NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
-
Xuejiao Lv
, Wenfeng Zhang
und Yan Meng
Abstract
We intended to explore the potential molecular mechanisms underlying the cardiac conduction block inducted by urea transporter (UT)-B deletion at the transcriptome level. The heart tissues were harvested from UT-B null mice and age-matched wild-type mice for lncRNA sequencing analysis. Based on the sequencing data, the differentially expressed mRNAs (DEMs) and lncRNAs (DELs) between UT-B knockout and control groups were identified, followed by function analysis and mRNA–lncRNA co-expression analysis. The miRNAs were predicted, and then the competing endogenous RNA (ceRNA) network was constructed. UT-B deletion results in the aberrant expression of 588 lncRNAs and 194 mRNAs. These DEMs were significantly enriched in the inflammation-related pathway. A lncRNA–mRNA co-expression network and a ceRNA network were constructed on the basis of the DEMs and DELs. The complement 7 (C7)–NONMMUT137216.1 co-expression pair had the highest correlation coefficient in the co-expression network. NONMMUT140591.1 had the highest degree in the ceRNA network and was involved in the ceRNA of NONMMUT140591.1-mmu-miR-298-5p-Gata5 (GATA binding protein 5). UT-B deletion may promote cardiac conduction block via inflammatory process. The ceRNA NONMMUT140591.1-mmu-miR-298-5p-Gata5 may be a potential molecular mechanism of UT-B knockout-induced cardiac conduction block.
Graphical abstract

1 Introduction
Cardiac conduction disease, characterized by the impaired integrity of the conduction system, is a serious, life threatening disease of the heart [1,2]. An impaired conduction system can result in slowed or even blocked impulse conduction, eventually leading to life-threatening rhythm disturbances [3]. The pathogenesis of the cardiac conduction disease is diverse, which can be caused by an acquired injury such as drug toxicity or ischemia, and is associated with heart diseases and neuromuscular diseases [4]. In the last decade, some ion channels associated genes that are responsible for the inherited cardiac conduction disease have been detected, such as KCNJ2, SCN5A, and HCN4 [5].
Urea transporters (UTs) are a family of small membrane proteins with specific permeability to urea, which include two types in mammals: UT-A and UT-B [6]. UT-A is mainly expressed in the kidney epithelial cells, while UT-B is widely distributed in the brain, heart, kidney, testis, bone marrow, urinary tract, and other tissues [7]. UT-B has a high expression in heart. Our previous study has found that UT-B deletion mice have prolonged P–R intervals from 6 to 52 weeks, indicating a delayed conduction from the atria to the ventricles [8]. Meng et al. [9] reported that the progressive heart block in UT-B null mice may be associated with the accumulated urea in cells. Urea has been considered to have negligible toxicity for a long time. Elevated blood urea in chronic renal failure is thought to have no influence on survival in patients [10]. High urea might have an important role in accelerated atherosclerosis in chronic dialysis patients [11]. Nevertheless, the role of urea accumulation in heart disease is still controversial.
In this study, the urea transporter B null mice and all urea transporters null mice provide us with proper tools to eliminate other interference factors. We have subtly controlled the concentration of urea through the knockout of the urea transporters to explore the potential molecular mechanisms underlying the cardiac conduction block induced by UT-B deletion at the transcriptome level. The RNA expression profiles of the heart tissues in UT-B knockout mice were analyzed by comparing with age-matched wild-type mice. The differentially expressed mRNAs (DEMs) and lncRNAs (DELs) between UT-B knockout and control groups were identified. Based on these DEMs and DELs, the competing endogenous RNA (ceRNA) network was constructed to explore the possible mechanism. The results may improve our understanding of the progression of cardiac conduction defect.
2 Materials and methods
2.1 Animals and tissue preparation
UT-B null mice were kindly provided by the University of California, San Francisco, School of Medicine. All the animals were kept in a standard experimental animal laboratory at the animal experiment center of Jilin University. Mice were raised with plenty of food and water under a 12:12 h light:dark cycle at 22 ± 2°C. In order to obtain stable UT-B null mice, the mice were mated with wild-type mice to obtain a heterozygous offspring. Then the heterozygous offspring were mated to obtain stable UT-B knockout mice, and wild-type mice from the same litter were used as control. At 16 weeks of age, three wild and null mice were respectively selected and anesthetized with 1% pentobarbital sodium. The hearts were harvested through thoracotomy and frozen in liquid nitrogen for sequencing analysis. The expression of UT-B in the UT-B knockout mice was verified, as shown in Figure 1.

The expression of UT-B in UT-B null mice detected by reverse-transcription (RT)-PCR (a) and western blot (b).
-
Ethical approval: The research related to animal use has complied with all the relevant national regulations and institutional policies for the care and use of animals, and were approved by the animal ethics committee of the Basic Medical Sciences unit of Jilin University (No. [2021]202014).
2.2 RNA extraction and RNA library construction for sequencing
Total RNA was extracted from heart tissue using TRIzol (Invitrogen; Carlsbad, CA, United States). The RNA quality was detected through gel electrophoresis. The concentration of RNA was detected using a NanoDrop ND-1000 spectrophotometer.
After removal of rRNA from the total RNA, the RNA samples were randomly cut into short fragments which were used to synthesize the first-strand cDNA using random hexamers. The synthesis of second-strand cDNA was performed through the mixing of first-strand cDNA with dNTPs, RNase H, and DNA polymerase I. After purification, the cDNA was degraded with uracil-N-glycosylase. The RNA fragments were separated using agarose gel electrophoresis, followed by amplification with polymerase chain reaction (PCR). The PCR products were sequenced on an Illumina HiSeq™ 2000 instrument (Illumina, Inc.; San Diego, CA, United States). The raw data were uploaded to the Gene Expression Omnibus (GSE168717, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168717).
2.3 Correlation analysis and principal component analysis (PCA) among samples
The expression level correlation between samples is an important index to test the reliability of the experiment and the rationality of sample selection. Using the cor function in R3.4.1, the Pearson correlation coefficient (p) between the two samples was calculated. For PCA, the prcomp function was used for data dimension reduction, and the ggfortify package was used to draw the PCA map.
2.4 DEMs and DELs identification
DESeq [12] software was used to conduct normalization processing on the counts of the genes of each sample (expression quantity was estimated using basemean value), and to calculate the fold change (FC). The negative binomial distribution test was used to conduct the difference significance test on reads. The differential genes (DEMs and DELs) were screened according to the FC and the results of the difference significance test. Here, the screening conditions were p < 0.05 and the FC >2.
2.5 Function and pathway enrichment analyses
After getting the differentially expressed genes, gene ontology (GO) (molecular function [MF], biological process [BP], and cellular component [CC]), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed by combining the GO [13] and KEGG [14] databases. The significance of DEMs enrichment in each GO term or pathway was calculated by the hypergeometric distribution test. The p value was adjusted using the Benjamini and Hochberg method. The threshold was set as count ≥3 and adjusted p value was <0.05.
2.6 Co-expression analysis of DELs and DEMs and function analysis of DELs
The Pearson correlation test was used to calculate the expression correlation between DELs and DEMs based on the gene expression data. The threshold values were set as |cor| >0.8 and p value <0.05. For each of the DELs, their co-expressed mRNAs were calculated, and the significance of DEMs enriched in each GO (or pathway) terms was calculated using the hypergeometric distribution test. The p value was adjusted using the Benjamini and Hochberg method. The threshold was set as count ≥3 and adjusted p value <0.05.
2.7 Cis of lncRNA and its adjacent coding gene analysis and lncRNA trans analysis
All the coding genes within the range of 100k in the upstream and downstream of DELs were searched, which were intersected with the genes that were significantly co-expressed with the lncRNAs (Pearson correlation calculation). These genes that were close to each other on the genome and had co-expression patterns were likely to be regulated by this lncRNA.
Based on the co-expression analysis results, the lncRNAs and mRNAs that were not on the same chromosome were screened out as candidate targets and the candidate sequences were extracted. The binding of candidate lncRNAs and genes at the nucleic acid level was predicted using RIsearch-2.0. The screening conditions were: the number of bases on which the two nucleic acid molecules interacted directly must be ≥10 and the base binding free energy ≤−50.
2.8 miRNA prediction and ceRNA network analysis
On the basis of the co-expression relationship of lncRNA–mRNA, the upstream miRNAs of mRNAs were predicted using the validated target module in the miRWalk2.0 [13] tool [15]. For lncRNAs in the lncRNA–mRNA co-expression relationship, we predicted the miRNA binding sites of the lncRNAs through miranda (v3.3a) [16] with parameters of score ≥140 and energy ≤−20, and obtained the miRNA–lncRNA relationship pairs. Based on the obtained lncRNA–miRNA and mRNA–miRNA relationships, the lncRNA–miRNA–mRNA relation pairs were screened, which were further screened according to the positive co-expression relationship between mRNA and lncRNA (correlation coefficient >0.95). Cytoscape (version 3.4.0) [17] was used for network building, namely the ceRNA network. The lncRNAs and mRNAs that had a positive co-expression relationship and were regulated by the same miRNA in the ceRNA network were considered as ceRNA. Finally, the node connection degree was analyzed using the Cytoscape plugin CytoNCA (version 2.1.6) [18].
3 Results
3.1 Correlation analysis and PCA among samples
Based on the mRNA and lncRNA expression matrices of each sample, the correlation between the samples was evaluated through the Pearson correlation coefficient. The closer p was to 1, the higher was the similarity of expression patterns between the samples (Figure 2a). The PCA map is shown in Figure 2b, indicating that the samples in the two groups were completely separated.
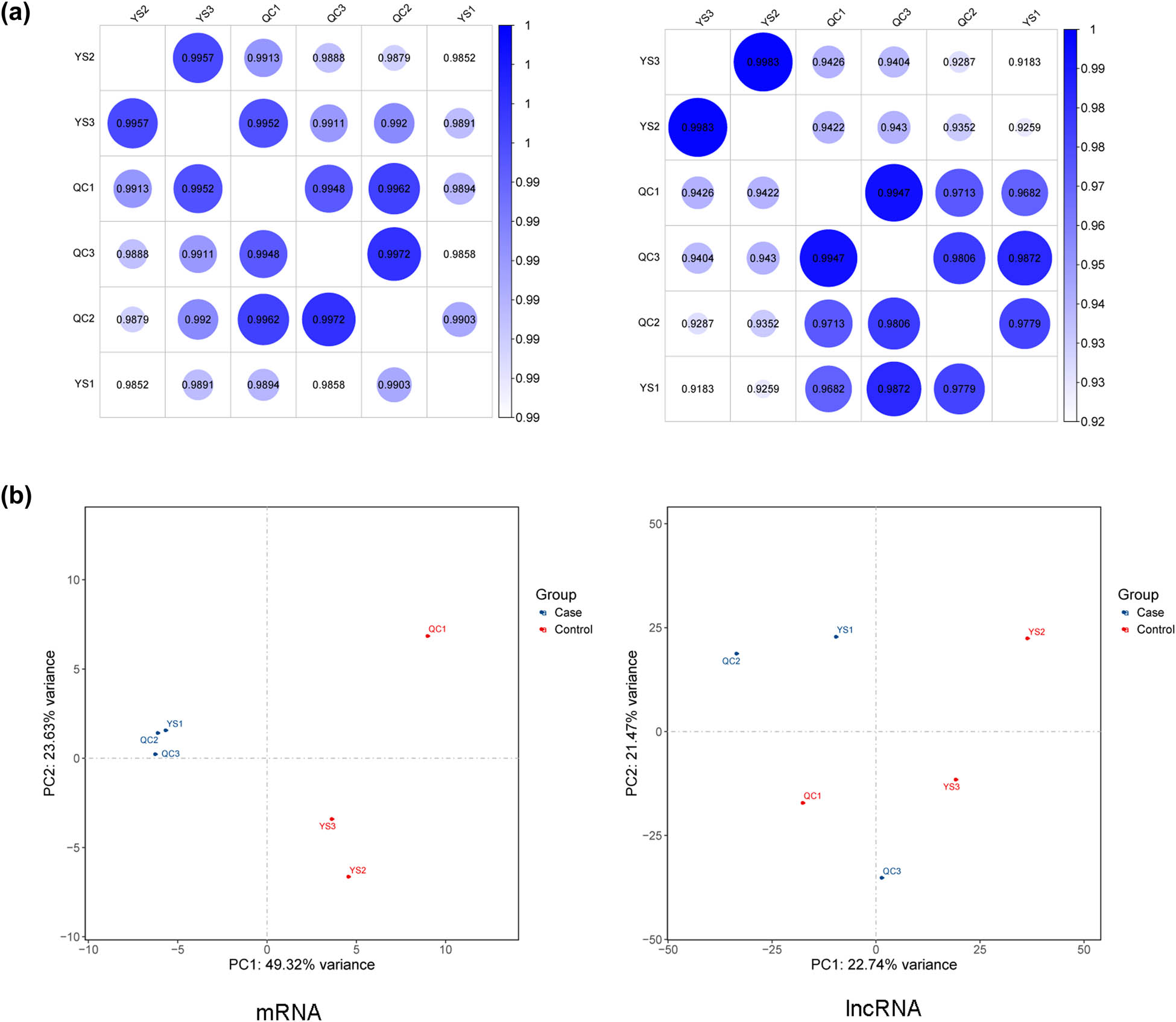
(a) Sample correlation heatmaps based on the expressive abundance of mRNA and lncRNA and (b) PCA maps for mRNA and lncRNA.
3.2 Differential expression analysis
According to the thresholds of p value <0.05 and |log2FC| >1,588 (278 up- and 310 downregulated) DELs and 194 (48 up- and 194 downregulated) the DEMs were identified. There were a greater number of downregulated than upregulated DELs and DEMs, and the NONMMUT140591.1 was a downregulated lncRNA. Bidirectional hierarchical clustering heatmaps are shown in Figure 3, which suggested that DELs and DEMs were well distinguished between the samples from the samples in the two groups.
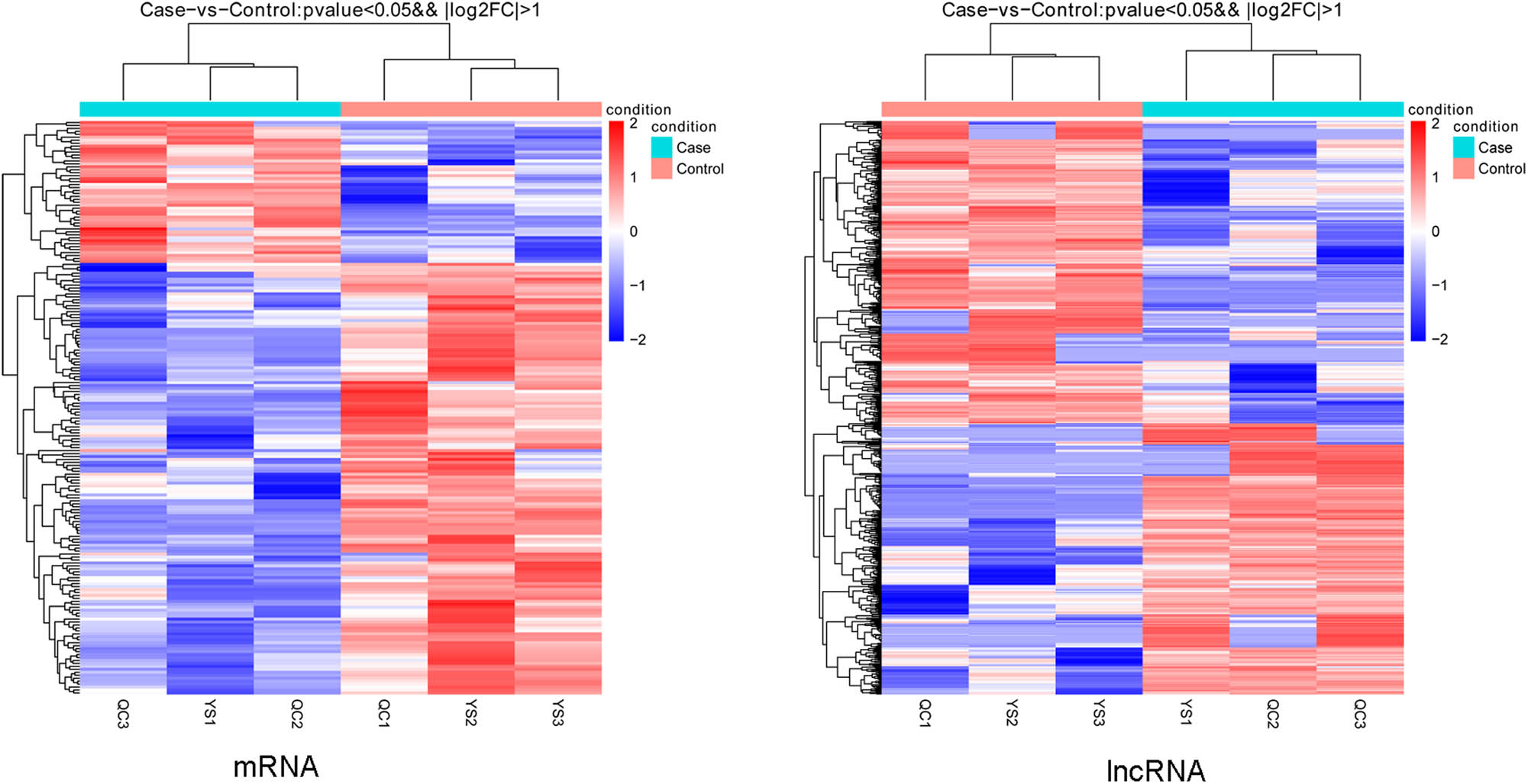
The clustering heatmaps of mRNAs and lncRNAs.
3.3 Functional enrichment analysis of up- and downregulated DEMs
The GO (BP, CC, and MF) functional enrichment analysis and the KEGG pathway enrichment analysis were performed on the up- and downregulated mRNAs, respectively. The results showed that seven BPs (such as DNA repair, nervous system development, and ion transport), three CCs (chromosome, nucleus, and nucleoplasm), three MFs (microtubule binding, ATPase activity, and DNA binding) (Figure 4a), and one KEGG pathway (viral carcinogenesis) (Figure 4c) were significantly enriched on the up-regulated mRNAs. Downregulated mRNAs were significantly enriched in 64 BPs (such as neutrophil chemotaxis, chemotaxis, and regulation of membrane depolarization), 19 CCs (such as extracellular region and extracellular space), 21 MFs (insulin-like growth factor binding and calcium ion binding) (Figure 4b), and 35 KEGG pathways (such as cytokine–cytokine receptor interaction, chemokine signaling pathway, interleukin (IL)-17 signaling pathway, and mitogen‑activated protein kinase signaling pathway) (Figure 4d).

The functions (a and b) and pathways (c and d) enriched by up- and downregulated mRNAs.
3.4 lncRNA–mRNA co-expression analysis
A total of 27,640 lncRNA–mRNA co-expression relation pairs, including 587 lncRNAs and 194 mRNAs, were obtained under |cor| >0.8 and p value <0.05. The top 500 pairs according to the p values were selected to construct the co-expression network (Figure 5), and the C7–NONMMUT137216.1 co-expression pair had the highest correlation coefficient in the co-expression network.

mRNA–lncRNA co-expression network (red triangle represents upregulated mRNA; the green inverted triangle represents downregulated mRNA; pink ellipse represents the upregulated lncRNA; the blue rectangle represents the downregulated lncRNA; the yellow line represents the positive co-expression relation; and the gray line represents the negative co-expression relation).
3.5 Function analysis of DELs
The top 10 lncRNAs, including NONMMUT040916.2, NONMMUT048938.2, NONMMUT065231.2, etc., with the largest number of GO enrichment (>5) were selected, and the top 30 of the GO terms were displayed (Supplementary file 1). For example, the positive regulation of the transforming growth factor beta production, extracellular region, and heparin binding were significantly enriched on the NONMMUT040916.2. Additionally, the top 10 lncRNAs, containing NONMMUT001827.2, NONMMUT019380.2, NONMMUT026967.2, etc., with the largest number of KEGG enrichment (no less than 2) were selected, and the top 20 of the pathways were displayed (Supplementary file 2). For example, malaria, TNF signaling pathway, and IL-17 signaling pathway were significantly enriched on the NONMMUT001827.2.
3.6 Cis and trans analyses
As mentioned in the method section, we found that Kdm5d, Eif2s3y, and Uty were regulated by NONMMUT074750.2; Adcy1 was regulated by NONMMUT140337.1; LOC101055758 was regulated by NONMMUT001777.2; Ddx3y and Uty were regulated by NONMMUT074763.2; Cacna2d3 was regulated by NONMMUT092640.1; Igfbp2 was regulated by NONMMUT001471.2; and Uty was regulated by NONMMUT074758.2.
The binding free energy was ranked from small to large, and the top 200 were selected to construct the network, as shown in Figure 6. There were 16 mRNAs in this network. Opcml, Pax9, Csf3r, Apol6, Gpr75, Slc12a8, and Cxcr2 interacted with numerous lncRNAs.
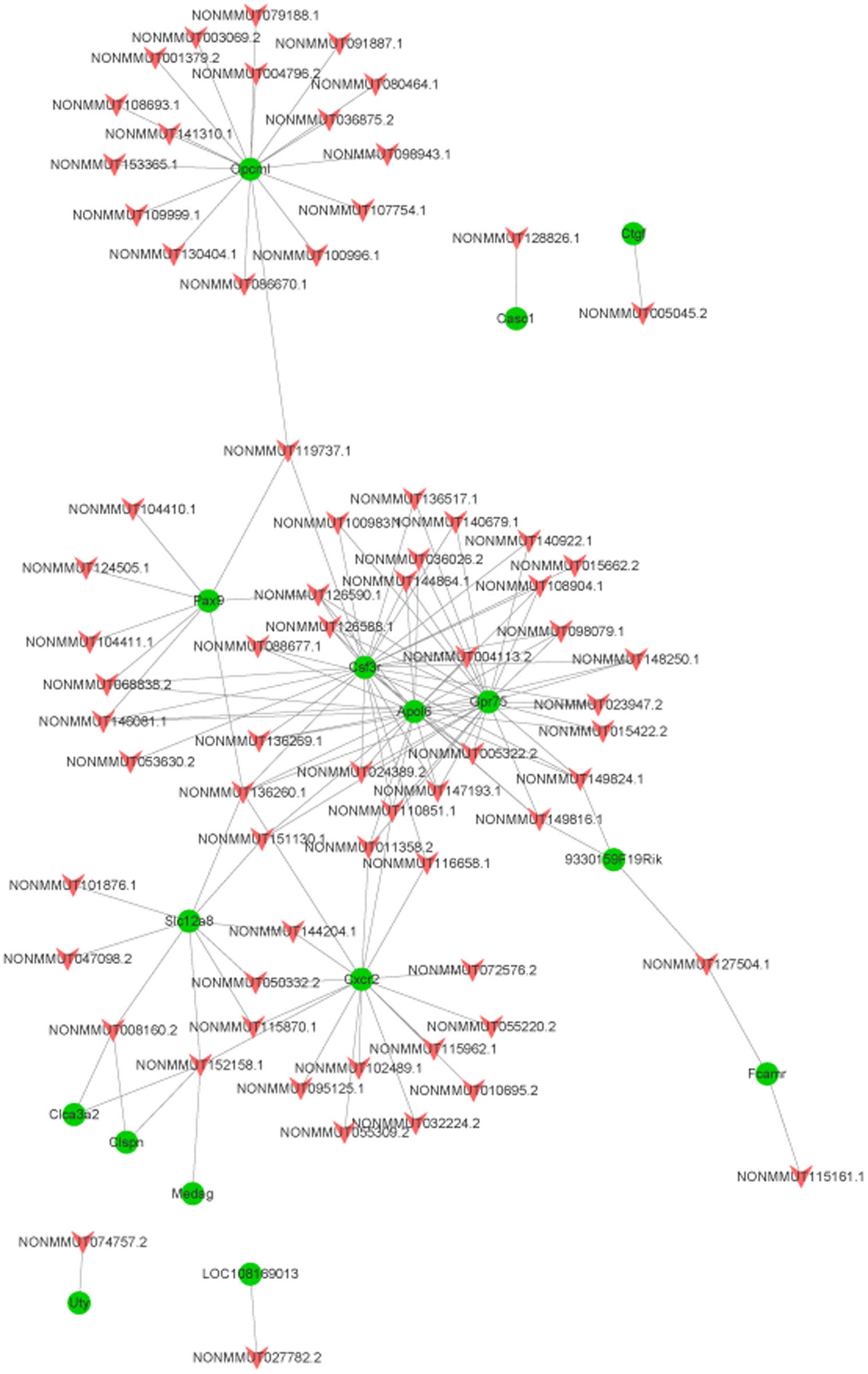
Network of lncRNA and gene trans analysis (red arrow nodes represent differentially expressed lncRNAs, while green circular nodes represent DEMs).
3.7 miRNA prediction and ceRNA network construction
A total of 396 miRNA–mRNA regulatory pairs were predicted for mRNAs in the lncRNA–mRNA co-expression relationships. Additionally, 54,916 lncRNA–miRNA pairs were predicted. The lncRNA–miRNA–mRNA pairs were then screened. Combining with the positive co-expression relationship between mRNA and lncRNA (correlation coefficient >0.8), 1,575 lncRNA–miRNA–mRNA pairs were obtained, including 349 lncRNAs, 150 miRNAs, and 68 mRNAs. Due to the large number of nodes, we focused on screening the lncRNA–miRNA–mRNA pairs with a positive co-expression correlation coefficient of >0.95 of mRNA–lncRNA, and constructed the ceRNA network, as shown in Figure 7. The network contained 129 regulatory pairs of miRNA–mRNA, 206 regulatory pairs of lncRNA–miRNA, and 134 co-expression pairs of lncRNA–mRNA. The hub nodes (top 5) in the network are shown in Table 1. NONMMUT140591.1 had the highest degree in the ceRNA network and was involved in the ceRNA of NONMMUT140591.1-mmu-miR-298-5p-Gata5 (GATA binding protein 5).
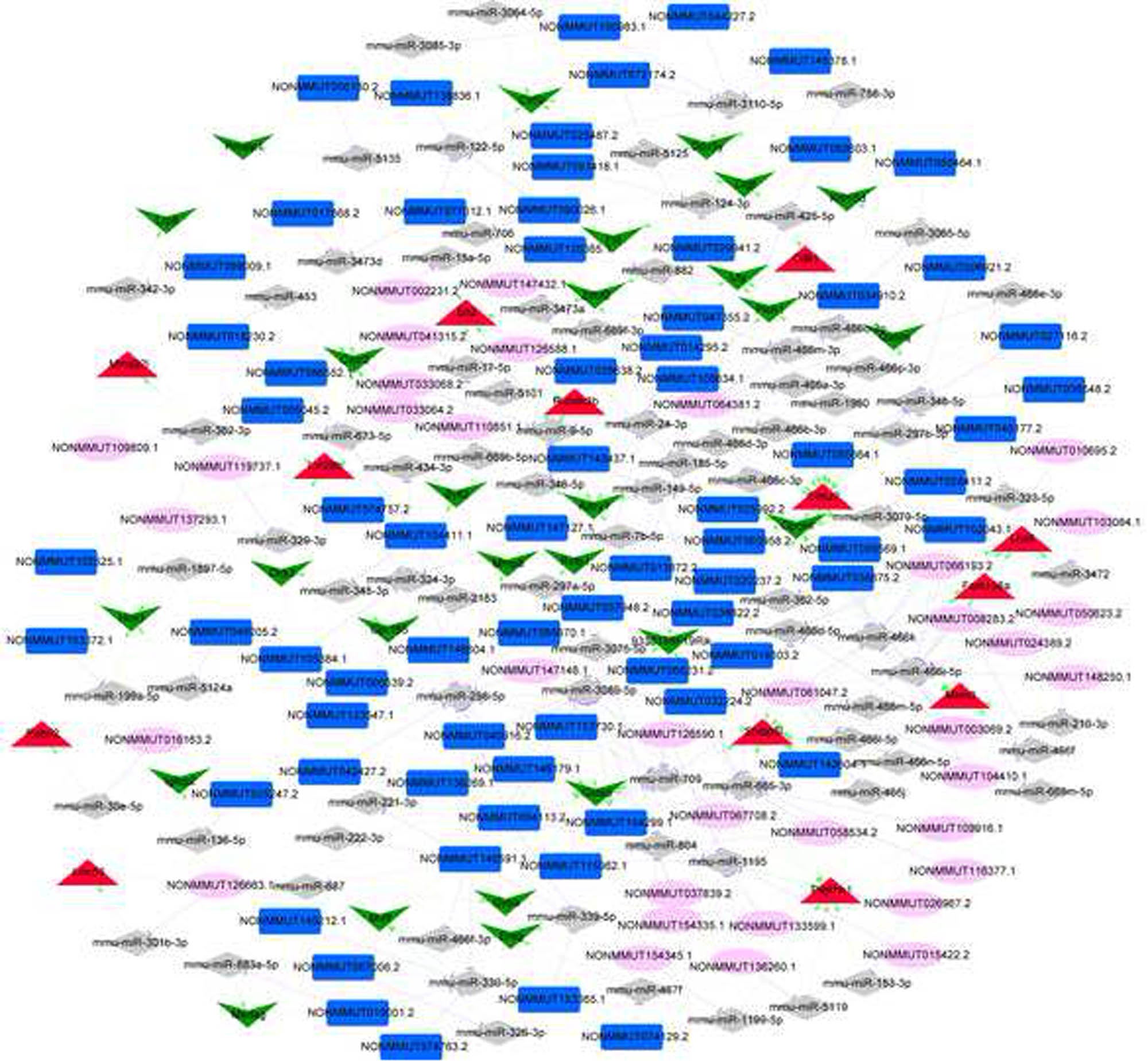
ceRNA network (red triangle represents upregulated mRNA; green inverted triangle represents downregulated mRNA; gray rhomboid represents miRNA; dark blue rectangle represents downregulated lncRNAs; pink ellipses represent upregulated lncRNAs; the green arrow represents the miRNA–mRNA regulatory relationship; the purple T-line represents the lncRNA–miRNA regulatory relationship; the yellow line represents the lncRNA–mRNA co-expression relationship).
Node connectivity degree ranking in ceRNA network (top 5)
| Node | Degree | Type |
|---|---|---|
| NONMMUT140591.1 | 15 | lncRNA_Down |
| NONMMUT064381.2 | 15 | lncRNA_Up |
| NONMMUT061047.2 | 11 | lncRNA_Up |
| NONMMUT047355.2 | 9 | lncRNA_Down |
| NONMMUT057948.2 | 8 | lncRNA_Down |
| Gata5 | 19 | mRNA_Down |
| Opcml | 18 | mRNA_Down |
| Plekhh1 | 18 | mRNA_Up |
| Ccna2 | 15 | mRNA_Up |
| Stxbp5l | 14 | mRNA_Up |
| mmu-miR-466i-5p | 17 | miRNA |
| mmu-miR-298-5p | 16 | miRNA |
| mmu-miR-466k | 15 | miRNA |
| mmu-miR-665-3p | 12 | miRNA |
| mmu-miR-709 | 10 | miRNA |
4 Discussion
In this study, urea accumulation by UT-B deletion results in the aberrant expression of 588 lncRNAs and 194 mRNAs. These DEMs were significantly enriched in the inflammation-related pathway. A lncRNA–mRNA co-expression network and a ceRNA network were built based on the DEMs and DELs. C7 (complement C7)–NONMMUT137216.1 co-expression pair had the highest correlation coefficient. NONMMUT140591.1 had the highest degree in the ceRNA network and was involved in the ceRNA of NONMMUT140591.1-mmu-miR-298-5p-Gata5 (GATA binding protein 5).
This study identified more downregulated genes than upregulated genes. The downregulated mRNAs were significantly involved in the IL-17 signaling pathway, cytokine–cytokine receptor interaction, and the chemokine signaling pathway. Interestingly, these pathways are inflammation related. Studies have reported that chronic inflammatory diseases have come into focus as a risk factor for the development of cardiovascular dysfunction [19,20]. Previous studies have focused more on the fact that urea exacerbates the progression of the disease or plays a role in certain complications. It has also been reported that high urea have toxicity in other cells and tissues. Urea induces the expression of proapoptotic proteins in human aortic endothelial cells [21], resulting in increased mitochondrial reactive oxygen species (ROS) production and activation of pro-inflammatory pathways which deteriorates the quality of life of patients with chronic kidney disease [22]. A previous study reported that congenital complete heart block is considered as an inflammatory process in patients with a structurally normal heart, which is due to transplacental transfer of maternal autoantibodies [23]. Clinical evidence suggests that tissue injury in both acute kidney injury and heart failure has immune-mediated inflammatory consequences that can initiate remote organ dysfunction [24]. The cardiovascular system is the main route of urea transportation. Therefore, urea specifically transmits organ crosstalk information from the kidney to the heart. The urea level is very stable in almost all conditions, which makes urea a reliable crosstalk language that only the kidney speaks. Wang et al. found that urea should be considered as an independent factor causing disease directly, and urea toxicity plays an important role at the cellular and systemic level [25]. Urea perseprobably participates in the pathogenesis of cardiovascular disease, insulin resistance, intestinal disease, and contributes to an overall accelerated ageing phenotype [26]. Our study may further support the report above. We speculated that urea accumulation by UT-B deletion may promote cardiac conduction block via the inflammatory process.
Co-expression analysis has been highly successful in the function of revealing genes, and has contributed greatly to the understanding of gene regulation systems [27–29]. Our study constructed a lncRNA–mRNA co-expression network. The C7–NONMMUT137216.1 co-expression pair had the highest correlation coefficient. C7 is a component of the terminal complement cascade [30]. The complement system is part of the humoral innate immune response, which forms a cascade of more than 30 proteins bound to the target surface or present in the plasma [31]. It has been suggested that the complement system plays a key role in the inflammatory response after acute myocardial infarction [32]. However, no one has reported that it has a role in cardiac conduction block to the best of our knowledge. As we mentioned above, cardiac conduction block may be associated with the inflammatory process. So, we speculated that urea accumulation by UT-B deletion may promote the co-expression of C7 and NONMMUT137216.1 to participate in the progression of cardiac conduction block.
NONMMUT140591.1 was a downregulated lncRNA in UT-B knockout mice. Additionally, it had the highest degree in the ceRNA network and was involved in the ceRNA of NONMMUT140591.1-mmu-miR-298-5p-Gata5. Gata5 belongs to the zinc finger transcription factor GATA family (GATA1–6), which is abundantly expressed in various endoderm- and mesoderm-derived tissues, predominantly in the embryonic heart [33]. Gata5, having partially overlapped function with Gata4 and Gata6, has been reported to have an important role in cardiovascular development [34]. Previous studies have suggested that Gata5 is associated with the bicuspid aortic valve [35,36]. Given the important role of Gata5 in cardiovascular development, we speculated that UT-B knockout may be involved in the cardiac conduction block trough downregulation of NONMMUT140591.1 as a ceRNA to regulated Gata5 expression.
However, this study had several limitations. First, the sample size of this study is relatively small. A large sample size is needed. In addition, the results identified from bioinformatics analyses have to be verified through in vitro and in vivo experiments.
5 Conclusion
To conclude, we found that urea accumulation by UT-B deletion may promote cardiac conduction block via inflammatory process. The ceRNA NONMMUT140591.1-mmu-miR-298-5p-Gata5 may be a potential molecular mechanism of UT-B knockout-induced cardiac conduction block.
Acknowledgments
We specially thank Professor Baoxue Yang of Peking University for the gift of mice.
-
Funding information: This study was supported by the National Natural Science Foundation of China (No. 81600207, No. 81370240), the Excellent Youth Fund of Jilin Provincial Science and Technology Department (No. 20190103090JH), the “13th five year plan” science and technology research project of the Jilin Provincial Department of Education (No. JJKH20200898KJ), the Health Technology Innovation Project of Jilin Province (No. 2017J059), the Science and Technology Department of Jilin Province (No. 20190304054YY), the “Xinglin scholar project” of the Changchun University of Traditional Chinese Medicine (No. 2019RS01), College Project Grant (No. 2018KJ01), and the National College Students’ Innovation and Entrepreneurship Training Program (No. 201910199001).
-
Author contributions: W.Z. and Y.M. conceived and designed the project. T.L., W.Y., L.Y., and X.Q. collected the data. S.F., Y.S., and N.S. interpreted the data. X.L. and W.T. performed the statistical analysis. Y.D. and S.F. wrote the manuscript. W.Z. and Y.M. revised the paper. All authors read and approved the final manuscript.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during this study are available from the corresponding author on reasonable request.
References
[1] Hoeker GS , James CC , Tegge AN , Gourdie RG , Smyth JW , Poelzing S . Attenuating loss of cardiac conduction during no-flow ischemia through changes in perfusate sodium and calcium. Am J Physiol Heart Circ Physiol. 2020;319(2):H396–409.10.1152/ajpheart.00112.2020Suche in Google Scholar PubMed PubMed Central
[2] Coluccia G , Oddone D , Maggi R , Corallo S , Senes J , Donateo P , et al. Left bundle branch area pacing in a young athlete with progressive cardiac conduction (Lev-Lenegre) disease. J Electrocardiol. 2021;64:95–8.10.1016/j.jelectrocard.2020.12.011Suche in Google Scholar PubMed
[3] Hsu L-A , Ko Y-S , Yeh Y-H , Chang C-J , Chan Y-H , Kuo C-T , et al. A novel DES L115F mutation identified by whole exome sequencing is associated with inherited cardiac conduction disease. Int J Mol Sci. 2019;20(24):6227.10.3390/ijms20246227Suche in Google Scholar PubMed PubMed Central
[4] Liu J , Liu D , Li M , Wu K , Liu N , Zhao C , et al. Identification of a nonsense mutation in TNNI3K associated with cardiac conduction disease. J Clin Lab Anal. 2020;34(9):e23418.10.1002/jcla.23418Suche in Google Scholar PubMed PubMed Central
[5] Wolf CM , Berul CI . Inherited conduction system abnormalities – one group of diseases, many genes. J Cardiovasc Electrophysiol. 2006;17(4):446–55.10.1111/j.1540-8167.2006.00427.xSuche in Google Scholar PubMed
[6] Yu L , Liu T , Fu S , Li L , Meng X , Su X , et al. Physiological functions of urea transporter B. Pflug Archiv Eur J Physiol. 2019;471(11–12):1359–68.10.1007/s00424-019-02323-xSuche in Google Scholar PubMed PubMed Central
[7] Li M , Zhang S , Yang B . Urea transporters identified as novel diuretic drug targets. Curr Drug Targets. 2020;21(3):279–87.10.2174/1389450120666191129101915Suche in Google Scholar PubMed
[8] Li X , Chen G , Yang B . Urea transporter physiology studied in knockout mice. Front Physiol. 2012;3:217.10.3389/fphys.2012.00217Suche in Google Scholar PubMed PubMed Central
[9] Meng Y , Zhao C , Zhang X , Zhao H , Guo L , Lü B , et al. Surface electrocardiogram and action potential in mice lacking urea transporter UT-B. Sci China Ser C Life Sci. 2009;52(5):474–8.10.1007/s11427-009-0047-ySuche in Google Scholar PubMed
[10] He L , Fu M , Chen X , Liu H , Chen X , Peng X , et al. Effect of dialysis dose and membrane flux on hemoglobin cycling in hemodialysis patients. Hemodial Int Int Symposium Home Hemodial. 2015 Apr;19(2):263–9.10.1111/hdi.12215Suche in Google Scholar PubMed
[11] Ivanovski O , Szumilak D , Nguyen-Khoa T , Ruellan N , Phan O , Lacour B , et al. The antioxidant N-acetylcysteine prevents accelerated atherosclerosis in uremic apolipoprotein E knockout mice. Kidney international. 2005 Jun;67(6):2288–94.10.1111/j.1523-1755.2005.00332.xSuche in Google Scholar PubMed
[12] Anders S , Huber W . Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL); 2012.Suche in Google Scholar
[13] Al-Mubaid H . Gene multifunctionality scoring using gene ontology. J Bioinf Comput Biol. 2018;16(5):1840018.10.1142/S0219720018400188Suche in Google Scholar PubMed
[14] Kanehisa M , Furumichi M , Tanabe M , Sato Y , Morishima K . KEGG: new perspectives on genomes, pathways, diseases, and drugs. Nucleic Acids Res. 2017;45(D1):D353–61.10.1093/nar/gkw1092Suche in Google Scholar PubMed PubMed Central
[15] Parveen A , Gretz N , Dweep H . Obtaining miRNA-target interaction information from miRWalk2.0. Curr Protoc Bioinforma. 2016;55:12.5.1–27.10.1002/cpbi.14Suche in Google Scholar PubMed
[16] Enright AJ , John B , Gaul U , Tuschl T , Sander C , Marks DS . MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1.10.1186/gb-2003-5-1-r1Suche in Google Scholar PubMed PubMed Central
[17] Doncheva NT , Morris JH , Gorodkin J , Jensen LJ . Cytoscape stringapp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18(2):623–32.10.1021/acs.jproteome.8b00702Suche in Google Scholar PubMed PubMed Central
[18] Liu X , Liu X , Qiao T , Chen W . Identification of crucial genes and pathways associated with colorectal cancer by bioinformatics analysis. Oncol Lett. 2020;19(3):1881–9.10.3892/ol.2020.11278Suche in Google Scholar PubMed PubMed Central
[19] Naseem Z , Iqbal MA , Ahmad S , Roohi N . Inflammatory markers as prognosticators of cardiovascular dysfunction in hypothyroid patients. J Biol Regul Homeost Agents. 2019;33(6):1891–5.Suche in Google Scholar
[20] Ruscica M , Corsini A , Ferri N , Banach M , Sirtori CR . Clinical approach to the inflammatory etiology of cardiovascular diseases. Pharmacol Res. 2020;159:104916.10.1016/j.phrs.2020.104916Suche in Google Scholar PubMed PubMed Central
[21] Caravaca F , Martín MV , Barroso S , Ruiz B , Hernández-Gallego R. Do inflammatory markers add predictive information of death beyond that provided by age and comorbidity in chronic renal failure patients?. Nephrol Dial Transplant. 2006 Jun;21(6):1575–81.10.1093/ndt/gfl033Suche in Google Scholar PubMed
[22] D'Apolito M , Du X , Pisanelli D , Pettoello-Mantovani M , Campanozzi A , Giacco F , et al. Urea-induced ROS cause endothelial dysfunction in chronic renal failure. Atherosclerosis 2015 Apr;239(2):393–400.10.1016/j.atherosclerosis.2015.01.034Suche in Google Scholar PubMed PubMed Central
[23] Scott JS , Maddison PJ , Taylor PV , Esscher E , Scott O , Skinner RP . Connective-tissue disease, antibodies to ribonucleoprotein, and congenital heart block. N Engl J Med. 1983;309(4):209–12.10.1056/NEJM198307283090403Suche in Google Scholar PubMed
[24] Virzì G , Day S , de Cal M , Vescovo G , Ronco C. Heart-kidney crosstalk and role of humoral signaling in critical illness. Critical Care (London, England). 2014 Jan 6;18(1):201.10.1186/cc13177Suche in Google Scholar PubMed PubMed Central
[25] Wang H , Huang B , Wang W , Li J , Chen Y , Flynn T , et al. High urea induces depression and LTP impairment through mTOR signalling suppression caused by carbamylation. EBioMedicine 2019 Oct;48:478–90.10.1016/j.ebiom.2019.09.049Suche in Google Scholar PubMed PubMed Central
[26] White WE , Yaqoob MM , Harwood SM. Aging and uremia: Is there cellular and molecular crossover? World J Nephrol. 2015 Feb 6;4(1):19–30.10.5527/wjn.v4.i1.19Suche in Google Scholar PubMed PubMed Central
[27] Mantini G , Vallés AM , Le Large TYS , Capula M , Funel N , Pham TV , et al. Co-expression analysis of pancreatic cancer proteome reveals biology and prognostic biomarkers. Cell Oncol (Dordr). 2020;43(6):1147–59.10.1007/s13402-020-00548-ySuche in Google Scholar PubMed PubMed Central
[28] Liu B , Zhan Y , Chen X , Hu X , Wu B , Pan S . Weighted gene co-expression network analysis can sort cancer-associated fibroblast-specific markers promoting bladder cancer progression. J Cell Physiol. 2021;236(2):1321–31.10.1002/jcp.29939Suche in Google Scholar PubMed
[29] Bai Q , Liu H , Guo H , Lin H , Song X , Jin Y , et al. Identification of hub genes associated with development and microenvironment of hepatocellular carcinoma by weighted gene co-expression network analysis and differential gene expression analysis. Front Genet. 2020;11:615308.10.3389/fgene.2020.615308Suche in Google Scholar PubMed PubMed Central
[30] Zhang D-F , Fan Y , Xu M , Wang G , Wang D , Li J , et al. Complement C7 is a novel risk gene for Alzheimer’s disease in Han Chinese. Natl Sci Rev. 2019;6(2):257–74.10.1093/nsr/nwy127Suche in Google Scholar PubMed PubMed Central
[31] Emmens RW , Wouters D , Zeerleder S , Van Ham SM , Niessen HW , Krijnen PA . On the value of therapeutic interventions targeting the complement system in acute myocardial infarction. Transl Res. 2017;182:103–22.10.1016/j.trsl.2016.10.005Suche in Google Scholar PubMed
[32] Bavia L , Lidani KCF , Andrade FA , Sobrinho MIAH , Nisihara RM , de Messias-Reason IJ . Complement activation in acute myocardial infarction: an early marker of inflammation and tissue injury? Immunol Lett. 2018;200:18–25.10.1016/j.imlet.2018.06.006Suche in Google Scholar PubMed
[33] Song Z , Chen L , Pang S , Yan B . Molecular genetic study on GATA5 gene promoter in acute myocardial infarction. PLoS One. 2021;16(3):e0248203.10.1371/journal.pone.0248203Suche in Google Scholar PubMed PubMed Central
[34] Wen B , Yuan H , Liu X , Wang H , Chen S , Chen Z , et al. GATA5 SUMOylation is indispensable for zebrafish cardiac development. Biochim Biophys Acta Gen Subj. 2017;1861(7):1691–701.10.1016/j.bbagen.2017.03.005Suche in Google Scholar PubMed
[35] Alonso-Montes C , Martín M , Martínez-Arias L , Coto E , Naves-Díaz M , Morís C , et al. Variants in cardiac GATA genes associated with bicuspid aortic valve. Eur J Clin Invest. 2018;48(12):e13027.10.1111/eci.13027Suche in Google Scholar PubMed
[36] Lo Presti F , Guzzardi DG , Bancone C , Fedak PWM , Della Corte A . The science of BAV aortopathy. Prog Cardiovasc Dis. 2020;63(4):465–74.10.1016/j.pcad.2020.06.009Suche in Google Scholar PubMed
© 2021 Xuejiao Lv et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells
Artikel in diesem Heft
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells

