Abstract
Many cancers exhibit resistance to chemotherapy, resulting in a poor prognosis. The transcription factor NRF2, activated in response to cellular antioxidants, plays a crucial role in cell survival, proliferation, and resistance to chemotherapy. This factor may serve as a promising target for therapeutic interventions in esophageal carcinoma. Recent research suggests that NRF2 activity is modulated by ubiquitination mediated by the KEAP1-CUL3 E3 ligase complex, highlighting the importance of deubiquitination. However, the specific deubiquitinase responsible for regulating NRF2 in esophageal cancer remains unknown. In this study, a novel regulator of the NRF2 protein, Ubiquitin-Specific Protease 35 (USP35), has been identified. Mechanistically, USP35 modulates NRF2 stability through enzymatic deubiquitination. USP35 interacts with NRF2 and facilitates its deubiquitination. Knockdown of USP35 leads to a notable increase in NRF2 levels and enhances the sensitivity of cells to chemotherapy. These findings suggest that the USP35-NRF2 axis is a key player in the regulation of therapeutic strategies for esophageal cancer.
1 Introduction
NRF2 is recognized as a key transcription factor that orchestrates the cellular antioxidant response. Recent studies have revealed multiple functions of NRF2 beyond redox regulation. Over the past two decades, research on NRF2 has increasingly focused on its potential as a primary target for cancer prevention and treatment. The expanding understanding of NRF2’s role in cancer therapy has presented novel challenges and opportunities in the field. NRF2, a Cap’n’collar (CNC) leucine zipper (bZIP) transcription factor, is composed of seven Neh domains, each serving a distinct function [1]. Through heterodimerization with small MAF proteins, the NRF2 protein regulates the expression of over 200 genes containing antioxidant response elements (AREs) [2]. NRF2 plays a critical role in controlling a wide array of biochemical processes and metabolic functions, including drug metabolism and excretion, iron metabolism, energetic metabolism, amino acid metabolism, autophagy, proliferation, survival, DNA repair, mitochondrial physiology, and proteasomal degradation [1,3,4].
The ubiquitin-proteasome system (UPS) plays a critical role in the regulation of numerous cellular functions, including cell growth, proliferation, and DNA repair [5,6]. Recent research has highlighted the significant impact of the UPS on various human diseases [7,8]. Post-translational modifications, such as ubiquitination, are involved in the modulation of protein stability and activity, influencing a wide range of biological processes, including tumorigenesis and tumor development [9–11]. Ubiquitination is catalyzed by the concertive actions of E1 activating enzymes, E2 conjugating enzymes, and E3 ligating enzymes, which covalently add ubiquitin to target proteins, leading to diverse biological outcomes, especially proteasomal degradation [12–14]. While NRF2 is ubiquitously expressed in all cell types, its basal protein levels typically remain low in the absence of stress. Three E3 ubiquitin ligases, including KEAP1-CUL3-RBX1 [15–19], β-TrCP-SKP1-CUL1-RBX1 [20,21], and HRD1 [22], have been identified as regulators of NRF2 ubiquitylation and degradation. Despite the extensive research on E3 ubiquitin ligases, the deubiquitinating enzymes (DUBs) responsible for NRF2 regulation have not been thoroughly investigated. Therefore, identifying the DUB(s) of NRF2 is crucial for a more efficient and comprehensive exploration of the NRF2-ARE signaling regulatory network.
A screening of an expression library comprising 56 DUBs was conducted to determine the enzyme responsible for regulating NRF2 stability. Our study revealed that ubiquitin-specific protease 35 (USP35) modulates NRF2 protein stability through its enzymatic activity by interacting with NRF2 and facilitating its deubiquitination. Furthermore, silencing of USP35 resulted in enhanced NRF2 turnover and increased cellular susceptibility to chemotherapy. Based on our findings, USP35 is a new type of DUB that is essential in sensitivity to chemotherapy, and may offer a potential clinical treatment for esophageal cancer.
2 Materials and methods
2.1 Cell culture
As for the HEK293T and TE-1 cell lines, they were from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and the KYSE-410 cell line was from Procell Life Science & Technology Co., Ltd (Hunan, China). A cell line called ECA-109 was purchased from iCell Bioscience. In addition to DMEM (HyClone), the cells were maintained in a solution containing 10% fetal bovine serum, 100 mg/ml streptomycin, 100 IU penicillin, and 4 mM l-glutamine. Cells are cultured at 37°C in 5% CO2 humidified incubators.
2.2 Reagents
We obtained anti-USP35 (24559-1-AP), anti-NRE2L2 (NRF2) (16396-1-AP), anti-Flag (20543-1-AP), anti-HA (51064-2-AP), rabbit anti-MYC (16286-1-AP), anti-GAPDH (60004-1-Ig), and anti-actin antibodies (66009-1-Ig) from Proteintech. Promega Company provided a CellTiter-Glo Luminescent Cell Viability Assay Kit and Caspa-Glo 3/7 Assay Kit. Selleck Company provided cycloheximide (CHX). MCE provided MG-132 and Cisplatin. Beyotime Company provided TRIzol reagent, TIANGEN Company provided qPCR kit (SYBR Green), Transgene supplied EL transfection reagent, Takara provided exonuclease III used for gene cloning.
2.3 Plasmids
Specifically, for lentivirus-based shRNA expression vectors, DNA oligonucleotides containing shRNA sequences were designed and cloned into the pLV-H1-EF1α-puro expression vector. The shRNA sequences targeting USP35 that were used in the study were as follows:
USP35-sh1: GGATAGAGAGGGAGGAAGA;
USP35-sh2; GTGGAGAAGGAGACAGAAA.
2.4 CRISPR/Cas9 knockout (KO) cell line
An online tool called CRISPR Design Tool was used to design single-guide RNA (sgRNA) that was then cloned into lentiCRISPRv2 (Addgene, #52961) using the CRISPR Design Tool. The sgRNA sequence for NRF2 was 5ʹ-TAGTTGTAACTGAGCGAAAA-3ʹ.
2.5 Transfection and DUBs screening
During the screening of DUBs, every DUB expression plasmid was co-transfected with NRF2 into 293T cells using the EL Transfection Reagent from transgene company. Next, the cells were lysed and examined using antibodies against HA (NRF2), Flag (DUBS), and ACTIN.
2.6 RNA extraction and real-time PCR analysis
In the same manner as described previously, RNA was extracted and real-time PCR was carried out [23,24]. Isolate total RNA from cells using TRIzol reagent according to the manufacturer’s instructions. With FastKing gRNA Dispelling RT SuperMix (TIANGEN), cDNA was synthesized from each RNA sample. The levels of RNA expression were normalized to GAPDH, a control gene. FastFire qPCR PreMix SYBR Green (TIANGEN) was used for the real-time PCR.
2.7 Ubiquitination assays
NRF2 ubiquitylation was determined by co-transfecting 293T cells with NRF2-HA and Flag-Ub, in the presence or absence of USP35 and its mutants. MG132 (10 M) was added to the cells for 6 h prior to harvesting, followed by RIPA buffer lysis, followed by immunoprecipitation of NRF2 with anti-HA beads. SDS-PAGE was performed on immunoprecipitated NRF2–HA, and anti-Flag antibodies were used to determine NRF2 ubiquitylation.
2.8 Immunoprecipitation
Protein interactions can be assessed using immunoprecipitation. After washing with cold PBS and lysing with protease inhibitor rich lysis buffer, cells were centrifuged. Centrifuging the supernatant of the cell lysate at 4°C for 30 min at 11,000g removed the supernatant from the cell lysate. As mentioned earlier [23,24], a M2 bead or an anti-HA bead was used in the immunoprecipitation step.
2.9 Cell viability assay
To determine the cell viability, approximately 2.5 × 104 cells were seeded into 96-well plates and incubated overnight according to the kit’s instructions. The cells were processed for an appropriate period of time whether they contain or do not contain related compounds. The 96-well plate was then incubated for 30 min at room temperature, followed by addition of equal volumes of CellTiter-Glo reagent and shaking for 2 min. After 15 min of further incubation at room temperature, the luminescence value was recorded.
3 Results
3.1 USP35 regulates the stability of NRF2 protein
In order to investigate the regulation of NRF2 stability by DUBs, we conducted experiments to determine if NRF2 protein degradation occurs in an ubiquitination/proteasome-dependent manner. Our findings, illustrated in Figure 1a, demonstrate that treatment with the proteasome inhibitor MG-132 led to a significant increase in NRF2 protein levels in esophageal cancer cell lines. To identify specific DUBs involved in the regulation of NRF2 protein degradation, we co-transfected NRF2 with expression plasmids encoding individual DUBs in 293T cells and assessed their impact on NRF2 protein levels. Our results indicate that the expression of USP35, a DUB, significantly enhanced NRF2 protein levels (Figure 1b). Furthermore, the upregulation of USP35 was found to elevate endogenous NRF2 protein levels while having no impact on NRF2 RNA levels (Figure 1c and d). Conversely, downregulation of USP35 resulted in decreased NRF2 protein levels (Figure 1e). To validate the role of USP35 in modulating NRF2 protein stability, TE-1 cells were co-transfected with USP35 and NRF2 and treated with the eukaryotic protein synthesis inhibitor CHX. The results depicted in Figure 1f demonstrate that USP35 expression significantly enhances NRF2 protein stability.

USP35 regulates the stability of NRF2 protein. (a) The proteasome inhibitor MG132 (10 μM) was applied to KYSE-410, TE-1, and ECA-109 cell lines for a duration of 6 h, followed by SDS-PAGE and western blot analysis using appropriate antibodies. (b) Plasmid expressing NRF2-HA was co-transfected with individual DUB expression plasmids into 293T cells. After 36 h, the cells were collected and analyzed using relevant antibodies. (c) Lentivirus expressing either empty or USP35-Flag was utilized to infect ECA-109 cells, which were subsequently harvested and analyzed with the appropriate antibodies. (d) NRF2 RNA levels were evaluated in ECA-109 cells infected with lentivirus carrying either empty or USP35-Flag constructs. (e) Lentivirus containing scramble or USP35-shRNAs was employed to infect ECA-109 cells, followed by cell harvesting and analysis using specific antibodies. (f) In TE-1 cells, co-transfection of a plasmid expressing NRF2-HA with either an empty vector or USP35-Flag expression plasmids was performed. Subsequently, the cells were treated with CHX (40 µg/mL) for a specified period, followed by cell harvesting and analysis using the designated antibodies after 24 h.
3.2 Enzymatic activity of USP35 is significant in its regulation of NRF2
Subsequently, our investigation focused on the role of USP35 in regulating the protein stability of NRF2. Given the importance of a DUB’s enzymatic activity for its function, we conducted experiments to determine if USP35’s enzymatic activity impacts the stability of NRF2 protein. Specifically, we examined the effects of USP35-C450S, an enzymatically inactive mutant of USP35 characterized by a serine mutation at amino acid position 450. As shown in Figure 2a, only wild-type USP35, and not its enzymatically inactive mutant, was able to increase NRF2 protein levels. Subsequently, our investigation focused on determining the specific domain of USP35 responsible for regulating the stability of NRF2. Our findings indicate that only the USP35 construct containing its enzymatic domain (amino acids 421-1018), as opposed to the construct containing amino acids 1-420, exhibited a significant increase in NRF2-protein levels compared to full-length USP35 (Figure 2b). Furthermore, the stability of the NRF2 protein was enhanced by USP35, but not by its enzymatically inactive mutant (Figure 2c). Collectively, these results suggest that USP35 influences NRF2 protein stability through enzymatic mechanisms.
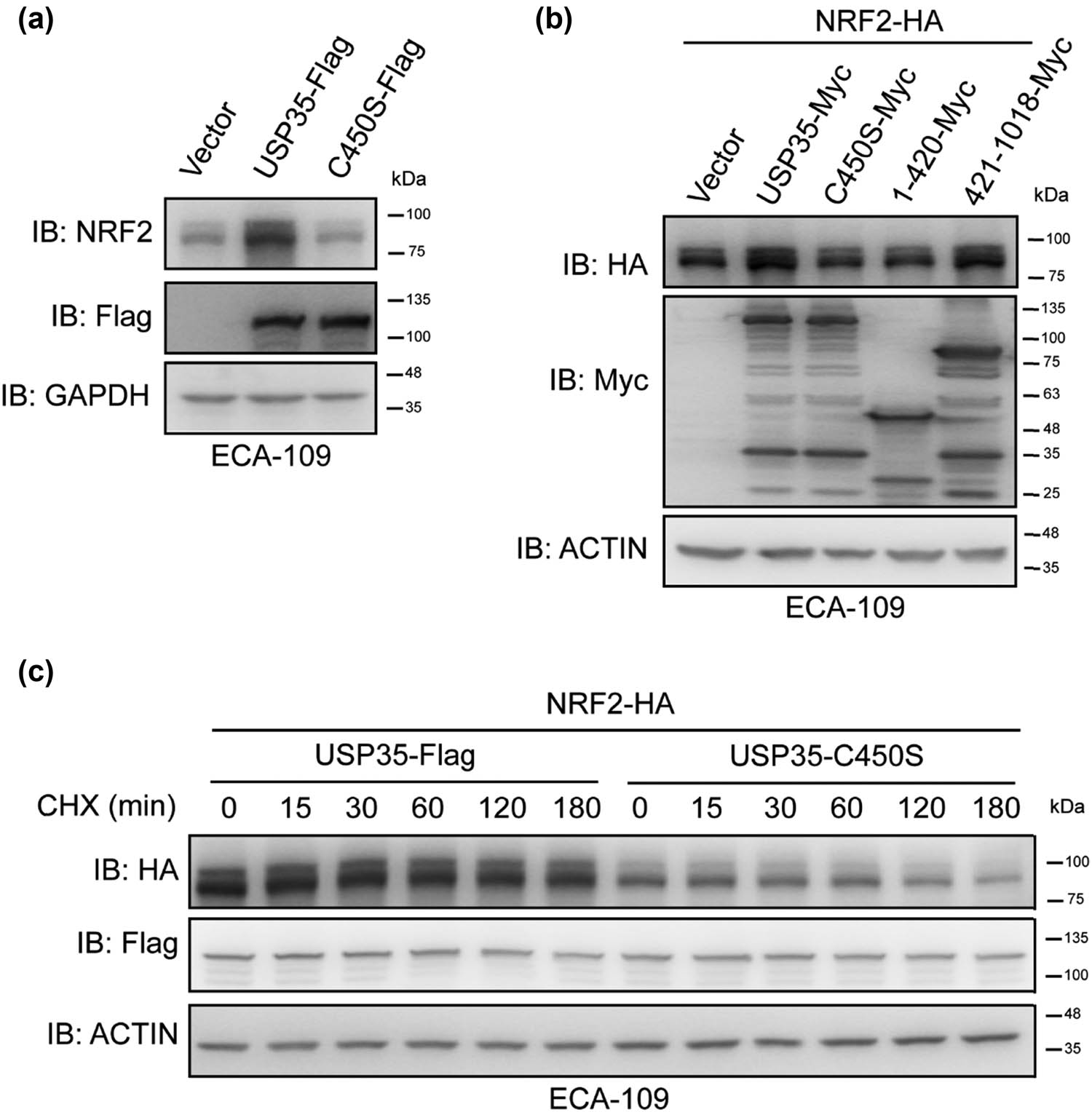
Enzymatic activity of USP35 is important in its regulation of NRF2. (a) Lentivirus expressing Vector, USP35-Flag, or USP35-C450S-Flag was utilized to transduce ECA-109 cells, followed by cell harvesting and analysis using specific antibodies. (b) Plasmid expressing NRF2-HA was co-transfected with plasmids expressing an empty vector, USP35, or its variants into ECA-109 cells. Subsequently, the cells were harvested after 36 h and subjected to analysis using the appropriate antibodies. (c) Plasmid encoding NRF2-HA was co-transfected with plasmids encoding an empty vector, USP35-Flag, or USP35-C450S-Flag into ECA-109 cells. Following a 24 h incubation period, the cells were exposed to CHX at a concentration of 40 µg/mL for designated durations. Subsequently, the cells were harvested and subjected to immunoblot analysis using the specified antibodies.
3.3 USP35 interacts with NRF2 and promotes NRF2 deubiquitination
Subsequently, an investigation was conducted to determine the potential interaction between USP35 and NRF2 in order to elucidate the mechanism by which USP35 regulates the protein stability of NRF2. The results presented in Figure 3a and b demonstrate a mutual interaction between USP35 and NRF2. Further analysis was carried out to ascertain the specific protein domains through which USP35 and NRF2 interact. As illustrated in Figure 3c, it was observed that NRF2 is capable of interacting with enzymatic domains of USP35 (421-1018). As numerous DUBs are known to modulate protein stability by cleaving ubiquitin chains from substrates, our study investigated the potential impact of USP35 on the ubiquitination of NRF2. Our findings, as depicted in Figure 3d, demonstrate that USP35 specifically targets the ubiquitin chain of NRF2, but not that of its inactive mutant. In conclusion, our results suggest that USP35 interacts with NRF2 and deubiquitinates it, thereby regulating its protein stability.
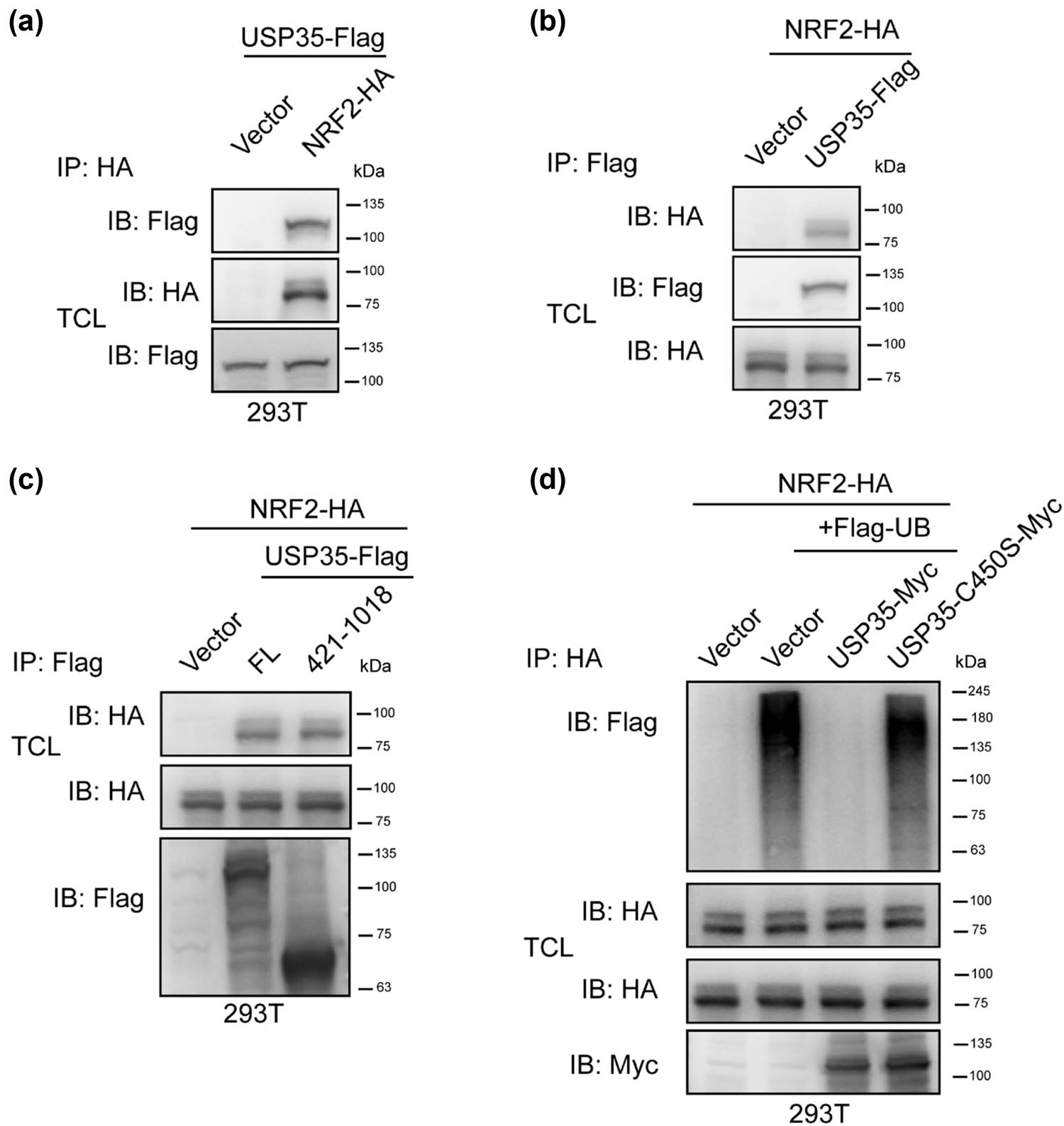
USP35 interacted with and promoted NRF2 deubiquitination. (a) An empty vector or NRF2-HA plasmid was co-transfected with a USP35-Flag plasmid into 293T cells. Prior to cell harvesting, the cells were treated with MG132 (10 µM) for a duration of 6 h. Subsequently, the cell lysates underwent immunoprecipitation utilizing the designated antibodies. Immunoblotting was carried out on both the total cell lysates and the immunoprecipitates using anti-HA or Flag antibodies. (b) Similarly to (a), with the exception that an empty vector or USP35-Flag plasmid was co-transfected with an NRF2-HA plasmid into 293T cells. (c) In a manner analogous to (a), the co-transfection of empty vector, USP35-Flag, or the enzymatic domain (aa 421-1018) plasmid with NRF2-HA plasmid was conducted in 293T cells. (d) Co-transfection of empty vector, USP35-MYC plasmid, or USP35 enzymatic mutant C450S-MYC plasmid with NRF2-HA and Flag-ubiquitin (Flag-Ub) plasmids was carried out in 293T cells. Subsequently, the cells were treated with MG132 (10 µM) for 6 h prior to harvesting. Immunoprecipitation was performed on the cell lysates using anti-HA antibody, followed by immunoblotting using Flag (Flag-UB), HA (NRF2-HA), and Myc (USP35-Myc) antibodies on both the lysates.
3.4 USP35 causes chemotherapy resistance by upregulating NRF2 in ECA-109 cells
A key emphasis of this research is the examination of drug resistance within the realm of cancer therapeutics. Previous research has demonstrated that the expression of the NRF2 protein is linked to the development of resistance to chemotherapy in cancer cells. The data presented indicate that USP35 acts as a novel deubiquitinase of NRF2. Notably, the upregulation of USP35 in various cancers implies a potential relationship between USP35 and NRF2 in the context of resistance to chemotherapeutic agents. To investigate this hypothesis, three stable ECA-109 cell lines were utilized, each expressing either an empty vector, NRF2, or USP35. Through the utilization of sell viability assays, our study examined the impact of Cisplatin, a chemotherapy agent, on esophageal cancer cells. Our findings indicate that the upregulation of NRF2 or USP35 diminishes the cytotoxic effects of Cisplatin on esophageal cancer cells (Figure 4a). Furthermore, a notable enhancement in susceptibility to Cisplatin treatment was observed in ECA-109 cells with NRF2 KOs and USP35 knockdowns (Figure 4b and c). The sensitization effects induced by NRF2 KO were reversible upon NRF2 re-expression, but not by USP35 (Figure 4d), suggesting that USP35 contributes to resistance against Cisplatin.
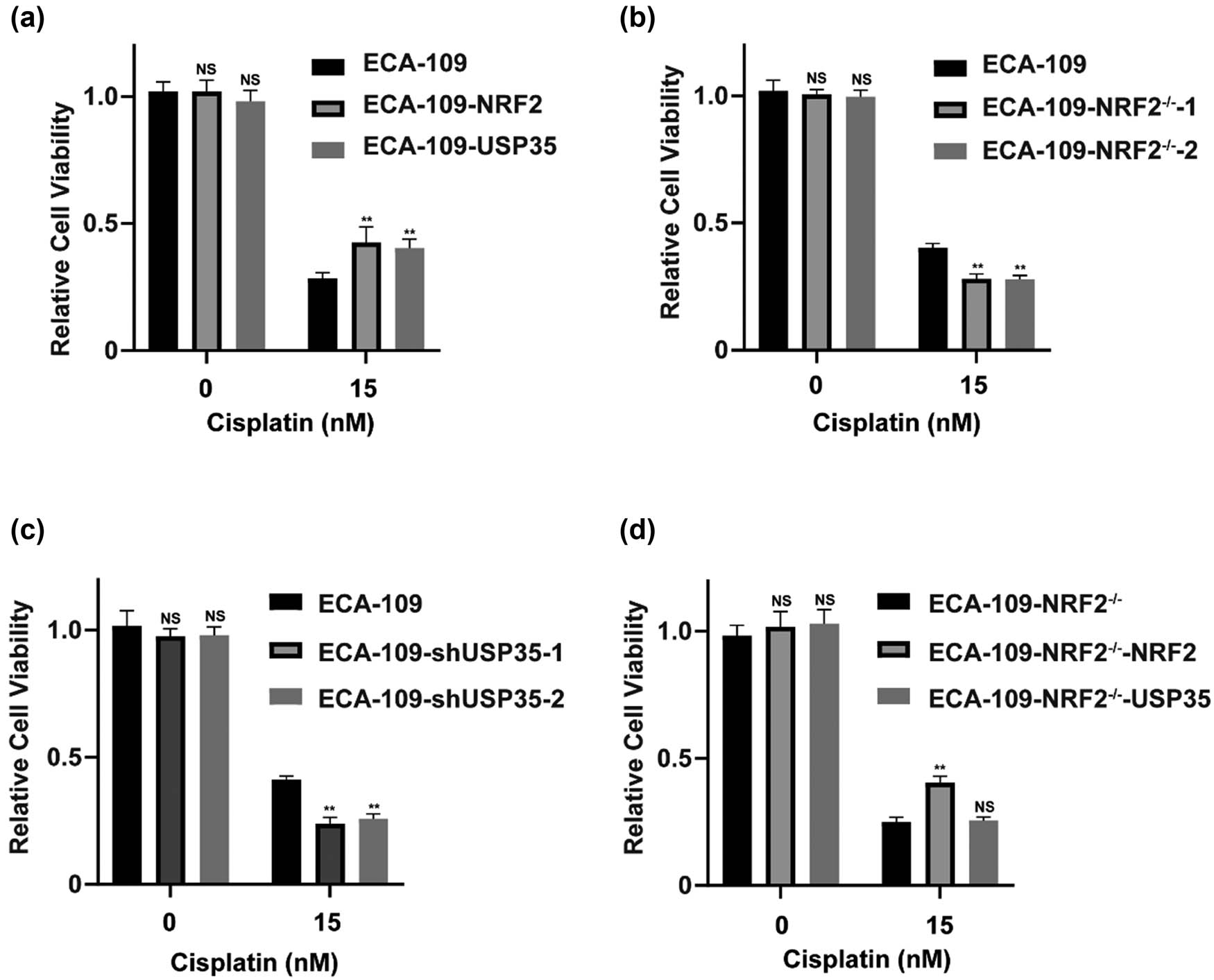
USP35 causes chemotherapy resistance by upregulating NRF2 in ECA-109 cells. (a)–(d) USP35 has been identified as a contributor to Cisplatin resistance through the upregulation of NRF2. To investigate this mechanism, we generated stable cell lines expressing ECA-109-vector, ECA-109-NRF2, and ECA-109-USP35, as well as KO cell lines ECA-109-NRF2−/−-1 and ECA-109-NRF2−/−-2, and knockdown cell lines ECA-109-shNC, ECA-109-shUSP35-1, and ECA-109-shUSP35-2. The ECA-109-NRF2−/− cell line was transduced with lentivirus containing NRF2 or USP35 to establish stably rescued cell lines designated as ECA-109-NRF2−/−-NRF2 and ECA-109-NRF2−/−-USP35. These cell lines were then subjected to treatment with varying doses of Cisplatin for 48 h in a 96-well plate, followed by measurement of cell viability.
4 Discussion
A significant portion of the ARE signaling pathway is mediated by NRF2 and its downstream effector genes, leading to the activation of numerous transcription factors. The NRF2-ARE signaling pathways are closely linked to various pathophysiological processes, such as metabolism and oxidative stress. Our study identifies USP35 as a prominent candidate in the screening process and highlights its extensive investigation into the molecular mechanisms governing its regulatory role in the NRF2-ARE signaling pathway in esophageal cancer cells. In our research, we found that USP35 interacts with NRF2 and aids in its deubiquitination, leading to the inhibition of its proteasomal degradation.
In numerous cellular processes, ubiquitination-proteasome degradation and the DUB system play vital roles in regulating transcription, DNA repair, signal transduction, cell cycle control, protein degradation, and cancer development. Nevertheless, additional research is needed to fully understand the relationship between the deubiquitinase USP35 and cancer chemotherapy. Given the association between NRF2 and chemoresistance, our study aimed to illustrate the role of USP35 in regulating chemoresistance through deubiquitination and stabilization of NRF2. Our findings indicate that depletion of USP35 significantly enhances Cisplatin-induced cell death in esophageal cancer cell lines. Consequently, our research will pave the way for further exploration into the deubiquitination mechanisms and regulatory pathways of NRF2. In conclusion, our results suggest that USP35 enhances chemotherapy resistance by deubiquitinating and stabilizing NRF2, offering a promising avenue for addressing chemoresistance in esophageal cancer through NRF2 activation.
Numerous antioxidant pathways, such as the NRF2-ARE antioxidant response pathway, are crucial in cancer therapy and chemoprevention. Thus, a stringent regulatory framework is necessary to impede the onset and advancement of cancer. Research indicates that cancer cells exhibiting elevated NRF2 levels demonstrate reduced sensitivity to chemotherapy [25]. The NRF2 protein is a transcription factor, and previous studies have shown that NRF2 is deubiquitinated by a variety of DUBs, such as DUB3, USP11, and USP35 [26–28]. Diverse DUBs play a key role in various tumors, which implies that different DUBs may be active in different tumors to regulate the stability of the NRF2 protein. For instance, the heterotopic expression of DUB3 has been shown to result in NRF2-dependent chemotherapy resistance in colon cancer cell lines [26]. Depletion of USP11 has been found to inhibit cell proliferation and induce ROS-mediated stress-induced iron cell death [27]. Silencing of USP35 leads to a reduction in NRF2 levels, thereby increasing the sensitivity of renal clear cell cancer cells to iron death induction [28]. Our research indicates that knockdown of USP35 results in a significant elevation of NRF2 levels and enhances the sensitivity of esophageal cancer cells to chemotherapy. Previous research has indicated that USP11 plays a role in stabilizing the NRF2 protein. However, our investigation into the regulation of NRF2 protein stability did not find evidence of USP11’s involvement in this process. This discrepancy may be attributed to the use of foreign expression vectors or screening methods. Both our study and the study by Wang et al. identified USP35 as a deubiquitination enzyme present in various tumors, suggesting that this enzyme may stabilize the same substrate protein in different tumor types to facilitate its function. Our study primarily investigates chemotherapy resistance as a significant barrier to effective treatment of esophageal cancer, with the aim of identifying key factors contributing to drug resistance. In our functional analysis, cells that overexpressed NRF2 and USP35 exhibited a survival rate approximately 10% lower than cells that did not overexpress these proteins. This is attributed to the presence of additional DUBs and drug-resistant molecules within esophageal cancer cell lines, which are implicated in the resistance to chemotherapy drugs. This observation further indicates the existence of untapped targets for addressing tumor resistance.
-
Funding information: This work was supported by the Henan Provincial Bureau of Science and Technology (Grant no. 20232048) and Luoyang Medical and Health Research (Grant no. 2001025A).
-
Author contributions: The majority of the biochemical experiments were conducted by Dian Zhang and Jiawen Li, with Chao Zhang, Jinliang Xue, Peihao Li, Kai Shang, and Xiao Zhang contributing to select cell experiments. Baoping Lang was responsible for the research design and the article composition. The final manuscript was reviewed and approved by all authors.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Smolková K, Mikó E, Kovács T, Leguina-Ruzzi A, Sipos A, Bai P. Nuclear factor erythroid 2-related factor 2 in regulating cancer metabolism. Antioxid Redox Signal. 2020;33(13):966–97.10.1089/ars.2020.8024Search in Google Scholar PubMed PubMed Central
[2] Fakhri S, Moradi SZ, Nouri Z, Cao H, Wang H, Khan H, et al. Modulation of integrin receptor by polyphenols: downstream Nrf2-Keap1/ARE and associated cross-talk mediators in cardiovascular diseases. Crit Rev Food Sci Nutr. 2024;64(6):1592–616.10.1080/10408398.2022.2118226Search in Google Scholar PubMed
[3] Crisman E, Duarte P, Dauden E, Cuadrado A, Rodríguez-Franco MI, López MG, et al. KEAP1-NRF2 protein–protein interaction inhibitors: design, pharmacological properties and therapeutic potential. Med Res Rev. 2023;43(1):237–87.10.1002/med.21925Search in Google Scholar PubMed PubMed Central
[4] Niu Z, Jin R, Zhang Y, Li H. Signaling pathways and targeted therapies in lung squamous cell carcinoma: mechanisms and clinical trials. Signal Transduct Target Ther. 2022;7(1):353.10.1038/s41392-022-01200-xSearch in Google Scholar PubMed PubMed Central
[5] Liu B, Ruan J, Chen M, Li Z, Manjengwa G, Schlüter D, et al. Deubiquitinating enzymes (DUBs): decipher underlying basis of neurodegenerative diseases. Mol Psychiatry. 2022;27(1):259–68.10.1038/s41380-021-01233-8Search in Google Scholar PubMed
[6] Olguín HC. The gentle side of the UPS: ubiquitin-proteasome system and the regulation of the myogenic program. Front Cell Dev Biol. 2022;9:821839.10.3389/fcell.2021.821839Search in Google Scholar PubMed PubMed Central
[7] Sivinski J, Zhang DD, Chapman E. Targeting NRF2 to treat cancer. Semin Cancer Biol. 2021;76:61–73.10.1016/j.semcancer.2021.06.003Search in Google Scholar PubMed PubMed Central
[8] Hanna J, Guerra-Moreno A, Ang J, Micoogullari Y. Protein degradation and the pathologic basis of disease. Am J Pathol. 2019;189(1):94–103.10.1016/j.ajpath.2018.09.004Search in Google Scholar PubMed PubMed Central
[9] Wang Y, Liu X, Huang W, Liang J, Chen Y. The intricate interplay between HIFs, ROS, and the ubiquitin system in the tumor hypoxic microenvironment. Pharmacol Ther. 2022;240:108303.10.1016/j.pharmthera.2022.108303Search in Google Scholar PubMed
[10] Kumar P, Kumar P, Mandal D, Velayutham R. The emerging role of Deubiquitinases (DUBs) in parasites: a foresight review. Front Cell Infect Microbiol. 2022;12:985178.10.3389/fcimb.2022.985178Search in Google Scholar PubMed PubMed Central
[11] Suresh B, Lee J, Kim H, Ramakrishna S. Regulation of pluripotency and differentiation by deubiquitinating enzymes. Cell Death Differ. 2016;23(8):1257–64.10.1038/cdd.2016.53Search in Google Scholar PubMed PubMed Central
[12] Martens S, Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020;6:23.10.1038/s41421-020-0155-1Search in Google Scholar PubMed PubMed Central
[13] Chang HM, Yeh ETH. SUMO: from bench to bedside. Physiol Rev. 2020;100(4):1599–1619.10.1152/physrev.00025.2019Search in Google Scholar PubMed PubMed Central
[14] Paiva SL, da Silva SR, de Araujo ED, Gunning PT. Regulating the master regulator: controlling ubiquitination by thinking outside the active site. J Med Chem. 2018;61(2):405–21.10.1021/acs.jmedchem.6b01346Search in Google Scholar PubMed
[15] McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281(34):24756–68.10.1074/jbc.M601119200Search in Google Scholar PubMed
[16] Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–53.10.1128/MCB.24.24.10941-10953.2004Search in Google Scholar PubMed PubMed Central
[17] Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–9.10.1128/MCB.24.16.7130-7139.2004Search in Google Scholar PubMed PubMed Central
[18] Tao S, Liu P, Luo G, Rojo de la Vega M, Chen H, Wu T, et al. p97 negatively regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex. Mol Cell Biol. 2017;37(8):e00660-16.10.1128/MCB.00660-16Search in Google Scholar PubMed PubMed Central
[19] Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23(22):8137–51.10.1128/MCB.23.22.8137-8151.2003Search in Google Scholar PubMed PubMed Central
[20] Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32(32):3765–81.10.1038/onc.2012.388Search in Google Scholar PubMed PubMed Central
[21] Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31(6):1121–33.10.1128/MCB.01204-10Search in Google Scholar PubMed PubMed Central
[22] Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T, et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28(7):708–22.10.1101/gad.238246.114Search in Google Scholar PubMed PubMed Central
[23] Huang F, Liang J, Lin Y, Chen Y, Hu F, Feng J, et al. Repurposing of Ibrutinib and Quizartinib as potent inhibitors of necroptosis. Commun Biol. 2023;6(1):972.10.1038/s42003-023-05353-5Search in Google Scholar PubMed PubMed Central
[24] Feng J, Liu P, Li X, Zhang D, Lin H, Hou Z, et al. The deubiquitinating enzyme USP20 regulates the stability of the MCL1 protein. Biochem Biophys Res Commun. 2022;593:122–8.10.1016/j.bbrc.2022.01.019Search in Google Scholar PubMed
[25] Cruz-Gregorio A, Aranda-Rivera AK, Pedraza-Chaverri J. Nuclear factor erythroid 2-related factor 2 in human papillomavirus-related cancers. Rev Med Virol. 2022;32(3):e2308.10.1002/rmv.2308Search in Google Scholar PubMed
[26] Zhang Q, Zhang ZY, Du H, Li SZ, Tu R, Jia YF, et al. DUB3 deubiquitinates and stabilizes NRF2 in chemotherapy resistance of colorectal cancer. Cell Death Differ. 2019;26(11):2300–13.10.1038/s41418-019-0303-zSearch in Google Scholar PubMed PubMed Central
[27] Meng C, Zhan J, Chen D, Shao G, Zhang H, Gu W, et al. The deubiquitinase USP11 regulates cell proliferation and ferroptotic cell death via stabilization of NRF2 USP11 deubiquitinates and stabilizes NRF2. Oncogene. 2021;40(9):1706–20.10.1038/s41388-021-01660-5Search in Google Scholar PubMed
[28] Wang S, Wang T, Zhang X, Cheng S, Chen C, Yang G, et al. The deubiquitylating enzyme USP35 restricts regulated cell death to promote survival of renal clear cell carcinoma. Cell Death Differ. 2023;30(7):1757–70.10.1038/s41418-023-01176-3Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Constitutive and evoked release of ATP in adult mouse olfactory epithelium
- LARP1 knockdown inhibits cultured gastric carcinoma cell cycle progression and metastatic behavior
- PEGylated porcine–human recombinant uricase: A novel fusion protein with improved efficacy and safety for the treatment of hyperuricemia and renal complications
- Research progress on ocular complications caused by type 2 diabetes mellitus and the function of tears and blepharons
- The role and mechanism of esketamine in preventing and treating remifentanil-induced hyperalgesia based on the NMDA receptor–CaMKII pathway
- Brucella infection combined with Nocardia infection: A case report and literature review
- Detection of serum interleukin-18 level and neutrophil/lymphocyte ratio in patients with antineutrophil cytoplasmic antibody-associated vasculitis and its clinical significance
- Ang-1, Ang-2, and Tie2 are diagnostic biomarkers for Henoch-Schönlein purpura and pediatric-onset systemic lupus erythematous
- PTTG1 induces pancreatic cancer cell proliferation and promotes aerobic glycolysis by regulating c-myc
- Role of serum B-cell-activating factor and interleukin-17 as biomarkers in the classification of interstitial pneumonia with autoimmune features
- Effectiveness and safety of a mumps containing vaccine in preventing laboratory-confirmed mumps cases from 2002 to 2017: A meta-analysis
- Low levels of sex hormone-binding globulin predict an increased breast cancer risk and its underlying molecular mechanisms
- A case of Trousseau syndrome: Screening, detection and complication
- Application of the integrated airway humidification device enhances the humidification effect of the rabbit tracheotomy model
- Preparation of Cu2+/TA/HAP composite coating with anti-bacterial and osteogenic potential on 3D-printed porous Ti alloy scaffolds for orthopedic applications
- Aquaporin-8 promotes human dermal fibroblasts to counteract hydrogen peroxide-induced oxidative damage: A novel target for management of skin aging
- Current research and evidence gaps on placental development in iron deficiency anemia
- Single-nucleotide polymorphism rs2910829 in PDE4D is related to stroke susceptibility in Chinese populations: The results of a meta-analysis
- Pheochromocytoma-induced myocardial infarction: A case report
- Kaempferol regulates apoptosis and migration of neural stem cells to attenuate cerebral infarction by O‐GlcNAcylation of β-catenin
- Sirtuin 5 regulates acute myeloid leukemia cell viability and apoptosis by succinylation modification of glycine decarboxylase
- Apigenin 7-glucoside impedes hypoxia-induced malignant phenotypes of cervical cancer cells in a p16-dependent manner
- KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1
- Current state of research on copper complexes in the treatment of breast cancer
- Exploring antioxidant strategies in the pathogenesis of ALS
- Helicobacter pylori causes gastric dysbacteriosis in chronic gastritis patients
- IL-33/soluble ST2 axis is associated with radiation-induced cardiac injury
- The predictive value of serum NLR, SII, and OPNI for lymph node metastasis in breast cancer patients with internal mammary lymph nodes after thoracoscopic surgery
- Carrying SNP rs17506395 (T > G) in TP63 gene and CCR5Δ32 mutation associated with the occurrence of breast cancer in Burkina Faso
- P2X7 receptor: A receptor closely linked with sepsis-associated encephalopathy
- Probiotics for inflammatory bowel disease: Is there sufficient evidence?
- Identification of KDM4C as a gene conferring drug resistance in multiple myeloma
- Microbial perspective on the skin–gut axis and atopic dermatitis
- Thymosin α1 combined with XELOX improves immune function and reduces serum tumor markers in colorectal cancer patients after radical surgery
- Highly specific vaginal microbiome signature for gynecological cancers
- Sample size estimation for AQP4-IgG seropositive optic neuritis: Retinal damage detection by optical coherence tomography
- The effects of SDF-1 combined application with VEGF on femoral distraction osteogenesis in rats
- Fabrication and characterization of gold nanoparticles using alginate: In vitro and in vivo assessment of its administration effects with swimming exercise on diabetic rats
- Mitigating digestive disorders: Action mechanisms of Mediterranean herbal active compounds
- Distribution of CYP2D6 and CYP2C19 gene polymorphisms in Han and Uygur populations with breast cancer in Xinjiang, China
- VSP-2 attenuates secretion of inflammatory cytokines induced by LPS in BV2 cells by mediating the PPARγ/NF-κB signaling pathway
- Factors influencing spontaneous hypothermia after emergency trauma and the construction of a predictive model
- Long-term administration of morphine specifically alters the level of protein expression in different brain regions and affects the redox state
- Application of metagenomic next-generation sequencing technology in the etiological diagnosis of peritoneal dialysis-associated peritonitis
- Clinical diagnosis, prevention, and treatment of neurodyspepsia syndrome using intelligent medicine
- Case report: Successful bronchoscopic interventional treatment of endobronchial leiomyomas
- Preliminary investigation into the genetic etiology of short stature in children through whole exon sequencing of the core family
- Cystic adenomyoma of the uterus: Case report and literature review
- Mesoporous silica nanoparticles as a drug delivery mechanism
- Dynamic changes in autophagy activity in different degrees of pulmonary fibrosis in mice
- Vitamin D deficiency and inflammatory markers in type 2 diabetes: Big data insights
- Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells
- Meta-analysis on the efficacy of allogeneic hematopoietic stem cell transplantation to treat malignant lymphoma
- Mitochondrial DNA drives neuroinflammation through the cGAS-IFN signaling pathway in the spinal cord of neuropathic pain mice
- Application value of artificial intelligence algorithm-based magnetic resonance multi-sequence imaging in staging diagnosis of cervical cancer
- Embedded monitoring system and teaching of artificial intelligence online drug component recognition
- Investigation into the association of FNDC1 and ADAMTS12 gene expression with plumage coloration in Muscovy ducks
- Yak meat content in feed and its impact on the growth of rats
- A rare case of Richter transformation with breast involvement: A case report and literature review
- First report of Nocardia wallacei infection in an immunocompetent patient in Zhejiang province
- Rhodococcus equi and Brucella pulmonary mass in immunocompetent: A case report and literature review
- Downregulation of RIP3 ameliorates the left ventricular mechanics and function after myocardial infarction via modulating NF-κB/NLRP3 pathway
- Evaluation of the role of some non-enzymatic antioxidants among Iraqi patients with non-alcoholic fatty liver disease
- The role of Phafin proteins in cell signaling pathways and diseases
- Ten-year anemia as initial manifestation of Castleman disease in the abdominal cavity: A case report
- Coexistence of hereditary spherocytosis with SPTB P.Trp1150 gene variant and Gilbert syndrome: A case report and literature review
- Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells
- Exploratory evaluation supported by experimental and modeling approaches of Inula viscosa root extract as a potent corrosion inhibitor for mild steel in a 1 M HCl solution
- Imaging manifestations of ductal adenoma of the breast: A case report
- Gut microbiota and sleep: Interaction mechanisms and therapeutic prospects
- Isomangiferin promotes the migration and osteogenic differentiation of rat bone marrow mesenchymal stem cells
- Prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer
- Circular RNAs as potential biomarkers for male severe sepsis
- Knockdown of Stanniocalcin-1 inhibits growth and glycolysis in oral squamous cell carcinoma cells
- The expression and biological role of complement C1s in esophageal squamous cell carcinoma
- A novel GNAS mutation in pseudohypoparathyroidism type 1a with articular flexion deformity: A case report
- Predictive value of serum magnesium levels for prognosis in patients with non-small cell lung cancer undergoing EGFR-TKI therapy
- HSPB1 alleviates acute-on-chronic liver failure via the P53/Bax pathway
- IgG4-related disease complicated by PLA2R-associated membranous nephropathy: A case report
- Baculovirus-mediated endostatin and angiostatin activation of autophagy through the AMPK/AKT/mTOR pathway inhibits angiogenesis in hepatocellular carcinoma
- Metformin mitigates osteoarthritis progression by modulating the PI3K/AKT/mTOR signaling pathway and enhancing chondrocyte autophagy
- Evaluation of the activity of antimicrobial peptides against bacterial vaginosis
- Atypical presentation of γ/δ mycosis fungoides with an unusual phenotype and SOCS1 mutation
- Analysis of the microecological mechanism of diabetic kidney disease based on the theory of “gut–kidney axis”: A systematic review
- Omega-3 fatty acids prevent gestational diabetes mellitus via modulation of lipid metabolism
- Refractory hypertension complicated with Turner syndrome: A case report
- Interaction of ncRNAs and the PI3K/AKT/mTOR pathway: Implications for osteosarcoma
- Association of low attenuation area scores with pulmonary function and clinical prognosis in patients with chronic obstructive pulmonary disease
- Long non-coding RNAs in bone formation: Key regulators and therapeutic prospects
- The deubiquitinating enzyme USP35 regulates the stability of NRF2 protein
- Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as potential diagnostic markers for rebleeding in patients with esophagogastric variceal bleeding
- G protein-coupled receptor 1 participating in the mechanism of mediating gestational diabetes mellitus by phosphorylating the AKT pathway
- LL37-mtDNA regulates viability, apoptosis, inflammation, and autophagy in lipopolysaccharide-treated RLE-6TN cells by targeting Hsp90aa1
- The analgesic effect of paeoniflorin: A focused review
- Chemical composition’s effect on Solanum nigrum Linn.’s antioxidant capacity and erythrocyte protection: Bioactive components and molecular docking analysis
- Knockdown of HCK promotes HREC cell viability and inner blood–retinal barrier integrity by regulating the AMPK signaling pathway
- The role of rapamycin in the PINK1/Parkin signaling pathway in mitophagy in podocytes
- Laryngeal non-Hodgkin lymphoma: Report of four cases and review of the literature
- Clinical value of macrogenome next-generation sequencing on infections
- Overview of dendritic cells and related pathways in autoimmune uveitis
- TAK-242 alleviates diabetic cardiomyopathy via inhibiting pyroptosis and TLR4/CaMKII/NLRP3 pathway
- Hypomethylation in promoters of PGC-1α involved in exercise-driven skeletal muscular alterations in old age
- Profile and antimicrobial susceptibility patterns of bacteria isolated from effluents of Kolladiba and Debark hospitals
- The expression and clinical significance of syncytin-1 in serum exosomes of hepatocellular carcinoma patients
- A histomorphometric study to evaluate the therapeutic effects of biosynthesized silver nanoparticles on the kidneys infected with Plasmodium chabaudi
- PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
- Analysis of MDA, SOD, TAOC, MNCV, SNCV, and TSS scores in patients with diabetes peripheral neuropathy
- SLIT3 deficiency promotes non-small cell lung cancer progression by modulating UBE2C/WNT signaling
- The relationship between TMCO1 and CALR in the pathological characteristics of prostate cancer and its effect on the metastasis of prostate cancer cells
- Heterogeneous nuclear ribonucleoprotein K is a potential target for enhancing the chemosensitivity of nasopharyngeal carcinoma
- PHB2 alleviates retinal pigment epithelium cell fibrosis by suppressing the AGE–RAGE pathway
- Anti-γ-aminobutyric acid-B receptor autoimmune encephalitis with syncope as the initial symptom: Case report and literature review
- Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua
- Human umbilical cord mesenchymal stem cells regulate glutathione metabolism depending on the ERK–Nrf2–HO-1 signal pathway to repair phosphoramide mustard-induced ovarian cancer cells
- Electroacupuncture on GB acupoints improves osteoporosis via the estradiol–PI3K–Akt signaling pathway
- Renalase protects against podocyte injury by inhibiting oxidative stress and apoptosis in diabetic nephropathy
- Review: Dicranostigma leptopodum: A peculiar plant of Papaveraceae
- Combination effect of flavonoids attenuates lung cancer cell proliferation by inhibiting the STAT3 and FAK signaling pathway
- Renal microangiopathy and immune complex glomerulonephritis induced by anti-tumour agents: A case report
- Correlation analysis of AVPR1a and AVPR2 with abnormal water and sodium and potassium metabolism in rats
- Gastrointestinal health anti-diarrheal mixture relieves spleen deficiency-induced diarrhea through regulating gut microbiota
- Myriad factors and pathways influencing tumor radiotherapy resistance
- Exploring the effects of culture conditions on Yapsin (YPS) gene expression in Nakaseomyces glabratus
- Screening of prognostic core genes based on cell–cell interaction in the peripheral blood of patients with sepsis
- Coagulation factor II thrombin receptor as a promising biomarker in breast cancer management
- Ileocecal mucinous carcinoma misdiagnosed as incarcerated hernia: A case report
- Methyltransferase like 13 promotes malignant behaviors of bladder cancer cells through targeting PI3K/ATK signaling pathway
- The debate between electricity and heat, efficacy and safety of irreversible electroporation and radiofrequency ablation in the treatment of liver cancer: A meta-analysis
- ZAG promotes colorectal cancer cell proliferation and epithelial–mesenchymal transition by promoting lipid synthesis
- Baicalein inhibits NLRP3 inflammasome activation and mitigates placental inflammation and oxidative stress in gestational diabetes mellitus
- Impact of SWCNT-conjugated senna leaf extract on breast cancer cells: A potential apoptotic therapeutic strategy
- MFAP5 inhibits the malignant progression of endometrial cancer cells in vitro
- Major ozonated autohemotherapy promoted functional recovery following spinal cord injury in adult rats via the inhibition of oxidative stress and inflammation
- Axodendritic targeting of TAU and MAP2 and microtubule polarization in iPSC-derived versus SH-SY5Y-derived human neurons
- Differential expression of phosphoinositide 3-kinase/protein kinase B and Toll-like receptor/nuclear factor kappa B signaling pathways in experimental obesity Wistar rat model
- The therapeutic potential of targeting Oncostatin M and the interleukin-6 family in retinal diseases: A comprehensive review
- BA inhibits LPS-stimulated inflammatory response and apoptosis in human middle ear epithelial cells by regulating the Nf-Kb/Iκbα axis
- Role of circRMRP and circRPL27 in chronic obstructive pulmonary disease
- Investigating the role of hyperexpressed HCN1 in inducing myocardial infarction through activation of the NF-κB signaling pathway
- Characterization of phenolic compounds and evaluation of anti-diabetic potential in Cannabis sativa L. seeds: In vivo, in vitro, and in silico studies
- Quantitative immunohistochemistry analysis of breast Ki67 based on artificial intelligence
- Ecology and Environmental Science
- Screening of different growth conditions of Bacillus subtilis isolated from membrane-less microbial fuel cell toward antimicrobial activity profiling
- Degradation of a mixture of 13 polycyclic aromatic hydrocarbons by commercial effective microorganisms
- Evaluation of the impact of two citrus plants on the variation of Panonychus citri (Acari: Tetranychidae) and beneficial phytoseiid mites
- Prediction of present and future distribution areas of Juniperus drupacea Labill and determination of ethnobotany properties in Antalya Province, Türkiye
- Population genetics of Todarodes pacificus (Cephalopoda: Ommastrephidae) in the northwest Pacific Ocean via GBS sequencing
- A comparative analysis of dendrometric, macromorphological, and micromorphological characteristics of Pistacia atlantica subsp. atlantica and Pistacia terebinthus in the middle Atlas region of Morocco
- Macrofungal sporocarp community in the lichen Scots pine forests
- Assessing the proximate compositions of indigenous forage species in Yemen’s pastoral rangelands
- Food Science
- Gut microbiota changes associated with low-carbohydrate diet intervention for obesity
- Reexamination of Aspergillus cristatus phylogeny in dark tea: Characteristics of the mitochondrial genome
- Differences in the flavonoid composition of the leaves, fruits, and branches of mulberry are distinguished based on a plant metabolomics approach
- Investigating the impact of wet rendering (solventless method) on PUFA-rich oil from catfish (Clarias magur) viscera
- Non-linear associations between cardiovascular metabolic indices and metabolic-associated fatty liver disease: A cross-sectional study in the US population (2017–2020)
- Knockdown of USP7 alleviates atherosclerosis in ApoE-deficient mice by regulating EZH2 expression
- Utility of dairy microbiome as a tool for authentication and traceability
- Agriculture
- Enhancing faba bean (Vicia faba L.) productivity through establishing the area-specific fertilizer rate recommendation in southwest Ethiopia
- Impact of novel herbicide based on synthetic auxins and ALS inhibitor on weed control
- Perspectives of pteridophytes microbiome for bioremediation in agricultural applications
- Fertilizer application parameters for drip-irrigated peanut based on the fertilizer effect function established from a “3414” field trial
- Improving the productivity and profitability of maize (Zea mays L.) using optimum blended inorganic fertilization
- Application of leaf multispectral analyzer in comparison to hyperspectral device to assess the diversity of spectral reflectance indices in wheat genotypes
- Animal Sciences
- Knockdown of ANP32E inhibits colorectal cancer cell growth and glycolysis by regulating the AKT/mTOR pathway
- Development of a detection chip for major pathogenic drug-resistant genes and drug targets in bovine respiratory system diseases
- Exploration of the genetic influence of MYOT and MB genes on the plumage coloration of Muscovy ducks
- Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism
- Comparison of nutritional value of the wild and cultivated spiny loaches at three growth stages
- Transcriptomic analysis of liver immune response in Chinese spiny frog (Quasipaa spinosa) infected with Proteus mirabilis
- Disruption of BCAA degradation is a critical characteristic of diabetic cardiomyopathy revealed by integrated transcriptome and metabolome analysis
- Plant Sciences
- Effect of long-term in-row branch covering on soil microorganisms in pear orchards
- Photosynthetic physiological characteristics, growth performance, and element concentrations reveal the calcicole–calcifuge behaviors of three Camellia species
- Transcriptome analysis reveals the mechanism of NaHCO3 promoting tobacco leaf maturation
- Bioinformatics, expression analysis, and functional verification of allene oxide synthase gene HvnAOS1 and HvnAOS2 in qingke
- Water, nitrogen, and phosphorus coupling improves gray jujube fruit quality and yield
- Improving grape fruit quality through soil conditioner: Insights from RNA-seq analysis of Cabernet Sauvignon roots
- Role of Embinin in the reabsorption of nucleus pulposus in lumbar disc herniation: Promotion of nucleus pulposus neovascularization and apoptosis of nucleus pulposus cells
- Revealing the effects of amino acid, organic acid, and phytohormones on the germination of tomato seeds under salinity stress
- Combined effects of nitrogen fertilizer and biochar on the growth, yield, and quality of pepper
- Comprehensive phytochemical and toxicological analysis of Chenopodium ambrosioides (L.) fractions
- Impact of “3414” fertilization on the yield and quality of greenhouse tomatoes
- Exploring the coupling mode of water and fertilizer for improving growth, fruit quality, and yield of the pear in the arid region
- Metagenomic analysis of endophytic bacteria in seed potato (Solanum tuberosum)
- Antibacterial, antifungal, and phytochemical properties of Salsola kali ethanolic extract
- Exploring the hepatoprotective properties of citronellol: In vitro and in silico studies on ethanol-induced damage in HepG2 cells
- Enhanced osmotic dehydration of watermelon rind using honey–sucrose solutions: A study on pre-treatment efficacy and mass transfer kinetics
- Effects of exogenous 2,4-epibrassinolide on photosynthetic traits of 53 cowpea varieties under NaCl stress
- Comparative transcriptome analysis of maize (Zea mays L.) seedlings in response to copper stress
- An optimization method for measuring the stomata in cassava (Manihot esculenta Crantz) under multiple abiotic stresses
- Fosinopril inhibits Ang II-induced VSMC proliferation, phenotype transformation, migration, and oxidative stress through the TGF-β1/Smad signaling pathway
- Antioxidant and antimicrobial activities of Salsola imbricata methanolic extract and its phytochemical characterization
- Bioengineering and Biotechnology
- Absorbable calcium and phosphorus bioactive membranes promote bone marrow mesenchymal stem cells osteogenic differentiation for bone regeneration
- New advances in protein engineering for industrial applications: Key takeaways
- An overview of the production and use of Bacillus thuringiensis toxin
- Research progress of nanoparticles in diagnosis and treatment of hepatocellular carcinoma
- Bioelectrochemical biosensors for water quality assessment and wastewater monitoring
- PEI/MMNs@LNA-542 nanoparticles alleviate ICU-acquired weakness through targeted autophagy inhibition and mitochondrial protection
- Unleashing of cytotoxic effects of thymoquinone-bovine serum albumin nanoparticles on A549 lung cancer cells
- Erratum
- Erratum to “Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM”
- Erratum to “Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation”
- Retraction
- Retraction to “MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB”
- Retraction to “A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis”
- Special Issue on Advances in Neurodegenerative Disease Research and Treatment
- Transplantation of human neural stem cell prevents symptomatic motor behavior disability in a rat model of Parkinson’s disease
- Special Issue on Multi-omics
- Inflammasome complex genes with clinical relevance suggest potential as therapeutic targets for anti-tumor drugs in clear cell renal cell carcinoma
- Gastroesophageal varices in primary biliary cholangitis with anti-centromere antibody positivity: Early onset?
Articles in the same Issue
- Biomedical Sciences
- Constitutive and evoked release of ATP in adult mouse olfactory epithelium
- LARP1 knockdown inhibits cultured gastric carcinoma cell cycle progression and metastatic behavior
- PEGylated porcine–human recombinant uricase: A novel fusion protein with improved efficacy and safety for the treatment of hyperuricemia and renal complications
- Research progress on ocular complications caused by type 2 diabetes mellitus and the function of tears and blepharons
- The role and mechanism of esketamine in preventing and treating remifentanil-induced hyperalgesia based on the NMDA receptor–CaMKII pathway
- Brucella infection combined with Nocardia infection: A case report and literature review
- Detection of serum interleukin-18 level and neutrophil/lymphocyte ratio in patients with antineutrophil cytoplasmic antibody-associated vasculitis and its clinical significance
- Ang-1, Ang-2, and Tie2 are diagnostic biomarkers for Henoch-Schönlein purpura and pediatric-onset systemic lupus erythematous
- PTTG1 induces pancreatic cancer cell proliferation and promotes aerobic glycolysis by regulating c-myc
- Role of serum B-cell-activating factor and interleukin-17 as biomarkers in the classification of interstitial pneumonia with autoimmune features
- Effectiveness and safety of a mumps containing vaccine in preventing laboratory-confirmed mumps cases from 2002 to 2017: A meta-analysis
- Low levels of sex hormone-binding globulin predict an increased breast cancer risk and its underlying molecular mechanisms
- A case of Trousseau syndrome: Screening, detection and complication
- Application of the integrated airway humidification device enhances the humidification effect of the rabbit tracheotomy model
- Preparation of Cu2+/TA/HAP composite coating with anti-bacterial and osteogenic potential on 3D-printed porous Ti alloy scaffolds for orthopedic applications
- Aquaporin-8 promotes human dermal fibroblasts to counteract hydrogen peroxide-induced oxidative damage: A novel target for management of skin aging
- Current research and evidence gaps on placental development in iron deficiency anemia
- Single-nucleotide polymorphism rs2910829 in PDE4D is related to stroke susceptibility in Chinese populations: The results of a meta-analysis
- Pheochromocytoma-induced myocardial infarction: A case report
- Kaempferol regulates apoptosis and migration of neural stem cells to attenuate cerebral infarction by O‐GlcNAcylation of β-catenin
- Sirtuin 5 regulates acute myeloid leukemia cell viability and apoptosis by succinylation modification of glycine decarboxylase
- Apigenin 7-glucoside impedes hypoxia-induced malignant phenotypes of cervical cancer cells in a p16-dependent manner
- KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1
- Current state of research on copper complexes in the treatment of breast cancer
- Exploring antioxidant strategies in the pathogenesis of ALS
- Helicobacter pylori causes gastric dysbacteriosis in chronic gastritis patients
- IL-33/soluble ST2 axis is associated with radiation-induced cardiac injury
- The predictive value of serum NLR, SII, and OPNI for lymph node metastasis in breast cancer patients with internal mammary lymph nodes after thoracoscopic surgery
- Carrying SNP rs17506395 (T > G) in TP63 gene and CCR5Δ32 mutation associated with the occurrence of breast cancer in Burkina Faso
- P2X7 receptor: A receptor closely linked with sepsis-associated encephalopathy
- Probiotics for inflammatory bowel disease: Is there sufficient evidence?
- Identification of KDM4C as a gene conferring drug resistance in multiple myeloma
- Microbial perspective on the skin–gut axis and atopic dermatitis
- Thymosin α1 combined with XELOX improves immune function and reduces serum tumor markers in colorectal cancer patients after radical surgery
- Highly specific vaginal microbiome signature for gynecological cancers
- Sample size estimation for AQP4-IgG seropositive optic neuritis: Retinal damage detection by optical coherence tomography
- The effects of SDF-1 combined application with VEGF on femoral distraction osteogenesis in rats
- Fabrication and characterization of gold nanoparticles using alginate: In vitro and in vivo assessment of its administration effects with swimming exercise on diabetic rats
- Mitigating digestive disorders: Action mechanisms of Mediterranean herbal active compounds
- Distribution of CYP2D6 and CYP2C19 gene polymorphisms in Han and Uygur populations with breast cancer in Xinjiang, China
- VSP-2 attenuates secretion of inflammatory cytokines induced by LPS in BV2 cells by mediating the PPARγ/NF-κB signaling pathway
- Factors influencing spontaneous hypothermia after emergency trauma and the construction of a predictive model
- Long-term administration of morphine specifically alters the level of protein expression in different brain regions and affects the redox state
- Application of metagenomic next-generation sequencing technology in the etiological diagnosis of peritoneal dialysis-associated peritonitis
- Clinical diagnosis, prevention, and treatment of neurodyspepsia syndrome using intelligent medicine
- Case report: Successful bronchoscopic interventional treatment of endobronchial leiomyomas
- Preliminary investigation into the genetic etiology of short stature in children through whole exon sequencing of the core family
- Cystic adenomyoma of the uterus: Case report and literature review
- Mesoporous silica nanoparticles as a drug delivery mechanism
- Dynamic changes in autophagy activity in different degrees of pulmonary fibrosis in mice
- Vitamin D deficiency and inflammatory markers in type 2 diabetes: Big data insights
- Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells
- Meta-analysis on the efficacy of allogeneic hematopoietic stem cell transplantation to treat malignant lymphoma
- Mitochondrial DNA drives neuroinflammation through the cGAS-IFN signaling pathway in the spinal cord of neuropathic pain mice
- Application value of artificial intelligence algorithm-based magnetic resonance multi-sequence imaging in staging diagnosis of cervical cancer
- Embedded monitoring system and teaching of artificial intelligence online drug component recognition
- Investigation into the association of FNDC1 and ADAMTS12 gene expression with plumage coloration in Muscovy ducks
- Yak meat content in feed and its impact on the growth of rats
- A rare case of Richter transformation with breast involvement: A case report and literature review
- First report of Nocardia wallacei infection in an immunocompetent patient in Zhejiang province
- Rhodococcus equi and Brucella pulmonary mass in immunocompetent: A case report and literature review
- Downregulation of RIP3 ameliorates the left ventricular mechanics and function after myocardial infarction via modulating NF-κB/NLRP3 pathway
- Evaluation of the role of some non-enzymatic antioxidants among Iraqi patients with non-alcoholic fatty liver disease
- The role of Phafin proteins in cell signaling pathways and diseases
- Ten-year anemia as initial manifestation of Castleman disease in the abdominal cavity: A case report
- Coexistence of hereditary spherocytosis with SPTB P.Trp1150 gene variant and Gilbert syndrome: A case report and literature review
- Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells
- Exploratory evaluation supported by experimental and modeling approaches of Inula viscosa root extract as a potent corrosion inhibitor for mild steel in a 1 M HCl solution
- Imaging manifestations of ductal adenoma of the breast: A case report
- Gut microbiota and sleep: Interaction mechanisms and therapeutic prospects
- Isomangiferin promotes the migration and osteogenic differentiation of rat bone marrow mesenchymal stem cells
- Prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer
- Circular RNAs as potential biomarkers for male severe sepsis
- Knockdown of Stanniocalcin-1 inhibits growth and glycolysis in oral squamous cell carcinoma cells
- The expression and biological role of complement C1s in esophageal squamous cell carcinoma
- A novel GNAS mutation in pseudohypoparathyroidism type 1a with articular flexion deformity: A case report
- Predictive value of serum magnesium levels for prognosis in patients with non-small cell lung cancer undergoing EGFR-TKI therapy
- HSPB1 alleviates acute-on-chronic liver failure via the P53/Bax pathway
- IgG4-related disease complicated by PLA2R-associated membranous nephropathy: A case report
- Baculovirus-mediated endostatin and angiostatin activation of autophagy through the AMPK/AKT/mTOR pathway inhibits angiogenesis in hepatocellular carcinoma
- Metformin mitigates osteoarthritis progression by modulating the PI3K/AKT/mTOR signaling pathway and enhancing chondrocyte autophagy
- Evaluation of the activity of antimicrobial peptides against bacterial vaginosis
- Atypical presentation of γ/δ mycosis fungoides with an unusual phenotype and SOCS1 mutation
- Analysis of the microecological mechanism of diabetic kidney disease based on the theory of “gut–kidney axis”: A systematic review
- Omega-3 fatty acids prevent gestational diabetes mellitus via modulation of lipid metabolism
- Refractory hypertension complicated with Turner syndrome: A case report
- Interaction of ncRNAs and the PI3K/AKT/mTOR pathway: Implications for osteosarcoma
- Association of low attenuation area scores with pulmonary function and clinical prognosis in patients with chronic obstructive pulmonary disease
- Long non-coding RNAs in bone formation: Key regulators and therapeutic prospects
- The deubiquitinating enzyme USP35 regulates the stability of NRF2 protein
- Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as potential diagnostic markers for rebleeding in patients with esophagogastric variceal bleeding
- G protein-coupled receptor 1 participating in the mechanism of mediating gestational diabetes mellitus by phosphorylating the AKT pathway
- LL37-mtDNA regulates viability, apoptosis, inflammation, and autophagy in lipopolysaccharide-treated RLE-6TN cells by targeting Hsp90aa1
- The analgesic effect of paeoniflorin: A focused review
- Chemical composition’s effect on Solanum nigrum Linn.’s antioxidant capacity and erythrocyte protection: Bioactive components and molecular docking analysis
- Knockdown of HCK promotes HREC cell viability and inner blood–retinal barrier integrity by regulating the AMPK signaling pathway
- The role of rapamycin in the PINK1/Parkin signaling pathway in mitophagy in podocytes
- Laryngeal non-Hodgkin lymphoma: Report of four cases and review of the literature
- Clinical value of macrogenome next-generation sequencing on infections
- Overview of dendritic cells and related pathways in autoimmune uveitis
- TAK-242 alleviates diabetic cardiomyopathy via inhibiting pyroptosis and TLR4/CaMKII/NLRP3 pathway
- Hypomethylation in promoters of PGC-1α involved in exercise-driven skeletal muscular alterations in old age
- Profile and antimicrobial susceptibility patterns of bacteria isolated from effluents of Kolladiba and Debark hospitals
- The expression and clinical significance of syncytin-1 in serum exosomes of hepatocellular carcinoma patients
- A histomorphometric study to evaluate the therapeutic effects of biosynthesized silver nanoparticles on the kidneys infected with Plasmodium chabaudi
- PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
- Analysis of MDA, SOD, TAOC, MNCV, SNCV, and TSS scores in patients with diabetes peripheral neuropathy
- SLIT3 deficiency promotes non-small cell lung cancer progression by modulating UBE2C/WNT signaling
- The relationship between TMCO1 and CALR in the pathological characteristics of prostate cancer and its effect on the metastasis of prostate cancer cells
- Heterogeneous nuclear ribonucleoprotein K is a potential target for enhancing the chemosensitivity of nasopharyngeal carcinoma
- PHB2 alleviates retinal pigment epithelium cell fibrosis by suppressing the AGE–RAGE pathway
- Anti-γ-aminobutyric acid-B receptor autoimmune encephalitis with syncope as the initial symptom: Case report and literature review
- Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua
- Human umbilical cord mesenchymal stem cells regulate glutathione metabolism depending on the ERK–Nrf2–HO-1 signal pathway to repair phosphoramide mustard-induced ovarian cancer cells
- Electroacupuncture on GB acupoints improves osteoporosis via the estradiol–PI3K–Akt signaling pathway
- Renalase protects against podocyte injury by inhibiting oxidative stress and apoptosis in diabetic nephropathy
- Review: Dicranostigma leptopodum: A peculiar plant of Papaveraceae
- Combination effect of flavonoids attenuates lung cancer cell proliferation by inhibiting the STAT3 and FAK signaling pathway
- Renal microangiopathy and immune complex glomerulonephritis induced by anti-tumour agents: A case report
- Correlation analysis of AVPR1a and AVPR2 with abnormal water and sodium and potassium metabolism in rats
- Gastrointestinal health anti-diarrheal mixture relieves spleen deficiency-induced diarrhea through regulating gut microbiota
- Myriad factors and pathways influencing tumor radiotherapy resistance
- Exploring the effects of culture conditions on Yapsin (YPS) gene expression in Nakaseomyces glabratus
- Screening of prognostic core genes based on cell–cell interaction in the peripheral blood of patients with sepsis
- Coagulation factor II thrombin receptor as a promising biomarker in breast cancer management
- Ileocecal mucinous carcinoma misdiagnosed as incarcerated hernia: A case report
- Methyltransferase like 13 promotes malignant behaviors of bladder cancer cells through targeting PI3K/ATK signaling pathway
- The debate between electricity and heat, efficacy and safety of irreversible electroporation and radiofrequency ablation in the treatment of liver cancer: A meta-analysis
- ZAG promotes colorectal cancer cell proliferation and epithelial–mesenchymal transition by promoting lipid synthesis
- Baicalein inhibits NLRP3 inflammasome activation and mitigates placental inflammation and oxidative stress in gestational diabetes mellitus
- Impact of SWCNT-conjugated senna leaf extract on breast cancer cells: A potential apoptotic therapeutic strategy
- MFAP5 inhibits the malignant progression of endometrial cancer cells in vitro
- Major ozonated autohemotherapy promoted functional recovery following spinal cord injury in adult rats via the inhibition of oxidative stress and inflammation
- Axodendritic targeting of TAU and MAP2 and microtubule polarization in iPSC-derived versus SH-SY5Y-derived human neurons
- Differential expression of phosphoinositide 3-kinase/protein kinase B and Toll-like receptor/nuclear factor kappa B signaling pathways in experimental obesity Wistar rat model
- The therapeutic potential of targeting Oncostatin M and the interleukin-6 family in retinal diseases: A comprehensive review
- BA inhibits LPS-stimulated inflammatory response and apoptosis in human middle ear epithelial cells by regulating the Nf-Kb/Iκbα axis
- Role of circRMRP and circRPL27 in chronic obstructive pulmonary disease
- Investigating the role of hyperexpressed HCN1 in inducing myocardial infarction through activation of the NF-κB signaling pathway
- Characterization of phenolic compounds and evaluation of anti-diabetic potential in Cannabis sativa L. seeds: In vivo, in vitro, and in silico studies
- Quantitative immunohistochemistry analysis of breast Ki67 based on artificial intelligence
- Ecology and Environmental Science
- Screening of different growth conditions of Bacillus subtilis isolated from membrane-less microbial fuel cell toward antimicrobial activity profiling
- Degradation of a mixture of 13 polycyclic aromatic hydrocarbons by commercial effective microorganisms
- Evaluation of the impact of two citrus plants on the variation of Panonychus citri (Acari: Tetranychidae) and beneficial phytoseiid mites
- Prediction of present and future distribution areas of Juniperus drupacea Labill and determination of ethnobotany properties in Antalya Province, Türkiye
- Population genetics of Todarodes pacificus (Cephalopoda: Ommastrephidae) in the northwest Pacific Ocean via GBS sequencing
- A comparative analysis of dendrometric, macromorphological, and micromorphological characteristics of Pistacia atlantica subsp. atlantica and Pistacia terebinthus in the middle Atlas region of Morocco
- Macrofungal sporocarp community in the lichen Scots pine forests
- Assessing the proximate compositions of indigenous forage species in Yemen’s pastoral rangelands
- Food Science
- Gut microbiota changes associated with low-carbohydrate diet intervention for obesity
- Reexamination of Aspergillus cristatus phylogeny in dark tea: Characteristics of the mitochondrial genome
- Differences in the flavonoid composition of the leaves, fruits, and branches of mulberry are distinguished based on a plant metabolomics approach
- Investigating the impact of wet rendering (solventless method) on PUFA-rich oil from catfish (Clarias magur) viscera
- Non-linear associations between cardiovascular metabolic indices and metabolic-associated fatty liver disease: A cross-sectional study in the US population (2017–2020)
- Knockdown of USP7 alleviates atherosclerosis in ApoE-deficient mice by regulating EZH2 expression
- Utility of dairy microbiome as a tool for authentication and traceability
- Agriculture
- Enhancing faba bean (Vicia faba L.) productivity through establishing the area-specific fertilizer rate recommendation in southwest Ethiopia
- Impact of novel herbicide based on synthetic auxins and ALS inhibitor on weed control
- Perspectives of pteridophytes microbiome for bioremediation in agricultural applications
- Fertilizer application parameters for drip-irrigated peanut based on the fertilizer effect function established from a “3414” field trial
- Improving the productivity and profitability of maize (Zea mays L.) using optimum blended inorganic fertilization
- Application of leaf multispectral analyzer in comparison to hyperspectral device to assess the diversity of spectral reflectance indices in wheat genotypes
- Animal Sciences
- Knockdown of ANP32E inhibits colorectal cancer cell growth and glycolysis by regulating the AKT/mTOR pathway
- Development of a detection chip for major pathogenic drug-resistant genes and drug targets in bovine respiratory system diseases
- Exploration of the genetic influence of MYOT and MB genes on the plumage coloration of Muscovy ducks
- Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism
- Comparison of nutritional value of the wild and cultivated spiny loaches at three growth stages
- Transcriptomic analysis of liver immune response in Chinese spiny frog (Quasipaa spinosa) infected with Proteus mirabilis
- Disruption of BCAA degradation is a critical characteristic of diabetic cardiomyopathy revealed by integrated transcriptome and metabolome analysis
- Plant Sciences
- Effect of long-term in-row branch covering on soil microorganisms in pear orchards
- Photosynthetic physiological characteristics, growth performance, and element concentrations reveal the calcicole–calcifuge behaviors of three Camellia species
- Transcriptome analysis reveals the mechanism of NaHCO3 promoting tobacco leaf maturation
- Bioinformatics, expression analysis, and functional verification of allene oxide synthase gene HvnAOS1 and HvnAOS2 in qingke
- Water, nitrogen, and phosphorus coupling improves gray jujube fruit quality and yield
- Improving grape fruit quality through soil conditioner: Insights from RNA-seq analysis of Cabernet Sauvignon roots
- Role of Embinin in the reabsorption of nucleus pulposus in lumbar disc herniation: Promotion of nucleus pulposus neovascularization and apoptosis of nucleus pulposus cells
- Revealing the effects of amino acid, organic acid, and phytohormones on the germination of tomato seeds under salinity stress
- Combined effects of nitrogen fertilizer and biochar on the growth, yield, and quality of pepper
- Comprehensive phytochemical and toxicological analysis of Chenopodium ambrosioides (L.) fractions
- Impact of “3414” fertilization on the yield and quality of greenhouse tomatoes
- Exploring the coupling mode of water and fertilizer for improving growth, fruit quality, and yield of the pear in the arid region
- Metagenomic analysis of endophytic bacteria in seed potato (Solanum tuberosum)
- Antibacterial, antifungal, and phytochemical properties of Salsola kali ethanolic extract
- Exploring the hepatoprotective properties of citronellol: In vitro and in silico studies on ethanol-induced damage in HepG2 cells
- Enhanced osmotic dehydration of watermelon rind using honey–sucrose solutions: A study on pre-treatment efficacy and mass transfer kinetics
- Effects of exogenous 2,4-epibrassinolide on photosynthetic traits of 53 cowpea varieties under NaCl stress
- Comparative transcriptome analysis of maize (Zea mays L.) seedlings in response to copper stress
- An optimization method for measuring the stomata in cassava (Manihot esculenta Crantz) under multiple abiotic stresses
- Fosinopril inhibits Ang II-induced VSMC proliferation, phenotype transformation, migration, and oxidative stress through the TGF-β1/Smad signaling pathway
- Antioxidant and antimicrobial activities of Salsola imbricata methanolic extract and its phytochemical characterization
- Bioengineering and Biotechnology
- Absorbable calcium and phosphorus bioactive membranes promote bone marrow mesenchymal stem cells osteogenic differentiation for bone regeneration
- New advances in protein engineering for industrial applications: Key takeaways
- An overview of the production and use of Bacillus thuringiensis toxin
- Research progress of nanoparticles in diagnosis and treatment of hepatocellular carcinoma
- Bioelectrochemical biosensors for water quality assessment and wastewater monitoring
- PEI/MMNs@LNA-542 nanoparticles alleviate ICU-acquired weakness through targeted autophagy inhibition and mitochondrial protection
- Unleashing of cytotoxic effects of thymoquinone-bovine serum albumin nanoparticles on A549 lung cancer cells
- Erratum
- Erratum to “Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM”
- Erratum to “Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation”
- Retraction
- Retraction to “MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB”
- Retraction to “A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis”
- Special Issue on Advances in Neurodegenerative Disease Research and Treatment
- Transplantation of human neural stem cell prevents symptomatic motor behavior disability in a rat model of Parkinson’s disease
- Special Issue on Multi-omics
- Inflammasome complex genes with clinical relevance suggest potential as therapeutic targets for anti-tumor drugs in clear cell renal cell carcinoma
- Gastroesophageal varices in primary biliary cholangitis with anti-centromere antibody positivity: Early onset?


