Abstract
The limitations of conventional urine culture methods can be avoided by using culture-independent approaches like polymerase chain reaction (PCR) and next-generation sequencing (NGS). However, the efficacy of these approaches in this setting is still subject to contention. PRISMA-compliant searches were performed on MEDLINE/PubMed, EMBASE, Web of Sciences, and the Cochrane Database until March 2023. The included articles compared PCR or NGS to conventional urine culture for the detection of urinary tract infections (UTIs). RevMan performed meta-analysis, and the Cochrane Risk of Bias Assessment Tool assessed study quality. A total of 10 selected studies that involved 1,291 individuals were included in this meta-analysis. The study found that PCR has a 99% sensitivity and a 94% specificity for diagnosing UTIs. Furthermore, NGS was shown to have a sensitivity of 90% for identifying UTIs and a specificity of 86%. The odds ratio (OR) for PCR to detect Gram-positive bacteria is 0.50 (95% confidence interval [CI] 0.41–0.61), while the OR for NGS to detect Gram-negative bacteria is 0.23 [95% CI 0.09–0.59]. UTIs are typically caused by Gram-negative bacteria like Escherichia coli and Gram-positive bacteria like Staphylococci and Streptococci. PCR and NGS are reliable, culture-free molecular diagnostic methods that, despite being expensive, are essential for UTI diagnosis and prevention due to their high sensitivity and specificity.
1 Introduction
Urinary tract infection (UTI) is an infectious condition that is frequently encountered in the adult population. Usually, these infections appear in the bladder or urethra. However, in more severe instances, they might impact the kidney [1]. Women exhibit a greater vulnerability to UTIs in comparison to men. Around 50–60% of women are projected to experience at least one UTI during their lifetime [2].
Bacterial infections are responsible for the majority of UTIs, and the standard therapy usually involves the use of antibiotics [3,4]. The healthcare industry bears substantial expenses for the treatment and management of UTIs, totaling billions of dollars annually, across both outpatient and inpatient settings [5]. The application of molecular testing techniques, such as next-generation sequencing (NGS) and polymerase chain reaction (PCR), for the identification and diagnosis of UTIs, has experienced substantial progress in recent years. The increase in popularity can be ascribed to the discontentment associated with the traditional method of exclusively depending on urine culture [6,7]. The accuracy of traditional culture methods in identifying acute UTIs is approximately 60%. The traditional approach of urine culture predominantly promotes the proliferation of rapidly growing aerobic bacteria, such as Escherichia coli, Enterococcus, and Staphylococcus species. Nevertheless, it is unable to adequately foster the majority of human commensal bacteria, which are distinguished by their slow growth, anaerobic nature, fastidiousness, or limited development in traditional cultures [8]. Therefore, the exact role of these bacteria in the development of UTIs is yet unknown. Molecular tools, such as NGS and PCR, have uncovered that the bladder has a diverse array of bacterial inhabitants, even in asymptomatic individuals who are in good health. PCR and NGS are culture-independent techniques used to identify and analyze microorganisms in a sample, thereby bypassing the limitations of standard urine culture methods [9]. The PCR test utilizes an advanced approach to duplicate a specific portion of DNA obtained from the patient’s urine sample. This replication process facilitates the identification of the particular pathogen accountable for the UTI, the determination of the most appropriate drugs for UTI treatment, and the evaluation of the bacteria’s resistance to different antibiotics. Qualitative PCR is used to determine the presence or absence of a pathogen, whereas quantitative PCR is performed to measure the amount of pathogen present [10,11]. NGS offers a thorough and detailed evaluation of the urine microbiome. Unlike PCR, which can only detect a limited number of organisms, NGS analyzes the complete microbial DNA in a urine sample and compares it to a comprehensive species database [12]. The application of these approaches has greatly improved our understanding of the urine microbiome and implicated these complex bacterial communities in the genesis of UTI symptoms. Recent studies have emphasized the use of molecular diagnostic tools like PCR and NGS to identify UTIs that are resistant to conventional urine culture techniques; as a result, clinical applications of commercial culture-independent diagnostic services like NGS and PCR are now widely available [13,14,15]. However, molecular diagnostic techniques are advocated for their enhanced sensitivity in detecting urine infections, but the efficacy of these strategies in this specific setting remains unknown. So, the aim of this meta-analysis is to compare how well culture-independent molecular diagnostic technologies like PCR and NGS work for diagnosing UTIs versus traditional urine culture. For this, relevant papers [15,16,17,18,19,20,21,22,23,24,25] selected as per the specific inclusion and exclusion criteria were used in this systematic review and meta-analysis.
2 Methods
The present study complied with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) recommendations [26].
2.1 Eligibility criteria
The current study conducted an extensive examination of relevant academic articles published between 2000 and 2023. The PICO structure was employed to formulate specific selection criteria. In this context, P represented individuals with UTIs; I stood for the application of PCR and NGS for the detection of UTIs. The letter C represents the use of conventional urine culture for UTI detection, while the letter O encompasses clinical outcomes, the total number of positive UTI cases, and the microorganisms responsible for the infections. The studies examined and compared the diagnosis results of UTIs in patients using traditional urine culture methods and advanced molecular diagnostic methods such as PCR or NGS techniques. The researchers prioritized the inclusion of (1) full-text papers and (2) articles published in English in this meta-analysis. The abstracts were only included in the meta-analysis if sufficient information was provided. The analysis excluded papers that had inadequate data, lacked relevance to UTIs, or were published before 2000.
2.2 Information sources
The researchers performed a comprehensive and methodical examination of pertinent literature by searching the databases of MEDLINE/PubMed, EMBASE, Web of Sciences, and the Cochrane database, adhering to the guidelines specified in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA).
2.3 Search strategy
The search was conducted using the following terms: “Urinary tract infections” or “UTI”; “Polymerase chain reaction” or “PCR”; “Next generation sequencing” or “NGS”; “meta-analysis”; “Causative agent of UTIs”; “Gram negative bacteria”; “Gram positive bacteria”; “Fungi”; “Protozoa”; “Conventional urine culture”; and “Molecular diagnostic methods.” The researcher conducted a comprehensive review of scholarly literature by utilizing the databases of PubMed and Cochrane libraries. In the context of searching Scopus, the title (ti)–abstract (abs)–keyword (key) field was utilized with the aforementioned keywords (Table A1). The key phrases “UTIs,” “conventional urine culture,” and “PCR and NGS for detection of UTI” were utilized in the Cochrane database. The integration of the Medical Subject Headings and textual keywords was accomplished by employing the Boolean operator “AND” within the context of the search strategy.
2.4 Selection process
The authors, MZ and SQ, conducted a comprehensive literature review to identify relevant studies. The researchers utilized inclusion criteria to exclude references that were outdated and to incorporate studies of significant relevance. In addition, two researchers conducted a thorough bibliographic search to identify pertinent and influential scholarly articles. A rigorous methodology was utilized to identify and incorporate pertinent studies published between the years 2010 and 2023
2.5 Data collection process and data items
The researchers MZ and SQ independently collected the demographic summary and event data from the studies included in this research. The main results were as follows: The study includes the following information: (1) the overall count of confirmed instances of UTI identified through traditional urine culture; (2) the overall count of confirmed instances of UTI identified through the molecular diagnostic techniques of PCR or NGS; (3) the types of Gram-positive and Gram-negative microorganisms implicated in UTI cases.
2.6 Sources of heterogeneity
The calculation of heterogeneity was performed among the experiments that were included. The Cochran Q statistic and the I 2 index were used in a random bivariate mode [27] and the RevMan software [28] was used to check for heterogeneity. Multiple sources of heterogeneity were investigated, encompassing the utilization of complete textual publications as opposed to abstracts, differences in age cohorts and sample sizes, variances in the bacteria evaluated, and differences in the outcomes of studies. Two reviewers, MZ and SQ, independently evaluated the methodological validity of the studies included in the analysis. The author XZ successfully settled any disputes that emerged between MZ and SQ through discussions and meticulous examination of data.
2.7 Risk of bias assessment
A pre-established, standardized questionnaire was used to assess the risk of bias in the articles that were considered for the analysis. The investigators utilized the Cochrane Risk of Bias: Robvis Tool [29] to produce a concise summary and visual representation illustrating the risk of bias.
2.8 Meta-analysis
The meta-analysis was performed utilizing the RevMan software (Review Manager, RevMan, Version 5, Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2020). The group exhibiting a degree of heterogeneity exceeding 50% opted to employ the random effect, while the subgroup with heterogeneity below 50% utilized the fixed effect. The primary methodology utilized in this study involved the application of the Mantel-Haenszel technique, which incorporated random bivariate effects. The aforementioned methodology was primarily utilized to calculate statistical measures such as sensitivity, specificity, and odds ratio (OR), along with a 95% confidence interval (CI) [30,31]. In addition, forest plots were generated to visually depict the aforementioned findings. The metrics used by the researchers to evaluate the extent of heterogeneity in the analyzed studies included tau2, chi2, I 2, and z values. Statistical significance was determined by considering a p-value below the predetermined threshold of 0.05. The DerSimonian and Lair method was utilized to compute the diagnostic OR using a 2 × 2 contingency table [32]. The evaluation of publication bias in the studies that were included in the analysis was performed utilizing Begg’s test [33] and Deek’s funnel plot [34]. The Deek’s funnel plot was constructed by plotting the logarithm of the OR for each individual study against its corresponding standard error, utilizing the MedCalc software [35].
-
Statement of ethics: An ethics statement is not applicable because this study is based exclusively on published literature.
-
Study approval statement: This study protocol was reviewed and approved by First Affiliated Hospital, Heilongjiang University of Chinese Medicine.
3 Results
3.1 Literature search results
The flowchart depicted in Figure 1 illustrates the utilization of the PRISMA framework in the process of selecting research studies. Following an extensive examination of online databases, a total of 398 studies were identified. After eliminating duplicate entries, a comprehensive set of 304 studies underwent a screening process based on the evaluation of their abstracts and titles. A thorough assessment was conducted on a total of 168 studies that met the predetermined criteria for inclusion. The present meta-analysis consisted of a total of ten studies, which were chosen according to pre-established criteria for inclusion and exclusion. The analysis included ten studies, of which five [16,17,18,19,20] evaluated the relative effectiveness of conventional urine culture and PCR in detecting UTIs, while the remaining studies examined the comparative efficacy of conventional urine culture and NGS for UTI detection. Table 1 provides a comprehensive summary of the relevant attributes of the studies that were included in the analysis pertaining to PCR. Conversely, Table 2 offers a comprehensive summary of the relevant attributes of the studies that were included in the analysis pertaining to NGS. The attributes encompass the identification of the studies, including their publication years, journals of publication, total number of UTI cases, age of patients, details of instruments and techniques employed, type of infection, molecular diagnostic method utilized, and identification of Gram-positive and Gram-negative bacteria responsible for the UTI.

PRISMA flowchart of selection of studies.
Characteristics of the included studies comparing PCR with conventional urine culture method
| Study ID | Year of publication | Journal of publication | Total number of participants | Age of patients (years) | Type of infection | Molecular diagnostic method used | Instrument details | Conventional method used | Micro-organisms identified | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gram positive | Gram negative | |||||||||
| Heytens et al. [16] | 2017 | Clinical Microbiology and Infection | 220 | ≥18 | UTI | PCR | DNA extraction: using Abbott Real-Time CT/NG assay (Abbott Laboratories) and Diagenode S-DiaMGTV (Diagenode Diagnostics, Seraing, Belgium) kit | Urine culture method | S. saprophyticus, Trichomonas vaginalis | E. coli, Mycoplasma genitalium |
| PCR process: Abbott | ||||||||||
| m2000sp/rt instrument | ||||||||||
| Ibraheam et al. [17] | 2016 | Pak. J. Biotechnol | 30 | ≥18 | UTI | PCR | DNA extraction: genomic DNA kit (Geneaid, China) | Urine culture method | S. saprophyticus, Enterobacter | E. coli, P aeruginosa, Proteus mirabilis |
| PCR process: thermocycler Eppendorf programmed cycler | ||||||||||
| Lehmann et al. [18] | 2011 | PLoS One | 81 | ≥18 | UTI | PCR | DNA extraction and PCR process: SeptiFastH, Roche Diagnostics | Urine culture method | Staphylococci, Streptococcus | Escherichia coli, P. aeruginosa, Acinetobacter, Proteus mirabilis |
| GmbH, Penzberg, Germany) | ||||||||||
| Wojno et al. [19] | 2020 | Infectious Diseases | 582 | ≥18 | UTI | PCR | DNA extraction: MagMAX DNA Multi-Sample Ultra Kit (ThermoFisher, Carlsbad, CA) | Urine culture method | Streptococcus, Enterococcus, Actinobaculum schaali | Escherichia coli, Proteus species, Citrobacter species, P. aeruginosa |
| PCR process: Life Technologies | ||||||||||
| 12K Flex OpenArray System. | ||||||||||
| van der Zee et al. [20] | 2016 | PLoS One | 211 | ≥18 | UTI | PCR | DNA extraction: using E7526 kit, Sigma-Aldrich, Munich, Germany | Urine culture method | Staphylococcus, Streptococcus | Escherichia coli, P. aeruginosa, Enterobacteriaceae |
| PCR process: ABI 7500 Real-Time PCR system (Applied Biosystems (ABI), Life Tech, Glasgow, UK) | ||||||||||
PCR: polymerase chain reaction; UTI: urinary tract infection.
Characteristics of the Included studies comparing NGS with Conventional urine culture method
| Study ID | Year of publication | Journal of Publication | Total number of cases | Type of Infection | Molecular diagnostic method used | Instrument Details | Conventional method used | Micro-organisms identified | |
|---|---|---|---|---|---|---|---|---|---|
| Gram positive | Gram negative | ||||||||
| Hasman et al. [21] | 2014 | Journal of Clinical Microbiology | 35 | UTI | NGS | Sequence analysis performed using MG-RAST. | Urine culture method | E. faecalis, Lactobacillus, or Bifidobacterium | E. coli, Prevotella, Gardnerella |
| Ishihara et al. [22] | 2020 | Drug Discoveries & Therapeutics | 10 | UTI | NGS | Sequence analysis performed using Genome Search Toolkit (GSTK) | Urine culture method | Enterococcus faecalis, Aerococcus urinae | Escherichia coli, Proteus mirabilis |
| McDonald et al. [23] | 2017 | Reviews in Urology | 57 | UTI | NGS | Sequence analysis performed using MicroGen DX (Orlando, FL) | Urine culture method | Staphylococcus, Streptococcus, Enterococcus, Aerococcus urinae, Corynebacterium urealyticum, Enterobacter | Escherichia coli, Proteus species, Citrobacter species, P. aeruginosa |
| Sabat et al. [24] | 2017 | Nature Scientific Reports | 23 | UTI | NGS | Sequence analysis performed using Nextera XT | Urine culture method | Staphylococcus, Bacillus | Pseudomonas fluorescens, E. coli, Chryseobacterium, Enhydrobacter, Paracoccus |
| DNA Sample Preparation Kit (Illumina) | |||||||||
| Yoo et al. [25] | 2021 | Journal of Clinical Medicine | 42 | UTI | NGS | Sequence analysis performed using NEXTflex 16S V4 | Urine culture method | Staphylococcus, Streptococcus, Rothia, Enterobacteriaceae | Pseudomonas, Acinetobacter, Sphingomonas |
| Amplicon-Seq (BioO Scientific, Austin, TX, USA) | |||||||||
UTI: urinary tract infection; NGS: next-generation sequencing.
3.2 Quality assessment of the included studies
Table 3 displays the evaluation of the quality of the studies that were incorporated into this meta-analysis. Figure 2 provides a concise overview of the risk of bias assessment conducted for studies pertaining to NGS, while Figure 3 presents a succinct summary of the risk of bias analysis conducted for studies pertaining to PCR. The analysis for NGS incorporated five studies, of which one demonstrated a critical risk of bias resulting from confounding factors, while another exhibited a minimal risk of bias due to the classification of interventions. In a similar vein, the analysis for PCR included five studies, of which two were found to have a low risk of bias attributed to missing data and biased selection of reported results. The plot depicted in Figure 4 exhibits an inverted funnel shape for both PCR and NGS, suggesting the absence of publication bias [36]. This observation is further supported by the lack of statistical significance (p > 0.05) in the Begg’s tests for both NGS (p = 0.342) and PCR (p = 0.417) [37].
Assessment of bias for included studies
| Heytens et al. [16] | Ibraheam et al. [17] | Lehmann et al. [18] | Wojno et al. [19] | van der Zee et al. [20] | Hasman et al. [21] | Ishihara et al. [22] | McDonald et al. [23] | Sabat et al. [24] | Yoo et al. [25] | |
|---|---|---|---|---|---|---|---|---|---|---|
| Did the study avoid inappropriate exclusions | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Did all patients receive the same reference standard | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Were all patients included in the analysis | N | N | N | N | N | N | N | N | N | N |
| Was the sample frame appropriate to address the target population? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Were study participants sampled in an appropriate way? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Were the study subjects and the setting described in detail? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Were valid methods used for the identification of the condition? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Was the condition measured in a standard, reliable way for all participants? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
Y –Yes; N – No.

Risk of Bias analysis for studies related to NGS. (a) Risk of bias graph. (b) Risk of bias summary.

Risk of Bias analysis for studies related to PCR. (a) Risk of bias graph. (b) Risk of bias summary.

Funnel Plot for publication bias. (a) NGS; (b) PCR.
3.3 Statistical analysis of the primary outcomes
The current meta-analysis comprised 10 research papers, which included a collective sample size of 1,291 individuals. The statistical analysis was conducted on the primary outcomes of the studies included in order to evaluate the effectiveness of culture-independent molecular diagnostic technologies, such as PCR and NGS, compared to conventional urine culture for diagnosing UTIs.
3.3.1 Sensitivity and specificity of PCR for detection of UTI
Figure 5 illustrates the sensitivity and specificity of PCR in detecting UTIs, as determined by analyzing data from five specific studies. These studies provided information on the number of positive UTI cases identified through both conventional urine culture and PCR, allowing for a comparative analysis. The combined sensitivity of PCR is 0.99 with a 95% CI of 0.82–1.0. Additionally, the combined specificity of PCR is 0.94 with a 95% CI of 0.55–1.0. The findings of this study indicate that PCR has a higher likelihood of detecting pathogens with greater specificity and accuracy when compared to the conventional culture method.

Forest plot for sensitivity and specificity of PCR for detection of UTI.
3.3.2 Sensitivity and specificity of NGS for detection of UTI
Figure 6 depicts the Sensitivity and specificity of NGS in the detection of UTIs, as determined through the analysis of data from five selected studies. These studies yielded data regarding the prevalence of positive UTI cases detected using both traditional urine culture methods and NGS, enabling a comparative examination. The aggregate sensitivity of NGS is 0.90, accompanied by a 95% CI ranging from 0.45 to 1.0. Moreover, the collective specificity of NGS is 0.86, accompanied by a 95% CI ranging from 0.35 to 1.0. The results of this study suggest that NGS exhibits a higher probability of identifying pathogens with increased specificity and accuracy in comparison to the traditional culture technique.

Forest plot for sensitivity and specificity of NGS for detection of UTI.
3.3.3 UTI detection rate of PCR
Figure 7 depicts a Box and Whisker plot that demonstrates a discernibly higher rate of UTI detection using PCR method in comparison to the conventional urine culture method. The statistical parameters associated with the PCR technique encompass a minimum value of 20, a first quartile (Q1) value of 51.5, a median value of 67, a third quartile (Q3) value of 243.5, and a maximum value of 326. Additionally, the mean value is calculated to be 138.2, while the skewness coefficient is estimated to be 0.090, suggesting a distribution that may exhibit symmetry (p-value = 0.32). Furthermore, the distribution is characterized by a mesokurtic tail. In contrast, the traditional method of urine culture exhibits a minimum value of 18, a first quartile (Q1) of 37.5, a median of 61, a third quartile (Q3) of 266, a maximum value of 431, a mean of 153, a skewness of 1.38 suggesting a possibly symmetrical distribution (p-value = 0.128), and a mesokurtic tail. In an analogous way, the forest plot depicted in Figure 8 demonstrates that the likelihood of detecting UTIs through PCR is higher, as indicated by an adjusted odds ratio (AOR) of 0.50 [95% CI 0.41–0.61]. The findings exhibited heterogeneity, as indicated by the values of tau2 (0.00), chi2 (2.91), df (4), I 2 (75%), z (6.75), and p < 0.00001.

Box and Whisker plot comparing UTI detection rate by conventional urine culture vs PCR.
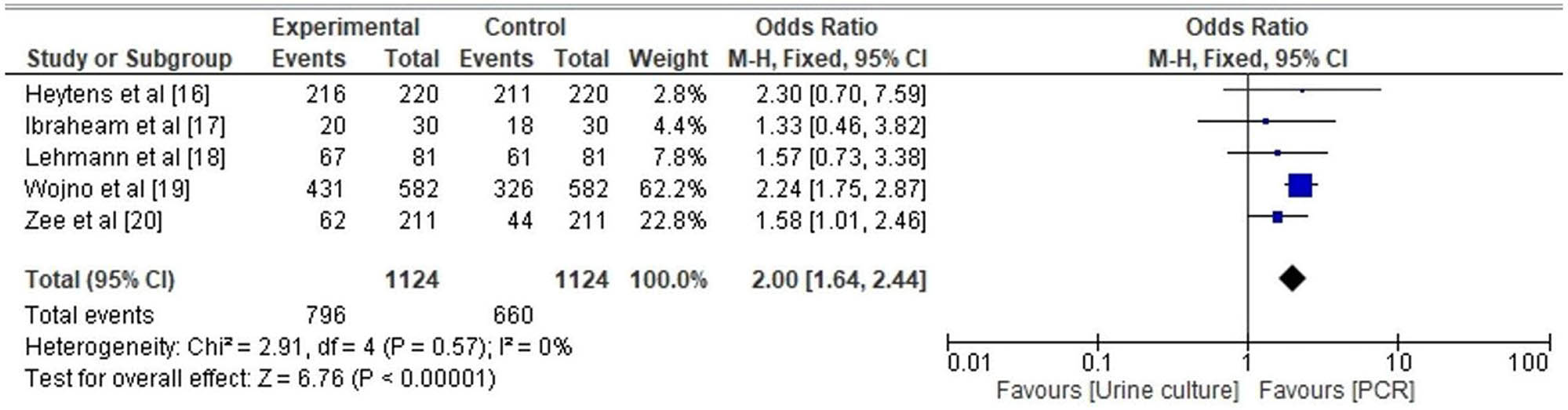
Forest plot for OR of detection of UTI by PCR.
3.3.4 UTI detection rate of NGS
The Box and Whisker plot presented in Figure 9 illustrates that the NGS method exhibits a notably higher rate of UTI detection compared to the conventional urine culture method. The statistical measures for the NGS method include a minimum value of 10, a first quartile (Q1) of 16.75, a median of 22, a third quartile (Q3) of 32.75, a maximum value of 44, a mean of 24.8, a skewness of 0.748371 indicating a potentially symmetrical distribution (p-value = 0.412), and a mesokurtic tail. On the other hand, the conventional urine culture method has a minimum value of 7, a Q1 of 8.5, a median of 13, a Q3 of 17.5, a maximum value of 19, a mean of 13, a skewness of 0.0 indicating a potentially symmetrical distribution (p-value = 1), and a mesokurtic tail. The forest plot displayed in Figure 10 illustrates that the likelihood of identifying UTIs using NGS is greater, as evidenced by an AOR of 0.23 [95% CI 0.09–0.59]. The results demonstrated heterogeneity, as evidenced by tau2 (0.68), chi2 (10.96), df (4), I 2 (64%), z (3.01), and p < 0.003.

Box and Whisker plot comparing UTI detection rate by conventional urine culture vs NGS.

Forest plot for OR of detection of UTI by NGS.
3.3.5 Pathogens detected in UTI by conventional urine culture, PCR, and NGS
The findings from conventional urine culture, PCR, and NGS analyses revealed that UTIs can be attributed to both Gram-positive and Gram-negative bacteria. The Gram-positive bacteria that have been identified include Staphylococcus saprophyticus, Enterobacter, Staphylococci, Streptococcus, Enterococcus, Actinobaculum schaali, Lactobacillus, or Bifidobacterium, Enterococcus faecalis, Aerococcus urinae, Corynebacterium urealyticum, Bacillus, and Rothia. Gram-positive bacteria, such as Staphylococcus and Streptococcus, have been found to be prevalent in the majority of UTIs. The Gram-negative bacteria included in this list are Escherichia coli, Mycoplasma genitalium, Pseudomonas aeruginosa, Proteus mirabilis, Acinetobacter, Citrobacter, Prevotella, Gardnerella, Pseudomonas fluorescens, Chryseobacterium, Enhydrobacter, Paracoccus, Acinetobacter, Enterobacteriaceae, and Sphingomonas. E. coli is the most commonly identified Gram-negative bacterium in the majority of UTIs. Other than this, the protozoa Trichomonas vaginalis was also responsible for causing UTIs (Table 4).
UTIs causing microorganisms detected by PCR and NGS
| Type of micro-organism | Name of micro-organisms detected |
|---|---|
| Gram-positive bacteria | S. saprophyticus, Enterobacter, Staphylococci, Streptococcus, Enterococcus, Actinobaculum schaali, Lactobacillus, or Bifidobacterium, Enterococcus faecalis, Aerococcus urinae, Corynebacterium urealyticum, Bacillus, and Rothia |
| Gram-negative bacteria | Escherichia coli, Mycoplasma genitalium, Pseudomonas aeruginosa, Proteus mirabilis, Acinetobacter, Citrobacter, Prevotella, Gardnerella, Pseudomonas fluorescens, Chryseobacterium, Enhydrobacter, Paracoccus, Acinetobacter, Enterobacteriaceae, and Sphingomonas |
| Protozoa | Trichomonas vaginalis |
4 Discussion
In recent years, the utilization of molecular-based microbial profiling in the evaluation of UTIs has gained a substantial amount of relevance. PCR and NGS offer valuable diagnostic tools that have the potential to alleviate the ongoing cycle of aggravation and discomfort experienced by patients suffering from persistent UTIs [38]. NGS provides a highly complete assessment of the urine microbiome and examines the whole of microbial DNA present in a urine sample and subsequently compares it to a comprehensive database of species [39]. Similarly, the Urine PCR test distinguishes the existence of bacteria in a distinct manner. The utilization of a multiplex PCR) test enables the identification of a greater number of microbial species compared to the conventional urine culture method in individuals displaying symptoms indicative of a UTI [40]. The misapplication and misuse of antibiotics accelerates the development of antibiotic resistance [41]. PCR and NGS tests are characterized by their rapid detection rate, heightened sensitivity, and remarkable accuracy in identifying the bacteria responsible for UTIs. Consequently, these tests effectively tackle the problem of antibiotic resistance, enabling healthcare practitioners to provide informed recommendations regarding the appropriate choice of antibiotics and their optimal length of administration [42]. PCR possesses the capability to identify the presence of a pathogen responsible for symptoms, as opposed to genetic material of a microorganism that is clinically insignificant [43]. By amplifying specific segments of DNA, PCR enables the detection and characterization of target microorganisms at the species, strain, and serovar/pathovar levels. The method may also be employed to characterize whole populations of microorganisms in samples [44]. NGS offers in-depth strain genotyping and antibiotic resistance surveillance in some pathogens, such as Streptococcus pneumoniae, and is a very reliable predictor of antimicrobial-resistant status in others. Unlike PCR, NGS gives information about a sample’s entire set of genetic, regulatory, and biological properties. A urine culture can take up to seven days to provide results, whereas a PCR urine test has a quick turnaround time; results are often available in a day, and PCR costs roughly $5 per test. Simultaneously, the total turnaround time for identifying pathogens by mNGS testing is approximately 4 h, which is substantially faster than normal urine culture testing but slightly more expensive at $200 per test. Nevertheless, the utilization of PCR and NGS techniques remains valuable. However, it is important to acknowledge that traditional urine culture methods depend on the growth of live bacteria to identify species, whereas molecular-based techniques such as NGS and PCR do not [45]. The purpose of this meta-analysis was to investigate whether or not NGS and PCR are more accurate than the traditional approach of urine culture in identifying UTIs. Based on the findings of our study, it was found that both NGS and PCR techniques exhibit high levels of accuracy, sensitivity, and specificity in the diagnosis of UTIs. The study determined that the sensitivity of PCR in detecting UTIs was 99%, with a specificity of 94%. In contrast, the sensitivity of NGS in detecting UTIs was found to be 90%, with a specificity of 86%. The culture-free molecular-based methods have been found to exhibit a higher likelihood of detecting various types of Gram-positive and Gram-negative bacteria. This includes the accurate detection of Gram-positive bacteria such as S. saprophyticus, Enterobacter, Staphylococci, Streptococcus, Aerococcus urinae, Corynebacterium urealyticum, among others, as well as Gram-negative bacteria such as Escherichia coli, Mycoplasma genitalium, Pseudomonas, Proteus mirabilis, Chryseobacterium, and others. The prevalence of UTIs has been observed to be primarily associated with Gram-positive bacteria, including Staphylococcus and Streptococcus, as well as Gram-negative Escherichia coli. In line with our findings, Gasiorek et al. (2020) [46] conducted a review study in which they observed that molecular-based techniques, such as NGS and PCR, offer the potential to enhance patient assessment and management by effectively evaluating the urinary microbiome. In addition, Xu et al. (2021) [47], Dixon et al. (2020) [48], Szlachta-McGinn et al. (2022) [49], and Behzadi et al. (2019) [50] also advocate for the use of NGS and PCR techniques in the identification of UTIs). Nevertheless, there remains a need for novel methodologies to assess the merits and drawbacks of these contemporary and emerging diagnostic techniques, as well as to supplant the conventional urine culture method, which presently serves as the benchmark for diagnosing UTIs.
5 Limitations
The present investigation is limited by the diversity of PCR and NGS instruments as well as the variability of DNA extraction kit tools. Additionally, the involvement of different technicians introduces the potential for human error, thereby increasing the likelihood of false-negative outcomes. The present study exclusively focused on English-language publications, which potentially introduces a selection bias. Furthermore, the present analysis was executed with meticulous adherence to scientific protocols, and it is important to acknowledge that the findings are constrained due to the utilization of only 10 comparative studies characterized by varying degrees of heterogeneity, ranging from moderate to high levels. In the context of this meta-analysis, it would be advantageous to possess the capability to examine a diverse range of study-specific attributes that may be linked to the observed variations in reported outcomes and could provide further elucidation on the importance and effectiveness of culture-free molecular diagnostic-based PCR and NGS in the identification of UTIs.
6 Conclusion
Culture-independent molecular technologies such as NGS and PCR are widely utilized in the commercial diagnosis of UTIs due to reports of limited sensitivity of urine cultures. Increased sensitivity and specificity in the detection of urine bacteria are supported by moderate evidence. Therefore, in order to further substantiate these pieces of evidence, we conducted a comparative analysis between culture-independent molecular methods and conventional urine culture. The findings of a meta-analysis suggest that PCR and NGS exhibit considerable sensitivity in the detection of UTIs. However, further research is required to ascertain whether their routine application is supported by clinical implications and to compare patient symptoms and cure rates subsequent to antibiotic selection guided by molecular methods versus traditional urine culture.
Abbreviations
- UTIs
-
Urinary tract infections
- PCR
-
Polymerase chain reaction
- NGS
-
Next-generation sequencing
- AOR
-
Adjusted odds ratio
Acknowledgement
None.
-
Funding information: The funding is based on PI3K/Akt/PSD-95 Fund, i.e., Signaling Pathway to Explore the Protective Mechanism of Electronic Twisting Needling Method on Hippocampal Synaptic Plasticity in Rats with Epilepsy and Depression.
-
Author contributions: MZ: concept and designed the study; SQ: analyzed data and drafting of the manuscript; YS: collected the data and helped in data analysis; XZ: proofreading and final editing along with guarantor of the manuscript.
-
Conflict of interest: The authors have no conflicts of interest to declare.
-
Data availability statement: Upon a reasonable request, the corresponding author will provide access to the requested information.
Appendix
Database search strategy
| Database | Search strategy |
|---|---|
| PubMed | #1 “Urinary tract infections” OR “UTI” OR “Polymerase chain reaction” [MeSH Terms] # OR “PCR” OR “Next generation sequencing” [All Fields] OR “NGS” OR “Gram positive bacteria” OR “Gram negative bacteria” [All Fields]” OR “Conventional urine culture” OR “Molecular diagnostic methods” [All Fields] OR “meta-analysis” OR “systematic review” [All Fields] |
| #2 “Positive UTI cases,” OR “type of bacteria,” [MeSH Terms] OR “sensitivity,” OR “specificity,” OR “diagnostic odds ratio,” [All Fields] | |
| #3 #1 AND #2 | |
| Embase | #1 “Urinary tract infections” / exp$ OR “UTI”/ exp OR “Polymerase chain reaction”/exp OR “PCR”/exp OR “Next generation sequencing”/ exp OR “NGS”/ exp OR “Gram positive bacteria” /exp OR “Gram negative bacteria”/ exp OR “Conventional urine culture” / exp OR “Molecular diagnostic methods”/exp OR “meta-analysis”/exp OR “systematic review”/exp |
| #2 “Positive UTI cases” / exp OR “type of bacteria”/ exp OR “sensitivity”/ exp OR “specificity” /exp OR “diagnostic odds ratio” exp | |
| #3 #1 AND #2 | |
| Cochrane library | #1 ( Urinary tract infections): ti, ab, kw@ OR ( UTI): ti, ab, kw OR ( Polymerase chain reaction): ti, ab, kw OR ( PCR): ti, ab, kw OR ( Next generation sequencing): ti, ab, kw OR ( NGS): ti, ab, kw OR ( Gram positive bacteria): ti, ab, kw OR ( Gram negative bacteria): ti, ab, kw OR ( Conventional urine culture):OR ( Molecular diagnostic methods): ti, ab, kw OR (meta-analysis) ti, ab, kw (Word variations have been searched) |
| #2 (Positive UTI cases): ti, ab, kw OR (type of bacteria): ti, ab, kw OR (sensitivity): ti, ab, kw or (specificity): ti, ab, kw or (diagnostic odds ratio): ti, ab, kw (Word variations have been searched) | |
| #3 #1 AND #2 |
#MeSH terms: Medical Subject Headings; $ exp: explosion in Emtree- searching of selected subject terms and related subjects; @ ti, ab, kw: either title or abstract or keyword fields.
References
[1] Gupta K, Grigoryan L, Trautner B. Urinary tract infection. Ann Intern Med. 2017;167(7):ITC49–64. 10.7326/AITC201710030.Search in Google Scholar PubMed
[2] Czajkowski K, Broś-Konopielko M, Teliga-Czajkowska J. Urinary tract infection in women. Prz Menopauzalny. 2021;20(1):40–7. 10.5114/pm.2021.105382.Search in Google Scholar PubMed PubMed Central
[3] Bader MS, Loeb M, Leto D, Brooks AA. Treatment of urinary tract infections in the era of antimicrobial resistance and new antimicrobial agents. Postgrad Med. 2020;132(3):234–50. 10.1080/00325481.2019.1680052.Search in Google Scholar PubMed
[4] Chu CM, Lowder JL. Diagnosis and treatment of urinary tract infections across age groups. Am J Obstet Gynecol. 2018;219(1):40–51. 10.1016/j.ajog.2017.12.231.Search in Google Scholar PubMed
[5] Jancel T, Dudas V. Management of uncomplicated urinary tract infections. West J Med. 2002;176(1):51–5. 10.1136/ewjm.176.1.51.Search in Google Scholar PubMed PubMed Central
[6] Mouraviev V, McDonald M. An implementation of next generation sequencing for prevention and diagnosis of urinary tract infection in urology. Can J Urol. 2018;25(3):9349–56.Search in Google Scholar
[7] Pirkani GS, Awan MA, Abbas F, Din M. Culture and PCR based detection of bacteria causing urinary tract infection in urine specimen. Pak J Med Sci. 2020;36(3):391–5. 10.12669/pjms.36.3.1577.Search in Google Scholar PubMed PubMed Central
[8] Sharma B, Mohan B, Sharma R, Lakhanpal V, Shankar P, Singh SK, et al. Evaluation of an automated rapid urine culture method for urinary tract infection: Comparison with gold standard conventional culture method. Indian J Med Microbiol. 2023;42:19–24. 10.1016/j.ijmmb.2023.01.003.Search in Google Scholar PubMed
[9] Almas S, Carpenter RE, Rowan C, Tamrakar VK, Bishop J, Sharma R. Advantage of precision metagenomics for urinary tract infection diagnostics. Front Cell Infect Microbiol. 2023;13:1221289. 10.3389/fcimb.2023.1221289.Search in Google Scholar PubMed PubMed Central
[10] Hinata N, Shirakawa T, Okada H, Shigemura K, Kamidono S, Gotoh A. Quantitative detection of Escherichia coli from urine of patients with bacteriuria by real-time PCR. Mol Diagn. 2004;8(3):179–84. 10.1007/BF03260062.Search in Google Scholar PubMed
[11] Kelly BN. UTI detection by PCR: Improving patient outcomes. J Mass Spectrom Adv Clin Lab. 2023;28:60–2. 10.1016/j.jmsacl.2023.02.006.Search in Google Scholar PubMed PubMed Central
[12] Huang R, Yuan Q, Gao J, Liu Y, Jin X, Tang L, et al. Application of metagenomic next-generation sequencing in the diagnosis of urinary tract infection in patients undergoing cutaneous ureterostomy. Front Cell Infect Microbiol. 2023;13:991011. 10.3389/fcimb.2023.991011.Search in Google Scholar PubMed PubMed Central
[13] Kotaskova I, Obrucova H, Malisova B, Videnska P, Zwinsova B, Peroutkova T, et al. Molecular techniques complement culture-based assessment of bacteria composition in mixed biofilms of urinary tract catheter-related samples. Front Microbiol. 2019;10:462. 10.3389/fmicb.2019.00462.Search in Google Scholar PubMed PubMed Central
[14] Harris M, Fasolino T. New and emerging technologies for the diagnosis of urinary tract infections. J Labor Med. 2022;46(1):3–15. 10.1515/labmed-2021-0085.Search in Google Scholar
[15] Korman HJ, Baunoch D, Luke N, Wang D, Zhao X, Levin M, et al. A diagnostic test combining molecular testing with phenotypic pooled antibiotic susceptibility improved the clinical outcomes of patients with non-E. coli or polymicrobial complicated urinary tract infections. Res Rep Urol. 2023;15:141–7. 10.2147/RRU.S404260.Search in Google Scholar PubMed PubMed Central
[16] Heytens S, De Sutter A, Coorevits L, Cools P, Boelens J, Van Simaey L, et al. Women with symptoms of a urinary tract infection but a negative urine culture: PCR-based quantification of Escherichia coli suggests infection in most cases. Clin Microbiol Infect. 2017;23(9):647–52. 10.1016/j.cmi.2017.04.004.Search in Google Scholar PubMed
[17] Ibraheam IA, Saleh IA, Kadhim HJ, Al-Mahdi ZK. Molecular diagnostic of Escherichia Coli among urninary tract infection patients using Polymerase chain reaction (PCR). Pak J Biotechnol. 2016;13(4):275–8.Search in Google Scholar
[18] Lehmann LE, Hauser S, Malinka T, Klaschik S, Weber SU, Schewe JC, et al. Rapid qualitative urinary tract infection pathogen identification by SeptiFast real-time PCR. PLoS One. 2011;6(2):e17146. 10.1371/journal.pone.0017146.Search in Google Scholar PubMed PubMed Central
[19] Wojno KJ, Baunoch D, Luke N, Opel M, Korman H, Kelly C, et al. Multiplex PCR based urinary tract infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology. 2020;136:119–26. 10.1016/j.urology.2019.10.018.Search in Google Scholar PubMed
[20] van der Zee A, Roorda L, Bosman G, Ossewaarde JM. Molecular diagnosis of urinary tract infections by semi-quantitative detection of uropathogens in a routine clinical hospital setting. PLoS One. 2016;11(3):e0150755. 10.1371/journal.pone.0150755.Search in Google Scholar PubMed PubMed Central
[21] Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, Frimodt-Møller N, et al. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol. 2014;52(1):139–46. 10.1128/JCM.02452-13.Search in Google Scholar PubMed PubMed Central
[22] Ishihara T, Watanabe N, Inoue S, Aoki H, Tsuji T, Yamamoto B, et al. Usefulness of next-generation DNA sequencing for the diagnosis of urinary tract infection. Drug Discov Ther. 2020;14(1):42–9. 10.5582/ddt.2020.01000.Search in Google Scholar PubMed
[23] McDonald M, Kameh D, Johnson ME, Johansen TEB, Albala D, Mouraviev V. A head-to-head comparative phase II study of standard urine culture and sensitivity versus DNA next-generation sequencing testing for urinary tract infections. Rev Urol. 2017;19(4):213–20. 10.3909/riu0780.Search in Google Scholar PubMed PubMed Central
[24] Sabat AJ, van Zanten E, Akkerboom V, Wisselink G, van Slochteren K, de Boer RF, et al. Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification - increased discrimination of closely related species. Sci Rep. 2017;7(1):3434. 10.1038/s41598-017-03458-6.Search in Google Scholar PubMed PubMed Central
[25] Yoo JJ, Shin HB, Song JS, Kim M, Yun J, Kim Z, et al. Urinary microbiome characteristics in female patients with acute uncomplicated cystitis and recurrent cystitis. J Clin Med. 2021 Mar;10(5):1097. 10.3390/jcm10051097.Search in Google Scholar PubMed PubMed Central
[26] Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. 10.1136/bmj.n71.Search in Google Scholar PubMed PubMed Central
[27] Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11(2):193–206. 10.1037/1082-989X.11.2.193.Search in Google Scholar PubMed
[28] Schmidt L, Shokraneh F, Steinhausen K, Adams CE. Introducing RAPTOR: RevMan parsing tool for reviewers. Syst Rev. 2019;8(1):151. 10.1186/s13643-019-1070-0.Search in Google Scholar PubMed PubMed Central
[29] McGuinness LA, Higgins JPT. Risk-of-bias visualization (Robvis): An R package and Shiny web app for visualizing risk-of-bias assessments. Res Syn Meth. 2021;12(1):55–61. 10.1002/jrsm.1411.Search in Google Scholar PubMed
[30] Lachin JM. Power of the Mantel-Haenszel and other tests for discrete or grouped time-to-event data under a chained binomial model. Stat Med. 2013;32(2):220–9. 10.1002/sim.5480.Search in Google Scholar PubMed PubMed Central
[31] Trevethan R. Sensitivity, specificity, and predictive values: Foundations, pliabilities, and pitfalls in research and practice. Front Public Health. 2017;5:307. 10.3389/fpubh.2017.00307.Search in Google Scholar PubMed PubMed Central
[32] George BJ, Aban IB. An application of meta-analysis based on DerSimonian and Laird method. J Nucl Cardiol. 2016;23:690–2. 10.1007/s12350-015-0249-6.Search in Google Scholar PubMed
[33] van Enst WA, Ochodo E, Scholten RJ, Hooft L, Leeflang MM. Investigation of publication bias in meta-analyses of diagnostic test accuracy: a meta-epidemiological study. BMC Med Res Methodol. 2014;14:70. 10.1186/1471-2288-14-70.Search in Google Scholar PubMed PubMed Central
[34] Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. 2001;54(10):1046–55. 10.1016/s0895-4356(01)00377-8.Search in Google Scholar PubMed
[35] Elovic A, Pourmand A. MDCalc medical calculator app review. J Digit Imaging. 2019 Oct;32(5):682–4. 10.1007/s10278-019-00218-y.Search in Google Scholar PubMed PubMed Central
[36] Godavitarne C, Robertson A, Ricketts DM, Rogers BA. Understanding and interpreting funnel plots for the clinician. Br J Hosp Med (Lond). 2018;79(10):578–83. 10.12968/hmed.2018.79.10.578.Search in Google Scholar PubMed
[37] Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.10.2307/2533446Search in Google Scholar
[38] Guo Z, Tian S, Wang W, Zhang Y, Li J, Lin R. Correlation analysis among genotype resistance, phenotype resistance, and eradication effect after resistance-guided quadruple therapies in refractory Helicobacter pylori infections. Front Microbiol. 2022;13:861626. 10.3389/fmicb.2022.861626.Search in Google Scholar PubMed PubMed Central
[39] Cason C, D’Accolti M, Soffritti I, Mazzacane S, Comar M, Caselli E. Next-generation sequencing and PCR technologies in monitoring the hospital microbiome and its drug resistance. Front Microbiol. 2022;13:969863. 10.3389/fmicb.2022.969863.Search in Google Scholar PubMed PubMed Central
[40] Gleicher S, Karram M, Wein AJ, Dmochowski RR. Recurrent and complicated urinary tract infections in women: Utility of advanced testing to enhance care. Neurourol Urodyn. 2024 Jan;43(1):161–6. 10.1002/nau.25280.Search in Google Scholar PubMed
[41] Bader MS, Loeb M, Leto D, Brooks AA. Treatment of urinary tract infections in the era of antimicrobial resistance and new antimicrobial agents. Postgrad Med. 2020 Apr;132(3):234–50. 10.1080/00325481.2019.1680052.Search in Google Scholar PubMed
[42] Ward DG, Baxter L, Gordon NS, Ott S, Savage RS, Beggs AD, et al. Multiplex PCR and next generation sequencing for the non-invasive detection of bladder cancer. PLoS One. 2016 Feb;11(2):e0149756. 10.1371/journal.pone.0149756.Search in Google Scholar PubMed PubMed Central
[43] Melnyk AI, Toal C, Glass Clark S, Bradley M. Home urinary tract infection testing: patient experience and satisfaction with polymerase chain reaction kit. Int Urogynecol J. 2023 May;34(5):1055–60. 10.1007/s00192-022-05309-z.Search in Google Scholar PubMed PubMed Central
[44] Hansen WLJ, van der Donk CFM, Bruggeman CA, Stobberingh EE, Wolffs PFG. A real-time PCR-based semi-quantitative breakpoint to aid in molecular identification of urinary tract infections. PLoS ONE. 2013;8(4):e61439. 10.1371/journal.pone.0061439.Search in Google Scholar PubMed PubMed Central
[45] Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, García-Cobos S, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017 Feb;243:16–24. 10.1016/j.jbiotec.2016.12.022.Search in Google Scholar PubMed
[46] Gasiorek M, Hsieh MH, Forster CS. Utility of DNA next-generation sequencing and expanded quantitative urine culture in diagnosis and management of chronic or persistent lower urinary tract symptoms. J Clin Microbiol. 2019;58(1):e00204-19. 10.1128/JCM.00204-19.Search in Google Scholar PubMed PubMed Central
[47] Xu R, Deebel N, Casals R, Dutta R, Mirzazadeh M. A new gold rush: A review of current and developing diagnostic tools for urinary tract infections. Diagnostics (Basel). 2021;11(3):479. 10.3390/diagnostics11030479.Search in Google Scholar PubMed PubMed Central
[48] Dixon M, Stefil M, McDonald M, Bjerklund-Johansen TE, Naber K, Wagenlehner F, et al. Metagenomics in diagnosis and improved targeted treatment of UTI. World J Urol. 2020;38(1):35–43. 10.1007/s00345-019-02731-9.Search in Google Scholar PubMed
[49] Szlachta-McGinn A, Douglass KM, Chung UYR, Jackson NJ, Nickel JC, Ackerman AL. Molecular diagnostic methods versus conventional urine culture for diagnosis and treatment of urinary tract infection: A systematic review and meta-analysis. Eur Urol Open Sci. 2022;44:113–24. 10.1016/j.euros.2022.08.009.Search in Google Scholar PubMed PubMed Central
[50] Behzadi P, Behzadi E, Pawlak-Adamska EA. Urinary tract infections (UTIs) or genital tract infections (GTIs)? It’s the diagnostics that count. GMS Hyg Infect Control. 2019;14:Doc14. 10.3205/dgkh000320.Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- EDNRB inhibits the growth and migration of prostate cancer cells by activating the cGMP-PKG pathway
- STK11 (LKB1) mutation suppresses ferroptosis in lung adenocarcinoma by facilitating monounsaturated fatty acid synthesis
- Association of SOX6 gene polymorphisms with Kashin-Beck disease risk in the Chinese Han population
- The pyroptosis-related signature predicts prognosis and influences the tumor immune microenvironment in dedifferentiated liposarcoma
- METTL3 attenuates ferroptosis sensitivity in lung cancer via modulating TFRC
- Identification and validation of molecular subtypes and prognostic signature for stage I and stage II gastric cancer based on neutrophil extracellular traps
- Novel lumbar plexus block versus femoral nerve block for analgesia and motor recovery after total knee arthroplasty
- Correlation between ABCB1 and OLIG2 polymorphisms and the severity and prognosis of patients with cerebral infarction
- Study on the radiotherapy effect and serum neutral granulocyte lymphocyte ratio and inflammatory factor expression of nasopharyngeal carcinoma
- Transcriptome analysis of effects of Tecrl deficiency on cardiometabolic and calcium regulation in cardiac tissue
- Aflatoxin B1 induces infertility, fetal deformities, and potential therapies
- Serum levels of HMW adiponectin and its receptors are associated with cytokine levels and clinical characteristics in chronic obstructive pulmonary disease
- METTL3-mediated methylation of CYP2C19 mRNA may aggravate clopidogrel resistance in ischemic stroke patients
- Understand how machine learning impact lung cancer research from 2010 to 2021: A bibliometric analysis
- Pressure ulcers in German hospitals: Analysis of reimbursement and length of stay
- Metformin plus L-carnitine enhances brown/beige adipose tissue activity via Nrf2/HO-1 signaling to reduce lipid accumulation and inflammation in murine obesity
- Downregulation of carbonic anhydrase IX expression in mouse xenograft nasopharyngeal carcinoma model via doxorubicin nanobubble combined with ultrasound
- Feasibility of 3-dimensional printed models in simulated training and teaching of transcatheter aortic valve replacement
- miR-335-3p improves type II diabetes mellitus by IGF-1 regulating macrophage polarization
- The analyses of human MCPH1 DNA repair machinery and genetic variations
- Activation of Piezo1 increases the sensitivity of breast cancer to hyperthermia therapy
- Comprehensive analysis based on the disulfidptosis-related genes identifies hub genes and immune infiltration for pancreatic adenocarcinoma
- Changes of serum CA125 and PGE2 before and after high-intensity focused ultrasound combined with GnRH-a in treatment of patients with adenomyosis
- The clinical value of the hepatic venous pressure gradient in patients undergoing hepatic resection for hepatocellular carcinoma with or without liver cirrhosis
- Development and validation of a novel model to predict pulmonary embolism in cardiology suspected patients: A 10-year retrospective analysis
- Downregulation of lncRNA XLOC_032768 in diabetic patients predicts the occurrence of diabetic nephropathy
- Circ_0051428 targeting miR-885-3p/MMP2 axis enhances the malignancy of cervical cancer
- Effectiveness of ginkgo diterpene lactone meglumine on cognitive function in patients with acute ischemic stroke
- The construction of a novel prognostic prediction model for glioma based on GWAS-identified prognostic-related risk loci
- Evaluating the impact of childhood BMI on the risk of coronavirus disease 2019: A Mendelian randomization study
- Lactate dehydrogenase to albumin ratio is associated with in-hospital mortality in patients with acute heart failure: Data from the MIMIC-III database
- CD36-mediated podocyte lipotoxicity promotes foot process effacement
- Efficacy of etonogestrel subcutaneous implants versus the levonorgestrel-releasing intrauterine system in the conservative treatment of adenomyosis
- FLRT2 mediates chondrogenesis of nasal septal cartilage and mandibular condyle cartilage
- Challenges in treating primary immune thrombocytopenia patients undergoing COVID-19 vaccination: A retrospective study
- Let-7 family regulates HaCaT cell proliferation and apoptosis via the ΔNp63/PI3K/AKT pathway
- Phospholipid transfer protein ameliorates sepsis-induced cardiac dysfunction through NLRP3 inflammasome inhibition
- Postoperative cognitive dysfunction in elderly patients with colorectal cancer: A randomized controlled study comparing goal-directed and conventional fluid therapy
- Long-pulsed ultrasound-mediated microbubble thrombolysis in a rat model of microvascular obstruction
- High SEC61A1 expression predicts poor outcome of acute myeloid leukemia
- Comparison of polymerase chain reaction and next-generation sequencing with conventional urine culture for the diagnosis of urinary tract infections: A meta-analysis
- Secreted frizzled-related protein 5 protects against renal fibrosis by inhibiting Wnt/β-catenin pathway
- Pan-cancer and single-cell analysis of actin cytoskeleton genes related to disulfidptosis
- Overexpression of miR-532-5p restrains oxidative stress response of chondrocytes in nontraumatic osteonecrosis of the femoral head by inhibiting ABL1
- Autologous liver transplantation for unresectable hepatobiliary malignancies in enhanced recovery after surgery model
- Clinical analysis of incomplete rupture of the uterus secondary to previous cesarean section
- Abnormal sleep duration is associated with sarcopenia in older Chinese people: A large retrospective cross-sectional study
- No genetic causality between obesity and benign paroxysmal vertigo: A two-sample Mendelian randomization study
- Identification and validation of autophagy-related genes in SSc
- Long non-coding RNA SRA1 suppresses radiotherapy resistance in esophageal squamous cell carcinoma by modulating glycolytic reprogramming
- Evaluation of quality of life in patients with schizophrenia: An inpatient social welfare institution-based cross-sectional study
- The possible role of oxidative stress marker glutathione in the assessment of cognitive impairment in multiple sclerosis
- Compilation of a self-management assessment scale for postoperative patients with aortic dissection
- Left atrial appendage closure in conjunction with radiofrequency ablation: Effects on left atrial functioning in patients with paroxysmal atrial fibrillation
- Effect of anterior femoral cortical notch grade on postoperative function and complications during TKA surgery: A multicenter, retrospective study
- Clinical characteristics and assessment of risk factors in patients with influenza A-induced severe pneumonia after the prevalence of SARS-CoV-2
- Analgesia nociception index is an indicator of laparoscopic trocar insertion-induced transient nociceptive stimuli
- High STAT4 expression correlates with poor prognosis in acute myeloid leukemia and facilitates disease progression by upregulating VEGFA expression
- Factors influencing cardiovascular system-related post-COVID-19 sequelae: A single-center cohort study
- HOXD10 regulates intestinal permeability and inhibits inflammation of dextran sulfate sodium-induced ulcerative colitis through the inactivation of the Rho/ROCK/MMPs axis
- Mesenchymal stem cell-derived exosomal miR-26a induces ferroptosis, suppresses hepatic stellate cell activation, and ameliorates liver fibrosis by modulating SLC7A11
- Endovascular thrombectomy versus intravenous thrombolysis for primary distal, medium vessel occlusion in acute ischemic stroke
- ANO6 (TMEM16F) inhibits gastrointestinal stromal tumor growth and induces ferroptosis
- Prognostic value of EIF5A2 in solid tumors: A meta-analysis and bioinformatics analysis
- The role of enhanced expression of Cx43 in patients with ulcerative colitis
- Choosing a COVID-19 vaccination site might be driven by anxiety and body vigilance
- Role of ICAM-1 in triple-negative breast cancer
- Cost-effectiveness of ambroxol in the treatment of Gaucher disease type 2
- HLA-DRB5 promotes immune thrombocytopenia via activating CD8+ T cells
- Efficacy and factors of myofascial release therapy combined with electrical and magnetic stimulation in the treatment of chronic pelvic pain syndrome
- Efficacy of tacrolimus monotherapy in primary membranous nephropathy
- Mechanisms of Tripterygium wilfordii Hook F on treating rheumatoid arthritis explored by network pharmacology analysis and molecular docking
- FBXO45 levels regulated ferroptosis renal tubular epithelial cells in a model of diabetic nephropathy by PLK1
- Optimizing anesthesia strategies to NSCLC patients in VATS procedures: Insights from drug requirements and patient recovery patterns
- Alpha-lipoic acid upregulates the PPARγ/NRF2/GPX4 signal pathway to inhibit ferroptosis in the pathogenesis of unexplained recurrent pregnancy loss
- Correlation between fat-soluble vitamin levels and inflammatory factors in paediatric community-acquired pneumonia: A prospective study
- CD1d affects the proliferation, migration, and apoptosis of human papillary thyroid carcinoma TPC-1 cells via regulating MAPK/NF-κB signaling pathway
- miR-let-7a inhibits sympathetic nerve remodeling after myocardial infarction by downregulating the expression of nerve growth factor
- Immune response analysis of solid organ transplantation recipients inoculated with inactivated COVID-19 vaccine: A retrospective analysis
- The H2Valdien derivatives regulate the epithelial–mesenchymal transition of hepatoma carcinoma cells through the Hedgehog signaling pathway
- Clinical efficacy of dexamethasone combined with isoniazid in the treatment of tuberculous meningitis and its effect on peripheral blood T cell subsets
- Comparison of short-segment and long-segment fixation in treatment of degenerative scoliosis and analysis of factors associated with adjacent spondylolisthesis
- Lycopene inhibits pyroptosis of endothelial progenitor cells induced by ox-LDL through the AMPK/mTOR/NLRP3 pathway
- Methylation regulation for FUNDC1 stability in childhood leukemia was up-regulated and facilitates metastasis and reduces ferroptosis of leukemia through mitochondrial damage by FBXL2
- Correlation of single-fiber electromyography studies and functional status in patients with amyotrophic lateral sclerosis
- Risk factors of postoperative airway obstruction complications in children with oral floor mass
- Expression levels and clinical significance of serum miR-19a/CCL20 in patients with acute cerebral infarction
- Physical activity and mental health trends in Korean adolescents: Analyzing the impact of the COVID-19 pandemic from 2018 to 2022
- Evaluating anemia in HIV-infected patients using chest CT
- Ponticulus posticus and skeletal malocclusion: A pilot study in a Southern Italian pre-orthodontic court
- Causal association of circulating immune cells and lymphoma: A Mendelian randomization study
- Assessment of the renal function and fibrosis indexes of conventional western medicine with Chinese medicine for dredging collaterals on treating renal fibrosis: A systematic review and meta-analysis
- Comprehensive landscape of integrator complex subunits and their association with prognosis and tumor microenvironment in gastric cancer
- New target-HMGCR inhibitors for the treatment of primary sclerosing cholangitis: A drug Mendelian randomization study
- Population pharmacokinetics of meropenem in critically ill patients
- Comparison of the ability of newly inflammatory markers to predict complicated appendicitis
- Comparative morphology of the cruciate ligaments: A radiological study
- Immune landscape of hepatocellular carcinoma: The central role of TP53-inducible glycolysis and apoptosis regulator
- Serum SIRT3 levels in epilepsy patients and its association with clinical outcomes and severity: A prospective observational study
- SHP-1 mediates cigarette smoke extract-induced epithelial–mesenchymal transformation and inflammation in 16HBE cells
- Acute hyper-hypoxia accelerates the development of depression in mice via the IL-6/PGC1α/MFN2 signaling pathway
- The GJB3 correlates with the prognosis, immune cell infiltration, and therapeutic responses in lung adenocarcinoma
- Physical fitness and blood parameters outcomes of breast cancer survivor in a low-intensity circuit resistance exercise program
- Exploring anesthetic-induced gene expression changes and immune cell dynamics in atrial tissue post-coronary artery bypass graft surgery
- Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism
- Analysis of the risk factors of the radiation-induced encephalopathy in nasopharyngeal carcinoma: A retrospective cohort study
- Reproductive outcomes in women with BRCA 1/2 germline mutations: A retrospective observational study and literature review
- Evaluation of upper airway ultrasonographic measurements in predicting difficult intubation: A cross-section of the Turkish population
- Prognostic and diagnostic value of circulating IGFBP2 in pancreatic cancer
- Postural stability after operative reconstruction of the AFTL in chronic ankle instability comparing three different surgical techniques
- Research trends related to emergence agitation in the post-anaesthesia care unit from 2001 to 2023: A bibliometric analysis
- Frequency and clinicopathological correlation of gastrointestinal polyps: A six-year single center experience
- ACSL4 mediates inflammatory bowel disease and contributes to LPS-induced intestinal epithelial cell dysfunction by activating ferroptosis and inflammation
- Affibody-based molecular probe 99mTc-(HE)3ZHER2:V2 for non-invasive HER2 detection in ovarian and breast cancer xenografts
- Effectiveness of nutritional support for clinical outcomes in gastric cancer patients: A meta-analysis of randomized controlled trials
- The relationship between IFN-γ, IL-10, IL-6 cytokines, and severity of the condition with serum zinc and Fe in children infected with Mycoplasma pneumoniae
- Paraquat disrupts the blood–brain barrier by increasing IL-6 expression and oxidative stress through the activation of PI3K/AKT signaling pathway
- Sleep quality associate with the increased prevalence of cognitive impairment in coronary artery disease patients: A retrospective case–control study
- Dioscin protects against chronic prostatitis through the TLR4/NF-κB pathway
- Association of polymorphisms in FBN1, MYH11, and TGF-β signaling-related genes with susceptibility of sporadic thoracic aortic aneurysm and dissection in the Zhejiang Han population
- Application value of multi-parameter magnetic resonance image-transrectal ultrasound cognitive fusion in prostate biopsy
- Laboratory variables‐based artificial neural network models for predicting fatty liver disease: A retrospective study
- Decreased BIRC5-206 promotes epithelial–mesenchymal transition in nasopharyngeal carcinoma through sponging miR-145-5p
- Sepsis induces the cardiomyocyte apoptosis and cardiac dysfunction through activation of YAP1/Serpine1/caspase-3 pathway
- Assessment of iron metabolism and iron deficiency in incident patients on incident continuous ambulatory peritoneal dialysis
- Tibial periosteum flap combined with autologous bone grafting in the treatment of Gustilo-IIIB/IIIC open tibial fractures
- The application of intravenous general anesthesia under nasopharyngeal airway assisted ventilation undergoing ureteroscopic holmium laser lithotripsy: A prospective, single-center, controlled trial
- Long intergenic noncoding RNA for IGF2BP2 stability suppresses gastric cancer cell apoptosis by inhibiting the maturation of microRNA-34a
- Role of FOXM1 and AURKB in regulating keratinocyte function in psoriasis
- Parental control attitudes over their pre-school children’s diet
- The role of auto-HSCT in extranodal natural killer/T cell lymphoma
- Significance of negative cervical cytology and positive HPV in the diagnosis of cervical lesions by colposcopy
- Echinacoside inhibits PASMCs calcium overload to prevent hypoxic pulmonary artery remodeling by regulating TRPC1/4/6 and calmodulin
- ADAR1 plays a protective role in proximal tubular cells under high glucose conditions by attenuating the PI3K/AKT/mTOR signaling pathway
- The risk of cancer among insulin glargine users in Lithuania: A retrospective population-based study
- The unusual location of primary hydatid cyst: A case series study
- Intraoperative changes in electrophysiological monitoring can be used to predict clinical outcomes in patients with spinal cavernous malformation
- Obesity and risk of placenta accreta spectrum: A meta-analysis
- Shikonin alleviates asthma phenotypes in mice via an airway epithelial STAT3-dependent mechanism
- NSUN6 and HTR7 disturbed the stability of carotid atherosclerotic plaques by regulating the immune responses of macrophages
- The effect of COVID-19 lockdown on admission rates in Maternity Hospital
- Temporal muscle thickness is not a prognostic predictor in patients with high-grade glioma, an experience at two centers in China
- Luteolin alleviates cerebral ischemia/reperfusion injury by regulating cell pyroptosis
- Therapeutic role of respiratory exercise in patients with tuberculous pleurisy
- Effects of CFTR-ENaC on spinal cord edema after spinal cord injury
- Irisin-regulated lncRNAs and their potential regulatory functions in chondrogenic differentiation of human mesenchymal stem cells
- DMD mutations in pediatric patients with phenotypes of Duchenne/Becker muscular dystrophy
- Combination of C-reactive protein and fibrinogen-to-albumin ratio as a novel predictor of all-cause mortality in heart failure patients
- Significant role and the underly mechanism of cullin-1 in chronic obstructive pulmonary disease
- Ferroptosis-related prognostic model of mantle cell lymphoma
- Observation of choking reaction and other related indexes in elderly painless fiberoptic bronchoscopy with transnasal high-flow humidification oxygen therapy
- A bibliometric analysis of Prader-Willi syndrome from 2002 to 2022
- The causal effects of childhood sunburn occasions on melanoma: A univariable and multivariable Mendelian randomization study
- Oxidative stress regulates glycogen synthase kinase-3 in lymphocytes of diabetes mellitus patients complicated with cerebral infarction
- Role of COX6C and NDUFB3 in septic shock and stroke
- Trends in disease burden of type 2 diabetes, stroke, and hypertensive heart disease attributable to high BMI in China: 1990–2019
- Purinergic P2X7 receptor mediates hyperoxia-induced injury in pulmonary microvascular endothelial cells via NLRP3-mediated pyroptotic pathway
- Investigating the role of oviductal mucosa–endometrial co-culture in modulating factors relevant to embryo implantation
- Analgesic effect of external oblique intercostal block in laparoscopic cholecystectomy: A retrospective study
- Elevated serum miR-142-5p correlates with ischemic lesions and both NSE and S100β in ischemic stroke patients
- Correlation between the mechanism of arteriopathy in IgA nephropathy and blood stasis syndrome: A cohort study
- Risk factors for progressive kyphosis after percutaneous kyphoplasty in osteoporotic vertebral compression fracture
- Predictive role of neuron-specific enolase and S100-β in early neurological deterioration and unfavorable prognosis in patients with ischemic stroke
- The potential risk factors of postoperative cognitive dysfunction for endovascular therapy in acute ischemic stroke with general anesthesia
- Fluoxetine inhibited RANKL-induced osteoclastic differentiation in vitro
- Detection of serum FOXM1 and IGF2 in patients with ARDS and their correlation with disease and prognosis
- Rhein promotes skin wound healing by activating the PI3K/AKT signaling pathway
- Differences in mortality risk by levels of physical activity among persons with disabilities in South Korea
- Review Articles
- Cutaneous signs of selected cardiovascular disorders: A narrative review
- XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis
- A narrative review on adverse drug reactions of COVID-19 treatments on the kidney
- Emerging role and function of SPDL1 in human health and diseases
- Adverse reactions of piperacillin: A literature review of case reports
- Molecular mechanism and intervention measures of microvascular complications in diabetes
- Regulation of mesenchymal stem cell differentiation by autophagy
- Molecular landscape of borderline ovarian tumours: A systematic review
- Advances in synthetic lethality modalities for glioblastoma multiforme
- Investigating hormesis, aging, and neurodegeneration: From bench to clinics
- Frankincense: A neuronutrient to approach Parkinson’s disease treatment
- Sox9: A potential regulator of cancer stem cells in osteosarcoma
- Early detection of cardiovascular risk markers through non-invasive ultrasound methodologies in periodontitis patients
- Advanced neuroimaging and criminal interrogation in lie detection
- Maternal factors for neural tube defects in offspring: An umbrella review
- The chemoprotective hormetic effects of rosmarinic acid
- CBD’s potential impact on Parkinson’s disease: An updated overview
- Progress in cytokine research for ARDS: A comprehensive review
- Utilizing reactive oxygen species-scavenging nanoparticles for targeting oxidative stress in the treatment of ischemic stroke: A review
- NRXN1-related disorders, attempt to better define clinical assessment
- Lidocaine infusion for the treatment of complex regional pain syndrome: Case series and literature review
- Trends and future directions of autophagy in osteosarcoma: A bibliometric analysis
- Iron in ventricular remodeling and aneurysms post-myocardial infarction
- Case Reports
- Sirolimus potentiated angioedema: A case report and review of the literature
- Identification of mixed anaerobic infections after inguinal hernia repair based on metagenomic next-generation sequencing: A case report
- Successful treatment with bortezomib in combination with dexamethasone in a middle-aged male with idiopathic multicentric Castleman’s disease: A case report
- Complete heart block associated with hepatitis A infection in a female child with fatal outcome
- Elevation of D-dimer in eosinophilic gastrointestinal diseases in the absence of venous thrombosis: A case series and literature review
- Four years of natural progressive course: A rare case report of juvenile Xp11.2 translocations renal cell carcinoma with TFE3 gene fusion
- Advancing prenatal diagnosis: Echocardiographic detection of Scimitar syndrome in China – A case series
- Outcomes and complications of hemodialysis in patients with renal cancer following bilateral nephrectomy
- Anti-HMGCR myopathy mimicking facioscapulohumeral muscular dystrophy
- Recurrent opportunistic infections in a HIV-negative patient with combined C6 and NFKB1 mutations: A case report, pedigree analysis, and literature review
- Letter to the Editor
- Letter to the Editor: Total parenteral nutrition-induced Wernicke’s encephalopathy after oncologic gastrointestinal surgery
- Erratum
- Erratum to “Bladder-embedded ectopic intrauterine device with calculus”
- Retraction
- Retraction of “XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis”
- Corrigendum
- Corrigendum to “Investigating hormesis, aging, and neurodegeneration: From bench to clinics”
- Corrigendum to “Frankincense: A neuronutrient to approach Parkinson’s disease treatment”
- Special Issue The evolving saga of RNAs from bench to bedside - Part II
- Machine-learning-based prediction of a diagnostic model using autophagy-related genes based on RNA sequencing for patients with papillary thyroid carcinoma
- Unlocking the future of hepatocellular carcinoma treatment: A comprehensive analysis of disulfidptosis-related lncRNAs for prognosis and drug screening
- Elevated mRNA level indicates FSIP1 promotes EMT and gastric cancer progression by regulating fibroblasts in tumor microenvironment
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part I
- Ultrasound-guided transperineal vs transrectal prostate biopsy: A meta-analysis of diagnostic accuracy and complication rates
- Assessment of diagnostic value of unilateral systematic biopsy combined with targeted biopsy in detecting clinically significant prostate cancer
- SENP7 inhibits glioblastoma metastasis and invasion by dissociating SUMO2/3 binding to specific target proteins
- MARK1 suppress malignant progression of hepatocellular carcinoma and improves sorafenib resistance through negatively regulating POTEE
- Analysis of postoperative complications in bladder cancer patients
- Carboplatin combined with arsenic trioxide versus carboplatin combined with docetaxel treatment for LACC: A randomized, open-label, phase II clinical study
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part I
- Comprehensive pan-cancer investigation of carnosine dipeptidase 1 and its prospective prognostic significance in hepatocellular carcinoma
- Identification of signatures associated with microsatellite instability and immune characteristics to predict the prognostic risk of colon cancer
- Single-cell analysis identified key macrophage subpopulations associated with atherosclerosis
Articles in the same Issue
- Research Articles
- EDNRB inhibits the growth and migration of prostate cancer cells by activating the cGMP-PKG pathway
- STK11 (LKB1) mutation suppresses ferroptosis in lung adenocarcinoma by facilitating monounsaturated fatty acid synthesis
- Association of SOX6 gene polymorphisms with Kashin-Beck disease risk in the Chinese Han population
- The pyroptosis-related signature predicts prognosis and influences the tumor immune microenvironment in dedifferentiated liposarcoma
- METTL3 attenuates ferroptosis sensitivity in lung cancer via modulating TFRC
- Identification and validation of molecular subtypes and prognostic signature for stage I and stage II gastric cancer based on neutrophil extracellular traps
- Novel lumbar plexus block versus femoral nerve block for analgesia and motor recovery after total knee arthroplasty
- Correlation between ABCB1 and OLIG2 polymorphisms and the severity and prognosis of patients with cerebral infarction
- Study on the radiotherapy effect and serum neutral granulocyte lymphocyte ratio and inflammatory factor expression of nasopharyngeal carcinoma
- Transcriptome analysis of effects of Tecrl deficiency on cardiometabolic and calcium regulation in cardiac tissue
- Aflatoxin B1 induces infertility, fetal deformities, and potential therapies
- Serum levels of HMW adiponectin and its receptors are associated with cytokine levels and clinical characteristics in chronic obstructive pulmonary disease
- METTL3-mediated methylation of CYP2C19 mRNA may aggravate clopidogrel resistance in ischemic stroke patients
- Understand how machine learning impact lung cancer research from 2010 to 2021: A bibliometric analysis
- Pressure ulcers in German hospitals: Analysis of reimbursement and length of stay
- Metformin plus L-carnitine enhances brown/beige adipose tissue activity via Nrf2/HO-1 signaling to reduce lipid accumulation and inflammation in murine obesity
- Downregulation of carbonic anhydrase IX expression in mouse xenograft nasopharyngeal carcinoma model via doxorubicin nanobubble combined with ultrasound
- Feasibility of 3-dimensional printed models in simulated training and teaching of transcatheter aortic valve replacement
- miR-335-3p improves type II diabetes mellitus by IGF-1 regulating macrophage polarization
- The analyses of human MCPH1 DNA repair machinery and genetic variations
- Activation of Piezo1 increases the sensitivity of breast cancer to hyperthermia therapy
- Comprehensive analysis based on the disulfidptosis-related genes identifies hub genes and immune infiltration for pancreatic adenocarcinoma
- Changes of serum CA125 and PGE2 before and after high-intensity focused ultrasound combined with GnRH-a in treatment of patients with adenomyosis
- The clinical value of the hepatic venous pressure gradient in patients undergoing hepatic resection for hepatocellular carcinoma with or without liver cirrhosis
- Development and validation of a novel model to predict pulmonary embolism in cardiology suspected patients: A 10-year retrospective analysis
- Downregulation of lncRNA XLOC_032768 in diabetic patients predicts the occurrence of diabetic nephropathy
- Circ_0051428 targeting miR-885-3p/MMP2 axis enhances the malignancy of cervical cancer
- Effectiveness of ginkgo diterpene lactone meglumine on cognitive function in patients with acute ischemic stroke
- The construction of a novel prognostic prediction model for glioma based on GWAS-identified prognostic-related risk loci
- Evaluating the impact of childhood BMI on the risk of coronavirus disease 2019: A Mendelian randomization study
- Lactate dehydrogenase to albumin ratio is associated with in-hospital mortality in patients with acute heart failure: Data from the MIMIC-III database
- CD36-mediated podocyte lipotoxicity promotes foot process effacement
- Efficacy of etonogestrel subcutaneous implants versus the levonorgestrel-releasing intrauterine system in the conservative treatment of adenomyosis
- FLRT2 mediates chondrogenesis of nasal septal cartilage and mandibular condyle cartilage
- Challenges in treating primary immune thrombocytopenia patients undergoing COVID-19 vaccination: A retrospective study
- Let-7 family regulates HaCaT cell proliferation and apoptosis via the ΔNp63/PI3K/AKT pathway
- Phospholipid transfer protein ameliorates sepsis-induced cardiac dysfunction through NLRP3 inflammasome inhibition
- Postoperative cognitive dysfunction in elderly patients with colorectal cancer: A randomized controlled study comparing goal-directed and conventional fluid therapy
- Long-pulsed ultrasound-mediated microbubble thrombolysis in a rat model of microvascular obstruction
- High SEC61A1 expression predicts poor outcome of acute myeloid leukemia
- Comparison of polymerase chain reaction and next-generation sequencing with conventional urine culture for the diagnosis of urinary tract infections: A meta-analysis
- Secreted frizzled-related protein 5 protects against renal fibrosis by inhibiting Wnt/β-catenin pathway
- Pan-cancer and single-cell analysis of actin cytoskeleton genes related to disulfidptosis
- Overexpression of miR-532-5p restrains oxidative stress response of chondrocytes in nontraumatic osteonecrosis of the femoral head by inhibiting ABL1
- Autologous liver transplantation for unresectable hepatobiliary malignancies in enhanced recovery after surgery model
- Clinical analysis of incomplete rupture of the uterus secondary to previous cesarean section
- Abnormal sleep duration is associated with sarcopenia in older Chinese people: A large retrospective cross-sectional study
- No genetic causality between obesity and benign paroxysmal vertigo: A two-sample Mendelian randomization study
- Identification and validation of autophagy-related genes in SSc
- Long non-coding RNA SRA1 suppresses radiotherapy resistance in esophageal squamous cell carcinoma by modulating glycolytic reprogramming
- Evaluation of quality of life in patients with schizophrenia: An inpatient social welfare institution-based cross-sectional study
- The possible role of oxidative stress marker glutathione in the assessment of cognitive impairment in multiple sclerosis
- Compilation of a self-management assessment scale for postoperative patients with aortic dissection
- Left atrial appendage closure in conjunction with radiofrequency ablation: Effects on left atrial functioning in patients with paroxysmal atrial fibrillation
- Effect of anterior femoral cortical notch grade on postoperative function and complications during TKA surgery: A multicenter, retrospective study
- Clinical characteristics and assessment of risk factors in patients with influenza A-induced severe pneumonia after the prevalence of SARS-CoV-2
- Analgesia nociception index is an indicator of laparoscopic trocar insertion-induced transient nociceptive stimuli
- High STAT4 expression correlates with poor prognosis in acute myeloid leukemia and facilitates disease progression by upregulating VEGFA expression
- Factors influencing cardiovascular system-related post-COVID-19 sequelae: A single-center cohort study
- HOXD10 regulates intestinal permeability and inhibits inflammation of dextran sulfate sodium-induced ulcerative colitis through the inactivation of the Rho/ROCK/MMPs axis
- Mesenchymal stem cell-derived exosomal miR-26a induces ferroptosis, suppresses hepatic stellate cell activation, and ameliorates liver fibrosis by modulating SLC7A11
- Endovascular thrombectomy versus intravenous thrombolysis for primary distal, medium vessel occlusion in acute ischemic stroke
- ANO6 (TMEM16F) inhibits gastrointestinal stromal tumor growth and induces ferroptosis
- Prognostic value of EIF5A2 in solid tumors: A meta-analysis and bioinformatics analysis
- The role of enhanced expression of Cx43 in patients with ulcerative colitis
- Choosing a COVID-19 vaccination site might be driven by anxiety and body vigilance
- Role of ICAM-1 in triple-negative breast cancer
- Cost-effectiveness of ambroxol in the treatment of Gaucher disease type 2
- HLA-DRB5 promotes immune thrombocytopenia via activating CD8+ T cells
- Efficacy and factors of myofascial release therapy combined with electrical and magnetic stimulation in the treatment of chronic pelvic pain syndrome
- Efficacy of tacrolimus monotherapy in primary membranous nephropathy
- Mechanisms of Tripterygium wilfordii Hook F on treating rheumatoid arthritis explored by network pharmacology analysis and molecular docking
- FBXO45 levels regulated ferroptosis renal tubular epithelial cells in a model of diabetic nephropathy by PLK1
- Optimizing anesthesia strategies to NSCLC patients in VATS procedures: Insights from drug requirements and patient recovery patterns
- Alpha-lipoic acid upregulates the PPARγ/NRF2/GPX4 signal pathway to inhibit ferroptosis in the pathogenesis of unexplained recurrent pregnancy loss
- Correlation between fat-soluble vitamin levels and inflammatory factors in paediatric community-acquired pneumonia: A prospective study
- CD1d affects the proliferation, migration, and apoptosis of human papillary thyroid carcinoma TPC-1 cells via regulating MAPK/NF-κB signaling pathway
- miR-let-7a inhibits sympathetic nerve remodeling after myocardial infarction by downregulating the expression of nerve growth factor
- Immune response analysis of solid organ transplantation recipients inoculated with inactivated COVID-19 vaccine: A retrospective analysis
- The H2Valdien derivatives regulate the epithelial–mesenchymal transition of hepatoma carcinoma cells through the Hedgehog signaling pathway
- Clinical efficacy of dexamethasone combined with isoniazid in the treatment of tuberculous meningitis and its effect on peripheral blood T cell subsets
- Comparison of short-segment and long-segment fixation in treatment of degenerative scoliosis and analysis of factors associated with adjacent spondylolisthesis
- Lycopene inhibits pyroptosis of endothelial progenitor cells induced by ox-LDL through the AMPK/mTOR/NLRP3 pathway
- Methylation regulation for FUNDC1 stability in childhood leukemia was up-regulated and facilitates metastasis and reduces ferroptosis of leukemia through mitochondrial damage by FBXL2
- Correlation of single-fiber electromyography studies and functional status in patients with amyotrophic lateral sclerosis
- Risk factors of postoperative airway obstruction complications in children with oral floor mass
- Expression levels and clinical significance of serum miR-19a/CCL20 in patients with acute cerebral infarction
- Physical activity and mental health trends in Korean adolescents: Analyzing the impact of the COVID-19 pandemic from 2018 to 2022
- Evaluating anemia in HIV-infected patients using chest CT
- Ponticulus posticus and skeletal malocclusion: A pilot study in a Southern Italian pre-orthodontic court
- Causal association of circulating immune cells and lymphoma: A Mendelian randomization study
- Assessment of the renal function and fibrosis indexes of conventional western medicine with Chinese medicine for dredging collaterals on treating renal fibrosis: A systematic review and meta-analysis
- Comprehensive landscape of integrator complex subunits and their association with prognosis and tumor microenvironment in gastric cancer
- New target-HMGCR inhibitors for the treatment of primary sclerosing cholangitis: A drug Mendelian randomization study
- Population pharmacokinetics of meropenem in critically ill patients
- Comparison of the ability of newly inflammatory markers to predict complicated appendicitis
- Comparative morphology of the cruciate ligaments: A radiological study
- Immune landscape of hepatocellular carcinoma: The central role of TP53-inducible glycolysis and apoptosis regulator
- Serum SIRT3 levels in epilepsy patients and its association with clinical outcomes and severity: A prospective observational study
- SHP-1 mediates cigarette smoke extract-induced epithelial–mesenchymal transformation and inflammation in 16HBE cells
- Acute hyper-hypoxia accelerates the development of depression in mice via the IL-6/PGC1α/MFN2 signaling pathway
- The GJB3 correlates with the prognosis, immune cell infiltration, and therapeutic responses in lung adenocarcinoma
- Physical fitness and blood parameters outcomes of breast cancer survivor in a low-intensity circuit resistance exercise program
- Exploring anesthetic-induced gene expression changes and immune cell dynamics in atrial tissue post-coronary artery bypass graft surgery
- Empagliflozin improves aortic injury in obese mice by regulating fatty acid metabolism
- Analysis of the risk factors of the radiation-induced encephalopathy in nasopharyngeal carcinoma: A retrospective cohort study
- Reproductive outcomes in women with BRCA 1/2 germline mutations: A retrospective observational study and literature review
- Evaluation of upper airway ultrasonographic measurements in predicting difficult intubation: A cross-section of the Turkish population
- Prognostic and diagnostic value of circulating IGFBP2 in pancreatic cancer
- Postural stability after operative reconstruction of the AFTL in chronic ankle instability comparing three different surgical techniques
- Research trends related to emergence agitation in the post-anaesthesia care unit from 2001 to 2023: A bibliometric analysis
- Frequency and clinicopathological correlation of gastrointestinal polyps: A six-year single center experience
- ACSL4 mediates inflammatory bowel disease and contributes to LPS-induced intestinal epithelial cell dysfunction by activating ferroptosis and inflammation
- Affibody-based molecular probe 99mTc-(HE)3ZHER2:V2 for non-invasive HER2 detection in ovarian and breast cancer xenografts
- Effectiveness of nutritional support for clinical outcomes in gastric cancer patients: A meta-analysis of randomized controlled trials
- The relationship between IFN-γ, IL-10, IL-6 cytokines, and severity of the condition with serum zinc and Fe in children infected with Mycoplasma pneumoniae
- Paraquat disrupts the blood–brain barrier by increasing IL-6 expression and oxidative stress through the activation of PI3K/AKT signaling pathway
- Sleep quality associate with the increased prevalence of cognitive impairment in coronary artery disease patients: A retrospective case–control study
- Dioscin protects against chronic prostatitis through the TLR4/NF-κB pathway
- Association of polymorphisms in FBN1, MYH11, and TGF-β signaling-related genes with susceptibility of sporadic thoracic aortic aneurysm and dissection in the Zhejiang Han population
- Application value of multi-parameter magnetic resonance image-transrectal ultrasound cognitive fusion in prostate biopsy
- Laboratory variables‐based artificial neural network models for predicting fatty liver disease: A retrospective study
- Decreased BIRC5-206 promotes epithelial–mesenchymal transition in nasopharyngeal carcinoma through sponging miR-145-5p
- Sepsis induces the cardiomyocyte apoptosis and cardiac dysfunction through activation of YAP1/Serpine1/caspase-3 pathway
- Assessment of iron metabolism and iron deficiency in incident patients on incident continuous ambulatory peritoneal dialysis
- Tibial periosteum flap combined with autologous bone grafting in the treatment of Gustilo-IIIB/IIIC open tibial fractures
- The application of intravenous general anesthesia under nasopharyngeal airway assisted ventilation undergoing ureteroscopic holmium laser lithotripsy: A prospective, single-center, controlled trial
- Long intergenic noncoding RNA for IGF2BP2 stability suppresses gastric cancer cell apoptosis by inhibiting the maturation of microRNA-34a
- Role of FOXM1 and AURKB in regulating keratinocyte function in psoriasis
- Parental control attitudes over their pre-school children’s diet
- The role of auto-HSCT in extranodal natural killer/T cell lymphoma
- Significance of negative cervical cytology and positive HPV in the diagnosis of cervical lesions by colposcopy
- Echinacoside inhibits PASMCs calcium overload to prevent hypoxic pulmonary artery remodeling by regulating TRPC1/4/6 and calmodulin
- ADAR1 plays a protective role in proximal tubular cells under high glucose conditions by attenuating the PI3K/AKT/mTOR signaling pathway
- The risk of cancer among insulin glargine users in Lithuania: A retrospective population-based study
- The unusual location of primary hydatid cyst: A case series study
- Intraoperative changes in electrophysiological monitoring can be used to predict clinical outcomes in patients with spinal cavernous malformation
- Obesity and risk of placenta accreta spectrum: A meta-analysis
- Shikonin alleviates asthma phenotypes in mice via an airway epithelial STAT3-dependent mechanism
- NSUN6 and HTR7 disturbed the stability of carotid atherosclerotic plaques by regulating the immune responses of macrophages
- The effect of COVID-19 lockdown on admission rates in Maternity Hospital
- Temporal muscle thickness is not a prognostic predictor in patients with high-grade glioma, an experience at two centers in China
- Luteolin alleviates cerebral ischemia/reperfusion injury by regulating cell pyroptosis
- Therapeutic role of respiratory exercise in patients with tuberculous pleurisy
- Effects of CFTR-ENaC on spinal cord edema after spinal cord injury
- Irisin-regulated lncRNAs and their potential regulatory functions in chondrogenic differentiation of human mesenchymal stem cells
- DMD mutations in pediatric patients with phenotypes of Duchenne/Becker muscular dystrophy
- Combination of C-reactive protein and fibrinogen-to-albumin ratio as a novel predictor of all-cause mortality in heart failure patients
- Significant role and the underly mechanism of cullin-1 in chronic obstructive pulmonary disease
- Ferroptosis-related prognostic model of mantle cell lymphoma
- Observation of choking reaction and other related indexes in elderly painless fiberoptic bronchoscopy with transnasal high-flow humidification oxygen therapy
- A bibliometric analysis of Prader-Willi syndrome from 2002 to 2022
- The causal effects of childhood sunburn occasions on melanoma: A univariable and multivariable Mendelian randomization study
- Oxidative stress regulates glycogen synthase kinase-3 in lymphocytes of diabetes mellitus patients complicated with cerebral infarction
- Role of COX6C and NDUFB3 in septic shock and stroke
- Trends in disease burden of type 2 diabetes, stroke, and hypertensive heart disease attributable to high BMI in China: 1990–2019
- Purinergic P2X7 receptor mediates hyperoxia-induced injury in pulmonary microvascular endothelial cells via NLRP3-mediated pyroptotic pathway
- Investigating the role of oviductal mucosa–endometrial co-culture in modulating factors relevant to embryo implantation
- Analgesic effect of external oblique intercostal block in laparoscopic cholecystectomy: A retrospective study
- Elevated serum miR-142-5p correlates with ischemic lesions and both NSE and S100β in ischemic stroke patients
- Correlation between the mechanism of arteriopathy in IgA nephropathy and blood stasis syndrome: A cohort study
- Risk factors for progressive kyphosis after percutaneous kyphoplasty in osteoporotic vertebral compression fracture
- Predictive role of neuron-specific enolase and S100-β in early neurological deterioration and unfavorable prognosis in patients with ischemic stroke
- The potential risk factors of postoperative cognitive dysfunction for endovascular therapy in acute ischemic stroke with general anesthesia
- Fluoxetine inhibited RANKL-induced osteoclastic differentiation in vitro
- Detection of serum FOXM1 and IGF2 in patients with ARDS and their correlation with disease and prognosis
- Rhein promotes skin wound healing by activating the PI3K/AKT signaling pathway
- Differences in mortality risk by levels of physical activity among persons with disabilities in South Korea
- Review Articles
- Cutaneous signs of selected cardiovascular disorders: A narrative review
- XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis
- A narrative review on adverse drug reactions of COVID-19 treatments on the kidney
- Emerging role and function of SPDL1 in human health and diseases
- Adverse reactions of piperacillin: A literature review of case reports
- Molecular mechanism and intervention measures of microvascular complications in diabetes
- Regulation of mesenchymal stem cell differentiation by autophagy
- Molecular landscape of borderline ovarian tumours: A systematic review
- Advances in synthetic lethality modalities for glioblastoma multiforme
- Investigating hormesis, aging, and neurodegeneration: From bench to clinics
- Frankincense: A neuronutrient to approach Parkinson’s disease treatment
- Sox9: A potential regulator of cancer stem cells in osteosarcoma
- Early detection of cardiovascular risk markers through non-invasive ultrasound methodologies in periodontitis patients
- Advanced neuroimaging and criminal interrogation in lie detection
- Maternal factors for neural tube defects in offspring: An umbrella review
- The chemoprotective hormetic effects of rosmarinic acid
- CBD’s potential impact on Parkinson’s disease: An updated overview
- Progress in cytokine research for ARDS: A comprehensive review
- Utilizing reactive oxygen species-scavenging nanoparticles for targeting oxidative stress in the treatment of ischemic stroke: A review
- NRXN1-related disorders, attempt to better define clinical assessment
- Lidocaine infusion for the treatment of complex regional pain syndrome: Case series and literature review
- Trends and future directions of autophagy in osteosarcoma: A bibliometric analysis
- Iron in ventricular remodeling and aneurysms post-myocardial infarction
- Case Reports
- Sirolimus potentiated angioedema: A case report and review of the literature
- Identification of mixed anaerobic infections after inguinal hernia repair based on metagenomic next-generation sequencing: A case report
- Successful treatment with bortezomib in combination with dexamethasone in a middle-aged male with idiopathic multicentric Castleman’s disease: A case report
- Complete heart block associated with hepatitis A infection in a female child with fatal outcome
- Elevation of D-dimer in eosinophilic gastrointestinal diseases in the absence of venous thrombosis: A case series and literature review
- Four years of natural progressive course: A rare case report of juvenile Xp11.2 translocations renal cell carcinoma with TFE3 gene fusion
- Advancing prenatal diagnosis: Echocardiographic detection of Scimitar syndrome in China – A case series
- Outcomes and complications of hemodialysis in patients with renal cancer following bilateral nephrectomy
- Anti-HMGCR myopathy mimicking facioscapulohumeral muscular dystrophy
- Recurrent opportunistic infections in a HIV-negative patient with combined C6 and NFKB1 mutations: A case report, pedigree analysis, and literature review
- Letter to the Editor
- Letter to the Editor: Total parenteral nutrition-induced Wernicke’s encephalopathy after oncologic gastrointestinal surgery
- Erratum
- Erratum to “Bladder-embedded ectopic intrauterine device with calculus”
- Retraction
- Retraction of “XRCC1 and hOGG1 polymorphisms and endometrial carcinoma: A meta-analysis”
- Corrigendum
- Corrigendum to “Investigating hormesis, aging, and neurodegeneration: From bench to clinics”
- Corrigendum to “Frankincense: A neuronutrient to approach Parkinson’s disease treatment”
- Special Issue The evolving saga of RNAs from bench to bedside - Part II
- Machine-learning-based prediction of a diagnostic model using autophagy-related genes based on RNA sequencing for patients with papillary thyroid carcinoma
- Unlocking the future of hepatocellular carcinoma treatment: A comprehensive analysis of disulfidptosis-related lncRNAs for prognosis and drug screening
- Elevated mRNA level indicates FSIP1 promotes EMT and gastric cancer progression by regulating fibroblasts in tumor microenvironment
- Special Issue Advancements in oncology: bridging clinical and experimental research - Part I
- Ultrasound-guided transperineal vs transrectal prostate biopsy: A meta-analysis of diagnostic accuracy and complication rates
- Assessment of diagnostic value of unilateral systematic biopsy combined with targeted biopsy in detecting clinically significant prostate cancer
- SENP7 inhibits glioblastoma metastasis and invasion by dissociating SUMO2/3 binding to specific target proteins
- MARK1 suppress malignant progression of hepatocellular carcinoma and improves sorafenib resistance through negatively regulating POTEE
- Analysis of postoperative complications in bladder cancer patients
- Carboplatin combined with arsenic trioxide versus carboplatin combined with docetaxel treatment for LACC: A randomized, open-label, phase II clinical study
- Special Issue Exploring the biological mechanism of human diseases based on MultiOmics Technology - Part I
- Comprehensive pan-cancer investigation of carnosine dipeptidase 1 and its prospective prognostic significance in hepatocellular carcinoma
- Identification of signatures associated with microsatellite instability and immune characteristics to predict the prognostic risk of colon cancer
- Single-cell analysis identified key macrophage subpopulations associated with atherosclerosis

