Abstract
C 17 H 17 Cl 2 N 5 , monoclinic, P21/c (no. 14), a = 14.603(2) Å, b = 24.059(4) Å, c = 10.1380(16) Å, β = 93.141(3)∘, V = 3556.4(10) Å 3 , Z = 8, R gt (F) = 0.0512, wR ref (F2) = 0.1416, T = 296(2) K.
Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
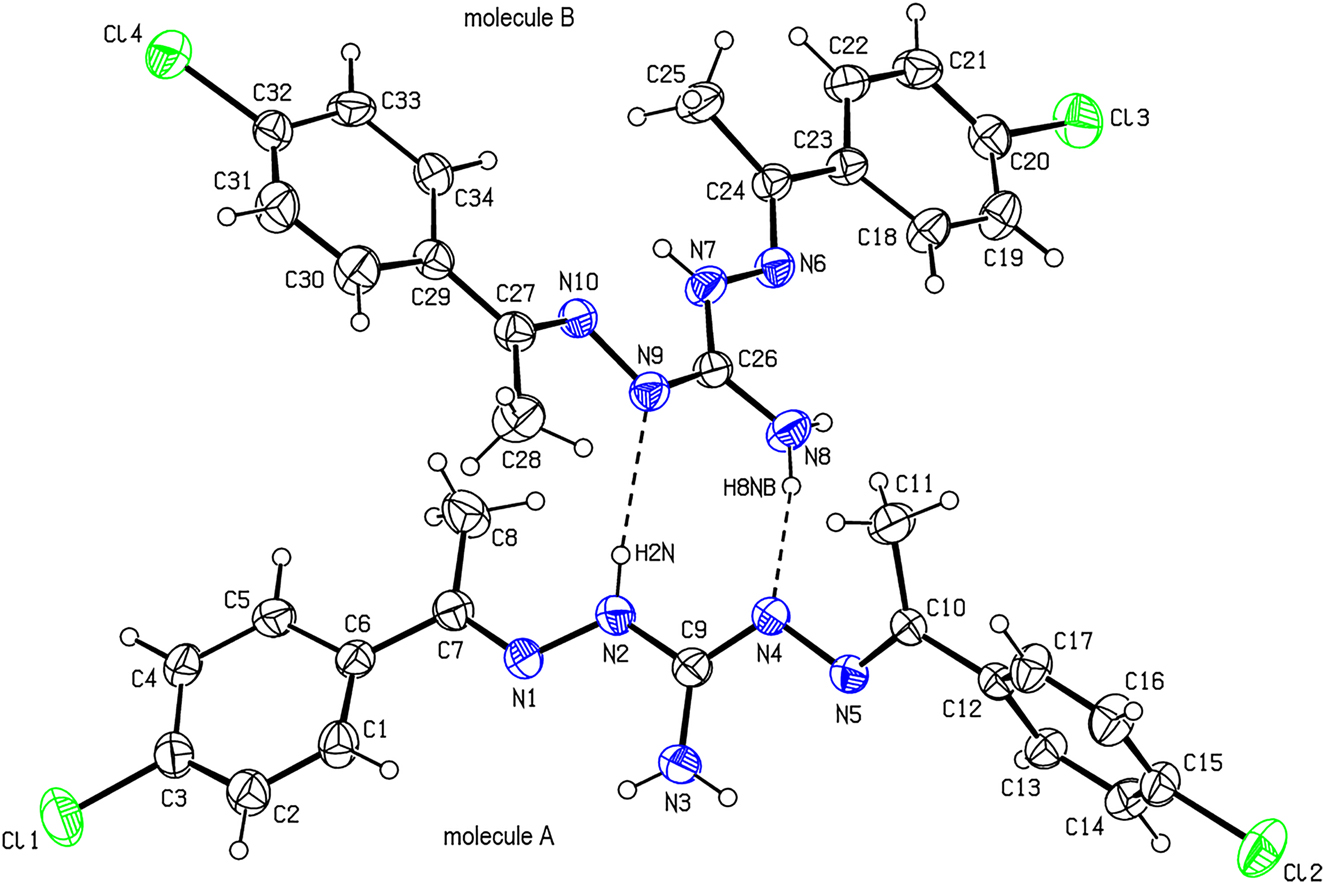
A perspective view of the asymmetric unit of the title compound. Displacement ellipsoids are drawn at the 30% probability level. The two molecules (A and B) in the asymmetric unit are linked by N—H···N hydrogen bonds shown as dashed lines. Only the major component of the disordered atoms is shown for clarity.
Data collection and handling.
| Crystal: | Yellow irregular |
| Size: | 0.32 × 0.22 × 0.12 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.37 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θmax, completeness: | 27.6°, >99 % |
| N(hkl)measured, N(hkl)unique, Rint: | 30263, 8240, 0.049 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 4576 |
| N(param)refined: | 474 |
| Programs: | Bruker [1], SHELX [2, 3], WinGX/ORTEP [4], PLATON [5] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| C1 | 0.98245 (18) | 0.17607 (12) | 0.4007 (3) | 0.0594 (7) |
| H1 | 0.936791 | 0.161750 | 0.342835 | 0.071* |
| C2 | 1.06576 (19) | 0.14891 (13) | 0.4150 (3) | 0.0655 (8) |
| H2 | 1.075867 | 0.116554 | 0.367751 | 0.079* |
| C3 | 1.13303 (18) | 0.17007 (13) | 0.4992 (3) | 0.0602 (7) |
| C4a | 1.1233 (6) | 0.2205 (4) | 0.5598 (10) | 0.0640 (19) |
| H4a | 1.172001 | 0.236487 | 0.609086 | 0.077* |
| C5a | 1.0390 (8) | 0.2472 (5) | 0.5458 (12) | 0.062 (2) |
| H5a | 1.031192 | 0.281168 | 0.587349 | 0.074* |
| C4Ab | 1.1079 (18) | 0.2084 (12) | 0.587 (3) | 0.0640 (19) |
| H4Ab | 1.147163 | 0.216109 | 0.659733 | 0.077* |
| C5Ab | 1.026 (2) | 0.2365 (14) | 0.572 (4) | 0.062 (2) |
| H5Ab | 1.012441 | 0.264172 | 0.631923 | 0.074* |
| C6 | 0.96532 (16) | 0.22390 (10) | 0.4702 (2) | 0.0473 (6) |
| C7 | 0.87451 (17) | 0.25136 (11) | 0.4591 (3) | 0.0517 (6) |
| C8 | 0.8611 (2) | 0.30554 (13) | 0.5271 (3) | 0.0796 (10) |
| H8A | 0.888836 | 0.334761 | 0.478693 | 0.119* |
| H8B | 0.889033 | 0.303935 | 0.614943 | 0.119* |
| H8C | 0.796630 | 0.312765 | 0.531419 | 0.119* |
| C9 | 0.65586 (17) | 0.21923 (11) | 0.3180 (3) | 0.0514 (6) |
| C10 | 0.45869 (15) | 0.24199 (10) | 0.1447 (2) | 0.0455 (6) |
| C11 | 0.4755 (2) | 0.30013 (12) | 0.1001 (3) | 0.0724 (9) |
| H11A | 0.528470 | 0.300823 | 0.048014 | 0.109* |
| H11B | 0.485903 | 0.323731 | 0.175833 | 0.109* |
| H11C | 0.423063 | 0.313192 | 0.047919 | 0.109* |
| C12 | 0.37824 (15) | 0.21137 (10) | 0.0876 (2) | 0.0442 (6) |
| C13 | 0.33074 (17) | 0.17375 (11) | 0.1612 (3) | 0.0512 (6) |
| H13 | 0.349573 | 0.167554 | 0.249076 | 0.061* |
| C14 | 0.25601 (19) | 0.14530 (11) | 0.1066 (3) | 0.0594 (7) |
| H14 | 0.224226 | 0.120529 | 0.157699 | 0.071* |
| C15 | 0.22890 (18) | 0.15369 (12) | −0.0229 (3) | 0.0592 (7) |
| C16 | 0.2755 (2) | 0.18980 (13) | −0.0987 (3) | 0.0684 (8) |
| H16 | 0.257431 | 0.194755 | −0.187287 | 0.082* |
| C17 | 0.34912 (18) | 0.21879 (13) | −0.0436 (3) | 0.0611 (7) |
| H17 | 0.379916 | 0.243824 | −0.095220 | 0.073* |
| N1 | 0.81069 (14) | 0.22430 (9) | 0.3944 (2) | 0.0522 (5) |
| N2 | 0.72570 (15) | 0.24858 (10) | 0.3799 (2) | 0.0584 (6) |
| H2N | 0.721 (2) | 0.2824 (12) | 0.370 (3) | 0.070* |
| N3 | 0.66766 (18) | 0.16488 (10) | 0.3000 (3) | 0.0710 (8) |
| H3NA | 0.716 (2) | 0.1505 (14) | 0.324 (3) | 0.085* |
| H3NB | 0.624 (2) | 0.1462 (14) | 0.265 (3) | 0.085* |
| N4 | 0.58347 (13) | 0.24812 (8) | 0.2838 (2) | 0.0541 (5) |
| N5 | 0.51198 (13) | 0.21556 (8) | 0.2278 (2) | 0.0496 (5) |
| Cl1 | 1.23700 (5) | 0.13522 (4) | 0.52022 (10) | 0.0941 (3) |
| Cl2 | 0.13562 (6) | 0.11783 (4) | −0.09404 (10) | 0.0943 (3) |
| C18a | 0.4264 (7) | 0.4648 (5) | 0.7502 (11) | 0.0581 (19) |
| H18a | 0.422294 | 0.434609 | 0.693014 | 0.070* |
| C19a | 0.3583 (7) | 0.4732 (5) | 0.8365 (10) | 0.0617 (19) |
| H19a | 0.308137 | 0.449396 | 0.836773 | 0.074* |
| C18Aa | 0.4481 (19) | 0.4565 (15) | 0.778 (3) | 0.0581 (19) |
| H18Aa | 0.457363 | 0.421667 | 0.740926 | 0.070* |
| C19Aa | 0.3810 (18) | 0.4639 (14) | 0.864 (3) | 0.0617 (19) |
| H19Aa | 0.344311 | 0.434008 | 0.885106 | 0.074* |
| C20 | 0.36573 (18) | 0.51709 (12) | 0.9218 (3) | 0.0567 (7) |
| C21 | 0.4329 (2) | 0.55541 (12) | 0.9122 (3) | 0.0657 (8) |
| H21 | 0.433523 | 0.587146 | 0.964623 | 0.079* |
| C22 | 0.50043 (19) | 0.54719 (11) | 0.8245 (3) | 0.0611 (7) |
| H22 | 0.546093 | 0.573863 | 0.818411 | 0.073* |
| C23 | 0.50231 (17) | 0.50082 (10) | 0.7460 (2) | 0.0476 (6) |
| C24 | 0.57422 (16) | 0.49163 (10) | 0.6518 (2) | 0.0468 (6) |
| C25 | 0.6417 (2) | 0.53660 (12) | 0.6260 (3) | 0.0659 (8) |
| H25A | 0.698988 | 0.528675 | 0.673364 | 0.099* |
| H25B | 0.618511 | 0.571593 | 0.655117 | 0.099* |
| H25C | 0.651020 | 0.538387 | 0.533120 | 0.099* |
| C26 | 0.63407 (16) | 0.38669 (10) | 0.4322 (2) | 0.0466 (6) |
| C27 | 0.82185 (17) | 0.40787 (11) | 0.2563 (2) | 0.0504 (6) |
| C28 | 0.8205 (2) | 0.35922 (13) | 0.1632 (3) | 0.0767 (9) |
| H28A | 0.760575 | 0.342657 | 0.158847 | 0.115* |
| H28B | 0.864980 | 0.332252 | 0.194557 | 0.115* |
| H28C | 0.834984 | 0.371678 | 0.076882 | 0.115* |
| C29 | 0.89736 (17) | 0.44853 (10) | 0.2558 (2) | 0.0485 (6) |
| C30 | 0.9702 (2) | 0.44238 (13) | 0.1747 (3) | 0.0699 (8) |
| H30 | 0.968988 | 0.413300 | 0.114229 | 0.084* |
| C31 | 1.04427 (19) | 0.47792 (13) | 0.1810 (3) | 0.0709 (8) |
| H31 | 1.092401 | 0.472538 | 0.125936 | 0.085* |
| C32 | 1.04652 (17) | 0.52066 (11) | 0.2675 (3) | 0.0552 (7) |
| C33a | 0.9718 (8) | 0.5331 (4) | 0.3408 (12) | 0.064 (2) |
| H33a | 0.971449 | 0.564752 | 0.393490 | 0.076* |
| C34a | 0.8958 (7) | 0.4963 (5) | 0.3333 (11) | 0.0593 (19) |
| H34a | 0.844527 | 0.504121 | 0.380481 | 0.071* |
| C33Ab | 0.983 (3) | 0.5188 (11) | 0.361 (4) | 0.064 (2) |
| H33Ab | 0.989021 | 0.543121 | 0.432118 | 0.076* |
| C34Ab | 0.9169 (18) | 0.4868 (13) | 0.356 (3) | 0.0593 (19) |
| H34Ab | 0.877315 | 0.488219 | 0.424784 | 0.071* |
| N6 | 0.57311 (14) | 0.44386 (9) | 0.5944 (2) | 0.0499 (5) |
| N7 | 0.63867 (15) | 0.43353 (9) | 0.5060 (2) | 0.0549 (6) |
| H7N | 0.6796 (19) | 0.4547 (12) | 0.494 (3) | 0.066* |
| N8 | 0.56333 (17) | 0.35217 (11) | 0.4463 (3) | 0.0610 (6) |
| H8NA | 0.530 (2) | 0.3569 (12) | 0.506 (3) | 0.073* |
| H8NB | 0.5619 (19) | 0.3212 (13) | 0.397 (3) | 0.073* |
| N9 | 0.69490 (14) | 0.37425 (8) | 0.3462 (2) | 0.0501 (5) |
| N10 | 0.76155 (13) | 0.41545 (8) | 0.3417 (2) | 0.0470 (5) |
| Cl3 | 0.28131 (5) | 0.52635 (4) | 1.03409 (8) | 0.0800 (3) |
| Cl4 | 1.14287 (5) | 0.56361 (3) | 0.28543 (9) | 0.0797 (3) |
-
aOccupancy: 0.73(2), bOccupancy: 0.27(2).
Source of material
Weigh 2 g (0.0159 mol) 1,3–diaminoguanidine hydrochloride salt and put into a 100 mL flask. It is dissolved in a mixture of purified water (15 mL) and methanol (15 mL), and 4.13 mL (2.2 eq) of 4-methylbenzaldehyde is added. After mixing at room for 24 h in a magnetic stirrer, it is refluxed for 2 h, then the reaction is stopped, cooled to room temperature and neutralized with 0.64 g (1 eq) NaOH. For precipitation the mixture is kept in the refrigerator for 30–45 min, the precipitated material is filtered with a crucible, washed with distilled water (3 × 5 mL) and dried under vacuum. After waiting for one day, crystallization is performed in ethanol (EtOH), kept in the refrigerator for 24 h, the crystalline substance formed is filtered, washed in cold ethanol, the impurity of the substance is checked with TLC (Thin Layer Chromatogram) paper, then dried in a vacuum oven. 5.05 g and the yield was 94%.
Experimental details
The hydrogen atoms of the NH and NH 2 groups of molecules A and B were located from a difference–Fourier map and refined freely with U iso (H) = 1.5U eq (N). All C-bound H atoms were refined using a riding model with d(C—H) = 0.93 Å, U iso (H) = 1.2U eq (C) for aromatic and 0.96 Å, U iso (H) = 1.5U eq (C) for methyl H atoms. Atoms C4, C5, C18, C19, C33 and C34 of the chlorobenzene rings of molecules A and B were refined as disordered over two sets of sites with site occupancies of 0.73 (2) and 0.27 (2). Ellipsoid displacement (EADP) constraints were applied to disordered atom groups C4/C4A, C5/C5A, C18/C18A, C19/C19A, C33/C33A and C34/C34A.
Comment
Aminoguanidine is a guanidine derivative known for centuries. It is a compound that is structurally similar to the amino acid L-arginine, which contains a guanidine structure. In addition, aminoguanidine is an important molecule in the formation of nitric oxide by the catalytic effect of nitric oxide synthase [6]. In addition, aminoguanidine was administered to rats with cataracts and it was stated that aminoguanidine had cataract-preventing activity in this application. In recent years, aminoguanidine has been found to be a new promising compound that delays aging. At the same time, it has been observed that atherosclerosis, which increases with age, decreases with the application of aminoguanidine [7].
Moreover, compounds containing an imidazole core are structural isosteres of naturally occurring nucleotides, allowing them to readily interact with biopolymers responsible for numerous biological activities and functions of the living system [8].
There is no symmetry relationship between two molecules A (with Cl1) and B (with Cl3) of the title compound in the asymmetric unit Figure 1. The dihedral angles between the aromatic rings (C1—C6, C12—C17 and C18—C22, C29—C34) of the major components of the disordered molecules A and B are 86.6(2) and 6.0(3) ∘ for molecule A and B, respectively. The dihedral angles between the aromatic rings [(C1—C3,C4A,C5A,C6), (C12—C17) and (C18A,C19A,C20—C22), (C29—C32,C33A,C34A)] of the minor components of the disordered molecules A and B are 88.3(7) and 19.2(9) ∘ for molecule A and B, respectively. The conformations of the molecules A and B are different. This may be attributed to the different steric interactions that occur due to having the different environments in the crystal.
The bond length values in the title compound can be compared with those of the related compounds which are 1,2-bis((pentafluorobenzylidene)amino)guanidine methanol solvate (CSD refcode EMOCIT [9]), 1,3-bis((pentafluorobenzylidene)amino)guanidinium benzoate benzoic acid solvate (EMOCUF [9]) and 1,3-bis((1-naphthylmethylene) amino)guanidinium benzoate benzoic acid solvate (EMOCOZ [9]).
In the crystal, molecules A and B are connected by N—H⃛N hydrogen bonds [N2—H2N/N9: H2N/N9 = 2.25(3) Å, N2/N9 = 3.073(3) Å with angle at H2N = 175(3) ∘ and N8—H8NB/N4: H8NB/N4 = 2.13(3) Å, N8/N4 = 3.020(3) Å with angle at H8NB = 170(3) ∘ ], forming a R22(8) ring motif parallel to the (001) plane. Furthermore, molecules are linked by C—H⃛Cl contacts C11—H11C/Cl1i: H11C/Cl1i = 2.99 Å, C11/Cl1i = 3.860(3) Å with angle at H11C = 151.9 ∘ for symmetry operation (i): −1 + x, 1/2 − y, − 1/2 + z; C19—H19/Cl1ii: H19/Cl1ii = 2.98 Å, C19/Cl1ii = 3.711(10) Å with angle at H19 = 136.3 ∘ for (ii): −1 + x, 1/2 − y, 1/2 + z; C19A—H19A/Cl1ii: H19A/Cl1ii = 2.71 Å, C19A/Cl1ii = 3.60(3) Å with angle at H19A = 161.8 ∘ for (ii): −1 + x, 1/2 − y, 1/2 + z and C33—H33/Cl2iii: H33/Cl2iii = 2.93 Å, C33/Cl2iii = 3.694(11) Å with angle at H33 = 140.9 ∘ for (iii): 1 − x, 1/2 + y, 1/2 − z], C—Cl ⃛ π interactions [C15—Cl2/Cg(C29—C34)iv: Cl2/Cg(C29—C34)iv = 3.729(3) Å with angle at Cl2 = 167.09(11) ∘ ; C15—Cl2/Cg(C29—C32/C33A,C34A)iv: Cl2/Cg(C29—C32/C33A,C34A)iv = 3.590(8) Å with angle at Cl2 = 167.21(16) ∘ for (iv) −1 + x, 1/2 − y, − 1/2 + z] and π − π stacking interactions [Cg1(C18—C23)⃛Cg1(C18—C23)v = 3.892(4) Å, angle = 0.0(3) ∘ and slippage = 1.68 Å; Cg2(C20—C23/C18A,C19A)⃛Cg2(C20—C23/C18A,C19A)v = 3.618(10) Å, angle = 0.0(9) ∘ and slippage = 1.15 Å for (v) 1 − x, 1 − y, 2 − z], forming a three-dimensional network.
-
Author contributions: The author has accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: None declared.
-
Conflict of interest statement: The author declares no conflicts of interest regarding this article.
References
1. Bruker. APEX2 and SAINT. Bruker AXS Inc.: Madison, WI, USA, 2012.Search in Google Scholar
2. Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122; https://doi.org/10.1107/s0108767307043930.Search in Google Scholar PubMed
3. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr.. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
4. Farrugia, L. J. WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854; https://doi.org/10.1107/s0021889812029111.Search in Google Scholar
5. Spek, A. L. Structure validation in chemical crystallography. Acta Crystallogr 2009, D65, 148–155; https://doi.org/10.1107/s090744490804362x.Search in Google Scholar
6. Parlakpınar, H., Örüm, M., Acet, A. Aminoguanidine and cardiovascular system. Ann. Health Sci. Res. 2012, 2, 9–14.Search in Google Scholar
7. Özgüneş, H., Atasayar, S. Aminoguanidine and the its significance in diseases: review. Türkiye Klinikleri J. Med. Sci., 29, 976–986.Search in Google Scholar
8. Çağlar Yavuz, S. Synthesis and in Vitro Cytotoxic Activity Studies of Compounds Containing Pyrimidine and Imidazole Nucleus. PhD Thesis, Erciyes University, Graduate School of Natural and Applied Sciences, Department of Chemistry, 2019; pp. 1–275.Search in Google Scholar
9. Bose, P., Ahamed, B. N., Ghosh, P. Functionalized guanidinium chloride based colourimetric sensors for fluoride and acetate: single crystal X-ray structural evidence of –NH deprotonation and complexation. Org. Biomol. Chem. 2011, 9, 1972–1979; https://doi.org/10.1039/c0ob00947d.Search in Google Scholar PubMed
© 2021 Zeliha Atioğlu, published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Redetermination of the crystal structure of 3-bromonitrobenzene at 200 K, C6H4BrNO2 – temperature effects on cell constants
- Crystal structure of (E)-ethyl 2-((4-oxo-4H-chromen-3-yl)methyleneaminooxy)acetate, C14H13NO5
- Crystal structure of (8R,10R,14R, Z)-2-((3–Fluoropyridin-4-yl) methylene)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6, 6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-3H-cyclopenta[a] phenanthren-3-one, C36H52FNO3
- Crystal structure of [6,6′-((1E,1′E)-(propane-1,3- diylbis(azaneylylidene))bis(methaneylylidene)) bis(3-chlorophenol)-κ4N,N′,O,O′] copper(II), C17H14Cl2CuN2O2
- The crystal structure of 6-amino-2-carboxypyridin-1-ium bromide, C6H7BrN2O2
- Redetermination of the crystal structure of bis[N,N′-ethylenebis(acetylacetoniminato)nickel(II)] sodium perchlorate, C24H36ClN4NaNi2O8
- The crystal structure of 3-methyl-2,6-dinitrophenol, C7H6N2O5
- The crystal structure of 5-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione, C17H9ClN2O2
- Crystal structure of trans-tetraaqua-bis{2-carboxy-4-((5-carboxypyridin-3-yl)oxy)benzoato-κ1 N}cobalt(II) dihydrate C28H28O20N2Co
- Crystal structure of 3-allyl-4-(2-bromoethyl)-5-(4-methoxyphenyl)-2-(p-tolyl)furan, C23H23BrO2
- The crystal structure of 6,6′-(((2-(dimethylamino)ethyl)azanediyl)bis(methylene))bis(benzo[d][1,3]dioxol-5-ol ato-κ4N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)-titanium(IV)-dichloromethane(1/1), C27H25N3O10Ti
- Crystal structure of (((1E,1′E)-1,2-phenylenebis(methaneylylidene))bis(hydrazin-1-yl-2-ylidene))bis(aminomethaniminium) dinitrate C10H16N10O6
- Crystal structure of catena-poly[triaqua-(μ 2-1,3-di(1H-imidazol-1-yl)propane-κ 2 N:N′)-(4,4′-(1H-1,2,4-triazole-3,5-diyl)dibenzoato-κ 1 O)nickel(II)]N,N′-dimethylformamide (1/1), C28H35N8O8Ni
- The crystal structure of 3,3′-[1,4-phenylenebis(methylene)]bis(1-ethenyl-1H-imidazol-3-ium) dichloride – dichloromethane – water (1/1/1), C19H24Cl4N4O1
- Crystal structure of 1,1′-(methane-1,1-diyl)bis(3-propyl-1H-imidazol-3-ium) bis(hexafluoridophosphate), C13H22F12N4P2
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)tin(IV), C12H8Cl4Sn
- Synthesis and crystal structure of 4-acetylpyrene, C18H12O
- Crystal structure of 2,2′-(butane-1,4-diylbis(azanylylidene))bis(methanylylidene))bis(4-methoxyphenol), C20H24N2O4
- The crystal structure of (E)-2-(((5-((triphenylstannyl)thio)-1,3,4-thiadiazol-2-yl)imino)methyl)phenol, C27H21N3OS2Sn
- Crystal structure of diaqua-bis(μ2-6-phenylpyridine-2-carboxylate-κ3N,O:O)-bis(6-phenylpyridine-2-carboxylato-κ2N,O)lead(II) – N,N-dimethylformamide – water (1/2/4), C54H58N6O16Pb2
- Crystal structure of methyl 4-acetoxy-3-methoxybenzoate, C11H12O5
- Crystal structure of 2,2′-(propane-1,3-dilylbis(azaneylylidene))bis(methanylylidene)bis(4-methylphenol), C19H22N2O2
- Crystal structure of dichlorido-bis(4-methylphenyl-κC1)tin(IV), C14H14Cl2Sn
- Crystal structure of methyl (E)-3-(4-acetoxyphenyl)acrylate, C12H12O4
- The crystal structure of bis(benzoato-κ2 O,O′)-(2,9-dimethyl-1,10-phenanthroline-κ2 N,N′)-copper(II), C28H22CuN2O4
- Crystal structure of (8R,10R,14R,Z)-12-hydroxy-2-((6-methoxypyridin-2-yl)methylene)-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl)hexadecahydro-3H-cyclopenta[a]phenanthren-3-one–water (2/1), C37H56NO4.5
- Crystal structure of dimethyl-bis(4-bromophenyl-κC1)tin(IV), C14H14Br2Sn
- The crystal structure of the cocrystal di-μ2-chlorido-octamethyl-di-μ3-oxido-bis(2,3,4,5-tetrafluorobenzoato-κ2 O,O′)tetratin(IV) ─ octamethyl-di-μ3-oxido-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O′)-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O;O′)tetratin(IV) C58H54Cl2F24O16Sn8
- Crystal structure of 3-iodo-N 2-(2-methyl-1-(methylsulfonyl)propan-2-yl)-N 1-(2-methyl-4-(perfluoropropan-2-yl)phenyl)phthalamide, C23H22F7I1N2O4S1
- Crystal structure of 1-(2-(4-bromophenyl)-2,3-dihydro-1H-benzo[e]indol-1-yl)-naphthalen-2-ol – dichloromethane – dimethyl sulfoxide (1/1/1), C28H18BrNO·CH2Cl2·C2H6SO
- Crystal structure of [meso-5,7,7,12,14,14,-hexamethyl-1,4,8,11-tetraazacyclotetradecane]nickel(II) diperchlorate – dimethylsulphoxide (1/2), C20H48Cl2N4NiO10S2
- Crystal structure of 1,1′-(1,3-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S:S) palladium(II), C26H18N6PdS4
- The crystal structure of bis(6-phenylpyridine-2-carboxylato-κ2 N,O)copper(II), C24H16N2O4Cu
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC)-bis(triphenylarsine oxide-κO)tin(IV), C48H38As2Cl4O2Sn
- Crystal structure of (4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane-κ 8 N 2, O 6) potassium cyclopentadienide, [K([2.2.2]crypt)]Cp, C23H41KN2O6
- The crystal structure of bis(2-oxidopyridin-1-ium-3-carboxylato-κ2O,O′)-(phenantroline-κ2N,N′)manganese(II) - methanol (1/3), C27H28N4O9Mn
- Crystal structure of 4-(dimethylamino)pyridinium dibromido-tris(4-chlorophenyl-κC)stannate(IV), C25H23Br2Cl3N2Sn
- Crystal structure of (3E,5E)-1-(4-cyanobenzenesulfonyl)-3,5-bis(3-fluorobenzylidene)piperidin-4-one-dichloromethane (1/1), C27H20Cl2F2N2O3S
- Crystal structure of (3E,5E)-3,5-bis(4-fluorobenzylidene)-1-((4-trifluoromethyl)benzenesulfonyl)piperidin-4-one, C26H18F5NO3S
- Crystal structure of chlorido-(4-methyl-2-((phenylimino)methyl)phenolato-κ2 N,O)-(pyridine-κ1 N)platinum(II), C19H17ClN2OPt
- Crystal structure of (4-methylbenzyl)(triphenyl)phosphonium chloride dihydrate, C26H28ClO2P
- The crystal structure of poly[μ2-chlorido-(μ2-1,2-bis(4-pyridyl)ethane-κ2N:N′silver(I)], C12H12AgClN2
- Crystal structure of poly[(μ4-benzene-1,2,4,5-tetracarboxylato)-bis(μ2-adipohydrazide)dicadmium], C11H15N4O6Cd
- The crystal structure of (E)-N′-(butan-2-ylidene)isonicotinohydrazide 0.5 hydrate C10H13N3O·0.5H2O
- The crystal structure of bis(6-phenylpyridine-2-carboxylate-κ2 N,O)-(2,2′-bipyridine-κ2 N,N′)zinc(II) monohydrate, C34H26N4O5Zn
- The crystal structure of (1R *,2S *)-1,2-bis(2-fluorophenyl)-3,8-dimethoxyacenaphthene-1,2-diol, C26H20F2O4
- Crystal structure of catena-poly[(μ2-1-((2-ethyl-4-methyl-1H-imidazol-1-yl)methyl)-1H-benzotriazole-κ2N:N′)-(nitrato-κ2O,O′)silver (I)], C13H15Ag1N6O3
- The crystal structure of [(phenantroline-κ2 N,N′)-bis(6-phenylpyridine-2-carboxylate-κ2 N,O)cobalt(II)]monohydrate, C36H26N4O5Co
- Crystal structure of (1E)-N′-[(1E)-1-(4-chlorophenyl)ethylidene]-2-[1-(4-chlorophenyl)ethylidene]hydrazine-1-carbohydrazonamide, C 17 H 17 Cl 2 N 5
- The crystal structure of (E)-2-((tert-butylimino)methyl)-4-chlorophenol, C11H14ClNO
- Crystal structure of all-cis-2,4,6-trihydroxycyclohexane- 1,3,5-triaminium chloride sulfate, C6H18ClN3O7S
- Crystal structure of dichlorido-bis(dimethyl sulfoxide-κO)bis(4-methylphenyl-κC 1)tin(IV), C18H26Cl2O2S2Sn
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)(2,2′-bipyridyl-κ 2 N,N′)tin(IV), C22H16Cl4N2Sn
- Redetermination of the crystal structure of (E)-5-bromo-2-hydroxybenzaldehyde oxime, C 7 H 6 BrNO 2
- The crystal structure of (E)-amino(2-(4-methylbenzylidene)hydrazineyl)methaniminium 4-methylbenzoate, C9H13N4 + C8H7O2 −
- Crystal structure of 2-chloro-3-(isopentylamino)naphthalene-1,4-dione, C 15 H 16 ClNO 2
- The crystal structure of bis(2-acetyl-5-methoxyphenyl)carbonate 1.5 hydrate, C19H18O7
- The crystal structure of poly[(μ 4-4,4′-(azanediylbis(methylene))dibenzoato-κ 4 O:N:O′:Oʺ)zinc(II)], C16H13NO4Zn
- The crystal structure of catena-poly[(1,10-phenanthroline-k2N,N′)-(μ3-tetraoxidomoybdato(VI)-k3O:O′:O″)manganese(II)] C12H8N2O4MoMn
- Crystal structure of catena-poly[(4-hydroxyl-5-(methoylcarbonyl)thiophene-2-carboxylato-κ1 O)-(μ2-piperazine-1,4-diylbis(pyridin-4-ylmethanone)-κ2 N:N′)silver(I)] monohydrate, C23H23AgN4O8S
- Crystal structure of bis(4-bromo-2-(((3-bromopropyl)imino)methyl)phenolato-κ2N,O)-oxido-vanadium(IV), C20H20Br4N2O3V
- The crystal structure of (2a′S,2a1′S,3R,5a′S,7′R)-5-(furan-3-yl)-2a′,2a1′-dihydroxy-7′-methyldecahydro-2H-spiro[furan-3,6′-naphtho[1,8-bc]furan]-2,2′(2a′H)-dione, C19H22O7
- The crystal structure of 3-bromopicolinic acid, C6H4BrNO2
- Crystal structure of 1,1′-(1,4-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S,S) platinum(II), C26H18N6PtS4
- Synthesis and crystal structure of 5-(8-((3-carboxyazetidin-1-ium-1-yl)methyl)-7-hydroxy-4-oxo-4H-chromen-3-yl)-2-hydroxybenzenesulfonate monohydrate, C20H19NO10S
- The crystal structure of 3-amino-5-carboxypyridin-1-ium bromide, C6H7BrN2O2
- The crystal structure of (2-hydroxy-5-methyl-phenyl)-(1H-pyrazol-4-yl)-methanone hemihydrate, C11H10.5N2O2.5
- Crystal structure of tetraaqua-(2-(4-formylphenoxy)acetato-k1O)cadmium(II), C18H22O12Cd
- Crystal structure of diethyl 6,12-dimethyl-3,9-di-p-tolyl-3,9-diazapentacyclo[6.4.0.02,7.04,11.05,10]dodecane-1,5-dicarboxylate, C32H38N2O4
- Crystal structure of (E)-N′-(1-(3-chloro-4-fluorophenyl)ethylidene)-4-hydroxy – tetrahydrofuran (2/1), C17H16ClFN2O2.5
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Redetermination of the crystal structure of 3-bromonitrobenzene at 200 K, C6H4BrNO2 – temperature effects on cell constants
- Crystal structure of (E)-ethyl 2-((4-oxo-4H-chromen-3-yl)methyleneaminooxy)acetate, C14H13NO5
- Crystal structure of (8R,10R,14R, Z)-2-((3–Fluoropyridin-4-yl) methylene)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6, 6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-3H-cyclopenta[a] phenanthren-3-one, C36H52FNO3
- Crystal structure of [6,6′-((1E,1′E)-(propane-1,3- diylbis(azaneylylidene))bis(methaneylylidene)) bis(3-chlorophenol)-κ4N,N′,O,O′] copper(II), C17H14Cl2CuN2O2
- The crystal structure of 6-amino-2-carboxypyridin-1-ium bromide, C6H7BrN2O2
- Redetermination of the crystal structure of bis[N,N′-ethylenebis(acetylacetoniminato)nickel(II)] sodium perchlorate, C24H36ClN4NaNi2O8
- The crystal structure of 3-methyl-2,6-dinitrophenol, C7H6N2O5
- The crystal structure of 5-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione, C17H9ClN2O2
- Crystal structure of trans-tetraaqua-bis{2-carboxy-4-((5-carboxypyridin-3-yl)oxy)benzoato-κ1 N}cobalt(II) dihydrate C28H28O20N2Co
- Crystal structure of 3-allyl-4-(2-bromoethyl)-5-(4-methoxyphenyl)-2-(p-tolyl)furan, C23H23BrO2
- The crystal structure of 6,6′-(((2-(dimethylamino)ethyl)azanediyl)bis(methylene))bis(benzo[d][1,3]dioxol-5-ol ato-κ4N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)-titanium(IV)-dichloromethane(1/1), C27H25N3O10Ti
- Crystal structure of (((1E,1′E)-1,2-phenylenebis(methaneylylidene))bis(hydrazin-1-yl-2-ylidene))bis(aminomethaniminium) dinitrate C10H16N10O6
- Crystal structure of catena-poly[triaqua-(μ 2-1,3-di(1H-imidazol-1-yl)propane-κ 2 N:N′)-(4,4′-(1H-1,2,4-triazole-3,5-diyl)dibenzoato-κ 1 O)nickel(II)]N,N′-dimethylformamide (1/1), C28H35N8O8Ni
- The crystal structure of 3,3′-[1,4-phenylenebis(methylene)]bis(1-ethenyl-1H-imidazol-3-ium) dichloride – dichloromethane – water (1/1/1), C19H24Cl4N4O1
- Crystal structure of 1,1′-(methane-1,1-diyl)bis(3-propyl-1H-imidazol-3-ium) bis(hexafluoridophosphate), C13H22F12N4P2
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)tin(IV), C12H8Cl4Sn
- Synthesis and crystal structure of 4-acetylpyrene, C18H12O
- Crystal structure of 2,2′-(butane-1,4-diylbis(azanylylidene))bis(methanylylidene))bis(4-methoxyphenol), C20H24N2O4
- The crystal structure of (E)-2-(((5-((triphenylstannyl)thio)-1,3,4-thiadiazol-2-yl)imino)methyl)phenol, C27H21N3OS2Sn
- Crystal structure of diaqua-bis(μ2-6-phenylpyridine-2-carboxylate-κ3N,O:O)-bis(6-phenylpyridine-2-carboxylato-κ2N,O)lead(II) – N,N-dimethylformamide – water (1/2/4), C54H58N6O16Pb2
- Crystal structure of methyl 4-acetoxy-3-methoxybenzoate, C11H12O5
- Crystal structure of 2,2′-(propane-1,3-dilylbis(azaneylylidene))bis(methanylylidene)bis(4-methylphenol), C19H22N2O2
- Crystal structure of dichlorido-bis(4-methylphenyl-κC1)tin(IV), C14H14Cl2Sn
- Crystal structure of methyl (E)-3-(4-acetoxyphenyl)acrylate, C12H12O4
- The crystal structure of bis(benzoato-κ2 O,O′)-(2,9-dimethyl-1,10-phenanthroline-κ2 N,N′)-copper(II), C28H22CuN2O4
- Crystal structure of (8R,10R,14R,Z)-12-hydroxy-2-((6-methoxypyridin-2-yl)methylene)-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl)hexadecahydro-3H-cyclopenta[a]phenanthren-3-one–water (2/1), C37H56NO4.5
- Crystal structure of dimethyl-bis(4-bromophenyl-κC1)tin(IV), C14H14Br2Sn
- The crystal structure of the cocrystal di-μ2-chlorido-octamethyl-di-μ3-oxido-bis(2,3,4,5-tetrafluorobenzoato-κ2 O,O′)tetratin(IV) ─ octamethyl-di-μ3-oxido-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O′)-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O;O′)tetratin(IV) C58H54Cl2F24O16Sn8
- Crystal structure of 3-iodo-N 2-(2-methyl-1-(methylsulfonyl)propan-2-yl)-N 1-(2-methyl-4-(perfluoropropan-2-yl)phenyl)phthalamide, C23H22F7I1N2O4S1
- Crystal structure of 1-(2-(4-bromophenyl)-2,3-dihydro-1H-benzo[e]indol-1-yl)-naphthalen-2-ol – dichloromethane – dimethyl sulfoxide (1/1/1), C28H18BrNO·CH2Cl2·C2H6SO
- Crystal structure of [meso-5,7,7,12,14,14,-hexamethyl-1,4,8,11-tetraazacyclotetradecane]nickel(II) diperchlorate – dimethylsulphoxide (1/2), C20H48Cl2N4NiO10S2
- Crystal structure of 1,1′-(1,3-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S:S) palladium(II), C26H18N6PdS4
- The crystal structure of bis(6-phenylpyridine-2-carboxylato-κ2 N,O)copper(II), C24H16N2O4Cu
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC)-bis(triphenylarsine oxide-κO)tin(IV), C48H38As2Cl4O2Sn
- Crystal structure of (4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane-κ 8 N 2, O 6) potassium cyclopentadienide, [K([2.2.2]crypt)]Cp, C23H41KN2O6
- The crystal structure of bis(2-oxidopyridin-1-ium-3-carboxylato-κ2O,O′)-(phenantroline-κ2N,N′)manganese(II) - methanol (1/3), C27H28N4O9Mn
- Crystal structure of 4-(dimethylamino)pyridinium dibromido-tris(4-chlorophenyl-κC)stannate(IV), C25H23Br2Cl3N2Sn
- Crystal structure of (3E,5E)-1-(4-cyanobenzenesulfonyl)-3,5-bis(3-fluorobenzylidene)piperidin-4-one-dichloromethane (1/1), C27H20Cl2F2N2O3S
- Crystal structure of (3E,5E)-3,5-bis(4-fluorobenzylidene)-1-((4-trifluoromethyl)benzenesulfonyl)piperidin-4-one, C26H18F5NO3S
- Crystal structure of chlorido-(4-methyl-2-((phenylimino)methyl)phenolato-κ2 N,O)-(pyridine-κ1 N)platinum(II), C19H17ClN2OPt
- Crystal structure of (4-methylbenzyl)(triphenyl)phosphonium chloride dihydrate, C26H28ClO2P
- The crystal structure of poly[μ2-chlorido-(μ2-1,2-bis(4-pyridyl)ethane-κ2N:N′silver(I)], C12H12AgClN2
- Crystal structure of poly[(μ4-benzene-1,2,4,5-tetracarboxylato)-bis(μ2-adipohydrazide)dicadmium], C11H15N4O6Cd
- The crystal structure of (E)-N′-(butan-2-ylidene)isonicotinohydrazide 0.5 hydrate C10H13N3O·0.5H2O
- The crystal structure of bis(6-phenylpyridine-2-carboxylate-κ2 N,O)-(2,2′-bipyridine-κ2 N,N′)zinc(II) monohydrate, C34H26N4O5Zn
- The crystal structure of (1R *,2S *)-1,2-bis(2-fluorophenyl)-3,8-dimethoxyacenaphthene-1,2-diol, C26H20F2O4
- Crystal structure of catena-poly[(μ2-1-((2-ethyl-4-methyl-1H-imidazol-1-yl)methyl)-1H-benzotriazole-κ2N:N′)-(nitrato-κ2O,O′)silver (I)], C13H15Ag1N6O3
- The crystal structure of [(phenantroline-κ2 N,N′)-bis(6-phenylpyridine-2-carboxylate-κ2 N,O)cobalt(II)]monohydrate, C36H26N4O5Co
- Crystal structure of (1E)-N′-[(1E)-1-(4-chlorophenyl)ethylidene]-2-[1-(4-chlorophenyl)ethylidene]hydrazine-1-carbohydrazonamide, C 17 H 17 Cl 2 N 5
- The crystal structure of (E)-2-((tert-butylimino)methyl)-4-chlorophenol, C11H14ClNO
- Crystal structure of all-cis-2,4,6-trihydroxycyclohexane- 1,3,5-triaminium chloride sulfate, C6H18ClN3O7S
- Crystal structure of dichlorido-bis(dimethyl sulfoxide-κO)bis(4-methylphenyl-κC 1)tin(IV), C18H26Cl2O2S2Sn
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)(2,2′-bipyridyl-κ 2 N,N′)tin(IV), C22H16Cl4N2Sn
- Redetermination of the crystal structure of (E)-5-bromo-2-hydroxybenzaldehyde oxime, C 7 H 6 BrNO 2
- The crystal structure of (E)-amino(2-(4-methylbenzylidene)hydrazineyl)methaniminium 4-methylbenzoate, C9H13N4 + C8H7O2 −
- Crystal structure of 2-chloro-3-(isopentylamino)naphthalene-1,4-dione, C 15 H 16 ClNO 2
- The crystal structure of bis(2-acetyl-5-methoxyphenyl)carbonate 1.5 hydrate, C19H18O7
- The crystal structure of poly[(μ 4-4,4′-(azanediylbis(methylene))dibenzoato-κ 4 O:N:O′:Oʺ)zinc(II)], C16H13NO4Zn
- The crystal structure of catena-poly[(1,10-phenanthroline-k2N,N′)-(μ3-tetraoxidomoybdato(VI)-k3O:O′:O″)manganese(II)] C12H8N2O4MoMn
- Crystal structure of catena-poly[(4-hydroxyl-5-(methoylcarbonyl)thiophene-2-carboxylato-κ1 O)-(μ2-piperazine-1,4-diylbis(pyridin-4-ylmethanone)-κ2 N:N′)silver(I)] monohydrate, C23H23AgN4O8S
- Crystal structure of bis(4-bromo-2-(((3-bromopropyl)imino)methyl)phenolato-κ2N,O)-oxido-vanadium(IV), C20H20Br4N2O3V
- The crystal structure of (2a′S,2a1′S,3R,5a′S,7′R)-5-(furan-3-yl)-2a′,2a1′-dihydroxy-7′-methyldecahydro-2H-spiro[furan-3,6′-naphtho[1,8-bc]furan]-2,2′(2a′H)-dione, C19H22O7
- The crystal structure of 3-bromopicolinic acid, C6H4BrNO2
- Crystal structure of 1,1′-(1,4-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S,S) platinum(II), C26H18N6PtS4
- Synthesis and crystal structure of 5-(8-((3-carboxyazetidin-1-ium-1-yl)methyl)-7-hydroxy-4-oxo-4H-chromen-3-yl)-2-hydroxybenzenesulfonate monohydrate, C20H19NO10S
- The crystal structure of 3-amino-5-carboxypyridin-1-ium bromide, C6H7BrN2O2
- The crystal structure of (2-hydroxy-5-methyl-phenyl)-(1H-pyrazol-4-yl)-methanone hemihydrate, C11H10.5N2O2.5
- Crystal structure of tetraaqua-(2-(4-formylphenoxy)acetato-k1O)cadmium(II), C18H22O12Cd
- Crystal structure of diethyl 6,12-dimethyl-3,9-di-p-tolyl-3,9-diazapentacyclo[6.4.0.02,7.04,11.05,10]dodecane-1,5-dicarboxylate, C32H38N2O4
- Crystal structure of (E)-N′-(1-(3-chloro-4-fluorophenyl)ethylidene)-4-hydroxy – tetrahydrofuran (2/1), C17H16ClFN2O2.5


