Abstract
Radiation inactivation of enveloped viruses occurs as the result of damages at the molecular level of their genome. The rapidly emerging and ongoing coronavirus disease 2019 (COVID-19) pneumonia pandemic prompted by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is now a global health crisis and an economic devastation. The readiness of an active and safe vaccine against the COVID-19 has become a race against time in this unqualified global panic caused by this pandemic. In this review, which we hope will be helpful in the current situation of COVID-19, we analyze the potential use of γ-irradiation to inactivate this virus by damaging at the molecular level its genetic material. This inactivation is a vital step towards the design and development of an urgently needed, effective vaccine against this disease.
1 Introduction
Viruses (or virions) are subcellular particles, commonly spherical or rod-shaped, which composed of a protein capsid that contains their genetic material made of RNA or DNA. Sometimes the viral genome is protected by an additional outer envelope made of a lipid bilayer with spikes of glycoproteins inserted inside the viral envelope [1]. Viruses are classified based on their size, shape, envelope, and structure of their genome. Unlike bacteria, viruses lack cell organelles and thus have no metabolic activities on their own. To transcript and replicate, they entirely depend on the host biochemical machinery of eukaryotic or prokaryotic host cells [2]. Once inside the host cell, viruses can mutate through genome deletion, insertion, and/or substitution to novel strains of different virulence [3,4]. This viral mutation is the major obstacle for the development of new vaccines [5,6].
1.1 Key features of human coronavirus
Six human coronaviruses (HCoVs) were known before the COVID-19 outbreak: 229E and NL63 (alpha coronavirus), OC43, HKU1, SARS-CoV, and MERS-CoV (beta coronavirus) [7]. Severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) first emerged in South China in 2002–2003 to cause a large-scale epidemic with over 8,000 infections and more than 800 deaths [8]. The Middle East Respiratory Syndrome CoV (MERS-CoV) has caused a persistent epidemic in the Arabian Peninsula, especially in Saudi Arabia in 2012 [9]. SARS-CoV and MERS-CoV are enveloped positive-sense RNA viruses (size ranging from 70 to 90 nm) belonging to the Coronaviridae family. It was shown that rodents, avians, and mainly bats are reservoir host of these family viruses that can be potentially transmitted from animals to humans [10,11] due to the growing consumption of animal proteins including those from exotic wild mammals in China.
A novel strain of coronavirus, labeled as SARS-CoV-2 by the International Committee on Taxonomy of Viruses-Coronavirus Study Group [12], belonging to the beta-coronavirus lineage, shares around 80% identity to SARS-CoV [13,14]. This strain, believed to have started from a seafood market in the city of Wuhan, China, in December 2019, is now spreading and creating a chaos across the entire world [15,16]. Human-to-human transmission of this virus was officially confirmed on January 20th 2020 [10]. World Health Organization (WHO) declared this disease as a global pandemic on March 11, 2020 [17]. As of 29 January 2021, 27,901,760 cases are still active, 72,337,017 recovered, and 2,210,259 died in 235 countries or territories worldwide [18].
Similar to previous coronaviruses symptoms, SARS-CoV-2 is mainly affecting the lower respiratory track, ranging from mild respiratory disease to SARS and septic shock in advanced stages. Damages to the cardiovascular system, gut, kidneys, and brain have also been reported [13,19] along with vital organ failures in comorbid patients [20,21].
The genetic material of SARS-CoV-2 consists of single-stranded RNA backbone made of alternating 5-carbon sugar (ribose) and phosphate groups. Attached to this backbone are 29,891 to 29,903 adenine, uracil, cytosine, and guanine bases [22,23] encoding for 9,860 amino acids [10]. One third of the SARS-CoV-2 genome make up the four major structural proteins: spike glycoprotein (S), membrane protein (M), envelope protein (E), and nucleocapsid protein (N) accountable for some important functions in virus replication [24,25]. The remaining two‐thirds of its viral genome encode for 16 nonstructural proteins (nsp-1 to 16). Each of these nsps has a specific role in the life cycle of the virus and its pathogenicity [26,27]. For instance, nsp-1 is used by the virus to elude the host innate immune system [28], nsp-2 is indispensable for its replication, and nsp-9, in complex with nsp-8, is involved in RNA replication and virulence [29].
There is a 76.5% similarity in the amino acid sequences of the spike glycoprotein in SARS-CoV and SARS-CoV-2 [30]. SARS-CoV-2 seems to have greater binding affinity to the angiotensin converting enzyme 2 (ACE2) cell membrane receptor than the other SARS-CoV virus strains, suggesting a greater capacity of SARS-CoV-2 for human to human transmission [31,32]. There are speculations that cellular overexpression of human ACE2 (associated with the usage of medications such as ACE2 inhibitors, angiotensin II receptor blockers) could enhance the COVID-19 severity [21,33,34,35,36]. Among the four structural proteins of the SARS-CoV family, the spike glycoprotein S plays a key role in viral docking and cellular internalization [37]. It binds via the receptor-binding domain (RBD) in the S1 subunit to the ACE2 receptors [38,39], expressed especially on the plasma membrane of human respiratory epithelial host cells, and virtually in all other organs. The virus penetrates the host cell via endocytosis through its S2 subunit [40,41] and infects it by hijacking its molecular machinery to encode for RNA polymerase enzyme necessary for the replication of its own RNA genome [42,43]. Viral entry in cells is a critical phase in the course of the COVID-19 disease. Thus, inhibition of the viral binding and internalization in a host cell constitute a strategy for potential therapeutics against COVID-19 pandemic.
1.2 SARS-CoV-2 infectivity
Aerial transmission by expelled respiratory droplets is considered as the main direct transmission vector of the SARS-CoV-2 when in close contact with an infected person coughing, sneezing, or even talking [10,44,45,46]. Some findings have indicated that the virus may as well be airborne [31,47,48]. Indirect transmission may also occur via fomites when respiratory droplets from infected people land on object surfaces which can be touched by a receptive host [49,50].
The WHO recommends that SARS CoV-2 sample handling should be conducted in no less than a Biosafety Level 3 (BSL-3) laboratory using BSL-3 practices (WHO, 2020) [17,51]. The SARS-CoV-2 cytopathogenic effect is measured by its capability to infect a host cell. This is usually expressed by the tissue culture infectious dose, TCID50/mL, calculated using the method of Reed and Muench, [52], which is the viral titer at which 50% of the host cell lines are infected when inoculated in vitro with a diluted viral solution. However, because of some limitations with the in vitro tests (slow viral growth), the use of in vivo assays is taken after inoculated animals were sacrificed for within-host virus titering and pathological study [53].
Basic reproductive number
One of the key epidemiological factors in the COVID-19 pandemic is the incubation period, time elapsed between the exposure and the appearance of the first symptoms. Different pooled analysis of confirmed COVID-19 cases showed that the estimated incubation times were 5.1 days [57], 6.4 days (range 2.2–11.1) [58], and 5.0 days (range 2–14 days) [59]. Sanche and colleagues estimated, in late January 2020 in Mainland China, the average incubation period to 4.2 days. A time duration from symptoms onset to admission to hospital for treatment was estimated to 1.5 days in late January 2020 and the time from symptoms onset to death to 16.1 days [60].
Various in vitro studies showed that RNA viruses are less vulnerable to corruption due to their ability to promptly repair their genome damages by proof-reading, excision, and removing flawed RNA nucleotides that occur during their replication [61,62,63,64]. Exoribonuclease (ExoN) enzymes encoded by nsp-14 play a crucial role in maintaining the viral genome integrity [18,64,65,66]. Eckerle and workers reported that mutations in nsp-14 of SARS viruses lead to 15-fold increase in replication errors [67].
1.3 Vaccine strategy
To help prevent the spreading of the COVID-19 pandemic, most of the countries have adopted immediate measures consisting of global travel restrictions on movement, lockdowns, social distancing, patient self-isolation, and provision of medical care to infected people. Few curative methods using already known antiviral agents such as hydroxychloroquine and remdesivir were tested on patients [68,69,70]. However, results were not very encouraging for any of these agents to be considered as significant therapy yet. The best option for ending this pandemic and reestablishing a normal life remains, by far, the development of safe and effective prophylactic vaccines. This has triggered an extensive collaboration and a colossal mission between pharma companies and scientists to expedite vaccine development and production in less than a year instead of the normal 10-year period time. By the end of 2020, 259 COVID-19 vaccine projects were in the pipeline [71]. Frontrunning coronavirus vaccines, sharing the same purpose of stimulating the immune system against SARS-CoV-2, can be broadly categorized into three platforms:
the classical inactivated virus vectored vaccines based on disrupting the viral genome through chemical or physical alterations. These viruses are no longer able to replicate to cause infection, but able to trigger an immune memory response [72].
the full-length S glycoprotein- or RBD-based vaccines that generate target antigens in the infected cell [73].
the groundbreaking DNA-, mRNA-based vaccines that encode in the host cell the full-length S glycoprotein as target antigen [74].
As of 29 January 2021, five vaccines went through the necessary multiple phases of trial to ensure safety, showing more than 90% efficacy. They have been approved and licensed for use by national and international public health regulators and are being rolled out worldwide (Table 1).
COVID-19 vaccines currently available in the market (January 2021)
| Vaccine | Platform | Inactivation method | Developer | Reference |
|---|---|---|---|---|
| BNT162b2 | mRNA | — | Pfizer – BioNTech (USA, Germany) | [75,76] |
| mRNA-1273 | mRNA | — | Moderna (USA) | [77,78] |
| AZD1222 | Nonreplicating viral vector | Deletions in E1 and E3 genes in adenovirus vector to inhibit replications | University of Oxford AstraZeneca (UK) | [15,79,80] |
| CoronaVac | Inactivated SARS-CoV-2 | β-Propiolactone to inhibit replication | Sinovac (China) | [29,81] |
| Sputnik V | Heterologous recombinant adenovirus (rAd26 and rAd5) | Deletions in E1 and E3 genes in adenovirus vector to inhibit replications | Gamaleya (Russia) | [82,83] |
1.4 Different agents for SARS-CoV-2 inactivation
Virus inactivation for vaccine purposes was already known since the late 1800s [84]. In 1885, Pasteur laid the foundations of immunization with inactivated rabies virus cultured in rabbit spinal cords [85]. It was not until the discovery of the in vitro culture of viruses outside the host organism procedures that inactivated viral vaccine development was truly initiated. This allowed a large-scale production of viruses as source for inactivated vaccine purposes [86]. Vaccine producers are generally using virus growth on continuous cell lines to reduce production costs and increase vaccine safety. Once the virus has been purified, inactivation can be achieved using chemical or physical methods or a combination of the two. A wide range of chemical agents are used: ascorbic acid [87], derivatives of ethylenimine [88], and hydrogen peroxide [89]. However, formaldehyde [90] and β-propiolactone [91] are the most widely used for inactivation for decades. To avoid the extensive and time-consuming downstream processing to detoxify the virus cultures from chemical inactivators, the use of γ‐irradiation as a physical alternative method to chemical inactivation has been proposed by many authors. Preparation of experimental vaccines against several viral diseases using γ‐irradiation is reported in the literature: bluetongue [92], Venezuelan equine encephalitis [93], rabies [94], smallpox [95], influenza [96], HIV [97], Ebola [98], rotavirus [99], and polio [100].
The choice of an inactivation method preserving the viral epitope integrity is important since the damage of the envelop protein will lessen the efficacy of the vaccine [101]. Several studies showed that viral inactivation by formaldehyde, hydrogen peroxide, or binary ethylenimine derivatives is nonselective and can damage the envelope protein leading to a poor immune response [102,103]. Nevertheless, γ-irradiation has shown a superior inactivation method by preserving the viral antigens intact to trigger the immunogenicity while destroying nucleic acids to inhibit the viral replication in human cells [104,105]. This advantageous attribute of γ-irradiation can be ascribed to its high penetration depth that causes direct damage to nucleic acids without altering structural proteins [96,106,107].
Due to the potentially dangerous consequences of SARS-CoV-2 human infection, extreme attention should be paid to ensure that inactivation procedures are efficient. Effective inactivation of the SARS-CoV-2 is vital as it allows research, especially the development of new vaccines, to be conducted under safe conditions [108]. Various methods are already available for SARS-CoV effective inactivation [109] and could be tested on SARS-CoV-2 since these two viruses share a great deal of genome: ultraviolet radiation, thermal treatment, extreme pH values, and commonly used disinfectants offer an effective virus inactivation. Duan and colleagues reported an inactivation of SARS-CoV virus after 15 min exposure to ultraviolet C, whereas ultraviolet A and B had no effect on its viability, irrespective of the duration of the exposure [110]. Heat can denature SARS-CoV secondary structural proteins. A complete inactivation of this virus at 75°C for 45 min was reported by Darnell and colleagues [111]. For inactivation with detergents, Gerlach et al. showed that SARS-CoV-2 can be efficiently inactivated by 70% ethanol, 0.1% hydrogen peroxide, and 0.1% sodium sulphate, commonly available in hand soaps, within 60 s of exposure on various surfaces [112]. pH has a great effect on the viability of SARS-CoV-2. Chan et al. reported that the virus survived for up to 6 days in a medium with pH range [5–9], but lost between 2.9 and 5.33 log10 infectivity. At pH 4 and pH 11, it remained viable for 1–2 days. At extreme pHs (pH 2–3 and pH 11–12), the virus lost 5.25 log10 infectivity within only 1 day [16].
In this perspective, γ-inactivation of viruses could be an important and promising tool for SARS-CoV-2 vaccine development (Figure 1). In this review, we analyze the potential use of γ-irradiation to inactivate the SARS-CoV-2 by altering its genetic material while preserving its structural proteins.
Schematic diagram showing the development of SARS-CoV-2 vaccine using radiation-induced inactivation of live virus.
1.5 Radiolysis of water in biological matter: mechanisms of radiobiological action
The two major processes in the interaction of energetic photons or charged particles with aqueous biological medium are ionization and excitation of water molecules and biologically important macromolecules such as DNA, RNA, lipids, and proteins [113,114]. The electrons liberated in the ionization have enough energy to ionize further molecules in a manner that energy is deposited in spurs. When their energy falls below the ionization threshold of water, the electrons become solvated
1.6 Mechanism of viral inactivation
γ-irradiation disrupts viruses by altering mainly their RNA genetic material. The number of nitrogenous bases and their sequence in the RNA is crucial for determining the viral sensitivity towards γ-irradiation. The more target nucleotides, the more likely the nucleic acid genome will be damaged for a given absorbed dose [118]. The mechanism behind this damage falls into two types: direct and indirect. Direct damage is caused by the radiation-induced cleavage of the sugar-phosphate backbone or the cross-linking, deletion, substitution, and insertion in the sequence of nitrogen bases [119,120]. Indirect damage is attributed to the oxidative stress of the radiolytically produced ROS on the viral material, leading to its fragmentation and cross-linking [121,122]. A minimum energy deposition of 17.5 eV within a critical distance of 6 Å from the nucleotide (corresponding to a sensitive spherical volume of 0.596 nm3) can induce a lethal damage to the viral RNA [123]. Disruption to the protein capsid and the lipid bilayer envelope, by lipid and protein peroxidation chain reactions, may as well result in the reduction of viral pathogenicity [70,124,125]. It has been reported that, for viruses belonging to the enveloped Coronaviridae family, the conformational changes in the spike glycoprotein S block the viral binding to the host cell plasma membrane and prevent cellular internalization, the first stage in viral infection [111,126]. Studies suggested that genetic material rather than protein and lipid envelopes is likely to be the primary target for viral inactivation [107,127]. Nims et al. reported that the presence or absence of a viral envelope does not seem to be a major factor of inactivation by γ-irradiation [128].
Single-strand break (for single-stranded viruses) and double-strand break (for double-stranded viruses) are generally sufficient to inactivate the viral genome [129]. Based on the hypothesis of the single-hit-single-target (SHST) model [130,131], the inactivation of viruses is typically expressed by the following relationship [132]:
where
It should be noted that
Feldmann and colleagues showed that the inactivation was inversely correlated with genome size [133]. They measured the radiation doses for a 6 log10 reduction and found 2 Mrads for coronaviruses (∼29 kb genome size), 4 Mrads for filoviruses (∼19 kb), 8 Mrads for arenaviruses, bunyaviruses, orthomyxoviruses, and paramyxoviruses (∼13 kb) and 10 Mrads for flaviviruses (∼9 kb). Viruses having single-strand nucleic acid present the highest radiosensitivity. Hume and colleagues [127] reported that the three enveloped single-stranded RNA viruses of similar sizes, namely morbillivirus (90–150 nm), bunyavirus (90–120 nm), and rhabdovirus (70–150 nm), showed a comparable D 10 values (2.53, 2.61, and 2.71 kGy respectively) when irradiated under the same experimental protocol.
Leung and workers cultured SARS-CoV-2 on Vero cells in the presence of 1% fetal bovine serum and 1% l-glutamine. Virus-containing supernatants were titered after irradiation [134]. The D 10 was 1.6 kGy and the complete inactivation of the SARS-CoV-2 was attained with an absorbed dose of 10 kGy, value lower than the 20 kGy previously reported value for the similar SARS-CoV [133]. Even though the single-stranded RNA viruses may present a certain radioresistance, Nims and colleagues showed that no strong clue can explain the discrepancies in the log10 values for virus inactivation by γ-irradiation, as shown in Table 2 [135]. Multiple causative parameters may be involved in this discrepancy, including, but not limited to, sera matrices preparation variability from sample-to-sample, variability in γ-irradiation procedures (dose rate), and variability in the purity of the virus stock quality.
Viral properties of some enveloped virus families. Efficacy of γ-irradiation on the log10 reduction
| Virus | Family | Morphology (not to scale) | Size (nm) | Genome size (kb) * | Nucleic acid genome | D 10 (kGy) | log10 reduction/kGy c | References |
|---|---|---|---|---|---|---|---|---|
| IBRb | Herpesviridae |
 |
100–120 | 120–230 | Double-stranded DNA | 3.22 | 0.310 | [136,137] |
| APV | Poxviridae |
 |
240–300 | 130–260 | Double-stranded DNA | 2.20 | 0.456 | [138,139] |
| PI3b | Paramyxoviridae |
 |
100–200 | 13–19 | Single-stranded RNA | 4.78 | 0.209 | [128,140] |
| BVDVa | Flaviviridae |
 |
50–70 | 9–13 | Single-stranded RNA | 5.05 | 0.198 | [141,142] |
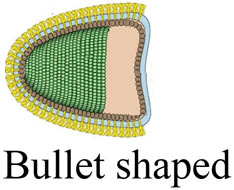
| SARS-CoV-2 | Coronaviridae |
 |
20–25 | 26–32 | Single-stranded RNA | 1.60 | 0.625 | [134,143] |
| BEFVa | Rhabdoviridae |
 |
75 × 150 | 10–16 | Single-stranded RNA | 2.94 | 0.340 | [142,144] |
| Akabanea | Bunyavuridae |
 |
80–120 | 11–23 | Single-stranded RNA | 2.50 | 0.400 | [142,145] |
| Ainoa | Bunyavuridae |
 |
80–120 | 11–23 | Single-stranded RNA | 3.45 | 0.290 | [142,146] |
APV: avian poxvirus, PI3: parainfluenza virus type 3, IBR: infectious bovine rhinotracheitis, BVDV: bovine viral diarrhea virus, BEFV: bovine ephemeral fever virus.
- *
A kilobase (kb) pair is a unit of genome size measurement equal to 1,000 base pairs of DNA or RNA.
- a
Viruses in bovine serum.
- b
Viruses spiked into fetal bovine serum.
- c
log10 reduction per kGy represents the slope of the viral inactivation (log10 reduction in titer) vs radiation dose in kGy.
In another study, Schmidt and workers [132] investigated the fractionated and continuous electron beam irradiation of four different types of viruses: the Human Immunodeficiency Virus-2 (Retroviridae enveloped HIV-2), the Hepatovirus A (Picornaviridae non-enveloped HAV), the Pseudorabies Virus (Herpesviridae, enveloped PRV), and the Porcine Parvovirus (Parvoviridae non-enveloped PPV). The cell lines for these viruses were respectively human T lymphocyte cells, mink-lung cells, embryonal rhesus monkey kidney cells, and pig kidney cells. The irradiation doses for the continuous beam were multiple of 3.4 kGy and up to 34 kGy, while for the fractionated beam, a dose of 3.4 Gy was applied up to 10 times. The
The presence of solutes in the irradiation culture of viruses renders the radiation-induced inactivation of viruses less efficient. de Roda Husman et al. [125] compared the inactivation of the respiratory feline calicivirus (FeCV) and the enteric canine calicivirus (CaCV) with the Escherichia bacteriophage MS2 in viral culture of different protein concentration. They reported a
2 Conclusion
The eruption of the SARS-CoV-2 new virus poses a real challenge and worries in a sense that its attributes are initially unknown and very limited viral data on COVID-19 infection is currently available. The virus is still infecting populations globally. Previous studies from other viruses and particularly the SARS-CoV and MERS-CoV have showed that damaging the genetic material can destroy infectivity while retaining immunogenicity. This review showed that ionizing radiation can potentially inactivate almost all the RNA viruses, by disrupting their genomic material. This could suggest a basis for developing a potential γ-inactivated virus-based vaccine against the spread of the COVID-19 pandemic. Improving its immunogenicity and preventing any potential undesired effects that could compromise the safety of the vaccine are of course other important factors which should also be taken into full consideration and thoroughly examined.
Acknowledgments
The authors gratefully acknowledge Deanship of Scientific Research at King Abdulaziz University for technical and financial support
-
Funding information: This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. RG-6-135-40.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
[1] Mäntynen S, Sundberg L-R, Oksanen H, Poranen M. Half a century of research on membrane-containing bacteriophages: bringing new concepts to modern virology. Viruses [Internet]. 2019 Jan 18;11(1):76. Available from: http://www.mdpi.com/1999-4915/11/1/7610.3390/v11010076Search in Google Scholar
[2] Menéndez-Arias L, Andino R. Viral polymerases. Virus Res [Internet]. 2017 Apr;234:1–3. Available from: https://linkinghub.elsevier.com/retrieve/pii/S016817021730128410.1016/j.virusres.2017.02.003Search in Google Scholar
[3] Islam MR, Hoque MN, Rahman MS, Alam ASMRU, Akther M, Puspo JA, et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci Rep [Internet]. 2020 Dec 1 [cited 2021 Mar 11];10(1):14004. Available from: https://doi.org/10.1038/s41598-020-70812-610.1038/s41598-020-70812-6Search in Google Scholar
[4] Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med [Internet]. 2020 Apr 22 [cited 2021 Mar 11];18(1):1–9. Available from: https://pubmed.ncbi.nlm.nih.gov/32321524/10.1186/s12967-020-02344-6Search in Google Scholar
[5] Servín-Blanco R, Zamora-Alvarado R, Gevorkian G, Manoutcharian K. Antigenic variability: obstacles on the road to vaccines against traditionally difficult targets. Hum Vaccin Immunother [Internet]. 2016 Oct 2;12(10):2640–8. Available from: https://www.tandfonline.com/doi/full/10.1080/21645515.2016.119171810.1080/21645515.2016.1191718Search in Google Scholar
[6] Carlson JM, Le AQ, Shahid A, Brumme ZL. HIV-1 adaptation to HLA: a window into virus–host immune interactions. Trends Microbiol [Internet]. 2015 Apr;23(4):212–24. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0966842X1400265010.1016/j.tim.2014.12.008Search in Google Scholar
[7] Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–66.10.1056/NEJMoa030781Search in Google Scholar
[8] Cheng VCC, Lau SKP, Woo PCY, Yuen Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev [Internet]. 2007 [cited 2020 Aug 9];20(4):660–94. Available from: http://cmr.asm.org/10.1128/CMR.00023-07Search in Google Scholar
[9] Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med [Internet]. 2012 Nov 8;367(19):1814–20. Available from: http://www.nejm.org/doi/abs/10.1056/NEJMoa121172110.1056/NEJMoa1211721Search in Google Scholar
[10] Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet [Internet]. 2020 Feb;39;5(10223):514–23. Available from: https://linkinghub.elsevier.com/retrieve/pii/S014067362030154910.1016/S0140-6736(20)30154-9Search in Google Scholar
[11] Lun Z-R, Qu L-H. Animal-to-human SARS-associated coronavirus transmission? Emerg Infect Dis [Internet]. 2004 May;10(5):959. Available from: http://wwwnc.cdc.gov/eid/article/10/5/04-0022_article.htm10.3201/eid1005.040022Search in Google Scholar
[12] Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Severe acute respiratory syndrome-related coronavirus: the species and its viruses-a statement of the coronavirus study group; 2020 [cited 2020 Aug 12];1:1–20. Available from: https://doi.org/10.1101/2020.02.07.93786210.1101/2020.02.07.937862Search in Google Scholar
[13] Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506.10.1016/S0140-6736(20)30183-5Search in Google Scholar
[14] Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez Med. 2020;28(2):174–84.Search in Google Scholar
[15] Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet [Internet]. 2020 Dec 19 [cited 2021 Feb 11];396(10267):1979–93.10.1016/S0140-6736(20)32466-1Search in Google Scholar
[16] Chan KH, Sridhar S, Zhang RR, Chu H, Fung AYF, Chan G, et al. Factors affecting stability and infectivity of SARS-CoV-2. J Hosp Infect. 2020 Oct 1;106(2):226–31.10.1016/j.jhin.2020.07.009Search in Google Scholar PubMed PubMed Central
[17] WHO. WHO coronavirus disease (COVID-19) dashboard. Geneva: World Health Organization; 2020.Search in Google Scholar
[18] Ojha R, Gupta N, Naik B, Singh S, Verma VK, Prusty D, et al. High throughput and comprehensive approach to develop multiepitope vaccine against minacious COVID-19. Eur J Pharm Sci [Internet]. 2020 Aug 1 [cited 2021 Feb 11];151:105375. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7224663/10.1016/j.ejps.2020.105375Search in Google Scholar PubMed PubMed Central
[19] Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China Supplemental content. JAMA Cardiol [Internet]. 2020 [cited 2020 Aug 9];5(7):802–10. Available from: https://jamanetwork.com/10.1001/jamacardio.2020.0950Search in Google Scholar PubMed PubMed Central
[20] Sanyaolu A, Okorie C, Marinkovic A, Patidar R, Younis K, Desai P, et al. Comorbidity and its impact on patients with COVID-19. SN Compr Clin Med [Internet]. 2020 Aug 25;2(8):1069–76. Available from: http://link.springer.com/10.1007/s42399-020-00363-410.1007/s42399-020-00363-4Search in Google Scholar PubMed PubMed Central
[21] Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, et al. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: a systematic review and meta-analysis. Int J Infect Dis [Internet]; 2020 [cited 2020 Aug 9]. Available from: https://www.sciencedirect.com/science/article/pii/S1201971220301363Search in Google Scholar
[22] Hassan SA, Sheikh FN, Jamal S, Ezeh JK, Akhtar A. Coronavirus (COVID-19): a review of clinical features, diagnosis, and treatment. Cureus. 2020 Mar 21;12:1–7.10.7759/cureus.7355Search in Google Scholar PubMed PubMed Central
[23] Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar 12;579(7798):265–9.10.1038/s41586-020-2008-3Search in Google Scholar PubMed PubMed Central
[24] Luan J, Lu Y, Jin X. Research LZ-B and biophysical, 2020 U. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun. 2020 May 21;526(1):165–9.10.1016/j.bbrc.2020.03.047Search in Google Scholar PubMed PubMed Central
[25] Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. 10.1016/bs.aivir.2019.08.002.Search in Google Scholar PubMed PubMed Central
[26] Raj R. Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing. Biochem Biophys Rep [Internet]. 2021 Mar [cited 2021 Mar 11];25:100847. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7750489/10.1016/j.bbrep.2020.100847Search in Google Scholar PubMed PubMed Central
[27] Silva SJR, Alves da Silva CT, Mendes RPG, Pena L. Role of nonstructural proteins in the pathogenesis of SARS‐CoV‐2. J Med Virol [Internet]. 2020 Sep 2 [cited 2021 Mar 11];92(9):1427–9. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.2585810.1002/jmv.25858Search in Google Scholar PubMed PubMed Central
[28] Clark LK, Green TJ, Petit CM. Structure of nonstructural protein 1 from SARS-CoV-2. J Virol [Internet]. 2020 Nov 24 [cited 2021 Mar 11];95(4):1–12. Available from: http://jvi.asm.org/10.1128/JVI.02019-20Search in Google Scholar PubMed PubMed Central
[29] Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (80) [Internet]. 2020 May 15 [cited 2021 Mar 11];368(6492):779–82. Available from: https://pubmed.ncbi.nlm.nih.gov/32277040/10.1126/science.abb7498Search in Google Scholar PubMed PubMed Central
[30] Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci [Internet]. 2020 Mar 21;63(3):457–60. Available from: http://link.springer.com/10.1007/s11427-020-1637-510.1007/s11427-020-1637-5Search in Google Scholar PubMed PubMed Central
[31] Yao M, Zhang L, Ma J, Zhou L. On airborne transmission and control of SARS-Cov-2. Sci Total Environ [Internet]. 2020 Aug;731:139178. Available from: https://linkinghub.elsevier.com/retrieve/pii/S004896972032695410.1016/j.scitotenv.2020.139178Search in Google Scholar PubMed PubMed Central
[32] Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. In: Gallagher T, editor. J Virol [Internet]. 2020 Jan 29;94(7):1–25. Available from: https://jvi.asm.org/content/94/7/e00127-2010.1128/JVI.00127-20Search in Google Scholar PubMed PubMed Central
[33] Yang X-H, Deng W, Tong Z, Liu Y-X, Zhang L-F, Zhu H, et al. Comparative medicine mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus Infection [Internet]. ingentaconnect.com; 2007 [cited 2020 Aug 9]. Available from: https://www.ingentaconnect.com/content/aalas/cm/2007/00000057/00000005/art00003Search in Google Scholar
[34] Devaux CA, Rolain J-M, Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect [Internet]. 2020 Jun;53(3):425–35. Available from: https://linkinghub.elsevier.com/retrieve/pii/S168411822030109210.1016/j.jmii.2020.04.015Search in Google Scholar PubMed PubMed Central
[35] Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. Mass Med Soc [Internet]. [cited 2020 Aug 9];382:1199–207. Available from: https://www.nejm.org/doi/full/10.1056/NEJMOa200131610.1056/NEJMoa2001316Search in Google Scholar PubMed PubMed Central
[36] Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. New Engl J Med. 2020;382:1199–207.10.1056/NEJMoa2001316Search in Google Scholar
[37] Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor. naturecom [Internet]. 2020 [cited 2020 Aug 12]. Available from: https://www.nature.com/articles/s41423-020-0400-4?report=reader10.1038/s41423-020-0400-4Search in Google Scholar PubMed PubMed Central
[38] Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 May 14;581(7807):221–4.10.1038/s41586-020-2179-ySearch in Google Scholar PubMed PubMed Central
[39] Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Int Med. 2020;76:14–20.10.1016/j.ejim.2020.04.037Search in Google Scholar PubMed PubMed Central
[40] Grove J, Marsh M. The cell biology of receptor-mediated virus entry. J Cell Biol. 2011;195:1071–82.10.1083/jcb.201108131Search in Google Scholar PubMed PubMed Central
[41] Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 Apr 16;181(2):281–92.10.1016/j.cell.2020.02.058Search in Google Scholar PubMed PubMed Central
[42] Sullivan CS, Ganem D. microRNAs and viral infection. Mol Cell. 2005;20(1):3–7.10.1016/j.molcel.2005.09.012Search in Google Scholar PubMed
[43] South AM, Diz DI, Chappell MC. COVID-19, ACE2, and the cardiovascular consequences. Am J Physiol Circ Physiol. 2020;318:1084–90.10.1152/ajpheart.00217.2020Search in Google Scholar PubMed PubMed Central
[44] Liu J, Liao X, Qian S, Yuan J, Wang F, Liu Y, et al. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis. 2020 Jun;26(6):1320–23. 10.3201/eid2606.200239.Search in Google Scholar PubMed PubMed Central
[45] Ghinai I, McPherson TD, Hunter JC, Kirking HL, Christiansen D, Joshi K, et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395:1137–44.10.1016/S0140-6736(20)30607-3Search in Google Scholar
[46] Lu J, Gu J, Li K, Xu C, Su W, Lai Z, et al. Early release-COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020; 2020.10.3201/eid2607.200764Search in Google Scholar PubMed PubMed Central
[47] Frontera A, Martin C, Vlachos K, Sgubin G. Regional air pollution persistence links to covid19 infection zoning. J Infect. 2020;81:318–56.10.1016/j.jinf.2020.03.045Search in Google Scholar PubMed PubMed Central
[48] Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int [Internet]. 2020 Jun;139:105730. Available from: https://linkinghub.elsevier.com/retrieve/pii/S016041202031254X10.1016/j.envint.2020.105730Search in Google Scholar PubMed PubMed Central
[49] Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun [Internet]. 2020 Dec 29;11(1):2800. Available from: http://www.nature.com/articles/s41467-020-16670-210.1038/s41467-020-16670-2Search in Google Scholar PubMed PubMed Central
[50] Guo Z-D, Wang Z-Y, Zhang S-F, Li X, Li L, Li C, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26(7):10–3201.10.3201/eid2607.200885Search in Google Scholar PubMed PubMed Central
[51] WHO. WHO-I guidance. Laboratory biosafety guidance related to the novel coronavirus (2019-nCoV). Geneva: World Health Organization; 2020.Search in Google Scholar
[52] Ramakrishnan MA. Determination of 50% endpoint titer using a simple formula. Artic World J Virol [Internet]. 2016 [cited 2020 Aug 12];5(2):85. Available from: http://dx.doi.org/10.5501/wjv.v5.i2.8510.5501/wjv.v5.i2.85Search in Google Scholar PubMed PubMed Central
[53] Gombold J, Karakasidis S, Niksa P, Podczasy J, Neumann K, Richardson J, et al. Systematic evaluation of in vitro and in vivo adventitious virus assays for the detection of viral contamination of cell banks and biological products. Vaccine [Internet]. 2014 May 19 [cited 2021 Mar 11];32(24):2916–26. Available from: https://pubmed.ncbi.nlm.nih.gov/24681273/10.1016/j.vaccine.2014.02.021Search in Google Scholar PubMed PubMed Central
[54] D’Arienzo M, Coniglio A. Assessment of the SARS-CoV-2 basic reproduction number, R0, based on the early phase of COVID-19 outbreak in Italy. Biosaf Heal. 2020;2:57–9.10.1016/j.bsheal.2020.03.004Search in Google Scholar PubMed PubMed Central
[55] Fanelli D, Piazza F. Analysis and forecast of COVID-19 spreading in China, Italy and France. Chaos Solitons Fractals [Internet]. 2020 May;134:109761. Available from: https://linkinghub.elsevier.com/retrieve/pii/S096007792030163610.1016/j.chaos.2020.109761Search in Google Scholar PubMed PubMed Central
[56] Alimohamadi Y, Taghdir M, Sepandi M. Estimate of the basic reproduction number for COVID-19: a systematic review and meta-analysis. J Prev Med Public Heal [Internet]. 2020 May 31;53(3):151–7. Available from: http://jpmph.org/journal/view.php?doi=10.3961/jpmph.20.07610.3961/jpmph.20.076Search in Google Scholar PubMed PubMed Central
[57] Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med [Internet]. 2020 May 5;172(9):577–82. Available from: https://www.acpjournals.org/doi/10.7326/M20-050410.7326/M20-0504Search in Google Scholar PubMed PubMed Central
[58] Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019- nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Euro Surveill. 2020;25(5):pii=2000062.10.2807/1560-7917.ES.2020.25.5.2000062Search in Google Scholar PubMed PubMed Central
[59] Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung S-M, et al. Clinical medicine incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. mdpi.com [Internet]; 2020 [cited 2020 Aug 9];538:1–9. Available from: www.mdpi.com/journal/jcm10.3390/jcm9020538Search in Google Scholar PubMed PubMed Central
[60] Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis [Internet]. 2020 Jul;26(7):1470–7. Available from: http://wwwnc.cdc.gov/eid/article/26/7/20-0282_article.htm10.3201/eid2607.200282Search in Google Scholar PubMed PubMed Central
[61] Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, et al. Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA [Internet]. 2006 Mar 28 [cited 2021 Feb 11];103(13):5108–13. Available from: www.pnas.orgcgidoi10.1073pnas.050820010310.1073/pnas.0508200103Search in Google Scholar PubMed PubMed Central
[62] Nagy PD, Carpenter CD, Simon AE. A novel 3’-end repair mechanism in an RNA virus. Proc Natl Acad Sci [Internet]. 1997 Feb 18;94(4):1113–8. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.94.4.111310.1073/pnas.94.4.1113Search in Google Scholar PubMed PubMed Central
[63] Chen P, Jiang M, Hu T, Liu Q, Chen XS, Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J Biochem Mol Biol [Internet]. 2007 [cited 2021 Feb 11];40(5):649–55. Available from: https://pubmed.ncbi.nlm.nih.gov/17927896/10.5483/BMBRep.2007.40.5.649Search in Google Scholar PubMed
[64] Barr JN, Fearns R. How RNA viruses maintain their genome integrity. J Gen Virol [Internet]. 2010 Jun 1;91(6):1373–87. Available from: https://www.microbiologyresearch.org/content/journal/jgv/10.1099/vir.0.020818-010.1099/vir.0.020818-0Search in Google Scholar PubMed
[65] Saunders RDC, Boubriak I, Clancy DJ, Cox LS. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell [Internet]. 2008 Jun;7(3):418–25. Available from: http://doi.wiley.com/10.1111/j.1474-9726.2008.00388.x10.1111/j.1474-9726.2008.00388.xSearch in Google Scholar PubMed PubMed Central
[66] Becares M, Pascual-Iglesias A, Nogales A, Sola I, Enjuanes L, Zuñiga S. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J Virol. 2016;90(11):5399–414.10.1128/JVI.03259-15Search in Google Scholar
[67] Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81(22):12135–44.10.1128/JVI.01296-07Search in Google Scholar
[68] Davies M, Osborne V, Lane S, Roy D, Dhanda S, Evans A, et al. Remdesivir in treatment of COVID-19: a systematic benefit–risk assessment. Drug Saf [Internet]. 2020 Jul 28;43(7):645–56. Available from: http://link.springer.com/10.1007/s40264-020-00952-110.1007/s40264-020-00952-1Search in Google Scholar
[69] Gautret P, Lagier J-C, Parola P, Hoang VT, Meddeb L, Sevestre J, et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med Infect Dis [Internet]. 2020 Mar;34:101663. Available from: https://linkinghub.elsevier.com/retrieve/pii/S147789392030131910.1016/j.tmaid.2020.101663Search in Google Scholar
[70] Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet [Internet]. 2020 Aug 15 [cited 2021 Feb 11];396(10249):479–88.10.1016/S0140-6736(20)31605-6Search in Google Scholar
[71] Haidere MF, Ratan ZA, Nowroz S, Zaman SB, Jung Y-J, Hosseinzadeh H, et al. COVID-19 vaccine: critical questions with complicated answers. Biomol Ther (Seoul). 2021 Jan;29(1):1–10.10.4062/biomolther.2020.178Search in Google Scholar PubMed PubMed Central
[72] Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 Feb 20;382(8):727–33.10.1056/NEJMoa2001017Search in Google Scholar PubMed PubMed Central
[73] Kuo TY, Lin MY, Coffman RL, Campbell JD, Traquina P, Lin YJ, et al. Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci Rep. 2020 Dec 1;10:1.10.1038/s41598-020-77077-zSearch in Google Scholar PubMed PubMed Central
[74] Wang F, Kream RM, Stefano GB. An evidence based perspective on mRNA-SARScov-2 vaccine development [Internet]. Med Sci Monit. 2020 [cited 2021 Feb 11];26:1–8. Available from: https://pubmed.ncbi.nlm.nih.gov/32366816/10.12659/MSM.924700Search in Google Scholar PubMed PubMed Central
[75] Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, et al. Immunogenic BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature [Internet]. 2021 Feb 1 [cited 2021 Feb 11];592:1–10. Available from: https://www.nature.com/articles/s41586-021-03275-ySearch in Google Scholar
[76] Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med [Internet]. 2020 Dec 17 [cited 2021 Feb 11];383(25):2439–50. Available from: http://www.nejm.org/doi/10.1056/NEJMoa202790610.1056/NEJMoa2027906Search in Google Scholar PubMed PubMed Central
[77] Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature [Internet]. 2020 Oct 22 [cited 2021 Feb 11];586(7830):567–71. Available from: https://doi.org/10.1038/s41586-020-2622-010.1038/s41586-020-2622-0Search in Google Scholar
[78] Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med [Internet]. 2020 Dec 17 [cited 2021 Feb 11];383(25):2427–38. Available from: https://pubmed.ncbi.nlm.nih.gov/32991794/10.1056/NEJMoa2028436Search in Google Scholar
[79] Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet [Internet]. 2020 Aug 15 [cited 2021 Feb 11];396(10249):467–78.10.1016/S0140-6736(20)31604-4Search in Google Scholar
[80] van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature [Internet]. 2020 Oct 22 [cited 2021 Feb 11];586(7830):578–82. Available from: https://doi.org/10.1038/s41586-020-2608-y10.1038/s41586-020-2608-ySearch in Google Scholar
[81] Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis [Internet]. 2021 Feb 1 [cited 2021 Mar 11];21(2):181–92. Available from: www.thelancet.com/infection10.1016/S1473-3099(20)30843-4Search in Google Scholar
[82] Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatullin AI, Shcheblyakov DV, Dzharullaeva AS, et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet [Internet]. 2020 Sep 26 [cited 2021 Feb 11];396(10255):887–97.10.1016/S0140-6736(20)31866-3Search in Google Scholar
[83] Lundstrom K. Viral vectors for COVID-19 vaccine development. Viruses [Internet]. 2021 Feb 19 [cited 2021 Mar 11];13(2):317. Available from: https://www.mdpi.com/1999-4915/13/2/31710.3390/v13020317Search in Google Scholar PubMed PubMed Central
[84] Sanders B, Koldijk M, Schuitemaker H. Inactivated viral vaccines. Vaccine analysis: strategies, principles, and control [Internet]. Berlin Heidelberg: Springer; 2015 [cited 2021 Mar 11]. p. 45–80. Available from: https://link.springer.com/chapter/10.1007/978-3-662-45024-6_210.1007/978-3-662-45024-6_2Search in Google Scholar
[85] Hicks DJ, Fooks AR, Johnson N. Developments in rabies vaccines [Internet]. Clin Exp Immunol. 2012 [cited 2021 Mar 11];169:199–204. Available from: https://pubmed.ncbi.nlm.nih.gov/22861358/10.1111/j.1365-2249.2012.04592.xSearch in Google Scholar PubMed PubMed Central
[86] Barrett PN, Mundt W, Kistner O, Howard MK. Vero cell platform in vaccine production: moving towards cell culture-based viral vaccines [Internet]. Expert Rev Vaccines. 2009 [cited 2021 Mar 11];8:607–18. Available from: https://pubmed.ncbi.nlm.nih.gov/19397417/10.1586/erv.09.19Search in Google Scholar PubMed
[87] Madhusudana SN, Shamsundar R, Seetharaman S. In vitro inactivation of the rabies virus by ascorbic acid. Int J Infect Dis [Internet]. 2004 [cited 2021 Mar 11];8(1):21–5. Available from: https://pubmed.ncbi.nlm.nih.gov/14690777/10.1016/j.ijid.2003.09.002Search in Google Scholar
[88] Bahnemann HG. Inactivation of viral antigens for vaccine preparation with particular reference to the application of binary ethylenimine [Internet]. Vaccine. 1990 [cited 2021 Mar 12];8:299–303. Available from: https://pubmed.ncbi.nlm.nih.gov/2204242/10.1016/0264-410X(90)90083-XSearch in Google Scholar
[89] Amanna IJ, Raué HP, Slifka MK. Development of a new hydrogen peroxide-based vaccine platform. Nat Med [Internet]. 2012 Jun [cited 2021 Mar 11];18(6):974–9. Available from: https://pubmed.ncbi.nlm.nih.gov/22635006/10.1038/nm.2763Search in Google Scholar PubMed PubMed Central
[90] Metz B, Kersten GFA, Hoogerhout P, Brugghe HF, Timmermans HAM, De Jong A, et al. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem [Internet]. 2004 Feb 20 [cited 2021 Mar 12];279(8):6235–43. Available from: https://pubmed.ncbi.nlm.nih.gov/14638685/10.1074/jbc.M310752200Search in Google Scholar PubMed
[91] Lawrence SA. beta-Propiolactone: viral inactivation in vaccines and plasma products. PDA J Pharm Sci Technol. 2000;54(3):209–17.Search in Google Scholar
[92] Campbell CH, Barber TL, Knudsen RC, Swaney LM. Immune response of mice and sheep to bluetongue virus inactivated by gamma irradiation. Prog Clin Biol Res [Internet]. 1985 Jan 1 [cited 2021 Mar 12];178:639–47. Available from: https://europepmc.org/article/med/2989913Search in Google Scholar
[93] Reitman M, Tribble HR, Green L. Gamma-irradiated Venezuelan equine encephalitis vaccines. Appl Microbiol [Internet]. 1970 [cited 2021 Mar 11];19(5):763–7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC376784/?report=abstract10.21236/AD0864450Search in Google Scholar
[94] Wiktor TJ, Aaslestad HG, Kaplan MM. Immunogenicity of rabies virus inactivated by -propiolactone, acetylethyleneimine, and ionizing irradiation. Appl Microbiol [Internet]. 1972 [cited 2021 Mar 11];23(5):914–8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC380470/?report=abstract10.1128/am.23.5.914-918.1972Search in Google Scholar PubMed PubMed Central
[95] Marennikova SS, Macevic GR. Experimental study of the role of inactivated vaccine in two step vaccination against smallpox bull world health organ [Internet]. 1975 Jan 1 [cited 2021 Mar 11];52(1):51–6. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/1082382/?tool=EBISearch in Google Scholar
[96] Mullbacher A, Ada GL, Tha Hla R. Gamma-irradiated influenza A virus can prime for a cross-reactive and cross-protective immune response against influenza A viruses. Immunol Cell Biol [Internet]. 1988 [cited 2021 Mar 11];66(2):153–7. Available from: https://pubmed.ncbi.nlm.nih.gov/2846435/10.1038/icb.1988.19Search in Google Scholar PubMed
[97] Kang CY, Gao Y. Killed whole-HIV vaccine, employing a well established strategy for antiviral vaccines [Internet]. AIDS Res Ther. 2017 [cited 2021 Feb 11];14:47. Available from: https://aidsrestherapy.biomedcentral.com/articles/10.1186/s12981-017-0176-510.1186/s12981-017-0176-5Search in Google Scholar PubMed PubMed Central
[98] Marzi A, Halfmann P, Hill-Batorski L, Feldmann F, Shupert WL, Neumann G, et al. An Ebola whole-virus vaccine is protective in nonhuman primates. Science (80) [Internet]. 2015 Apr 24 [cited 2021 Feb 11];348(6233):439–42. Available from: https://pubmed.ncbi.nlm.nih.gov/25814063/10.1126/science.aaa4919Search in Google Scholar PubMed PubMed Central
[99] Shahrudin S, Chen C, David SC, Singleton EV, Davies J, Kirkwood CD, et al. Gamma-irradiated rotavirus: a possible whole virus inactivated vaccine. PLoS One [Internet]. 2018 Jun 7 [cited 2021 Feb 11];13(6):e0198182. Available from: https://dx.plos.org/10.1371/journal.pone.019818210.1371/journal.pone.0198182Search in Google Scholar PubMed PubMed Central
[100] Tobin GJ, Tobin JK, Gaidamakova EK, Wiggins TJ, Bushnell RV, Lee W-M, et al. A novel gamma radiation-inactivated sabin-based polio vaccine. In: Alsharifi M, editor. PLoS One [Internet]. 2020 Jan 30 [cited 2021 Feb 11];15(1):e0228006. Available from: https://dx.plos.org/10.1371/journal.pone.022800610.1371/journal.pone.0228006Search in Google Scholar
[101] Feldmann F, Shupert WL, Haddock E, Twardoski B, Feldmann H. Gamma irradiation as an effective method for inactivation of emerging viral pathogens. Am J Trop Med Hyg [Internet]. 2019 [cited 2021 Feb 11];100(5):1275–7. Available from: https://pubmed.ncbi.nlm.nih.gov/30860018/10.4269/ajtmh.18-0937Search in Google Scholar
[102] Jahrling PB, Stephenson EH. Protective efficacies of live attenuated and formaldehyde-inactivated Venezuelan equine encephalitis virus vaccines against aerosol challenge in hamsters. J Clin Microbiol [Internet]. 1984 [cited 2021 Mar 12];19(3):429–31. Available from: https://pubmed.ncbi.nlm.nih.gov/pmc/articles/PMC271080/?report=abstract10.1128/jcm.19.3.429-431.1984Search in Google Scholar
[103] Delrue I, Verzele D, Madder A, Nauwynck HJ. Inactivated virus vaccines from chemistry to prophylaxis: merits, risks and challenges. Expert Rev Vaccines [Internet]. 2012 Jun 9 [cited 2021 Mar 12];11(6):695–719. Available from: http://www.tandfonline.com/doi/full/10.1586/erv.12.3810.1586/erv.12.38Search in Google Scholar
[104] Ohshima H, Iida Y, Matsuda A, Kuwabara M. Damage induced by hydroxyl radicals generated in the hydration layer of GAMMA-irradiated frozen aqueous solution of DNA. J Radiat Res [Internet]. 1996 Sep 1 [cited 2021 Mar 12];37(3):199–207. Available from: https://academic.oup.com/jrr/article-lookup/doi/10.1269/jrr.37.19910.1269/jrr.37.199Search in Google Scholar
[105] Alsharifi M, Müllbacher A. The γ-irradiated influenza vaccine and the prospect of producing safe vaccines in general. Immunol Cell Biol. 2010;88:103–4.10.1038/icb.2009.81Search in Google Scholar
[106] Mullbacher A, Marshall ID, Ferris P. Classification of Barmah Forest virus as an alphavirus using cytotoxic T cell assays. J Gen Virol [Internet]. 1986 [cited 2021 Mar 12];67(2):295–9. Available from: https://pubmed.ncbi.nlm.nih.gov/3003237/10.1099/0022-1317-67-2-295Search in Google Scholar
[107] Lowy RJ, Vavrina GA, LaBarre DD. Comparison of gamma and neutron radiation inactivation of influenza A virus. Antiviral Res [Internet]. 2001 [cited 2021 Feb 11];52(3):261–73. Available from: https://pubmed.ncbi.nlm.nih.gov/11675143/10.1016/S0166-3542(01)00169-3Search in Google Scholar
[108] Patterson EI, Prince T, Anderson ER, Casas-Sanchez A, Smith SL, Cansado-Utrilla C, et al. Methods of inactivation of SARS-CoV-2 for downstream biological assays. J Infect Dis [Internet]. 2020 Oct 1 [cited 2021 Feb 11];222(9):1462–7. Available from: https://academic.oup.com/jid/article/222/9/1462/589295110.1093/infdis/jiaa507Search in Google Scholar
[109] Koopmans M, Duizer E. Foodborne viruses: an emerging problem. Int J Food Microbiol. 2004 Jan;90(1):23–41.10.1016/S0168-1605(03)00169-7Search in Google Scholar
[110] Duan S-M, Zhao X-S, Wen R-F, Huang J-J, Pi G-H, Zhang S-X, et al. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed Environ Sci. 2003 Sep;16(3):246–55.Search in Google Scholar
[111] Darnell MER, Subbarao K, Feinstone SM, Taylor DR. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J Virol Methods [Internet]. 2004 Oct;121(1):85–91. Available from: https://linkinghub.elsevier.com/retrieve/pii/S016609340400179X10.1016/j.jviromet.2004.06.006Search in Google Scholar PubMed PubMed Central
[112] Gerlach M, Wolff S, Ludwig S, Schäfer W, Keiner B, Roth NJ, et al. Rapid SARS-CoV-2 inactivation by commonly available chemicals on inanimate surfaces [Internet]. J Hosp Infect. 2020 [cited 2021 Feb 11];106:633–4. Available from: https://doi.org/10.1016/j.jhin.2020.09.00110.1016/j.jhin.2020.09.001Search in Google Scholar PubMed PubMed Central
[113] Kumar A, Becker D, Adhikary A, Sevilla MD. Molecular sciences reaction of electrons with DNA: radiation damage to radiosensitization. mdpi.com [Internet]; 2019 [cited 2020 Aug 9]. Available from: www.mdpi.com/journal/ijms10.3390/ijms20163998Search in Google Scholar PubMed PubMed Central
[114] Alizadeh E, Orlando TM, Sanche L. Biomolecular damage induced by ionizing radiation: the direct and indirect effects of low-energy electrons on DNA. Annu Rev Phys Chem [Internet]. 2015 Apr;66(1):379–98. Available from: http://www.annualreviews.org/doi/10.1146/annurev-physchem-040513-10360510.1146/annurev-physchem-040513-103605Search in Google Scholar PubMed
[115] O’Neill P, Stevens DL, Garman E. Physical and chemical considerations of damage induced in protein crystals by synchrotron radiation: a radiation chemical perspective. J Synchrotron Radiat. 2002;9(6):329–32.10.1107/S0909049502014553Search in Google Scholar
[116] Bartels DM, Cook AR, Mudaliar M, Jonah CD. Spur decay of the solvated electron in picosecond radiolysis measured with time-correlated absorption spectroscopy. J Phys Chem A [Internet]. 2000 Mar;104(8):1686–91. Available from: https://pubs.acs.org/doi/10.1021/jp992723e10.1021/jp992723eSearch in Google Scholar
[117] Uehara S, Nikjoo H. Monte Carlo simulation of water radiolysis for low-energy charged particles [Internet]. J Radiat Res. 2006 [cited 2020 Aug 9];47:69–81. Available from: http://jrr.jstage.jst.go.jp10.1269/jrr.47.69Search in Google Scholar PubMed
[118] Lytle CD, Sagripanti J-L. Predicted inactivation of viruses of relevance to biodefense by solar radiation. J Virol. 2005;79(22):14244–52.10.1128/JVI.79.22.14244-14252.2005Search in Google Scholar PubMed PubMed Central
[119] Lomax ME, Folkes LK, O’neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol. 2013;25(10):578–85.10.1016/j.clon.2013.06.007Search in Google Scholar PubMed
[120] Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22(11):1747–60.10.1021/tx900242kSearch in Google Scholar PubMed PubMed Central
[121] Sobotta L, Skupin-Mrugalska P, Mielcarek J, Goslinski T, Balzarini J. Photosensitizers mediated photodynamic inactivation against virus particles mini-reviews. Med Chem [Internet]. 2015 Apr 18 [cited 2021 Feb 11];15(6):503–21. Available from: https://pubmed.ncbi.nlm.nih.gov/25877599/10.2174/1389557515666150415151505Search in Google Scholar
[122] Costa L, Faustino MAF, Neves MGPMS, Cunha Â, Almeida A. Photodynamic inactivation of mammalian viruses and bacteriophages [Internet]. Viruses. 2012 [cited 2021 Feb 11];4:1034–74. Available from: https://pubmed.ncbi.nlm.nih.gov/pmc/articles/PMC3407894/10.3390/v4071034Search in Google Scholar
[123] Lampe N, Karamitros M, Breton V, Brown JMC, Sakata D, Sarramia D, et al. Mechanistic DNA damage simulations in Geant4-DNA Part 2: electron and proton damage in a bacterial cell. Phys Med. 2018;48:146–55.10.1016/j.ejmp.2017.12.008Search in Google Scholar
[124] Sommer R, Pribil W, Appelt S, Gehringer P, Eschweiler H, Leth H, et al. Inactivation of bacteriophages in water by means of non-ionizing (UV-253.7 nm) and ionizing (gamma) radiation: a comparative approach. Water Res. 2001;35(13):3109–16.10.1016/S0043-1354(01)00030-6Search in Google Scholar
[125] de Roda Husman AM, Bijkerk P, Lodder W, van den Berg H, Pribil W, Cabaj A, et al. Calicivirus Inactivation by nonionizing (253.7-nanometer-wavelength [UV]) and ionizing (gamma) radiation. Appl Environ Microbiol [Internet]. 2004 Sep;70(9):5089–93. Available from: https://aem.asm.org/content/70/9/508910.1128/AEM.70.9.5089-5093.2004Search in Google Scholar PubMed PubMed Central
[126] Wang T-Y, Libardo MDJ, Angeles-Boza AM, Pellois J-P. Membrane oxidation in cell delivery and cell killing applications. ACS Chem Biol [Internet]. 2017 May 19;12(5):1170–82. Available from: https://pubs.acs.org/doi/10.1021/acschembio.7b0023710.1021/acschembio.7b00237Search in Google Scholar PubMed PubMed Central
[127] Hume AJ, Ames J, Rennick LJ, Duprex WP, Marzi A, Tonkiss J, et al. Inactivation of RNA viruses by gamma irradiation: a study on mitigating factors. Viruses. 2016;8(7):204.10.3390/v8070204Search in Google Scholar PubMed PubMed Central
[128] Nims RW, Gauvin G, Plavsic M. Gamma irradiation of animal sera for inactivation of viruses and mollicutes–a review. Biologicals. 2011;39(6):370–7.10.1016/j.biologicals.2011.05.003Search in Google Scholar PubMed
[129] Durante M, Schulze K, Incerti S, Francis Z, Zein S, Guzmán CA. Virus irradiation and COVID-19 disease. Front Phys [Internet]. 2020 Oct 20 [cited 2021 Feb 11];8:565861. Available from: https://www.frontiersin.org/articles/10.3389/fphy.2020.565861/full10.3389/fphy.2020.565861Search in Google Scholar
[130] Zhao L, Mi D, Hu B, Sun Y. A generalized target theory and its applications. Sci Rep [Internet]. 2015 Nov 28;5(1):14568. Available from: http://www.nature.com/articles/srep1456810.1038/srep14568Search in Google Scholar PubMed PubMed Central
[131] Nomiya T. Discussions on target theory: past and present. J Radiat Res [Internet]. 2013 Nov;54(6):1161–3. Available from: https://academic.oup.com/jrr/article-lookup/doi/10.1093/jrr/rrt07510.1093/jrr/rrt075Search in Google Scholar PubMed PubMed Central
[132] Pruß A, Schmidt T, Hoburg AT, Gohs U, Schumann W, Sim-Brandenburg J-W, et al. Original article originalarbeit inactivation effect of standard and fractionated electron beam irradiation on enveloped and non-enveloped viruses in a tendon transplant model. Transfus Med Hemother [Internet]. 2012 Feb [cited 2020 Aug 12];39(1):29–35. Available from: www.karger.com/tmh10.1159/000336380Search in Google Scholar PubMed PubMed Central
[133] Feldmann F, Shupert WL, Haddock E, Twardoski B, Feldmann H. Gamma irradiation as an effective method for inactivation of emerging viral pathogens. Am J Trop Med Hyg [Internet]. 2019 May 1;100(5):1275–7. Available from: http://www.ajtmh.org/content/journals/10.4269/ajtmh.18-093710.4269/ajtmh.18-0937Search in Google Scholar PubMed PubMed Central
[134] Leung A, Tran K, Audet J, Lavineway S, Bastien N, Krishnan J. In vitro Inactivation of SARS-CoV-2 using gamma radiation. Appl Biosaf [Internet]. 2020 Sep 1 [cited 2021 Feb 11];25(3):157–60. Available from: https://www.liebertpub.com/doi/10.1177/153567602093424210.1177/1535676020934242Search in Google Scholar PubMed PubMed Central
[135] Gauvin G. RN-P Journal of Pharmaceutical Science. Gamma-irradiation of serum for the inactivation of adventitious contaminants. journal.pda.org [Internet]; 2010 Undefined [cited 2020 Aug 9]. Available from: https://journal.pda.org/content/64/5/432.shortSearch in Google Scholar
[136] Davison AJ. Herpesvirus systematics. Vet Microbiol. 2010 Jun;143(1):52–69.10.1016/j.vetmic.2010.02.014Search in Google Scholar PubMed PubMed Central
[137] Plavsic MZ, Daley JP, Danner DJ, Weppner DJ. Gamma irradiation of bovine sera. Dev Biol Stand. 1999;99:95–109.Search in Google Scholar
[138] Moss B. Poxvirus DNA replication. cshperspectives.cshlp.org [Internet]. Vol. 5, 2013 Sep [cited 2020 Aug 14]. p. 9. Available from: http://cshperspectives.cshlp.org/10.1101/cshperspect.a010199Search in Google Scholar PubMed PubMed Central
[139] Thomas FC, Davies AG, Dulac GC, Willis NG, Papp-Vid G, Girard A. Gamma ray inactivation of some animal viruses. Can J Comp Med [Internet]. 1981 [cited 2021 Feb 11];45(4):397–9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1320171/?report=abstractSearch in Google Scholar
[140] Cox RM, Plemper RK. Structure and organization of paramyxovirus particles. Curr Opin Virol. 2017;24:105–14.10.1016/j.coviro.2017.05.004Search in Google Scholar PubMed PubMed Central
[141] Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. naturecom [Internet]. 2018 Feb 12 [cited 2020 Aug 14];16(3):125–42. Available from: www.nature.com/nrmicro10.1038/nrmicro.2017.170Search in Google Scholar PubMed PubMed Central
[142] House C, House JA, Yedloutschnig RJ. Inactivation of viral agents in bovine serum by gamma irradiation1. Can J Microbiol. 1990;36:737–40.10.1139/m90-126Search in Google Scholar PubMed
[143] Phan MVT, Ngo Tri T, Hong Anh P, Baker S, Kellam P, Cotten M. Identification and characterization of Coronaviridae genomes from Vietnamese bats and rats based on conserved protein domains. Virus Evol [Internet]. 2018 Jul 1 [cited 2020 Aug 14];4(2):1–12. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6295324/?report=abstract10.1093/ve/vey035Search in Google Scholar PubMed PubMed Central
[144] Dietzgen RG, Kondo H, Goodin MM, Kurath G, Vasilakis N. The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins [Internet]. Virus Res. 2017 [cited 2021 Feb 11];227:158–70. Available from: https://pubmed.ncbi.nlm.nih.gov/27773769/10.1016/j.virusres.2016.10.010Search in Google Scholar PubMed PubMed Central
[145] Klemm C, Reguera J, Cusack S, Zielecki F, Kochs G, Weber F. Systems to establish bunyavirus genome replication in the absence of transcription. J Virol [Internet]. 2013 Jul 15 [cited 2021 Feb 11];87(14):8205–12. Available from: http://jvi.asm.org/10.1128/JVI.00371-13Search in Google Scholar PubMed PubMed Central
[146] Akashi H, Onuma S, Nagano H, Ohta M, Fukutomi T. Detection and differentiation of Aino and Akabane Simbu serogroup bunyaviruses by nested polymerase chain reaction. Arch Virol [Internet]. 1999 [cited 2021 Feb 11];144(11):2101–9. Available from: https://pubmed.ncbi.nlm.nih.gov/10603165/10.1007/s007050050625Search in Google Scholar PubMed
© 2021 Fouad A. Abolaban and Fathi M. Djouider, published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells
Articles in the same Issue
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells

