Abstract
Background
Colorectal cancer (CRC) is one of the deadliest cancers in the world. Increasing evidence suggests that circular RNAs (circRNAs) are implicated in CRC pathogenesis. This study aimed to determine the role of circAPLP2 and explore a potential mechanism of circAPLP2 action in CRC.
Methods
The expression of circAPLP2, miR-335-5p and helicase lymphoid-specific (HELLS) mRNA in CRC tissues and cells was measured by quantitative real-time polymerase chain reaction (qPCR). The functional effects of circAPLP2 on cell cycle progression/cell apoptosis, colony formation, cell migration, invasion and glycolysis metabolism were investigated by flow cytometry assay, colony formation assay, wound healing assay, transwell assay and glycolysis stress test. Glycolysis metabolism was also assessed by the levels of glucose uptake and lactate production. The protein levels of HELLS and HK2 were detected by western blot. The interaction between circAPLP2 and miR-335-5p, or miR-335-5p and HELLS was verified by dual-luciferase reporter assay. The role of circAPLP2 on solid tumor growth in nude mice was investigated.
Results
circAPLP2 and HELLS were overexpressed, but miR-335-5p was downregulated in CRC tissues and cells. Functional analyses showed that circAPLP2 knockdown suppressed CRC cell cycle progression, colony formation, migration, invasion and glycolysis metabolism, induced cell apoptosis and blocked solid tumor growth in nude mice. Moreover, miR-335-5p was a target of circAPLP2, and miR-335-5p could also bind to HELLS. Rescue experiments presented that miR-335-5p inhibition reversed the effects of circAPLP2 knockdown, and HELLS overexpression abolished the role of miR-335-5p restoration. Importantly, circAPLP2 could positively regulate HELLS expression by mediating miR-335-5p.
Conclusion
circAPLP2 triggered CRC malignant development by increasing HELLS expression via targeting miR-335-5p, which might be a novel strategy to understand and treat CRC.
1 Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related death, second only to lung cancer, liver cancer and stomach cancer [1]. In gender, CRC is the second common cancer in females and the third most common cancer in males[2]. Most CRC cases are found in Western countries (55%), but due to the rapid development of other countries in the past, this trend is changing, making CRC a problem of globalization [3,4]. Mutations in specific genes contribute to CRC, just like other types of cancer. These mutations can appear in oncogenes, tumor suppressor genes and genes related to DNA repair mechanisms [1]. Recently, the dysregulation of circular RNAs (circRNAs) in cancers has aroused much concern [5]. The exploration in the function of CRC-specific circRNAs will help to further understand the pathogenesis of CRC.
circRNAs are a cluster of non-coding RNAs (ncRNAs) that harbor covalently closed-loop structures. Numerous circRNAs originate from the exon region of protein-coding genes, in the process of “back-splicing” [6,7]. Compared with linear RNAs, circRNAs are more stable because of the lack of 5′ caps and 3′ ends, and circRNAs are gradually acknowledged to be novel biomarkers in cancer [5]. circRNAs are shown to be differently expressed in a tissue-specific manner in tumors. For example, certain circRNAs were identified to be significantly upregulated or downregulated in CRC tissues compared with adjacent normal tissues by RNA-sequencing technology [8,9]. Increasing studies manifested that circRNAs participated in CRC development, functioning as oncogenes or tumor suppressors, such as circ_0079993 and circ_0026344 [10,11]. As for circAPLP2, RNA-sequencing data suggested that it was highly expressed in CRC tissues [12]. However, its function is not fully disclosed.
circRNAs regulate gene expression in various ways, acting as microRNA (miRNA) sponges, RNA-binding protein sponges or regulators of transcription and translation [13,14,15]. miRNAs are well-known to be crucial for the initiation, progression and dissemination of human cancers, and are defined as treasures for cancer diagnosis and therapy [16]. The interaction between circRNA and miRNA has been identified in several studies. For example, miR-335-5p was a target of circZMYM2, and circZMYM2 targeted miR-335-5p to promote pancreatic cancer development [17]. It is still unknown whether miR-335-5p can be targeted by circTLK1 and whether miR-335-5p is involved in the circTLK1 regulatory pathway in CRC.
Helicase lymphoid-specific (HELLS) is shown to be upregulated in CRC tissues in Gene Expression Profiling Interactive Analysis (GEPIA) database, hinting that HELLS may play functions in CRC development. Given that miRNAs regulate gene expression by binding to their 3′ untranslated region (3′UTR) [18], bioinformatics analysis presents that miR-335-5p can bind to HELLS 3′UTR. It is worth exploring whether miR-335-5p is involved in CRC progression through the suppression of HELLS.
In this study, we not only investigated the expression of circAPLP2 in CRC tissues and cells, but also explored the functions of circAPLP2 on the development of CRC both in vitro and in vivo. Besides, we provided a potential mechanism of circAPLP2 action in CRC associated with miR-335-5p and HELLS, aiming to further understand the progression of CRC from the perspective of circAPLP2.
2 Materials and methods
2.1 Clinical samples
CRC tissues (n = 33) and matched normal tissues (n = 33) were collected from CRC patients recruited from West Hospital of Zibo Central Hospital. The use of these tissues was approved by these subjects with the written informed consent. All samples frozen by liquid nitrogen were stored at −80°C freezer. This study was conducted with the approval of the Ethics Committee of West Hospital of Zibo Central Hospital.
2.2 Cell lines and cell culture
CRC cell lines, including SW480, HCT116, SW620, LOVO and HT29, and normal intestinal epithelial cells (HIEC6) were purchased from Bena Culture Collection (Beijing, China). According to the guideline, SW480, SW620 and HIEC6 cells were cultured in 90% DMEM (Bena Culture Collection) containing 10% FBS, and HCT116 cells were cultured in 90% RPMI-1640 (Bena Culture Collection) containing 10% FBS. LOVO cells were cultured in 90% F-12K medium (Bena Culture Collection) containing 10% FBS, and HT29 cells were cultured in 90% McCoy’s 5a medium (Bena Culture Collection) containing 10% FBS. These cells were placed in a 37°C incubator supplemented with 5% CO2 for cell culture.
2.3 Quantitative real-time polymerase chain reaction (qPCR)
First, total RNA was isolated using the Total RNA Extraction Kit (Novland BioPharma; Shanghai, China). The reverse transcription of circRNA and mRNA was performed using One Step Reverse Transcription PCR Kit (Novland BioPharma), and the reverse transcription of miRNA was performed using One Step Stemaim-it miR Reverse Transcription Quantitation Kit (Novland BioPharma). QPCR reactions were carried out using the SYBR Green Mix (Novland BioPharma) under a CFX 96-touch Real-Time PCR System (Bio-Rad; Hercules, CA, USA). GAPDH or U6 was used as the normalization control, and the relative expression was calculated using the 2−ΔΔCt method. The sequences of used primers were listed as follows:
circAPLP2, F: 5′-CTTTCAAGTGTCTCGGCTCT-3′ and R: 5′-TCTTTTGTTTCAAAGCAGCTCT-3′; APLP2, F: 5′-GCAACCGAATGGACAGGGTA-3′ and R: 5′-TCAGAGTCTGCCTCTCTGCT-3′; HELLS, F: 5′-GCCATGAGAGACCGAAATGC-3′ and R: 5′-ACTCCCTGATTAGACGGCAC-3′; GAPDH, F: 5′-CAATGACCCCTTCATTGACC-3′ and R: 5′-GACAAGCTTCCCGTTCTCAG-3′; miR-335-5p, F: 5′-TCAAGAGCAATAACGAAAAATGT-3′ and R: 5′-GCTGTCAACGATACGCTACGT-3′; U6, F: 5′-GGTCGGGCAGGAAAGAGGGC-3′ and R: 5′-GCTAATCTTCTCTGTATCGTTCC-3′.
2.4 RNase R treatment
Total RNA isolated from SW480 and HCT116 cells were exposed to RNase R (1 U/μg; Epicentre; Madison, WI, USA) at 37°C for 30 min. Then, the expression of circAPLP2 and linear APLP2 was detected by qPCR as mentioned above.
2.5 Cell transfection
We used small interference RNA (siRNA) to mediate circAPLP2 knockdown, and siRNA targeting circAPLP2 (si-circAPLP2) and siRNA negative control (si-NC) were assembled by Genepharma (Shanghai, China). Besides, miR-335-5p mimic, miR-335-5p inhibitor and their matched negative control (miR-NC mimic or miR-NC inhibitor) were purchased from Ribobio (Guangzhou, China). The pCD-ciR vector harboring circAPLP2 sequence was used for circAPLP2 overexpression (pCD-ciR-circAPLP2; Geneseed; Guangzhou, China), with pCD-ciR vector as the control. The pcDNA vector harboring HELLS sequence was used for HELLS overexpression (pcDNA-HELLS; Sangon; Shanghai, China), with pcDNA-NC as the negative control. Cells were transiently transfected with these oligonucleotides or plasmids using Lipofectamine 3000 (Invitrogen; Carlsbad, CA, USA).
2.6 Flow cytometry assay
For cell cycle distribution analysis, cells were harvested, digested with trypsin and centrifuged for 15 min. Cell pellets were resuspended with 70% ethanol for fixing at −20°C overnight. Afterward, cells were washed with PBS twice and incubated with RNase A (180 μg/mL; Invitrogen) for 15 min at 37°C. Then, cells were stained with propidium iodide (PI; 50 μg/mL; Invitrogen) for 30 min at room temperature in the dark. Cells were analyzed by flow cytometry system (BD Biosciences, San Jose, CA, USA).
The apoptotic cells were detected using the Bioscience™ Annexin V-FITC Apoptosis Detection Kit (Invitrogen). Cells were harvested and centrifuged for 10 min. The cells were then washed with PBS twice and resuspended in binding buffer, and then stained with Annexin V-FITC and PI in the dark at room temperature for 15 min. The stained cells for apoptosis detection were analyzed by a flow cytometry system (BD Biosciences).
2.7 Colony formation assay
SW480 and HCT116 cells were seeded in 6-well plates at a density of 200 cells per well and cultured for 12 days with replacement with fresh medium every 3 days in a 37°C incubator containing 5% CO2. Subsequently, colonies were washed with PBS, fixed with methanol and stained with 0.1% crystal violet (Sangon). The images of colonies were captured, and the number of colonies was counted under a microscope (Olympus; Tokyo, Japan).
2.8 Wound healing assay
SW480 and HCT116 cells were seeded into a 6-well plate with a density of 2.0 × 105 cells/well. Then, the monolayer was gently scratched using a 10 μL pipette tip, ensuring that the tip is perpendicular to the bottom of the well. Next, the monolayer with a wound was gently washed twice with PBS to remove the detached cells. After culturing for 24 h, the representative images of migration distance were recorded under a microscope (40×; Olympus), and the data were analyzed using the Image J software (NIH, Bethesda, MA, USA).
2.9 Transwell assay
For migration assay, cells (5 × 104) were resuspended in serum-free medium and plated on the top of uncoated transwell chambers (Corning Incorporated; Corning, NY, USA). For invasion assay, cells (5 × 104) were transferred on the top of transwell chambers pre-coated with Matrigel matrix (Corning Incorporated). Meanwhile, the bottom of chambers in these two assays was filled with culture medium containing 10% FBS. After incubation for 24 h, the remaining cells attached to the inserts were fixed with methanol and stained with 0.1% crystal violet (Sangon). Cells were then photographed and counted under a microscope (100×; Olympus).
2.10 Glycolysis stress test
Extracellular acidification rate (ECAR) was measured to monitor glycolysis metabolism using a Seahorse XF96 Analyzer Glycolysis (Seahorse Bioscience, Santa Clara, CA, USA) in line with the guidelines. Briefly, SW480 and HCT116 cells in culture medium containing 10% FBS were seeded in XF 96-well plates. For glycolysis stress test, D-glucose (10 mM), oligomycin (1 μM) and 2-deoxyglucose (2-DG; 100 mM) were seriatim added into the wells at the indicated time points, and the value of corresponding ECAR (mPH/min) was assessed.
2.11 Glucose uptake and lactate production
To monitor glycolysis metabolism, the levels of glucose uptake and lactate production were detected using the Glucose Uptake Assay Kit (Abcam; Cambridge, MA, USA) and Lactate Assay Kit (Abcam) according to the manuscript’s protocols.
2.12 Western blot
Total proteins were extracted using RIPA Lysis Buffer (Sangon) and quantified by BCA Protein Assay Kit (Sangon). The equal amount of protein was isolated by 12% SDS-PAGE and then transferred on PVDF membranes (Bio-Rad). The protein-included membranes were blocked by 5% skim milk and then incubated with the primary antibodies, including anti-hexokinase 2 (anti-HK2; ab227198; Abcam), anti-HELLS (ab3851; Abcam) and anti-GAPDH (ab9485; Abcam) overnight at 4°C. Then, the membranes were incubated with the secondary antibody (ab205718; Abcam) for 1.5 h at room temperature. The target proteins were detected by enhanced chemiluminescence (ECL; Sangon) reagents and quantified using Image J software.
2.13 Dual-luciferase reporter assay
The target relationship between miR-335-5p and circAPLP2 or HELLS was predicted by the bioinformatics tool starbase v3.0 (http://starbase.sysu.edu.cn/). To verify the predicted relationship, dual-luciferase reporter assay was performed. In brief, the fragment of wild-type circAPLP2, including the miR-335-5p binding site, was amplified and cloned into PGL4 reporter plasmid, naming as WT-circAPLP2. The fragment of mutant-type circAPLP2 containing mutants on the miR-335-5p binding site was amplified and cloned into PGL4 reporter plasmid, named as MUT-circAPLP2. Similarly, WT-HELLS and MUT-HELLS were also constructed. SW480 and HCT116 cells were cotransfected with miR-335-5p mimic or miR-NC mimic and WT-circAPLP2, MUT-circAPLP2, WT-HELLS or MUT-HELLS and incubated for 48 h. The luciferase activity in SW480 and HCT116 cells with different transfection was measured using the Dual-Luciferase Reporter Assay System (Promega; Madison, WI, USA).
2.14 Animal experiments
For stable circAPLP2 knockdown, short hairpin RNA targeting circAPLP2 (sh-circAPLP2; Genepharma) was packaged into lentiviral vector by Genepharma, with sh-NC as the negative control. Lentiviral vector harboring sh-circAPLP2 or sh-NC was transfected into 293FT cells. SW480 cells were infected with the viral supernatants, and sh-circAPLP2 or sh-NC was infected into SW480 cells. The experimental mice (Balb/c; Female; 6 weeks old) were purchased from SLAC Animal Lab. (Shanghai, China) and divided into two groups (sh-circAPLP2 group and sh-NC group; n = 6 per group). SW480 cells harboring sh-circAPLP2 or sh-NC were subcutaneously injected into nude mice to allow tumor growth. The status of tumor growth was observed every 3 days, and tumor volume (length × width2 × 0.5) was recorded. After 4 weeks, all mice were sacrificed by cervical dislocation, and tumor tissues were excised for weighting and expression detection. All procedures were approved by the Animal Care and Use of Committee of West Hospital of Zibo Central Hospital.
2.15 Statistical analysis
All experiments were repeated at least three times. All data were presented as mean ± standard deviation (SD). P < 0.05 was set as statistically significant. The difference was analyzed using the unpaired Student’s t-test for comparisons of two groups or ANOVA for multiple group comparisons. Pearson correlation coefficient was adopted to analyze the correlation between two parameters. All statistical analyses were conducted using GraphPad Prism 7.0 (GraphPad, San Diego, CA, USA).
3 Results
3.1 circAPLP2 abundantly existed in CRC tissues and cells and was resistant to RNase R
circAPLP2 (hsa_circ_0000372) was derived from the exon2-exon3 of APLP2 mRNA, and the schematic diagram of circAPLP2 formation is shown in Figure 1a. The expression pattern of circAPLP2 in CRC was unclear, and our result showed that the expression of circAPLP2 was remarkably increased in CRC tissues compared with normal tissues (Figure 1b). Besides, circAPLP2 expression was also shown to be elevated in CRC cell lines, including SW480, HCT116, SW620, LOVO and HT29 cells, compared with that in HIEC6 cells (non-cancer cells) (Figure 1c). Next, we used RNase R to examine the stability of circAPLP2, and the result presented that RNase R significantly reduced the expression of linear APLP2 mRNA but unaffected the expression of circAPLP2 compared with Mock treatment (Figure 1d). In short, circAPLP2 was upregulated in CRC, and circAPLP2 was stable and resistant to RNase R.

circAPLP2 was upregulated in CRC tissues and cells, and resistant to RNase R. (a) The schematic diagram of circAPLP2 formation. (b) The expression of circAPLP2 in CRC tissues and paired normal tissues detected by qPCR. (c) The expression of circAPLP2 in cancer cells (SW480, HCT116, SW620, LOVO and HT29) and non-cancer cells (HIEC6) detected by qPCR. (d) The effects of RNase R or Mock treatment on the expression of circAPLP2 and linear APLP2. *P < 0.05.
3.2 circAPLP2 knockdown induced cell cycle arrest and cell apoptosis but suppressed colony formation, migration, invasion and glycolysis metabolism
To explore the role of circAPLP2 in CRC, we used siRNA-mediated circAPLP2 knockdown to investigate the effects of circAPLP2 in CRC cells. The data showed that circAPLP2 expression level was strikingly decreased after si-circAPLP2 transfection (Figure 2a). In function, circAPLP2 knockdown noticeably induced cell cycle arrest at the G0/G1 stage in SW480 and HCT116 cells (Figure 2b). The ability of colony formation was notably suppressed by circAPLP2 knockdown (Figure 2c). Compared to si-NC transfection, si-circAPLP2 transfection significantly promoted cell apoptosis (Figure 2d). From wound healing and transwell assays, we monitored that the capacity of cell migration was strikingly inhibited in SW480 and HCT116 cells transfected with si-circAPLP2 compared with si-NC (Figure 2e and f), and the capacity of cell invasion was also inhibited by si-circAPLP2 transfection in SW480 and HCT116 cells by transwell assay (Figure 2g). Moreover, glycolysis stress test showed that the ECAR value was decreased in SW480 and HCT116 cells transfected with si-circAPLP2 compared with si-NC with sequential addition of Glucose, Oligomycin and 2-DG (Figure 2h), and the levels of glucose uptake and lactate production were notably decreased after circAPLP2 knockdown (Figure 2i and j). In addition, the expression of HK2 (a marker of glycolysis) was also significantly reduced in cells with circAPLP2 knockdown (Figure 2k). In short, circAPLP2 knockdown blocked a series of CRC cell malignant behaviors, including colony formation, cell cycle progression, migration, invasion and glycolysis metabolism.

circAPLP2 knockdown induced cell cycle arrest and apoptosis but suppressed colony formation, migration, invasion and glycolysis metabolism. (a) The efficiency of circAPLP2 knockdown was examined using qPCR. Then, the effects of circAPLP2 knockdown on (b) cell cycle progression, (c) colony formation, (d) cell apoptosis, (e and f) cell migration, (g) cell invasion and (h) glycolysis metabolism in SW480 and HCT116 cells were detected by flow cytometry assay, colony formation assay, flow cytometry assay, wound healing assay, transwell assay and glycolysis stress test. (i and j) Glucose uptake and lactate production were examined using detection kits. (k) The expression of a glycolysis-related protein, HK2, was measured by western blot. *P < 0.05.
3.3 circAPLP2 targeted miR-335-5p and suppressed its expression
We attempted to identify the target miRNAs of circAPLP2, thus providing a mechanism to elucidate the role of circAPLP2 in CRC. Bioinformatics analysis (starbase v3.0) showed that circAPLP2 harbored targeting site (miR-335-5p response element) with miR-335-5p (Figure 3a). For further validation, WT-circAPLP2 and MUT-circAPLP2 reporter plasmids were constructed, and the data showed that the luciferase activity was markedly impaired in SW480 and HCT116 cells with cotransfection of miR-335-5p mimic and WT-circAPLP2 but not MUT-circAPLP2 (Figure 3b and c), verifying the interaction between circAPLP2 and miR-335-5p. In addition, circAPLP2 expression was remarkably increased in SW480 and HCT116 cells transfected with pcDNA-circAPLP2 compared with pcDNA-NC (Figure 3d). Then, in pCDNA-circAPLP2-transfected cells, the expression of miR-335-5p was decreased, while the expression of miR-335-5p was remarkably increased in si-circAPLP2-transfected cells (Figure 3e). Furthermore, the expression of miR-335-5p was lower in CRC tissues and cell lines (SW480, HCT116 and SW620) than those in normal tissues and non-cancer cells (HIEC6), respectively (Figure 3f and g). Pearson correlation analysis showed that miR-335-5p expression was negatively associated with circAPLP2 expression in CRC tissues (Figure 3h). In short, circAPLP2 functioned as a molecular sponge of miR-335-5p and suppressed miR-335-5p expression.

circAPLP2 targeted miR-335-5p to suppress miR-335-5p expression. (a) The binding site between circAPLP2 and miR-335-5p was analyzed by starbase v3.0. (b and c) The relationship between circAPLP2 and miR-335-5p was verified by dual-luciferase reporter assay. (d) The efficiency of circAPLP2 overexpression was examined using qPCR. (e) The effects of circAPLP2 overexpression and knockdown on the expression of miR-335-5p were examined using qPCR. (f) The expression of miR-335-5p in CRC and normal tissues detected by qPCR. (g) The expression of miR-335-5p in cancer cells (SW480, HCT116 and SW620) and non-cancer cells (HIEC6) detected by qPCR. (h) The association between miR-335-5p expression and circAPLP2 expression in CRC tissues was analyzed by Pearson correlation coefficient. *P < 0.05.
3.4 miR-335-5p deficiency abolished the effects of circAPLP2 knockdown in CRC cells
We performed rescue experiments to verify whether circAPLP2 played functions in CRC by targeting miR-335-5p. The expression of miR-335-5p was visibly lessened in cells transfected with miR-335-5p inhibitor compared with miR-NC inhibitor (Figure 4a). In function, circAPLP2 knockdown-induced cell cycle arrest was alleviated by the reintroduction of miR-335-5p inhibitor (Figure 4b). The ability of colony formation was suppressed by circAPLP2 knockdown in SW480 and HCT116 cells but restored by miR-335-5p inhibitor (Figure 4c). circAPLP2 knockdown-induced cell apoptosis was partly blocked by the inhibition of miR-335-5p (Figure 4d). Besides, the migration distance in wound healing assay and the number of migrated cells in transwell assay were blocked in cells after si-circAPLP2 transfection but renovated in cells after si-circAPLP2 + miR-335-5p inhibitor cotransfection (Figure 4e and f). Also, the number of invaded cells was elevated in cells after si-circAPLP2 + miR-335-5p inhibitor cotransfection compared with si-circAPLP2 transfection (Figure 4g). Glycolysis stress test showed that the value of ECAR was weakened in cells transfected with si-circAPLP2 but recovered in cells transfected with si-circAPLP2 + miR-335-5p inhibitor (Figure 4h). The levels of glucose uptake and lactate production were also recovered by the reintroduction of miR-335-5p inhibitor (Figure 4i and j). The expression of HK2 impaired by circAPLP2 knockdown was enhanced by miR-335-5p inhibition (Figure 4k). In short, circAPLP2 knockdown blocked a series of CRC cell malignant behaviors by increasing miR-335-5p expression.

miR-335-5p inhibition reversed the effects of circAPLP2 knockdown. (a) The efficiency of miR-335-5p inhibitor was examined using qPCR. (b) Cell cycle progression, (c) the ability of colony formation, (d) cell apoptosis, (e and f) cell migration, (g) cell invasion, (h) glycolysis metabolism, (i and j) the levels of glucose uptake and lactate production, and (k) the expression of HK2 in SW480 and HCT116 cells transfected with si-circAPLP2, si-NC, si-circAPLP2 + miR-335-5p inhibitor or si-circAPLP2 + miR-NC inhibitor were examined using flow cytometry assay, colony formation assay, flow cytometry assay, wound healing assay, transwell assay, glycolysis stress test, detection kits and western blot. *P < 0.05.
3.5 HELLS was a target of miR-335-5p
Additionally, we analyzed the potential target mRNAs of miR-335-5p. Bioinformatics analysis predicted that miR-335-5p bound to HELLS 3′UTR through a special binding site (Figure 5a), indicating that HELLS might be a target of miR-335-5p, which was further verified by dual-luciferase reporter assay (Figure 5b). In the following assay, we detected that the expression of miR-335-5p was pronouncedly promoted in cells transfected with miR-335-5p mimic compared with miR-NC mimic (Figure 5c). Besides, miR-335-5p negatively regulated HELLS expression because HELLS expression was notably increased in SW480 and HCT116 cells transfected with miR-335-5p inhibitor but decreased in cells transfected with miR-335-5p mimic (Figure 5d). We next examined the expression pattern of HELLS in CRC. The result showed that HELLS was upregulated in CRC tissues and cell lines (SW480, HCT116 and SW620) compared with that in normal tissues and non-cancer cells (HIEC6), respectively (Figure 5e–g). Moreover, Pearson correlation analysis introduced that HELLS expression was negatively associated with miR-335-5p expression in CRC tissues (Figure 5h). In short, miR-335-5p bound to HELLS and suppressed its expression.

miR-335-5p bound to HELLS 3′UTR. (a) The binding site between miR-335-5p and HELLS 3′UTR was analyzed by starbase v3.0. (b) The relationship between miR-335-5p and HELLS was verified by dual-luciferase reporter assay. (c) The efficiency of miR-335-5p mimic was examined using qPCR. (d) The effects of miR-335-5p enrichment and inhibition on the expression of HELLS were examined using western blot. (e and f) The expression of HELLS in CRC and normal tissues was detected by qPCR and western blot. (g) The expression of HELLS in HIEC6, SW480, HCT116 and SW620 cells was detected by western blot. (h) The association between HELLS expression and miR-335-5p expression in CRC tissues was analyzed by Pearson correlation coefficient. *P < 0.05.
3.6 miR-335-5p overexpression induced cell cycle arrest and cell apoptosis but suppressed cell colony formation, migration, invasion and glycolysis metabolism by repressing HELLS expression
We next performed rescue experiments to determine whether miR-335-5p exerted its role by degrading HELLS. The detection of HELLS overexpression efficiency showed that the expression of HELLS was remarkably enhanced in SW480 and HCT116 cells transfected with pcDNA-HELLS compared with pcDNA-NC (Figure 6a). In function, miR-335-5p mimic transfection notably induced cell cycle arrest and cell apoptotic rate, which was alleviated by the reintroduction of pcDNA-HELLS (Figure 6b and d). The ability of colony formation in SW480 and HCT116 cells was markedly blocked by miR-335-5p overexpression but partly enhanced by synchronous HELLS overexpression (Figure 6c). Besides, the capacities of migration and invasion were suppressed in SW480 and HCT116 cells transfected with miR-335-5p mimic but recovered in cells transfected with miR-335-5p mimic + pcDNA-HELLS (Figure 6e–g). Moreover, the value of ECAR in glycolysis stress test and the levels of glucose uptake and lactate production were also weakened in miR-335-5p mimic-transfected cells but restored in miR-335-5p mimic + pcDNA-HELLS-transfected cells (Figure 6h–j). The expression of HK2 was decreased in SW480 and HCT116 cells transfected with miR-335-5p mimic but reinforced in cells transfected with miR-335-5p mimic + pcDNA-HELLS (Figure 6k). In short, miR-335-5p overexpression blocked CRC cell malignant behaviors by targeting HELLS.
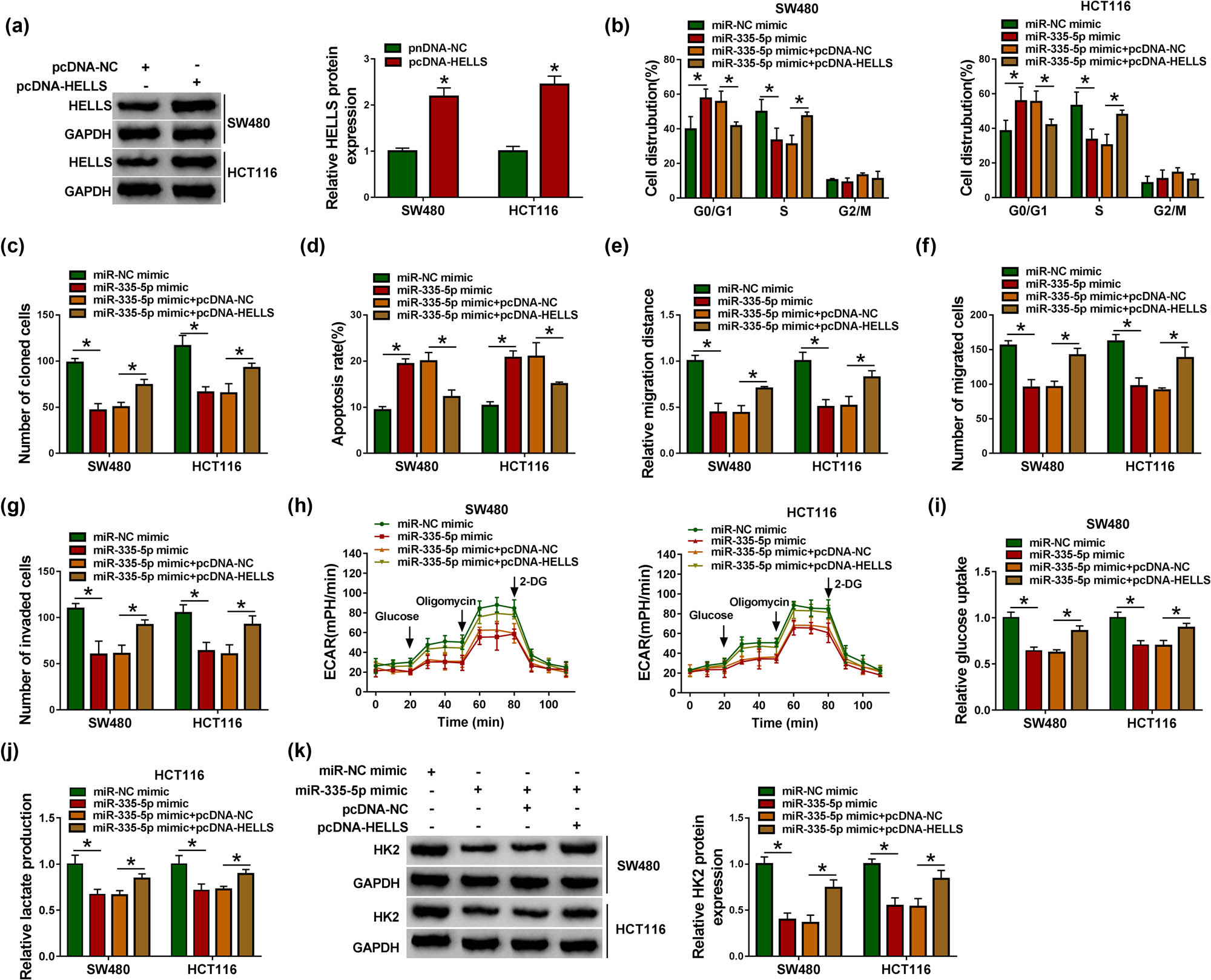
miR-335-5p restoration suppressed CRC development in vitro by targeting HELLS. (a) The efficiency of HELLS overexpression was examined using western blot. (b) Cell cycle progression, (c) the ability of colony formation, (d) cell apoptosis, (e and f) cell migration, (g) cell invasion, (h) glycolysis metabolism, (i and j) the levels of glucose uptake and lactate production, and (k) the expression of HK2 in SW480 and HCT116 cells transfected with miR-335-5p mimic, miR-NC mimic, miR-335-5p mimic + pcDNA-HELLS or miR-335-5p mimic + pcDNA-NC were examined using flow cytometry assay, colony formation assay, flow cytometry assay, wound healing assay, transwell assay, glycolysis stress test, detection kits and western blot. *P < 0.05.
3.7 circAPLP2 positively regulated HELLS expression by targeting miR-335-5p
Further dual-luciferase reporter assay discovered that the cotransfection of WT-HELLS 3′UTR and miR-335-5p remarkably reduced luciferase activity in SW480 and HCT116 cells, while further WT-circAPLP2 but not MUT-circAPLP2 reintroduction partly recovered luciferase activity (Figure 7a and b). Interestingly, we found that HELLS expression at the mRNA level in CRC tissues was positively associated with circAPLP2 expression (Figure 7c). Besides, the expression of HELLS was strikingly decreased in SW480 and HCT116 cells transfected with si-circAPLP2 compared with si-NC but partly recovered in cells transfected with si-circAPLP2 + miR-335-5p inhibitor compared with si-circAPLP2 + miR-NC inhibitor (Figure 7d). We deduced that circAPLP2 functioned as miR-335-5p sponge to increase the expression of HELLS.

circAPLP2 positively regulated HELLS expression by targeting miR-335-5p. (a and b) The interactions among circAPLP2, miR-335-5p and HELLS were further analyzed by dual-luciferase reporter assay. (c) The association between HELLS expression and circAPLP2 expression in CRC tissues was analyzed by Pearson correlation coefficient. (d) The expression of HELLS in SW480 and HCT116 cells transfected with si-circAPLP2, si-NC, si-circAPLP2 + miR-335-5p inhibitor or si-circAPLP2 + miR-NC inhibitor was detected by western blot. *P < 0.05.
3.8 circAPLP2 downregulation suppressed tumor growth in vivo
Animal experiments presented that tumor volume and tumor weight in the sh-circAPLP2 group were significantly lower than that in the sh-NC group (Figure 8a and b), suggesting that circAPLP2 downregulation inhibited tumor growth. Besides, the expression of circAPLP2 and HELLS was declined, while the expression of miR-335-5p was elevated in tumor tissues from the sh-circAPLP2 group relative to the sh-NC group (Figure 8c–e), indicating that circAPLP2 promoted tumor growth in vivo by mediating the miR-335-5p/HELLS axis.

circAPLP2 downregulation impaired tumor growth in vivo. (a) Nude mice were injected SW480 cells with sh-circAPLP2 or sh-NC, and tumor volume in each mouse was detected every 3 days. (b) Tumor weight was detected after 4 weeks when all mice were killed. (c and d) The expression of circAPLP2 and miR-335-5p in these tumor tissues was detected by qPCR. (e) The expression of HELLS in these tumor tissues was detected by western blot. *P < 0.05.
4 Discussion
The use of molecular markers for risk stratification, early detection and prognostic markers of CRC shows promise in molecular medicine [19]. circRNA satisfies a variety of suitable characteristics as biomarkers, including non-digestible and degraded, high abundance and wide distribution [20]. For example, circRNA expression profile revealed that circDDX17 was notably downregulated in CRC tissues, and low expression of circDDX17 contributed to CRC cell proliferation, migration and invasion [12]. Similarly, circBANP was presented to be upregulated in CRC tissues by circRNA expression profile, and circBANP knockdown suppressed CRC cell proliferation and served as a prognostic and therapeutic marker [21]. A circRNA profile from a previous study also monitored that circAPLP2 expression was strikingly enhanced in CRC tissues [12], while its detailed functions were not expounded. In our study, the expression of circAPLP2 was remarkably higher in 33 CRC tissues compared with that in matched normal tissues. Functional assays presented that underexpression of circAPLP2 weakened colony formation ability, the capacities of migration and invasion, and glycolysis metabolism, but promoted CRC cell cycle arrest and apoptosis. Also, in vivo assay showed that circAPLP2 knockdown blocked solid tumor growth. Our study provided sufficient evidence that circAPLP2 downregulation blocked CRC malignant development both in vitro and in vivo, suggesting that circAPLP2 was an oncogene in CRC.
It is a canonical phenomenon that circRNAs act as potent molecular sponges of miRNAs, sequestering miRNA activity and thus affecting the expression of downstream target genes [14]. In this paper, miR-335-5p was regarded as a target of circRNA through the prediction of bioinformatics analysis and the validation of dual-luciferase reporter assay. Interestingly, the role of miR-335-5p had been partly defined. Previous studies documented that miR-335-5p expression was remarkably declined in the plasma of patients with colorectal adenoma by high-depth small RNA sequencing [22]. In addition, miR-335-5p was markedly downregulated in CRC tissues and cells, and forced expression of miR-335-5p impaired CRC cell proliferation, migration and invasion, as well as lung and liver metastasis in vivo by mediating its target genes [23,24]. These studies strongly corroborated our observation of a lower abundance of miR-335-5p in clinical CRC tissues and cell lines, and miR-335-5p restoration induced cell cycle arrest and apoptosis but inhibited colony formation ability, migration, invasion and glycolysis metabolism.
Further study illustrated that HELLS was a target gene of miR-335-5p. HELLS was stated to regulate chromatin remodeling and mediate epigenetic silencing of tumor suppressor genes, thus aggravating the progression of cancers [25,26]. HELLS was demonstrated to be a downstream target of Forkhead Box M1 (FOXM1), and HELLS was used to be a biomarker for early cancer detection and an indicator for malignant progression [27]. Importantly, a study exposed that HELLS was highly expressed in CRC tissues, and high HELLS expression was associated with high TNM stage and low overall survival [28]. Besides, HELLS knockdown suppressed CRC cell proliferation and promoted cell cycle arrest [28]. Consistent with these findings, we also defined HELLS as an oncogene because its expression was strikingly increased in CRC tissues and cells. Moreover, HELLS overexpression abolished the role of miR-335-5p restoration and recovered CRC cell malignant activities.
In conclusion, higher expression of circAPLP2 was detected in CRC tissues and cells. circAPLP2 knockdown was associated with inhibitory colony formation ability, migration/invasion capacity and glycolysis metabolism. Besides, circAPLP2 knockdown also blocked solid tumor growth in vivo. Further mechanism analysis demonstrated that circAPLP2 played pro-cancer effects in CRC by upregulating HELLS via targeting miR-335-5p. Our study enriched the role of circAPLP2 in CRC and broadened the insights into CRC pathogenesis from the perspective of circAPLP2.
-
Disclosure of interest: The authors declare that they have no financial conflicts of interest.
References
[1] Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197.10.3390/ijms18010197Suche in Google Scholar
[2] Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065.10.1038/nrdp.2015.65Suche in Google Scholar
[3] Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–47.10.1016/S0140-6736(10)60353-4Suche in Google Scholar
[4] Brody H. Colorectal cancer. Nature. 2015;521:S1.10.1038/521S1aSuche in Google Scholar PubMed
[5] Bolha L, Ravnik-Glavac M, Glavac D. circular RNAs: biogenesis, function, and a role as possible cancer biomarkers. Int J Genomics. 2017;2017:6218353.10.1155/2017/6218353Suche in Google Scholar PubMed PubMed Central
[6] Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.10.1038/nature11928Suche in Google Scholar PubMed
[7] Barrett SP, Wang PL, Salzman J. circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540.10.7554/eLife.07540Suche in Google Scholar PubMed PubMed Central
[8] Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499–512.10.3892/or.2018.6372Suche in Google Scholar PubMed PubMed Central
[9] Ge J, Jin Y, Lv X, Liao Q, Luo C, Ye G, et al. Expression profiles of circular RNAs in human colorectal cancer based on RNA deep sequencing. J Clin Lab Anal. 2019;33:e22952.10.1002/jcla.22952Suche in Google Scholar PubMed PubMed Central
[10] Lu X, Yu Y, Liao F, Tan S. Homo sapiens circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 gene acts as an oncogene in colorectal cancer through the microRNA-203a-3p.1 and CREB1 axis. Med Sci Monit. 2019;25:6872–83.10.12659/MSM.916064Suche in Google Scholar PubMed PubMed Central
[11] Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S. circRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503:870–5.10.1016/j.bbrc.2018.06.089Suche in Google Scholar PubMed
[12] Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37:325.10.1186/s13046-018-1006-xSuche in Google Scholar PubMed PubMed Central
[13] Hentze MW, Preiss T. circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–5.10.1038/emboj.2013.53Suche in Google Scholar PubMed PubMed Central
[14] Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.10.1038/nature11993Suche in Google Scholar PubMed
[15] Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.10.1038/nsmb.2959Suche in Google Scholar PubMed
[16] Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–36.10.3322/caac.21244Suche in Google Scholar PubMed PubMed Central
[17] An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun D, et al. circZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. 2018;51:2224–36.10.1159/000495868Suche in Google Scholar PubMed
[18] Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–50.10.1038/nsmb.1552Suche in Google Scholar PubMed PubMed Central
[19] Grady WM, Pritchard CC. Molecular alterations and biomarkers in colorectal cancer. Toxicol Pathol. 2014;42:124–39.10.1177/0192623313505155Suche in Google Scholar PubMed PubMed Central
[20] Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90.10.1186/s12943-019-1002-6Suche in Google Scholar PubMed PubMed Central
[21] Zhu M, Xu Y, Chen Y, Yan F. circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–44.10.1016/j.biopha.2016.12.097Suche in Google Scholar PubMed
[22] Roberts BS, Hardigan AA, Moore DE, Ramaker RC, Jones AL, Fitz-Gerald MB, et al. Discovery and validation of circulating biomarkers of colorectal adenoma by high-depth small RNA sequencing. Clin Cancer Res. 2018;24:2092–9.10.1158/1078-0432.CCR-17-1960Suche in Google Scholar PubMed PubMed Central
[23] Zhang D, Yang N. miR-335-5p inhibits cell proliferation, migration and invasion in colorectal cancer through downregulating LDHB. J BUON. 2019;24:1128–36.Suche in Google Scholar
[24] Sun Z, Zhang Z, Liu Z, Qiu B, Liu K, Dong G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med Oncol. 2014;31:982.10.1007/s12032-014-0982-8Suche in Google Scholar PubMed
[25] Law CT, Wei L, Tsang FH, Chan CY, Xu IM, Lai RK, et al. HELLS regulates chromatin remodeling and epigenetic silencing of multiple tumor suppressor genes in human hepatocellular carcinoma. Hepatology. 2019;69:2013–30.10.1002/hep.30414Suche in Google Scholar PubMed
[26] Zocchi L, Mehta A, Wu SC, Wu J, Gu Y, Wang J, et al. Chromatin remodeling protein HELLS is critical for retinoblastoma tumor initiation and progression. Oncogenesis. 2020;9:25.10.1038/s41389-020-0210-7Suche in Google Scholar PubMed PubMed Central
[27] Waseem A, Ali M, Odell EW, Fortune F, Teh MT. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010;46:536–42.10.1016/j.oraloncology.2010.03.022Suche in Google Scholar PubMed
[28] Liu X, Hou X, Zhou Y, Li Q, Kong F, Yan S, et al. Downregulation of the helicase lymphoid-specific (HELLS) gene impairs cell proliferation and induces cell cycle arrest in colorectal cancer cells. Oncol Targets Ther. 2019;12:10153–63.10.2147/OTT.S223668Suche in Google Scholar PubMed PubMed Central
© 2021 Rui Xiang et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Research Articles
- Identification of ZG16B as a prognostic biomarker in breast cancer
- Behçet’s disease with latent Mycobacterium tuberculosis infection
- Erratum
- Erratum to “Suffering from Cerebral Small Vessel Disease with and without Metabolic Syndrome”
- Research Articles
- GPR37 promotes the malignancy of lung adenocarcinoma via TGF-β/Smad pathway
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Additional baricitinib loading dose improves clinical outcome in COVID-19
- The co-treatment of rosuvastatin with dapagliflozin synergistically inhibited apoptosis via activating the PI3K/AKt/mTOR signaling pathway in myocardial ischemia/reperfusion injury rats
- SLC12A8 plays a key role in bladder cancer progression and EMT
- LncRNA ATXN8OS enhances tamoxifen resistance in breast cancer
- Case Report
- Serratia marcescens as a cause of unfavorable outcome in the twin pregnancy
- Spleno-adrenal fusion mimicking an adrenal metastasis of a renal cell carcinoma: A case report and embryological background
- Research Articles
- TRIM25 contributes to the malignancy of acute myeloid leukemia and is negatively regulated by microRNA-137
- CircRNA circ_0004370 promotes cell proliferation, migration, and invasion and inhibits cell apoptosis of esophageal cancer via miR-1301-3p/COL1A1 axis
- LncRNA XIST regulates atherosclerosis progression in ox-LDL-induced HUVECs
- Potential role of IFN-γ and IL-5 in sepsis prediction of preterm neonates
- Rapid Communication
- COVID-19 vaccine: Call for employees in international transportation industries and international travelers as the first priority in global distribution
- Case Report
- Rare squamous cell carcinoma of the kidney with concurrent xanthogranulomatous pyelonephritis: A case report and review of the literature
- An infertile female delivered a baby after removal of primary renal carcinoid tumor
- Research Articles
- Hypertension, BMI, and cardiovascular and cerebrovascular diseases
- Case Report
- Coexistence of bilateral macular edema and pale optic disc in the patient with Cohen syndrome
- Research Articles
- Correlation between kinematic sagittal parameters of the cervical lordosis or head posture and disc degeneration in patients with posterior neck pain
- Review Articles
- Hepatoid adenocarcinoma of the lung: An analysis of the Surveillance, Epidemiology, and End Results (SEER) database
- Research Articles
- Thermography in the diagnosis of carpal tunnel syndrome
- Pemetrexed-based first-line chemotherapy had particularly prominent objective response rate for advanced NSCLC: A network meta-analysis
- Comparison of single and double autologous stem cell transplantation in multiple myeloma patients
- The influence of smoking in minimally invasive spinal fusion surgery
- Impact of body mass index on left atrial dimension in HOCM patients
- Expression and clinical significance of CMTM1 in hepatocellular carcinoma
- miR-142-5p promotes cervical cancer progression by targeting LMX1A through Wnt/β-catenin pathway
- Comparison of multiple flatfoot indicators in 5–8-year-old children
- Early MRI imaging and follow-up study in cerebral amyloid angiopathy
- Intestinal fatty acid-binding protein as a biomarker for the diagnosis of strangulated intestinal obstruction: A meta-analysis
- miR-128-3p inhibits apoptosis and inflammation in LPS-induced sepsis by targeting TGFBR2
- Dynamic perfusion CT – A promising tool to diagnose pancreatic ductal adenocarcinoma
- Biomechanical evaluation of self-cinching stitch techniques in rotator cuff repair: The single-loop and double-loop knot stitches
- Review Articles
- The ambiguous role of mannose-binding lectin (MBL) in human immunity
- Case Report
- Membranous nephropathy with pulmonary cryptococcosis with improved 1-year follow-up results: A case report
- Fertility problems in males carrying an inversion of chromosome 10
- Acute myeloid leukemia with leukemic pleural effusion and high levels of pleural adenosine deaminase: A case report and review of literature
- Metastatic renal Ewing’s sarcoma in adult woman: Case report and review of the literature
- Burkitt-like lymphoma with 11q aberration in a patient with AIDS and a patient without AIDS: Two cases reports and literature review
- Skull hemophilia pseudotumor: A case report
- Judicious use of low-dosage corticosteroids for non-severe COVID-19: A case report
- Adult-onset citrullinaemia type II with liver cirrhosis: A rare cause of hyperammonaemia
- Clinicopathologic features of Good’s syndrome: Two cases and literature review
- Fatal immune-related hepatitis with intrahepatic cholestasis and pneumonia associated with camrelizumab: A case report and literature review
- Research Articles
- Effects of hydroxyethyl starch and gelatin on the risk of acute kidney injury following orthotopic liver transplantation: A multicenter retrospective comparative clinical study
- Significance of nucleic acid positive anal swab in COVID-19 patients
- circAPLP2 promotes colorectal cancer progression by upregulating HELLS by targeting miR-335-5p
- Ratios between circulating myeloid cells and lymphocytes are associated with mortality in severe COVID-19 patients
- Risk factors of left atrial appendage thrombus in patients with non-valvular atrial fibrillation
- Clinical features of hypertensive patients with COVID-19 compared with a normotensive group: Single-center experience in China
- Surgical myocardial revascularization outcomes in Kawasaki disease: systematic review and meta-analysis
- Decreased chromobox homologue 7 expression is associated with epithelial–mesenchymal transition and poor prognosis in cervical cancer
- FGF16 regulated by miR-520b enhances the cell proliferation of lung cancer
- Platelet-rich fibrin: Basics of biological actions and protocol modifications
- Accurate diagnosis of prostate cancer using logistic regression
- miR-377 inhibition enhances the survival of trophoblast cells via upregulation of FNDC5 in gestational diabetes mellitus
- Prognostic significance of TRIM28 expression in patients with breast carcinoma
- Integrative bioinformatics analysis of KPNA2 in six major human cancers
- Exosomal-mediated transfer of OIP5-AS1 enhanced cell chemoresistance to trastuzumab in breast cancer via up-regulating HMGB3 by sponging miR-381-3p
- A four-lncRNA signature for predicting prognosis of recurrence patients with gastric cancer
- Knockdown of circ_0003204 alleviates oxidative low-density lipoprotein-induced human umbilical vein endothelial cells injury: Circulating RNAs could explain atherosclerosis disease progression
- Propofol postpones colorectal cancer development through circ_0026344/miR-645/Akt/mTOR signal pathway
- Knockdown of lncRNA TapSAKI alleviates LPS-induced injury in HK-2 cells through the miR-205/IRF3 pathway
- COVID-19 severity in relation to sociodemographics and vitamin D use
- Clinical analysis of 11 cases of nocardiosis
- Cis-regulatory elements in conserved non-coding sequences of nuclear receptor genes indicate for crosstalk between endocrine systems
- Four long noncoding RNAs act as biomarkers in lung adenocarcinoma
- Real-world evidence of cytomegalovirus reactivation in non-Hodgkin lymphomas treated with bendamustine-containing regimens
- Relation between IL-8 level and obstructive sleep apnea syndrome
- circAGFG1 sponges miR-28-5p to promote non-small-cell lung cancer progression through modulating HIF-1α level
- Nomogram prediction model for renal anaemia in IgA nephropathy patients
- Effect of antibiotic use on the efficacy of nivolumab in the treatment of advanced/metastatic non-small cell lung cancer: A meta-analysis
- NDRG2 inhibition facilitates angiogenesis of hepatocellular carcinoma
- A nomogram for predicting metabolic steatohepatitis: The combination of NAMPT, RALGDS, GADD45B, FOSL2, RTP3, and RASD1
- Clinical and prognostic features of MMP-2 and VEGF in AEG patients
- The value of miR-510 in the prognosis and development of colon cancer
- Functional implications of PABPC1 in the development of ovarian cancer
- Prognostic value of preoperative inflammation-based predictors in patients with bladder carcinoma after radical cystectomy
- Sublingual immunotherapy increases Treg/Th17 ratio in allergic rhinitis
- Prediction of improvement after anterior cruciate ligament reconstruction
- Effluent Osteopontin levels reflect the peritoneal solute transport rate
- circ_0038467 promotes PM2.5-induced bronchial epithelial cell dysfunction
- Significance of miR-141 and miR-340 in cervical squamous cell carcinoma
- Association between hair cortisol concentration and metabolic syndrome
- Microvessel density as a prognostic indicator of prostate cancer: A systematic review and meta-analysis
- Characteristics of BCR–ABL gene variants in patients of chronic myeloid leukemia
- Knee alterations in rheumatoid arthritis: Comparison of US and MRI
- Long non-coding RNA TUG1 aggravates cerebral ischemia and reperfusion injury by sponging miR-493-3p/miR-410-3p
- lncRNA MALAT1 regulated ATAD2 to facilitate retinoblastoma progression via miR-655-3p
- Development and validation of a nomogram for predicting severity in patients with hemorrhagic fever with renal syndrome: A retrospective study
- Analysis of COVID-19 outbreak origin in China in 2019 using differentiation method for unusual epidemiological events
- Laparoscopic versus open major liver resection for hepatocellular carcinoma: A case-matched analysis of short- and long-term outcomes
- Travelers’ vaccines and their adverse events in Nara, Japan
- Association between Tfh and PGA in children with Henoch–Schönlein purpura
- Can exchange transfusion be replaced by double-LED phototherapy?
- circ_0005962 functions as an oncogene to aggravate NSCLC progression
- Circular RNA VANGL1 knockdown suppressed viability, promoted apoptosis, and increased doxorubicin sensitivity through targeting miR-145-5p to regulate SOX4 in bladder cancer cells
- Serum intact fibroblast growth factor 23 in healthy paediatric population
- Algorithm of rational approach to reconstruction in Fournier’s disease
- A meta-analysis of exosome in the treatment of spinal cord injury
- Src-1 and SP2 promote the proliferation and epithelial–mesenchymal transition of nasopharyngeal carcinoma
- Dexmedetomidine may decrease the bupivacaine toxicity to heart
- Hypoxia stimulates the migration and invasion of osteosarcoma via up-regulating the NUSAP1 expression
- Long noncoding RNA XIST knockdown relieves the injury of microglia cells after spinal cord injury by sponging miR-219-5p
- External fixation via the anterior inferior iliac spine for proximal femoral fractures in young patients
- miR-128-3p reduced acute lung injury induced by sepsis via targeting PEL12
- HAGLR promotes neuron differentiation through the miR-130a-3p-MeCP2 axis
- Phosphoglycerate mutase 2 is elevated in serum of patients with heart failure and correlates with the disease severity and patient’s prognosis
- Cell population data in identifying active tuberculosis and community-acquired pneumonia
- Prognostic value of microRNA-4521 in non-small cell lung cancer and its regulatory effect on tumor progression
- Mean platelet volume and red blood cell distribution width is associated with prognosis in premature neonates with sepsis
- 3D-printed porous scaffold promotes osteogenic differentiation of hADMSCs
- Association of gene polymorphisms with women urinary incontinence
- Influence of COVID-19 pandemic on stress levels of urologic patients
- miR-496 inhibits proliferation via LYN and AKT pathway in gastric cancer
- miR-519d downregulates LEP expression to inhibit preeclampsia development
- Comparison of single- and triple-port VATS for lung cancer: A meta-analysis
- Fluorescent light energy modulates healing in skin grafted mouse model
- Silencing CDK6-AS1 inhibits LPS-induced inflammatory damage in HK-2 cells
- Predictive effect of DCE-MRI and DWI in brain metastases from NSCLC
- Severe postoperative hyperbilirubinemia in congenital heart disease
- Baicalin improves podocyte injury in rats with diabetic nephropathy by inhibiting PI3K/Akt/mTOR signaling pathway
- Clinical factors predicting ureteral stent failure in patients with external ureteral compression
- Novel H2S donor proglumide-ADT-OH protects HUVECs from ox-LDL-induced injury through NF-κB and JAK/SATA pathway
- Triple-Endobutton and clavicular hook: A propensity score matching analysis
- Long noncoding RNA MIAT inhibits the progression of diabetic nephropathy and the activation of NF-κB pathway in high glucose-treated renal tubular epithelial cells by the miR-182-5p/GPRC5A axis
- Serum exosomal miR-122-5p, GAS, and PGR in the non-invasive diagnosis of CAG
- miR-513b-5p inhibits the proliferation and promotes apoptosis of retinoblastoma cells by targeting TRIB1
- Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p
- The diagnostic and prognostic value of miR-92a in gastric cancer: A systematic review and meta-analysis
- Prognostic value of α2δ1 in hypopharyngeal carcinoma: A retrospective study
- No significant benefit of moderate-dose vitamin C on severe COVID-19 cases
- circ_0000467 promotes the proliferation, metastasis, and angiogenesis in colorectal cancer cells through regulating KLF12 expression by sponging miR-4766-5p
- Downregulation of RAB7 and Caveolin-1 increases MMP-2 activity in renal tubular epithelial cells under hypoxic conditions
- Educational program for orthopedic surgeons’ influences for osteoporosis
- Expression and function analysis of CRABP2 and FABP5, and their ratio in esophageal squamous cell carcinoma
- GJA1 promotes hepatocellular carcinoma progression by mediating TGF-β-induced activation and the epithelial–mesenchymal transition of hepatic stellate cells
- lncRNA-ZFAS1 promotes the progression of endometrial carcinoma by targeting miR-34b to regulate VEGFA expression
- Anticoagulation is the answer in treating noncritical COVID-19 patients
- Effect of late-onset hemorrhagic cystitis on PFS after haplo-PBSCT
- Comparison of Dako HercepTest and Ventana PATHWAY anti-HER2 (4B5) tests and their correlation with silver in situ hybridization in lung adenocarcinoma
- VSTM1 regulates monocyte/macrophage function via the NF-κB signaling pathway
- Comparison of vaginal birth outcomes in midwifery-led versus physician-led setting: A propensity score-matched analysis
- Treatment of osteoporosis with teriparatide: The Slovenian experience
- New targets of morphine postconditioning protection of the myocardium in ischemia/reperfusion injury: Involvement of HSP90/Akt and C5a/NF-κB
- Superenhancer–transcription factor regulatory network in malignant tumors
- β-Cell function is associated with osteosarcopenia in middle-aged and older nonobese patients with type 2 diabetes: A cross-sectional study
- Clinical features of atypical tuberculosis mimicking bacterial pneumonia
- Proteoglycan-depleted regions of annular injury promote nerve ingrowth in a rabbit disc degeneration model
- Effect of electromagnetic field on abortion: A systematic review and meta-analysis
- miR-150-5p affects AS plaque with ASMC proliferation and migration by STAT1
- MALAT1 promotes malignant pleural mesothelioma by sponging miR-141-3p
- Effects of remifentanil and propofol on distant organ lung injury in an ischemia–reperfusion model
- miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway
- Identification of LIG1 and LIG3 as prognostic biomarkers in breast cancer
- MitoQ inhibits hepatic stellate cell activation and liver fibrosis by enhancing PINK1/parkin-mediated mitophagy
- Dissecting role of founder mutation p.V727M in GNE in Indian HIBM cohort
- circATP2A2 promotes osteosarcoma progression by upregulating MYH9
- Prognostic role of oxytocin receptor in colon adenocarcinoma
- Review Articles
- The function of non-coding RNAs in idiopathic pulmonary fibrosis
- Efficacy and safety of therapeutic plasma exchange in stiff person syndrome
- Role of cesarean section in the development of neonatal gut microbiota: A systematic review
- Small cell lung cancer transformation during antitumor therapies: A systematic review
- Research progress of gut microbiota and frailty syndrome
- Recommendations for outpatient activity in COVID-19 pandemic
- Rapid Communication
- Disparity in clinical characteristics between 2019 novel coronavirus pneumonia and leptospirosis
- Use of microspheres in embolization for unruptured renal angiomyolipomas
- COVID-19 cases with delayed absorption of lung lesion
- A triple combination of treatments on moderate COVID-19
- Social networks and eating disorders during the Covid-19 pandemic
- Letter
- COVID-19, WHO guidelines, pedagogy, and respite
- Inflammatory factors in alveolar lavage fluid from severe COVID-19 pneumonia: PCT and IL-6 in epithelial lining fluid
- COVID-19: Lessons from Norway tragedy must be considered in vaccine rollout planning in least developed/developing countries
- What is the role of plasma cell in the lamina propria of terminal ileum in Good’s syndrome patient?
- Case Report
- Rivaroxaban triggered multifocal intratumoral hemorrhage of the cabozantinib-treated diffuse brain metastases: A case report and review of literature
- CTU findings of duplex kidney in kidney: A rare duplicated renal malformation
- Synchronous primary malignancy of colon cancer and mantle cell lymphoma: A case report
- Sonazoid-enhanced ultrasonography and pathologic characters of CD68 positive cell in primary hepatic perivascular epithelioid cell tumors: A case report and literature review
- Persistent SARS-CoV-2-positive over 4 months in a COVID-19 patient with CHB
- Pulmonary parenchymal involvement caused by Tropheryma whipplei
- Mediastinal mixed germ cell tumor: A case report and literature review
- Ovarian female adnexal tumor of probable Wolffian origin – Case report
- Rare paratesticular aggressive angiomyxoma mimicking an epididymal tumor in an 82-year-old man: Case report
- Perimenopausal giant hydatidiform mole complicated with preeclampsia and hyperthyroidism: A case report and literature review
- Primary orbital ganglioneuroblastoma: A case report
- Primary aortic intimal sarcoma masquerading as intramural hematoma
- Sustained false-positive results for hepatitis A virus immunoglobulin M: A case report and literature review
- Peritoneal loose body presenting as a hepatic mass: A case report and review of the literature
- Chondroblastoma of mandibular condyle: Case report and literature review
- Trauma-induced complete pacemaker lead fracture 8 months prior to hospitalization: A case report
- Primary intradural extramedullary extraosseous Ewing’s sarcoma/peripheral primitive neuroectodermal tumor (PIEES/PNET) of the thoracolumbar spine: A case report and literature review
- Computer-assisted preoperative planning of reduction of and osteosynthesis of scapular fracture: A case report
- High quality of 58-month life in lung cancer patient with brain metastases sequentially treated with gefitinib and osimertinib
- Rapid response of locally advanced oral squamous cell carcinoma to apatinib: A case report
- Retrieval of intrarenal coiled and ruptured guidewire by retrograde intrarenal surgery: A case report and literature review
- Usage of intermingled skin allografts and autografts in a senior patient with major burn injury
- Retraction
- Retraction on “Dihydromyricetin attenuates inflammation through TLR4/NF-kappa B pathway”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part I
- An artificial immune system with bootstrap sampling for the diagnosis of recurrent endometrial cancers
- Breast cancer recurrence prediction with ensemble methods and cost-sensitive learning
Artikel in diesem Heft
- Research Articles
- Identification of ZG16B as a prognostic biomarker in breast cancer
- Behçet’s disease with latent Mycobacterium tuberculosis infection
- Erratum
- Erratum to “Suffering from Cerebral Small Vessel Disease with and without Metabolic Syndrome”
- Research Articles
- GPR37 promotes the malignancy of lung adenocarcinoma via TGF-β/Smad pathway
- Expression and role of ABIN1 in sepsis: In vitro and in vivo studies
- Additional baricitinib loading dose improves clinical outcome in COVID-19
- The co-treatment of rosuvastatin with dapagliflozin synergistically inhibited apoptosis via activating the PI3K/AKt/mTOR signaling pathway in myocardial ischemia/reperfusion injury rats
- SLC12A8 plays a key role in bladder cancer progression and EMT
- LncRNA ATXN8OS enhances tamoxifen resistance in breast cancer
- Case Report
- Serratia marcescens as a cause of unfavorable outcome in the twin pregnancy
- Spleno-adrenal fusion mimicking an adrenal metastasis of a renal cell carcinoma: A case report and embryological background
- Research Articles
- TRIM25 contributes to the malignancy of acute myeloid leukemia and is negatively regulated by microRNA-137
- CircRNA circ_0004370 promotes cell proliferation, migration, and invasion and inhibits cell apoptosis of esophageal cancer via miR-1301-3p/COL1A1 axis
- LncRNA XIST regulates atherosclerosis progression in ox-LDL-induced HUVECs
- Potential role of IFN-γ and IL-5 in sepsis prediction of preterm neonates
- Rapid Communication
- COVID-19 vaccine: Call for employees in international transportation industries and international travelers as the first priority in global distribution
- Case Report
- Rare squamous cell carcinoma of the kidney with concurrent xanthogranulomatous pyelonephritis: A case report and review of the literature
- An infertile female delivered a baby after removal of primary renal carcinoid tumor
- Research Articles
- Hypertension, BMI, and cardiovascular and cerebrovascular diseases
- Case Report
- Coexistence of bilateral macular edema and pale optic disc in the patient with Cohen syndrome
- Research Articles
- Correlation between kinematic sagittal parameters of the cervical lordosis or head posture and disc degeneration in patients with posterior neck pain
- Review Articles
- Hepatoid adenocarcinoma of the lung: An analysis of the Surveillance, Epidemiology, and End Results (SEER) database
- Research Articles
- Thermography in the diagnosis of carpal tunnel syndrome
- Pemetrexed-based first-line chemotherapy had particularly prominent objective response rate for advanced NSCLC: A network meta-analysis
- Comparison of single and double autologous stem cell transplantation in multiple myeloma patients
- The influence of smoking in minimally invasive spinal fusion surgery
- Impact of body mass index on left atrial dimension in HOCM patients
- Expression and clinical significance of CMTM1 in hepatocellular carcinoma
- miR-142-5p promotes cervical cancer progression by targeting LMX1A through Wnt/β-catenin pathway
- Comparison of multiple flatfoot indicators in 5–8-year-old children
- Early MRI imaging and follow-up study in cerebral amyloid angiopathy
- Intestinal fatty acid-binding protein as a biomarker for the diagnosis of strangulated intestinal obstruction: A meta-analysis
- miR-128-3p inhibits apoptosis and inflammation in LPS-induced sepsis by targeting TGFBR2
- Dynamic perfusion CT – A promising tool to diagnose pancreatic ductal adenocarcinoma
- Biomechanical evaluation of self-cinching stitch techniques in rotator cuff repair: The single-loop and double-loop knot stitches
- Review Articles
- The ambiguous role of mannose-binding lectin (MBL) in human immunity
- Case Report
- Membranous nephropathy with pulmonary cryptococcosis with improved 1-year follow-up results: A case report
- Fertility problems in males carrying an inversion of chromosome 10
- Acute myeloid leukemia with leukemic pleural effusion and high levels of pleural adenosine deaminase: A case report and review of literature
- Metastatic renal Ewing’s sarcoma in adult woman: Case report and review of the literature
- Burkitt-like lymphoma with 11q aberration in a patient with AIDS and a patient without AIDS: Two cases reports and literature review
- Skull hemophilia pseudotumor: A case report
- Judicious use of low-dosage corticosteroids for non-severe COVID-19: A case report
- Adult-onset citrullinaemia type II with liver cirrhosis: A rare cause of hyperammonaemia
- Clinicopathologic features of Good’s syndrome: Two cases and literature review
- Fatal immune-related hepatitis with intrahepatic cholestasis and pneumonia associated with camrelizumab: A case report and literature review
- Research Articles
- Effects of hydroxyethyl starch and gelatin on the risk of acute kidney injury following orthotopic liver transplantation: A multicenter retrospective comparative clinical study
- Significance of nucleic acid positive anal swab in COVID-19 patients
- circAPLP2 promotes colorectal cancer progression by upregulating HELLS by targeting miR-335-5p
- Ratios between circulating myeloid cells and lymphocytes are associated with mortality in severe COVID-19 patients
- Risk factors of left atrial appendage thrombus in patients with non-valvular atrial fibrillation
- Clinical features of hypertensive patients with COVID-19 compared with a normotensive group: Single-center experience in China
- Surgical myocardial revascularization outcomes in Kawasaki disease: systematic review and meta-analysis
- Decreased chromobox homologue 7 expression is associated with epithelial–mesenchymal transition and poor prognosis in cervical cancer
- FGF16 regulated by miR-520b enhances the cell proliferation of lung cancer
- Platelet-rich fibrin: Basics of biological actions and protocol modifications
- Accurate diagnosis of prostate cancer using logistic regression
- miR-377 inhibition enhances the survival of trophoblast cells via upregulation of FNDC5 in gestational diabetes mellitus
- Prognostic significance of TRIM28 expression in patients with breast carcinoma
- Integrative bioinformatics analysis of KPNA2 in six major human cancers
- Exosomal-mediated transfer of OIP5-AS1 enhanced cell chemoresistance to trastuzumab in breast cancer via up-regulating HMGB3 by sponging miR-381-3p
- A four-lncRNA signature for predicting prognosis of recurrence patients with gastric cancer
- Knockdown of circ_0003204 alleviates oxidative low-density lipoprotein-induced human umbilical vein endothelial cells injury: Circulating RNAs could explain atherosclerosis disease progression
- Propofol postpones colorectal cancer development through circ_0026344/miR-645/Akt/mTOR signal pathway
- Knockdown of lncRNA TapSAKI alleviates LPS-induced injury in HK-2 cells through the miR-205/IRF3 pathway
- COVID-19 severity in relation to sociodemographics and vitamin D use
- Clinical analysis of 11 cases of nocardiosis
- Cis-regulatory elements in conserved non-coding sequences of nuclear receptor genes indicate for crosstalk between endocrine systems
- Four long noncoding RNAs act as biomarkers in lung adenocarcinoma
- Real-world evidence of cytomegalovirus reactivation in non-Hodgkin lymphomas treated with bendamustine-containing regimens
- Relation between IL-8 level and obstructive sleep apnea syndrome
- circAGFG1 sponges miR-28-5p to promote non-small-cell lung cancer progression through modulating HIF-1α level
- Nomogram prediction model for renal anaemia in IgA nephropathy patients
- Effect of antibiotic use on the efficacy of nivolumab in the treatment of advanced/metastatic non-small cell lung cancer: A meta-analysis
- NDRG2 inhibition facilitates angiogenesis of hepatocellular carcinoma
- A nomogram for predicting metabolic steatohepatitis: The combination of NAMPT, RALGDS, GADD45B, FOSL2, RTP3, and RASD1
- Clinical and prognostic features of MMP-2 and VEGF in AEG patients
- The value of miR-510 in the prognosis and development of colon cancer
- Functional implications of PABPC1 in the development of ovarian cancer
- Prognostic value of preoperative inflammation-based predictors in patients with bladder carcinoma after radical cystectomy
- Sublingual immunotherapy increases Treg/Th17 ratio in allergic rhinitis
- Prediction of improvement after anterior cruciate ligament reconstruction
- Effluent Osteopontin levels reflect the peritoneal solute transport rate
- circ_0038467 promotes PM2.5-induced bronchial epithelial cell dysfunction
- Significance of miR-141 and miR-340 in cervical squamous cell carcinoma
- Association between hair cortisol concentration and metabolic syndrome
- Microvessel density as a prognostic indicator of prostate cancer: A systematic review and meta-analysis
- Characteristics of BCR–ABL gene variants in patients of chronic myeloid leukemia
- Knee alterations in rheumatoid arthritis: Comparison of US and MRI
- Long non-coding RNA TUG1 aggravates cerebral ischemia and reperfusion injury by sponging miR-493-3p/miR-410-3p
- lncRNA MALAT1 regulated ATAD2 to facilitate retinoblastoma progression via miR-655-3p
- Development and validation of a nomogram for predicting severity in patients with hemorrhagic fever with renal syndrome: A retrospective study
- Analysis of COVID-19 outbreak origin in China in 2019 using differentiation method for unusual epidemiological events
- Laparoscopic versus open major liver resection for hepatocellular carcinoma: A case-matched analysis of short- and long-term outcomes
- Travelers’ vaccines and their adverse events in Nara, Japan
- Association between Tfh and PGA in children with Henoch–Schönlein purpura
- Can exchange transfusion be replaced by double-LED phototherapy?
- circ_0005962 functions as an oncogene to aggravate NSCLC progression
- Circular RNA VANGL1 knockdown suppressed viability, promoted apoptosis, and increased doxorubicin sensitivity through targeting miR-145-5p to regulate SOX4 in bladder cancer cells
- Serum intact fibroblast growth factor 23 in healthy paediatric population
- Algorithm of rational approach to reconstruction in Fournier’s disease
- A meta-analysis of exosome in the treatment of spinal cord injury
- Src-1 and SP2 promote the proliferation and epithelial–mesenchymal transition of nasopharyngeal carcinoma
- Dexmedetomidine may decrease the bupivacaine toxicity to heart
- Hypoxia stimulates the migration and invasion of osteosarcoma via up-regulating the NUSAP1 expression
- Long noncoding RNA XIST knockdown relieves the injury of microglia cells after spinal cord injury by sponging miR-219-5p
- External fixation via the anterior inferior iliac spine for proximal femoral fractures in young patients
- miR-128-3p reduced acute lung injury induced by sepsis via targeting PEL12
- HAGLR promotes neuron differentiation through the miR-130a-3p-MeCP2 axis
- Phosphoglycerate mutase 2 is elevated in serum of patients with heart failure and correlates with the disease severity and patient’s prognosis
- Cell population data in identifying active tuberculosis and community-acquired pneumonia
- Prognostic value of microRNA-4521 in non-small cell lung cancer and its regulatory effect on tumor progression
- Mean platelet volume and red blood cell distribution width is associated with prognosis in premature neonates with sepsis
- 3D-printed porous scaffold promotes osteogenic differentiation of hADMSCs
- Association of gene polymorphisms with women urinary incontinence
- Influence of COVID-19 pandemic on stress levels of urologic patients
- miR-496 inhibits proliferation via LYN and AKT pathway in gastric cancer
- miR-519d downregulates LEP expression to inhibit preeclampsia development
- Comparison of single- and triple-port VATS for lung cancer: A meta-analysis
- Fluorescent light energy modulates healing in skin grafted mouse model
- Silencing CDK6-AS1 inhibits LPS-induced inflammatory damage in HK-2 cells
- Predictive effect of DCE-MRI and DWI in brain metastases from NSCLC
- Severe postoperative hyperbilirubinemia in congenital heart disease
- Baicalin improves podocyte injury in rats with diabetic nephropathy by inhibiting PI3K/Akt/mTOR signaling pathway
- Clinical factors predicting ureteral stent failure in patients with external ureteral compression
- Novel H2S donor proglumide-ADT-OH protects HUVECs from ox-LDL-induced injury through NF-κB and JAK/SATA pathway
- Triple-Endobutton and clavicular hook: A propensity score matching analysis
- Long noncoding RNA MIAT inhibits the progression of diabetic nephropathy and the activation of NF-κB pathway in high glucose-treated renal tubular epithelial cells by the miR-182-5p/GPRC5A axis
- Serum exosomal miR-122-5p, GAS, and PGR in the non-invasive diagnosis of CAG
- miR-513b-5p inhibits the proliferation and promotes apoptosis of retinoblastoma cells by targeting TRIB1
- Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p
- The diagnostic and prognostic value of miR-92a in gastric cancer: A systematic review and meta-analysis
- Prognostic value of α2δ1 in hypopharyngeal carcinoma: A retrospective study
- No significant benefit of moderate-dose vitamin C on severe COVID-19 cases
- circ_0000467 promotes the proliferation, metastasis, and angiogenesis in colorectal cancer cells through regulating KLF12 expression by sponging miR-4766-5p
- Downregulation of RAB7 and Caveolin-1 increases MMP-2 activity in renal tubular epithelial cells under hypoxic conditions
- Educational program for orthopedic surgeons’ influences for osteoporosis
- Expression and function analysis of CRABP2 and FABP5, and their ratio in esophageal squamous cell carcinoma
- GJA1 promotes hepatocellular carcinoma progression by mediating TGF-β-induced activation and the epithelial–mesenchymal transition of hepatic stellate cells
- lncRNA-ZFAS1 promotes the progression of endometrial carcinoma by targeting miR-34b to regulate VEGFA expression
- Anticoagulation is the answer in treating noncritical COVID-19 patients
- Effect of late-onset hemorrhagic cystitis on PFS after haplo-PBSCT
- Comparison of Dako HercepTest and Ventana PATHWAY anti-HER2 (4B5) tests and their correlation with silver in situ hybridization in lung adenocarcinoma
- VSTM1 regulates monocyte/macrophage function via the NF-κB signaling pathway
- Comparison of vaginal birth outcomes in midwifery-led versus physician-led setting: A propensity score-matched analysis
- Treatment of osteoporosis with teriparatide: The Slovenian experience
- New targets of morphine postconditioning protection of the myocardium in ischemia/reperfusion injury: Involvement of HSP90/Akt and C5a/NF-κB
- Superenhancer–transcription factor regulatory network in malignant tumors
- β-Cell function is associated with osteosarcopenia in middle-aged and older nonobese patients with type 2 diabetes: A cross-sectional study
- Clinical features of atypical tuberculosis mimicking bacterial pneumonia
- Proteoglycan-depleted regions of annular injury promote nerve ingrowth in a rabbit disc degeneration model
- Effect of electromagnetic field on abortion: A systematic review and meta-analysis
- miR-150-5p affects AS plaque with ASMC proliferation and migration by STAT1
- MALAT1 promotes malignant pleural mesothelioma by sponging miR-141-3p
- Effects of remifentanil and propofol on distant organ lung injury in an ischemia–reperfusion model
- miR-654-5p promotes gastric cancer progression via the GPRIN1/NF-κB pathway
- Identification of LIG1 and LIG3 as prognostic biomarkers in breast cancer
- MitoQ inhibits hepatic stellate cell activation and liver fibrosis by enhancing PINK1/parkin-mediated mitophagy
- Dissecting role of founder mutation p.V727M in GNE in Indian HIBM cohort
- circATP2A2 promotes osteosarcoma progression by upregulating MYH9
- Prognostic role of oxytocin receptor in colon adenocarcinoma
- Review Articles
- The function of non-coding RNAs in idiopathic pulmonary fibrosis
- Efficacy and safety of therapeutic plasma exchange in stiff person syndrome
- Role of cesarean section in the development of neonatal gut microbiota: A systematic review
- Small cell lung cancer transformation during antitumor therapies: A systematic review
- Research progress of gut microbiota and frailty syndrome
- Recommendations for outpatient activity in COVID-19 pandemic
- Rapid Communication
- Disparity in clinical characteristics between 2019 novel coronavirus pneumonia and leptospirosis
- Use of microspheres in embolization for unruptured renal angiomyolipomas
- COVID-19 cases with delayed absorption of lung lesion
- A triple combination of treatments on moderate COVID-19
- Social networks and eating disorders during the Covid-19 pandemic
- Letter
- COVID-19, WHO guidelines, pedagogy, and respite
- Inflammatory factors in alveolar lavage fluid from severe COVID-19 pneumonia: PCT and IL-6 in epithelial lining fluid
- COVID-19: Lessons from Norway tragedy must be considered in vaccine rollout planning in least developed/developing countries
- What is the role of plasma cell in the lamina propria of terminal ileum in Good’s syndrome patient?
- Case Report
- Rivaroxaban triggered multifocal intratumoral hemorrhage of the cabozantinib-treated diffuse brain metastases: A case report and review of literature
- CTU findings of duplex kidney in kidney: A rare duplicated renal malformation
- Synchronous primary malignancy of colon cancer and mantle cell lymphoma: A case report
- Sonazoid-enhanced ultrasonography and pathologic characters of CD68 positive cell in primary hepatic perivascular epithelioid cell tumors: A case report and literature review
- Persistent SARS-CoV-2-positive over 4 months in a COVID-19 patient with CHB
- Pulmonary parenchymal involvement caused by Tropheryma whipplei
- Mediastinal mixed germ cell tumor: A case report and literature review
- Ovarian female adnexal tumor of probable Wolffian origin – Case report
- Rare paratesticular aggressive angiomyxoma mimicking an epididymal tumor in an 82-year-old man: Case report
- Perimenopausal giant hydatidiform mole complicated with preeclampsia and hyperthyroidism: A case report and literature review
- Primary orbital ganglioneuroblastoma: A case report
- Primary aortic intimal sarcoma masquerading as intramural hematoma
- Sustained false-positive results for hepatitis A virus immunoglobulin M: A case report and literature review
- Peritoneal loose body presenting as a hepatic mass: A case report and review of the literature
- Chondroblastoma of mandibular condyle: Case report and literature review
- Trauma-induced complete pacemaker lead fracture 8 months prior to hospitalization: A case report
- Primary intradural extramedullary extraosseous Ewing’s sarcoma/peripheral primitive neuroectodermal tumor (PIEES/PNET) of the thoracolumbar spine: A case report and literature review
- Computer-assisted preoperative planning of reduction of and osteosynthesis of scapular fracture: A case report
- High quality of 58-month life in lung cancer patient with brain metastases sequentially treated with gefitinib and osimertinib
- Rapid response of locally advanced oral squamous cell carcinoma to apatinib: A case report
- Retrieval of intrarenal coiled and ruptured guidewire by retrograde intrarenal surgery: A case report and literature review
- Usage of intermingled skin allografts and autografts in a senior patient with major burn injury
- Retraction
- Retraction on “Dihydromyricetin attenuates inflammation through TLR4/NF-kappa B pathway”
- Special Issue Computational Intelligence Methodologies Meets Recurrent Cancers - Part I
- An artificial immune system with bootstrap sampling for the diagnosis of recurrent endometrial cancers
- Breast cancer recurrence prediction with ensemble methods and cost-sensitive learning

