A machine learning-based prognostic model integrating mRNA stemness index, hypoxia, and glycolysis‑related biomarkers for colorectal cancer
-
Dan Liu
Abstract
Background
Cancer stemness, hypoxia, and glycolysis collectively influence colorectal cancer (CRC) progression. However, the intricate relationships among these factors remain incompletely understood.
Methods
This study (1) explored hypoxia and glycolysis-related genes (HGRGs) in CRC by mRNA stemness index (mRNAsi), analyzed the gene expression profiles from Gene Expression Omnibus and The Cancer Genome Atlas (TCGA) databases, (2) established a Cox-prognostic model based on single-sample gene set enrichment analysis, differentially expressed gene analysis, weighted gene co-expression network analysis, and Least Absolute Shrinkage and Selection Operator (LASSO) and Cox regression analyses, and (3) assessed the predictive accuracy of the model. Decision curve analysis (DCA) was employed to determine the clinical utility of the model.
Results
Ten HGRGs were selected based on mRNAsi to create the LASSO model. High-risk CRC patients in the TCGA dataset displayed unfavorable clinical outcomes and responses to immunotherapy. Consensus cluster analysis revealed two distinct colon adenocarcinoma/rectal adenocarcinoma clusters, with patients in cluster 2 having a worse prognosis compared to patients in cluster 1. A five-gene prognostic nomogram was developed through univariate and multivariate Cox regression analyses, with DCA confirming its accuracy.
Conclusions
This innovative prognostic model, incorporating ALDOB, AQP1, IL1A, PHGDH, and PTGIS, is highly accurate in predicting patient survival.
1 Introduction
Colorectal cancer (CRC) is the third most dominant global malignancy, profoundly impacting the gastrointestinal tract [1]. Despite multiple treatment modalities, including radiotherapy, targeted therapy, chemotherapy, surgery, immunotherapy, and traditional Chinese medicine, which have shown promise in extending patient survival, CRC continues to stand as the second most prominent contributor to cancer-related mortality, primarily attributing to early metastasis, late-stage diagnosis, and rapid progression [1,2,3,4]. Therefore, it is urgent to identify novel therapeutic targets and early diagnostic markers for CRC. CRC is a heterogeneous disease influenced by complex genomic variation and abnormal biological microenvironments [5]. Single-gene/factor prediction models often have limitations, whereas multigene-based models show excellent predictive power across a wide range of cancers [6,7,8]. Therefore, there is an urgent need to identify reliable genetic profiles for early CRC diagnosis, prognostic assessment, and targeted therapy.
Cancer stem cells (CSCs) represent a specialized and rare cell population present within tumor cell communities. They are self-renewing and play a key role in tumour formation, metastasis, recurrence, and resistance to therapeutic agents, with important implications for tumor progression and prognosis [9,10]. In CRC, CSCs are identified and defined by specific markers, including CD133, CD24, CD166, Lgr5, EpCAM, CD44, CD29, ALDH1, and β-catenin [11,12]. Recently, sequencing data analysis has emerged as a key approach to cancer prognosis assessment, which provides a new analytical perspective by characterizing cancer stemness using stemness-associated genes and the mRNA stemness index (mRNAsi) derived from machine learning algorithms [13]. Studies have explored the application of stemness scores in predicting prognosis, assessing immunotherapy response, and evaluating clinical outcomes in cancers, including glioblastoma, lung cancer, cutaneous melanoma, and CRC [14,15,16,17].
Hypoxia is a common feature of most solid tumors, resulting from increased oxygen consumption and vascular disturbances, and is frequently associated with a poor prognosis [18,19]. Hypoxia has profound effects on intracellular and extracellular metabolic processes, triggering the activation of hypoxia-inducible factors. These factors in turn stimulate the transcription of important genes involved in various processes such as angiogenesis, pH regulation, glucose metabolism, tumor invasion, and metastasis [20,21]. Moreover, recent research has shown that a hypoxic tumor microenvironment can influence the efficacy of chemotherapy in CRC patients [22,23]. In CRC cells, a modified energy metabolism, particularly abnormal activation of the glycolytic pathway, has been observed [24,25]. In the presence of sufficient oxygen, cancer cells rely mainly on glycolysis to produce energy [26]. In cancer cells, glycolysis accounts for the production of 50–60% of total adenosine triphosphate [27]. Aerobic glycolysis creates a specific microenvironment that favours the unrestricted growth and invasion of cancer cells [28]. Currently, investigations have developed hypoxia and glycolysis (HG)-related prognostic signatures for CRC patients [29,30], and hypoxia and glycolysis-related genes (HGRGs) are anticipated to serve as valuable prognostic biomarkers for CRC.
The main focus of the study was to identify HGRGs by analyzing tumor stemness in CRC patients, to study their pattern in normal and tumour tissues, and to use them as potential prognostic markers. We identified 30 HGRGs related to mRNAsi and validated their expression levels using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. Further insights into their functional implications were gained through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. A prognostic model comprising ten genes was developed using the Least Absolute Shrinkage and Selection Operator (LASSO) model. Consensus clustering of these genes identified two colon adenocarcinoma/rectal adenocarcinoma (COADREAD) subtypes and compared their survival, tumor mutational burden (TMB), and microsatellite instability (MSI) in the Tumour Immune Dysfunction and Rejection (TIDE) database. Finally, a prognostic nomogram incorporating five genes was introduced through LASSO–Cox regression analysis, and its predictive performance was verified using decision curve analysis (DCA). The DCA curve takes the sensitivity and specificity of the predictive model as the horizontal axis and the benefit as the vertical axis. If the decision curve of the predictive model is above the decision curve of the reference strategy, the clinical utility of the predictive model is higher; conversely, the clinical utility is lower. In conclusion, this study enriches our understanding of the pathogenesis of CRC and paves the way for personalized and tailored patient care.
2 Materials and methods
2.1 Data manipulation
The gene expression matrix data for COADREAD and relevant clinical details were retrieved from TCGA (https://portal.gdc.cancer.gov/) and UCSC Xena (http://genome.ucsc.edu), respectively [31,32]. The raw read counts were converted to Fragments Per Kilobase per Million using the R package limma [33]. Samples lacking key clinical data were eliminated, yielding in a final dataset comprising 644 cancer samples and 51 normal samples. Validation datasets for COADREAD were obtained from GSE14333 [34], GSE74602 [35], GSE87211 [36], and GSE161158 [37] from the GEO database [38] using the GEOquery package [39].
Hypoxia-related genes with a relevance score of >1 as the screening criterion were collected from GeneCards, a database containing extensive information about human genomes [40], using the term “hypoxia” as the primary search keyword, resulting in 2041 genes. We also collected 3147 hypoxia-related genes from 65 reference gene sets in MSigDB [41] using “hypoxia” as the keyword. Additionally, we searched the AmiGO2 website (http://amigo.geneontology.org/amigo) [42] using “hypoxia” as the keyword and obtained 298 hypoxia-related genes. By considering the common genes identified across these three sources, we collected 130 hypoxia-related genes. Similarly, we searched the GeneCards database, using the term “glycolysis” to compile a list of glycolysis-related genes and retrieved 851 genes with a relevance score of >1 as the screening criterion. Additionally, we found 753 glycolysis-related genes from 21 reference gene sets in MSigDB using “glycolysis” as the search term. By considering the common genes across these two sources, we collected 158 glycolysis-related genes. Ultimately, we obtained a subset of 273 HGRGs. Additional details can be found in Table S1.
In addition to gene expression data, we retrieved somatic mutation data, including single-nucleotide polymorphism (SNP), for COADREAD from TCGA and processed the data with the R package maftools [43]. These genomic characterizations were designed to explore the potential association between the pattern of genetic variation in HGRG-related genes and their expression levels and prognosis, and to assess the impact of genomic instability on immunotherapy response. Specifically, (1) analysis of somatic mutation types and frequencies by maftools can reveal the relationship between the mutational load of HGRG-related genes and patient prognosis; (2) copy number variation (CNV) analysis (GISTIC 2.0) can help to identify the genomic amplification/deletion events that drive the expression of HGRG; and (3) analysis of the MSI/TMB data in combination with the TIDE scores can assess the genomic instability’s role in the regulation of immune escape. Information regarding TMB and MSI in the COADREAD dataset was acquired from cBioPortal (https://www.cbioportal.org/) [44].
2.2 Dataset normalization and merging
We normalized COADREAD datasets from both TCGA and GEO using the R package limma. We then used the R package sva to remove the batch effects found in the COADREAD datasets (GSE14333, GSE74602, GSE87211, and GSE161158) from GEO and created a merged COADREAD dataset. The differences between the GEO datasets pre- and post-processing were illustrated in the distribution box and principal component analysis (PCA) plots.
2.3 Stemness index calculation
The mRNAsi for each sample was calculated using the stemness training dataset by single-sample gene set enrichment analysis (ssGSEA). The mRNAsi score for COADREAD patients was computed in accordance with the expression profile of the TCGA-COADREAD dataset with the R package GSVA [45]. The resulting score enabled the categorization of cancer samples into separate groups and facilitated correlation analysis.
2.4 Differentially expressed gene (DEG) analysis
The individuals afflicted with COADREAD from TCGA were stratified into two mRNAsi cohorts with a median mRNAsi score as the threshold. Subsequently, DEGs between the two groups were uncovered based on P < 0.05 and |log2 fold change (FC)| > 0. Genes with Log2 FC > 0 or < 0 are considered up-regulated and down-regulated genes, respectively. Finally, we obtained mRNAsi-related HGRGs for subsequent analysis by taking the intersection of DEGs and HGRGs.
2.5 Gene set enrichment analysis (GSEA)
We conducted GSEA [46] using the R package clusterProfiler [47] to examine DEGs distribution pattern and determine their impacts on phenotypes based on the c2.cp.v7.2 symbol reference in the MSigDB. The GSEA parameters were configured as: seed value of 2020, calculation number of 100,000, minimum of five genes per set, and a maximum of 500 genes. P-values were corrected using the Benjamini–Hochberg method to assess the statistical significance of enrichment. Gene sets with P < 0.05 and false discovery rate (FDR) (q value) <0.25 were considered significant enrichment.
2.6 HGRG score calculation
The abundance of individual genes within the dataset was assessed using the ssGSEA method. The HGRG score for each sample was computed according to the TCGA-COADREAD levels and mRNAsi-related HGRGs using the ssGSEA algorithm from the R package GSVA and used for sample grouping and correlation analysis. COADREAD patients were divided into two groups, using the median HGRGs score as the cut-off value.
2.7 Weighted gene co-expression network analysis (WGCNA)
WGCNA [48] was employed to delineate gene correlation patterns across multiple samples. Highly coordinated gene sets and potential biomarkers or therapeutic targets were identified by examining their connectivity and correlations with phenotypes using the R package WGCNA [49]. Correlation coefficients were first calculated for any two genes, and then weighted correlation coefficients were applied to ensure a scale-free distribution of gene connections within the network. Subsequently, a hierarchical clustering tree between genes was built based on the correlation between genes. Different gene modules were used to describe different branches of the tree based on unique colors, and the importance of each module was assessed. The top 25% of genes with the most pronounced differences in gene expression in the ARDS dataset were entered with a minimum number of 50 genes, a soft power threshold of 10, no module merging (shear height of 0), and a minimum distance of 0.2. Following this, the correlation between the HGRGs scores and various modules was computed. Simultaneously, genes in each module were considered signature genes. Prominent modules exhibiting strong correlation coefficients were selected, and their intersection with mRNAsi-related HGRGs was used as hypoxia and glycolysis-related module DEGs (HGRMDEGs) for further analysis.
2.8 GO and KEGG annotation analyses
The expression levels of HGRMDEGs were validated using a validation set, and genes with inconsistent validation results were excluded. The HGRMDEGs were annotated by GO [50] and KEGG [51] analyses using the R package clusterProfiler based on P < 0.05 and an FDR value (q value) of <0.25 and the Benjamini–Hochberg method.
2.9 LASSO model
LASSO regression was used to construct prognostic models to address the risk of overfitting in high-dimensional genetic data. Tenfold cross-validation (seed = 2,020) assessed model stability through repeated sampling, and a penalty term of λ = 0.01 prevented model complexity while preserving key genes. A median risk score grouping strategy maximized survival differences between the two groups to ensure clinical translational value.
2.10 Immunotherapy analysis
Gene expression profile was analyzed based on the TIDE database [52] to determine the likelihood of tumor immune evasion. Elevated TIDE scores indicate an increased likelihood of immunosurveillance evasion and reduced success of immunotherapy. Sample matrices from the TCGA-COADREAD dataset were uploaded to obtain TIDE scores for each COADREAD patient and analyzed for their association with HGRGs.
2.11 Protein–protein interaction (PPI) analysis
PPI network of HGRMDEGs between known and predicted proteins was predicted by searching the STRING [53] with a minimum interaction score of 0.4 to ensure moderate confidence. Local regions with close connections were identified by visualizing the PPI network using Cytoscape software (version 3.9.1) [54]. The GeneMANIA website [55] was utilized to predict genes functionally similar to the target genes and to construct an interaction network of HGRMDEGs from the LASSO model. miRNA interactions with HGRMDEGs were predicted using the ENCORI database [56]. Subsequently, miRNAs with supported database counts greater than three were filtered out, and the mRNA–miRNA interaction was analyzed using Cytoscape software.
2.12 Consensus clustering
Consensus clustering [57], a method based on resampling, was utilized to establish consensus and evaluate the stability of clusters identified using a clustering algorithm. Using the R package ConsensusClusterPlus, different COADREAD disease subtypes were identified based on HGRMDEG [58]. In this procedure, the cluster counts ranged from 2 to 8, and the entire sample set was repeated 1,000 times at a sampling rate of 80%. clusterAlg parameter was set to “km” (k-means algorithm), and the distance parameter was set to “euclidean.”
2.13 Drug sensitivity analysis
To assess the potential sensitivity of CRC patients in the TCGA cohort to commonly used anticancer drugs, a computational pharmacology approach was used. The Genomics of Drug Sensitivity in Cancer (GDSC) database v2.0 (https://www.cancerrxgene.org/) was first used as a training set, which contains drug dose-response data (IC50 values) and gene expression profiles of human cancer cell lines. Then, a drug sensitivity prediction model was constructed by a ridge regression algorithm based on gene expression data (RNA-seq or microarray) from the GDSC training set with IC50 values using the pRophetic R software package. For each drug, pRophetic performed feature selection (screening the most relevant genes based on the correlation between gene expression and IC50), model training (constructing a ridge regression model using the screened gene features and cell line IC50 values), and cross-validation (evaluating the model performance with Pearson’s correlation coefficient and RMSE using LOOCV or k-fold cross-validation on the GDSC training set and recording the average metrics). Subsequently, the trained model was applied to the gene expression profiles of TCGA samples (which should be consistent with the detection platform and normalization method of the GDSC training set), and the predicted IC50 values of each sample for different drugs were output by pRophetic (the lower the IC50, the higher the sensitivity). Finally, the TCGA samples were grouped according to HGRG scores (or variables such as risk group), and the Wilcoxon rank-sum test was used to compare the differences in predicted IC50 between the groups, screen for significant drugs (P < 0.05), and visualize the IC50 distribution.
2.14 Immune activity analysis
Estimating Stromal and Immune Cells in Malignant Tissue Using Expression Data (ESTIMATE) uses gene expression profiles of tumor samples to detect the relative proportions of tumor cells and infiltrating normal cells. It also quantifies immune activity to derive an immune score. To gain insight into the prognostic impact of genes associated with stromal and immune cells, we analyzed the immune response of tumors using expression profiles from the TCGA-COADREAD dataset and using the R package ESTIMATE [59]. In addition, we quantified the immune activity in the cancer samples using gene expression profiling to derive an immune score for each cancer sample. Subsequently, we assessed the association between the immune infiltration profile of COADREAD patients and the HGRG score.
2.15 Immune infiltration analysis
CIBERSORTx is an online tool for analyzing immune cells utilizing a deconvolution algorithm to examine the relative fraction of immune cells in cancer tissues. We provided the grouped gene expression data to the CIBERSORTx website, which was then combined with a signature gene matrix called LM22 to produce immune cell infiltration matrix data. We further analyzed the transcriptome data and accurately quantified the immune and stromal cells in the tissue using the R package MCPcounter [60]. In addition, we calculated an enrichment score for each sample using the ssGSEA algorithm to reflect the overall infiltration level of 28 types of immune cells.
2.16 Cox model
To examine the clinical prognostic value of HGRMDEGs obtained from the LASSO prognostic model in COADREAD, we performed univariate Cox regressions using the TCGA-COADREAD dataset to examine the expression pattern in relation to clinical variables. Multivariate Cox regression was examined for variables with P < 0.1, and the optimal set of variables was determined by stepwise regression. Based on the Cox model, we generated forest plots and nomograms to estimate 1-, 3-, and 5-year survival for COADREAD patients and built calibration curves using the R package rms to determine their accuracy. We built DCA plots using the R package ggDCA [61] to assess the performance of the nomogram model in predicting survival in COADREAD patients.
2.17 Statistical analysis
Drug sensitivity and all other data were assessed using R software. Continuous variables were shown as the median ± quartile interval, and the differences between the two groups were examined using the Wilcoxon rank sum test. The statistical significance in normally distributed variables and categorical variables between two groups was analyzed using the Chi-square or Fisher’s exact tests. Differences among different groups were examined with the Kruskal–Wallis test. LASSO regression analysis was conducted with the R package glmnet [62]. Receiver operating characteristic curve (ROC) and the time-dependent ROC [63] were established with the R package pROC and timeROC, respectively. Survival curves were analyzed with the R package survival and visualized with the survminer package. Significant variations in survival time between the two groups were evaluated with the log-rank test. Spearman’s rank correlation analysis was used to evaluate correlation coefficients between two numerators or scores, unless specified otherwise. All P values were two-tailed, and P < 0.05 was determined as statistical significance.
3 Results
3.1 Flow chart
The workflow is shown in Figure 1.
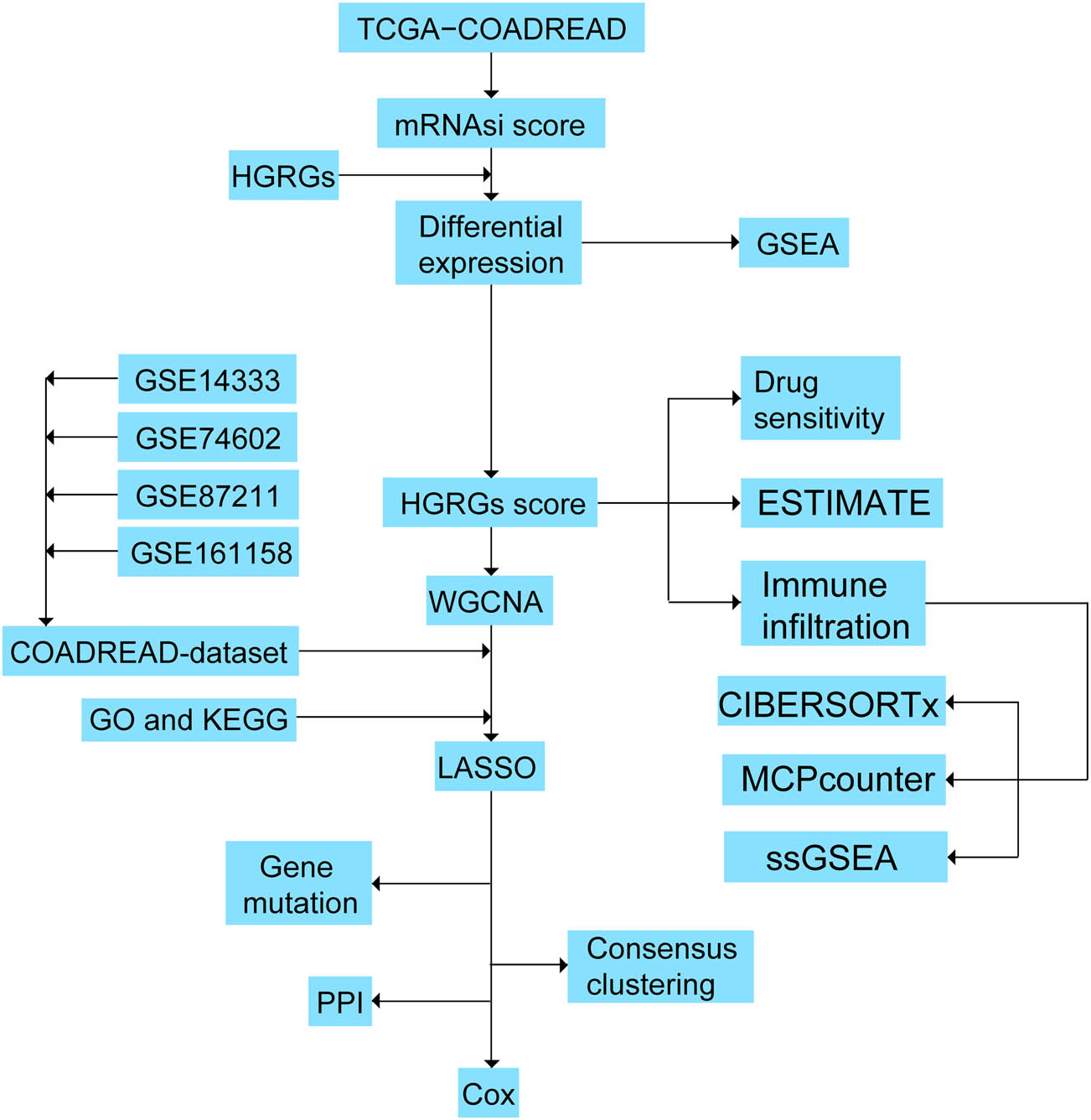
Study flow chart.
3.2 Stemness-related gene screen reveals 198 HGRGs
We first created a merged COADREAD dataset by eliminating the batch effect in the GEO-COADREAD datasets (GSE14333, GSE74602, GSE87211, and GSE161158). We then used distribution box plots and PCA plots to compare the datasets before and after correction (Figure S1a–d). These visual illustrations clearly show that the samples in the COADREAD dataset successfully eliminate the batch effect.
We calculated mRNAsi values for all patients in the TCGA-COADREAD dataset using the ssGSEA algorithm to assess their stemness levels. Subsequently, we divided these patients into two groups based on the median mRNAsi value. We plotted Kaplan–Meier curves (Figure 2a) and found a statistically significant difference in the prognosis of these two groups of patients, with the low mRNAsi group having a worse prognosis. Among the 19,572 genes in the TCGA-COADREAD dataset, we identified 11,934 DEGs, of which 6,388 genes were overexpressed and 5,546 genes were underexpressed in the high mRNAsi group (Figure 2b). We intersected DEGs associated with mRNAsi with HGRGs and obtained 198 HGRGs associated with mRNAsi, as shown in the Venn diagram (Figure 2c). These genes are shown in Table S2, and the first 40 genes are shown in the heatmap.

Screening of mRNAsi-related HGRGs in COADREAD patients. (a) The Kaplan–Meier survival curve of the two mRNAsi groups. (b) Volcano plot of DEGs between the two mRNAsi groups. (c) The overlaps of HGRGs and DEGs. (d) The heat map of the top 40 DEGs.
3.3 Hypoxia, glycolysis, and oncogenic pathways drive CRC stemness
We identified biological functions and pathways associated with these DEGs by GSEA. Several significantly enriched pathways can be seen in the pathway map (Figure 3a–i) and are listed in Table S3. Among them, cellular response to hypoxia (Figure 3a) and glycolytic pathway (Figure 3b) were associated with HG phenotypes, whereas Notch (Figure 3c), JAK-STAT (Figure 3d), Wnt (Figure 3e), MAPK (Figure 3f), PI3K-AKT (Figure 3g), Hedgehog (Figure 3h), and FcεRI-mediated NF-кB activation pathway (Figure 3i) were associated with hotspot molecules.

Hypoxia, glycolysis, and oncogenic pathways drive CRC stemness. Genes in the TCGA-COADREAD dataset were significantly enriched in the cellular response to hypoxia (a), glycolysis (b), Notch (c), JAK-STAT (d), Wnt (e), MAPK (f), PI3K-AKT (g), Hedgehog (h), and FcεRI-mediated NF-кB (i) signaling pathways.
3.4 WGCNA analysis and HGRG score cross-validation reveal 50 core stemness-regulated genes in CRC
Based on the levels of the 198 mRNAsi-associated HGRGs (Table S2), we calculated HGRG scores for all cancer samples in the TCGA-COADREAD dataset using the ssGSEA algorithm. We created a stacked bar chart to visualize the distribution of all clinical stages in the high and low HGRGs subgroups (Figure 4a–d). Our analyses showed no significant differences between the two groups, except for some changes observed in the T stage relative to the N and M stages. We further performed WGCNA analysis on all genes in the dataset and identified the top 25% of genes with the most significant differences between patients by setting the screening threshold criterion to 0.85 (Figure 4e). These genes were further clustered into nine modules by setting the optimal soft threshold to 10 (Figure 4f and g) and the branch merge cutoff height to 0, i.e., no modules were merged (Figure 4f). Subsequently, we analyzed the correlation between their expression patterns in these modules and the HGRGs scores of COADREAD patients (Figure 4g). Four modules with robust correlations (P < 0.05, |r| ≥ 0.30), namely MEred (r = 0.37, P = 9 × 10−23), MEyellow (r = 0.49, P = 9 × 10−41), MEbrown (r = 0.36, P = 1 × 10−20), and MEgrey (r = −0.38, P = 7 × 10−24), were selected and further analyzed. We then intersected the 198 mRNAsi-related HGRGs with the genes in the MEred module (Figure 4h), MEyellow module (Figure 4i), MEbrown module (Figure 4j), and MEgrey module (Figure 4k) of the TCGA-COADREAD dataset. We identified 50 HGRMDEGs and presented them in Venn diagrams (Table S4).

WGCNA analysis and HGRG score cross-validation reveal 50 core stemness-regulated genes in CRC. The proportions of the clinical-pathological (a), as well as clinical T (b), M (c), and N (d) stages in the two HGRG score groups. (e) The scale-free network of optimal soft thresholding power screening in the TCGA-COADREAD dataset. (f) Gene dendrogram and module colors. (g) The relationship between module eigengenes and HGRGs scores in the TCGA-COADREAD dataset. The Venn diagrams showing the overlaps between HGRMDEGs and the genes in the MEred module (h), MEyellow module (i), MEbrown module (j), and MEgrey module (k) of TCGA-COADREAD dataset.
3.5 Chromosomal localization and PPI network analysis reveal functional clusters among differentially expressed 30 HGRMDEGs in CRC
We compared the expression levels of 50 HGRMDEGs (Table S4) between the cancer and normal groups in the COADREAD and TCGA-COADREAD datasets (Figure 5a and b). The analyses revealed 30 HGRMDEGs with consistent expression trend in both datasets, including ADM, ALDOB, ALDOC, ANG, AQP1, CBFA2T3, CCNA2, CD44, ENO1, EPAS1, HK3, HMOX1, IL1A, IRS2, JUND, KCNMB1, LDHB, MMP14, MYCN, NDRG1, NUP210, PCK1, PHGDH, PLAT, PMAIP1, PRKACB, PTGIS, STC1, STC2, and VEGFA. We further explored the chromosomal localization analysis of these 30 HGRMDEGs to observe their distributions. The chromosome localization map (Figure 5c) showed that these genes were primarily located on chromosomes 1, 2, 5, and 8, with each hosting three genes. The genes on the same chromosome might have functional connections. We then analyzed the PPI of these 30 HGRMDEGs in the STRING database by setting the required minimum interaction score to medium confidence (0.4). Subsequently, a PPI network comprising 27 genes was established and visualized using Cytoscape software (Figure 5d). Notably, at the lowest interaction score of 0.4, all genes interacted with at least one HGRMDEG, except CBFA2T3, JUND, and NUP210, which did not interact with other HGRMDEGs.

Chromosomal localization and PPI network analysis reveal functional clusters among differentially expressed 30 HGRMDEGs in CRC. The levels of HGRMDEGs in the TCGA-COADREAD (a) and COADREAD datasets (b). (c) Chromosomal localizations of HGRMDEGs. (d) PPI network of HGRMDEGs. NS: P ≥ 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001.
3.6 A 10-gene hypoxia–glycolysis signature predicts poor prognosis in CRC and unveils a core metabolic mechanism
GO enrichment (Table S5) unveiled 30 HGRMDEGs with predominant association with biological processes (BPs), such as response to hypoxia (GO: 0001666), cellular response to hypoxia (GO: 0071456), and glycolytic process (GO: 0006096) (Figure 6a), and molecular functions (MFs) including lyase activity (GO: 0016829), receptor-ligand activity (GO: 0048018) and carbon-carbon lyase activity (GO: 0016830) (Figure 6b). Further, KEGG enrichment analysis (Table S5) demonstrated their significant enrichment in the HIF-1α signaling pathway (hsa04066), glycolysis/gluconeogenesis (hsa00010), and type II diabetes mellitus (hsa04930) pathways. We further performed GO and KEGG enrichment analyses on these 30 HGRMDEGs based on the logFC values between the two mRNAsi groups in the TCGA-COADREAD dataset and calculated the corresponding Z-scores for each gene. The results are presented in the form of chord plots (Figure 6c) and circle plots (Figure 6d) with logFC. The circle plot shows that carbon-carbon cleavage enzyme activity (GO: 0016830) was significantly upregulated. The glycolysis/gluconeogenesis (hsa00010) pathway was selected to illustrate the KEGG pathway (Figure 6e).

A 10-gene hypoxia-glycolysis signature predicts poor prognosis in CRC and unveils a core metabolic mechanism. Significantly enriched GO and KEGG terms of HGRMDEGs with bar graph (a) and circular network diagram (b). GO and KEGG enrichment analyses with logFC by chord diagram (c) and circle diagram (d). (e) The KEGG pathway diagram of Glycolysis/Gluconeogenesis (hsa00010). In (b), the blue dots and red squares represent specific genes and pathways, respectively. In (d), red and blue dots represent upregulated (logFC > 0) and down-regulated genes (logFC < 0), respectively. The screening criteria for GO and KEGG enrichment terms were P < 0.05 and FDR (q value) < 0.25.
3.7 The LASSO model stratifies CRC risk with survival prediction and accuracy
To examine the prognostic value of the 30 HGRMDEGs (ADM, ALDOB, ALDOC, ANG, AQP, CBFA2T3, CCNA2, CD44, ENO1, EPAS1, HK3, HMOX1, IL1A, IRS2, JUND, KCNMB1, LDHB, MMP14, MYCN, NDRG1, NUP210, PCK1, PHGDH, PLAT, PMAIP1, PRKACB, PTGIS, STC1, STC2, VEGFA) in the TCGA-COADREAD dataset, we conducted a LASSO regression analysis and developed a prognostic model consisting of 10 HGRMDEGs (ALDOB, ALDOC, AQP1, CCNA2, IL1A, JUND, NDRG1, PHGDH, PTGIS, VEGFA) (Figure 7a and b). The risk factor plot (Figure 7c) visually depicts the risk groupings derived from the LASSO model. We categorized the cancer samples into two risk groups using the median risk score as the threshold. Subsequently, we plotted a prognostic Kaplan–Meier curve to compare survival outcomes between the two risk groups (Figure 7d). The analysis showed that the prognosis was significantly worse in the higher risk group (log-rank P < 0.001). We then plotted the prognostic time-dependent ROC curves for the risk scores (Figure 7e). The area under the curve (AUC) of the ROC curves for 1-, 3-, and 5-year survival was all greater than 0.6, suggesting that higher-risk scores were associated with worse prognosis in COADREAD. We further compared the levels of 10 HGRMDEGs in the LASSO prognostic model between the two groups (Figure 7f) and observed significant differences in the expression of these genes between the two groups (P < 0.01). To investigate the association between risk scores and HGRGs scores in COADREAD patients, we visualized the correlation between the two using a scatterplot (Figure 7g), which showed a significant (P < 0.05) but very weak (|r| < 0.3) linear correlation. In addition, ALDOB (AUC = 0.711), CCNA2 (AUC = 0.770), and PHGDH (AUC = 0.705) had some accuracy in predicting COADREAD high- and low-risk patients (Figure 7h–k).

LASSO model stratifies CRC risk with survival prediction and accuracy. (a) Optimization parameter lambda determined by the LASSO regression model of HGRMDEGs. LASSO coefficient profile of the 10 HGRMDEGs (b) and risk factor plot (c) of the LASSO regression model. (d) Kaplan–Meier curves comparing the two risk groups in the TCGA-COADREAD dataset. (e) Time-dependent ROC curves illustrating the risk score for predicting COADREAD prognosis. (f) Comparison of the 10 HGRMDEGs levels between the two risk groups in the TCGA-COADREAD dataset. (g) Correlation between the risk score and the HGRG score of COADREAD patients. (h–k) The ROC curves of the 10 HGRMDEGs in the TCGA-COADREAD dataset. In the LASSO regression model plot (a), the y-axis represents the likelihood deviation, and the x-axis shows the log (λ), representing the penalty term in the LASSO regression. The numbers on the upper x-axis represent nonzero coefficients under each lambda. In the risk factor plot (c), red dots and blue dots represent the deceased and surviving patients, respectively. **P < 0.01 and ***P < 0.001.
3.8 Correlation and functional analysis reveal VEGFA dominance and mRNAsi-linked genes in CRC
Based on the levels of 10 HGRMDEGs (ALDOB, ALDOC, AQP1, CCNA2, IL1A, JUND, NDRG1, PHGDH, PTGIS, VEGFA) in cancer samples in the TCGA-COADREAD dataset, we first investigated the correlation between them using the Spearman algorithm and presented the results in the form of correlation heatmaps (Figure 8a) and correlation chord plots (Figure 8b). The results showed a linear correlation of ALDOC with NDRG1, AQP1 with CCNA2, IL1A, and PTGIS, and CCNA2 with PTGIS (|r| > 0.3, P < 0.05). Subsequently, a functional similarity analysis to assess the similarities among the 10 HGRMDEGs (Figure 8c) revealed that VEGFA exhibited the highest functional similarity score. We further investigated the association of the levels of these ten genes with the corresponding mRNAsi in the cancer samples (Figure 8d–m) and showed that CCNA2 (r = 0.608, P < 0.001, Figure 8g) and PTGIS (r = −0.347, P < 0.001, Figure 8i) were significantly and linearly correlated with mRNAsi.

Correlation and functional analysis reveal VEGFA dominance and mRNAsi-linked genes in CRC. Correlation heatmap (a) and correlation chord diagram (b) displaying the correlation of HGRMDEGs in the TCGA-COADREAD dataset. (c) Box plot showing the functional similarity of HGRMDEGs. (d–m) Scatter plots exhibiting the association between HGRMDEGs and mRNAsi. *P < 0.05 and **P < 0.01. The absolute correlation coefficient (r) value in the scatter plots denotes a strong correlation if it is above 0.8, a moderate correlation if it is between 0.5 and 0.8, a weak correlation if it is between 0.3 and 0.5, and little or no correlation if it is below 0.3.
3.9 High-risk CRC genomic profiling links to immune evasion and ICB resistance
Given that somatic genomic alterations (including mutations, CNVs, MSIs, and TMBs) are central mechanisms for cancer development, driving key pathways and influencing the immune microenvironment and therapeutic response, we further explored the somatic mutational profiles of 10 key HGRMDEGs and their association with risk models. The results showed that the TCGA-COADREAD dataset contains four types of somatic mutations: missense and nonsense substitutions and frame-shift deletions and insertions, with missense substitutions being the most prevalent (Figure 9a). Furthermore, the mutation types of the 10 HGRMDEGs in COADREAD patients primarily included SNPs along with some insertions and deletions (INS and DEL). Among SNVs, C > T was dominant, followed by C > A (Figure 8a). Among the 10 HGRMDEGs, ALDOB had the most somatic mutations, including missense, frame-shift deletion, and nonsense mutations (Figure 9b).

High-risk CRC genomic profiling links to immune evasion and ICB resistance. (a) Mutation type distributions of 10 HGRMDEGs in COADREAD patients. (b) Waterfall plot of somatic mutations. (c) CNV of 10 HGRMDEGs in COADREAD patients. (d) Genes with significant copy number amplification in COADREAD patients. (e) Genes with significant copy number deletion in COADREAD patients. Comparison of MSI (f), TMB (g), and TIDE score (h) between the two risk groups of COADREAD patients. Scatter plots showing the association of MSI (i), TMB (j), and TIDE score (k) with the risk score in COADREAD patients. *P < 0.05, **P < 0.01, and ***P < 0.001. In the scatter plot, the absolute correlation coefficient (r) > 0.8 indicates a strong correlation; 0.5–0.8 indicates a moderate correlation; 0.3–0.5 indicates a weak correlation, and <0.3 indicates little or no correlation.
We also examined CNVs of the 10 HGRMDEGs in COADREAD patients from the TCGA-COADREAD dataset using GISTIC 2.0 (Figure 9c–e) and identified significant amplifications and deletions in the 10 HGRMDEGs among the COADREAD patient samples. Among these genes, PTGIS, AQP1, and NDRG1 had the highest amplification frequency, while ALDOC, CCNA2, and ALDOB had the highest deletion frequency (Figure 9c).
Subsequently, we processed MSI and TMB data corresponding to COADREAD patients from the TCGA-COADREAD dataset, evaluated the TIDE scores using the TIDE algorithm, and generated grouped comparison charts (Figure 9f–h) and correlation scatter plots (Figure 9i–k) with respect to the corresponding risk scores. Our findings indicated statistically significant variations in MSI, TMB, and TIDE scores between the two risk groups (P < 0.05). Specifically, the high-risk group displayed lower MSI and TMB levels but higher TIDE scores. This signifies that the high-risk group is susceptible to immune escape and benefits less from immune checkpoint inhibitor therapy. Correlation scatter plots showed that MSI and TMB were weakly linearly associated with risk scores in COADREAD patients.
3.10 Gene-miRNA networks and consensus clustering identify prognostic subtypes in CRC
We established an interaction network of functionally similar genes related to the 10 HGRMDEGs using the GeneMANIA website (Figure 10) to observe shared protein domains, co-expression, pathways, co-localization, and gene interactions. To identify miRNAs interacting with these 10 HGRMDEGs, mRNA–miRNA data from the ENCORI database were utilized. Subsequently, miRNAs were screened against more than three databases, and the mRNA–miRNA interaction network consisting of 6 mRNAs and 38 miRNAs was depicted using Cytoscape software (Figure 10) and Table S6.

PPI network. (a) The PPI network of functionally similar genes related to the 10 HGRMDEGs. (b) The mRNA-miRNA regulatory network of the 10 HGRMDEGs, with blue rectangles representing mRNA and pink ellipses representing miRNA. The co-expression network (a) was generated using the GeneMANIA website. Black circles with white diagonal lines represent input RNAMRGs, while other black circles represent predicted functionally similar genes. Yellow-green lines represent shared protein domain relationships between genes; purple lines represent co-expression relationships between genes; light blue lines represent pathway-related connections between genes; dark blue lines represent co-localization relationships between genes; and green lines represent genetic interactions between genes.
To identify clinically significant molecular subtypes and to explore the association of the 10 HGRMDEG levels with COADREAD subtypes, we used consensus cluster analysis. The analysis categorized COADREAD disease into two subtypes: cluster 1 contained 308 samples and cluster 2 contained 336 samples (Figure 11a–c). In the TCGA-COADREAD dataset, 6 out of 10 HGRMDEGs showed significant expression differences between the two subtypes (P < 0.05, Figure 11d). The further constructed Kaplan–Meier survival curves showed that there was a significant difference in prognosis between the two groups (log-rank P = 0.017), with cluster 2 patients having a worse prognosis than cluster 1 (Figure 11e).

Consensus clustering identifies prognostic subtypes in CRC. (a) The COADREAD patients were divided into two clusters based on the consensus clustering matrix (K = 2). (b) Consensus clustering model with cumulative distribution function (CDF) for k values ranging from 2 to 9. (c) The delta area under CDF curves for k values ranging from 2 to 9. (d) The expression levels of the 10 HGRMDEGs between cluster 1 and cluster 2 in the TCGA-COADREAD dataset. (e) Kaplan–Meier curves between cluster 1 and cluster 2 in the TCGA-COADREAD dataset. ns: P ≥ 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001.
3.11 HGRG score stratification guides precision medication in CRC-high scoring patients have significantly increased sensitivity to 20 targeted agents
To explore the sensitivity of COADREAD patients to commonly used anticancer drugs in the TCGA-COADREAD dataset and to investigate potential treatment strategies for patients with different HGRG scores, we studied drug sensitivity using the GDSC database as a training set. We compared the IC50 of anticancer drugs in the two groups with different HGRG scores and used violin plots to show the top 20 drugs with significant differences in IC50 (Figure S2a–t). Among these 20 drugs, patients with higher HGRG scores had lower IC50 values compared to those with lower HGRG scores, suggesting that individuals with higher HGRG scores may be more sensitive to these drugs. This further emphasizes the importance of personalized treatment for cancer patients.
3.12 HGRG score and significant association with tumor microenvironmental characteristics
We investigated the expression profiles of cancer samples in the TCGA-COADREAD dataset, including stromal, immune, ESTIMATE, and tumor purity scores, as well as their distributions with respect to HGRGs scores, using the R package ESTIMATE. Our results showed that there were substantial differences (P < 0.001) in stroma (Figure S3a), immune (Figure S3b), ESTIMATE (Figure S3c), and tumor purity (Figure S3d) scores between the two groups of patients.
We further analyzed the association between HGRG scores and stromal, immune, ESTIMATE, and tumor purity scores of COADREAD patients and found that HGRG scores were weakly positively associated with stromal (r = 0.151, P < 0.001, Figure S3e), immune (r = 0.192, P < 0.001, Figure S3f), and ESTIMATE scores (r = 0.184, P < 0.001, Figure S3g) and weakly negatively correlated with tumor purity score (r = −0.184, P < 0.001, Figure S3h).
3.13 Characteristics of immune cell enrichment and key gene regulatory hubs in high HGRG-scoring groups
To examine differences in immune infiltration between the 2 HGRG score groups, we used multiple algorithms to calculate the abundance of 28 immune cell types in cancer patients from the TCGA-COADREAD dataset. We first applied the CIBERSORTx algorithm (Figure S4a) to visualize the distribution of immune cell types; subsequently, group comparisons were plotted by other algorithms (CIBERSORTx, MCPcounter, and ssGSEA) to demonstrate the changes in abundance between the two HGRG score groups (Figure S4b–d). The results showed that the high HGRG group presented significant enrichment in 25 out of 28 immune cell types (P < 0.05), suggesting the presence of extensive enhanced immune infiltration in the high HGRG group.
The findings exhibited that (1) when using the CIBERSORTx algorithm, 9 out of 22 immune cells demonstrated statistically significant differences (P < 0.05), 2) when using the MCPcounter algorithm, 8 out of 10 immune cells and stromal cells manifested significant differences (P < 0.05), 3) when with the ssGSEA algorithm, 25 out of 28 immune cells presented statistically significant differences (P < 0.05).
We then selected immune cells with differential infiltration abundance (P < 0.05) between the two HGRG score groups to analyze their relationships with the 10 HGRMDEGs. The results (Figure 12a) showed that (1) under both MCPcounter and ssGSEA algorithms, immune cells exhibited positive linear correlations with AQP1 and PTGIS immune cells but negative linear correlations with PHGDH and (2) immune cells exhibited a negative linear correlation with VEGFA under all three algorithms. We also plotted a complex heatmap to show the infiltration abundance of all immune cells in the two HGRG score groups under the three algorithms (Figure 12b). Notably, the high HGRGsscore group presented significantly increased immune cell infiltration abundance under both MCPcounter and ssGSEA algorithms.

Characteristics of immune cell enrichment and key gene regulatory hubs in high HGRG scoring groups. (a) The associations between the 10 HGRMDEGs and the immune cell infiltration abundance under the CIBERSORTx algorithm, MCPcounter algorithm, and ssGSEA algorithm. (b) The infiltration abundance of all immune cells under the CIBERSORTx algorithm, MCPcounter algorithm, and ssGSEA algorithm in the two HGRGs score groups. *P < 0.05 and **P < 0.01.
3.14 HGRMDEGs identified as prognostic biomarkers linking TNM staging to clinical outcomes in CRC
We further comparatively analyzed the levels of the 10 HGRMDEGs in patients at different clinical substages, including T1, T2, T3, T4, N0, N1, N2, N3, M0, M1, and pathological stages I, II, III, and IV. The outcomes displayed a potential association between N stage subtype and the genes ALDOB (Figure 13a), AQP1 (Figure 13d), CCNA2 (Figure 13g), IL1A (Figure 13i), NDRG1 (Figure 13j), PHGDH (Figure 13m), and VEGFA (Figure 13o). Furthermore, the expression of genes AQP1 (Figure 13c), CCNA2 (Figure 13f), PHGDH (Figure 13m), and VEGFA (Figure 13n) showed a potential association with M stage subtype. Lastly, the genes ALDOC (Figure 13b), AQP1 (Figure 13e), CCNA2 (Figure 13h), NDRG1 (Figure 13k), and VEGFA (Figure 13p) exhibited a potential association with clinical pathologic stage.

HGRMDEGs identified as prognostic biomarkers linking TNM staging to clinical outcomes in CRC. (a–p) The correlations of HGRMDEGs with N, M, and pathological stages. *P < 0.05, **P < 0.01, and ***P < 0.001.
3.15 The prediction model for CRC shows high prediction accuracy and high net benefit
At last, we analyzed the clinical traits of COADREAD patients in the TCGA-COADREAD dataset (Table 1) and executed univariate Cox regression analyses to examine their association with the 10 HGRMDEGs. Variables with P < 0.1, including T, N, M, and pathological stages, as well as age and BMI, were subjected to multivariate Cox regression analysis. The optimal variables, including T, N, and M stages, as well as age, were further identified with stepwise regression and chosen to create the multivariate Cox regression model (Table S7). The findings were visualized in a forest plot (Figure 14a). Furthermore, we performed nomogram analyses to judge the predictive value of the genes and clinical traits included in the multivariate Cox regression model (Figure 14b). Additionally, we generated calibration curves for the 1-, 3-, and 5-year survival (Figure 14c–e) and demonstrated that the red lines corresponding to the 1- and 3-year survival were near the ideal gray line, suggesting strong predictive performance of the model. Lastly, we evaluated the clinical application of the multivariate Cox regression model using DCA (Figure 14f–h). The results revealed that the red lines representing the model were significantly distant from both the blue lines (all positive) and the gray line (all negative), indicating its high accuracy.
Clinicopathological features of patients with CRC
| Characteristic | Levels | Overall |
|---|---|---|
| N | 644 | |
| T stage, n (%) | T1 | 20 (3.1%) |
| T2 | 111 (17.3%) | |
| T3 | 436 (68%) | |
| T4 | 74 (11.5%) | |
| N stage, n (%) | N0 | 368 (57.5%) |
| N1 | 153 (23.9%) | |
| N2 | 119 (18.6%) | |
| M stage, n (%) | M0 | 475 (84.2%) |
| M1 | 89 (15.8%) | |
| Pathologic stage, n (%) | Stage I | 111 (17.8%) |
| Stage II | 238 (38.2%) | |
| Stage III | 184 (29.5%) | |
| Stage IV | 90 (14.4%) | |
| Gender, n (%) | Female | 301 (46.7%) |
| Male | 343 (53.3%) | |
| Age, n (%) | ≤65 | 276 (42.9%) |
| >65 | 368 (57.1%) | |
| BMI, n (%) | <25 | 107 (32.5%) |
| ≥25 | 222 (67.5%) | |
| OS event, n (%) | Alive | 515 (80%) |
| Dead | 129 (20%) | |
| Age, median (IQR) | 68 (58, 76) |

The prediction model for CRC shows high prediction accuracy and high net benefit. The forest plot (a) and the nomogram (b) of the multivariate Cox regression model. The calibration curves of the nomogram at 1-year (c), 3-year (d), and 5-year (e) intervals. DCA plots of the nomograms at 1-year (f), 3-year (g), and 5-year (h) intervals. The x-axis in the DCA plot represents the threshold probability, while the y-axis represents the net benefit.
4 Discussion
As a global, highly prevalent malignant tumour, the molecular heterogeneity of CRC has led to the insufficient predictive efficacy of existing biomarkers, which restricts the development of precision diagnosis and treatment. In this study, we broke through the limitations of a single molecular dimension and constructed a 10-gene prognostic model and disease subtype classification system through mRNAsi, hypoxia–glycolysis-related genes, and multi-omics analysis (DEG/WGCNA/LASSO). The key findings indicated that (1) patients in the high-risk group had significantly lower survival and limited response to immunotherapy; (2) subtype two (Cluster2) identified by consensus clustering had a worse prognosis; and (3) a column-line graph model based on TNM stage-age-core genes (C-index = 0.76–0.79) and DCA validation provided a quantitative tool for clinical decision-making. This multimodal analysis framework provides a new paradigm for molecular typing and individualized treatment of CRC.
The LASSO risk score model consists of ten genes: ALDOB, ALDOC, AQP1, CCNA2, IL1A, JUND, NDRG1, PHGDH, PTGIS, and VEGFA. As far as we know, both ALDOB and ALDOC belong to the aldolase family. Aldolase, as the fourth enzyme of the glycolysis pathway, is vital in glycolysis and gluconeogenesis. Previous explorations have revealed a correlation between high ALDOB levels and unfavorable survival outcomes for CRC patients. Additionally, increased ALDOB expression serves as a pivotal trigger for metabolic reprogramming [64,65]. ALDOC overexpression acts as an independent prognostic factor for CRC patients. Moreover, ALDOC downregulation inhibits cell proliferation, migration, and spheroid formation [66]. AQP1 belongs to the aquaporin family and is responsible for the rapid passive transport of water across biological membranes. It has been shown that AQP9 promotes cancer cell invasion and motility through the AKT signaling pathway [67]. Similarly, AQP1 expression in CRC cells may enhance their ability to invade surrounding tissues and enter the bloodstream, thereby promoting spread to distal organs such as the liver. AQP1 level is a poor prognostic factor for advanced colon cancer [68]. CCNA2, a vital member of the conserved cyclin family and referred to as cyclin A2, is critical in regulating the cell cycle. Additionally, CCNA2 demonstrates a strong association with the survival rate of CRC patients. Silencing CCNA2 suppresses CRC cell proliferation and induces apoptosis [69]. IL1A encodes IL-1α, which belongs to the IL-1 protein cluster, serves as a pro-inflammatory factor, and is considered a potential prognostic biomarker related to T-cell infiltration in CRC patients [70]. JUND, an AP-1 transcription factor, regulates cell differentiation, proliferation, and apoptosis [71]. NDRG1 is involved in stress response and many BPs, such as hypoxia, cell proliferation, lipid metabolism, and chemotherapy resistance [72]. NDRG1 downregulation and TNM stage are important independent factors affecting the OS of CRC patients [73]. Downregulation of NDRG1 might promote epithelial–mesenchymal transition (EMT) progression in CRC through the NF-κB signaling [74]. PHGDH is an essential enzyme in synthesizing serine, a metabolic substrate for producing NADPH and glycine [75]. PHGDH-mediated serine metabolism promotes DNA hypermethylation in CRC stem cells by maintaining redox homeostasis and providing single-carbon units [76]. Inhibitors targeting PHGDH, such as NCT-503, have shown potential in cancer therapy. NCT-503 effectively inhibits PHGDH activity, thereby blocking the serine synthesis pathway, which in turn affects cancer cell growth and survival [77]. PHGDH overexpression is linked to the TNM stage and tumor size and independently predicts a poor prognosis in CRC patients [78]. PTGIS is an enzyme that converts prostaglandin H2 into prostaglandin I2 (PGI2). In CRC, PGE2 promotes tumor metastasis by upregulating miR-675-5p, a process that involves the regulation of p53 expression [79]. Furthermore, PGE2 regulates cell proliferation and signaling in CRC cells by inhibiting the expression of TIG1 and GRK5 and affecting the β-linker protein/TCF and cAMP signaling pathways [80]. Its expression is related to tumor growth and progression [81]. PTGIS overexpression is closely linked to liver metastasis and predicts a poor prognosis in colon cancer patients [82]. VEGFA is in mediating tumor angiogenesis, with its expression being controlled by oncogenes, diverse growth factors, and hypoxia. Nevertheless, the influence of JUND on the prognosis among CRC patients remains unexplored, and its effects on CRC patient prognosis have not been well explored. Therefore, from the initial ensemble of 273 HGRGs, a 10-gene prognostic model was constructed through multi-step screening such as DEG intersection, WGCNA modular association, and LASSO regression. The core advantages of this model are as follows: (1) pathway synergy can be captured, and the selected genes (e.g., glycolysis-related ALDOB, ALDOC, PHGDH, hypoxia-responsive VEGFA, and NDRG1) together constitute the core network of hypoxia-glycolysis-proliferation, avoiding single-gene misclassification. (2) Predictive robustness is improved, with multigene combinations significantly improving prognostic stratification (time-dependent ROC AUC > 0.6) and risk prediction accuracy (C-index 0.76–0.79) over single markers. (3) Regulatory hubs are revealed, and PPI network and functional analyses show that the 10 genes form a tight functional module with VEGFA and other hubs, providing a basis for multi-target combinations for co-targeted therapies.
The results obtained from GSEA indicated significant enrichment of all DEGs between the two mRNAsi groups in various pathways, including the cellular response to hypoxia, glycolysis, as well as Notch, JAK-STAT, Wnt, PI3K-AKT, MAPK, Hedgehog, and FcεRI-mediated NF-κB signaling pathways. GO analysis unveiled that the 30 HGRMDEGs were enriched in BPs related to response to hypoxia, cellular response to hypoxia, and glycolysis, as well as MFs involving lyase activity, receptor–ligand activity, and carbon–carbon lyase activity. The KEGG analysis of the 30 HGRMDEGs mainly revealed enrichment in the HIF-1α signaling pathway, glycolysis/gluconeogenesis, and type II diabetes mellitus pathway. Most of the enriched terms were associated with cancer hallmarks, including CSCs, hypoxia, and glycolysis. Wnt/β-catenin, Notch, and hypoxia signaling pathways are crucial in regulating CSC self-renewal and participating in tumorigenesis [83]. In addition, the WNT pathway is connected to the poor prognosis of CRC [84]. The PI3K-AKT pathway is vital in maintaining CSCs in multiple cancers, including CRC, promoting CSCs’ proliferation, migration, EMT, and invasion [85]. The AKT/mTOR/HIF-1α signaling pathway is crucial in promoting glycolysis and lactation, thus contributing to the “metabolic reprogramming” of cancer cells [86]. Epidemiological studies have established that type 2 diabetes mellitus (T2DM) is a risk factor for CRC [87]. Interestingly, our KEGG enrichment results included T2DM pathways, suggesting a potential shared risk factor between T2DM and CRC.
The multi-omics analysis in this study not only revealed the functions of key genes but also provided an in-depth portrayal of the tumor microenvironment shaped by them. Comprehensive analysis of the tumor microenvironment (including immuno-score, stroma-score, ESTIMATE-score, tumor purity, immune-cell infiltration as assessed by multiple algorithms, and the TIDE-score) showed that the high-risk-score group (based on the 10-gene signature) and/or high HGRG score groups exhibited significant alterations in tumor microenvironmental characteristics. Specifically, these groups were characterized by enhanced immunosuppression: e.g., ALDOB overexpression may inhibit CD8+ T-cell function through the WNT/PD-L1 axis [88], and IL1A promotes TAM-mediated immunosuppressive microenvironment formation [89]. High expression of the key gene VEGFA drives significant angiogenesis, which has been shown to be a major driver of tumor angiogenesis in the high-scoring group in animal models [90]. Combined CIBERSORTx, MCPcounter, and ssGSEA algorithm evaluations found that while the high-risk/high-HGRG group showed higher overall immune cell infiltration abundance (especially under ssGSEA and MCPcounter evaluations), this infiltration was accompanied by specific cellular subset disproportionality and altered functional status. MSI and TMB levels were significantly lower in the high-risk group compared to the low-risk group, while the TIDE score, which reflects immune escape potential, was significantly higher. Higher HGRG scores were associated with higher matrix scores, immune scores, and ESTIMATE scores, accompanied by reduced tumor purity. The synergistic effect of hypoxia-glycolysis-dryness features (characterized by HGRG scores and 10-gene risk scores) shapes a tumor microenvironment with immunosuppressive, angiogenically active, matrix-enriched features and a high potential for immune escape, which may be one of the core drivers of poor prognosis in high-risk patients and directly affects their response to immunotherapy. Therefore, risk scores derived from the analysis of 10 HGRGs can effectively identify high-risk patients with the aforementioned unfavorable tumor microenvironmental features and help select the most appropriate immunotherapy candidates.
The multivariate Cox regression model established using stepwise regression by incorporating ALDOB, AQP1, IL1A, PHGDH, and PTGIS, TNM stage, and age indicated that PHGDH could be an independent prognostic factor for CRC, in line with a previous report with a smaller sample size. Notably, our study, for the first time, identified a significant correlation between PHGDH and age. To enhance its clinical applicability, we established a prognostic nomogram by integrating ALDOB, AQP1, IL1A, PHGDH, and PTGIS with various clinicopathological features. Its accuracy was validated in the TCGA dataset, revealing a good predictive power. Notably, this is the first prognostic nomogram based on mRNAsi-related HGRGs, and these five important genes have yet to be included in other predictive models for cancers, including CRC. The utilization of this model could aid in molecular typing and screening of differential patient subgroups, ultimately optimizing personalized therapy.
Despite the valuable insights, this study had certain limitations. First, in terms of data sources, although multi-omics data (e.g., GEO and TCGA) have been integrated, the samples are mainly derived from Western populations, especially the TCGA database, which has more than 85% of Western populations. This data heterogeneity may limit the generalisability and accuracy of the constructed model in Asian populations. Second, in terms of computational drug sensitivity prediction, the pRophetic algorithm integrates GDSC and TCGA data to predict drug sensitivity in CRC and identify potential therapeutic clues for people with high HGRG scores. However, the model has three limitations: the biological level does not incorporate tumor microenvironment components (e.g., fibroblasts, Treg cells, tumor-associated macrophages) and dynamic interactions, the pharmacological mechanism only reflects in vitro cellular effects but ignores in vivo pharmacokinetics, and the heterogeneity is not sufficiently characterized to recapitulate the intra-tumor genetic/spatial heterogeneity. The performance of the algorithm is limited by the feature matching between the training set and the test set, and lacks prospective validation with clinical efficacy indexes. Therefore, the current drug sensitivity analysis should be regarded as an exploratory hypothesis to guide subsequent studies, and in the future, it should be deepened through the functional validation of patient-derived organoid/xenograft model, biomarker-based clinical drug tracking, and mechanistic studies (e.g., analysis of the association between drug targets and metabolic pathways), in order to provide a reliable basis for translational decision-making in the clinic. Furthermore, in terms of mechanistic studies, the functional regulatory networks of key genes (e.g., PHGDH and VEGFA) rely mainly on bioinformatics inference, while functional validation by gene editing (e.g., CRISPR) or organoid models is lacking. This lack of experimental validation makes the understanding of gene function insufficiently deep and accurate. Finally, in terms of the clinical application of the prognostic model, although the column-line diagram model has been validated by DCA, the supporting test kits (e.g., NGS panel) have not yet been developed, which limits the on-the-ground application and promotion of the model in clinical practice. In summary, the current study faces certain limitations in terms of data sources, computational drug sensitivity prediction, mechanism study, and clinical application of the prognostic model, which need to be further explored and addressed in future studies. Our future research interests include multi-centre cohort validation, organoid drug screening, functional genome analysis, and development of clinical translational tools. We plan to incorporate more than 2,000 samples from multiple centers in Asia to correct for population variation, analyze the tumour microenvironment using single-cell transcriptome technology, and develop race-adaptive algorithms. Meanwhile, the CRC organoid library is established to verify drug synergy, detect serine metabolism, and screen drug resistance genes. In terms of the functional genome, we validate the PHGDH regulatory mechanism, locate HGRG gene expression, and analyze the PHGDH transcriptional network. Finally, a portable gene testing kit and AI decision-making system are developed to prospectively validate the guiding value for chemotherapy regimen selection.
5 Conclusion
By integrating multi-omics data and systems biology analysis, this study deeply analyzed the interaction network between stemness characteristics and hypoxia–glycolysis pathway in CRC, and successfully established a molecular typing and prognosis prediction system with clinical translational value. The main conclusions include: the identification of metabolic-microenvironmental regulatory axes centered on PHGDH and VEGFA; the classification of CRC into two molecular subtypes with different therapeutic sensitivities, which provides a potential strategy for targeted metabolism in combination with antivascular therapies; and the construction of a columnar graph model that significantly optimizes prognosis prediction of patients with CRC, which is particularly valuable in individualized survival assessment of patients with advanced disease.
Abbreviations
- AUC
-
area under the curve
- BP
-
biological process
- CDF
-
cumulative distribution function
- CNV
-
copy number variation
- COADREAD
-
colon adenocarcinoma/rectal adenocarcinoma
- DEG
-
differentially expressed gene
- GDSC
-
Genomics of Drug Sensitivity in Cancer
- GEO
-
Gene Expression Omnibus
- GO
-
Gene Ontology
- GSEA
-
gene set enrichment analysis
- HGRGs
-
hypoxia and glycolysis-related genes
- HGRMDEGs
-
hypoxia and glycolysis-related module DEGs
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- LASSO
-
Least Absolute Shrinkage and Selection Operator
- MF
-
molecular function
- mRNAsi
-
mRNA stemness index
- MSI
-
microsatellite instability
- PCA
-
principal component analysis
- PPI
-
protein–protein interaction
- ROC
-
receiver operating characteristic curve
- SNP
-
single-nucleotide polymorphism
- ssGSEA
-
single-sample GSEA
- DCA
-
decision curve analysis
- TCGA
-
The Cancer Genome Atlas
- TIDE
-
Tumor Immune Dysfunction and Exclusion
- TMB
-
tumor mutational burden
- WGCNA
-
weighted gene co-expression network analysis
Acknowledgments
We thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
-
Funding information: This work was sponsored by the Education Department of Heilongjiang Province and Qiqihar Bureau of Science and Technology (2019-KYYWF-1227, 2018-KYYWF-0112, 2020-KYYWF-0013, 2019-KYYWF-1231, and LHYD-202005).
-
Author contributions: Conceptualization and supervision, Q.M.; resources, software formal analysis, and validation, D.L., X.L., M.Z., and W.L.; writing – original draft preparation, D.L.; writing – review and editing, L.Y. and Y.N.; and visualization, L.W., X.M., and P.L. All authors approved the current version.
-
Conflict of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
-
Data availability statement: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.10.3322/caac.21660Suche in Google Scholar PubMed
[2] Qin F, Xu H, Wei G, Ji Y, Yu J, Hu C, et al. A prognostic model based on the immune-related lncRNAs in colorectal cancer. Front Genet. 2021;12:658736.10.3389/fgene.2021.658736Suche in Google Scholar PubMed PubMed Central
[3] Hissong E, Pittman ME. Colorectal carcinoma screening: Established methods and emerging technology. Crit Rev Clin Lab Sci. 2020;57(1):22–36.10.1080/10408363.2019.1670614Suche in Google Scholar PubMed
[4] Sun Y, Yang B, Lin M, Yu H, Chen H, Zhang Z. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019;24(3):299–305.10.3233/CBM-182129Suche in Google Scholar PubMed
[5] Xu H, Wong CC, Li W, Zhou Y, Li Y, Wang L, et al. RING-finger protein 6 promotes colorectal tumorigenesis by transcriptionally activating SF3B2. Oncogene. 2021;40(47):6513–26.10.1038/s41388-021-01872-9Suche in Google Scholar PubMed PubMed Central
[6] Yang B, Fan Y, Liang R, Wu Y, Gu A. Identification of a prognostic six-immune-gene signature and a nomogram model for uveal melanoma. BMC Ophthalmol. 2023;23(1):2.10.1186/s12886-022-02723-1Suche in Google Scholar PubMed PubMed Central
[7] Liu J, Wu Z, Sun R, Nie S, Meng H, Zhong Y, et al. Using mRNAsi to identify prognostic-related genes in endometrial carcinoma based on WGCNA. Life Sci. 2020;258:118231.10.1016/j.lfs.2020.118231Suche in Google Scholar PubMed
[8] Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7(1):71.10.1038/s41420-021-00451-xSuche in Google Scholar PubMed PubMed Central
[9] De Francesco EM, Sotgia F, Lisanti MP. Cancer stem cells (CSCs): Metabolic strategies for their identification and eradication. Biochem J. 2018;475(9):1611–34.10.1042/BCJ20170164Suche in Google Scholar PubMed PubMed Central
[10] Koh SY, Moon JY, Unno T, Cho SK. Baicalein suppresses stem cell-like characteristics in radio- and chemoresistant MDA-MB-231 human breast cancer cells through Up-regulation of IFIT2. Nutrients. 2019;11(3):624.10.3390/nu11030624Suche in Google Scholar PubMed PubMed Central
[11] Zhang R, Zhang X, Zhang W, Cui W, Xiao Y, Liu L, et al. Sohlh2 regulates the stemness and differentiation of colon cancer stem cells by downregulating LncRNA-H19 transcription. Mol Cancer Res. 2023;21(2):115–26.10.1158/1541-7786.MCR-22-0134Suche in Google Scholar PubMed
[12] Peng L, Xiong Y, Wang R, Xiang L, Zhou H, Fu Z. The critical role of peroxiredoxin-2 in colon cancer stem cells. Aging (Albany NY). 2021;13(8):11170–87.10.18632/aging.202784Suche in Google Scholar PubMed PubMed Central
[13] Bai KH, He SY, Shu LL, Wang WD, Lin SY, Zhang QY, et al. Identification of cancer stem cell characteristics in liver hepatocellular carcinoma by WGCNA analysis of transcriptome stemness index. Cancer Med. 2020;9(12):4290–8.10.1002/cam4.3047Suche in Google Scholar PubMed PubMed Central
[14] Liao Y, Wang Y, Cheng M, Huang C, Fan X. Weighted gene coexpression network analysis of features that control cancer stem cells reveals prognostic biomarkers in lung adenocarcinoma. Front Genet. 2020;11:311.10.3389/fgene.2020.00311Suche in Google Scholar PubMed PubMed Central
[15] Wang Z, Wang Y, Yang T, Xing H, Wang Y, Gao L, et al. Machine learning revealed stemness features and a novel stemness-based classification with appealing implications in discriminating the prognosis, immunotherapy and temozolomide responses of 906 glioblastoma patients. Brief Bioinform. 2021;22(5):bbab032.10.1093/bib/bbab032Suche in Google Scholar PubMed PubMed Central
[16] Wang X, Wan Q, Jin L, Liu C, Liu C, Cheng Y, et al. The integrative analysis identifies three cancer subtypes and stemness features in cutaneous melanoma. Front Mol Biosci. 2020;7:598725.10.3389/fmolb.2020.598725Suche in Google Scholar PubMed PubMed Central
[17] Weng M, Li T, Zhao J, Guo M, Zhao W, Gu W, et al. mRNAsi-related metabolic risk score model identifies poor prognosis, immunoevasive contexture, and low chemotherapy response in colorectal cancer patients through machine learning. Front Immunol. 2022;13:950782.10.3389/fimmu.2022.950782Suche in Google Scholar PubMed PubMed Central
[18] Favaro E, Lord S, Harris AL, Buffa FM. Gene expression and hypoxia in breast cancer. Genome Med. 2011;3(8):55.10.1186/gm271Suche in Google Scholar PubMed PubMed Central
[19] Jing X, Yang F, Shao C, Wei K, Xie M, Shen H, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18(1):157.10.1186/s12943-019-1089-9Suche in Google Scholar PubMed PubMed Central
[20] Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–32.10.1038/nrc1187Suche in Google Scholar PubMed
[21] Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–69.10.1182/blood-2004-07-2958Suche in Google Scholar PubMed
[22] Wen YA, Stevens PD, Gasser ML, Andrei R, Gao T. Downregulation of PHLPP expression contributes to hypoxia-induced resistance to chemotherapy in colon cancer cells. Mol Cell Biol. 2013;33(22):4594–605.10.1128/MCB.00695-13Suche in Google Scholar PubMed PubMed Central
[23] Tang YA, Chen YF, Bao Y, Mahara S, Yatim S, Oguz G, et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote chemoresistance in colorectal cancer. Proc Natl Acad Sci U S A. 2018;115(26):E5990–e9.10.1073/pnas.1801348115Suche in Google Scholar PubMed PubMed Central
[24] Nenkov M, Ma Y, Gaßler N, Chen Y. Metabolic reprogramming of colorectal cancer cells and the microenvironment: Implication for therapy. Int J Mol Sci. 2021;22(12):6262.10.3390/ijms22126262Suche in Google Scholar PubMed PubMed Central
[25] Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, et al. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J Cell Physiol. 2019;234(10):17023–49.10.1002/jcp.28436Suche in Google Scholar PubMed
[26] Chen X, Qian Y, Wu S. The Warburg effect: Evolving interpretations of an established concept. Free Radic Biol Med. 2015;79:253–63.10.1016/j.freeradbiomed.2014.08.027Suche in Google Scholar PubMed PubMed Central
[27] Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol Lett. 2012;4(6):1151–7.10.3892/ol.2012.928Suche in Google Scholar PubMed PubMed Central
[28] Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–9.10.1038/nrc1478Suche in Google Scholar PubMed
[29] Zhu J, Wang S, Bai H, Wang K, Hao J, Zhang J, et al. Identification of five glycolysis-related gene signature and risk score model for colorectal cancer. Front Oncol. 2021;11:588811.10.3389/fonc.2021.588811Suche in Google Scholar PubMed PubMed Central
[30] Lee JH, Jung S, Park WS, Choe EK, Kim E, Shin R, et al. Prognostic nomogram of hypoxia-related genes predicting overall survival of colorectal cancer-Analysis of TCGA database. Sci Rep. 2019;9(1):1803.10.1038/s41598-018-38116-ySuche in Google Scholar PubMed PubMed Central
[31] Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–8.10.1038/s41587-020-0546-8Suche in Google Scholar PubMed PubMed Central
[32] Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71.10.1093/nar/gkv1507Suche in Google Scholar PubMed PubMed Central
[33] Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.10.1093/nar/gkv007Suche in Google Scholar PubMed PubMed Central
[34] Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15(24):7642–51.10.1158/1078-0432.CCR-09-1431Suche in Google Scholar PubMed PubMed Central
[35] Gao P, He M, Zhang C, Geng C. Integrated analysis of gene expression signatures associated with colon cancer from three datasets. Gene. 2018;654:95–102.10.1016/j.gene.2018.02.007Suche in Google Scholar PubMed
[36] Hu Y, Gaedcke J, Emons G, Beissbarth T, Grade M, Jo P, et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer. 2018;57(3):140–9.10.1002/gcc.22512Suche in Google Scholar PubMed PubMed Central
[37] Szeglin BC, Wu C, Marco MR, Park HS, Zhang Z, Zhang B, et al. A SMAD4-modulated gene profile predicts disease-free survival in stage II and III colorectal cancer. Cancer Rep (Hoboken). 2022;5(1):e1423.10.1002/cnr2.1423Suche in Google Scholar PubMed PubMed Central
[38] Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–5.10.1093/nar/gks1193Suche in Google Scholar PubMed PubMed Central
[39] Davis S, Meltzer PS. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–7.10.1093/bioinformatics/btm254Suche in Google Scholar PubMed
[40] Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The genecards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinforma. 2016;54:1.30.1-1.3.10.1002/cpbi.5Suche in Google Scholar PubMed
[41] Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25.10.1016/j.cels.2015.12.004Suche in Google Scholar PubMed PubMed Central
[42] Balsa-Canto E, Henriques D, Gábor A, Banga JR. AMIGO2, a toolbox for dynamic modeling, optimization and control in systems biology. Bioinformatics. 2016;32(21):3357–9.10.1093/bioinformatics/btw411Suche in Google Scholar PubMed PubMed Central
[43] Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56.10.1101/gr.239244.118Suche in Google Scholar PubMed PubMed Central
[44] Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.10.1126/scisignal.2004088Suche in Google Scholar PubMed PubMed Central
[45] Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7.10.1186/1471-2105-14-7Suche in Google Scholar PubMed PubMed Central
[46] Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.10.1073/pnas.0506580102Suche in Google Scholar PubMed PubMed Central
[47] Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7.10.1089/omi.2011.0118Suche in Google Scholar PubMed PubMed Central
[48] Liu W, Li L, Ye H, Tu W. Weighted gene co-expression network analysis in biomedicine research. Sheng Wu Gong Cheng Xue Bao. 2017;33(11):1791–801.Suche in Google Scholar
[49] Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559.10.1186/1471-2105-9-559Suche in Google Scholar PubMed PubMed Central
[50] Gene ontology consortium: Going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56.10.1093/nar/gku1179Suche in Google Scholar PubMed PubMed Central
[51] Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.10.1093/nar/28.1.27Suche in Google Scholar PubMed PubMed Central
[52] Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12(1):21.10.1186/s13073-020-0721-zSuche in Google Scholar PubMed PubMed Central
[53] Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d13.10.1093/nar/gky1131Suche in Google Scholar PubMed PubMed Central
[54] Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.10.1101/gr.1239303Suche in Google Scholar PubMed PubMed Central
[55] Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–w4.10.1093/nar/gky311Suche in Google Scholar PubMed PubMed Central
[56] Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7.10.1093/nar/gkt1248Suche in Google Scholar PubMed PubMed Central
[57] Brière G, Darbo É, Thébault P, Uricaru R. Consensus clustering applied to multi-omics disease subtyping. BMC Bioinf. 2021;22(1):361.10.1186/s12859-021-04279-1Suche in Google Scholar PubMed PubMed Central
[58] Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3.10.1093/bioinformatics/btq170Suche in Google Scholar PubMed PubMed Central
[59] Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.10.1038/ncomms3612Suche in Google Scholar PubMed PubMed Central
[60] Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218.10.1186/s13059-016-1070-5Suche in Google Scholar PubMed PubMed Central
[61] Tataranni T, Piccoli C. Dichloroacetate (DCA) and cancer: An overview towards clinical applications. Oxid Med Cell Longev. 2019;2019:8201079.10.1155/2019/8201079Suche in Google Scholar PubMed PubMed Central
[62] Engebretsen S, Bohlin J. Statistical predictions with glmnet. Clin Epigenetics. 2019;11(1):123.10.1186/s13148-019-0730-1Suche in Google Scholar PubMed PubMed Central
[63] Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5(9):1315–6.10.1097/JTO.0b013e3181ec173dSuche in Google Scholar PubMed
[64] Li Q, Li Y, Xu J, Wang S, Xu Y, Li X, et al. Aldolase B overexpression is associated with poor prognosis and promotes tumor progression by epithelial-mesenchymal transition in colorectal adenocarcinoma. Cell Physiol Biochem. 2017;42(1):397–406.10.1159/000477484Suche in Google Scholar PubMed
[65] Huang HC, Lin WR, Lim SN, Yeh CT, Yen TH, Alison MR, et al. Aldolase triggers metabolic reprogramming in colorectal cancer in hypoxia and stiff desmoplastic microenvironments. Colloids Surf B Biointerfaces. 2020;190:110969.10.1016/j.colsurfb.2020.110969Suche in Google Scholar PubMed
[66] Maruyama R, Nagaoka Y, Ishikawa A, Akabane S, Fujiki Y, Taniyama D, et al. Overexpression of aldolase, fructose-bisphosphate C and its association with spheroid formation in colorectal cancer. Pathol Int. 2022;72(3):176–86.10.1111/pin.13200Suche in Google Scholar PubMed
[67] Lv Y, Huang Q, Dai W, Jie Y, Yu G, Fan X, et al. AQP9 promotes astrocytoma cell invasion and motility via the AKT pathway. Oncol Lett. 2018;16(5):6059–64.10.3892/ol.2018.9361Suche in Google Scholar PubMed PubMed Central
[68] Yoshida T, Hojo S, Sekine S, Sawada S, Okumura T, Nagata T, et al. Expression of aquaporin-1 is a poor prognostic factor for stage II and III colon cancer. Mol Clin Oncol. 2013;1(6):953–8.10.3892/mco.2013.165Suche in Google Scholar PubMed PubMed Central
[69] Gan Y, Li Y, Li T, Shu G, Yin G. CCNA2 acts as a novel biomarker in regulating the growth and apoptosis of colorectal cancer. Cancer Manag Res. 2018;10:5113–24.10.2147/CMAR.S176833Suche in Google Scholar PubMed PubMed Central
[70] Liao R, Ma QZ, Zhou CY, Li JJ, Weng NN, Yang Y, et al. Identification of biomarkers related to tumor-infiltrating lymphocytes (TILs) infiltration with gene co-expression network in colorectal cancer. Bioengineered. 2021;12(1):1676–88.10.1080/21655979.2021.1921551Suche in Google Scholar PubMed PubMed Central
[71] Hirai SI, Ryseck RP, Mechta F, Bravo R, Yaniv M. Characterization of junD: A new member of the jun proto-oncogene family. Embo J. 1989;8(5):1433–9.10.1002/j.1460-2075.1989.tb03525.xSuche in Google Scholar PubMed PubMed Central
[72] Villodre ES, Gong Y, Hu X, Huo L, Yoon EC, Ueno NT, et al. NDRG1 expression is an independent prognostic factor in inflammatory breast cancer. Cancers (Basel). 2020;12(12):3711.10.3390/cancers12123711Suche in Google Scholar PubMed PubMed Central
[73] Strzelczyk B, Szulc A, Rzepko R, Kitowska A, Skokowski J, Szutowicz A, et al. Identification of high-risk stage II colorectal tumors by combined analysis of the NDRG1 gene expression and the depth of tumor invasion. Ann Surg Oncol. 2009;16(5):1287–94.10.1245/s10434-009-0381-0Suche in Google Scholar PubMed
[74] Ma J, Gao Q, Zeng S, Shen H. Knockdown of NDRG1 promote epithelial-mesenchymal transition of colorectal cancer via NF-κB signaling. J Surg Oncol. 2016;114(4):520–7.10.1002/jso.24348Suche in Google Scholar PubMed
[75] Shen L, Zhang J, Zheng Z, Yang F, Liu S, Wu Y, et al. PHGDH inhibits ferroptosis and promotes malignant progression by upregulating SLC7A11 in bladder cancer. Int J Biol Sci. 2022;18(14):5459–74.10.7150/ijbs.74546Suche in Google Scholar PubMed PubMed Central
[76] Li HM, Li X, Xia R, Zhang X, Jin TZ, Zhang HS. PHGDH knockdown increases sensitivity to SR1, an aryl hydrocarbon receptor antagonist, in colorectal cancer by activating the autophagy pathway. Febs J. 2024;291(8):1780–94.10.1111/febs.17080Suche in Google Scholar PubMed
[77] Zhao JY, Feng KR, Wang F, Zhang JW, Cheng JF, Lin GQ, et al. A retrospective overview of PHGDH and its inhibitors for regulating cancer metabolism. Eur J Med Chem. 2021;217:113379.10.1016/j.ejmech.2021.113379Suche in Google Scholar PubMed
[78] Jia XQ, Zhang S, Zhu HJ, Wang W, Zhu JH, Wang XD, et al. Increased expression of PHGDH and prognostic significance in colorectal cancer. Transl Oncol. 2016;9(3):191–6.10.1016/j.tranon.2016.03.006Suche in Google Scholar PubMed PubMed Central
[79] Cen B, Lang JD, Du Y, Wei J, Xiong Y, Bradley N, et al. Prostaglandin E(2) induces miR675-5p to promote colorectal tumor metastasis via modulation of p53 expression. Gastroenterology. 2020;158(4):971–84.e10.10.1053/j.gastro.2019.11.013Suche in Google Scholar PubMed PubMed Central
[80] Tsai FM, Wu CC, Shyu RY, Wang CH, Jiang SY. Tazarotene-induced gene 1 inhibits prostaglandin E2-stimulated HCT116 colon cancer cell growth. J Biomed Sci. 2011;18(1):88.10.1186/1423-0127-18-88Suche in Google Scholar PubMed PubMed Central
[81] Pan XY, Yang Y, Meng HW, Li HD, Chen X, Huang HM, et al. DNA methylation of PTGIS enhances hepatic stellate cells activation and liver fibrogenesis. Front Pharmacol. 2018;9:553.10.3389/fphar.2018.00553Suche in Google Scholar PubMed PubMed Central
[82] Lichao S, Liang P, Chunguang G, Fang L, Zhihua Y, Yuliang R. Overexpression of PTGIS could predict liver metastasis and is correlated with poor prognosis in colon cancer patients. Pathol Oncol Res. 2012;18(3):563–9.10.1007/s12253-011-9478-4Suche in Google Scholar PubMed
[83] Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381–95.10.1002/jcp.27740Suche in Google Scholar PubMed
[84] Menck K, Wlochowitz D, Wachter A, Conradi LC, Wolff A, Scheel AH, et al. High-throughput profiling of colorectal cancer liver metastases reveals intra- and inter-patient heterogeneity in the EGFR and WNT pathways associated with clinical outcome. Cancers (Basel). 2022;14(9):2084.10.3390/cancers14092084Suche in Google Scholar PubMed PubMed Central
[85] Wang XC, Liu Y, Long FW, Liu LR, Fan CW. Identification of a lncRNA prognostic signature-related to stem cell index and its significance in colorectal cancer. Future Oncol. 2021;17(23):3087–100.10.2217/fon-2020-1163Suche in Google Scholar PubMed
[86] Weng ML, Chen WK, Chen XY, Lu H, Sun ZR, Yu Q, et al. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat Commun. 2020;11(1):1869.10.1038/s41467-020-15795-8Suche in Google Scholar PubMed PubMed Central
[87] González N, Prieto I, Del Puerto-Nevado L, Portal-Nuñez S, Ardura JA, Corton M, et al. 2017 update on the relationship between diabetes and colorectal cancer: epidemiology, potential molecular mechanisms and therapeutic implications. Oncotarget. 2017;8(11):18456–85.10.18632/oncotarget.14472Suche in Google Scholar PubMed PubMed Central
[88] Liu J, Hu C, Jin Y. ALDOB suppresses the activity of CD8(+) T cells in colorectal cancer via the WNT signaling pathway. Immunol Cell Biol. 2025;103(3):307–16.10.1111/imcb.12853Suche in Google Scholar PubMed
[89] Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer’s inflammatory roots. Clin Cancer Res. 2011;17(19):6125–9.10.1158/1078-0432.CCR-10-2743Suche in Google Scholar PubMed PubMed Central
[90] Moens S, Goveia J, Stapor PC, Cantelmo AR, Carmeliet P. The multifaceted activity of VEGF in angiogenesis - Implications for therapy responses. Cytokine Growth Factor Rev. 2014;25(4):473–82.10.1016/j.cytogfr.2014.07.009Suche in Google Scholar PubMed
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Network pharmacological analysis and in vitro testing of the rutin effects on triple-negative breast cancer
- Impact of diabetes on long-term survival in elderly liver cancer patients: A retrospective study
- Knockdown of CCNB1 alleviates high glucose-triggered trophoblast dysfunction during gestational diabetes via Wnt/β-catenin signaling pathway
- Risk factors for severe adverse drug reactions in hospitalized patients
- Analysis of the effect of ALA-PDT on macrophages in footpad model of mice infected with Fonsecaea monophora based on single-cell sequencing
- Development and validation of headspace gas chromatography with a flame ionization detector method for the determination of ethanol in the vitreous humor
- CMSP exerts anti-tumor effects on small cell lung cancer cells by inducing mitochondrial dysfunction and ferroptosis
- Predictive value of plasma sB7-H3 and YKL-40 in pediatric refractory Mycoplasma pneumoniae pneumonia
- Antiangiogenic potential of Elaeagnus umbellata extracts and molecular docking study by targeting VEGFR-2 pathway
- Comparison of the effectiveness of nurse-led preoperative counseling and postoperative follow-up care vs standard care for patients with gastric cancer
- Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis
- Adhered macrophages as an additional marker of cardiomyocyte injury in biopsies of patients with dilated cardiomyopathy
- Association between statin administration and outcome in patients with sepsis: A retrospective study
- Exploration of the association between estimated glucose disposal rate and osteoarthritis in middle-aged and older adults: An analysis of NHANES data from 2011 to 2018
- A comparative analysis of the binary and multiclass classified chest X-ray images of pneumonia and COVID-19 with ML and DL models
- Lysophosphatidic acid 2 alleviates deep vein thrombosis via protective endothelial barrier function
- Transcription factor A, mitochondrial promotes lymph node metastasis and lymphangiogenesis in epithelial ovarian carcinoma
- Serum PM20D1 levels are associated with nutritional status and inflammatory factors in gastric cancer patients undergoing early enteral nutrition
- Hydromorphone reduced the incidence of emergence agitation after adenotonsillectomy in children with obstructive sleep apnea: A randomized, double-blind study
- Vitamin D replacement therapy may regulate sleep habits in patients with restless leg syndrome
- The first-line antihypertensive nitrendipine potentiated the therapeutic effect of oxaliplatin by downregulating CACNA1D in colorectal cancer
- Health literacy and health-related quality of life: The mediating role of irrational happiness
- Modulatory effects of Lycium barbarum polysaccharide on bone cell dynamics in osteoporosis
- Mechanism research on inhibition of gastric cancer in vitro by the extract of Pinellia ternata based on network pharmacology and cellular metabolomics
- Examination of the causal role of immune cells in non-alcoholic fatty liver disease by a bidirectional Mendelian randomization study
- Clinical analysis of ten cases of HIV infection combined with acute leukemia
- Investigating the cardioprotective potential of quercetin against tacrolimus-induced cardiotoxicity in Wistar rats: A mechanistic insights
- Clinical observation of probiotics combined with mesalazine and Yiyi Baitouweng Decoction retention enema in treating mild-to-moderate ulcerative colitis
- Diagnostic value of ratio of blood inflammation to coagulation markers in periprosthetic joint infection
- Sex-specific associations of sex hormone binding globulin and risk of bladder cancer
- Core muscle strength and stability-oriented breathing training reduces inter-recti distance in postpartum women
- The ERAS nursing care strategy for patients undergoing transsphenoidal endoscopic pituitary tumor resection: A randomized blinded controlled trial
- The serum IL-17A levels in patients with traumatic bowel rupture post-surgery and its predictive value for patient prognosis
- Impact of Kolb’s experiential learning theory-based nursing on caregiver burden and psychological state of caregivers of dementia patients
- Analysis of serum NLR combined with intraoperative margin condition to predict the prognosis of cervical HSIL patients undergoing LEEP surgery
- Commiphora gileadensis ameliorate infertility and erectile dysfunction in diabetic male mice
- The correlation between epithelial–mesenchymal transition classification and MMP2 expression of circulating tumor cells and prognosis of advanced or metastatic nasopharyngeal carcinoma
- Tetrahydropalmatine improves mitochondrial function in vascular smooth muscle cells of atherosclerosis in vitro by inhibiting Ras homolog gene family A/Rho-associated protein kinase-1 signaling pathway
- A cross-sectional study: Relationship between serum oxidative stress levels and arteriovenous fistula maturation in maintenance dialysis patients
- A comparative analysis of the impact of repeated administration of flavan 3-ol on brown, subcutaneous, and visceral adipose tissue
- Identifying early screening factors for depression in middle-aged and older adults: A cohort study
- Perform tumor-specific survival analysis for Merkel cell carcinoma patients undergoing surgical resection based on the SEER database by constructing a nomogram chart
- Unveiling the role of CXCL10 in pancreatic cancer progression: A novel prognostic indicator
- High-dose preoperative intraperitoneal erythropoietin and intravenous methylprednisolone in acute traumatic spinal cord injuries following decompression surgeries
- RAB39B: A novel biomarker for acute myeloid leukemia identified via multi-omics and functional validation
- Impact of peripheral conditioning on reperfusion injury following primary percutaneous coronary intervention in diabetic and non-diabetic STEMI patients
- Clinical efficacy of azacitidine in the treatment of middle- and high-risk myelodysplastic syndrome in middle-aged and elderly patients: A retrospective study
- The effect of ambulatory blood pressure load on mitral regurgitation in continuous ambulatory peritoneal dialysis patients
- Expression and clinical significance of ITGA3 in breast cancer
- Single-nucleus RNA sequencing reveals ARHGAP28 expression of podocytes as a biomarker in human diabetic nephropathy
- rSIG combined with NLR in the prognostic assessment of patients with multiple injuries
- Toxic metals and metalloids in collagen supplements of fish and jellyfish origin: Risk assessment for daily intake
- Exploring causal relationship between 41 inflammatory cytokines and marginal zone lymphoma: A bidirectional Mendelian randomization study
- Gender beliefs and legitimization of dating violence in adolescents
- Effect of serum IL-6, CRP, and MMP-9 levels on the efficacy of modified preperitoneal Kugel repair in patients with inguinal hernia
- Effect of smoking and smoking cessation on hematological parameters in polycythemic patients
- Pathogen surveillance and risk factors for pulmonary infection in patients with lung cancer: A retrospective single-center study
- Necroptosis of hippocampal neurons in paclitaxel chemotherapy-induced cognitive impairment mediates microglial activation via TLR4/MyD88 signaling pathway
- Celastrol suppresses neovascularization in rat aortic vascular endothelial cells stimulated by inflammatory tenocytes via modulating the NLRP3 pathway
- Cord-lamina angle and foraminal diameter as key predictors of C5 palsy after anterior cervical decompression and fusion surgery
- GATA1: A key biomarker for predicting the prognosis of patients with diffuse large B-cell lymphoma
- Influencing factors of false lumen thrombosis in type B aortic dissection: A single-center retrospective study
- MZB1 regulates the immune microenvironment and inhibits ovarian cancer cell migration
- Integrating experimental and network pharmacology to explore the pharmacological mechanisms of Dioscin against glioblastoma
- Trends in research on preterm birth in twin pregnancy based on bibliometrics
- Four-week IgE/baseline IgE ratio combined with tryptase predicts clinical outcome in omalizumab-treated children with moderate-to-severe asthma
- Single-cell transcriptomic analysis identifies a stress response Schwann cell subtype
- Acute pancreatitis risk in the diagnosis and management of inflammatory bowel disease: A critical focus
- Effect of subclinical esketamine on NLRP3 and cognitive dysfunction in elderly ischemic stroke patients
- Interleukin-37 mediates the anti-oral tumor activity in oral cancer through STAT3
- CA199 and CEA expression levels, and minimally invasive postoperative prognosis analysis in esophageal squamous carcinoma patients
- Efficacy of a novel drainage catheter in the treatment of CSF leak after posterior spine surgery: A retrospective cohort study
- Comprehensive biomedicine assessment of Apteranthes tuberculata extracts: Phytochemical analysis and multifaceted pharmacological evaluation in animal models
- Relation of time in range to severity of coronary artery disease in patients with type 2 diabetes: A cross-sectional study
- Dopamine attenuates ethanol-induced neuronal apoptosis by stimulating electrical activity in the developing rat retina
- Correlation between albumin levels during the third trimester and the risk of postpartum levator ani muscle rupture
- Factors associated with maternal attention and distraction during breastfeeding and childcare: A cross-sectional study in the west of Iran
- Mechanisms of hesperetin in treating metabolic dysfunction-associated steatosis liver disease via network pharmacology and in vitro experiments
- The law on oncological oblivion in the Italian and European context: How to best uphold the cancer patients’ rights to privacy and self-determination?
- The prognostic value of the neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and prognostic nutritional index for survival in patients with colorectal cancer
- Factors affecting the measurements of peripheral oxygen saturation values in healthy young adults
- Comparison and correlations between findings of hysteroscopy and vaginal color Doppler ultrasonography for detection of uterine abnormalities in patients with recurrent implantation failure
- The effects of different types of RAGT on balance function in stroke patients with low levels of independent walking in a convalescent rehabilitation hospital
- Causal relationship between asthma and ankylosing spondylitis: A bidirectional two-sample univariable and multivariable Mendelian randomization study
- Correlations of health literacy with individuals’ understanding and use of medications in Southern Taiwan
- Correlation of serum calprotectin with outcome of acute cerebral infarction
- Comparison of computed tomography and guided bronchoscopy in the diagnosis of pulmonary nodules: A systematic review and meta-analysis
- Curdione protects vascular endothelial cells and atherosclerosis via the regulation of DNMT1-mediated ERBB4 promoter methylation
- The identification of novel missense variant in ChAT gene in a patient with gestational diabetes denotes plausible genetic association
- Molecular genotyping of multi-system rare blood types in foreign blood donors based on DNA sequencing and its clinical significance
- Exploring the role of succinyl carnitine in the association between CD39⁺ CD4⁺ T cell and ulcerative colitis: A Mendelian randomization study
- Dexmedetomidine suppresses microglial activation in postoperative cognitive dysfunction via the mmu-miRNA-125/TRAF6 signaling axis
- Analysis of serum metabolomics in patients with different types of chronic heart failure
- Diagnostic value of hematological parameters in the early diagnosis of acute cholecystitis
- Pachymaran alleviates fat accumulation, hepatocyte degeneration, and injury in mice with nonalcoholic fatty liver disease
- Decrease in CD4 and CD8 lymphocytes are predictors of severe clinical picture and unfavorable outcome of the disease in patients with COVID-19
- METTL3 blocked the progression of diabetic retinopathy through m6A-modified SOX2
- The predictive significance of anti-RO-52 antibody in patients with interstitial pneumonia after treatment of malignant tumors
- Exploring cerebrospinal fluid metabolites, cognitive function, and brain atrophy: Insights from Mendelian randomization
- Development and validation of potential molecular subtypes and signatures of ocular sarcoidosis based on autophagy-related gene analysis
- Widespread venous thrombosis: Unveiling a complex case of Behçet’s disease with a literature perspective
- Uterine fibroid embolization: An analysis of clinical outcomes and impact on patients’ quality of life
- Discovery of lipid metabolism-related diagnostic biomarkers and construction of diagnostic model in steroid-induced osteonecrosis of femoral head
- Serum-derived exomiR-188-3p is a promising novel biomarker for early-stage ovarian cancer
- Enhancing chronic back pain management: A comparative study of ultrasound–MRI fusion guidance for paravertebral nerve block
- Peptide CCAT1-70aa promotes hepatocellular carcinoma proliferation and invasion via the MAPK/ERK pathway
- Electroacupuncture-induced reduction of myocardial ischemia–reperfusion injury via FTO-dependent m6A methylation modulation
- Hemorrhoids and cardiovascular disease: A bidirectional Mendelian randomization study
- Cell-free adipose extract inhibits hypertrophic scar formation through collagen remodeling and antiangiogenesis
- HALP score in Demodex blepharitis: A case–control study
- Assessment of SOX2 performance as a marker for circulating cancer stem-like cells (CCSCs) identification in advanced breast cancer patients using CytoTrack system
- Risk and prognosis for brain metastasis in primary metastatic cervical cancer patients: A population-based study
- Comparison of the two intestinal anastomosis methods in pediatric patients
- Factors influencing hematological toxicity and adverse effects of perioperative hyperthermic intraperitoneal vs intraperitoneal chemotherapy in gastrointestinal cancer
- Endotoxin tolerance inhibits NLRP3 inflammasome activation in macrophages of septic mice by restoring autophagic flux through TRIM26
- Lateral transperitoneal laparoscopic adrenalectomy: A single-centre experience of 21 procedures
- Petunidin attenuates lipopolysaccharide-induced retinal microglia inflammatory response in diabetic retinopathy by targeting OGT/NF-κB/LCN2 axis
- Procalcitonin and C-reactive protein as biomarkers for diagnosing and assessing the severity of acute cholecystitis
- Factors determining the number of sessions in successful extracorporeal shock wave lithotripsy patients
- Development of a nomogram for predicting cancer-specific survival in patients with renal pelvic cancer following surgery
- Inhibition of ATG7 promotes orthodontic tooth movement by regulating the RANKL/OPG ratio under compression force
- A machine learning-based prognostic model integrating mRNA stemness index, hypoxia, and glycolysis‑related biomarkers for colorectal cancer
- Glutathione attenuates sepsis-associated encephalopathy via dual modulation of NF-κB and PKA/CREB pathways
- FAHD1 prevents neuronal ferroptosis by modulating R-loop and the cGAS–STING pathway
- Association of placenta weight and morphology with term low birth weight: A case–control study
- Investigation of the pathogenic variants induced Sjogren’s syndrome in Turkish population
- Nucleotide metabolic abnormalities in post-COVID-19 condition and type 2 diabetes mellitus patients and their association with endocrine dysfunction
- TGF-β–Smad2/3 signaling in high-altitude pulmonary hypertension in rats: Role and mechanisms via macrophage M2 polarization
- Ultrasound-guided unilateral versus bilateral erector spinae plane block for postoperative analgesia of patients undergoing laparoscopic cholecystectomy
- Profiling gut microbiome dynamics in subacute thyroiditis: Implications for pathogenesis, diagnosis, and treatment
- Delta neutrophil index, CRP/albumin ratio, procalcitonin, immature granulocytes, and HALP score in acute appendicitis: Best performing biomarker?
- Anticancer activity mechanism of novelly synthesized and characterized benzofuran ring-linked 3-nitrophenyl chalcone derivative on colon cancer cells
- H2valdien3 arrests the cell cycle and induces apoptosis of gastric cancer
- Prognostic relevance of PRSS2 and its immune correlates in papillary thyroid carcinoma
- Association of SGLT2 inhibition with psychiatric disorders: A Mendelian randomization study
- Motivational interviewing for alcohol use reduction in Thai patients
- 10.1515/med-2025-1198
- Polyphyllin II inhibits thyroid cancer cell growth by simultaneously inhibiting glycolysis and oxidative phosphorylation
- Relationship between the expression of copper death promoting factor SLC31A1 in papillary thyroid carcinoma and clinicopathological indicators and prognosis
- CSF2 polarized neutrophils and invaded renal cancer cells in vitro influence
- Proton pump inhibitors-induced thrombocytopenia: A systematic literature analysis of case reports
- The current status and influence factors of research ability among community nurses: A sequential qualitative–quantitative study
- OKAIN: A comprehensive oncology knowledge base for the interpretation of clinically actionable alterations
- The relationship between serum CA50, CA242, and SAA levels and clinical pathological characteristics and prognosis in patients with pancreatic cancer
- Identification and external validation of a prognostic signature based on hypoxia–glycolysis-related genes for kidney renal clear cell carcinoma
- Engineered RBC-derived nanovesicles functionalized with tumor-targeting ligands: A comparative study on breast cancer targeting efficiency and biocompatibility
- Relationship of resting echocardiography combined with serum micronutrients to the severity of low-gradient severe aortic stenosis
- Review Articles
- The effects of enhanced external counter-pulsation on post-acute sequelae of COVID-19: A narrative review
- Diabetes-related cognitive impairment: Mechanisms, symptoms, and treatments
- Microscopic changes and gross morphology of placenta in women affected by gestational diabetes mellitus in dietary treatment: A systematic review
- Review of mechanisms and frontier applications in IL-17A-induced hypertension
- Research progress on the correlation between islet amyloid peptides and type 2 diabetes mellitus
- The safety and efficacy of BCG combined with mitomycin C compared with BCG monotherapy in patients with non-muscle-invasive bladder cancer: A systematic review and meta-analysis
- The application of augmented reality in robotic general surgery: A mini-review
- The effect of Greek mountain tea extract and wheat germ extract on peripheral blood flow and eicosanoid metabolism in mammals
- Neurogasobiology of migraine: Carbon monoxide, hydrogen sulfide, and nitric oxide as emerging pathophysiological trinacrium relevant to nociception regulation
- Plant polyphenols, terpenes, and terpenoids in oral health
- Laboratory medicine between technological innovation, rights safeguarding, and patient safety: A bioethical perspective
- End-of-life in cancer patients: Medicolegal implications and ethical challenges in Europe
- The maternal factors during pregnancy for intrauterine growth retardation: An umbrella review
- Intra-abdominal hypertension/abdominal compartment syndrome of pediatric patients in critical care settings
- PI3K/Akt pathway and neuroinflammation in sepsis-associated encephalopathy
- Screening of Group B Streptococcus in pregnancy: A systematic review for the laboratory detection
- Giant borderline ovarian tumours – review of the literature
- Leveraging artificial intelligence for collaborative care planning: Innovations and impacts in shared decision-making – A systematic review
- Cholera epidemiology analysis through the experience of the 1973 Naples epidemic
- Risk factors of frailty/sarcopenia in community older adults: Meta-analysis
- Supplement strategies for infertility in overweight women: Evidence and legal insights
- Scurvy, a not obsolete disorder: Clinical report in eight young children and literature review
- A meta-analysis of the effects of DBS on cognitive function in patients with advanced PD
- Protective role of selenium in sepsis: Mechanisms and potential therapeutic strategies
- Strategies for hyperkalemia management in dialysis patients: A systematic review
- Case Reports
- Delayed graft function after renal transplantation
- Semaglutide treatment for type 2 diabetes in a patient with chronic myeloid leukemia: A case report and review of the literature
- Diverse electrophysiological demyelinating features in a late-onset glycogen storage disease type IIIa case
- Giant right atrial hemangioma presenting with ascites: A case report
- Laser excision of a large granular cell tumor of the vocal cord with subglottic extension: A case report
- EsoFLIP-assisted dilation for dysphagia in systemic sclerosis: Highlighting the role of multimodal esophageal evaluation
- Molecular hydrogen-rhodiola as an adjuvant therapy for ischemic stroke in internal carotid artery occlusion: A case report
- Coronary artery anomalies: A case of the “malignant” left coronary artery and its surgical management
- Rapid Communication
- Biological properties of valve materials using RGD and EC
-
A single oral administration of flavanols enhances short
-term memory in mice along with increased brain-derived neurotrophic factor - Letter to the Editor
- Role of enhanced external counterpulsation in long COVID
- Expression of Concern
- Expression of concern “A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma”
- Expression of concern “Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway”
- Expression of concern “circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8”
- Corrigendum
- Corrigendum to “Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism”
- Corrigendum to “Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis”
- Corrigendum to “The progress of autoimmune hepatitis research and future challenges”
- Retraction
- Retraction of “miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway”
- Retraction of: “LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through downregulating SP-A by sponging to miR-424”
- Retraction of: “SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways”
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part II
- Unveiling novel biomarkers for platinum chemoresistance in ovarian cancer
- Lathyrol affects the expression of AR and PSA and inhibits the malignant behavior of RCC cells
- The era of increasing cancer survivorship: Trends in fertility preservation, medico-legal implications, and ethical challenges
- Bone scintigraphy and positron emission tomography in the early diagnosis of MRONJ
- Meta-analysis of clinical efficacy and safety of immunotherapy combined with chemotherapy in non-small cell lung cancer
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part IV
- Exploration of mRNA-modifying METTL3 oncogene as momentous prognostic biomarker responsible for colorectal cancer development
- Special Issue The evolving saga of RNAs from bench to bedside - Part III
- Interaction and verification of ferroptosis-related RNAs Rela and Stat3 in promoting sepsis-associated acute kidney injury
- The mRNA MOXD1: Link to oxidative stress and prognostic significance in gastric cancer
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part II
- Dynamic changes in lactate-related genes in microglia and their role in immune cell interactions after ischemic stroke
- A prognostic model correlated with fatty acid metabolism in Ewing’s sarcoma based on bioinformatics analysis
- Red cell distribution width predicts early kidney injury: A NHANES cross-sectional study
- Special Issue Diabetes mellitus: pathophysiology, complications & treatment
- Nutritional risk assessment and nutritional support in children with congenital diabetes during surgery
- Correlation of the differential expressions of RANK, RANKL, and OPG with obesity in the elderly population in Xinjiang
- A discussion on the application of fluorescence micro-optical sectioning tomography in the research of cognitive dysfunction in diabetes
- A review of brain research on T2DM-related cognitive dysfunction
- Metformin and estrogen modulation in LABC with T2DM: A 36-month randomized trial
- Special Issue Innovative Biomarker Discovery and Precision Medicine in Cancer Diagnostics
- CircASH1L-mediated tumor progression in triple-negative breast cancer: PI3K/AKT pathway mechanisms
Artikel in diesem Heft
- Research Articles
- Network pharmacological analysis and in vitro testing of the rutin effects on triple-negative breast cancer
- Impact of diabetes on long-term survival in elderly liver cancer patients: A retrospective study
- Knockdown of CCNB1 alleviates high glucose-triggered trophoblast dysfunction during gestational diabetes via Wnt/β-catenin signaling pathway
- Risk factors for severe adverse drug reactions in hospitalized patients
- Analysis of the effect of ALA-PDT on macrophages in footpad model of mice infected with Fonsecaea monophora based on single-cell sequencing
- Development and validation of headspace gas chromatography with a flame ionization detector method for the determination of ethanol in the vitreous humor
- CMSP exerts anti-tumor effects on small cell lung cancer cells by inducing mitochondrial dysfunction and ferroptosis
- Predictive value of plasma sB7-H3 and YKL-40 in pediatric refractory Mycoplasma pneumoniae pneumonia
- Antiangiogenic potential of Elaeagnus umbellata extracts and molecular docking study by targeting VEGFR-2 pathway
- Comparison of the effectiveness of nurse-led preoperative counseling and postoperative follow-up care vs standard care for patients with gastric cancer
- Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis
- Adhered macrophages as an additional marker of cardiomyocyte injury in biopsies of patients with dilated cardiomyopathy
- Association between statin administration and outcome in patients with sepsis: A retrospective study
- Exploration of the association between estimated glucose disposal rate and osteoarthritis in middle-aged and older adults: An analysis of NHANES data from 2011 to 2018
- A comparative analysis of the binary and multiclass classified chest X-ray images of pneumonia and COVID-19 with ML and DL models
- Lysophosphatidic acid 2 alleviates deep vein thrombosis via protective endothelial barrier function
- Transcription factor A, mitochondrial promotes lymph node metastasis and lymphangiogenesis in epithelial ovarian carcinoma
- Serum PM20D1 levels are associated with nutritional status and inflammatory factors in gastric cancer patients undergoing early enteral nutrition
- Hydromorphone reduced the incidence of emergence agitation after adenotonsillectomy in children with obstructive sleep apnea: A randomized, double-blind study
- Vitamin D replacement therapy may regulate sleep habits in patients with restless leg syndrome
- The first-line antihypertensive nitrendipine potentiated the therapeutic effect of oxaliplatin by downregulating CACNA1D in colorectal cancer
- Health literacy and health-related quality of life: The mediating role of irrational happiness
- Modulatory effects of Lycium barbarum polysaccharide on bone cell dynamics in osteoporosis
- Mechanism research on inhibition of gastric cancer in vitro by the extract of Pinellia ternata based on network pharmacology and cellular metabolomics
- Examination of the causal role of immune cells in non-alcoholic fatty liver disease by a bidirectional Mendelian randomization study
- Clinical analysis of ten cases of HIV infection combined with acute leukemia
- Investigating the cardioprotective potential of quercetin against tacrolimus-induced cardiotoxicity in Wistar rats: A mechanistic insights
- Clinical observation of probiotics combined with mesalazine and Yiyi Baitouweng Decoction retention enema in treating mild-to-moderate ulcerative colitis
- Diagnostic value of ratio of blood inflammation to coagulation markers in periprosthetic joint infection
- Sex-specific associations of sex hormone binding globulin and risk of bladder cancer
- Core muscle strength and stability-oriented breathing training reduces inter-recti distance in postpartum women
- The ERAS nursing care strategy for patients undergoing transsphenoidal endoscopic pituitary tumor resection: A randomized blinded controlled trial
- The serum IL-17A levels in patients with traumatic bowel rupture post-surgery and its predictive value for patient prognosis
- Impact of Kolb’s experiential learning theory-based nursing on caregiver burden and psychological state of caregivers of dementia patients
- Analysis of serum NLR combined with intraoperative margin condition to predict the prognosis of cervical HSIL patients undergoing LEEP surgery
- Commiphora gileadensis ameliorate infertility and erectile dysfunction in diabetic male mice
- The correlation between epithelial–mesenchymal transition classification and MMP2 expression of circulating tumor cells and prognosis of advanced or metastatic nasopharyngeal carcinoma
- Tetrahydropalmatine improves mitochondrial function in vascular smooth muscle cells of atherosclerosis in vitro by inhibiting Ras homolog gene family A/Rho-associated protein kinase-1 signaling pathway
- A cross-sectional study: Relationship between serum oxidative stress levels and arteriovenous fistula maturation in maintenance dialysis patients
- A comparative analysis of the impact of repeated administration of flavan 3-ol on brown, subcutaneous, and visceral adipose tissue
- Identifying early screening factors for depression in middle-aged and older adults: A cohort study
- Perform tumor-specific survival analysis for Merkel cell carcinoma patients undergoing surgical resection based on the SEER database by constructing a nomogram chart
- Unveiling the role of CXCL10 in pancreatic cancer progression: A novel prognostic indicator
- High-dose preoperative intraperitoneal erythropoietin and intravenous methylprednisolone in acute traumatic spinal cord injuries following decompression surgeries
- RAB39B: A novel biomarker for acute myeloid leukemia identified via multi-omics and functional validation
- Impact of peripheral conditioning on reperfusion injury following primary percutaneous coronary intervention in diabetic and non-diabetic STEMI patients
- Clinical efficacy of azacitidine in the treatment of middle- and high-risk myelodysplastic syndrome in middle-aged and elderly patients: A retrospective study
- The effect of ambulatory blood pressure load on mitral regurgitation in continuous ambulatory peritoneal dialysis patients
- Expression and clinical significance of ITGA3 in breast cancer
- Single-nucleus RNA sequencing reveals ARHGAP28 expression of podocytes as a biomarker in human diabetic nephropathy
- rSIG combined with NLR in the prognostic assessment of patients with multiple injuries
- Toxic metals and metalloids in collagen supplements of fish and jellyfish origin: Risk assessment for daily intake
- Exploring causal relationship between 41 inflammatory cytokines and marginal zone lymphoma: A bidirectional Mendelian randomization study
- Gender beliefs and legitimization of dating violence in adolescents
- Effect of serum IL-6, CRP, and MMP-9 levels on the efficacy of modified preperitoneal Kugel repair in patients with inguinal hernia
- Effect of smoking and smoking cessation on hematological parameters in polycythemic patients
- Pathogen surveillance and risk factors for pulmonary infection in patients with lung cancer: A retrospective single-center study
- Necroptosis of hippocampal neurons in paclitaxel chemotherapy-induced cognitive impairment mediates microglial activation via TLR4/MyD88 signaling pathway
- Celastrol suppresses neovascularization in rat aortic vascular endothelial cells stimulated by inflammatory tenocytes via modulating the NLRP3 pathway
- Cord-lamina angle and foraminal diameter as key predictors of C5 palsy after anterior cervical decompression and fusion surgery
- GATA1: A key biomarker for predicting the prognosis of patients with diffuse large B-cell lymphoma
- Influencing factors of false lumen thrombosis in type B aortic dissection: A single-center retrospective study
- MZB1 regulates the immune microenvironment and inhibits ovarian cancer cell migration
- Integrating experimental and network pharmacology to explore the pharmacological mechanisms of Dioscin against glioblastoma
- Trends in research on preterm birth in twin pregnancy based on bibliometrics
- Four-week IgE/baseline IgE ratio combined with tryptase predicts clinical outcome in omalizumab-treated children with moderate-to-severe asthma
- Single-cell transcriptomic analysis identifies a stress response Schwann cell subtype
- Acute pancreatitis risk in the diagnosis and management of inflammatory bowel disease: A critical focus
- Effect of subclinical esketamine on NLRP3 and cognitive dysfunction in elderly ischemic stroke patients
- Interleukin-37 mediates the anti-oral tumor activity in oral cancer through STAT3
- CA199 and CEA expression levels, and minimally invasive postoperative prognosis analysis in esophageal squamous carcinoma patients
- Efficacy of a novel drainage catheter in the treatment of CSF leak after posterior spine surgery: A retrospective cohort study
- Comprehensive biomedicine assessment of Apteranthes tuberculata extracts: Phytochemical analysis and multifaceted pharmacological evaluation in animal models
- Relation of time in range to severity of coronary artery disease in patients with type 2 diabetes: A cross-sectional study
- Dopamine attenuates ethanol-induced neuronal apoptosis by stimulating electrical activity in the developing rat retina
- Correlation between albumin levels during the third trimester and the risk of postpartum levator ani muscle rupture
- Factors associated with maternal attention and distraction during breastfeeding and childcare: A cross-sectional study in the west of Iran
- Mechanisms of hesperetin in treating metabolic dysfunction-associated steatosis liver disease via network pharmacology and in vitro experiments
- The law on oncological oblivion in the Italian and European context: How to best uphold the cancer patients’ rights to privacy and self-determination?
- The prognostic value of the neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, and prognostic nutritional index for survival in patients with colorectal cancer
- Factors affecting the measurements of peripheral oxygen saturation values in healthy young adults
- Comparison and correlations between findings of hysteroscopy and vaginal color Doppler ultrasonography for detection of uterine abnormalities in patients with recurrent implantation failure
- The effects of different types of RAGT on balance function in stroke patients with low levels of independent walking in a convalescent rehabilitation hospital
- Causal relationship between asthma and ankylosing spondylitis: A bidirectional two-sample univariable and multivariable Mendelian randomization study
- Correlations of health literacy with individuals’ understanding and use of medications in Southern Taiwan
- Correlation of serum calprotectin with outcome of acute cerebral infarction
- Comparison of computed tomography and guided bronchoscopy in the diagnosis of pulmonary nodules: A systematic review and meta-analysis
- Curdione protects vascular endothelial cells and atherosclerosis via the regulation of DNMT1-mediated ERBB4 promoter methylation
- The identification of novel missense variant in ChAT gene in a patient with gestational diabetes denotes plausible genetic association
- Molecular genotyping of multi-system rare blood types in foreign blood donors based on DNA sequencing and its clinical significance
- Exploring the role of succinyl carnitine in the association between CD39⁺ CD4⁺ T cell and ulcerative colitis: A Mendelian randomization study
- Dexmedetomidine suppresses microglial activation in postoperative cognitive dysfunction via the mmu-miRNA-125/TRAF6 signaling axis
- Analysis of serum metabolomics in patients with different types of chronic heart failure
- Diagnostic value of hematological parameters in the early diagnosis of acute cholecystitis
- Pachymaran alleviates fat accumulation, hepatocyte degeneration, and injury in mice with nonalcoholic fatty liver disease
- Decrease in CD4 and CD8 lymphocytes are predictors of severe clinical picture and unfavorable outcome of the disease in patients with COVID-19
- METTL3 blocked the progression of diabetic retinopathy through m6A-modified SOX2
- The predictive significance of anti-RO-52 antibody in patients with interstitial pneumonia after treatment of malignant tumors
- Exploring cerebrospinal fluid metabolites, cognitive function, and brain atrophy: Insights from Mendelian randomization
- Development and validation of potential molecular subtypes and signatures of ocular sarcoidosis based on autophagy-related gene analysis
- Widespread venous thrombosis: Unveiling a complex case of Behçet’s disease with a literature perspective
- Uterine fibroid embolization: An analysis of clinical outcomes and impact on patients’ quality of life
- Discovery of lipid metabolism-related diagnostic biomarkers and construction of diagnostic model in steroid-induced osteonecrosis of femoral head
- Serum-derived exomiR-188-3p is a promising novel biomarker for early-stage ovarian cancer
- Enhancing chronic back pain management: A comparative study of ultrasound–MRI fusion guidance for paravertebral nerve block
- Peptide CCAT1-70aa promotes hepatocellular carcinoma proliferation and invasion via the MAPK/ERK pathway
- Electroacupuncture-induced reduction of myocardial ischemia–reperfusion injury via FTO-dependent m6A methylation modulation
- Hemorrhoids and cardiovascular disease: A bidirectional Mendelian randomization study
- Cell-free adipose extract inhibits hypertrophic scar formation through collagen remodeling and antiangiogenesis
- HALP score in Demodex blepharitis: A case–control study
- Assessment of SOX2 performance as a marker for circulating cancer stem-like cells (CCSCs) identification in advanced breast cancer patients using CytoTrack system
- Risk and prognosis for brain metastasis in primary metastatic cervical cancer patients: A population-based study
- Comparison of the two intestinal anastomosis methods in pediatric patients
- Factors influencing hematological toxicity and adverse effects of perioperative hyperthermic intraperitoneal vs intraperitoneal chemotherapy in gastrointestinal cancer
- Endotoxin tolerance inhibits NLRP3 inflammasome activation in macrophages of septic mice by restoring autophagic flux through TRIM26
- Lateral transperitoneal laparoscopic adrenalectomy: A single-centre experience of 21 procedures
- Petunidin attenuates lipopolysaccharide-induced retinal microglia inflammatory response in diabetic retinopathy by targeting OGT/NF-κB/LCN2 axis
- Procalcitonin and C-reactive protein as biomarkers for diagnosing and assessing the severity of acute cholecystitis
- Factors determining the number of sessions in successful extracorporeal shock wave lithotripsy patients
- Development of a nomogram for predicting cancer-specific survival in patients with renal pelvic cancer following surgery
- Inhibition of ATG7 promotes orthodontic tooth movement by regulating the RANKL/OPG ratio under compression force
- A machine learning-based prognostic model integrating mRNA stemness index, hypoxia, and glycolysis‑related biomarkers for colorectal cancer
- Glutathione attenuates sepsis-associated encephalopathy via dual modulation of NF-κB and PKA/CREB pathways
- FAHD1 prevents neuronal ferroptosis by modulating R-loop and the cGAS–STING pathway
- Association of placenta weight and morphology with term low birth weight: A case–control study
- Investigation of the pathogenic variants induced Sjogren’s syndrome in Turkish population
- Nucleotide metabolic abnormalities in post-COVID-19 condition and type 2 diabetes mellitus patients and their association with endocrine dysfunction
- TGF-β–Smad2/3 signaling in high-altitude pulmonary hypertension in rats: Role and mechanisms via macrophage M2 polarization
- Ultrasound-guided unilateral versus bilateral erector spinae plane block for postoperative analgesia of patients undergoing laparoscopic cholecystectomy
- Profiling gut microbiome dynamics in subacute thyroiditis: Implications for pathogenesis, diagnosis, and treatment
- Delta neutrophil index, CRP/albumin ratio, procalcitonin, immature granulocytes, and HALP score in acute appendicitis: Best performing biomarker?
- Anticancer activity mechanism of novelly synthesized and characterized benzofuran ring-linked 3-nitrophenyl chalcone derivative on colon cancer cells
- H2valdien3 arrests the cell cycle and induces apoptosis of gastric cancer
- Prognostic relevance of PRSS2 and its immune correlates in papillary thyroid carcinoma
- Association of SGLT2 inhibition with psychiatric disorders: A Mendelian randomization study
- Motivational interviewing for alcohol use reduction in Thai patients
- 10.1515/med-2025-1198
- Polyphyllin II inhibits thyroid cancer cell growth by simultaneously inhibiting glycolysis and oxidative phosphorylation
- Relationship between the expression of copper death promoting factor SLC31A1 in papillary thyroid carcinoma and clinicopathological indicators and prognosis
- CSF2 polarized neutrophils and invaded renal cancer cells in vitro influence
- Proton pump inhibitors-induced thrombocytopenia: A systematic literature analysis of case reports
- The current status and influence factors of research ability among community nurses: A sequential qualitative–quantitative study
- OKAIN: A comprehensive oncology knowledge base for the interpretation of clinically actionable alterations
- The relationship between serum CA50, CA242, and SAA levels and clinical pathological characteristics and prognosis in patients with pancreatic cancer
- Identification and external validation of a prognostic signature based on hypoxia–glycolysis-related genes for kidney renal clear cell carcinoma
- Engineered RBC-derived nanovesicles functionalized with tumor-targeting ligands: A comparative study on breast cancer targeting efficiency and biocompatibility
- Relationship of resting echocardiography combined with serum micronutrients to the severity of low-gradient severe aortic stenosis
- Review Articles
- The effects of enhanced external counter-pulsation on post-acute sequelae of COVID-19: A narrative review
- Diabetes-related cognitive impairment: Mechanisms, symptoms, and treatments
- Microscopic changes and gross morphology of placenta in women affected by gestational diabetes mellitus in dietary treatment: A systematic review
- Review of mechanisms and frontier applications in IL-17A-induced hypertension
- Research progress on the correlation between islet amyloid peptides and type 2 diabetes mellitus
- The safety and efficacy of BCG combined with mitomycin C compared with BCG monotherapy in patients with non-muscle-invasive bladder cancer: A systematic review and meta-analysis
- The application of augmented reality in robotic general surgery: A mini-review
- The effect of Greek mountain tea extract and wheat germ extract on peripheral blood flow and eicosanoid metabolism in mammals
- Neurogasobiology of migraine: Carbon monoxide, hydrogen sulfide, and nitric oxide as emerging pathophysiological trinacrium relevant to nociception regulation
- Plant polyphenols, terpenes, and terpenoids in oral health
- Laboratory medicine between technological innovation, rights safeguarding, and patient safety: A bioethical perspective
- End-of-life in cancer patients: Medicolegal implications and ethical challenges in Europe
- The maternal factors during pregnancy for intrauterine growth retardation: An umbrella review
- Intra-abdominal hypertension/abdominal compartment syndrome of pediatric patients in critical care settings
- PI3K/Akt pathway and neuroinflammation in sepsis-associated encephalopathy
- Screening of Group B Streptococcus in pregnancy: A systematic review for the laboratory detection
- Giant borderline ovarian tumours – review of the literature
- Leveraging artificial intelligence for collaborative care planning: Innovations and impacts in shared decision-making – A systematic review
- Cholera epidemiology analysis through the experience of the 1973 Naples epidemic
- Risk factors of frailty/sarcopenia in community older adults: Meta-analysis
- Supplement strategies for infertility in overweight women: Evidence and legal insights
- Scurvy, a not obsolete disorder: Clinical report in eight young children and literature review
- A meta-analysis of the effects of DBS on cognitive function in patients with advanced PD
- Protective role of selenium in sepsis: Mechanisms and potential therapeutic strategies
- Strategies for hyperkalemia management in dialysis patients: A systematic review
- Case Reports
- Delayed graft function after renal transplantation
- Semaglutide treatment for type 2 diabetes in a patient with chronic myeloid leukemia: A case report and review of the literature
- Diverse electrophysiological demyelinating features in a late-onset glycogen storage disease type IIIa case
- Giant right atrial hemangioma presenting with ascites: A case report
- Laser excision of a large granular cell tumor of the vocal cord with subglottic extension: A case report
- EsoFLIP-assisted dilation for dysphagia in systemic sclerosis: Highlighting the role of multimodal esophageal evaluation
- Molecular hydrogen-rhodiola as an adjuvant therapy for ischemic stroke in internal carotid artery occlusion: A case report
- Coronary artery anomalies: A case of the “malignant” left coronary artery and its surgical management
- Rapid Communication
- Biological properties of valve materials using RGD and EC
-
A single oral administration of flavanols enhances short
-term memory in mice along with increased brain-derived neurotrophic factor - Letter to the Editor
- Role of enhanced external counterpulsation in long COVID
- Expression of Concern
- Expression of concern “A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma”
- Expression of concern “Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway”
- Expression of concern “circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8”
- Corrigendum
- Corrigendum to “Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism”
- Corrigendum to “Comparing the therapeutic efficacy of endoscopic minimally invasive surgery and traditional surgery for early-stage breast cancer: A meta-analysis”
- Corrigendum to “The progress of autoimmune hepatitis research and future challenges”
- Retraction
- Retraction of “miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway”
- Retraction of: “LncRNA CASC15 inhibition relieves renal fibrosis in diabetic nephropathy through downregulating SP-A by sponging to miR-424”
- Retraction of: “SCARA5 inhibits oral squamous cell carcinoma via inactivating the STAT3 and PI3K/AKT signaling pathways”
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part II
- Unveiling novel biomarkers for platinum chemoresistance in ovarian cancer
- Lathyrol affects the expression of AR and PSA and inhibits the malignant behavior of RCC cells
- The era of increasing cancer survivorship: Trends in fertility preservation, medico-legal implications, and ethical challenges
- Bone scintigraphy and positron emission tomography in the early diagnosis of MRONJ
- Meta-analysis of clinical efficacy and safety of immunotherapy combined with chemotherapy in non-small cell lung cancer
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part IV
- Exploration of mRNA-modifying METTL3 oncogene as momentous prognostic biomarker responsible for colorectal cancer development
- Special Issue The evolving saga of RNAs from bench to bedside - Part III
- Interaction and verification of ferroptosis-related RNAs Rela and Stat3 in promoting sepsis-associated acute kidney injury
- The mRNA MOXD1: Link to oxidative stress and prognostic significance in gastric cancer
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part II
- Dynamic changes in lactate-related genes in microglia and their role in immune cell interactions after ischemic stroke
- A prognostic model correlated with fatty acid metabolism in Ewing’s sarcoma based on bioinformatics analysis
- Red cell distribution width predicts early kidney injury: A NHANES cross-sectional study
- Special Issue Diabetes mellitus: pathophysiology, complications & treatment
- Nutritional risk assessment and nutritional support in children with congenital diabetes during surgery
- Correlation of the differential expressions of RANK, RANKL, and OPG with obesity in the elderly population in Xinjiang
- A discussion on the application of fluorescence micro-optical sectioning tomography in the research of cognitive dysfunction in diabetes
- A review of brain research on T2DM-related cognitive dysfunction
- Metformin and estrogen modulation in LABC with T2DM: A 36-month randomized trial
- Special Issue Innovative Biomarker Discovery and Precision Medicine in Cancer Diagnostics
- CircASH1L-mediated tumor progression in triple-negative breast cancer: PI3K/AKT pathway mechanisms

