Abstract
Backgrounds
The integrator complex (INT) is a multiprotein assembly in gene transcription. Although several subunits of INT complex have been implicated in multiple cancers, the complex’s role in gastric cancer (GC) is poorly understood.
Methods
The gene expressions, prognostic values, and the associations with microsatellite instability (MSI) of INT subunits were confirmed by GEO and The Cancer Genome Atlas (TCGA) databases. cBioPortal, GeneMANIA, TISIDB, and MCPcounter algorithm were adopted to investigate the mutation frequency, protein–protein interaction network, and the association with immune cells of INT subunits in GC. Additionally, in vitro experiments were performed to confirm the role of INTS11 in pathogenesis of GC.
Results
The mRNA expression levels of INTS2/4/5/7/8/9/10/11/12/13/14 were significantly elevated both in GSE183904 and TCGA datasets. Through functional enrichment analysis, the functions of INT subunits were mainly associated with snRNA processing, INT, and DNA-directed 5′–3′ RNA polymerase activity. Moreover, these INT subunit expressions were associated with tumor-infiltrating lymphocytes and MSI in GC. In vitro experiments demonstrated that knockdown of the catalytic core INTS11 in GC cells inhibits cell proliferation ability. INTS11 overexpression showed opposite effects.
Conclusions
Our data demonstrate that the INT complex might act as an oncogene and can be used as a prognosis biomarker for GC.
1 Introduction
Gastric cancer (GC) remains the leading cause of cancer-associated death worldwide [1,2]. Due to low rate of routine screening and high metastatic potential of GC, most patients are diagnosed in the advanced stage, with a median survival time of less than 15 months [3]. The present therapeutic strategies, including radiotherapy, biomarker-directed therapy, surgical resection, immunotherapy, and chemotherapy, are insufficient to improve the survival rates of advanced GC patients [1,4]. Hence, new optimal predictive biomarkers and therapeutic targets are badly required for early diagnosis and to prolong the survival of advanced GC.
The integrator complex (INT) is a polyprotein complex consisting of at least 14 subunits, INTS1 to INTS14, with a molecular weight greater than 1.4 MDa [5]. This protein complex binds to the C-terminal domain (CTD) of RNA polymerase II and exerts biological functions in small nuclear RNA processing [6]. INT was first identified as a complex involved in the formation of U-rich small nuclear RNAs [7]. Notably, given its major role in transcriptional regulation and snRNAs processing, it is feasible that some subunits of INT are also involved in the initiation and progression of human tumors [8]. INTS6 was first identified as a candidate tumor suppressor gene in 1999 because it was absent or highly downregulated in non-small cell lung cancer [9]. However, in colorectal cancer (CRC), Ding et al. observed INTS6 was upregulated in CRC and mediated CRC cells proliferation and metastasis by regulating AKT/ERK signaling pathway [10]. Similarly, INTS7 expression was found to be upregulated in tumor tissues of lung adenocarcinoma patients, compared with the adjacent normal tissues [11]. A previous study revealed that INTS8 participates epithelial-to-mesenchymal transition in hepatocellular carcinoma (HCC) [12]. It also functions as an oncogene in intrahepatic cholangiocarcinoma [13].
On the whole, these studies indicated that the INT subunits may be promising therapeutic targets for a variety of tumors. However, the roles of INT subunits in carcinogenesis have not been fully clarified. In particular, there are few reports about INT subunits in GC. It is well known that the systematic bioinformatics analysis of biological functions is one of the most important methods in cancer research. Therefore, in this study, we analyzed the differential expressions and mutation patterns, biological functions, the association with immune cells, protein–protein interactions, and different prognostic values of INT subunits in GC patients based on public databases. Furthermore, INTS11 is the catalytic core subunit of the INT complex and loss of INTS11 would impair the ability of processing U1 and U2 primary transcripts [14]. Therefore, targeting INTS11 may obstruct the function of INT complex, leading to dysregulation of the target genes, and ultimately affecting GC tumorigenesis. Therefore, in our study, a series of experiments were conducted to explore its potential biological functions in GC cells.
2 Methods
2.1 The Cancer Genome Atlas (TCGA) data analysis
TCGA is a public cancer genomic project supported by the National Cancer Institute that can be used to analyze differences in gene expression and detect co-expressed genes. The RNA-seq transcriptome data of 375 GC tissues and 32 adjacent normal tissues were obtained from TCGA website (https://www.cancer.gov/ccg/research/genome-sequencing/tcga).
2.2 Kaplan–Meier (K–M) survival analysis
To explore the potential prognostic values of INT subunits, we analyzed the relationship between INT subunit expressions and overall survival (OS) based on by K–M plotter website (http://kmplot.com/analysis/).
2.3 cBioPortal data analysis
cBioPortal (http://cbioportal.org) is a web-based platform for the detection, visualization, and analysis of large-scale cancer genomics data, including epigenetic, gene expression profiling, and proteomics data. We used cBioPortal (Stomach Adenocarcinoma [TCGA, PanCancer Atlas]) to analyze alterations, copy number alterations in the raw RNA-seq data.
2.4 Single-cell RNA sequencing analysis
GC scRNA-seq dataset (GSE183904) comprising 11 normal tissues and 29 gastric tumor tissues was downloaded from GEO database. Quality control was performed using the Seurat R package (V 5.0.1), filtering out cells with poor quality (expressed genes <200 or >7,000, mitochondrial gene content >20%). The remaining expressed gene data were normalized using “LogNormalize” scaling and the top 2,000 variable genes were identified using the FindVariableFeatures function (method = vst). Then, data scaling was conducted using ScaleData function, followed by principal component analysis for dimension reduction. Batch effects were corrected using Harmony package (V 1.2.0). Cell types were identified based on marker gene expressions, such as epithelial cells (EPCAM, CDH1, KRT7), T (CD3D, CD8A, CD4), NK cells (NCAM1), endothelial cells (VWF, PECAM1), fibroblasts (COL1A1, PDGFRA), mast cell (CPA3), B cell (CD19, CD79A), plasma B cell (JCHAIN), and myeloid cell (FCGR3A). Data visualization was performed using the UMAP methods.
2.5 Relationship between INT subunits and immune cell infiltration in GC
To investigate the specific relationship of INT subunits with immune cells, we utilized TISIDB website (http://cis.hku.hk/TISIDB/index.php) for evaluating whether INT subunits were related to immune subtypes in GC patients. Furthermore, we applied the MCPcounter algorithm for analysis of the relationship between INT subunits and infiltrating immune cells including endothelial cell, neutrophil, cancer-associated fibroblast, monocyte, macrophage monocyte, T cell, T cell CD8, B cell, NK cell, and myeloid dendritic cell.
2.6 Correlational research on INT subunits and microsatellite instability (MSI)
MSI is caused by functional defects in DNA mismatch repair and leads to gene duplication disorder, tumor heterogeneity, tumor progression, causing drug resistance or more immune epitopes, and affect tumor prognosis [15]. MSI score was obtained from the TCGA database. Finally, the results were visualized in the form of box plots.
2.7 Cell culture
The GC cell lines HGC-27 and AGS were obtained from Cell Bank of Academy of Sciences (Shanghai, China). These cells were cultured in RPMI-1640 medium (Gibco, Life technologies, USA) with 10% fetal bovine serum. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. All cell lines were authenticated by short tandem repeat profiling.
2.8 Proliferation assay
For cell proliferation assay, 1 × 103 transfected cells were seeded into 96-well plates per well. Then, at the indicated time-point, 10 μL of CCK-8 solution (EpiZyme Inc., Shanghai, China) was added into each well, and the plates were incubated for 2 h at 37℃. The optical density values were measured at 450 nm (OD450) by an enzyme labeling instrument.
In parallel, for colony formation assays, 1 × 103 transfected cells were seeded into six-well plates per well. After 14 days, we fixed the colonies with 4% paraformaldehyde and stained the colonies with 0.1% crystal violet. Subsequently, colonies were photographed and counted to evaluate the colony-forming capability of the transfected cells.
2.9 Interference of INTS11 expression in GC cell lines and validation
To knockdown INTS11, two shRNA sequences: shINTS11 #1 and shINTS11 #2 and a scrambled Mock shRNA were chemically synthesized at Sangon Biotech (Shanghai, China). Subsequently, the shRNAs were cloned into pLKO.1-shRNA-puromycin vector and co-transfected with packaging plasmids psPAX2 and pMD2G into 293T cells using Lipofectamine 2000 (Invitrogen). The appropriate concentration of virus was added into tumor cells to generate and screen stable INTS11-knockdown GC cells with 1 µg/mL puromycin for 2 days. Then the cells were used for subsequent assays after verifying the INTS11 expression by quantitative real-time polymerase chain reaction (qPCR) and western blotting. shRNAs sequence targeting INTS11 are summarized in Table A1.
2.10 INTS11 overexpression in GC cell lines and validation
To investigate the effect of INTS11 overexpression, GC cell lines were transfected with a recombinant plasmid pcDNA3.1-INTS11 (Sangon Biotech, Shanghai, China), which contains the full length cDNA sequence of human INTS11. Both GC cell lines, HGC-27 and AGS, were transfected with the recombinant plasmids. qPCR and western blotting were used to verify the successful overexpression of INTS11 in these cell lines.
2.11 Western blotting
Total protein was extracted using RIPA lysis buffer and separated by 10% SDS-PAGE. Subsequently, proteins were transferred onto PVDF membranes. After blocking for 1 h in 5% BSA solution, the membranes were first treated overnight with primary antibodies: INTS11 (1:1,000; 15860-1-AP, Proteintech) and GAPDH (1:1,000; 81640-5-RR, Proteintech). The following day, membranes were treated with HRP-conjugated secondary antibody at room temperature for 1 h at 1:5,000 dilutions. Protein bands were visualized using enhanced chemiluminescence on a luminescent image analyzer (Bio-Rad, Marnes la Coquette, France).
2.12 qPCR
Total RNA was extracted using TRIzol reagent and 1 μg of total RNA was reverse transcribed into cDNA using a PrimeScript RT Reagent Kit (Takara Bio, Nojihigashi, Kusatsu, Japan). qPCR analysis of INTS11 and GAPDH gene was performed on a QuantStudio5 real-time PCR system (Applied Biosystems, Life Technologies, USA) using SYBR Green PCR Kit (Takara Bio, Shiga, Japan) according to the recommended protocol. The primer sequences are summarized in Table A1.
-
Ethical approval: This study was based on guidelines of the Declaration of Helsinki and approved by Zhongshan Hospital Qingpu Branch of Fudan University.
3 Results
3.1 Expression patterns of INT subunits in GC patients
The research strategy is presented in Figure 1. The TCGA dataset was used to investigate the mRNA expression differences of INT subunits between 375 GC tissues and 32 normal gastric tissues. INTS1–INTS14 expression levels were remarkably elevated in GC samples than in normal gastric specimens, both in unpaired tumor-adjacent normal GC samples and in the paired tumor-normal GC samples (Figure 2a and b). To further verify the expressions of INT subunits in GC patients, single-cell sequencing analysis was conducted. Except for INTS1, INTS3, and INTS6, the heat maps and dot plots showed that the other INTS subunits had higher gene expressions in gastric tissues compared to normal tissues, the heat maps and the dot plots indicate the average expression of the gene in all cell clusters (Figure 3a and b).

Work flow and the databases used in this study.
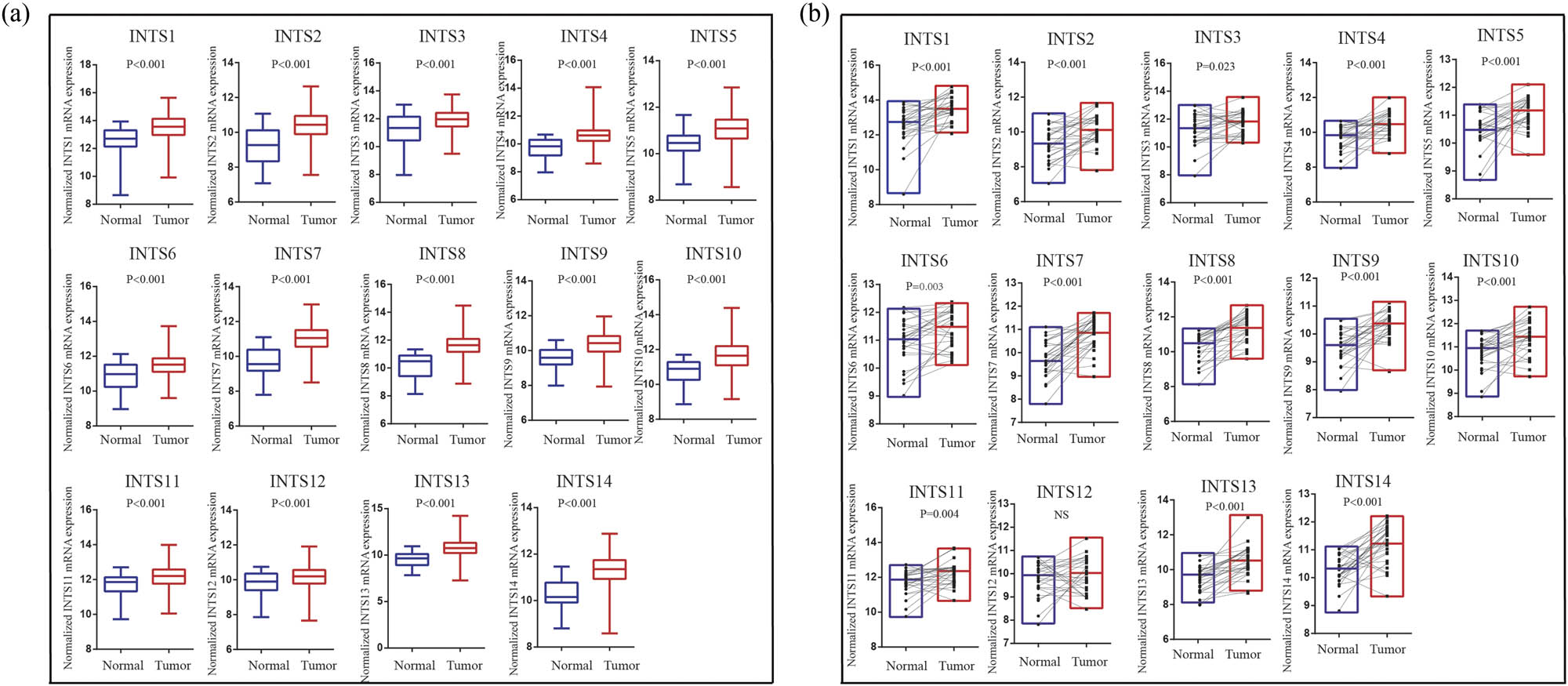
Differences of INT subunits gene expression levels. INT subunits mRNA expression levels were significantly different in the unpaired (a) and paired (b) tumor-adjacent normal GC samples.
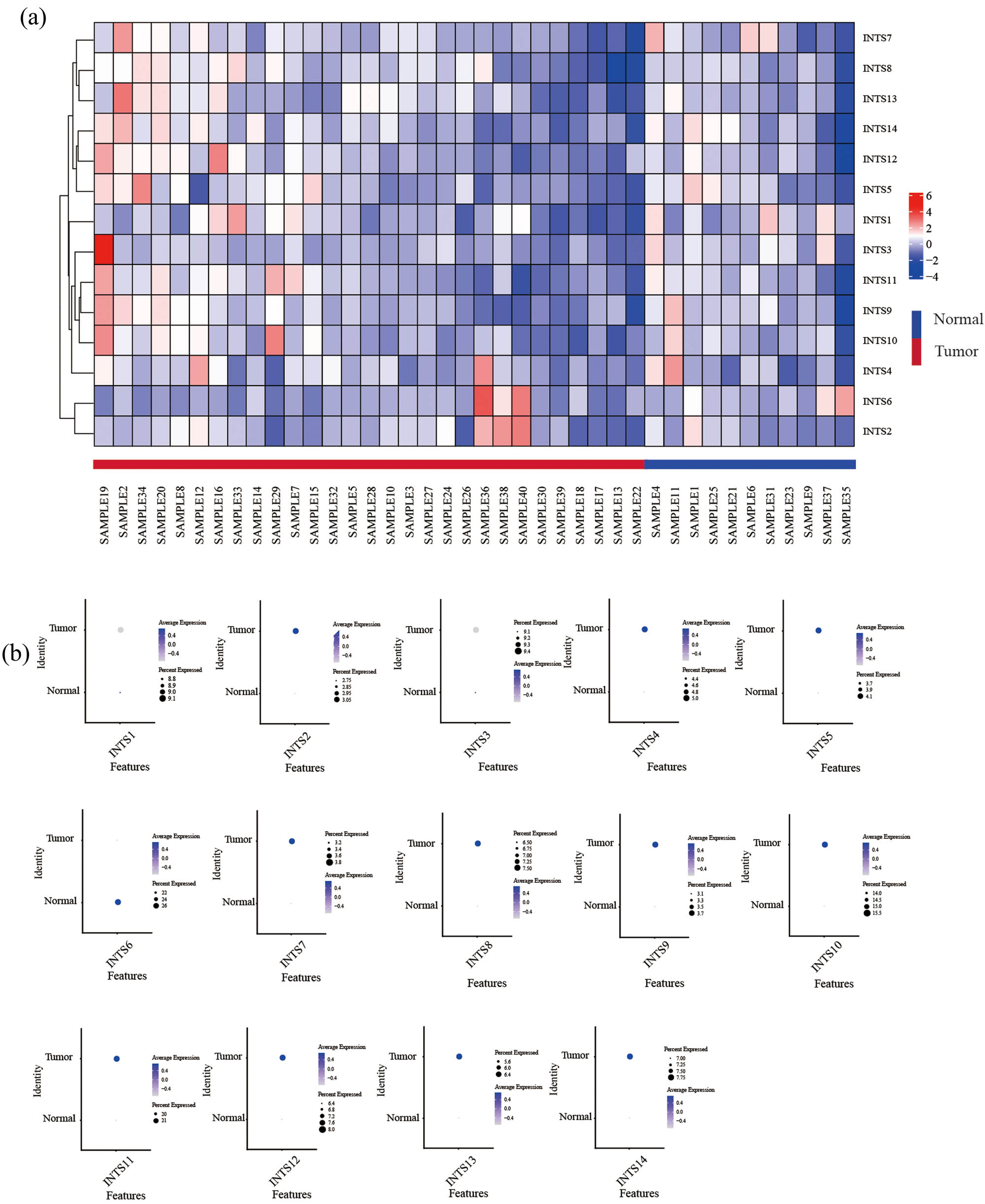
INT subunit expressions in GC tissues and normal tissues were examined by single-cell sequencing analysis. (a) Heat map and (b) dot plot of INT subunit expression profiles identified in gastric normal and cancer tissues.
3.2 Prognostic values of INT subunits in GC patients
Next, we continued to identify the prognostic values of the INT subunits for GC patients using K–M plotter. The expression levels of INT subunits were divided into high expression group and low expression group according to the best cut-off value provided by the K–M plotter. The K–M curves revealed that higher expression levels of INTS1/3/4/5/9/11 were related to shorter OS (Figure 4a). Conversely, the low expression levels of INTS2/6/7/8/10/12/13/14 indicated poorer OS in all GC patients (Figure 4a).

Prognostic analysis of INT subunits in GC patients. K–M analysis of prognostic effects of INT subunits with (a) OS in GC patients. OS, overall survival.
3.3 Gene mutation of INT subunits in GC patients
cBioPortal website was applied to access the frequency of genetic variation of INT subunits in GC patients. Our findings showed that INTS1 (7%), INTS2 (6%), INTS3 (6%), INTS4 (7%), INTS5 (2.3%), INTS6 (4%), INTS7 (2.5%), INTS8 (6%), INTS9 (1.8%), INTS10 (2.5%), INTS11 (5%), INTS12 (2.5%), INTS13 (5%), and INTS14 (1.1%) were altered in GC samples (Figure 5a). Of the 440 cases, 39.09% exhibited alterations in INT subunits, including mutation (15.68%; 69 cases), structural variant (0.23%, 1 cases), amplification (18.41%; 81 cases), deep deletion (2.5%; 11 cases), and multiple alterations (2.27%; 10 cases) (Figure 5b). In addition, the results of K–M plotter and log-rank test indicated that alterations of INT subunits in GC patients were not significantly associated with OS (log-rank test p-value = 0.169) and progression free survival (PFS; log-rank test p-value = 0.332) (Figure 5c and d).
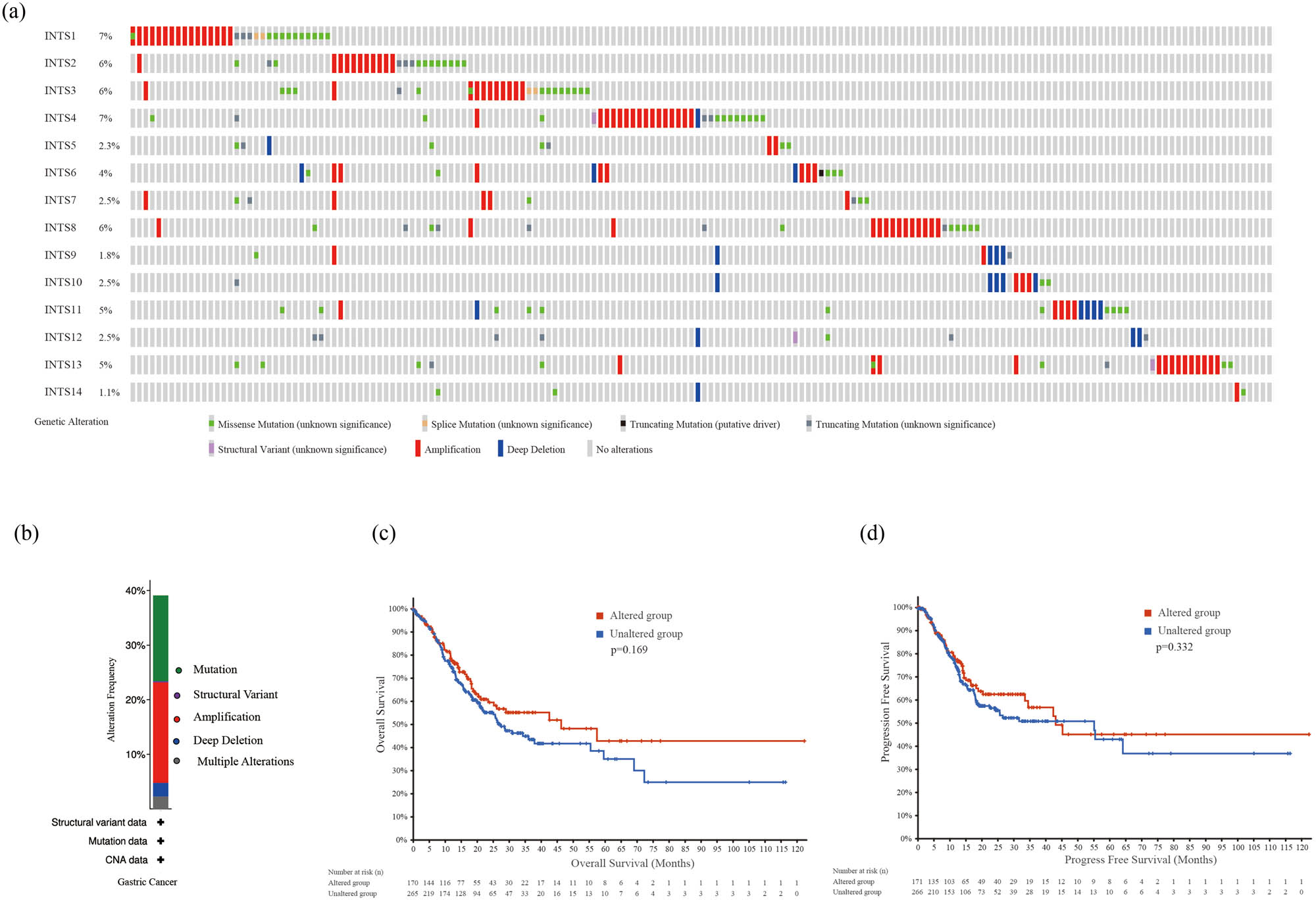
INT subunits gene mutation analysis in GC (cBioPortal). (a) Mutation patterns summary of INT subunits. (b) Mutation frequency of INT complex in GC. (c) K–M plot comparing OS in patients with/without INT subunits gene alterations. (d) K–M plot comparing PFS in patients with/without INT subunits gene alterations. PFS: progression free survival.
3.4 Correlation and functional enrichment analysis of INT subunits in GC patients
To investigate potential association of INT subunits in GC patients, we performed Pearson correlation analysis to evaluate the correlation between these subunits in GC using TCGA dataset. The results showed that the expressions of these subunits were positively inter-correlated in GC (Figure 6a). In addition, we adopted GeneMANIA to draw gene–gene interaction network between INT subunits and their functionally related genes. The results demonstrated that, except for INT subunits, there were 19 genes, including INTS6L, C7orf26, POLR2B, POLR2G, POLR2L, POLR21B, POLR2J, PPP2CB, ZMYND8, POLR2H, POLR2D, POLR2C, ZNF592, NAIF1, ZNF687, CTDP1, INIP, NABP1, and NABP2, associated with regulatory functions of aberrantly expressed INT subunits in GC patients (Figure 6b). Subsequently, functional and enrichment analyses of the above genes were performed using DAVID and presented in bubble diagrams. We found that the main biological process (BP) of these genes was snRNA processing and the main cellular component (CC) of these genes corresponds to INT (Figure 6c and d). The main molecular function (MF) of these genes was DNA-directed 5′–3′ RNA polymerase activity (Figure 6e). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was adopted to clarify pathways related to INT subunits and related genes, and the results revealed that these genes were mainly involved in RNA polymerase (Figure 6f).
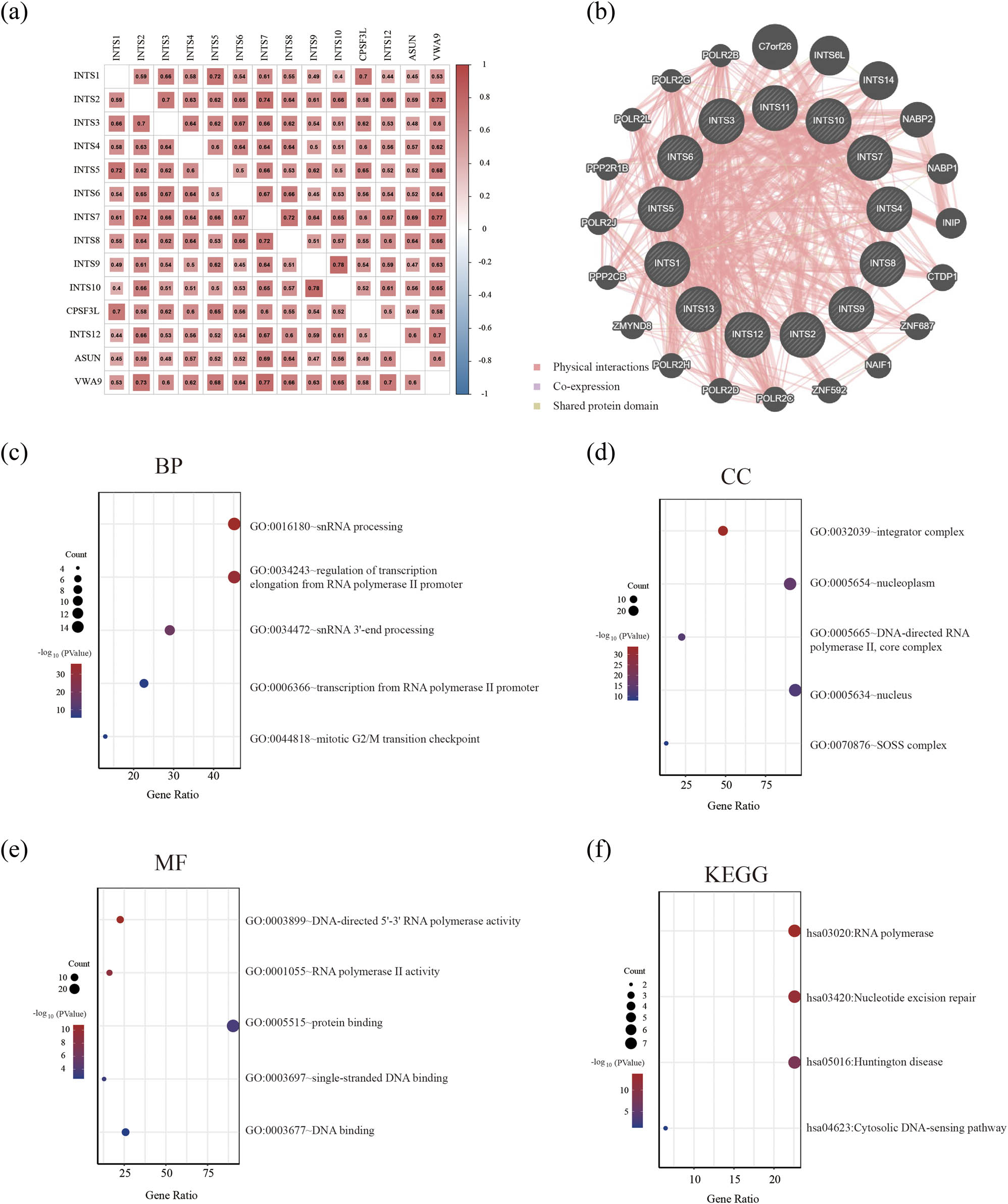
Correlations between INT subunits and related genes functional enrichment analysis in GC patients. (a) Gene correlations between INT subunits were analyzed using the Pearson test in TCGA dataset. (b) Co-expression network of INT subunits and related genes based on GeneMANIA. (c)–(e) Display the top five BP, CC, and MF for INT subunits and related genes, respectively. (f) KEGG pathways of the genes involved. KEGG: Kyoto Encyclopedia of Genes and Genomes; BP: biological process; CC: cellular component; MF: molecular function.
3.5 Relationship between INT subunits and immune cell infiltration in GC
To further explore the expression of INT subunits across immune and molecular subtypes, the TISIDB website was applied to perform an integrated analysis. Interestingly, we found that the expression levels of INT subunits were significantly different across different immune subtypes (Figure 7a–n). Subsequently, we further investigated the association between INT subunit expressions and immune cell infiltration in GC using MCPcounter algorithm. Our results showed that most INT subunit expressions were strongly positively related to neutrophil and endothelial cells (Figure 8a–n). Infiltration and polarization of neutrophils are mainly mediated by ELR + CXC (CXCL) chemokines and regulated by various stimulant-cell interactions [16–18]. So we further verified the correlation between INT subunits and CXCL chemokines and our results indicated the INT subunit expressions were positively associated with most of the CXCL chemokines (Figure 8o). Vascular endothelial growth factor (VEGF) is a special class of cytokines, which is only involved in regulating the biological activity of endothelial cells [19,20]. Functionally, VEGF can specifically induce interstitial production and promote vascular endothelial growth [21]. So we also probed the correlation between INT subunits and three VEGFs. The correlation analysis showed that INT subunits had a strong positive correlation with these three VEGFs (Figure 8o).

Effect of INT subunits on immunological status in GC using the TISIDB. (a)–(n) INT subunits mRNA expressions in different immune subtypes in GC, C1 (would healing), C2 (IFN-gamma dominant), C3 (inflammatory), C4 (lymphocyte depleted), and C6 (TGF-β dominant).

Correlations between the expression of INT subunits and immune infiltrating cells, CXCL chemokines, and VEGFs (a)–(n) Correlations between the expression of INTS11-14 subunits and immune infiltrating cells, respectively. (o) Correlations between INT subunits and CXCL chemokines, VEGFs.
3.6 Association of INT subunits with MSI
Numerous studies have demonstrated the value of MSI as indicators of the tumor immune response and linked with immune checkpoint inhibitor sensitivity. We thus performed MSI investigations. Our results found that most of the INT subunit expressions were positively associated with MSI in GC (Figure 9a–n), indicating that INT subunits may play important roles in the immune regulation and could predict the immunotherapy effect in GC. All these data together indicated that most of INT subunit expressions are widely associated with immunity in GC.

Correlations between MSI and the expression of INT subunit genes. (a)–(n) INTS1–14, respectively. MSI: microsatellite instability.
3.7 INTS11 distribution in GC were examined by single-cell analysis
INTS11 is the catalytic core subunit of the INT complex and the loss of INTS11 would impair the ability of processing U1 and U2 primary transcripts [22]. It is essential for the eviction of paused RNAPII and transcriptional elongation [23]. Therefore, targeting INTS11 may obstruct the function of INT complex, leading to deregulation of the target genes, and ultimately affecting GC tumorigenesis. To further determine the oncogenic role of INT complex in GC, we analyzed GSE183904 to examine the association between the expression of catalytic core subunit INTS11 and different types of cell. Based on the marker gene expression, we identified eight clusters of cell types, including epithelial cells, fibroblasts, myeloid cells, B and plasma B cells, T cells, mast cells, NK cells, and endothelial cells (Figure 10a). The UMAP plots showed that INTS11 could express in all cell types, especially in epithelial cells (Figure 10b–d). The UMAP plots and the expression of others INT subunits in different cell types are shown in Figure A1. Moreover, GSEA was conducted and found that there were positive correlations between high INTS11 expression and hallmark pathway of tumors, such as DNA replication and cell cycle pathways (Figure 10e and f).

INTS11 expression in different types of cell in GC was examined by single-cell analysis. (a) Cell clusters of GSE183904 of 29 GC patients. (b)–(d) INTS11 expression and distribution in eight clusters of cell types. (e) and (f) Hallmark pathways of tumors associated with INTS11.
3.8 Knockdown of INTS11 inhibited proliferation of GC cells
Next, we continued to investigate the role of INTS11 in GC cell proliferation. The result of INTS11 down expression was verified by qRT-CR and western blotting assays independently (Figure 11a and b). Then we performed CCK-8 assays to investigate the role of this complex in GC cell proliferation and the results showed that knockdown of INTS11 significantly inhibited cell proliferation of AGS and HGC-27 cells compared to its corresponding controls (Figure 11c and d). The effect of INTS11 on cell viability was further verified in colony formation assays. Results showed that knockdown of INTS11 produced less colonies compared to the mock groups (Figure 11e–h).

Knockdown of INTS11 inhibited GC cell proliferation. (a) qPCR and (b) western blotting analysis verified successful decreased expression of INTS11 in HGC-27 and AGS cells. (c) and (d) In HGC-27 and AGS cells, CCK8 assays showed knockdown of INTS11 significantly inhibited cell growth. (e)–(h) In HGC-27 and AGS cells, colony formation assays showed knockdown of INTS11 significantly inhibited cell colony formation abilities.
3.9 INTS11 promoted cell proliferation in GC cell lines
We subsequently enhanced the expression of INTS11 protein in HGC-27 and AGS cell lines via transfecting with a pcDNA3.1-INTS11 overexpression vector. The efficacy of INTS11 overexpression was independently confirmed through qPCR and western blotting assays (Figure 12a and b). The CCK8 assays revealed that INTS11 overexpression significantly enhanced the proliferative abilities of HGC-27 and AGS cells (Figure 12c and d). Additionally, the enhanced expression of INTS11 was observed to promote colony formation in these cell lines (Figure 12e–h).

Overexpression of INTS11 promoted GC cell proliferation. (a) qPCR and (b) western blotting analysis verified successful increased expression of INTS11 in HGC-27 and AGS cells. (c) and (d) In HGC-27 and AGS cells, CCK8 assays showed overexpression of INTS11 significantly promoted cell growth. (e)–(h) In HGC-27 and AGS cells, colony formation assays showed overexpression of INTS11 significantly promoted cell colony formation abilities.
4 Discussion
This is the first study to provide a systematic and comprehensive overview of genetic alterations, prognostic values, and the expression patterns of all genes encoding INT complex subunits in GC. INT complex is metazoan specific protein group composed of at least 14 subunits with a variety of biological functions, like impacting the transcriptional activation, small nuclear RNA production and processing, blast lineage development, and nucleic acid metabolism [24].
Many studies have shown that dysregulation of INT subunits is involved in occurrence and development of multiple cancers [7,25]. Although several INT subunits have been shown to play critical regulatory roles in tumors, the distinct roles of INT subunits in GC remain to be elucidated. In this study, bioinformatics tools were applied to identify mutations, mRNA expressions, prognostic values, the association with MSI, and biological significance of different INT subunits in GC.
INT complex is associated with the biogenesis of enhancer RNAs development and small nuclear RNA transcription regulation [23,26]. Teng found that mutations in INTS1 exerted their function during carcinogenesis mainly via post-transcriptional mechanisms [27]. Also, INTS1 gene mutations were associated with rare recessive human neurodevelopmental syndromes [6]. INTS6, also named as DICE1, is a member of the ATP-dependent helicases and related proteins [9]. It showed loss or downregulation of protein expression in the great mass of NSCLC tested [28]. In prostate cancer (PC), hypermethylation of the DICE1 promoter was observed in PC cell lines and in four of the eight tested PC patients [29]. INTS7 was involved in ATR pathway activation to promote viral genome replication [30]. INTS7 also worked with BAG3 to regulate bone marrow mesenchymal stem cell proliferation and migration [31]. As for INTS8, Yin et al. found that it was stably upregulated in HCC and Tong et al. demonstrated that INTS8 can accelerate the epithelial-mesenchymal transition in HCC by participating in the TGF-β signaling pathway [12]. INTS11 was a kind of catalytic nuclease and it was also the catalytic subunit of the INT complex [22]. The structure analysis revealed that INTS11 binds to INTS9 to constitute the catalytic core of INT complex, and INTS4 plays a key role in stabilizing nuclease domains and other components [22,32].
This study indicated that 14 INT subunits were all significantly increased in GC tissues compared with those normal gastric tissues based on TCGA databases. We further verified the 14 INT subunits mRNA expressions in single-cell sequencing dataset. The results showed that except for INTS1, INTS3, and INTS6, other 11 INT subunits in GC tissue cells were upregulated compared with normal tissue cells. To explore the cause of the abnormal expression of INT subunits in GC, we further assessed the genetic variations, constructed co-expression network, and applied functional enrichment analysis in GC. These results confirm that INT subunits were altered. Amplification and mutation of these subunits may be the main cause of their abnormal expression. Previous studies had demonstrated that INT subunits play important roles in RNA production and processing. In this study, GO analysis also revealed that INT subunits and those related genes were mainly involved in snRNA processing, INT, and DNA-directed 5′–3′ RNA polymerase activity. KEGG analysis of these subunits revealed RNA polymerase, nucleotide excision repair, and Huntington disease. Moreover, according to the results of TISIDB website analysis, there were close correlations between INT subunits and immune cell infiltration in GC. Our study also found that most of INT subunit expressions were positively related to the MSI, which indicated that INT subunits may play important roles in immune regulation in GC. Subsequently, MCPcounter algorithm showed that most INT subunit expressions were strongly positively related to neutrophil and endothelial cells. Neutrophils, as the most abundant leukocytes in peripheral blood, may influence tumor progression through the paracrine release of cytokines and chemokines with pro-tumor functions [33]. Also, it can form neutrophil extracellular traps to promote metastasis in GC patients and tumor-associated neutrophils were considered as a strong predictor for poor prognosis across human cancers [34,35]. The endothelial cells can line blood vessels and are especially prevalent in tumors [36]. The crosstalk between endothelial cells and tumor cells has been characterized as one of the key cell–cell interactions within the tumor microenvironment [37]. Endothelial cell-secreted epidermal growth factor induces epithelial-to-mesenchymal transition in head and neck cancer cells [37]. Tumor endothelial cells also can perform angiogenesis to support the growth, establishment, and dissemination of tumors to distant organs [38]. Our findings suggested that INT subunits involved in development of GC maybe be by influencing the proportion of neutrophil and endothelial cells.
5 Conclusion
In the present study, we systematically and comprehensively analyzed differential expression and mutation patterns, biological functions, protein–protein interactions, different prognostic values, and immune cell infiltration of INT subunits in GC patients based on public databases. Our analysis showed that most INT subunits were expressed at significantly elevated levels in GC tissues compared with normal tissues, suggesting that INT subunits may be potential predictors of prognosis for GC patients. Moreover, functional enrichment analysis indicated that differentially expressed INT subunits were mainly involved in snRNA processing, INT, and DNA-directed 5′–3′ RNA polymerase activity, and most INT subunit expressions were strongly positively related to MSI, neutrophil, and endothelial cells. Considering that INTS11 is the catalytic core subunit of the INT complex, knockdown or overexpression of the INTS11 subunit would influence the function of the INT complex. Therefore, we performed various experiments to assess the effect of INTS11 in GC cells. In conclusion, INT subunits could be effective markers with prognostic and expression significance for GC. Our results can help to better understand the pathogenesis of GC and develop more effective clinical treatments in the future.
Acknowledgements
The authors wish to thank all the authors for the useful suggestions.
-
Funding information: This work was supported by Qingpu District Health Commission Youth program (QWJ2023-16).
-
Author contributions: X.X.T. was responsible for data analysis, financial support, and molecular and cellular experiments. X.X.T. was responsible for writing this article. L.M., D.W., and Y.B.L. were responsible for revising the article. Y.L.L. was responsible for supervision and revising the article. All authors read and approved the final manuscript.
-
Conflict of interest: The authors declare that they have no competing interests.
-
Data availability statement: Publicly available datasets were analyzed in this study. This data can be found in online repositories: TCGA (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and GEO (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE183904.
Appendix

UMAP plots and the expression of INT subunits in different cell types.
The sequence of qRT-PCR primers and shRNAs used in this study
| Name | Sequence(5′–3′) | |||
|---|---|---|---|---|
| qRT-PCR primers | ||||
| INTS11 | Forward | GGAGCGCATGAACCTGAAG | ||
| Reverse | GTCCAGGGGATGAACAGCTT | |||
| GAPDH | Forward | GTCGCCAGCCGAGCCACATC | ||
| Reverse | CCAGGCGCCCAA TACGACCA | |||
| shRNAs targeting INTS11 | ||||
| shRNA #1 | sense | CCGGTCCTGGACTGTGTGATCATTA CTCGAGTAATGATCACACAGTC CAGGATTTTTG | ||
| antisense | AATTCAAAAATCCTGGACTGTGTGATCATT ACTCGAGTAATGATCACACAGTCCAGGA | |||
| shRNA #2 | sense | CCGGCCAGAAGAT CCGCAAGACTTTCTCGAGAAA GTCTTGCGGATCTTCTGGTTTTTG | ||
| antisense | AATTCAAAAACCAGAAGA TCCGCAAGACTTTCTCGAGAAA GTCTTGCGGATCTTCTGG | |||
References
[1] Guan WL, He Y, Xu RH. Gastric cancer treatment: recent progress and future perspectives. J Hematol Oncol. 2023;16(1):57.10.1186/s13045-023-01451-3Search in Google Scholar PubMed PubMed Central
[2] Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–48.10.1016/S0140-6736(20)31288-5Search in Google Scholar PubMed
[3] Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol. 2023;20(3):155–70.10.1038/s41575-022-00703-wSearch in Google Scholar PubMed
[4] Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric cancer: Off J Int Gastric Cancer Assoc Japanese Gastric Cancer Assoc. 2020;23(4):565–78.10.1007/s10120-020-01090-4Search in Google Scholar PubMed
[5] Zheng H, Qi Y, Hu S, Cao X, Xu C, Yin Z, et al. Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science. 2020;370:6520.10.1126/science.abb5872Search in Google Scholar PubMed
[6] Oegema R, Baillat D, Schot R, van Unen LM, Brooks A, Kia SK, et al. Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet. 2017;13(5):e1006809.10.1371/journal.pgen.1006809Search in Google Scholar PubMed PubMed Central
[7] Federico A, Rienzo M, Abbondanza C, Costa V, Ciccodicola A, Casamassimi A. Pan-cancer mutational and transcriptional analysis of the integrator complex. Int J Mol Sci. 2017;18:5.10.3390/ijms18050936Search in Google Scholar PubMed PubMed Central
[8] Li Z, Zhu P, Wang M, Fang C, Ji H. Correlation between oncogene integrator complex subunit 7 and a poor prognosis in lung adenocarcinoma. J Thorac Dis. 2022;14(12):4815–27.10.21037/jtd-22-1533Search in Google Scholar PubMed PubMed Central
[9] Wieland I, Arden KC, Michels D, Klein-Hitpass L, Bohm M, Viars CS, et al. Isolation of DICE1: a gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene. 1999;18(32):4530–7.10.1038/sj.onc.1202806Search in Google Scholar PubMed
[10] Ding X, Chen T, Shi Q, Nan P, Wang X, Xie D, et al. INTS6 promotes colorectal cancer progression by activating of AKT and ERK signaling. Exp Cell Res. 2021;407(2):112826.10.1016/j.yexcr.2021.112826Search in Google Scholar PubMed
[11] Li X, Yao Y, Qian J, Jin G, Zeng G, Zhao H. Overexpression and diagnostic significance of INTS7 in lung adenocarcinoma and its effects on tumor microenvironment. Int Immunopharmacol. 2021;101(Pt B):108346.10.1016/j.intimp.2021.108346Search in Google Scholar PubMed
[12] Tong H, Liu X, Li T, Qiu W, Peng C, Shen B, et al. INTS8 accelerates the epithelial-to-mesenchymal transition in hepatocellular carcinoma by upregulating the TGF-beta signaling pathway. Cancer Manag Res. 2019;11:1869–79.10.2147/CMAR.S184392Search in Google Scholar PubMed PubMed Central
[13] Zhou Q, Ji L, Shi X, Deng D, Guo F, Wang Z, et al. INTS8 is a therapeutic target for intrahepatic cholangiocarcinoma via the integration of bioinformatics analysis and experimental validation. Sci Rep. 2021;11(1):23649.10.1038/s41598-021-03017-0Search in Google Scholar PubMed PubMed Central
[14] Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123(2):265–76.10.1016/j.cell.2005.08.019Search in Google Scholar PubMed
[15] Liu T, Yang K, Chen J, Qi L, Zhou X, Wang P. Comprehensive pan-cancer analysis of KIF18A as a marker for prognosis and immunity. Biomolecules. 2023;13:2.10.3390/biom13020326Search in Google Scholar PubMed PubMed Central
[16] Cambier S, Gouwy M, Proost P. The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell Mol Immunol. 2023;20(3):217–51.10.1038/s41423-023-00974-6Search in Google Scholar PubMed PubMed Central
[17] Kobayashi Y. Neutrophil infiltration and chemokines. Crit Rev Immunol. 2006;26(4):307–16.10.1615/CritRevImmunol.v26.i4.20Search in Google Scholar
[18] Shen J, Wang R, Chen Y, Fang Z, Tang J, Yao J, et al. Prognostic significance and mechanisms of CXCL genes in clear cell renal cell carcinoma. Aging (Albany NY). 2023;15(16):7974–96.10.18632/aging.204922Search in Google Scholar PubMed PubMed Central
[19] Perez-Gutierrez L, Ferrara N. Biology and therapeutic targeting of vascular endothelial growth factor A. Nat Rev Mol Cell Biol. 2023;24(11):816–34.10.1038/s41580-023-00631-wSearch in Google Scholar PubMed
[20] Wu L, Su C, Yang C, Liu J, Ye Y. TBX3 regulates the transcription of VEGFA to promote osteoblasts proliferation and microvascular regeneration. PeerJ. 2022;10:e13722.10.7717/peerj.13722Search in Google Scholar PubMed PubMed Central
[21] Brogowska KK, Zajkowska M, Mroczko B. Vascular endothelial growth factor ligands and receptors in breast cancer. J Clin Med. 2023;12(6):2412.10.3390/jcm12062412Search in Google Scholar PubMed PubMed Central
[22] Pfleiderer MM, Galej WP. Structure of the catalytic core of the integrator complex. Mol Cell. 2021;81(6):1246–59.10.1016/j.molcel.2021.01.005Search in Google Scholar PubMed PubMed Central
[23] Wang H, Fan Z, Shliaha PV, Miele M, Hendrickson RC, Jiang X, et al. H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature. 2023;615(7951):339–48.10.1038/s41586-023-05780-8Search in Google Scholar PubMed PubMed Central
[24] Zhang Y, Koe CT, Tan YS, Ho J, Tan P, Yu F, et al. The integrator complex prevents dedifferentiation of intermediate neural progenitors back into neural stem cells. Cell Rep. 2019;27(4):987–96.10.1016/j.celrep.2019.03.089Search in Google Scholar PubMed
[25] Vervoort SJ, Welsh SA, Devlin JR, Barbieri E, Knight DA, Offley S, et al. The PP2A-integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell. 2021;184(12):3143–62.10.1016/j.cell.2021.04.022Search in Google Scholar PubMed PubMed Central
[26] Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, et al. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015;25(3):288–305.10.1038/cr.2015.19Search in Google Scholar PubMed PubMed Central
[27] Teng H, Wei W, Li Q, Xue M, Shi X, Li X, et al. Prevalence and architecture of posttranscriptionally impaired synonymous mutations in 8,320 genomes across 22 cancer types. Nucleic Acids Res. 2020;48(3):1192–205.10.1093/nar/gkaa019Search in Google Scholar PubMed PubMed Central
[28] Wieland I, Sell C, Weidle UH, Wieacker P. Ectopic expression of DICE1 suppresses tumor cell growth. Oncol Rep. 2004;12(2):207–11.10.3892/or.12.2.207Search in Google Scholar
[29] Ropke A, Buhtz P, Bohm M, Seger J, Wieland I, Allhoff EP, et al. Promoter CpG hypermethylation and downregulation of DICE1 expression in prostate cancer. Oncogene. 2005;24(44):6667–75.10.1038/sj.onc.1208824Search in Google Scholar PubMed
[30] Postigo A, Ramsden AE, Howell M, Way M. Cytoplasmic ATR activation promotes vaccinia virus genome replication. Cell Rep. 2017;19(5):1022–32.10.1016/j.celrep.2017.04.025Search in Google Scholar PubMed PubMed Central
[31] Liu Y, Xu R, Xu J, Wu T, Zhang X. BAG3 regulates bone marrow mesenchymal stem cell proliferation by targeting INTS7. PeerJ. 2023;11:e15828.10.7717/peerj.15828Search in Google Scholar PubMed PubMed Central
[32] Albrecht TR, Shevtsov SP, Wu Y, Mascibroda LG, Peart NJ, Huang KL, et al. Integrator subunit 4 is a ‘Symplekin-like’ scaffold that associates with INTS9/11 to form the Integrator cleavage module. Nucleic Acids Res. 2018;46(8):4241–55.10.1093/nar/gky100Search in Google Scholar PubMed PubMed Central
[33] Li S, Cong X, Gao H, Lan X, Li Z, Wang W, et al. Tumor-associated neutrophils induce EMT by IL-17a to promote migration and invasion in gastric cancer cells. J Exp Clin Cancer Research: CR. 2019;38(1):6.10.1186/s13046-018-1003-0Search in Google Scholar PubMed PubMed Central
[34] Xia X, Zhang Z, Zhu C, Ni B, Wang S, Yang S, et al. Neutrophil extracellular traps promote metastasis in gastric cancer patients with postoperative abdominal infectious complications. Nat Commun. 2022;13(1):1017.10.1038/s41467-022-28492-5Search in Google Scholar PubMed PubMed Central
[35] Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–45.10.1038/nm.3909Search in Google Scholar PubMed PubMed Central
[36] Franses JW, Baker AB, Chitalia VC, Edelman ER. Stromal endothelial cells directly influence cancer progression. Sci Transl Med. 2011;3(66):66ra5.10.1126/scitranslmed.3001542Search in Google Scholar PubMed PubMed Central
[37] Zhang Z, Dong Z, Lauxen IS, Filho MS, Nor JE. Endothelial cell-secreted EGF induces epithelial to mesenchymal transition and endows head and neck cancer cells with stem-like phenotype. Cancer Res. 2014;74(10):2869–81.10.1158/0008-5472.CAN-13-2032Search in Google Scholar PubMed PubMed Central
[38] Annan DA, Kikuchi H, Maishi N, Hida Y, Hida K. Tumor endothelial cell - a biological tool for translational cancer research. Int J Mol Sci. 2020;21(9):3238.10.3390/ijms21093238Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- EDNRB inhibits the growth and migration of prostate cancer cells by activating the cGMP-PKG pathway
- STK11 (LKB1) mutation suppresses ferroptosis in lung adenocarcinoma by facilitating monounsaturated fatty acid synthesis
- Association of SOX6 gene polymorphisms with Kashin-Beck disease risk in the Chinese Han population
- The pyroptosis-related signature predicts prognosis and influences the tumor immune microenvironment in dedifferentiated liposarcoma
- METTL3 attenuates ferroptosis sensitivity in lung cancer via modulating TFRC
- Identification and validation of molecular subtypes and prognostic signature for stage I and stage II gastric cancer based on neutrophil extracellular traps
- Novel lumbar plexus block versus femoral nerve block for analgesia and motor recovery after total knee arthroplasty
- Correlation between ABCB1 and OLIG2 polymorphisms and the severity and prognosis of patients with cerebral infarction
- Study on the radiotherapy effect and serum neutral granulocyte lymphocyte ratio and inflammatory factor expression of nasopharyngeal carcinoma
- Transcriptome analysis of effects of Tecrl deficiency on cardiometabolic and calcium regulation in cardiac tissue
- Aflatoxin B1 induces infertility, fetal deformities, and potential therapies
- Serum levels of HMW adiponectin and its receptors are associated with cytokine levels and clinical characteristics in chronic obstructive pulmonary disease
- METTL3-mediated methylation of CYP2C19 mRNA may aggravate clopidogrel resistance in ischemic stroke patients
- Understand how machine learning impact lung cancer research from 2010 to 2021: A bibliometric analysis
- Pressure ulcers in German hospitals: Analysis of reimbursement and length of stay
- Metformin plus L-carnitine enhances brown/beige adipose tissue activity via Nrf2/HO-1 signaling to reduce lipid accumulation and inflammation in murine obesity
- Downregulation of carbonic anhydrase IX expression in mouse xenograft nasopharyngeal carcinoma model via doxorubicin nanobubble combined with ultrasound
- Feasibility of 3-dimensional printed models in simulated training and teaching of transcatheter aortic valve replacement
- miR-335-3p improves type II diabetes mellitus by IGF-1 regulating macrophage polarization
- The analyses of human MCPH1 DNA repair machinery and genetic variations
- Activation of Piezo1 increases the sensitivity of breast cancer to hyperthermia therapy
- Comprehensive analysis based on the disulfidptosis-related genes identifies hub genes and immune infiltration for pancreatic adenocarcinoma
- Changes of serum CA125 and PGE2 before and after high-intensity focused ultrasound combined with GnRH-a in treatment of patients with adenomyosis
- The clinical value of the hepatic venous pressure gradient in patients undergoing hepatic resection for hepatocellular carcinoma with or without liver cirrhosis
- Development and validation of a novel model to predict pulmonary embolism in cardiology suspected patients: A 10-year retrospective analysis
- Downregulation of lncRNA XLOC_032768 in diabetic patients predicts the occurrence of diabetic nephropathy
- Circ_0051428 targeting miR-885-3p/MMP2 axis enhances the malignancy of cervical cancer
- Effectiveness of ginkgo diterpene lactone meglumine on cognitive function in patients with acute ischemic stroke
- The construction of a novel prognostic prediction model for glioma based on GWAS-identified prognostic-related risk loci
- Evaluating the impact of childhood BMI on the risk of coronavirus disease 2019: A Mendelian randomization study
- Lactate dehydrogenase to albumin ratio is associated with in-hospital mortality in patients with acute heart failure: Data from the MIMIC-III database
- CD36-mediated podocyte lipotoxicity promotes foot process effacement
- Efficacy of etonogestrel subcutaneous implants versus the levonorgestrel-releasing intrauterine system in the conservative treatment of adenomyosis
- FLRT2 mediates chondrogenesis of nasal septal cartilage and mandibular condyle cartilage
- Challenges in treating primary immune thrombocytopenia patients undergoing COVID-19 vaccination: A retrospective study
- Let-7 family regulates HaCaT cell proliferation and apoptosis via the ΔNp63/PI3K/AKT pathway
- Phospholipid transfer protein ameliorates sepsis-induced cardiac dysfunction through NLRP3 inflammasome inhibition
- Postoperative cognitive dysfunction in elderly patients with colorectal cancer: A randomized controlled study comparing goal-directed and conventional fluid therapy
- Long-pulsed ultrasound-mediated microbubble thrombolysis in a rat model of microvascular obstruction
- High SEC61A1 expression predicts poor outcome of acute myeloid leukemia
- Comparison of polymerase chain reaction and next-generation sequencing with conventional urine culture for the diagnosis of urinary tract infections: A meta-analysis
- Secreted frizzled-related protein 5 protects against renal fibrosis by inhibiting Wnt/β-catenin pathway
- Pan-cancer and single-cell analysis of actin cytoskeleton genes related to disulfidptosis
- Overexpression of miR-532-5p restrains oxidative stress response of chondrocytes in nontraumatic osteonecrosis of the femoral head by inhibiting ABL1
- Autologous liver transplantation for unresectable hepatobiliary malignancies in enhanced recovery after surgery model
- Clinical analysis of incomplete rupture of the uterus secondary to previous cesarean section
- Abnormal sleep duration is associated with sarcopenia in older Chinese people: A large retrospective cross-sectional study
- No genetic causality between obesity and benign paroxysmal vertigo: A two-sample Mendelian randomization study
- Identification and validation of autophagy-related genes in SSc
- Long non-coding RNA SRA1 suppresses radiotherapy resistance in esophageal squamous cell carcinoma by modulating glycolytic reprogramming
- Evaluation of quality of life in patients with schizophrenia: An inpatient social welfare institution-based cross-sectional study
- The possible role of oxidative stress marker glutathione in the assessment of cognitive impairment in multiple sclerosis
- Compilation of a self-management assessment scale for postoperative patients with aortic dissection
- Left atrial appendage closure in conjunction with radiofrequency ablation: Effects on left atrial functioning in patients with paroxysmal atrial fibrillation
- Effect of anterior femoral cortical notch grade on postoperative function and complications during TKA surgery: A multicenter, retrospective study
- Clinical characteristics and assessment of risk factors in patients with influenza A-induced severe pneumonia after the prevalence of SARS-CoV-2
- Analgesia nociception index is an indicator of laparoscopic trocar insertion-induced transient nociceptive stimuli
- High STAT4 expression correlates with poor prognosis in acute myeloid leukemia and facilitates disease progression by upregulating VEGFA expression
- Factors influencing cardiovascular system-related post-COVID-19 sequelae: A single-center cohort study
- HOXD10 regulates intestinal permeability and inhibits inflammation of dextran sulfate sodium-induced ulcerative colitis through the inactivation of the Rho/ROCK/MMPs axis
- Mesenchymal stem cell-derived exosomal miR-26a induces ferroptosis, suppresses hepatic stellate cell activation, and ameliorates liver fibrosis by modulating SLC7A11
- Endovascular thrombectomy versus intravenous thrombolysis for primary distal, medium vessel occlusion in acute ischemic stroke
- ANO6 (TMEM16F) inhibits gastrointestinal stromal tumor growth and induces ferroptosis
- Prognostic value of EIF5A2 in solid tumors: A meta-analysis and bioinformatics analysis
- The role of enhanced expression of Cx43 in patients with ulcerative colitis
- Choosing a COVID-19 vaccination site might be driven by anxiety and body vigilance
- Role of ICAM-1 in triple-negative breast cancer
- Cost-effectiveness of ambroxol in the treatment of Gaucher disease type 2
- HLA-DRB5 promotes immune thrombocytopenia via activating CD8+ T cells
- Efficacy and factors of myofascial release therapy combined with electrical and magnetic stimulation in the treatment of chronic pelvic pain syndrome
- Efficacy of tacrolimus monotherapy in primary membranous nephropathy
- Mechanisms of Tripterygium wilfordii Hook F on treating rheumatoid arthritis explored by network pharmacology analysis and molecular docking
- FBXO45 levels regulated ferroptosis renal tubular epithelial cells in a model of diabetic nephropathy by PLK1
- Optimizing anesthesia strategies to NSCLC patients in VATS procedures: Insights from drug requirements and patient recovery patterns
- Alpha-lipoic acid upregulates the PPARγ/NRF2/GPX4 signal pathway to inhibit ferroptosis in the pathogenesis of unexplained recurrent pregnancy loss
- Correlation between fat-soluble vitamin levels and inflammatory factors in paediatric community-acquired pneumonia: A prospective study
- CD1d affects the proliferation, migration, and apoptosis of human papillary thyroid carcinoma TPC-1 cells via regulating MAPK/NF-κB signaling pathway
- miR-let-7a inhibits sympathetic nerve remodeling after myocardial infarction by downregulating the expression of nerve growth factor
- Immune response analysis of solid organ transplantation recipients inoculated with inactivated COVID-19 vaccine: A retrospective analysis
- The H2Valdien derivatives regulate the epithelial–mesenchymal transition of hepatoma carcinoma cells through the Hedgehog signaling pathway
- Clinical efficacy of dexamethasone combined with isoniazid in the treatment of tuberculous meningitis and its effect on peripheral blood T cell subsets
- Comparison of short-segment and long-segment fixation in treatment of degenerative scoliosis and analysis of factors associated with adjacent spondylolisthesis
- Lycopene inhibits pyroptosis of endothelial progenitor cells induced by ox-LDL through the AMPK/mTOR/NLRP3 pathway
- Methylation regulation for FUNDC1 stability in childhood leukemia was up-regulated and facilitates metastasis and reduces ferroptosis of leukemia through mitochondrial damage by FBXL2
- Correlation of single-fiber electromyography studies and functional status in patients with amyotrophic lateral sclerosis
- Risk factors of postoperative airway obstruction complications in children with oral floor mass
- Expression levels and clinical significance of serum miR-19a/CCL20 in patients with acute cerebral infarction
- Physical activity and mental health trends in Korean adolescents: Analyzing the impact of the COVID-19 pandemic from 2018 to 2022
- Evaluating anemia in HIV-infected patients using chest CT
- Ponticulus posticus and skeletal malocclusion: A pilot study in a Southern Italian pre-orthodontic court
- Causal association of circulating immune cells and lymphoma: A Mendelian randomization study
- Assessment of the renal function and fibrosis indexes of conventional western medicine with Chinese medicine for dredging collaterals on treating renal fibrosis: A systematic review and meta-analysis
- Comprehensive landscape of integrator complex subunits and their association with prognosis and tumor microenvironment in gastric cancer
- New target-HMGCR inhibitors for the treatment of primary sclerosing cholangitis: A drug Mendelian randomization study
- Population pharmacokinetics of meropenem in critically ill patients
- Comparison of the ability of newly inflammatory markers to predict complicated appendicitis
- Comparative morphology of the cruciate ligaments: A radiological study
- Immune landscape of hepatocellular carcinoma: The central role of TP53-inducible glycolysis and apoptosis regulator
- Serum SIRT3 levels in epilepsy patients and its association with clinical outcomes and severity: A prospective observational study
- SHP-1 mediates cigarette smoke extract-induced epithelial–mesenchymal transformation and inflammation in 16HBE cells
- Acute hyper-hypoxia accelerates the development of depression in mice via the IL-6/PGC1α/MFN2 signaling pathway
- The GJB3 correlates with the prognosis, immune cell infiltration, and therapeutic responses in lung adenocarcinoma
- Physical fitness and blood parameters outcomes of breast cancer survivor in a low-intensity circuit resistance exercise program
- Exploring anesthetic-induced gene expression changes and immune cell dynamics in atrial tissue post-coronary artery bypass graft surgery
- Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism
- Analysis of the risk factors of the radiation-induced encephalopathy in nasopharyngeal carcinoma: A retrospective cohort study
- Reproductive outcomes in women with BRCA 1/2 germline mutations: A retrospective observational study and literature review
- Evaluation of upper airway ultrasonographic measurements in predicting difficult intubation: A cross-section of the Turkish population
- Prognostic and diagnostic value of circulating IGFBP2 in pancreatic cancer
- Postural stability after operative reconstruction of the AFTL in chronic ankle instability comparing three different surgical techniques
- Research trends related to emergence agitation in the post-anaesthesia care unit from 2001 to 2023: A bibliometric analysis
- Frequency and clinicopathological correlation of gastrointestinal polyps: A six-year single center experience
- ACSL4 mediates inflammatory bowel disease and contributes to LPS-induced intestinal epithelial cell dysfunction by activating ferroptosis and inflammation
- Affibody-based molecular probe 99mTc-(HE)3ZHER2:V2 for non-invasive HER2 detection in ovarian and breast cancer xenografts
- Effectiveness of nutritional support for clinical outcomes in gastric cancer patients: A meta-analysis of randomized controlled trials
- The relationship between IFN-γ, IL-10, IL-6 cytokines, and severity of the condition with serum zinc and Fe in children infected with Mycoplasma pneumoniae
- Paraquat disrupts the blood–brain barrier by increasing IL-6 expression and oxidative stress through the activation of PI3K/AKT signaling pathway
- Sleep quality associate with the increased prevalence of cognitive impairment in coronary artery disease patients: A retrospective case–control study
- Dioscin protects against chronic prostatitis through the TLR4/NF-κB pathway
- Association of polymorphisms in FBN1, MYH11, and TGF-β signaling-related genes with susceptibility of sporadic thoracic aortic aneurysm and dissection in the Zhejiang Han population
- Application value of multi-parameter magnetic resonance image-transrectal ultrasound cognitive fusion in prostate biopsy
- Laboratory variables‐based artificial neural network models for predicting fatty liver disease: A retrospective study
- Decreased BIRC5-206 promotes epithelial–mesenchymal transition in nasopharyngeal carcinoma through sponging miR-145-5p
- Sepsis induces the cardiomyocyte apoptosis and cardiac dysfunction through activation of YAP1/Serpine1/caspase-3 pathway
- Assessment of iron metabolism and iron deficiency in incident patients on incident continuous ambulatory peritoneal dialysis
- Tibial periosteum flap combined with autologous bone grafting in the treatment of Gustilo-IIIB/IIIC open tibial fractures
- The application of intravenous general anesthesia under nasopharyngeal airway assisted ventilation undergoing ureteroscopic holmium laser lithotripsy: A prospective, single-center, controlled trial
- Long intergenic noncoding RNA for IGF2BP2 stability suppresses gastric cancer cell apoptosis by inhibiting the maturation of microRNA-34a
- Role of FOXM1 and AURKB in regulating keratinocyte function in psoriasis
- Parental control attitudes over their pre-school children’s diet
- The role of auto-HSCT in extranodal natural killer/T cell lymphoma
- Significance of negative cervical cytology and positive HPV in the diagnosis of cervical lesions by colposcopy
- Echinacoside inhibits PASMCs calcium overload to prevent hypoxic pulmonary artery remodeling by regulating TRPC1/4/6 and calmodulin
- ADAR1 plays a protective role in proximal tubular cells under high glucose conditions by attenuating the PI3K/AKT/mTOR signaling pathway
- The risk of cancer among insulin glargine users in Lithuania: A retrospective population-based study
- The unusual location of primary hydatid cyst: A case series study
- Intraoperative changes in electrophysiological monitoring can be used to predict clinical outcomes in patients with spinal cavernous malformation
- Obesity and risk of placenta accreta spectrum: A meta-analysis
- Shikonin alleviates asthma phenotypes in mice via an airway epithelial STAT3-dependent mechanism
- NSUN6 and HTR7 disturbed the stability of carotid atherosclerotic plaques by regulating the immune responses of macrophages
- The effect of COVID-19 lockdown on admission rates in Maternity Hospital
- Temporal muscle thickness is not a prognostic predictor in patients with high-grade glioma, an experience at two centers in China
- Luteolin alleviates cerebral ischemia/reperfusion injury by regulating cell pyroptosis
- Therapeutic role of respiratory exercise in patients with tuberculous pleurisy
- Effects of CFTR-ENaC on spinal cord edema after spinal cord injury
- Irisin-regulated lncRNAs and their potential regulatory functions in chondrogenic differentiation of human mesenchymal stem cells
- DMD mutations in pediatric patients with phenotypes of Duchenne/Becker muscular dystrophy
- Combination of C-reactive protein and fibrinogen-to-albumin ratio as a novel predictor of all-cause mortality in heart failure patients
- Significant role and the underly mechanism of cullin-1 in chronic obstructive pulmonary disease
- Ferroptosis-related prognostic model of mantle cell lymphoma
- Observation of choking reaction and other related indexes in elderly painless fiberoptic bronchoscopy with transnasal high-flow humidification oxygen therapy
- A bibliometric analysis of Prader-Willi syndrome from 2002 to 2022
- The causal effects of childhood sunburn occasions on melanoma: A univariable and multivariable Mendelian randomization study
- Oxidative stress regulates glycogen synthase kinase-3 in lymphocytes of diabetes mellitus patients complicated with cerebral infarction
- Role of COX6C and NDUFB3 in septic shock and stroke
- Trends in disease burden of type 2 diabetes, stroke, and hypertensive heart disease attributable to high BMI in China: 1990–2019
- Purinergic P2X7 receptor mediates hyperoxia-induced injury in pulmonary microvascular endothelial cells via NLRP3-mediated pyroptotic pathway
- Investigating the role of oviductal mucosa–endometrial co-culture in modulating factors relevant to embryo implantation
- Analgesic effect of external oblique intercostal block in laparoscopic cholecystectomy: A retrospective study
- Elevated serum miR-142-5p correlates with ischemic lesions and both NSE and S100β in ischemic stroke patients
- Correlation between the mechanism of arteriopathy in IgA nephropathy and blood stasis syndrome: A cohort study
- Risk factors for progressive kyphosis after percutaneous kyphoplasty in osteoporotic vertebral compression fracture
- Predictive role of neuron-specific enolase and S100-β in early neurological deterioration and unfavorable prognosis in patients with ischemic stroke
- The potential risk factors of postoperative cognitive dysfunction for endovascular therapy in acute ischemic stroke with general anesthesia
- Fluoxetine inhibited RANKL-induced osteoclastic differentiation in vitro
- Detection of serum FOXM1 and IGF2 in patients with ARDS and their correlation with disease and prognosis
- Rhein promotes skin wound healing by activating the PI3K/AKT signaling pathway
- Differences in mortality risk by levels of physical activity among persons with disabilities in South Korea
- Review Articles
- Cutaneous signs of selected cardiovascular disorders: A narrative review
- XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis
- A narrative review on adverse drug reactions of COVID-19 treatments on the kidney
- Emerging role and function of SPDL1 in human health and diseases
- Adverse reactions of piperacillin: A literature review of case reports
- Molecular mechanism and intervention measures of microvascular complications in diabetes
- Regulation of mesenchymal stem cell differentiation by autophagy
- Molecular landscape of borderline ovarian tumours: A systematic review
- Advances in synthetic lethality modalities for glioblastoma multiforme
- Investigating hormesis, aging, and neurodegeneration: From bench to clinics
- Frankincense: A neuronutrient to approach Parkinson’s disease treatment
- Sox9: A potential regulator of cancer stem cells in osteosarcoma
- Early detection of cardiovascular risk markers through non-invasive ultrasound methodologies in periodontitis patients
- Advanced neuroimaging and criminal interrogation in lie detection
- Maternal factors for neural tube defects in offspring: An umbrella review
- The chemoprotective hormetic effects of rosmarinic acid
- CBD’s potential impact on Parkinson’s disease: An updated overview
- Progress in cytokine research for ARDS: A comprehensive review
- Utilizing reactive oxygen species-scavenging nanoparticles for targeting oxidative stress in the treatment of ischemic stroke: A review
- NRXN1-related disorders, attempt to better define clinical assessment
- Lidocaine infusion for the treatment of complex regional pain syndrome: Case series and literature review
- Trends and future directions of autophagy in osteosarcoma: A bibliometric analysis
- Iron in ventricular remodeling and aneurysms post-myocardial infarction
- Case Reports
- Sirolimus potentiated angioedema: A case report and review of the literature
- Identification of mixed anaerobic infections after inguinal hernia repair based on metagenomic next-generation sequencing: A case report
- Successful treatment with bortezomib in combination with dexamethasone in a middle-aged male with idiopathic multicentric Castleman’s disease: A case report
- Complete heart block associated with hepatitis A infection in a female child with fatal outcome
- Elevation of D-dimer in eosinophilic gastrointestinal diseases in the absence of venous thrombosis: A case series and literature review
- Four years of natural progressive course: A rare case report of juvenile Xp11.2 translocations renal cell carcinoma with TFE3 gene fusion
- Advancing prenatal diagnosis: Echocardiographic detection of Scimitar syndrome in China – A case series
- Outcomes and complications of hemodialysis in patients with renal cancer following bilateral nephrectomy
- Anti-HMGCR myopathy mimicking facioscapulohumeral muscular dystrophy
- Recurrent opportunistic infections in a HIV-negative patient with combined C6 and NFKB1 mutations: A case report, pedigree analysis, and literature review
- Letter to the Editor
- Letter to the Editor: Total parenteral nutrition-induced Wernicke’s encephalopathy after oncologic gastrointestinal surgery
- Erratum
- Erratum to “Bladder-embedded ectopic intrauterine device with calculus”
- Retraction
- Retraction of “XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis”
- Corrigendum
- Corrigendum to “Investigating hormesis, aging, and neurodegeneration: From bench to clinics”
- Corrigendum to “Frankincense: A neuronutrient to approach Parkinson’s disease treatment”
- Special Issue The evolving saga of RNAs from bench to bedside - Part II
- Machine-learning-based prediction of a diagnostic model using autophagy-related genes based on RNA sequencing for patients with papillary thyroid carcinoma
- Unlocking the future of hepatocellular carcinoma treatment: A comprehensive analysis of disulfidptosis-related lncRNAs for prognosis and drug screening
- Elevated mRNA level indicates FSIP1 promotes EMT and gastric cancer progression by regulating fibroblasts in tumor microenvironment
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part I
- Ultrasound-guided transperineal vs transrectal prostate biopsy: A meta-analysis of diagnostic accuracy and complication rates
- Assessment of diagnostic value of unilateral systematic biopsy combined with targeted biopsy in detecting clinically significant prostate cancer
- SENP7 inhibits glioblastoma metastasis and invasion by dissociating SUMO2/3 binding to specific target proteins
- MARK1 suppress malignant progression of hepatocellular carcinoma and improves sorafenib resistance through negatively regulating POTEE
- Analysis of postoperative complications in bladder cancer patients
- Carboplatin combined with arsenic trioxide versus carboplatin combined with docetaxel treatment for LACC: A randomized, open-label, phase II clinical study
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part I
- Comprehensive pan-cancer investigation of carnosine dipeptidase 1 and its prospective prognostic significance in hepatocellular carcinoma
- Identification of signatures associated with microsatellite instability and immune characteristics to predict the prognostic risk of colon cancer
- Single-cell analysis identified key macrophage subpopulations associated with atherosclerosis
Articles in the same Issue
- Research Articles
- EDNRB inhibits the growth and migration of prostate cancer cells by activating the cGMP-PKG pathway
- STK11 (LKB1) mutation suppresses ferroptosis in lung adenocarcinoma by facilitating monounsaturated fatty acid synthesis
- Association of SOX6 gene polymorphisms with Kashin-Beck disease risk in the Chinese Han population
- The pyroptosis-related signature predicts prognosis and influences the tumor immune microenvironment in dedifferentiated liposarcoma
- METTL3 attenuates ferroptosis sensitivity in lung cancer via modulating TFRC
- Identification and validation of molecular subtypes and prognostic signature for stage I and stage II gastric cancer based on neutrophil extracellular traps
- Novel lumbar plexus block versus femoral nerve block for analgesia and motor recovery after total knee arthroplasty
- Correlation between ABCB1 and OLIG2 polymorphisms and the severity and prognosis of patients with cerebral infarction
- Study on the radiotherapy effect and serum neutral granulocyte lymphocyte ratio and inflammatory factor expression of nasopharyngeal carcinoma
- Transcriptome analysis of effects of Tecrl deficiency on cardiometabolic and calcium regulation in cardiac tissue
- Aflatoxin B1 induces infertility, fetal deformities, and potential therapies
- Serum levels of HMW adiponectin and its receptors are associated with cytokine levels and clinical characteristics in chronic obstructive pulmonary disease
- METTL3-mediated methylation of CYP2C19 mRNA may aggravate clopidogrel resistance in ischemic stroke patients
- Understand how machine learning impact lung cancer research from 2010 to 2021: A bibliometric analysis
- Pressure ulcers in German hospitals: Analysis of reimbursement and length of stay
- Metformin plus L-carnitine enhances brown/beige adipose tissue activity via Nrf2/HO-1 signaling to reduce lipid accumulation and inflammation in murine obesity
- Downregulation of carbonic anhydrase IX expression in mouse xenograft nasopharyngeal carcinoma model via doxorubicin nanobubble combined with ultrasound
- Feasibility of 3-dimensional printed models in simulated training and teaching of transcatheter aortic valve replacement
- miR-335-3p improves type II diabetes mellitus by IGF-1 regulating macrophage polarization
- The analyses of human MCPH1 DNA repair machinery and genetic variations
- Activation of Piezo1 increases the sensitivity of breast cancer to hyperthermia therapy
- Comprehensive analysis based on the disulfidptosis-related genes identifies hub genes and immune infiltration for pancreatic adenocarcinoma
- Changes of serum CA125 and PGE2 before and after high-intensity focused ultrasound combined with GnRH-a in treatment of patients with adenomyosis
- The clinical value of the hepatic venous pressure gradient in patients undergoing hepatic resection for hepatocellular carcinoma with or without liver cirrhosis
- Development and validation of a novel model to predict pulmonary embolism in cardiology suspected patients: A 10-year retrospective analysis
- Downregulation of lncRNA XLOC_032768 in diabetic patients predicts the occurrence of diabetic nephropathy
- Circ_0051428 targeting miR-885-3p/MMP2 axis enhances the malignancy of cervical cancer
- Effectiveness of ginkgo diterpene lactone meglumine on cognitive function in patients with acute ischemic stroke
- The construction of a novel prognostic prediction model for glioma based on GWAS-identified prognostic-related risk loci
- Evaluating the impact of childhood BMI on the risk of coronavirus disease 2019: A Mendelian randomization study
- Lactate dehydrogenase to albumin ratio is associated with in-hospital mortality in patients with acute heart failure: Data from the MIMIC-III database
- CD36-mediated podocyte lipotoxicity promotes foot process effacement
- Efficacy of etonogestrel subcutaneous implants versus the levonorgestrel-releasing intrauterine system in the conservative treatment of adenomyosis
- FLRT2 mediates chondrogenesis of nasal septal cartilage and mandibular condyle cartilage
- Challenges in treating primary immune thrombocytopenia patients undergoing COVID-19 vaccination: A retrospective study
- Let-7 family regulates HaCaT cell proliferation and apoptosis via the ΔNp63/PI3K/AKT pathway
- Phospholipid transfer protein ameliorates sepsis-induced cardiac dysfunction through NLRP3 inflammasome inhibition
- Postoperative cognitive dysfunction in elderly patients with colorectal cancer: A randomized controlled study comparing goal-directed and conventional fluid therapy
- Long-pulsed ultrasound-mediated microbubble thrombolysis in a rat model of microvascular obstruction
- High SEC61A1 expression predicts poor outcome of acute myeloid leukemia
- Comparison of polymerase chain reaction and next-generation sequencing with conventional urine culture for the diagnosis of urinary tract infections: A meta-analysis
- Secreted frizzled-related protein 5 protects against renal fibrosis by inhibiting Wnt/β-catenin pathway
- Pan-cancer and single-cell analysis of actin cytoskeleton genes related to disulfidptosis
- Overexpression of miR-532-5p restrains oxidative stress response of chondrocytes in nontraumatic osteonecrosis of the femoral head by inhibiting ABL1
- Autologous liver transplantation for unresectable hepatobiliary malignancies in enhanced recovery after surgery model
- Clinical analysis of incomplete rupture of the uterus secondary to previous cesarean section
- Abnormal sleep duration is associated with sarcopenia in older Chinese people: A large retrospective cross-sectional study
- No genetic causality between obesity and benign paroxysmal vertigo: A two-sample Mendelian randomization study
- Identification and validation of autophagy-related genes in SSc
- Long non-coding RNA SRA1 suppresses radiotherapy resistance in esophageal squamous cell carcinoma by modulating glycolytic reprogramming
- Evaluation of quality of life in patients with schizophrenia: An inpatient social welfare institution-based cross-sectional study
- The possible role of oxidative stress marker glutathione in the assessment of cognitive impairment in multiple sclerosis
- Compilation of a self-management assessment scale for postoperative patients with aortic dissection
- Left atrial appendage closure in conjunction with radiofrequency ablation: Effects on left atrial functioning in patients with paroxysmal atrial fibrillation
- Effect of anterior femoral cortical notch grade on postoperative function and complications during TKA surgery: A multicenter, retrospective study
- Clinical characteristics and assessment of risk factors in patients with influenza A-induced severe pneumonia after the prevalence of SARS-CoV-2
- Analgesia nociception index is an indicator of laparoscopic trocar insertion-induced transient nociceptive stimuli
- High STAT4 expression correlates with poor prognosis in acute myeloid leukemia and facilitates disease progression by upregulating VEGFA expression
- Factors influencing cardiovascular system-related post-COVID-19 sequelae: A single-center cohort study
- HOXD10 regulates intestinal permeability and inhibits inflammation of dextran sulfate sodium-induced ulcerative colitis through the inactivation of the Rho/ROCK/MMPs axis
- Mesenchymal stem cell-derived exosomal miR-26a induces ferroptosis, suppresses hepatic stellate cell activation, and ameliorates liver fibrosis by modulating SLC7A11
- Endovascular thrombectomy versus intravenous thrombolysis for primary distal, medium vessel occlusion in acute ischemic stroke
- ANO6 (TMEM16F) inhibits gastrointestinal stromal tumor growth and induces ferroptosis
- Prognostic value of EIF5A2 in solid tumors: A meta-analysis and bioinformatics analysis
- The role of enhanced expression of Cx43 in patients with ulcerative colitis
- Choosing a COVID-19 vaccination site might be driven by anxiety and body vigilance
- Role of ICAM-1 in triple-negative breast cancer
- Cost-effectiveness of ambroxol in the treatment of Gaucher disease type 2
- HLA-DRB5 promotes immune thrombocytopenia via activating CD8+ T cells
- Efficacy and factors of myofascial release therapy combined with electrical and magnetic stimulation in the treatment of chronic pelvic pain syndrome
- Efficacy of tacrolimus monotherapy in primary membranous nephropathy
- Mechanisms of Tripterygium wilfordii Hook F on treating rheumatoid arthritis explored by network pharmacology analysis and molecular docking
- FBXO45 levels regulated ferroptosis renal tubular epithelial cells in a model of diabetic nephropathy by PLK1
- Optimizing anesthesia strategies to NSCLC patients in VATS procedures: Insights from drug requirements and patient recovery patterns
- Alpha-lipoic acid upregulates the PPARγ/NRF2/GPX4 signal pathway to inhibit ferroptosis in the pathogenesis of unexplained recurrent pregnancy loss
- Correlation between fat-soluble vitamin levels and inflammatory factors in paediatric community-acquired pneumonia: A prospective study
- CD1d affects the proliferation, migration, and apoptosis of human papillary thyroid carcinoma TPC-1 cells via regulating MAPK/NF-κB signaling pathway
- miR-let-7a inhibits sympathetic nerve remodeling after myocardial infarction by downregulating the expression of nerve growth factor
- Immune response analysis of solid organ transplantation recipients inoculated with inactivated COVID-19 vaccine: A retrospective analysis
- The H2Valdien derivatives regulate the epithelial–mesenchymal transition of hepatoma carcinoma cells through the Hedgehog signaling pathway
- Clinical efficacy of dexamethasone combined with isoniazid in the treatment of tuberculous meningitis and its effect on peripheral blood T cell subsets
- Comparison of short-segment and long-segment fixation in treatment of degenerative scoliosis and analysis of factors associated with adjacent spondylolisthesis
- Lycopene inhibits pyroptosis of endothelial progenitor cells induced by ox-LDL through the AMPK/mTOR/NLRP3 pathway
- Methylation regulation for FUNDC1 stability in childhood leukemia was up-regulated and facilitates metastasis and reduces ferroptosis of leukemia through mitochondrial damage by FBXL2
- Correlation of single-fiber electromyography studies and functional status in patients with amyotrophic lateral sclerosis
- Risk factors of postoperative airway obstruction complications in children with oral floor mass
- Expression levels and clinical significance of serum miR-19a/CCL20 in patients with acute cerebral infarction
- Physical activity and mental health trends in Korean adolescents: Analyzing the impact of the COVID-19 pandemic from 2018 to 2022
- Evaluating anemia in HIV-infected patients using chest CT
- Ponticulus posticus and skeletal malocclusion: A pilot study in a Southern Italian pre-orthodontic court
- Causal association of circulating immune cells and lymphoma: A Mendelian randomization study
- Assessment of the renal function and fibrosis indexes of conventional western medicine with Chinese medicine for dredging collaterals on treating renal fibrosis: A systematic review and meta-analysis
- Comprehensive landscape of integrator complex subunits and their association with prognosis and tumor microenvironment in gastric cancer
- New target-HMGCR inhibitors for the treatment of primary sclerosing cholangitis: A drug Mendelian randomization study
- Population pharmacokinetics of meropenem in critically ill patients
- Comparison of the ability of newly inflammatory markers to predict complicated appendicitis
- Comparative morphology of the cruciate ligaments: A radiological study
- Immune landscape of hepatocellular carcinoma: The central role of TP53-inducible glycolysis and apoptosis regulator
- Serum SIRT3 levels in epilepsy patients and its association with clinical outcomes and severity: A prospective observational study
- SHP-1 mediates cigarette smoke extract-induced epithelial–mesenchymal transformation and inflammation in 16HBE cells
- Acute hyper-hypoxia accelerates the development of depression in mice via the IL-6/PGC1α/MFN2 signaling pathway
- The GJB3 correlates with the prognosis, immune cell infiltration, and therapeutic responses in lung adenocarcinoma
- Physical fitness and blood parameters outcomes of breast cancer survivor in a low-intensity circuit resistance exercise program
- Exploring anesthetic-induced gene expression changes and immune cell dynamics in atrial tissue post-coronary artery bypass graft surgery
- Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism
- Analysis of the risk factors of the radiation-induced encephalopathy in nasopharyngeal carcinoma: A retrospective cohort study
- Reproductive outcomes in women with BRCA 1/2 germline mutations: A retrospective observational study and literature review
- Evaluation of upper airway ultrasonographic measurements in predicting difficult intubation: A cross-section of the Turkish population
- Prognostic and diagnostic value of circulating IGFBP2 in pancreatic cancer
- Postural stability after operative reconstruction of the AFTL in chronic ankle instability comparing three different surgical techniques
- Research trends related to emergence agitation in the post-anaesthesia care unit from 2001 to 2023: A bibliometric analysis
- Frequency and clinicopathological correlation of gastrointestinal polyps: A six-year single center experience
- ACSL4 mediates inflammatory bowel disease and contributes to LPS-induced intestinal epithelial cell dysfunction by activating ferroptosis and inflammation
- Affibody-based molecular probe 99mTc-(HE)3ZHER2:V2 for non-invasive HER2 detection in ovarian and breast cancer xenografts
- Effectiveness of nutritional support for clinical outcomes in gastric cancer patients: A meta-analysis of randomized controlled trials
- The relationship between IFN-γ, IL-10, IL-6 cytokines, and severity of the condition with serum zinc and Fe in children infected with Mycoplasma pneumoniae
- Paraquat disrupts the blood–brain barrier by increasing IL-6 expression and oxidative stress through the activation of PI3K/AKT signaling pathway
- Sleep quality associate with the increased prevalence of cognitive impairment in coronary artery disease patients: A retrospective case–control study
- Dioscin protects against chronic prostatitis through the TLR4/NF-κB pathway
- Association of polymorphisms in FBN1, MYH11, and TGF-β signaling-related genes with susceptibility of sporadic thoracic aortic aneurysm and dissection in the Zhejiang Han population
- Application value of multi-parameter magnetic resonance image-transrectal ultrasound cognitive fusion in prostate biopsy
- Laboratory variables‐based artificial neural network models for predicting fatty liver disease: A retrospective study
- Decreased BIRC5-206 promotes epithelial–mesenchymal transition in nasopharyngeal carcinoma through sponging miR-145-5p
- Sepsis induces the cardiomyocyte apoptosis and cardiac dysfunction through activation of YAP1/Serpine1/caspase-3 pathway
- Assessment of iron metabolism and iron deficiency in incident patients on incident continuous ambulatory peritoneal dialysis
- Tibial periosteum flap combined with autologous bone grafting in the treatment of Gustilo-IIIB/IIIC open tibial fractures
- The application of intravenous general anesthesia under nasopharyngeal airway assisted ventilation undergoing ureteroscopic holmium laser lithotripsy: A prospective, single-center, controlled trial
- Long intergenic noncoding RNA for IGF2BP2 stability suppresses gastric cancer cell apoptosis by inhibiting the maturation of microRNA-34a
- Role of FOXM1 and AURKB in regulating keratinocyte function in psoriasis
- Parental control attitudes over their pre-school children’s diet
- The role of auto-HSCT in extranodal natural killer/T cell lymphoma
- Significance of negative cervical cytology and positive HPV in the diagnosis of cervical lesions by colposcopy
- Echinacoside inhibits PASMCs calcium overload to prevent hypoxic pulmonary artery remodeling by regulating TRPC1/4/6 and calmodulin
- ADAR1 plays a protective role in proximal tubular cells under high glucose conditions by attenuating the PI3K/AKT/mTOR signaling pathway
- The risk of cancer among insulin glargine users in Lithuania: A retrospective population-based study
- The unusual location of primary hydatid cyst: A case series study
- Intraoperative changes in electrophysiological monitoring can be used to predict clinical outcomes in patients with spinal cavernous malformation
- Obesity and risk of placenta accreta spectrum: A meta-analysis
- Shikonin alleviates asthma phenotypes in mice via an airway epithelial STAT3-dependent mechanism
- NSUN6 and HTR7 disturbed the stability of carotid atherosclerotic plaques by regulating the immune responses of macrophages
- The effect of COVID-19 lockdown on admission rates in Maternity Hospital
- Temporal muscle thickness is not a prognostic predictor in patients with high-grade glioma, an experience at two centers in China
- Luteolin alleviates cerebral ischemia/reperfusion injury by regulating cell pyroptosis
- Therapeutic role of respiratory exercise in patients with tuberculous pleurisy
- Effects of CFTR-ENaC on spinal cord edema after spinal cord injury
- Irisin-regulated lncRNAs and their potential regulatory functions in chondrogenic differentiation of human mesenchymal stem cells
- DMD mutations in pediatric patients with phenotypes of Duchenne/Becker muscular dystrophy
- Combination of C-reactive protein and fibrinogen-to-albumin ratio as a novel predictor of all-cause mortality in heart failure patients
- Significant role and the underly mechanism of cullin-1 in chronic obstructive pulmonary disease
- Ferroptosis-related prognostic model of mantle cell lymphoma
- Observation of choking reaction and other related indexes in elderly painless fiberoptic bronchoscopy with transnasal high-flow humidification oxygen therapy
- A bibliometric analysis of Prader-Willi syndrome from 2002 to 2022
- The causal effects of childhood sunburn occasions on melanoma: A univariable and multivariable Mendelian randomization study
- Oxidative stress regulates glycogen synthase kinase-3 in lymphocytes of diabetes mellitus patients complicated with cerebral infarction
- Role of COX6C and NDUFB3 in septic shock and stroke
- Trends in disease burden of type 2 diabetes, stroke, and hypertensive heart disease attributable to high BMI in China: 1990–2019
- Purinergic P2X7 receptor mediates hyperoxia-induced injury in pulmonary microvascular endothelial cells via NLRP3-mediated pyroptotic pathway
- Investigating the role of oviductal mucosa–endometrial co-culture in modulating factors relevant to embryo implantation
- Analgesic effect of external oblique intercostal block in laparoscopic cholecystectomy: A retrospective study
- Elevated serum miR-142-5p correlates with ischemic lesions and both NSE and S100β in ischemic stroke patients
- Correlation between the mechanism of arteriopathy in IgA nephropathy and blood stasis syndrome: A cohort study
- Risk factors for progressive kyphosis after percutaneous kyphoplasty in osteoporotic vertebral compression fracture
- Predictive role of neuron-specific enolase and S100-β in early neurological deterioration and unfavorable prognosis in patients with ischemic stroke
- The potential risk factors of postoperative cognitive dysfunction for endovascular therapy in acute ischemic stroke with general anesthesia
- Fluoxetine inhibited RANKL-induced osteoclastic differentiation in vitro
- Detection of serum FOXM1 and IGF2 in patients with ARDS and their correlation with disease and prognosis
- Rhein promotes skin wound healing by activating the PI3K/AKT signaling pathway
- Differences in mortality risk by levels of physical activity among persons with disabilities in South Korea
- Review Articles
- Cutaneous signs of selected cardiovascular disorders: A narrative review
- XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis
- A narrative review on adverse drug reactions of COVID-19 treatments on the kidney
- Emerging role and function of SPDL1 in human health and diseases
- Adverse reactions of piperacillin: A literature review of case reports
- Molecular mechanism and intervention measures of microvascular complications in diabetes
- Regulation of mesenchymal stem cell differentiation by autophagy
- Molecular landscape of borderline ovarian tumours: A systematic review
- Advances in synthetic lethality modalities for glioblastoma multiforme
- Investigating hormesis, aging, and neurodegeneration: From bench to clinics
- Frankincense: A neuronutrient to approach Parkinson’s disease treatment
- Sox9: A potential regulator of cancer stem cells in osteosarcoma
- Early detection of cardiovascular risk markers through non-invasive ultrasound methodologies in periodontitis patients
- Advanced neuroimaging and criminal interrogation in lie detection
- Maternal factors for neural tube defects in offspring: An umbrella review
- The chemoprotective hormetic effects of rosmarinic acid
- CBD’s potential impact on Parkinson’s disease: An updated overview
- Progress in cytokine research for ARDS: A comprehensive review
- Utilizing reactive oxygen species-scavenging nanoparticles for targeting oxidative stress in the treatment of ischemic stroke: A review
- NRXN1-related disorders, attempt to better define clinical assessment
- Lidocaine infusion for the treatment of complex regional pain syndrome: Case series and literature review
- Trends and future directions of autophagy in osteosarcoma: A bibliometric analysis
- Iron in ventricular remodeling and aneurysms post-myocardial infarction
- Case Reports
- Sirolimus potentiated angioedema: A case report and review of the literature
- Identification of mixed anaerobic infections after inguinal hernia repair based on metagenomic next-generation sequencing: A case report
- Successful treatment with bortezomib in combination with dexamethasone in a middle-aged male with idiopathic multicentric Castleman’s disease: A case report
- Complete heart block associated with hepatitis A infection in a female child with fatal outcome
- Elevation of D-dimer in eosinophilic gastrointestinal diseases in the absence of venous thrombosis: A case series and literature review
- Four years of natural progressive course: A rare case report of juvenile Xp11.2 translocations renal cell carcinoma with TFE3 gene fusion
- Advancing prenatal diagnosis: Echocardiographic detection of Scimitar syndrome in China – A case series
- Outcomes and complications of hemodialysis in patients with renal cancer following bilateral nephrectomy
- Anti-HMGCR myopathy mimicking facioscapulohumeral muscular dystrophy
- Recurrent opportunistic infections in a HIV-negative patient with combined C6 and NFKB1 mutations: A case report, pedigree analysis, and literature review
- Letter to the Editor
- Letter to the Editor: Total parenteral nutrition-induced Wernicke’s encephalopathy after oncologic gastrointestinal surgery
- Erratum
- Erratum to “Bladder-embedded ectopic intrauterine device with calculus”
- Retraction
- Retraction of “XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis”
- Corrigendum
- Corrigendum to “Investigating hormesis, aging, and neurodegeneration: From bench to clinics”
- Corrigendum to “Frankincense: A neuronutrient to approach Parkinson’s disease treatment”
- Special Issue The evolving saga of RNAs from bench to bedside - Part II
- Machine-learning-based prediction of a diagnostic model using autophagy-related genes based on RNA sequencing for patients with papillary thyroid carcinoma
- Unlocking the future of hepatocellular carcinoma treatment: A comprehensive analysis of disulfidptosis-related lncRNAs for prognosis and drug screening
- Elevated mRNA level indicates FSIP1 promotes EMT and gastric cancer progression by regulating fibroblasts in tumor microenvironment
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part I
- Ultrasound-guided transperineal vs transrectal prostate biopsy: A meta-analysis of diagnostic accuracy and complication rates
- Assessment of diagnostic value of unilateral systematic biopsy combined with targeted biopsy in detecting clinically significant prostate cancer
- SENP7 inhibits glioblastoma metastasis and invasion by dissociating SUMO2/3 binding to specific target proteins
- MARK1 suppress malignant progression of hepatocellular carcinoma and improves sorafenib resistance through negatively regulating POTEE
- Analysis of postoperative complications in bladder cancer patients
- Carboplatin combined with arsenic trioxide versus carboplatin combined with docetaxel treatment for LACC: A randomized, open-label, phase II clinical study
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part I
- Comprehensive pan-cancer investigation of carnosine dipeptidase 1 and its prospective prognostic significance in hepatocellular carcinoma
- Identification of signatures associated with microsatellite instability and immune characteristics to predict the prognostic risk of colon cancer
- Single-cell analysis identified key macrophage subpopulations associated with atherosclerosis

