Abstract
C10H8BrN3, orthorhombic,
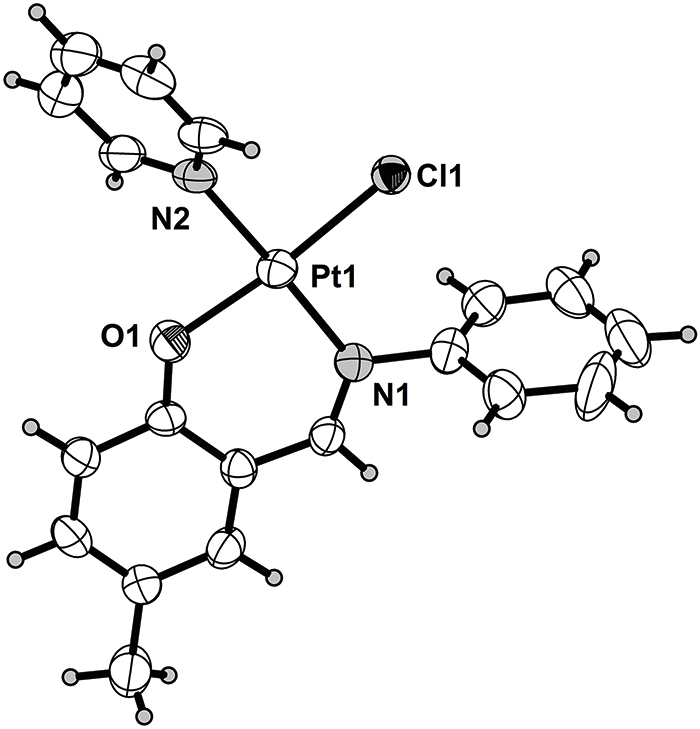
One of the two crystallographically independent tile complexes is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Yellow block |
| Size: | 0.55 × 0.40 × 0.20 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 7.74 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θ max, completeness: | 27.6°, 99% |
| N(hkl)measured, N(hkl)unique, R int: | 13,405, 8311, 0.042 |
| Criterion for I obs, N(hkl)gt: | I obs > 2 σ(I obs), 5880 |
| N(param)refined: | 435 |
| Programs: | Bruker [1], SHELX [2], Diamond [3] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| Pt1 | 0.18009 (3) | 0.03568 (3) | 0.19872 (3) | 0.03592 (13) |
| Pt2 | 0.68252 (3) | 0.33125 (3) | 0.19834 (3) | 0.03727 (13) |
| Cl1 | 0.1784 (2) | −0.1114 (3) | 0.1260 (3) | 0.0605 (8) |
| Cl2 | 0.6815 (2) | 0.5276 (2) | 0.1190 (3) | 0.0655 (8) |
| N1 | 0.0221 (7) | 0.0089 (7) | 0.2603 (7) | 0.0377 (17) |
| N2 | 0.3379 (7) | 0.0727 (8) | 0.1339 (7) | 0.044 (2) |
| N3 | 0.5231 (7) | 0.3216 (7) | 0.2591 (7) | 0.0383 (18) |
| N4 | 0.8425 (7) | 0.3338 (7) | 0.1330 (7) | 0.044 (2) |
| O1 | 0.2002 (5) | 0.1639 (7) | 0.2583 (7) | 0.055 (2) |
| O2 | 0.7023 (6) | 0.1616 (6) | 0.2605 (6) | 0.0460 (17) |
| C1 | 0.1246 (8) | 0.2057 (9) | 0.3107 (8) | 0.041 (2) |
| C2 | 0.0142 (8) | 0.1610 (8) | 0.3430 (8) | 0.038 (2) |
| C3 | −0.0624 (9) | 0.2186 (9) | 0.3918 (9) | 0.047 (2) |
| H3 | −0.1352 | 0.1901 | 0.4083 | 0.056* |
| C4 | −0.0347 (9) | 0.3181 (9) | 0.4173 (9) | 0.047 (2) |
| C5 | 0.0748 (10) | 0.3574 (11) | 0.3896 (10) | 0.059 (3) |
| H5 | 0.0968 | 0.4210 | 0.4075 | 0.070* |
| C6 | 0.1511 (9) | 0.3073 (10) | 0.3377 (10) | 0.051 (3) |
| H6 | 0.2224 | 0.3394 | 0.3189 | 0.061* |
| C7 | −0.0295 (8) | 0.0678 (8) | 0.3133 (8) | 0.039 (2) |
| H7 | −0.1039 | 0.0481 | 0.3352 | 0.047* |
| C8 | −0.0496 (8) | −0.0800 (8) | 0.2439 (10) | 0.045 (2) |
| C9 | −0.0956 (9) | −0.0643 (11) | 0.1505 (11) | 0.055 (3) |
| H9 | −0.0782 | 0.0042 | 0.0955 | 0.066* |
| C10 | −0.1643 (11) | −0.1397 (12) | 0.1311 (13) | 0.066 (4) |
| H10 | −0.1923 | −0.1263 | 0.0650 | 0.079* |
| C11 | −0.1918 (13) | −0.2434 (15) | 0.2204 (17) | 0.089 (6) |
| H11 | −0.2404 | −0.2981 | 0.2127 | 0.106* |
| C12 | −0.1478 (14) | −0.2616 (14) | 0.3145 (18) | 0.104 (6) |
| H12 | −0.1648 | −0.3295 | 0.3704 | 0.125* |
| C13 | −0.0761 (11) | −0.1785 (9) | 0.3287 (10) | 0.057 (3) |
| H13 | −0.0472 | −0.1899 | 0.3941 | 0.068* |
| C14 | −0.1162 (12) | 0.3737 (13) | 0.4744 (16) | 0.092 (6) |
| H14A | −0.1250 | 0.3299 | 0.5492 | 0.137* |
| H14B | −0.1862 | 0.3770 | 0.4449 | 0.137* |
| H14C | −0.0903 | 0.4502 | 0.4660 | 0.137* |
| C15 | 0.3699 (9) | 0.1831 (11) | 0.0767 (9) | 0.053 (3) |
| H15 | 0.3157 | 0.2385 | 0.0673 | 0.063* |
| C16 | 0.4750 (11) | 0.2192 (11) | 0.0318 (11) | 0.062 (3) |
| H16 | 0.4933 | 0.2953 | −0.0077 | 0.074* |
| C17 | 0.5541 (10) | 0.1302 (17) | 0.0509 (13) | 0.083 (5) |
| H17 | 0.6273 | 0.1480 | 0.0221 | 0.099* |
| C18 | 0.5244 (9) | 0.0173 (12) | 0.1117 (10) | 0.059 (3) |
| H18 | 0.5769 | −0.0398 | 0.1255 | 0.071* |
| C19 | 0.4198 (9) | −0.0059 (10) | 0.1490 (8) | 0.046 (2) |
| H19 | 0.4002 | −0.0816 | 0.1886 | 0.056* |
| C20 | 0.6213 (8) | 0.0837 (8) | 0.3105 (7) | 0.036 (2) |
| C21 | 0.5102 (7) | 0.1110 (7) | 0.3312 (7) | 0.0316 (18) |
| C22 | 0.4337 (8) | 0.0211 (10) | 0.3808 (9) | 0.050 (3) |
| H22 | 0.3612 | 0.0392 | 0.3978 | 0.059* |
| C23 | 0.4591 (9) | −0.0885 (12) | 0.4046 (10) | 0.058 (3) |
| C24 | 0.5723 (9) | −0.1174 (10) | 0.3859 (9) | 0.052 (3) |
| H24 | 0.5927 | −0.1940 | 0.4060 | 0.062* |
| C25 | 0.6505 (9) | −0.0315 (9) | 0.3381 (9) | 0.048 (3) |
| H25 | 0.7235 | −0.0503 | 0.3240 | 0.057* |
| C26 | 0.4711 (8) | 0.2236 (8) | 0.3078 (8) | 0.037 (2) |
| H26 | 0.3968 | 0.2290 | 0.3305 | 0.044* |
| C27 | 0.4550 (9) | 0.4239 (10) | 0.2484 (11) | 0.053 (3) |
| C28 | 0.4387 (17) | 0.4658 (13) | 0.3326 (16) | 0.100 (6) |
| H28 | 0.4726 | 0.4329 | 0.3952 | 0.120* |
| C29 | 0.3717 (18) | 0.5569 (15) | 0.3225 (18) | 0.112 (6) |
| H29 | 0.3622 | 0.5886 | 0.3781 | 0.135* |
| C30 | 0.3179 (18) | 0.6029 (16) | 0.2331 (19) | 0.112 (6) |
| H30 | 0.2704 | 0.6639 | 0.2282 | 0.134* |
| C31 | 0.3351 (14) | 0.5581 (13) | 0.1516 (15) | 0.088 (5) |
| H31 | 0.3002 | 0.5900 | 0.0893 | 0.106* |
| C32 | 0.4021 (9) | 0.4678 (11) | 0.1590 (11) | 0.061 (3) |
| H32 | 0.4116 | 0.4364 | 0.1032 | 0.073* |
| C33 | 0.3727 (10) | −0.1838 (9) | 0.4537 (11) | 0.058 (3) |
| H33A | 0.3303 | −0.1695 | 0.5132 | 0.088* |
| H33B | 0.4088 | −0.2561 | 0.4782 | 0.088* |
| H33C | 0.3242 | −0.1863 | 0.4007 | 0.088* |
| C34 | 0.8717 (9) | 0.2678 (11) | 0.0737 (11) | 0.060 (3) |
| H34 | 0.8171 | 0.2211 | 0.0620 | 0.072* |
| C35 | 0.9775 (10) | 0.2635 (11) | 0.0279 (12) | 0.073 (4) |
| H35 | 0.9937 | 0.2153 | −0.0132 | 0.088* |
| C36 | 1.0565 (11) | 0.3303 (13) | 0.0439 (12) | 0.077 (4) |
| H36 | 1.1284 | 0.3290 | 0.0131 | 0.092* |
| C37 | 1.0331 (10) | 0.4000 (12) | 0.1046 (11) | 0.066 (4) |
| H37 | 1.0877 | 0.4476 | 0.1147 | 0.079* |
| C38 | 0.9214 (9) | 0.3987 (10) | 0.1529 (10) | 0.064 (4) |
| H38 | 0.9040 | 0.4427 | 0.1979 | 0.077* |
Source of materials
The complex was synthesized in quantitate yield from our previously reported method [4]. Block-shaped yellow crystals of the title complex were grown by slow evaporation from their CH2Cl2 solution.
Experimental details
The hydrogen atoms were placed in calculated positions and refined using a riding mode. Some non-fitting reflections were removed manually. The structural model is not affected by this procedure.
Comment
Platinum metal-based complexes (cisplatin, carboplatin, and oxaliplatin) draw immense attention owing to their anticancer potentials. The interest in Pt-based complexes growing worldwide interest due to their ease of synthesis, stability, favorable therapeutic indices, lower toxicity, etc. [4]. The chemistry of platinum complexes has been extensively studied owing to their antitumor/anticancer properties [5], [6], [7], [8], [9], [10], [11], [12], [13] and as a sensor [14, 15].
The asymmetric unit of the title structure contains two Pt-complexes. Each Pt is coordinated by nitrogen and oxygen atom to form a six-member ring and exhibited a square planar environment because the angles of OPtCl and N1PtN2 were ca. 180° respectively. The crystal structures is assembled by collaborative π–π interactions (3.345 Å between pyridine and benzene ring) and CH⃛π bonds (3.110 Å) to form a linear chain along the c-axis.
Funding source: Sichuan University of Science & Engineering
Funding source: Taif University http://dx.doi.org/10.13039/501100006261
Award Identifier / Grant number: TURSP-2020/01
Funding source: Inner Mongolia University
Award Identifier / Grant number: No.10000-21311201/092
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: This research was financially supported by the Scientific Research Foundation for Talents, Sichuan University of Science & Engineering, Taif University Researches Supporting Project number (TURSP-2020/01), Taif University, Taif, Saudi Arabia. Inner Mongolia University funding under the title Academic Backbone (No.10000-21311201/092).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. BRUKER. SAINT, APEX2 and SADABS; Bruker AXS Inc.: Madison, Wisconsin, USA, 2009.Search in Google Scholar
2. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
3. Brandenburg, K. DIAMOND. Visual Crystal Structure Information System (Ver. 4.0); Crystal Impact: Bonn, Germany, 2015.Search in Google Scholar
4. Rahman, F.-U., Ali, A., Guo, R., Wang, W.-K., Wang, H., Li, Z.-T., Lin, Y., Zhang, D.-W. Efficient one-pot synthesis of trans- Pt(ii)(salicylaldimine)(4-picoline)Cl complexes: effective agents for enhanced expression of p53 tumor suppressor genes. Dalton Trans. 2015, 44, 9872–9880; https://doi.org/10.1039/c5dt01098e.Search in Google Scholar PubMed
5. Rahman, F.-U., Ali, A., Guo, R., Zhang, Y.-C., Wang, H., Li, Z.-T., Zhang, D.-W. Synthesis and anticancer activities of a novel class of mono- and di- metallic Pt(ii)(salicylaldiminato)(DMSO or Picolino)Cl complexes. Dalton Trans. 2015, 44, 2166–2175; https://doi.org/10.1039/c4dt03018d.Search in Google Scholar PubMed
6. Rahman, F.-U., Ali, A., Khan, I. U., Duong, H.-Q., Yu, S.-B., Lin, Y.-J., Wang, H., Li, Z.-T., Zhang, D.-W. Morpholine or methylpiperazine and salicylaldimine based heteroleptic square planner platinum (II) complexes: in vitro anticancer study and growth retardation effect on E. coli. Eur. J. Med. Chem. 2017, 131, 263–274; https://doi.org/10.1016/j.ejmech.2017.03.014.Search in Google Scholar PubMed
7. Rahman, F.-U., Ali, A., Duong, H.-Q., Khan, I. U., Bhatti, M. Z., Li, Z.-T., Wang, H., Zhang, D.-W. ONS-donor ligand based Pt (II) complexes display extremely high anticancer potency through autophagic cell death pathway. Eur. J. Med. Chem. 2019, 164, 546–561; https://doi.org/10.1016/j.ejmech.2018.12.052.Search in Google Scholar PubMed
8. Rahman, F.-U., Bhatti, M. Z., Ali, A., Duong, H.-Q., Zhang, Y., Yang, B., Koppireddi, S., Lin, Y., Wang, H., Li, Z.-T. Homo-and heteroleptic Pt (II) complexes of ONN donor hydrazone and 4-picoline: a synthetic, structural and detailed mechanistic anticancer investigation. Eur. J. Med. Chem. 2018, 143, 1039–1052; https://doi.org/10.1016/j.ejmech.2017.11.044.Search in Google Scholar PubMed
9. Wilson, J. J., Lippard, S. J. Synthetic methods for the preparation of platinum anticancer complexes. Chem. Rev. 2014, 114, 4470–4495; https://doi.org/10.1021/cr4004314.Search in Google Scholar PubMed PubMed Central
10. Bai, X., Ali, A., Lv, Z., Wang, N., Zhao, X., Hao, H., Zhang, Y., Rahman, F.-U. Platinum complexes inhibit HER-2 enriched and triple-negative breast cancer cells metabolism to suppress growth, stemness and migration by targeting PKM/LDHA and CCND1/BCL2/ATG3 signaling pathways. Eur. J. Med. Chem. 2021, 224, 113689; https://doi.org/10.1016/j.ejmech.2021.113689.Search in Google Scholar PubMed
11. Rahman, F. U., Ali, A., Guo, R., Wang, W. K., Wang, H., Li, Z. T., Lin, Y., Zhang, D. W. Efficient one-pot synthesis of trans-Pt(II)(salicylaldimine)(4-picoline)Cl complexes: effective agents for enhanced expression of p53 tumor suppressor genes. Dalton Trans. 2015, 44, 9872–9880; https://doi.org/10.1039/c5dt01098e.Search in Google Scholar
12. Rahman, F.-U., Ali, A., Khan, I., Guo, R., Chen, L., Wang, H., Li, Z.-T., Lin, Y., Zhang, D.-W. Synthesis and characterization of trans- Pt(II)(salicylaldimine)(pyridine/pyridine-4-carbinol)Cl complexes: in vivo inhibition of E. coli growth and in vitro anticancer activities. Polyhedron 2015, 100, 264–270; https://doi.org/10.1016/j.poly.2015.08.034.Search in Google Scholar
13. Rahman, F.-U., Wang, H., Zhang, D.-W., Li, Z.-T. 42 members new hydroquinone bridged supramolecular macrocycle and its tetra-nuclear mixed ligands Pt(II) complex: a synthetic, structural and spectroscopic investigation. Inorg. Chem. Commun. 2018, 97, 157–165; https://doi.org/10.1016/j.inoche.2018.09.026.Search in Google Scholar
14. Wong, Y.-S., Ng, M., Yeung, M. C.-L., Yam, V. W.-W. Platinum(II)-based host–guest coordination-driven supramolecular co-assembly assisted by Pt⃛Pt and pi–pi stacking interactions: a dual-selective luminescence sensor for cations and anions. J. Am. Chem. Soc. 2021, 143, 973–982; https://doi.org/10.1021/jacs.0c11162.Search in Google Scholar PubMed
15. Kumar, P., Pachisia, S., Gupta, R. Turn-on detection of assorted phosphates by luminescent chemosensors. Inorg. Chem. Front. 2021, 8, 3587–3607; https://doi.org/10.1039/d1qi00032b.Search in Google Scholar
© 2021 Arshad Khan et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Redetermination of the crystal structure of 3-bromonitrobenzene at 200 K, C6H4BrNO2 – temperature effects on cell constants
- Crystal structure of (E)-ethyl 2-((4-oxo-4H-chromen-3-yl)methyleneaminooxy)acetate, C14H13NO5
- Crystal structure of (8R,10R,14R, Z)-2-((3–Fluoropyridin-4-yl) methylene)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6, 6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-3H-cyclopenta[a] phenanthren-3-one, C36H52FNO3
- Crystal structure of [6,6′-((1E,1′E)-(propane-1,3- diylbis(azaneylylidene))bis(methaneylylidene)) bis(3-chlorophenol)-κ4N,N′,O,O′] copper(II), C17H14Cl2CuN2O2
- The crystal structure of 6-amino-2-carboxypyridin-1-ium bromide, C6H7BrN2O2
- Redetermination of the crystal structure of bis[N,N′-ethylenebis(acetylacetoniminato)nickel(II)] sodium perchlorate, C24H36ClN4NaNi2O8
- The crystal structure of 3-methyl-2,6-dinitrophenol, C7H6N2O5
- The crystal structure of 5-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione, C17H9ClN2O2
- Crystal structure of trans-tetraaqua-bis{2-carboxy-4-((5-carboxypyridin-3-yl)oxy)benzoato-κ1 N}cobalt(II) dihydrate C28H28O20N2Co
- Crystal structure of 3-allyl-4-(2-bromoethyl)-5-(4-methoxyphenyl)-2-(p-tolyl)furan, C23H23BrO2
- The crystal structure of 6,6′-(((2-(dimethylamino)ethyl)azanediyl)bis(methylene))bis(benzo[d][1,3]dioxol-5-ol ato-κ4N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)-titanium(IV)-dichloromethane(1/1), C27H25N3O10Ti
- Crystal structure of (((1E,1′E)-1,2-phenylenebis(methaneylylidene))bis(hydrazin-1-yl-2-ylidene))bis(aminomethaniminium) dinitrate C10H16N10O6
- Crystal structure of catena-poly[triaqua-(μ 2-1,3-di(1H-imidazol-1-yl)propane-κ 2 N:N′)-(4,4′-(1H-1,2,4-triazole-3,5-diyl)dibenzoato-κ 1 O)nickel(II)]N,N′-dimethylformamide (1/1), C28H35N8O8Ni
- The crystal structure of 3,3′-[1,4-phenylenebis(methylene)]bis(1-ethenyl-1H-imidazol-3-ium) dichloride – dichloromethane – water (1/1/1), C19H24Cl4N4O1
- Crystal structure of 1,1′-(methane-1,1-diyl)bis(3-propyl-1H-imidazol-3-ium) bis(hexafluoridophosphate), C13H22F12N4P2
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)tin(IV), C12H8Cl4Sn
- Synthesis and crystal structure of 4-acetylpyrene, C18H12O
- Crystal structure of 2,2′-(butane-1,4-diylbis(azanylylidene))bis(methanylylidene))bis(4-methoxyphenol), C20H24N2O4
- The crystal structure of (E)-2-(((5-((triphenylstannyl)thio)-1,3,4-thiadiazol-2-yl)imino)methyl)phenol, C27H21N3OS2Sn
- Crystal structure of diaqua-bis(μ2-6-phenylpyridine-2-carboxylate-κ3N,O:O)-bis(6-phenylpyridine-2-carboxylato-κ2N,O)lead(II) – N,N-dimethylformamide – water (1/2/4), C54H58N6O16Pb2
- Crystal structure of methyl 4-acetoxy-3-methoxybenzoate, C11H12O5
- Crystal structure of 2,2′-(propane-1,3-dilylbis(azaneylylidene))bis(methanylylidene)bis(4-methylphenol), C19H22N2O2
- Crystal structure of dichlorido-bis(4-methylphenyl-κC1)tin(IV), C14H14Cl2Sn
- Crystal structure of methyl (E)-3-(4-acetoxyphenyl)acrylate, C12H12O4
- The crystal structure of bis(benzoato-κ2 O,O′)-(2,9-dimethyl-1,10-phenanthroline-κ2 N,N′)-copper(II), C28H22CuN2O4
- Crystal structure of (8R,10R,14R,Z)-12-hydroxy-2-((6-methoxypyridin-2-yl)methylene)-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl)hexadecahydro-3H-cyclopenta[a]phenanthren-3-one–water (2/1), C37H56NO4.5
- Crystal structure of dimethyl-bis(4-bromophenyl-κC1)tin(IV), C14H14Br2Sn
- The crystal structure of the cocrystal di-μ2-chlorido-octamethyl-di-μ3-oxido-bis(2,3,4,5-tetrafluorobenzoato-κ2 O,O′)tetratin(IV) ─ octamethyl-di-μ3-oxido-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O′)-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O;O′)tetratin(IV) C58H54Cl2F24O16Sn8
- Crystal structure of 3-iodo-N 2-(2-methyl-1-(methylsulfonyl)propan-2-yl)-N 1-(2-methyl-4-(perfluoropropan-2-yl)phenyl)phthalamide, C23H22F7I1N2O4S1
- Crystal structure of 1-(2-(4-bromophenyl)-2,3-dihydro-1H-benzo[e]indol-1-yl)-naphthalen-2-ol – dichloromethane – dimethyl sulfoxide (1/1/1), C28H18BrNO·CH2Cl2·C2H6SO
- Crystal structure of [meso-5,7,7,12,14,14,-hexamethyl-1,4,8,11-tetraazacyclotetradecane]nickel(II) diperchlorate – dimethylsulphoxide (1/2), C20H48Cl2N4NiO10S2
- Crystal structure of 1,1′-(1,3-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S:S) palladium(II), C26H18N6PdS4
- The crystal structure of bis(6-phenylpyridine-2-carboxylato-κ2 N,O)copper(II), C24H16N2O4Cu
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC)-bis(triphenylarsine oxide-κO)tin(IV), C48H38As2Cl4O2Sn
- Crystal structure of (4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane-κ 8 N 2, O 6) potassium cyclopentadienide, [K([2.2.2]crypt)]Cp, C23H41KN2O6
- The crystal structure of bis(2-oxidopyridin-1-ium-3-carboxylato-κ2O,O′)-(phenantroline-κ2N,N′)manganese(II) - methanol (1/3), C27H28N4O9Mn
- Crystal structure of 4-(dimethylamino)pyridinium dibromido-tris(4-chlorophenyl-κC)stannate(IV), C25H23Br2Cl3N2Sn
- Crystal structure of (3E,5E)-1-(4-cyanobenzenesulfonyl)-3,5-bis(3-fluorobenzylidene)piperidin-4-one-dichloromethane (1/1), C27H20Cl2F2N2O3S
- Crystal structure of (3E,5E)-3,5-bis(4-fluorobenzylidene)-1-((4-trifluoromethyl)benzenesulfonyl)piperidin-4-one, C26H18F5NO3S
- Crystal structure of chlorido-(4-methyl-2-((phenylimino)methyl)phenolato-κ2 N,O)-(pyridine-κ1 N)platinum(II), C19H17ClN2OPt
- Crystal structure of (4-methylbenzyl)(triphenyl)phosphonium chloride dihydrate, C26H28ClO2P
- The crystal structure of poly[μ2-chlorido-(μ2-1,2-bis(4-pyridyl)ethane-κ2N:N′silver(I)], C12H12AgClN2
- Crystal structure of poly[(μ4-benzene-1,2,4,5-tetracarboxylato)-bis(μ2-adipohydrazide)dicadmium], C11H15N4O6Cd
- The crystal structure of (E)-N′-(butan-2-ylidene)isonicotinohydrazide 0.5 hydrate C10H13N3O·0.5H2O
- The crystal structure of bis(6-phenylpyridine-2-carboxylate-κ2 N,O)-(2,2′-bipyridine-κ2 N,N′)zinc(II) monohydrate, C34H26N4O5Zn
- The crystal structure of (1R *,2S *)-1,2-bis(2-fluorophenyl)-3,8-dimethoxyacenaphthene-1,2-diol, C26H20F2O4
- Crystal structure of catena-poly[(μ2-1-((2-ethyl-4-methyl-1H-imidazol-1-yl)methyl)-1H-benzotriazole-κ2N:N′)-(nitrato-κ2O,O′)silver (I)], C13H15Ag1N6O3
- The crystal structure of [(phenantroline-κ2 N,N′)-bis(6-phenylpyridine-2-carboxylate-κ2 N,O)cobalt(II)]monohydrate, C36H26N4O5Co
- Crystal structure of (1E)-N′-[(1E)-1-(4-chlorophenyl)ethylidene]-2-[1-(4-chlorophenyl)ethylidene]hydrazine-1-carbohydrazonamide, C 17 H 17 Cl 2 N 5
- The crystal structure of (E)-2-((tert-butylimino)methyl)-4-chlorophenol, C11H14ClNO
- Crystal structure of all-cis-2,4,6-trihydroxycyclohexane- 1,3,5-triaminium chloride sulfate, C6H18ClN3O7S
- Crystal structure of dichlorido-bis(dimethyl sulfoxide-κO)bis(4-methylphenyl-κC 1)tin(IV), C18H26Cl2O2S2Sn
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)(2,2′-bipyridyl-κ 2 N,N′)tin(IV), C22H16Cl4N2Sn
- Redetermination of the crystal structure of (E)-5-bromo-2-hydroxybenzaldehyde oxime, C 7 H 6 BrNO 2
- The crystal structure of (E)-amino(2-(4-methylbenzylidene)hydrazineyl)methaniminium 4-methylbenzoate, C9H13N4 + C8H7O2 −
- Crystal structure of 2-chloro-3-(isopentylamino)naphthalene-1,4-dione, C 15 H 16 ClNO 2
- The crystal structure of bis(2-acetyl-5-methoxyphenyl)carbonate 1.5 hydrate, C19H18O7
- The crystal structure of poly[(μ 4-4,4′-(azanediylbis(methylene))dibenzoato-κ 4 O:N:O′:Oʺ)zinc(II)], C16H13NO4Zn
- The crystal structure of catena-poly[(1,10-phenanthroline-k2N,N′)-(μ3-tetraoxidomoybdato(VI)-k3O:O′:O″)manganese(II)] C12H8N2O4MoMn
- Crystal structure of catena-poly[(4-hydroxyl-5-(methoylcarbonyl)thiophene-2-carboxylato-κ1 O)-(μ2-piperazine-1,4-diylbis(pyridin-4-ylmethanone)-κ2 N:N′)silver(I)] monohydrate, C23H23AgN4O8S
- Crystal structure of bis(4-bromo-2-(((3-bromopropyl)imino)methyl)phenolato-κ2N,O)-oxido-vanadium(IV), C20H20Br4N2O3V
- The crystal structure of (2a′S,2a1′S,3R,5a′S,7′R)-5-(furan-3-yl)-2a′,2a1′-dihydroxy-7′-methyldecahydro-2H-spiro[furan-3,6′-naphtho[1,8-bc]furan]-2,2′(2a′H)-dione, C19H22O7
- The crystal structure of 3-bromopicolinic acid, C6H4BrNO2
- Crystal structure of 1,1′-(1,4-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S,S) platinum(II), C26H18N6PtS4
- Synthesis and crystal structure of 5-(8-((3-carboxyazetidin-1-ium-1-yl)methyl)-7-hydroxy-4-oxo-4H-chromen-3-yl)-2-hydroxybenzenesulfonate monohydrate, C20H19NO10S
- The crystal structure of 3-amino-5-carboxypyridin-1-ium bromide, C6H7BrN2O2
- The crystal structure of (2-hydroxy-5-methyl-phenyl)-(1H-pyrazol-4-yl)-methanone hemihydrate, C11H10.5N2O2.5
- Crystal structure of tetraaqua-(2-(4-formylphenoxy)acetato-k1O)cadmium(II), C18H22O12Cd
- Crystal structure of diethyl 6,12-dimethyl-3,9-di-p-tolyl-3,9-diazapentacyclo[6.4.0.02,7.04,11.05,10]dodecane-1,5-dicarboxylate, C32H38N2O4
- Crystal structure of (E)-N′-(1-(3-chloro-4-fluorophenyl)ethylidene)-4-hydroxy – tetrahydrofuran (2/1), C17H16ClFN2O2.5
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Redetermination of the crystal structure of 3-bromonitrobenzene at 200 K, C6H4BrNO2 – temperature effects on cell constants
- Crystal structure of (E)-ethyl 2-((4-oxo-4H-chromen-3-yl)methyleneaminooxy)acetate, C14H13NO5
- Crystal structure of (8R,10R,14R, Z)-2-((3–Fluoropyridin-4-yl) methylene)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6, 6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-3H-cyclopenta[a] phenanthren-3-one, C36H52FNO3
- Crystal structure of [6,6′-((1E,1′E)-(propane-1,3- diylbis(azaneylylidene))bis(methaneylylidene)) bis(3-chlorophenol)-κ4N,N′,O,O′] copper(II), C17H14Cl2CuN2O2
- The crystal structure of 6-amino-2-carboxypyridin-1-ium bromide, C6H7BrN2O2
- Redetermination of the crystal structure of bis[N,N′-ethylenebis(acetylacetoniminato)nickel(II)] sodium perchlorate, C24H36ClN4NaNi2O8
- The crystal structure of 3-methyl-2,6-dinitrophenol, C7H6N2O5
- The crystal structure of 5-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione, C17H9ClN2O2
- Crystal structure of trans-tetraaqua-bis{2-carboxy-4-((5-carboxypyridin-3-yl)oxy)benzoato-κ1 N}cobalt(II) dihydrate C28H28O20N2Co
- Crystal structure of 3-allyl-4-(2-bromoethyl)-5-(4-methoxyphenyl)-2-(p-tolyl)furan, C23H23BrO2
- The crystal structure of 6,6′-(((2-(dimethylamino)ethyl)azanediyl)bis(methylene))bis(benzo[d][1,3]dioxol-5-ol ato-κ4N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)-titanium(IV)-dichloromethane(1/1), C27H25N3O10Ti
- Crystal structure of (((1E,1′E)-1,2-phenylenebis(methaneylylidene))bis(hydrazin-1-yl-2-ylidene))bis(aminomethaniminium) dinitrate C10H16N10O6
- Crystal structure of catena-poly[triaqua-(μ 2-1,3-di(1H-imidazol-1-yl)propane-κ 2 N:N′)-(4,4′-(1H-1,2,4-triazole-3,5-diyl)dibenzoato-κ 1 O)nickel(II)]N,N′-dimethylformamide (1/1), C28H35N8O8Ni
- The crystal structure of 3,3′-[1,4-phenylenebis(methylene)]bis(1-ethenyl-1H-imidazol-3-ium) dichloride – dichloromethane – water (1/1/1), C19H24Cl4N4O1
- Crystal structure of 1,1′-(methane-1,1-diyl)bis(3-propyl-1H-imidazol-3-ium) bis(hexafluoridophosphate), C13H22F12N4P2
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)tin(IV), C12H8Cl4Sn
- Synthesis and crystal structure of 4-acetylpyrene, C18H12O
- Crystal structure of 2,2′-(butane-1,4-diylbis(azanylylidene))bis(methanylylidene))bis(4-methoxyphenol), C20H24N2O4
- The crystal structure of (E)-2-(((5-((triphenylstannyl)thio)-1,3,4-thiadiazol-2-yl)imino)methyl)phenol, C27H21N3OS2Sn
- Crystal structure of diaqua-bis(μ2-6-phenylpyridine-2-carboxylate-κ3N,O:O)-bis(6-phenylpyridine-2-carboxylato-κ2N,O)lead(II) – N,N-dimethylformamide – water (1/2/4), C54H58N6O16Pb2
- Crystal structure of methyl 4-acetoxy-3-methoxybenzoate, C11H12O5
- Crystal structure of 2,2′-(propane-1,3-dilylbis(azaneylylidene))bis(methanylylidene)bis(4-methylphenol), C19H22N2O2
- Crystal structure of dichlorido-bis(4-methylphenyl-κC1)tin(IV), C14H14Cl2Sn
- Crystal structure of methyl (E)-3-(4-acetoxyphenyl)acrylate, C12H12O4
- The crystal structure of bis(benzoato-κ2 O,O′)-(2,9-dimethyl-1,10-phenanthroline-κ2 N,N′)-copper(II), C28H22CuN2O4
- Crystal structure of (8R,10R,14R,Z)-12-hydroxy-2-((6-methoxypyridin-2-yl)methylene)-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl)hexadecahydro-3H-cyclopenta[a]phenanthren-3-one–water (2/1), C37H56NO4.5
- Crystal structure of dimethyl-bis(4-bromophenyl-κC1)tin(IV), C14H14Br2Sn
- The crystal structure of the cocrystal di-μ2-chlorido-octamethyl-di-μ3-oxido-bis(2,3,4,5-tetrafluorobenzoato-κ2 O,O′)tetratin(IV) ─ octamethyl-di-μ3-oxido-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O′)-bis(μ2-2,3,4,5-tetrafluorobenzoato-κ2 O:O;O′)tetratin(IV) C58H54Cl2F24O16Sn8
- Crystal structure of 3-iodo-N 2-(2-methyl-1-(methylsulfonyl)propan-2-yl)-N 1-(2-methyl-4-(perfluoropropan-2-yl)phenyl)phthalamide, C23H22F7I1N2O4S1
- Crystal structure of 1-(2-(4-bromophenyl)-2,3-dihydro-1H-benzo[e]indol-1-yl)-naphthalen-2-ol – dichloromethane – dimethyl sulfoxide (1/1/1), C28H18BrNO·CH2Cl2·C2H6SO
- Crystal structure of [meso-5,7,7,12,14,14,-hexamethyl-1,4,8,11-tetraazacyclotetradecane]nickel(II) diperchlorate – dimethylsulphoxide (1/2), C20H48Cl2N4NiO10S2
- Crystal structure of 1,1′-(1,3-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S:S) palladium(II), C26H18N6PdS4
- The crystal structure of bis(6-phenylpyridine-2-carboxylato-κ2 N,O)copper(II), C24H16N2O4Cu
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC)-bis(triphenylarsine oxide-κO)tin(IV), C48H38As2Cl4O2Sn
- Crystal structure of (4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane-κ 8 N 2, O 6) potassium cyclopentadienide, [K([2.2.2]crypt)]Cp, C23H41KN2O6
- The crystal structure of bis(2-oxidopyridin-1-ium-3-carboxylato-κ2O,O′)-(phenantroline-κ2N,N′)manganese(II) - methanol (1/3), C27H28N4O9Mn
- Crystal structure of 4-(dimethylamino)pyridinium dibromido-tris(4-chlorophenyl-κC)stannate(IV), C25H23Br2Cl3N2Sn
- Crystal structure of (3E,5E)-1-(4-cyanobenzenesulfonyl)-3,5-bis(3-fluorobenzylidene)piperidin-4-one-dichloromethane (1/1), C27H20Cl2F2N2O3S
- Crystal structure of (3E,5E)-3,5-bis(4-fluorobenzylidene)-1-((4-trifluoromethyl)benzenesulfonyl)piperidin-4-one, C26H18F5NO3S
- Crystal structure of chlorido-(4-methyl-2-((phenylimino)methyl)phenolato-κ2 N,O)-(pyridine-κ1 N)platinum(II), C19H17ClN2OPt
- Crystal structure of (4-methylbenzyl)(triphenyl)phosphonium chloride dihydrate, C26H28ClO2P
- The crystal structure of poly[μ2-chlorido-(μ2-1,2-bis(4-pyridyl)ethane-κ2N:N′silver(I)], C12H12AgClN2
- Crystal structure of poly[(μ4-benzene-1,2,4,5-tetracarboxylato)-bis(μ2-adipohydrazide)dicadmium], C11H15N4O6Cd
- The crystal structure of (E)-N′-(butan-2-ylidene)isonicotinohydrazide 0.5 hydrate C10H13N3O·0.5H2O
- The crystal structure of bis(6-phenylpyridine-2-carboxylate-κ2 N,O)-(2,2′-bipyridine-κ2 N,N′)zinc(II) monohydrate, C34H26N4O5Zn
- The crystal structure of (1R *,2S *)-1,2-bis(2-fluorophenyl)-3,8-dimethoxyacenaphthene-1,2-diol, C26H20F2O4
- Crystal structure of catena-poly[(μ2-1-((2-ethyl-4-methyl-1H-imidazol-1-yl)methyl)-1H-benzotriazole-κ2N:N′)-(nitrato-κ2O,O′)silver (I)], C13H15Ag1N6O3
- The crystal structure of [(phenantroline-κ2 N,N′)-bis(6-phenylpyridine-2-carboxylate-κ2 N,O)cobalt(II)]monohydrate, C36H26N4O5Co
- Crystal structure of (1E)-N′-[(1E)-1-(4-chlorophenyl)ethylidene]-2-[1-(4-chlorophenyl)ethylidene]hydrazine-1-carbohydrazonamide, C 17 H 17 Cl 2 N 5
- The crystal structure of (E)-2-((tert-butylimino)methyl)-4-chlorophenol, C11H14ClNO
- Crystal structure of all-cis-2,4,6-trihydroxycyclohexane- 1,3,5-triaminium chloride sulfate, C6H18ClN3O7S
- Crystal structure of dichlorido-bis(dimethyl sulfoxide-κO)bis(4-methylphenyl-κC 1)tin(IV), C18H26Cl2O2S2Sn
- Crystal structure of dichlorido-bis(4-chlorophenyl-κC 1)(2,2′-bipyridyl-κ 2 N,N′)tin(IV), C22H16Cl4N2Sn
- Redetermination of the crystal structure of (E)-5-bromo-2-hydroxybenzaldehyde oxime, C 7 H 6 BrNO 2
- The crystal structure of (E)-amino(2-(4-methylbenzylidene)hydrazineyl)methaniminium 4-methylbenzoate, C9H13N4 + C8H7O2 −
- Crystal structure of 2-chloro-3-(isopentylamino)naphthalene-1,4-dione, C 15 H 16 ClNO 2
- The crystal structure of bis(2-acetyl-5-methoxyphenyl)carbonate 1.5 hydrate, C19H18O7
- The crystal structure of poly[(μ 4-4,4′-(azanediylbis(methylene))dibenzoato-κ 4 O:N:O′:Oʺ)zinc(II)], C16H13NO4Zn
- The crystal structure of catena-poly[(1,10-phenanthroline-k2N,N′)-(μ3-tetraoxidomoybdato(VI)-k3O:O′:O″)manganese(II)] C12H8N2O4MoMn
- Crystal structure of catena-poly[(4-hydroxyl-5-(methoylcarbonyl)thiophene-2-carboxylato-κ1 O)-(μ2-piperazine-1,4-diylbis(pyridin-4-ylmethanone)-κ2 N:N′)silver(I)] monohydrate, C23H23AgN4O8S
- Crystal structure of bis(4-bromo-2-(((3-bromopropyl)imino)methyl)phenolato-κ2N,O)-oxido-vanadium(IV), C20H20Br4N2O3V
- The crystal structure of (2a′S,2a1′S,3R,5a′S,7′R)-5-(furan-3-yl)-2a′,2a1′-dihydroxy-7′-methyldecahydro-2H-spiro[furan-3,6′-naphtho[1,8-bc]furan]-2,2′(2a′H)-dione, C19H22O7
- The crystal structure of 3-bromopicolinic acid, C6H4BrNO2
- Crystal structure of 1,1′-(1,4-phenylenebis(methylene))bis(pyridin-1-ium) bis(1,2-dicyanoethene-1,2-dithiolato-κ2 S,S) platinum(II), C26H18N6PtS4
- Synthesis and crystal structure of 5-(8-((3-carboxyazetidin-1-ium-1-yl)methyl)-7-hydroxy-4-oxo-4H-chromen-3-yl)-2-hydroxybenzenesulfonate monohydrate, C20H19NO10S
- The crystal structure of 3-amino-5-carboxypyridin-1-ium bromide, C6H7BrN2O2
- The crystal structure of (2-hydroxy-5-methyl-phenyl)-(1H-pyrazol-4-yl)-methanone hemihydrate, C11H10.5N2O2.5
- Crystal structure of tetraaqua-(2-(4-formylphenoxy)acetato-k1O)cadmium(II), C18H22O12Cd
- Crystal structure of diethyl 6,12-dimethyl-3,9-di-p-tolyl-3,9-diazapentacyclo[6.4.0.02,7.04,11.05,10]dodecane-1,5-dicarboxylate, C32H38N2O4
- Crystal structure of (E)-N′-(1-(3-chloro-4-fluorophenyl)ethylidene)-4-hydroxy – tetrahydrofuran (2/1), C17H16ClFN2O2.5

