Abstract
This study aimed to determine the prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer (HER2+ BC). Transcriptome sequencing and clinicopathological data were downloaded from the Cancer Genome Atlas. The 10X single cell sequencing dataset was downloaded from the National Center for Biotechnology Information Gene Expression Omnibus. Exosomes-Related Genes were extracted from the ExoCarta and Gene Set Enrichment Analysis databases. FGF9, SF3B4, and EPCAM were found and deemed the most accurate predictive signatures. Patients with HER2+ BC were subtyped into three groupings by exosome prognostic gene (EPGs). The expression of SF3B4 was positively linked with the infiltration of macrophages, neutrophils, and CD4+ T cells. The expression characteristics of EPGs were associated with the biological process of “response to xenobiotic stimuli.” Interactions were relatively high between malignant epithelial cells and fibroblasts, endothelial cells, monocytes, and macrophages. Malignant epithelial cells interact more with fibroblasts and endothelial cells. The migration inhibitory factor pathway was the primary outgoing signaling pattern, while the C-C motif chemokine ligand pathway was the primary incoming signaling pattern for communication between malignant epithelial cells and macrophages. This study described the role of exosome signatures in the prognosis and microenvironment of HER2+ BC and provided a basis for future research.
1 Introduction
Breast cancer is the most prevalent female malignant tumor worldwide. It is predicted that there will be 300,590 new cases of breast cancer and 43,700 deaths in the United States alone in 2023 [1]. The number of new breast cancer cases in China in 2022 was estimated to be 420,000, with breast cancer ranking first and fourth in morbidity and mortality, respectively, among the Chinese population [2]. The overexpressed human epidermal growth factor receptor 2 positive breast cancer (HER2+ BC) comprises 15–20% of breast cancer cases and displays sensitivity to anti-HER2-targeted medicines, such as transtuzumab (binding specifically to the HER2 receptor and preventing its activation), pertuzumab (blocking the dimerization of HER2 with other growth factor receptors), lapatinib (inhibiting directly the internal kinase activity of HER2 and epidermal growth factor receptor), and T-DM1 (allowing tertuzumab to deliver chemotherapy directly to HER2-overexpressing tumor cells) [3]. Despite the efficacy of these drugs, primary and secondary resistance frequently develop.
Since HER2+ BC is characterized by dense immunological cell infiltration and reduced immune exhaustion [4], immune checkpoint inhibitors combined with standard anti-HER2 drugs have demonstrated success in the treatment of HER2+ BC [5]. Exosomes are extracellular vesicles that are 30–150 nm in diameter and originate from the multivesicular endosome pathway. Exosomes are composed of a lipid bilayer and contain DNA, RNA, proteins, and lipids from their parent cells. Exosomes are involved in the internal signal interaction, tumor metastasis, extracellular matrix reconstruction, tumor microenvironment control (cell communication and signaling, immunoregulation, extracellular matrix remodeling, angiogenesis, promoting tumor drug resistance), and drug resistance [6]. Exosome-related genes (ERG) have shown important value in the diagnosis and prognosis evaluation of breast cancer. They affect the tumor microenvironment and cancer progression by transmitting information between cells. HER2+ BC is a specific subtype characterized by overexpression of HER2, which is associated with specific clinicopathological features and treatment response. Understanding the role of ERGs in HER2+ BC has potential implications for developing new therapeutic strategies and improving patient outcomes. Ciravolo et al. found that HER2-overexpressing breast cancer cell lines exhibited resistance to trastuzumab by releasing HER2-expressed exosomes into the extracellular environment [7]. Barok et al. discovered that T-DM1 may bind to HER2+ BC-derived exosomes and be transferred to additional cancer cells by the exosomes, thereby inhibiting the survival of target cells [8]. Limoni et al. created a drug delivery method by engineering exosomes and delivering siRNA to HER2+ BC cells [9].
Thus, we hypothesize that exosomes may serve as a predictive biomarker for HER2+ BC and play a crucial role in the interaction between HER2+ BC and the microenvironment. However, there are insufficient reports on this topic.
In this study, we identified the differentially expressed genes (DEGs) in HER2+ BC and ERGs, determined the exosome prognostic genes (EPGs) via Kaplan–Meier (KM) analysis, LASSO Cox regression, and random forest model, performed the clustering analysis, mutation feature analysis, enrichment analysis, and determined the relationship between EPGs and the immune microenvironment using a single cell RNA-sequencing dataset containing five clinical samples. Our findings not only validated the markers that predict the clinical prognosis of HER2+ BC from the perspective of exosomes, but also provide a research foundation for establishing the link between exosome signatures and the immunological microenvironment in HER2+ BC.
2 Methods
2.1 Raw data acquisition
We acquired the breast cancer transcriptome sequencing data and clinicopathological information from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) database [10] through the R program TCGAbiolink [11]. All downloaded sample data met the following criteria: (a) cases with complete mRNA expression data and clinical data: ensure that each case selected for the study has sufficient information for subsequent statistical analysis and biological interpretation; (b) by pathological diagnosis of breast cancer: ensure that the research focuses on a specific disease group, and to enhance the pertinence and accuracy of the results; (c) cases accepted standardized treatment of breast cancer, including surgery, chemotherapy, and radiotherapy: select standard treatment cases, helps to reduce the deviation caused by different treatment results; (d) cases of survival time for more than 30 days: eliminate survival cases in a very short time, these cases may be unrepresentative due to special circumstances; and (e) HER2+ BC samples can be extracted from all datasets: focus on HER2-positive breast cancer, a patient population with specific biological characteristics and treatment response. The transcriptome data were reannotated using an annotation file from UCSC Xena (https://xenabrowser.net/datapages) for future analysis [12]. Finally, 74 cases of HER2+ BC and 99 cases of precancerous normal samples were included. Somatic mutation data on HER2+ BC (n = 74) were collected from “Masked Somatic Mutation” on the TCGA GDC website (https://portal.gdc.cancer.gov/), preprocessed with VarScan software, and visualized using maftools R package [13]. The single cell sequencing (scRNA seq) dataset for combined analysis was downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) [14]. GSE176078 was derived from human DNA and detected on the GPL18573 IIIumina NextSeq 500. The data derived from the primary lesion of 26 patients with breast cancer, including five patients with HER2+ BC were assessed in this study (Table 1). ERGs were referenced from the ExoCarta database [15] and the gene set enrichment analysis (GSEA, https://www.gsea-msigdb.org/) [16]. We enlisted a total of 121 ERGs after combination and de-duplication (Table S1).
Published transcriptome database information
| TCGA-BRCA (HER2+) | GSE176078 | |
|---|---|---|
| Platform | Illumina | GPL18573 |
| Species | Homo sapiens | Homo sapiens |
| Tissue | Breast cancer | Breast cancer |
| Samples in cancer group | 74 | 5 |
| Samples in normal group | 99 | 0 |
| Reference | — | PMID: 34493872 |
2.2 Gene set variation analysis (GSVA)
GSVA was performed on HER2+ BC from TCGA-BRCA using the R GSVA package (version 1.42.0) [17].
2.3 Identification of exosome-related differentially expressed genes (ER-DEGs) in HER2+ BC
The R limma package was used to screen DEGs between HER2+ BC samples and precancerous TCGA-BRCA samples. The absolute value of Log2 [Fold change] (Log2 FC) > 1 and p < 0.05 were set as the threshold of DEGs. The genes with Log2 FC > 1 and p < 0.05 were DEGs with up-regulated expression, while the genes with Log2 FC < −1 and p < 0.05 were DEGs with down-regulated expression. The DEGs were displayed using volcano plots and heat maps.
2.4 Identification of EPGs in HER2+ BC
Univariate and multivariate Cox regression analyses were first conducted to determine ER-DEGs, followed by the Least Absolute Shrinkage and Selection Operator (LASSO) regression and random forest model to filter the findings of multivariate Cox regression. LASSO Cox model is a statistical technique that combines the LASSO method with the Cox proportional hazard model, which is widely used in survival analysis, especially when dealing with high-dimensional data (such as gene expression data), aiming to identify variables (such as genes or biomarkers) that are significantly correlated with patient survival time. The candidates identified by the two models were plotted and intersected using a Venn Diagram. The dimensionality reduction screening was performed by 1,000 iterations with ten folds to cross-validate the LASSO Cox model [18]. The random forest model was simulated 1,000 times, and the significance of the final results was determined and then ranked. ERGs with an importance greater than 0.2 were chosen as the results. The intersection of genes screened by the two preceding models was considered as the EPGs in HER2+ BC. We created a Spearman’s analysis, a heat map, a scatter plot, and a correlation curve drawing using the R package cowplot. Correlation analysis of key genes was performed using the R package GOSemSim [19]. The chromosome localization map was drawn and displayed using the R package RCircos [20].
2.5 Mutation analysis of EPGs in HER2+ BC
The breast cancer MuTect2 file for Whole Exome Sequencing Mutation Annotation Format was obtained from the TCGA GDC website (https://portal.gdc.cancer.gov/). All non-synonymous mutations were chosen for downstream analysis. EPG mutations in HER2+ BC were exhibited through R package maftools [21].
2.6 Construction of prognostic predictive model by EPGs in HER2+ BC
The nomogram graphic, calibration analysis, and calibration curve drawing were completed using the R package rms [22]. Next, a calibration analysis was run and a calibration curve built to evaluate the accuracy and resolution of the nomogram. We created the time-dependent receiver operative characteristic (ROC) curves to assess the accuracy of the prognostic model for the 1-, 3-, and 5-year survival outcomes for HER2+ BC using the R package survivalROC [23]. Decision curve analysis (DCA) maps were created to evaluate the accuracy and resolution of Cox regression models using the R package ggDA [24].
2.7 Determination of exosome-related subgroups of HER2+ BC
Populations with varied exosome functional phenotypic characteristics based on previous EPGs in HER2+ BC were found using the consensus clustering method of CONSENSUSClusterPlus in R [25]. Principal coordinates analysis (PCoA) was performed to confirm the consistency clustering results [26]. The box charts were created using the R package ggpubr to group the sample clustering labels. The significant differences between the groups were discovered using the Wilcoxon rank-sum test, with p-values less than 0.05 considered statistically significant.
2.8 Evaluation of biological characteristics and tumor microenvironment of functional phenotypes of different exosomes in HER2+ BC
The Wilcoxon rank-sum test was performed to compare the difference in tumor mutational burden and microsatellite instability (MSI) between different exosome functional phenotypes in HER2+ BC, and the results are displayed in a box graph. The TIMER algorithm was used to assess the entrance status of immune cells, and the CIBERSORT algorithm was utilized to assess the difference in immune cell infiltration between HER2+ BC samples and normal samples [27]. The TIMER algorithm evaluates the status and activity of immune cells by estimating the proportion of each immune cell in the tumor microenvironment through a deconvolution method using specific immune cell marker genes. The CIBERSORT algorithm provides a powerful tool to explore immune infiltration and immune microenvironment in HER2+ BC and other cancers by analyzing a set of gene expression signatures linked to specific cell types.
Single sample gene set enrichment analysis (ssGSEA) was used to perform GSEA and estimate the relative abundance of each immune cell infiltration [28]. The distribution of immune cell infiltration in different risk groups and disease subtypes of HER2+ BC was displayed using the R package ggplot2. Correlation heatmaps were created to show the relationship between EPGs, key genes connected to exosomes, and immune cells in different risk categories using the R package heatmap.
2.9 Quality control, cluster analysis, cell annotation of single cell data, scoring of ERG sets, and analysis of intercellular communication networks
The Seurat objects for this analysis were created, and quality check was performed by importing the count matrix of five samples from the single cell dataset GSE176078 using the R package Seurat (version 4.0). The standards are listed as follows: (1) The number of genes expressed in cells could not be less than 250. (2) 20% or more unique molecular identifiers (UMIs) were located on mitochondrial or ribosome gene cells. (3) The outlier was filtered by cutting the gene expression to 2 × (mean ± standard deviation). If they met one of the criteria, they were eliminated. To check double cells, the R program DoubletFinder was used with the default parameter settings [29]. The batch effect between samples was corrected using the R package Harmony [30]. After quality standardization and conversion to Seurat objects, the first 2,000 genes with significant variations in expression levels were extracted using the FindVariableFeature function. Dimensionality reduction analysis was implemented using principal component analysis (PCA) based on the expression levels of the 2,000 previously selected genes. Cell clustering analysis was conducted using the FindClusters (with the resolution parameter set to 0.8) and FindAllMarkers functions. To identify the marker genes for each cluster, a cutoff threshold of p value <0.05 and a multiple of differences >0.5 were applied. Following the preceding stages, we obtained 48,785 cells. Standard cell types were annotated using the HumanPrimaryCellAtlasData dataset from the R package SingleR (version 1.8.1) [31]. Nine cell types were identified such as T cells, B cells, endothelial cells, epithelial cells, fibroblasts, monocytes, NK cells, dendritic cells (DC), and macrophages by referencing the expression patterns of the marker gene in the GSE176078 and CellMarker datasets [32,33]. Malignant epithelial cells were identified by changes in copy number variation using the CopyKat R program [34]. “Malignant epithelial cells” appraisal aims to identify cancer cells in single cell data and that for us to study the tumor microenvironment, the heterogeneity of cancer cells and immune cells around the interaction is of great significance. Exosomes based on the GSE176078 dataset for scoring were obtained using the Area Under the Curve Cell (AUCell) R package (version 1.12.0) [35]. The set of exosome genes was organized as input data for determining the value of the area under the curve (AUC). A gene expression ranking was established for each cell based on the AUC value. The AUC was used to estimate the proportion of highly expressed genes in each cell. The AUCell_ explore threshold function was used to generate a threshold for evaluating the activity of the present gene set. Then, based on the AUC score of each cell, Uniform Manifold Approximation and Projection (UMAP) encapsulation of the cell clusters was stained to reveal which cell clusters were active in the ERGs. Intercellular communication networks from single-cell RNA sequencing data were inferred statistically and examined using the R package CellChat [36]. Intended cell contact refers to the expected or potential cell-to-cell contact in the analysis of single-cell data, which is usually mediated by specific receptor–ligand pairings. Receptor and the ligand matching is one of the basic mechanisms of intercellular communication, through the combination of cell surface receptors and ligands, transmit signals to promote interaction between cells, the cell behavior and the regulation of physiological status is of great significance. The cell contact of HER2+ BC single cell subsets was depicted using a circle diagram. All receptor–ligand pairings during intercellular signal transduction were counted using bubble plots.
2.10 Statistical analysis
All data processing and analysis in this study were conducted using R (Version 4.2.0). Continuous variables are presented as the mean ± standard deviation. The comparison between the two groups was conducted using the Wilcoxon rank-sum test. Three or more groups were compared using the Kruskal–Wallis test. The Kruskal–Wallis test is a generalization of the Wilcoxon rank-sum test on groups of three or more independent samples. It was used to test multiple sample group median whether there was a statistically significant difference, which did not have to require the same data of normal distribution. This method is a non-parametric approach to assess whether multiple groups have the same distribution on a continuous variable. The chi-squared test or the Fisher’s exact test was utilized to compare and analyze the statistical significance between two sets of categorical variables. GSVA analysis was performed on the TCGA-BRCA samples obtained after integration through the EPGs. Exosome function-related scores of HER2+ BC in TCGA-BRCA were calculated. To investigate the association between exosome function and clinical phenotype, Wilcoxon analysis was used. KM analysis was used to evaluate the prognosis. Spearman’s correlation analysis was used to determine correlation coefficients between different genes. p Value of less than 0.05 was considered as the criterion for statistically significant differences.
3 Results
3.1 Identifying EPGs in patients with HER2+ BC
The flowchart of this study is depicted in Figure 1. In cases with HER2+ BC from the TCGA-BRCA dataset, we found 6,259 DEGs, of which, 3,252 were upregulated and 3,006 were downregulated between breast cancer and precancerous normal samples. The blue scatter points indicate highly expressed genes, whereas the red scatter points indicate low expressed genes in HER2+ BC (Figure 2a). Heat maps (Figure 2b) were used to visually display the expression of each gene in HER2+ BC, where blue indicates low expression and red indicates high expression. Through this gradient of color, the heat map revealed significant differences in gene expression levels in HER2+ BC compared to precancerous normal samples. Based on the intersection of ERGs and DEGs, we found 42 ER-DEGs, which are represented in a Venn diagram (Figure 2c). We performed survival analyses on patients with HER2+ BC and discovered that 8 of the 42 ER-DEGs had significant survival differences – they were EPGAM (p = 0.041), FGF9 (p = 0.017), POLR2K (p = 0.015), HRNR (p = 0.002), RANBP1 (p = 0.003), SF3B4 (p = 0.003), TAC1 (p = 0.024), and FZD5 (p = 0.001) (Figure 2d). To determine the association between ER-DEGs and overall survival of patients with HER2+ BC, we performed the univariate Cox regression in the subgroup of HER2+ BC from TCGA-BRCA and discovered that 9 of the 42 ER-DEGs were associated with clinical prognosis (p < 0.1). We conducted additional research on the nine genes using Lasso Cox regression and the random forest algorithm. The optimal Lambda value was employed in the LASSO model (Figure 2e). To display the LASSO Cox regression outcomes, the LASSO variable trace plot was used (Figure 2f). The nine ER-DEGs were ranked using the random forest variable importance values, indicating that the error rate reflecting survival reduced with different genes (Figure 2g). To discover the genes most associated with the prognosis of HER2+ BC, we chose genes with an importance greater than 0.2 as HER2+ BC prognosis-related genes (Figure 2h). Then, we integrated the two screening models, took their intersection, and discovered three HER2+ BC-related prognostic genes (FGF9, SF3B4, and EPCAM) (Figure 2i) that we dubbed EPGs. We drew a chromosomal map to reveal their location on a chromosome. FGF9 is found on chromosome 13, SF3B4 is found on chromosome 1, and EPCAM is found on chromosome 2 (Figure 2j).

Work flowchart of this study.
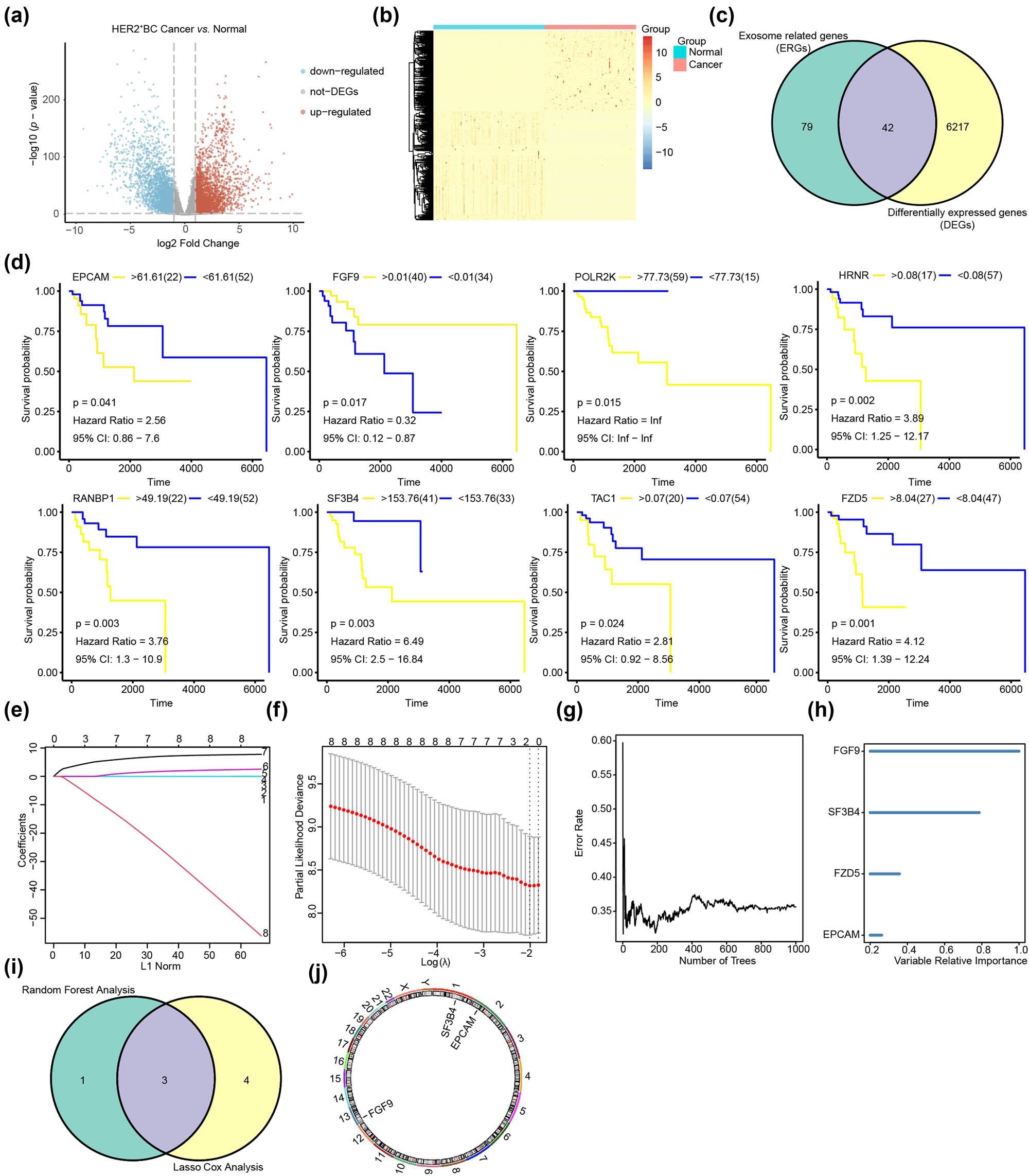
Screening of critical prognostic genes related to exosomes in patients with HER2+ BC. (a) DEGs in HER2+ BC. The abscissa of the volcanic map is log2FoldChange and the ordinate is log10 (p-value). The red node indicates up-regulated DEGs, the blue node represents down-regulated DEGs, and the gray node represents genes that are not significantly differentially expressed. (b) A heat map reveals DEGs in HER2+ BC. The blue bar indicates HER2+ BC samples, the pink bar indicates normal control samples, the red bar indicates high gene expression, and the blue bar indicates low gene expression. (c) The number of intersecting genes between ERGs and DEGs in HER2+ BC. (d) KM plots show eight ER-DEGs that exhibit significant differences in survival. (e) Diagnostic model diagram of ER-DEGs in the TCGA-BRCA dataset. (f) Variable trace plot of ER-DEGs in the LASSO-COX regression model. (g) Error rate of ER-DEGs in the random forest model. (h) Importance ranking diagram of ERGs in random forest model. (i) The Venn diagram depicting the EPGs identified in the HER2+ BC using random forest analysis and Lasso cox analysis. (j) Chromosomal mapping of EPGs.
3.2 Constructing a prognostic model based on EPGs in HER2+ BC
The correlation analysis results revealed that the correlation between EPCAM and FGF9 was −0.18, while the correlation between EPCAM and SF3B4 was 0.2. (Figure 3a). The survival of patients with HER2+ BC was associated with the EPGs (Figure 3b). To assess the prognostic efficacy of EPGs in patients with HER2+ BC, we developed a predictive prognostic model using multivariate Cox regression. We evaluated the prognostic value of the Cox regression model and illustrated the nomogram (Figure 3c), plotted the curve through calibration analysis, and evaluated the prediction effect of the model on the actual results based on the model’s fitting, to predict the 1-, 3-, and 5-year actual survival probabilities of patients with HER2+ BC (Figure 3d). We demonstrated the differentiation of the model in predicting the 1-, 3-, and 5-year survival probability of patients with HER2+ BC using time-dependent ROC curves. The results indicated that the model was optimal for predicting the 1-, 3-, and 5-year survival probabilities, with AUC values of 0.84 (70.77–96.78), 0.77 (61.62–93.11), and 0.78 (60.32–94.75). (Figure 3e). The role of the Cox regression model in clinical utility was examined using DCA (Figure 3f). When the line of the model stabilizes above All Positive and All Negative within a given range, the broader the range, the greater the net income and the better the performance of model.
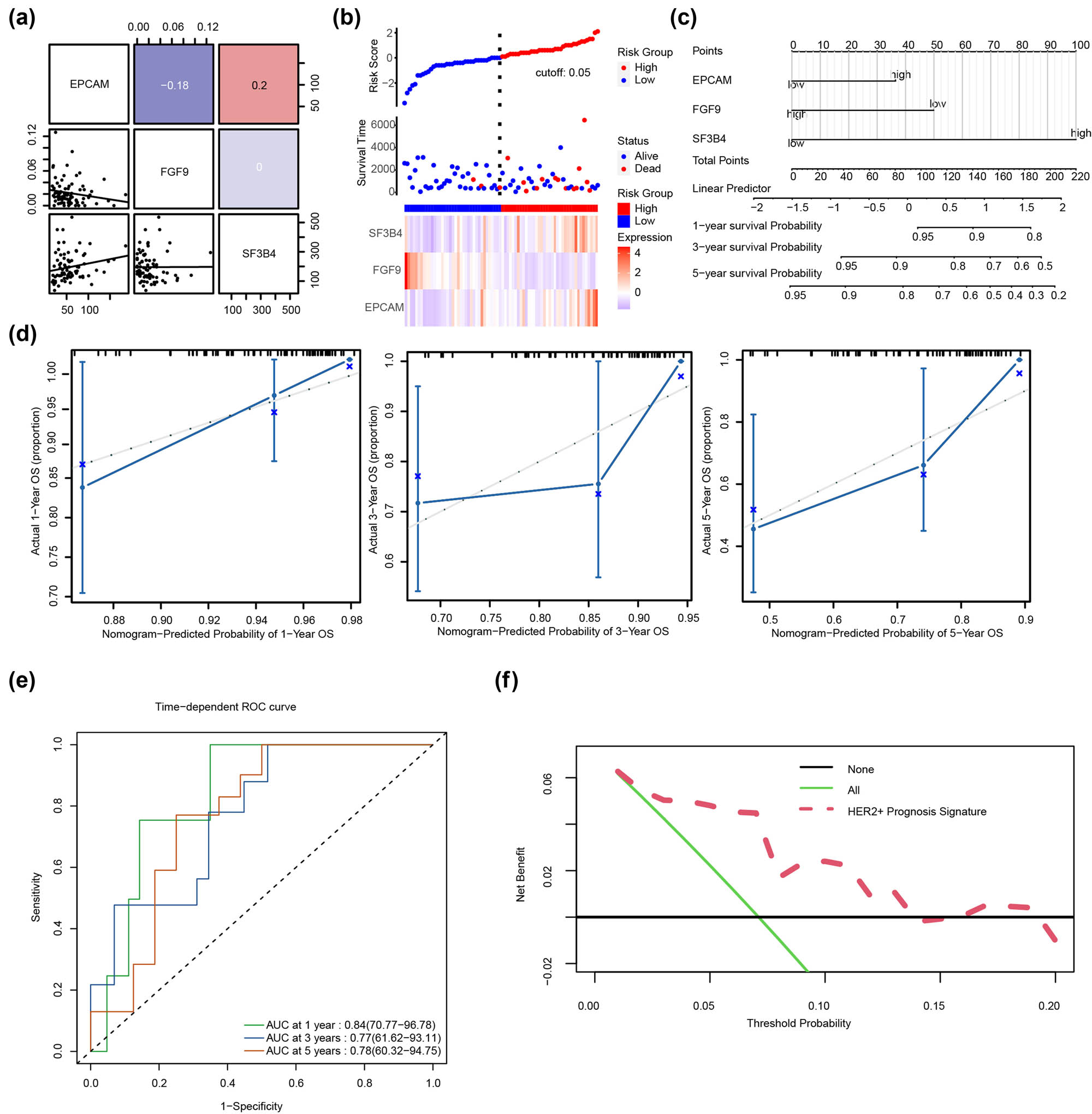
Constructing the predictive model using EPGs. (a) The link between EPGs in HER2+ BC. Red indicates a positive correlation, whereas blue indicates a negative correlation. (b) The relationship between EPGs and survival of patients with HER2+ BC. (c) The nomogram was created using Cox regression analysis of EPGs from the TCGA-BRCA dataset, specifically for HER2+ BC. The middle section reflects the expression levels of FGF9, SF3B4, and EPCAM, while the total score represents the total score of the patient based on the three gene scores. The total score correlates with the chance of occurrence listed below it. (d) The 1-, 3-, and 5-year recall curves revealed that the black diagonal dashed line representing the true sample situation and the blue solid line representing the predicted patient’s disease status by the prediction model overlapped in most cases. (e) The time-dependent ROC curve demonstrated the model’s ability to predict 1-, 3-, and 5-year survival. (f) The plot DCA of ERGs in the Cox regression model. The vertical axis indicates net income, and the horizontal axis represents probability threshold or threshold probability.
3.3 Three subtypes recognized by EPGs in HER2+ BC
We discovered three subgroups based on FGF9, SF3B4, and EPCAM using a consensus clustering approach (Figure 4a). The screen plot revealed that the optimal classification parameters could be identified using inflection points (Figure 4b). The cumulative distribution function indicated that three categories were more logical, as the rise slope of the consistency index was the lowest when consensus clustering was divided into three groups (Figure 4c). PCoA validated the features of three subtypes, and the results indicated the optimal classification stability of the three disease subtypes (Figure 4d). KM analysis revealed that the prognosis of patients with Cluster A subtype was better than that of patients with Cluster B or Cluster C with the p-value of 0.038 (Figure 4e). The Wilcoxon rank-sum test indicated that the degree of MSI in Cluster C was substantially greater than its degree in Cluster A (p < 0.05). (Figure 4f). ssGSEA analyses revealed substantially enriched biological signaling pathways in three subtypes of exonemes. The results showed that the PANCREAS BETA CELLS and COAGULATION pathways were highly enriched in Cluster A (Figure 4g). ALLOGRAFT REJECTION, IL6 JAK STAT3 SIGNALING, INTERFERON GAMMA RESPONSE, E2F TARGETS, and INFLAMMATORY RESPONSE were enriched in Cluster B (Figure 4h). P53 PATHWAY, XENOBIOTIC METABOLISM, OXIDATIVE PHOSPHORYLATION, MTORC1 SIGNALING, and UV RESPONSE UP were enriched in Cluster C (Figure 4i).
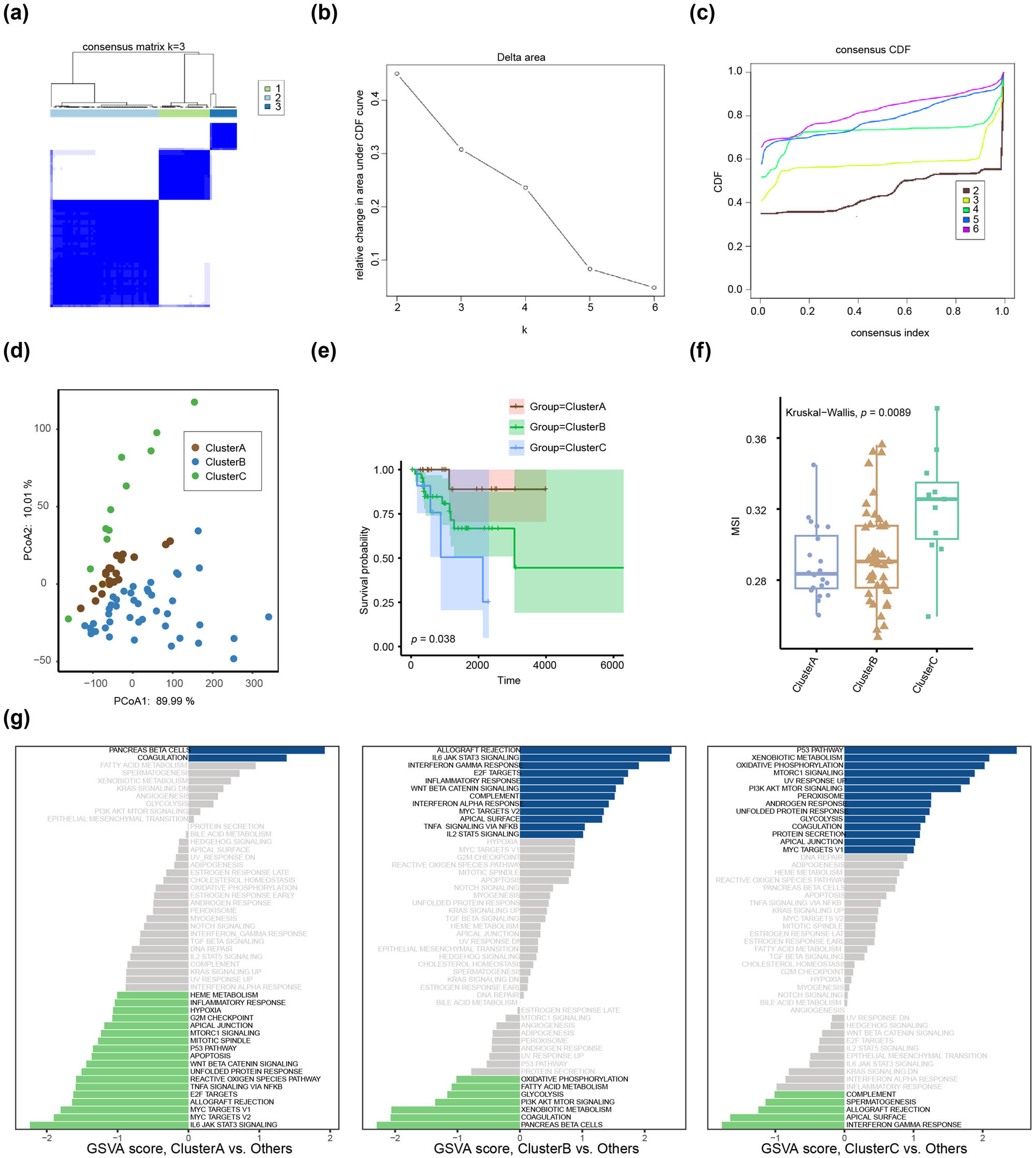
Unsupervised cluster analysis for TCGA-BRCA. (a) Consensus clustering based on EPGs (FGF9, SF3B4, and EPCAM TCGA) in the HER2+ BC dataset. (b) The scree plot according to cluster analysis. (c) Cumulative distribution function showed the consensus index under different K values. (d) PCoA analysis among the three clusters. (e) Survival curves showed the survival probability of patients in the three clusters. (f) The difference in MSI of patients in the three clusters. (g) Kyoto Encyclopedia of Genes and Genomes bifunctional enrichment analysis for Cluster A, Cluster B, and Cluster C.
3.4 Mutational signatures and gene druggability of EPGs in HER2+ BC subgroup
The mutation features of the exosome-related HER2+ BC subgroups were studied using the HER2+ BC in the TCGA-BRCA dataset and the R package maftools. The results showed that TP53, PIK3CA, TTN, and MUC16 had a high mutation frequency in HER2+ BC, of which TP53 was 79% (Figure 5a) in Cluster A, 67% (Figure 5b) in Cluster B, and 33% (Figure 5c) in Cluster C. The biological functional alterations caused by mutations in Cluster A and Cluster B were mainly concentrated in the TP53 signaling pathway, while they were mainly concentrated in the RTK-RAS signaling pathway in Cluster C subtype (Figure 5d). We explored the gene druggability and the interactions between drugs and genes based on the Drug Gene Interaction database and found that the gene predicting the possible action of drugs on patients in the Cluster A was TRANSCRIPTION FACTOR COMPLEX (TP53 et al.) (Figure 5e). The gene that predicted the potential effect of drugs on Cluster B was PHOSPHOLIPASSE (LRP1 et al.) (Figure 5f). The gene that predicted the potential effect of the drug on patients in Cluster C was SHORT CHAIN DEDROGENASE REDUCTASE (FASN et al.) (Figure 5g).
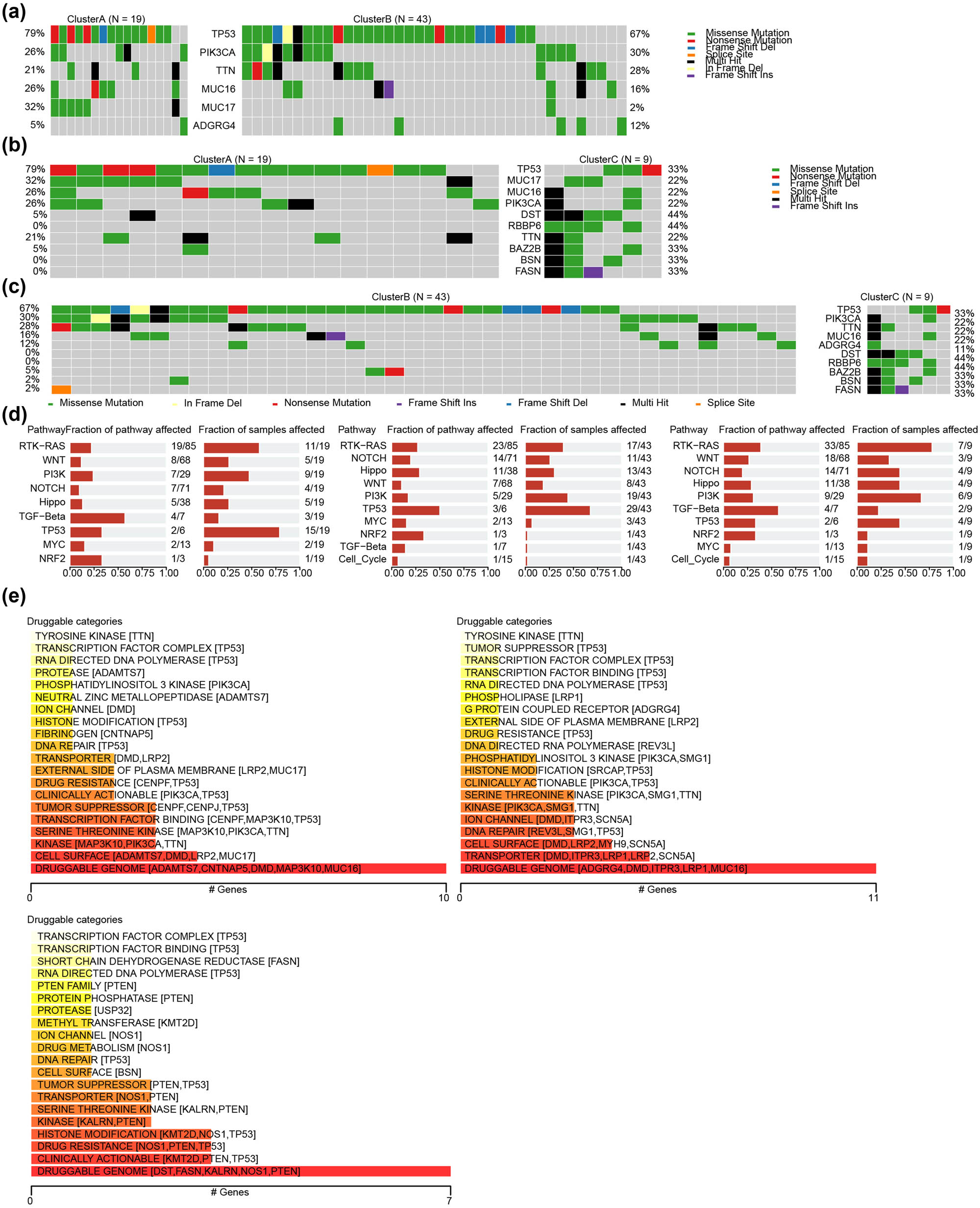
Mutation and gene druggability analysis for EPGs in the HER2+ BC subgroup. Comparing the landscape of mutations in ERGs among HER2+ BC subgroup in (a) Cluster A vs Cluster B, (b) Cluster A vs Cluster C), (c) Cluster B vs Cluster C. (d) Biological functional analysis affected by mutations in exosome-related HER2+ BC subgroup (left: Cluster A, middle: Cluster B; right: Cluster C). (e) The categories of potential role of druggability.
3.5 Immune microenvironment characteristics of EPGs in HER2+ BC subgroup
Based on the TCGA-BRCA dataset for HER2+ BC, we assessed the status of six types of immune cell infiltration and the immune cell infiltration among three subtypes using TIMER analysis and its algorithm. The results exhibited statistically significant differences in DC infiltration among three subtypes (p = 0.0001, Figure 6a). We analyzed the three subtypes in pairs and found that Cluster A differed significantly from Cluster B (p = 0.0032, Figure 6b). We performed correlation analyses between ERGs and the infiltration of various immune cells in HER2+ BC and discovered that DCs were positively correlated with the presence of FGF9-expressed instances (r = 0.37, p < 0.001, Figure 6c). Macrophages were positively correlated with the content of EPCAM expressed cases (r = 0.31, p = 0.01, Figure 6d). Macrophages (r = 0.29, p = 0.01, Figure 6e), neutrophils (r = 0.31, p = 0.01, Figure 6f), and CD4+ T cells (r = 0.35, p < 0.001, Figure 6g) were positively correlated with the content of SF3B4 expressed cases.
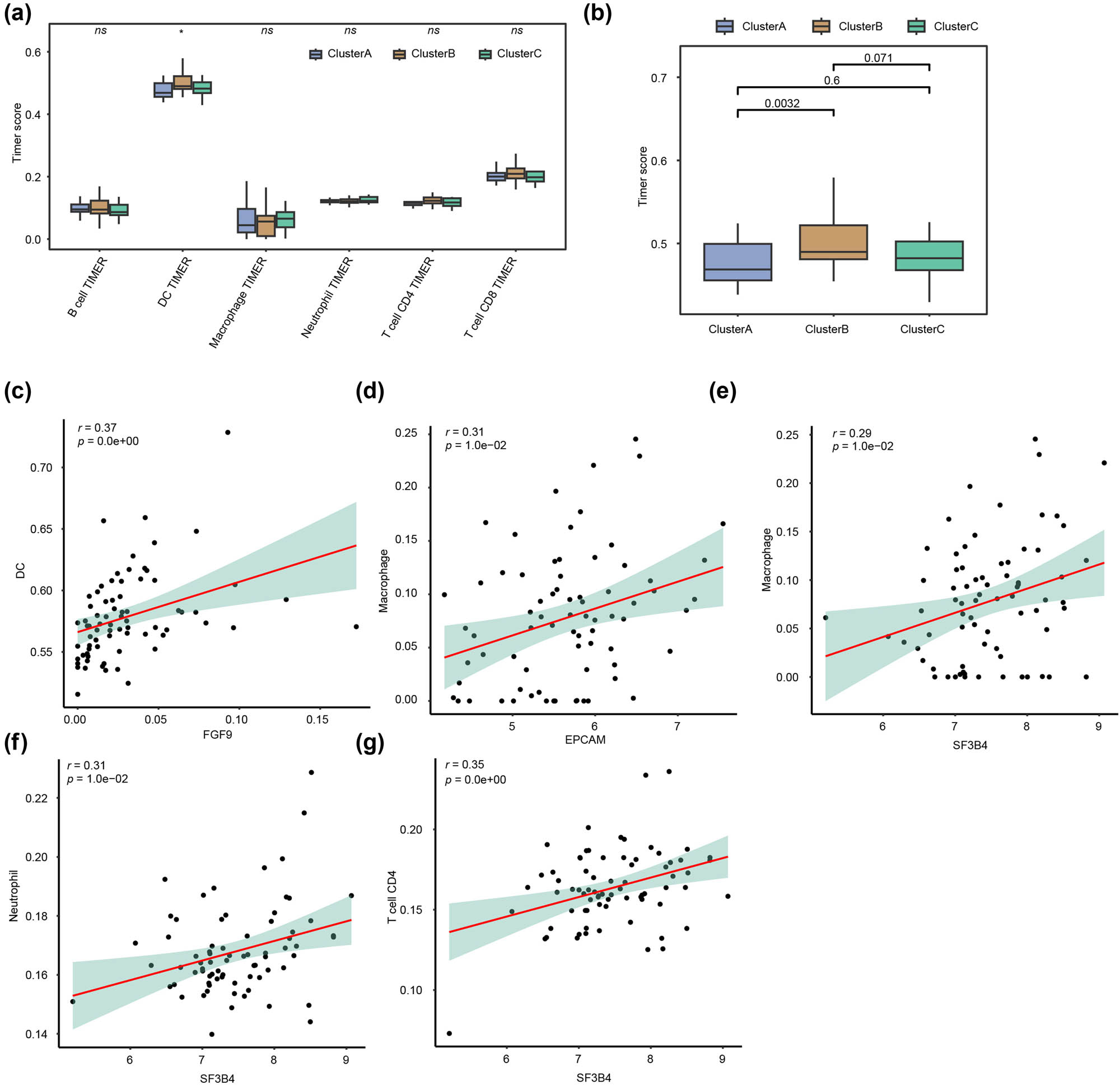
Immune infiltration analysis for EPGs in the HER2+ BC subgroup. (a). The box diagram of immune cell infiltration abundance in the exosome-related HER2+ BC subgroup. (b) Pair-wise comparisons of the box plot among the three subgroups. (c) Correlation analysis between FGF9 and DC infiltration abundance. (d) Correlation analysis between EPCAM and macrophage infiltration abundance. (e) Correlation analysis between SF3B4 and macrophage infiltration abundance. (f) Correlation analysis between SF3B4 and neutrophil infiltration abundance. (g) Correlation analysis between SF3B4 and CD4+ T cells infiltration abundance.
3.6 Validating the EPGs in 10X genomics single-cell data of BC
To validate the role of the three EPGs (EPCAM, FGF9, and SF3B4) in HER2+ BC in various cell types, we conducted validation analysis using 10× single cell dataset-GSE176078. Initially, we imported the count matrix of five samples from the single cell dataset using the Seurat (version 4.0.2) R package to generate the Seurat object for analysis. Based on the preliminary quality of the Cell Ranger, we did a second round of data quality control by preserving the following high-quality cells: gene numbers greater than 250, UMI numbers greater than 500, log10 genes per UMI greater than 0.8, mitochondrial UMI ratio less than 20%, and red blood cell gene ratio less than 5%. Then, we removed the doublet using the R package DoubletFinder. Following the previous steps, we obtained a total of 19,311 cells (Figure S1). We then standardized the sequencing depth of GSE176078 using the “Normalize Data” function, which defaults to “Log Normalize.” We identified 2,000 variable features of the dataset using the “vst” approach using the “Find Variable Features” function. Then, we used “Scale Data” to scale the data to exclude the effect of sequencing depth and used PCA with variable genes as input to identify significant principal components based on the Elbow Plot function. The top 20 principal components were chosen as statistically significant UMAP inputs.
3.7 Cluster analysis and cell type annotation in the single-cell dataset and scoring of EPGs in HER2+ BC subtypes
Following quality control of the HER2+ BC single cell dataset of GSE176078, we effectively categorized 19,311 cells into 29 independent cell clusters (Figure 7a). We annotated cells into nine distinct cell types through singleR and sample sources. Clusters 0, 1, 2, 3, 6, 10, 14, 15, and 28 were annotated as T cells (10,576, 54.77%); Clusters 12, 24, and 26 were annotated as B cells (919, 4.76%); Cluster 4 was annotated as endothelial cells (1128, 5.84%); Clusters 9, 13, 16, 17, 19, and 23 were annotated as epithelial cells (2821, 14.61%); Clusters 5, 7, 22, and 25 were annotated as fibroblasts (883, 4.57%); Cluster 27 was annotated as monocytes (477, 2.47%); Clusters 11, 20, and 21 were annotated as NK cells (1613, 8.35%); Cluster 8 was annotated as DCs (124, 0.64%); Clusters 8 and 18 were annotated as macrophages (882, 4.57%) (Figure 7b). Each sample appeared as a homogenous distribution (Figure 7c). The proportions of cell types were shown in each sample, with T cells accounting for a high proportion in each sample (Figure 7d). We screened the main marker genes of each cell type using the Cellmarker dataset and displayed them in dot plot format. EPCAM and KRT19 represent epithelial cells. PECAM1 and VWF represent endothelial cells. CD68 and CD163 represent macrophages. CD14 represents monocytes. CD3D and CD3E represent T cells. NKG7 represents NK cells. CD79A and CD79B represent B cells (Figure 7e). The DEGs between cell subtypes are listed in Table 2. Epithelial cells were classified as either malignant or non-malignant using CopyKat analysis, as depicted in Figure S2. To gain an understanding of the expression characteristics of ERGs in HER2+ BC, three identified EPGs were scored using the R program AUCell to compute the activity of exosome function in each cell based on the proportion of the highly expressed gene set in each cell. Cells expressing more genes from the gene set exhibit higher AUC values than those cells expressing fewer genes. We found two peaks in the AUC values of all cells. When the AUC threshold was set to 0.2, 3,434 cells showed relatively high AUC and 12,774 cells showed relatively low AUC (Figure 7f), through UMAP clustering analysis (Figure 7g) or box plot AUC comparison (Figure 7h; Figure S2, these cells are mainly distributed in malignant epithelial cells). Using gene ontology and pathway enrichment analysis to determine the functional characteristics of cell subsets in these malignant epithelial cells, we discovered that the expression properties of GRGs in HER2+ BC were primarily associated with RESPONSE TO XENOBIOTIC STIMULUS. The screening criteria were p < 0.05, and the top ten pathways were sorted by ES score (Figure 7i, Table 3)
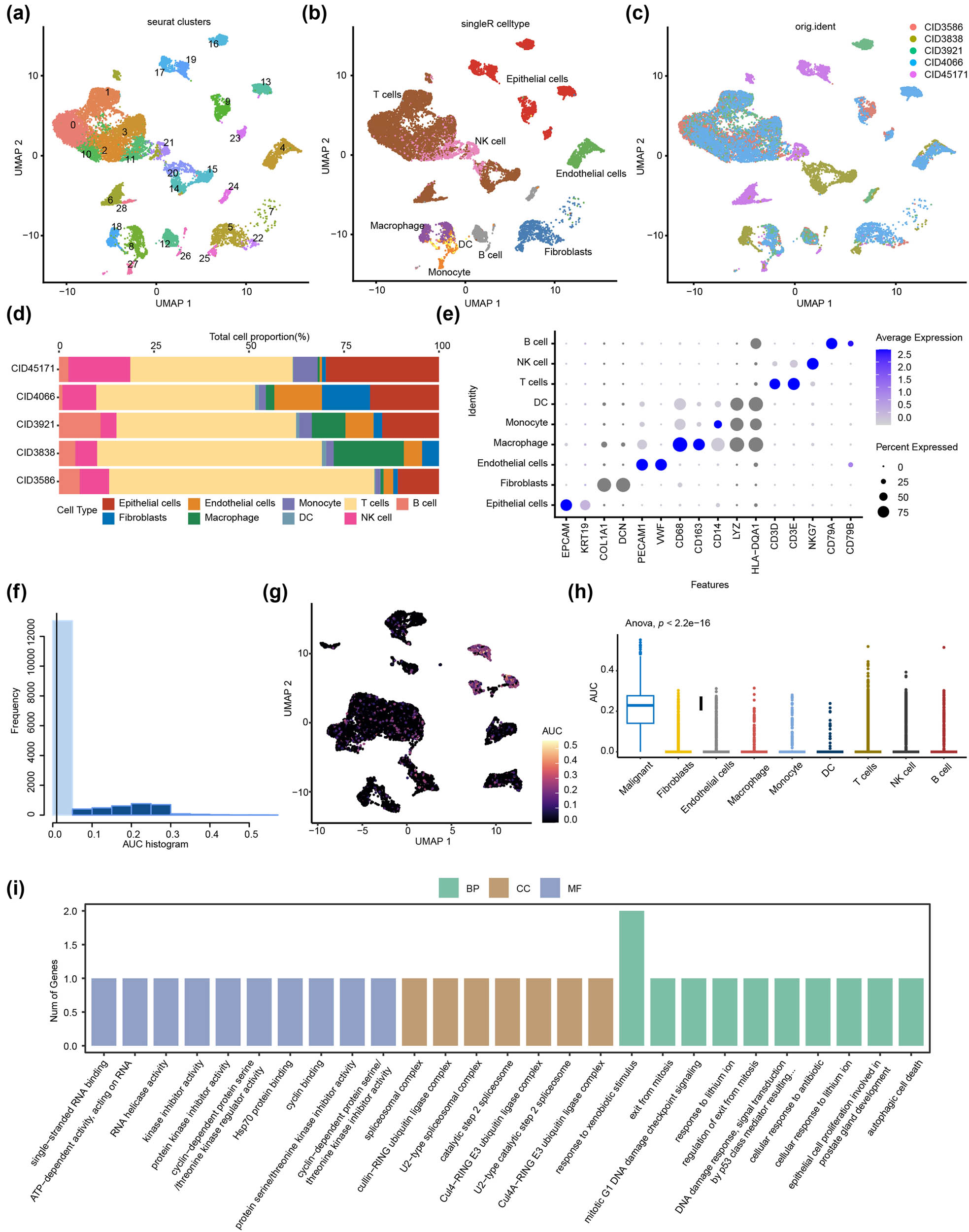
Single-cell RNA sequencing revealed the complexity of the microenvironment in HER2+ BC. (a) Cells were clustered into 29 clusters through UMAP based on the single cell dataset (GSE176078). (b) Cells were annotated into nine cell types through singleR and sample sources, including T cells, B cells, endothelial cells, epithelial cells, fibroblasts, monocytes, NK cells, DCs, and macrophages. (c) The UMAP diagram showed the distribution of nine cell types. (d) The bar chart displayed the proportion of each cell type in the nine cell types. (e) The heat map suggested the expression of genes specifically expressed for each cell type in the cell, with circles ranging from small to large representing the average expression value of characteristic genes. (f) The scores of the EPGs. (g) UMAP analysis in immune cells. (h) AUC box plot of EPG scores in different cell subtypes. (i) Pathway enrichment analysis of EPGs in HER2+ BC.
Characteristic gene information of cell type
| p_val | avg_log2FC | pct.1 | pct.2 | p_val_adj | Cluster | Gene |
|---|---|---|---|---|---|---|
| 0 | 3.5008097 | 0.975 | 0.085 | 0 | Endothelial cells | IGFBP7 |
| 0 | 2.7874423 | 0.9 | 0.007 | 0 | Endothelial cells | RAMP2 |
| 0 | 2.6477013 | 0.83 | 0.003 | 0 | Endothelial cells | PLVAP |
| 0 | 2.6393137 | 0.87 | 0.051 | 0 | Endothelial cells | SPARCL1 |
| 0 | 2.5954879 | 0.99 | 0.302 | 0 | Endothelial cells | IFITM3 |
| 0 | 2.5215686 | 0.51 | 0.002 | 0 | Endothelial cells | ACKR1 |
| 0 | 2.5190479 | 0.943 | 0.1 | 0 | Endothelial cells | IFI27 |
| 0 | 2.1214615 | 0.875 | 0.035 | 0 | Endothelial cells | RNASE1 |
| 0 | 2.0813175 | 0.836 | 0.027 | 0 | Endothelial cells | HSPG2 |
| 0 | 2.0796495 | 0.897 | 0.114 | 0 | Endothelial cells | IGFBP4 |
| 0 | 3.0812194 | 0.625 | 0.047 | 0 | NK_cell | GNLY |
| 0 | 2.2828379 | 0.759 | 0.141 | 0 | NK_cell | NKG7 |
| 0 | 1.8853469 | 0.525 | 0.066 | 0 | NK_cell | XCL1 |
| 0 | 1.8804565 | 0.485 | 0.05 | 0 | NK_cell | XCL2 |
| 0 | 1.5454339 | 0.42 | 0.052 | 0 | NK_cell | GZMB |
| 0 | 1.2911218 | 0.809 | 0.33 | 0 | NK_cell | CCL5 |
| 0 | 1.2029094 | 0.672 | 0.144 | 0 | NK_cell | CTSW |
| 0 | 1.1969762 | 0.592 | 0.057 | 0 | NK_cell | KLRD1 |
| 0 | 1.0595189 | 0.678 | 0.205 | 0 | NK_cell | CST7 |
| 0 | 1.777129 | 0.607 | 0.189 | 0 | NK_cell | CCL4 |
| 0 | 5.3961354 | 0.925 | 0.021 | 0 | Fibroblasts | COL1A1 |
| 0 | 5.1014286 | 0.946 | 0.02 | 0 | Fibroblasts | COL1A2 |
| 0 | 4.8643579 | 0.931 | 0.014 | 0 | Fibroblasts | COL3A1 |
| 0 | 4.6323219 | 0.909 | 0.007 | 0 | Fibroblasts | SFRP2 |
| 0 | 4.2776688 | 0.93 | 0.008 | 0 | Fibroblasts | LUM |
| 0 | 4.0859518 | 0.94 | 0.07 | 0 | Fibroblasts | SPARC |
| 0 | 4.0236418 | 0.946 | 0.007 | 0 | Fibroblasts | DCN |
| 0 | 3.9159311 | 0.809 | 0.013 | 0 | Fibroblasts | POSTN |
| 0 | 3.8171428 | 0.66 | 0.006 | 0 | Fibroblasts | MMP11 |
| 0 | 3.5036856 | 0.853 | 0.022 | 0 | Fibroblasts | CTHRC1 |
| 0 | 5.5917694 | 0.971 | 0.08 | 0 | Macrophage | APOE |
| 0 | 4.4926105 | 0.933 | 0.029 | 0 | Macrophage | APOC1 |
| 0 | 4.1681647 | 0.542 | 0.024 | 0 | Macrophage | SPP1 |
| 0 | 4.0036169 | 0.999 | 0.261 | 0 | Macrophage | HLA-DRA |
| 0 | 4.0025266 | 0.977 | 0.019 | 0 | Macrophage | C1QB |
| 0 | 3.9784419 | 1 | 0.958 | 0 | Macrophage | FTL |
| 0 | 3.8855317 | 0.985 | 0.02 | 0 | Macrophage | C1QA |
| 0 | 3.782816 | 0.98 | 0.017 | 0 | Macrophage | C1QC |
| 0 | 3.6845234 | 0.981 | 0.208 | 0 | Macrophage | CTSB |
| 0 | 3.680925 | 0.957 | 0.039 | 0 | Macrophage | LYZ |
| 0 | 4.5888552 | 0.305 | 0.023 | 0 | B_cell | IGHG1 |
| 0 | 3.6141401 | 0.168 | 0.003 | 0 | B_cell | IGHG2 |
| 0 | 2.822074 | 0.34 | 0.007 | 0 | B_cell | JCHAIN |
| 0 | 4.5251031 | 0.119 | 0.003 | 0 | B_cell | IGHV3-23 |
| 0 | 4.3981597 | 0.265 | 0.031 | 0 | B_cell | IGHG3 |
| 0 | 3.1499148 | 0.245 | 0.032 | 0 | B_cell | IGHA1 |
| 0 | 5.7093188 | 0.26 | 0.038 | 0 | B_cell | IGLC2 |
| 0 | 5.2700108 | 0.357 | 0.075 | 0 | B_cell | IGKC |
| 0 | 6.6128799 | 0.134 | 0.014 | 0 | B_cell | IGLV2-14 |
| 0 | 4.5139674 | 0.167 | 0.025 | 0 | B_cell | IGLC3 |
| 0 | 1.6121143 | 0.706 | 0.101 | 0 | T_cells | IL7R |
| 0 | 1.4663435 | 0.857 | 0.36 | 0 | T_cells | RGCC |
| 0 | 1.3521761 | 0.987 | 0.695 | 0 | T_cells | BTG1 |
| 0 | 1.3504311 | 0.808 | 0.269 | 0 | T_cells | TNFAIP3 |
| 0 | 1.2750278 | 0.859 | 0.451 | 0 | T_cells | TSC22D3 |
| 0 | 1.2696695 | 0.901 | 0.577 | 0 | T_cells | SARAF |
| 0 | 1.2689862 | 0.775 | 0.086 | 0 | T_cells | CD3E |
| 0 | 1.1577115 | 0.577 | 0.108 | 0 | T_cells | LTB |
| 0 | 1.145201 | 0.684 | 0.049 | 0 | T_cells | CD3D |
| 0 | 1.1788459 | 0.634 | 0.472 | 0 | T_cells | ANXA1 |
| 0 | 3.2032857 | 0.868 | 0.061 | 0 | Monocyte | LYZ |
| 0 | 2.7284108 | 0.979 | 0.277 | 0 | Monocyte | HLA-DRA |
| 0 | 2.6981768 | 0.977 | 0.216 | 0 | Monocyte | CST3 |
| 0 | 2.5125578 | 0.589 | 0.021 | 0 | Monocyte | IL1B |
| 0 | 2.426797 | 0.927 | 0.109 | 0 | Monocyte | TYROBP |
| 0 | 2.3950507 | 0.374 | 0.035 | 0 | Monocyte | S100A8 |
| 0 | 2.8961996 | 0.479 | 0.062 | 0 | Monocyte | S100A9 |
| 0 | 2.6940977 | 0.881 | 0.301 | 0 | Monocyte | HLA-DPA1 |
| 0 | 2.6823252 | 0.886 | 0.312 | 0 | Monocyte | HLA-DPB1 |
| 0 | 2.5512792 | 0.984 | 0.662 | 0 | Monocyte | CD74 |
| 0 | 3.0065951 | 0.83 | 0.076 | 0 | DC | LYZ |
| 0 | 2.872482 | 0.955 | 0.195 | 0 | DC | HLA-DQA1 |
| 0 | 3.1076022 | 0.955 | 0.23 | 0 | DC | CST3 |
| 0 | 3.3514534 | 0.991 | 0.29 | 0 | DC | HLA-DRA |
| 0 | 3.3879318 | 0.982 | 0.312 | 0 | DC | HLA-DPA1 |
| 0 | 3.3525582 | 0.982 | 0.322 | 0 | DC | HLA-DPB1 |
| 0 | 3.1123536 | 0.991 | 0.36 | 0 | DC | HLA-DRB1 |
| 0 | 2.6186613 | 0.429 | 0.05 | 0 | DC | G0S2 |
| 0 | 3.1815724 | 1 | 0.668 | 0 | DC | CD74 |
| 0 | 2.7433919 | 0.705 | 0.192 | 0 | DC | HLA-DRB5 |
| 0 | 4.899681 | 0.439 | 0.077 | 0 | Epithelial cells | MUCL1 |
| 0 | 4.1936505 | 0.314 | 0.028 | 0 | Epithelial cells | SCGB2A2 |
| 0 | 3.2109667 | 0.519 | 0.02 | 0 | Epithelial cells | CALML5 |
| 0 | 2.8713105 | 0.62 | 0.143 | 0 | Epithelial cells | MIEN1 |
| 0 | 2.8496767 | 0.871 | 0.031 | 0 | Epithelial cells | CD24 |
| 0 | 2.826804 | 0.875 | 0.027 | 0 | Epithelial cells | KRT7 |
| 0 | 2.8203469 | 0.607 | 0.01 | 0 | Epithelial cells | KRT19 |
| 0 | 2.723252 | 0.548 | 0.111 | 0 | Epithelial cells | MGP |
| 0 | 2.6634161 | 0.796 | 0.395 | 0 | Epithelial cells | DBI |
| 0 | 5.4093452 | 0.272 | 0.088 | 0 | Epithelial cells | SCGB1B2P |
Enrichment analysis results of DEGs in cell clusters of high malignant epithelial cell
| Ontology | ID | Description | Gene ratio | Bg ratio | p value | p.adjust | q value | gene ID | Count |
|---|---|---|---|---|---|---|---|---|---|
| CC | GO:0005743 | Mitochondrial inner membrane | 8/67 | 491/19594 | 0.0002619 | 0.0335883 | 0.0263503 | NME4/TIMM13/MRPS34/NDUFB9/COX6C/MRPL14/SLC25A39/UQCRQ | 8 |
| CC | GO:0098798 | Mitochondrial protein-containing complex | 6/67 | 281/19594 | 0.0003954 | 0.0335883 | 0.0263503 | TIMM13/MRPS34/NDUFB9/COX6C/MRPL14/UQCRQ | 6 |
| CC | GO:0071005 | U2-type precatalytic spliceosome | 3/67 | 50/19594 | 0.0006677 | 0.0335883 | 0.0263503 | SF3B4/LSM4/SNRPE | 3 |
| CC | GO:1990204 | Oxidoreductase complex | 4/67 | 120/19594 | 0.0007615 | 0.0335883 | 0.0263503 | NDUFB9/ETFA/UQCRQ/P4HB | 4 |
| CC | GO:0071011 | Precatalytic spliceosome | 3/67 | 53/19594 | 0.0007922 | 0.0335883 | 0.0263503 | SF3B4/LSM4/SNRPE | 3 |
| CC | GO:0016328 | Lateral plasma membrane | 3/67 | 64/19594 | 0.0013717 | 0.0484654 | 0.0380215 | EPCAM/TACSTD2/CLDN4 | 3 |
| MF | GO:0016209 | Antioxidant activity | 4/64 | 85/18410 | 0.0002177 | 0.042997 | 0.0370511 | MGST1/TXN/PRDX2/PRDX4 | 4 |
| MF | GO:0016860 | Intramolecular oxidoreductase activity | 3/64 | 50/18410 | 0.0006989 | 0.042997 | 0.0370511 | MIF/P4HB/DDT | 3 |
| MF | GO:0004601 | Peroxidase activity | 3/64 | 52/18410 | 0.0007842 | 0.042997 | 0.0370511 | MGST1/PRDX2/PRDX4 | 3 |
| MF | GO:0016684 | Oxidoreductase activity, acting on peroxide as acceptor | 3/64 | 56/18410 | 0.0009739 | 0.042997 | 0.0370511 | MGST1/PRDX2/PRDX4 | 3 |
| MF | GO:0016863 | Intramolecular oxidoreductase activity, transposing C═C bonds | 2/64 | 14/18410 | 0.0010538 | 0.042997 | 0.0370511 | MIF/DDT | 2 |
3.8 Cell–cell communication analysis
We inferred and quantified communication between nine cell types using CellChat, and visualized cell communication intensity and quantity using heat maps (Figure 8a) and circular plots (Figure 8b), which indicated a relatively high interaction intensity between malignant epithelial cells and fibroblasts or endothelial cells or monocytes, as well as a relatively high number of interactions between malignant epithelial cells and fibroblasts or endothelial cells. We also systematically investigated the cell–cell communication network to identify the two pathways that contribute the most to the outgoing or incoming signals of malignant epithelial cells (Figure 8c). Migration inhibitory factors (MIF) pathway contributes most to input, whereas C-C motif chemokine ligand (CCL) pathway contributes most to output. We demonstrated the cellular communication received by malignant epithelial cells (Figure 8d) and emitted by malignant epithelial cells (Figure 8e) via the MIF pathway and CCL pathway through bubble diagrams. In the cellular communication received by malignant epithelial cells, macrophages were the major immune cell in both, in the MIF pathway engaged in incoming cell–cell communication and in the CCL pathway engaged in outgoing cell–cell communication.
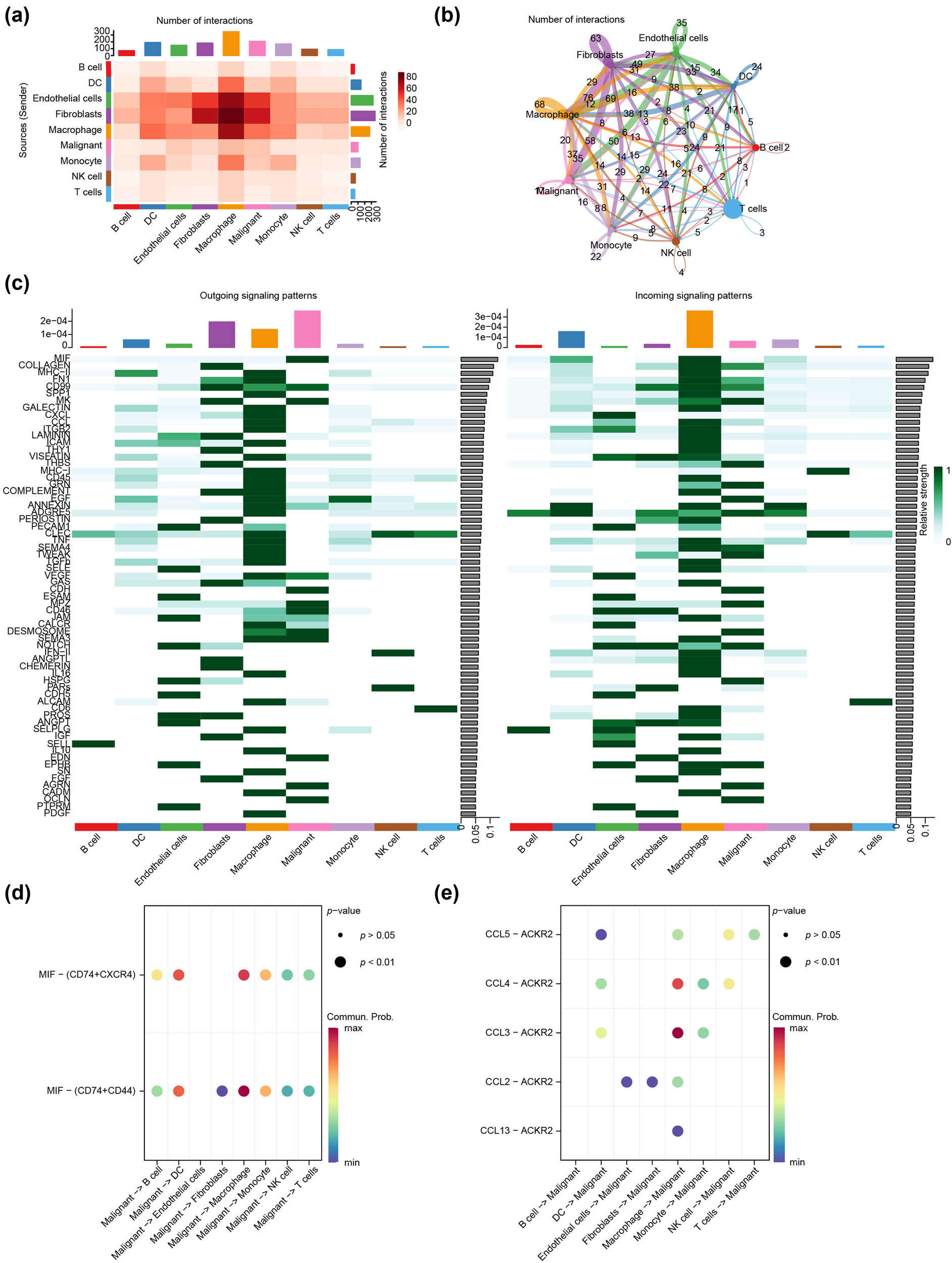
Cell–cell communication analysis. (a) The heatmap revealed the intensity of cell–cell communication including T cells, B cells, endothelial cells, epithelial cells, fibroblasts, monocytes, NK cells, DCs, and macrophages based on the GSE176078 dataset. (b) Circular graph exhibits cell communication numbers. (c) The heat map indicating contribution values of pathway involved in output and input, with the green color representing the intensity of correlation between the cells with each other. (d) and (e) Bubble diagram showing the cell–cell communication (d) received and (e) emitted by malignant epithelial cells. The bubble size represents the p-value, and the color represents the cell–cell communication probability.
4 Discussion
Although ERGs have been identified as diagnostic and prognostic indicators for breast cancer [37], their potential clinical importance in HER2+ BC is uncertain. Herein, eight genes were extracted via deferentially expressed genes (normal vs tumor) of TCGA BRCA dataset and ERGs were merged and KM analysis was utilized to obtain eight prognostic genes. As higher infiltration of immune cells and expression of immune activation markers are seen in HER2+ BC primary lesions compared to metastatic tissues [38], we attempted to determine the relationship between exosomes and prognosis or microenvironment in patients with HER2+ BC. By combining diverse expression analysis, prognostic relevance, and ERGs, we determined the candidates for EPGs. After LASSO Cox analysis and random forest cox analysis, we finally identified FGF9, SF3B4, and EPCAM as the EPGs for HER2+ BC.
In the context of examining the expression profile of EPGs in HER2+ BC, “EPGs” specifically refers to the three genes we identified as critically important: FGF9, SF3B4, and EPCAM. These genes showed a clear association with the immune response to foreign stimuli, underscoring their potential role in regulating the tumor microenvironment in HER2+ BC through an exosome-mediated mechanism. FGF9, the protein produced by the fibroblast growth factor 9, acts as a secreted factor that binds fibroblast growth factor receptor and inhibits growth stimulation in the microenvironment [39]. In contrast to our findings, Xu et al. discovered that patients with ovarian cancer had decreased FGF9 expression and displayed favorable prognostic values. Abnormally produced FGF9 was associated with immunological markers, such as immunoinhibitory and chemokine receptors in ovarian cancer [40]. Our results indicated that patients with HER2+ BC and low FGF9 expression experienced poor survival. EPCAM encodes an antigen related to carcinoma called epithelial cell adhesion molecule, which is expressed in malignant epithelial tumors. EPCAM is used as a target for immunotherapy in the treatment of various carcinomas [41]. Although EPCAM serves as the marker protein for enriching tumor-derived exosomes, Rupp et al. discovered a reduction of EPCAM expression in serum exosomes produced from patients with breast cancer [42]. Leblanc et al. demonstrated that immunocapture of breast cell exosomes by targeting EPCAM was blocked by syntenin [43]. We discovered a negative correlation between EPCAM and FGF9 expression in HER2+ BC. SF3B4 encodes one of four subunits of the splicing factor 3B and is involved in pre-mRNA splicing [44]. Its function in exosomes and clinical importance in breast cancer are well understood. Our research demonstrated that SF3B4 had prognostic value in HER2+ BC.
We subtyped all HER2+ BC cases into three clusters and discovered that Cluster C had a relatively high level of MSI and p53 signaling pathway genes. Pizzi et al. demonstrated that p53 was downregulated in breast cancer characterized by MSI [45]. Dutta et al. discovered that breast cancer cells stimulate DNA damage repair and phosphorylation of p53 in mammary epithelial cells by secreting exosomes that are taken up by these cells, thereby creating a favorable environment for breast cancer [46]. The results of our study suggests that patients with HER2+ BC subtyped into Cluster C have a high frequency of TP53 mutations, whereas the biological functional abnormalities produced by mutations in Cluster A and Cluster B were primarily centered in the TP53 signaling pathway. Beis et al. proposed that potential new drug targets included genes that were differentially expressed in patients with various prognostic profiles or drug administration [47]. Domenis et al. discovered that the activation of pro-inflammatory cytokines increased the transfer of mutated p53 from tumor cells to epithelial cells via exosomes, thereby inserting the effect on malignant transformation and tumor progression [48]. Our research demonstrated that p53 may be the target that Cluster A drugs act upon. Zhou et al. reported that exosomal LRP1 promoted the migration of ovarian cancer cells, whereas Ricklefs discovered that FASN was elevated in malignant glioma cells and extracellular vesicles of plasma [49,50]. We believe that LRP1 and FASN are the drug-related targets of Cluster B and Cluster C.
DCs are antigen-presenting cells with multiple functions that contribute to the initiation and modulation of innate and adaptive immune responses. Romagnoli et al. suggested that CD3+ T cells cultured with DCs derived exosome-treated breast cancer cells, which exhibited a significantly higher proportion of IFN-γ-secreting cells, indicating that incorporation of DC-derived exosomes by the breast cancer cells could activate T-cells for a potentially more effective cancer immunotherapy [51]. It is generally recognized that DC-based treatments, particularly DC vaccination, have a therapeutic effect on breast cancer [52]. Our research revealed that the infiltration of DCs varied over three clusters, and that the expression of FGF9 was strongly linked with DCs.
Wu performed scRNA Seq (Chromium, 10X Genomics) on 26 primary tumors from three major clinical subtypes of breast cancer, including 11 ER+ BC, 5 HER2+ BC, and 10 triple-negative breast cancer; this allowed us the opportunity to investigate the involvement of EPGs in the microenvironment for subtypes of HER2+ BC [53]. The functions of EPGs may be primarily associated with RESPONSE TO XENOBIOTIC STIMULUS, indicating that exosomes play a vital role in the cellular response to external stimulation.
CCL family and macrophage MIFs play a significant role in breast cancer progression and its interaction with the microenvironment. Breast cancer cells promote tumor-derived CCL20 and upregulate PD-L1 expression on neutrophils to exacerbate T cell immunosuppression [54]. Liu et al. discovered that exosomes produced from mesenchymal stem cells mediate heart healing following myocardial infarction through the upregulation of MIF [55]. Li et al. found that adipose-derived stem cells interact with macrophages and accelerate bone repair via downregulation of macrophage MIF in exosomes [56]. Our research suggests that HER2+ BC cells interacted mostly with macrophages via MIF and CCL signaling pathway by exosomes. However, our study contains some limitations. The detected EPGs necessitate functional investigation in the context of the immunological microenvironment of HER2+ BC, and the HER2+ BC clinical samples must be confirmed using scRNA sequencing. There are also intrinsic biases in bioinformatics analyses. Despite these limitations, our approach provides bioinformatic hints for further research.
5 Conclusion
Our study reveals that exosomes from HER2+ BC patients have significant prognostic value and successfully identified EPGs (FGF9, SF3B4, and EPCAM), which were significantly associated with patient survival. In addition, our analysis highlighted the complex interactions between tumor and microenvironment, revealing the mechanism of information transmission through exosomes, which are not only critical for tumor growth and progression, but also have a profound impact on treatment response and patient prognosis.
Acknowledgment
We thank Xiang Xiao for his support for our study.
-
Funding information: The study was supported by funds as follows: Natural Science Foundation of Shanghai (No. 21ZR1412500), National Natural Science Foundation of China (No. 82172925), and Shanghai Changning “Quality-Balance” Research Talent Development Fund (CNJH10).
-
Author contributions: Conception and design of the research: Li Liang and Ji Zhao; acquisition of data: Feng Shen and Yue-Mei Hu; analysis and interpretation of the data: Feng Shen and Qun-chao Hu; statistical analysis: Yue-Mei Hu, Yan-Jie Chen, Kai Yin, and Ying Chen; obtaining funding: Li Liang and Qun-chao Hu; writing of the manuscript: Yan-Jie Chen and Ji Zhao; critical revision of the manuscript for intellectual content: Ying Chen and Kai Yin.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48.10.3322/caac.21763Search in Google Scholar PubMed
[2] Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135(5):584–90.10.1097/CM9.0000000000002108Search in Google Scholar PubMed PubMed Central
[3] Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov. 2023;22(2):101–26.10.1038/s41573-022-00579-0Search in Google Scholar PubMed PubMed Central
[4] Solek J, Chrzanowski J, Cieslak A, Zielinska A, Piasecka D, Braun M, et al. Subtype-specific tumour immune microenvironment in risk of recurrence of ductal carcinoma in situ: prognostic value of HER2. Biomedicines. 2022;10(5):1061.10.3390/biomedicines10051061Search in Google Scholar PubMed PubMed Central
[5] Vos H, Lambein K, Richard F, Marien B, Nevelsteen I, Punie K, et al. Comparison of the tumor immune microenvironment of primary hormone receptor-negative HER2-positive and triple negative breast cancer. NPJ Breast Cancer. 2021;7(1):128.10.1038/s41523-021-00332-7Search in Google Scholar PubMed PubMed Central
[6] Wang X, Tian L, Lu J, Ng IO. Exosomes and cancer – diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis. 2022;11(1):54.10.1038/s41389-022-00431-5Search in Google Scholar PubMed PubMed Central
[7] Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, Campiglio M, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227(2):658–67.10.1002/jcp.22773Search in Google Scholar PubMed
[8] Barok M, Puhka M, Vereb G, Szollosi J, Isola J, Joensuu H. Cancer-derived exosomes from HER2-positive cancer cells carry trastuzumab-emtansine into cancer cells leading to growth inhibition and caspase activation. BMC Cancer. 2018;18(1):504.10.1186/s12885-018-4418-2Search in Google Scholar PubMed PubMed Central
[9] Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol. 2019;187(1):352–64.10.1007/s12010-018-2813-4Search in Google Scholar PubMed
[10] Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1A):A68–77.10.5114/wo.2014.47136Search in Google Scholar PubMed PubMed Central
[11] Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71.10.1093/nar/gkv1507Search in Google Scholar
[12] Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–8.10.1038/s41587-020-0546-8Search in Google Scholar
[13] Xu Q, Chen S, Hu Y, Huang W. Landscape of immune microenvironment under immune cell infiltration pattern in breast cancer. Front Immunol. 2021;12:711433.10.3389/fimmu.2021.711433Search in Google Scholar
[14] Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets – update. Nucleic Acids Res. 2013;41:D991–5. (Database issue).10.1093/nar/gks1193Search in Google Scholar
[15] Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9(6):1324–38.10.1074/mcp.M000063-MCP201Search in Google Scholar
[16] Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40.10.1093/bioinformatics/btr260Search in Google Scholar
[17] Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7.10.1186/1471-2105-14-7Search in Google Scholar
[18] Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16(4):385–95.10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3Search in Google Scholar
[19] Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26(7):976–8.10.1093/bioinformatics/btq064Search in Google Scholar
[20] Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244.10.1186/1471-2105-14-244Search in Google Scholar
[21] Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56.10.1101/gr.239244.118Search in Google Scholar PubMed PubMed Central
[22] Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–70.10.1200/JCO.2007.12.9791Search in Google Scholar PubMed
[23] Cao R, Lopez-de-Ullibarri I. ROC curves for the statistical analysis of microarray data. Methods Mol Biol. 2019;1986:245–53.10.1007/978-1-4939-9442-7_11Search in Google Scholar PubMed
[24] Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. JAMA. 2015;313(4):409–10.10.1001/jama.2015.37Search in Google Scholar PubMed
[25] Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3.10.1093/bioinformatics/btq170Search in Google Scholar PubMed PubMed Central
[26] Shi Y, Zhang L, Do KA, Peterson CB, Jenq RR. aPCoA: covariate adjusted principal coordinates analysis. Bioinformatics. 2020;36(13):4099–101.10.1093/bioinformatics/btaa276Search in Google Scholar PubMed PubMed Central
[27] Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14.10.1093/nar/gkaa407Search in Google Scholar PubMed PubMed Central
[28] Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–62.10.1016/j.celrep.2016.12.019Search in Google Scholar PubMed
[29] McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–37e324.10.1016/j.cels.2019.03.003Search in Google Scholar PubMed PubMed Central
[30] Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–96.10.1038/s41592-019-0619-0Search in Google Scholar PubMed PubMed Central
[31] Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72.10.1038/s41590-018-0276-ySearch in Google Scholar PubMed PubMed Central
[32] Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184(3):792–809.10.1016/j.cell.2021.01.010Search in Google Scholar PubMed
[33] Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–8.10.1093/nar/gky900Search in Google Scholar PubMed PubMed Central
[34] Gao R, Bai S, Henderson YC, Lin Y, Schalck A, Yan Y, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39(5):599–608.10.1038/s41587-020-00795-2Search in Google Scholar PubMed PubMed Central
[35] Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–6.10.1038/nmeth.4463Search in Google Scholar PubMed PubMed Central
[36] Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell–cell communication using CellChat. Nat Commun. 2021;12(1):1088.10.1038/s41467-021-21246-9Search in Google Scholar PubMed PubMed Central
[37] Alagundagi DB, Ghate SD, Rajendra VKJ, Gollapalli P, Shetty VV, D’Souza C, et al. Exploring breast cancer exosomes for novel biomarkers of potential diagnostic and prognostic importance. 3 Biotech. 2023;13(1):7.10.1007/s13205-022-03422-wSearch in Google Scholar PubMed PubMed Central
[38] Quaranta M, Scaramella F, Costanzo G. Biocompatibility of new porcelains in dentistry. Dent Cadmos. 1987;55(18):49–53.Search in Google Scholar
[39] Lindner M, Thummler K, Arthur A, Brunner S, Elliott C, McElroy D, et al. Fibroblast growth factor signalling in multiple sclerosis: inhibition of myelination and induction of pro-inflammatory environment by FGF9. Brain. 2015;138(Pt 7):1875–93.10.1093/brain/awv102Search in Google Scholar PubMed PubMed Central
[40] Xu Z, Cai Y, Liu W, Kang F, He Q, Hong Q, et al. Downregulated exosome-associated gene FGF9 as a novel diagnostic and prognostic target for ovarian cancer and its underlying roles in immune regulation. Aging (Albany NY). 2022;14(4):1822–35.10.18632/aging.203905Search in Google Scholar PubMed PubMed Central
[41] Gires O, Bauerle PA. EpCAM as a target in cancer therapy. J Clin Oncol. 2010;28(15):e239–40.10.1200/JCO.2009.26.8540Search in Google Scholar PubMed
[42] Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122(2):437–46.10.1016/j.ygyno.2011.04.035Search in Google Scholar PubMed
[43] Leblanc R, Kashyap R, Barral K, Egea-Jimenez AL, Kovalskyy D, Feracci M, et al. Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loading with syndecan and EpCAM cargo. J Extracell Vesicles. 2020;10(2):e12039.10.1002/jev2.12039Search in Google Scholar PubMed PubMed Central
[44] Yan L, Yang X, Yang X, Yuan X, Wei L, Si Y, et al. The role of splicing factor SF3B4 in congenital diseases and tumors. Discov Med. 2021;32(167):123–32.Search in Google Scholar
[45] Pizzi C, Panico L, De Marchis L, Mastranzo P, Di Maio M, D’Amico C, et al. p53 expression is decreased in primary breast carcinomas with microsatellite instability. Breast Cancer Res Treat. 2002;73(3):257–66.10.1023/A:1015806530091Search in Google Scholar
[46] Dutta S, Warshall C, Bandyopadhyay C, Dutta D, Chandran B. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS One. 2014;9(5):e97580.10.1371/journal.pone.0097580Search in Google Scholar PubMed PubMed Central
[47] Beis G, Serafeim AP, Papasotiriou I. Data-driven analysis and druggability assessment methods to accelerate the identification of novel cancer targets. Comput Struct Biotechnol J. 2023;21:46–57.10.1016/j.csbj.2022.11.042Search in Google Scholar PubMed PubMed Central
[48] Domenis R, Cifu A, Mio C, Fabris M, Curcio F. Pro-inflammatory microenvironment modulates the transfer of mutated TP53 mediated by tumor exosomes. Int J Mol Sci. 2021;22(12):6258.10.3390/ijms22126258Search in Google Scholar PubMed PubMed Central
[49] Zhou W, Ma J, Zhao H, Wang Q, Guo X, Chen L, et al. Serum exosomes from epithelial ovarian cancer patients contain LRP1, which promotes the migration of epithelial ovarian cancer cell. Mol Cell Proteomics. 2023;22(4):100520.10.1016/j.mcpro.2023.100520Search in Google Scholar PubMed PubMed Central
[50] Ricklefs FL, Maire CL, Matschke J, Duhrsen L, Sauvigny T, Holz M, et al. FASN is a biomarker enriched in malignant glioma-derived extracellular vesicles. Int J Mol Sci. 2020;21(6):1931.10.3390/ijms21061931Search in Google Scholar PubMed PubMed Central
[51] Romagnoli GG, Zelante BB, Toniolo PA, Migliori IK, Barbuto JA. Dendritic cell-derived exosomes may be a tool for cancer immunotherapy by converting tumor cells into immunogenic targets. Front Immunol. 2014;5:692.10.3389/fimmu.2014.00692Search in Google Scholar PubMed PubMed Central
[52] Mejias Sosa L, Lopez-Janeiro A, Cordoba Iturriagagoitia A, Sala P, Solans BP, Hato L, et al. Modification of breast cancer milieu with chemotherapy plus dendritic cell vaccine: an approach to select best therapeutic strategies. Biomedicines. 2023;11(2):238.10.3390/biomedicines11020238Search in Google Scholar PubMed PubMed Central
[53] Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–47.10.1038/s41588-021-00911-1Search in Google Scholar PubMed PubMed Central
[54] Kwantwi LB, Wang S, Zhang W, Peng W, Cai Z, Sheng Y, et al. Tumor-associated neutrophils activated by tumor-derived CCL20 (C-C motif chemokine ligand 20) promote T cell immunosuppression via programmed death-ligand 1 (PD-L1) in breast cancer. Bioengineered. 2021;12(1):6996–7006.10.1080/21655979.2021.1977102Search in Google Scholar PubMed PubMed Central
[55] Liu X, Li X, Zhu W, Zhang Y, Hong Y, Liang X, et al. Exosomes from mesenchymal stem cells overexpressing MIF enhance myocardial repair. J Cell Physiol. 2020;235(11):8010–22.10.1002/jcp.29456Search in Google Scholar PubMed
[56] Li R, Li D, Wang H, Chen K, Wang S, Xu J, et al. Exosomes from adipose-derived stem cells regulate M1/M2 macrophage phenotypic polarization to promote bone healing via miR-451a/MIF. Stem Cell Res Ther. 2022;13(1):149.10.1186/s13287-022-02823-1Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Constitutive and evoked release of ATP in adult mouse olfactory epithelium
- LARP1 knockdown inhibits cultured gastric carcinoma cell cycle progression and metastatic behavior
- PEGylated porcine–human recombinant uricase: A novel fusion protein with improved efficacy and safety for the treatment of hyperuricemia and renal complications
- Research progress on ocular complications caused by type 2 diabetes mellitus and the function of tears and blepharons
- The role and mechanism of esketamine in preventing and treating remifentanil-induced hyperalgesia based on the NMDA receptor–CaMKII pathway
- Brucella infection combined with Nocardia infection: A case report and literature review
- Detection of serum interleukin-18 level and neutrophil/lymphocyte ratio in patients with antineutrophil cytoplasmic antibody-associated vasculitis and its clinical significance
- Ang-1, Ang-2, and Tie2 are diagnostic biomarkers for Henoch-Schönlein purpura and pediatric-onset systemic lupus erythematous
- PTTG1 induces pancreatic cancer cell proliferation and promotes aerobic glycolysis by regulating c-myc
- Role of serum B-cell-activating factor and interleukin-17 as biomarkers in the classification of interstitial pneumonia with autoimmune features
- Effectiveness and safety of a mumps containing vaccine in preventing laboratory-confirmed mumps cases from 2002 to 2017: A meta-analysis
- Low levels of sex hormone-binding globulin predict an increased breast cancer risk and its underlying molecular mechanisms
- A case of Trousseau syndrome: Screening, detection and complication
- Application of the integrated airway humidification device enhances the humidification effect of the rabbit tracheotomy model
- Preparation of Cu2+/TA/HAP composite coating with anti-bacterial and osteogenic potential on 3D-printed porous Ti alloy scaffolds for orthopedic applications
- Aquaporin-8 promotes human dermal fibroblasts to counteract hydrogen peroxide-induced oxidative damage: A novel target for management of skin aging
- Current research and evidence gaps on placental development in iron deficiency anemia
- Single-nucleotide polymorphism rs2910829 in PDE4D is related to stroke susceptibility in Chinese populations: The results of a meta-analysis
- Pheochromocytoma-induced myocardial infarction: A case report
- Kaempferol regulates apoptosis and migration of neural stem cells to attenuate cerebral infarction by O‐GlcNAcylation of β-catenin
- Sirtuin 5 regulates acute myeloid leukemia cell viability and apoptosis by succinylation modification of glycine decarboxylase
- Apigenin 7-glucoside impedes hypoxia-induced malignant phenotypes of cervical cancer cells in a p16-dependent manner
- KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1
- Current state of research on copper complexes in the treatment of breast cancer
- Exploring antioxidant strategies in the pathogenesis of ALS
- Helicobacter pylori causes gastric dysbacteriosis in chronic gastritis patients
- IL-33/soluble ST2 axis is associated with radiation-induced cardiac injury
- The predictive value of serum NLR, SII, and OPNI for lymph node metastasis in breast cancer patients with internal mammary lymph nodes after thoracoscopic surgery
- Carrying SNP rs17506395 (T > G) in TP63 gene and CCR5Δ32 mutation associated with the occurrence of breast cancer in Burkina Faso
- P2X7 receptor: A receptor closely linked with sepsis-associated encephalopathy
- Probiotics for inflammatory bowel disease: Is there sufficient evidence?
- Identification of KDM4C as a gene conferring drug resistance in multiple myeloma
- Microbial perspective on the skin–gut axis and atopic dermatitis
- Thymosin α1 combined with XELOX improves immune function and reduces serum tumor markers in colorectal cancer patients after radical surgery
- Highly specific vaginal microbiome signature for gynecological cancers
- Sample size estimation for AQP4-IgG seropositive optic neuritis: Retinal damage detection by optical coherence tomography
- The effects of SDF-1 combined application with VEGF on femoral distraction osteogenesis in rats
- Fabrication and characterization of gold nanoparticles using alginate: In vitro and in vivo assessment of its administration effects with swimming exercise on diabetic rats
- Mitigating digestive disorders: Action mechanisms of Mediterranean herbal active compounds
- Distribution of CYP2D6 and CYP2C19 gene polymorphisms in Han and Uygur populations with breast cancer in Xinjiang, China
- VSP-2 attenuates secretion of inflammatory cytokines induced by LPS in BV2 cells by mediating the PPARγ/NF-κB signaling pathway
- Factors influencing spontaneous hypothermia after emergency trauma and the construction of a predictive model
- Long-term administration of morphine specifically alters the level of protein expression in different brain regions and affects the redox state
- Application of metagenomic next-generation sequencing technology in the etiological diagnosis of peritoneal dialysis-associated peritonitis
- Clinical diagnosis, prevention, and treatment of neurodyspepsia syndrome using intelligent medicine
- Case report: Successful bronchoscopic interventional treatment of endobronchial leiomyomas
- Preliminary investigation into the genetic etiology of short stature in children through whole exon sequencing of the core family
- Cystic adenomyoma of the uterus: Case report and literature review
- Mesoporous silica nanoparticles as a drug delivery mechanism
- Dynamic changes in autophagy activity in different degrees of pulmonary fibrosis in mice
- Vitamin D deficiency and inflammatory markers in type 2 diabetes: Big data insights
- Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells
- Meta-analysis on the efficacy of allogeneic hematopoietic stem cell transplantation to treat malignant lymphoma
- Mitochondrial DNA drives neuroinflammation through the cGAS-IFN signaling pathway in the spinal cord of neuropathic pain mice
- Application value of artificial intelligence algorithm-based magnetic resonance multi-sequence imaging in staging diagnosis of cervical cancer
- Embedded monitoring system and teaching of artificial intelligence online drug component recognition
- Investigation into the association of FNDC1 and ADAMTS12 gene expression with plumage coloration in Muscovy ducks
- Yak meat content in feed and its impact on the growth of rats
- A rare case of Richter transformation with breast involvement: A case report and literature review
- First report of Nocardia wallacei infection in an immunocompetent patient in Zhejiang province
- Rhodococcus equi and Brucella pulmonary mass in immunocompetent: A case report and literature review
- Downregulation of RIP3 ameliorates the left ventricular mechanics and function after myocardial infarction via modulating NF-κB/NLRP3 pathway
- Evaluation of the role of some non-enzymatic antioxidants among Iraqi patients with non-alcoholic fatty liver disease
- The role of Phafin proteins in cell signaling pathways and diseases
- Ten-year anemia as initial manifestation of Castleman disease in the abdominal cavity: A case report
- Coexistence of hereditary spherocytosis with SPTB P.Trp1150 gene variant and Gilbert syndrome: A case report and literature review
- Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells
- Exploratory evaluation supported by experimental and modeling approaches of Inula viscosa root extract as a potent corrosion inhibitor for mild steel in a 1 M HCl solution
- Imaging manifestations of ductal adenoma of the breast: A case report
- Gut microbiota and sleep: Interaction mechanisms and therapeutic prospects
- Isomangiferin promotes the migration and osteogenic differentiation of rat bone marrow mesenchymal stem cells
- Prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer
- Circular RNAs as potential biomarkers for male severe sepsis
- Knockdown of Stanniocalcin-1 inhibits growth and glycolysis in oral squamous cell carcinoma cells
- The expression and biological role of complement C1s in esophageal squamous cell carcinoma
- A novel GNAS mutation in pseudohypoparathyroidism type 1a with articular flexion deformity: A case report
- Predictive value of serum magnesium levels for prognosis in patients with non-small cell lung cancer undergoing EGFR-TKI therapy
- HSPB1 alleviates acute-on-chronic liver failure via the P53/Bax pathway
- IgG4-related disease complicated by PLA2R-associated membranous nephropathy: A case report
- Baculovirus-mediated endostatin and angiostatin activation of autophagy through the AMPK/AKT/mTOR pathway inhibits angiogenesis in hepatocellular carcinoma
- Metformin mitigates osteoarthritis progression by modulating the PI3K/AKT/mTOR signaling pathway and enhancing chondrocyte autophagy
- Evaluation of the activity of antimicrobial peptides against bacterial vaginosis
- Atypical presentation of γ/δ mycosis fungoides with an unusual phenotype and SOCS1 mutation
- Analysis of the microecological mechanism of diabetic kidney disease based on the theory of “gut–kidney axis”: A systematic review
- Omega-3 fatty acids prevent gestational diabetes mellitus via modulation of lipid metabolism
- Refractory hypertension complicated with Turner syndrome: A case report
- Interaction of ncRNAs and the PI3K/AKT/mTOR pathway: Implications for osteosarcoma
- Association of low attenuation area scores with pulmonary function and clinical prognosis in patients with chronic obstructive pulmonary disease
- Long non-coding RNAs in bone formation: Key regulators and therapeutic prospects
- The deubiquitinating enzyme USP35 regulates the stability of NRF2 protein
- Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as potential diagnostic markers for rebleeding in patients with esophagogastric variceal bleeding
- G protein-coupled receptor 1 participating in the mechanism of mediating gestational diabetes mellitus by phosphorylating the AKT pathway
- LL37-mtDNA regulates viability, apoptosis, inflammation, and autophagy in lipopolysaccharide-treated RLE-6TN cells by targeting Hsp90aa1
- The analgesic effect of paeoniflorin: A focused review
- Chemical composition’s effect on Solanum nigrum Linn.’s antioxidant capacity and erythrocyte protection: Bioactive components and molecular docking analysis
- Knockdown of HCK promotes HREC cell viability and inner blood–retinal barrier integrity by regulating the AMPK signaling pathway
- The role of rapamycin in the PINK1/Parkin signaling pathway in mitophagy in podocytes
- Laryngeal non-Hodgkin lymphoma: Report of four cases and review of the literature
- Clinical value of macrogenome next-generation sequencing on infections
- Overview of dendritic cells and related pathways in autoimmune uveitis
- TAK-242 alleviates diabetic cardiomyopathy via inhibiting pyroptosis and TLR4/CaMKII/NLRP3 pathway
- Hypomethylation in promoters of PGC-1α involved in exercise-driven skeletal muscular alterations in old age
- Profile and antimicrobial susceptibility patterns of bacteria isolated from effluents of Kolladiba and Debark hospitals
- The expression and clinical significance of syncytin-1 in serum exosomes of hepatocellular carcinoma patients
- A histomorphometric study to evaluate the therapeutic effects of biosynthesized silver nanoparticles on the kidneys infected with Plasmodium chabaudi
- PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
- Analysis of MDA, SOD, TAOC, MNCV, SNCV, and TSS scores in patients with diabetes peripheral neuropathy
- SLIT3 deficiency promotes non-small cell lung cancer progression by modulating UBE2C/WNT signaling
- The relationship between TMCO1 and CALR in the pathological characteristics of prostate cancer and its effect on the metastasis of prostate cancer cells
- Heterogeneous nuclear ribonucleoprotein K is a potential target for enhancing the chemosensitivity of nasopharyngeal carcinoma
- PHB2 alleviates retinal pigment epithelium cell fibrosis by suppressing the AGE–RAGE pathway
- Anti-γ-aminobutyric acid-B receptor autoimmune encephalitis with syncope as the initial symptom: Case report and literature review
- Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua
- Human umbilical cord mesenchymal stem cells regulate glutathione metabolism depending on the ERK–Nrf2–HO-1 signal pathway to repair phosphoramide mustard-induced ovarian cancer cells
- Electroacupuncture on GB acupoints improves osteoporosis via the estradiol–PI3K–Akt signaling pathway
- Renalase protects against podocyte injury by inhibiting oxidative stress and apoptosis in diabetic nephropathy
- Review: Dicranostigma leptopodum: A peculiar plant of Papaveraceae
- Combination effect of flavonoids attenuates lung cancer cell proliferation by inhibiting the STAT3 and FAK signaling pathway
- Renal microangiopathy and immune complex glomerulonephritis induced by anti-tumour agents: A case report
- Correlation analysis of AVPR1a and AVPR2 with abnormal water and sodium and potassium metabolism in rats
- Gastrointestinal health anti-diarrheal mixture relieves spleen deficiency-induced diarrhea through regulating gut microbiota
- Myriad factors and pathways influencing tumor radiotherapy resistance
- Exploring the effects of culture conditions on Yapsin (YPS) gene expression in Nakaseomyces glabratus
- Screening of prognostic core genes based on cell–cell interaction in the peripheral blood of patients with sepsis
- Coagulation factor II thrombin receptor as a promising biomarker in breast cancer management
- Ileocecal mucinous carcinoma misdiagnosed as incarcerated hernia: A case report
- Methyltransferase like 13 promotes malignant behaviors of bladder cancer cells through targeting PI3K/ATK signaling pathway
- The debate between electricity and heat, efficacy and safety of irreversible electroporation and radiofrequency ablation in the treatment of liver cancer: A meta-analysis
- ZAG promotes colorectal cancer cell proliferation and epithelial–mesenchymal transition by promoting lipid synthesis
- Baicalein inhibits NLRP3 inflammasome activation and mitigates placental inflammation and oxidative stress in gestational diabetes mellitus
- Impact of SWCNT-conjugated senna leaf extract on breast cancer cells: A potential apoptotic therapeutic strategy
- MFAP5 inhibits the malignant progression of endometrial cancer cells in vitro
- Major ozonated autohemotherapy promoted functional recovery following spinal cord injury in adult rats via the inhibition of oxidative stress and inflammation
- Axodendritic targeting of TAU and MAP2 and microtubule polarization in iPSC-derived versus SH-SY5Y-derived human neurons
- Differential expression of phosphoinositide 3-kinase/protein kinase B and Toll-like receptor/nuclear factor kappa B signaling pathways in experimental obesity Wistar rat model
- The therapeutic potential of targeting Oncostatin M and the interleukin-6 family in retinal diseases: A comprehensive review
- BA inhibits LPS-stimulated inflammatory response and apoptosis in human middle ear epithelial cells by regulating the Nf-Kb/Iκbα axis
- Role of circRMRP and circRPL27 in chronic obstructive pulmonary disease
- Investigating the role of hyperexpressed HCN1 in inducing myocardial infarction through activation of the NF-κB signaling pathway
- Characterization of phenolic compounds and evaluation of anti-diabetic potential in Cannabis sativa L. seeds: In vivo, in vitro, and in silico studies
- Quantitative immunohistochemistry analysis of breast Ki67 based on artificial intelligence
- Ecology and Environmental Science
- Screening of different growth conditions of Bacillus subtilis isolated from membrane-less microbial fuel cell toward antimicrobial activity profiling
- Degradation of a mixture of 13 polycyclic aromatic hydrocarbons by commercial effective microorganisms
- Evaluation of the impact of two citrus plants on the variation of Panonychus citri (Acari: Tetranychidae) and beneficial phytoseiid mites
- Prediction of present and future distribution areas of Juniperus drupacea Labill and determination of ethnobotany properties in Antalya Province, Türkiye
- Population genetics of Todarodes pacificus (Cephalopoda: Ommastrephidae) in the northwest Pacific Ocean via GBS sequencing
- A comparative analysis of dendrometric, macromorphological, and micromorphological characteristics of Pistacia atlantica subsp. atlantica and Pistacia terebinthus in the middle Atlas region of Morocco
- Macrofungal sporocarp community in the lichen Scots pine forests
- Assessing the proximate compositions of indigenous forage species in Yemen’s pastoral rangelands
- Food Science
- Gut microbiota changes associated with low-carbohydrate diet intervention for obesity
- Reexamination of Aspergillus cristatus phylogeny in dark tea: Characteristics of the mitochondrial genome
- Differences in the flavonoid composition of the leaves, fruits, and branches of mulberry are distinguished based on a plant metabolomics approach
- Investigating the impact of wet rendering (solventless method) on PUFA-rich oil from catfish (Clarias magur) viscera
- Non-linear associations between cardiovascular metabolic indices and metabolic-associated fatty liver disease: A cross-sectional study in the US population (2017–2020)
- Knockdown of USP7 alleviates atherosclerosis in ApoE-deficient mice by regulating EZH2 expression
- Utility of dairy microbiome as a tool for authentication and traceability
- Agriculture
- Enhancing faba bean (Vicia faba L.) productivity through establishing the area-specific fertilizer rate recommendation in southwest Ethiopia
- Impact of novel herbicide based on synthetic auxins and ALS inhibitor on weed control
- Perspectives of pteridophytes microbiome for bioremediation in agricultural applications
- Fertilizer application parameters for drip-irrigated peanut based on the fertilizer effect function established from a “3414” field trial
- Improving the productivity and profitability of maize (Zea mays L.) using optimum blended inorganic fertilization
- Application of leaf multispectral analyzer in comparison to hyperspectral device to assess the diversity of spectral reflectance indices in wheat genotypes
- Animal Sciences
- Knockdown of ANP32E inhibits colorectal cancer cell growth and glycolysis by regulating the AKT/mTOR pathway
- Development of a detection chip for major pathogenic drug-resistant genes and drug targets in bovine respiratory system diseases
- Exploration of the genetic influence of MYOT and MB genes on the plumage coloration of Muscovy ducks
- Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism
- Comparison of nutritional value of the wild and cultivated spiny loaches at three growth stages
- Transcriptomic analysis of liver immune response in Chinese spiny frog (Quasipaa spinosa) infected with Proteus mirabilis
- Disruption of BCAA degradation is a critical characteristic of diabetic cardiomyopathy revealed by integrated transcriptome and metabolome analysis
- Plant Sciences
- Effect of long-term in-row branch covering on soil microorganisms in pear orchards
- Photosynthetic physiological characteristics, growth performance, and element concentrations reveal the calcicole–calcifuge behaviors of three Camellia species
- Transcriptome analysis reveals the mechanism of NaHCO3 promoting tobacco leaf maturation
- Bioinformatics, expression analysis, and functional verification of allene oxide synthase gene HvnAOS1 and HvnAOS2 in qingke
- Water, nitrogen, and phosphorus coupling improves gray jujube fruit quality and yield
- Improving grape fruit quality through soil conditioner: Insights from RNA-seq analysis of Cabernet Sauvignon roots
- Role of Embinin in the reabsorption of nucleus pulposus in lumbar disc herniation: Promotion of nucleus pulposus neovascularization and apoptosis of nucleus pulposus cells
- Revealing the effects of amino acid, organic acid, and phytohormones on the germination of tomato seeds under salinity stress
- Combined effects of nitrogen fertilizer and biochar on the growth, yield, and quality of pepper
- Comprehensive phytochemical and toxicological analysis of Chenopodium ambrosioides (L.) fractions
- Impact of “3414” fertilization on the yield and quality of greenhouse tomatoes
- Exploring the coupling mode of water and fertilizer for improving growth, fruit quality, and yield of the pear in the arid region
- Metagenomic analysis of endophytic bacteria in seed potato (Solanum tuberosum)
- Antibacterial, antifungal, and phytochemical properties of Salsola kali ethanolic extract
- Exploring the hepatoprotective properties of citronellol: In vitro and in silico studies on ethanol-induced damage in HepG2 cells
- Enhanced osmotic dehydration of watermelon rind using honey–sucrose solutions: A study on pre-treatment efficacy and mass transfer kinetics
- Effects of exogenous 2,4-epibrassinolide on photosynthetic traits of 53 cowpea varieties under NaCl stress
- Comparative transcriptome analysis of maize (Zea mays L.) seedlings in response to copper stress
- An optimization method for measuring the stomata in cassava (Manihot esculenta Crantz) under multiple abiotic stresses
- Fosinopril inhibits Ang II-induced VSMC proliferation, phenotype transformation, migration, and oxidative stress through the TGF-β1/Smad signaling pathway
- Antioxidant and antimicrobial activities of Salsola imbricata methanolic extract and its phytochemical characterization
- Bioengineering and Biotechnology
- Absorbable calcium and phosphorus bioactive membranes promote bone marrow mesenchymal stem cells osteogenic differentiation for bone regeneration
- New advances in protein engineering for industrial applications: Key takeaways
- An overview of the production and use of Bacillus thuringiensis toxin
- Research progress of nanoparticles in diagnosis and treatment of hepatocellular carcinoma
- Bioelectrochemical biosensors for water quality assessment and wastewater monitoring
- PEI/MMNs@LNA-542 nanoparticles alleviate ICU-acquired weakness through targeted autophagy inhibition and mitochondrial protection
- Unleashing of cytotoxic effects of thymoquinone-bovine serum albumin nanoparticles on A549 lung cancer cells
- Erratum
- Erratum to “Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM”
- Erratum to “Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation”
- Retraction
- Retraction to “MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB”
- Retraction to “A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis”
- Special Issue on Advances in Neurodegenerative Disease Research and Treatment
- Transplantation of human neural stem cell prevents symptomatic motor behavior disability in a rat model of Parkinson’s disease
- Special Issue on Multi-omics
- Inflammasome complex genes with clinical relevance suggest potential as therapeutic targets for anti-tumor drugs in clear cell renal cell carcinoma
- Gastroesophageal varices in primary biliary cholangitis with anti-centromere antibody positivity: Early onset?
Articles in the same Issue
- Biomedical Sciences
- Constitutive and evoked release of ATP in adult mouse olfactory epithelium
- LARP1 knockdown inhibits cultured gastric carcinoma cell cycle progression and metastatic behavior
- PEGylated porcine–human recombinant uricase: A novel fusion protein with improved efficacy and safety for the treatment of hyperuricemia and renal complications
- Research progress on ocular complications caused by type 2 diabetes mellitus and the function of tears and blepharons
- The role and mechanism of esketamine in preventing and treating remifentanil-induced hyperalgesia based on the NMDA receptor–CaMKII pathway
- Brucella infection combined with Nocardia infection: A case report and literature review
- Detection of serum interleukin-18 level and neutrophil/lymphocyte ratio in patients with antineutrophil cytoplasmic antibody-associated vasculitis and its clinical significance
- Ang-1, Ang-2, and Tie2 are diagnostic biomarkers for Henoch-Schönlein purpura and pediatric-onset systemic lupus erythematous
- PTTG1 induces pancreatic cancer cell proliferation and promotes aerobic glycolysis by regulating c-myc
- Role of serum B-cell-activating factor and interleukin-17 as biomarkers in the classification of interstitial pneumonia with autoimmune features
- Effectiveness and safety of a mumps containing vaccine in preventing laboratory-confirmed mumps cases from 2002 to 2017: A meta-analysis
- Low levels of sex hormone-binding globulin predict an increased breast cancer risk and its underlying molecular mechanisms
- A case of Trousseau syndrome: Screening, detection and complication
- Application of the integrated airway humidification device enhances the humidification effect of the rabbit tracheotomy model
- Preparation of Cu2+/TA/HAP composite coating with anti-bacterial and osteogenic potential on 3D-printed porous Ti alloy scaffolds for orthopedic applications
- Aquaporin-8 promotes human dermal fibroblasts to counteract hydrogen peroxide-induced oxidative damage: A novel target for management of skin aging
- Current research and evidence gaps on placental development in iron deficiency anemia
- Single-nucleotide polymorphism rs2910829 in PDE4D is related to stroke susceptibility in Chinese populations: The results of a meta-analysis
- Pheochromocytoma-induced myocardial infarction: A case report
- Kaempferol regulates apoptosis and migration of neural stem cells to attenuate cerebral infarction by O‐GlcNAcylation of β-catenin
- Sirtuin 5 regulates acute myeloid leukemia cell viability and apoptosis by succinylation modification of glycine decarboxylase
- Apigenin 7-glucoside impedes hypoxia-induced malignant phenotypes of cervical cancer cells in a p16-dependent manner
- KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1
- Current state of research on copper complexes in the treatment of breast cancer
- Exploring antioxidant strategies in the pathogenesis of ALS
- Helicobacter pylori causes gastric dysbacteriosis in chronic gastritis patients
- IL-33/soluble ST2 axis is associated with radiation-induced cardiac injury
- The predictive value of serum NLR, SII, and OPNI for lymph node metastasis in breast cancer patients with internal mammary lymph nodes after thoracoscopic surgery
- Carrying SNP rs17506395 (T > G) in TP63 gene and CCR5Δ32 mutation associated with the occurrence of breast cancer in Burkina Faso
- P2X7 receptor: A receptor closely linked with sepsis-associated encephalopathy
- Probiotics for inflammatory bowel disease: Is there sufficient evidence?
- Identification of KDM4C as a gene conferring drug resistance in multiple myeloma
- Microbial perspective on the skin–gut axis and atopic dermatitis
- Thymosin α1 combined with XELOX improves immune function and reduces serum tumor markers in colorectal cancer patients after radical surgery
- Highly specific vaginal microbiome signature for gynecological cancers
- Sample size estimation for AQP4-IgG seropositive optic neuritis: Retinal damage detection by optical coherence tomography
- The effects of SDF-1 combined application with VEGF on femoral distraction osteogenesis in rats
- Fabrication and characterization of gold nanoparticles using alginate: In vitro and in vivo assessment of its administration effects with swimming exercise on diabetic rats
- Mitigating digestive disorders: Action mechanisms of Mediterranean herbal active compounds
- Distribution of CYP2D6 and CYP2C19 gene polymorphisms in Han and Uygur populations with breast cancer in Xinjiang, China
- VSP-2 attenuates secretion of inflammatory cytokines induced by LPS in BV2 cells by mediating the PPARγ/NF-κB signaling pathway
- Factors influencing spontaneous hypothermia after emergency trauma and the construction of a predictive model
- Long-term administration of morphine specifically alters the level of protein expression in different brain regions and affects the redox state
- Application of metagenomic next-generation sequencing technology in the etiological diagnosis of peritoneal dialysis-associated peritonitis
- Clinical diagnosis, prevention, and treatment of neurodyspepsia syndrome using intelligent medicine
- Case report: Successful bronchoscopic interventional treatment of endobronchial leiomyomas
- Preliminary investigation into the genetic etiology of short stature in children through whole exon sequencing of the core family
- Cystic adenomyoma of the uterus: Case report and literature review
- Mesoporous silica nanoparticles as a drug delivery mechanism
- Dynamic changes in autophagy activity in different degrees of pulmonary fibrosis in mice
- Vitamin D deficiency and inflammatory markers in type 2 diabetes: Big data insights
- Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells
- Meta-analysis on the efficacy of allogeneic hematopoietic stem cell transplantation to treat malignant lymphoma
- Mitochondrial DNA drives neuroinflammation through the cGAS-IFN signaling pathway in the spinal cord of neuropathic pain mice
- Application value of artificial intelligence algorithm-based magnetic resonance multi-sequence imaging in staging diagnosis of cervical cancer
- Embedded monitoring system and teaching of artificial intelligence online drug component recognition
- Investigation into the association of FNDC1 and ADAMTS12 gene expression with plumage coloration in Muscovy ducks
- Yak meat content in feed and its impact on the growth of rats
- A rare case of Richter transformation with breast involvement: A case report and literature review
- First report of Nocardia wallacei infection in an immunocompetent patient in Zhejiang province
- Rhodococcus equi and Brucella pulmonary mass in immunocompetent: A case report and literature review
- Downregulation of RIP3 ameliorates the left ventricular mechanics and function after myocardial infarction via modulating NF-κB/NLRP3 pathway
- Evaluation of the role of some non-enzymatic antioxidants among Iraqi patients with non-alcoholic fatty liver disease
- The role of Phafin proteins in cell signaling pathways and diseases
- Ten-year anemia as initial manifestation of Castleman disease in the abdominal cavity: A case report
- Coexistence of hereditary spherocytosis with SPTB P.Trp1150 gene variant and Gilbert syndrome: A case report and literature review
- Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells
- Exploratory evaluation supported by experimental and modeling approaches of Inula viscosa root extract as a potent corrosion inhibitor for mild steel in a 1 M HCl solution
- Imaging manifestations of ductal adenoma of the breast: A case report
- Gut microbiota and sleep: Interaction mechanisms and therapeutic prospects
- Isomangiferin promotes the migration and osteogenic differentiation of rat bone marrow mesenchymal stem cells
- Prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer
- Circular RNAs as potential biomarkers for male severe sepsis
- Knockdown of Stanniocalcin-1 inhibits growth and glycolysis in oral squamous cell carcinoma cells
- The expression and biological role of complement C1s in esophageal squamous cell carcinoma
- A novel GNAS mutation in pseudohypoparathyroidism type 1a with articular flexion deformity: A case report
- Predictive value of serum magnesium levels for prognosis in patients with non-small cell lung cancer undergoing EGFR-TKI therapy
- HSPB1 alleviates acute-on-chronic liver failure via the P53/Bax pathway
- IgG4-related disease complicated by PLA2R-associated membranous nephropathy: A case report
- Baculovirus-mediated endostatin and angiostatin activation of autophagy through the AMPK/AKT/mTOR pathway inhibits angiogenesis in hepatocellular carcinoma
- Metformin mitigates osteoarthritis progression by modulating the PI3K/AKT/mTOR signaling pathway and enhancing chondrocyte autophagy
- Evaluation of the activity of antimicrobial peptides against bacterial vaginosis
- Atypical presentation of γ/δ mycosis fungoides with an unusual phenotype and SOCS1 mutation
- Analysis of the microecological mechanism of diabetic kidney disease based on the theory of “gut–kidney axis”: A systematic review
- Omega-3 fatty acids prevent gestational diabetes mellitus via modulation of lipid metabolism
- Refractory hypertension complicated with Turner syndrome: A case report
- Interaction of ncRNAs and the PI3K/AKT/mTOR pathway: Implications for osteosarcoma
- Association of low attenuation area scores with pulmonary function and clinical prognosis in patients with chronic obstructive pulmonary disease
- Long non-coding RNAs in bone formation: Key regulators and therapeutic prospects
- The deubiquitinating enzyme USP35 regulates the stability of NRF2 protein
- Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as potential diagnostic markers for rebleeding in patients with esophagogastric variceal bleeding
- G protein-coupled receptor 1 participating in the mechanism of mediating gestational diabetes mellitus by phosphorylating the AKT pathway
- LL37-mtDNA regulates viability, apoptosis, inflammation, and autophagy in lipopolysaccharide-treated RLE-6TN cells by targeting Hsp90aa1
- The analgesic effect of paeoniflorin: A focused review
- Chemical composition’s effect on Solanum nigrum Linn.’s antioxidant capacity and erythrocyte protection: Bioactive components and molecular docking analysis
- Knockdown of HCK promotes HREC cell viability and inner blood–retinal barrier integrity by regulating the AMPK signaling pathway
- The role of rapamycin in the PINK1/Parkin signaling pathway in mitophagy in podocytes
- Laryngeal non-Hodgkin lymphoma: Report of four cases and review of the literature
- Clinical value of macrogenome next-generation sequencing on infections
- Overview of dendritic cells and related pathways in autoimmune uveitis
- TAK-242 alleviates diabetic cardiomyopathy via inhibiting pyroptosis and TLR4/CaMKII/NLRP3 pathway
- Hypomethylation in promoters of PGC-1α involved in exercise-driven skeletal muscular alterations in old age
- Profile and antimicrobial susceptibility patterns of bacteria isolated from effluents of Kolladiba and Debark hospitals
- The expression and clinical significance of syncytin-1 in serum exosomes of hepatocellular carcinoma patients
- A histomorphometric study to evaluate the therapeutic effects of biosynthesized silver nanoparticles on the kidneys infected with Plasmodium chabaudi
- PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
- Analysis of MDA, SOD, TAOC, MNCV, SNCV, and TSS scores in patients with diabetes peripheral neuropathy
- SLIT3 deficiency promotes non-small cell lung cancer progression by modulating UBE2C/WNT signaling
- The relationship between TMCO1 and CALR in the pathological characteristics of prostate cancer and its effect on the metastasis of prostate cancer cells
- Heterogeneous nuclear ribonucleoprotein K is a potential target for enhancing the chemosensitivity of nasopharyngeal carcinoma
- PHB2 alleviates retinal pigment epithelium cell fibrosis by suppressing the AGE–RAGE pathway
- Anti-γ-aminobutyric acid-B receptor autoimmune encephalitis with syncope as the initial symptom: Case report and literature review
- Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua
- Human umbilical cord mesenchymal stem cells regulate glutathione metabolism depending on the ERK–Nrf2–HO-1 signal pathway to repair phosphoramide mustard-induced ovarian cancer cells
- Electroacupuncture on GB acupoints improves osteoporosis via the estradiol–PI3K–Akt signaling pathway
- Renalase protects against podocyte injury by inhibiting oxidative stress and apoptosis in diabetic nephropathy
- Review: Dicranostigma leptopodum: A peculiar plant of Papaveraceae
- Combination effect of flavonoids attenuates lung cancer cell proliferation by inhibiting the STAT3 and FAK signaling pathway
- Renal microangiopathy and immune complex glomerulonephritis induced by anti-tumour agents: A case report
- Correlation analysis of AVPR1a and AVPR2 with abnormal water and sodium and potassium metabolism in rats
- Gastrointestinal health anti-diarrheal mixture relieves spleen deficiency-induced diarrhea through regulating gut microbiota
- Myriad factors and pathways influencing tumor radiotherapy resistance
- Exploring the effects of culture conditions on Yapsin (YPS) gene expression in Nakaseomyces glabratus
- Screening of prognostic core genes based on cell–cell interaction in the peripheral blood of patients with sepsis
- Coagulation factor II thrombin receptor as a promising biomarker in breast cancer management
- Ileocecal mucinous carcinoma misdiagnosed as incarcerated hernia: A case report
- Methyltransferase like 13 promotes malignant behaviors of bladder cancer cells through targeting PI3K/ATK signaling pathway
- The debate between electricity and heat, efficacy and safety of irreversible electroporation and radiofrequency ablation in the treatment of liver cancer: A meta-analysis
- ZAG promotes colorectal cancer cell proliferation and epithelial–mesenchymal transition by promoting lipid synthesis
- Baicalein inhibits NLRP3 inflammasome activation and mitigates placental inflammation and oxidative stress in gestational diabetes mellitus
- Impact of SWCNT-conjugated senna leaf extract on breast cancer cells: A potential apoptotic therapeutic strategy
- MFAP5 inhibits the malignant progression of endometrial cancer cells in vitro
- Major ozonated autohemotherapy promoted functional recovery following spinal cord injury in adult rats via the inhibition of oxidative stress and inflammation
- Axodendritic targeting of TAU and MAP2 and microtubule polarization in iPSC-derived versus SH-SY5Y-derived human neurons
- Differential expression of phosphoinositide 3-kinase/protein kinase B and Toll-like receptor/nuclear factor kappa B signaling pathways in experimental obesity Wistar rat model
- The therapeutic potential of targeting Oncostatin M and the interleukin-6 family in retinal diseases: A comprehensive review
- BA inhibits LPS-stimulated inflammatory response and apoptosis in human middle ear epithelial cells by regulating the Nf-Kb/Iκbα axis
- Role of circRMRP and circRPL27 in chronic obstructive pulmonary disease
- Investigating the role of hyperexpressed HCN1 in inducing myocardial infarction through activation of the NF-κB signaling pathway
- Characterization of phenolic compounds and evaluation of anti-diabetic potential in Cannabis sativa L. seeds: In vivo, in vitro, and in silico studies
- Quantitative immunohistochemistry analysis of breast Ki67 based on artificial intelligence
- Ecology and Environmental Science
- Screening of different growth conditions of Bacillus subtilis isolated from membrane-less microbial fuel cell toward antimicrobial activity profiling
- Degradation of a mixture of 13 polycyclic aromatic hydrocarbons by commercial effective microorganisms
- Evaluation of the impact of two citrus plants on the variation of Panonychus citri (Acari: Tetranychidae) and beneficial phytoseiid mites
- Prediction of present and future distribution areas of Juniperus drupacea Labill and determination of ethnobotany properties in Antalya Province, Türkiye
- Population genetics of Todarodes pacificus (Cephalopoda: Ommastrephidae) in the northwest Pacific Ocean via GBS sequencing
- A comparative analysis of dendrometric, macromorphological, and micromorphological characteristics of Pistacia atlantica subsp. atlantica and Pistacia terebinthus in the middle Atlas region of Morocco
- Macrofungal sporocarp community in the lichen Scots pine forests
- Assessing the proximate compositions of indigenous forage species in Yemen’s pastoral rangelands
- Food Science
- Gut microbiota changes associated with low-carbohydrate diet intervention for obesity
- Reexamination of Aspergillus cristatus phylogeny in dark tea: Characteristics of the mitochondrial genome
- Differences in the flavonoid composition of the leaves, fruits, and branches of mulberry are distinguished based on a plant metabolomics approach
- Investigating the impact of wet rendering (solventless method) on PUFA-rich oil from catfish (Clarias magur) viscera
- Non-linear associations between cardiovascular metabolic indices and metabolic-associated fatty liver disease: A cross-sectional study in the US population (2017–2020)
- Knockdown of USP7 alleviates atherosclerosis in ApoE-deficient mice by regulating EZH2 expression
- Utility of dairy microbiome as a tool for authentication and traceability
- Agriculture
- Enhancing faba bean (Vicia faba L.) productivity through establishing the area-specific fertilizer rate recommendation in southwest Ethiopia
- Impact of novel herbicide based on synthetic auxins and ALS inhibitor on weed control
- Perspectives of pteridophytes microbiome for bioremediation in agricultural applications
- Fertilizer application parameters for drip-irrigated peanut based on the fertilizer effect function established from a “3414” field trial
- Improving the productivity and profitability of maize (Zea mays L.) using optimum blended inorganic fertilization
- Application of leaf multispectral analyzer in comparison to hyperspectral device to assess the diversity of spectral reflectance indices in wheat genotypes
- Animal Sciences
- Knockdown of ANP32E inhibits colorectal cancer cell growth and glycolysis by regulating the AKT/mTOR pathway
- Development of a detection chip for major pathogenic drug-resistant genes and drug targets in bovine respiratory system diseases
- Exploration of the genetic influence of MYOT and MB genes on the plumage coloration of Muscovy ducks
- Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism
- Comparison of nutritional value of the wild and cultivated spiny loaches at three growth stages
- Transcriptomic analysis of liver immune response in Chinese spiny frog (Quasipaa spinosa) infected with Proteus mirabilis
- Disruption of BCAA degradation is a critical characteristic of diabetic cardiomyopathy revealed by integrated transcriptome and metabolome analysis
- Plant Sciences
- Effect of long-term in-row branch covering on soil microorganisms in pear orchards
- Photosynthetic physiological characteristics, growth performance, and element concentrations reveal the calcicole–calcifuge behaviors of three Camellia species
- Transcriptome analysis reveals the mechanism of NaHCO3 promoting tobacco leaf maturation
- Bioinformatics, expression analysis, and functional verification of allene oxide synthase gene HvnAOS1 and HvnAOS2 in qingke
- Water, nitrogen, and phosphorus coupling improves gray jujube fruit quality and yield
- Improving grape fruit quality through soil conditioner: Insights from RNA-seq analysis of Cabernet Sauvignon roots
- Role of Embinin in the reabsorption of nucleus pulposus in lumbar disc herniation: Promotion of nucleus pulposus neovascularization and apoptosis of nucleus pulposus cells
- Revealing the effects of amino acid, organic acid, and phytohormones on the germination of tomato seeds under salinity stress
- Combined effects of nitrogen fertilizer and biochar on the growth, yield, and quality of pepper
- Comprehensive phytochemical and toxicological analysis of Chenopodium ambrosioides (L.) fractions
- Impact of “3414” fertilization on the yield and quality of greenhouse tomatoes
- Exploring the coupling mode of water and fertilizer for improving growth, fruit quality, and yield of the pear in the arid region
- Metagenomic analysis of endophytic bacteria in seed potato (Solanum tuberosum)
- Antibacterial, antifungal, and phytochemical properties of Salsola kali ethanolic extract
- Exploring the hepatoprotective properties of citronellol: In vitro and in silico studies on ethanol-induced damage in HepG2 cells
- Enhanced osmotic dehydration of watermelon rind using honey–sucrose solutions: A study on pre-treatment efficacy and mass transfer kinetics
- Effects of exogenous 2,4-epibrassinolide on photosynthetic traits of 53 cowpea varieties under NaCl stress
- Comparative transcriptome analysis of maize (Zea mays L.) seedlings in response to copper stress
- An optimization method for measuring the stomata in cassava (Manihot esculenta Crantz) under multiple abiotic stresses
- Fosinopril inhibits Ang II-induced VSMC proliferation, phenotype transformation, migration, and oxidative stress through the TGF-β1/Smad signaling pathway
- Antioxidant and antimicrobial activities of Salsola imbricata methanolic extract and its phytochemical characterization
- Bioengineering and Biotechnology
- Absorbable calcium and phosphorus bioactive membranes promote bone marrow mesenchymal stem cells osteogenic differentiation for bone regeneration
- New advances in protein engineering for industrial applications: Key takeaways
- An overview of the production and use of Bacillus thuringiensis toxin
- Research progress of nanoparticles in diagnosis and treatment of hepatocellular carcinoma
- Bioelectrochemical biosensors for water quality assessment and wastewater monitoring
- PEI/MMNs@LNA-542 nanoparticles alleviate ICU-acquired weakness through targeted autophagy inhibition and mitochondrial protection
- Unleashing of cytotoxic effects of thymoquinone-bovine serum albumin nanoparticles on A549 lung cancer cells
- Erratum
- Erratum to “Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM”
- Erratum to “Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation”
- Retraction
- Retraction to “MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB”
- Retraction to “A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis”
- Special Issue on Advances in Neurodegenerative Disease Research and Treatment
- Transplantation of human neural stem cell prevents symptomatic motor behavior disability in a rat model of Parkinson’s disease
- Special Issue on Multi-omics
- Inflammasome complex genes with clinical relevance suggest potential as therapeutic targets for anti-tumor drugs in clear cell renal cell carcinoma
- Gastroesophageal varices in primary biliary cholangitis with anti-centromere antibody positivity: Early onset?

