Abstract
C30H46O3·1/6H2O, monoclinic, C2 (no. 5), a = 29.4307(4) Å, b = 15.42797(17) Å, c = 18.4667(2) Å, β = 104.4652(12)°, V = 8119.11(17) Å3, Z = 12, Rgt(F) = 0.0432, wRref(F2) = 0.1197, T = 293(2) K.
CCDC no.: 2016692
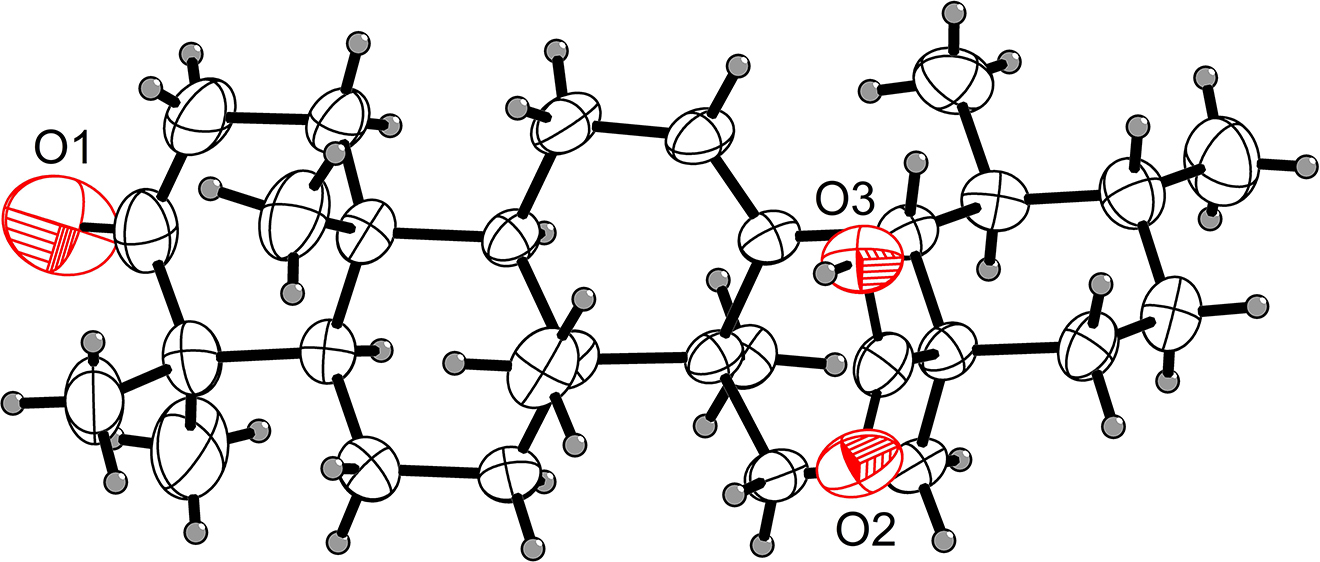
One of three independent molecules of the title crystal structure is shown in the Figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colorless block |
| Size: | 0.29 × 0.24 × 0.20 mm |
| Wavelength: | Cu Kα radiation (1.54184 Å) |
| μ: | 0.54 mm−1 |
| Diffractometer, scan mode: | Xcalibur, ω |
| θmax, completeness: | 66.8°, 99% |
| N(hkl)measured, N(hkl)unique, Rint: | 34,354, 12,647, 0.029 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 11372 |
| N(param)refined: | 917 |
| Programs: | CrysAlisPRO [1], Olex2 [2], SUPERFLIP [3], [4], [5], SHELX [6] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| O1 | 0.5867 (2) | 0.0091 (5) | 0.6690 (3) | 0.206 (3) |
| O2 | 0.81273 (9) | 0.5601 (2) | 0.58312 (15) | 0.0939 (8) |
| O3 | 0.82126 (8) | 0.5673 (2) | 0.70414 (13) | 0.0832 (7) |
| H3 | 0.845625 | 0.541511 | 0.704072 | 0.125* |
| C1 | 0.64593 (16) | 0.2046 (3) | 0.7422 (2) | 0.0847 (10) |
| H1A | 0.617945 | 0.240355 | 0.729465 | 0.102* |
| H1B | 0.663778 | 0.220992 | 0.791858 | 0.102* |
| C2 | 0.6315 (2) | 0.1089 (3) | 0.7428 (3) | 0.1146 (17) |
| H2A | 0.658852 | 0.075973 | 0.768576 | 0.137* |
| H2B | 0.608486 | 0.104143 | 0.772169 | 0.137* |
| C3 | 0.61238 (15) | 0.0684 (3) | 0.6719 (3) | 0.0934 (11) |
| C4 | 0.62633 (18) | 0.0975 (3) | 0.6011 (2) | 0.0941 (12) |
| C5 | 0.64576 (12) | 0.1921 (2) | 0.60814 (18) | 0.0678 (8) |
| H5 | 0.617630 | 0.228598 | 0.599046 | 0.081* |
| C6 | 0.66939 (15) | 0.2177 (2) | 0.5464 (2) | 0.0802 (9) |
| H6A | 0.700742 | 0.193304 | 0.557064 | 0.096* |
| H6B | 0.651618 | 0.194471 | 0.498942 | 0.096* |
| C7 | 0.67217 (12) | 0.3153 (2) | 0.54113 (17) | 0.0668 (7) |
| H7A | 0.640574 | 0.338578 | 0.526466 | 0.080* |
| H7B | 0.687709 | 0.329975 | 0.502202 | 0.080* |
| C8 | 0.69863 (9) | 0.3591 (2) | 0.61457 (15) | 0.0566 (6) |
| C9 | 0.68082 (10) | 0.3223 (2) | 0.68112 (15) | 0.0555 (6) |
| H9 | 0.648855 | 0.344946 | 0.673465 | 0.067* |
| C10 | 0.67558 (11) | 0.2219 (2) | 0.68597 (18) | 0.0661 (7) |
| C11 | 0.70835 (14) | 0.3634 (3) | 0.75465 (18) | 0.0828 (11) |
| H11E | 0.689882 | 0.358446 | 0.791221 | 0.099* |
| H11F | 0.737060 | 0.330730 | 0.773263 | 0.099* |
| C12 | 0.72067 (12) | 0.4562 (2) | 0.74834 (16) | 0.0702 (8) |
| H12 | 0.735000 | 0.484809 | 0.792481 | 0.084* |
| C13 | 0.71313 (9) | 0.5019 (2) | 0.68574 (14) | 0.0537 (6) |
| C14 | 0.68919 (9) | 0.46032 (18) | 0.61035 (14) | 0.0508 (6) |
| C15 | 0.70782 (11) | 0.5008 (2) | 0.54577 (15) | 0.0614 (7) |
| H15E | 0.736816 | 0.471730 | 0.544320 | 0.074* |
| H15F | 0.685220 | 0.488969 | 0.498742 | 0.074* |
| C16 | 0.71689 (10) | 0.5977 (2) | 0.55143 (15) | 0.0601 (7) |
| H16E | 0.687273 | 0.628033 | 0.544898 | 0.072* |
| H16F | 0.730921 | 0.615634 | 0.511564 | 0.072* |
| C17 | 0.74956 (9) | 0.6230 (2) | 0.62708 (15) | 0.0561 (6) |
| C18 | 0.72583 (9) | 0.5979 (2) | 0.68990 (15) | 0.0549 (6) |
| H18 | 0.749599 | 0.605955 | 0.737117 | 0.066* |
| C19 | 0.68422 (11) | 0.6589 (2) | 0.69336 (18) | 0.0664 (7) |
| H19 | 0.659969 | 0.652305 | 0.646483 | 0.080* |
| C20 | 0.70001 (13) | 0.7542 (3) | 0.6998 (2) | 0.0784 (9) |
| H20 | 0.724955 | 0.760179 | 0.745886 | 0.094* |
| C21 | 0.72040 (14) | 0.7786 (3) | 0.6351 (2) | 0.0824 (9) |
| H21E | 0.730882 | 0.838343 | 0.640779 | 0.099* |
| H21F | 0.696154 | 0.774052 | 0.588762 | 0.099* |
| C22 | 0.76128 (12) | 0.7208 (2) | 0.6307 (2) | 0.0702 (8) |
| H22E | 0.787027 | 0.731529 | 0.674103 | 0.084* |
| H22F | 0.771793 | 0.736495 | 0.586657 | 0.084* |
| C23 | 0.5831 (3) | 0.0931 (4) | 0.5358 (4) | 0.179 (4) |
| H23G | 0.562858 | 0.141183 | 0.538260 | 0.268* |
| H23H | 0.566652 | 0.039829 | 0.538243 | 0.268* |
| H23I | 0.592488 | 0.095336 | 0.489651 | 0.268* |
| C24 | 0.6624 (3) | 0.0291 (3) | 0.5896 (4) | 0.142 (2) |
| H24G | 0.671938 | 0.042124 | 0.544782 | 0.214* |
| H24H | 0.648328 | −0.027414 | 0.585423 | 0.214* |
| H24I | 0.689323 | 0.030138 | 0.631619 | 0.214* |
| C25 | 0.72265 (17) | 0.1732 (3) | 0.7114 (3) | 0.1086 (15) |
| H25G | 0.737438 | 0.170223 | 0.670655 | 0.163* |
| H25H | 0.717037 | 0.115634 | 0.726841 | 0.163* |
| H25I | 0.742767 | 0.203515 | 0.752559 | 0.163* |
| C26 | 0.75182 (11) | 0.3422 (3) | 0.6267 (2) | 0.0842 (10) |
| H26G | 0.761864 | 0.362608 | 0.584149 | 0.126* |
| H26H | 0.757866 | 0.281143 | 0.632920 | 0.126* |
| H26I | 0.768729 | 0.372342 | 0.670717 | 0.126* |
| C27 | 0.63612 (9) | 0.4818 (2) | 0.59340 (17) | 0.0617 (7) |
| H27G | 0.624045 | 0.464180 | 0.634833 | 0.093* |
| H27H | 0.619764 | 0.451455 | 0.549165 | 0.093* |
| H27I | 0.631684 | 0.543072 | 0.585686 | 0.093* |
| C28 | 0.79677 (10) | 0.5790 (2) | 0.63502 (17) | 0.0612 (7) |
| C29 | 0.6608 (2) | 0.8166 (4) | 0.7045 (4) | 0.1164 (16) |
| H29G | 0.634197 | 0.806733 | 0.663189 | 0.175* |
| H29H | 0.671565 | 0.875160 | 0.702828 | 0.175* |
| H29I | 0.652009 | 0.807346 | 0.750596 | 0.175* |
| C30 | 0.66287 (15) | 0.6325 (3) | 0.7575 (2) | 0.0904 (11) |
| H30G | 0.650918 | 0.574491 | 0.749372 | 0.136* |
| H30H | 0.637762 | 0.671421 | 0.759515 | 0.136* |
| H30I | 0.686531 | 0.635082 | 0.803846 | 0.136* |
| O1B | 0.59041 (9) | 0.9870 (2) | 0.28189 (19) | 0.1010 (9) |
| O2B | 0.54190 (7) | 0.33317 (19) | 0.45594 (10) | 0.0734 (6) |
| O3B | 0.46502 (7) | 0.33191 (19) | 0.40640 (11) | 0.0744 (6) |
| H3B | 0.464788 | 0.331844 | 0.450717 | 0.112* |
| C1B | 0.62096 (10) | 0.7771 (2) | 0.3542 (2) | 0.0767 (10) |
| H1BA | 0.647531 | 0.749705 | 0.388332 | 0.092* |
| H1BB | 0.622085 | 0.762033 | 0.303707 | 0.092* |
| C2B | 0.62565 (11) | 0.8759 (3) | 0.3637 (3) | 0.0932 (12) |
| H2BA | 0.629581 | 0.890400 | 0.416018 | 0.112* |
| H2BB | 0.653561 | 0.894924 | 0.349259 | 0.112* |
| C3B | 0.58429 (11) | 0.9234 (2) | 0.3182 (2) | 0.0773 (9) |
| C4B | 0.53508 (10) | 0.8917 (2) | 0.3183 (2) | 0.0680 (8) |
| C5B | 0.53382 (8) | 0.79018 (17) | 0.31693 (16) | 0.0508 (6) |
| H5B | 0.535805 | 0.775022 | 0.266279 | 0.061* |
| C6B | 0.48726 (9) | 0.7517 (2) | 0.32311 (19) | 0.0615 (7) |
| H6BA | 0.461565 | 0.785991 | 0.293759 | 0.074* |
| H6BB | 0.485475 | 0.753141 | 0.374840 | 0.074* |
| C7B | 0.48276 (8) | 0.65834 (19) | 0.29499 (17) | 0.0556 (6) |
| H7BA | 0.481680 | 0.658359 | 0.242059 | 0.067* |
| H7BB | 0.453267 | 0.634803 | 0.300602 | 0.067* |
| C8B | 0.52298 (8) | 0.59835 (17) | 0.33584 (13) | 0.0468 (5) |
| C9B | 0.57078 (8) | 0.64406 (18) | 0.34279 (14) | 0.0476 (5) |
| H9B | 0.574751 | 0.645445 | 0.291673 | 0.057* |
| C10B | 0.57543 (9) | 0.74087 (19) | 0.36895 (15) | 0.0552 (6) |
| C11B | 0.61063 (10) | 0.5858 (2) | 0.3860 (2) | 0.0698 (8) |
| H11C | 0.639428 | 0.602085 | 0.372967 | 0.084* |
| H11D | 0.615063 | 0.596225 | 0.439121 | 0.084* |
| C12B | 0.60226 (9) | 0.4917 (2) | 0.37120 (16) | 0.0593 (7) |
| H12B | 0.626584 | 0.454661 | 0.393829 | 0.071* |
| C13B | 0.56415 (9) | 0.45500 (18) | 0.32950 (13) | 0.0489 (6) |
| C14B | 0.52257 (8) | 0.51176 (16) | 0.28929 (12) | 0.0451 (5) |
| C15B | 0.47503 (9) | 0.46460 (18) | 0.28050 (15) | 0.0537 (6) |
| H15C | 0.464744 | 0.472816 | 0.325997 | 0.064* |
| H15D | 0.451987 | 0.491810 | 0.240058 | 0.064* |
| C16B | 0.47576 (11) | 0.3679 (2) | 0.26465 (15) | 0.0591 (7) |
| H16C | 0.480601 | 0.359238 | 0.215088 | 0.071* |
| H16D | 0.445574 | 0.343028 | 0.265228 | 0.071* |
| C17B | 0.51444 (11) | 0.32051 (19) | 0.32204 (14) | 0.0556 (6) |
| C18B | 0.56276 (10) | 0.35620 (18) | 0.31921 (14) | 0.0544 (6) |
| H18B | 0.585265 | 0.332419 | 0.363030 | 0.065* |
| C19B | 0.57919 (13) | 0.3246 (2) | 0.24972 (17) | 0.0692 (8) |
| H19B | 0.558515 | 0.350779 | 0.205165 | 0.083* |
| C20B | 0.57500 (19) | 0.2251 (2) | 0.2412 (2) | 0.0909 (13) |
| H20B | 0.596529 | 0.199173 | 0.285020 | 0.109* |
| C21B | 0.5265 (2) | 0.1947 (2) | 0.2400 (2) | 0.0948 (13) |
| H21C | 0.525310 | 0.132054 | 0.235467 | 0.114* |
| H21D | 0.504577 | 0.218718 | 0.196451 | 0.114* |
| C22B | 0.51171 (16) | 0.2212 (2) | 0.30996 (18) | 0.0760 (9) |
| H22C | 0.531876 | 0.192839 | 0.353045 | 0.091* |
| H22D | 0.479796 | 0.201921 | 0.305877 | 0.091* |
| C23B | 0.50085 (12) | 0.9262 (2) | 0.2475 (2) | 0.0823 (10) |
| H23D | 0.506217 | 0.896874 | 0.204512 | 0.123* |
| H23E | 0.505710 | 0.987295 | 0.243032 | 0.123* |
| H23F | 0.469205 | 0.915993 | 0.250516 | 0.123* |
| C24B | 0.52183 (15) | 0.9326 (3) | 0.3865 (3) | 0.0970 (13) |
| H24D | 0.491303 | 0.912688 | 0.388685 | 0.145* |
| H24E | 0.521457 | 0.994612 | 0.381799 | 0.145* |
| H24F | 0.544481 | 0.916040 | 0.431404 | 0.145* |
| C25B | 0.57787 (15) | 0.7520 (3) | 0.45288 (19) | 0.0862 (10) |
| H25D | 0.595310 | 0.803457 | 0.471237 | 0.129* |
| H25E | 0.593148 | 0.702575 | 0.479977 | 0.129* |
| H25F | 0.546674 | 0.756898 | 0.459642 | 0.129* |
| C26B | 0.51576 (12) | 0.5764 (2) | 0.41406 (16) | 0.0665 (8) |
| H26D | 0.488777 | 0.539683 | 0.408583 | 0.100* |
| H26E | 0.511025 | 0.628951 | 0.439025 | 0.100* |
| H26F | 0.543010 | 0.546940 | 0.443041 | 0.100* |
| C27B | 0.52876 (10) | 0.52852 (19) | 0.20915 (13) | 0.0548 (6) |
| H27D | 0.560363 | 0.546872 | 0.212276 | 0.082* |
| H27E | 0.507323 | 0.572923 | 0.185388 | 0.082* |
| H27F | 0.522416 | 0.476104 | 0.180376 | 0.082* |
| C28B | 0.50774 (10) | 0.33101 (19) | 0.40069 (14) | 0.0549 (6) |
| C29B | 0.5894 (3) | 0.1922 (3) | 0.1712 (3) | 0.131 (2) |
| H29D | 0.569430 | 0.218049 | 0.127367 | 0.197* |
| H29E | 0.586204 | 0.130332 | 0.168194 | 0.197* |
| H29F | 0.621390 | 0.207911 | 0.174615 | 0.197* |
| C30B | 0.62901 (15) | 0.3562 (3) | 0.2549 (2) | 0.0923 (12) |
| H30D | 0.629253 | 0.418355 | 0.252853 | 0.139* |
| H30E | 0.639808 | 0.333010 | 0.213956 | 0.139* |
| H30F | 0.649341 | 0.337082 | 0.301290 | 0.139* |
| O1C | 0.6814 (2) | 1.0565 (4) | 0.0633 (3) | 0.190 (3) |
| O2C | 0.53260 (7) | 0.39302 (18) | −0.06581 (10) | 0.0709 (6) |
| H2C | 0.507221 | 0.385396 | −0.055970 | 0.106* |
| O3C | 0.55522 (7) | 0.40157 (18) | 0.05711 (11) | 0.0691 (6) |
| C1C | 0.65497 (12) | 0.8612 (2) | 0.13312 (17) | 0.0681 (8) |
| H1CA | 0.651996 | 0.837028 | 0.180221 | 0.082* |
| H1CB | 0.685728 | 0.845543 | 0.127055 | 0.082* |
| C2C | 0.65174 (16) | 0.9599 (3) | 0.1368 (2) | 0.0863 (10) |
| H2CA | 0.622572 | 0.975939 | 0.148669 | 0.104* |
| H2CB | 0.677438 | 0.981642 | 0.176271 | 0.104* |
| C3C | 0.6537 (2) | 1.0003 (3) | 0.0645 (3) | 0.1052 (14) |
| C4C | 0.62014 (19) | 0.9665 (3) | −0.0066 (2) | 0.0942 (12) |
| C5C | 0.62088 (12) | 0.8650 (2) | −0.00414 (16) | 0.0638 (7) |
| H5C | 0.652207 | 0.849611 | −0.009347 | 0.077* |
| C6C | 0.58735 (13) | 0.8216 (2) | −0.07124 (17) | 0.0741 (9) |
| H6CA | 0.555715 | 0.822060 | −0.064434 | 0.089* |
| H6CB | 0.587265 | 0.853588 | −0.116509 | 0.089* |
| C7C | 0.60290 (12) | 0.7286 (2) | −0.07864 (15) | 0.0631 (7) |
| H7CA | 0.633553 | 0.729433 | −0.089271 | 0.076* |
| H7CB | 0.581147 | 0.701801 | −0.120931 | 0.076* |
| C8C | 0.60562 (8) | 0.67255 (18) | −0.00883 (13) | 0.0481 (5) |
| C9C | 0.63137 (8) | 0.72287 (18) | 0.06254 (13) | 0.0481 (5) |
| H9C | 0.664287 | 0.724597 | 0.060335 | 0.058* |
| C10C | 0.61752 (9) | 0.8205 (2) | 0.06941 (15) | 0.0554 (6) |
| C11C | 0.63144 (12) | 0.6691 (2) | 0.13221 (15) | 0.0636 (7) |
| H11A | 0.656685 | 0.689191 | 0.173228 | 0.076* |
| H11B | 0.602127 | 0.678748 | 0.145998 | 0.076* |
| C12C | 0.63733 (9) | 0.5737 (2) | 0.12191 (14) | 0.0548 (6) |
| H12C | 0.640257 | 0.538955 | 0.163990 | 0.066* |
| C13C | 0.63878 (8) | 0.53371 (18) | 0.05877 (13) | 0.0466 (5) |
| C14C | 0.63410 (8) | 0.58604 (18) | −0.01343 (13) | 0.0466 (5) |
| C15C | 0.60845 (10) | 0.53319 (19) | −0.08299 (14) | 0.0528 (6) |
| H15A | 0.574908 | 0.538001 | −0.088342 | 0.063* |
| H15B | 0.615247 | 0.559169 | −0.126956 | 0.063* |
| C16C | 0.62120 (10) | 0.43798 (19) | −0.08125 (15) | 0.0547 (6) |
| H16A | 0.653508 | 0.432250 | −0.084184 | 0.066* |
| H16B | 0.601417 | 0.409439 | −0.124604 | 0.066* |
| C17C | 0.61534 (9) | 0.39313 (18) | −0.01042 (14) | 0.0528 (6) |
| C18C | 0.64763 (9) | 0.43686 (19) | 0.05929 (15) | 0.0523 (6) |
| H18C | 0.638108 | 0.413759 | 0.102702 | 0.063* |
| C19C | 0.70031 (10) | 0.4120 (2) | 0.07052 (18) | 0.0688 (8) |
| H19C | 0.711065 | 0.436300 | 0.028665 | 0.083* |
| C20C | 0.70609 (13) | 0.3134 (3) | 0.0691 (2) | 0.0835 (11) |
| H20C | 0.695616 | 0.289301 | 0.111276 | 0.100* |
| C21C | 0.67651 (14) | 0.2738 (3) | −0.0016 (2) | 0.0857 (11) |
| H21A | 0.680729 | 0.211367 | 0.000406 | 0.103* |
| H21B | 0.686904 | 0.295380 | −0.044055 | 0.103* |
| C22C | 0.62449 (13) | 0.2948 (2) | −0.0123 (2) | 0.0704 (8) |
| H22A | 0.613190 | 0.266771 | 0.026899 | 0.084* |
| H22B | 0.606956 | 0.271531 | −0.059855 | 0.084* |
| C23C | 0.6372 (3) | 0.9963 (4) | −0.0741 (3) | 0.146 (3) |
| H23A | 0.664807 | 0.964324 | −0.076263 | 0.220* |
| H23B | 0.644382 | 1.057004 | −0.069634 | 0.220* |
| H23C | 0.613032 | 0.986147 | −0.119016 | 0.220* |
| C24C | 0.5720 (2) | 1.0073 (4) | −0.0115 (4) | 0.138 (2) |
| H24A | 0.549252 | 0.982564 | −0.052887 | 0.207* |
| H24B | 0.573691 | 1.068719 | −0.018592 | 0.207* |
| H24C | 0.562760 | 0.996114 | 0.034001 | 0.207* |
| C25C | 0.56941 (11) | 0.8324 (3) | 0.0877 (2) | 0.0766 (9) |
| H25A | 0.544909 | 0.829813 | 0.042354 | 0.115* |
| H25B | 0.568552 | 0.887660 | 0.111262 | 0.115* |
| H25C | 0.564902 | 0.787146 | 0.120911 | 0.115* |
| C26C | 0.55512 (9) | 0.6495 (2) | −0.00511 (19) | 0.0658 (8) |
| H26A | 0.556273 | 0.612664 | 0.037199 | 0.099* |
| H26B | 0.539353 | 0.619882 | −0.050088 | 0.099* |
| H26C | 0.538385 | 0.701687 | −0.000139 | 0.099* |
| C27C | 0.68417 (9) | 0.6046 (2) | −0.02168 (16) | 0.0578 (6) |
| H27A | 0.702940 | 0.627728 | 0.024378 | 0.087* |
| H27B | 0.682677 | 0.645765 | −0.061160 | 0.087* |
| H27C | 0.697998 | 0.551687 | −0.033379 | 0.087* |
| C28C | 0.56441 (9) | 0.39821 (18) | −0.00600 (14) | 0.0493 (5) |
| C29C | 0.75723 (18) | 0.2862 (5) | 0.0781 (4) | 0.130 (2) |
| H29A | 0.768697 | 0.310667 | 0.038248 | 0.195* |
| H29B | 0.759133 | 0.224177 | 0.076484 | 0.195* |
| H29C | 0.775937 | 0.306812 | 0.125222 | 0.195* |
| C30C | 0.72983 (12) | 0.4517 (3) | 0.1424 (2) | 0.0923 (12) |
| H30A | 0.727594 | 0.513758 | 0.139260 | 0.138* |
| H30B | 0.761986 | 0.434576 | 0.149252 | 0.138* |
| H30C | 0.718545 | 0.431914 | 0.183993 | 0.138* |
Source of material
3-Oxo-urs-12-en-28-oic acid was synthesized by oxidation of ursolic acid (UA) with the Sarett reagent. Specifically, the Sarett reagent was added to a solution of UA in dichloromethane (DCM) at room temperature over a period till the brown color persisted. The resulting solution was stirred for further 6 h. The progress of the reaction was monitored by TLC. After completion of the reaction, isopropanol was added and stirred at r.t. for 30 min. The DCM was evaporated in vacuo to yield a black solid, which was purified using silica-gel column chromatography (petroleum ether:ethyl acetate = 20:1, v/v) to afford a white powder. Suitable crystals were obtained by recrystallization in ethyl acetate solution.
Experimental details
All hydrogen atoms were placed in idealized positions with the carrier atom–H distances = 0.93 Å for alkene, 0.96 Å for methyl, 0.97 Å for methylene and 0.98 Å for methine. The Uiso values of the hydrogen atoms of methyl groups were set to 1.5 Ueq and the Uiso values of all other hydrogen atoms were set to 1.2Ueq(C). The absolute configuration was derived from the synthesis. The solvent molecule was presumed to be two water molecule on the basis of the 43 Å3 voids.
Comment
Ursolic acid (UA), a pentacyclic triterpenoid compound, is widely distributed in food products such as apples, basil, as well as in more than 120 plant species, many of which are applied as medicinal plants in traditional formulations [7]. UA has been reported to have multiple pharmacological activities like anti-inflammatory [8], antitumor [9], antioxidative [10], antiviral [11]. The pentacyclic triterpene system of title compound molecule consists of condensed six-membered rings with a ketone functional group at C-3 and a carboxyl moiety at C-28. It has been reported that 3-oxo-ursolic acid derivatives exhibit significant activity in inhibiting the production of nitric oxide induced by interferon-γ in mouse macrophages [12], [13]. And other study also showed that its derivatives more potently inhibit the growth of MCF-7 and THP-1 cancer cell lines [14]. Accordingly, the title compound was synthesized according to reported procedures [15]. In the molecule of the title compound, bond lengths and angles within the six-membered rings are similar to those given in the literature for tetracyclic triterpeneoids [16], [17], [18].
Single-crystal structure analysis indicated that there are three molecules in the asymmetric unit of the title crystal structure. All cyclohexane rings are in the usually observed chair conformation, cyclohexene is in a conformation intermediate between sofa and half-chair: the C8 and C9 atoms deviate in different directions with respect to the planar C11–C12=C13–C14 fragment by 0.66 and 0.12 Å, respectively, in one molecule, by 0.70 and 0.84 Å in the second molecule and by 0.66 and 0.13 Å in the third molecule.
Funding source: National Natural Science Foundation of China
Award Identifier / Grant number: 81473104
Award Identifier / Grant number: 81773563
Funding source: Science and Technology Innovation Development Plan of Yantai
Award Identifier / Grant number: 2020XDRH105
Acknowledgments
X-ray data were collected at Institute of Medical Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College.
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: This work was supported by Science and Technology Innovation Development Plan of Yantai (No. 2020XDRH105) and the National Natural Science Foundation of China (No. 81473104 and 81773563).
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Rigaku Oxford Diffraction: CrysAlisPRO; Rigaku Corporation: Oxford, UK, 2018.Search in Google Scholar
2. Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., Puschmann, H. Olex2: a complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
3. Palatinus, L., Chapuis, G. SUPERFLIP – a computer program for the solution of crystal structures by charge flipping in arbitrary dimensions. J. Appl. Cryst. 2007, 40, 786–790; https://doi.org/10.1107/s0021889807029238.Search in Google Scholar
4. Palatinus, L., van der Lee, A. Symmetry determination following structure solution in P1. J. Appl. Cryst. 2008, 41, 975–984; https://doi.org/10.1107/s0021889808028185.Search in Google Scholar
5. Palatinus, L., Prathapa, S. J., van Smaalen, S. EDMA: a computer program for topological analysis of discrete electron densities. J. Appl. Cryst. 2012, 45, 575–580; https://doi.org/10.1107/s0021889812016068.Search in Google Scholar
6. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
7. Wang, X. T., Gong, Y., Zhou, B., Yang, J. J., Cheng, Y., Zhao, J. G., Qi, M. Y. Ursolic acid ameliorates oxidative stress, inflammation and fibrosis in diabetic cardiomyopathy rats. Biomed. Pharmacother. 2018, 97, 1461–1467; https://doi.org/10.1016/j.biopha.2017.11.032.Search in Google Scholar
8. Wei, Z. Y., Chi, K. Q., Wang, K. S., Wu, J., Liu, L. P., Piao, H. R. Design, synthesis, evaluation, and molecular docking of ursolic acid derivatives containing a nitrogen heterocycle as anti-inflammatory agents. Bioorg. Med. Chem. Lett. 2018, 28, 1797–1803; https://doi.org/10.1016/j.bmcl.2018.04.021.Search in Google Scholar
9. Jiang, W., Huang, R. Z., Zhang, J., Guo, T., Zhang, M. T., Huang, X. C., Zhang, B., Liao, Z. X., Sun, J., Wang, H. S. Discovery of antitumor ursolic acid long-chain diamine derivatives as potent inhibitors of NF-\kB. Bioorg. Chem. 2018, 79, 265–276; https://doi.org/10.1016/j.bioorg.2018.05.005.Search in Google Scholar
10. Wang, X. T., Gong, Y., Zhou, B., Yang, J. J., Cheng, Y., Zhao, J. G., Qi, M. Y. Ursolic acid ameliorates oxidative stress, inflammation and fibrosis in diabetic cardiomyopathy rats. Biomed. Pharmacother. 2018, 97, 1461–1467; https://doi.org/10.1016/j.biopha.2017.11.032.Search in Google Scholar
11. Tohmé, M. J., Giménez, M. C., Peralta, A., Colombo, M. I., Delgui, L. R. Ursolic acid: a novel antiviral compound inhibiting rotavirus infection in vitro. Int. J. Antimicrob. Agents 2019, 54, 601–609; https://doi.org/10.1016/j.ijantimicag.2019.07.015.Search in Google Scholar
12. Honda, T., Gribble, G. W., Suh, N., Finlay, H. J., Rounds, B. V., Bore, L., Favaloro, F. G., Wang, Y. P., Sporn, M. B. Novel synthetic oleanane and ursane triterpenoids with various enone functionalities in ring a as inhibitors of nitric oxide production in mouse macrophages. J. Med. Chem. 2000, 41, 1866–1877; https://doi.org/10.1021/jm000008j.Search in Google Scholar
13. Honda, T., Rounds, B. V., Bore, L., Finlay, H. J., Favaloro, F. G.Jr., Suh, N., Wang, Y. P., Sporn, M. B., Gribble, G. W. Synthetic oleanane and ursane triterpenoids with modified rings A and C: a series of highly active inhibitors of nitric oxide production in mouse macrophages. J. Med. Chem. 2000, 41, 4233–4246; https://doi.org/10.1021/jm0002230.Search in Google Scholar
14. Wang, W. Z., Lei, L., Liu, Z., Wang, H. B., Meng, Q. G. Design, synthesis, and biological evaluation of novel nitrogen heterocycle-containing ursolic acid analogs as antitumor agents. Molecules 2019, 41, 877; https://doi.org/10.3390/molecules24050877.Search in Google Scholar
15. Zhao, R. L., Wang, H. Y., Luan, M. Z., Zheng, X., Zhao, F. L., Meng, Q. G. Crystal structure of (3R,5R,8R,9R,10R,12R,13R,14R)-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-1H-cyclopenta[a]phenanthrene-3,12-diol, C30H52O3. Z. Kristallogr. NCS 2020, 235, 129–131.10.1515/ncrs-2019-0533Search in Google Scholar
16. Liu, J., Xu, Y. R., Yang, J. J., Wang, W. Z., Zhang, J. Q., Zhang, R. M., Meng, Q. G. Discovery, semisynthesis, biological activities, and metabolism of ocotillol-type saponins. J. Ginseng Res. 2017, 41, 373–378; https://doi.org/10.1016/j.jgr.2017.01.001.Search in Google Scholar
17. Wang, C. M., Liu, J., Deng, J. Q., Wang, J. Z., Weng, W. Z., Chu, H. X., Meng, Q. G. Advances in the chemistry, pharmacological diversity, and metabolism of 20(R)-ginseng saponins. J. Ginseng Res. 2020, 44, 14–23; https://doi.org/10.1016/j.jgr.2019.01.005.Search in Google Scholar
18. Zhang, J. Q., Xu, Y. R., Li, H. X., Zhao, F. L., Wang, C. M., Liu, Z., Liu, P., Liu, Y. N., Meng, Q. G., Zhao, F. Synthesis and in vitro anti-inflammatory activity of C20 epimeric ocotillol-type triterpenes and protopanaxadiol. Planta Med. 2019, 85, 292–301; https://doi.org/10.1055/a-0770-0994.Search in Google Scholar
© 2020 Xiao-Hui Wang et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of 3-oxo-urs-12-en-28-oic acid, C30H46O3·1/6H2O
- The crystal structure of (3S,12R,20R,24R)-3,12-diacetyl-20,24-epoxy-dammarane-3,12,25–triol–ethyl acetate (4/1), C34H56O6⋅ 0.25(C4H8O2)
- A new polymorph of tetrakis(dimethylammonium) catena-poly[tetrakis(μ2-sulfato-κ2O:O′)dizinc(II)], Zn2C8H32N4O16S4
- Crystal structure of 10-oxysophoridine, C15H22N2O2
- The crystal structure of (5R,8R,9R,10R,12R,13R,14R)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl)tetradecahydro-3H-cyclopenta[a]phenanthrene-3,6(2H)-dione, C30H48O4
- Synthesis, crystal structure and optical property of 1,6-bis(p-tolylthio)pyrene, C30H22S2
- The crystal structure of hexakis(2-(pyridin-2-ylamino)pyridin-1-ium) decavanadate(V) dihydrate, C60H64N18O30V10
- Preparation and crystal structure of a cationic olefin polymerization precatalyst: (1,7-bis(2,6–dichlorophenyl)-1,7-di-aza-4-oxo-heptan-1,4,7-triyl)dimethyl zirconium(IV), C18H20Cl4N2OZr
- The crystal structure of fac-tricarbonyl(4,4-dimethyl-2,2-dipyridyl-κ2N,N′)- (pyrazole-κN)rhenium(I) nitrate, C18H16O3N4Re
- Synthesis and crystal structure of hexaaquacopper(II) 2,5-dicarboxyterephthalate, C10H16O14Cu
- The crystal structure of (8R,10R,12R,14R)- 12-hydroxy-16-(5-(2-hydroxypropan-2-yl)-2-methyltetrahydrofuran-2-yl)- 4,4,8,10,14-pentamethyltetradecahydro-3H- cyclopenta[a]phenanthrene-3,6(2H)-dione, C30H48O5
- Structure of the mixed crystal (S)-(6-(bromo/chloro)-2-methoxy-2,6-dihydroquinolin-3-yl)(phenyl)methanol, C17H14Br0.5Cl0.5NO2
- The crystal structure of trans-tetraaqua-bis(4-acetylphenoxyacetato-κ1O)manganese(II), C20H26O12Mn
- Crystal structure of (E)-2-(4-fluoro-3-(trifluoromethyl)benzylidene)-7-methoxy-3,4-dihydronaphthalen-1(2H)-one, C19H14F4O2
- Crystal structure of DL-α-(methylaminomethyl)benzyl alcohol, C9H13NO
- The crystal structure of dipentaerthritol hexanitrate, C10H16N6O19
- Crystal structure of N,N-diphenylformamide, C13H11NO
- Crystal structure of (E)-2-(3,5-bis(trifluoromethyl)benzylidene)-7-methoxy-3,4-dihydronaphthalen- 1(2H)-one, C20H14F6O2
- Crystal structure of ortho-methoxy benzaldehyde, C8H8O2 – a second polymorph and deposition of 3D coordinates
- Crystal structure of catena-poly[diaqua-bis(μ2-2-(4-(2,4,4-trimethylpentan-2-yl)phenoxy)propanoato-κ2O:O')-(2-(4-(2,4,4-trimethylpentan-2-yl)phenoxy)propanoato-κ2O,O')yttrium(III)], C51H79O11Y
- Crystal structure of benzylthiouronium chloride, C8H11ClN2S
- Synthesis and crystal structure of tert-butyl (2′R,3R,3′R,4a′R,9a′S)-1-acetyl-5-chloro-3″-methyl-2,5″,9′-trioxo-1″-phenyl-1″,4a′,5″,9a′-tetrahydro-1′H,3′H,9′H-dispiro[indoline-3,4′-xanthene-2′,4″-pyrazole]-3′-carboxylate, C36H32ClN3O7
- Crystal structure of 2-hydroxy-4-methoxy benzaldehyde, C8H8O3
- Crystal structure of poly[diaqua-(m3-3′,5′-dicarboxy-[1,1′-biphenyl]-3,4-dicarboxylato-K4O,O′:O″:O‴) cadmium(II)], C16H11O10Cd
- Crystal structure of {tetraaqua-bis(1-(4-hydroxy-2-oxotetrahydrofuran-3-yl)-2-((4aS,6R,8aS)-6-hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylenedecahydronaphthalen-1-yl)ethane-1-sulfonato-k2O,O') calcium(II)}-{triaqua-bis(1-(4-hydroxy-2-oxotetrahydrofuran-3-yl)-2-((4aS,6R,8aS)-6-hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylenedecahydronaphthalen-1-yl)ethane-1-sulfonato-k2O,O') calcium(II)} – water – acetone (1/1/8/2)
- Synthesis and crystal structure of bis{2-bromo-6-((E)-((4-((E)-1-(methoxy-imino)ethyl)phenyl)imino)methyl)phenolato- κ2N,O}zinc(II)-methanol(1/2), C65H60Br4N8O9Zn2
- Crystal structure of benzenesulphonic acid
- Crystal structure of N-benzyl-N-nicotinoyl-nicotine amide C19H15N3O2
- Crystal structure of poly[aqua(μ3-2,4-diamino-benzenesulfonato-κ4N:N′,O:O′)silver(I)], C12H18O8N4S2Ag2
- Crystal structure of 1,4-bis(methylpyridinium benzene) bis(1,2-dicyanoethene-1,2-dithiolato-κ2S:S)nickel(II), C26H18N6NiS4
- Crystal structure of the Cu(II) complex chlorido-(6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxylato-k2N,O)-(phenanthroline-k2N,N')copper(II), C23H15ClCuN4O3
- Crystal structure of phenarsazine chloride acetic acid solvate, C14H13AsClNO2
- Crystal structure of poly[aqua-(μ2-3,3′,4,5′-biphenyl tetracarboxylate- κ3O,O′:O′′) -(μ2-4,4′-bis(pyrid-4-yl)biphenyl-κ2N:N′)zinc(II)], C27H18NO9Zn
- Crystal structure of catena-poly[(μ2-3-amino-benzenedisulfonato-κ2N:O)-bis (3-methyl-isoquinoline-κN)silver(I)], C26H24N3O3SAg
- Crystal structure of 2-((4-Aminophenyl)thio)acetic acid, C8H9NO2S
- Crystal structure of phenarsazine chloride dimethylsulfoxide solvate, C14H15AsClNOS
- Synthesis and crystal structure of 2-azido-N-phenylacetamide, C8H8N4O
- Crystal structure of chlorido{hydridotris[3-phenyl-5-methylpyrazol-1-yl-κN3]borato}copper(II), C30H28BClCuN6
- Crystal structure of benzanthrone – a redetermination for correct molecular geometry and localization of hydrogen atoms
- Crystal structure of 4-bromobenzaldehyde – complete redetermination at 200 K, C7H5BrO
- Crystal structure and spectroscopic properties of chlorido{hydridotris[3-,5-dimethylpyrazol-1-yl-κN3]borato}(3-,5-dimethylpyrazol-1-yl-κN)copper(II), C20H30BClCuN8
- The crystal structure of 4-((2-hydroxynaphthalen-1-yl)(pyrrolidin-1-yl)methyl)benzonitrile, C22H20N2O
- Crystal structure of 4-ethyl-3-phenylisoquinolin-1(2H)-one, C17H15NO
- Crystal structure of (tricyclohexylphosphane-κP)-[(Z)-N-(3-fluorophenyl)-O-methylthiocarbamato-k1S]gold(I), C26H40AuFNOPS
- Crystal structure of (3S,8R,10R,12R,14R)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-1H-cyclopenta[a]phenanthren-3-yl acetate, C32H54O4
- The crystal structure of 2-[(S)-1-(naphthalen-1-yl)ethyl]-2,3,7,7a- tetrahydro-3a,6-epoxyisoindol-1(6H)-one, C19H20NO2
- Crystal structure of {hydridotris[3-(t-butyl)-5-isopropylpyrazol-1-yl-κN3]borato}thallium(I), C30H52BN6Tl
- Synthesis and crystal structure of 1-octyl-3-phenylquinoxalin-2(1H)-one, C22H26N2O
- The crystal structure of 2,6-difluorophenol, C6H4F2O
- 4-(9H-Fluoren-9-yl)-4-methylmorpholin-4-ium bromide, C18H20BrNO
- The crystal structure of 2,4-dimethylimidazole monohydrate, C5H10N2O
- The crystal structure of 1,2-dimethylimidazole, C5H8N2
- The crystal structure of 3-ammonio-4-aminobenzoate, C7H8N2O2 – a second polymorph
- The crystal structure of 4-hydroxy-2,5-bis(1-methyl-1H-imidazol-3-ium-2-ylthio)-3,6-dioxocyclohexa-1,4-dienolate chloride monohydrate, C14H15N4O5S2Cl
- The crystal structure of butyrylferrocene, C14H16FeO
- The crystal structure of bi-1,1′-cyclopentane-1,1′-diol, C10H18O2
- The crystal structure of 2-iso-propylimidazole, C6H10N2
- The crystal structure of aqua-tris (1,3-diphenylpropane-1,3-dionato-κ2O,O′)-lanthanum(III), C45H35LaO7
- Crystal structure of (3E,5E)-3,5-bis-4-methoxy-3-(trifluoromethyl)benzylidene)-1-methylpiperidin-4-one, C24H21F6NO3
- The crystal structure of 3,5-dichloro-6-diazo-2,4-dinitrocyclohexa-2,4-dien-1-one, C6Cl2N4O5
- Crystal structure of carbonyl(2-methylquinolin-8-olato-κ2N,O)(triphenylarsine-κAs)rhodium(I), C29H23AsNO2Rh
- Crystal structure of (1aS,1a1S,2S)-4a-butoxy-1a,1a1,2,4a,5,6-hexahydro-1H-cyclobuta[de]naphthalen-2-yl-4-nitrobenzoate, C22H25NO5
- Crystal structure of carbonyl(2-oxopyridin-1(2H)-olato-k2O,O′)(triphenylarsine-κAs)rhodium(I), C24H19AsNO3Rh
- Crystal structure of catena-poly[triqua-bis(μ2-4-carboxy-2-(1H-tetrazol-1-yl)-1H-imidazole-5-carboxylato-k3N,O:O′)barium(II)] tetrahydrate, C14H14BaN12O15
- Crystal structure of (E)-3′,6′-bis(ethylamino)-2-((quinoxalin-2-ylmethylene)amino)spiro[isoindoline-1,9′-xanthen]-3-one, C35H32N6O2
- Crystal structure of diaqua-bis(μ2-5-chloro-salicylato-κ3O,O′:O′)-bis(5-chloro-salicylato-κ2O,O′)-bis(1,10-phenanthroline-κ2N,N′) dilead(II) – water (1/2), C52H36C14N4O14Pb2·2(H2O)
- Crystal structure of (E)-2-(4-ethoxycarbonyl-3,5-dimethyl-2-(pyrrole-2-ylmethyleneamino)-3′,6′-dihydroxylspiro[isoindoline-1,9′-xanthen]-3-one-methanol (1/1), C31H29N3O7
- The crystal structure of 5H-dibenzo[b,e]azepine-6,11-dione, C14H9NO2
- Crystal structure of (E)-2-(4-fluoro-2-(trifluoromethyl)benzylidene)-7-methoxy-3,4-dihydronaphthalen-1(2H)-one, C19H14F4O2
- The crystal structure of N-(2-methoxy-4,5-bis[phenylselanyl]phenyl)picolinamide, C25H20N2O2Se2
- The crystal structure of (E)-2-(5-bromo-2-hydroxybenzylidene)-N-phenylhydrazine-1- carboxamide monohydrate, C14H14BrN3O3
- Crystal structure of fac-tricarbonyl-(nitrato-k1O)-bis(pyridine-κN)-rhenium, C13H10O6N3Re
- Crystal structure of (E)-2-(((1H-pyrrol-2-yl)methylene)amino)-3′,6′-dihydroxyspiro[isoindoline-1,9′-xanthen]-3-one — methanol (1/2), C27H25N3O6
- The crystal structure of 4-amino-N′-(4-aminobenzoyl)benzohydrazide monohydrate, C14H16N4O3
- Crystal structure of bis(amino(carbamothioylamino)methaniminium) 5-hydroxyisophthalate monohydrate, C12H20N8O6S2
- The crystal structure of 2-(chloromethyl)pyridine, C6H6ClN
- The crystal structure of 1-bromo-4-iodo-benzene, C6H4BrI
- The crystal structure of 2,6-dimethyl-4-nitro-phenol, C8H9NO3
- The crystal structure of 3-chloropropionic acid, C3H5ClO2
- The crystal structure of 2-(2-methoxyphenyl)acetic acid, C9H10O3
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of 3-oxo-urs-12-en-28-oic acid, C30H46O3·1/6H2O
- The crystal structure of (3S,12R,20R,24R)-3,12-diacetyl-20,24-epoxy-dammarane-3,12,25–triol–ethyl acetate (4/1), C34H56O6⋅ 0.25(C4H8O2)
- A new polymorph of tetrakis(dimethylammonium) catena-poly[tetrakis(μ2-sulfato-κ2O:O′)dizinc(II)], Zn2C8H32N4O16S4
- Crystal structure of 10-oxysophoridine, C15H22N2O2
- The crystal structure of (5R,8R,9R,10R,12R,13R,14R)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl)tetradecahydro-3H-cyclopenta[a]phenanthrene-3,6(2H)-dione, C30H48O4
- Synthesis, crystal structure and optical property of 1,6-bis(p-tolylthio)pyrene, C30H22S2
- The crystal structure of hexakis(2-(pyridin-2-ylamino)pyridin-1-ium) decavanadate(V) dihydrate, C60H64N18O30V10
- Preparation and crystal structure of a cationic olefin polymerization precatalyst: (1,7-bis(2,6–dichlorophenyl)-1,7-di-aza-4-oxo-heptan-1,4,7-triyl)dimethyl zirconium(IV), C18H20Cl4N2OZr
- The crystal structure of fac-tricarbonyl(4,4-dimethyl-2,2-dipyridyl-κ2N,N′)- (pyrazole-κN)rhenium(I) nitrate, C18H16O3N4Re
- Synthesis and crystal structure of hexaaquacopper(II) 2,5-dicarboxyterephthalate, C10H16O14Cu
- The crystal structure of (8R,10R,12R,14R)- 12-hydroxy-16-(5-(2-hydroxypropan-2-yl)-2-methyltetrahydrofuran-2-yl)- 4,4,8,10,14-pentamethyltetradecahydro-3H- cyclopenta[a]phenanthrene-3,6(2H)-dione, C30H48O5
- Structure of the mixed crystal (S)-(6-(bromo/chloro)-2-methoxy-2,6-dihydroquinolin-3-yl)(phenyl)methanol, C17H14Br0.5Cl0.5NO2
- The crystal structure of trans-tetraaqua-bis(4-acetylphenoxyacetato-κ1O)manganese(II), C20H26O12Mn
- Crystal structure of (E)-2-(4-fluoro-3-(trifluoromethyl)benzylidene)-7-methoxy-3,4-dihydronaphthalen-1(2H)-one, C19H14F4O2
- Crystal structure of DL-α-(methylaminomethyl)benzyl alcohol, C9H13NO
- The crystal structure of dipentaerthritol hexanitrate, C10H16N6O19
- Crystal structure of N,N-diphenylformamide, C13H11NO
- Crystal structure of (E)-2-(3,5-bis(trifluoromethyl)benzylidene)-7-methoxy-3,4-dihydronaphthalen- 1(2H)-one, C20H14F6O2
- Crystal structure of ortho-methoxy benzaldehyde, C8H8O2 – a second polymorph and deposition of 3D coordinates
- Crystal structure of catena-poly[diaqua-bis(μ2-2-(4-(2,4,4-trimethylpentan-2-yl)phenoxy)propanoato-κ2O:O')-(2-(4-(2,4,4-trimethylpentan-2-yl)phenoxy)propanoato-κ2O,O')yttrium(III)], C51H79O11Y
- Crystal structure of benzylthiouronium chloride, C8H11ClN2S
- Synthesis and crystal structure of tert-butyl (2′R,3R,3′R,4a′R,9a′S)-1-acetyl-5-chloro-3″-methyl-2,5″,9′-trioxo-1″-phenyl-1″,4a′,5″,9a′-tetrahydro-1′H,3′H,9′H-dispiro[indoline-3,4′-xanthene-2′,4″-pyrazole]-3′-carboxylate, C36H32ClN3O7
- Crystal structure of 2-hydroxy-4-methoxy benzaldehyde, C8H8O3
- Crystal structure of poly[diaqua-(m3-3′,5′-dicarboxy-[1,1′-biphenyl]-3,4-dicarboxylato-K4O,O′:O″:O‴) cadmium(II)], C16H11O10Cd
- Crystal structure of {tetraaqua-bis(1-(4-hydroxy-2-oxotetrahydrofuran-3-yl)-2-((4aS,6R,8aS)-6-hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylenedecahydronaphthalen-1-yl)ethane-1-sulfonato-k2O,O') calcium(II)}-{triaqua-bis(1-(4-hydroxy-2-oxotetrahydrofuran-3-yl)-2-((4aS,6R,8aS)-6-hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylenedecahydronaphthalen-1-yl)ethane-1-sulfonato-k2O,O') calcium(II)} – water – acetone (1/1/8/2)
- Synthesis and crystal structure of bis{2-bromo-6-((E)-((4-((E)-1-(methoxy-imino)ethyl)phenyl)imino)methyl)phenolato- κ2N,O}zinc(II)-methanol(1/2), C65H60Br4N8O9Zn2
- Crystal structure of benzenesulphonic acid
- Crystal structure of N-benzyl-N-nicotinoyl-nicotine amide C19H15N3O2
- Crystal structure of poly[aqua(μ3-2,4-diamino-benzenesulfonato-κ4N:N′,O:O′)silver(I)], C12H18O8N4S2Ag2
- Crystal structure of 1,4-bis(methylpyridinium benzene) bis(1,2-dicyanoethene-1,2-dithiolato-κ2S:S)nickel(II), C26H18N6NiS4
- Crystal structure of the Cu(II) complex chlorido-(6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxylato-k2N,O)-(phenanthroline-k2N,N')copper(II), C23H15ClCuN4O3
- Crystal structure of phenarsazine chloride acetic acid solvate, C14H13AsClNO2
- Crystal structure of poly[aqua-(μ2-3,3′,4,5′-biphenyl tetracarboxylate- κ3O,O′:O′′) -(μ2-4,4′-bis(pyrid-4-yl)biphenyl-κ2N:N′)zinc(II)], C27H18NO9Zn
- Crystal structure of catena-poly[(μ2-3-amino-benzenedisulfonato-κ2N:O)-bis (3-methyl-isoquinoline-κN)silver(I)], C26H24N3O3SAg
- Crystal structure of 2-((4-Aminophenyl)thio)acetic acid, C8H9NO2S
- Crystal structure of phenarsazine chloride dimethylsulfoxide solvate, C14H15AsClNOS
- Synthesis and crystal structure of 2-azido-N-phenylacetamide, C8H8N4O
- Crystal structure of chlorido{hydridotris[3-phenyl-5-methylpyrazol-1-yl-κN3]borato}copper(II), C30H28BClCuN6
- Crystal structure of benzanthrone – a redetermination for correct molecular geometry and localization of hydrogen atoms
- Crystal structure of 4-bromobenzaldehyde – complete redetermination at 200 K, C7H5BrO
- Crystal structure and spectroscopic properties of chlorido{hydridotris[3-,5-dimethylpyrazol-1-yl-κN3]borato}(3-,5-dimethylpyrazol-1-yl-κN)copper(II), C20H30BClCuN8
- The crystal structure of 4-((2-hydroxynaphthalen-1-yl)(pyrrolidin-1-yl)methyl)benzonitrile, C22H20N2O
- Crystal structure of 4-ethyl-3-phenylisoquinolin-1(2H)-one, C17H15NO
- Crystal structure of (tricyclohexylphosphane-κP)-[(Z)-N-(3-fluorophenyl)-O-methylthiocarbamato-k1S]gold(I), C26H40AuFNOPS
- Crystal structure of (3S,8R,10R,12R,14R)-12-hydroxy-4,4,8,10,14-pentamethyl-17-((R)-2,6,6-trimethyltetrahydro-2H-pyran-2-yl) hexadecahydro-1H-cyclopenta[a]phenanthren-3-yl acetate, C32H54O4
- The crystal structure of 2-[(S)-1-(naphthalen-1-yl)ethyl]-2,3,7,7a- tetrahydro-3a,6-epoxyisoindol-1(6H)-one, C19H20NO2
- Crystal structure of {hydridotris[3-(t-butyl)-5-isopropylpyrazol-1-yl-κN3]borato}thallium(I), C30H52BN6Tl
- Synthesis and crystal structure of 1-octyl-3-phenylquinoxalin-2(1H)-one, C22H26N2O
- The crystal structure of 2,6-difluorophenol, C6H4F2O
- 4-(9H-Fluoren-9-yl)-4-methylmorpholin-4-ium bromide, C18H20BrNO
- The crystal structure of 2,4-dimethylimidazole monohydrate, C5H10N2O
- The crystal structure of 1,2-dimethylimidazole, C5H8N2
- The crystal structure of 3-ammonio-4-aminobenzoate, C7H8N2O2 – a second polymorph
- The crystal structure of 4-hydroxy-2,5-bis(1-methyl-1H-imidazol-3-ium-2-ylthio)-3,6-dioxocyclohexa-1,4-dienolate chloride monohydrate, C14H15N4O5S2Cl
- The crystal structure of butyrylferrocene, C14H16FeO
- The crystal structure of bi-1,1′-cyclopentane-1,1′-diol, C10H18O2
- The crystal structure of 2-iso-propylimidazole, C6H10N2
- The crystal structure of aqua-tris (1,3-diphenylpropane-1,3-dionato-κ2O,O′)-lanthanum(III), C45H35LaO7
- Crystal structure of (3E,5E)-3,5-bis-4-methoxy-3-(trifluoromethyl)benzylidene)-1-methylpiperidin-4-one, C24H21F6NO3
- The crystal structure of 3,5-dichloro-6-diazo-2,4-dinitrocyclohexa-2,4-dien-1-one, C6Cl2N4O5
- Crystal structure of carbonyl(2-methylquinolin-8-olato-κ2N,O)(triphenylarsine-κAs)rhodium(I), C29H23AsNO2Rh
- Crystal structure of (1aS,1a1S,2S)-4a-butoxy-1a,1a1,2,4a,5,6-hexahydro-1H-cyclobuta[de]naphthalen-2-yl-4-nitrobenzoate, C22H25NO5
- Crystal structure of carbonyl(2-oxopyridin-1(2H)-olato-k2O,O′)(triphenylarsine-κAs)rhodium(I), C24H19AsNO3Rh
- Crystal structure of catena-poly[triqua-bis(μ2-4-carboxy-2-(1H-tetrazol-1-yl)-1H-imidazole-5-carboxylato-k3N,O:O′)barium(II)] tetrahydrate, C14H14BaN12O15
- Crystal structure of (E)-3′,6′-bis(ethylamino)-2-((quinoxalin-2-ylmethylene)amino)spiro[isoindoline-1,9′-xanthen]-3-one, C35H32N6O2
- Crystal structure of diaqua-bis(μ2-5-chloro-salicylato-κ3O,O′:O′)-bis(5-chloro-salicylato-κ2O,O′)-bis(1,10-phenanthroline-κ2N,N′) dilead(II) – water (1/2), C52H36C14N4O14Pb2·2(H2O)
- Crystal structure of (E)-2-(4-ethoxycarbonyl-3,5-dimethyl-2-(pyrrole-2-ylmethyleneamino)-3′,6′-dihydroxylspiro[isoindoline-1,9′-xanthen]-3-one-methanol (1/1), C31H29N3O7
- The crystal structure of 5H-dibenzo[b,e]azepine-6,11-dione, C14H9NO2
- Crystal structure of (E)-2-(4-fluoro-2-(trifluoromethyl)benzylidene)-7-methoxy-3,4-dihydronaphthalen-1(2H)-one, C19H14F4O2
- The crystal structure of N-(2-methoxy-4,5-bis[phenylselanyl]phenyl)picolinamide, C25H20N2O2Se2
- The crystal structure of (E)-2-(5-bromo-2-hydroxybenzylidene)-N-phenylhydrazine-1- carboxamide monohydrate, C14H14BrN3O3
- Crystal structure of fac-tricarbonyl-(nitrato-k1O)-bis(pyridine-κN)-rhenium, C13H10O6N3Re
- Crystal structure of (E)-2-(((1H-pyrrol-2-yl)methylene)amino)-3′,6′-dihydroxyspiro[isoindoline-1,9′-xanthen]-3-one — methanol (1/2), C27H25N3O6
- The crystal structure of 4-amino-N′-(4-aminobenzoyl)benzohydrazide monohydrate, C14H16N4O3
- Crystal structure of bis(amino(carbamothioylamino)methaniminium) 5-hydroxyisophthalate monohydrate, C12H20N8O6S2
- The crystal structure of 2-(chloromethyl)pyridine, C6H6ClN
- The crystal structure of 1-bromo-4-iodo-benzene, C6H4BrI
- The crystal structure of 2,6-dimethyl-4-nitro-phenol, C8H9NO3
- The crystal structure of 3-chloropropionic acid, C3H5ClO2
- The crystal structure of 2-(2-methoxyphenyl)acetic acid, C9H10O3


