Abstract
The purpose of this study is to observe the potential value and underlying mechanism of the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)/miR-143 axis in ISR. A total of 150 participants were enrolled, including 100 patients (observation group) with coronary heart disease who underwent stent implantation in the Department of Cardiology of our hospital between January 2018 and January 2020, and 50 healthy people (control group) concurrently underwent a physical examination. Serum MALAT1 and miR-143 levels were detected by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Tumor necrosis factor-α (TNF-α; 10 ng/mL) induced human vascular smooth muscle cells (HVSMCs). MALAT1 increased while miR-143 decreased in the observation group versus the control group (P < 0.001). The non-restenosis group had significantly elevated MALAT1 expression while decreased miR-143 expression than the restenosis group (P < 0.001). The areas under the curves of the expression of MALAT1 and miR-143 in predicting restenosis were 0.917 and 0.881, respectively. Following si-MALAT1 transfection, HVSMC multiplication and invasiveness decreased significantly (P < 0.05). miR-143-inhibitor was observed to upregulate the luciferase activity of MALAT1-WT (P < 0.05). MALAT1 is highly expressed in patients with ISR while miR-143 is decreased, and the MALAT1/miR-143 axis is a potential pathway to modulate the multiplication and invasiveness of HVSMCs.
1 Introduction
Coronary heart disease (CHD), also known as coronary atherosclerotic heart disease, is a common cardiovascular disease and a global public health safety issue at present [1,2]. The latest statistics reveal a global incidence of CHD as high as 36.37% [3]. Coronary stent implantation, as an important intervention for heart rate rhythm, is mainly to improve the myocardial blood supply by reconstructing the narrow lumen to achieve the purpose of treatment [4]. However, in-stent restenosis (ISR) after stent implantation has always been one of the difficulties in coronary interventional therapy. Despite the significantly reduced incidence of ISR, thanks to the advent of drug-coated stents in recent years, ISR cannot be avoided entirely [5,6].
Long-chain noncoding RNAs (lncRNAs) are noncoding RNAs of over 200 nt in length, which cannot directly encode proteins [7,8]. Although the bulk of the human genome is translated into RNA molecules, only around 2% of these transcripts encode proteins. As a result, scientists have begun to investigate the vast universe of noncoding RNAs (ncRNAs) as critical physiological and pathological cell activity regulators. Numerous studies have shown the role of small noncoding RNAs (ncRNAs) such as microRNAs (miRNAs) in cardiovascular illness. Still, the role of long noncoding RNAs (lncRNAs) as important regulators in the genesis and progression of cardiac disorders remains unknown. Because all ncRNAs with a length of more than 200 nucleotides are arbitrarily classed as lncRNAs, this class of molecules is highly diverse, performing a range of biological activities and interacting with a variety of other RNAs and proteins [9]. Nonetheless, recent studies have found that lncRNAs are involved in many malignant tumors [10,11] and interfere with the occurrence and progression of a broad spectrum of cardiovascular diseases and nervous system diseases [12,13]. Importantly, lncRNAs have also been shown to play essential roles in embryonic development and cardiac lineage commitment (such as Fendrr and Braveheart [14]), and are important regulators of proper heart functionality. More details on lncRNAs in organogenesis and heart development have been extensively reviewed elsewhere [15]. Of these, metabolism associated lung adenocarcinoma transcript 1 (MALAT1), a highly conserved lncRNA located on chromosome 11q13.1 [14], is of predictive value in vascular stenosis of CHD patients [15], despite the fact that it cannot directly encode proteins as reported by the previous literature [16]. In addition, in recent years, lncRNAs have been revealed to be able to regulate microRNAs (miRs) by acting as competitive endogenous RNAs (ceRNAs) to participate in a variety of diseases [17]. For example, the MALAT1 has vital functions in nuclear speckles and the regulation of genes expressions. Recent research studies have proved that MALAT1 was overexpressed and oncogenic in some tumors, including lung, colorectal, bladder, and laryngeal cancers [18]. MALAT1/miR-15b-5p/MAPK1 axis mediates endothelial progenitor cell autophagy via the mTOR pathway and affects coronary atherosclerotic heart disease [19]. miR-31 promotes Th22 differentiation in patients with CHR by targeting Bach2 [20], and lncRNA ANRIL and miR-181b mediate the interaction of the NF-kappaB axis in inflammation-related coronary artery disease [21]. miR is a kind of 22 nt-length long noncoding short-chain RNAs in eukaryotes, which can bind to 3′UTRs and 5′UTRs of downstream target genes and coding area to regulate the expression of target genes and participate in many intercellular signal regulation [22]. miR-143, a classic miR, has been revealed to be pivotal in the occurrence and development of CHR via modulating KLLN and its mechanism [23]. Another literature study has pointed out that miR-143 is an independent, prognostic, and diagnostic marker of CHR [24]. In this study, we found that there were potential targeting sites between lncRNA MALAT1 and miR-143 through online prediction.
Hence, we speculate that the two may be potential predictors of ISR, with certain regulatory mechanisms between them.
2 Methods and materials
2.1 Clinical data
One hundred CHR patients (observation group, hereinafter referred to as OG) who received stent implantation in the Department of Cardiology of our hospital from January 2018 to January 2020 and 50 healthy controls (control group, hereinafter referred to as CG) without CHR who concurrently underwent physical examination were collected as research participants. Gender and age were similar in the two cohorts (P > 0.05).
2.1.1 Inclusion criteria for patients
Patients between 55 and 75 years of age who were tolerant to stent implantation were included. Patients in OG were all diagnosed with CHR by coronary angiography.
2.1.2 Exclusion criteria for patients
Exclusion criteria included severe liver and kidney dysfunction, other infectious or immune diseases, communication disorder, cognitive dysfunction, and malignant tumor(s).
-
Informed consent: Informed consent has been obtained from all individuals included in this study.
-
Ethical approval: The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance with the tenets of the Helsinki declaration, and has been approved by the author's institutional review board or equivalent committee.
2.2 Cell source
Human vascular smooth muscle cells (HVSMCs) were purchased from Beijing Bena Culture Collection (BNCC340087). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), in an incubator supplemented with 5% CO2 at 37°C and were passaged when the confluence reached 80%.
2.3 Cell transfection
HVSMCs were transfected with si-MALAT1 (inhibitory sequence), si-NC (independent sequence; control), miR-143-inhibitor (inhibitory sequence), and miR-NC (control) with the Lipofectamine 2000 transfection kit according to the kit’s instructions. All the sequences were designed and synthesized by Shanghai Sangon Biological Co., Ltd. Then, the cells were transfected by tumor necrosis factor-α (TNF-α; 10 ng/mL) for 24 h. The transfection effect was verified by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
2.4 qRT-PCR detection
The total RNA was separated from collected serum and cells with TRIzol reagent and subjected to purity, concentration, and integrity determination using an ultraviolet spectrophotometer and gel electrophoresis. cDNA was then obtained using the reverse transcription kit. Subsequently, PCR amplification of miRs was carried out with an all-in-one miRNA first-chain cDNA synthesis kit and all-in-one miR qPCR kit, while that of lncRNA and mRNA was done by SYBR Premix Ex Taq kits. RT-qPCR (ABI 7500, ABI) instrument was used for detection. Amplification conditions were as follows: predenaturation at 95°C for 1 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min; 95°C for 15 s, 60°C 1 min, and finally, 95°C for 15 s for melting curve analysis. The amplification system was configured according to the kit instructions. U6 and GAPDH were used as an internal reference for miR and lncRNA, respectively, and the target genes’ expression profiles were calculated using the 2−ΔΔCT method [25]. The amplified primer sequences are detailed in Table 1.
Primer sequences
| Gene name | Upstream sequence 5-3 | Downstream sequence 5-3 |
|---|---|---|
| MALAT1 | GCCTGGAAGCTGAAAAACGG | TGGAAAACGCCTCAATCCCA |
| miR-143 | AGTGCGTGTCGTGGTGT | GCCTGAGATGAAGCACGTG |
| GAPDH | CTGACTTCAACAGCGACACC | GTGGTCCAGGGGTCTTACTC |
| U6 | TGCGGGTGCTCGCTTCGGCAGC | CCAGTGCAGGGTCCGAGGT |
2.5 Determination of cell multiplication
This study analyzed cell viability using the cell counting kit 8 (CCK-8) (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Cells (4 × 103 cells per well) seeded into the wells of a 96-well plate were cultivated at 37°C with 5% CO2 in the air and then immersed in a CCK-8 reagent (10 µL) at 0, 24, 48, and 72 h after cultivation, respectively. After another 2 h of incubation at 37°C, the cells were processed with a microplate reader for optical density (450 nm) determination.
2.6 Detection of cell invasiveness
Transwell test was used to detect the changes in cell invasiveness capacity. After 48 h of transfection, 200 μL of cell suspension (1 × 108/L) was put into the upper chamber containing Matrigel (Corning, USA), and 500 μL of DMEM containing 10% FBS was added into the lower chamber for 24 h of culture in the cell incubator. Then, the chamber was subjected to 3 PBS rinses, 10 min of fixing with formaldehyde, and 15 min of dyeing with 0.1% crystal violet solution. After washing 3 times with PSB, the cells inside the ventricular membrane were gently wiped off with cotton swabs, and the chamber was taken off and laid on the slide to count the invasive cells in three visual fields under a microscope (200×).
2.7 Double luciferase reporter assay (DLRA)
miRs that could bound to MALAT1 were predicted using online prediction websites StarBase v3.0 and LncBase Predicted v.2, and it was found that there were targeted binding sites between MALAT1 and miR-143. Then, the wild-type (WT) and mutant (MUT) MALAT1 luciferase reporter plasmid vectors were constructed. HVSMCs were inoculated into a 24-well plate with 1 × 105 cells per well and co-transfected with either miR-NC or miR-143-inhibitor after 24 h. DLRA was conducted 24 h later to detect the activity of Renilla and firefly luciferases, with Renilla luciferase as an internal reference. The experiment was repeated 3 times.
2.8 Statistical analysis
In this study, GraphPad 8 software package was adopted to draw the required pictures, and the SPSS20.0 software package was used to conduct statistical analysis on the collected data. Inter-group comparisons employed independent sample t-test, multigroup comparisons adopted one-way ANOVA (expressed as F) and LSD/t posthoc test, and multitime point expression comparisons used repeated measures ANOVA (expressed as F) and Bonferroni posthoc test. The correlation analysis was performed by Pearson correlation coefficient, and the diagnostic value of lncRNA and miR in restenosis patients was visualized using receiver operating characteristic (ROC) curves. A P value less than 0.05 was considered statistically significant.
3 Results
3.1 Expression and predictive values of MALAT1 and miR-143 in patients with restenosis
To explore the expression of MALAT1 and miR-143 in patients with stenosis, we first analyzed their levels in CHD patients. It was found that MALAT1 increased while miR-143 decreased in OG compared with CG (Figure 1a and b, P < 0.001), indicating that the two interfered with the development of CHD. To further study the correlation of MALAT1 and miR-143 with restenosis in CHD patients, we divided 100 patients into the restenosis group (n = 27) and nonrestenosis group (n = 73) according to the stenosis status after treatment to further detect the expression of MALAT1 and miR-143 in patients. By testing, we found that MALAT1 was significantly increased, and miR-143 was decreased in the restenosis group compared with the nonrestenosis group (Figure 2a and b, P < 0.001). Then, we analyzed the predictive value of the two in patients with restenosis using the ROC curve. It showed that the areas under the curves (AUCs) of MALAT1 and miR-143 were 0.917 and 0.881, respectively (Figure 3 and Table 2), suggesting that the two were of high predictive value in patients with restenosis.

Expression of MALAT1 and miR-143 in CHD patients. (a) QRT-PCR was used to detect the expression level of MALAT1 in the serum of healthy people and CHD patients (n = 50). (b) QRT-PCR was used to detect the expression level of miR-143 in the serum of healthy people and CHD patients (n = 50). ***P < 0.001.
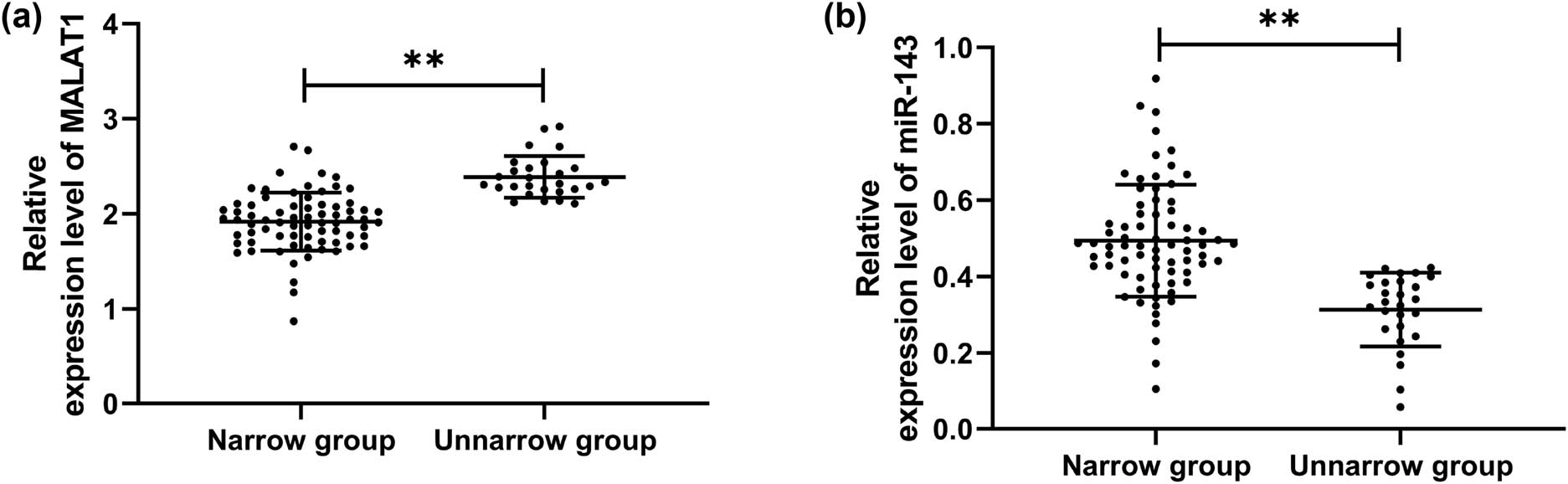
Expression of MALAT1 and miR-143 in CHD patients with restenosis. (a) Expression of MALAT1 in CHD patients with restenosis by qRT-PCR (restenosis group = 27, nonrestenosis group = 73). (b) Expression of miR-143 in CHD patients with restenosis by qRT-PCR (restenosis group = 27, nonrestenosis group = 73). **P < 0.01.

Predictive value of MALAT1 and miR-143 in CHD patients with restenosis. (a) ROC curve analysis of the AUC of MALAT1 in predicting restenosis in patients with CHD. (b) ROC curve analysis of the AUC of miR-143 in predicting restenosis in patients with CHD.
ROC parameters
| Variables | AUC | 95 CI% | Specificity (%) | Sensitivity | Youden index (%) | Cutoff value |
|---|---|---|---|---|---|---|
| MALAT1 | 0.917 | 0.864–0.989 | 79.45 | 100.00% | 79.45 | >2.108 |
| miR-143 | 0.881 | 0.818–0.945 | 72.60 | 100.00 | 72.60 | <0.426 |
Note: AUC: area under the curve, 95 CI%: 95% confidence interval.
3.2 Downregulation of MALAT1 suppresses the multiplication and invasiveness of HVSMCs
Through the above, we confirmed the clinical value of MALAT1 and miR-143 in patients with restenosis. However, we are not clear about the related mechanism, so we treated HVSMCs with TNF-α for exploration. Through qRT-PCR detection, we found that MALAT1 elevated significantly in HVSMCs after intervention (Figure 4a, P < 0.01). Subsequently, si-MALAT1 was transfected into the intervened HVSMCs and remarkably reduced MALAT1 was determined (Figure 4b, P < 0.01). In addition, compared with HVSMCs treated with TNF-α after transfection of si-NC, the multiplication and invasiveness of HVSMCs treated with TNF-α after transfection of si-MALAT1 reduced evidently, as demonstrated by CCK-8 and Transwell assays (Figures 4c and d, P < 0.05).
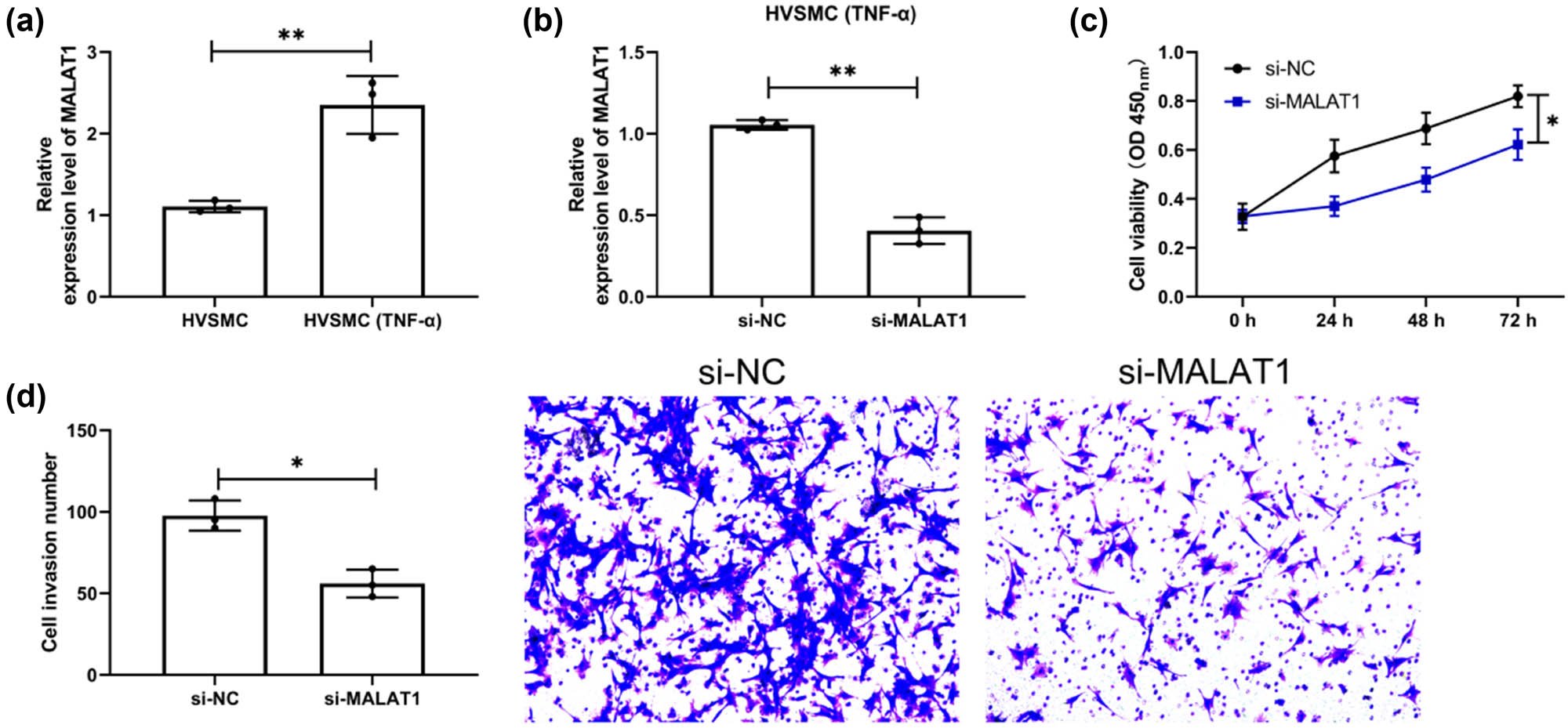
Downregulation of MALAT1 inhibits the multiplication and invasiveness of HVSMCs. (a) QRT-PCR was used to detect the expression level of MALAT1 in the cells after TNF-α-induced HVSMCs. (n = 3). (b) QRT-PCR was used to detect the expression level of MALAT1 in HVSMCs cells induced by TNF-α after si-MALAT1 transfection (n = 3). (c) Changes in multiplication ability of si-MALAT1-transfected HVSMCs after TNF-α induction by CCK-8 assay (n = 3). (d) Changes in invasiveness ability of si-MALAT1-transfected HVSMCs after TNF-α induction by Transwell assay (n = 3). *P < 0.05 and **P < 0.01.
3.3 MALAT1 has a targeted binding relationship with miR-143
CeRNA is an important way for lncRNA to participate in various functions of the body. To further determine the correlation between MALAT1 and miR-143, we conducted the DLRA. The experimental results showed that the miR-143-inhibitor could upregulate the luciferase activity of MALAT1-WT (Figure 5a, P < 0.05). Furthermore, qRT-PCR showed that miR-143 in cells transfected with si-MALAT1 was significantly increased but that in cells co-transfected with miR-143-inhibitor and si-MALAT1 was reversed (Figure 5b, P < 0.05). Moreover, an inverse association was determined between MALAT1 and miR-143 in CHD patients (Figure 5c). These experimental results indicate that MALAT1 has a targeted binding relationship with miR-143.
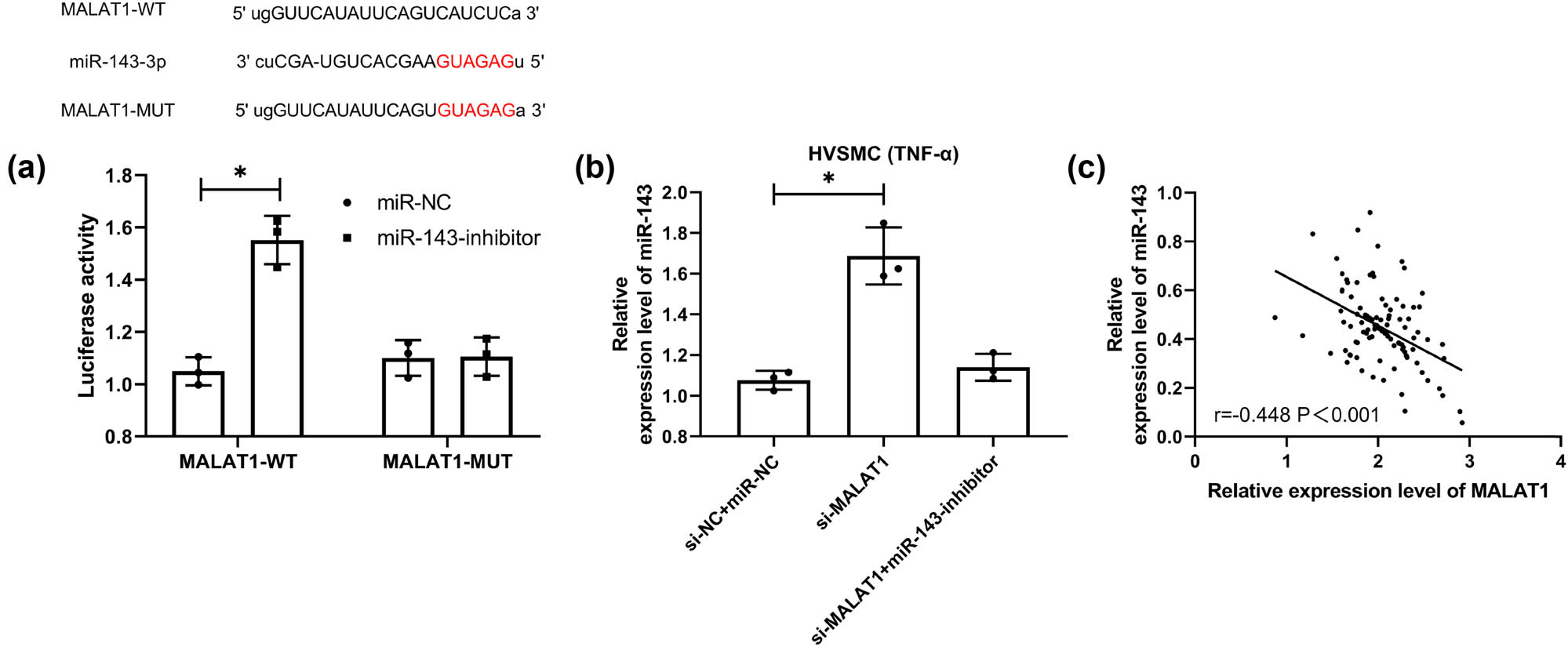
Targeted binding relationship between MALAT1 and miR-143. (a) The online prediction website verifies the target prediction of MALAT1 and miR-143 and the dual-luciferase report (n = 3). (b) qRT-PCR was used to detect the expression level of miR-143 in co-transfected si-MALAT1 cells (n = 3). (c) Correlation analysis of MALAT1 and miR-143 in CHD patients by Pearson correlation coefficient (n = 50). *P < 0.05.
3.4 Downregulation of miR-143 reverses the inhibitory action of si-MALAT1 on multiplication and invasiveness of HVSMCs
At the end of the study, the relationship between MALAT1 and miR-143 was further verified. The changes of multiplication and invasiveness of TNF-α-treated HVSMCs were analyzed by the co-transfection experiment. CCK-8 and Transwell experiments demonstrated that, after co-transfection of miR-143-inhibitor and si-MALAT1, the inhibitory action of si-MALAT1 on HVSMC multiplication and invasiveness was reversed (Figure 6a and b, P < 0.05). Therefore, the MALAT1/miR-143 axis can regulate the multiplication and invasiveness of HVSMCs and may be a potential therapeutic target.

Downregulation of miR-143 reverses the inhibition of si-MALAT1 on the multiplication and invasiveness of HVSMCs. (a) Changes of cell multiplication ability after co-transfection by CCK-8 assay (n = 3). (b) Changes in cell invasiveness ability after co-transfection by Transwell assay (n = 3). *P < 0.05.
4 Discussion
Recent years have witnessed the great progress made in the treatment of CHD, a commonly seen cardiovascular disease in clinical practice [26]. Currently, coronary stent implantation is the mainstay of treatment for CHR [27]. However, some patients still suffer from vascular restenosis after treatment, which is one of the causes of death in patients [28]. In this study, we found that MALAT1 and miR-143 were differentially expressed in CHD patients with restenosis, and MALAT1 can participate in the multiplication and invasiveness of HVSMCs via modulating miR-143.
lncRNAs have been reported to play an important regulatory role in many diseases, especially in metabolic diseases and cardiovascular diseases. For example, the research by Liu et al. proposed that insulin resistance of type 2 diabetes can be effectively reduced by the lncRNA MALAT1/miRNA-382-3p/resistin axis [29]. In addition, lncRNA CAIF is shown to suppress autophagy and alleviate myocardial infarction by blocking p53-mediated cardiac troponin transcription [30], and lncRNA H19 can reduce myocardial injury and maladaptive cardiac remodeling caused by myocardial infarction by regulating KDM3A [31]. MALAT1, an early discovered lncRNA, is found to be highly expressed in many cancers such as lung cancer [32], gastric carcinoma [33], and breast cancer [34]. CeRNA is currently one of the important ways for lncRNA to participate in the life process [35]. lncRNAs can also participate in the occurrence of acute myocardial infarction by regulating the downstream binding of miRs. For example, MALAT1 knockout can alleviate acute myocardial infarction through the miR-320/Pten axis [36], and lncRNA Gas5 targets the miR-525-5p/CALM2 axis to regulate myocardial infarction [37]. Earlier, a literature study has reported that miR-143 is involved in the development of CHD [38]. In this paper, through the database prediction analysis, we found the binding target sites between miR-143 and MALAT1, indicating a regulatory relationship between the two.
We first analyzed the expression of miR-143 and MALAT1 in CHD patients and found decreased expression of miR-143 and increased expression of MALAT1. Further, we detected miR-143 and MALAT1 in patients with restenosis and observed that MALAT1 was increased while miR-143 was decreased. Similarly, early studies found that MALAT1 was increased in patients with ISR, which verified the consistency of MALAT1 expression [14]. Furthermore, we detected the expression of miR-143. It was found for the first time that miR-143 was decreased in patients with ISR; moreover, ROC curve analysis showed that miR-143 had clinical value in predicting ISR. The above results suggest that miR-143 has a high clinical value for ISR in CHD patients but the underlying mechanism needs further verification.
The primary mechanism of ISR is an aggregation of platelets, leukocytes, and macrophages, which results in the migration and proliferation of medial smooth muscle cells [39,40]. Recently, researchers discovered that around one-third of ISR events were caused by in-stent neoatherosclerosis (ISNA), a mechanism that is clearly distinct from the traditional process [41]. However, there is a common phenomenon that occurs in each of these mechanisms: ISR and ISNA inflammation. Mir-143/145 has been demonstrated to be abundantly and selectively expressed in vascular smooth muscle cells (VSMCs) and to have a critical role in the proliferation and migration of VSMCs and the function of arteries, in earlier research [42,43]. Additional research established a direct link between miR-143/145 and cardiovascular illness in humans and experimental animal models [44,45]. miR-143/145 was demonstrated to be downregulated during carotid artery neointimal development in rats. Researchers discovered a substantial reduction in miR-143/145 expression in animal models of damaged or atherosclerotic arteries, including ApoE mutant mice’s atherosclerotic aortas, mouse ligation-induced carotid artery, and rat carotid balloon-injured carotid artery [44]. These data imply that miR-143/145 plays a role in developing atherosclerosis and ISR induced by VSMC migration and proliferation. Additionally, a few recent studies discovered that miR-143/145 might play some inflammatory disorders [46]. From the above experiments, we can see that the relative expression of MALAT1 and miR-143 was opposite in CHD patients with ISR, so we speculated that there might be a targeted regulation relationship between the two. It was identified by DLRA that MALAT1-WT fluorescence activity could be upregulated by miR-143-inhibit, and correlation analysis revealed an inverse connection between MALAT1 and miR-143 in CHD patients, which suggested a targeted regulatory relationship between them. Then, we found through experiments that the multiplication and invasiveness of TNF-α interfered HVSMCs was significantly inhibited after MALAT1 knockdown. However, after the co-transfection of the miR-143-inhibitor and si-MALAT1 into HVSMCs, the multiplication and invasiveness of cells were not significantly different from those transfected with miR-NC + si-NC. This suggests that the MALAT1/miR-143 axis may be a potential pathway for restenosis.
Experiments have determined the clinical value and relevant mechanism of the MALAT1/miR-143 axis in CHD patients with ISR. However, this study still shows margins of improvement. First, an in-depth investigation of the MALAT1/miR-143 axis mechanism in CHD patients with ISR is warranted. Second, more experiments are needed to verify the results since we only tested the impacts of the MALAT1/miR-143 axis on HVSMC multiplication and invasiveness. Therefore, we hope that we will refine our experimental design to improve our research conclusions in future research.
To sum up, MALAT1 is highly expressed in CHD patients with ISR while miR-143 expression is decreased. Besides, the MALAT1/miR-143 axis can be used as a potential pathway to modulate the multiplication and invasiveness of HVSMCs.
There are several limitations to this study. We used a limited number of cell lines as well as we did not evaluate the effect on the primary cells. However, some restrictions should garner more attention. For instance, the mTOR signaling pathway was not the only one active in CAD blood samples, as seen by the dot plot of the KEGG pathway. Although most of the genes involved in the mTOR signaling pathway were discovered in CAD, the other two pathways require more investigation. Additional in vivo investigations are required.
The more vigorous study needs to include numerous cells lines and human primary cell lines. The area of lncRNAs as possible therapeutic targets is still in its infancy. It is not implausible that lncRNAs may emerge as useful new tools for the treatment of a variety of illnesses, including CVDs, in the near future, which is worthy of future investigation. The discovery of lncRNAs revolutionized our knowledge of disease-regulating circuits and accelerated the development of technological breakthroughs in molecular interrogation methods. As additional lncRNAs were discovered to be associated with cardiovascular physiology, critical concerns remained unresolved before we could fully exploit the therapeutic potential of these new biological modulators. On the other hand, the mechanism by which lncRNAs are targeted to specific genomic loci remains unknown. Indeed, the temporal-spatial specificity of lncRNA effects is particularly perplexing in light of the fact that a substantial number appear to interact with highly promiscuous variables, such as PRC2 (polycomb repressive complex 2). The biochemical and structural reason for retaining specific lncRNAs that mimic mRNAs in the nucleus is also unknown. Finally, a critical bottom-line issue is: Can we utilize lncRNA’s diagnostic or therapeutic potential to enhance human health? Although this has been a difficult aim to achieve, promising translational initiatives are underway.
-
Funding information: The authors state no funding involved.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Khamis RY, Ammari T, Mikhail GW. Gender differences in coronary heart disease. Heart. 2016;102:1142–9.10.1136/heartjnl-2014-306463Suche in Google Scholar PubMed
[2] Wirtz PH, von Kanel R. Psychological stress, inflammation, and coronary heart disease. Curr Cardiol Rep. 2017;19:111.10.1007/s11886-017-0919-xSuche in Google Scholar PubMed
[3] Lao XQ, Liu X, Deng HB, Chan TC, Ho KF, Wang F, et al. Sleep quality, sleep duration, and the risk of coronary heart disease: a prospective cohort study with 60,586 adults. J Clin Sleep Med. 2018;14:109–17.10.5664/jcsm.6894Suche in Google Scholar PubMed PubMed Central
[4] Sukovatykh BS, Sukovatykh MB, Polianskii DV. Effect of the type of a coronary stent on dynamics of quality of life in patients with ischaemic heart disease. Angiol Sosud Khir. 2020;26:43–8.10.33529/ANGIO2020426Suche in Google Scholar PubMed
[5] Byrne RA, Joner M, Kastrati A. Stent thrombosis and restenosis: what have we learned and where are we going? The Andreas Gruntzig Lecture ESC 2014. Eur Heart J. 2015;36:3320–31.10.1093/eurheartj/ehv511Suche in Google Scholar PubMed PubMed Central
[6] Nicolais C, Lakhter V, Virk HUH, Sardar P, Bavishi C, O’Murchu B, et al. Therapeutic options for in-stent restenosis. Curr Cardiol Rep. 2018;20:7.10.1007/s11886-018-0952-4Suche in Google Scholar PubMed
[7] Bhan A, Soleimani M, Mandal SS. Long non-coding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81.10.1158/0008-5472.CAN-16-2634Suche in Google Scholar PubMed PubMed Central
[8] Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y, et al. lncRNA-ATB: an indispensable cancer-related long non-coding RNA. Cell Prolif. 2017;50:e12381.10.1111/cpr.12381Suche in Google Scholar PubMed PubMed Central
[9] Hobuß L, Bär C, Thum T. Long non-coding RNAs: at the heart of cardiac dysfunction? Front Physiol. 2019;10:30.10.3389/fphys.2019.00030Suche in Google Scholar PubMed PubMed Central
[10] Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: a new regulator in cancer signaling. Cancer Lett. 2018;419:27–9.10.1016/j.canlet.2018.01.008Suche in Google Scholar PubMed
[11] Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, et al. lncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–33.10.1016/j.cca.2018.07.004Suche in Google Scholar PubMed
[12] Liao J, Wang J, Liu Y, Li J, Duan L. Transcriptome sequencing of lncRNA, miRNA, mRNA and interaction network constructing in coronary heart disease. BMC Med Genomics. 2019;12:124.10.1186/s12920-019-0570-zSuche in Google Scholar PubMed PubMed Central
[13] Sun Y, Lin J, Huang S, Xu X, Cai Y, Yang L, et al. Preliminary verification of lncRNA ENST00000609755.1 potential ceRNA regulatory network in coronary heart disease. Int J Cardiol. 2021;328:165–75.10.1016/j.ijcard.2020.11.064Suche in Google Scholar PubMed
[14] Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–68.10.2147/CMAR.S169406Suche in Google Scholar PubMed PubMed Central
[15] Devaux Y. Braveheart, a long non-coding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–83; Devaux Y, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, et al. Long non-coding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12(7):415–25.10.1016/j.cell.2013.01.003Suche in Google Scholar PubMed PubMed Central
[16] Qiu S, Sun J. lncRNA-MALAT1 expression in patients with coronary atherosclerosis and its predictive value for in-stent restenosis. Exp Ther Med. 2020;20:129.10.3892/etm.2020.9258Suche in Google Scholar PubMed PubMed Central
[17] Zhang X, Hamblin MH, Yin KJ. The long non-coding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14.10.1080/15476286.2017.1358347Suche in Google Scholar PubMed PubMed Central
[18] Zhu J, Zhang X, Gao W, Hu H, Wang X, Hao D. lncRNA/circRNAmiRNAmRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20:3160–74.10.3892/mmr.2019.10569Suche in Google Scholar
[19] Zhu Y, Yang T, Duan J, Mu N, Zhang T. MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging (Albany NY). 2019;11:1089–109.10.18632/aging.101766Suche in Google Scholar PubMed PubMed Central
[20] Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel non-coding RNA, and thymosin β 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41.10.1038/sj.onc.1206928Suche in Google Scholar PubMed
[21] Huang R, Chen X, Long Y, Chen R. MiR-31 promotes Th22 differentiation through targeting Bach2 in coronary heart disease. Biosci Rep. 2019;39:BSR20190986.10.1042/BSR20190986Suche in Google Scholar PubMed PubMed Central
[22] Guo F, Tang C, Li Y, Liu Y, Lv P, Wang W, et al. The interplay of lncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-kappaB signalling pathway. J Cell Mol Med. 2018;22:5062–75.10.1111/jcmm.13790Suche in Google Scholar PubMed PubMed Central
[23] Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–7.10.1016/j.jaci.2017.08.034Suche in Google Scholar PubMed PubMed Central
[24] Liu H, Xiong W, Liu F, Lin F, He J, Liu C, et al. Significant role and mechanism of microRNA-143-3p/KLLN axis in the development of coronary heart disease. Am J Transl Res. 2019;11:3610–9.Suche in Google Scholar
[25] Lin DC, Lin JB, Chen Z, Chen R, Wan CY, Lin SW, et al. Independent and combined effects of environmental factors and miR-126, miR-143, and miR-145 on the risk of coronary heart disease. J Geriatr Cardiol. 2017;14:688–95.Suche in Google Scholar
[26] Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.10.1006/meth.2001.1262Suche in Google Scholar PubMed
[27] Malakar AK, Choudhury D, Halder B, Paul P, Uddin A, Chakraborty S. A review on coronary artery disease, its risk factors, and therapeutics. J Cell Physiol. 2019;234:16812–23.10.1002/jcp.28350Suche in Google Scholar PubMed
[28] Cheng HM, Chiou LJ, Chen TC, Sung SH, Chen CH, Lang HC. Real-world cost-effectiveness of drug-eluting stents vs. bare-metal stents for coronary heart disease-a five-year follow-up study. Health Policy. 2019;123:229–34.10.1016/j.healthpol.2018.11.010Suche in Google Scholar PubMed
[29] Kokkinidis DG, Waldo SW, Armstrong EJ. Treatment of coronary artery in-stent restenosis. Expert Rev Cardiovasc Ther. 2017;15:191–202.10.1080/14779072.2017.1284588Suche in Google Scholar PubMed
[30] Liu SX, Zheng F, Xie KL, Xie MR, Jiang LJ, Cai Y. Exercise reduces insulin resistance in type 2 diabetes mellitus via mediating the lncRNA MALAT1/MicroRNA-382-3p/resistin axis. Mol Ther Nucleic Acids. 2019;18:34–44.10.1016/j.omtn.2019.08.002Suche in Google Scholar PubMed PubMed Central
[31] Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, et al. lncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29.10.1038/s41467-017-02280-ySuche in Google Scholar PubMed PubMed Central
[32] Zhang BF, Jiang H, Chen J, Hu Q, Yang S, Liu XP, et al. lncRNA H19 ameliorates myocardial infarction-induced myocardial injury and maladaptive cardiac remodelling by regulating KDM3A. J Cell Mol Med. 2020;24:1099–115.10.1111/jcmm.14846Suche in Google Scholar PubMed PubMed Central
[33] Feng C, Zhao Y, Li Y, Zhang T, Ma Y, Liu Y. lncRNA MALAT1 promotes lung cancer proliferation and gefitinib resistance by acting as a miR-200a sponge. Arch Bronconeumol (Engl Ed). 2019;55:627–33.10.1016/j.arbr.2019.03.018Suche in Google Scholar
[34] YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long non-coding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174.10.1186/s12943-017-0743-3Suche in Google Scholar PubMed PubMed Central
[35] Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long non-coding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–15.10.1038/s41588-018-0252-3Suche in Google Scholar PubMed PubMed Central
[36] Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–8.10.1136/jmedgenet-2015-103334Suche in Google Scholar PubMed
[37] Hu H, Wu J, Li D, Zhou J, Yu H, Ma L. Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed Pharmacother. 2018;106:738–46.10.1016/j.biopha.2018.06.122Suche in Google Scholar PubMed
[38] Zhang Y, Hou YM, Gao F, Xiao JW, Li CC, Tang Y. lncRNA GAS5 regulates myocardial infarction by targeting the miR-525-5p/CALM2 axis. J Cell Biochem. 2019;120:18678–88.10.1002/jcb.29156Suche in Google Scholar PubMed
[39] Minha S, Pichard AD, Waksman R. In-stent restenosis of drug-eluting stents. Future Cardiol. 2013;9(5):721–31.10.2217/fca.13.45Suche in Google Scholar PubMed
[40] Zhou H, Zhang S, Sun X, Yang D, Zhuang X, Guo Y, et al. Lipid management for coronary heart disease patients: an appraisal of updated international guidelines applying Appraisal of Guidelines for Research and Evaluation II-clinical practice guideline appraisal for lipid management in coronary heart disease. J Thorac Dis. 2019;11(8):3534–46.10.21037/jtd.2019.07.71Suche in Google Scholar PubMed PubMed Central
[41] Park SJ, Kang SJ, Virmani R, Nakano M, Ueda Y. In-stent neoatherosclerosis: a final common pathway of late stent failure. J Am Coll Cardiol. 2012;59(23):2051–7.10.1016/j.jacc.2011.10.909Suche in Google Scholar PubMed
[42] Kohlstedt K, Trouvain C, Boettger T, Shi L, Fisslthaler B, Fleming I. AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145. Circ Res. 2013;112(8):1150–8.10.1161/CIRCRESAHA.113.301282Suche in Google Scholar PubMed
[43] Norata GD, Pinna C, Zappella F, Elia L, Sala A, Condorelli G, et al. MicroRNA 143–145 deficiency impairs vascular function. Int J Immunopathol Pharmacol. 2012;25(2):467–74.10.1177/039463201202500216Suche in Google Scholar PubMed
[44] Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100(11):1579–88.10.1161/CIRCRESAHA.106.141986Suche in Google Scholar PubMed
[45] Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–10.10.1038/nature08195Suche in Google Scholar PubMed PubMed Central
[46] Pekow JR, Dougherty U, Mustafi R, Zhu H, Kocherginsky M, Rubin DT, et al. miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflamm Bowel Dis. 2012;18(1):94–100.10.1002/ibd.21742Suche in Google Scholar PubMed PubMed Central
© 2021 Chen Cao et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells
Artikel in diesem Heft
- Biomedical Sciences
- Research progress on the mechanism of orexin in pain regulation in different brain regions
- Adriamycin-resistant cells are significantly less fit than adriamycin-sensitive cells in cervical cancer
- Exogenous spermidine affects polyamine metabolism in the mouse hypothalamus
- Iris metastasis of diffuse large B-cell lymphoma misdiagnosed as primary angle-closure glaucoma: A case report and review of the literature
- LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression
- Two new inflammatory markers related to the CURB-65 score for disease severity in patients with community-acquired pneumonia: The hypersensitive C-reactive protein to albumin ratio and fibrinogen to albumin ratio
- Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis
- Silencing XIST mitigated lipopolysaccharide (LPS)-induced inflammatory injury in human lung fibroblast WI-38 cells through modulating miR-30b-5p/CCL16 axis and TLR4/NF-κB signaling pathway
- Protocatechuic acid attenuates cerebral aneurysm formation and progression by inhibiting TNF-alpha/Nrf-2/NF-kB-mediated inflammatory mechanisms in experimental rats
- ABCB1 polymorphism in clopidogrel-treated Montenegrin patients
- Metabolic profiling of fatty acids in Tripterygium wilfordii multiglucoside- and triptolide-induced liver-injured rats
- miR-338-3p inhibits cell growth, invasion, and EMT process in neuroblastoma through targeting MMP-2
- Verification of neuroprotective effects of alpha-lipoic acid on chronic neuropathic pain in a chronic constriction injury rat model
- Circ_WWC3 overexpression decelerates the progression of osteosarcoma by regulating miR-421/PDE7B axis
- Knockdown of TUG1 rescues cardiomyocyte hypertrophy through targeting the miR-497/MEF2C axis
- MiR-146b-3p protects against AR42J cell injury in cerulein-induced acute pancreatitis model through targeting Anxa2
- miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer
- Diabetes and COVID-19
- Discovery of novel potential KIT inhibitors for the treatment of gastrointestinal stromal tumor
- TEAD4 is a novel independent predictor of prognosis in LGG patients with IDH mutation
- circTLK1 facilitates the proliferation and metastasis of renal cell carcinoma by regulating miR-495-3p/CBL axis
- microRNA-9-5p protects liver sinusoidal endothelial cell against oxygen glucose deprivation/reperfusion injury
- Long noncoding RNA TUG1 regulates degradation of chondrocyte extracellular matrix via miR-320c/MMP-13 axis in osteoarthritis
- Duodenal adenocarcinoma with skin metastasis as initial manifestation: A case report
- Effects of Loofah cylindrica extract on learning and memory ability, brain tissue morphology, and immune function of aging mice
- Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways
- Blocking circ_UBR4 suppressed proliferation, migration, and cell cycle progression of human vascular smooth muscle cells in atherosclerosis
- Gene therapy in PIDs, hemoglobin, ocular, neurodegenerative, and hemophilia B disorders
- Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis
- Vitamin D deficiency and cardiovascular risk in type 2 diabetes population
- Circ_0013359 facilitates the tumorigenicity of melanoma by regulating miR-136-5p/RAB9A axis
- Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression
- lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis
- Identification of circRNA circ-CSPP1 as a potent driver of colorectal cancer by directly targeting the miR-431/LASP1 axis
- Hyperhomocysteinemia exacerbates ischemia-reperfusion injury-induced acute kidney injury by mediating oxidative stress, DNA damage, JNK pathway, and apoptosis
- Potential prognostic markers and significant lncRNA–mRNA co-expression pairs in laryngeal squamous cell carcinoma
- Gamma irradiation-mediated inactivation of enveloped viruses with conservation of genome integrity: Potential application for SARS-CoV-2 inactivated vaccine development
- ADHFE1 is a correlative factor of patient survival in cancer
- The association of transcription factor Prox1 with the proliferation, migration, and invasion of lung cancer
- Is there a relationship between the prevalence of autoimmune thyroid disease and diabetic kidney disease?
- Immunoregulatory function of Dictyophora echinovolvata spore polysaccharides in immunocompromised mice induced by cyclophosphamide
- T cell epitopes of SARS-CoV-2 spike protein and conserved surface protein of Plasmodium malariae share sequence homology
- Anti-obesity effect and mechanism of mesenchymal stem cells influence on obese mice
- Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis
- Glucocorticoids protect HEI-OC1 cells from tunicamycin-induced cell damage via inhibiting endoplasmic reticulum stress
- Prognostic value of the neutrophil-to-lymphocyte ratio in acute organophosphorus pesticide poisoning
- Gastroprotective effects of diosgenin against HCl/ethanol-induced gastric mucosal injury through suppression of NF-κβ and myeloperoxidase activities
- Silencing of LINC00707 suppresses cell proliferation, migration, and invasion of osteosarcoma cells by modulating miR-338-3p/AHSA1 axis
- Successful extracorporeal membrane oxygenation resuscitation of patient with cardiogenic shock induced by phaeochromocytoma crisis mimicking hyperthyroidism: A case report
- Effects of miR-185-5p on replication of hepatitis C virus
- Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis
- Primary localized cutaneous nodular amyloidosis presenting as lymphatic malformation: A case report
- Multimodal magnetic resonance imaging analysis in the characteristics of Wilson’s disease: A case report and literature review
- Therapeutic potential of anticoagulant therapy in association with cytokine storm inhibition in severe cases of COVID-19: A case report
- Neoadjuvant immunotherapy combined with chemotherapy for locally advanced squamous cell lung carcinoma: A case report and literature review
- Rufinamide (RUF) suppresses inflammation and maintains the integrity of the blood–brain barrier during kainic acid-induced brain damage
- Inhibition of ADAM10 ameliorates doxorubicin-induced cardiac remodeling by suppressing N-cadherin cleavage
- Invasive ductal carcinoma and small lymphocytic lymphoma/chronic lymphocytic leukemia manifesting as a collision breast tumor: A case report and literature review
- Clonal diversity of the B cell receptor repertoire in patients with coronary in-stent restenosis and type 2 diabetes
- CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression
- WDR74 promotes proliferation and metastasis in colorectal cancer cells through regulating the Wnt/β-catenin signaling pathway
- Down-regulation of IGHG1 enhances Protoporphyrin IX accumulation and inhibits hemin biosynthesis in colorectal cancer by suppressing the MEK-FECH axis
- Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis
- Scutellarin-induced A549 cell apoptosis depends on activation of the transforming growth factor-β1/smad2/ROS/caspase-3 pathway
- lncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis
- A two-microRNA signature predicts the progression of male thyroid cancer
- Isolation of microglia from retinas of chronic ocular hypertensive rats
- Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study
- Calcineurin Aβ gene knockdown inhibits transient outward potassium current ion channel remodeling in hypertrophic ventricular myocyte
- Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma
- Clinical significance of activated Wnt/β-catenin signaling in apoptosis inhibition of oral cancer
- circ_CHFR regulates ox-LDL-mediated cell proliferation, apoptosis, and EndoMT by miR-15a-5p/EGFR axis in human brain microvessel endothelial cells
- Resveratrol pretreatment mitigates LPS-induced acute lung injury by regulating conventional dendritic cells’ maturation and function
- Ubiquitin-conjugating enzyme E2T promotes tumor stem cell characteristics and migration of cervical cancer cells by regulating the GRP78/FAK pathway
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso
- Protective effect of Lactobacillus-containing probiotics on intestinal mucosa of rats experiencing traumatic hemorrhagic shock
- Glucocorticoids induce osteonecrosis of the femoral head through the Hippo signaling pathway
- Endothelial cell-derived SSAO can increase MLC20 phosphorylation in VSMCs
- Downregulation of STOX1 is a novel prognostic biomarker for glioma patients
- miR-378a-3p regulates glioma cell chemosensitivity to cisplatin through IGF1R
- The molecular mechanisms underlying arecoline-induced cardiac fibrosis in rats
- TGF-β1-overexpressing mesenchymal stem cells reciprocally regulate Th17/Treg cells by regulating the expression of IFN-γ
- The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia
- Red blood cell distribution width-standard deviation but not red blood cell distribution width-coefficient of variation as a potential index for the diagnosis of iron-deficiency anemia in mid-pregnancy women
- Small cell neuroendocrine carcinoma expressing alpha fetoprotein in the endometrium
- Superoxide dismutase and the sigma1 receptor as key elements of the antioxidant system in human gastrointestinal tract cancers
- Molecular characterization and phylogenetic studies of Echinococcus granulosus and Taenia multiceps coenurus cysts in slaughtered sheep in Saudi Arabia
- ITGB5 mutation discovered in a Chinese family with blepharophimosis-ptosis-epicanthus inversus syndrome
- ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting
- Facilitation of mouse skin-derived precursor growth and yield by optimizing plating density
- 3,4-Dihydroxyphenylethanol ameliorates lipopolysaccharide-induced septic cardiac injury in a murine model
- Downregulation of PITX2 inhibits the proliferation and migration of liver cancer cells and induces cell apoptosis
- Expression of CDK9 in endometrial cancer tissues and its effect on the proliferation of HEC-1B
- Novel predictor of the occurrence of DKA in T1DM patients without infection: A combination of neutrophil/lymphocyte ratio and white blood cells
- Investigation of molecular regulation mechanism under the pathophysiology of subarachnoid hemorrhage
- miR-25-3p protects renal tubular epithelial cells from apoptosis induced by renal IRI by targeting DKK3
- Bioengineering and Biotechnology
- Green fabrication of Co and Co3O4 nanoparticles and their biomedical applications: A review
- Agriculture
- Effects of inorganic and organic selenium sources on the growth performance of broilers in China: A meta-analysis
- Crop-livestock integration practices, knowledge, and attitudes among smallholder farmers: Hedging against climate change-induced shocks in semi-arid Zimbabwe
- Food Science and Nutrition
- Effect of food processing on the antioxidant activity of flavones from Polygonatum odoratum (Mill.) Druce
- Vitamin D and iodine status was associated with the risk and complication of type 2 diabetes mellitus in China
- Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”
- Comparison between voltammetric detection methods for abalone-flavoring liquid
- Composition of low-molecular-weight glutenin subunits in common wheat (Triticum aestivum L.) and their effects on the rheological properties of dough
- Application of culture, PCR, and PacBio sequencing for determination of microbial composition of milk from subclinical mastitis dairy cows of smallholder farms
- Investigating microplastics and potentially toxic elements contamination in canned Tuna, Salmon, and Sardine fishes from Taif markets, KSA
- From bench to bar side: Evaluating the red wine storage lesion
- Establishment of an iodine model for prevention of iodine-excess-induced thyroid dysfunction in pregnant women
- Plant Sciences
- Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose
- Comparative analysis of the SPL gene family in five Rosaceae species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia
- Identification of leaf rust resistance genes Lr34 and Lr46 in common wheat (Triticum aestivum L. ssp. aestivum) lines of different origin using multiplex PCR
- Investigation of bioactivities of Taxus chinensis, Taxus cuspidata, and Taxus × media by gas chromatography-mass spectrometry
- Morphological structures and histochemistry of roots and shoots in Myricaria laxiflora (Tamaricaceae)
- Transcriptome analysis of resistance mechanism to potato wart disease
- In silico analysis of glycosyltransferase 2 family genes in duckweed (Spirodela polyrhiza) and its role in salt stress tolerance
- Comparative study on growth traits and ions regulation of zoysiagrasses under varied salinity treatments
- Role of MS1 homolog Ntms1 gene of tobacco infertility
- Biological characteristics and fungicide sensitivity of Pyricularia variabilis
- In silico/computational analysis of mevalonate pyrophosphate decarboxylase gene families in Campanulids
- Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa)
- How photoautotrophy, photomixotrophy, and ventilation affect the stomata and fluorescence emission of pistachios rootstock?
- Apoplastic histochemical features of plant root walls that may facilitate ion uptake and retention
- Ecology and Environmental Sciences
- The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield
- Domestication of wild animals may provide a springboard for rapid variation of coronavirus
- Response of benthic invertebrate assemblages to seasonal and habitat condition in the Wewe River, Ashanti region (Ghana)
- Molecular record for the first authentication of Isaria cicadae from Vietnam
- Twig biomass allocation of Betula platyphylla in different habitats in Wudalianchi Volcano, northeast China
- Animal Sciences
- Supplementation of probiotics in water beneficial growth performance, carcass traits, immune function, and antioxidant capacity in broiler chickens
- Predators of the giant pine scale, Marchalina hellenica (Gennadius 1883; Hemiptera: Marchalinidae), out of its natural range in Turkey
- Honey in wound healing: An updated review
- NONMMUT140591.1 may serve as a ceRNA to regulate Gata5 in UT-B knockout-induced cardiac conduction block
- Radiotherapy for the treatment of pulmonary hydatidosis in sheep
- Retraction
- Retraction of “Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating microRNA-34a-5p/NOTCH1 signaling pathway”
- Special Issue on Reuse of Agro-Industrial By-Products
- An effect of positional isomerism of benzoic acid derivatives on antibacterial activity against Escherichia coli
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part II
- Relationship of Gensini score with retinal vessel diameter and arteriovenous ratio in senile CHD
- Effects of different enantiomers of amlodipine on lipid profiles and vasomotor factors in atherosclerotic rabbits
- Establishment of the New Zealand white rabbit animal model of fatty keratopathy associated with corneal neovascularization
- lncRNA MALAT1/miR-143 axis is a potential biomarker for in-stent restenosis and is involved in the multiplication of vascular smooth muscle cells

