Abstract
The circ_UBR4 (hsa_circ_0010283) is a novel abnormally overexpressed circRNA in oxidized low-density lipoprotein (ox-LDL)-induced model of atherosclerosis (AS) in human vascular smooth muscle cells (VSMCs). However, its role in the dysfunction of VSMCs remains to be further investigated. Here, we attempted to explore its role in ox-LDL-induced excessive proliferation and migration in VSMCs by regulating Rho/Rho-associated coiled-coil containing kinase 1 (ROCK1), a therapeutic target of AS. Expression of circ_UBR4 and ROCK1 was upregulated, whereas miR-107 was downregulated in human AS serum and ox-LDL-induced VSMCs. Depletion of circ_UBR4 arrested cell cycle, suppressed cell viability, colony-forming ability, and migration ability, and depressed expression of proliferating cell nuclear antigen and matrix metalloproteinase 2 in VSMCs in spite of the opposite effects of ox-LDL. Notably, ROCK1 upregulation mediated by plasmid transfection or miR-107 deletion could counteract the suppressive role of circ_UBR4 knockdown in ox-LDL-induced VSMCs proliferation, migration, and cell cycle progression. In mechanism, miR-107 was identified as a target of circ_UBR4 to mediate the regulatory effect of circ_UBR4 on ROCK1. circ_UBR4 might be a contributor in human AS partially by regulating VSMCs’ cell proliferation, migration, and cell cycle progression via circ_UBR4/miR-107/ROCK1 pathway.
1 Introduction
Atherosclerosis (AS), a common cause of cardiovascular and cerebrovascular diseases, is a chronic inflammatory disease in blood vessels [1]. AS is initially distinguished by the formation of lipid-rich plaques in the wall of medium-to-large sized arteries [2]. Because AS is a progressive process, circulating oxidized lipids accumulate in inner layers of artery walls and then internalized by resident macrophages, eventually leading to the establishment of atheromatic plaque core [3]. Endothelial dysfunction followed by inflammation of the vessel wall leads to atherosclerotic lesion formation [4]. The excessive migration, proliferation, and inflammation of vascular smooth muscle cells (VSMCs), one essential component of blood vessels, are important pathological features after vascular injury in AS [5,6].
Circular RNAs (circRNAs) are endogenous and stable transcripts formed by back-splicing events. Emerging evidence demonstrates that circRNAs have been implicated in multiple cardiovascular diseases (CVDs) including AS [7], even supposed as potential theranostics [8]. VSMCs have long been considered as one key cellular target in preventing AS [9], and circRNA biology in VSMCs has also been extensively overviewed [10]. The circ_UBR4 (hsa_circ_0010283), located in the chromosome 1 and derived from exons 45 to 66 of UBR4, is one novel highly expressed circRNA in abnormal lipid metabolism in VSMCs stimulated by oxidized low-density lipoprotein (ox-LDL) [11]. ox-LDL is a biomarker of CVDs [12], and ox-LDL force could mimic inflammatory environment and lipid deposition in arterial wall cells. However, the specific molecular functions and biological effects of circ_UBR4 remain to be further defined.
circRNAs and competing endogenous RNAs (ceRNAs) are described as the “professional” microRNAs (miRNAs) “inhibitors/sponges” to regulate their cellular concentrations [13,14]. As miRNA-target interaction is strongly concentration dependent, circRNAs could depress the expression of functional genes and then participate in diverse cell progressions [15]. Rho-associated coiled-coil containing kinase 1 (ROCK1) is one effector of small GTP-binding protein Rho, and Rho/ROCK1 pathway has been developed as therapeutic agent for CVDs [16], as well as therapeutic targets for statins in AS [17]. The facilitating role of ROCK1 has been well documented in dysfunction of VSMCs and vascular endothelial cells in AS models in vitro and in vivo [18,19,20]. Yet, miRNA regulatory mechanism on ROCK1 in AS is poorly illustrated, especially the interplay among circRNAs, miRNAs, and ROCK1.
During atherogenesis, big epigenetic changes in different noncoding RNAs, especially miRNAs, are discovered in VSMCs, vascular endothelial cells, and macrophages [21]. miRNA (miR)-107, belonging to miR-15/107 family [22], has been found to be implicated in the pathogenesis of some diseases [23], including in vascular endothelial cells dysfunctions [24,25,26]; however, its role in VSMCs remains largely uncovered. In this study, we aimed to investigate the expression and role of circ_UBR4 in human VSMCs induced by ox-LDL; furthermore, the relationship and reciprocal role of circ_UBR4, ROCK1, and miR-107 in proliferation, migration, and cell cycle progression of this model cells were validated.
2 Materials and methods
2.1 Clinical serum samples
A group of 41 AS patients and 41 normal control people were recruited from Zhongshan Affiliated Hospital, Dalian University. The blood samples were harvested, and the serum was extracted by centrifugation at 3,000 g for 10 min. The control volunteers were without cancer, infection, or autoimmune diseases. No significant differences had been found in age and sex among patients and healthy volunteers, and more clinicopathological data of all participators are presented in Table 1.
Clinicopathological data of AS patients and healthy volunteers
| Clinical characteristics | Groups | P | |
|---|---|---|---|
| Normal (N = 41) | AS (N = 41) | ||
| Gender, % (male) | 63.41 | 68.29 | >0.05 |
| Age (years) | 53 ± 7 | 55 ± 11 | >0.05 |
| Hypertension, % (n) | 43.91 (18) | 46.34 (19) | >0.05 |
| Diabetes, % (n) | 26.83 (11) | 21.95 (9) | >0.05 |
| Mean heart rate (bpm) | 75.34 ± 11.63 | 78.26 ± 2.67 | >0.05 |
| Current cigarette user, % (n) | 55.11 | 62.3 | >0.05 |
| Current alcohol user, % (n) | 31.2 | 33.4 | >0.05 |
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations and institutional policies and in accordance with the tenets of the Helsinki Declaration, and has been approved by the Ethics Committee of Zhongshan Affiliated Hospital, Dalian University.
2.2 Cell culture
Human VSMCs (BNCC340087) were obtained from BeNa Culture Collection (Beijing, China) and cultivated in Dulbecco’s Modified Eagle’s Medium, high glucose (M22650; R&D SYSTEMS, Shanghai, China) equipped with 10% fetal bovine serum (S11150H; R&D SYSTEMS). For AS model establishment in vitro, VSMCs at 80% confluence were administrated with 20, 40, 80, or 100 μg/mL ox-LDL (YB-002; Yiyuanbiotech, Guangzhou, China) for 24 h. Control cells were VSMCs without ox-LDL treatment. All cells were incubated in a humidified 5% CO2 incubator at 37°C.
2.3 Reverse transcriptase quantitative PCR (RT-qPCR)
Total RNA was isolated in TRIzol LS (for serum; Invitrogen, Carlsbad, CA, USA) and TRIzol (for cells; Invitrogen) according to its manufacturer protocol. The RNA samples were subjected to RT-qPCR with SuperScript First-strand Synthesis System (Invitrogen) with gDNA remover for cDNA synthesis and SYBR Green qPCR Master Mix (Invitrogen) for cDNA amplification. The special qPCR primers for circ_UBR4, UBR4, miR-107, ROCK1, glyceraldehyde-phosphate dehydrogenase (GAPDH), and U6 were provided by GENEWIZ (Beijing, China), and the sequence of primers is presented in Table 2. Cycle threshold (Ct) values were determined on 7500 real-time PCR System (Applied Biosystems, Foster City, MA, USA), and the condition of running RT-qPCR is presented in Table 3. Relative expression was calculated using the 2−ΔΔCt method. GAPDH (for circRNA and mRNA) and U6 (for miRNA) were used as endogenous controls.
The primers of RT-qPCR
| Name/length | Sequence (5′–3′) | Specificity (≤length) |
|---|---|---|
| circ_UBR4 (112nt) | 5′-ACACTCATCAGCCTGTTCCA-3′ forward and 5′-CCCTGTAGTTTGCTGGACAC-3′ reverse | Yes https://www.ncbi.nlm.nih.gov/tools/primer-blast/circ_UBR4 FR |
| UBR4 (118nt) | TTCCCCTCGAAGCAACACTC forward | Yes https://www.ncbi.nlm.nih.gov/tools/primer-blast/UBR4 FR |
| CTGGCTTTCCTGACGGATGT reverse | ||
| miR-107 (74nt) | 5′-CAGCATTGTACAGGGCT-3′ forward and 5′-GAACATGTCTGCGTATCTC-3′ reverse | Yes https://www.ncbi.nlm.nih.gov/tools/primer-blast/miR-107 FR |
| ROCK1 (136nt) | AAGAGGGCATTGTCACAGCA forward | Yes https://www.ncbi.nlm.nih.gov/tools/primer-blast/ROCK1 FR |
| AGCATCCAATCCATCCAGCA reverse | ||
| GAPDH (101nt) | ACAACTTTGGTATCGTGGAAGG forward | Yes https://www.ncbi.nlm.nih.gov/tools/primer-blast/GAPDH FR |
| GCCATCACGCCACAGTTTC reverse | ||
| U6 (87nt) | 5′-CGCTTCGGCAGCACATATAC-3′ forward and 5′-TTCACGAATTTGCGTGTCATC-3′ reverse | Yes https://www.ncbi.nlm.nih.gov/tools/primer-blast/U6 FR |
The conditions of running RT-qPCR
| Procedure | Temperature (°C) | Time (s) |
|---|---|---|
| Pre-denaturation | 95 | 30 |
| Denaturation (for 40 cycles) | 95 | 5 |
| Annealing/extension (for 40 cycles) | 62 | 20 |
2.4 Ribonuclease R (RNase R) and actinomycin D (ActD) treatment
RNase R is an exoribonuclease that degraded linear RNA but left circular transcripts intact [27]. The extracted RNA in 100 μg/mL ox-LDL-treated VSMCs was treated with RNase R (Geneseed, Guangzhou, China) with a ratio of 3 U enzyme/μg RNA for 30 min at 37°C or treated with the 1× Reaction Buffer (as Mock group). Ethanol precipitation was carried out to remove the enzyme and salts. After that, RT-qPCR was used to examine circ_UBR4 and UBR4 expression. ActD was used to block transcription, and 100 μg/mL ox-LDL-treated VSMCs in 24-well plate were added to 2 μg/mL ActD (Amyjet, Wuhan, China) for 0, 4, 8, 16, and 24 h before total RNA isolation and RT-qPCR for the detection of circ_UBR4 and UBR4 expression.
2.5 Cell transfection
VSMCs in 60% confluence were performed for cell transfection with the aid of Lipofectamine 3000 (Invitrogen) following the manufacturer’s protocols. The nucleotides included siRNA targeting circ_UBR4 (si-circ_UBR4), miR-107 mimic (miR-107), miR-107 inhibitor (anti-miR-107), and the negative controls (si-NC, miR-NC, and anti-miR-NC), as well as pcDNA plasmid (pcDNA) and pcDNA-ROCK1 plasmid (ROCK1). Post-transfection for 24 h, VSMCs were subjected to 100 μg/mL ox-LDL treatment for 24 h. The sequence of oligonucleotides was listed, including si-circ_UBR4, 5′-CAGCCTGTTCCAATGGTGGCA-3′; miR-107, 5′-AGCAGCAUUGUACAGGGCUAUCA-3′; anti-miR-107, 5′-TGATAGCCCTGTACAATGCTGCT-3′; si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-NC, 5′-GGUUCGUACGUACACUGUUCA-3′; anti-miR-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′.
2.6 Flow cytometry (FCM)
Cell cycle distribution of VSMCs in control group and ox-LDL group was determined using propidium iodide (PI; Sigma-Aldrich, St. Louis, MO, USA) staining and FCM analysis. VSMCs were collected and prepared before testing. Briefly, 1 × 106 cells were digested with trypsin free from EDTA and then fixed in 70% ethanol overnight at 4°C. After RNase A treatment (100 μg/mL, 1 h, 37°C), the cells were stained with 50 μg/mL PI at room temperature for 20 min. Promptly, the DNA contents of stained cells were analyzed on FlowJo software 7.6 (Tree Star, San Carlos, CA, USA).
2.7 Colony formation assay and cell counting kit (CCK)-8 assay
Control VSMCs and ox-LDL-treated VSMCs were seeded into 6-well plate at a density of 300 cells/well, and these cells were cultured in complete medium for another 14 days. Fresh medium was replaced per 3 days. Then, cells were fixed with 70% ethanol for 30 min and stained with 0.2% crystal violet for 30 min at the room temperature. Images of the colonies formed were captured by Quantity One (Bio-Rad, Hercules, CA, USA).
Cell viability of VSMCs before and after ox-LDL treatment was measured by CCK-8 assay; 20 μL of CCK-8 solution (Sigma-Aldrich) was added to the medium of each well, and five repeated wells were set up in each group. After incubation for 2 h at 37°C, the optical density (OD) value at 570 nm was measured using an automatic multi-well spectrophotometer (Bio-Rad).
2.8 Transwell assay
Cell migration ability was evaluated by transwell assay with transwell chamber (6.5 μm pore; Corning, NY, USA). Control VSMCs and ox-LDL-treated VSMCs were re-suspended in serum-free medium at a density of 1 × 104 cells/chamber, followed by transferring in the top chamber. Another 400 μL of complete medium was placed into the lower chamber. Transwell chambers were maintained at 37°C for 48 h. The migrated cells on the basolateral chamber were fixed with 70% ethanol for 30 min and stained with 0.2% crystal violet for 30 min at room temperature. The stained cells were captured under an inverted microscope (Olympus, Tokyo, Japan).
2.9 Western blotting
Total cellular proteins of VSMCs were lysed in RIPA buffer supplemented with protease inhibitors, phosphatase inhibitors, and phenylmethanesulfonyl fluoride (PMSF). With centrifugation at 12,000 g for 20 min at 4°C, protein samples were harvested. The concentration of obtained proteins was quantified using Bradford reagent (Sigma-Aldrich), and 20 μg proteins were separated, transferred, and exposed following the standard procedures [28]. The primary antibodies against ROCK1 (ab45171, 1:10,000), proliferating cell nuclear antigen (PCNA; ab18197, 1:1,000), matrix metalloproteinase 2 (MMP2; ab97779, 1:2,500), and GAPDH (ab9485, 1:2,500) were from Abcam (Cambridge, UK), as well as HRP-conjugated secondary antibody against rabbit (ab205718, 1:50,000). Staining intensity of the bands was detected on Integrated chemiluminescence apparatus (ChemiScope 5300 Pro, Clinx, Shanghai, China). GAPDH was the internal control.
2.10 Dual-luciferase reporter assay
Artificially synthesized fragments of circ_UBR4 and ROCK1 3′ untranslated region (3′UTR) containing miR-107-binding sites were, respectively, inserted into the luciferase report vector pmirGLO (Promega, Madison, WI, USA), named as circ_UBR4-wt and ROCK1 3′UTR-wt. A mutation of these sites was performed and inserted into pmirGLO (Promega) as well, named as circ_UBR4-mut and ROCK1 3′UTR-mut. VSMCs seeded in 24-well plate were co-transfected with pmirGLO vectors carrying circ_UBR4-wt, ROCK1 3′UTR-wt, circ_UBR4-mut, or ROCK1 3′UTR-mut together with miR-107 or miR-NC. The transfection was performed in triplicate. Post transfection for 48 h, the Firefly luciferase activity was examined by Dual Luciferase Assay System (Promega) and normalized to the Renilla luciferase activity.
2.11 Pearson’s correlation analysis
The correlation among circ_UBR4, miR-107, and ROCK1 mRNA levels in serum samples from AS patients (n = 41) was identified using Pearson’s correlation analysis. The correlation coefficient was greater than 0.8.
2.12 Statistical analysis
All data were analyzed with software GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA), and data were presented as mean ± standard deviation for at least three times from biological level. Statistical analyses were performed using unpaired Student’s t-test or one-way analysis of variance (ANOVA). Tukey’s post hoc analysis was performed after ANOVA. P < 0.05 was deemed as significantly different.
3 Results
3.1 circ_UBR4 was upregulated in human AS serum and ox-LDL-induced human VSMCs with a stable structure
To screen circ_UBR4 expression in AS patients, serum samples were collected. The level of circ_UBR4 in human AS serum was 3.1-fold of that in normal people (Figure 1a). In vitro, human VSMCs were disposed to ox-LDL to stimuli AS model, and circ_UBR4 level was increased in ox-LDL-treated VSMCs (Figure 1b). Besides, compared with its host gene UBR4, circ_UBR4 expression was unaffected with RNase R and ActD treatment (Figure 1c and d). These results suggested a potential role of circ_UBR4 underlying the pathogenesis of AS, especially in VSMCs.
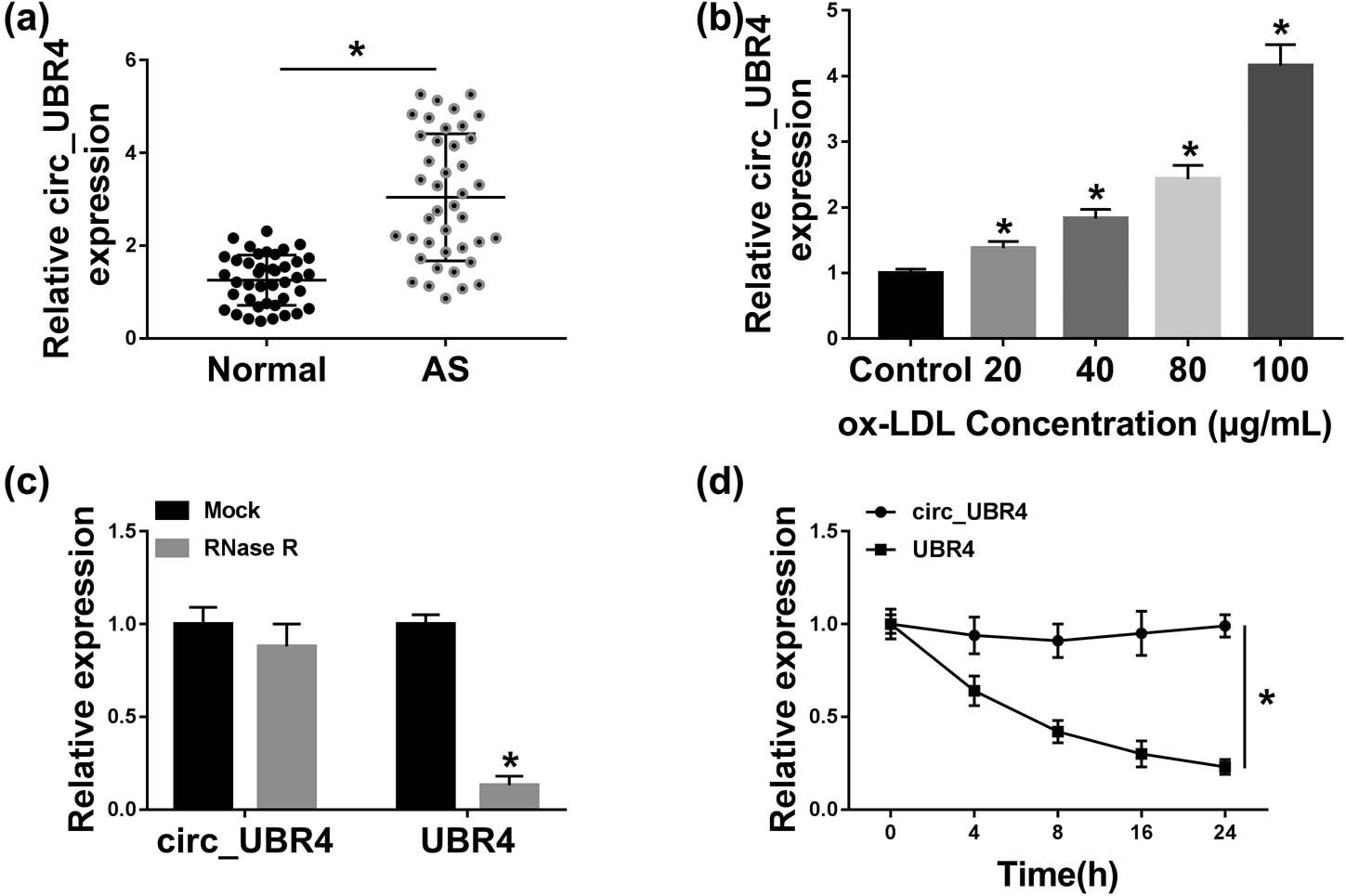
Expression of circ_UBR4 (hsa_circ_0010283) was upregulated in human AS serum and ox-LDL-induced VSMCs. RT-qPCR detected circ_UBR4 expression level in (a) serum samples from AS patients (AS; n = 41) and normal control people (normal; n = 41), and (b) human VSMCs treated with different concentrations of ox-LDL for 24 h. (c and d) RT-qPCR measured levels of circ_UBR4 and its host gene UBR4 in (c) extracted RNA treated with RNase R or with Mock, and (d) 100 μg/mL ox-LDL-treated VSMCs treated with ActD. *P < 0.05.
3.2 The blockage of circ_UBR4 restrained proliferation, migration, and cell cycle progression of ox-LDL-induced human VSMCs
To investigate the cellular function of circ_UBR4 in human VSMCs, circ_UBR4 was artificially silenced using siRNA transfection, and RT-qPCR identified a high knockdown efficiency of si-circ_UBR4 in VSMCs (Figure 2a). Loss-of-function experiments were carried out in ox-LDL-treated VSMCs with siRNA transfection. ox-LDL treatment induced higher cell distribution in S-phase and lower cell distribution in G0/G1 phase (Figure 2b), whereas these cell distributions were reversed with si-circ_UBR4 pre-transfection. Colony-forming ability of VSMCs was facilitated in response to ox-LDL treatment, which was attenuated by introducing si-circ_UBR4 (Figure 2c). CCK-8 assay showed cell viability promotion in ox-LDL-treated VSMCs (Figure 2d), accompanied with elevated expression of PCNA (proliferation marker) (Figure 2e). Moreover, si-circ_UBR4-mediated circ_UBR4 downregulation led to diminishment of cell viability and PCNA expression in ox-LDL-challenged VSMCs (Figure 2d and e). Transwell migration ability of VSMCs along with expression of MMP2 (migration marker) was promoted under ox-LDL stress, and this promotion was cancelled with pre-transfection of si-circ_UBR4 (Figure 2f and g). Above outcomes indicated a suppressive effect of circ_UBR4 blockage on ox-LDL-induced excessive proliferation, migration, and cell cycle progression of human VSMCs in vitro.
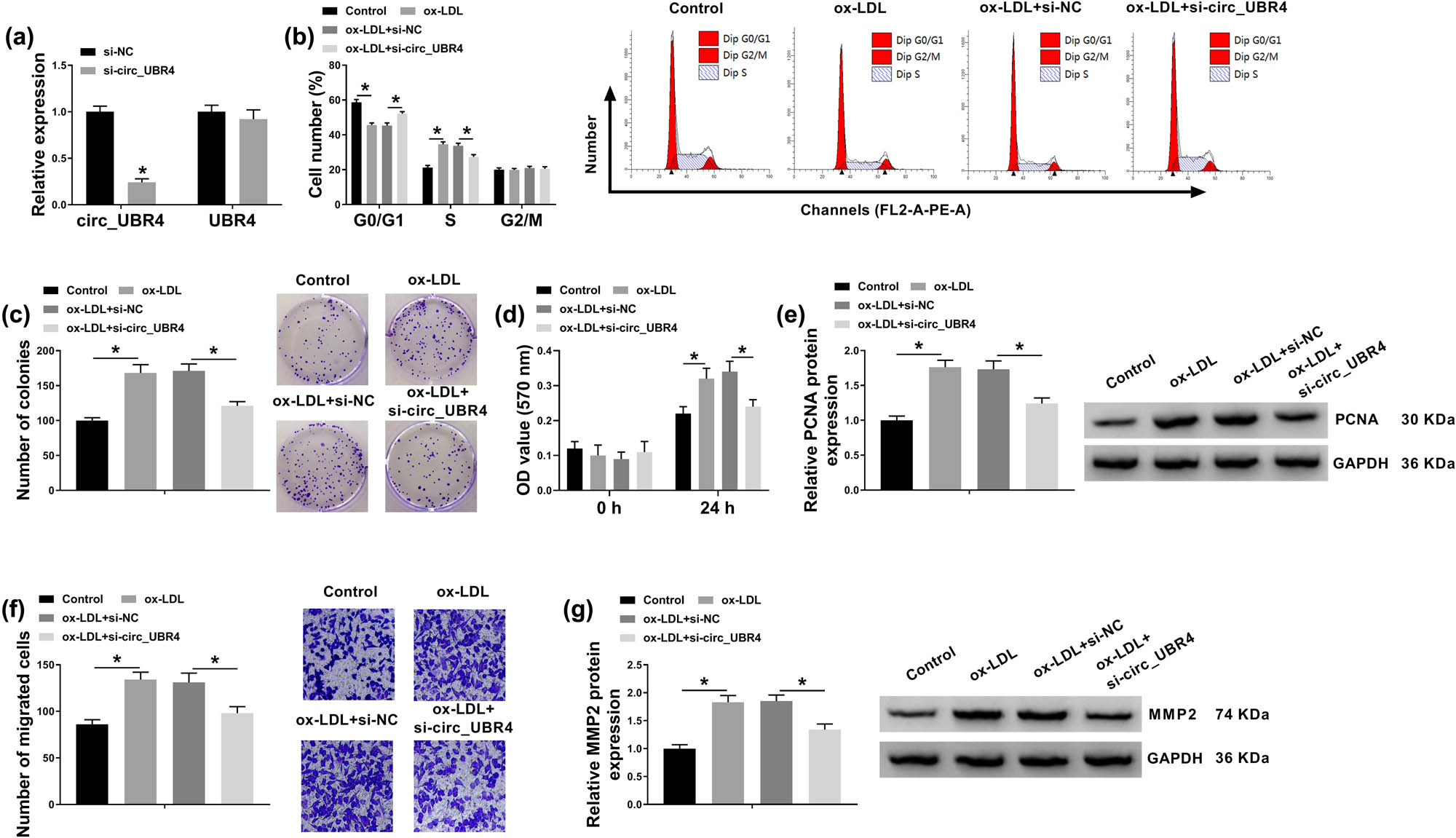
Suppressive role of circ_UBR4 depletion in cell cycle progression, proliferation, and migration of ox-LDL-induced human VSMCs. (a) RT-qPCR detected circ_UBR4 and UBR4 expression levels in VSMCs transfected with siRNA targeting circ_UBR4 (si-circ_UBR4), normalized to its negative control (si-NC) transfection. (b–g) VSMCs were co-treated with siRNA transfection for 24 h and ox-LDL (100 μg/mL) treatment for another 24 h. (b) FCM analyzed cell distributions in different cell cycle phases (G0/G1, S, and G2/M) by counting cell number. (c) Colony formation assay measured colony-forming ability by determining number of colonies. (d) CCK-8 assay monitored cell viability by analyzing OD value at 570 nm. (e) Western blotting detected expression of proliferation marker, PCNA. (f) Transwell assay evaluated cell migration ability by counting number of migrated cells. (g) Western blotting detected expression of migration marker, matrix MMP2. *P < 0.05.
3.3 ROCK1 overexpression counteracted the suppressive role of circ_UBR4 depletion in ox-LDL-induced human VSMCs
ROCK1 plasmid transfection restored the expression of ROCK1 on both mRNA level and protein level in VSMCs transfected with si-circ_UBR4 (Figure 3a and b). Moreover, ROCK1 plasmid also abrogated the suppression of circ_UBR4 silence on cell cycle progression, as depicted by decreased G0/G1 cells and increased S-cells (Figure 3c). Colony number of ox-LDL-treated VSMCs was reduced by silencing circ_UBR4, and this reduction was mitigated with the presence of ROCK1 plasmid (Figure 3d). Under ox-LDL stimuli, si-circ_UBR4 lowered cell viability and PCNA expression of VSMCs, which was rescued because of co-administration of ROCK1 plasmid (Figure 3e and f). Anti-migration role of circ_UBR4 knockdown in ox-LDL-disposed VSMCs was augmented by restoring ROCK1 expression via plasmid transfection, as evidenced by increased transwell migrated cells and higher MMP2 protein level (Figure 3g and h). These findings demonstrated that ROCK1 upregulation could counteract the suppression of circ_UBR4 knockdown on ox-LDL-induced VSMCs excessive proliferation, migration, and cell cycle progression.
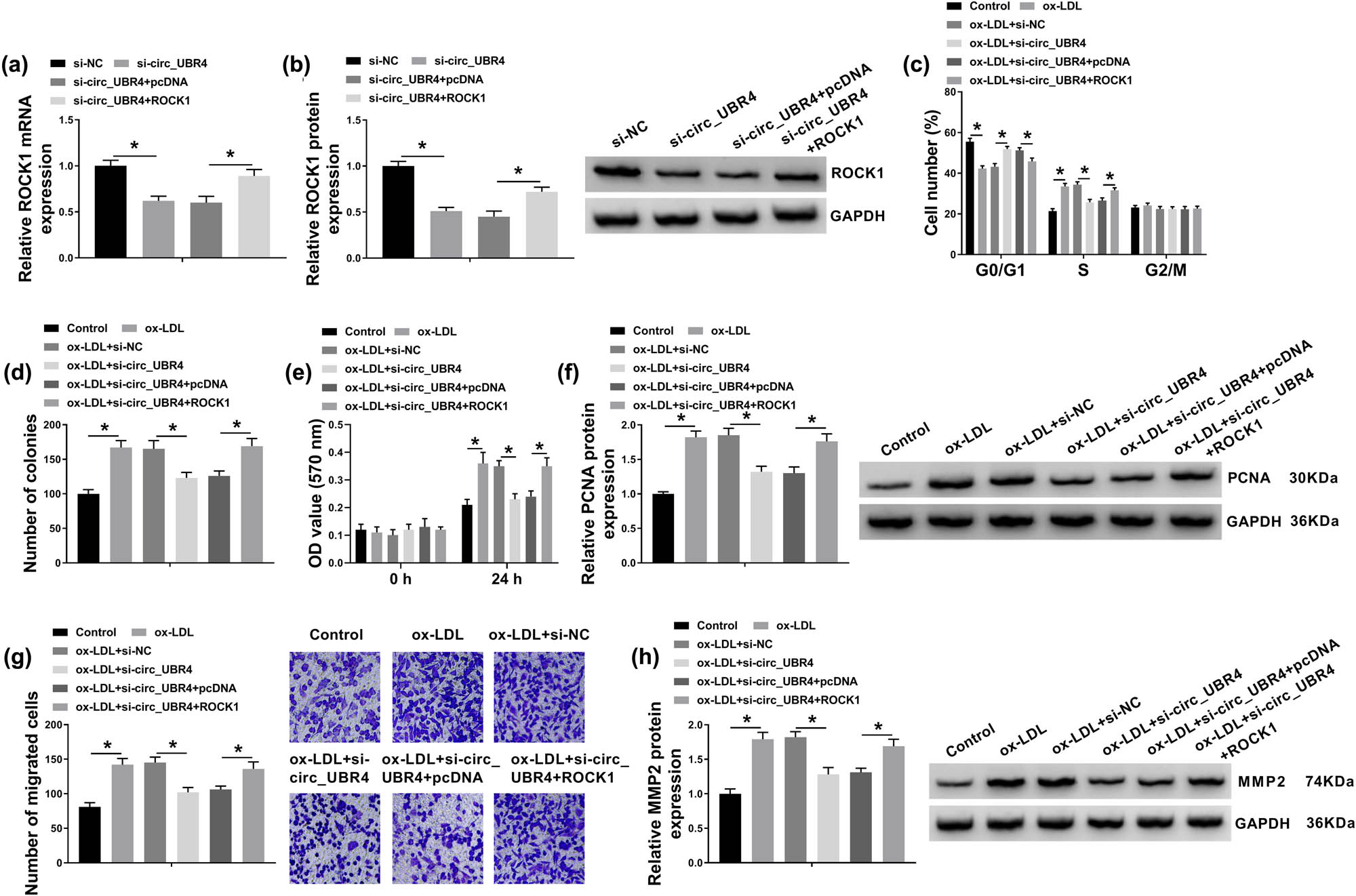
Rho/Rho-associated coiled-coil containing kinase 1 (ROCK1) counteracted the role of circ_UBR4 depletion in ox-LDL-induced human VSMCs. (a and b) RT-qPCR and western blotting examined ROCK mRNA expression level and protein expression level in VSMCs transfected with si-NC, si-circ_UBR4, si-circ_UBR4 along with pcDNA plasmid (pcDNA), si-circ_UBR4 along with pcDNA-ROCK1 plasmid (ROCK1). (c–h) VSMCs were co-treated with above transfections for 24 h and ox-LDL (100 μg/mL) treatment for another 24 h. (c) FCM analyzed cell numbers in G0/G1, S, and G2/M phases. (d) Colony formation assay determined number of colonies. (e) CCK-8 assay monitored OD value at 570 nm. (f) Western blotting detected PCNA protein expression level. (g) Transwell assay evaluated number of migrated cells. (h) Western blotting detected MMP2 protein expression level. *P < 0.05.
3.4 miR-107 directly interacted with both circ_UBR4 and ROCK1 in human VSMCs
With the in silico data on starbase3.0 software (http://starbase/circRNA&miRNA/=UBR4/=miR-107, http://starbase/mRNA&miRNA/=ROCK1/=miR-107), miR-107 was complementary to both circ_UBR4 and ROCK1. According to the predicted miR-107-binding site in circ_UBR4-wt and ROCK1 3′UTR-wt (Figure 4a and g), circ_UBR4-mut and ROCK1 3′UTR-mut were constructed. Dual-luciferase reporter assay revealed that luciferase activities of vectors containing circ_UBR4-wt and ROCK1 3′UTR-wt were declined in VSMCs transfected with miR-107 compared to miR-NC transfection (Figure 4b and h). These data hinted a target relationship between miR-107 and circ_UBR4 or ROCK1. In terms of miR-107 expression, its level was upregulated by si-circ_UBR4 transfection and miR-107 mimic transfection (Figure 4c and i), whereas downregulated by anti-miR-107 transfection (Figure 4i). In AS samples, miR-107 expression was lower in human AS serum and VSMCs under ox-LDL stress (Figure 4d and f). Moreover, miR-107 expression was negatively correlated with circ_UBR4 in AS patients’ serum (Figure 4e). On the contrary, ROCK1 expression on both mRNA level and protein level was inversely regulated by miR-107, as described by increased ROCK1 level in anti-miR-107-transfected VSMCs and decreased ROCK1 level in miR-107-transfected VSMCs (Figure 4j and k). In AS patients, ROCK1 expression level was higher in the serum samples comparing to normal people (Figure 4l and o), and its mRNA expression was negatively correlated with miR-107 and positively correlated with circ_UBR4 (Figure 4m and n). In addition, ROCK1 expression was highly expressed in ox-LDL-induced human VSMCs (Figure 4p and q). Taken together, miR-107 was abnormally expressed in AS probably through directly interacting with circ_UBR4 and ROCK1.
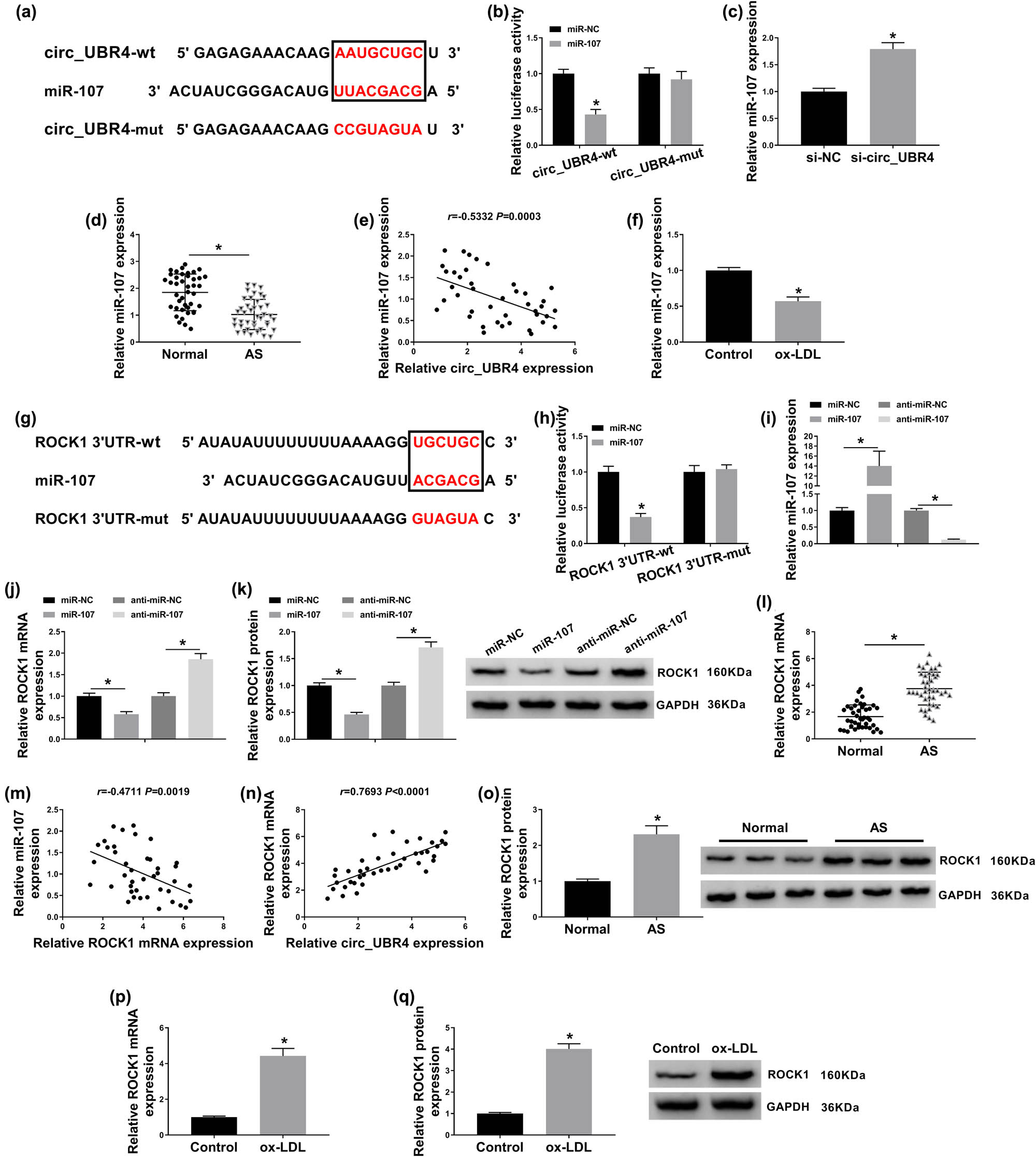
miRNA (miR)-107 directly interacted with circ_UBR4 and ROCK1. (a and g) Starbase3.0 software predicted the potential complementary sites of miR-107 on (a) wild type of circ_UBR4 (circ_UBR4-wt) and (g) wild type of ROCK1 in 3′untranslated region (ROCR1 3′UTR-wt). The red in black box was the binding sites, and the red out of black box was the mutated sites in the mutant type of circ_UBR4 (circ_UBR4-mut) and mutant type of ROCK1 (ROCK1 3′UTR-mut). (b and h) Dual-luciferase reporter assay measured luciferase activity of vector carrying (b) circ_UBR4-wt or circ_UBR4-mut, and (h) ROCR1-wt or ROCR1-mut in VSMCs transfected with miR-107 mimic (miR-107) or its negative control (miR-NC). (c and d) RT-qPCR detected miR-107 expression level in VSMCs transfected with si-NC or si-circ_UBR4, and serum samples in AS and normal groups. (e) Pearson’s correlation analysis determined the association between circ_UBR4 and miR-107 expression in human AS serum. (f) RT-qPCR detected miR-107 expression level in ox-LDL (100 μg/mL)-treated human VSMCs. (i–k) VSMCs were transfected with miR-NC, miR-107, miR-107-5p inhibitor (anti-miR-107), or the negative control (anti-miR-NC), and then (i and j) RT-qPCR measured miR-107 expression level and ROCK1 mRNA expression level, and (k) western blotting examined ROCK1 protein expression level. (l) RT-qPCR examined ROCK1 mRNA expression level in serum samples in AS and normal groups. (m and n) Pearson’s correlation analysis determined the association between ROCK1 mRNA and either miR-107 or circ_UBR4 expression in human AS serum. (o) Western blotting detected ROCK1 protein expression level in three groups of serum samples from AS patients and normal people. (p and q) RT-qPCR and western blotting measured ROCK1 expression in ox-LDL (100 μg/mL)-treated human VSMCs both on mRNA level and protein level. *P < 0.05.
3.5 Silencing miR-107 abolished the suppressive role of circ_UBR4 depletion in ox-LDL-induced human VSMCs through upregulating ROCK1
The effect of miR-107 on the role of circ_UBR4 depletion in VSMCs in AS using rescue experiment was further explored. Blocking circ_UBR4 resulted in higher level of miR-107 in ox-LDL-treated human VSMCs, and anti-miR-107 presence could downregulate this upregulation (Figure 5a). Moreover, anti-miR-107 transfection caused a series of promoting effects on excessive cell progressions of ox-LDL-induced VSMCs with circ_UBR4 knockdown, including cell entrance to S phase (Figure 5b), colony-formation ability (Figure 5c), cell viability (Figure 5d), migration ability (Figure 5f), as well as PCNA and MMP2 expression (Figure 5e and g). Furthermore, in ox-LDL-induced VSMCs transfected with si-circ_UBR4, anti-miR-107 co-transfection not only suppressed miR-107 expression (Figure 5a) but also enhanced ROCK1 expression (Figure 6a and b). These outcomes illuminated that silencing miR-107 accompanied with ROCK1 upregulation could abolish the role of circ_UBR4 knockdown in human VSMCs under ox-LDL condition, suggesting a circ_UBR4/miR-107/ROCK1 regulatory axis in VSMCs underlying AS pathogenesis.
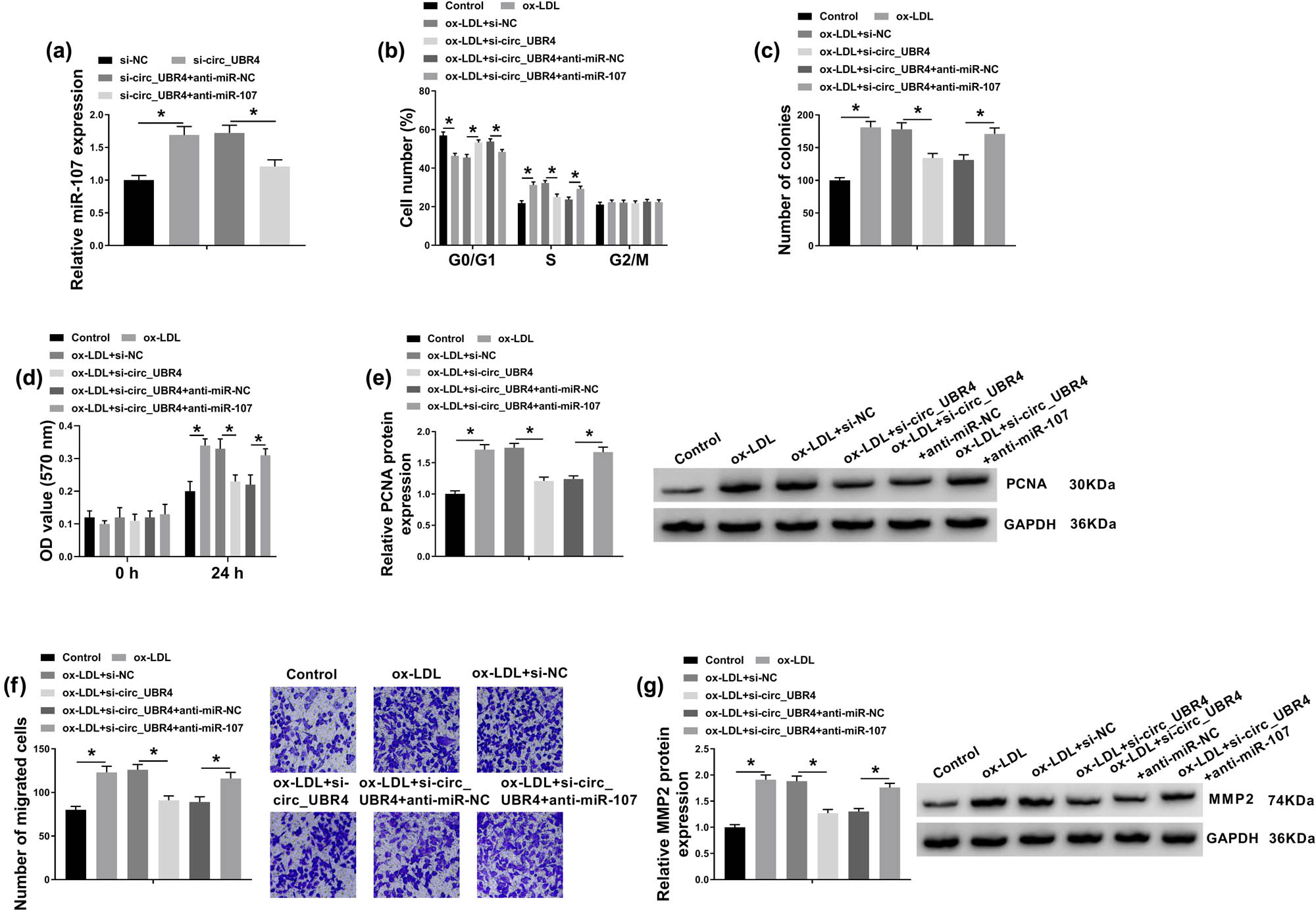
Silencing miR-107 abolished the role of circ_UBR4 depletion in ox-LDL-induced human VSMCs. (a) RT-qPCR examined miR-107 expression level in VSMCs transfected with si-NC alone, si-circ_UBR4 alone, or together with anti-NC or anti-miR-107. (b–g) VSMCs were co-treated with above transfections for 24 h and ox-LDL (100 μg/mL) treatment for another 24 h. (b) FCM analyzed cell numbers in G0/G1, S, and G2/M phases. (c) Colony formation assay determined number of colonies. (d) CCK-8 assay monitored OD value at 570 nm. (e) Western blotting detected PCNA protein expression level. (f) Transwell assay evaluated number of migrated cells. (g) Western blotting detected MMP2 protein expression level. *P < 0.05.
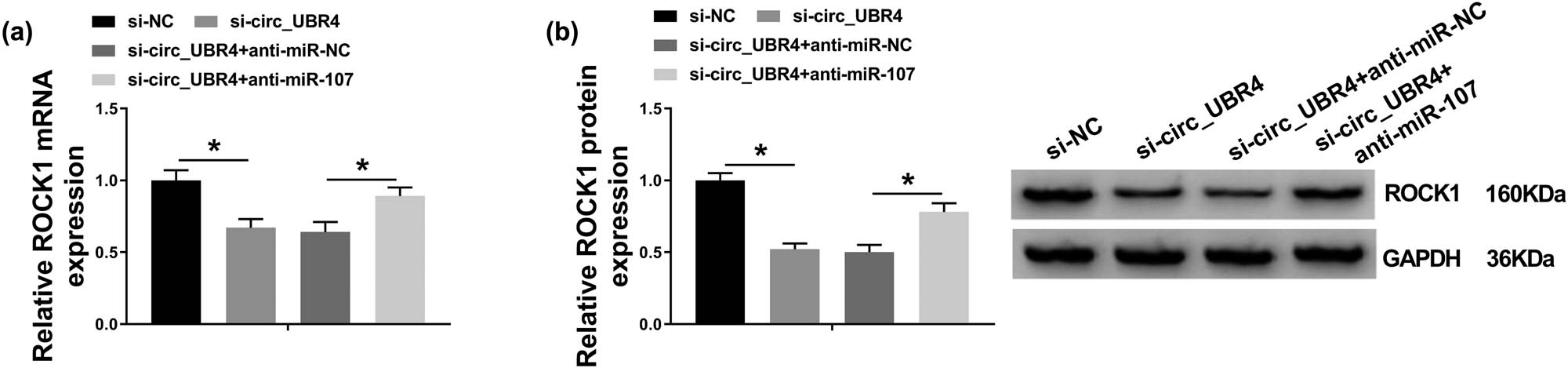
circ_UBR4 modulated ROCK1 expression in human VSMCs via miR-107 regulation. (a and b) RT-qPCR and western blotting measured ROCK1 expression levels in VSMCs transfected with si-NC alone, si-circ_UBR4 alone, or together with anti-NC or anti-miR-107. *P < 0.05.
4 Discussion
Recently, differential expression and bioinformatics analysis of circRNA in AS tissues and cell models in VSMCs had been disclosed, both under platelet-derived growth factor (PDGF-BB) stress and ox-LDL pressure [11,29,30]. There were several circRNAs involved in the dysfunction of VSMCs and the pathogenesis of AS. For example, circ_CHFR depletion weakened ox-LDL-induced promotion of cell growth, migration, and inflammation in VSMCs through targeting miR-370/FOXO1/Cyclin D1 and miR-214-3p/Wnt3/β-catenin pathways [11,31]. Besides, VSMCs phenotypic differentiation, proliferation, apoptosis, and migration were regulated by circ_SATB2 and circ_RUSC2 via sponging miRNAs [32,33]. However, comprehensive analysis of the role of circRNAs in the dysfunction of VSMCs remained largely unclear. Thus, we selected a novel circRNA circ_UBR4 that was abnormally expressed in ox-LDL-stimulated VSMCs to further explore its role in VSMCs during AS.
Here, circ_UBR4 was upregulated in human AS serum and ox-LDL-induced VSMCs, which was consistent with the initial study [11,34]. Besides, the expression of circ_UBR4 was resistant to RNase R and ActD treatment, suggesting a stable structure of circ_UBR4 and its host gene UBR4. As for the function, ox-LDL-induced VSMCs’ excessive proliferation, colony-formation ability, migration, and cell cycle progression were attenuated by blocking circ_UBR4. Moreover, the suppressive role of circUBR4 knockdown in ox-LDL-induced proliferation, migration, and invasion in VSMCs had been previously demonstrated [34,35]. Molecularly, expression of PCNA and MMP2 was also depressed by blocking circ_UBR4 [35]; furthermore, circ_UBR4 modulated ox-LDL-evoked dysfunction of VSMCs through serving as a ceRNA for miRNAs, such as miR-133a-3p and miR-370-3p. In this study, we discovered that miR-107 was sponged by circUBR4 as well. These findings suggested a circ_UBR4-miR-107/133a-3p/370-3p-ROCK1/pregnancy-associated plasma protein A/high mobility group box 1 ceRNA network in modulating VSMCs proliferation and migration under ox-LDL pressure in AS.
In this present study, miR-107 was downregulated in human AS serum, and this finding was in favor of the previous outcomes [36]. Here, we also demonstrated the low level of miR-107 in ox-LDL-induced VSMCs, and this downregulation was also discovered in PDGF-BB-induced pulmonary arterial smooth muscle cells (PASMCs) [37]. In terms of cellular functions, we observed that miR-107 downregulation was associated with promotion of proliferation, migration, and cell cycle progression in ox-LDL-challenged human VSMCs in spite of the anti-AS role of circ_UBR4 deficiency. Our findings supported the notion that miR-107 blockage exhibited facilitating effect on proliferation and migration of PASMCs stimulated by PDGF-BB [37]. Moreover, Shen et al. [36] pointed out the link of miR-107 silence in VSMCs proliferation and invasion but also stated its involvement in proliferation and invasion of vascular endothelial cells under stimuli of lipopolysaccharide, a potentially important stimulator and AS risk factor. Besides, Gao et al. [24] indicated a pharmacological effect of miR-107 in coronary AS model in mouse, and miR-107 upregulation reduced blood lipid level and atherosclerotic index by suppressing vascular endothelial cells apoptosis, inflammation, and endoplasmic reticulum stress. Taken together, miR-107 might represent a new approach for pharmacological treatment of diseases including AS.
Except for KRT1 and NOR1 [24,37], ROCK1 was confirmed as a novel target gene of miR-107. ROCK1 was upregulated in the serum of AS patients, which was positively correlated with circ_UBR4 expression and negatively correlated with miR-107 expression. ROCK1 upregulation mediated the promotion of ox-LDL-induced excessive proliferation, migration, and cell cycle progression in VSMCs despite circ_UBR4 silencing exerting the opposite effects. Chen et al. [19] also noted similar facilitating role of ROCK1 in proliferation and migration of VSMCs induced by high glucose, also an in vitro model of AS. It was reported that ROCK1/2 suppression mediated by the inhibitor Y-27632 or siRNAs transfection could reduce expression of MMP2, belonging to MMP family that played a crucial role in the pathogenesis of CVDs [38]. Similar to this announcement, circ_UBR4 depletion accompanied ROCK1 downregulation to depress MMP2 expression. The suppressive effect of miR-107 on MMP2 expression might be first documented in this study. By the way, starbase software also predicted a potential binding between miR-107 and ROCK2, and ROCK2 was also involved in Rho/ROCKs pathway in regulating AS endothelial function and vascular inflammation [17]. This hypothesis should be further validated.
In terms of PCNA expression regulation, we noticed a suppression of miR-107, as well as a promotion of ROCK1 on PCNA expression in ox-LDL-induced VSMCs. Furthermore, PCNA together with cyclin D1 could be significantly decreased with miR-107 ectopic expression in lung cancer cells and colorectal cancer cells [39,40], suggesting that miR-107 was an important regulator of cell proliferation. MMP2, MMP9, PCNA, ROCK1, and ROCK2 were increased in a mouse model of AS, and the receptor antagonist of IL-8, playing a key part in VSMCs proliferation and migration, could suppress the development of AS by inhibiting ROCKs [41]. Intriguingly, Lu et al. [42] expounded that knockdown of PCNA inhibited ROCK1 expression in colon cancer HCT116 cells, whereas ROCK1 knockdown did not affect PCNA expression. This finding hinted that ROCK1 silence could be downregulating PCNA expression in a content-dependent manner, such as in VSMCs with high ox-LDL level.
Collectively, we demonstrated that silencing of circ_UBR4 could attenuate ox-LDL-induced excessive proliferation, migration, and cell cycle progression in human VSMCs via circ_UBR4/miR-107/ROCK1 ceRNA regulatory axis. This result might suggest circ_UBR4, miR-107, and ROCK1 as potential targets in therapeutic schemes of AS.
-
Funding information: The authors state no funding involved.
-
Author contributions: Y.Z. designed the experiments. C.Z. along with Z.C. and M.W. carried them out. C.Z. and Z.C. prepared the manuscript with contributions from all co-authors. Y.Z. and Z.H. interpreted and discussed the results and conclusion. The authors applied the SDC approach for the sequence of authors.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Fasolo F, Di Gregoli K, Maegdefessel L, Johnson JL. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res. 2019;115(12):1732–56.10.1093/cvr/cvz203Search in Google Scholar PubMed PubMed Central
[2] Grover SP, Mackman N. Tissue factor in atherosclerosis and atherothrombosis. Atherosclerosis. 2020;307:80–6.10.1016/j.atherosclerosis.2020.06.003Search in Google Scholar PubMed
[3] Avgerinos NA, Neofytou P. Mathematical modelling and simulation of atherosclerosis formation and progress: a review. Ann Biomed Eng. 2019;47(8):1764–85.10.1007/s10439-019-02268-3Search in Google Scholar PubMed
[4] Zhao TC, Wang Z, Zhao TY. The important role of histone deacetylases in modulating vascular physiology and arteriosclerosis. Atherosclerosis. 2020;303:36–42.10.1016/j.atherosclerosis.2020.04.020Search in Google Scholar PubMed PubMed Central
[5] Li L, Li Y, Tang C. The role of microRNAs in the involvement of vascular smooth muscle cells in the development of atherosclerosis. Cell Biol Int. 2019;43(10):1102–12.10.1002/cbin.11164Search in Google Scholar PubMed
[6] Basatemur GL, Jorgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol. 2019;16(12):727–44.10.1038/s41569-019-0227-9Search in Google Scholar PubMed
[7] Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588–600.10.1002/jcp.27384Search in Google Scholar PubMed
[8] Bei Y, Yang T, Wang L, Holvoet P, Das S, Sluijter JPG, et al. Circular RNAs as potential theranostics in the cardiovascular system. Mol Ther Nucleic Acids. 2018;13:407–18.10.1016/j.omtn.2018.09.022Search in Google Scholar PubMed PubMed Central
[9] Cao Q, Guo Z, Du S, Ling H, Song C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020;255:117837.10.1016/j.lfs.2020.117837Search in Google Scholar PubMed
[10] Maguire EM, Xiao Q. Noncoding RNAs in vascular smooth muscle cell function and neointimal hyperplasia. FEBS J. 2020;287:5260–83.10.1111/febs.15357Search in Google Scholar PubMed
[11] Yang L, Yang F, Zhao H, Wang M, Zhang Y. Circular RNA circCHFR Facilitates the Proliferation and Migration of Vascular Smooth Muscle via miR-370/FOXO1/Cyclin D1 Pathway. Mol Ther Nucleic Acids. 2019;16:434–41.10.1016/j.omtn.2019.02.028Search in Google Scholar PubMed PubMed Central
[12] Trpkovic A, Resanovic I, Stanimirovic J, Radak D, Mousa SA, Cenic-Milosevic D, et al. Oxidized low-density lipoprotein as a biomarker of cardiovascular diseases. Crit Rev Clin Lab Sci. 2015;52(2):70–85.10.3109/10408363.2014.992063Search in Google Scholar PubMed
[13] Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–83.10.1038/nrg.2016.20Search in Google Scholar PubMed
[14] Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.10.1007/978-981-13-1426-1_6Search in Google Scholar PubMed
[15] Navarro E, Mallen A, Cruzado JM, Torras J, Hueso M. Unveiling ncRNA regulatory axes in atherosclerosis progression. Clin Transl Med. 2020;9(1):5.10.1186/s40169-020-0256-3Search in Google Scholar PubMed PubMed Central
[16] Liao JK, Seto M, Noma K. Rho kinase (ROCK) inhibitors. J Cardiovasc Pharmacol. 2007;50(1):17–24.10.1097/FJC.0b013e318070d1bdSearch in Google Scholar PubMed PubMed Central
[17] Sawada N, Liao JK. Rho/Rho-associated coiled-coil forming kinase pathway as therapeutic targets for statins in atherosclerosis. Antioxid Redox Signal. 2014;20(8):1251–67.10.1089/ars.2013.5524Search in Google Scholar PubMed PubMed Central
[18] Bu DX, Rai V, Shen X, Rosario R, Lu Y, D’Agati V, et al. Activation of the ROCK1 branch of the transforming growth factor-beta pathway contributes to RAGE-dependent acceleration of atherosclerosis in diabetic ApoE-null mice. Circ Res. 2010;106(6):1040–51.10.1161/CIRCRESAHA.109.201103Search in Google Scholar PubMed PubMed Central
[19] Chen M, Zhang Y, Li W, Yang J. MicroRNA-145 alleviates high glucose-induced proliferation and migration of vascular smooth muscle cells through targeting ROCK1. Biomed Pharmacother. 2018;99:81–6.10.1016/j.biopha.2018.01.014Search in Google Scholar PubMed
[20] Tang H, Zhu M, Zhao G, Fu W, Shi Z, Ding Y, et al. Loss of CLOCK under high glucose upregulates ROCK1-mediated endothelial to mesenchymal transition and aggravates plaque vulnerability. Atherosclerosis. 2018;275:58–67.10.1016/j.atherosclerosis.2018.05.046Search in Google Scholar PubMed
[21] Schiano C, Benincasa G, Franzese M, Della Mura N, Pane K, Salvatore M, et al. Epigenetic-sensitive pathways in personalized therapy of major cardiovascular diseases. Pharmacol Ther. 2020;210:107514.10.1016/j.pharmthera.2020.107514Search in Google Scholar PubMed
[22] Turco C, Donzelli S, Fontemaggi G. miR-15/107 microRNA gene group: characteristics and functional implications in cancer. Front Cell Dev Biol. 2020;8:427.10.3389/fcell.2020.00427Search in Google Scholar PubMed PubMed Central
[23] Jiang ZP, Zhou TB. Role of miR-107 and its signaling pathways in diseases. J Recept Signal Transduct Res. 2014;34(5):338–41.10.3109/10799893.2014.896383Search in Google Scholar PubMed
[24] Gao ZF, Ji XL, Gu J, Wang XY, Ding L, Zhang H. microRNA-107 protects against inflammation and endoplasmic reticulum stress of vascular endothelial cells via KRT1-dependent Notch signaling pathway in a mouse model of coronary atherosclerosis. J Cell Physiol. 2019;234(7):12029–41.10.1002/jcp.27864Search in Google Scholar PubMed
[25] Khor ES, Wong PF. Endothelial replicative senescence delayed by the inhibition of MTORC1 signaling involves MicroRNA-107. Int J Biochem Cell Biol. 2018;101:64–73.10.1016/j.biocel.2018.05.016Search in Google Scholar PubMed
[26] Zhuang Y, Peng H, Mastej V, Chen W. MicroRNA regulation of endothelial junction proteins and clinical consequence. Mediators Inflamm. 2016;2016:5078627.10.1155/2016/5078627Search in Google Scholar PubMed PubMed Central
[27] Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63.10.1093/nar/gkl151Search in Google Scholar PubMed PubMed Central
[28] Chen Z, Wang M, Huang K, He Q, Li H, Chang G. MicroRNA-125b affects vascular smooth muscle cell function by targeting serum response factor. Cell Physiol Biochem. 2018;46(4):1566–80.10.1159/000489203Search in Google Scholar PubMed
[29] Zhang F, Zhang R, Zhang X, Wu Y, Li X, Zhang S, et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging. 2018;10(9):2266–83.10.18632/aging.101541Search in Google Scholar PubMed PubMed Central
[30] Tian J, Fu Y, Li Q, Xu Y, Xi X, Zheng Y, et al. Differential expression and bioinformatics analysis of circRNA in PDGF-BB-induced vascular smooth muscle cells. Front Genet. 2020;11:530.10.3389/fgene.2020.00530Search in Google Scholar PubMed PubMed Central
[31] Zhuang JB, Li T, Hu XM, Ning M, Gao WQ, Lang YH, et al. circ_CHFR expedites cell growth, migration and inflammation in ox-LDL-treated human vascular smooth muscle cells via the miR-214-3p/Wnt3/beta-catenin pathway. Eur Rev Med Pharmacol Sci. 2020;24(6):3282–92.Search in Google Scholar
[32] Mao YY, Wang JQ, Guo XX, Bi Y, Wang CX. circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochem Biophys Res Commun. 2018;505(1):119–25.10.1016/j.bbrc.2018.09.069Search in Google Scholar PubMed
[33] Sun J, Zhang Z, Yang S. circ_RUSC2 upregulates the expression of miR-661 target gene SYK and regulates the function of vascular smooth muscle cells. Biochem Cell Biol. 2019;97(6):709–14.10.1139/bcb-2019-0031Search in Google Scholar PubMed
[34] Feng Z, Zhu Y, Zhang J, Yang W, Chen Z, Li B. Hsa-circ_0010283 regulates oxidized low-density lipoprotein-induced proliferation and migration of vascular smooth muscle cells by targeting the miR-133a-3p/pregnancy-associated plasma protein A axis. Circ J. 2020;84(12):2259–69.10.1253/circj.CJ-20-0345Search in Google Scholar PubMed
[35] Ding P, Ding Y, Tian Y, Lei X. Circular RNA circ_0010283 regulates the viability and migration of oxidized low density lipoprotein induced vascular smooth muscle cells via an miR3703p/HMGB1 axis in atherosclerosis. Int J Mol Med. 2020;46(4):1399–408.10.3892/ijmm.2020.4703Search in Google Scholar PubMed PubMed Central
[36] Shen L, Hu Y, Lou J, Yin S, Wang W, Wang Y, et al. circRNA0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting miR107. Mol Med Rep. 2019;19(5):3923–32.10.3892/mmr.2019.10011Search in Google Scholar
[37] Chen S, Yu C, Lu R, Song T, Wang X, Tang W, et al. miR-107 inhibits PDGF-BB-induced proliferation of human pulmonary arterial smooth muscle cells and migration through targeting NOR1. Int J Clin Exp Pathol. 2019;12(5):1599–608.Search in Google Scholar
[38] Cui Y, Sun YW, Lin HS, Su WM, Fang Y, Zhao Y, et al. Platelet-derived growth factor-BB induces matrix metalloproteinase-2 expression and rat vascular smooth muscle cell migration via ROCK and ERK/p38 MAPK pathways. Mol Cell Biochem. 2014;393(1–2):255–63.10.1007/s11010-014-2068-5Search in Google Scholar PubMed
[39] Cai P, Li J, Chen G, Peng B, Yu L, Zhao B, et al. MicroRNA-107 may regulate lung cancer cell proliferation and apoptosis by targeting TP53 regulated inhibitor of apoptosis 1. Oncol Lett. 2020;19(3):1958–66.10.3892/ol.2020.11248Search in Google Scholar PubMed PubMed Central
[40] Zhang W, Li W, Han X. Skullcapflavone I inhibits proliferation of human colorectal cancer cells via down-regulation of miR-107 expression. Neoplasma. 2019;66(2):203–10.10.4149/neo_2018_180427N279Search in Google Scholar PubMed
[41] Qin Y, Mao W, Pan L, Sun Y, Fan F, Zhao Y, et al. Inhibitory effect of recombinant human CXCL8(3-72)K11R/G31P on atherosclerotic plaques in a mouse model of atherosclerosis. Immunopharmacol Immunotoxicol. 2019;41(3):446–54.10.1080/08923973.2019.1616753Search in Google Scholar PubMed
[42] Lu YC, Wang P, Wang J, Ma R, Lee SC. PCNA and JNK1-Stat3 pathways respectively promotes and inhibits diabetes-associated centrosome amplification by targeting at the ROCK1/14-3-3sigma complex in human colon cancer HCT116 cells. J Cell Physiol. 2019;234(7):11511–23.10.1002/jcp.27813Search in Google Scholar PubMed PubMed Central
© 2021 Ying Zhang et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells
Articles in the same Issue
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells

