Abstract
Disease development and progression are very complex processes which make clinical decision making non-trivial. On the one hand, examination results that are stored in multiple formats and data types in clinical information systems need to be considered. Beyond, biological or molecular-biological processes can influence clinical decision making. So far, biological knowledge and patient data is separated from each other. This complicates inclusion of all relevant knowledge and information into the decision making. In this paper, we describe a concept of model-based decision support that links knowledge about biological processes, treatment decisions and clinical data. It consists of three models: 1) a biological model, 2) a decision model encompassing medical knowledge about the treatment workflow and decision parameters, and 3) a patient data model generated from clinical data. Requirements and future steps for realizing the concept will be presented and it will be shown how the concept can support the clinical decision making.
1 Introduction
Individualized patient treatment requires considering all relevant clinical information items and weighting them according to the patient-specific situation. Additionally, knowledge and information on the development of diseases as well as on influence factors including the underlying biological processes are required. Further, knowledge and experiences about treatment and treatment plans, risk factors, problems and complications etc. need to be accessible. However, the challenges we face nowadays in clinical settings are that (1) clinical data is distributed in several systems and available in multiple formats, and (2) best practices and information about treatment plans and options are captured in biomedical literature or clinical practice guidelines, but are not formalized. Not only accessing the relevant data during diagnosis and treatment among the large set of available patient data is a challenge, also its mental integration, analysis and interpretation. Understanding the dependencies among examination results, biological processes and medical conditions is crucial for making proper decisions. For these reasons, support in evidence-based medicine is important to ensure high quality treatment in the different pre-, intra and perioperative stages, and also beyond the domain of surgery.
The concept of model-based therapy [1, 2] addresses the challenges outlined before by relying upon knowledge and data models that provide an evidence-based, reproducible basis for decision making. Among others biological processes can be modelled such as the effect of hormones on disease development. A digital patient model integrates various patient-specific information entities that need to be considered within therapy planning and clinical decision making. It further describes relations between clinical parameters and sociological factors related to a specific clinical pathology. In this work, we will introduce a concept of model-based decision support that links knowledge about biological processes, treatment decisions and clinical data. It consists of three models: 1) a biological model, 2) a decision model encompassing medical knowledge about the treatment workflow and decision parameters and 3) a patient data model generated from clinical data. We will collect the requirements for the realization of the concept.
2 Three-part modelling concept
Our concept of model-based decision support relies upon three types of models (see Fig. 1):
– A biological model describes the disease mechanisms and underlying biological processes and parameters (e.g. hormone-disease effect, eye functions, latitude of a tumor).
– A decision model encompasses the medical knowledge on the diagnostic of a disease, the therapy decision including the therapy options, their risks for the patient and their relations to single information items relevant in decision making.
– A patient specific data model includes and integrates the relevant patient data (laboratory tests, examination results, medical report, signal and image data).
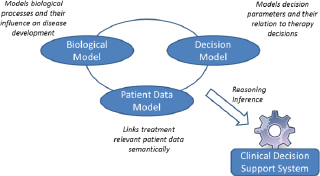
By integrating information on biological processes, decision processes and clinical data, a model-based decision support system can be built. Information from one model is relevant for calculations in another one.
The decision model reflects the intellectual reasoning performed by clinicians during diagnosis, treatment and monitoring of the disease. It includes decision parameters and their relations to therapy options. Further, the patient model integrates the relevant clinical patient data of different types. The biological model can also contain information on the efficacy of drugs given some biological circumstances of patients for a personalized therapy plan. The models clearly contain information relevant for each other. For example, predictions about cell growth in the context of tumor diseases impact the predictions on treatment options or expectancy of life. However, to reduce complexity and to enable their re-use (in particular re-use of models on biological processes), it is necessary to separate biological models from the other model types even though they are related to the same disease.
We suggest building a clinical decision support system on top of a model that integrates these three model types in the context of a specific medical condition. All relevant information items of a decision making process together with the relevant decision parameters and the dependencies among biological and clinical aspects of a disease are linked in an integrated model. The resulting decision support system will be capable of providing a prognosis of treatment outcome, providing hints to missing examination results or just providing an integrated view on data and therapeutic options with the relationships among them. Further, it helps to learn dependencies between biological processes and their influence on factors relevant for patient treatment.
3 Use case
In this section, we want to describe a use case for our three-model concept and specify the concrete requirements based on the general collection from the section before. To demonstrate the application of the three models as basis for a model-based clinical decision support system, consider the following use case of treatment and monitoring of patients suffering from pituitary adenoma.
Pituitary adenomas are common benign tumours of the pituitary gland. Some tumours secrete one or more hormones in excess. Such so-called secretory pituitary adenomas are usually found due to hormonal imbalances that affect bodily functions. The treatment of these tumours requires the competences of interdisciplinary clinicians, endocrinologists, neurosurgeons and radiologists. The treatment decision is usually performed by considering a broad range of patient data such as clinical symptoms (infertility, acromegaly, deterioration of the visual functions) or results from clinical examinations (hormone test, image data acquisition, ophthalmological examination).
A model-based decision support system for supporting in pituitary adenoma treatment comprises a biological model reflecting the healthy biological processes of the pituitary gland, a decision model on the treatment options and their dependencies to clinical and biological parameter. When applied to a specific patient record, the patient data model provides the patient-specific data. Based on the decision model, our model-based decision support system sends notifications when relevant patient data for deciding for a treatment is missing. Once all relevant data is available to the system, it compares the healthy processes to the given pathological data of patient, suggests possible treatments (surgery, medical treatment, radiotherapy or simple observation) and predicts the outcome and risks for the single treatment options. The calculation considers the severity of the clinical symptoms, the findings of the clinical examination (size and location of the tumours). Together with the general state of the patient this leads to the choice of the most relevant therapy for the patient.
From this use case, it becomes clear that for its realisation among others a biological model on hormone effects is required as well as a therapy decision model for pituitary adenoma, and a standardized representation and storage of the patient-specific data.
4 Requirements
More specifically, in order to realize a clinical decision support system based on the three models, methods and challenges in three main areas need to be considered:
– Modelling of biological processes, diseases and clinical decision making: Methods for semi-automatic modelling of diseases, biological processes and clinical decision processes need to be developed.
– Semantic linking : Methods for linking data and biological models with patient models through ontologies including methods for integrating the underlying ontologies are necessary. Semantic annotations can be produced by extracting relevant terms from texts or classifying image data automatically.
– Ontology developing : Ontologies and classification systems for describing the concepts of the models and providing the relations for inferencing are required.
More details on these requirements are provided in the following.
Requirement 1: Model development. For generating biological and decision model, mathematical modelling techniques such as Multi-Entity Bayesian Networks (MEBN) can be exploited. MEBNs are based on the Bayesian probability theory with first-order logic [3]. In general, MEBN are similar to Bayesian Networks (BN) with nodes, edges and conditional probabilities. MEBN have been used to model and simulate abstractions of real-life situations and processes [3]. They are based on both, mathematical correlations between random variables (representing uncertainty and what is known with reasonable certainty) and a graph, denoting the conditional dependency structure between these random variables. Information entities from the domain of interest, e.g. results of clinical examinations, medical imaging, patient behavior and patient characteristics (e.g. age, gender, and tobacco and alcohol consumption) are integrated in the model as random variables [4].
However, developing decision models or biological models requires expertise in the areas to be modelled which results in a very time-consuming process. In our experience, it took a team of two persons one year to build a therapy decision model for laryngeal carcinoma [5]. Methods need to be developed for learning relevant information items and their correlations from clinical practice guidelines, biomedical literature and learned from patient data to build semi-automatically a graph-like structure that connects the relevant information items for a concrete disease or organ. In this context methods for information and relation extraction can be exploited to collect relevant information items and their relations from textual documents (see requirement 2). Given the fact, that not all decision relevant information is described in literature and other sources (e.g. experiences), methods are necessary to support physicians in validating the generated model and in easily adding missing information items or relations. For example, starting from an ontology restricted to a specific subdomain, concepts could be suggested to the biologist or physician to be selected and included into the model.
For generating patient data models, relevant clinical data have to be collected, the semantic information extracted and stored in appropriate data bases. The set of relevant patient data for the treatment of a given disease is generally determined from observations in the clinic and from interviews with physicians. In order to be able to process the patient data, the semantic information has to be extracted (see requirement 2), using automatic techniques if possible, and stored in appropriate databases. These last ones enable to represent the information in a structured and standard way in order to facilitate the information access and interpretation. For example, the openEHR Foundation proposes appropriate tools for the patient data modelling [6, 7].
Requirement 2: Methods for collecting information items and extracting information.
For disease modelling, parameter, diagnoses, and other important information normally hidden in free text need to be extracted, analysed automatically and mapped to an underlying ontology. For these purposes, natural language processing techniques are required which typically involve tokenization and sentence splitting, stop-list filtering, stemming, lemmatization, POS tagging, chunking and shallow or deep parsing. For these basic tasks open source software is available, for example natural language processing tools provided by the Stanford language processing group (http://nlp.stanford.edu/software/indexs.html, accessed: 22.02.2015). Named-entity recognition aims at identifying within a collection of text all of the instances of a name for a specific type of thing [8]. Examples of named entity categories in the medical domain include diseases and illnesses, symptoms, procedures or drugs. Domain-specific named entity recognition can be enabled through tools such as MetaMap (http://metamap. nlm.nih.gov/, accessed: 22.02.2015) or OpenCalais (http://www.opencalais.com/, accessed: 22.02.2015). Moreover, segmentation methods are appropriate to extract the semantic information from image data, in general represented by the anatomical structure to be examined such as a tumour [9, 10].
The biological and clinical data relevant for model-generation ranges from text-based content, to medical images, numerical data, and genomic data whose processing requires a variety of methods due to different data characteristics and content types. An optimized data analysis system for the target domain is necessary that provides structured data or information surrogates with semantic annotations. Besides the relevant information items, it is crucial for semi-automatic model generation, to determine the dependencies of entities as described in texts. For this purpose, relationship extraction methods are required that make use of semantic knowledge and natural language processing.
Requirement 3: Ontologies. Mapping of extracted terms to concepts of ontologies is necessary to get a standardized representation and to assign semantic categories to terms. Further, the use of ontologies and formal concepts of a domain is necessary for adapting inference functionalities to various situations and application scenarios, but also to make unstructured text automatically processable within the context of reasoning. In the domain of medicine, there are already ontologies available that describe medical concepts such as disease or agents (viruses) including MeSH, SNOMED CT or UMLS. They can be considered as basis to identify and formally describe relevant clinical concepts.
Requirement 4: Data and model linking. For realizing the concept, methods are required to link the models or to exploit the output from one model in another one. In particular in the patient model, data linking is necessary. Patient information is currently stored in multiple information systems. Semantics are unavailable, i.e. data items are not semantically annotated which complicates annotation, analysis and linking.
Integration of the patient specific model with the biological and decision model is crucial for interpreting patient data, making predictions, evaluating risks and of course for decision making. Remember that the decision model comprises the information entities that should be considered in decision making. Instantiating these entities with real patient values would lead to a patient-individual decision model. A semantically integrated model is necessary to answer questions such as:
– Which patient data are pathological? How do the parameters influence each other?
– Which therapies may be envisaged with a given patient data set?
– Which are the risks and success rates of the therapies for a particular patient?
Answers to these questions allow to better understand the disease mechanisms.
The most straight forward approach for integrating the models is to identify output from one model and exploit it for calculation of the other model. To realize this, it is essential to base upon the same semantics and ontologies (see requirement 2). On the basis of the integrated models, a clinical decision support system can be developed. The system can provide answers to the questions mentioned. Further, clinicians can be assisted in diagnosing and categorizing a disease or even in the choice of an optimal therapy. The integration of the three model types also supports in controlling the success of the therapy for a given patient: For example, questions can be answered such as which is the success of each therapy given the individual patient condition? In order to suggest therapies, methods are required for reasoning and inferencing. Integration of statistical methods into the system will allow for epidemiological studies or analysis of treatment outcome and thus goes beyond clinical decision support by supporting also research and quality management.
5 Discussion and conclusion
The main benefit of the three-model based approach is that functionalities of current information or clinical decision support systems are extended by:
bridging the gap between system biological models and patient and clinical data (patient model) as well as medical knowledge data (decision model) – in order to make justified, well-grounded therapeutic decisions, and by
improved semantic annotation and linking of the heterogeneous data – in order to support users with an integrated view on patient data and on knowledge about disease development and treatment.
There are still many research problems to address to realize the concept completely, for example: (1) How can we build semi-automatically a digital patient model, including biological models and therapy decision models? and (2) How can the models be linked and interpreted together in order to provide a decision support system? We are currently focusing on developing tools to support expert-based modelling of decision processes.
Funding
This research is part of the Digital Patient Modelling group partially funded by the BMBF.
Author's Statement
Conflict of interest: Authors state no conflict of interest. Material and Methods: Informed consent: Informed consent has been obtained from all individuals included in this study. Ethical approval: The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance the tenets of the Helsinki Declaration, and has been approved by the authors’ institutional review board or equivalent committee.
References
[1] Denecke K. Model-based Decision Support: Requirements and Future for its Application in Surgery. Biomed Tech (Berl) . 2013 Sep 7.10.1515/bmt-2013-4300Search in Google Scholar PubMed
[2] Berliner L, Lemke H, vanSonnenberg E, Ashamalla H, Mattes MD, Dosik D, Hazin H, Shah S, Mohanty S, Verma S, Esposito G, Bargellini I, Battaglia V, Caramella D, Bartolozzi C, Morrison P. Model-guided therapy for hepatocellular carcinoma: a role for information technology in predictive, preventive and personalized medicine.The EPMA Journal 2014:5(16).10.1186/1878-5085-5-16Search in Google Scholar PubMed PubMed Central
[3] Laskey KB. MEBN: A language for first-order Bayesian knowledge bases.Artificial Intelligence, 2008;Volume 172, Issues 2–3: 140-178.10.1016/j.artint.2007.09.006Search in Google Scholar
[4] Lemke HU, Cypko M, Berliner L, Information integration for patient specific modelling using MEBNs: example of laryngeal carcinoma,Int J CARS (2012), Volume 7 (Suppl 1): 246-248.Search in Google Scholar
[5] Stöhr M, Cypko M, Denecke K, Lemke HU, Dietz A: A model of the decision-making process: therapy of laryngeal cancer.Int J CARS (2014), Volume 9 (Suppl 1)Search in Google Scholar
[6] Beale T, Heard S. openEHR Architecture Overview.http://www. openehr.org/releases/1.0.2/architecture/overview.pdf2008.Search in Google Scholar
[7] Garde S, Knaup P, Hovenga EJS, Heard S. Towards semantic interoperability for electronic health records. Methods Inf Med 2007: 46: 332-343.10.1160/ME5001Search in Google Scholar PubMed
[8] Cohen A and Hersh W. A survey of current work in biomedical text mining.Brief Bioin-form, 6(1):57–71, Jan. 2005.10.1093/bib/6.1.57Search in Google Scholar PubMed
[9] Chalopin C, Krissian K, Meixensberger J, Müns A, Arlt F, Lind-ner. Evaluation of a semi-automatic segmentation algorithm in 3D intraoperative ultrasound brain angiography.Biomed Tech . 2013; 58 (3): 293-302.10.1515/bmt-2012-0089Search in Google Scholar PubMed
[10] Daenzer S, Freitag S, von Sachsen S, Steinke H, Groll M, Meixensberger J, Leimert M.: VolHOG: A volumetric object recognition approach based on bivariate histograms of oriented gradients for vertebra detection in cervical spine MRI.Med Phys . 2014 Aug;41(8):08230510.1118/1.4890587Search in Google Scholar PubMed
© 2015 by Walter de Gruyter GmbH, Berlin/Boston
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Articles in the same Issue
- Research Article
- Development and characterization of superparamagnetic coatings
- Research Article
- The development of an experimental setup to measure acousto-electric interaction signal
- Research Article
- Stability analysis of ferrofluids
- Research Article
- Investigation of endothelial growth using a sensors-integrated microfluidic system to simulate physiological barriers
- Research Article
- Energy harvesting for active implants: powering a ruminal pH-monitoring system
- Research Article
- New type of fluxgate magnetometer for the heart’s magnetic fields detection
- Research Article
- Field mapping of ballistic pressure pulse sources
- Research Article
- Development of a new homecare sleep monitor using body sounds and motion tracking
- Research Article
- Noise properties of textile, capacitive EEG electrodes
- Research Article
- Detecting phase singularities and rotor center trajectories based on the Hilbert transform of intraatrial electrograms in an atrial voxel model
- Research Article
- Spike sorting: the overlapping spikes challenge
- Research Article
- Separating the effect of respiration from the heart rate variability for cases of constant harmonic breathing
- Research Article
- Locating regions of arrhythmogenic substrate by analyzing the duration of triggered atrial activities
- Research Article
- Combining different ECG derived respiration tracking methods to create an optimal reconstruction of the breathing pattern
- Research Article
- Atrial and ventricular signal averaging electrocardiography in pacemaker and cardiac resynchronization therapy
- Research Article
- Estimation of a respiratory signal from a single-lead ECG using the 4th order central moments
- Research Article
- Compressed sensing of multi-lead ECG signals by compressive multiplexing
- Research Article
- Heart rate monitoring in ultra-high-field MRI using frequency information obtained from video signals of the human skin compared to electrocardiography and pulse oximetry
- Research Article
- Synchronization in wireless biomedical-sensor networks with Bluetooth Low Energy
- Research Article
- Automated classification of stages of anaesthesia by populations of evolutionary optimized fuzzy rules
- Research Article
- Effects of sampling rate on automated fatigue recognition in surface EMG signals
- Research Article
- Closed-loop transcranial alternating current stimulation of slow oscillations
- Research Article
- Cardiac index in atrio- and interventricular delay optimized cardiac resynchronization therapy and cardiac contractility modulation
- Research Article
- The role of expert evaluation for microsleep detection
- Research Article
- The impact of baseline wander removal techniques on the ST segment in simulated ischemic 12-lead ECGs
- Research Article
- Metal artifact reduction by projection replacements and non-local prior image integration
- Research Article
- A novel coaxial nozzle for in-process adjustment of electrospun scaffolds’ fiber diameter
- Research Article
- Processing of membranes for oxygenation using the Bellhouse-effect
- Research Article
- Inkjet printing of viable human dental follicle stem cells
- Research Article
- The use of an icebindingprotein out of the snowflea Hypogastrura harveyi as a cryoprotectant in the cryopreservation of mesenchymal stem cells
- Research Article
- New NIR spectroscopy based method to determine ischemia in vivo in liver – a first study on rats
- Research Article
- QRS and QT ventricular conduction times and permanent pacemaker therapy after transcatheter aortic valve implantation
- Research Article
- Adopting oculopressure tonometry as a transient in vivo rabbit glaucoma model
- Research Article
- Next-generation vision testing: the quick CSF
- Research Article
- Improving tactile sensation in laparoscopic surgery by overcoming size restrictions
- Research Article
- Design and control of a 3-DOF hydraulic driven surgical instrument
- Research Article
- Evaluation of endourological tools to improve the diagnosis and therapy of ureteral tumors – from model development to clinical application
- Research Article
- Frequency based assessment of surgical activities
- Research Article
- “Hands free for intervention”, a new approach for transoral endoscopic surgery
- Research Article
- Pseudo-haptic feedback in medical teleoperation
- Research Article
- Feasibility of interactive gesture control of a robotic microscope
- Research Article
- Towards structuring contextual information for workflow-driven surgical assistance functionalities
- Research Article
- Towards a framework for standardized semantic workflow modeling and management in the surgical domain
- Research Article
- Closed-loop approach for situation awareness of medical devices and operating room infrastructure
- Research Article
- Kinect based physiotherapy system for home use
- Research Article
- Evaluating the microsoft kinect skeleton joint tracking as a tool for home-based physiotherapy
- Research Article
- Integrating multimodal information for intraoperative assistance in neurosurgery
- Research Article
- Respiratory motion tracking using Microsoft’s Kinect v2 camera
- Research Article
- Using smart glasses for ultrasound diagnostics
- Research Article
- Measurement of needle susceptibility artifacts in magnetic resonance images
- Research Article
- Dimensionality reduction of medical image descriptors for multimodal image registration
- Research Article
- Experimental evaluation of different weighting schemes in magnetic particle imaging reconstruction
- Research Article
- Evaluation of CT capability for the detection of thin bone structures
- Research Article
- Towards contactless optical coherence elastography with acoustic tissue excitation
- Research Article
- Development and implementation of algorithms for automatic and robust measurement of the 2D:4D digit ratio using image data
- Research Article
- Automated high-throughput analysis of B cell spreading on immobilized antibodies with whole slide imaging
- Research Article
- Tissue segmentation from head MRI: a ground truth validation for feature-enhanced tracking
- Research Article
- Video tracking of swimming rodents on a reflective water surface
- Research Article
- MR imaging of model drug distribution in simulated vitreous
- Research Article
- Studying the extracellular contribution to the double wave vector diffusion-weighted signal
- Research Article
- Artifacts in field free line magnetic particle imaging in the presence of inhomogeneous and nonlinear magnetic fields
- Research Article
- Introducing a frequency-tunable magnetic particle spectrometer
- Research Article
- Imaging of aortic valve dynamics in 4D OCT
- Research Article
- Intravascular optical coherence tomography (OCT) as an additional tool for the assessment of stent structures
- Research Article
- Simple concept for a wide-field lensless digital holographic microscope using a laser diode
- Research Article
- Intraoperative identification of somato-sensory brain areas using optical imaging and standard RGB camera equipment – a feasibility study
- Research Article
- Respiratory surface motion measurement by Microsoft Kinect
- Research Article
- Improving image quality in EIT imaging by measurement of thorax excursion
- Research Article
- A clustering based dual model framework for EIT imaging: first experimental results
- Research Article
- Three-dimensional anisotropic regularization for limited angle tomography
- Research Article
- GPU-based real-time generation of large ultrasound volumes from freehand 3D sweeps
- Research Article
- Experimental computer tomograph
- Research Article
- US-tracked steered FUS in a respiratory ex vivo ovine liver phantom
- Research Article
- Contribution of brownian rotation and particle assembly polarisation to the particle response in magnetic particle spectrometry
- Research Article
- Preliminary investigations of magnetic modulated nanoparticles for microwave breast cancer detection
- Research Article
- Construction of a device for magnetic separation of superparamagnetic iron oxide nanoparticles
- Research Article
- An IHE-conform telecooperation platform supporting the treatment of dementia patients
- Research Article
- Automated respiratory therapy system based on the ARDSNet protocol with systemic perfusion control
- Research Article
- Identification of surgical instruments using UHF-RFID technology
- Research Article
- A generic concept for the development of model-guided clinical decision support systems
- Research Article
- Evaluation of local alterations in femoral bone mineral density measured via quantitative CT
- Research Article
- Creating 3D gelatin phantoms for experimental evaluation in biomedicine
- Research Article
- Influence of short-term fixation with mixed formalin or ethanol solution on the mechanical properties of human cortical bone
- Research Article
- Analysis of the release kinetics of surface-bound proteins via laser-induced fluorescence
- Research Article
- Tomographic particle image velocimetry of a water-jet for low volume harvesting of fat tissue for regenerative medicine
- Research Article
- Wireless medical sensors – context, robustness and safety
- Research Article
- Sequences for real-time magnetic particle imaging
- Research Article
- Speckle-based off-axis holographic detection for non-contact photoacoustic tomography
- Research Article
- A machine learning approach for planning valve-sparing aortic root reconstruction
- Research Article
- An in-ear pulse wave velocity measurement system using heart sounds as time reference
- Research Article
- Measuring different oxygenation levels in a blood perfusion model simulating the human head using NIRS
- Research Article
- Multisegmental fusion of the lumbar spine a curse or a blessing?
- Research Article
- Numerical analysis of the biomechanical complications accompanying the total hip replacement with NANOS-Prosthetic: bone remodelling and prosthesis migration
- Research Article
- A muscle model for hybrid muscle activation
- Research Article
- Mathematical, numerical and in-vitro investigation of cooling performance of an intra-carotid catheter for selective brain hypothermia
- Research Article
- An ideally parameterized unscented Kalman filter for the inverse problem of electrocardiography
- Research Article
- Interactive visualization of cardiac anatomy and atrial excitation for medical diagnosis and research
- Research Article
- Virtualizing clinical cases of atrial flutter in a fast marching simulation including conduction velocity and ablation scars
- Research Article
- Mesh structure-independent modeling of patient-specific atrial fiber orientation
- Research Article
- Accelerating mono-domain cardiac electrophysiology simulations using OpenCL
- Research Article
- Understanding the cellular mode of action of vernakalant using a computational model: answers and new questions
- Research Article
- A java based simulator with user interface to simulate ventilated patients
- Research Article
- Evaluation of an algorithm to choose between competing models of respiratory mechanics
- Research Article
- Numerical simulation of low-pulsation gerotor pumps for use in the pharmaceutical industry and in biomedicine
- Research Article
- Numerical and experimental flow analysis in centifluidic systems for rapid allergy screening tests
- Research Article
- Biomechanical parameter determination of scaffold-free cartilage constructs (SFCCs) with the hyperelastic material models Yeoh, Ogden and Demiray
- Research Article
- FPGA controlled artificial vascular system
- Research Article
- Simulation based investigation of source-detector configurations for non-invasive fetal pulse oximetry
- Research Article
- Test setup for characterizing the efficacy of embolic protection devices
- Research Article
- Impact of electrode geometry on force generation during functional electrical stimulation
- Research Article
- 3D-based visual physical activity assessment of children
- Research Article
- Realtime assessment of foot orientation by Accelerometers and Gyroscopes
- Research Article
- Image based reconstruction for cystoscopy
- Research Article
- Image guided surgery innovation with graduate students - a new lecture format
- Research Article
- Multichannel FES parameterization for controlling foot motion in paretic gait
- Research Article
- Smartphone supported upper limb prosthesis
- Research Article
- Use of quantitative tremor evaluation to enhance target selection during deep brain stimulation surgery for essential tremor
- Research Article
- Evaluation of adhesion promoters for Parylene C on gold metallization
- Research Article
- The influence of metallic ions from CoCr28Mo6 on the osteogenic differentiation and cytokine release of human osteoblasts
- Research Article
- Increasing the visibility of thin NITINOL vascular implants
- Research Article
- Possible reasons for early artificial bone failure in biomechanical tests of ankle arthrodesis systems
- Research Article
- Development of a bending test procedure for the characterization of flexible ECoG electrode arrays
- Research Article
- Tubular manipulators: a new concept for intracochlear positioning of an auditory prosthesis
- Research Article
- Investigation of the dynamic diameter deformation of vascular stents during fatigue testing with radial loading
- Research Article
- Electrospun vascular grafts with anti-kinking properties
- Research Article
- Integration of temperature sensors in polyimide-based thin-film electrode arrays
- Research Article
- Use cases and usability challenges for head-mounted displays in healthcare
- Research Article
- Device- and service profiles for integrated or systems based on open standards
- Research Article
- Risk management for medical devices in research projects
- Research Article
- Simulation of varying femoral attachment sites of medial patellofemoral ligament using a musculoskeletal multi-body model
- Research Article
- Does enhancing consciousness for strategic planning processes support the effectiveness of problem-based learning concepts in biomedical education?
- Research Article
- SPIO processing in macrophages for MPI: The breast cancer MPI-SNLB-concept
- Research Article
- Numerical simulations of airflow in the human pharynx of OSAHS patients
Articles in the same Issue
- Research Article
- Development and characterization of superparamagnetic coatings
- Research Article
- The development of an experimental setup to measure acousto-electric interaction signal
- Research Article
- Stability analysis of ferrofluids
- Research Article
- Investigation of endothelial growth using a sensors-integrated microfluidic system to simulate physiological barriers
- Research Article
- Energy harvesting for active implants: powering a ruminal pH-monitoring system
- Research Article
- New type of fluxgate magnetometer for the heart’s magnetic fields detection
- Research Article
- Field mapping of ballistic pressure pulse sources
- Research Article
- Development of a new homecare sleep monitor using body sounds and motion tracking
- Research Article
- Noise properties of textile, capacitive EEG electrodes
- Research Article
- Detecting phase singularities and rotor center trajectories based on the Hilbert transform of intraatrial electrograms in an atrial voxel model
- Research Article
- Spike sorting: the overlapping spikes challenge
- Research Article
- Separating the effect of respiration from the heart rate variability for cases of constant harmonic breathing
- Research Article
- Locating regions of arrhythmogenic substrate by analyzing the duration of triggered atrial activities
- Research Article
- Combining different ECG derived respiration tracking methods to create an optimal reconstruction of the breathing pattern
- Research Article
- Atrial and ventricular signal averaging electrocardiography in pacemaker and cardiac resynchronization therapy
- Research Article
- Estimation of a respiratory signal from a single-lead ECG using the 4th order central moments
- Research Article
- Compressed sensing of multi-lead ECG signals by compressive multiplexing
- Research Article
- Heart rate monitoring in ultra-high-field MRI using frequency information obtained from video signals of the human skin compared to electrocardiography and pulse oximetry
- Research Article
- Synchronization in wireless biomedical-sensor networks with Bluetooth Low Energy
- Research Article
- Automated classification of stages of anaesthesia by populations of evolutionary optimized fuzzy rules
- Research Article
- Effects of sampling rate on automated fatigue recognition in surface EMG signals
- Research Article
- Closed-loop transcranial alternating current stimulation of slow oscillations
- Research Article
- Cardiac index in atrio- and interventricular delay optimized cardiac resynchronization therapy and cardiac contractility modulation
- Research Article
- The role of expert evaluation for microsleep detection
- Research Article
- The impact of baseline wander removal techniques on the ST segment in simulated ischemic 12-lead ECGs
- Research Article
- Metal artifact reduction by projection replacements and non-local prior image integration
- Research Article
- A novel coaxial nozzle for in-process adjustment of electrospun scaffolds’ fiber diameter
- Research Article
- Processing of membranes for oxygenation using the Bellhouse-effect
- Research Article
- Inkjet printing of viable human dental follicle stem cells
- Research Article
- The use of an icebindingprotein out of the snowflea Hypogastrura harveyi as a cryoprotectant in the cryopreservation of mesenchymal stem cells
- Research Article
- New NIR spectroscopy based method to determine ischemia in vivo in liver – a first study on rats
- Research Article
- QRS and QT ventricular conduction times and permanent pacemaker therapy after transcatheter aortic valve implantation
- Research Article
- Adopting oculopressure tonometry as a transient in vivo rabbit glaucoma model
- Research Article
- Next-generation vision testing: the quick CSF
- Research Article
- Improving tactile sensation in laparoscopic surgery by overcoming size restrictions
- Research Article
- Design and control of a 3-DOF hydraulic driven surgical instrument
- Research Article
- Evaluation of endourological tools to improve the diagnosis and therapy of ureteral tumors – from model development to clinical application
- Research Article
- Frequency based assessment of surgical activities
- Research Article
- “Hands free for intervention”, a new approach for transoral endoscopic surgery
- Research Article
- Pseudo-haptic feedback in medical teleoperation
- Research Article
- Feasibility of interactive gesture control of a robotic microscope
- Research Article
- Towards structuring contextual information for workflow-driven surgical assistance functionalities
- Research Article
- Towards a framework for standardized semantic workflow modeling and management in the surgical domain
- Research Article
- Closed-loop approach for situation awareness of medical devices and operating room infrastructure
- Research Article
- Kinect based physiotherapy system for home use
- Research Article
- Evaluating the microsoft kinect skeleton joint tracking as a tool for home-based physiotherapy
- Research Article
- Integrating multimodal information for intraoperative assistance in neurosurgery
- Research Article
- Respiratory motion tracking using Microsoft’s Kinect v2 camera
- Research Article
- Using smart glasses for ultrasound diagnostics
- Research Article
- Measurement of needle susceptibility artifacts in magnetic resonance images
- Research Article
- Dimensionality reduction of medical image descriptors for multimodal image registration
- Research Article
- Experimental evaluation of different weighting schemes in magnetic particle imaging reconstruction
- Research Article
- Evaluation of CT capability for the detection of thin bone structures
- Research Article
- Towards contactless optical coherence elastography with acoustic tissue excitation
- Research Article
- Development and implementation of algorithms for automatic and robust measurement of the 2D:4D digit ratio using image data
- Research Article
- Automated high-throughput analysis of B cell spreading on immobilized antibodies with whole slide imaging
- Research Article
- Tissue segmentation from head MRI: a ground truth validation for feature-enhanced tracking
- Research Article
- Video tracking of swimming rodents on a reflective water surface
- Research Article
- MR imaging of model drug distribution in simulated vitreous
- Research Article
- Studying the extracellular contribution to the double wave vector diffusion-weighted signal
- Research Article
- Artifacts in field free line magnetic particle imaging in the presence of inhomogeneous and nonlinear magnetic fields
- Research Article
- Introducing a frequency-tunable magnetic particle spectrometer
- Research Article
- Imaging of aortic valve dynamics in 4D OCT
- Research Article
- Intravascular optical coherence tomography (OCT) as an additional tool for the assessment of stent structures
- Research Article
- Simple concept for a wide-field lensless digital holographic microscope using a laser diode
- Research Article
- Intraoperative identification of somato-sensory brain areas using optical imaging and standard RGB camera equipment – a feasibility study
- Research Article
- Respiratory surface motion measurement by Microsoft Kinect
- Research Article
- Improving image quality in EIT imaging by measurement of thorax excursion
- Research Article
- A clustering based dual model framework for EIT imaging: first experimental results
- Research Article
- Three-dimensional anisotropic regularization for limited angle tomography
- Research Article
- GPU-based real-time generation of large ultrasound volumes from freehand 3D sweeps
- Research Article
- Experimental computer tomograph
- Research Article
- US-tracked steered FUS in a respiratory ex vivo ovine liver phantom
- Research Article
- Contribution of brownian rotation and particle assembly polarisation to the particle response in magnetic particle spectrometry
- Research Article
- Preliminary investigations of magnetic modulated nanoparticles for microwave breast cancer detection
- Research Article
- Construction of a device for magnetic separation of superparamagnetic iron oxide nanoparticles
- Research Article
- An IHE-conform telecooperation platform supporting the treatment of dementia patients
- Research Article
- Automated respiratory therapy system based on the ARDSNet protocol with systemic perfusion control
- Research Article
- Identification of surgical instruments using UHF-RFID technology
- Research Article
- A generic concept for the development of model-guided clinical decision support systems
- Research Article
- Evaluation of local alterations in femoral bone mineral density measured via quantitative CT
- Research Article
- Creating 3D gelatin phantoms for experimental evaluation in biomedicine
- Research Article
- Influence of short-term fixation with mixed formalin or ethanol solution on the mechanical properties of human cortical bone
- Research Article
- Analysis of the release kinetics of surface-bound proteins via laser-induced fluorescence
- Research Article
- Tomographic particle image velocimetry of a water-jet for low volume harvesting of fat tissue for regenerative medicine
- Research Article
- Wireless medical sensors – context, robustness and safety
- Research Article
- Sequences for real-time magnetic particle imaging
- Research Article
- Speckle-based off-axis holographic detection for non-contact photoacoustic tomography
- Research Article
- A machine learning approach for planning valve-sparing aortic root reconstruction
- Research Article
- An in-ear pulse wave velocity measurement system using heart sounds as time reference
- Research Article
- Measuring different oxygenation levels in a blood perfusion model simulating the human head using NIRS
- Research Article
- Multisegmental fusion of the lumbar spine a curse or a blessing?
- Research Article
- Numerical analysis of the biomechanical complications accompanying the total hip replacement with NANOS-Prosthetic: bone remodelling and prosthesis migration
- Research Article
- A muscle model for hybrid muscle activation
- Research Article
- Mathematical, numerical and in-vitro investigation of cooling performance of an intra-carotid catheter for selective brain hypothermia
- Research Article
- An ideally parameterized unscented Kalman filter for the inverse problem of electrocardiography
- Research Article
- Interactive visualization of cardiac anatomy and atrial excitation for medical diagnosis and research
- Research Article
- Virtualizing clinical cases of atrial flutter in a fast marching simulation including conduction velocity and ablation scars
- Research Article
- Mesh structure-independent modeling of patient-specific atrial fiber orientation
- Research Article
- Accelerating mono-domain cardiac electrophysiology simulations using OpenCL
- Research Article
- Understanding the cellular mode of action of vernakalant using a computational model: answers and new questions
- Research Article
- A java based simulator with user interface to simulate ventilated patients
- Research Article
- Evaluation of an algorithm to choose between competing models of respiratory mechanics
- Research Article
- Numerical simulation of low-pulsation gerotor pumps for use in the pharmaceutical industry and in biomedicine
- Research Article
- Numerical and experimental flow analysis in centifluidic systems for rapid allergy screening tests
- Research Article
- Biomechanical parameter determination of scaffold-free cartilage constructs (SFCCs) with the hyperelastic material models Yeoh, Ogden and Demiray
- Research Article
- FPGA controlled artificial vascular system
- Research Article
- Simulation based investigation of source-detector configurations for non-invasive fetal pulse oximetry
- Research Article
- Test setup for characterizing the efficacy of embolic protection devices
- Research Article
- Impact of electrode geometry on force generation during functional electrical stimulation
- Research Article
- 3D-based visual physical activity assessment of children
- Research Article
- Realtime assessment of foot orientation by Accelerometers and Gyroscopes
- Research Article
- Image based reconstruction for cystoscopy
- Research Article
- Image guided surgery innovation with graduate students - a new lecture format
- Research Article
- Multichannel FES parameterization for controlling foot motion in paretic gait
- Research Article
- Smartphone supported upper limb prosthesis
- Research Article
- Use of quantitative tremor evaluation to enhance target selection during deep brain stimulation surgery for essential tremor
- Research Article
- Evaluation of adhesion promoters for Parylene C on gold metallization
- Research Article
- The influence of metallic ions from CoCr28Mo6 on the osteogenic differentiation and cytokine release of human osteoblasts
- Research Article
- Increasing the visibility of thin NITINOL vascular implants
- Research Article
- Possible reasons for early artificial bone failure in biomechanical tests of ankle arthrodesis systems
- Research Article
- Development of a bending test procedure for the characterization of flexible ECoG electrode arrays
- Research Article
- Tubular manipulators: a new concept for intracochlear positioning of an auditory prosthesis
- Research Article
- Investigation of the dynamic diameter deformation of vascular stents during fatigue testing with radial loading
- Research Article
- Electrospun vascular grafts with anti-kinking properties
- Research Article
- Integration of temperature sensors in polyimide-based thin-film electrode arrays
- Research Article
- Use cases and usability challenges for head-mounted displays in healthcare
- Research Article
- Device- and service profiles for integrated or systems based on open standards
- Research Article
- Risk management for medical devices in research projects
- Research Article
- Simulation of varying femoral attachment sites of medial patellofemoral ligament using a musculoskeletal multi-body model
- Research Article
- Does enhancing consciousness for strategic planning processes support the effectiveness of problem-based learning concepts in biomedical education?
- Research Article
- SPIO processing in macrophages for MPI: The breast cancer MPI-SNLB-concept
- Research Article
- Numerical simulations of airflow in the human pharynx of OSAHS patients

