Abstract
Background
Malignant tumors were considered as the leading causes of cancer-related mortality globally. More and more studies found that dysregulated genes played an important role in carcinogenesis. The aim of this study was to explore the significance of KPNA2 in human six major cancers including non-small cell lung cancer (NSCLC), gastric cancer, colorectal cancer, breast cancer, hepatocellular carcinoma, and bladder cancer based on bioinformatics analysis.
Methods
The data were collected and comprehensively analyzed based on multiple databases. KPNA2 mRNA expression in six major cancers was investigated in Oncomine, the human protein atlas, and GEPIA databases. The mutation status of KPNA2 in the six major cancers was evaluated by online data analysis tool Catalog of Somatic Mutations in Cancer (COSMIC) and cBioPortal. Co-expressed genes with KPNA2 were identified by using LinkedOmics and made pairwise correlation by Cancer Regulome tools. Protein-protein interaction (PPI) network relevant to KPNA2 was constructed by STRING database and KEGG pathway of the included proteins of the PPI network was explored and demonstrated by circus plot. Survival analysis-relevant KPNA2 of the six cancers was performed by GEPIA online data analysis tool based on TCGA database.
Results
Compared with paired normal tissue, KPNA2 mRNA was upregulated in all of the six types of cancers. KPNA2 mutations, especially missense substitution, were widely identified in six cancers and interact with different genes in different cancer types. Genes involved in PPI network were mainly enriched in p53 signaling pathway, cell cycle, viral carcinogenesis, and Foxo signaling pathway. KPNA2 protein was mainly expressed in nucleoplasm and cytosol in cancer cells. Immunohistochemistry assay indicated that KPNA2 protein was also positively expressed in nucleoplasm with brownish yellow staining. Overall survival (OS) and progression free survival (PFS) were different between KPNA2 high and low expression groups.
Conclusions
KPNA2 was widely dysregulated and mutated in carcinomas and correlated with the patients prognosis which may be potential target for cancer treatment and biomarker for prognosis.
1 Introduction
Cancer is the leading cause of death globally. According to the cancer statistical analysis in year 2018, 9.6 million deaths and 18.1 million new cases of all types of cancers were estimated [1]. Cancer had posed a heavy burden not only for the human beings, but also for the governments and health provides. Lung cancer especially non-small cell lung cancer (NSCLC), gastric cancer, colorectal cancer, breast cancer, liver hepatic cancer, and bladder cancer were the most common carcinomas diagnosed clinically with high incidence and mortality [2,3]. Although the above six cancers had high incidence and poor prognosis, the general carcinogenesis was not completely cleart. In recent years, with the development of biology and life science, more and more evidence had made it clear that the driving genes had played an important role in the carcinogenesis and pathways [4]. The driving genes including oncogenes and tumor suppressor genes involved in the cell division, apoptosis [5], proliferation [6], and migration may act as the general phenotype of malignant carcinoma.
Karyopherin subunit alpha 2 (KPNA2) is one of the important members of karyophenin family [7]. It has three functional domains: N-terminal Importin β binding domain, central domain, and a short acid C-terminal. The central domain contains the nuclear localization signal (NLS) binding site and CAS binding site [8,9]. In the cytoplasm, Karyopherin subunit alpha 2 can recognize and bind nucleophilic NLS, while Importin β will combine with karyophenin to form NLS-α/β complex, and then enter the nucleus through the nuclear pore complex under the energy provided by RanGTP enzyme [8]. KPNA2 is not expressed or low expressed in normal tissues, but it is upregulated in some types of carcinoma such as breast cancer [10,11] and ovarian cancer [12]. However, seldom studies focused on the KPNA2 expression, mutation, and as prognostic marker for pan-cancer. In the present work, we investigated KPNA2 mRNA expression, mutation, and prognostic significance in six major carcinomas through well-known online databases.
2 Methods
2.1 KPNA2 mRNA expression analysis
KPNA2 mRNA expression level of human normal tissues and multiple cancers was identified in the human protein atlas database (https://www.proteinatlas.org/) with data original from HPA, GTEx, and FANTOM5 project. KPNA2 mRNA expression level between cancer tissue and paired normal tissue was further validated by Oncomine database (https://www.oncomine.org/resource/login.html) [13] and GEPIA online data analysis tool (http://gepia.cancer-pku.cn/detail.php) with data origin from the TCGA database.
2.2 KPNA2 gene mutation analysis
KPNA2 gene mutation was analyzed through the cBio cancer genomics portal (http://cbioportal.org) with the data origin from the TCGA database. The mutation frequency of nonsense substitution, missense substitution, synonymous substitution, inframe insertion, frameshift, etc. was identified and expressed by pie plot. The single nucleotide mutation of KPNA2 was also screened by catalogue of somatic mutation in cancer (COSMIC) (https://cancer.sanger.ac.uk/cosmic/) online data analysis tool [14].
2.3 Genome-wide association of KPNA2 mRNA in cancer analysis
The expression of KPNA2 gene and its correlation with other genes of the six cancer types was expressed by the circus plots generated from the Cancer Regulome tools and data (http://explorer.cancerregulome.org/). Co-expressed genes were clustered and demonstrated by the heat map generated from LinkedOmics database (http://www.linkedomics.org/login.php) [15]. The top positive and negative correlated genes with KPNA2 were identified and made Pearson correlations test.
2.4 PPI network construction and KEGG pathway enrichment
The protein–protein interaction (PPI) network relevant to KPNA2 was constructed by the STRINIG database (http://string-db.org/cgi/input.pl) [16]. The PPI network was constructed under the condition of: network type: full STRING network type; meaning of network edges: evidence; minimum required interaction score: 0.40. The genes included in the PPI network were identified and made KEGG pathway enrichment demonstrated by circus plot.
2.5 Survival analysis
According to the median expression level of KPNA2 mRNA, cancer patients were divided into high expression (≥median expression) group and low expression group. The progression free survival (PFS) and overall survival (OS) were compared between KPNA2 high and low expression groups of the six cancer types and demonstrated by survival curve [17].
2.6 KPNA2 protein expression analysis
KPNA2 protein expression in tumor cell lines and cancer tissues was detected by immunofluorescent staining and immunohistochemistry assay in the human protein atlas database (https://www.proteinatlas.org/).
2.7 Statistical analysis
The data were analyzed based on the relevant databases or online data analysis tool.
3 Results
3.1 KPNA2 mRNA expression in normal and tumor tissue
KPNA2 mRNA expression in all human body tissues is demonstrated in Figure 1a. The expression level was quite different across different tissues. KPNA2 mRNA expression levels in different type cancers are showed in Figure 1b, which indicated that the expression level across different cancers was not obviously different. KPNA2 was upregulated in cancer tissue compared with paired normal tissue in all the six major cancers based on Oncomine database (Figure 1c) and GEPIA with statistical difference (p < 0.05) (Figure 2).
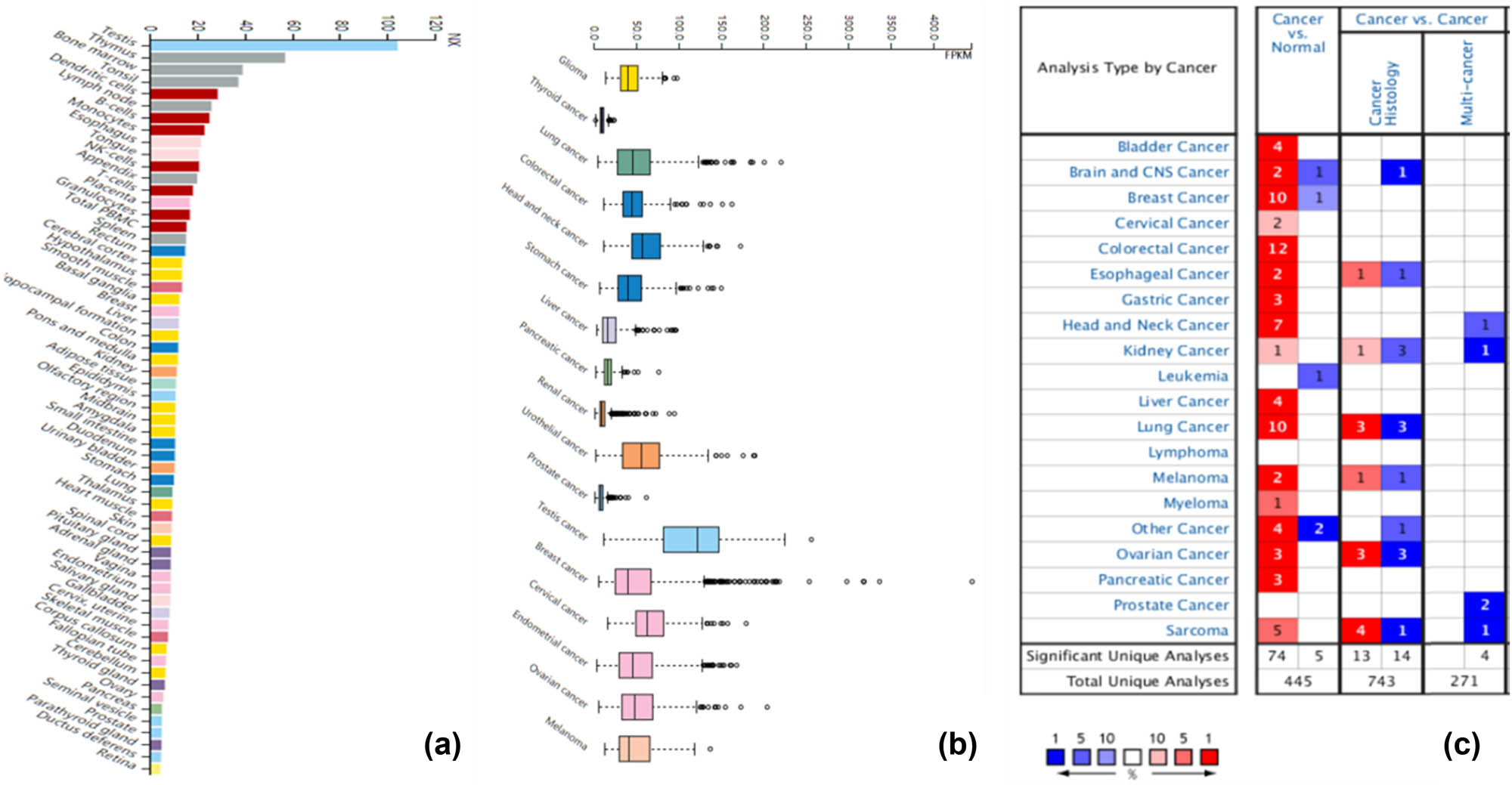
KPNA2 expression analysis. (a) KPNA2 expression in across human body tissues, (b) KPNS2 expression across carcinomas, (c) KPNA2 expression between cancer tissue and paired normal tissues based on oncoming database.
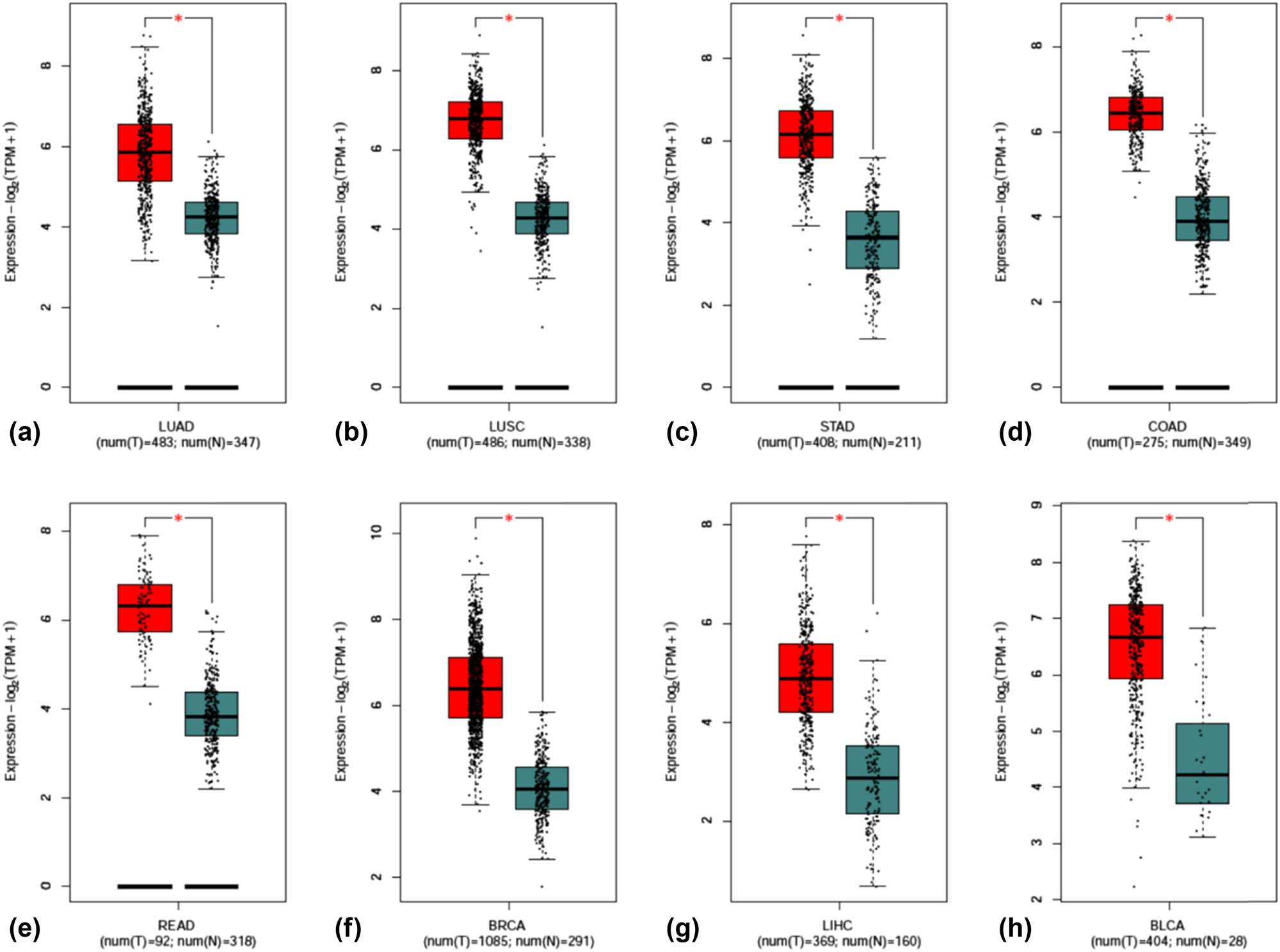
Scatter plot of KPNA2 mRNA expression between paired normal tissue and cancer tissue. (a) Lung adenocarcinoma, (b) lung squamous cell carcinoma, (c) gastric cancer, (d) colon cancer, (e) Rectal cancer, (f) breast cancer, (g) hepatocellular carcinoma, (h) bladder cancer.
3.2 KPNA2 mutation analysis
KPNA2 mutation status of the six cancers was evaluated by online data analysis tool Catalog of Somatic Mutations in Cancer (COSMIC) and cBioPortal. KPNA2 mutations were widely identified in six major cancers and interact with different genes in different cancer types. Missense substitution was found in lung cancer (84.21%), gastric cancer (48.15%), colorectal cancer (46.94%), breast cancer (30.43%), liver hepatic cancer (38.46%), and bladder cancer (87.50%). Other major mutations including nonsense substitution and synonymous substitution were also identified in the six major cancers (Figure 3a). For pan-cancers analysis, KPNA2 highly mutated in uterine carcinoma, stomach cancer, cervical cancer, breast cancer, etc. based on TCGA database (Figure 3b). For single nucleotide mutation, C > T and G > T were most common in the KPNA2 coding strand, both of which were identified in the six major cancer types. And other kinds of single nucleotide mutations were rare in TCGA cancer samples of the six cancer types (Figure 4).
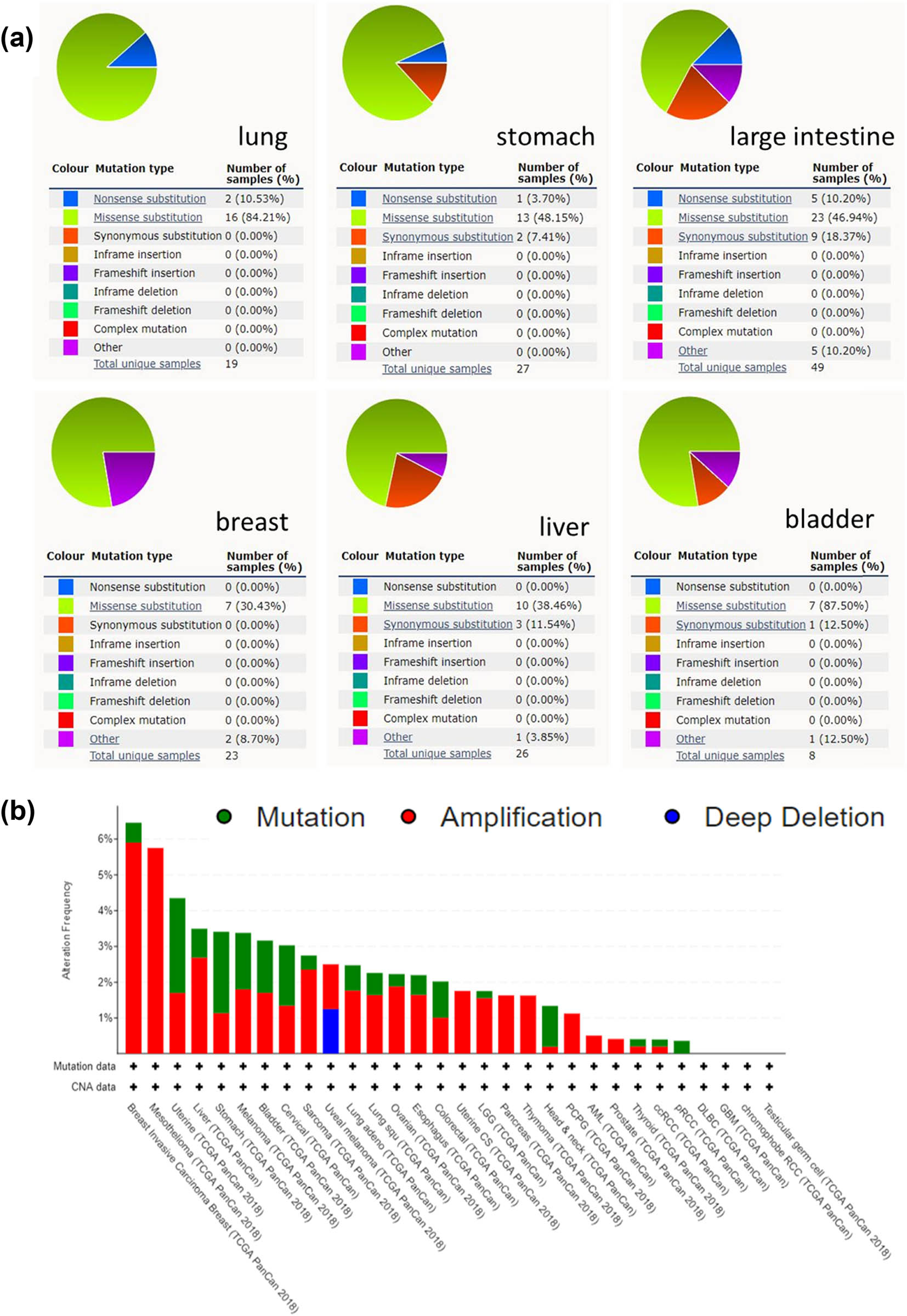
KPNA2 mutation analysis. (a) Pie plot of the KPNA2 mutation frequency of the six major cancers. (b) Bar chart of KPNA2 mutation in pan-cancers based on TCGA database.
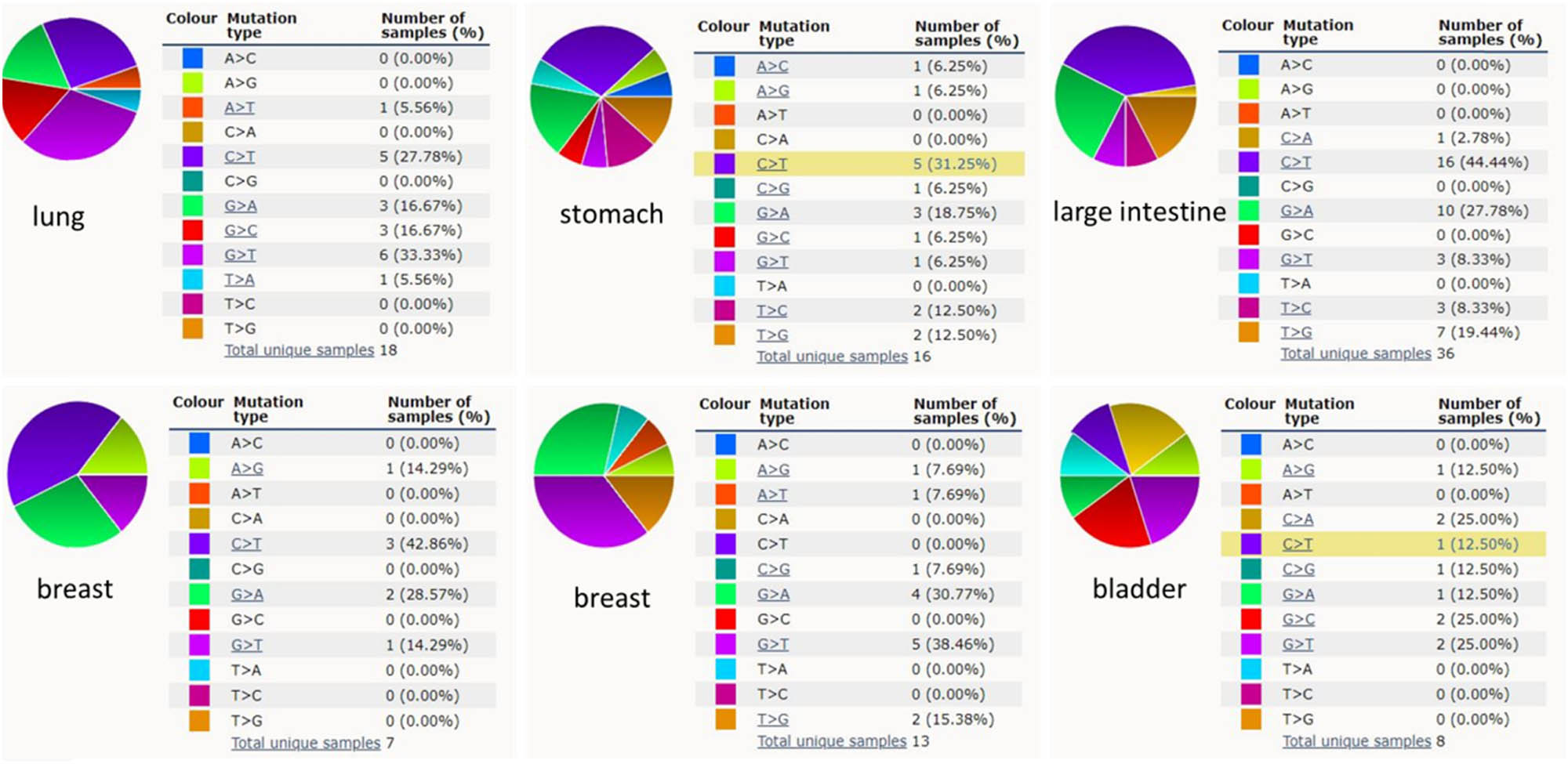
KPNA2 single nucleotide mutation analysis.
3.3 Genome-wide association of KPNA2 in cancer
Based on the association among genes, DNA methylation, somatic copy number, somatic mutation, and protein level, circus plots were drawn to display the interrelation between KPNA2 and other genes. According to the data from TCGA, KPNA2 was associated with other genes that could be detected in NSCLC, gastric cancer, colorectal cancer, liver hepatic cancer, and bladder cancer (Figure 5).
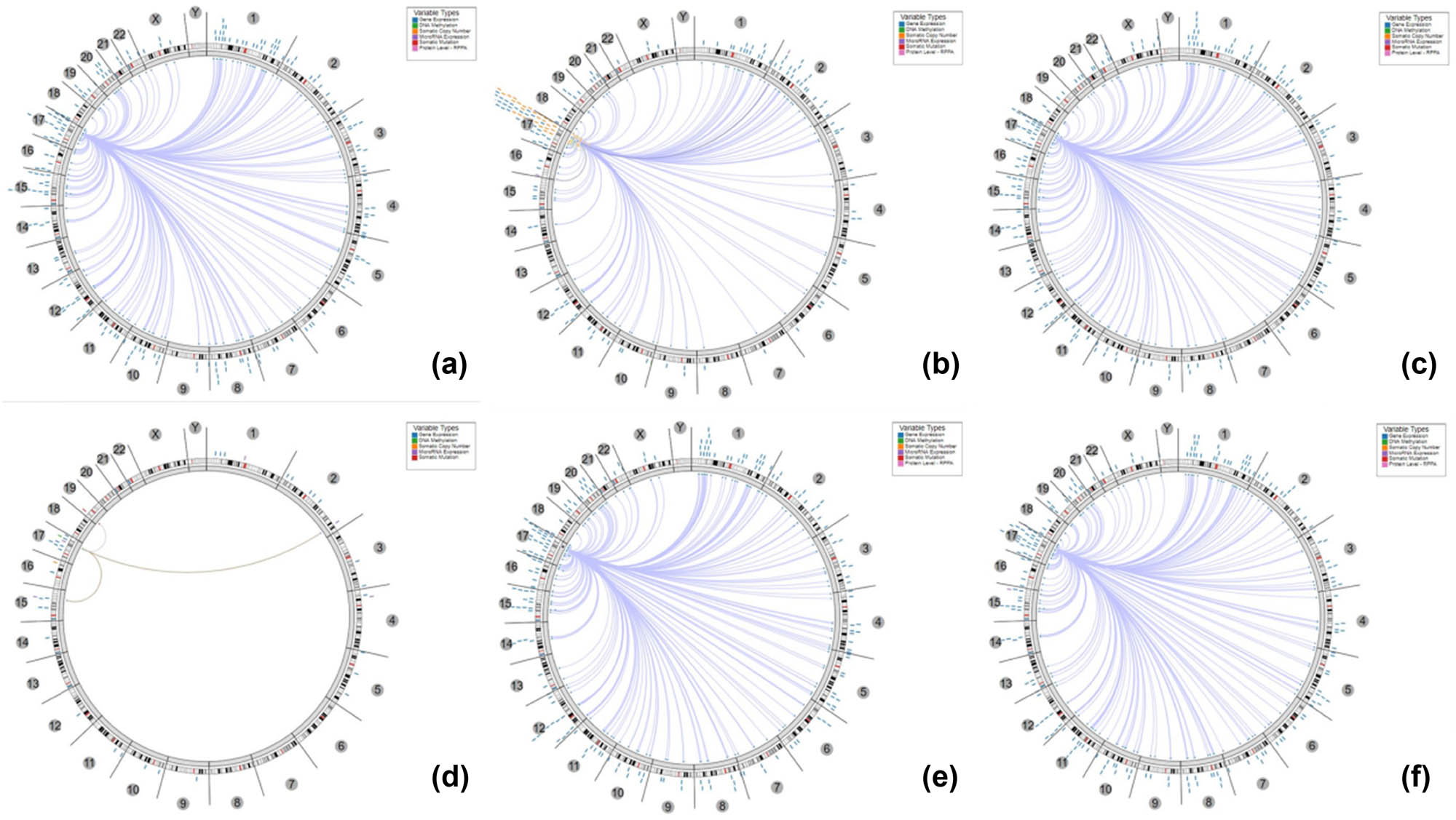
The circus plot demonstrated the associations between KPNA2 and other genes. The edges in the center connecting the features (with genomic coordinates) displayed around the perimeter. The outer ring displays cytogenetic bands. The inner ring displays associations that contain features lacking genomic coordinates. (a) Non-small cell lung cancer, (b) gastric cancer, (c) colorectal cancer, (d) breast cancer, (e) liver hepatic cancer, (f) bladder cancer.
3.4 Co-expressed genes with KPAN2 in six major cancers
The co-expressed genes with KPAN2 in six cancers were demonstrated with the heatmap (Figure 6). The top positive and negative correlated genes with KPAN2 in six cancers are showed in Figure 7.
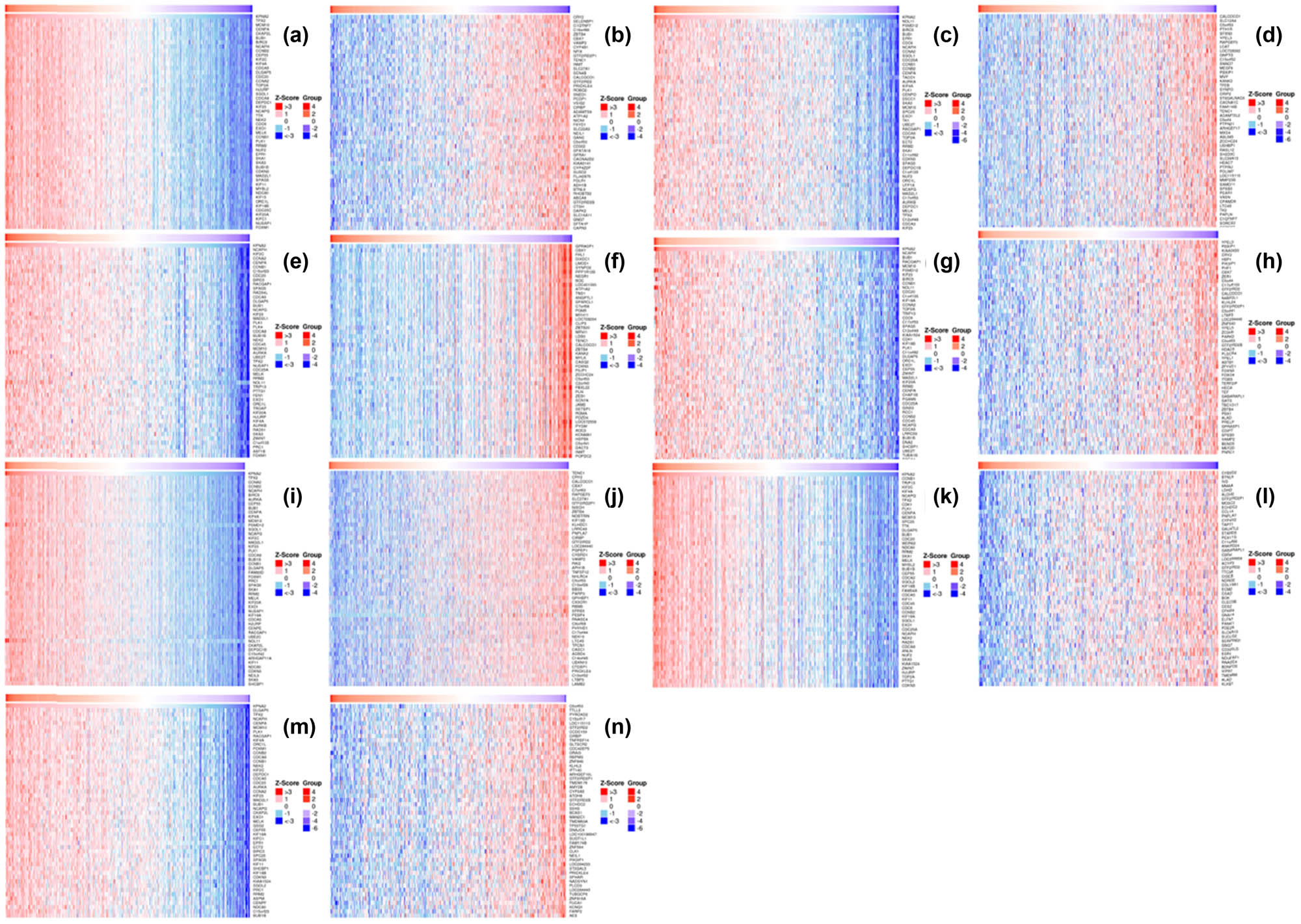
Heat map of co-expressed genes with KPAN2 in six major cancers. (a) Positive co-expressed genes with KPAN2 in lung adenocarcinoma, (b) negative co-expressed genes with KPAN2 in lung adenocarcinoma, (c) positive co-expressed genes with KPAN2 in lung squamous carcinoma, (d) negative co-expressed genes with KPAN2 in lung squamous carcinoma, (e) positive co-expressed genes with KPAN2 in gastric cancer, (f) negative co-expressed genes with KPAN2 in gastric cancer, (g) positive co-expressed genes with KPAN2 in colorectal cancer, (h) negative-expressed genes with KPAN2 in colorectal cancer, (i) positive co-expressed genes with KPAN2 in breast cancer, (j) negative co-expressed genes with KPAN2 in breast cancer, (k) positive co-expressed genes with KPAN2 in liver hepatic cancer, (l) negative co-expressed genes with KPAN2 in breast cancer, (m) positive co-expressed genes with KPAN2 in bladder cancer, (n) negative co-expressed genes with KPAN2 in bladder cancer.
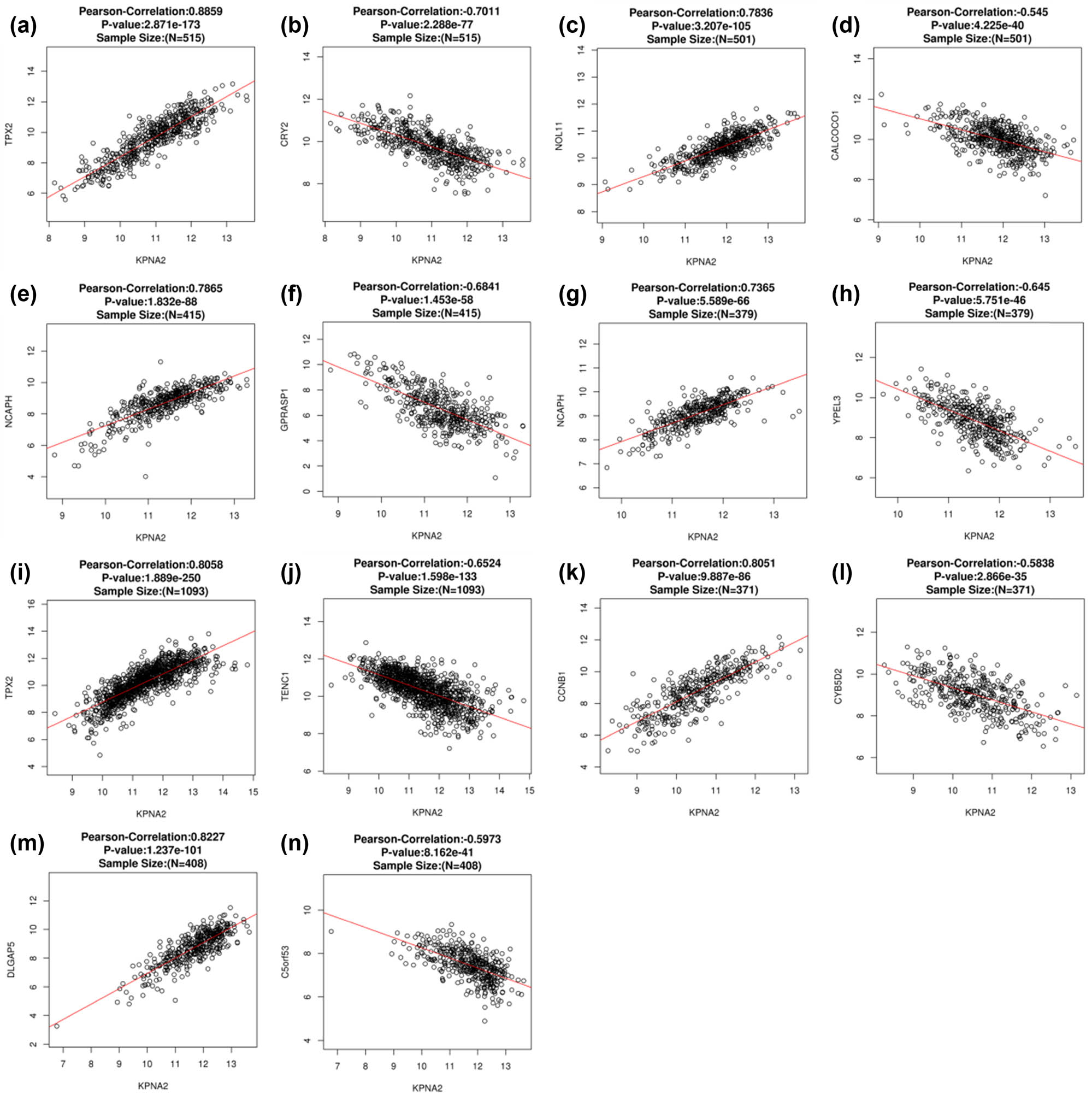
The top positive and negative correlated genes with KPAN2 in six major cancers. (a) Positive co-expressed genes with KPAN2 in lung adenocarcinoma, (b) negative co-expressed genes with KPAN2 in lung adenocarcinoma, (c) positive co-expressed genes with KPAN2 in lung squamous carcinoma, (d) negative co-expressed genes with KPAN2 in lung squamous carcinoma, (e) positive co-expressed genes with KPAN2 in gastric cancer, (f) negative co-expressed genes with KPAN2 in gastric cancer, (g) positive co-expressed genes with KPAN2 in colorectal cancer, (h) negative-expressed genes with KPAN2 in colorectal cancer, (i) positive co-expressed genes with KPAN2 in breast cancer, (j) negative co-expressed genes with KPAN2 in breast cancer, (k) positive co-expressed genes with KPAN2 in liver hepatic cancer, (l) negative co-expressed genes with KPAN2 in breast cancer, (m) positive co-expressed genes with KPAN2 in bladder cancer, (n) negative co-expressed genes with KPAN2 in bladder cancer.
3.5 PPI network of KPNA2
Twenty genes were included in the PPI network with the edges of 105 and local clustering coefficient of 0.713, which indicated that the PPI enrichment was significant with statistical difference (p < 0.001) (Figure 8).
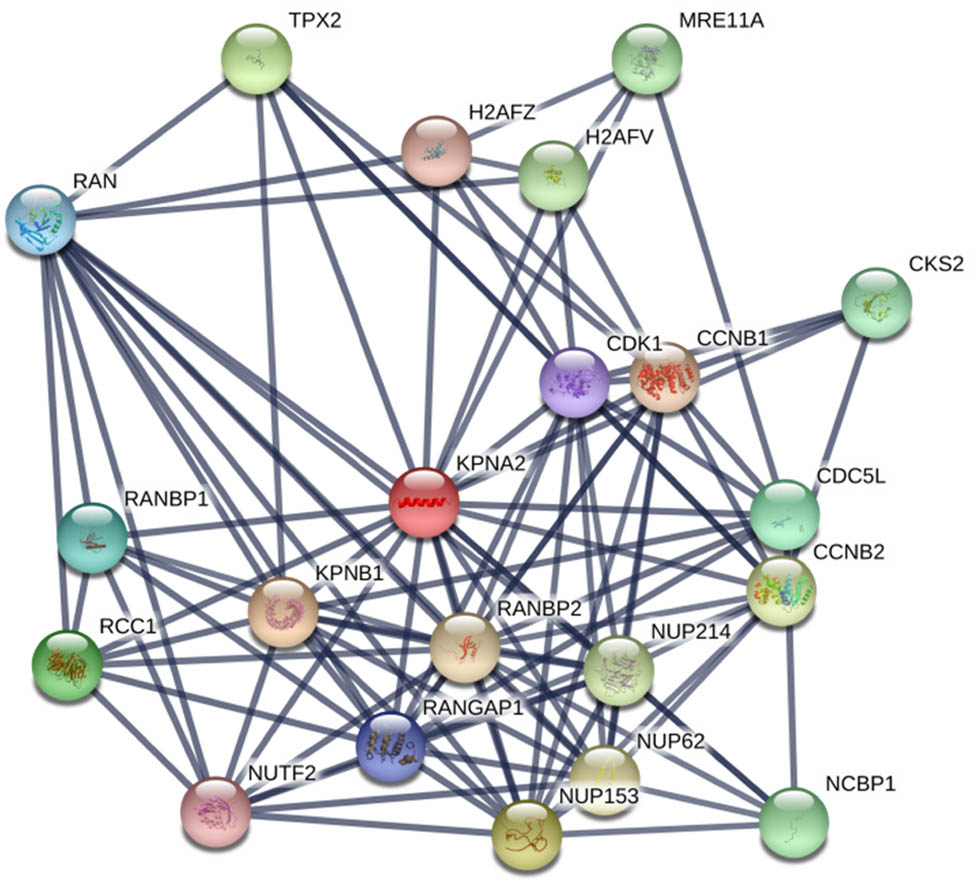
The PPI network included KPNA2 and relevant genes
3.6 KEGG pathway relevant KPNA2
Genes involved in PPI network were mainly enriched in p53 signaling pathway, cell cycle, viral carcinogenesis, Foxo signaling pathway, etc. (Figure 9).
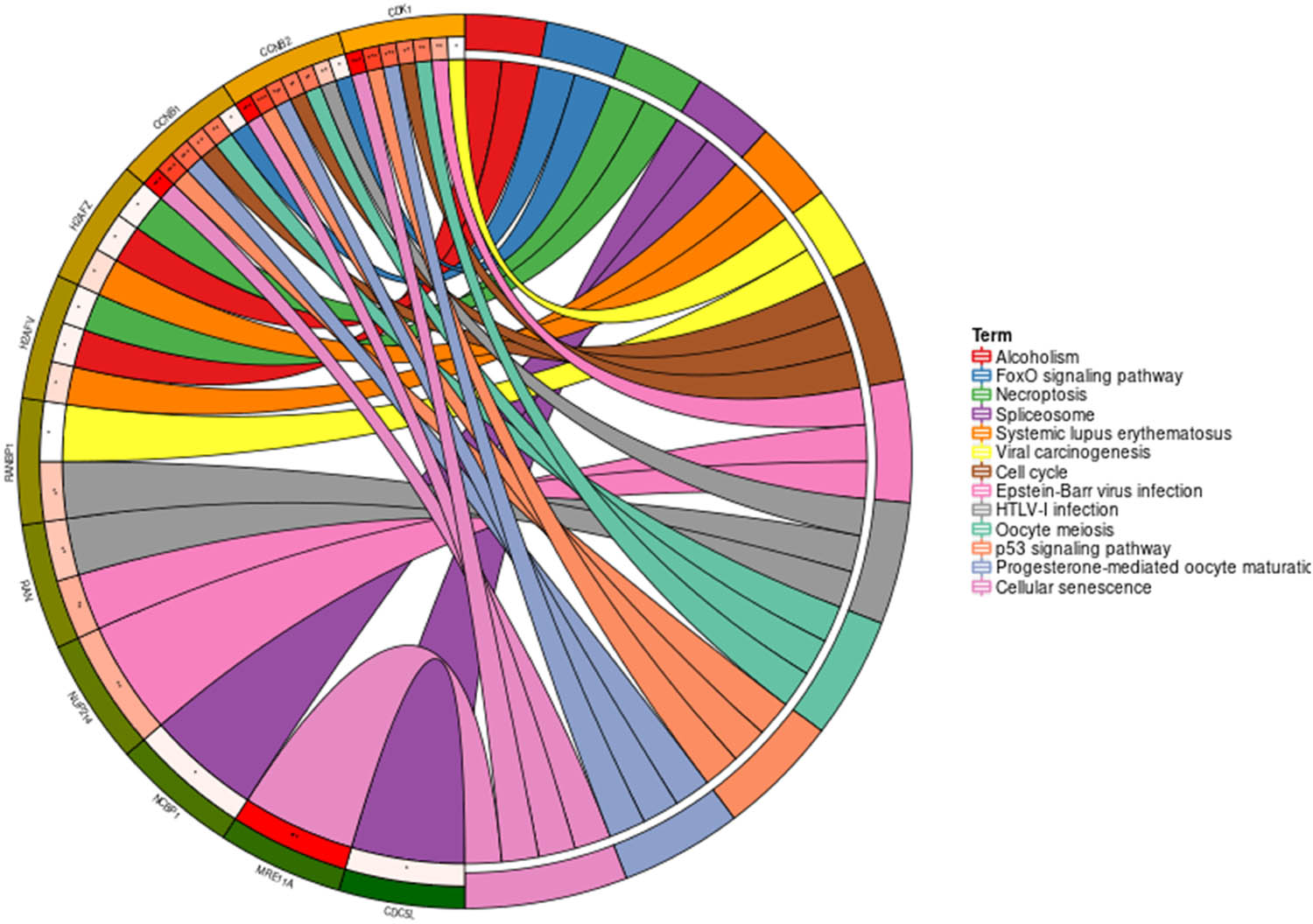
Circus plot of KEGG pathway enrichment of genes that are relevant to KPNA2
3.7 KPNA2 mRNA level and patients’ prognosis
OS was statistically different between KPNA2 mRNA high and low expression groups in NSCLC (HR = 1.2, p < 0.05), colorectal cancer (HR = 0.51, p < 0.01), and liver hepatic carcinoma (HR = 2.1, p < 0.001) (Figure 10). For disease free survival (DFS), the statistical difference was found in gastric cancer (HR = 0.67, p < 0.05) and liver hepatic cancer (HR = 1.9, p < 0.001) (Figure 11).
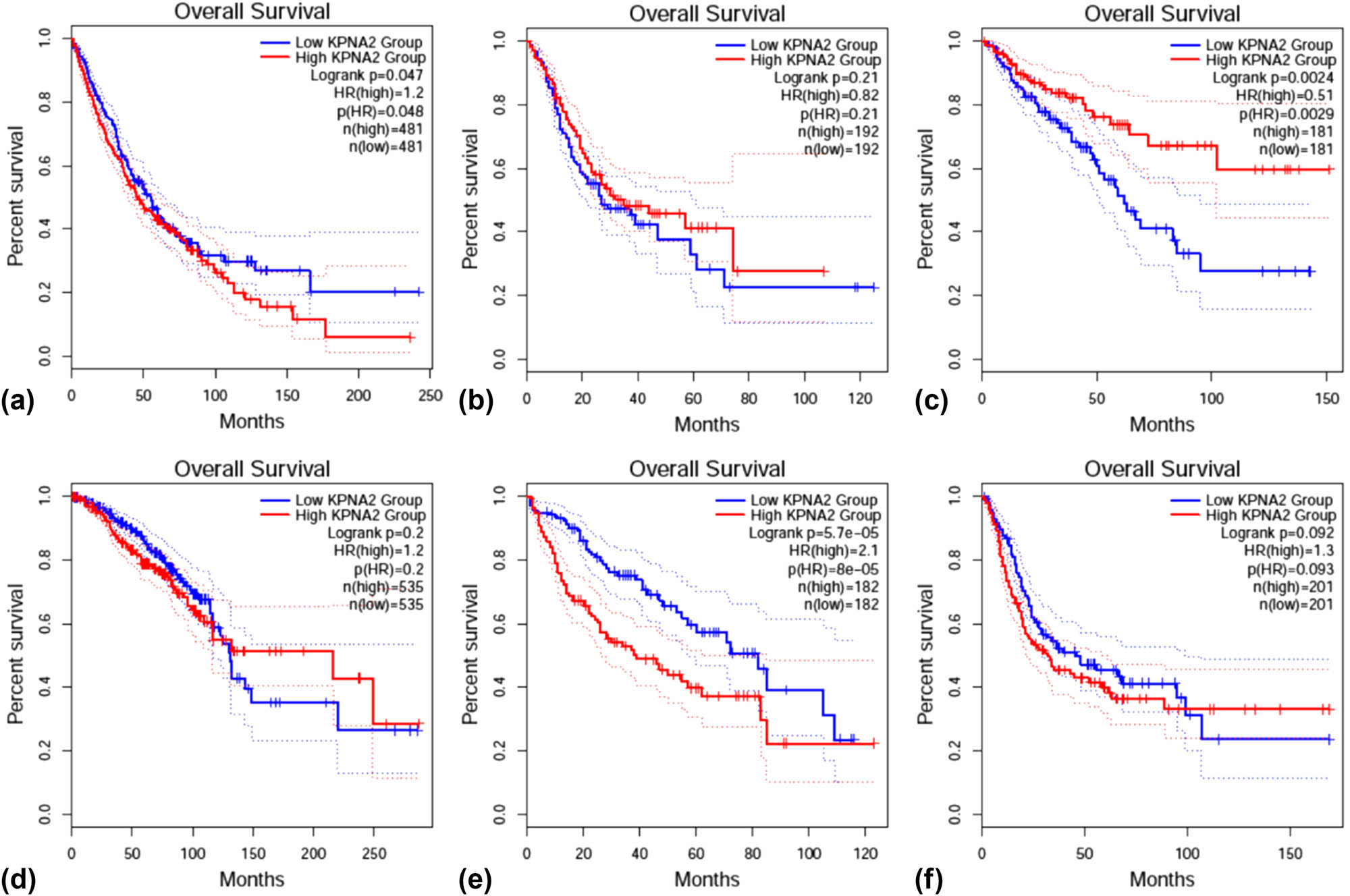
Overall survival (OS) between KPNA2 between high and low expression groups in six major cancers. (a) NSCLC, (b) gastric cancer, (c) colorectal cancer, (d) breast cancer, (e) liver hepatic cancer, (f) bladder cancer.
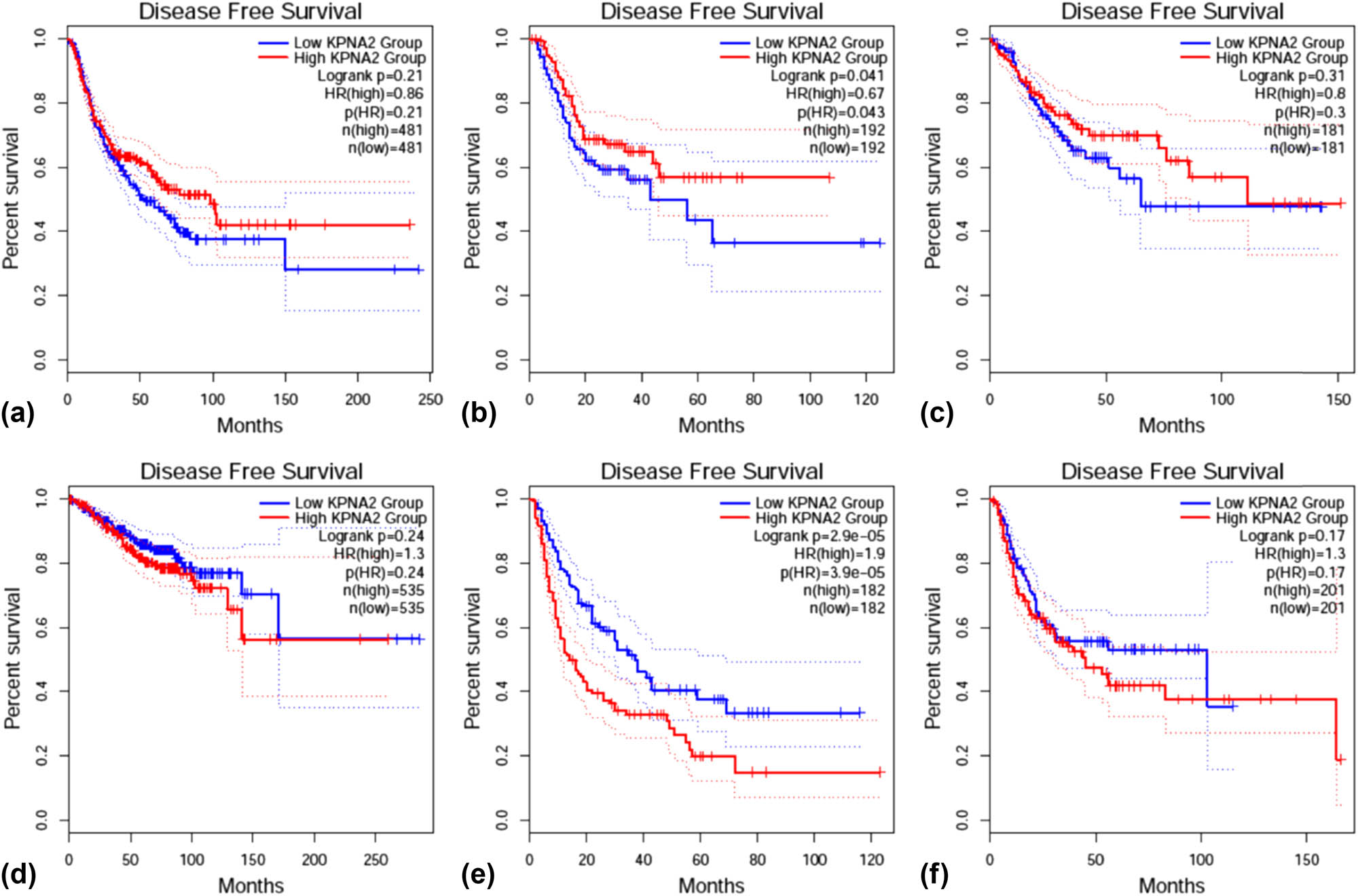
Disease free survival (DFS) between KPNA2 between high and low expression groups in six major cancers. (a) NSCLC, (b) gastric cancer, (c) colorectal cancer, (d) breast cancer, (e) liver hepatic cancer, (f) bladder cancer.
3.8 KPNA2 protein expression
KPNA2 protein was mainly expressed in nucleoplasm and cytosol in cancer cells detected by immunofluorescence assay (Figure 12). Immunohistochemistry assay indicated that KPNA2 protein was also positively expressed in nucleoplasm with Brownish yellow staining (Figure 13).
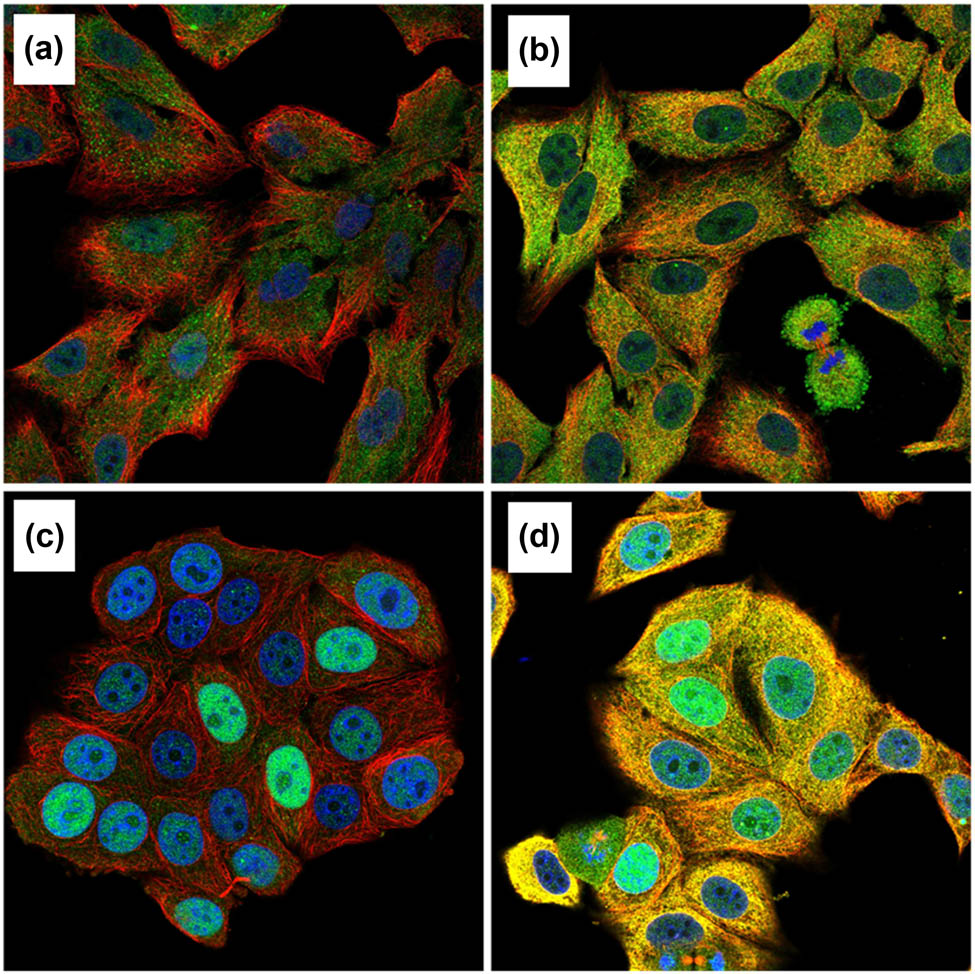
KPNA2 protein was mainly localized to the nucleoplasm and cytosol in cancer cells with blue staining. (a) Immunofluorescent staining of human lung cancer cell line A549 shows localization to nucleoplasm in antibody + nucleus + microtubule channels. (b) Immunofluorescent staining of human lung cancer cell line A549 shows localization to nucleoplasm in antibody + nucleus + microtubule + ER channels. (c) Immunofluorescent staining of human breast cancer cell line MCF7 shows localization to nucleoplasm in antibody + nucleus + microtubule channels. (d) Immunofluorescent staining of human breast cancer cell line MCF7 shows localization to nucleoplasm in antibody + nucleus + microtubule + ER channels.
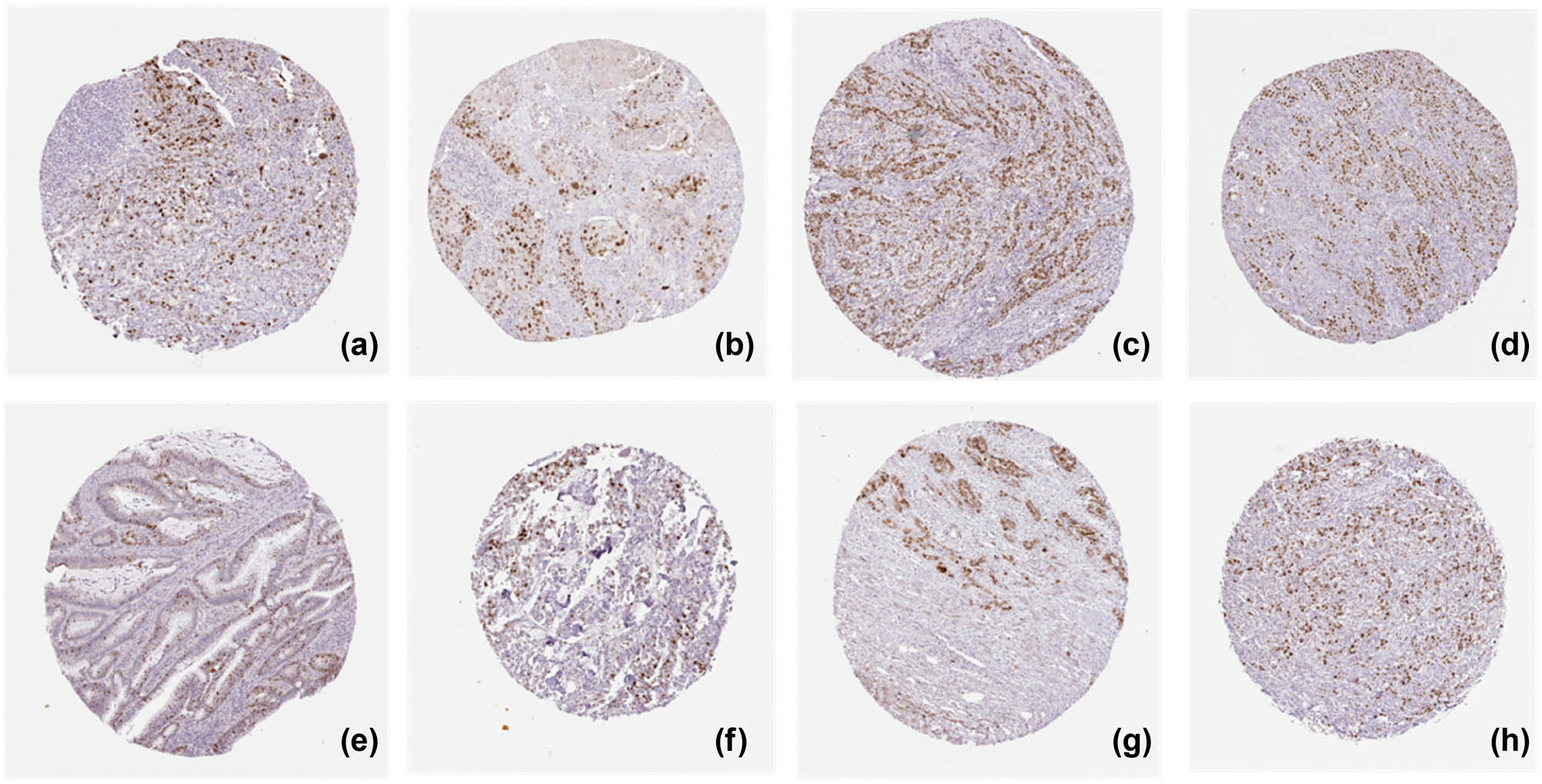
KPNA2 protein positive expression in six major cancers detected by Immunohistochemistry assay. (a) Lung adenocarcinoma, (b) lung squamous cell carcinoma, (c) gastric cancer, (d) colon cancer, (e) rectal cancer, (f) breast cancer, (g) liver hepatic carcinoma, (h) bladder cancer.
4 Discussion
The structural nuclear transporter family of KPNA2 includes the input protein family and the output protein family [18]. It mainly mediates proteins with molecular weight greater than 40 kDa through nuclear pore complexes (NPC). The input protein family includes karyophenin α family and import β family. There are seven members in karyophenin α family, of which karyophenin α 2 (KPNA2) is one of the most important members. KPNA2 gene is located in chromosome 17q23-q24 in human being and its encoded protein contains 529 amino acids, with a molecular weight of 58 kDa [9,19]. The N-terminal is the Importin β binding domain, which has self-inhibition function, so that KPNA2 can only bind to importin at the same time β and cargo molecules can only be translocated to the nucleus [20,21]. The central region is composed of ten arm repeat sequences, including 2 NLS binding sites, which can bind to the nucleoprotein with NLS, and the 10th arm sequence can bind to CAS, which is responsible for kpna2 nucleoplasm recycling; the function of C-terminal is not completely clear yet (Figure 14a).
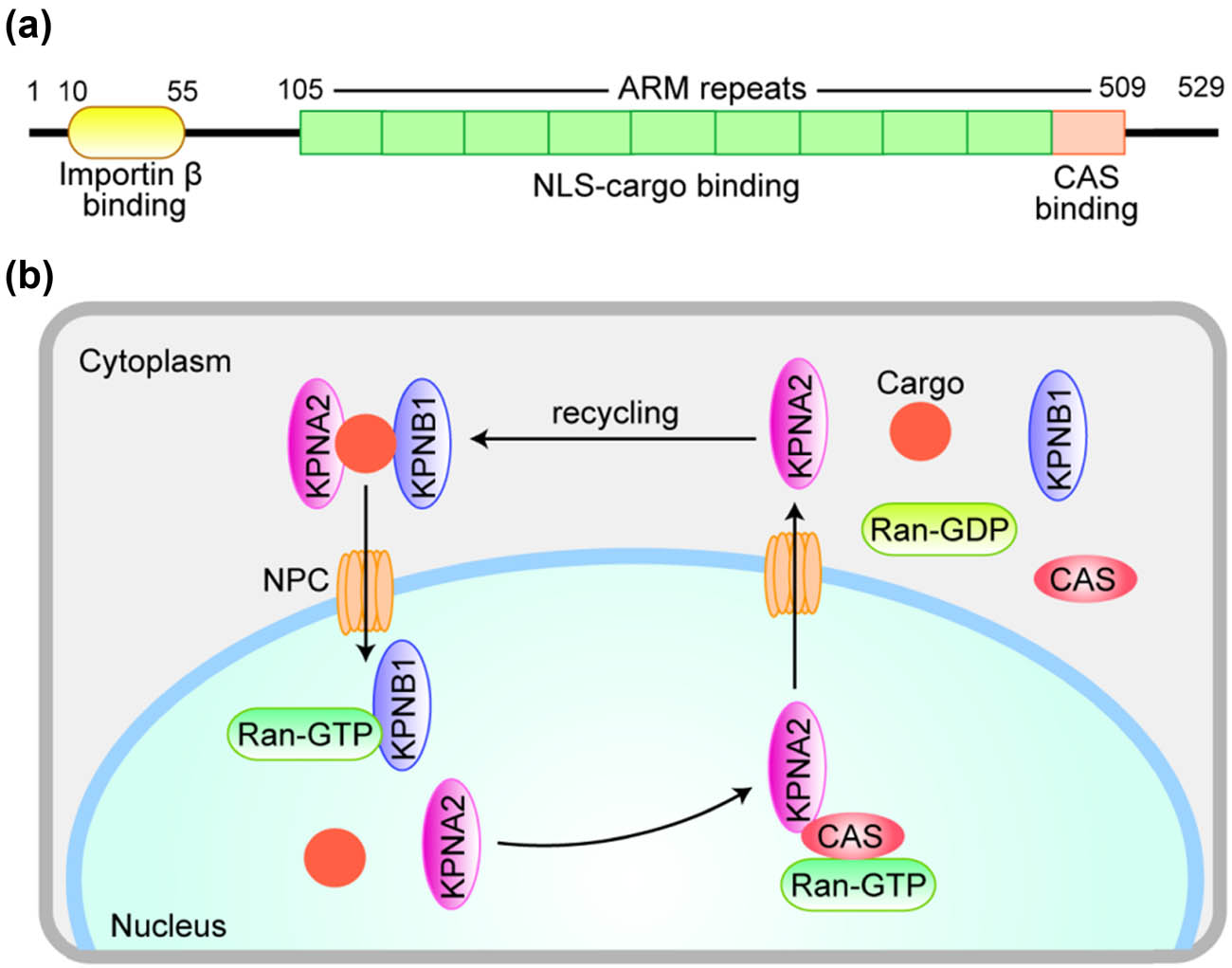
A diagrammatic representation of KPNA2 protein structure (a) and the molecular mechanism of KPNA2 nucleoplasmic recirculation (b). (a) The N-terminus is the Importin β (KPNB1) binding domain, ensuring that KPNA2 can only be translocated into the nucleus while simultaneously combining KPNB1 and cargo protein. The central region consists of 10 armadillo (ARM) repeats, including two NLS-cargo binding sites. The last ARM repeat mediates CAS binding. (b) KPNB1 brings a complex of KPNA2 and Cargo protein into the nucleus via NPC and binds to RanGTP to release KPNA2 and cargo proteins into the nucleus. Then KPNB1 returns directly to the cytoplasm. KNPA2 returns to the cytoplasm with the help of another transporter, CAS, for the next cycle.
The classical nuclear protein input is regulated by heterodimer composed of importin β and karyophenin α. Karyophenin α protein can recognize and bind NLS of cargo protein. Importin β brings the complex composed of karyophenin α and nucleoprotein into the nucleus through NPC and combines with RanGTP to form protein complex in the nucleus, so as to release karyophenin α and nucleoprotein into the nucleus, and then importin β returns directly to the cytoplasm, while karyophenin α returns to the cytoplasm with the help of CAS [22] (Figure 14b).
KPNA2 is a member of the karyopherin family. Given its function in nucleocytoplasmic transport, KPNA2 mediates the translocation of various proteins and is involved in numerous cellular processes, such as cellular differentiation, proliferation, apoptosis, transcriptional regulation, immune response, and viral infection. Recently, several studies have demonstrated that KPNA2 was upregulated in multiple malignancies [23,24,25]. Its aberrant expression was often associated with adverse outcomes in affected patients, indicating that KPNA2 played a significant role in carcinogenesis and tumor progression [23,26,27]. These findings were supported by previous studies, which reported that KPNA2 may have a functional role in the malignant transformation of cells.
As an intranuclear transporter, KPNA2 is involved in cell differentiation, proliferation and apoptosis, transcriptional regulation, immune response, and virus infection. More importantly, many studies have found that Pna2 was involved in tumor progression by regulating the nuclear translocation of tumor-related proteins. It has been reported that: (1) KPNA2 was involved in the nuclear translocation of many tumor-related transcription factors, including E2F1 and pleomorphic adenoma, two members of the zinc finger plag family Gene1, plag1, [28] and lot1 [29] and BTB/POZ transcription factor kaiso [30], etc.; (2) KPNA2 mediates Rac-1 nuclear translocation [31]; Rac-1 participates in tumor formation by participating in cell cycle, cell adhesion, and migration; (3) KPNA2 mediates the entry of cell cycle regulatory protein CHK2 into the nucleus, and overexpression of KPNA2 leads to increased nuclear input of CHK2 [32]; (4) KPNA2 participates in breast cancer suppression – BRCA1 has the function of DNA repair and cell cycle monitoring, which affects the process of tumor formation; (5) KPNA2 participates in the entry of NBS1, which is a kind of DNA repair complex protein and also involved in tumor formation [33].
In our present work, we noticed that KPNA2 mRNA was upregulated in all of the six types of cancers compared with paired normal tissue. KPNA2 mutations, especially missense substitution, were widely identified in six cancers and interact with different genes in different cancer types. Genes involved in PPI network were mainly enriched in p53 signaling pathway, cell cycle, viral carcinogenesis, and Foxo signaling pathway. Immunohistochemistry assay indicated that KPNA2 protein was also positively expressed in nucleoplasm with brownish yellow staining. OS and PFS were generally different between KPNA2 high and low expression groups, which may be a potential biomarker for cancer prognosis. However, there are still limitations for the present work. First, there may be potential heterogeneity across the six cancer types and the results need further validation. Second, the results of the present were mainly based on data mining from relevant databases which should be further validated by local data or experiments.
5 Conclusion
KPNA2, as an intranuclear transport protein, participates in a variety of biological activities through the transport function of nucleoplasm, and its role in tumor development has attracted more and more attention. In view of the abnormal expression of KPNA2 in most cancer tissues and serum of the cancer patients, and related to the proliferation, migration, and invasion of tumor cells, KPNA2 can be used as a potential biomarker for prognosis. However, the mechanism of KPNA2 in tumorigenesis and progression is not completely clear yet and needs further investigation.
-
Ethics approval and consent to participate: Not applicable.
-
Author contributions: Xu C. B. analyzed the data and prepared the manuscript. Liu M. designed the work.
-
Conflict of interest statement: Authors state no conflict of interest.
-
Data availability statement: The analyzed data sets generated during the study are available from the corresponding author on reasonable request. The data that support the findings of this study are available at the relevant databases (https://www.proteinatlas.org/); (http://cbioportal.org); (https://cancer.sanger.ac.uk/cosmic/).
References
[1] Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.10.3322/caac.21492Suche in Google Scholar PubMed
[2] Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.10.3322/caac.21551Suche in Google Scholar PubMed
[3] Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.10.3322/caac.21590Suche in Google Scholar PubMed
[4] Johnson LA, June CH. Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 2017;27:38–58.10.1038/cr.2016.154Suche in Google Scholar PubMed PubMed Central
[5] Zhang KJ, Qian J, Wang SB, Yang Y. Targeting gene-viro-therapy with AFP driving apoptin gene shows potent antitumor effect in hepatocarcinoma. J Biomed Sci. 2012;19:20.10.1186/1423-0127-19-20Suche in Google Scholar PubMed PubMed Central
[6] Cui Y, Zhang C, Ma S, Guo W, Cao W, Guan F. CASC5 is a potential tumour driving gene in lung adenocarcinoma. Cell Biochem Funct. 2020;38(6):733–42.10.1002/cbf.3540Suche in Google Scholar PubMed
[7] Kelley JB, Talley AM, Spencer A, Gioeli D, Paschal BM. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11:63.10.1186/1471-2121-11-63Suche in Google Scholar PubMed PubMed Central
[8] Yasuhara N, Kumar PK. Aptamers that bind specifically to human KPNA2 (importin-α1) and efficiently interfere with nuclear transport. J Biochem. 2016;160:259–68.10.1093/jb/mvw032Suche in Google Scholar PubMed PubMed Central
[9] Dörr SN, Schlicker MN, Hansmann IN. Genomic structure of karyopherin alpha2 ( KPNA2) within a low-copy repeat on chromosome 17q23-q24 and mutation analysis in patients with Russell-Silver syndrome. Hum Genet. 2001;109:479–86.10.1007/s004390100605Suche in Google Scholar PubMed
[10] Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N, et al. A KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Br J Cancer. 2015;112:1929–37.10.1038/bjc.2015.165Suche in Google Scholar PubMed PubMed Central
[11] Dankof A, Fritzsche FR, Dahl E, Pahl S, Wild P, Dietel M, et al. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Arch. 2007;451:877–81.10.1007/s00428-007-0513-5Suche in Google Scholar
[12] Huang L, Zhou Y, Cao XP, Lin JX, Zhang L, Huang ST, et al. KPNA2 is a potential diagnostic serum biomarker for epithelial ovarian cancer and correlates with poor prognosis. Tumour Biol. 2017;39:1010428317706289.10.1177/1010428317706289Suche in Google Scholar
[13] Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6.10.1016/S1476-5586(04)80047-2Suche in Google Scholar
[14] Li QT, Huang ZZ, Chen YB, Yao HY, Ke ZH, He XX, et al. Integrative analysis of Siglec-15 mRNA in human cancers based on data mining. J Cancer. 2020;11:2453–64.10.7150/jca.38747Suche in Google Scholar PubMed PubMed Central
[15] Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–63.10.1093/nar/gkx1090Suche in Google Scholar PubMed PubMed Central
[16] Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13.10.1093/nar/gky1131Suche in Google Scholar PubMed PubMed Central
[17] Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102.10.1093/nar/gkx247Suche in Google Scholar PubMed PubMed Central
[18] Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406.10.1074/mcp.M113.035600Suche in Google Scholar PubMed PubMed Central
[19] Mai M, Qian C, Yokomizo A, Smith DI, Liu W. Cloning of the human homolog of conductin (AXIN2), a gene mapping to chromosome 17q23-q24. Genomics. 1999;55:341–4.10.1006/geno.1998.5650Suche in Google Scholar PubMed
[20] Zu YL, Ai Y, Huang CK. Characterization of an autoinhibitory domain in human mitogen-activated protein kinase-activated protein kinase 2. J Biol Chem. 1995;270:202–6.10.1074/jbc.270.1.202Suche in Google Scholar PubMed
[21] Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–15.10.1016/S0959-440X(01)00264-0Suche in Google Scholar
[22] Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–14.10.1016/j.tcb.2004.07.016Suche in Google Scholar PubMed
[23] Zhou J, Dong D, Cheng R, Wang Y, Jiang S, Zhu Y, et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to OCT4 nuclear transportation in bladder cancer. Oncotarget. 2016;7:72767–76.10.18632/oncotarget.11889Suche in Google Scholar PubMed PubMed Central
[24] Takada T, Tsutsumi S, Takahashi R, Ohsone K, Tatsuki H, Suto T, et al. KPNA2 over-expression is a potential marker of prognosis and therapeutic sensitivity in colorectal cancer patients. J Surg Oncol. 2016;113:213–7.10.1002/jso.24114Suche in Google Scholar PubMed
[25] Li XL, Jia LL, Shi MM, Li X, Li ZH, Li HF, et al. Downregulation of KPNA2 in non-small-cell lung cancer is associated with Oct4 expression. J Transl Med. 2013;11:232.10.1186/1479-5876-11-232Suche in Google Scholar PubMed PubMed Central
[26] Shi B, Su B, Fang D, Tang Y, Xiong G, Guo Z, et al. High expression of KPNA2 defines poor prognosis in patients with upper tract urothelial carcinoma treated with radical nephroureterectomy. BMC Cancer. 2015;15:380.10.1186/s12885-015-1369-8Suche in Google Scholar PubMed PubMed Central
[27] Jiang P, Tang Y, He L, Tang H, Liang M, Mai C, et al. Aberrant expression of nuclear KPNA2 is correlated with early recurrence and poor prognosis in patients with small hepatocellular carcinoma after hepatectomy. Med Oncol. 2014;31:131.10.1007/s12032-014-0131-4Suche in Google Scholar PubMed
[28] Braem CV, Kas K, Meyen E, Debiec-Rychter M, Van De Ven WJ, Voz ML. Identification of a karyopherin alpha 2 recognition site in PLAG1, which functions as a nuclear localization signal. J Biol Chem. 2002;277:19673–8.10.1074/jbc.M112112200Suche in Google Scholar PubMed
[29] Huang SM, Huang SP, Wang SL, Liu PY. Importin alpha1 is involved in the nuclear localization of Zac1 and the induction of p21WAF1/CIP1 by Zac1. Biochem J. 2007;402:359–66.10.1042/BJ20061295Suche in Google Scholar PubMed PubMed Central
[30] Kelly KF, Otchere AA, Graham M, Daniel JM. Nuclear import of the BTB/POZ transcriptional regulator Kaiso. J Cell Sci. 2004;117:6143–52.10.1242/jcs.01541Suche in Google Scholar PubMed
[31] Sandrock K, Bielek H, Schradi K, Schmidt G, Klugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic. 2010;11:198–209.10.1111/j.1600-0854.2009.01015.xSuche in Google Scholar PubMed
[32] Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, et al. Karyopherin-alpha2 protein interacts with Chk2 and contributes to its nuclear import. J Biol Chem. 2003;278:42346–51.10.1074/jbc.M303304200Suche in Google Scholar PubMed
[33] Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J Biol Chem. 2005;280:39594–600.10.1074/jbc.M508425200Suche in Google Scholar PubMed
© 2021 Chaobo Xu and Ming Liu, published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Identification of ZG16B as a prognostic biomarker in breast cancer
- Behçet’s disease with latent Mycobacterium tuberculosis infection
- Erratum
- Erratum to “Suffering from Cerebral Small Vessel Disease with and without Metabolic Syndrome”
- Research Articles
- GPR37 promotes the malignancy of lung adenocarcinoma via TGF-β/Smad pathway
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Additional baricitinib loading dose improves clinical outcome in COVID-19
- The co-treatment of rosuvastatin with dapagliflozin synergistically inhibited apoptosis via activating the PI3K/AKt/mTOR signaling pathway in myocardial ischemia/reperfusion injury rats
- SLC12A8 plays a key role in bladder cancer progression and EMT
- LncRNA ATXN8OS enhances tamoxifen resistance in breast cancer
- Case Report
- Serratia marcescens as a cause of unfavorable outcome in the twin pregnancy
- Spleno-adrenal fusion mimicking an adrenal metastasis of a renal cell carcinoma: A case report and embryological background
- Research Articles
- TRIM25 contributes to the malignancy of acute myeloid leukemia and is negatively regulated by microRNA-137
- CircRNA circ_0004370 promotes cell proliferation, migration, and invasion and inhibits cell apoptosis of esophageal cancer via miR-1301-3p/COL1A1 axis
- LncRNA XIST regulates atherosclerosis progression in ox-LDL-induced HUVECs
- Potential role of IFN-γ and IL-5 in sepsis prediction of preterm neonates
- Rapid Communication
- COVID-19 vaccine: Call for employees in international transportation industries and international travelers as the first priority in global distribution
- Case Report
- Rare squamous cell carcinoma of the kidney with concurrent xanthogranulomatous pyelonephritis: A case report and review of the literature
- An infertile female delivered a baby after removal of primary renal carcinoid tumor
- Research Articles
- Hypertension, BMI, and cardiovascular and cerebrovascular diseases
- Case Report
- Coexistence of bilateral macular edema and pale optic disc in the patient with Cohen syndrome
- Research Articles
- Correlation between kinematic sagittal parameters of the cervical lordosis or head posture and disc degeneration in patients with posterior neck pain
- Review Articles
- Hepatoid adenocarcinoma of the lung: An analysis of the Surveillance, Epidemiology, and End Results (SEER) database
- Research Articles
- Thermography in the diagnosis of carpal tunnel syndrome
- Pemetrexed-based first-line chemotherapy had particularly prominent objective response rate for advanced NSCLC: A network meta-analysis
- Comparison of single and double autologous stem cell transplantation in multiple myeloma patients
- The influence of smoking in minimally invasive spinal fusion surgery
- Impact of body mass index on left atrial dimension in HOCM patients
- Expression and clinical significance of CMTM1 in hepatocellular carcinoma
- miR-142-5p promotes cervical cancer progression by targeting LMX1A through Wnt/β-catenin pathway
- Comparison of multiple flatfoot indicators in 5–8-year-old children
- Early MRI imaging and follow-up study in cerebral amyloid angiopathy
- Intestinal fatty acid-binding protein as a biomarker for the diagnosis of strangulated intestinal obstruction: A meta-analysis
- miR-128-3p inhibits apoptosis and inflammation in LPS-induced sepsis by targeting TGFBR2
- Dynamic perfusion CT – A promising tool to diagnose pancreatic ductal adenocarcinoma
- Biomechanical evaluation of self-cinching stitch techniques in rotator cuff repair: The single-loop and double-loop knot stitches
- Review Articles
- The ambiguous role of mannose-binding lectin (MBL) in human immunity
- Case Report
- Membranous nephropathy with pulmonary cryptococcosis with improved 1-year follow-up results: A case report
- Fertility problems in males carrying an inversion of chromosome 10
- Acute myeloid leukemia with leukemic pleural effusion and high levels of pleural adenosine deaminase: A case report and review of literature
- Metastatic renal Ewing’s sarcoma in adult woman: Case report and review of the literature
- Burkitt-like lymphoma with 11q aberration in a patient with AIDS and a patient without AIDS: Two cases reports and literature review
- Skull hemophilia pseudotumor: A case report
- Judicious use of low-dosage corticosteroids for non-severe COVID-19: A case report
- Adult-onset citrullinaemia type II with liver cirrhosis: A rare cause of hyperammonaemia
- Clinicopathologic features of Good’s syndrome: Two cases and literature review
- Fatal immune-related hepatitis with intrahepatic cholestasis and pneumonia associated with camrelizumab: A case report and literature review
- Research Articles
- Effects of hydroxyethyl starch and gelatin on the risk of acute kidney injury following orthotopic liver transplantation: A multicenter retrospective comparative clinical study
- Significance of nucleic acid positive anal swab in COVID-19 patients
- circAPLP2 promotes colorectal cancer progression by upregulating HELLS by targeting miR-335-5p
- Ratios between circulating myeloid cells and lymphocytes are associated with mortality in severe COVID-19 patients
- Risk factors of left atrial appendage thrombus in patients with non-valvular atrial fibrillation
- Clinical features of hypertensive patients with COVID-19 compared with a normotensive group: Single-center experience in China
- Surgical myocardial revascularization outcomes in Kawasaki disease: systematic review and meta-analysis
- Decreased chromobox homologue 7 expression is associated with epithelial–mesenchymal transition and poor prognosis in cervical cancer
- FGF16 regulated by miR-520b enhances the cell proliferation of lung cancer
- Platelet-rich fibrin: Basics of biological actions and protocol modifications
- Accurate diagnosis of prostate cancer using logistic regression
- miR-377 inhibition enhances the survival of trophoblast cells via upregulation of FNDC5 in gestational diabetes mellitus
- Prognostic significance of TRIM28 expression in patients with breast carcinoma
- Integrative bioinformatics analysis of KPNA2 in six major human cancers
- Exosomal-mediated transfer of OIP5-AS1 enhanced cell chemoresistance to trastuzumab in breast cancer via up-regulating HMGB3 by sponging miR-381-3p
- A four-lncRNA signature for predicting prognosis of recurrence patients with gastric cancer
- Knockdown of circ_0003204 alleviates oxidative low-density lipoprotein-induced human umbilical vein endothelial cells injury: Circulating RNAs could explain atherosclerosis disease progression
- Propofol postpones colorectal cancer development through circ_0026344/miR-645/Akt/mTOR signal pathway
- Knockdown of lncRNA TapSAKI alleviates LPS-induced injury in HK-2 cells through the miR-205/IRF3 pathway
- COVID-19 severity in relation to sociodemographics and vitamin D use
- Clinical analysis of 11 cases of nocardiosis
- Cis-regulatory elements in conserved non-coding sequences of nuclear receptor genes indicate for crosstalk between endocrine systems
- Four long noncoding RNAs act as biomarkers in lung adenocarcinoma
- Real-world evidence of cytomegalovirus reactivation in non-Hodgkin lymphomas treated with bendamustine-containing regimens
- Relation between IL-8 level and obstructive sleep apnea syndrome
- circAGFG1 sponges miR-28-5p to promote non-small-cell lung cancer progression through modulating HIF-1α level
- Nomogram prediction model for renal anaemia in IgA nephropathy patients
- Effect of antibiotic use on the efficacy of nivolumab in the treatment of advanced/metastatic non-small cell lung cancer: A meta-analysis
- NDRG2 inhibition facilitates angiogenesis of hepatocellular carcinoma
- A nomogram for predicting metabolic steatohepatitis: The combination of NAMPT, RALGDS, GADD45B, FOSL2, RTP3, and RASD1
- Clinical and prognostic features of MMP-2 and VEGF in AEG patients
- The value of miR-510 in the prognosis and development of colon cancer
- Functional implications of PABPC1 in the development of ovarian cancer
- Prognostic value of preoperative inflammation-based predictors in patients with bladder carcinoma after radical cystectomy
- Sublingual immunotherapy increases Treg/Th17 ratio in allergic rhinitis
- Prediction of improvement after anterior cruciate ligament reconstruction
- Effluent Osteopontin levels reflect the peritoneal solute transport rate
- circ_0038467 promotes PM2.5-induced bronchial epithelial cell dysfunction
- Significance of miR-141 and miR-340 in cervical squamous cell carcinoma
- Association between hair cortisol concentration and metabolic syndrome
- Microvessel density as a prognostic indicator of prostate cancer: A systematic review and meta-analysis
- Characteristics of BCR–ABL gene variants in patients of chronic myeloid leukemia
- Knee alterations in rheumatoid arthritis: Comparison of US and MRI
- Long non-coding RNA TUG1 aggravates cerebral ischemia and reperfusion injury by sponging miR-493-3p/miR-410-3p
- lncRNA MALAT1 regulated ATAD2 to facilitate retinoblastoma progression via miR-655-3p
- Development and validation of a nomogram for predicting severity in patients with hemorrhagic fever with renal syndrome: A retrospective study
- Analysis of COVID-19 outbreak origin in China in 2019 using differentiation method for unusual epidemiological events
- Laparoscopic versus open major liver resection for hepatocellular carcinoma: A case-matched analysis of short- and long-term outcomes
- Travelers’ vaccines and their adverse events in Nara, Japan
- Association between Tfh and PGA in children with Henoch–Schönlein purpura
- Can exchange transfusion be replaced by double-LED phototherapy?
- circ_0005962 functions as an oncogene to aggravate NSCLC progression
- Circular RNA VANGL1 knockdown suppressed viability, promoted apoptosis, and increased doxorubicin sensitivity through targeting miR-145-5p to regulate SOX4 in bladder cancer cells
- Serum intact fibroblast growth factor 23 in healthy paediatric population
- Algorithm of rational approach to reconstruction in Fournier’s disease
- A meta-analysis of exosome in the treatment of spinal cord injury
- Src-1 and SP2 promote the proliferation and epithelial–mesenchymal transition of nasopharyngeal carcinoma
- Dexmedetomidine may decrease the bupivacaine toxicity to heart
- Hypoxia stimulates the migration and invasion of osteosarcoma via up-regulating the NUSAP1 expression
- Long noncoding RNA XIST knockdown relieves the injury of microglia cells after spinal cord injury by sponging miR-219-5p
- External fixation via the anterior inferior iliac spine for proximal femoral fractures in young patients
- miR-128-3p reduced acute lung injury induced by sepsis via targeting PEL12
- HAGLR promotes neuron differentiation through the miR-130a-3p-MeCP2 axis
- Phosphoglycerate mutase 2 is elevated in serum of patients with heart failure and correlates with the disease severity and patient’s prognosis
- Cell population data in identifying active tuberculosis and community-acquired pneumonia
- Prognostic value of microRNA-4521 in non-small cell lung cancer and its regulatory effect on tumor progression
- Mean platelet volume and red blood cell distribution width is associated with prognosis in premature neonates with sepsis
- 3D-printed porous scaffold promotes osteogenic differentiation of hADMSCs
- Association of gene polymorphisms with women urinary incontinence
- Influence of COVID-19 pandemic on stress levels of urologic patients
- miR-496 inhibits proliferation via LYN and AKT pathway in gastric cancer
- miR-519d downregulates LEP expression to inhibit preeclampsia development
- Comparison of single- and triple-port VATS for lung cancer: A meta-analysis
- Fluorescent light energy modulates healing in skin grafted mouse model
- Silencing CDK6-AS1 inhibits LPS-induced inflammatory damage in HK-2 cells
- Predictive effect of DCE-MRI and DWI in brain metastases from NSCLC
- Severe postoperative hyperbilirubinemia in congenital heart disease
- Baicalin improves podocyte injury in rats with diabetic nephropathy by inhibiting PI3K/Akt/mTOR signaling pathway
- Clinical factors predicting ureteral stent failure in patients with external ureteral compression
- Novel H2S donor proglumide-ADT-OH protects HUVECs from ox-LDL-induced injury through NF-κB and JAK/SATA pathway
- Triple-Endobutton and clavicular hook: A propensity score matching analysis
- Long noncoding RNA MIAT inhibits the progression of diabetic nephropathy and the activation of NF-κB pathway in high glucose-treated renal tubular epithelial cells by the miR-182-5p/GPRC5A axis
- Serum exosomal miR-122-5p, GAS, and PGR in the non-invasive diagnosis of CAG
- miR-513b-5p inhibits the proliferation and promotes apoptosis of retinoblastoma cells by targeting TRIB1
- Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p
- The diagnostic and prognostic value of miR-92a in gastric cancer: A systematic review and meta-analysis
- Prognostic value of α2δ1 in hypopharyngeal carcinoma: A retrospective study
- No significant benefit of moderate-dose vitamin C on severe COVID-19 cases
- circ_0000467 promotes the proliferation, metastasis, and angiogenesis in colorectal cancer cells through regulating KLF12 expression by sponging miR-4766-5p
- Downregulation of RAB7 and Caveolin-1 increases MMP-2 activity in renal tubular epithelial cells under hypoxic conditions
- Educational program for orthopedic surgeons’ influences for osteoporosis
- Expression and function analysis of CRABP2 and FABP5, and their ratio in esophageal squamous cell carcinoma
- GJA1 promotes hepatocellular carcinoma progression by mediating TGF-β-induced activation and the epithelial–mesenchymal transition of hepatic stellate cells
- lncRNA-ZFAS1 promotes the progression of endometrial carcinoma by targeting miR-34b to regulate VEGFA expression
- Anticoagulation is the answer in treating noncritical COVID-19 patients
- Effect of late-onset hemorrhagic cystitis on PFS after haplo-PBSCT
- Comparison of Dako HercepTest and Ventana PATHWAY anti-HER2 (4B5) tests and their correlation with silver in situ hybridization in lung adenocarcinoma
- VSTM1 regulates monocyte/macrophage function via the NF-κB signaling pathway
- Comparison of vaginal birth outcomes in midwifery-led versus physician-led setting: A propensity score-matched analysis
- Treatment of osteoporosis with teriparatide: The Slovenian experience
- New targets of morphine postconditioning protection of the myocardium in ischemia/reperfusion injury: Involvement of HSP90/Akt and C5a/NF-κB
- Superenhancer–transcription factor regulatory network in malignant tumors
- β-Cell function is associated with osteosarcopenia in middle-aged and older nonobese patients with type 2 diabetes: A cross-sectional study
- Clinical features of atypical tuberculosis mimicking bacterial pneumonia
- Proteoglycan-depleted regions of annular injury promote nerve ingrowth in a rabbit disc degeneration model
- Effect of electromagnetic field on abortion: A systematic review and meta-analysis
- miR-150-5p affects AS plaque with ASMC proliferation and migration by STAT1
- MALAT1 promotes malignant pleural mesothelioma by sponging miR-141-3p
- Effects of remifentanil and propofol on distant organ lung injury in an ischemia–reperfusion model
- miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway
- Identification of LIG1 and LIG3 as prognostic biomarkers in breast cancer
- MitoQ inhibits hepatic stellate cell activation and liver fibrosis by enhancing PINK1/parkin-mediated mitophagy
- Dissecting role of founder mutation p.V727M in GNE in Indian HIBM cohort
- circATP2A2 promotes osteosarcoma progression by upregulating MYH9
- Prognostic role of oxytocin receptor in colon adenocarcinoma
- Review Articles
- The function of non-coding RNAs in idiopathic pulmonary fibrosis
- Efficacy and safety of therapeutic plasma exchange in stiff person syndrome
- Role of cesarean section in the development of neonatal gut microbiota: A systematic review
- Small cell lung cancer transformation during antitumor therapies: A systematic review
- Research progress of gut microbiota and frailty syndrome
- Recommendations for outpatient activity in COVID-19 pandemic
- Rapid Communication
- Disparity in clinical characteristics between 2019 novel coronavirus pneumonia and leptospirosis
- Use of microspheres in embolization for unruptured renal angiomyolipomas
- COVID-19 cases with delayed absorption of lung lesion
- A triple combination of treatments on moderate COVID-19
- Social networks and eating disorders during the Covid-19 pandemic
- Letter
- COVID-19, WHO guidelines, pedagogy, and respite
- Inflammatory factors in alveolar lavage fluid from severe COVID-19 pneumonia: PCT and IL-6 in epithelial lining fluid
- COVID-19: Lessons from Norway tragedy must be considered in vaccine rollout planning in least developed/developing countries
- What is the role of plasma cell in the lamina propria of terminal ileum in Good’s syndrome patient?
- Case Report
- Rivaroxaban triggered multifocal intratumoral hemorrhage of the cabozantinib-treated diffuse brain metastases: A case report and review of literature
- CTU findings of duplex kidney in kidney: A rare duplicated renal malformation
- Synchronous primary malignancy of colon cancer and mantle cell lymphoma: A case report
- Sonazoid-enhanced ultrasonography and pathologic characters of CD68 positive cell in primary hepatic perivascular epithelioid cell tumors: A case report and literature review
- Persistent SARS-CoV-2-positive over 4 months in a COVID-19 patient with CHB
- Pulmonary parenchymal involvement caused by Tropheryma whipplei
- Mediastinal mixed germ cell tumor: A case report and literature review
- Ovarian female adnexal tumor of probable Wolffian origin – Case report
- Rare paratesticular aggressive angiomyxoma mimicking an epididymal tumor in an 82-year-old man: Case report
- Perimenopausal giant hydatidiform mole complicated with preeclampsia and hyperthyroidism: A case report and literature review
- Primary orbital ganglioneuroblastoma: A case report
- Primary aortic intimal sarcoma masquerading as intramural hematoma
- Sustained false-positive results for hepatitis A virus immunoglobulin M: A case report and literature review
- Peritoneal loose body presenting as a hepatic mass: A case report and review of the literature
- Chondroblastoma of mandibular condyle: Case report and literature review
- Trauma-induced complete pacemaker lead fracture 8 months prior to hospitalization: A case report
- Primary intradural extramedullary extraosseous Ewing’s sarcoma/peripheral primitive neuroectodermal tumor (PIEES/PNET) of the thoracolumbar spine: A case report and literature review
- Computer-assisted preoperative planning of reduction of and osteosynthesis of scapular fracture: A case report
- High quality of 58-month life in lung cancer patient with brain metastases sequentially treated with gefitinib and osimertinib
- Rapid response of locally advanced oral squamous cell carcinoma to apatinib: A case report
- Retrieval of intrarenal coiled and ruptured guidewire by retrograde intrarenal surgery: A case report and literature review
- Usage of intermingled skin allografts and autografts in a senior patient with major burn injury
- Retraction
- Retraction on “Dihydromyricetin attenuates inflammation through TLR4/NF-kappa B pathway”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part I
- An artificial immune system with bootstrap sampling for the diagnosis of recurrent endometrial cancers
- Breast cancer recurrence prediction with ensemble methods and cost-sensitive learning
Artikel in diesem Heft
- Research Articles
- Identification of ZG16B as a prognostic biomarker in breast cancer
- Behçet’s disease with latent Mycobacterium tuberculosis infection
- Erratum
- Erratum to “Suffering from Cerebral Small Vessel Disease with and without Metabolic Syndrome”
- Research Articles
- GPR37 promotes the malignancy of lung adenocarcinoma via TGF-β/Smad pathway
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Additional baricitinib loading dose improves clinical outcome in COVID-19
- The co-treatment of rosuvastatin with dapagliflozin synergistically inhibited apoptosis via activating the PI3K/AKt/mTOR signaling pathway in myocardial ischemia/reperfusion injury rats
- SLC12A8 plays a key role in bladder cancer progression and EMT
- LncRNA ATXN8OS enhances tamoxifen resistance in breast cancer
- Case Report
- Serratia marcescens as a cause of unfavorable outcome in the twin pregnancy
- Spleno-adrenal fusion mimicking an adrenal metastasis of a renal cell carcinoma: A case report and embryological background
- Research Articles
- TRIM25 contributes to the malignancy of acute myeloid leukemia and is negatively regulated by microRNA-137
- CircRNA circ_0004370 promotes cell proliferation, migration, and invasion and inhibits cell apoptosis of esophageal cancer via miR-1301-3p/COL1A1 axis
- LncRNA XIST regulates atherosclerosis progression in ox-LDL-induced HUVECs
- Potential role of IFN-γ and IL-5 in sepsis prediction of preterm neonates
- Rapid Communication
- COVID-19 vaccine: Call for employees in international transportation industries and international travelers as the first priority in global distribution
- Case Report
- Rare squamous cell carcinoma of the kidney with concurrent xanthogranulomatous pyelonephritis: A case report and review of the literature
- An infertile female delivered a baby after removal of primary renal carcinoid tumor
- Research Articles
- Hypertension, BMI, and cardiovascular and cerebrovascular diseases
- Case Report
- Coexistence of bilateral macular edema and pale optic disc in the patient with Cohen syndrome
- Research Articles
- Correlation between kinematic sagittal parameters of the cervical lordosis or head posture and disc degeneration in patients with posterior neck pain
- Review Articles
- Hepatoid adenocarcinoma of the lung: An analysis of the Surveillance, Epidemiology, and End Results (SEER) database
- Research Articles
- Thermography in the diagnosis of carpal tunnel syndrome
- Pemetrexed-based first-line chemotherapy had particularly prominent objective response rate for advanced NSCLC: A network meta-analysis
- Comparison of single and double autologous stem cell transplantation in multiple myeloma patients
- The influence of smoking in minimally invasive spinal fusion surgery
- Impact of body mass index on left atrial dimension in HOCM patients
- Expression and clinical significance of CMTM1 in hepatocellular carcinoma
- miR-142-5p promotes cervical cancer progression by targeting LMX1A through Wnt/β-catenin pathway
- Comparison of multiple flatfoot indicators in 5–8-year-old children
- Early MRI imaging and follow-up study in cerebral amyloid angiopathy
- Intestinal fatty acid-binding protein as a biomarker for the diagnosis of strangulated intestinal obstruction: A meta-analysis
- miR-128-3p inhibits apoptosis and inflammation in LPS-induced sepsis by targeting TGFBR2
- Dynamic perfusion CT – A promising tool to diagnose pancreatic ductal adenocarcinoma
- Biomechanical evaluation of self-cinching stitch techniques in rotator cuff repair: The single-loop and double-loop knot stitches
- Review Articles
- The ambiguous role of mannose-binding lectin (MBL) in human immunity
- Case Report
- Membranous nephropathy with pulmonary cryptococcosis with improved 1-year follow-up results: A case report
- Fertility problems in males carrying an inversion of chromosome 10
- Acute myeloid leukemia with leukemic pleural effusion and high levels of pleural adenosine deaminase: A case report and review of literature
- Metastatic renal Ewing’s sarcoma in adult woman: Case report and review of the literature
- Burkitt-like lymphoma with 11q aberration in a patient with AIDS and a patient without AIDS: Two cases reports and literature review
- Skull hemophilia pseudotumor: A case report
- Judicious use of low-dosage corticosteroids for non-severe COVID-19: A case report
- Adult-onset citrullinaemia type II with liver cirrhosis: A rare cause of hyperammonaemia
- Clinicopathologic features of Good’s syndrome: Two cases and literature review
- Fatal immune-related hepatitis with intrahepatic cholestasis and pneumonia associated with camrelizumab: A case report and literature review
- Research Articles
- Effects of hydroxyethyl starch and gelatin on the risk of acute kidney injury following orthotopic liver transplantation: A multicenter retrospective comparative clinical study
- Significance of nucleic acid positive anal swab in COVID-19 patients
- circAPLP2 promotes colorectal cancer progression by upregulating HELLS by targeting miR-335-5p
- Ratios between circulating myeloid cells and lymphocytes are associated with mortality in severe COVID-19 patients
- Risk factors of left atrial appendage thrombus in patients with non-valvular atrial fibrillation
- Clinical features of hypertensive patients with COVID-19 compared with a normotensive group: Single-center experience in China
- Surgical myocardial revascularization outcomes in Kawasaki disease: systematic review and meta-analysis
- Decreased chromobox homologue 7 expression is associated with epithelial–mesenchymal transition and poor prognosis in cervical cancer
- FGF16 regulated by miR-520b enhances the cell proliferation of lung cancer
- Platelet-rich fibrin: Basics of biological actions and protocol modifications
- Accurate diagnosis of prostate cancer using logistic regression
- miR-377 inhibition enhances the survival of trophoblast cells via upregulation of FNDC5 in gestational diabetes mellitus
- Prognostic significance of TRIM28 expression in patients with breast carcinoma
- Integrative bioinformatics analysis of KPNA2 in six major human cancers
- Exosomal-mediated transfer of OIP5-AS1 enhanced cell chemoresistance to trastuzumab in breast cancer via up-regulating HMGB3 by sponging miR-381-3p
- A four-lncRNA signature for predicting prognosis of recurrence patients with gastric cancer
- Knockdown of circ_0003204 alleviates oxidative low-density lipoprotein-induced human umbilical vein endothelial cells injury: Circulating RNAs could explain atherosclerosis disease progression
- Propofol postpones colorectal cancer development through circ_0026344/miR-645/Akt/mTOR signal pathway
- Knockdown of lncRNA TapSAKI alleviates LPS-induced injury in HK-2 cells through the miR-205/IRF3 pathway
- COVID-19 severity in relation to sociodemographics and vitamin D use
- Clinical analysis of 11 cases of nocardiosis
- Cis-regulatory elements in conserved non-coding sequences of nuclear receptor genes indicate for crosstalk between endocrine systems
- Four long noncoding RNAs act as biomarkers in lung adenocarcinoma
- Real-world evidence of cytomegalovirus reactivation in non-Hodgkin lymphomas treated with bendamustine-containing regimens
- Relation between IL-8 level and obstructive sleep apnea syndrome
- circAGFG1 sponges miR-28-5p to promote non-small-cell lung cancer progression through modulating HIF-1α level
- Nomogram prediction model for renal anaemia in IgA nephropathy patients
- Effect of antibiotic use on the efficacy of nivolumab in the treatment of advanced/metastatic non-small cell lung cancer: A meta-analysis
- NDRG2 inhibition facilitates angiogenesis of hepatocellular carcinoma
- A nomogram for predicting metabolic steatohepatitis: The combination of NAMPT, RALGDS, GADD45B, FOSL2, RTP3, and RASD1
- Clinical and prognostic features of MMP-2 and VEGF in AEG patients
- The value of miR-510 in the prognosis and development of colon cancer
- Functional implications of PABPC1 in the development of ovarian cancer
- Prognostic value of preoperative inflammation-based predictors in patients with bladder carcinoma after radical cystectomy
- Sublingual immunotherapy increases Treg/Th17 ratio in allergic rhinitis
- Prediction of improvement after anterior cruciate ligament reconstruction
- Effluent Osteopontin levels reflect the peritoneal solute transport rate
- circ_0038467 promotes PM2.5-induced bronchial epithelial cell dysfunction
- Significance of miR-141 and miR-340 in cervical squamous cell carcinoma
- Association between hair cortisol concentration and metabolic syndrome
- Microvessel density as a prognostic indicator of prostate cancer: A systematic review and meta-analysis
- Characteristics of BCR–ABL gene variants in patients of chronic myeloid leukemia
- Knee alterations in rheumatoid arthritis: Comparison of US and MRI
- Long non-coding RNA TUG1 aggravates cerebral ischemia and reperfusion injury by sponging miR-493-3p/miR-410-3p
- lncRNA MALAT1 regulated ATAD2 to facilitate retinoblastoma progression via miR-655-3p
- Development and validation of a nomogram for predicting severity in patients with hemorrhagic fever with renal syndrome: A retrospective study
- Analysis of COVID-19 outbreak origin in China in 2019 using differentiation method for unusual epidemiological events
- Laparoscopic versus open major liver resection for hepatocellular carcinoma: A case-matched analysis of short- and long-term outcomes
- Travelers’ vaccines and their adverse events in Nara, Japan
- Association between Tfh and PGA in children with Henoch–Schönlein purpura
- Can exchange transfusion be replaced by double-LED phototherapy?
- circ_0005962 functions as an oncogene to aggravate NSCLC progression
- Circular RNA VANGL1 knockdown suppressed viability, promoted apoptosis, and increased doxorubicin sensitivity through targeting miR-145-5p to regulate SOX4 in bladder cancer cells
- Serum intact fibroblast growth factor 23 in healthy paediatric population
- Algorithm of rational approach to reconstruction in Fournier’s disease
- A meta-analysis of exosome in the treatment of spinal cord injury
- Src-1 and SP2 promote the proliferation and epithelial–mesenchymal transition of nasopharyngeal carcinoma
- Dexmedetomidine may decrease the bupivacaine toxicity to heart
- Hypoxia stimulates the migration and invasion of osteosarcoma via up-regulating the NUSAP1 expression
- Long noncoding RNA XIST knockdown relieves the injury of microglia cells after spinal cord injury by sponging miR-219-5p
- External fixation via the anterior inferior iliac spine for proximal femoral fractures in young patients
- miR-128-3p reduced acute lung injury induced by sepsis via targeting PEL12
- HAGLR promotes neuron differentiation through the miR-130a-3p-MeCP2 axis
- Phosphoglycerate mutase 2 is elevated in serum of patients with heart failure and correlates with the disease severity and patient’s prognosis
- Cell population data in identifying active tuberculosis and community-acquired pneumonia
- Prognostic value of microRNA-4521 in non-small cell lung cancer and its regulatory effect on tumor progression
- Mean platelet volume and red blood cell distribution width is associated with prognosis in premature neonates with sepsis
- 3D-printed porous scaffold promotes osteogenic differentiation of hADMSCs
- Association of gene polymorphisms with women urinary incontinence
- Influence of COVID-19 pandemic on stress levels of urologic patients
- miR-496 inhibits proliferation via LYN and AKT pathway in gastric cancer
- miR-519d downregulates LEP expression to inhibit preeclampsia development
- Comparison of single- and triple-port VATS for lung cancer: A meta-analysis
- Fluorescent light energy modulates healing in skin grafted mouse model
- Silencing CDK6-AS1 inhibits LPS-induced inflammatory damage in HK-2 cells
- Predictive effect of DCE-MRI and DWI in brain metastases from NSCLC
- Severe postoperative hyperbilirubinemia in congenital heart disease
- Baicalin improves podocyte injury in rats with diabetic nephropathy by inhibiting PI3K/Akt/mTOR signaling pathway
- Clinical factors predicting ureteral stent failure in patients with external ureteral compression
- Novel H2S donor proglumide-ADT-OH protects HUVECs from ox-LDL-induced injury through NF-κB and JAK/SATA pathway
- Triple-Endobutton and clavicular hook: A propensity score matching analysis
- Long noncoding RNA MIAT inhibits the progression of diabetic nephropathy and the activation of NF-κB pathway in high glucose-treated renal tubular epithelial cells by the miR-182-5p/GPRC5A axis
- Serum exosomal miR-122-5p, GAS, and PGR in the non-invasive diagnosis of CAG
- miR-513b-5p inhibits the proliferation and promotes apoptosis of retinoblastoma cells by targeting TRIB1
- Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p
- The diagnostic and prognostic value of miR-92a in gastric cancer: A systematic review and meta-analysis
- Prognostic value of α2δ1 in hypopharyngeal carcinoma: A retrospective study
- No significant benefit of moderate-dose vitamin C on severe COVID-19 cases
- circ_0000467 promotes the proliferation, metastasis, and angiogenesis in colorectal cancer cells through regulating KLF12 expression by sponging miR-4766-5p
- Downregulation of RAB7 and Caveolin-1 increases MMP-2 activity in renal tubular epithelial cells under hypoxic conditions
- Educational program for orthopedic surgeons’ influences for osteoporosis
- Expression and function analysis of CRABP2 and FABP5, and their ratio in esophageal squamous cell carcinoma
- GJA1 promotes hepatocellular carcinoma progression by mediating TGF-β-induced activation and the epithelial–mesenchymal transition of hepatic stellate cells
- lncRNA-ZFAS1 promotes the progression of endometrial carcinoma by targeting miR-34b to regulate VEGFA expression
- Anticoagulation is the answer in treating noncritical COVID-19 patients
- Effect of late-onset hemorrhagic cystitis on PFS after haplo-PBSCT
- Comparison of Dako HercepTest and Ventana PATHWAY anti-HER2 (4B5) tests and their correlation with silver in situ hybridization in lung adenocarcinoma
- VSTM1 regulates monocyte/macrophage function via the NF-κB signaling pathway
- Comparison of vaginal birth outcomes in midwifery-led versus physician-led setting: A propensity score-matched analysis
- Treatment of osteoporosis with teriparatide: The Slovenian experience
- New targets of morphine postconditioning protection of the myocardium in ischemia/reperfusion injury: Involvement of HSP90/Akt and C5a/NF-κB
- Superenhancer–transcription factor regulatory network in malignant tumors
- β-Cell function is associated with osteosarcopenia in middle-aged and older nonobese patients with type 2 diabetes: A cross-sectional study
- Clinical features of atypical tuberculosis mimicking bacterial pneumonia
- Proteoglycan-depleted regions of annular injury promote nerve ingrowth in a rabbit disc degeneration model
- Effect of electromagnetic field on abortion: A systematic review and meta-analysis
- miR-150-5p affects AS plaque with ASMC proliferation and migration by STAT1
- MALAT1 promotes malignant pleural mesothelioma by sponging miR-141-3p
- Effects of remifentanil and propofol on distant organ lung injury in an ischemia–reperfusion model
- miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway
- Identification of LIG1 and LIG3 as prognostic biomarkers in breast cancer
- MitoQ inhibits hepatic stellate cell activation and liver fibrosis by enhancing PINK1/parkin-mediated mitophagy
- Dissecting role of founder mutation p.V727M in GNE in Indian HIBM cohort
- circATP2A2 promotes osteosarcoma progression by upregulating MYH9
- Prognostic role of oxytocin receptor in colon adenocarcinoma
- Review Articles
- The function of non-coding RNAs in idiopathic pulmonary fibrosis
- Efficacy and safety of therapeutic plasma exchange in stiff person syndrome
- Role of cesarean section in the development of neonatal gut microbiota: A systematic review
- Small cell lung cancer transformation during antitumor therapies: A systematic review
- Research progress of gut microbiota and frailty syndrome
- Recommendations for outpatient activity in COVID-19 pandemic
- Rapid Communication
- Disparity in clinical characteristics between 2019 novel coronavirus pneumonia and leptospirosis
- Use of microspheres in embolization for unruptured renal angiomyolipomas
- COVID-19 cases with delayed absorption of lung lesion
- A triple combination of treatments on moderate COVID-19
- Social networks and eating disorders during the Covid-19 pandemic
- Letter
- COVID-19, WHO guidelines, pedagogy, and respite
- Inflammatory factors in alveolar lavage fluid from severe COVID-19 pneumonia: PCT and IL-6 in epithelial lining fluid
- COVID-19: Lessons from Norway tragedy must be considered in vaccine rollout planning in least developed/developing countries
- What is the role of plasma cell in the lamina propria of terminal ileum in Good’s syndrome patient?
- Case Report
- Rivaroxaban triggered multifocal intratumoral hemorrhage of the cabozantinib-treated diffuse brain metastases: A case report and review of literature
- CTU findings of duplex kidney in kidney: A rare duplicated renal malformation
- Synchronous primary malignancy of colon cancer and mantle cell lymphoma: A case report
- Sonazoid-enhanced ultrasonography and pathologic characters of CD68 positive cell in primary hepatic perivascular epithelioid cell tumors: A case report and literature review
- Persistent SARS-CoV-2-positive over 4 months in a COVID-19 patient with CHB
- Pulmonary parenchymal involvement caused by Tropheryma whipplei
- Mediastinal mixed germ cell tumor: A case report and literature review
- Ovarian female adnexal tumor of probable Wolffian origin – Case report
- Rare paratesticular aggressive angiomyxoma mimicking an epididymal tumor in an 82-year-old man: Case report
- Perimenopausal giant hydatidiform mole complicated with preeclampsia and hyperthyroidism: A case report and literature review
- Primary orbital ganglioneuroblastoma: A case report
- Primary aortic intimal sarcoma masquerading as intramural hematoma
- Sustained false-positive results for hepatitis A virus immunoglobulin M: A case report and literature review
- Peritoneal loose body presenting as a hepatic mass: A case report and review of the literature
- Chondroblastoma of mandibular condyle: Case report and literature review
- Trauma-induced complete pacemaker lead fracture 8 months prior to hospitalization: A case report
- Primary intradural extramedullary extraosseous Ewing’s sarcoma/peripheral primitive neuroectodermal tumor (PIEES/PNET) of the thoracolumbar spine: A case report and literature review
- Computer-assisted preoperative planning of reduction of and osteosynthesis of scapular fracture: A case report
- High quality of 58-month life in lung cancer patient with brain metastases sequentially treated with gefitinib and osimertinib
- Rapid response of locally advanced oral squamous cell carcinoma to apatinib: A case report
- Retrieval of intrarenal coiled and ruptured guidewire by retrograde intrarenal surgery: A case report and literature review
- Usage of intermingled skin allografts and autografts in a senior patient with major burn injury
- Retraction
- Retraction on “Dihydromyricetin attenuates inflammation through TLR4/NF-kappa B pathway”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part I
- An artificial immune system with bootstrap sampling for the diagnosis of recurrent endometrial cancers
- Breast cancer recurrence prediction with ensemble methods and cost-sensitive learning

