Mendelian randomization study
- Interleukin 33, soluble suppression of tumorigenicity 2, interleukin 27, and galectin 3 as predictors for outcome in patients admitted to intensive care units
- Identification of diagnostic immune-related gene biomarkers for predicting heart failure after acute myocardial infarction
- Long-term administration of probiotics prevents gastrointestinal mucosal barrier dysfunction in septic mice partly by upregulating the 5-HT degradation pathway
- miR-192 inhibits the activation of hepatic stellate cells by targeting Rictor
- Diagnostic and prognostic value of MR-pro ADM, procalcitonin, and copeptin in sepsis
- Review Articles
- Prenatal diagnosis of fetal defects and its implications on the delivery mode
- Electromagnetic fields exposure on fetal and childhood abnormalities: Systematic review and meta-analysis
- Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae
- Saddle pulmonary embolism in the setting of COVID-19 infection: A systematic review of case reports and case series
- Vitamin C and epigenetics: A short physiological overview
- Ebselen: A promising therapy protecting cardiomyocytes from excess iron in iron-overloaded thalassemia patients
- Aspirin versus LMWH for VTE prophylaxis after orthopedic surgery
- Mechanism of rhubarb in the treatment of hyperlipidemia: A recent review
- Surgical management and outcomes of traumatic global brachial plexus injury: A concise review and our center approach
- The progress of autoimmune hepatitis research and future challenges
- METTL16 in human diseases: What should we do next?
- New insights into the prevention of ureteral stents encrustation
- VISTA as a prospective immune checkpoint in gynecological malignant tumors: A review of the literature
- Case Reports
- Mycobacterium xenopi infection of the kidney and lymph nodes: A case report
- Genetic mutation of SLC6A20 (c.1072T > C) in a family with nephrolithiasis: A case report
- Chronic hepatitis B complicated with secondary hemochromatosis was cured clinically: A case report
- Liver abscess complicated with multiple organ invasive infection caused by hematogenous disseminated hypervirulent Klebsiella pneumoniae: A case report
- Urokinase-based lock solutions for catheter salvage: A case of an upcoming kidney transplant recipient
- Two case reports of maturity-onset diabetes of the young type 3 caused by the hepatocyte nuclear factor 1α gene mutation
- Immune checkpoint inhibitor-related pancreatitis: What is known and what is not
- Does total hip arthroplasty result in intercostal nerve injury? A case report and literature review
- Clinicopathological characteristics and diagnosis of hepatic sinusoidal obstruction syndrome caused by Tusanqi – Case report and literature review
- Synchronous triple primary gastrointestinal malignant tumors treated with laparoscopic surgery: A case report
- CT-guided percutaneous microwave ablation combined with bone cement injection for the treatment of transverse metastases: A case report
- Malignant hyperthermia: Report on a successful rescue of a case with the highest temperature of 44.2°C
- Anesthetic management of fetal pulmonary valvuloplasty: A case report
- Rapid Communication
- Impact of COVID-19 lockdown on glycemic levels during pregnancy: A retrospective analysis
- Erratum
- Erratum to “Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway”
- Erratum to: “Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p”
- Retraction
- Retraction of “Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients”
- Retraction of “circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis”
- Retraction of “miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells”
- Retraction of “SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis”
- Retraction of “circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury”
- Retraction of “lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells”
- Special issue Linking Pathobiological Mechanisms to Clinical Application for cardiovascular diseases
- Effect of cardiac rehabilitation therapy on depressed patients with cardiac insufficiency after cardiac surgery
- Special issue The evolving saga of RNAs from bench to bedside - Part I
- FBLIM1 mRNA is a novel prognostic biomarker and is associated with immune infiltrates in glioma
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part III
- Development of a machine learning-based signature utilizing inflammatory response genes for predicting prognosis and immune microenvironment in ovarian cancer
Articles in the same Issue
- Research Articles
- Exosomes derived from mesenchymal stem cells overexpressing miR-210 inhibits neuronal inflammation and contribute to neurite outgrowth through modulating microglia polarization
- Current situation of acute ST-segment elevation myocardial infarction in a county hospital chest pain center during an epidemic of novel coronavirus pneumonia
- circ-IARS depletion inhibits the progression of non-small-cell lung cancer by circ-IARS/miR-1252-5p/HDGF ceRNA pathway
- circRNA ITGA7 restrains growth and enhances radiosensitivity by up-regulating SMAD4 in colorectal carcinoma
- WDR79 promotes aerobic glycolysis of pancreatic ductal adenocarcinoma (PDAC) by the suppression of SIRT4
- Up-regulation of collagen type V alpha 2 (COL5A2) promotes malignant phenotypes in gastric cancer cell via inducing epithelial–mesenchymal transition (EMT)
- Inhibition of TERC inhibits neural apoptosis and inflammation in spinal cord injury through Akt activation and p-38 inhibition via the miR-34a-5p/XBP-1 axis
- 3D-printed polyether-ether-ketone/n-TiO2 composite enhances the cytocompatibility and osteogenic differentiation of MC3T3-E1 cells by downregulating miR-154-5p
- Propofol-mediated circ_0000735 downregulation restrains tumor growth by decreasing integrin-β1 expression in non-small cell lung cancer
- PVT1/miR-16/CCND1 axis regulates gastric cancer progression
- Silencing of circ_002136 sensitizes gastric cancer to paclitaxel by targeting the miR-16-5p/HMGA1 axis
- Short-term outcomes after simultaneous gastrectomy plus cholecystectomy in gastric cancer: A pooling up analysis
- SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways
- Molecular mechanism by which the Notch signaling pathway regulates autophagy in a rat model of pulmonary fibrosis in pigeon breeder’s lung
- lncRNA TPT1-AS1 promotes cell migration and invasion in esophageal squamous-cell carcinomas by regulating the miR-26a/HMGA1 axis
- SIRT1/APE1 promotes the viability of gastric cancer cells by inhibiting p53 to suppress ferroptosis
- Glycoprotein non-metastatic melanoma B interacts with epidermal growth factor receptor to regulate neural stem cell survival and differentiation
- Treatments for brain metastases from EGFR/ALK-negative/unselected NSCLC: A network meta-analysis
- Association of osteoporosis and skeletal muscle loss with serum type I collagen carboxyl-terminal peptide β glypeptide: A cross-sectional study in elder Chinese population
- circ_0000376 knockdown suppresses non-small cell lung cancer cell tumor properties by the miR-545-3p/PDPK1 pathway
- Delivery in a vertical birth chair supported by freedom of movement during labor: A randomized control trial
- UBE2J1 knockdown promotes cell apoptosis in endometrial cancer via regulating PI3K/AKT and MDM2/p53 signaling
- Metabolic resuscitation therapy in critically ill patients with sepsis and septic shock: A pilot prospective randomized controlled trial
- Lycopene ameliorates locomotor activity and urinary frequency induced by pelvic venous congestion in rats
- UHRF1-induced connexin26 methylation is involved in hearing damage triggered by intermittent hypoxia in neonatal rats
- LINC00511 promotes melanoma progression by targeting miR-610/NUCB2
- Ultra-high-performance liquid chromatography-tandem mass spectrometry analysis of serum metabolomic characteristics in people with different vitamin D levels
- Role of Jumonji domain-containing protein D3 and its inhibitor GSK-J4 in Hashimoto’s thyroiditis
- circ_0014736 induces GPR4 to regulate the biological behaviors of human placental trophoblast cells through miR-942-5p in preeclampsia
- Monitoring of sirolimus in the whole blood samples from pediatric patients with lymphatic anomalies
- Effects of osteogenic growth peptide C-terminal pentapeptide and its analogue on bone remodeling in an osteoporosis rat model
- A novel autophagy-related long non-coding RNAs signature predicting progression-free interval and I-131 therapy benefits in papillary thyroid carcinoma
- WGCNA-based identification of potential targets and pathways in response to treatment in locally advanced breast cancer patients
- Radiomics model using preoperative computed tomography angiography images to differentiate new from old emboli of acute lower limb arterial embolism
- Dysregulated lncRNAs are involved in the progress of myocardial infarction by constructing regulatory networks
- Single-arm trial to evaluate the efficacy and safety of baclofen in treatment of intractable hiccup caused by malignant tumor chemotherapy
- Genetic polymorphisms of MRPS30-DT and NINJ2 may influence lung cancer risk
- Efficacy of immune checkpoint inhibitors in patients with KRAS-mutant advanced non-small cell lung cancer: A retrospective analysis
- Pyroptosis-based risk score predicts prognosis and drug sensitivity in lung adenocarcinoma
- Upregulation of lncRNA LANCL1-AS1 inhibits the progression of non-small-cell lung cancer via the miR-3680-3p/GMFG axis
- CircRANBP17 modulated KDM1A to regulate neuroblastoma progression by sponging miR-27b-3p
- Exosomal miR-93-5p regulated the progression of osteoarthritis by targeting ADAMTS9
- Downregulation of RBM17 enhances cisplatin sensitivity and inhibits cell invasion in human hypopharyngeal cancer cells
- HDAC5-mediated PRAME regulates the proliferation, migration, invasion, and EMT of laryngeal squamous cell carcinoma via the PI3K/AKT/mTOR signaling pathway
- The association between sleep duration, quality, and nonalcoholic fatty liver disease: A cross-sectional study
- Myostatin silencing inhibits podocyte apoptosis in membranous nephropathy through Smad3/PKA/NOX4 signaling pathway
- A novel long noncoding RNA AC125257.1 facilitates colorectal cancer progression by targeting miR-133a-3p/CASC5 axis
- Impact of omicron wave and associated control measures in Shanghai on health management and psychosocial well-being of patients with chronic conditions
- Clinicopathological characteristics and prognosis of young patients aged ≤45 years old with non-small cell lung cancer
- TMT-based comprehensive proteomic profiling identifies serum prognostic signatures of acute myeloid leukemia
- The dose limits of teeth protection for patients with nasopharyngeal carcinoma undergoing radiotherapy based on the early oral health-related quality of life
- miR-30b-5p targeting GRIN2A inhibits hippocampal damage in epilepsy
- Long non-coding RNA AL137789.1 promoted malignant biological behaviors and immune escape of pancreatic carcinoma cells
- IRF6 and FGF1 polymorphisms in non-syndromic cleft lip with or without cleft palate in the Polish population
- Comprehensive analysis of the role of SFXN family in breast cancer
- Efficacy of bronchoscopic intratumoral injection of endostar and cisplatin in lung squamous cell carcinoma patients underwent conventional chemoradiotherapy
- Silencing of long noncoding RNA MIAT inhibits the viability and proliferation of breast cancer cells by promoting miR-378a-5p expression
- AG1024, an IGF-1 receptor inhibitor, ameliorates renal injury in rats with diabetic nephropathy via the SOCS/JAK2/STAT pathway
- Downregulation of KIAA1199 alleviated the activation, proliferation, and migration of hepatic stellate cells by the inhibition of epithelial–mesenchymal transition
- Exendin-4 regulates the MAPK and WNT signaling pathways to alleviate the osteogenic inhibition of periodontal ligament stem cells in a high glucose environment
- Inhibition of glycolysis represses the growth and alleviates the endoplasmic reticulum stress of breast cancer cells by regulating TMTC3
- The function of lncRNA EMX2OS/miR-653-5p and its regulatory mechanism in lung adenocarcinoma
- Tectorigenin alleviates the apoptosis and inflammation in spinal cord injury cell model through inhibiting insulin-like growth factor-binding protein 6
- Ultrasound examination supporting CT or MRI in the evaluation of cervical lymphadenopathy in patients with irradiation-treated head and neck cancer
- F-box and WD repeat domain containing 7 inhibits the activation of hepatic stellate cells by degrading delta-like ligand 1 to block Notch signaling pathway
- Knockdown of circ_0005615 enhances the radiosensitivity of colorectal cancer by regulating the miR-665/NOTCH1 axis
- Long noncoding RNA Mhrt alleviates angiotensin II-induced cardiac hypertrophy phenotypes by mediating the miR-765/Wnt family member 7B pathway
- Effect of miR-499-5p/SOX6 axis on atrial fibrosis in rats with atrial fibrillation
- Cholesterol induces inflammation and reduces glucose utilization
- circ_0004904 regulates the trophoblast cell in preeclampsia via miR-19b-3p/ARRDC3 axis
- NECAB3 promotes the migration and invasion of liver cancer cells through HIF-1α/RIT1 signaling pathway
- The poor performance of cardiovascular risk scores in identifying patients with idiopathic inflammatory myopathies at high cardiovascular risk
- miR-2053 inhibits the growth of ovarian cancer cells by downregulating SOX4
- Nucleophosmin 1 associating with engulfment and cell motility protein 1 regulates hepatocellular carcinoma cell chemotaxis and metastasis
- α-Hederin regulates macrophage polarization to relieve sepsis-induced lung and liver injuries in mice
- Changes of microbiota level in urinary tract infections: A meta-analysis
- Identification of key enzalutamide-resistance-related genes in castration-resistant prostate cancer and verification of RAD51 functions
- Falls during oxaliplatin-based chemotherapy for gastrointestinal malignancies – (lessons learned from) a prospective study
- Outcomes of low-risk birth care during the Covid-19 pandemic: A cohort study from a tertiary care center in Lithuania
- Vitamin D protects intestines from liver cirrhosis-induced inflammation and oxidative stress by inhibiting the TLR4/MyD88/NF-κB signaling pathway
- Integrated transcriptome analysis identifies APPL1/RPS6KB2/GALK1 as immune-related metastasis factors in breast cancer
- Genomic analysis of immunogenic cell death-related subtypes for predicting prognosis and immunotherapy outcomes in glioblastoma multiforme
- Circular RNA Circ_0038467 promotes the maturation of miRNA-203 to increase lipopolysaccharide-induced apoptosis of chondrocytes
- An economic evaluation of fine-needle cytology as the primary diagnostic tool in the diagnosis of lymphadenopathy
- Midazolam impedes lung carcinoma cell proliferation and migration via EGFR/MEK/ERK signaling pathway
- Network pharmacology combined with molecular docking and experimental validation to reveal the pharmacological mechanism of naringin against renal fibrosis
- PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma
- miR-141-3p accelerates ovarian cancer progression and promotes M2-like macrophage polarization by targeting the Keap1-Nrf2 pathway
- lncRNA OIP5-AS1 attenuates the osteoarthritis progression in IL-1β-stimulated chondrocytes
- Overexpression of LINC00607 inhibits cell growth and aggressiveness by regulating the miR-1289/EFNA5 axis in non-small-cell lung cancer
- Subjective well-being in informal caregivers during the COVID-19 pandemic
- Nrf2 protects against myocardial ischemia-reperfusion injury in diabetic rats by inhibiting Drp1-mediated mitochondrial fission
- Unfolded protein response inhibits KAT2B/MLKL-mediated necroptosis of hepatocytes by promoting BMI1 level to ubiquitinate KAT2B
- Bladder cancer screening: The new selection and prediction model
- circNFATC3 facilitated the progression of oral squamous cell carcinoma via the miR-520h/LDHA axis
- Prone position effect in intensive care patients with SARS-COV-2 pneumonia
- Clinical observation on the efficacy of Tongdu Tuina manipulation in the treatment of primary enuresis in children
- Dihydroartemisinin ameliorates cerebral I/R injury in rats via regulating VWF and autophagy-mediated SIRT1/FOXO1 pathway
- Knockdown of circ_0113656 assuages oxidized low-density lipoprotein-induced vascular smooth muscle cell injury through the miR-188-3p/IGF2 pathway
- Low Ang-(1–7) and high des-Arg9 bradykinin serum levels are correlated with cardiovascular risk factors in patients with COVID-19
- Effect of maternal age and body mass index on induction of labor with oral misoprostol for premature rupture of membrane at term: A retrospective cross-sectional study
- Potential protective effects of Huanglian Jiedu Decoction against COVID-19-associated acute kidney injury: A network-based pharmacological and molecular docking study
- Clinical significance of serum MBD3 detection in girls with central precocious puberty
- Clinical features of varicella-zoster virus caused neurological diseases detected by metagenomic next-generation sequencing
- Collagen treatment of complex anorectal fistula: 3 years follow-up
- LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through down-regulating SP-A by sponging to miR-424
- Efficacy analysis of empirical bismuth quadruple therapy, high-dose dual therapy, and resistance gene-based triple therapy as a first-line Helicobacter pylori eradication regimen – An open-label, randomized trial
- SMOC2 plays a role in heart failure via regulating TGF-β1/Smad3 pathway-mediated autophagy
- A prospective cohort study of the impact of chronic disease on fall injuries in middle-aged and older adults
- circRNA THBS1 silencing inhibits the malignant biological behavior of cervical cancer cells via the regulation of miR-543/HMGB2 axis
- hsa_circ_0000285 sponging miR-582-3p promotes neuroblastoma progression by regulating the Wnt/β-catenin signaling pathway
- Long non-coding RNA GNAS-AS1 knockdown inhibits proliferation and epithelial–mesenchymal transition of lung adenocarcinoma cells via the microRNA-433-3p/Rab3A axis
- lncRNA UCA1 regulates miR-132/Lrrfip1 axis to promote vascular smooth muscle cell proliferation
- Twenty-four-color full spectrum flow cytometry panel for minimal residual disease detection in acute myeloid leukemia
- Hsa-miR-223-3p participates in the process of anthracycline-induced cardiomyocyte damage by regulating NFIA gene
- Anti-inflammatory effect of ApoE23 on Salmonella typhimurium-induced sepsis in mice
- Analysis of somatic mutations and key driving factors of cervical cancer progression
- Hsa_circ_0028007 regulates the progression of nasopharyngeal carcinoma through the miR-1179/SQLE axis
- Variations in sexual function after laparoendoscopic single-site hysterectomy in women with benign gynecologic diseases
- Effects of pharmacological delay with roxadustat on multi-territory perforator flap survival in rats
- Analysis of heroin effects on calcium channels in rat cardiomyocytes based on transcriptomics and metabolomics
- Risk factors of recurrent bacterial vaginosis among women of reproductive age: A cross-sectional study
- Alkbh5 plays indispensable roles in maintaining self-renewal of hematopoietic stem cells
- Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients
- Correlation between microvessel maturity and ISUP grades assessed using contrast-enhanced transrectal ultrasonography in prostate cancer
- The protective effect of caffeic acid phenethyl ester in the nephrotoxicity induced by α-cypermethrin
- Norepinephrine alleviates cyclosporin A-induced nephrotoxicity by enhancing the expression of SFRP1
- Effect of RUNX1/FOXP3 axis on apoptosis of T and B lymphocytes and immunosuppression in sepsis
- The function of Foxp1 represses β-adrenergic receptor transcription in the occurrence and development of bladder cancer through STAT3 activity
- Risk model and validation of carbapenem-resistant Klebsiella pneumoniae infection in patients with cerebrovascular disease in the ICU
- Calycosin protects against chronic prostatitis in rats via inhibition of the p38MAPK/NF-κB pathway
- Pan-cancer analysis of the PDE4DIP gene with potential prognostic and immunotherapeutic values in multiple cancers including acute myeloid leukemia
- The safety and immunogenicity to inactivated COVID-19 vaccine in patients with hyperlipemia
- Circ-UBR4 regulates the proliferation, migration, inflammation, and apoptosis in ox-LDL-induced vascular smooth muscle cells via miR-515-5p/IGF2 axis
- Clinical characteristics of current COVID-19 rehabilitation outpatients in China
- Luteolin alleviates ulcerative colitis in rats via regulating immune response, oxidative stress, and metabolic profiling
- miR-199a-5p inhibits aortic valve calcification by targeting ATF6 and GRP78 in valve interstitial cells
- The application of iliac fascia space block combined with esketamine intravenous general anesthesia in PFNA surgery of the elderly: A prospective, single-center, controlled trial
- Elevated blood acetoacetate levels reduce major adverse cardiac and cerebrovascular events risk in acute myocardial infarction
- The effects of progesterone on the healing of obstetric anal sphincter damage in female rats
- Identification of cuproptosis-related genes for predicting the development of prostate cancer
- Lumican silencing ameliorates β-glycerophosphate-mediated vascular smooth muscle cell calcification by attenuating the inhibition of APOB on KIF2C activity
- Targeting PTBP1 blocks glutamine metabolism to improve the cisplatin sensitivity of hepatocarcinoma cells through modulating the mRNA stability of glutaminase
- A single center prospective study: Influences of different hip flexion angles on the measurement of lumbar spine bone mineral density by dual energy X-ray absorptiometry
- Clinical analysis of AN69ST membrane continuous venous hemofiltration in the treatment of severe sepsis
- Antibiotics therapy combined with probiotics administered intravaginally for the treatment of bacterial vaginosis: A systematic review and meta-analysis
- Construction of a ceRNA network to reveal a vascular invasion associated prognostic model in hepatocellular carcinoma
- A pan-cancer analysis of STAT3 expression and genetic alterations in human tumors
- A prognostic signature based on seven T-cell-related cell clustering genes in bladder urothelial carcinoma
- Pepsin concentration in oral lavage fluid of rabbit reflux model constructed by dilating the lower esophageal sphincter
- The antihypertensive felodipine shows synergistic activity with immune checkpoint blockade and inhibits tumor growth via NFAT1 in LUSC
- Tanshinone IIA attenuates valvular interstitial cells’ calcification induced by oxidized low density lipoprotein via reducing endoplasmic reticulum stress
- AS-IV enhances the antitumor effects of propofol in NSCLC cells by inhibiting autophagy
- Establishment of two oxaliplatin-resistant gallbladder cancer cell lines and comprehensive analysis of dysregulated genes
- Trial protocol: Feasibility of neuromodulation with connectivity-guided intermittent theta-burst stimulation for improving cognition in multiple sclerosis
- LncRNA LINC00592 mediates the promoter methylation of WIF1 to promote the development of bladder cancer
- Factors associated with gastrointestinal dysmotility in critically ill patients
- Mechanisms by which spinal cord stimulation intervenes in atrial fibrillation: The involvement of the endothelin-1 and nerve growth factor/p75NTR pathways
- Analysis of two-gene signatures and related drugs in small-cell lung cancer by bioinformatics
- Silencing USP19 alleviates cigarette smoke extract-induced mitochondrial dysfunction in BEAS-2B cells by targeting FUNDC1
- Menstrual irregularities associated with COVID-19 vaccines among women in Saudi Arabia: A survey during 2022
- Ferroptosis involves in Schwann cell death in diabetic peripheral neuropathy
- The effect of AQP4 on tau protein aggregation in neurodegeneration and persistent neuroinflammation after cerebral microinfarcts
- Activation of UBEC2 by transcription factor MYBL2 affects DNA damage and promotes gastric cancer progression and cisplatin resistance
- Analysis of clinical characteristics in proximal and distal reflux monitoring among patients with gastroesophageal reflux disease
- Exosomal circ-0020887 and circ-0009590 as novel biomarkers for the diagnosis and prediction of short-term adverse cardiovascular outcomes in STEMI patients
- Upregulated microRNA-429 confers endometrial stromal cell dysfunction by targeting HIF1AN and regulating the HIF1A/VEGF pathway
- Bibliometrics and knowledge map analysis of ultrasound-guided regional anesthesia
- Knockdown of NUPR1 inhibits angiogenesis in lung cancer through IRE1/XBP1 and PERK/eIF2α/ATF4 signaling pathways
- D-dimer trends predict COVID-19 patient’s prognosis: A retrospective chart review study
- WTAP affects intracranial aneurysm progression by regulating m6A methylation modification
- Using of endoscopic polypectomy in patients with diagnosed malignant colorectal polyp – The cross-sectional clinical study
- Anti-S100A4 antibody administration alleviates bronchial epithelial–mesenchymal transition in asthmatic mice
- Prognostic evaluation of system immune-inflammatory index and prognostic nutritional index in double expressor diffuse large B-cell lymphoma
- Prevalence and antibiogram of bacteria causing urinary tract infection among patients with chronic kidney disease
- Reactive oxygen species within the vaginal space: An additional promoter of cervical intraepithelial neoplasia and uterine cervical cancer development?
- Identification of disulfidptosis-related genes and immune infiltration in lower-grade glioma
- A new technique for uterine-preserving pelvic organ prolapse surgery: Laparoscopic rectus abdominis hysteropexy for uterine prolapse by comparing with traditional techniques
- Self-isolation of an Italian long-term care facility during COVID-19 pandemic: A comparison study on care-related infectious episodes
- A comparative study on the overlapping effects of clinically applicable therapeutic interventions in patients with central nervous system damage
- Low intensity extracorporeal shockwave therapy for chronic pelvic pain syndrome: Long-term follow-up
- The diagnostic accuracy of touch imprint cytology for sentinel lymph node metastases of breast cancer: An up-to-date meta-analysis of 4,073 patients
- Mortality associated with Sjögren’s syndrome in the United States in the 1999–2020 period: A multiple cause-of-death study
- CircMMP11 as a prognostic biomarker mediates miR-361-3p/HMGB1 axis to accelerate malignant progression of hepatocellular carcinoma
- Analysis of the clinical characteristics and prognosis of adult de novo acute myeloid leukemia (none APL) with PTPN11 mutations
- KMT2A maintains stemness of gastric cancer cells through regulating Wnt/β-catenin signaling-activated transcriptional factor KLF11
- Evaluation of placental oxygenation by near-infrared spectroscopy in relation to ultrasound maturation grade in physiological term pregnancies
- The role of ultrasonographic findings for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative breast cancer
- Construction of immunogenic cell death-related molecular subtypes and prognostic signature in colorectal cancer
- Long-term prognostic value of high-sensitivity cardiac troponin-I in patients with idiopathic dilated cardiomyopathy
- Establishing a novel Fanconi anemia signaling pathway-associated prognostic model and tumor clustering for pediatric acute myeloid leukemia patients
- Integrative bioinformatics analysis reveals STAT2 as a novel biomarker of inflammation-related cardiac dysfunction in atrial fibrillation
- Adipose-derived stem cells repair radiation-induced chronic lung injury via inhibiting TGF-β1/Smad 3 signaling pathway
- Real-world practice of idiopathic pulmonary fibrosis: Results from a 2000–2016 cohort
- lncRNA LENGA sponges miR-378 to promote myocardial fibrosis in atrial fibrillation
- Diagnostic value of urinary Tamm-Horsfall protein and 24 h urine osmolality for recurrent calcium oxalate stones of the upper urinary tract: Cross-sectional study
- The value of color Doppler ultrasonography combined with serum tumor markers in differential diagnosis of gastric stromal tumor and gastric cancer
- The spike protein of SARS-CoV-2 induces inflammation and EMT of lung epithelial cells and fibroblasts through the upregulation of GADD45A
- Mycophenolate mofetil versus cyclophosphamide plus in patients with connective tissue disease-associated interstitial lung disease: Efficacy and safety analysis
- MiR-1278 targets CALD1 and suppresses the progression of gastric cancer via the MAPK pathway
- Metabolomic analysis of serum short-chain fatty acid concentrations in a mouse of MPTP-induced Parkinson’s disease after dietary supplementation with branched-chain amino acids
- Cimifugin inhibits adipogenesis and TNF-α-induced insulin resistance in 3T3-L1 cells
- Predictors of gastrointestinal complaints in patients on metformin therapy
- Prescribing patterns in patients with chronic obstructive pulmonary disease and atrial fibrillation
- A retrospective analysis of the effect of latent tuberculosis infection on clinical pregnancy outcomes of in vitro fertilization–fresh embryo transferred in infertile women
- Appropriateness and clinical outcomes of short sustained low-efficiency dialysis: A national experience
- miR-29 regulates metabolism by inhibiting JNK-1 expression in non-obese patients with type 2 diabetes mellitus and NAFLD
- Clinical features and management of lymphoepithelial cyst
- Serum VEGF, high-sensitivity CRP, and cystatin-C assist in the diagnosis of type 2 diabetic retinopathy complicated with hyperuricemia
- ENPP1 ameliorates vascular calcification via inhibiting the osteogenic transformation of VSMCs and generating PPi
- Significance of monitoring the levels of thyroid hormone antibodies and glucose and lipid metabolism antibodies in patients suffer from type 2 diabetes
- The causal relationship between immune cells and different kidney diseases: A Mendelian randomization study
- Interleukin 33, soluble suppression of tumorigenicity 2, interleukin 27, and galectin 3 as predictors for outcome in patients admitted to intensive care units
- Identification of diagnostic immune-related gene biomarkers for predicting heart failure after acute myocardial infarction
- Long-term administration of probiotics prevents gastrointestinal mucosal barrier dysfunction in septic mice partly by upregulating the 5-HT degradation pathway
- miR-192 inhibits the activation of hepatic stellate cells by targeting Rictor
- Diagnostic and prognostic value of MR-pro ADM, procalcitonin, and copeptin in sepsis
- Review Articles
- Prenatal diagnosis of fetal defects and its implications on the delivery mode
- Electromagnetic fields exposure on fetal and childhood abnormalities: Systematic review and meta-analysis
- Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae
- Saddle pulmonary embolism in the setting of COVID-19 infection: A systematic review of case reports and case series
- Vitamin C and epigenetics: A short physiological overview
- Ebselen: A promising therapy protecting cardiomyocytes from excess iron in iron-overloaded thalassemia patients
- Aspirin versus LMWH for VTE prophylaxis after orthopedic surgery
- Mechanism of rhubarb in the treatment of hyperlipidemia: A recent review
- Surgical management and outcomes of traumatic global brachial plexus injury: A concise review and our center approach
- The progress of autoimmune hepatitis research and future challenges
- METTL16 in human diseases: What should we do next?
- New insights into the prevention of ureteral stents encrustation
- VISTA as a prospective immune checkpoint in gynecological malignant tumors: A review of the literature
- Case Reports
- Mycobacterium xenopi infection of the kidney and lymph nodes: A case report
- Genetic mutation of SLC6A20 (c.1072T > C) in a family with nephrolithiasis: A case report
- Chronic hepatitis B complicated with secondary hemochromatosis was cured clinically: A case report
- Liver abscess complicated with multiple organ invasive infection caused by hematogenous disseminated hypervirulent Klebsiella pneumoniae: A case report
- Urokinase-based lock solutions for catheter salvage: A case of an upcoming kidney transplant recipient
- Two case reports of maturity-onset diabetes of the young type 3 caused by the hepatocyte nuclear factor 1α gene mutation
- Immune checkpoint inhibitor-related pancreatitis: What is known and what is not
- Does total hip arthroplasty result in intercostal nerve injury? A case report and literature review
- Clinicopathological characteristics and diagnosis of hepatic sinusoidal obstruction syndrome caused by Tusanqi – Case report and literature review
- Synchronous triple primary gastrointestinal malignant tumors treated with laparoscopic surgery: A case report
- CT-guided percutaneous microwave ablation combined with bone cement injection for the treatment of transverse metastases: A case report
- Malignant hyperthermia: Report on a successful rescue of a case with the highest temperature of 44.2°C
- Anesthetic management of fetal pulmonary valvuloplasty: A case report
- Rapid Communication
- Impact of COVID-19 lockdown on glycemic levels during pregnancy: A retrospective analysis
- Erratum
- Erratum to “Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway”
- Erratum to: “Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p”
- Retraction
- Retraction of “Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients”
- Retraction of “circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis”
- Retraction of “miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells”
- Retraction of “SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis”
- Retraction of “circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury”
- Retraction of “lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells”
- Special issue Linking Pathobiological Mechanisms to Clinical Application for cardiovascular diseases
- Effect of cardiac rehabilitation therapy on depressed patients with cardiac insufficiency after cardiac surgery
- Special issue The evolving saga of RNAs from bench to bedside - Part I
- FBLIM1 mRNA is a novel prognostic biomarker and is associated with immune infiltrates in glioma
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part III
- Development of a machine learning-based signature utilizing inflammatory response genes for predicting prognosis and immune microenvironment in ovarian cancer

