Abstract
Osteoporosis is a major health concern worldwide. The present study aimed to identify effective biomarkers for osteoporosis detection. In osteoporosis, 559 differentially expressed genes (DEGs) were enriched in PI3K-Akt signaling pathway and Foxo signaling pathway. Weighted gene co-expression network analysis showed that green, pink, and tan modules were clinically significant modules, and that six genes (VEGFA, DDX5, SOD2, HNRNPD, EIF5B, and HSP90B1) were identified as “real” hub genes in the protein–protein interaction network, co-expression network, and 559 DEGs. The sensitivity and specificity of the support vector machine (SVM) for identifying patients with osteoporosis was 100%, with an area under curve of 1 in both training and validation datasets. Our results indicated that the current system using the SVM method could identify patients with osteoporosis.
1 Introduction
Osteoporosis is a systemic metabolic disease characterized by a decrease in bone mass and destruction of bone tissue microstructure, which could lead to bone fragility and susceptibility to fracture. As a common bone disease syndrome [1], osteoporosis has been considered as an aging disease in which bone density decreases and further aggravates the condition [2,3]. In normal physiological state, human bones are in constant metabolism that osteoblasts form new bone tissue and osteoclasts resorb old or damaged bone tissue. The dynamic balance of bone volume is maintained by continuous bone formation and bone resorption [4]. However, when the body is affected by injury, aging, estrogen deficiency, and/or other factors, the balance will be disrupted, causing osteoporosis [5,6].
As a supervised learning method in machine learning, support vector machine (SVM) algorithm [7] was first proposed in 1995, and has been developed more maturely and widely applied in the mid-1990s. Later on, a series of improved and extended algorithms, including multi-classification SVM, support vector regression, least squares SVM, support vector clustering, semi-supervised SVM, etc., have been developed.
Machine learning tools could display key features from complex datasets [8,9]. SVM is widely used in disease research for building predictive models, and could produce effective and predictable models [10–12]. For example, a study has shown that applying SVM approach helps identify postmenopausal women with low bone density [13]. Moreover, a gene signature associated with postmenopausal osteoporosis was developed and validated by SVM [14]. Chen et al. reported that a diagnostic model established based on nine key genes could reliably separate osteoporosis patients from healthy subjects [15]. Chen et al. have identified a group of circulating miRNAs as non-invasive biomarkers for the detection of postmenopausal and mechanical unloading osteoporosis through a large-scale screening based on microarray [16]. Xia et al. showed that genes such as VPS35, FCGR2A, TBCA, HIRA, TYROBP, JUND, PHF20, NFKB2, RPL35A, and BICD2 may be considered to be potential pathogenic genes of osteoporosis and may be useful for further study of the mechanisms underlying osteoporosis [17]. Although a large number of biomarkers have been identified in different ways, there is still a lack of model analyses based on multiple key genes that are valid in osteoporosis for clinical application.
The Gene Expression Omnibus (GEO) [18], an online public database made available by National Center for Biotechnology Information (NCBI) in 2000, is currently one of the most comprehensive gene expression databases. We systematically analyzed the expression patterns of genes associated with osteoporosis samples from this database at a transcriptional level. In addition, a risk prediction model was developed for osteoporosis patients based on SVM.
2 Materials and methods
2.1 Data acquisition
MINiML formatted family file(s) of osteoporosis-related microarray datasets GSE35959 [19], GSE7158 [20], GSE7429 [21], and GSE13850 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13850) and GSE62402 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62402) were downloaded from GEO.
From the GEO dataset, osteoporosis samples and control samples were retained. The probes were converted to Gene Symbol. One probe corresponding to multiple genes was removed. Median expressions of genes corresponding to multiple Gene Symbols were taken. The clinical statistics of the processed samples are shown in Table 1.
Clinical information of the samples
| Dataset | Expression | Platforms |
|---|---|---|
| GSE35959 | ||
| Control | 14 | GPL570 |
| Osteoporosis | 5 | |
| GSE7158 | ||
| High PBM | 14 | GPL570 |
| Low PBM | 12 | |
| GSE62402 | ||
| High BMD | 5 | GPL5175 |
| Low BMD | 5 | |
| GSE13850 | ||
| High BMD | 20 | GPL96 |
| Low BMD | 20 | |
| GSE7429 | ||
| High BMD | 10 | GPL96 |
| Low BMD | 10 |
PBM (Peak bone mass) is an important determinant of osteoporosis. BMD (bone mineral density) can be reliably and accurately measured and has high genetic determination with heritability of 0.5–0.9, indicating that genetic factors play an important role in risk of osteoporosis.
2.2 Identification of differentially expressed genes (DEGs)
The “Limma” package [22] in the latest R language was used to screen DEGs between osteoporosis tissues and normal samples under the interception criteria |log2FC| > 1 and FDR < 0.05.
2.3 Functional enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and gene ontology (GO) functional enrichment analysis (cellular component [CC], biological process [BP], and molecular function [MF]) on the DEGs were performed using the R software package clusterProfiler [23]. p < 0.05 was the threshold for significant enrichment.
2.4 Weighted gene co-expression network analysis (WGCNA)
WGCNA was performed on the DEGs using the R language “WGCNA” package [24]. The expression profile in the osteoporosis samples was constructed as a sample clustering dendrogram using the function hclust, and sample clustering analysis was performed. If the gene expression value of a sample was significantly higher than the average, the sample was considered as an outlier. The vertical coordinate was the height of the sample clustering tree, with a higher height indicating a higher gene expression of the sample. The soft threshold was selected using the function pick soft threshold, a criterion based on the approximate scale-free network that allows the constructed network to be more consistent with the power-law distribution. In this study, the correlation coefficient was 0.85.
Based on the topological overlap measure, the genes were clustered using the average linkage hierarchical clustering method under the criteria of hybrid dynamic shear tree. The minimum number of genes per gene network module was 100, and 0.25 was the threshold of cut height.
2.5 Protein–protein interaction (PPI) networks
PPI network could help discover new drug targets and study the molecular mechanisms of a disease from a system perspective. The PPI network was constructed based on the STRING database, which is a database used for searching interactions between known proteins and predicting protein interactions. The PPI network of the module genes was developed by importing the module genes into the STRING database. Generally, the importance of a network node is positively related to greater connections to the genes in the network. Four algorithms of Degree, Maximal Clique Centrality (MCC), Closeness, and Betweenness of Cytoscape (version: 3.7.2) [25] software plug-in cytoHubba [26] were used to calculate each point in the PPI network for screening the module pivot genes.
2.6 Construction of SVM classifier
SVM is a supervised machine learning classification algorithm that distinguishes sample types by estimating the degree of a sample belonging to a certain class. For the training set, the SVM classifier was constructed using the SVM method based on the optimal mRNA set in R package e1071 (version 1.6-8, http://cran.r-project.org/web/packages/e1071).
The performance of the SVM classifier was evaluated in the training and validation sets with the area under curve (AUC) of the receiver operating characteristic (ROC) curve as an evaluation metric.
GitHub page
3 Results
3.1 Identification and functional annotation of DEGs
The analysis flow chart of this study is shown in Figure 1. The Limma package was used to calculate the DEGs between osteoporosis and control samples from GSE35959 dataset. A total of 559 DEGs were obtained incorporating 152 upregulated and 407 downregulated genes in osteoporosis samples (Figure 2a and b). Furthermore, KEGG pathway analysis and GO function enrichment analysis on the 559 DEGs were performed using R software package Clusterprofiler. The top ten significantly enriched BPs, MFs, and CCs are shown in Figure 2c–e. Based on the KEGG annotation, eight pathways including PI3K-Akt signaling pathway, Foxo signaling pathway, and osteoclast differentiation (Figure 2f) were obtained.
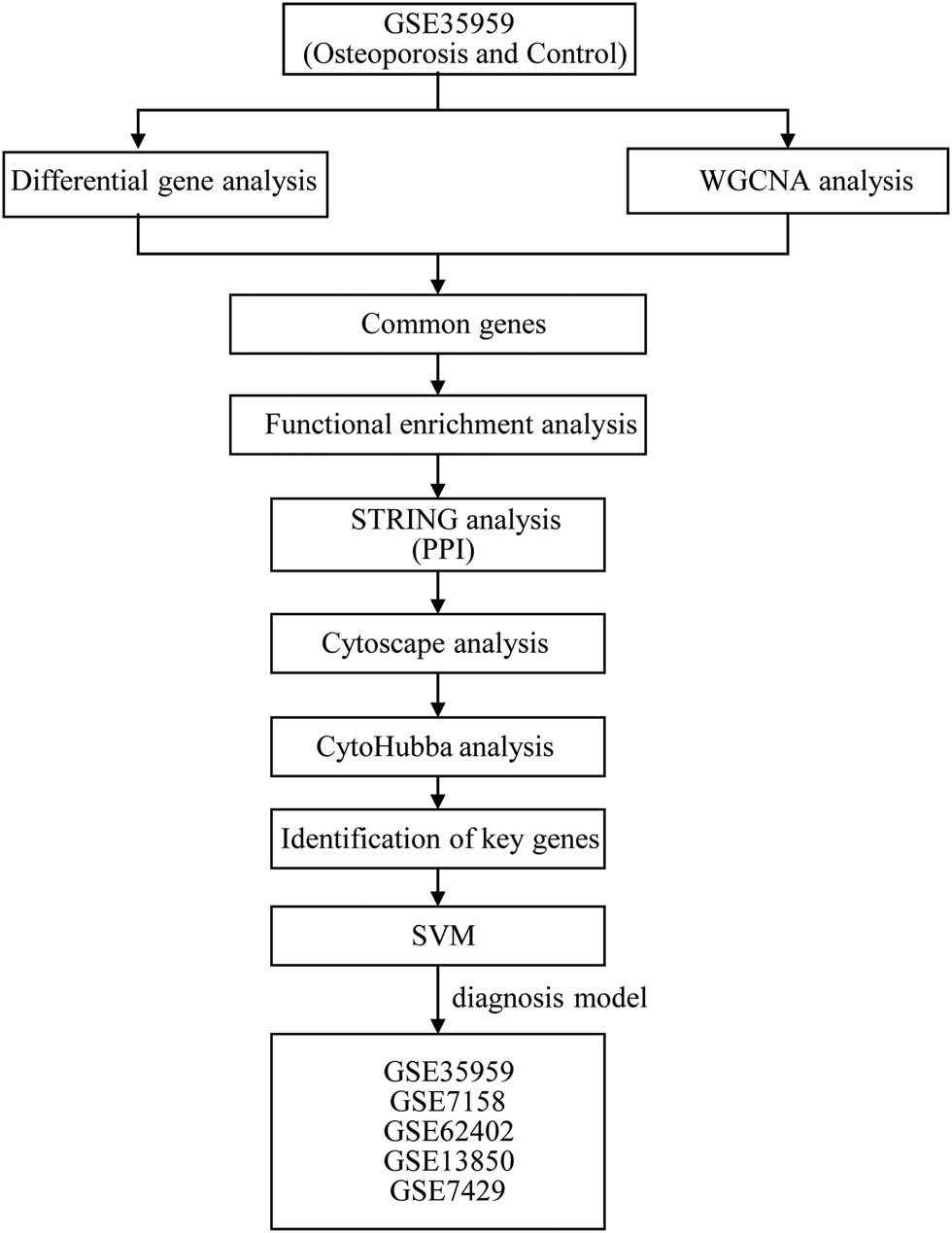
Work flow chart.
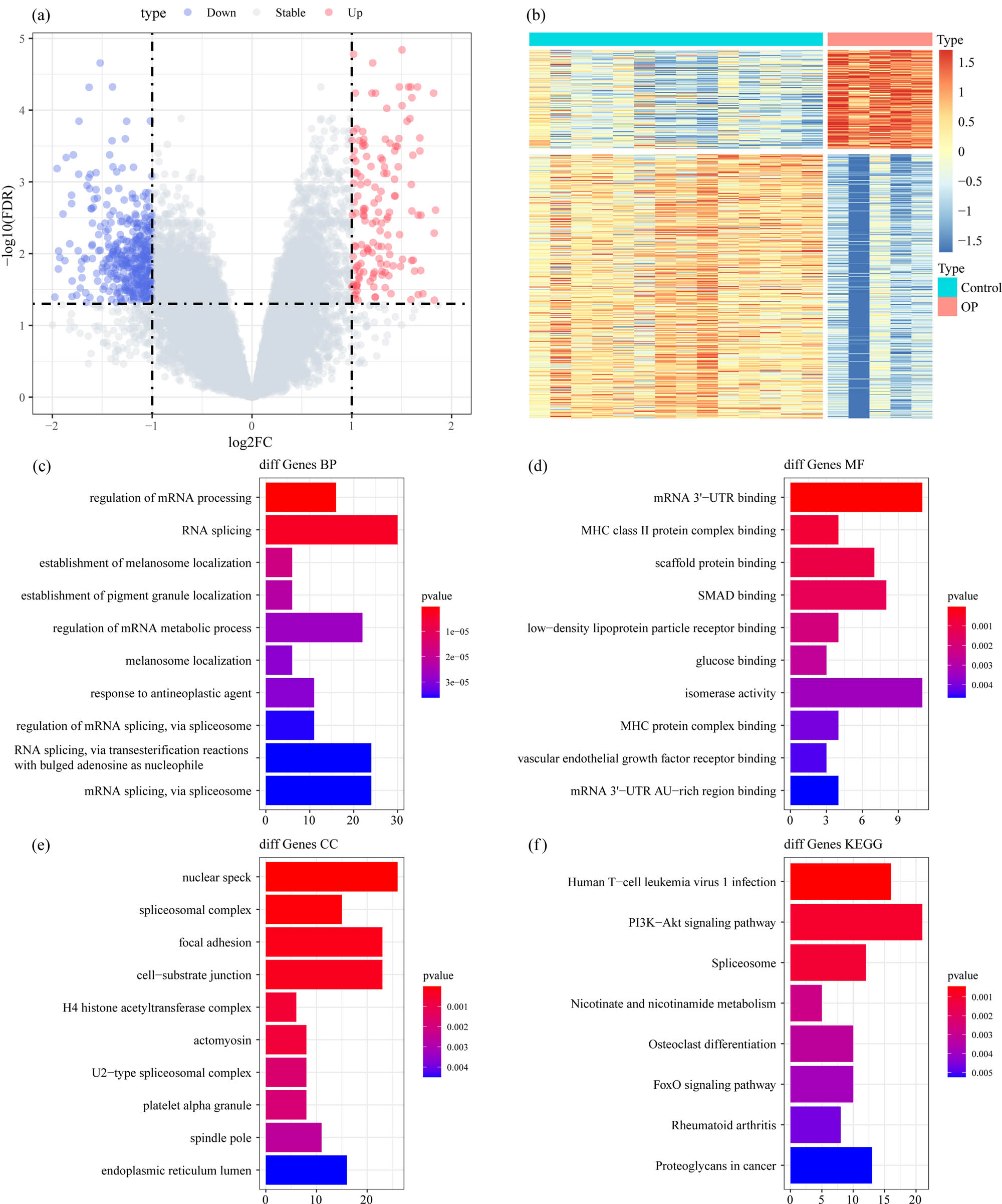
Identification of DEGs: (a) the volcano map of DEGs in GSE35959 dataset, (b) the heat map of DEGs in GSE35959 dataset, (c) the BP annotation map of DEGs, (d) the CC annotation map of DEGs, (e) the MF annotation map of DEGs, and (f) the KEGG annotation map of DEGs.
3.2 WGCNA
WGCNA was used to construct a co-expression network and identify co-expression modules. The hierarchical clustering of samples showed that 18 samples met the requirements (Figure 3a). The study of sufficient soft threshold validation converged to a scale-free topology with a value of 13 (Figure 3b). The first group of modules was obtained by Dynamic Tree Cut algorithm, and the related modules (height = 0.25, deepSplit = 2, and minModuleSize = 100) were merged together (merged Dynamic). Here, we identified a total of 15 modules (Figure 3c). The correlation between each module and the sample type (osteoporosis and control) was further analyzed. The results demonstrated that the green and pink modules were significantly positively correlated with osteoporosis, and that the tan module was obviously negatively correlated with osteoporosis (Figure 3d). Furthermore, using the R software package Clusterprofile, 1,719 genes, 1,143 genes, and 225 genes obtained, respectively, from green, pink, and tan modules of WGCNA co-expression were analyzed for KEGG pathway analysis and GO function enrichment. For the 1,719 genes in the green model, and the top ten significantly enriched BPs, MFs, and CCs are shown in Figure 4a–c. Based on KEGG annotation, nine pathways including osteoclast differentiation, cell adhesion molecules, Th17 cell differentiation, and Epstein-Barr virus infection were obtained (Figure 4d).
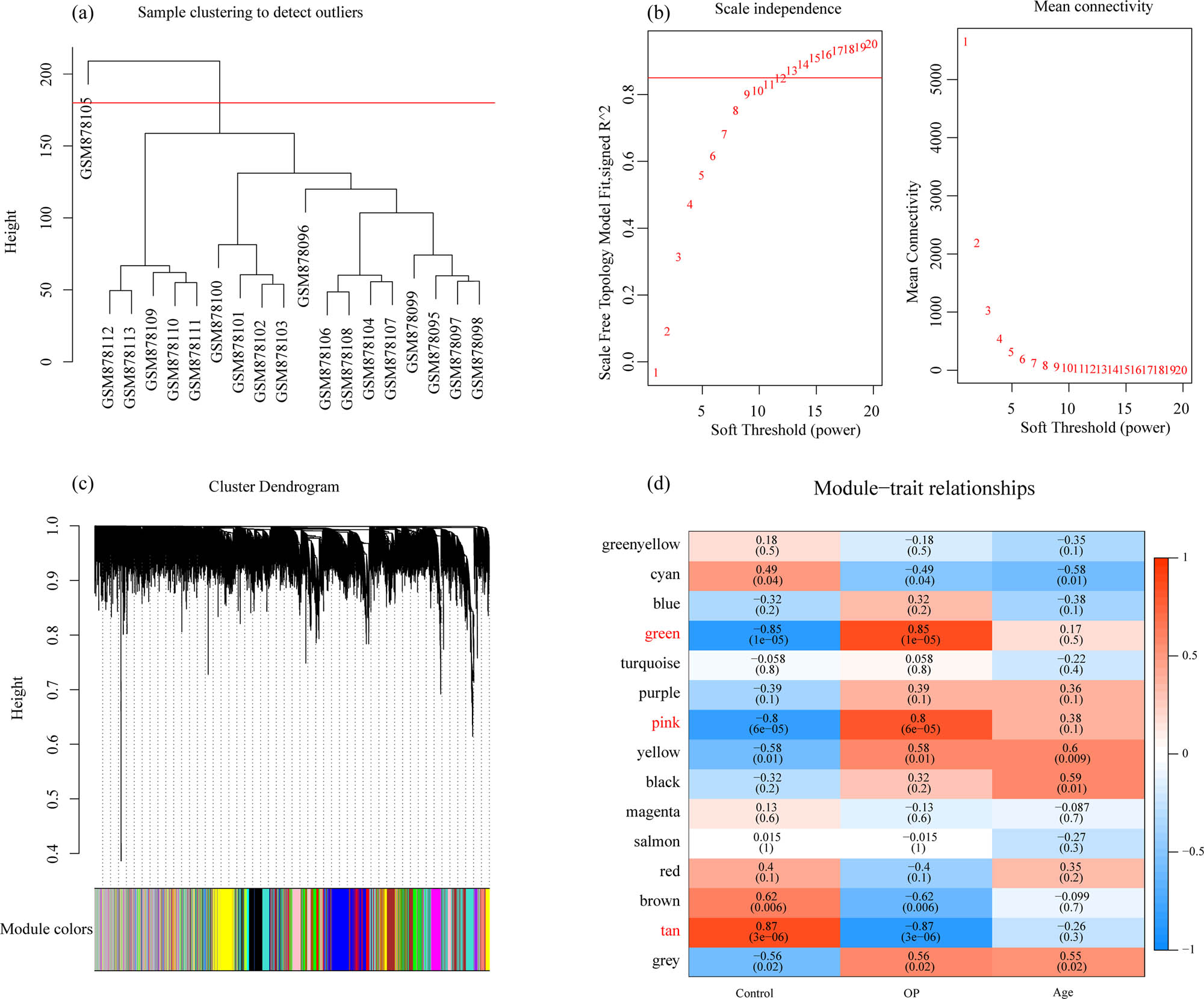
WGCNA: (a) cluster analysis of samples, (b) analysis of network topology for various soft-thresholding powers, (c) gene dendrogram and module colors, and (d) correlation results between 15 modules and clinical phenotypes.
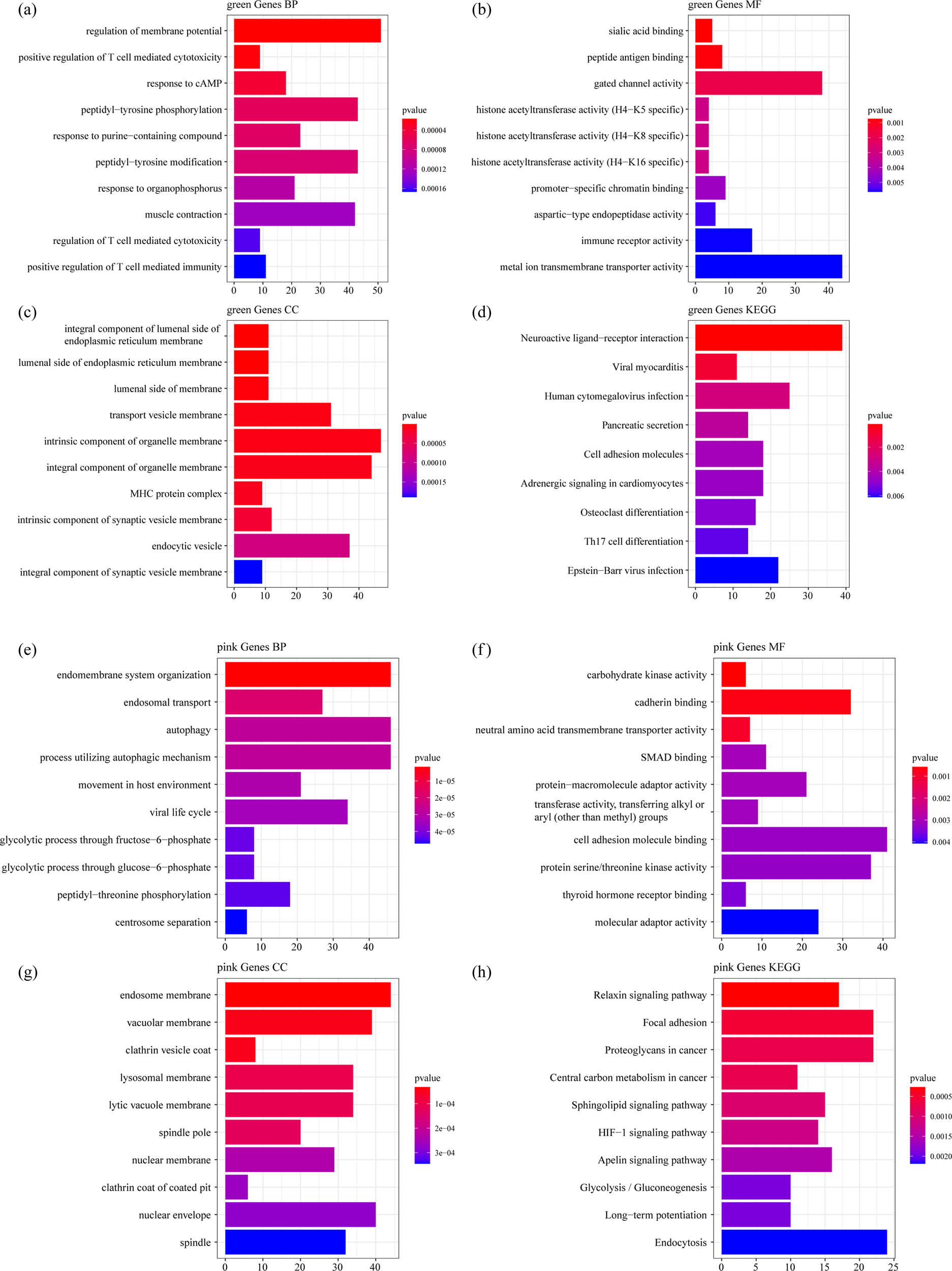
Functional enrichment analysis of DEGs in green and pink model: (a) the BP annotation map of DEGs in green model, (b) the CC annotation map of DEGs in green model, (c) the MF annotation map of DEGs in green model, (d) the KEGG annotation map of DEGs in green model, (e) the BP annotation map of DEGs in pink model, (f) the CC annotation map of DEGs in pink model, (g) the MF annotation map of DEGs in pink model, and (h) the KEGG annotation map of DEGs in pink model.
For the 1,143 genes in pink model, the top ten significantly enriched BPs, MFs, and CCs are shown in Figure 4e–g. Based on the KEGG annotation, 73 pathways including HIF-1 signaling pathway, Apelin signaling pathway, mTOR signaling pathway, and MAPK signaling pathway were obtained (Figure 4h).
For the 225 genes in tan model, the top ten significantly enriched BPs, MFs, and CCs are shown in Figure 5a–c. There were no pathways obtained based on the KEGG annotation.
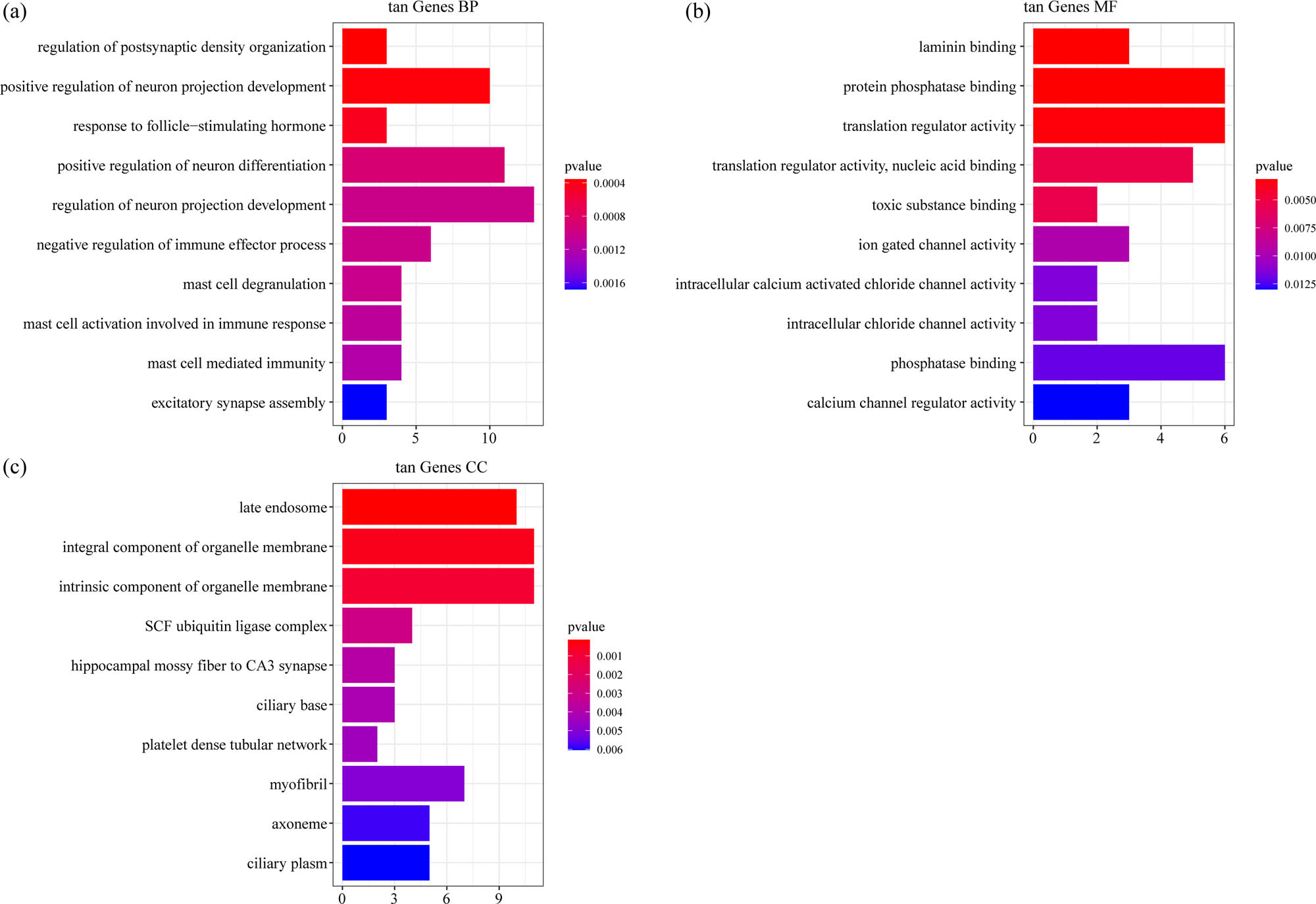
Functional enrichment analysis of DEGs in tan model: (a) the BP annotation map of DEGs in tan model, (b) the CC annotation map of DEGs in tan model, and (c) the MF annotation map of DEGs in tan model.
3.3 Identification of co-expressed differential genes
The DEGs in GSE35959 dataset and green, pink, and tan co-expressed by WGCNA showed 109 intersected genes between the green module and 559 DEGs. Among the 559 DEGs, the pink module gene had 115 intersected genes, 58 intersected genes were obtained between the tan module and 559 DEGs, and a total of 282 genes were in the intersection (Figure 6a). The KEGG pathway analysis and GO function enrichment analysis on the 282 DEGs were performed by software package Clusterprofiler. The top ten significantly enriched BPs, MFs, and CCs are shown in Figure 6b–d. Based on the KEGG annotation, six pathways including PI3K-Akt signaling pathway, osteoclast differentiation, and focal adhesion were obtained (Figure 6e).
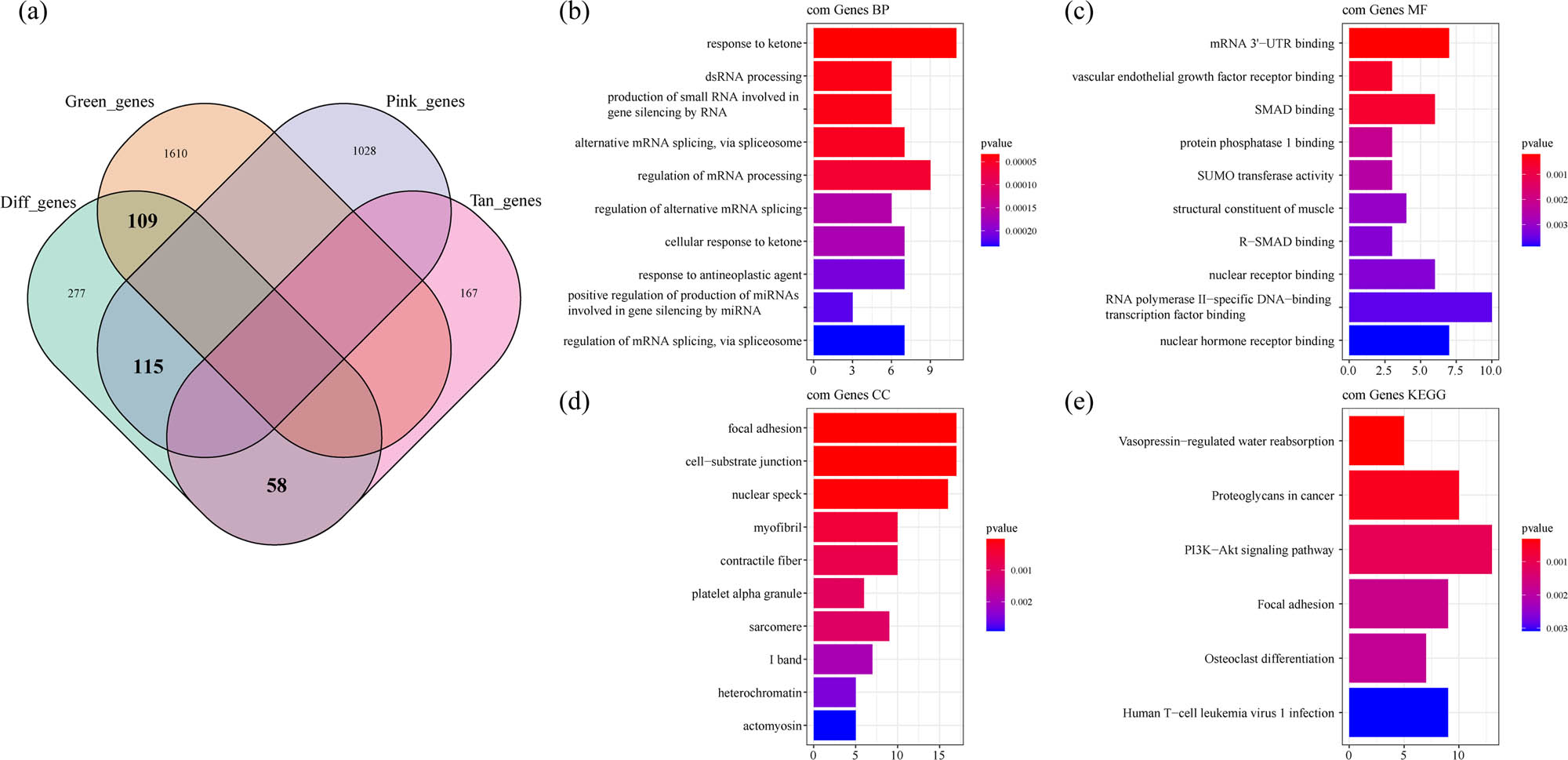
Identification of co-expression of DEGs: (a) Venn diagram of co-expressed genes and DEGs, (b) the BP annotation map of co-expression of DEGs, (c) the CC annotation map of co-expression of DEGs, (d) the MF annotation map of co-expression of DEGs, and (e) the KEGG annotation map of co-expression of DEGs.
3.4 PPI network
The PPI network of the 282 DEGs was further developed using the STRING database (Figure 7). For the PPI network developed with the 282 DEGs, the four algorithms of Degree, MCC, Closeness, and Betweenness of cytoHubba, a plug-in of Cytoscape (version: 3.7.2) software were used for calculations. Here, the top 15 genes were selected as the key genes (Figure 8a–d). The hub genes obtained by the four algorithms were taken as intersection, and finally six genes (VEGFA, DDX5, SOD2, HNRNPD, EIF5B, and HSP90B1) were acquired (Figure 8e).
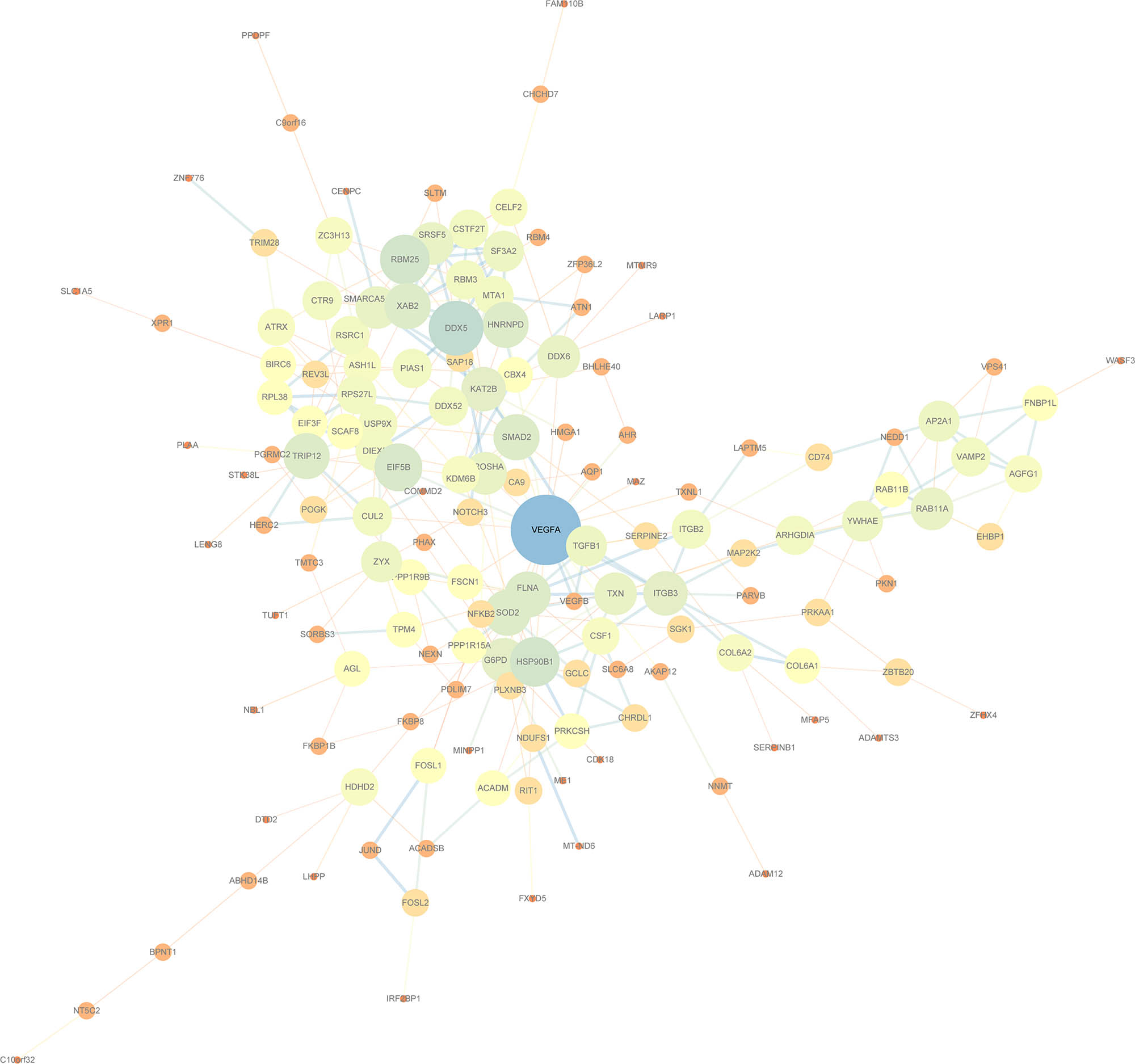
PPI diagram of STRING protein network visualized by Cytoscape.
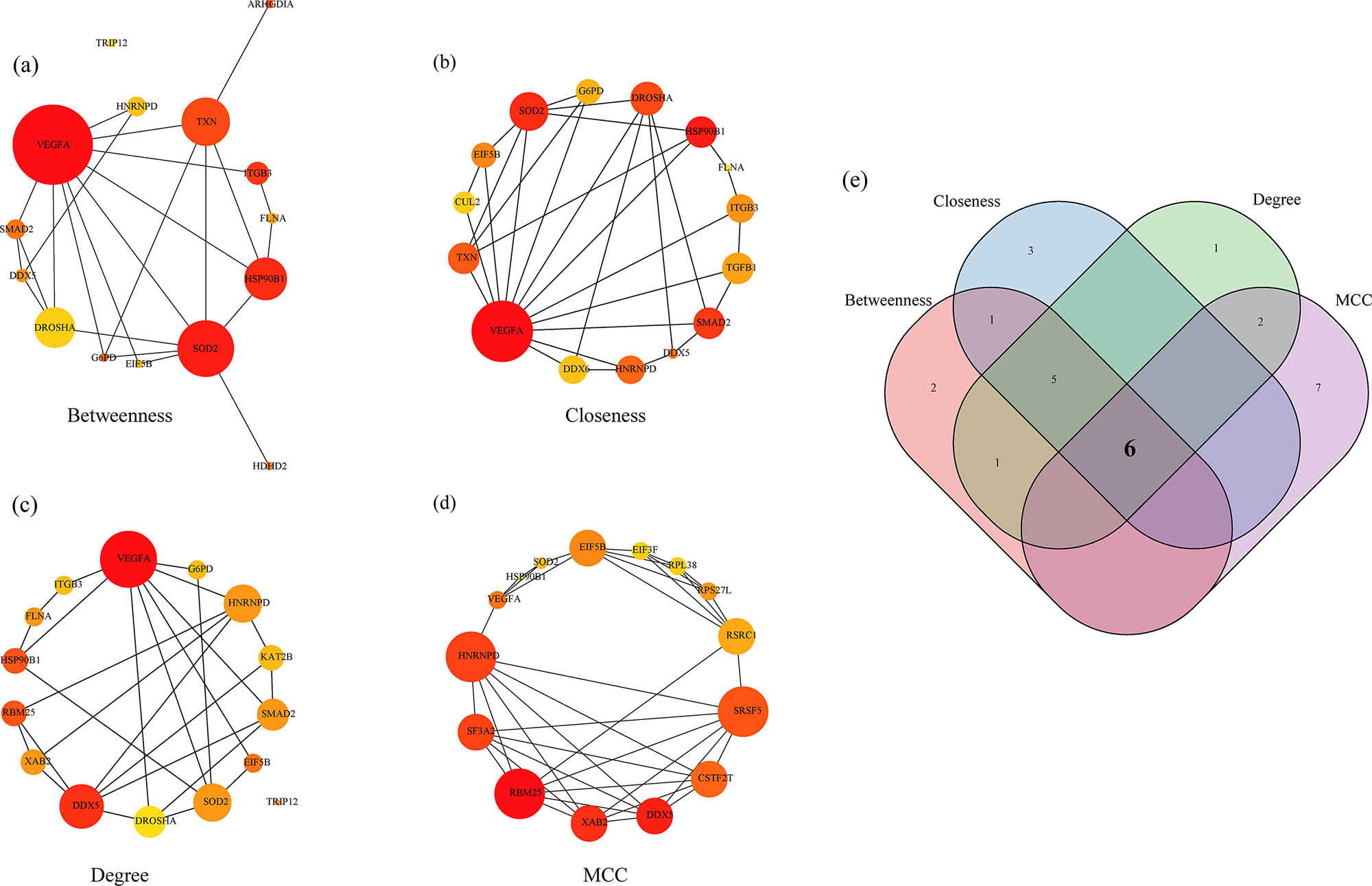
Identification of Hub genes: (a) PPI network of hub genes obtained by Betweenness algorithm, (b) PPI network of hub genes obtained by Closeness algorithm, (c) PPI network of hub genes obtained by Degree algorithm, (d) PPI network of hub genes obtained by MCC algorithm, and (e) Venn diagram of the Hub genes obtained by the four algorithms.
3.5 Construction and validation of the diagnostic model
GSE35959 was used as train dataset, and datasets (GSE7429, GSE7158, GSE13850, and GSE62402) were used as validation datasets. Six hub genes served as features in the training dataset, their corresponding expression profiles were obtained, and the SVM classification model was constructed. The classification accuracy of GSE35959 dataset was 100%, as 19 out of 19 samples were correctly classified; moreover, the sensitivity and specificity of the models were all 100%, with the area under the ROC curve (AUC) of 1 (Figure 9a). Furthermore, the GSE7429, GSE7158, GSE13850, and GSE62402 datasets verified that all the samples in the dataset were correctly classified. The classification accuracy was 100%, and the sensitivity and specificity of the models were 100%, with an area under the ROC curve of 1 (Figure 9b–e).
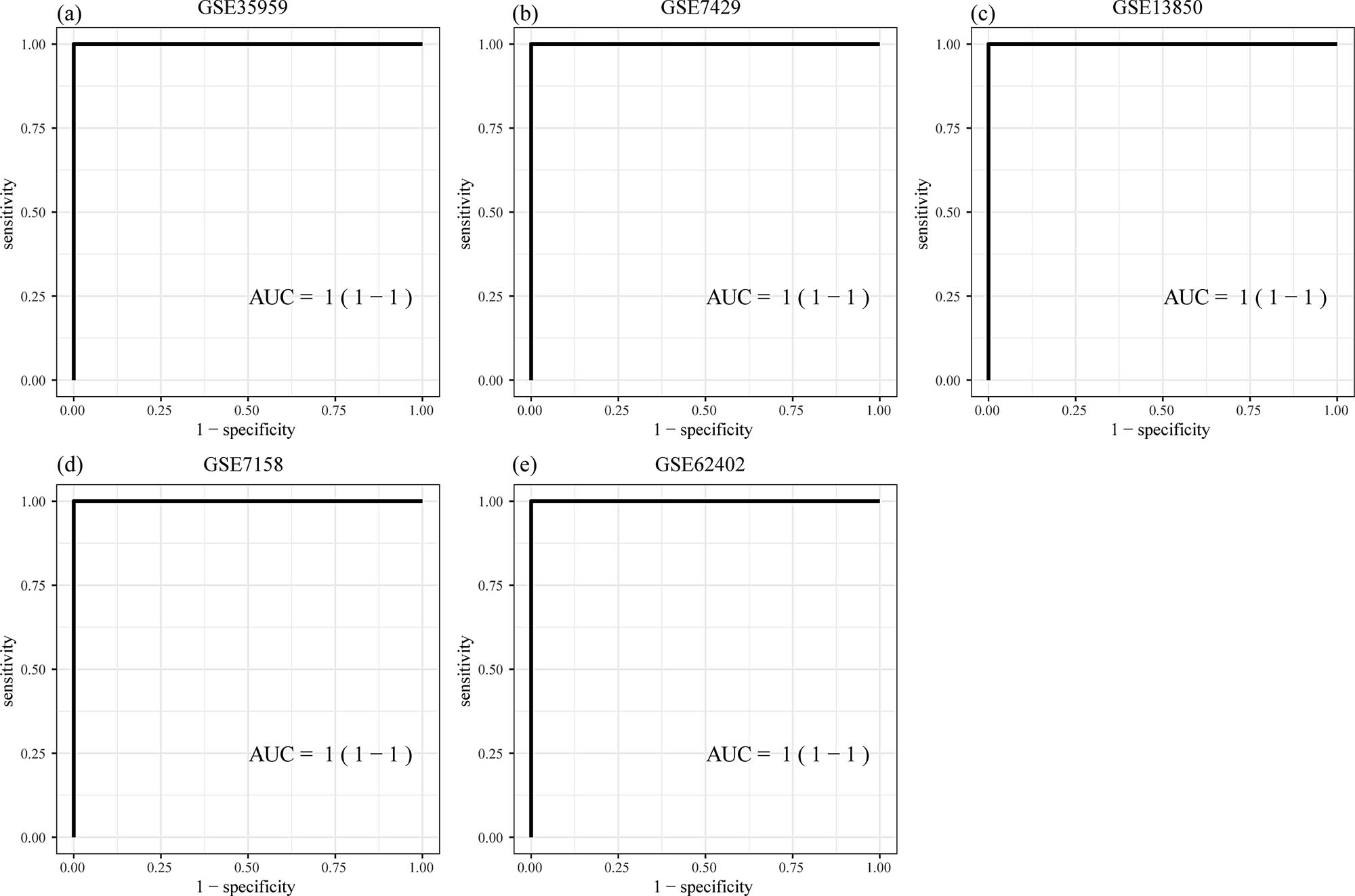
Construction and validation of diagnostic models: (a) ROC curves of the classification results of the diagnostic model on sample of GSE35959 dataset, (b) ROC curves of the classification results of the diagnostic model on the samples of GSE7429 dataset, (c) ROC curves of the classification results of the diagnostic model on the samples of GSE13850 dataset, (d) ROC curves of the classification results of the diagnostic model on the samples of GSE7158 dataset, and (e) ROC curves of the classification results of the diagnostic model on the samples of GSE62402 dataset.
4 Discussion
Osteoporotic fractures can be prevented by pharmacological treatment. The current available osteoporosis treatments are anti-resorptive (inhibiting the osteoclasts), bone forming (stimulating the osteoblasts), or dual acting (simultaneously stimulating the osteoblasts and inhibiting the osteoclasts) [27]. The anti-resorptive treatments are bisphosphonates, receptor activator of nuclear factor κ-B ligand (RANKL) antibody, and selective estrogen receptor modulators (SERMs) that either cause osteoclast apoptosis (bisphosphonates) or inhibit osteoclast recruitment (RANKL antibody and SERMs). Teriparatide (parathyroid hormone, amino acids 1–34) and abaloparatide are bone-forming treatments of which abaloparatide currently is only available in the United States [27]. Romosozumab is a dual-acting treatment that stimulates bone formation and inhibits bone resorption at the same time [28].
In the present study, 559 DEGs between osteoporosis patients and normal controls were identified from GSE35959 dataset. WGCNA identified the green, pink, and tan modules as clinically significant. Subsequent analysis revealed six genes (VEGFA, DDX5, SOD2, HNRNPD, EIF5B, and HSP90B1) overlapped in PPI, 559 DEGs and co-expression analysis were the “real” hub genes. Based on SVM, the six-gene signature achieved a 100% prediction accuracy in distinguishing patients with osteoporosis from normal controls, with 100% sensitivity, 100% specificity, and AUC of 1. The results obtained using other four datasets (GSE7158, GSE62402, GSE13850, and GSE7429) further supported such findings.
SVM methods have the feasibility of extracting higher order statistics, and have been widely used in classification and prediction. Lin et al. used different classifiers to evaluate fractures from X-ray images, and reported a high classification accuracy of 98.2% based on a combination of SVM classifiers [29]. Caligiuri et al. showed that SVM is highly effective in distinguishing healthy bones with high Az values from fractured bones [30]. Based on the SVM, the 11-gene combination achieved a 94% prediction accuracy in distinguishing patients with postmenopausal osteoporosis from healthy controls [14]. Chen et al. identified six hub genes as features to build a predictive prognostic model for osteoporosis [31]. Hu et al. constructed a five-feature gene model by SVM for classifying osteoporosis samples [32]. Liu et al. screened three key pathways associated with the development of osteoporosis [33]. In this work, SVM was also applied to identify osteoporosis patients from normal samples based on a six-gene signature with 100% sensitivity.
As for VEGFA, Yu et al. reported that miR-16a-5p and VEGFA contributed to the postmenopausal osteoporosis [34]. A study demonstrated that in the osteoporosis group, VEGFA gene showed a significant association with osteoporosis [35]. VEGFA is a pro-angiogenic factor upregulated when responding to uniaxial cyclic tensile strain in human adipose-derived stem cells (hASCs) and hMSCs from osteoporotic donors [36]. DDX5 is differentially expressed in bone marrow microenvironment of osteoporosis patients [37]. Melatonin promotes osteogenesis by inducing oxidative stress in mitochondria via the SIRT3/SOD2 signaling pathway [38]. Deng et al. reported that SOD2 is a susceptibility gene for osteoporosis among Chinese population [39]. HnRNPL inhibits osteogenic differentiation of PDLCs through downregulating the H3K36me3-specific methyltransferase Setd2. However, there are no reports of HNRNPD in osteoporosis. According to previous study, EIF5B and HSP90B1 have also not been reported in osteoporosis. Still, further research is required to explore the roles of six genes in osteoporosis.
In summary, the present study applied weighted gene co-expression analysis combined with PPI analysis to identify hub genes associated with osteoporosis. In addition, the six-gene combinations may serve as potential biomarkers for osteoporosis to guide clinical treatment for different patients. However, lack of biological investigation and experimental validation with a larger sample size was considered as a limitation of this study. Further studies are therefore needed to verify the diagnostic ability of this gene signature for osteoporosis before clinical application.
Acknowledgments
None.
-
Funding information: This study was supported by Science and Technology Project of Jiangsu Provincial Bureau of Traditional Chinese Medicine (YB201964), Lianyungang Health Science and Technology Project (201915 and 201917), Growth Foundation for middle-aged and young medical talents of The Second People’s Hospital of Lianyungang (TQ201901 and TQ202002), and the major scientific and technological projects of Bengbu Medical College (2020byzd339).
-
Author contributions: Nanning Lv and Mingming Liu concepted and designed the research; Mingming Liu drafted the manuscript and got agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved; Zhangzhe Zhou and Shuangjun He contributed to data acquisition; Xiaofeng Shao and Xinfeng Zhou analyzed the data; Xiaoxiao Feng and Nanning Lv interpreted the data; Zhonglai Qian and Yijian Zhang also revised the manuscript for important intellectual content. All authors approved the final version to be published.
-
Conflict of interest: The authors declare that they have no competing interests.
-
Data availability statement: The datasets generated and/or analyzed during the current study are available in the GSE35959, GSE7158, GSE7429, GSE13850, and GSE62402 datasets.
References
[1] Russell LA. Management of difficult osteoporosis. Best Pract Res Clin Rheumatol. 2018;32(6):835–47.10.1016/j.berh.2019.04.002Suche in Google Scholar PubMed
[2] Curtis EM, Moon RJ, Harvey NC, Cooper C. The impact of fragility fracture and approaches to osteoporosis risk assessment worldwide. Bone. 2017;104:29–38.10.1016/j.bone.2017.01.024Suche in Google Scholar PubMed PubMed Central
[3] Murray TM, Ste-Marie LG. Prevention and management of osteoporosis: consensus statements from the scientific advisory board of the osteoporosis society of Canada. 7. Fluoride therapy for osteoporosis. CMAJ: Canadian Med Assoc J. 1996;155(7):949–54.Suche in Google Scholar
[4] Alford AI, Kozloff KM, Hankenson KD. Extracellular matrix networks in bone remodeling. Int J Biochem Cell Biol. 2015;65:20–31.10.1016/j.biocel.2015.05.008Suche in Google Scholar PubMed
[5] Schürer C, Wallaschofski H, Nauck M, Völzke H, Schober HC, Hannemann A. Fracture risk and risk factors for osteoporosis. Dtsch Arzteblatt Int. 2015;112(21–22):365–71.10.3238/arztebl.2015.0365Suche in Google Scholar PubMed PubMed Central
[6] Leder BZ, Tsai JN, Uihlein AV, Burnett-Bowie SA, Zhu Y, Foley K, et al. Two years of denosumab and teriparatide administration in postmenopausal women with osteoporosis (The DATA Extension Study): a randomized controlled trial. J Clin Endocrinol Metab. 2014;99(5):1694–700.10.1210/jc.2013-4440Suche in Google Scholar PubMed PubMed Central
[7] Idicula-Thomas S, Kulkarni AJ, Kulkarni BD, Jayaraman VK, Balaji PV. A support vector machine-based method for predicting the propensity of a protein to be soluble or to form inclusion body on overexpression in Escherichia coli. Bioinformatics. 2006;22(3):278–84.10.1093/bioinformatics/bti810Suche in Google Scholar PubMed
[8] Qiu Y, Li H, Xie J, Qiao X, Wu J. Identification of ABCC5 among ATP-binding cassette transporter family as a new biomarker for hepatocellular carcinoma based on bioinformatics analysis. Int J Gen Med. 2021;14:7235–46.10.2147/IJGM.S333904Suche in Google Scholar PubMed PubMed Central
[9] Xie J, Li H, Chen L, Cao Y, Hu Y, Zhu Z, et al. A novel pyroptosis-related lncRNA signature for predicting the prognosis of skin cutaneous melanoma. Int J Gen Med. 2021;14:6517–27.10.2147/IJGM.S335396Suche in Google Scholar PubMed PubMed Central
[10] Sapthagirivasan V, Anburajan M. Diagnosis of osteoporosis by extraction of trabecular features from hip radiographs using support vector machine: an investigation panorama with DXA. Comput Biol Med. 2013;43(11):1910–9.10.1016/j.compbiomed.2013.09.002Suche in Google Scholar PubMed
[11] Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteom. 2018;15(1):41–51.10.21873/cgp.20063Suche in Google Scholar
[12] Lynch CM, Abdollahi B, Fuqua JD, de Carlo AR, Bartholomai JA, Balgemann RN, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inform. 2017;108:1–8.10.1016/j.ijmedinf.2017.09.013Suche in Google Scholar PubMed PubMed Central
[13] Kavitha MS, Asano A, Taguchi A, Kurita T, Sanada M. Diagnosis of osteoporosis from dental panoramic radiographs using the support vector machine method in a computer-aided system. BMC Med Imaging. 2012;12:1.10.1186/1471-2342-12-1Suche in Google Scholar PubMed PubMed Central
[14] Yang C, Ren J, Li B, Jin C, Ma C, Cheng C, et al. Identification of gene biomarkers in patients with postmenopausal osteoporosis. Mol Med Rep. 2019;19(2):1065–73.10.3892/mmr.2018.9752Suche in Google Scholar PubMed PubMed Central
[15] Chen X, Liu G, Wang S, Zhang H, Xue P. Machine learning analysis of gene expression profile reveals a novel diagnostic signature for osteoporosis. J Orthopaedic Surg Res. 2021;16(1):189.10.1186/s13018-021-02329-1Suche in Google Scholar PubMed PubMed Central
[16] Chen J, Li K, Pang Q, Yang C, Zhang H, Wu F, et al. Identification of suitable reference gene and biomarkers of serum miRNAs for osteoporosis. Sci Rep. 2016;6:36347.10.1038/srep36347Suche in Google Scholar PubMed PubMed Central
[17] Xia B, Li Y, Zhou J, Tian B, Feng L. Identification of potential pathogenic genes associated with osteoporosis. Bone Jt Res. 2017;6(12):640–8.10.1302/2046-3758.612.BJR-2017-0102.R1Suche in Google Scholar PubMed PubMed Central
[18] Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets – update. Nucleic Acids Res. 2013;41(Database issue):D991–5.10.1093/nar/gks1193Suche in Google Scholar PubMed PubMed Central
[19] Benisch P, Schilling T, Klein-Hitpass L, Frey SP, Seefried L, Raaijmakers N, et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One. 2012;7(9):e45142.10.1371/journal.pone.0045142Suche in Google Scholar PubMed PubMed Central
[20] Cheishvili D, Parashar S, Mahmood N, Arakelian A, Kremer R, Goltzman D, et al. Identification of an epigenetic signature of osteoporosis in blood DNA of postmenopausal women. J Bone Min Res. 2018;33(11):1980–9.10.1002/jbmr.3527Suche in Google Scholar PubMed
[21] Xiao P, Chen Y, Jiang H, Liu YZ, Pan F, Yang TL, et al. In vivo genome-wide expression study on human circulating B cells suggests a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J Bone Min Res. 2008;23(5):644–54.10.1359/jbmr.080105Suche in Google Scholar PubMed PubMed Central
[22] Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.10.1093/nar/gkv007Suche in Google Scholar PubMed PubMed Central
[23] Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7.10.1089/omi.2011.0118Suche in Google Scholar PubMed PubMed Central
[24] Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559.10.1186/1471-2105-9-559Suche in Google Scholar PubMed PubMed Central
[25] Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.10.1101/gr.1239303Suche in Google Scholar PubMed PubMed Central
[26] Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(4):S11.10.1186/1752-0509-8-S4-S11Suche in Google Scholar PubMed PubMed Central
[27] Langdahl BL, Harsløf T. Medical treatment of osteoporotic vertebral fractures. Therapeutic Adv Musculoskelet Dis. 2011;3(1):17–29.10.1177/1759720X10392105Suche in Google Scholar PubMed PubMed Central
[28] Langdahl BL. Overview of treatment approaches to osteoporosis. Br J Pharmacol. 2021;178(9):1891–906.10.1111/bph.15024Suche in Google Scholar PubMed
[29] Kim KC, Cho HC, Jang TJ, Choi JM, Seo JK. Automatic detection and segmentation of lumbar vertebrae from X-ray images for compression fracture evaluation. Comput Methods Prog Biomed. 2021;200:105833.10.1016/j.cmpb.2020.105833Suche in Google Scholar PubMed
[30] Caligiuri P, Giger ML, Favus M. Multifractal radiographic analysis of osteoporosis. Med Phys. 1994;21(4):503–8.10.1118/1.597390Suche in Google Scholar PubMed
[31] Chen Y, Zou L, Lu J, Hu M, Yang Z, Sun C. Identification and validation of novel gene markers of osteoporosis by weighted co expression analysis. Ann Transl Med. 2022;10(4):210.10.21037/atm-22-229Suche in Google Scholar PubMed PubMed Central
[32] Hu M, Zou L, Lu J, Yang Z, Chen Y, Xu Y, et al. Construction of a 5-feature gene model by support vector machine for classifying osteoporosis samples. Bioengineered. 2021;12(1):6821–30.10.1080/21655979.2021.1971026Suche in Google Scholar PubMed PubMed Central
[33] Liu Y, Wang Y, Zhang Y, Liu Z, Xiang H, Peng X, et al. Screening for key pathways associated with the development of osteoporosis by bioinformatics analysis. BioMed Res Int. 2017;2017:8589347.10.1155/2017/8589347Suche in Google Scholar PubMed PubMed Central
[34] Yu T, You X, Zhou H, He W, Li Z, Li B, et al. MiR-16-5p regulates postmenopausal osteoporosis by directly targeting VEGFA. Aging. 2020;12(10):9500–14.10.18632/aging.103223Suche in Google Scholar PubMed PubMed Central
[35] Lee KH, Kim SH, Kim CH, Min BJ, Kim GJ, Lim Y, et al. Identifying genetic variants underlying medication-induced osteonecrosis of the jaw in cancer and osteoporosis: a case control study. J Transl Med. 2019;17(1):381.10.1186/s12967-019-2129-3Suche in Google Scholar PubMed PubMed Central
[36] Charoenpanich A, Wall ME, Tucker CJ, Andrews DM, Lalush DS, Dirschl DR, et al. Cyclic tensile strain enhances osteogenesis and angiogenesis in mesenchymal stem cells from osteoporotic donors. Tissue Eng Part A. 2014;20(1–2):67–78.10.1089/ten.tea.2013.0006Suche in Google Scholar
[37] Zhou Q, Xie F, Zhou B, Wang J, Wu B, Li L, et al. Differentially expressed proteins identified by TMT proteomics analysis in bone marrow microenvironment of osteoporotic patients. Osteoporos Int. 2019;30(5):1089–98.10.1007/s00198-019-04884-0Suche in Google Scholar PubMed
[38] Zhou W, Liu Y, Shen J, Yu B, Bai J, Lin J, et al. Melatonin increases bone mass around the prostheses of OVX rats by ameliorating mitochondrial oxidative stress via the SIRT3/SOD2 signaling pathway. Oxid Med Cell Longev. 2019;2019:4019619.10.1155/2019/4019619Suche in Google Scholar PubMed PubMed Central
[39] Deng FY, Lei SF, Chen XD, Tan LJ, Zhu XZ, Deng HW. An integrative study ascertained SOD2 as a susceptibility gene for osteoporosis in Chinese. J Bone Miner Res. 2011;26(11):2695–701.10.1002/jbmr.471Suche in Google Scholar PubMed PubMed Central
© 2022 Nanning Lv et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- AMBRA1 attenuates the proliferation of uveal melanoma cells
- A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma
- Differences in complications between hepatitis B-related cirrhosis and alcohol-related cirrhosis
- Effect of gestational diabetes mellitus on lipid profile: A systematic review and meta-analysis
- Long noncoding RNA NR2F1-AS1 stimulates the tumorigenic behavior of non-small cell lung cancer cells by sponging miR-363-3p to increase SOX4
- Promising novel biomarkers and candidate small-molecule drugs for lung adenocarcinoma: Evidence from bioinformatics analysis of high-throughput data
- Plasmapheresis: Is it a potential alternative treatment for chronic urticaria?
- The biomarkers of key miRNAs and gene targets associated with extranodal NK/T-cell lymphoma
- Gene signature to predict prognostic survival of hepatocellular carcinoma
- Effects of miRNA-199a-5p on cell proliferation and apoptosis of uterine leiomyoma by targeting MED12
- Does diabetes affect paraneoplastic thrombocytosis in colorectal cancer?
- Is there any effect on imprinted genes H19, PEG3, and SNRPN during AOA?
- Leptin and PCSK9 concentrations are associated with vascular endothelial cytokines in patients with stable coronary heart disease
- Pericentric inversion of chromosome 6 and male fertility problems
- Staple line reinforcement with nebulized cyanoacrylate glue in laparoscopic sleeve gastrectomy: A propensity score-matched study
- Retrospective analysis of crescent score in clinical prognosis of IgA nephropathy
- Expression of DNM3 is associated with good outcome in colorectal cancer
- Activation of SphK2 contributes to adipocyte-induced EOC cell proliferation
- CRRT influences PICCO measurements in febrile critically ill patients
- SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis
- lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells
- circ_AKT3 knockdown suppresses cisplatin resistance in gastric cancer
- Prognostic value of nicotinamide N-methyltransferase in human cancers: Evidence from a meta-analysis and database validation
- GPC2 deficiency inhibits cell growth and metastasis in colon adenocarcinoma
- A pan-cancer analysis of the oncogenic role of Holliday junction recognition protein in human tumors
- Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer
- Association between preventable risk factors and metabolic syndrome
- miR-29c-5p knockdown reduces inflammation and blood–brain barrier disruption by upregulating LRP6
- Cardiac contractility modulation ameliorates myocardial metabolic remodeling in a rabbit model of chronic heart failure through activation of AMPK and PPAR-α pathway
- Quercitrin protects human bronchial epithelial cells from oxidative damage
- Smurf2 suppresses the metastasis of hepatocellular carcinoma via ubiquitin degradation of Smad2
- circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury
- Sonoclot’s usefulness in prediction of cardiopulmonary arrest prognosis: A proof of concept study
- Four drug metabolism-related subgroups of pancreatic adenocarcinoma in prognosis, immune infiltration, and gene mutation
- Decreased expression of miR-195 mediated by hypermethylation promotes osteosarcoma
- LMO3 promotes proliferation and metastasis of papillary thyroid carcinoma cells by regulating LIMK1-mediated cofilin and the β-catenin pathway
- Cx43 upregulation in HUVECs under stretch via TGF-β1 and cytoskeletal network
- Evaluation of menstrual irregularities after COVID-19 vaccination: Results of the MECOVAC survey
- Histopathologic findings on removed stomach after sleeve gastrectomy. Do they influence the outcome?
- Analysis of the expression and prognostic value of MT1-MMP, β1-integrin and YAP1 in glioma
- Optimal diagnosis of the skin cancer using a hybrid deep neural network and grasshopper optimization algorithm
- miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells
- Clinical value of SIRT1 as a prognostic biomarker in esophageal squamous cell carcinoma, a systematic meta-analysis
- circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8
- miR-22-5p regulates the self-renewal of spermatogonial stem cells by targeting EZH2
- hsa-miR-340-5p inhibits epithelial–mesenchymal transition in endometriosis by targeting MAP3K2 and inactivating MAPK/ERK signaling
- circ_0085296 inhibits the biological functions of trophoblast cells to promote the progression of preeclampsia via the miR-942-5p/THBS2 network
- TCD hemodynamics findings in the subacute phase of anterior circulation stroke patients treated with mechanical thrombectomy
- Development of a risk-stratification scoring system for predicting risk of breast cancer based on non-alcoholic fatty liver disease, non-alcoholic fatty pancreas disease, and uric acid
- Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway
- circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis
- Human amniotic fluid as a source of stem cells
- lncRNA NONRATT013819.2 promotes transforming growth factor-β1-induced myofibroblastic transition of hepatic stellate cells by miR24-3p/lox
- NORAD modulates miR-30c-5p-LDHA to protect lung endothelial cells damage
- Idiopathic pulmonary fibrosis telemedicine management during COVID-19 outbreak
- Risk factors for adverse drug reactions associated with clopidogrel therapy
- Serum zinc associated with immunity and inflammatory markers in Covid-19
- The relationship between night shift work and breast cancer incidence: A systematic review and meta-analysis of observational studies
- LncRNA expression in idiopathic achalasia: New insight and preliminary exploration into pathogenesis
- Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway
- Moscatilin suppresses the inflammation from macrophages and T cells
- Zoledronate promotes ECM degradation and apoptosis via Wnt/β-catenin
- Epithelial-mesenchymal transition-related genes in coronary artery disease
- The effect evaluation of traditional vaginal surgery and transvaginal mesh surgery for severe pelvic organ prolapse: 5 years follow-up
- Repeated partial splenic artery embolization for hypersplenism improves platelet count
- Low expression of miR-27b in serum exosomes of non-small cell lung cancer facilitates its progression by affecting EGFR
- Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis
- miR-455-5p enhances 5-fluorouracil sensitivity in colorectal cancer cells by targeting PIK3R1 and DEPDC1
- The effect of tranexamic acid on the reduction of intraoperative and postoperative blood loss and thromboembolic risk in patients with hip fracture
- Isocitrate dehydrogenase 1 mutation in cholangiocarcinoma impairs tumor progression by sensitizing cells to ferroptosis
- Artemisinin protects against cerebral ischemia and reperfusion injury via inhibiting the NF-κB pathway
- A 16-gene signature associated with homologous recombination deficiency for prognosis prediction in patients with triple-negative breast cancer
- Lidocaine ameliorates chronic constriction injury-induced neuropathic pain through regulating M1/M2 microglia polarization
- MicroRNA 322-5p reduced neuronal inflammation via the TLR4/TRAF6/NF-κB axis in a rat epilepsy model
- miR-1273h-5p suppresses CXCL12 expression and inhibits gastric cancer cell invasion and metastasis
- Clinical characteristics of pneumonia patients of long course of illness infected with SARS-CoV-2
- circRNF20 aggravates the malignancy of retinoblastoma depending on the regulation of miR-132-3p/PAX6 axis
- Linezolid for resistant Gram-positive bacterial infections in children under 12 years: A meta-analysis
- Rack1 regulates pro-inflammatory cytokines by NF-κB in diabetic nephropathy
- Comprehensive analysis of molecular mechanism and a novel prognostic signature based on small nuclear RNA biomarkers in gastric cancer patients
- Smog and risk of maternal and fetal birth outcomes: A retrospective study in Baoding, China
- Let-7i-3p inhibits the cell cycle, proliferation, invasion, and migration of colorectal cancer cells via downregulating CCND1
- β2-Adrenergic receptor expression in subchondral bone of patients with varus knee osteoarthritis
- Possible impact of COVID-19 pandemic and lockdown on suicide behavior among patients in Southeast Serbia
- In vitro antimicrobial activity of ozonated oil in liposome eyedrop against multidrug-resistant bacteria
- Potential biomarkers for inflammatory response in acute lung injury
- A low serum uric acid concentration predicts a poor prognosis in adult patients with candidemia
- Antitumor activity of recombinant oncolytic vaccinia virus with human IL2
- ALKBH5 inhibits TNF-α-induced apoptosis of HUVECs through Bcl-2 pathway
- Risk prediction of cardiovascular disease using machine learning classifiers
- Value of ultrasonography parameters in diagnosing polycystic ovary syndrome
- Bioinformatics analysis reveals three key genes and four survival genes associated with youth-onset NSCLC
- Identification of autophagy-related biomarkers in patients with pulmonary arterial hypertension based on bioinformatics analysis
- Protective effects of glaucocalyxin A on the airway of asthmatic mice
- Overexpression of miR-100-5p inhibits papillary thyroid cancer progression via targeting FZD8
- Bioinformatics-based analysis of SUMOylation-related genes in hepatocellular carcinoma reveals a role of upregulated SAE1 in promoting cell proliferation
- Effectiveness and clinical benefits of new anti-diabetic drugs: A real life experience
- Identification of osteoporosis based on gene biomarkers using support vector machine
- Tanshinone IIA reverses oxaliplatin resistance in colorectal cancer through microRNA-30b-5p/AVEN axis
- miR-212-5p inhibits nasopharyngeal carcinoma progression by targeting METTL3
- Association of ST-T changes with all-cause mortality among patients with peripheral T-cell lymphomas
- LINC00665/miRNAs axis-mediated collagen type XI alpha 1 correlates with immune infiltration and malignant phenotypes in lung adenocarcinoma
- The perinatal factors that influence the excretion of fecal calprotectin in premature-born children
- Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study
- Does the use of 3D-printed cones give a chance to postpone the use of megaprostheses in patients with large bone defects in the knee joint?
- lncRNA HAGLR modulates myocardial ischemia–reperfusion injury in mice through regulating miR-133a-3p/MAPK1 axis
- Protective effect of ghrelin on intestinal I/R injury in rats
- In vivo knee kinematics of an innovative prosthesis design
- Relationship between the height of fibular head and the incidence and severity of knee osteoarthritis
- lncRNA WT1-AS attenuates hypoxia/ischemia-induced neuronal injury during cerebral ischemic stroke via miR-186-5p/XIAP axis
- Correlation of cardiac troponin T and APACHE III score with all-cause in-hospital mortality in critically ill patients with acute pulmonary embolism
- LncRNA LINC01857 reduces metastasis and angiogenesis in breast cancer cells via regulating miR-2052/CENPQ axis
- Endothelial cell-specific molecule 1 (ESM1) promoted by transcription factor SPI1 acts as an oncogene to modulate the malignant phenotype of endometrial cancer
- SELENBP1 inhibits progression of colorectal cancer by suppressing epithelial–mesenchymal transition
- Visfatin is negatively associated with coronary artery lesions in subjects with impaired fasting glucose
- Treatment and outcomes of mechanical complications of acute myocardial infarction during the Covid-19 era: A comparison with the pre-Covid-19 period. A systematic review and meta-analysis
- Neonatal stroke surveillance study protocol in the United Kingdom and Republic of Ireland
- Oncogenic role of TWF2 in human tumors: A pan-cancer analysis
- Mean corpuscular hemoglobin predicts the length of hospital stay independent of severity classification in patients with acute pancreatitis
- Association of gallstone and polymorphisms of UGT1A1*27 and UGT1A1*28 in patients with hepatitis B virus-related liver failure
- TGF-β1 upregulates Sar1a expression and induces procollagen-I secretion in hypertrophic scarring fibroblasts
- Antisense lncRNA PCNA-AS1 promotes esophageal squamous cell carcinoma progression through the miR-2467-3p/PCNA axis
- NK-cell dysfunction of acute myeloid leukemia in relation to the renin–angiotensin system and neurotransmitter genes
- The effect of dilution with glucose and prolonged injection time on dexamethasone-induced perineal irritation – A randomized controlled trial
- miR-146-5p restrains calcification of vascular smooth muscle cells by suppressing TRAF6
- Role of lncRNA MIAT/miR-361-3p/CCAR2 in prostate cancer cells
- lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2
- Noninvasive diagnosis of AIH/PBC overlap syndrome based on prediction models
- lncRNA FAM230B is highly expressed in colorectal cancer and suppresses the maturation of miR-1182 to increase cell proliferation
- circ-LIMK1 regulates cisplatin resistance in lung adenocarcinoma by targeting miR-512-5p/HMGA1 axis
- LncRNA SNHG3 promoted cell proliferation, migration, and metastasis of esophageal squamous cell carcinoma via regulating miR-151a-3p/PFN2 axis
- Risk perception and affective state on work exhaustion in obstetrics during the COVID-19 pandemic
- lncRNA-AC130710/miR-129-5p/mGluR1 axis promote migration and invasion by activating PKCα-MAPK signal pathway in melanoma
- SNRPB promotes cell cycle progression in thyroid carcinoma via inhibiting p53
- Xylooligosaccharides and aerobic training regulate metabolism and behavior in rats with streptozotocin-induced type 1 diabetes
- Serpin family A member 1 is an oncogene in glioma and its translation is enhanced by NAD(P)H quinone dehydrogenase 1 through RNA-binding activity
- Silencing of CPSF7 inhibits the proliferation, migration, and invasion of lung adenocarcinoma cells by blocking the AKT/mTOR signaling pathway
- Ultrasound-guided lumbar plexus block versus transversus abdominis plane block for analgesia in children with hip dislocation: A double-blind, randomized trial
- Relationship of plasma MBP and 8-oxo-dG with brain damage in preterm
- Identification of a novel necroptosis-associated miRNA signature for predicting the prognosis in head and neck squamous cell carcinoma
- Delayed femoral vein ligation reduces operative time and blood loss during hip disarticulation in patients with extremity tumors
- The expression of ASAP3 and NOTCH3 and the clinicopathological characteristics of adult glioma patients
- Longitudinal analysis of factors related to Helicobacter pylori infection in Chinese adults
- HOXA10 enhances cell proliferation and suppresses apoptosis in esophageal cancer via activating p38/ERK signaling pathway
- Meta-analysis of early-life antibiotic use and allergic rhinitis
- Marital status and its correlation with age, race, and gender in prognosis of tonsil squamous cell carcinomas
- HPV16 E6E7 up-regulates KIF2A expression by activating JNK/c-Jun signal, is beneficial to migration and invasion of cervical cancer cells
- Amino acid profiles in the tissue and serum of patients with liver cancer
- Pain in critically ill COVID-19 patients: An Italian retrospective study
- Immunohistochemical distribution of Bcl-2 and p53 apoptotic markers in acetamiprid-induced nephrotoxicity
- Estradiol pretreatment in GnRH antagonist protocol for IVF/ICSI treatment
- Long non-coding RNAs LINC00689 inhibits the apoptosis of human nucleus pulposus cells via miR-3127-5p/ATG7 axis-mediated autophagy
- The relationship between oxygen therapy, drug therapy, and COVID-19 mortality
- Monitoring hypertensive disorders in pregnancy to prevent preeclampsia in pregnant women of advanced maternal age: Trial mimicking with retrospective data
- SETD1A promotes the proliferation and glycolysis of nasopharyngeal carcinoma cells by activating the PI3K/Akt pathway
- The role of Shunaoxin pills in the treatment of chronic cerebral hypoperfusion and its main pharmacodynamic components
- TET3 governs malignant behaviors and unfavorable prognosis of esophageal squamous cell carcinoma by activating the PI3K/AKT/GSK3β/β-catenin pathway
- Associations between morphokinetic parameters of temporary-arrest embryos and the clinical prognosis in FET cycles
- Long noncoding RNA WT1-AS regulates trophoblast proliferation, migration, and invasion via the microRNA-186-5p/CADM2 axis
- The incidence of bronchiectasis in chronic obstructive pulmonary disease
- Integrated bioinformatics analysis shows integrin alpha 3 is a prognostic biomarker for pancreatic cancer
- Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway
- Comparison of hospitalized patients with severe pneumonia caused by COVID-19 and influenza A (H7N9 and H1N1): A retrospective study from a designated hospital
- lncRNA ZFAS1 promotes intervertebral disc degeneration by upregulating AAK1
- Pathological characteristics of liver injury induced by N,N-dimethylformamide: From humans to animal models
- lncRNA ELFN1-AS1 enhances the progression of colon cancer by targeting miR-4270 to upregulate AURKB
- DARS-AS1 modulates cell proliferation and migration of gastric cancer cells by regulating miR-330-3p/NAT10 axis
- Dezocine inhibits cell proliferation, migration, and invasion by targeting CRABP2 in ovarian cancer
- MGST1 alleviates the oxidative stress of trophoblast cells induced by hypoxia/reoxygenation and promotes cell proliferation, migration, and invasion by activating the PI3K/AKT/mTOR pathway
- Bifidobacterium lactis Probio-M8 ameliorated the symptoms of type 2 diabetes mellitus mice by changing ileum FXR-CYP7A1
- circRNA DENND1B inhibits tumorigenicity of clear cell renal cell carcinoma via miR-122-5p/TIMP2 axis
- EphA3 targeted by miR-3666 contributes to melanoma malignancy via activating ERK1/2 and p38 MAPK pathways
- Pacemakers and methylprednisolone pulse therapy in immune-related myocarditis concomitant with complete heart block
- miRNA-130a-3p targets sphingosine-1-phosphate receptor 1 to activate the microglial and astrocytes and to promote neural injury under the high glucose condition
- Review Articles
- Current management of cancer pain in Italy: Expert opinion paper
- Hearing loss and brain disorders: A review of multiple pathologies
- The rationale for using low-molecular weight heparin in the therapy of symptomatic COVID-19 patients
- Amyotrophic lateral sclerosis and delayed onset muscle soreness in light of the impaired blink and stretch reflexes – watch out for Piezo2
- Interleukin-35 in autoimmune dermatoses: Current concepts
- Recent discoveries in microbiota dysbiosis, cholangiocytic factors, and models for studying the pathogenesis of primary sclerosing cholangitis
- Advantages of ketamine in pediatric anesthesia
- Congenital adrenal hyperplasia. Role of dentist in early diagnosis
- Migraine management: Non-pharmacological points for patients and health care professionals
- Atherogenic index of plasma and coronary artery disease: A systematic review
- Physiological and modulatory role of thioredoxins in the cellular function
- Case Reports
- Intrauterine Bakri balloon tamponade plus cervical cerclage for the prevention and treatment of postpartum haemorrhage in late pregnancy complicated with acute aortic dissection: Case series
- A case of successful pembrolizumab monotherapy in a patient with advanced lung adenocarcinoma: Use of multiple biomarkers in combination for clinical practice
- Unusual neurological manifestations of bilateral medial medullary infarction: A case report
- Atypical symptoms of malignant hyperthermia: A rare causative mutation in the RYR1 gene
- A case report of dermatomyositis with the missed diagnosis of non-small cell lung cancer and concurrence of pulmonary tuberculosis
- A rare case of endometrial polyp complicated with uterine inversion: A case report and clinical management
- Spontaneous rupturing of splenic artery aneurysm: Another reason for fatal syncope and shock (Case report and literature review)
- Fungal infection mimicking COVID-19 infection – A case report
- Concurrent aspergillosis and cystic pulmonary metastases in a patient with tongue squamous cell carcinoma
- Paraganglioma-induced inverted takotsubo-like cardiomyopathy leading to cardiogenic shock successfully treated with extracorporeal membrane oxygenation
- Lineage switch from lymphoma to myeloid neoplasms: First case series from a single institution
- Trismus during tracheal extubation as a complication of general anaesthesia – A case report
- Simultaneous treatment of a pubovesical fistula and lymph node metastasis secondary to multimodal treatment for prostate cancer: Case report and review of the literature
- Two case reports of skin vasculitis following the COVID-19 immunization
- Ureteroiliac fistula after oncological surgery: Case report and review of the literature
- Synchronous triple primary malignant tumours in the bladder, prostate, and lung harbouring TP53 and MEK1 mutations accompanied with severe cardiovascular diseases: A case report
- Huge mucinous cystic neoplasms with adhesion to the left colon: A case report and literature review
- Commentary
- Commentary on “Clinicopathological features of programmed cell death-ligand 1 expression in patients with oral squamous cell carcinoma”
- Rapid Communication
- COVID-19 fear, post-traumatic stress, growth, and the role of resilience
- Erratum
- Erratum to “Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway”
- Erratum to “Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study”
- Erratum to “lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2”
- Retraction
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Retraction to “miR-519d downregulates LEP expression to inhibit preeclampsia development”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part II
- Usefulness of close surveillance for rectal cancer patients after neoadjuvant chemoradiotherapy
Artikel in diesem Heft
- Research Articles
- AMBRA1 attenuates the proliferation of uveal melanoma cells
- A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma
- Differences in complications between hepatitis B-related cirrhosis and alcohol-related cirrhosis
- Effect of gestational diabetes mellitus on lipid profile: A systematic review and meta-analysis
- Long noncoding RNA NR2F1-AS1 stimulates the tumorigenic behavior of non-small cell lung cancer cells by sponging miR-363-3p to increase SOX4
- Promising novel biomarkers and candidate small-molecule drugs for lung adenocarcinoma: Evidence from bioinformatics analysis of high-throughput data
- Plasmapheresis: Is it a potential alternative treatment for chronic urticaria?
- The biomarkers of key miRNAs and gene targets associated with extranodal NK/T-cell lymphoma
- Gene signature to predict prognostic survival of hepatocellular carcinoma
- Effects of miRNA-199a-5p on cell proliferation and apoptosis of uterine leiomyoma by targeting MED12
- Does diabetes affect paraneoplastic thrombocytosis in colorectal cancer?
- Is there any effect on imprinted genes H19, PEG3, and SNRPN during AOA?
- Leptin and PCSK9 concentrations are associated with vascular endothelial cytokines in patients with stable coronary heart disease
- Pericentric inversion of chromosome 6 and male fertility problems
- Staple line reinforcement with nebulized cyanoacrylate glue in laparoscopic sleeve gastrectomy: A propensity score-matched study
- Retrospective analysis of crescent score in clinical prognosis of IgA nephropathy
- Expression of DNM3 is associated with good outcome in colorectal cancer
- Activation of SphK2 contributes to adipocyte-induced EOC cell proliferation
- CRRT influences PICCO measurements in febrile critically ill patients
- SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis
- lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells
- circ_AKT3 knockdown suppresses cisplatin resistance in gastric cancer
- Prognostic value of nicotinamide N-methyltransferase in human cancers: Evidence from a meta-analysis and database validation
- GPC2 deficiency inhibits cell growth and metastasis in colon adenocarcinoma
- A pan-cancer analysis of the oncogenic role of Holliday junction recognition protein in human tumors
- Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer
- Association between preventable risk factors and metabolic syndrome
- miR-29c-5p knockdown reduces inflammation and blood–brain barrier disruption by upregulating LRP6
- Cardiac contractility modulation ameliorates myocardial metabolic remodeling in a rabbit model of chronic heart failure through activation of AMPK and PPAR-α pathway
- Quercitrin protects human bronchial epithelial cells from oxidative damage
- Smurf2 suppresses the metastasis of hepatocellular carcinoma via ubiquitin degradation of Smad2
- circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury
- Sonoclot’s usefulness in prediction of cardiopulmonary arrest prognosis: A proof of concept study
- Four drug metabolism-related subgroups of pancreatic adenocarcinoma in prognosis, immune infiltration, and gene mutation
- Decreased expression of miR-195 mediated by hypermethylation promotes osteosarcoma
- LMO3 promotes proliferation and metastasis of papillary thyroid carcinoma cells by regulating LIMK1-mediated cofilin and the β-catenin pathway
- Cx43 upregulation in HUVECs under stretch via TGF-β1 and cytoskeletal network
- Evaluation of menstrual irregularities after COVID-19 vaccination: Results of the MECOVAC survey
- Histopathologic findings on removed stomach after sleeve gastrectomy. Do they influence the outcome?
- Analysis of the expression and prognostic value of MT1-MMP, β1-integrin and YAP1 in glioma
- Optimal diagnosis of the skin cancer using a hybrid deep neural network and grasshopper optimization algorithm
- miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells
- Clinical value of SIRT1 as a prognostic biomarker in esophageal squamous cell carcinoma, a systematic meta-analysis
- circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8
- miR-22-5p regulates the self-renewal of spermatogonial stem cells by targeting EZH2
- hsa-miR-340-5p inhibits epithelial–mesenchymal transition in endometriosis by targeting MAP3K2 and inactivating MAPK/ERK signaling
- circ_0085296 inhibits the biological functions of trophoblast cells to promote the progression of preeclampsia via the miR-942-5p/THBS2 network
- TCD hemodynamics findings in the subacute phase of anterior circulation stroke patients treated with mechanical thrombectomy
- Development of a risk-stratification scoring system for predicting risk of breast cancer based on non-alcoholic fatty liver disease, non-alcoholic fatty pancreas disease, and uric acid
- Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway
- circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis
- Human amniotic fluid as a source of stem cells
- lncRNA NONRATT013819.2 promotes transforming growth factor-β1-induced myofibroblastic transition of hepatic stellate cells by miR24-3p/lox
- NORAD modulates miR-30c-5p-LDHA to protect lung endothelial cells damage
- Idiopathic pulmonary fibrosis telemedicine management during COVID-19 outbreak
- Risk factors for adverse drug reactions associated with clopidogrel therapy
- Serum zinc associated with immunity and inflammatory markers in Covid-19
- The relationship between night shift work and breast cancer incidence: A systematic review and meta-analysis of observational studies
- LncRNA expression in idiopathic achalasia: New insight and preliminary exploration into pathogenesis
- Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway
- Moscatilin suppresses the inflammation from macrophages and T cells
- Zoledronate promotes ECM degradation and apoptosis via Wnt/β-catenin
- Epithelial-mesenchymal transition-related genes in coronary artery disease
- The effect evaluation of traditional vaginal surgery and transvaginal mesh surgery for severe pelvic organ prolapse: 5 years follow-up
- Repeated partial splenic artery embolization for hypersplenism improves platelet count
- Low expression of miR-27b in serum exosomes of non-small cell lung cancer facilitates its progression by affecting EGFR
- Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis
- miR-455-5p enhances 5-fluorouracil sensitivity in colorectal cancer cells by targeting PIK3R1 and DEPDC1
- The effect of tranexamic acid on the reduction of intraoperative and postoperative blood loss and thromboembolic risk in patients with hip fracture
- Isocitrate dehydrogenase 1 mutation in cholangiocarcinoma impairs tumor progression by sensitizing cells to ferroptosis
- Artemisinin protects against cerebral ischemia and reperfusion injury via inhibiting the NF-κB pathway
- A 16-gene signature associated with homologous recombination deficiency for prognosis prediction in patients with triple-negative breast cancer
- Lidocaine ameliorates chronic constriction injury-induced neuropathic pain through regulating M1/M2 microglia polarization
- MicroRNA 322-5p reduced neuronal inflammation via the TLR4/TRAF6/NF-κB axis in a rat epilepsy model
- miR-1273h-5p suppresses CXCL12 expression and inhibits gastric cancer cell invasion and metastasis
- Clinical characteristics of pneumonia patients of long course of illness infected with SARS-CoV-2
- circRNF20 aggravates the malignancy of retinoblastoma depending on the regulation of miR-132-3p/PAX6 axis
- Linezolid for resistant Gram-positive bacterial infections in children under 12 years: A meta-analysis
- Rack1 regulates pro-inflammatory cytokines by NF-κB in diabetic nephropathy
- Comprehensive analysis of molecular mechanism and a novel prognostic signature based on small nuclear RNA biomarkers in gastric cancer patients
- Smog and risk of maternal and fetal birth outcomes: A retrospective study in Baoding, China
- Let-7i-3p inhibits the cell cycle, proliferation, invasion, and migration of colorectal cancer cells via downregulating CCND1
- β2-Adrenergic receptor expression in subchondral bone of patients with varus knee osteoarthritis
- Possible impact of COVID-19 pandemic and lockdown on suicide behavior among patients in Southeast Serbia
- In vitro antimicrobial activity of ozonated oil in liposome eyedrop against multidrug-resistant bacteria
- Potential biomarkers for inflammatory response in acute lung injury
- A low serum uric acid concentration predicts a poor prognosis in adult patients with candidemia
- Antitumor activity of recombinant oncolytic vaccinia virus with human IL2
- ALKBH5 inhibits TNF-α-induced apoptosis of HUVECs through Bcl-2 pathway
- Risk prediction of cardiovascular disease using machine learning classifiers
- Value of ultrasonography parameters in diagnosing polycystic ovary syndrome
- Bioinformatics analysis reveals three key genes and four survival genes associated with youth-onset NSCLC
- Identification of autophagy-related biomarkers in patients with pulmonary arterial hypertension based on bioinformatics analysis
- Protective effects of glaucocalyxin A on the airway of asthmatic mice
- Overexpression of miR-100-5p inhibits papillary thyroid cancer progression via targeting FZD8
- Bioinformatics-based analysis of SUMOylation-related genes in hepatocellular carcinoma reveals a role of upregulated SAE1 in promoting cell proliferation
- Effectiveness and clinical benefits of new anti-diabetic drugs: A real life experience
- Identification of osteoporosis based on gene biomarkers using support vector machine
- Tanshinone IIA reverses oxaliplatin resistance in colorectal cancer through microRNA-30b-5p/AVEN axis
- miR-212-5p inhibits nasopharyngeal carcinoma progression by targeting METTL3
- Association of ST-T changes with all-cause mortality among patients with peripheral T-cell lymphomas
- LINC00665/miRNAs axis-mediated collagen type XI alpha 1 correlates with immune infiltration and malignant phenotypes in lung adenocarcinoma
- The perinatal factors that influence the excretion of fecal calprotectin in premature-born children
- Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study
- Does the use of 3D-printed cones give a chance to postpone the use of megaprostheses in patients with large bone defects in the knee joint?
- lncRNA HAGLR modulates myocardial ischemia–reperfusion injury in mice through regulating miR-133a-3p/MAPK1 axis
- Protective effect of ghrelin on intestinal I/R injury in rats
- In vivo knee kinematics of an innovative prosthesis design
- Relationship between the height of fibular head and the incidence and severity of knee osteoarthritis
- lncRNA WT1-AS attenuates hypoxia/ischemia-induced neuronal injury during cerebral ischemic stroke via miR-186-5p/XIAP axis
- Correlation of cardiac troponin T and APACHE III score with all-cause in-hospital mortality in critically ill patients with acute pulmonary embolism
- LncRNA LINC01857 reduces metastasis and angiogenesis in breast cancer cells via regulating miR-2052/CENPQ axis
- Endothelial cell-specific molecule 1 (ESM1) promoted by transcription factor SPI1 acts as an oncogene to modulate the malignant phenotype of endometrial cancer
- SELENBP1 inhibits progression of colorectal cancer by suppressing epithelial–mesenchymal transition
- Visfatin is negatively associated with coronary artery lesions in subjects with impaired fasting glucose
- Treatment and outcomes of mechanical complications of acute myocardial infarction during the Covid-19 era: A comparison with the pre-Covid-19 period. A systematic review and meta-analysis
- Neonatal stroke surveillance study protocol in the United Kingdom and Republic of Ireland
- Oncogenic role of TWF2 in human tumors: A pan-cancer analysis
- Mean corpuscular hemoglobin predicts the length of hospital stay independent of severity classification in patients with acute pancreatitis
- Association of gallstone and polymorphisms of UGT1A1*27 and UGT1A1*28 in patients with hepatitis B virus-related liver failure
- TGF-β1 upregulates Sar1a expression and induces procollagen-I secretion in hypertrophic scarring fibroblasts
- Antisense lncRNA PCNA-AS1 promotes esophageal squamous cell carcinoma progression through the miR-2467-3p/PCNA axis
- NK-cell dysfunction of acute myeloid leukemia in relation to the renin–angiotensin system and neurotransmitter genes
- The effect of dilution with glucose and prolonged injection time on dexamethasone-induced perineal irritation – A randomized controlled trial
- miR-146-5p restrains calcification of vascular smooth muscle cells by suppressing TRAF6
- Role of lncRNA MIAT/miR-361-3p/CCAR2 in prostate cancer cells
- lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2
- Noninvasive diagnosis of AIH/PBC overlap syndrome based on prediction models
- lncRNA FAM230B is highly expressed in colorectal cancer and suppresses the maturation of miR-1182 to increase cell proliferation
- circ-LIMK1 regulates cisplatin resistance in lung adenocarcinoma by targeting miR-512-5p/HMGA1 axis
- LncRNA SNHG3 promoted cell proliferation, migration, and metastasis of esophageal squamous cell carcinoma via regulating miR-151a-3p/PFN2 axis
- Risk perception and affective state on work exhaustion in obstetrics during the COVID-19 pandemic
- lncRNA-AC130710/miR-129-5p/mGluR1 axis promote migration and invasion by activating PKCα-MAPK signal pathway in melanoma
- SNRPB promotes cell cycle progression in thyroid carcinoma via inhibiting p53
- Xylooligosaccharides and aerobic training regulate metabolism and behavior in rats with streptozotocin-induced type 1 diabetes
- Serpin family A member 1 is an oncogene in glioma and its translation is enhanced by NAD(P)H quinone dehydrogenase 1 through RNA-binding activity
- Silencing of CPSF7 inhibits the proliferation, migration, and invasion of lung adenocarcinoma cells by blocking the AKT/mTOR signaling pathway
- Ultrasound-guided lumbar plexus block versus transversus abdominis plane block for analgesia in children with hip dislocation: A double-blind, randomized trial
- Relationship of plasma MBP and 8-oxo-dG with brain damage in preterm
- Identification of a novel necroptosis-associated miRNA signature for predicting the prognosis in head and neck squamous cell carcinoma
- Delayed femoral vein ligation reduces operative time and blood loss during hip disarticulation in patients with extremity tumors
- The expression of ASAP3 and NOTCH3 and the clinicopathological characteristics of adult glioma patients
- Longitudinal analysis of factors related to Helicobacter pylori infection in Chinese adults
- HOXA10 enhances cell proliferation and suppresses apoptosis in esophageal cancer via activating p38/ERK signaling pathway
- Meta-analysis of early-life antibiotic use and allergic rhinitis
- Marital status and its correlation with age, race, and gender in prognosis of tonsil squamous cell carcinomas
- HPV16 E6E7 up-regulates KIF2A expression by activating JNK/c-Jun signal, is beneficial to migration and invasion of cervical cancer cells
- Amino acid profiles in the tissue and serum of patients with liver cancer
- Pain in critically ill COVID-19 patients: An Italian retrospective study
- Immunohistochemical distribution of Bcl-2 and p53 apoptotic markers in acetamiprid-induced nephrotoxicity
- Estradiol pretreatment in GnRH antagonist protocol for IVF/ICSI treatment
- Long non-coding RNAs LINC00689 inhibits the apoptosis of human nucleus pulposus cells via miR-3127-5p/ATG7 axis-mediated autophagy
- The relationship between oxygen therapy, drug therapy, and COVID-19 mortality
- Monitoring hypertensive disorders in pregnancy to prevent preeclampsia in pregnant women of advanced maternal age: Trial mimicking with retrospective data
- SETD1A promotes the proliferation and glycolysis of nasopharyngeal carcinoma cells by activating the PI3K/Akt pathway
- The role of Shunaoxin pills in the treatment of chronic cerebral hypoperfusion and its main pharmacodynamic components
- TET3 governs malignant behaviors and unfavorable prognosis of esophageal squamous cell carcinoma by activating the PI3K/AKT/GSK3β/β-catenin pathway
- Associations between morphokinetic parameters of temporary-arrest embryos and the clinical prognosis in FET cycles
- Long noncoding RNA WT1-AS regulates trophoblast proliferation, migration, and invasion via the microRNA-186-5p/CADM2 axis
- The incidence of bronchiectasis in chronic obstructive pulmonary disease
- Integrated bioinformatics analysis shows integrin alpha 3 is a prognostic biomarker for pancreatic cancer
- Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway
- Comparison of hospitalized patients with severe pneumonia caused by COVID-19 and influenza A (H7N9 and H1N1): A retrospective study from a designated hospital
- lncRNA ZFAS1 promotes intervertebral disc degeneration by upregulating AAK1
- Pathological characteristics of liver injury induced by N,N-dimethylformamide: From humans to animal models
- lncRNA ELFN1-AS1 enhances the progression of colon cancer by targeting miR-4270 to upregulate AURKB
- DARS-AS1 modulates cell proliferation and migration of gastric cancer cells by regulating miR-330-3p/NAT10 axis
- Dezocine inhibits cell proliferation, migration, and invasion by targeting CRABP2 in ovarian cancer
- MGST1 alleviates the oxidative stress of trophoblast cells induced by hypoxia/reoxygenation and promotes cell proliferation, migration, and invasion by activating the PI3K/AKT/mTOR pathway
- Bifidobacterium lactis Probio-M8 ameliorated the symptoms of type 2 diabetes mellitus mice by changing ileum FXR-CYP7A1
- circRNA DENND1B inhibits tumorigenicity of clear cell renal cell carcinoma via miR-122-5p/TIMP2 axis
- EphA3 targeted by miR-3666 contributes to melanoma malignancy via activating ERK1/2 and p38 MAPK pathways
- Pacemakers and methylprednisolone pulse therapy in immune-related myocarditis concomitant with complete heart block
- miRNA-130a-3p targets sphingosine-1-phosphate receptor 1 to activate the microglial and astrocytes and to promote neural injury under the high glucose condition
- Review Articles
- Current management of cancer pain in Italy: Expert opinion paper
- Hearing loss and brain disorders: A review of multiple pathologies
- The rationale for using low-molecular weight heparin in the therapy of symptomatic COVID-19 patients
- Amyotrophic lateral sclerosis and delayed onset muscle soreness in light of the impaired blink and stretch reflexes – watch out for Piezo2
- Interleukin-35 in autoimmune dermatoses: Current concepts
- Recent discoveries in microbiota dysbiosis, cholangiocytic factors, and models for studying the pathogenesis of primary sclerosing cholangitis
- Advantages of ketamine in pediatric anesthesia
- Congenital adrenal hyperplasia. Role of dentist in early diagnosis
- Migraine management: Non-pharmacological points for patients and health care professionals
- Atherogenic index of plasma and coronary artery disease: A systematic review
- Physiological and modulatory role of thioredoxins in the cellular function
- Case Reports
- Intrauterine Bakri balloon tamponade plus cervical cerclage for the prevention and treatment of postpartum haemorrhage in late pregnancy complicated with acute aortic dissection: Case series
- A case of successful pembrolizumab monotherapy in a patient with advanced lung adenocarcinoma: Use of multiple biomarkers in combination for clinical practice
- Unusual neurological manifestations of bilateral medial medullary infarction: A case report
- Atypical symptoms of malignant hyperthermia: A rare causative mutation in the RYR1 gene
- A case report of dermatomyositis with the missed diagnosis of non-small cell lung cancer and concurrence of pulmonary tuberculosis
- A rare case of endometrial polyp complicated with uterine inversion: A case report and clinical management
- Spontaneous rupturing of splenic artery aneurysm: Another reason for fatal syncope and shock (Case report and literature review)
- Fungal infection mimicking COVID-19 infection – A case report
- Concurrent aspergillosis and cystic pulmonary metastases in a patient with tongue squamous cell carcinoma
- Paraganglioma-induced inverted takotsubo-like cardiomyopathy leading to cardiogenic shock successfully treated with extracorporeal membrane oxygenation
- Lineage switch from lymphoma to myeloid neoplasms: First case series from a single institution
- Trismus during tracheal extubation as a complication of general anaesthesia – A case report
- Simultaneous treatment of a pubovesical fistula and lymph node metastasis secondary to multimodal treatment for prostate cancer: Case report and review of the literature
- Two case reports of skin vasculitis following the COVID-19 immunization
- Ureteroiliac fistula after oncological surgery: Case report and review of the literature
- Synchronous triple primary malignant tumours in the bladder, prostate, and lung harbouring TP53 and MEK1 mutations accompanied with severe cardiovascular diseases: A case report
- Huge mucinous cystic neoplasms with adhesion to the left colon: A case report and literature review
- Commentary
- Commentary on “Clinicopathological features of programmed cell death-ligand 1 expression in patients with oral squamous cell carcinoma”
- Rapid Communication
- COVID-19 fear, post-traumatic stress, growth, and the role of resilience
- Erratum
- Erratum to “Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway”
- Erratum to “Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study”
- Erratum to “lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2”
- Retraction
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Retraction to “miR-519d downregulates LEP expression to inhibit preeclampsia development”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part II
- Usefulness of close surveillance for rectal cancer patients after neoadjuvant chemoradiotherapy


