Abstract
Background
Circular RNAs (circRNAs) are associated with cisplatin resistance in gastric cancer (GC). This study aims to explore the role of circRNA AKT serine/threonine kinase 3 (circ_AKT3) in the resistance of GC to cisplatin.
Methods
42 sensitive and 23 resistant GC patients were recruited for tissue collection. The cisplatin-resistant GC cells MKN-7/DDP and HGC-27/DDP were used for in vitro study. circ_AKT3, microRNA-206 (miR-206) and protein tyrosine phosphatase non-receptor type 14 (PTPN14) levels were detected via quantitative reverse transcription real-time PCR (qPCR) and Western blot. Cisplatin resistance was assessed by detecting P-glycoprotein (P-gp) level, half maximal inhibitory concentration (IC50) of cisplatin and cell apoptosis. The target relationship between miR-206 and circ_AKT3 or PTPN14 was analyzed via dual-luciferase reporter and RNA pull-down assays. The role of circ_AKT3 in vivo was assessed using xenograft model.
Results
circ_AKT3 level was increased, but miR-206 was declined in cisplatin-resistant GC tissues and cells. circ_AKT3 knockdown or miR-206 overexpression decreased the level of P-gp and IC50 of cisplatin and increased apoptosis of MKN-7/DDP and HGC-27/DDP cells. Additionally, circ_AKT3 targeted miR-206, and regulated cisplatin resistance by interacting with miR-206. PTPN14 was regulated by circ_AKT3 through miR-206 as a bridge. Also, circ_AKT3 knockdown decreased xenograft tumor growth.
Conclusion
circ_AKT3 knockdown suppressed cisplatin resistance using miR-206/PTPN14 axis in cisplatin-resistant GC cells.
1 Introduction
Gastric cancer (GC) is one of the main malignancies with high mortality and incidence [1]. Surgery is the main therapeutic strategy of GC, while the clinical outcomes of advanced GC patients remain poor. The combined treatment of surgery and chemotherapy or radiotherapy has been implemented in GC [2]. Cisplatin (DDP) is a major known chemotherapy drug, whereas development of drug resistance limits its therapeutic efficacy [3]. Hence, it is necessary to urgently explore novel strategies to improve cisplatin sensitivity in GC treatment.
Noncoding RNAs, such as circular RNAs (circRNAs) and microRNAs (miRNAs), are not able to code proteins and play important roles in development and therapeutics of cancers [4]. circRNAs have a covalently closed continuous loop structure and play essential roles in cancers [5]. Moreover, circRNAs are associated with occurrence of chemoresistance in human cancers [6]. circRNA AKT serine/threonine kinase 3 (circ_AKT3) is derived from AKT3, which has been regarded to have a vital role in glioblastoma and clear cell renal cell carcinoma [7,8]. More importantly, circ_AKT3 could increase chemoresistance in GC by suppressing miR-198 [9]. However, the mechanism involving circ_AKT3 is complicated and requires further investigation, particularly with respect to cisplatin resistance.
miRNAs regulate eukaryotic gene expression and exhibit pivotal roles in diagnosis, therapy and prognosis of GC [10]. Former works suggested that miR-206 could inhibit resistance to drugs, such as paclitaxel, euthyrox, gefitinib and cisplatin [11,12,13,14]. Moreover, miR-206 is reported to play an anti-cancer role and could inhibit cisplatin resistance in GC [15]. Nevertheless, whether miR-206 is responsible for circ_AKT3-meidated cisplatin resistance is unknown. Furthermore, previous studies report that protein tyrosine phosphatase non-receptor type 14 (PTPN14) is an oncogene and associated with drug resistance in GC [16,17]. Through bioinformatics analysis, we found the predicted complementary sites of miR-206 with circ_AKT3 and PTPN14. Thus, we hypothesized that miR-206 and PTPN14 might be associated with the mechanism addressed by circ_AKT3. In this study, we investigated the influence of circ_AKT3 on cisplatin resistance in GC and explored whether it was regulated by miR-206/PTPN14 axis.
2 Materials and methods
2.1 Patients and tissues
42 cisplatin-sensitive and 23 cisplatin-resistant patients with advanced GC were recruited for this study. Patients had undergone two cycles of cisplatin-based chemotherapy and were divided into sensitive or resistant group according to the efficacy of the therapy [18]. The cancer tissues were collected and stored at −80°C for RNA extraction. Written informed consents were obtained. This study was approved by the ethics committee of the Changchun University of Chinese Medicine.
2.2 Cell culture and transfection
MKN-7 and HGC-27 cells were purchased from BeNa Culture Collection (Beijing, China). The corresponding cisplatin-resistant variants MKN-7/DDP and HGC-27/DDP were obtained through exposure to increased doses of cisplatin. All cells were grown in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco). To maintain the resistant phenotype, the resistant cells were cultured in a medium containing 1 μg/mL cisplatin (Sigma, St. Louis, MO, USA).
Small interfering RNA (siRNA) for circ_AKT3 (si-circ_AKT3), negative control (si-NC), circ_AKT3 overexpression vector, pcDNA circRNA mini vector (vector), PTPN14 overexpression vector, pcDNA3.1 empty vector (pcDNA), miR-206 mimic, miRNA negative control (miR-NC), miR-206 inhibitor (anti-miR-206) and inhibitor negative control (anti-miR-NC) were synthesized by RiboBio (Guangzhou, China). Upon reaching 60% confluence, MKN-7/DDP and HGC-27/DDP cells were transfected with 40 nM oligonucleotides or 500 ng vectors via LipoFiterTM Liposomal Transfection Reagent (Hanbio, Shanghai, China) and cultured for 24 h.
2.3 RNA isolation and quantitative reverse transcription PCR (RT-qPCR)
RNA isolation was performed through Trizol-based methods. The RNA was employed for reverse transcription and RT-qPCR using All-in-OneTM First-Strand cDNA Synthesis Kit and RT-qPCR Detection Kit (FulenGen, Guangzhou, China). The specific primers used in this study were shown as follows: circ_AKT3 (Forward, 5′-TTGGTGGAGGACCAGATGAT-3′ and Reverse, 5′-ATAGAAACGTGTGCGGTCCT-3′); PTPN14 (Forward, 5′-TCTAGGATGTGTGCCAGGGA-3′ and Reverse, 5′-TGTGGTGCTTCCGGAAATGT-3′); and GAPDH (Forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and Reverse, 5′-GCCATCACGCCACAGTTTC-3′). The primers for miR-206 and U6 were provided by FulenGen with a universal adaptor PCR primer. The relative expression levels of circ_AKT3, PTPN14 and miR-206 were analyzed according to the 2−ΔΔCt method with GAPDH or U6 as a reference [19].
2.4 Cisplatin cytotoxicity assay
Cisplatin cytotoxicity was investigated by a MTT Cell Cytotoxicity Assay Kit (Beyotime, Shanghai, China). MKN-7/DDP and HGC-27/DDP cells (10,000 cells per well) were placed into 96-well plates overnight and then incubated with a series of concentrations (0–20 μg/mL) of cisplatin for 48 h. Then, the cells were incubated with MTT reagent for 4 h and then incubated with formazan dissolving solution. Subsequently, the output of the absorbance at OD570 nm was determined with a microplate reader (Bio-Rad, Hercules, CA, USA). The half maximal inhibitory concentration (IC50) of cisplatin was estimated according to the survival curve.
2.5 Western blot
MKN-7/DDP and HGC-27/DDP cells were lysed for total protein isolation using RIPA buffer with 1% phenylmethylsulfonyl fluoride (Solarbio, Beijing, China). After the high-speed centrifugation (12,000g, 5 min) at 4°C, the protein in supernatant was quantified and used for SDS-PAGE. Transmembrane was performed using nitrocellulose membranes (Solarbio) and western blot transfer buffer (10×, Solarbio). QuickBlockTM Blocking Buffer (Beyotime) was used for blocking the nonspecific binding sites. Subsequently, the membranes were incubated with primary antibodies overnight and then interacted with special secondary antibody. The antibodies against p-glycoprotein (P-gp) (ab235954), PTPN14 (ab204321), GAPDH (ab181602) and immunoglobulin G (ab205718) were obtained from Abcam (Cambridge, MA, USA). Protein signals were developed using BeyoECL Plus (Beyotime). The relative protein expression was half-quantified with Quantity One software (Bio-Rad) and normalized to GAPDH level.
2.6 Cell apoptosis assay
Cell apoptosis was measured through flow cytometry using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Solarbio). MKN-7/DDP and HGC-27/DDP cells (100,000 cells per well) were added into 6-well plates and cultured for 48 h. Subsequently, cells were incubated with 10 μL of Annexin V-FITC and PI. The apoptotic rate was determined using a flow cytometer (Agilent, Hangzhou, China) displaying the percentage of cells labelled by Annexin V-FITC + and PI +/−.
2.7 Bioinformatics analysis, luciferase reporter analysis and RNA pull-down
Bioinformatics analysis was used to search potential binding sites of miR-206 and circ_AKT3 or PTPN14 by using starBase. For luciferase reporter assay, firefly luciferase-expressing pmirGLO vectors (Promega, Madison, WI, USA) were exploited for establishment of wild-type (Wt) or mutant (Mut) luciferase reporter vectors targeting circ_AKT3 or PTPN14, named as circ_AKT3-Wt, circ_AKT3-Mut, PTPN14 3′UTR-Wt or PTPN14 3′UTR-Mut, respectively. MKN-7/DDP and HGC-27/DDP cells were co-transfected with miR-206 mimic or miR-NC and these constructs, along with renilla vector. After 24 h post-transfection, luciferase activity was detected through a luciferase reporter assay kit (Promega).
For RNA pull-down, the Wt or Mut sequences of miR-206 and circ_AKT3 were labeled with biotin, and named as bio-miR-206, bio-miR-206-Mut, bio-circ_AKT3 and bio-circ_AKT3-Mut, respectively. The bio-miR-NC and bio-NC were used as the control. MKN-7/DDP and HGC-27/DDP cells were lysed in RIPA buffer with RNase inhibitor (Invitrogen, Carlsbad, CA, USA) and then incubated with biotinylated products for 2 h, followed by incubation with streptavidin beads for 1 h. After extraction of bound RNAs, the expressions of circ_AKT3 and miR-206 enriched in precipitates were detected by RT-qPCR analysis.
2.8 Xenograft model
The 5-week-old male BALB/c nude mice were obtained from Beijing Laboratory Animal Center (Beijing, China). The lentiviral vector of short hairpin RNA targeting circ_AKT3 (sh-circ_AKT3) or negative control (sh-NC) was generated by RiboBio. MKN-7/DDP cells (2 × 106 cells per mouse) stably transfected with sh-circ_AKT3 or sh-NC were subcutaneously injected into the flanks of the mice (n = 6/group). Tumor volume was examined every 4 days after 7 days and calculated as: 0.5 × length (mm) × width2 (mm2). After 31 days, the mice were killed by cervical dislocation. Tumor weight was measured and tumor samples were collected for the abundances of circ_AKT3, miR-206, PTPN14 and P-gp. The animal experiments were approved by the Animal Care and Use Committee of the Changchun University of Chinese Medicine.
2.9 Statistical analysis
Statistical analysis of every experiment was conducted by GraphPad Prism 6 (GraphPad Inc., La Jolla, CA, USA). The data from three independent experiments were shown as mean value ± standard deviation. Student’s t test or Mann-Whitney test was employed for the comparison between the two groups, while ANOVA with Tukey test was employed for the comparison between three or more groups. The potential linear relationship between miR-206 and circ_AKT3 or PTPN14 level was analyzed by spearman’s correlation assay. P < 0.05 was considered as significant difference.
-
Research involving human participants and/or animals: This study was approved by the Institutional Animal Care and Use Committee of the Changchun University of Chinese Medicine. We have strictly carried out the relevant research work protecting the rights and interests of animals under the supervision of the Institutional Animal Care and Use Committee to ensure that the research is in line with the relevant provisions of the Institutional Animal Care and Use Committee.
-
Informed consent: Informed consent was obtained from all individual participants included in the study.
-
Ethics approval: All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
-
Data availability statement: The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
3 Results
3.1 circ_AKT3 level is upregulated and miR-206 level is downregulated in cisplatin-resistant GC
To probe the role of circ_AKT3 and miR-206 in cisplatin resistance, their levels were detected in cisplatin-sensitive or resistant GC tissues. Compared with sensitive samples (n = 42), circ_AKT3 level was evidently enhanced in cisplatin-resistant GC tissues (n = 23) (Figure 1a). On the contrary, the abundance of miR-206 was abnormally reduced in resistant tissues in comparison to that in sensitive group (Figure 1b). Moreover, the data from Figure 1c and d displayed that IC50 of cisplatin in MKN-7/DDP and HGC-27/DDP cells was higher than that in MKN-7 and HGC-27 cells, which suggested the high resistance of both MKN-7/DDP and HGC-27/DDP cells to cisplatin. The data of RT-qPCR assay showed that circ_AKT3 expression was obviously increased in MKN-7/DDP and HGC-27/DDP cells in comparison to sensitive cells, while miR-206 abundance showed an opposite trend (Figure 1e and f). The above data suggested that cisplatin resistance might be associated with circ_AKT3 and miR-206.
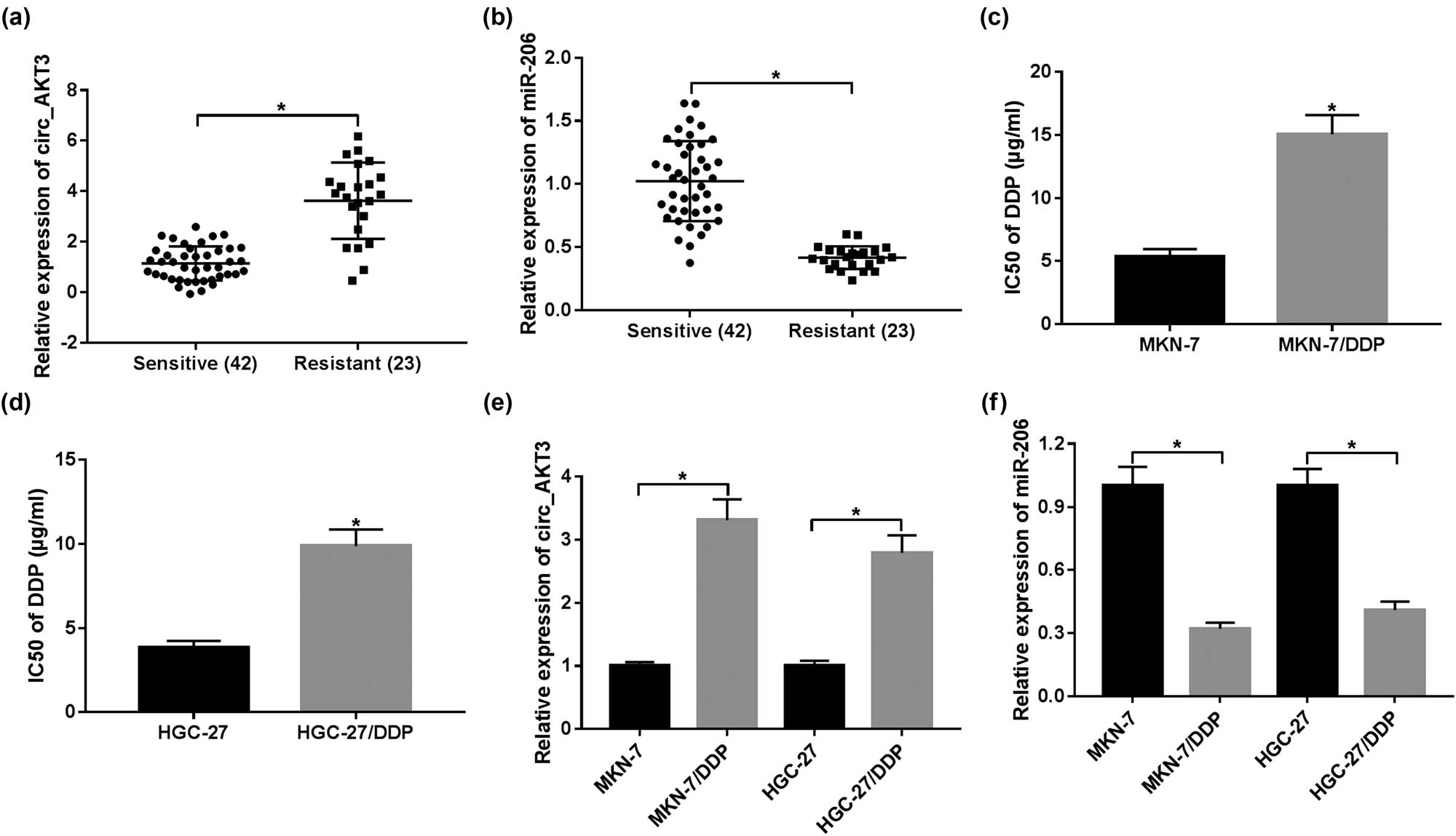
circ_AKT3 expression is elevated and miR-206 level is decreased in resistant GC. (a and b) The levels of circ_AKT3 and miR-206 were measured in sensitive (n = 42) and resistant (n = 23) GC tissues via RT-qPCR. (c and d) The IC50 of cisplatin was analyzed in MKN-7, MKN-7/DDP, HGC-27 and HGC-27/DDP cells by MTT assay. (e and f) The levels of circ_AKT3 and miR-206 were detected in DDP-sensitive and DDP-resistant GC cells via RT-qPCR. *P < 0.05.
3.2 Interference of circ_AKT3 reduces cisplatin resistance in resistant GC cells
To assess the biological role of circ_AKT3 in cisplatin resistance, circ_AKT3 was silenced in MKN-7/DDP and HGC-27/DDP cells by a siRNA. As displayed in Figure 2a, circ_AKT3 expression was effectively reduced in MKN-7/DDP and HGC-27/DDP cells after transfection of si-circ_AKT3, showing the high efficiency of si-circ_AKT3 in reducing circ_AKT3 expression. MTT assay displayed that circ_AKT3 knockdown significantly decreased the IC50 of cisplatin in the two cell lines (Figure 2b). Moreover, downregulation of circ_AKT3 inhibited the expression of P-gp protein level in MKN-7/DDP and HGC-27/DDP cells (Figure 2c and d). In addition, the analysis of flow cytometry showed higher apoptotic rate in the two resistant cell lines after silencing circ_AKT3 as compared with the two sensitive cell lines (Figure 2e and f). The above findings manifested that circ_AKT3 knockdown improved sensitivity of GC cells to cisplatin.

Silence of circ_AKT3 inhibits cisplatin resistance of resistant GC cells. MKN-7/DDP and HGC-27/DDP cells were transfected with si-circ_AKT3 or s-NC. Then, circ_AKT3 expression (a), IC50 of cisplatin (b), P-gp protein level (c and d) and apoptosis (e and f) were measured via RT-qPCR, MTT, Western blot and flow cytometry, respectively. *P < 0.05.
3.3 Overexpression of miR-206 represses cisplatin resistance of resistant GC cells
The role of miR-206 in cisplatin resistance was also explored in MKN-7/DDP and HGC-27/DDP cells by overexpressing miR-206 using miRNA mimic. After the transfection of miR-206 mimic, miR-206 abundance was markedly increased in MKN-7/DDP and HGC-27/DDP cells (Figure 3a). Then, miR-206 overexpression induced obvious reduction in IC50 of cisplatin in the two types of resistant cells (Figure 3b). Meanwhile, the results of Western blot demonstrated that miR-206 overexpression led to a decline in P-gp protein production (Figure 3c and d). Besides, the apoptotic rate of MKN-7/DDP and HGC-27/DDP cells was significantly increased by miR-206 overexpression (Figure 3e and f). Taken together, these results explained that miR-206 was able to improve cisplatin sensitivity in GC cells.
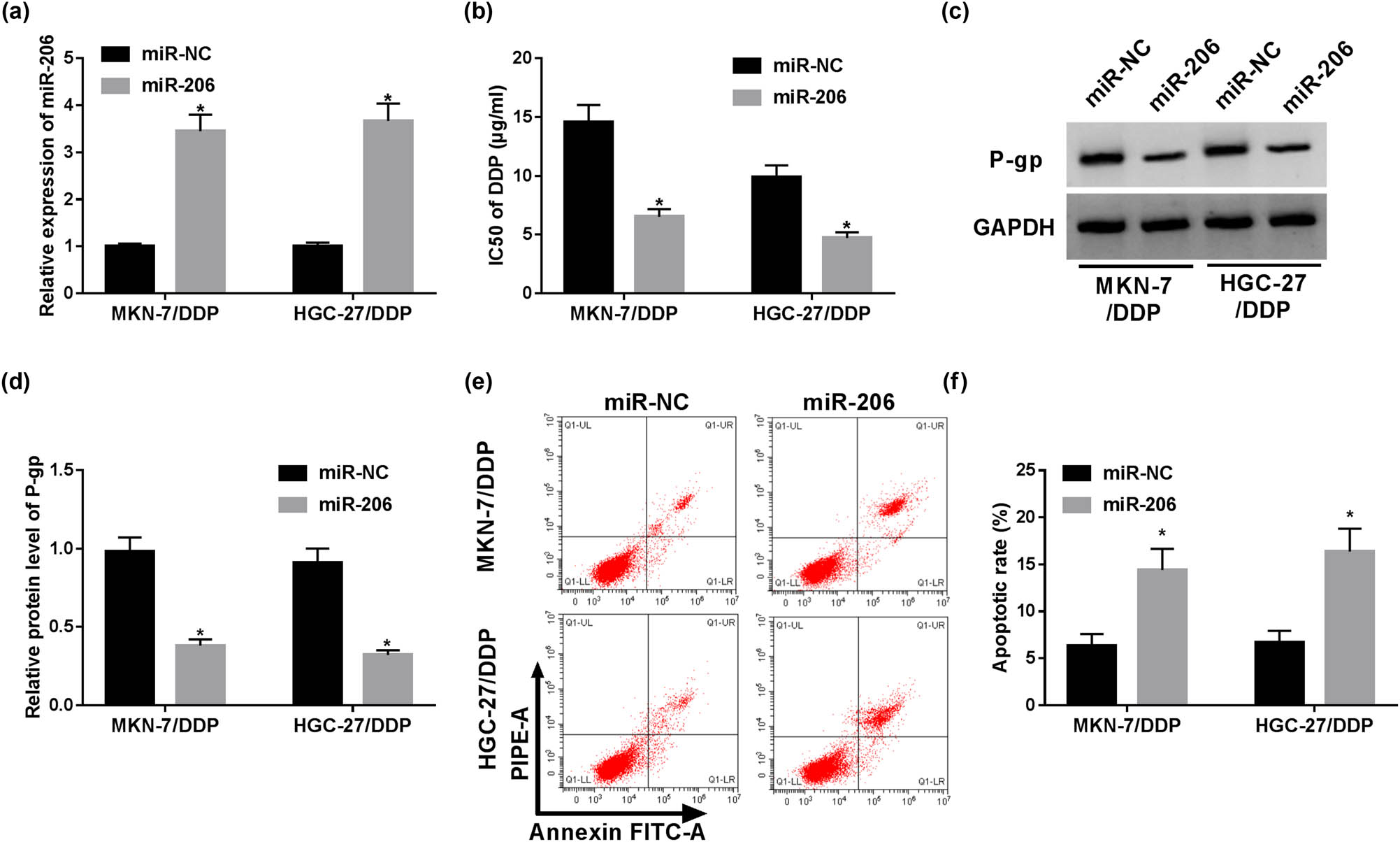
Overexpression of miR-206 suppresses cisplatin resistance of resistant GC cells. The expression of miR-206 (a), IC50 of cisplatin (b), P-gp protein level (c and d) and apoptosis (e and f) were detected in MKN-7/DDP and HGC-27/DDP cells with transfection of miR-206 mimic or miR-NC via RT-qPCR, MTT, Western blot and flow cytometry, respectively. *P < 0.05.
3.4 circ_AKT3 regulates cisplatin resistance of resistant GC cells by sponging miR-206
Results of bioinformatics analysis displayed that circ_AKT3 carried the complementary sites of miR-206 (Figure 4a), suggesting the potential correlation between them. To validate this interaction, the Wt or Mut luciferase reporter vectors of circ_AKT3 were constructed and transfected into MKN-7/DDP and HGC-27/DDP cells. Results first showed that miR-206 overexpression induced decrease in luciferase activity in circ_AKT3-Wt-transfected cells, but the luciferase activity of circ_AKT3-Mut had no response to miR-206 overexpression (Figure 4b and c). Moreover, RNA pull-down assay revealed the binding ability of circ_AKT3 and miR-206 by biotinylated miR-206 or circ_AKT3, respectively (Figure 4d and e). According to spearman’s correlation analysis, miR-206 expression in GC tissues was inversely associated with circ_AKT3 level (r = −0.629 and P < 0.001) (Figure 4f). Subsequently, the effect of circ_AKT3 on miR-206 level was investigated in MKN-7/DDP and HGC-27/DDP cells by overexpressing or silencing circ_AKT3. As exhibited in Figure 4g and h, miR-206 abundance was obviously inhibited by circ_AKT3 overexpression, but enhanced by circ_AKT3 interference. To explore whether the effect of circ_AKT3 on cisplatin resistance was modulated via miR-206, MKN-7/DDP and HGC-27/DDP cells were transfected with si-NC, si-circ_AKT3, si-circ_AKT3 + anti-miR-NC or anti-miR-206. The data of RT-qPCR demonstrated that transfection of anti-miR-206 obviously abolished the promoting impact of circ_AKT3 interference on miR-206 expression level (Figure 5a). Besides, the rescue experiments displayed that miR-206 deficiency attenuated the inhibitive role of circ_AKT3 silence in IC50 of cisplatin and P-gp protein level as well as the promoting effect of circ_AKT3 on apoptosis (Figure 5b–f). Thus, all these findings explained that circ_AKT3 regulated cisplatin resistance by interacting with miR-206 in GC cells.

circ_AKT3 is a decoy of miR-206. (a) The potential complementary sites of circ_AKT3 and miR-206 were analyzed via starBase. (b and c) Luciferase activity was detected in MKN-7/DDP and HGC-27/DDP cells co-transfected with miR-NC or miR-206 mimic and circ_AKT3-Wt or circ_AKT3-Mut. (d and e) The levels of circ_AKT3 and miR-206 were detected in MKN-7/DDP and HGC-27/DDP cells after biotin pull-down. (f) The linear correlation between expression of circ_AKT3 and miR-206 in GC tissues was analyzed. (g and h) miR-206 level was detected in MKN-7/DDP and HGC-27/DDP cells transfected with vector, circ_AKT3 overexpression vector, si-NC or si-circ_AKT3 via RT-qPCR. *P < 0.05.

Down-regulation of miR-206 attenuates the effect of circ_AKT3 knockdown on cisplatin resistance of resistant GC cells. MKN-7/DDP and HGC-27/DDP cells were transfected with si-NC, si-circ_AKT3, si-circ_AKT3 + anti-miR-NC or anti-miR-206. Then, miR-206 level (a), IC50 of cisplatin (b), P-gp protein level (c–e) and apoptotic rate (f) were investigated by RT-qPCR, MTT, Western blot and flow cytometry, respectively. *P < 0.05.
3.5 miR-206 inhibits cisplatin resistance of resistant GC cells via targeting PTPN14
The mRNA and protein levels of PTPN14 were evidently enhanced in cisplatin-resistant GC tissues and cells (Figure 6a–d). Meanwhile, PTPN14 level was negatively associated with miR-206 level in GC tissues (Figure 6e). To further elucidate the mechanism, bioinformatics analysis showed that miR-206 was able to bind to PTPN14 (Figure 6f). Subsequent results displayed that the luciferase activity was notably reduced in PTPN14 3′UTR-Wt + miR-206 group in MKN-7/DDP and HGC-27/DDP cells, but it was not changed in PTPN14 3′UTR-Mut + miR-206 group (Figure 6f). Moreover, restoration of PTPN14 weakened the effect of miR-206 overexpression on IC50 of cisplatin, P-gp expression and cell apoptosis in MKN-7/DDP and HGC-27/DDP cells (Figure 6g–i). By large, all the above observations demonstrated that PTPN14 could regulate cisplatin resistance through interaction with miR-206.

miR-206 inhibits cisplatin resistance of resistant GC cells via targeting PTPN14. (a and b) PTPN14 expression was measured in sensitive (n = 42) and resistant (n = 23) GC tissues via RT-qPCR and Western blot. (c and d) PTPN14 expression was detected in sensitive and resistant GC cells by RT-qPCR and Western blot. (e) The linear correlation between levels of PTPN14 and miR-206 in GC tissues was analyzed. (f) The binding sites of miR-206 and PTPN14 were searched via starBase, and luciferase activity was detected in MKN-7/DDP and HGC-27/DDP cells co-transfected with miR-NC or miR-206 mimic and PTPN14 3′UTR-Wt or PTPN14 3′UTR-Mut. IC50 of cisplatin (g), P-gp protein level (h) and apoptotic rate (i) were measured in MKN-7/DDP and HGC-27/DDP cells transfected with miR-NC, miR-206 mimic, miR-206 mimic + pcDNA or PTPN14 overexpression vector via MTT, Western blot and flow cytometry, respectively. *P < 0.05.
3.6 circ_AKT3 modulates PTPN14 expression by miR-206
To explore whether circ_AKT3 could regulate PTPN14 in DDP-resistant GC cells, MKN-7/DDP and HGC-27/DDP cells were transfected with si-NC, si-circ_AKT3, si-circ_AKT3 + anti-miR-NC or anti-miR-206. As displayed in Figure 7a–c, PTPN14 expression at mRNA and protein levels were notably decreased via knockdown of circ_AKT3, which were restored by inhibition of miR-206, indicating that circ_AKT3 regulated PTPN14 by associating with miR-206.

circ_AKT3 regulates PTPN14 expression via miR-206 in resistant GC cells. (a–c) PTPN14 expression was detected in MKN-7/DDP and HGC-27/DDP cells transfected with si-NC, si-circ_AKT3, si-circ_AKT3 + anti-miR-NC or anti-miR-206 via RT-qPCR and Western blot. *P < 0.05.
3.7 circ_AKT3 knockdown decreases xenograft tumor growth
The study continued to explore the effect of circ_AKT3 in vivo. As shown in Figure 8a and b, the volume and weight of tumors were obviously declined in sh-circ_AKT3 group in comparison to sh-NC group. In addition, circ_AKT3, PTPN14 and P-gp levels were reduced and miR-206 abundance was increased in tumor samples from sh-circ_AKT3 group in comparison to sh-NC group (Figure 8c–e). The above data ascertained that circ_AKT3 knockdown inhibited MKN-7/DDP cell malignancy in vivo.

Knockdown of circ_AKT3 reduces xenograft tumor growth. MKN-7/DDP cells stably transfected with sh-circ_AKT3 or sh-NC were used for establishment of xenograft model. (a) Tumor volume was detected every 4 days. (b) Weight of tumor tissues was measured at the end point. (c–e) The levels of circ_AKT3, miR-206, PTPN14 and P-gp were detected in each group. *P < 0.05.
4 Discussion
Previous studies demonstrated that circRNAs could act as miRNA sponges or competing endogenous RNAs (ceRNAs) to further modulate drug resistance in GC [20,21]. Moreover, it was reported that circ_AKT3 contributed to cisplatin resistance in GC [9]. Nevertheless, more insights in the mechanism of circ_AKT3 regulating cisplatin resistance in GC are needed. In the current research, circ_AKT3 was highly expressed in GC tissues and cells with cisplatin resistance. Knockdown of circ_AKT3 decreased cisplatin resistance by downregulating P-gp expression. For the first time, we demonstrated a ceRNA network of circ_AKT3/miR-206/PTPN14 that was associated with cisplatin resistance.
In this study, we found high level of circ_AKT3 might be associated with cisplatin resistance in GC, indicating the positive role of circ_AKT3 in GC, which was opposite to that in other cancers [7,8]. We hypothesized that the exact roles of circRNAs in cancers might be induced due to the alteration of tumor microenvironment. By using MKN-7/DDP and HGC-27/DDP cells, we found that silencing circ_AKT3 decreased cisplatin resistance as reduction in IC50 of cisplatin and increase in apoptosis after circ_AKT3 knockdown, which was consistent with the findings of previous study [9]. P-gp is one of the key proteins associated with drug resistance and can trigger multidrug resistance in GC [22,23]. This study found that circ_AKT3 knockdown inhibited P-gp protein expression in GC, suggesting the inimical role of circ_AKT3 in chemotherapy. The ceRNA regulatory network, including circRNA, miRNA and mRNA, is one of the main mechanisms associated with tumorigenesis and pathogenesis of GC [24]. To explore whether circ_AKT3 regulated cisplatin resistance by functioning as a ceRNA, its potential target miRNAs were searched. As a result, we confirmed circ_AKT3 acted as a sponge for miR-206 in GC cells.
miR-206 has been reported to negatively modulate drug resistance in human cancers [11,12,13,14]. In the current work, low level of miR-206 was involved in cisplatin resistance and its overexpression attenuated the cisplatin resistance by regulating P-gp. This was also consistent with previous studies [14,15]. Moreover, the effect of circ_AKT3 silence on cisplatin resistance was weakened by miR-206 exhaustion, indicating that circ_AKT3 regulated drug resistance by sponging miR-206 in GC. To further explore the ceRNA crosstalk of circ_AKT3, the target mRNA of miR-206 was searched. Previous studies focusing on the role of miR-206 in human cancers have showed some targets, such as cyclinD2, CXC chemokine receptor 4 and mitogen activating protein kinase 3 (MAPK3) [15,25,26]. This research validated PTPN14 as a functional target of miR-206 using luciferase reporter analysis.
Previous studies suggested PTPN14 served as an oncogene in GC by regulating proliferation, migration and epithelial-to-mesenchymal transition [16,27,28]. Moreover, PTPN14 was implicated in paclitaxel and doxorubicin resistance in GC [17]. Our study showed that PTPN14 expression was elevated in cisplatin-resistant GC, indicating that PTPN14 might also contribute to cisplatin resistance. Furthermore, we found that PTPN14 weakened the effect of miR-206 on cisplatin resistance, and it was regulated by circ_AKT3 through miR-206. Collectively, circ_AKT3 induced PTPN14 to participate in the regulation of cisplatin resistance by sponging miR-206 in GC cells. To better understand the underlying mechanism, in vivo experiments were performed. And our data confirmed the suppressive effect of circ_AKT3 knockdown on growth of cisplatin-resistant GC cells in vivo.
5 Conclusion
In conclusion, our findings described the sensitization role of circ_AKT3 knockdown in GC, uncovered by decrease in IC50 of cisplatin and P-gp expression and increase in apoptosis. The inner mechanism was that circ_AKT3 regulated miR-206/PTPN14 axis in a ceRNA-based manner. This work first confirmed the ceRNA network of circ_AKT3/miR-206/PTPN14, providing a new idea for ameliorating cisplatin sensitivity in GC treatment.
Abbreviations
- anti-miR-206
-
miR-206 inhibitor
- anti-miR-NC
-
miR-206 inhibitor negative control
- circRNAs
-
circular RNAs
- circ_AKT3
-
circRNA AKT serine/threonine kinase 3
- GC
-
gastric cancer
- IC50
-
half maximal inhibitory concentration
- miRNAs
-
microRNAs
- miR-NC
-
miRNA negative control
- P-gp
-
p-glycoprotein
- TPN14
-
protein tyrosine phosphatase non-receptor type 14
- si-NC
-
siRNA for negative control
- siRNA
-
small interfering RNA
- RT-qPCR
-
quantitative reverse transcription PCR
- vector
-
pcDNA circRNA mini vector
Acknowledgement
Not applicable
-
Funding information: This work was funded by the department of Liaoning Science and Technology (Grant No. 20170540361).
-
Author contributions: Wenting Shi and Fang Wang made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agreed to be accountable for all aspects of the work.
-
Conflict of interest: The authors declare that they have no competing interests.
References
[1] Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64. 10.1016/S0140-6736(16)30354-3.Suche in Google Scholar
[2] Rawicz-Pruszynski K, van Sandick JW, Mielko J, Cisel B, Polkowski WP. Current challenges in gastric cancer surgery: European perspective. Surg Oncol. 2018;27:650–6. 10.1016/j.suronc.2018.08.004.Suche in Google Scholar PubMed
[3] Abbas M, Habib M, Naveed M, Karthik K, Dhama K, Shi M, et al. The relevance of gastric cancer biomarkers in prognosis and pre- and post- chemotherapy in clinical practice. Biomed Pharmacother. 2017;95:1082–90. 10.1016/j.biopha.2017.09.032.Suche in Google Scholar PubMed
[4] Zhang M, Du X. Noncoding RNAs in gastric cancer: research progress and prospects. World J Gastroenterol. 2016;22:6610–8. 10.3748/wjg.v22.i29.6610.Suche in Google Scholar PubMed PubMed Central
[5] Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015–21. 10.7150/jca.30828.Suche in Google Scholar PubMed PubMed Central
[6] Hua X, Sun Y, Chen J, Wu Y, Sha J, Han S, et al. Circular RNAs in drug resistant tumors. Biomed Pharmacother. 2019;118:109233. 10.1016/j.biopha.2019.109233.Suche in Google Scholar PubMed
[7] Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131. 10.1186/s12943-019-1056-5.Suche in Google Scholar PubMed PubMed Central
[8] Xue D, Wang H, Chen Y, Shen D, Lu J, Wang M, et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol Cancer. 2019;18:151. 10.1186/s12943-019-1072-5.Suche in Google Scholar PubMed PubMed Central
[9] Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. 10.1186/s12943-019-0969-3.Suche in Google Scholar PubMed PubMed Central
[10] Yuan HL, Wang T, Zhang KH. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Onco Targets Ther. 2018;11:3891–900. 10.2147/OTT.S156921.Suche in Google Scholar PubMed PubMed Central
[11] Wang R, Zhang T, Yang Z, Jiang C, Seng J. Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J Cell Mol Med. 2018;22:4068–75. 10.1111/jcmm.13679.Suche in Google Scholar PubMed PubMed Central
[12] Liu F, Yin R, Chen X, Chen W, Qian Y, Zhao Y, et al. Overexpression of miR-206 decreases the Euthyrox-resistance by targeting MAP4K3 in papillary thyroid carcinoma. Biomed Pharmacother. 2019;114:108605. 10.1016/j.biopha.2019.108605.Suche in Google Scholar PubMed
[13] Wu K, Li J, Qi Y, Zhang C, Zhu D, Liu D, et al. SNHG14 confers gefitinib resistance in non-small cell lung cancer by upregulating ABCB1 via sponging miR-206-3p. Biomed Pharmacother. 2019;116:108995. 10.1016/j.biopha.2019.108995.Suche in Google Scholar PubMed
[14] Chen QY, Jiao DM, Wang J, Hu H, Tang X, Chen J, et al. miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget. 2016;7:24510–26. 10.18632/oncotarget.8229.Suche in Google Scholar PubMed PubMed Central
[15] Chen Z, Gao YJ, Hou RZ, Ding DY, Song DF, Wang DY, et al. MicroRNA-206 facilitates gastric cancer cell apoptosis and suppresses cisplatin resistance by targeting MAPK2 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:171–80. 10.26355/eurrev_201901_16761.Suche in Google Scholar PubMed
[16] Han X, Sun T, Hong J, Wei R, Dong Y, Huang D, et al. Nonreceptor tyrosine phosphatase 14 promotes proliferation and migration through regulating phosphorylation of YAP of Hippo signaling pathway in gastric cancer cells. J Cell Biochem. 2019;120:17723–30. 10.1002/jcb.29038.Suche in Google Scholar PubMed
[17] Wang H, Qin R, Guan A, Yao Y, Huang Y, Jia H, et al. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem. 2018;119:7226–34. 10.1002/jcb.26901.Suche in Google Scholar PubMed
[18] Peng C, Huang K, Liu G, Li Y, Yu C. MiR-876-3p regulates cisplatin resistance and stem cell-like properties of gastric cancer cells by targeting TMED3. J Gastroenterol Hepatol. 2019;34:1711–9. 10.1111/jgh.14649.Suche in Google Scholar PubMed
[19] Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. 10.1006/meth.2001.1262.Suche in Google Scholar PubMed
[20] Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124-3p. Biosci Rep. 2019;39:BSR20193045. 10.1042/BSR20193045.Suche in Google Scholar PubMed PubMed Central
[21] Huang XX, Zhang Q, Hu H, Jin Y, Zeng AL, Xia YB, et al. A novel circular RNA circFN1 enhances cisplatin resistance in gastric cancer via sponging miR-182-5p. J Cell Biochem. 2020; 10.1002/jcb.29641.Suche in Google Scholar PubMed
[22] Breier A, Gibalova L, Seres M, Barancik M, Sulova Z. New insight into p-glycoprotein as a drug target. Anticancer Agents Med Chem. 2013;13:159–70.10.2174/187152013804487380Suche in Google Scholar
[23] Xu HW, Xu L, Hao JH, Qin CY, Liu H. Expression of P-glycoprotein and multidrug resistance-associated protein is associated with multidrug resistance in gastric cancer. J Int Med Res. 2010;38:34–42. 10.1177/147323001003800104.Suche in Google Scholar PubMed
[24] Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–7. 10.3748/wjg.v21.i41.11680.Suche in Google Scholar PubMed PubMed Central
[25] Zhang L, Liu X, Jin H, Guo X, Xia L, Chen Z, et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 2013;332:94–101. 10.1016/j.canlet.2013.01.023.Suche in Google Scholar PubMed
[26] Li H, Yao G, Feng B, Lu X, Fan Y. Circ_0056618 and CXCR4 act as competing endogenous in gastric cancer by regulating miR-206. J Cell Biochem. 2018;119:9543–51. 10.1002/jcb.27271.Suche in Google Scholar PubMed
[27] Liu YP, Sun XH, Cao XL, Jiang WW, Wang XX, Zhang YF, et al. MicroRNA-217 suppressed epithelial-to-mesenchymal transition in gastric cancer metastasis through targeting PTPN14. Eur Rev Med Pharmacol Sci. 2017;21:1759–67.Suche in Google Scholar
[28] Chen G, Yang Z, Feng M, Wang Z, Ye Q. microRNA-217 suppressed epithelial-to-mesenchymal transition through targeting PTPN14 in gastric cancer. Biosci Rep. 2019;40:BSR20193176. 10.1042/BSR20193176.Suche in Google Scholar PubMed PubMed Central
© 2022 Wenting Shi and Fang Wang, published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- AMBRA1 attenuates the proliferation of uveal melanoma cells
- A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma
- Differences in complications between hepatitis B-related cirrhosis and alcohol-related cirrhosis
- Effect of gestational diabetes mellitus on lipid profile: A systematic review and meta-analysis
- Long noncoding RNA NR2F1-AS1 stimulates the tumorigenic behavior of non-small cell lung cancer cells by sponging miR-363-3p to increase SOX4
- Promising novel biomarkers and candidate small-molecule drugs for lung adenocarcinoma: Evidence from bioinformatics analysis of high-throughput data
- Plasmapheresis: Is it a potential alternative treatment for chronic urticaria?
- The biomarkers of key miRNAs and gene targets associated with extranodal NK/T-cell lymphoma
- Gene signature to predict prognostic survival of hepatocellular carcinoma
- Effects of miRNA-199a-5p on cell proliferation and apoptosis of uterine leiomyoma by targeting MED12
- Does diabetes affect paraneoplastic thrombocytosis in colorectal cancer?
- Is there any effect on imprinted genes H19, PEG3, and SNRPN during AOA?
- Leptin and PCSK9 concentrations are associated with vascular endothelial cytokines in patients with stable coronary heart disease
- Pericentric inversion of chromosome 6 and male fertility problems
- Staple line reinforcement with nebulized cyanoacrylate glue in laparoscopic sleeve gastrectomy: A propensity score-matched study
- Retrospective analysis of crescent score in clinical prognosis of IgA nephropathy
- Expression of DNM3 is associated with good outcome in colorectal cancer
- Activation of SphK2 contributes to adipocyte-induced EOC cell proliferation
- CRRT influences PICCO measurements in febrile critically ill patients
- SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis
- lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells
- circ_AKT3 knockdown suppresses cisplatin resistance in gastric cancer
- Prognostic value of nicotinamide N-methyltransferase in human cancers: Evidence from a meta-analysis and database validation
- GPC2 deficiency inhibits cell growth and metastasis in colon adenocarcinoma
- A pan-cancer analysis of the oncogenic role of Holliday junction recognition protein in human tumors
- Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer
- Association between preventable risk factors and metabolic syndrome
- miR-29c-5p knockdown reduces inflammation and blood–brain barrier disruption by upregulating LRP6
- Cardiac contractility modulation ameliorates myocardial metabolic remodeling in a rabbit model of chronic heart failure through activation of AMPK and PPAR-α pathway
- Quercitrin protects human bronchial epithelial cells from oxidative damage
- Smurf2 suppresses the metastasis of hepatocellular carcinoma via ubiquitin degradation of Smad2
- circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury
- Sonoclot’s usefulness in prediction of cardiopulmonary arrest prognosis: A proof of concept study
- Four drug metabolism-related subgroups of pancreatic adenocarcinoma in prognosis, immune infiltration, and gene mutation
- Decreased expression of miR-195 mediated by hypermethylation promotes osteosarcoma
- LMO3 promotes proliferation and metastasis of papillary thyroid carcinoma cells by regulating LIMK1-mediated cofilin and the β-catenin pathway
- Cx43 upregulation in HUVECs under stretch via TGF-β1 and cytoskeletal network
- Evaluation of menstrual irregularities after COVID-19 vaccination: Results of the MECOVAC survey
- Histopathologic findings on removed stomach after sleeve gastrectomy. Do they influence the outcome?
- Analysis of the expression and prognostic value of MT1-MMP, β1-integrin and YAP1 in glioma
- Optimal diagnosis of the skin cancer using a hybrid deep neural network and grasshopper optimization algorithm
- miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells
- Clinical value of SIRT1 as a prognostic biomarker in esophageal squamous cell carcinoma, a systematic meta-analysis
- circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8
- miR-22-5p regulates the self-renewal of spermatogonial stem cells by targeting EZH2
- hsa-miR-340-5p inhibits epithelial–mesenchymal transition in endometriosis by targeting MAP3K2 and inactivating MAPK/ERK signaling
- circ_0085296 inhibits the biological functions of trophoblast cells to promote the progression of preeclampsia via the miR-942-5p/THBS2 network
- TCD hemodynamics findings in the subacute phase of anterior circulation stroke patients treated with mechanical thrombectomy
- Development of a risk-stratification scoring system for predicting risk of breast cancer based on non-alcoholic fatty liver disease, non-alcoholic fatty pancreas disease, and uric acid
- Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway
- circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis
- Human amniotic fluid as a source of stem cells
- lncRNA NONRATT013819.2 promotes transforming growth factor-β1-induced myofibroblastic transition of hepatic stellate cells by miR24-3p/lox
- NORAD modulates miR-30c-5p-LDHA to protect lung endothelial cells damage
- Idiopathic pulmonary fibrosis telemedicine management during COVID-19 outbreak
- Risk factors for adverse drug reactions associated with clopidogrel therapy
- Serum zinc associated with immunity and inflammatory markers in Covid-19
- The relationship between night shift work and breast cancer incidence: A systematic review and meta-analysis of observational studies
- LncRNA expression in idiopathic achalasia: New insight and preliminary exploration into pathogenesis
- Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway
- Moscatilin suppresses the inflammation from macrophages and T cells
- Zoledronate promotes ECM degradation and apoptosis via Wnt/β-catenin
- Epithelial-mesenchymal transition-related genes in coronary artery disease
- The effect evaluation of traditional vaginal surgery and transvaginal mesh surgery for severe pelvic organ prolapse: 5 years follow-up
- Repeated partial splenic artery embolization for hypersplenism improves platelet count
- Low expression of miR-27b in serum exosomes of non-small cell lung cancer facilitates its progression by affecting EGFR
- Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis
- miR-455-5p enhances 5-fluorouracil sensitivity in colorectal cancer cells by targeting PIK3R1 and DEPDC1
- The effect of tranexamic acid on the reduction of intraoperative and postoperative blood loss and thromboembolic risk in patients with hip fracture
- Isocitrate dehydrogenase 1 mutation in cholangiocarcinoma impairs tumor progression by sensitizing cells to ferroptosis
- Artemisinin protects against cerebral ischemia and reperfusion injury via inhibiting the NF-κB pathway
- A 16-gene signature associated with homologous recombination deficiency for prognosis prediction in patients with triple-negative breast cancer
- Lidocaine ameliorates chronic constriction injury-induced neuropathic pain through regulating M1/M2 microglia polarization
- MicroRNA 322-5p reduced neuronal inflammation via the TLR4/TRAF6/NF-κB axis in a rat epilepsy model
- miR-1273h-5p suppresses CXCL12 expression and inhibits gastric cancer cell invasion and metastasis
- Clinical characteristics of pneumonia patients of long course of illness infected with SARS-CoV-2
- circRNF20 aggravates the malignancy of retinoblastoma depending on the regulation of miR-132-3p/PAX6 axis
- Linezolid for resistant Gram-positive bacterial infections in children under 12 years: A meta-analysis
- Rack1 regulates pro-inflammatory cytokines by NF-κB in diabetic nephropathy
- Comprehensive analysis of molecular mechanism and a novel prognostic signature based on small nuclear RNA biomarkers in gastric cancer patients
- Smog and risk of maternal and fetal birth outcomes: A retrospective study in Baoding, China
- Let-7i-3p inhibits the cell cycle, proliferation, invasion, and migration of colorectal cancer cells via downregulating CCND1
- β2-Adrenergic receptor expression in subchondral bone of patients with varus knee osteoarthritis
- Possible impact of COVID-19 pandemic and lockdown on suicide behavior among patients in Southeast Serbia
- In vitro antimicrobial activity of ozonated oil in liposome eyedrop against multidrug-resistant bacteria
- Potential biomarkers for inflammatory response in acute lung injury
- A low serum uric acid concentration predicts a poor prognosis in adult patients with candidemia
- Antitumor activity of recombinant oncolytic vaccinia virus with human IL2
- ALKBH5 inhibits TNF-α-induced apoptosis of HUVECs through Bcl-2 pathway
- Risk prediction of cardiovascular disease using machine learning classifiers
- Value of ultrasonography parameters in diagnosing polycystic ovary syndrome
- Bioinformatics analysis reveals three key genes and four survival genes associated with youth-onset NSCLC
- Identification of autophagy-related biomarkers in patients with pulmonary arterial hypertension based on bioinformatics analysis
- Protective effects of glaucocalyxin A on the airway of asthmatic mice
- Overexpression of miR-100-5p inhibits papillary thyroid cancer progression via targeting FZD8
- Bioinformatics-based analysis of SUMOylation-related genes in hepatocellular carcinoma reveals a role of upregulated SAE1 in promoting cell proliferation
- Effectiveness and clinical benefits of new anti-diabetic drugs: A real life experience
- Identification of osteoporosis based on gene biomarkers using support vector machine
- Tanshinone IIA reverses oxaliplatin resistance in colorectal cancer through microRNA-30b-5p/AVEN axis
- miR-212-5p inhibits nasopharyngeal carcinoma progression by targeting METTL3
- Association of ST-T changes with all-cause mortality among patients with peripheral T-cell lymphomas
- LINC00665/miRNAs axis-mediated collagen type XI alpha 1 correlates with immune infiltration and malignant phenotypes in lung adenocarcinoma
- The perinatal factors that influence the excretion of fecal calprotectin in premature-born children
- Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study
- Does the use of 3D-printed cones give a chance to postpone the use of megaprostheses in patients with large bone defects in the knee joint?
- lncRNA HAGLR modulates myocardial ischemia–reperfusion injury in mice through regulating miR-133a-3p/MAPK1 axis
- Protective effect of ghrelin on intestinal I/R injury in rats
- In vivo knee kinematics of an innovative prosthesis design
- Relationship between the height of fibular head and the incidence and severity of knee osteoarthritis
- lncRNA WT1-AS attenuates hypoxia/ischemia-induced neuronal injury during cerebral ischemic stroke via miR-186-5p/XIAP axis
- Correlation of cardiac troponin T and APACHE III score with all-cause in-hospital mortality in critically ill patients with acute pulmonary embolism
- LncRNA LINC01857 reduces metastasis and angiogenesis in breast cancer cells via regulating miR-2052/CENPQ axis
- Endothelial cell-specific molecule 1 (ESM1) promoted by transcription factor SPI1 acts as an oncogene to modulate the malignant phenotype of endometrial cancer
- SELENBP1 inhibits progression of colorectal cancer by suppressing epithelial–mesenchymal transition
- Visfatin is negatively associated with coronary artery lesions in subjects with impaired fasting glucose
- Treatment and outcomes of mechanical complications of acute myocardial infarction during the Covid-19 era: A comparison with the pre-Covid-19 period. A systematic review and meta-analysis
- Neonatal stroke surveillance study protocol in the United Kingdom and Republic of Ireland
- Oncogenic role of TWF2 in human tumors: A pan-cancer analysis
- Mean corpuscular hemoglobin predicts the length of hospital stay independent of severity classification in patients with acute pancreatitis
- Association of gallstone and polymorphisms of UGT1A1*27 and UGT1A1*28 in patients with hepatitis B virus-related liver failure
- TGF-β1 upregulates Sar1a expression and induces procollagen-I secretion in hypertrophic scarring fibroblasts
- Antisense lncRNA PCNA-AS1 promotes esophageal squamous cell carcinoma progression through the miR-2467-3p/PCNA axis
- NK-cell dysfunction of acute myeloid leukemia in relation to the renin–angiotensin system and neurotransmitter genes
- The effect of dilution with glucose and prolonged injection time on dexamethasone-induced perineal irritation – A randomized controlled trial
- miR-146-5p restrains calcification of vascular smooth muscle cells by suppressing TRAF6
- Role of lncRNA MIAT/miR-361-3p/CCAR2 in prostate cancer cells
- lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2
- Noninvasive diagnosis of AIH/PBC overlap syndrome based on prediction models
- lncRNA FAM230B is highly expressed in colorectal cancer and suppresses the maturation of miR-1182 to increase cell proliferation
- circ-LIMK1 regulates cisplatin resistance in lung adenocarcinoma by targeting miR-512-5p/HMGA1 axis
- LncRNA SNHG3 promoted cell proliferation, migration, and metastasis of esophageal squamous cell carcinoma via regulating miR-151a-3p/PFN2 axis
- Risk perception and affective state on work exhaustion in obstetrics during the COVID-19 pandemic
- lncRNA-AC130710/miR-129-5p/mGluR1 axis promote migration and invasion by activating PKCα-MAPK signal pathway in melanoma
- SNRPB promotes cell cycle progression in thyroid carcinoma via inhibiting p53
- Xylooligosaccharides and aerobic training regulate metabolism and behavior in rats with streptozotocin-induced type 1 diabetes
- Serpin family A member 1 is an oncogene in glioma and its translation is enhanced by NAD(P)H quinone dehydrogenase 1 through RNA-binding activity
- Silencing of CPSF7 inhibits the proliferation, migration, and invasion of lung adenocarcinoma cells by blocking the AKT/mTOR signaling pathway
- Ultrasound-guided lumbar plexus block versus transversus abdominis plane block for analgesia in children with hip dislocation: A double-blind, randomized trial
- Relationship of plasma MBP and 8-oxo-dG with brain damage in preterm
- Identification of a novel necroptosis-associated miRNA signature for predicting the prognosis in head and neck squamous cell carcinoma
- Delayed femoral vein ligation reduces operative time and blood loss during hip disarticulation in patients with extremity tumors
- The expression of ASAP3 and NOTCH3 and the clinicopathological characteristics of adult glioma patients
- Longitudinal analysis of factors related to Helicobacter pylori infection in Chinese adults
- HOXA10 enhances cell proliferation and suppresses apoptosis in esophageal cancer via activating p38/ERK signaling pathway
- Meta-analysis of early-life antibiotic use and allergic rhinitis
- Marital status and its correlation with age, race, and gender in prognosis of tonsil squamous cell carcinomas
- HPV16 E6E7 up-regulates KIF2A expression by activating JNK/c-Jun signal, is beneficial to migration and invasion of cervical cancer cells
- Amino acid profiles in the tissue and serum of patients with liver cancer
- Pain in critically ill COVID-19 patients: An Italian retrospective study
- Immunohistochemical distribution of Bcl-2 and p53 apoptotic markers in acetamiprid-induced nephrotoxicity
- Estradiol pretreatment in GnRH antagonist protocol for IVF/ICSI treatment
- Long non-coding RNAs LINC00689 inhibits the apoptosis of human nucleus pulposus cells via miR-3127-5p/ATG7 axis-mediated autophagy
- The relationship between oxygen therapy, drug therapy, and COVID-19 mortality
- Monitoring hypertensive disorders in pregnancy to prevent preeclampsia in pregnant women of advanced maternal age: Trial mimicking with retrospective data
- SETD1A promotes the proliferation and glycolysis of nasopharyngeal carcinoma cells by activating the PI3K/Akt pathway
- The role of Shunaoxin pills in the treatment of chronic cerebral hypoperfusion and its main pharmacodynamic components
- TET3 governs malignant behaviors and unfavorable prognosis of esophageal squamous cell carcinoma by activating the PI3K/AKT/GSK3β/β-catenin pathway
- Associations between morphokinetic parameters of temporary-arrest embryos and the clinical prognosis in FET cycles
- Long noncoding RNA WT1-AS regulates trophoblast proliferation, migration, and invasion via the microRNA-186-5p/CADM2 axis
- The incidence of bronchiectasis in chronic obstructive pulmonary disease
- Integrated bioinformatics analysis shows integrin alpha 3 is a prognostic biomarker for pancreatic cancer
- Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway
- Comparison of hospitalized patients with severe pneumonia caused by COVID-19 and influenza A (H7N9 and H1N1): A retrospective study from a designated hospital
- lncRNA ZFAS1 promotes intervertebral disc degeneration by upregulating AAK1
- Pathological characteristics of liver injury induced by N,N-dimethylformamide: From humans to animal models
- lncRNA ELFN1-AS1 enhances the progression of colon cancer by targeting miR-4270 to upregulate AURKB
- DARS-AS1 modulates cell proliferation and migration of gastric cancer cells by regulating miR-330-3p/NAT10 axis
- Dezocine inhibits cell proliferation, migration, and invasion by targeting CRABP2 in ovarian cancer
- MGST1 alleviates the oxidative stress of trophoblast cells induced by hypoxia/reoxygenation and promotes cell proliferation, migration, and invasion by activating the PI3K/AKT/mTOR pathway
- Bifidobacterium lactis Probio-M8 ameliorated the symptoms of type 2 diabetes mellitus mice by changing ileum FXR-CYP7A1
- circRNA DENND1B inhibits tumorigenicity of clear cell renal cell carcinoma via miR-122-5p/TIMP2 axis
- EphA3 targeted by miR-3666 contributes to melanoma malignancy via activating ERK1/2 and p38 MAPK pathways
- Pacemakers and methylprednisolone pulse therapy in immune-related myocarditis concomitant with complete heart block
- miRNA-130a-3p targets sphingosine-1-phosphate receptor 1 to activate the microglial and astrocytes and to promote neural injury under the high glucose condition
- Review Articles
- Current management of cancer pain in Italy: Expert opinion paper
- Hearing loss and brain disorders: A review of multiple pathologies
- The rationale for using low-molecular weight heparin in the therapy of symptomatic COVID-19 patients
- Amyotrophic lateral sclerosis and delayed onset muscle soreness in light of the impaired blink and stretch reflexes – watch out for Piezo2
- Interleukin-35 in autoimmune dermatoses: Current concepts
- Recent discoveries in microbiota dysbiosis, cholangiocytic factors, and models for studying the pathogenesis of primary sclerosing cholangitis
- Advantages of ketamine in pediatric anesthesia
- Congenital adrenal hyperplasia. Role of dentist in early diagnosis
- Migraine management: Non-pharmacological points for patients and health care professionals
- Atherogenic index of plasma and coronary artery disease: A systematic review
- Physiological and modulatory role of thioredoxins in the cellular function
- Case Reports
- Intrauterine Bakri balloon tamponade plus cervical cerclage for the prevention and treatment of postpartum haemorrhage in late pregnancy complicated with acute aortic dissection: Case series
- A case of successful pembrolizumab monotherapy in a patient with advanced lung adenocarcinoma: Use of multiple biomarkers in combination for clinical practice
- Unusual neurological manifestations of bilateral medial medullary infarction: A case report
- Atypical symptoms of malignant hyperthermia: A rare causative mutation in the RYR1 gene
- A case report of dermatomyositis with the missed diagnosis of non-small cell lung cancer and concurrence of pulmonary tuberculosis
- A rare case of endometrial polyp complicated with uterine inversion: A case report and clinical management
- Spontaneous rupturing of splenic artery aneurysm: Another reason for fatal syncope and shock (Case report and literature review)
- Fungal infection mimicking COVID-19 infection – A case report
- Concurrent aspergillosis and cystic pulmonary metastases in a patient with tongue squamous cell carcinoma
- Paraganglioma-induced inverted takotsubo-like cardiomyopathy leading to cardiogenic shock successfully treated with extracorporeal membrane oxygenation
- Lineage switch from lymphoma to myeloid neoplasms: First case series from a single institution
- Trismus during tracheal extubation as a complication of general anaesthesia – A case report
- Simultaneous treatment of a pubovesical fistula and lymph node metastasis secondary to multimodal treatment for prostate cancer: Case report and review of the literature
- Two case reports of skin vasculitis following the COVID-19 immunization
- Ureteroiliac fistula after oncological surgery: Case report and review of the literature
- Synchronous triple primary malignant tumours in the bladder, prostate, and lung harbouring TP53 and MEK1 mutations accompanied with severe cardiovascular diseases: A case report
- Huge mucinous cystic neoplasms with adhesion to the left colon: A case report and literature review
- Commentary
- Commentary on “Clinicopathological features of programmed cell death-ligand 1 expression in patients with oral squamous cell carcinoma”
- Rapid Communication
- COVID-19 fear, post-traumatic stress, growth, and the role of resilience
- Erratum
- Erratum to “Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway”
- Erratum to “Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study”
- Erratum to “lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2”
- Retraction
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Retraction to “miR-519d downregulates LEP expression to inhibit preeclampsia development”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part II
- Usefulness of close surveillance for rectal cancer patients after neoadjuvant chemoradiotherapy
Artikel in diesem Heft
- Research Articles
- AMBRA1 attenuates the proliferation of uveal melanoma cells
- A ceRNA network mediated by LINC00475 in papillary thyroid carcinoma
- Differences in complications between hepatitis B-related cirrhosis and alcohol-related cirrhosis
- Effect of gestational diabetes mellitus on lipid profile: A systematic review and meta-analysis
- Long noncoding RNA NR2F1-AS1 stimulates the tumorigenic behavior of non-small cell lung cancer cells by sponging miR-363-3p to increase SOX4
- Promising novel biomarkers and candidate small-molecule drugs for lung adenocarcinoma: Evidence from bioinformatics analysis of high-throughput data
- Plasmapheresis: Is it a potential alternative treatment for chronic urticaria?
- The biomarkers of key miRNAs and gene targets associated with extranodal NK/T-cell lymphoma
- Gene signature to predict prognostic survival of hepatocellular carcinoma
- Effects of miRNA-199a-5p on cell proliferation and apoptosis of uterine leiomyoma by targeting MED12
- Does diabetes affect paraneoplastic thrombocytosis in colorectal cancer?
- Is there any effect on imprinted genes H19, PEG3, and SNRPN during AOA?
- Leptin and PCSK9 concentrations are associated with vascular endothelial cytokines in patients with stable coronary heart disease
- Pericentric inversion of chromosome 6 and male fertility problems
- Staple line reinforcement with nebulized cyanoacrylate glue in laparoscopic sleeve gastrectomy: A propensity score-matched study
- Retrospective analysis of crescent score in clinical prognosis of IgA nephropathy
- Expression of DNM3 is associated with good outcome in colorectal cancer
- Activation of SphK2 contributes to adipocyte-induced EOC cell proliferation
- CRRT influences PICCO measurements in febrile critically ill patients
- SLCO4A1-AS1 mediates pancreatic cancer development via miR-4673/KIF21B axis
- lncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells
- circ_AKT3 knockdown suppresses cisplatin resistance in gastric cancer
- Prognostic value of nicotinamide N-methyltransferase in human cancers: Evidence from a meta-analysis and database validation
- GPC2 deficiency inhibits cell growth and metastasis in colon adenocarcinoma
- A pan-cancer analysis of the oncogenic role of Holliday junction recognition protein in human tumors
- Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer
- Association between preventable risk factors and metabolic syndrome
- miR-29c-5p knockdown reduces inflammation and blood–brain barrier disruption by upregulating LRP6
- Cardiac contractility modulation ameliorates myocardial metabolic remodeling in a rabbit model of chronic heart failure through activation of AMPK and PPAR-α pathway
- Quercitrin protects human bronchial epithelial cells from oxidative damage
- Smurf2 suppresses the metastasis of hepatocellular carcinoma via ubiquitin degradation of Smad2
- circRNA_0001679/miR-338-3p/DUSP16 axis aggravates acute lung injury
- Sonoclot’s usefulness in prediction of cardiopulmonary arrest prognosis: A proof of concept study
- Four drug metabolism-related subgroups of pancreatic adenocarcinoma in prognosis, immune infiltration, and gene mutation
- Decreased expression of miR-195 mediated by hypermethylation promotes osteosarcoma
- LMO3 promotes proliferation and metastasis of papillary thyroid carcinoma cells by regulating LIMK1-mediated cofilin and the β-catenin pathway
- Cx43 upregulation in HUVECs under stretch via TGF-β1 and cytoskeletal network
- Evaluation of menstrual irregularities after COVID-19 vaccination: Results of the MECOVAC survey
- Histopathologic findings on removed stomach after sleeve gastrectomy. Do they influence the outcome?
- Analysis of the expression and prognostic value of MT1-MMP, β1-integrin and YAP1 in glioma
- Optimal diagnosis of the skin cancer using a hybrid deep neural network and grasshopper optimization algorithm
- miR-223-3p alleviates TGF-β-induced epithelial-mesenchymal transition and extracellular matrix deposition by targeting SP3 in endometrial epithelial cells
- Clinical value of SIRT1 as a prognostic biomarker in esophageal squamous cell carcinoma, a systematic meta-analysis
- circ_0020123 promotes cell proliferation and migration in lung adenocarcinoma via PDZD8
- miR-22-5p regulates the self-renewal of spermatogonial stem cells by targeting EZH2
- hsa-miR-340-5p inhibits epithelial–mesenchymal transition in endometriosis by targeting MAP3K2 and inactivating MAPK/ERK signaling
- circ_0085296 inhibits the biological functions of trophoblast cells to promote the progression of preeclampsia via the miR-942-5p/THBS2 network
- TCD hemodynamics findings in the subacute phase of anterior circulation stroke patients treated with mechanical thrombectomy
- Development of a risk-stratification scoring system for predicting risk of breast cancer based on non-alcoholic fatty liver disease, non-alcoholic fatty pancreas disease, and uric acid
- Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway
- circ_0062491 alleviates periodontitis via the miR-142-5p/IGF1 axis
- Human amniotic fluid as a source of stem cells
- lncRNA NONRATT013819.2 promotes transforming growth factor-β1-induced myofibroblastic transition of hepatic stellate cells by miR24-3p/lox
- NORAD modulates miR-30c-5p-LDHA to protect lung endothelial cells damage
- Idiopathic pulmonary fibrosis telemedicine management during COVID-19 outbreak
- Risk factors for adverse drug reactions associated with clopidogrel therapy
- Serum zinc associated with immunity and inflammatory markers in Covid-19
- The relationship between night shift work and breast cancer incidence: A systematic review and meta-analysis of observational studies
- LncRNA expression in idiopathic achalasia: New insight and preliminary exploration into pathogenesis
- Notoginsenoside R1 alleviates spinal cord injury through the miR-301a/KLF7 axis to activate Wnt/β-catenin pathway
- Moscatilin suppresses the inflammation from macrophages and T cells
- Zoledronate promotes ECM degradation and apoptosis via Wnt/β-catenin
- Epithelial-mesenchymal transition-related genes in coronary artery disease
- The effect evaluation of traditional vaginal surgery and transvaginal mesh surgery for severe pelvic organ prolapse: 5 years follow-up
- Repeated partial splenic artery embolization for hypersplenism improves platelet count
- Low expression of miR-27b in serum exosomes of non-small cell lung cancer facilitates its progression by affecting EGFR
- Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis
- miR-455-5p enhances 5-fluorouracil sensitivity in colorectal cancer cells by targeting PIK3R1 and DEPDC1
- The effect of tranexamic acid on the reduction of intraoperative and postoperative blood loss and thromboembolic risk in patients with hip fracture
- Isocitrate dehydrogenase 1 mutation in cholangiocarcinoma impairs tumor progression by sensitizing cells to ferroptosis
- Artemisinin protects against cerebral ischemia and reperfusion injury via inhibiting the NF-κB pathway
- A 16-gene signature associated with homologous recombination deficiency for prognosis prediction in patients with triple-negative breast cancer
- Lidocaine ameliorates chronic constriction injury-induced neuropathic pain through regulating M1/M2 microglia polarization
- MicroRNA 322-5p reduced neuronal inflammation via the TLR4/TRAF6/NF-κB axis in a rat epilepsy model
- miR-1273h-5p suppresses CXCL12 expression and inhibits gastric cancer cell invasion and metastasis
- Clinical characteristics of pneumonia patients of long course of illness infected with SARS-CoV-2
- circRNF20 aggravates the malignancy of retinoblastoma depending on the regulation of miR-132-3p/PAX6 axis
- Linezolid for resistant Gram-positive bacterial infections in children under 12 years: A meta-analysis
- Rack1 regulates pro-inflammatory cytokines by NF-κB in diabetic nephropathy
- Comprehensive analysis of molecular mechanism and a novel prognostic signature based on small nuclear RNA biomarkers in gastric cancer patients
- Smog and risk of maternal and fetal birth outcomes: A retrospective study in Baoding, China
- Let-7i-3p inhibits the cell cycle, proliferation, invasion, and migration of colorectal cancer cells via downregulating CCND1
- β2-Adrenergic receptor expression in subchondral bone of patients with varus knee osteoarthritis
- Possible impact of COVID-19 pandemic and lockdown on suicide behavior among patients in Southeast Serbia
- In vitro antimicrobial activity of ozonated oil in liposome eyedrop against multidrug-resistant bacteria
- Potential biomarkers for inflammatory response in acute lung injury
- A low serum uric acid concentration predicts a poor prognosis in adult patients with candidemia
- Antitumor activity of recombinant oncolytic vaccinia virus with human IL2
- ALKBH5 inhibits TNF-α-induced apoptosis of HUVECs through Bcl-2 pathway
- Risk prediction of cardiovascular disease using machine learning classifiers
- Value of ultrasonography parameters in diagnosing polycystic ovary syndrome
- Bioinformatics analysis reveals three key genes and four survival genes associated with youth-onset NSCLC
- Identification of autophagy-related biomarkers in patients with pulmonary arterial hypertension based on bioinformatics analysis
- Protective effects of glaucocalyxin A on the airway of asthmatic mice
- Overexpression of miR-100-5p inhibits papillary thyroid cancer progression via targeting FZD8
- Bioinformatics-based analysis of SUMOylation-related genes in hepatocellular carcinoma reveals a role of upregulated SAE1 in promoting cell proliferation
- Effectiveness and clinical benefits of new anti-diabetic drugs: A real life experience
- Identification of osteoporosis based on gene biomarkers using support vector machine
- Tanshinone IIA reverses oxaliplatin resistance in colorectal cancer through microRNA-30b-5p/AVEN axis
- miR-212-5p inhibits nasopharyngeal carcinoma progression by targeting METTL3
- Association of ST-T changes with all-cause mortality among patients with peripheral T-cell lymphomas
- LINC00665/miRNAs axis-mediated collagen type XI alpha 1 correlates with immune infiltration and malignant phenotypes in lung adenocarcinoma
- The perinatal factors that influence the excretion of fecal calprotectin in premature-born children
- Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study
- Does the use of 3D-printed cones give a chance to postpone the use of megaprostheses in patients with large bone defects in the knee joint?
- lncRNA HAGLR modulates myocardial ischemia–reperfusion injury in mice through regulating miR-133a-3p/MAPK1 axis
- Protective effect of ghrelin on intestinal I/R injury in rats
- In vivo knee kinematics of an innovative prosthesis design
- Relationship between the height of fibular head and the incidence and severity of knee osteoarthritis
- lncRNA WT1-AS attenuates hypoxia/ischemia-induced neuronal injury during cerebral ischemic stroke via miR-186-5p/XIAP axis
- Correlation of cardiac troponin T and APACHE III score with all-cause in-hospital mortality in critically ill patients with acute pulmonary embolism
- LncRNA LINC01857 reduces metastasis and angiogenesis in breast cancer cells via regulating miR-2052/CENPQ axis
- Endothelial cell-specific molecule 1 (ESM1) promoted by transcription factor SPI1 acts as an oncogene to modulate the malignant phenotype of endometrial cancer
- SELENBP1 inhibits progression of colorectal cancer by suppressing epithelial–mesenchymal transition
- Visfatin is negatively associated with coronary artery lesions in subjects with impaired fasting glucose
- Treatment and outcomes of mechanical complications of acute myocardial infarction during the Covid-19 era: A comparison with the pre-Covid-19 period. A systematic review and meta-analysis
- Neonatal stroke surveillance study protocol in the United Kingdom and Republic of Ireland
- Oncogenic role of TWF2 in human tumors: A pan-cancer analysis
- Mean corpuscular hemoglobin predicts the length of hospital stay independent of severity classification in patients with acute pancreatitis
- Association of gallstone and polymorphisms of UGT1A1*27 and UGT1A1*28 in patients with hepatitis B virus-related liver failure
- TGF-β1 upregulates Sar1a expression and induces procollagen-I secretion in hypertrophic scarring fibroblasts
- Antisense lncRNA PCNA-AS1 promotes esophageal squamous cell carcinoma progression through the miR-2467-3p/PCNA axis
- NK-cell dysfunction of acute myeloid leukemia in relation to the renin–angiotensin system and neurotransmitter genes
- The effect of dilution with glucose and prolonged injection time on dexamethasone-induced perineal irritation – A randomized controlled trial
- miR-146-5p restrains calcification of vascular smooth muscle cells by suppressing TRAF6
- Role of lncRNA MIAT/miR-361-3p/CCAR2 in prostate cancer cells
- lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2
- Noninvasive diagnosis of AIH/PBC overlap syndrome based on prediction models
- lncRNA FAM230B is highly expressed in colorectal cancer and suppresses the maturation of miR-1182 to increase cell proliferation
- circ-LIMK1 regulates cisplatin resistance in lung adenocarcinoma by targeting miR-512-5p/HMGA1 axis
- LncRNA SNHG3 promoted cell proliferation, migration, and metastasis of esophageal squamous cell carcinoma via regulating miR-151a-3p/PFN2 axis
- Risk perception and affective state on work exhaustion in obstetrics during the COVID-19 pandemic
- lncRNA-AC130710/miR-129-5p/mGluR1 axis promote migration and invasion by activating PKCα-MAPK signal pathway in melanoma
- SNRPB promotes cell cycle progression in thyroid carcinoma via inhibiting p53
- Xylooligosaccharides and aerobic training regulate metabolism and behavior in rats with streptozotocin-induced type 1 diabetes
- Serpin family A member 1 is an oncogene in glioma and its translation is enhanced by NAD(P)H quinone dehydrogenase 1 through RNA-binding activity
- Silencing of CPSF7 inhibits the proliferation, migration, and invasion of lung adenocarcinoma cells by blocking the AKT/mTOR signaling pathway
- Ultrasound-guided lumbar plexus block versus transversus abdominis plane block for analgesia in children with hip dislocation: A double-blind, randomized trial
- Relationship of plasma MBP and 8-oxo-dG with brain damage in preterm
- Identification of a novel necroptosis-associated miRNA signature for predicting the prognosis in head and neck squamous cell carcinoma
- Delayed femoral vein ligation reduces operative time and blood loss during hip disarticulation in patients with extremity tumors
- The expression of ASAP3 and NOTCH3 and the clinicopathological characteristics of adult glioma patients
- Longitudinal analysis of factors related to Helicobacter pylori infection in Chinese adults
- HOXA10 enhances cell proliferation and suppresses apoptosis in esophageal cancer via activating p38/ERK signaling pathway
- Meta-analysis of early-life antibiotic use and allergic rhinitis
- Marital status and its correlation with age, race, and gender in prognosis of tonsil squamous cell carcinomas
- HPV16 E6E7 up-regulates KIF2A expression by activating JNK/c-Jun signal, is beneficial to migration and invasion of cervical cancer cells
- Amino acid profiles in the tissue and serum of patients with liver cancer
- Pain in critically ill COVID-19 patients: An Italian retrospective study
- Immunohistochemical distribution of Bcl-2 and p53 apoptotic markers in acetamiprid-induced nephrotoxicity
- Estradiol pretreatment in GnRH antagonist protocol for IVF/ICSI treatment
- Long non-coding RNAs LINC00689 inhibits the apoptosis of human nucleus pulposus cells via miR-3127-5p/ATG7 axis-mediated autophagy
- The relationship between oxygen therapy, drug therapy, and COVID-19 mortality
- Monitoring hypertensive disorders in pregnancy to prevent preeclampsia in pregnant women of advanced maternal age: Trial mimicking with retrospective data
- SETD1A promotes the proliferation and glycolysis of nasopharyngeal carcinoma cells by activating the PI3K/Akt pathway
- The role of Shunaoxin pills in the treatment of chronic cerebral hypoperfusion and its main pharmacodynamic components
- TET3 governs malignant behaviors and unfavorable prognosis of esophageal squamous cell carcinoma by activating the PI3K/AKT/GSK3β/β-catenin pathway
- Associations between morphokinetic parameters of temporary-arrest embryos and the clinical prognosis in FET cycles
- Long noncoding RNA WT1-AS regulates trophoblast proliferation, migration, and invasion via the microRNA-186-5p/CADM2 axis
- The incidence of bronchiectasis in chronic obstructive pulmonary disease
- Integrated bioinformatics analysis shows integrin alpha 3 is a prognostic biomarker for pancreatic cancer
- Inhibition of miR-21 improves pulmonary vascular responses in bronchopulmonary dysplasia by targeting the DDAH1/ADMA/NO pathway
- Comparison of hospitalized patients with severe pneumonia caused by COVID-19 and influenza A (H7N9 and H1N1): A retrospective study from a designated hospital
- lncRNA ZFAS1 promotes intervertebral disc degeneration by upregulating AAK1
- Pathological characteristics of liver injury induced by N,N-dimethylformamide: From humans to animal models
- lncRNA ELFN1-AS1 enhances the progression of colon cancer by targeting miR-4270 to upregulate AURKB
- DARS-AS1 modulates cell proliferation and migration of gastric cancer cells by regulating miR-330-3p/NAT10 axis
- Dezocine inhibits cell proliferation, migration, and invasion by targeting CRABP2 in ovarian cancer
- MGST1 alleviates the oxidative stress of trophoblast cells induced by hypoxia/reoxygenation and promotes cell proliferation, migration, and invasion by activating the PI3K/AKT/mTOR pathway
- Bifidobacterium lactis Probio-M8 ameliorated the symptoms of type 2 diabetes mellitus mice by changing ileum FXR-CYP7A1
- circRNA DENND1B inhibits tumorigenicity of clear cell renal cell carcinoma via miR-122-5p/TIMP2 axis
- EphA3 targeted by miR-3666 contributes to melanoma malignancy via activating ERK1/2 and p38 MAPK pathways
- Pacemakers and methylprednisolone pulse therapy in immune-related myocarditis concomitant with complete heart block
- miRNA-130a-3p targets sphingosine-1-phosphate receptor 1 to activate the microglial and astrocytes and to promote neural injury under the high glucose condition
- Review Articles
- Current management of cancer pain in Italy: Expert opinion paper
- Hearing loss and brain disorders: A review of multiple pathologies
- The rationale for using low-molecular weight heparin in the therapy of symptomatic COVID-19 patients
- Amyotrophic lateral sclerosis and delayed onset muscle soreness in light of the impaired blink and stretch reflexes – watch out for Piezo2
- Interleukin-35 in autoimmune dermatoses: Current concepts
- Recent discoveries in microbiota dysbiosis, cholangiocytic factors, and models for studying the pathogenesis of primary sclerosing cholangitis
- Advantages of ketamine in pediatric anesthesia
- Congenital adrenal hyperplasia. Role of dentist in early diagnosis
- Migraine management: Non-pharmacological points for patients and health care professionals
- Atherogenic index of plasma and coronary artery disease: A systematic review
- Physiological and modulatory role of thioredoxins in the cellular function
- Case Reports
- Intrauterine Bakri balloon tamponade plus cervical cerclage for the prevention and treatment of postpartum haemorrhage in late pregnancy complicated with acute aortic dissection: Case series
- A case of successful pembrolizumab monotherapy in a patient with advanced lung adenocarcinoma: Use of multiple biomarkers in combination for clinical practice
- Unusual neurological manifestations of bilateral medial medullary infarction: A case report
- Atypical symptoms of malignant hyperthermia: A rare causative mutation in the RYR1 gene
- A case report of dermatomyositis with the missed diagnosis of non-small cell lung cancer and concurrence of pulmonary tuberculosis
- A rare case of endometrial polyp complicated with uterine inversion: A case report and clinical management
- Spontaneous rupturing of splenic artery aneurysm: Another reason for fatal syncope and shock (Case report and literature review)
- Fungal infection mimicking COVID-19 infection – A case report
- Concurrent aspergillosis and cystic pulmonary metastases in a patient with tongue squamous cell carcinoma
- Paraganglioma-induced inverted takotsubo-like cardiomyopathy leading to cardiogenic shock successfully treated with extracorporeal membrane oxygenation
- Lineage switch from lymphoma to myeloid neoplasms: First case series from a single institution
- Trismus during tracheal extubation as a complication of general anaesthesia – A case report
- Simultaneous treatment of a pubovesical fistula and lymph node metastasis secondary to multimodal treatment for prostate cancer: Case report and review of the literature
- Two case reports of skin vasculitis following the COVID-19 immunization
- Ureteroiliac fistula after oncological surgery: Case report and review of the literature
- Synchronous triple primary malignant tumours in the bladder, prostate, and lung harbouring TP53 and MEK1 mutations accompanied with severe cardiovascular diseases: A case report
- Huge mucinous cystic neoplasms with adhesion to the left colon: A case report and literature review
- Commentary
- Commentary on “Clinicopathological features of programmed cell death-ligand 1 expression in patients with oral squamous cell carcinoma”
- Rapid Communication
- COVID-19 fear, post-traumatic stress, growth, and the role of resilience
- Erratum
- Erratum to “Tollip promotes hepatocellular carcinoma progression via PI3K/AKT pathway”
- Erratum to “Effect of femoral head necrosis cystic area on femoral head collapse and stress distribution in femoral head: A clinical and finite element study”
- Erratum to “lncRNA NORAD promotes lung cancer progression by competitively binding to miR-28-3p with E2F2”
- Retraction
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Retraction to “miR-519d downregulates LEP expression to inhibit preeclampsia development”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part II
- Usefulness of close surveillance for rectal cancer patients after neoadjuvant chemoradiotherapy

