Abstract
This study aimed to investigate the presence of antibiotic susceptibility patterns and bacterial profiles of some multi-drug-resistant bacteria isolated from the effluents of Kolladiba and Debark Hospitals. Sixteen samples were collected from Kolladiba and Debark Hospitals in North Gondar, Ethiopia, to investigate the presence of multi-drug-resistant bacteria. To assess susceptibility patterns, well-isolated bacterial colonies were subjected to seven antibiotics. The selected resistant isolates were characterized using morphological and biochemical tests. Plasmid DNA analysis of the isolates was also performed. Out of a total of 28 bacterial isolates, 12 were found to be multi-drug resistant. Among the tested antibiotics, erythromycin was the most resistant antibiotic, while novobiocin was the most effective antibiotic. A plasmid profile study of the isolates revealed both the presence and absence of plasmids. The number of plasmids ranged from zero to four, with plasmid sizes of 100, 900, 1,000, 1,400, 1,500, and 1,800 base pairs. This study concluded that effluents from both hospitals have high number of multi-drug-resistant isolates. The genes responsible for multi-drug resistance in bacterial isolates under this study could be either plasmid-mediated or chromosomal DNA-mediated. The presence of multi-drug-resistant bacteria in these effluents should not be overlooked.
1 Introduction
Serious infections caused by pathogenic bacteria that have become resistant to commonly used broad-spectrum antibiotics have become major global healthcare issues in the current century, and also, the growth of antibiotic-resistant bacteria in hospital setups is becoming a serious problem. Such emerging antibiotic resistance is threatening the management and control of bacterial infections. Although antibiotic resistance as an environmental problem is increasing from time to time, it has largely been overlooked, and prevention and containment have received far too little attention. As a result, the increasing incidence of resistance to a wide range of antibiotics by a variety of pathogenic organisms is a major concern facing modern medicine. Furthermore, antibiotic resistance has resulted in increased healthcare costs, morbidity, and mortality among patients with infectious diseases [1].
In Africa, including Ethiopia, many hospitals and clinics are major sources/reservoirs for large numbers of disease-causing bacteria, and such bacterial strains are introduced into the public environment [2]. The spread of antibiotic-resistant bacteria from hospitals to the environment can occur through various mechanisms, such as discharged antibiotics that are excreted through feces and urine of patients, discharged patients and health-care workers, accidental spilling of different antibiotics, and discarding expired drugs without burning. Overuse and misuse of antibiotics have accelerated antibiotic resistance. Uncontrolled and unrestricted uses of different antibiotics by the community also cause a rise in antibiotic resistance and result in the spread of resistance genes in hospital effluents. These multiple antibiotic-resistant pathogenic bacteria entering into an environment mainly through effluents are causing a serious public health impact. Bacteria resistant to these antibiotics, which carry the transmissible gene, have the ability to transfer such transmissible genes to other normal non-resistant bacterial communities. Such infections caused by the transferred resistance gene of bacteria are habitually difficult to treat [3]. It also minimizes the power of the antibiotic pool for handling bacterial infections [3]. Hence, hospital effluents could raise a number of common antibiotic-resistant bacteria in the recipient environment both by the mechanisms of introduction and selection pressure [4]. It is also true that hospitals provide an environment conducive to multi-drug bacteria, making the treatment options limited and expensive [5].
Genes coding for antibiotic resistance and virulence may share common features of being located in the bacterial chromosome, as well as on plasmids. The genes they carry may be expressed under stressed conditions and are considered to be living organisms in their simple structure since they are the main elements of a continuous lineage with every individual’s evolutionary history [6]. Many pathogenic bacteria have plasmids that carry antibiotic resistance encoding genes, which contribute to the emergence of multi-drug resistance [7,8]. These plasmids also encode virulence factors.
In the study areas, there is a lack of information on whether hospital effluents carrying antibiotics could contribute to the development of antibiotic-resistant microorganisms in the environment or not. The current study was designed to assess the antimicrobial susceptibility pattern of bacteria isolated from the effluents of two hospitals in North Gondar, Amhara Region, Ethiopia, against commonly used antibiotics. The study also aimed to analyze the plasmid profile of the multi-drug-resistant isolates. The purpose of studying the plasmid profile was to check whether the resistance is plasmid-mediated or chromosome-mediated. The findings of this study therefore would help to make public health authorities aware of the dissemination of drug-resistant bacteria in the hospital environments for proper management of effluents.
2 Materials and methods
2.1 Description of the study areas
The study was carried out in Debark and Kolladiba hospitals of North Gondar, Amhara Region, Ethiopia. Debark is located in the northern part of Gondar town near Ras Dashen mountain. The town has 13°08′N latitude and 37°54′E longitude with an elevation of 2,850 m above sea level. Conversely, Kolladiba is 16 km far from Gondar town in the southwestern part. Kolladiba town has 12°40′N latitude and 37°10′E longitude with an elevation of 2,146 m above sea level. The distances from Addis Ababa to Kolladiba and Debark are 729 and 835.3 km, respectively. Maps of both study areas are depicted in Figures 1 and 2.
![Figure 1
Map showing study area – Debark district (Debark town) [9].](/document/doi/10.1515/biol-2022-0960/asset/graphic/j_biol-2022-0960_fig_001.jpg)
Map showing study area – Debark district (Debark town) [9].
Map showing Dembia district (Kolladiba town).
2.2 Sample collection and isolation of bacteria
A cross-sectional study was used to conduct this research. A total of 16 effluent samples were collected from Debark and Kolladiba hospitals in North Gondar, Amhara Region, Ethiopia. They were taken from different sites of the two hospitals (around the drug store, laboratory, shower, surgical room, and patient bedding room) and finally at the sites where all the hospital effluents collectively enter into the septic tank. The samples were collected using sterilized plastic bags. The collected samples were transported to the Microbial Laboratory, Department of Biotechnology, Institute of Biotechnology, University of Gondar. Then, the samples were preserved at 4°C in a refrigerator for further analysis.
Bacteria were isolated by a serial dilution method. Stock solution was prepared by diluting 1 mL of the effluent sample in 9 mL of distilled water and shaken using a vortex mixer. From the stock solution, 1 mL was used to prepare the final volume of 10−4, 10−5, 10−6, and 10−7 and distributed on Mueller Hinton agar medium aseptically. Then, the plates were incubated at 37°C for 48 h. Distinct bacterial colonies that were grown well on each plate were observed. Distinct representative colonies were sub-cultured on differential and selective media such as mannitol salt agar (Staphylococcus sp. confirmation test), MacConkey agar (Escherichia coli isolation test), Salmonela–Shigella agar (Salmonella and Shigella confirmation test), pseudomonas agar (Pseudomonas sp. confirmation test), and XLD (xylose lysine deoxycholate) agar media (Salmonella confirmation test) [10].
2.3 Morphological and biochemical characterization tests
The selected isolates underwent morphological and biochemical analysis [11].
2.4 Morphological tests
The morphology of growing bacterial colonies, such as size, color, shape, margin, elevation, texture, appearance, and opacity of each isolate, was tested and used for identification purposes [11].
2.5 Biochemical characterization
Isolates were characterized using various biochemical tests following the procedures set in Bergey’s Manual of Systematic Bacteriology and Determinative Bacteriology [11], including Gram straining, motility and catalase tests, citrate utilization tests, coagulase and urease production tests, indole production tests, oxidase tests, and Methyl Red–Voges Proskauer (MR–VP) tests.
2.6 Antibiotic susceptibility test
The antimicrobial susceptibility profile of isolates was assessed using the standard Kirby Bauer disk diffusion assay [12]. The inoculum of each bacterium was prepared by suspending the freshly grown bacteria in 5 mL sterile nutrient broth, and the turbidity was adjusted to an approximate value of 0.5 McFarland standard. Then, the antibiotic discs were placed over Mueller Hinton agar plates seeded with each bacterial isolate. The common antibiotics used for susceptibility tests were ampicillin (AMP, 10 µg), tetracycline (TET, 30 µg), gentamycin (GEN, 10 µg), amoxicillin (AMX, 10 µg), novobiocin (NV, 30 µg), erythromycin (E, 15 µg), and cotrimoxazole (COT, 25 µg). The Petri plates were incubated at 37°C for 24 h. The zone of growth inhibition around each disc was measured, and the results were interpreted according to the guidelines from the Clinical and Laboratory Standards Institute [13]. All experiments were done in triplicates. The experimental results were expressed as the mean of three replicates.
2.7 Plasmid DNA isolation and analysis
Those selected antibiotic-resistant isolates were subjected to plasmid analysis [14]. The isolates were grown in 10 mL of nutrient broth in test tubes and incubated at 37°C for 18–24 h. After incubation, 1,500 µL of each culture was transferred into 2 mL Eppendorf tubes for plasmid extraction, and glycerol was added to the remaining culture and stored at 4°C. The 1,500 µL bacterial culture in the Eppendorf tubes was centrifuged at 6,000 rpm for 8–10 min and placed in ice. Then, the supernatant was carefully removed using a fine-tip micropipette, and the pellet was thoroughly re-suspended in 100 µL of ice-cold solution I (containing sucrose and Tris–ethylene diamine tetra acetic acid). The suspension was vortexed gently, placed in ice for 5 min, and then shifted to room temperature.
After that, 200 µL of solution II (containing sodium hydroxide and sodium dodecyl sulfate) was added to the suspension containing pellets and solution I. Those pellet–suspension mixtures were mixed by inverting the mixture six to seven times, and the tube was maintained for 1 min at 0°C.
Then, 150 µL of solution III (containing acetic acid and potassium acetate) was added from the ice box to the sample containing test tubes. The content of each tube was gently mixed by inverting the tubes for five to six times and maintained at 0°C for 5 min. The tubes were centrifuged for 10 min at 6,000–8,000 rpm to yield a clear supernatant. This fractionation step separates the plasmid DNA from the cellular debris and chromosomal DNA in the pellet. Plasmid DNA renatures and remains in solution. Other components get precipitated. The supernatant was removed carefully without disturbing the pellet from each tube and transferred into new Eppendorf tubes, and 200 µL of ethanol was added to it. The precipitate was collected using centrifugation at 10,000 rpm for 20 min. Then, the supernatant was decanted, and the Eppendorf tube was inverted on bloating paper to drain left over, which was still stuck to the Eppendorf tube at the bottom side. The inverted tube was dried for about 10–15 min at room temperature. The dried pellet was suspended with 50 µL of 1× TE buffer and mixed by tapping by finger; the extracted plasmid DNA was quantified using nanodrop, and then the plasmid DNA in solution was applied to an agarose gel for electrophoresis. The samples were processed using horizontal gel electrophoresis to identify the number of plasmid copies present in different isolates of each cell. For this purpose, an agarose gel of 1% was used. Staining of DNA fragments was carried out using ethidium bromide (0.5 μg/mL), and they were visualized by UV-Trans illumination equipment. A standard DNA molecular weight marker was used to estimate the plasmid size. The standard DNA marker was used in this 1 kb ladder with 100 bp ladder gaps. This was done to check whether the isolated bacterial-resistant gene is plasmid-mediated or chromosomal DNA-mediated.
3 Results and discussion
3.1 Bacterial colony isolation and subculture and screening
In this study, a total of 16 effluent samples were collected from Kolladiba and Debark hospitals. Of these samples, 28 bacterial isolates were recovered. The bacterial isolates from Debark hospital effluent samples were designated as SD1, SD2, SD3, SD4, SD5, SD6, SD7, SD8, SD9, SD10, SD11, SD12, SD13, SD14, SD15, and SD16, while that of Kolladiba effluent samples were labeled as SK1, SK2, SK3, SK4, SK5, SK6, SK7, SK8, SK9, SK10, SK11, and SK12. The isolates were sub-cultured using differential and selective media. From these bacterial isolates, 12 (SD1, SD2, SD4, SD7, SD11, SD15, SD16, SK2, SK3, SK5, SK7, and SK8) were found to be multi-drug resistant. By the way, multi-drug resistance is defined as non-susceptibility to at least one agent in three or more classes of antibiotics.
3.2 Morphological and biochemical characteristics of multi-drug resistant bacteria
Colony morphology is the visual culture characteristics of a bacterial colony on an agar media. Observing colony morphology is an important skill used in the microbiology laboratory for the identification of microorganisms. Colonies need to be well isolated from other colonies to observe the characteristic shape, size, color, surface appearance, and texture. The different colony morphology characteristics, i.e., colony shape, size, texture, appearance, etc., were characterized for each of the 12 bacterial isolates. The details are described in Table 1.
Colony characteristics of selected bacterial isolate
| Isolate | Colony characteristics | |||||||
|---|---|---|---|---|---|---|---|---|
| Size | Color | Shape | Margin | Elevation | Texture | Appearance | Opacity | |
| SD1 | 1.0 mm | Bright yellow | Circular | Entire | Convex | Mucoid | Rough | Opaque |
| SD2 | 2 mm | Grey | Circular | Entire | Convex | Shiny | Smooth | Opaque |
| SD4 | 2.2 mm | White | Irregular | Entire | Raised | Shiny | Smooth | Transparent |
| SD7 | 1.5 mm | Yellow | Filamentous | Entire | Raised | Shiny | Smooth | Opaque |
| SD11 | 2 mm | Pale yellow | Circular | Entire | Convex | Shiny | Smooth | Transparent |
| SD15 | 1.8 mm | Colorless | Circular | Undulate | Raised | Shiny | Smooth | Opaque |
| SD16 | 1.9 mm | Yellow | Filamentous | Entire | Convex | Mucoid | Smooth | Transparent |
| SK2 | 0.8 m | Yellow | Spindle | Entire | Convex | Mucoid | Smooth | Transparent |
| SK3 | 2.6 mm | Colorless | Circular | Undulate | Raised | Shiny | Smooth | Opaque |
| SK5 | 1 mm | Golden yellow | Circular | Entire | Convex | Mucoid | Rough | Opaque |
| SK7 | 2.5 mm | White | Circular | Entire | Convex | Mucoid | Smooth | Opaque |
| SK8 | 2.0 m | White | Circular | Entire | Raised | Shiny | Smooth | Transparent |
In this study, ten biochemical tests were performed against each isolated multi-drug-resistant bacteria from effluent samples. Variations were observed for the different tests across the isolates. Each of the biochemical test results is presented in Table 2.
Biochemical test results for bacterial isolates from effluent Debark and Kolladiba hospital sample
| Tests | SD1 | SD2 | SD4 | SD7 | SD11 | SD15 | SD16 | SK2 | SK3 | SK5 | SK7 | SK8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram staining | + | − | − | + | − | − | − | − | − | + | − | − |
| Shape | Spherical | Rod | Rod | Spherical | Rod | Rod | Rod | Rod | Rod | Spherical | Rod | Rod |
| Indole test | − | − | + | − | − | − | − | − | − | − | − | + |
| Catalase test | + | + | + | + | + | + | + | + | + | + | + | + |
| Oxidase test | + | + | − | + | − | + | + | + | + | + | − | − |
| Urease test | − | − | − | − | − | + | − | − | + | − | − | − |
| Coagulase test | + | − | − | + | − | − | − | − | − | + | − | − |
| Citrate test | + | − | − | + | − | − | + | + | − | + | − | − |
| Motility test | − | + | + | − | − | + | − | − | + | − | + | + |
| Methyl red | + | − | + | + | + | − | − | − | − | + | + | + |
| VP test | + | − | − | − | − | − | + | + | − | + | − | − |
| Isolate name | Staphylococcus sp. | Citrobacter sp. | E. coli | Staphylococcus sp. | Shigella sp. | P. aeruginasa | K. pneumoniae | K. pneumoniae | P. aeruginosa | Staphylococcus sp. | Salmonella sp. | E. coli |
In this study, different bacterial colonies were isolated from hospital effluents and identified using morphological characteristics (shape, size, color, texture, margin, and elevation of the colony) and different biochemical tests (Gram staining, urease, catalase, motility, oxidase, indole production, coagulase, citrate, and MR-VP tests) [11]. Based on the morphological and biochemical test results, isolates were assigned as Staphylococcus sp. (SD1, SD7, and SK5), Citrobacter sp. (SD2), E. coli (SD4 and SK8), Shigella sp. (SD11), P. aeruginosa (SD15 and SK3), K. pneumoniae (SD16 and SK2), and Salmonella sp. (SK7).
3.3 Antibiotic susceptibility tests
The 28 bacterial isolates that were recovered from effluent samples were subjected to antibiotic susceptibility tests using seven commonly used antibiotics. Almost all of the commonly used antibiotics that were tested in this study failed to inhibit all 12 bacterial isolates, while the rest isolates were sensitive to all 7 antibiotics (Table 3).
Resistance rate of bacterial isolates from Debark and Kolladiba Hospital effluent samples against commonly used antibiotics
| Bacterial isolates | Antibiotic resistance N (%) | ||||||
|---|---|---|---|---|---|---|---|
| AMP | AMX | COT | GEN | NV | TET | E | |
| Staphylococcus sp. (n = 3) | 3(100) | 2(66.7) | 3(100) | 3(100) | 3(0) | 2(66.7) | 3(100) |
| E. coli (n = 2) | 1(50) | 1(50) | 1(100) | 1(50) | 1(50) | 2(100) | 2(100) |
| P. aeruginosa (n = 2) | 2(100) | 2(100) | 1(50) | 1(50) | 1(50) | 2(100) | 2(100) |
| Salmonella sp. (n = 1) | 1(0) | 1(0) | 1(0) | 1(100 | 1(0) | 1(100) | 1(100) |
| Citrobacter sp. (n = 1) | 1(0) | 1(100) | 1(100) | 1(0) | 1(100) | 1(0) | 1(100) |
| Shigella sp. (n = 1) | 1(100) | 1(100) | 1(100) | 1(100) | 1(0) | 1(0) | 1(100) |
| K. pneumoniae (n = 2) | 2(100) | 2(100) | 2(0) | 2(100) | 2(100) | 2(100) | 2(100) |
AMP: Ampicillin, TET: Tetracycline, GEN: Gentamycin, AMX: Amoxicillin, NV: Novobiocin, E: Erythromycin, COT: Cotrimoxazole.
Susceptibility to antibiotics result presented a varying degree of resistance to the different antibiotics tested against the tested bacteria. However, all isolates were sensitive to at least one of the seven antibiotics except against erythromycin (100% resistance). One isolate of E. coli (SK8) was 100% resistant to all the tested antibiotics. When we take into consideration about isolates showing resistant to more than five antibiotics, the distribution of multi-drug-resistant isolates in Debark and Kolladiba hospitals is still found to be high.
Antibiotic resistance is a major public health threat, and the presence of resistant organisms in environmental wastewaters/effluents is an emerging concern around the world. In this study, high incidences of antibiotics-resistant bacteria were isolated from Kolladiba and Debark Hospitals effluents. Out of 28 isolates, 12 were found to be resistant to at least one to seven tested antibiotics. This appeared to be analogous to what was predicted in many reports by researchers in Ethiopia that there is clear evidence of abuse of antibiotics for which the emergence of multi-drug-resistant bacteria is continuously increasing from time to time [15,16]. The indiscriminate use of antibiotics (misuse and overuse of antibiotics) currently has resulted in the increase of occurrence of antibiotic-resistant bacteria in the hospital effluents. The presence of a high rate of multi-drug-resistant bacteria in the hospital effluent is an alarm to take measures. This is because it may further result in disease spread and treatment failure. Similar results have been reported by Praveenkumarreddy et al. [17] in India and Molla et al. [18] in Gondar, Ethiopia. In this study, the antimicrobial susceptibility pattern shows that the isolates were highly resistant to the most commonly used antibiotics. One isolate of E. coli (SK8), which was isolated from the Kolladiba Hospital effluent sample, was 100% resistant to all the tested antibiotics (Table 3). Conversely, there was a high degree of resistance to erythromycin (100% resistance). The most effective antibiotic was novobiocin, with 58.3% inhibition effectiveness against the tested bacteria. A similar result was reported by Teshome et al. [19] in eastern Ethiopia, who isolated samples from hospital effluents. Study results by Osadebe and Okounim [20] also demonstrated that waste effluent from hospital environments contains a wide range of multi-drug-resistant bacteria.
3.4 Plasmid isolation and bacterial profile
Plasmid isolation and profiling of the 12 bacterial isolates that were resistant to the commonly used broad-spectrum antibiotics were performed. Plasmid profiles for the bacterial isolates revealed that some possessed four plasmids, just a few had single plasmid, and two of them had no plasmid. Isolates SD1 (L111), SK5 (L2), SD15 (L3), SK7(L5), SK3 (L6), SD11 (L9), SD2 (L10), SK2 (L11), and SD16 (L12) were having plasmid(s). Conversely, isolates SK8 (L4), SD4 (L7), and SD7 (L8) do not possess any plasmid when exposed to the horizontal gel electrophoresis, and it means that the multi-drug-resistant gene is not plasmid mediated but it may be chromosomal DNA-mediated. Therefore, the multi-drug resistance in these isolates may be plasmid or chromosomal DNA-mediated. The plasmid sizes for the isolates were found to have different base pair sizes (bps). SD1 (L1), SK7 (L5), and SK2 (L11) had one plasmid with a size of 100 bp, while SK5 (L2), SD15 (L3), SK3 (L6), and SD11 (L9) had two plasmid fragments of 100 bp and approximately 1,500 bp. SD16 (L12) had four plasmid sizes of 100, 900, 1,000, and approximately 1,500 bp, while SD2 (L10) contains two plasmids with sizes of 100 and approximately 1,800 bp. The result of the plasmid profile of the isolates is shown in Figure 3.
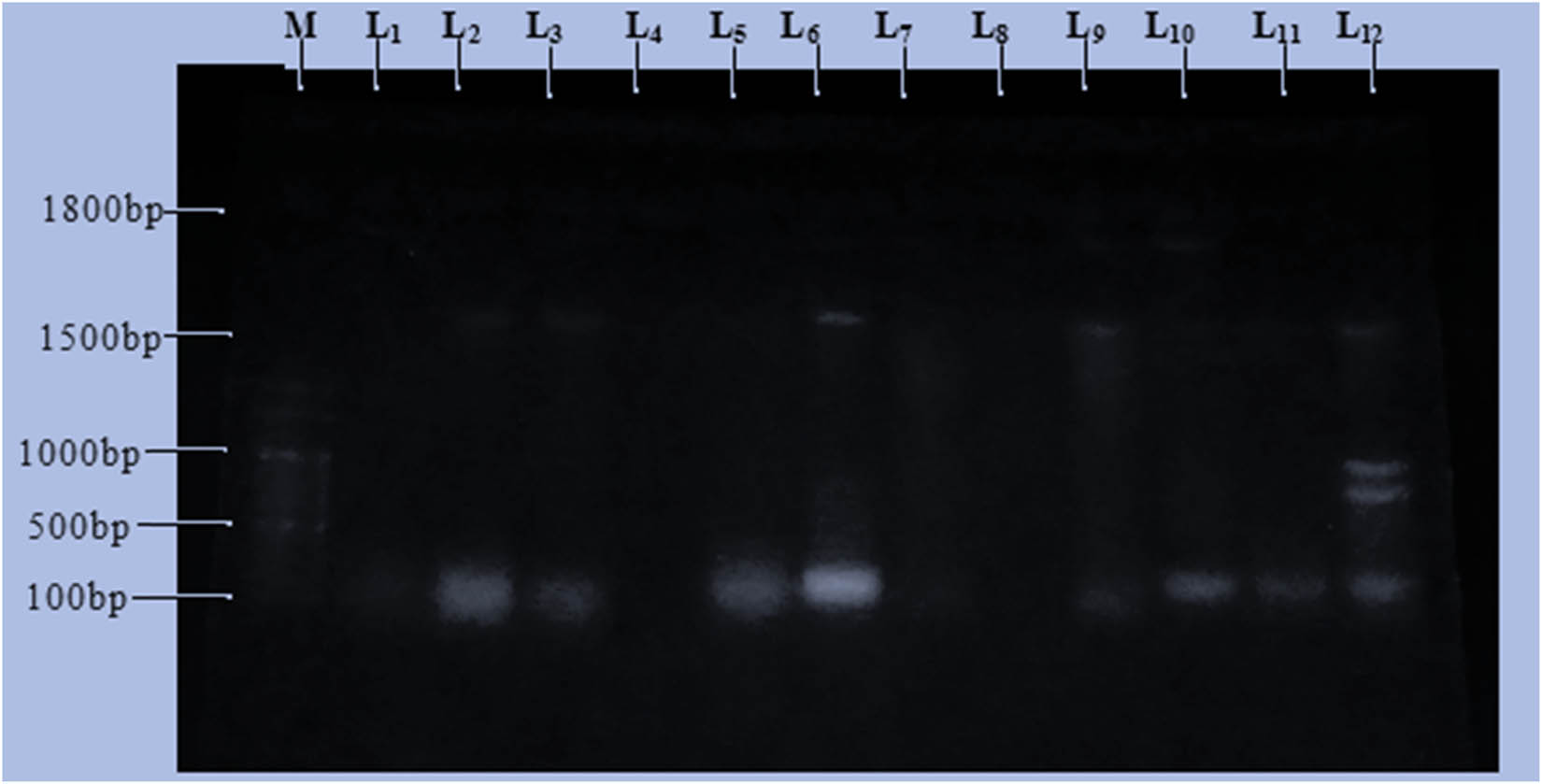
Plasmid size determination and a band size of 12 multi-drug-resistant bacteria. Key: M-Marker, L1-SD1, L2-SK5, L3-SD15, L4-SK8, L5-SK7, L6-SK3, L7-SD4, L8-SD7, L9-SD11, L10-SD2, L11-SK2, L12-SD16.
Plasmid profile studies are widely used for epidemiological studies, and the role of plasmids in drug resistance is well understood. In this study, the presence, number, and molecular size of plasmids were analyzed for the 12 multi-drug-resistant bacterial isolates. The isolates were found to contain various numbers of plasmids (1–4) with different sizes (from 100 to 1,800 bp) (Figure 1). The most common plasmid sizes encountered were 100 and 1,500 bps. These different-sized plasmids were responsible for the presence of multi-drug resistance. The isolates might acquire genetic elements (plasmids) from other bacteria. Large mobile genetic elements appear to encode both antibiotic-resistant factors and proteins that are responsible for the increase in virulence, thus giving the organism the ability to adapt to the selective pressure of antibiotics. This study correlates with the study done by Ojo et al. [21] and Olawale et al. [22]. The analysis of the plasmid profile makes it possible to estimate the dependence between antibiotic susceptibility and the presence of plasmids with respect to a particular isolate [20,22]. It can be said that resistance mediated by plasmid which has been observed in various studies such as Ojo et al. [21], Olawale et al. [22], Jaran [23], Agrahari et al. [24], Nmesirionye et al. [25], and Olorunfemi et al. [26] may be responsible for the differences observed in treatment by clinicians. The gene responsible for multi-drug resistance that occurred in bacterial isolates under this study may be grouped into chromosomal mediated [(SK8 (L4), SD4 (L7), and SD7 (L8)] and plasmid-mediated (the rest isolates).
3.5 Limitation of this study
The limitation of our study was the small number of sample sizes collected due to lack of resources.
4 Conclusions
This study aimed to investigate the presence of antibiotic resistance patterns and bacterial profiles of some multi-drug-resistant bacteria isolated from effluents of Kolladiba and Debark Hospitals. The findings reveal that effluents of Kolladiba and Debark Hospitals have a high incidence of multi-drug-resistant bacteria. From a total of 28 bacterial isolates, 12 were found to be multi-drug resistant. Among the tested antibiotics, erythromycin was the most resisted antibiotic, while novobiocin was the most effective antibiotic. A plasmid profile study of the isolates revealed both the presence and absence of plasmids. The number of plasmids ranged from zero to four, with plasmid sizes of 100, 900, 1,000, 1,400, 1,500, and 1,800 base pairs. Most of multi-drug resistances could be due to plasmid genes. To reduce the incidence of drug resistance, the unwise use of antibiotics by individuals whose health is impaired should be discouraged. In order to reduce the impact of hospital effluents on the spread of antimicrobial resistance in the community, a proper effluent treatment mechanism should be devised.
Acknowledgements
Both authors would like to acknowledge University of Gondar.
-
Funding information: Authors state no funding involved.
-
Author contributions: E.A. performed the experiments as part of his master’s thesis work. This work was carried out under the supervision of T.M.J. T.M.J. helped in editing the manuscript. Both authors read and approved the final manuscript.
-
Conflict of interest: Authors state no conflicts of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Warren C, Mespa M, Ephraim C, Justin C, Mathias T, Joseph K, et al. Evaluation of antibiotic susceptibility patterns of pathogens isolated from routine laboratory specimens at Ndola Teaching Hospital: A retrospective study. PLoS One. 2019;14(12):1–15. 10.1371/journal.pone.0226676.Search in Google Scholar PubMed PubMed Central
[2] Adugna C, Sivalingam KM. Prevalence of multiple drug-resistant bacteria in the main campus wastewater treatment plant of Wolaita Sodo University, Southern Ethiopia. Int J Microbiol. 2020;2020:1–9. 10.1155/2022/1781518.1781518.Search in Google Scholar
[3] Stanton IC, Bethel A, Leonard AFC, Gaze WH, Garside R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: a systematic map. Environ Evidence. 2022;11:8. 10.1186/s13750-022-00262-2.Search in Google Scholar PubMed PubMed Central
[4] Larsson DGJ, Flach C-F. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20(5):257–69. 10.1038/s41579-021-00649-x.Search in Google Scholar PubMed PubMed Central
[5] Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske GG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. 10.1111/j.1469-0691.2011.03570.x.Search in Google Scholar PubMed
[6] Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38. 10.1128/AAC.01707-08.Search in Google Scholar PubMed PubMed Central
[7] Li F, Son M, Xu L, Deng B, Zhu S, Li X. Risk factors for catheter-associated urinary tract infection among hospitalized patients: A systematic review and meta-analysis of observational studies. J Adv Nurs. 2019;75(3):517–27. 10.1111/jan.13863.Search in Google Scholar PubMed
[8] Mutai WC, Peter GW, Samuel K, Anne WTM. Plasmid profiling and incompatibility grouping of multi-drug resistant Salmonella enteric serovar typhi isolates in Nairobi, Kenya. BMC Res Notes. 2019;12:422. 10.1186/s13104-019-4468-9.Search in Google Scholar PubMed PubMed Central
[9] Tefera B, Ruelle ML, Asfaw Z, Tsegay BA. Woody plant diversity in an Afromontane agricultural landscape (Debark District, northern Ethiopia). Forests Trees Livelihoods. 2014;1–19. 10.1080/14728028.2014.942709.Search in Google Scholar
[10] Feleke M, Mengistu E, Andargachew M, Belay T, Yeshambel B, Yitayal S, et al. The growing challenges of antibacterial drug resistance in Ethiopia. J Glob Antimicrob Resist. 2014;2(3):148–54. 10.1016/j.jgar.2014.02.004.Search in Google Scholar PubMed
[11] Williams ST, Sharpe ME, Holt JG. Bergey’s manual of systematic bacteriology. Vol. 4, Baltimore: William and Wilkins Co.; 1989.Search in Google Scholar
[12] Bauer AW, Kirby WM, Sherirs JC, Turck M. Antibiotic susceptibility testing by standardized single disk method. Am J Clin Pathol. 1966;45:433–96. 10.1093/ajcp/45.4_ts.493.Search in Google Scholar
[13] Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. 30th edn. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.Search in Google Scholar
[14] He F. Plasmid DNA extraction from E. coli using alkaline lysis. Method Mol Biol. 2011;101:e30. 10.21769/BioProtoc.30.Search in Google Scholar
[15] Muhie OA. Antibiotic use and resistance pattern in Ethiopia: systematic review and meta analysis. Int J Microbiol. 2019;2019:1–8. 10.1155/2019/2489063.Search in Google Scholar PubMed PubMed Central
[16] Alemu B, Assefa A, Bedasa M, Amenu K, Wieleand KB. Antimicrobial resistance in Ethiopia: A systematic review and meta-analysis of prevalence in foods, food handlers, animals, and the environment. One Health. 2021;13:100286. 10.1016/j.onehlt.2021.100286.Search in Google Scholar PubMed PubMed Central
[17] Praveenkumarreddy Y, Akiba M, Guruge KS, Balakrishna K, Vandana KE, Kumar V. Occurrence of antimicrobial-resistant Escherichia coli in sewage treatment plants of South India. J Water, Sanit Hyg Dev. 2020;10(1):48–55. 10.2166/washdev.2020.051.Search in Google Scholar
[18] Molla R, Tiruneh M, Abebe W, Moges F. Bacterial profile and antimicrobial susceptibility patterns in chronic suppurative otitis media at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Res Notes. 2019;12:414. 10.1186/s13104-019-4452-4.Search in Google Scholar PubMed PubMed Central
[19] Teshome A, Alemayehu T, Deriba W, Ayele Y. Antibiotic resistance profile of bacteria isolated from wastewater systems in eastern ethiopia. J Environ Public Health. 2020;2020:1–10. 10.1155/2020/2796365.Search in Google Scholar PubMed PubMed Central
[20] Osadebe AU, Okounim B. Multidrug-resistant bacteria in the wastewater of the hospitals in Port Harcourt metropolis: Implications for environmental health. J Adv Environ Health Res. 2020;8:46–55. 10.22102/jaehr. 2020. 200910.1144.Search in Google Scholar
[21] Ojo SKS, Sargin BO, Esumeh FI. Plasmid curing analysis of antibiotic resistance in β-lactamase producing staphylococci from wounds and burns patients. Pak J Biol Sci. 2014;17:130–3. 10.3923/pjbs.2014.130.133.Search in Google Scholar PubMed
[22] Olawale SI, Busayo OOM, Olatunji OI, Mariam M, Olayinka OS. Plasmid profiles and antibiotic susceptibility patterns of bacteria isolated from abattoirs wastewater within Ilorin, Kwara, Nigeria. Iran J Microbiol. 2020;12(6):547–55. 10.18502/ijm.v12i6.5029.Search in Google Scholar PubMed PubMed Central
[23] Jaran AS. Antimicrobial resistance patterns and plasmid profiles of staphylococcus aureus isolated from different clinical specimens in Saudi Arabia. Eur Sci J. 2017;13(9):1–9.10.19044/esj.2017.v13n9p1Search in Google Scholar
[24] Agrahari G, Koirala A, Thapa R, Chaudhary MK, Tuladhar R. Antimicrobial resistance patterns and plasmid profiles of methicillin resistant Staphylococcus aureus isolated from clinical samples. Nepal J Biotechnol. 2019;7(1):8–14.10.3126/njb.v7i1.26945Search in Google Scholar
[25] Nmesirionye BU, Ugwu CT, Nworie KM. Antibiotic Susceptibility and Plasmid Profile of Multidrug resistant Uropathogenic Serratia marcescens. J Microbiol Infect Dis. 2022;12(1):12–8.10.5799/jmid.1085914Search in Google Scholar
[26] Olorunfemi PO, Damshit JD, Chris-Otubor GO. Antimicrobial susceptibility pattern and plasmid profiles of bacterial isolates from four hospitals in JOS Metropolis. Afr J Pharm Res Dev. 2023;15(2):23–35.Search in Google Scholar
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Constitutive and evoked release of ATP in adult mouse olfactory epithelium
- LARP1 knockdown inhibits cultured gastric carcinoma cell cycle progression and metastatic behavior
- PEGylated porcine–human recombinant uricase: A novel fusion protein with improved efficacy and safety for the treatment of hyperuricemia and renal complications
- Research progress on ocular complications caused by type 2 diabetes mellitus and the function of tears and blepharons
- The role and mechanism of esketamine in preventing and treating remifentanil-induced hyperalgesia based on the NMDA receptor–CaMKII pathway
- Brucella infection combined with Nocardia infection: A case report and literature review
- Detection of serum interleukin-18 level and neutrophil/lymphocyte ratio in patients with antineutrophil cytoplasmic antibody-associated vasculitis and its clinical significance
- Ang-1, Ang-2, and Tie2 are diagnostic biomarkers for Henoch-Schönlein purpura and pediatric-onset systemic lupus erythematous
- PTTG1 induces pancreatic cancer cell proliferation and promotes aerobic glycolysis by regulating c-myc
- Role of serum B-cell-activating factor and interleukin-17 as biomarkers in the classification of interstitial pneumonia with autoimmune features
- Effectiveness and safety of a mumps containing vaccine in preventing laboratory-confirmed mumps cases from 2002 to 2017: A meta-analysis
- Low levels of sex hormone-binding globulin predict an increased breast cancer risk and its underlying molecular mechanisms
- A case of Trousseau syndrome: Screening, detection and complication
- Application of the integrated airway humidification device enhances the humidification effect of the rabbit tracheotomy model
- Preparation of Cu2+/TA/HAP composite coating with anti-bacterial and osteogenic potential on 3D-printed porous Ti alloy scaffolds for orthopedic applications
- Aquaporin-8 promotes human dermal fibroblasts to counteract hydrogen peroxide-induced oxidative damage: A novel target for management of skin aging
- Current research and evidence gaps on placental development in iron deficiency anemia
- Single-nucleotide polymorphism rs2910829 in PDE4D is related to stroke susceptibility in Chinese populations: The results of a meta-analysis
- Pheochromocytoma-induced myocardial infarction: A case report
- Kaempferol regulates apoptosis and migration of neural stem cells to attenuate cerebral infarction by O‐GlcNAcylation of β-catenin
- Sirtuin 5 regulates acute myeloid leukemia cell viability and apoptosis by succinylation modification of glycine decarboxylase
- Apigenin 7-glucoside impedes hypoxia-induced malignant phenotypes of cervical cancer cells in a p16-dependent manner
- KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1
- Current state of research on copper complexes in the treatment of breast cancer
- Exploring antioxidant strategies in the pathogenesis of ALS
- Helicobacter pylori causes gastric dysbacteriosis in chronic gastritis patients
- IL-33/soluble ST2 axis is associated with radiation-induced cardiac injury
- The predictive value of serum NLR, SII, and OPNI for lymph node metastasis in breast cancer patients with internal mammary lymph nodes after thoracoscopic surgery
- Carrying SNP rs17506395 (T > G) in TP63 gene and CCR5Δ32 mutation associated with the occurrence of breast cancer in Burkina Faso
- P2X7 receptor: A receptor closely linked with sepsis-associated encephalopathy
- Probiotics for inflammatory bowel disease: Is there sufficient evidence?
- Identification of KDM4C as a gene conferring drug resistance in multiple myeloma
- Microbial perspective on the skin–gut axis and atopic dermatitis
- Thymosin α1 combined with XELOX improves immune function and reduces serum tumor markers in colorectal cancer patients after radical surgery
- Highly specific vaginal microbiome signature for gynecological cancers
- Sample size estimation for AQP4-IgG seropositive optic neuritis: Retinal damage detection by optical coherence tomography
- The effects of SDF-1 combined application with VEGF on femoral distraction osteogenesis in rats
- Fabrication and characterization of gold nanoparticles using alginate: In vitro and in vivo assessment of its administration effects with swimming exercise on diabetic rats
- Mitigating digestive disorders: Action mechanisms of Mediterranean herbal active compounds
- Distribution of CYP2D6 and CYP2C19 gene polymorphisms in Han and Uygur populations with breast cancer in Xinjiang, China
- VSP-2 attenuates secretion of inflammatory cytokines induced by LPS in BV2 cells by mediating the PPARγ/NF-κB signaling pathway
- Factors influencing spontaneous hypothermia after emergency trauma and the construction of a predictive model
- Long-term administration of morphine specifically alters the level of protein expression in different brain regions and affects the redox state
- Application of metagenomic next-generation sequencing technology in the etiological diagnosis of peritoneal dialysis-associated peritonitis
- Clinical diagnosis, prevention, and treatment of neurodyspepsia syndrome using intelligent medicine
- Case report: Successful bronchoscopic interventional treatment of endobronchial leiomyomas
- Preliminary investigation into the genetic etiology of short stature in children through whole exon sequencing of the core family
- Cystic adenomyoma of the uterus: Case report and literature review
- Mesoporous silica nanoparticles as a drug delivery mechanism
- Dynamic changes in autophagy activity in different degrees of pulmonary fibrosis in mice
- Vitamin D deficiency and inflammatory markers in type 2 diabetes: Big data insights
- Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells
- Meta-analysis on the efficacy of allogeneic hematopoietic stem cell transplantation to treat malignant lymphoma
- Mitochondrial DNA drives neuroinflammation through the cGAS-IFN signaling pathway in the spinal cord of neuropathic pain mice
- Application value of artificial intelligence algorithm-based magnetic resonance multi-sequence imaging in staging diagnosis of cervical cancer
- Embedded monitoring system and teaching of artificial intelligence online drug component recognition
- Investigation into the association of FNDC1 and ADAMTS12 gene expression with plumage coloration in Muscovy ducks
- Yak meat content in feed and its impact on the growth of rats
- A rare case of Richter transformation with breast involvement: A case report and literature review
- First report of Nocardia wallacei infection in an immunocompetent patient in Zhejiang province
- Rhodococcus equi and Brucella pulmonary mass in immunocompetent: A case report and literature review
- Downregulation of RIP3 ameliorates the left ventricular mechanics and function after myocardial infarction via modulating NF-κB/NLRP3 pathway
- Evaluation of the role of some non-enzymatic antioxidants among Iraqi patients with non-alcoholic fatty liver disease
- The role of Phafin proteins in cell signaling pathways and diseases
- Ten-year anemia as initial manifestation of Castleman disease in the abdominal cavity: A case report
- Coexistence of hereditary spherocytosis with SPTB P.Trp1150 gene variant and Gilbert syndrome: A case report and literature review
- Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells
- Exploratory evaluation supported by experimental and modeling approaches of Inula viscosa root extract as a potent corrosion inhibitor for mild steel in a 1 M HCl solution
- Imaging manifestations of ductal adenoma of the breast: A case report
- Gut microbiota and sleep: Interaction mechanisms and therapeutic prospects
- Isomangiferin promotes the migration and osteogenic differentiation of rat bone marrow mesenchymal stem cells
- Prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer
- Circular RNAs as potential biomarkers for male severe sepsis
- Knockdown of Stanniocalcin-1 inhibits growth and glycolysis in oral squamous cell carcinoma cells
- The expression and biological role of complement C1s in esophageal squamous cell carcinoma
- A novel GNAS mutation in pseudohypoparathyroidism type 1a with articular flexion deformity: A case report
- Predictive value of serum magnesium levels for prognosis in patients with non-small cell lung cancer undergoing EGFR-TKI therapy
- HSPB1 alleviates acute-on-chronic liver failure via the P53/Bax pathway
- IgG4-related disease complicated by PLA2R-associated membranous nephropathy: A case report
- Baculovirus-mediated endostatin and angiostatin activation of autophagy through the AMPK/AKT/mTOR pathway inhibits angiogenesis in hepatocellular carcinoma
- Metformin mitigates osteoarthritis progression by modulating the PI3K/AKT/mTOR signaling pathway and enhancing chondrocyte autophagy
- Evaluation of the activity of antimicrobial peptides against bacterial vaginosis
- Atypical presentation of γ/δ mycosis fungoides with an unusual phenotype and SOCS1 mutation
- Analysis of the microecological mechanism of diabetic kidney disease based on the theory of “gut–kidney axis”: A systematic review
- Omega-3 fatty acids prevent gestational diabetes mellitus via modulation of lipid metabolism
- Refractory hypertension complicated with Turner syndrome: A case report
- Interaction of ncRNAs and the PI3K/AKT/mTOR pathway: Implications for osteosarcoma
- Association of low attenuation area scores with pulmonary function and clinical prognosis in patients with chronic obstructive pulmonary disease
- Long non-coding RNAs in bone formation: Key regulators and therapeutic prospects
- The deubiquitinating enzyme USP35 regulates the stability of NRF2 protein
- Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as potential diagnostic markers for rebleeding in patients with esophagogastric variceal bleeding
- G protein-coupled receptor 1 participating in the mechanism of mediating gestational diabetes mellitus by phosphorylating the AKT pathway
- LL37-mtDNA regulates viability, apoptosis, inflammation, and autophagy in lipopolysaccharide-treated RLE-6TN cells by targeting Hsp90aa1
- The analgesic effect of paeoniflorin: A focused review
- Chemical composition’s effect on Solanum nigrum Linn.’s antioxidant capacity and erythrocyte protection: Bioactive components and molecular docking analysis
- Knockdown of HCK promotes HREC cell viability and inner blood–retinal barrier integrity by regulating the AMPK signaling pathway
- The role of rapamycin in the PINK1/Parkin signaling pathway in mitophagy in podocytes
- Laryngeal non-Hodgkin lymphoma: Report of four cases and review of the literature
- Clinical value of macrogenome next-generation sequencing on infections
- Overview of dendritic cells and related pathways in autoimmune uveitis
- TAK-242 alleviates diabetic cardiomyopathy via inhibiting pyroptosis and TLR4/CaMKII/NLRP3 pathway
- Hypomethylation in promoters of PGC-1α involved in exercise-driven skeletal muscular alterations in old age
- Profile and antimicrobial susceptibility patterns of bacteria isolated from effluents of Kolladiba and Debark hospitals
- The expression and clinical significance of syncytin-1 in serum exosomes of hepatocellular carcinoma patients
- A histomorphometric study to evaluate the therapeutic effects of biosynthesized silver nanoparticles on the kidneys infected with Plasmodium chabaudi
- PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
- Analysis of MDA, SOD, TAOC, MNCV, SNCV, and TSS scores in patients with diabetes peripheral neuropathy
- SLIT3 deficiency promotes non-small cell lung cancer progression by modulating UBE2C/WNT signaling
- The relationship between TMCO1 and CALR in the pathological characteristics of prostate cancer and its effect on the metastasis of prostate cancer cells
- Heterogeneous nuclear ribonucleoprotein K is a potential target for enhancing the chemosensitivity of nasopharyngeal carcinoma
- PHB2 alleviates retinal pigment epithelium cell fibrosis by suppressing the AGE–RAGE pathway
- Anti-γ-aminobutyric acid-B receptor autoimmune encephalitis with syncope as the initial symptom: Case report and literature review
- Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua
- Human umbilical cord mesenchymal stem cells regulate glutathione metabolism depending on the ERK–Nrf2–HO-1 signal pathway to repair phosphoramide mustard-induced ovarian cancer cells
- Electroacupuncture on GB acupoints improves osteoporosis via the estradiol–PI3K–Akt signaling pathway
- Renalase protects against podocyte injury by inhibiting oxidative stress and apoptosis in diabetic nephropathy
- Review: Dicranostigma leptopodum: A peculiar plant of Papaveraceae
- Combination effect of flavonoids attenuates lung cancer cell proliferation by inhibiting the STAT3 and FAK signaling pathway
- Renal microangiopathy and immune complex glomerulonephritis induced by anti-tumour agents: A case report
- Correlation analysis of AVPR1a and AVPR2 with abnormal water and sodium and potassium metabolism in rats
- Gastrointestinal health anti-diarrheal mixture relieves spleen deficiency-induced diarrhea through regulating gut microbiota
- Myriad factors and pathways influencing tumor radiotherapy resistance
- Exploring the effects of culture conditions on Yapsin (YPS) gene expression in Nakaseomyces glabratus
- Screening of prognostic core genes based on cell–cell interaction in the peripheral blood of patients with sepsis
- Coagulation factor II thrombin receptor as a promising biomarker in breast cancer management
- Ileocecal mucinous carcinoma misdiagnosed as incarcerated hernia: A case report
- Methyltransferase like 13 promotes malignant behaviors of bladder cancer cells through targeting PI3K/ATK signaling pathway
- The debate between electricity and heat, efficacy and safety of irreversible electroporation and radiofrequency ablation in the treatment of liver cancer: A meta-analysis
- ZAG promotes colorectal cancer cell proliferation and epithelial–mesenchymal transition by promoting lipid synthesis
- Baicalein inhibits NLRP3 inflammasome activation and mitigates placental inflammation and oxidative stress in gestational diabetes mellitus
- Impact of SWCNT-conjugated senna leaf extract on breast cancer cells: A potential apoptotic therapeutic strategy
- MFAP5 inhibits the malignant progression of endometrial cancer cells in vitro
- Major ozonated autohemotherapy promoted functional recovery following spinal cord injury in adult rats via the inhibition of oxidative stress and inflammation
- Axodendritic targeting of TAU and MAP2 and microtubule polarization in iPSC-derived versus SH-SY5Y-derived human neurons
- Differential expression of phosphoinositide 3-kinase/protein kinase B and Toll-like receptor/nuclear factor kappa B signaling pathways in experimental obesity Wistar rat model
- The therapeutic potential of targeting Oncostatin M and the interleukin-6 family in retinal diseases: A comprehensive review
- BA inhibits LPS-stimulated inflammatory response and apoptosis in human middle ear epithelial cells by regulating the Nf-Kb/Iκbα axis
- Role of circRMRP and circRPL27 in chronic obstructive pulmonary disease
- Investigating the role of hyperexpressed HCN1 in inducing myocardial infarction through activation of the NF-κB signaling pathway
- Characterization of phenolic compounds and evaluation of anti-diabetic potential in Cannabis sativa L. seeds: In vivo, in vitro, and in silico studies
- Quantitative immunohistochemistry analysis of breast Ki67 based on artificial intelligence
- Ecology and Environmental Science
- Screening of different growth conditions of Bacillus subtilis isolated from membrane-less microbial fuel cell toward antimicrobial activity profiling
- Degradation of a mixture of 13 polycyclic aromatic hydrocarbons by commercial effective microorganisms
- Evaluation of the impact of two citrus plants on the variation of Panonychus citri (Acari: Tetranychidae) and beneficial phytoseiid mites
- Prediction of present and future distribution areas of Juniperus drupacea Labill and determination of ethnobotany properties in Antalya Province, Türkiye
- Population genetics of Todarodes pacificus (Cephalopoda: Ommastrephidae) in the northwest Pacific Ocean via GBS sequencing
- A comparative analysis of dendrometric, macromorphological, and micromorphological characteristics of Pistacia atlantica subsp. atlantica and Pistacia terebinthus in the middle Atlas region of Morocco
- Macrofungal sporocarp community in the lichen Scots pine forests
- Assessing the proximate compositions of indigenous forage species in Yemen’s pastoral rangelands
- Food Science
- Gut microbiota changes associated with low-carbohydrate diet intervention for obesity
- Reexamination of Aspergillus cristatus phylogeny in dark tea: Characteristics of the mitochondrial genome
- Differences in the flavonoid composition of the leaves, fruits, and branches of mulberry are distinguished based on a plant metabolomics approach
- Investigating the impact of wet rendering (solventless method) on PUFA-rich oil from catfish (Clarias magur) viscera
- Non-linear associations between cardiovascular metabolic indices and metabolic-associated fatty liver disease: A cross-sectional study in the US population (2017–2020)
- Knockdown of USP7 alleviates atherosclerosis in ApoE-deficient mice by regulating EZH2 expression
- Utility of dairy microbiome as a tool for authentication and traceability
- Agriculture
- Enhancing faba bean (Vicia faba L.) productivity through establishing the area-specific fertilizer rate recommendation in southwest Ethiopia
- Impact of novel herbicide based on synthetic auxins and ALS inhibitor on weed control
- Perspectives of pteridophytes microbiome for bioremediation in agricultural applications
- Fertilizer application parameters for drip-irrigated peanut based on the fertilizer effect function established from a “3414” field trial
- Improving the productivity and profitability of maize (Zea mays L.) using optimum blended inorganic fertilization
- Application of leaf multispectral analyzer in comparison to hyperspectral device to assess the diversity of spectral reflectance indices in wheat genotypes
- Animal Sciences
- Knockdown of ANP32E inhibits colorectal cancer cell growth and glycolysis by regulating the AKT/mTOR pathway
- Development of a detection chip for major pathogenic drug-resistant genes and drug targets in bovine respiratory system diseases
- Exploration of the genetic influence of MYOT and MB genes on the plumage coloration of Muscovy ducks
- Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism
- Comparison of nutritional value of the wild and cultivated spiny loaches at three growth stages
- Transcriptomic analysis of liver immune response in Chinese spiny frog (Quasipaa spinosa) infected with Proteus mirabilis
- Disruption of BCAA degradation is a critical characteristic of diabetic cardiomyopathy revealed by integrated transcriptome and metabolome analysis
- Plant Sciences
- Effect of long-term in-row branch covering on soil microorganisms in pear orchards
- Photosynthetic physiological characteristics, growth performance, and element concentrations reveal the calcicole–calcifuge behaviors of three Camellia species
- Transcriptome analysis reveals the mechanism of NaHCO3 promoting tobacco leaf maturation
- Bioinformatics, expression analysis, and functional verification of allene oxide synthase gene HvnAOS1 and HvnAOS2 in qingke
- Water, nitrogen, and phosphorus coupling improves gray jujube fruit quality and yield
- Improving grape fruit quality through soil conditioner: Insights from RNA-seq analysis of Cabernet Sauvignon roots
- Role of Embinin in the reabsorption of nucleus pulposus in lumbar disc herniation: Promotion of nucleus pulposus neovascularization and apoptosis of nucleus pulposus cells
- Revealing the effects of amino acid, organic acid, and phytohormones on the germination of tomato seeds under salinity stress
- Combined effects of nitrogen fertilizer and biochar on the growth, yield, and quality of pepper
- Comprehensive phytochemical and toxicological analysis of Chenopodium ambrosioides (L.) fractions
- Impact of “3414” fertilization on the yield and quality of greenhouse tomatoes
- Exploring the coupling mode of water and fertilizer for improving growth, fruit quality, and yield of the pear in the arid region
- Metagenomic analysis of endophytic bacteria in seed potato (Solanum tuberosum)
- Antibacterial, antifungal, and phytochemical properties of Salsola kali ethanolic extract
- Exploring the hepatoprotective properties of citronellol: In vitro and in silico studies on ethanol-induced damage in HepG2 cells
- Enhanced osmotic dehydration of watermelon rind using honey–sucrose solutions: A study on pre-treatment efficacy and mass transfer kinetics
- Effects of exogenous 2,4-epibrassinolide on photosynthetic traits of 53 cowpea varieties under NaCl stress
- Comparative transcriptome analysis of maize (Zea mays L.) seedlings in response to copper stress
- An optimization method for measuring the stomata in cassava (Manihot esculenta Crantz) under multiple abiotic stresses
- Fosinopril inhibits Ang II-induced VSMC proliferation, phenotype transformation, migration, and oxidative stress through the TGF-β1/Smad signaling pathway
- Antioxidant and antimicrobial activities of Salsola imbricata methanolic extract and its phytochemical characterization
- Bioengineering and Biotechnology
- Absorbable calcium and phosphorus bioactive membranes promote bone marrow mesenchymal stem cells osteogenic differentiation for bone regeneration
- New advances in protein engineering for industrial applications: Key takeaways
- An overview of the production and use of Bacillus thuringiensis toxin
- Research progress of nanoparticles in diagnosis and treatment of hepatocellular carcinoma
- Bioelectrochemical biosensors for water quality assessment and wastewater monitoring
- PEI/MMNs@LNA-542 nanoparticles alleviate ICU-acquired weakness through targeted autophagy inhibition and mitochondrial protection
- Unleashing of cytotoxic effects of thymoquinone-bovine serum albumin nanoparticles on A549 lung cancer cells
- Erratum
- Erratum to “Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM”
- Erratum to “Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation”
- Retraction
- Retraction to “MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB”
- Retraction to “A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis”
- Special Issue on Advances in Neurodegenerative Disease Research and Treatment
- Transplantation of human neural stem cell prevents symptomatic motor behavior disability in a rat model of Parkinson’s disease
- Special Issue on Multi-omics
- Inflammasome complex genes with clinical relevance suggest potential as therapeutic targets for anti-tumor drugs in clear cell renal cell carcinoma
- Gastroesophageal varices in primary biliary cholangitis with anti-centromere antibody positivity: Early onset?
Articles in the same Issue
- Biomedical Sciences
- Constitutive and evoked release of ATP in adult mouse olfactory epithelium
- LARP1 knockdown inhibits cultured gastric carcinoma cell cycle progression and metastatic behavior
- PEGylated porcine–human recombinant uricase: A novel fusion protein with improved efficacy and safety for the treatment of hyperuricemia and renal complications
- Research progress on ocular complications caused by type 2 diabetes mellitus and the function of tears and blepharons
- The role and mechanism of esketamine in preventing and treating remifentanil-induced hyperalgesia based on the NMDA receptor–CaMKII pathway
- Brucella infection combined with Nocardia infection: A case report and literature review
- Detection of serum interleukin-18 level and neutrophil/lymphocyte ratio in patients with antineutrophil cytoplasmic antibody-associated vasculitis and its clinical significance
- Ang-1, Ang-2, and Tie2 are diagnostic biomarkers for Henoch-Schönlein purpura and pediatric-onset systemic lupus erythematous
- PTTG1 induces pancreatic cancer cell proliferation and promotes aerobic glycolysis by regulating c-myc
- Role of serum B-cell-activating factor and interleukin-17 as biomarkers in the classification of interstitial pneumonia with autoimmune features
- Effectiveness and safety of a mumps containing vaccine in preventing laboratory-confirmed mumps cases from 2002 to 2017: A meta-analysis
- Low levels of sex hormone-binding globulin predict an increased breast cancer risk and its underlying molecular mechanisms
- A case of Trousseau syndrome: Screening, detection and complication
- Application of the integrated airway humidification device enhances the humidification effect of the rabbit tracheotomy model
- Preparation of Cu2+/TA/HAP composite coating with anti-bacterial and osteogenic potential on 3D-printed porous Ti alloy scaffolds for orthopedic applications
- Aquaporin-8 promotes human dermal fibroblasts to counteract hydrogen peroxide-induced oxidative damage: A novel target for management of skin aging
- Current research and evidence gaps on placental development in iron deficiency anemia
- Single-nucleotide polymorphism rs2910829 in PDE4D is related to stroke susceptibility in Chinese populations: The results of a meta-analysis
- Pheochromocytoma-induced myocardial infarction: A case report
- Kaempferol regulates apoptosis and migration of neural stem cells to attenuate cerebral infarction by O‐GlcNAcylation of β-catenin
- Sirtuin 5 regulates acute myeloid leukemia cell viability and apoptosis by succinylation modification of glycine decarboxylase
- Apigenin 7-glucoside impedes hypoxia-induced malignant phenotypes of cervical cancer cells in a p16-dependent manner
- KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1
- Current state of research on copper complexes in the treatment of breast cancer
- Exploring antioxidant strategies in the pathogenesis of ALS
- Helicobacter pylori causes gastric dysbacteriosis in chronic gastritis patients
- IL-33/soluble ST2 axis is associated with radiation-induced cardiac injury
- The predictive value of serum NLR, SII, and OPNI for lymph node metastasis in breast cancer patients with internal mammary lymph nodes after thoracoscopic surgery
- Carrying SNP rs17506395 (T > G) in TP63 gene and CCR5Δ32 mutation associated with the occurrence of breast cancer in Burkina Faso
- P2X7 receptor: A receptor closely linked with sepsis-associated encephalopathy
- Probiotics for inflammatory bowel disease: Is there sufficient evidence?
- Identification of KDM4C as a gene conferring drug resistance in multiple myeloma
- Microbial perspective on the skin–gut axis and atopic dermatitis
- Thymosin α1 combined with XELOX improves immune function and reduces serum tumor markers in colorectal cancer patients after radical surgery
- Highly specific vaginal microbiome signature for gynecological cancers
- Sample size estimation for AQP4-IgG seropositive optic neuritis: Retinal damage detection by optical coherence tomography
- The effects of SDF-1 combined application with VEGF on femoral distraction osteogenesis in rats
- Fabrication and characterization of gold nanoparticles using alginate: In vitro and in vivo assessment of its administration effects with swimming exercise on diabetic rats
- Mitigating digestive disorders: Action mechanisms of Mediterranean herbal active compounds
- Distribution of CYP2D6 and CYP2C19 gene polymorphisms in Han and Uygur populations with breast cancer in Xinjiang, China
- VSP-2 attenuates secretion of inflammatory cytokines induced by LPS in BV2 cells by mediating the PPARγ/NF-κB signaling pathway
- Factors influencing spontaneous hypothermia after emergency trauma and the construction of a predictive model
- Long-term administration of morphine specifically alters the level of protein expression in different brain regions and affects the redox state
- Application of metagenomic next-generation sequencing technology in the etiological diagnosis of peritoneal dialysis-associated peritonitis
- Clinical diagnosis, prevention, and treatment of neurodyspepsia syndrome using intelligent medicine
- Case report: Successful bronchoscopic interventional treatment of endobronchial leiomyomas
- Preliminary investigation into the genetic etiology of short stature in children through whole exon sequencing of the core family
- Cystic adenomyoma of the uterus: Case report and literature review
- Mesoporous silica nanoparticles as a drug delivery mechanism
- Dynamic changes in autophagy activity in different degrees of pulmonary fibrosis in mice
- Vitamin D deficiency and inflammatory markers in type 2 diabetes: Big data insights
- Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells
- Meta-analysis on the efficacy of allogeneic hematopoietic stem cell transplantation to treat malignant lymphoma
- Mitochondrial DNA drives neuroinflammation through the cGAS-IFN signaling pathway in the spinal cord of neuropathic pain mice
- Application value of artificial intelligence algorithm-based magnetic resonance multi-sequence imaging in staging diagnosis of cervical cancer
- Embedded monitoring system and teaching of artificial intelligence online drug component recognition
- Investigation into the association of FNDC1 and ADAMTS12 gene expression with plumage coloration in Muscovy ducks
- Yak meat content in feed and its impact on the growth of rats
- A rare case of Richter transformation with breast involvement: A case report and literature review
- First report of Nocardia wallacei infection in an immunocompetent patient in Zhejiang province
- Rhodococcus equi and Brucella pulmonary mass in immunocompetent: A case report and literature review
- Downregulation of RIP3 ameliorates the left ventricular mechanics and function after myocardial infarction via modulating NF-κB/NLRP3 pathway
- Evaluation of the role of some non-enzymatic antioxidants among Iraqi patients with non-alcoholic fatty liver disease
- The role of Phafin proteins in cell signaling pathways and diseases
- Ten-year anemia as initial manifestation of Castleman disease in the abdominal cavity: A case report
- Coexistence of hereditary spherocytosis with SPTB P.Trp1150 gene variant and Gilbert syndrome: A case report and literature review
- Utilization of convolutional neural networks to analyze microscopic images for high-throughput screening of mesenchymal stem cells
- Exploratory evaluation supported by experimental and modeling approaches of Inula viscosa root extract as a potent corrosion inhibitor for mild steel in a 1 M HCl solution
- Imaging manifestations of ductal adenoma of the breast: A case report
- Gut microbiota and sleep: Interaction mechanisms and therapeutic prospects
- Isomangiferin promotes the migration and osteogenic differentiation of rat bone marrow mesenchymal stem cells
- Prognostic value and microenvironmental crosstalk of exosome-related signatures in human epidermal growth factor receptor 2 positive breast cancer
- Circular RNAs as potential biomarkers for male severe sepsis
- Knockdown of Stanniocalcin-1 inhibits growth and glycolysis in oral squamous cell carcinoma cells
- The expression and biological role of complement C1s in esophageal squamous cell carcinoma
- A novel GNAS mutation in pseudohypoparathyroidism type 1a with articular flexion deformity: A case report
- Predictive value of serum magnesium levels for prognosis in patients with non-small cell lung cancer undergoing EGFR-TKI therapy
- HSPB1 alleviates acute-on-chronic liver failure via the P53/Bax pathway
- IgG4-related disease complicated by PLA2R-associated membranous nephropathy: A case report
- Baculovirus-mediated endostatin and angiostatin activation of autophagy through the AMPK/AKT/mTOR pathway inhibits angiogenesis in hepatocellular carcinoma
- Metformin mitigates osteoarthritis progression by modulating the PI3K/AKT/mTOR signaling pathway and enhancing chondrocyte autophagy
- Evaluation of the activity of antimicrobial peptides against bacterial vaginosis
- Atypical presentation of γ/δ mycosis fungoides with an unusual phenotype and SOCS1 mutation
- Analysis of the microecological mechanism of diabetic kidney disease based on the theory of “gut–kidney axis”: A systematic review
- Omega-3 fatty acids prevent gestational diabetes mellitus via modulation of lipid metabolism
- Refractory hypertension complicated with Turner syndrome: A case report
- Interaction of ncRNAs and the PI3K/AKT/mTOR pathway: Implications for osteosarcoma
- Association of low attenuation area scores with pulmonary function and clinical prognosis in patients with chronic obstructive pulmonary disease
- Long non-coding RNAs in bone formation: Key regulators and therapeutic prospects
- The deubiquitinating enzyme USP35 regulates the stability of NRF2 protein
- Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as potential diagnostic markers for rebleeding in patients with esophagogastric variceal bleeding
- G protein-coupled receptor 1 participating in the mechanism of mediating gestational diabetes mellitus by phosphorylating the AKT pathway
- LL37-mtDNA regulates viability, apoptosis, inflammation, and autophagy in lipopolysaccharide-treated RLE-6TN cells by targeting Hsp90aa1
- The analgesic effect of paeoniflorin: A focused review
- Chemical composition’s effect on Solanum nigrum Linn.’s antioxidant capacity and erythrocyte protection: Bioactive components and molecular docking analysis
- Knockdown of HCK promotes HREC cell viability and inner blood–retinal barrier integrity by regulating the AMPK signaling pathway
- The role of rapamycin in the PINK1/Parkin signaling pathway in mitophagy in podocytes
- Laryngeal non-Hodgkin lymphoma: Report of four cases and review of the literature
- Clinical value of macrogenome next-generation sequencing on infections
- Overview of dendritic cells and related pathways in autoimmune uveitis
- TAK-242 alleviates diabetic cardiomyopathy via inhibiting pyroptosis and TLR4/CaMKII/NLRP3 pathway
- Hypomethylation in promoters of PGC-1α involved in exercise-driven skeletal muscular alterations in old age
- Profile and antimicrobial susceptibility patterns of bacteria isolated from effluents of Kolladiba and Debark hospitals
- The expression and clinical significance of syncytin-1 in serum exosomes of hepatocellular carcinoma patients
- A histomorphometric study to evaluate the therapeutic effects of biosynthesized silver nanoparticles on the kidneys infected with Plasmodium chabaudi
- PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
- Analysis of MDA, SOD, TAOC, MNCV, SNCV, and TSS scores in patients with diabetes peripheral neuropathy
- SLIT3 deficiency promotes non-small cell lung cancer progression by modulating UBE2C/WNT signaling
- The relationship between TMCO1 and CALR in the pathological characteristics of prostate cancer and its effect on the metastasis of prostate cancer cells
- Heterogeneous nuclear ribonucleoprotein K is a potential target for enhancing the chemosensitivity of nasopharyngeal carcinoma
- PHB2 alleviates retinal pigment epithelium cell fibrosis by suppressing the AGE–RAGE pathway
- Anti-γ-aminobutyric acid-B receptor autoimmune encephalitis with syncope as the initial symptom: Case report and literature review
- Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua
- Human umbilical cord mesenchymal stem cells regulate glutathione metabolism depending on the ERK–Nrf2–HO-1 signal pathway to repair phosphoramide mustard-induced ovarian cancer cells
- Electroacupuncture on GB acupoints improves osteoporosis via the estradiol–PI3K–Akt signaling pathway
- Renalase protects against podocyte injury by inhibiting oxidative stress and apoptosis in diabetic nephropathy
- Review: Dicranostigma leptopodum: A peculiar plant of Papaveraceae
- Combination effect of flavonoids attenuates lung cancer cell proliferation by inhibiting the STAT3 and FAK signaling pathway
- Renal microangiopathy and immune complex glomerulonephritis induced by anti-tumour agents: A case report
- Correlation analysis of AVPR1a and AVPR2 with abnormal water and sodium and potassium metabolism in rats
- Gastrointestinal health anti-diarrheal mixture relieves spleen deficiency-induced diarrhea through regulating gut microbiota
- Myriad factors and pathways influencing tumor radiotherapy resistance
- Exploring the effects of culture conditions on Yapsin (YPS) gene expression in Nakaseomyces glabratus
- Screening of prognostic core genes based on cell–cell interaction in the peripheral blood of patients with sepsis
- Coagulation factor II thrombin receptor as a promising biomarker in breast cancer management
- Ileocecal mucinous carcinoma misdiagnosed as incarcerated hernia: A case report
- Methyltransferase like 13 promotes malignant behaviors of bladder cancer cells through targeting PI3K/ATK signaling pathway
- The debate between electricity and heat, efficacy and safety of irreversible electroporation and radiofrequency ablation in the treatment of liver cancer: A meta-analysis
- ZAG promotes colorectal cancer cell proliferation and epithelial–mesenchymal transition by promoting lipid synthesis
- Baicalein inhibits NLRP3 inflammasome activation and mitigates placental inflammation and oxidative stress in gestational diabetes mellitus
- Impact of SWCNT-conjugated senna leaf extract on breast cancer cells: A potential apoptotic therapeutic strategy
- MFAP5 inhibits the malignant progression of endometrial cancer cells in vitro
- Major ozonated autohemotherapy promoted functional recovery following spinal cord injury in adult rats via the inhibition of oxidative stress and inflammation
- Axodendritic targeting of TAU and MAP2 and microtubule polarization in iPSC-derived versus SH-SY5Y-derived human neurons
- Differential expression of phosphoinositide 3-kinase/protein kinase B and Toll-like receptor/nuclear factor kappa B signaling pathways in experimental obesity Wistar rat model
- The therapeutic potential of targeting Oncostatin M and the interleukin-6 family in retinal diseases: A comprehensive review
- BA inhibits LPS-stimulated inflammatory response and apoptosis in human middle ear epithelial cells by regulating the Nf-Kb/Iκbα axis
- Role of circRMRP and circRPL27 in chronic obstructive pulmonary disease
- Investigating the role of hyperexpressed HCN1 in inducing myocardial infarction through activation of the NF-κB signaling pathway
- Characterization of phenolic compounds and evaluation of anti-diabetic potential in Cannabis sativa L. seeds: In vivo, in vitro, and in silico studies
- Quantitative immunohistochemistry analysis of breast Ki67 based on artificial intelligence
- Ecology and Environmental Science
- Screening of different growth conditions of Bacillus subtilis isolated from membrane-less microbial fuel cell toward antimicrobial activity profiling
- Degradation of a mixture of 13 polycyclic aromatic hydrocarbons by commercial effective microorganisms
- Evaluation of the impact of two citrus plants on the variation of Panonychus citri (Acari: Tetranychidae) and beneficial phytoseiid mites
- Prediction of present and future distribution areas of Juniperus drupacea Labill and determination of ethnobotany properties in Antalya Province, Türkiye
- Population genetics of Todarodes pacificus (Cephalopoda: Ommastrephidae) in the northwest Pacific Ocean via GBS sequencing
- A comparative analysis of dendrometric, macromorphological, and micromorphological characteristics of Pistacia atlantica subsp. atlantica and Pistacia terebinthus in the middle Atlas region of Morocco
- Macrofungal sporocarp community in the lichen Scots pine forests
- Assessing the proximate compositions of indigenous forage species in Yemen’s pastoral rangelands
- Food Science
- Gut microbiota changes associated with low-carbohydrate diet intervention for obesity
- Reexamination of Aspergillus cristatus phylogeny in dark tea: Characteristics of the mitochondrial genome
- Differences in the flavonoid composition of the leaves, fruits, and branches of mulberry are distinguished based on a plant metabolomics approach
- Investigating the impact of wet rendering (solventless method) on PUFA-rich oil from catfish (Clarias magur) viscera
- Non-linear associations between cardiovascular metabolic indices and metabolic-associated fatty liver disease: A cross-sectional study in the US population (2017–2020)
- Knockdown of USP7 alleviates atherosclerosis in ApoE-deficient mice by regulating EZH2 expression
- Utility of dairy microbiome as a tool for authentication and traceability
- Agriculture
- Enhancing faba bean (Vicia faba L.) productivity through establishing the area-specific fertilizer rate recommendation in southwest Ethiopia
- Impact of novel herbicide based on synthetic auxins and ALS inhibitor on weed control
- Perspectives of pteridophytes microbiome for bioremediation in agricultural applications
- Fertilizer application parameters for drip-irrigated peanut based on the fertilizer effect function established from a “3414” field trial
- Improving the productivity and profitability of maize (Zea mays L.) using optimum blended inorganic fertilization
- Application of leaf multispectral analyzer in comparison to hyperspectral device to assess the diversity of spectral reflectance indices in wheat genotypes
- Animal Sciences
- Knockdown of ANP32E inhibits colorectal cancer cell growth and glycolysis by regulating the AKT/mTOR pathway
- Development of a detection chip for major pathogenic drug-resistant genes and drug targets in bovine respiratory system diseases
- Exploration of the genetic influence of MYOT and MB genes on the plumage coloration of Muscovy ducks
- Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism
- Comparison of nutritional value of the wild and cultivated spiny loaches at three growth stages
- Transcriptomic analysis of liver immune response in Chinese spiny frog (Quasipaa spinosa) infected with Proteus mirabilis
- Disruption of BCAA degradation is a critical characteristic of diabetic cardiomyopathy revealed by integrated transcriptome and metabolome analysis
- Plant Sciences
- Effect of long-term in-row branch covering on soil microorganisms in pear orchards
- Photosynthetic physiological characteristics, growth performance, and element concentrations reveal the calcicole–calcifuge behaviors of three Camellia species
- Transcriptome analysis reveals the mechanism of NaHCO3 promoting tobacco leaf maturation
- Bioinformatics, expression analysis, and functional verification of allene oxide synthase gene HvnAOS1 and HvnAOS2 in qingke
- Water, nitrogen, and phosphorus coupling improves gray jujube fruit quality and yield
- Improving grape fruit quality through soil conditioner: Insights from RNA-seq analysis of Cabernet Sauvignon roots
- Role of Embinin in the reabsorption of nucleus pulposus in lumbar disc herniation: Promotion of nucleus pulposus neovascularization and apoptosis of nucleus pulposus cells
- Revealing the effects of amino acid, organic acid, and phytohormones on the germination of tomato seeds under salinity stress
- Combined effects of nitrogen fertilizer and biochar on the growth, yield, and quality of pepper
- Comprehensive phytochemical and toxicological analysis of Chenopodium ambrosioides (L.) fractions
- Impact of “3414” fertilization on the yield and quality of greenhouse tomatoes
- Exploring the coupling mode of water and fertilizer for improving growth, fruit quality, and yield of the pear in the arid region
- Metagenomic analysis of endophytic bacteria in seed potato (Solanum tuberosum)
- Antibacterial, antifungal, and phytochemical properties of Salsola kali ethanolic extract
- Exploring the hepatoprotective properties of citronellol: In vitro and in silico studies on ethanol-induced damage in HepG2 cells
- Enhanced osmotic dehydration of watermelon rind using honey–sucrose solutions: A study on pre-treatment efficacy and mass transfer kinetics
- Effects of exogenous 2,4-epibrassinolide on photosynthetic traits of 53 cowpea varieties under NaCl stress
- Comparative transcriptome analysis of maize (Zea mays L.) seedlings in response to copper stress
- An optimization method for measuring the stomata in cassava (Manihot esculenta Crantz) under multiple abiotic stresses
- Fosinopril inhibits Ang II-induced VSMC proliferation, phenotype transformation, migration, and oxidative stress through the TGF-β1/Smad signaling pathway
- Antioxidant and antimicrobial activities of Salsola imbricata methanolic extract and its phytochemical characterization
- Bioengineering and Biotechnology
- Absorbable calcium and phosphorus bioactive membranes promote bone marrow mesenchymal stem cells osteogenic differentiation for bone regeneration
- New advances in protein engineering for industrial applications: Key takeaways
- An overview of the production and use of Bacillus thuringiensis toxin
- Research progress of nanoparticles in diagnosis and treatment of hepatocellular carcinoma
- Bioelectrochemical biosensors for water quality assessment and wastewater monitoring
- PEI/MMNs@LNA-542 nanoparticles alleviate ICU-acquired weakness through targeted autophagy inhibition and mitochondrial protection
- Unleashing of cytotoxic effects of thymoquinone-bovine serum albumin nanoparticles on A549 lung cancer cells
- Erratum
- Erratum to “Investigating the association between dietary patterns and glycemic control among children and adolescents with T1DM”
- Erratum to “Activation of hypermethylated P2RY1 mitigates gastric cancer by promoting apoptosis and inhibiting proliferation”
- Retraction
- Retraction to “MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB”
- Retraction to “A data mining technique for detecting malignant mesothelioma cancer using multiple regression analysis”
- Special Issue on Advances in Neurodegenerative Disease Research and Treatment
- Transplantation of human neural stem cell prevents symptomatic motor behavior disability in a rat model of Parkinson’s disease
- Special Issue on Multi-omics
- Inflammasome complex genes with clinical relevance suggest potential as therapeutic targets for anti-tumor drugs in clear cell renal cell carcinoma
- Gastroesophageal varices in primary biliary cholangitis with anti-centromere antibody positivity: Early onset?

