Abstract
C13H17NO3, monoclinic, Cc (no. 9), a = 11.6502(8) Å, b = 13.8752(8) Å, c = 7.9644(5) Å, β = 106.051(2)°, V = 1237.25(14) Å3, Z = 4, R gt (F) = 0.0427, wR ref(F 2) = 0.1023, T = 170 K.
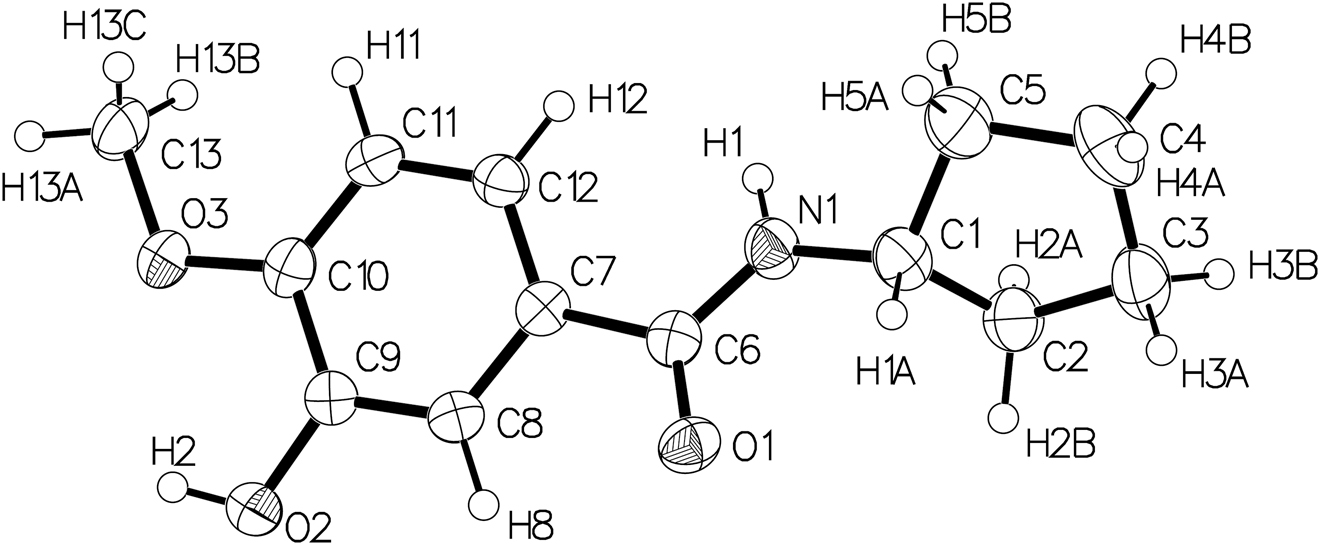
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colorless block |
| Size: | 0.28 × 0.15 × 0.12 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.09 mm−1 |
| Diffractometer, scan mode: | D8 VENTURE, φ and ω |
| θ max, completeness: | 26.4°, 98% |
| N(hkl)measured, N(hkl)unique, R int: | 5924, 2141, 0.045 |
| Criterion for I obs, N(hkl)gt: | I obs > 2 σ(I obs), 1747 |
| N(param)refined: | 157 |
| Programs: | Bruker [1], Olex2 [2], SHELX [3, 4] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| C1 | 0.7121 (3) | 0.5779 (2) | 0.8326 (4) | 0.0368 (9) |
| H1A | 0.708766 | 0.551263 | 0.947884 | 0.044* |
| C2 | 0.8348 (3) | 0.5581 (3) | 0.8098 (5) | 0.0426 (9) |
| H2A | 0.836531 | 0.570422 | 0.688133 | 0.051* |
| H2B | 0.859790 | 0.490786 | 0.841437 | 0.051* |
| C3 | 0.9146 (4) | 0.6297 (3) | 0.9369 (5) | 0.0503 (11) |
| H3A | 0.942702 | 0.601663 | 1.055539 | 0.060* |
| H3B | 0.984837 | 0.647802 | 0.896745 | 0.060* |
| C4 | 0.8349 (4) | 0.7172 (3) | 0.9366 (6) | 0.0570 (12) |
| H4A | 0.839466 | 0.736907 | 1.057803 | 0.068* |
| H4B | 0.860384 | 0.772170 | 0.876208 | 0.068* |
| C5 | 0.7080 (4) | 0.6871 (3) | 0.8407 (6) | 0.0526 (11) |
| H5A | 0.651019 | 0.708429 | 0.905182 | 0.063* |
| H5B | 0.683580 | 0.715006 | 0.721744 | 0.063* |
| C6 | 0.5604 (3) | 0.4543 (2) | 0.7103 (4) | 0.0301 (8) |
| C7 | 0.4537 (3) | 0.4270 (2) | 0.5658 (4) | 0.0286 (8) |
| C8 | 0.4214 (3) | 0.3299 (2) | 0.5526 (4) | 0.0337 (8) |
| H8 | 0.468580 | 0.284715 | 0.631995 | 0.040* |
| C9 | 0.3223 (3) | 0.2986 (2) | 0.4263 (5) | 0.0339 (8) |
| C10 | 0.2517 (3) | 0.3656 (2) | 0.3104 (4) | 0.0319 (8) |
| C11 | 0.2831 (3) | 0.4617 (2) | 0.3241 (4) | 0.0321 (8) |
| H11 | 0.235661 | 0.507130 | 0.245544 | 0.039* |
| C12 | 0.3835 (3) | 0.4925 (2) | 0.4517 (4) | 0.0311 (8) |
| H12 | 0.404043 | 0.558965 | 0.460637 | 0.037* |
| C13 | 0.0842 (3) | 0.3921 (3) | 0.0620 (5) | 0.0464 (10) |
| H13A | 0.018946 | 0.356452 | −0.017842 | 0.070* |
| H13B | 0.134297 | 0.421863 | −0.004277 | 0.070* |
| H13C | 0.050918 | 0.442449 | 0.121220 | 0.070* |
| N1 | 0.6129 (3) | 0.5381 (2) | 0.6962 (3) | 0.0357 (7) |
| H1 | 0.586283 | 0.571266 | 0.599029 | 0.043* |
| O1 | 0.5970 (2) | 0.40001 (16) | 0.8395 (3) | 0.0357 (6) |
| O2 | 0.2991 (2) | 0.20202 (16) | 0.4160 (4) | 0.0509 (8) |
| H2 | 0.225880 | 0.192950 | 0.370114 | 0.076* |
| O3 | 0.1550 (2) | 0.32725 (16) | 0.1895 (3) | 0.0384 (6) |
Source of material
A mixture of 10 mmol 3-hydroxy-4-methoxybenzoic acid, 10.4 mmol cyclopentylamine, and 20 mmol N,N-diisopropylethylamine (DIEA) were dissolved in 20 mL DMF. Then, 10.4 mmol of 2-(7-Azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) was added to the above solution. After 5 h stirring, the reaction was complete as monitored by Thin Layer Chromatography. Hundred milliliters of brine was added to the mixture, and the product was extracted with ethyl acetate (3 times with 500 mL). This product was washed with 5% citric acid, 5% NaHCO3, and H2O respectively. The mixture was dried over MgSO4, filtered, and concentrated under vacuum. The crystals were achieved through recrystallization in ethyl acetate, and dried at room temperature for seven days.
Experimental details
All H atoms bonded to their parent C atoms were treated as riding atoms in their geometrically idealized positions, with d(C–H) = 0.95 Å and U iso = 1.2 U eq.
Comment
Benzamide derivatives were studied to exhibit several biological properties, like antiviral [5], antipsychotic [6], anti-cancer [7], anti-oxidant [8], anti-inflammatory [9], anti-Alzheimer, anti-fatigue [10], and antidiabetic effects [11]. For example, several sulfamoylbenzamides and benzamide derivatives were verified to exhibit antiviral effects via significantly reducing intracellular HBV DNA in the cytoplasm [5, 12]. Some nitro benzamide analogs could significantly inhibit the expression of cyclooxygenase-2, interleukin-1β and tumor necrosis factor, and exhibited anti-inflammatory activities [13]. Additionally, some benzamide derivatives, like metoclopramide, exhibited anti-emetic properties through blocking dopaminergic and serotonergic receptors [6], and were also used for the treatment of gastroesophageal reflux disease in infants [14].
The molecular structure of the title compound is displayed in figure. All bond lengths and angles are normal, and agree with structural reports of 4-methoxybenzamide derivatives [15], [16], [17]. The methoxy-substituted benzene [C7–C12] forms a dihedral angle of 22.9° with the amide group [–N1–C6=O1–]. The cyclopentyl ring adopts a conformation between envelope and half-chair. The hydroxyl group donates its H atom to O1, which acts as acceptor in the strong intermolecular hydrogen bond (O2–H2···O1, 1.943(3) Å).
Funding source: National Natural Science Foundation of China http://dx.doi.org/10.13039/501100001809
Award Identifier / Grant number: 21902071
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: This work was financially supported by the National Natural Science Foundation of China (21902071).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Bruker. SMART APEX-II CCD; Bruker AXS Inc.: Madison, WI, USA, 2006.Search in Google Scholar
2. Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
3. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
4. Sheldrick, G. M. SHELXTL – integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8; https://doi.org/10.1107/s2053273314026370.Search in Google Scholar PubMed PubMed Central
5. Wu, S., Zhao, Q., Zhang, P., Kulp, J., Hu, L., Hwang, N., Zhang, J., Block, T. M., Xu, X., Du, Y., Chang, J., Guo, J. T. Discovery and mechanistic study of benzamide derivatives that modulate hepatitis B virus capsid assembly. J. Virol. 2017, 91, Art. no. e00519-17; https://doi.org/10.1128/JVI.00519-17.Search in Google Scholar PubMed PubMed Central
6. Oltmanns, D., Eisenhut, M., Mier, W., Haberkorn, U. Benzamides as melanotropic carriers for radioisotopes, metals, cytotoxic agents and as enzyme inhibitors. Curr. Med. Chem. 2009, 16, 2086–2094; https://doi.org/10.2174/092986709788612684.Search in Google Scholar PubMed
7. Mahdavi, M., Lavi, M. M., Yekta, R., Moosavi, M. A., Nobarani, M., Balalaei, S., Arami, S., Rashidi, M. R. Evaluation of the cytotoxic, apoptosis inducing activity and molecular docking of spiroquinazolinone benzamide derivatives in MCF-7 breast cancer cells. Chem. Biol. Interact. 2016, 260, 232–242; https://doi.org/10.1016/j.cbi.2016.10.004.Search in Google Scholar PubMed
8. Perin, N., Roškarić, P., Sović, I., Boček, I., Starčević, K., Hranjec, M., Vianello, R. Amino-substituted benzamide derivatives as promising antioxidant agents: a combined experimental and computational study. Chem. Res. Toxicol. 2018, 31, 974–984; https://doi.org/10.1021/acs.chemrestox.8b00175.Search in Google Scholar PubMed
9. Hussain, A. A., Abdulla, M. M., Amr, A. E.-G. E., Al-Omar, M. A., Shalaby, A. F. A. Anti-inflammatory activities of some newly synthesized pyridinyl- and indazolyl benzamide derivatives. Russ. J. Bioorg. Chem. 2015, 41, 87–96; https://doi.org/10.1134/s106816201405015x.Search in Google Scholar
10. Taj, M., Tirmizi, S., Raheel, A., Ali, H. M., Qureshi, S., Alshatir, H. Benzamide derivatives as potential candidates for anti-Alzheimer, anti-fatigue, anti-urease and anti-oxidant activity. J. Chil. Chem. Soc. 2017, 62, 3342–3344; https://doi.org/10.4067/s0717-97072017000100007.Search in Google Scholar
11. Singh, R., Lather, V., Pandita, D., Judge, V., Arumugam, K. N., Singh Grewal, A. Synthesis, docking and antidiabetic activity of some newer benzamide derivatives as potential glucokinase activators. Lett. Drug Des. Discov. 2017, 14, 540–553; https://doi.org/10.2174/1570180813666160819125342.Search in Google Scholar
12. Campagna, M. R., Liu, F., Mao, R., Mills, C., Cai, D., Guo, F., Zhao, X., Ye, H., Cuconati, A., Guo, H., Chang, J., Xu, X., Block, T. M., Guo, J.-T. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J. Virol. 2013, 87, 6931–6942; https://doi.org/10.1128/jvi.00582-13.Search in Google Scholar PubMed PubMed Central
13. Tumer, T. B., Onder, F. C., Ipek, H., Gungor, T., Savranoglu, S., Tok, T. T., Celik, A., Ay, M. Biological evaluation and molecular docking studies of nitro benzamide derivatives with respect to in vitro anti-inflammatory activity. Int. Immunopharm. 2017, 43, 129–139; https://doi.org/10.1016/j.intimp.2016.12.009.Search in Google Scholar PubMed
14. Hibbs, A. M., Lorch, S. A. Metoclopramide for the treatment of gastroesophageal reflux disease in infants: a systematic review. Pediatrics 2006, 118, 746–752; https://doi.org/10.1542/peds.2005-2664.Search in Google Scholar PubMed
15. Zhang, J., Zhao, X., Cai, C. The crystal structure of N-cyclohexyl-3-hydroxy-4-methoxybenzamide C14H19NO3. Z. Kristallogr. N. Cryst. Struct. 2022, 237, 543–545.10.1515/ncrs-2022-0066Search in Google Scholar
16. Kant, R., Sahi, S., Gupta, V. K., Kapoor, K., Paul, S. N-(4-Chlorophenyl)-4-methoxybenzamide. Acta Crystallogr. 2012, E68, o3077; https://doi.org/10.1107/s1600536812041384.Search in Google Scholar PubMed PubMed Central
17. Li, H. L., Zeng, H. S., Kang, S. S., Wang, H. B. 4-Methoxybenzamide oxime. Acta Crystallogr. 2007, E63, o4763; https://doi.org/10.1107/s1600536807059429.Search in Google Scholar
© 2022 Jingxiao Zhang et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of N-((3s,5s,7s)-adamantan-1-yl)-2-(3-benzoylphenyl)propanamide, C26H29NO2
- The crystal structure of bis(μ2-5-chloro-2-oxido-N-(1-oxidopropylidene)benzohydrazonato-κ5 N,O,O′:N′,O′′)-octakis(pyridine-κ1 N)trinickel(II) C60H56Cl2N12Ni3O6
- The crystal structure of 3-(4-chlorophenyl)-1,5-di-p-tolylpentane-1,5-dione, C25H23ClO2
- The crystal structure of 2,4,4-triphenyl-4H-benzo[b][1,4]oxaphosphinin-4-ium bromide – dichloromethane (1/1), C27H22BrCl2OP
- The crystal structure of 2-(3,6-di-tert-butyl-1,8-diiodo-9H-carbazol-9-yl)acetonitrile, C22H24I2N2
- Crystal structure of 3-phenylpropyl 2-(6-methoxynaphthalen-2-yl)propanoate, C23H24O3
- The crystal structure of (4-fluorophenyl)(5-(hydroxymethyl)furan-2-yl)methanol, C12H11FO3
- Crystal structure of the dihydrate of tetraethylammonium 1,3,5-thiadiazole-5-amido-2-carbamate, C11H27N5O4S
- Crystal structure of (Z)-4-[(p-tolylamino)(furan-2-yl)methylene]-3-phenyl-1-1-p-tolyl-1H-phenyl-1H-pyrazol-5(4H)-one, C28H23N3O2
- The crystal structure of (E)-3-(2-chlorophenyl)-1-ferrocenylprop-2-en-1-one, C19H15ClFeO
- The pseudosymmetric crystal structure of 3-((1R,2S)-1-methylpyrrolidin-1-ium-2-yl)pyridin-1-ium hexachloridostannate(IV), C10H16N2SnCl6
- Crystal structure of (2-(1-hydroxyheptyl)octahydro-8aH-chromene-5,8,8a-triol), C16H30O5
- The crystal structure of N-cyclohexyl-3-hydroxy-4-methoxybenzamide, C14H19NO3
- Crystal structure of 1-(4-hydroxybenzyl)-4-methoxy-9,10-dihydrophenanthrene-2,7-diol from Arundina graminifolia, C22H20O4
- The crystal structure of N-cyclopentyl-3-hydroxy-4-methoxybenzamide, C13H17NO3
- The crystal structure of 2,5,5-triphenyl-3,5-dihydro-4H-imidazol-4-one, C21H16N2O
- Crystal structure of 1H-1,2,3-Triazolo[4,5-b]-pyridin-4-ium nitrate, C5H5N5O3
- Crystal structure of (Z)-4-(((4-bromophenyl)amino)(furan-2-yl)methylene)-2,5-diphenyl-2,4-dihydro-3H-pyrazol-3-one, C26H18BrN3O2
- Crystal structure of 2-(4-methoxyphenyl)-3-methyl-1,8-naphthyridine, C16H14N2O
- The crystal structure of 3-([1,1′-biphenyl]-2-yl)-1,2-diphenylbenzo[b]phosphole-1-oxide, C32H23OP
- The crystal structure of ammonium (E)-4-((4-carboxyphenyl)diazenyl)benzoate, C14H13N3O4
- Crystal structure of bis(5-amino-1,2,4-triazol-4-ium-3-yl)methane sulfate, C5H10N8O4S
- The crystal structure of phenantroline-κ2 N,N′-bis(6-phenylpyridine-2-carboxylato-κ2 N,O)copper(II), C36H24N4O4Cu
- The crystal structure of tris(6-methylpyridin-2-yl)phosphine oxide, C18H18N3OP
- The crystal structure of N-(2′-hydroxymethyl-5′-phenyl-3′,4′-dihydro-[1,1′:3′,1″-terphenyl]- 1′(2′H)-yl)-P,P-diphenylphosphinic amide, C37H34NO2P
- Crystal structure of (E)-4-(6-(4-(2-(pyridin-4-yl)vinyl)phenoxy)pyrimidin-4-yl)morpholine, C21H20N4O2
- Crystal structure of 5-(adamantan-1-yl)-3-[(4-trifluoromethylanilino)methyl]-2,3-dihydro-1,3,4-oxadiazole-2-thione, C20H22F3N3OS
- Crystal structure of 2,2-dichloro-1-(4-chloro-1H-indol-1-yl)ethan-1-one, C10H6Cl3NO
- The crystal structure of 4-(((3-bromo-5-(trifluoromethyl)pyridin-2-yl)oxy)methyl)benzonitrile, C28H16Br2F6N4O2
- The crystal structure of 1H-benzimidazole-2-carboxamide, C8H7N3O
- The crystal structure of Histidinium hydrogensquarate, C10H11N3O6
- The crystal structure of 3-amino-5-carboxypyridin-1-ium iodide, C6H7IN2O2
- Crystal structure of (E)-amino(2-(3-ethoxy-4-hydroxybenzylidene)hydrazineyl)methaniminium nitrate hemihydrate C10H16N5O5.5
- Crystal structure of 1,2-bis(4,5-dinitro-1H-imidazol-1-yl)ethane, C8H6N8O8
- The crystal structure of diaqua-bis(pyrazolo[1,5-a]pyrimidine-3-carboxylato-κ2N,O)manganese(II), C14H12N6O6Mn
- The crystal structure of catena-poly[aqua-2,2′bipyridine-κ2N,N′-(μ2-5-ethoxyisophthalato-κ 4O,O′:Oʺ,O′ʺ)cadmium(II)] monohydrate, C20H20CdN2O7
- The crystal structure of (1S,3R)-1-(4-isopropylphenyl)-3-(methoxycarbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indol-2-iumchloride monohydrate, C22H27ClN2O3
- Crystal structure of 1-isopropyl-3-(prop-1-en-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, C11H15N5
- The crystal structure of (2,2′-bipyridine-κ2N,N′)- bis(6-phenylpyridine-2-carboxylate-κ2N,O)manganese(II)] monohydrate, C34H26N4O5Mn
- Crystal structure of the cocrystal 1,3,5,7-tetranitro-1,3,5,7-tetrazoctane ─ 2,3-dihydroindole (1/1), C12H17N9O8
- Crystal structure of 3-acetyl-6-hydroxy-2H-chromen-2-one monohydrate, C11H10O5
- Crystal structure of 6,9-diamino-2-ethoxyacridinium 3,5-dinitrobenozate — dimethylsulfoxide — water (1/1/1), C24H27N5O9S
- The crystal structure of 4,4′-bipyridinium bis-(2-hydroxy-3-methoxybenzoate), 2(C8H7.68O4)·C10H8.64N2
- Crystal structure of (Z)-4-(((4-fluorophenyl)amino)(furan-2-yl)methylene)-5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one
- The crystal structure of bis(4-chloro-2-(((2-chloroethyl)imino)methyl)phenolato-κ2N,O)-oxidovanadium(IV), C18H16Cl4N2O3V
- The crystal structure of 17-(bromoethynyl)-17-hydroxy-10, 13-dimethyl- 1,2,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-3H-cyclopenta[a]phenanthren-3-one, C21H27BrO2
- The crystal structure of 4-((6-fluoropyridin-2-yloxy)methyl)benzonitrile, C13H9FN2O
- Crystal structure of (Z)-2-(1-bromo-2-phenylvinyl)-5-ethyl-2-methyl-1,3-dioxane-5-carboxylic acid, C15H17Br1O4
- Crystal structure of catena-poly[tribenzyl-κ1C-(μ2-6-oxidopyridin-1-ium-3-carboxylato-κ2O:O’)tin(IV)-dichloromethane-methanol (1/1/1), C29H31Cl2NO4Sn
- Crystal structure of bis{2-(tert-butyl)-6-((E)-((4-((E)-1-(methoxyimino)ethyl)phenyl)imino)methyl)phenolato-κ2N,O}zinc(II), C40H46N4O4Zn
- Crystal structure of diaqua-bis(μ2-2-carboxy-3,4,5,6-tetrafluorobenzoato-κ2O:O′)-bis(phenanthroline-κ2N,N′)-bis(μ2-3,4,5,6-tetrafluorophthalato-κ3O:O,O′)dieuropium(III) – phenanthroline (1/2), C40H19EuF8N4O9
- The crystal structure of diaqua-bis(6-phenylpyridine-2-carboxylato-κ2N,O) manganese(II) — water — dimethylformamide (1/2/1), C27H31N3O9Mn
- The crystal structure of bis(pyrazolo[1,5-a]pyrimidine-3-carboxylato-κ2N,O)-copper(ii), C14H8N6O4Cu
- Crystal structure of poly[(μ2-1-(1-imidazolyl)-4-(imidazol-1-ylmethyl)benzene-κ2N:N′)-(μ3-pyridazine-4,5-dicarboxylate-κ3O:O′:N)]copper(II) hydrate, C19H16CuN6O5
- Crystal structure of acrinidinium tetrafluorohydrogenphthalate, C21H11F4NO4
- Crystal structure of 2-(1H-pyrazol-3-yl-κN)pyridine-κN-bis(2-(2,4-difluorophenyl)pyridinato-κ2C,N)iridium(III) sesquihydrate, C30H18F4IrN5·1.5[H2O]
- Crystal structure of 2-(2-hydroxy-5-nitrophenyl)-5-methyl-1,3-dioxane-5-carboxylic acid, C12H13N1O7
- The crystal structure of 1,2-bis(pyridinium-4-yl)ethane diperchlorate, C12H14N2·2ClO4 – a second polymorph
- The crystal structure of [(1,10-phenantroline-κ2N,N′)-bis(6-phenylpyridine-2-carboxylato-κ2N,O)manganese(II)] monohydrate, C36H26N4O5Mn
- Crystal structure of 1,2-bis(2,2,3,3,5,5,5-heptamethyl-1,1,4,4- tetrakis(trimethylsilyl)pentasilan-1-yl)ditellane, C38H114Si18Te2
- Crystal structure of 1,2-bis(2,4-dinitro-1H-imidazol-1-yl)ethane – dimethylformamide (1/1), C11H13N9O9
- Crystal structure of (Z)-3-((tert-butylamino) methylene)-2-(2-hydroxynaphthalen-1-yl) chroman-4-one, C24H23NO3
- Synthesis and crystal structure of (E)-1-(4-(((E)-3-(tert-butyl)-2-hydroxybenzylidene)amino)phenyl)ethan-1-one O-ethyl oxime, C21H26N2O2
- Crystal structure of the double salt bis(5-amino-1,2,4-triazol-4-ium-3-yl)methane hydrogen oxalate hemioxalate, C8H11N8O6
- Hydrothermal synthesis and crystal structure of catena-poly[diaqua-bis(μ2-4-[(4-pyridinylmethyl)amino]benzoato-κ2N:O)cobalt(II)]–1,2bi(4-pyridyl)ethene–water (1/1/1), C50H50N8O8Co
- Crystal structure of 3-(3-bromophenyl)-1′,3′-dimethyl-2′H,3H,4H-spiro[furo[3, 2-c]chromene-2,5′-pyrimidine]-2′,4,4′,6′(1′H,3′H) tetraone, C22H15BrN2O6
- The crystal structure of poly[aqua-(μ2-4,4′- bis(imidazolyl)biphenyl-κ2N:N′)-(μ2-3-nitrobenzene-1,2-dicarboxylato-κ2O:O′)]copper (II) hydrate, C26H21N5O8Cu
- The crystal structure of bis(4-(6-carboxy-8-ethyl-3-fluoro-5-oxo-5,8-dihydro-1,8-naphthyridin-2- yl)piperazin-1-ium) adipate tetrahydrate, C36H52F2N8O14
- Synthesis and crystal structure of poly[aqua(μ4-(1R,2S,4R)-4-hydroxy-1-((7-hydroxy-3-(4-hydroxy-3-sulfonatophenyl)-4-oxo-4H-chromen-8-yl)methyl)pyrrolidin-1-ium-2-carboxylate-κ4O:O′:O″:O‴)sodium(I)] monohydrate, C21H22NNaO12S
- Crystal structure of chlorido-(η6-toluene)(2,2′-bipyridine-κ2N,N′)ruthenium(II) hexafluorophosphate, C17H16ClN2RuPF6
- The crystal structure of (R)-6-hydroxy-8-methoxy-3-methylisochroman-1-one, C11H12O4
- Crystal structure of catena-poly[(5,5,7,12,12,14-hexamethyl -1,4,8,11-tetraazacyclotetradecane- κ4N,N′,Nʺ,N‴)nickel(II)-(μ2-perchlorato-κ2O:O′)] 3,5-dicarboxybenzoate – methanol (1/2), C27H49ClN4NiO12
- The crystal structure of 4-(chloromethyl)benzonitrile, C8H6ClN
- The crystal structure of dimethylammonium 8-[(7,9-dioxo-6,10-dioxaspiro[4.5]decan-8-ylidene)methyl]-9-oxo-6,10-dioxaspiro[4.5]dec-7-en-7-olate, C19H25NO8
- Crystal structure of (2R,3S,4S,5R,6S)-2-(acetoxymethyl)-6-((1-acetyl-5-bromo-4-chloro-1H-indol-3-yl)oxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate hemihydrate C24H25BrClNO11
- The crystal structure of the co-crystal tetrakis[2-(tris(4-methoxyphenyl)stannyl)ethyl]silane – tetrahydrofuran – toluene – tetrahydrofurane (1/1/1), C103H116O13SiSn4
- Crystal structure of methyl 3-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)propanoate, C16H13NO4
- Crystal structure of ethyl (Z)-3-amino-2-cyano-3-(2-oxo-2H-chromen-3-yl)acrylate, C15H12N2O4
- Crystal structure of methyl 2-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)acetate, C15H11NO4
- Crystal structure of catena-poly[diaqua-bis(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2N:N′)cobalt(II)] tetrafluoroterephthalate, C26H28N8O6F4Co
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of N-((3s,5s,7s)-adamantan-1-yl)-2-(3-benzoylphenyl)propanamide, C26H29NO2
- The crystal structure of bis(μ2-5-chloro-2-oxido-N-(1-oxidopropylidene)benzohydrazonato-κ5 N,O,O′:N′,O′′)-octakis(pyridine-κ1 N)trinickel(II) C60H56Cl2N12Ni3O6
- The crystal structure of 3-(4-chlorophenyl)-1,5-di-p-tolylpentane-1,5-dione, C25H23ClO2
- The crystal structure of 2,4,4-triphenyl-4H-benzo[b][1,4]oxaphosphinin-4-ium bromide – dichloromethane (1/1), C27H22BrCl2OP
- The crystal structure of 2-(3,6-di-tert-butyl-1,8-diiodo-9H-carbazol-9-yl)acetonitrile, C22H24I2N2
- Crystal structure of 3-phenylpropyl 2-(6-methoxynaphthalen-2-yl)propanoate, C23H24O3
- The crystal structure of (4-fluorophenyl)(5-(hydroxymethyl)furan-2-yl)methanol, C12H11FO3
- Crystal structure of the dihydrate of tetraethylammonium 1,3,5-thiadiazole-5-amido-2-carbamate, C11H27N5O4S
- Crystal structure of (Z)-4-[(p-tolylamino)(furan-2-yl)methylene]-3-phenyl-1-1-p-tolyl-1H-phenyl-1H-pyrazol-5(4H)-one, C28H23N3O2
- The crystal structure of (E)-3-(2-chlorophenyl)-1-ferrocenylprop-2-en-1-one, C19H15ClFeO
- The pseudosymmetric crystal structure of 3-((1R,2S)-1-methylpyrrolidin-1-ium-2-yl)pyridin-1-ium hexachloridostannate(IV), C10H16N2SnCl6
- Crystal structure of (2-(1-hydroxyheptyl)octahydro-8aH-chromene-5,8,8a-triol), C16H30O5
- The crystal structure of N-cyclohexyl-3-hydroxy-4-methoxybenzamide, C14H19NO3
- Crystal structure of 1-(4-hydroxybenzyl)-4-methoxy-9,10-dihydrophenanthrene-2,7-diol from Arundina graminifolia, C22H20O4
- The crystal structure of N-cyclopentyl-3-hydroxy-4-methoxybenzamide, C13H17NO3
- The crystal structure of 2,5,5-triphenyl-3,5-dihydro-4H-imidazol-4-one, C21H16N2O
- Crystal structure of 1H-1,2,3-Triazolo[4,5-b]-pyridin-4-ium nitrate, C5H5N5O3
- Crystal structure of (Z)-4-(((4-bromophenyl)amino)(furan-2-yl)methylene)-2,5-diphenyl-2,4-dihydro-3H-pyrazol-3-one, C26H18BrN3O2
- Crystal structure of 2-(4-methoxyphenyl)-3-methyl-1,8-naphthyridine, C16H14N2O
- The crystal structure of 3-([1,1′-biphenyl]-2-yl)-1,2-diphenylbenzo[b]phosphole-1-oxide, C32H23OP
- The crystal structure of ammonium (E)-4-((4-carboxyphenyl)diazenyl)benzoate, C14H13N3O4
- Crystal structure of bis(5-amino-1,2,4-triazol-4-ium-3-yl)methane sulfate, C5H10N8O4S
- The crystal structure of phenantroline-κ2 N,N′-bis(6-phenylpyridine-2-carboxylato-κ2 N,O)copper(II), C36H24N4O4Cu
- The crystal structure of tris(6-methylpyridin-2-yl)phosphine oxide, C18H18N3OP
- The crystal structure of N-(2′-hydroxymethyl-5′-phenyl-3′,4′-dihydro-[1,1′:3′,1″-terphenyl]- 1′(2′H)-yl)-P,P-diphenylphosphinic amide, C37H34NO2P
- Crystal structure of (E)-4-(6-(4-(2-(pyridin-4-yl)vinyl)phenoxy)pyrimidin-4-yl)morpholine, C21H20N4O2
- Crystal structure of 5-(adamantan-1-yl)-3-[(4-trifluoromethylanilino)methyl]-2,3-dihydro-1,3,4-oxadiazole-2-thione, C20H22F3N3OS
- Crystal structure of 2,2-dichloro-1-(4-chloro-1H-indol-1-yl)ethan-1-one, C10H6Cl3NO
- The crystal structure of 4-(((3-bromo-5-(trifluoromethyl)pyridin-2-yl)oxy)methyl)benzonitrile, C28H16Br2F6N4O2
- The crystal structure of 1H-benzimidazole-2-carboxamide, C8H7N3O
- The crystal structure of Histidinium hydrogensquarate, C10H11N3O6
- The crystal structure of 3-amino-5-carboxypyridin-1-ium iodide, C6H7IN2O2
- Crystal structure of (E)-amino(2-(3-ethoxy-4-hydroxybenzylidene)hydrazineyl)methaniminium nitrate hemihydrate C10H16N5O5.5
- Crystal structure of 1,2-bis(4,5-dinitro-1H-imidazol-1-yl)ethane, C8H6N8O8
- The crystal structure of diaqua-bis(pyrazolo[1,5-a]pyrimidine-3-carboxylato-κ2N,O)manganese(II), C14H12N6O6Mn
- The crystal structure of catena-poly[aqua-2,2′bipyridine-κ2N,N′-(μ2-5-ethoxyisophthalato-κ 4O,O′:Oʺ,O′ʺ)cadmium(II)] monohydrate, C20H20CdN2O7
- The crystal structure of (1S,3R)-1-(4-isopropylphenyl)-3-(methoxycarbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indol-2-iumchloride monohydrate, C22H27ClN2O3
- Crystal structure of 1-isopropyl-3-(prop-1-en-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, C11H15N5
- The crystal structure of (2,2′-bipyridine-κ2N,N′)- bis(6-phenylpyridine-2-carboxylate-κ2N,O)manganese(II)] monohydrate, C34H26N4O5Mn
- Crystal structure of the cocrystal 1,3,5,7-tetranitro-1,3,5,7-tetrazoctane ─ 2,3-dihydroindole (1/1), C12H17N9O8
- Crystal structure of 3-acetyl-6-hydroxy-2H-chromen-2-one monohydrate, C11H10O5
- Crystal structure of 6,9-diamino-2-ethoxyacridinium 3,5-dinitrobenozate — dimethylsulfoxide — water (1/1/1), C24H27N5O9S
- The crystal structure of 4,4′-bipyridinium bis-(2-hydroxy-3-methoxybenzoate), 2(C8H7.68O4)·C10H8.64N2
- Crystal structure of (Z)-4-(((4-fluorophenyl)amino)(furan-2-yl)methylene)-5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one
- The crystal structure of bis(4-chloro-2-(((2-chloroethyl)imino)methyl)phenolato-κ2N,O)-oxidovanadium(IV), C18H16Cl4N2O3V
- The crystal structure of 17-(bromoethynyl)-17-hydroxy-10, 13-dimethyl- 1,2,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-3H-cyclopenta[a]phenanthren-3-one, C21H27BrO2
- The crystal structure of 4-((6-fluoropyridin-2-yloxy)methyl)benzonitrile, C13H9FN2O
- Crystal structure of (Z)-2-(1-bromo-2-phenylvinyl)-5-ethyl-2-methyl-1,3-dioxane-5-carboxylic acid, C15H17Br1O4
- Crystal structure of catena-poly[tribenzyl-κ1C-(μ2-6-oxidopyridin-1-ium-3-carboxylato-κ2O:O’)tin(IV)-dichloromethane-methanol (1/1/1), C29H31Cl2NO4Sn
- Crystal structure of bis{2-(tert-butyl)-6-((E)-((4-((E)-1-(methoxyimino)ethyl)phenyl)imino)methyl)phenolato-κ2N,O}zinc(II), C40H46N4O4Zn
- Crystal structure of diaqua-bis(μ2-2-carboxy-3,4,5,6-tetrafluorobenzoato-κ2O:O′)-bis(phenanthroline-κ2N,N′)-bis(μ2-3,4,5,6-tetrafluorophthalato-κ3O:O,O′)dieuropium(III) – phenanthroline (1/2), C40H19EuF8N4O9
- The crystal structure of diaqua-bis(6-phenylpyridine-2-carboxylato-κ2N,O) manganese(II) — water — dimethylformamide (1/2/1), C27H31N3O9Mn
- The crystal structure of bis(pyrazolo[1,5-a]pyrimidine-3-carboxylato-κ2N,O)-copper(ii), C14H8N6O4Cu
- Crystal structure of poly[(μ2-1-(1-imidazolyl)-4-(imidazol-1-ylmethyl)benzene-κ2N:N′)-(μ3-pyridazine-4,5-dicarboxylate-κ3O:O′:N)]copper(II) hydrate, C19H16CuN6O5
- Crystal structure of acrinidinium tetrafluorohydrogenphthalate, C21H11F4NO4
- Crystal structure of 2-(1H-pyrazol-3-yl-κN)pyridine-κN-bis(2-(2,4-difluorophenyl)pyridinato-κ2C,N)iridium(III) sesquihydrate, C30H18F4IrN5·1.5[H2O]
- Crystal structure of 2-(2-hydroxy-5-nitrophenyl)-5-methyl-1,3-dioxane-5-carboxylic acid, C12H13N1O7
- The crystal structure of 1,2-bis(pyridinium-4-yl)ethane diperchlorate, C12H14N2·2ClO4 – a second polymorph
- The crystal structure of [(1,10-phenantroline-κ2N,N′)-bis(6-phenylpyridine-2-carboxylato-κ2N,O)manganese(II)] monohydrate, C36H26N4O5Mn
- Crystal structure of 1,2-bis(2,2,3,3,5,5,5-heptamethyl-1,1,4,4- tetrakis(trimethylsilyl)pentasilan-1-yl)ditellane, C38H114Si18Te2
- Crystal structure of 1,2-bis(2,4-dinitro-1H-imidazol-1-yl)ethane – dimethylformamide (1/1), C11H13N9O9
- Crystal structure of (Z)-3-((tert-butylamino) methylene)-2-(2-hydroxynaphthalen-1-yl) chroman-4-one, C24H23NO3
- Synthesis and crystal structure of (E)-1-(4-(((E)-3-(tert-butyl)-2-hydroxybenzylidene)amino)phenyl)ethan-1-one O-ethyl oxime, C21H26N2O2
- Crystal structure of the double salt bis(5-amino-1,2,4-triazol-4-ium-3-yl)methane hydrogen oxalate hemioxalate, C8H11N8O6
- Hydrothermal synthesis and crystal structure of catena-poly[diaqua-bis(μ2-4-[(4-pyridinylmethyl)amino]benzoato-κ2N:O)cobalt(II)]–1,2bi(4-pyridyl)ethene–water (1/1/1), C50H50N8O8Co
- Crystal structure of 3-(3-bromophenyl)-1′,3′-dimethyl-2′H,3H,4H-spiro[furo[3, 2-c]chromene-2,5′-pyrimidine]-2′,4,4′,6′(1′H,3′H) tetraone, C22H15BrN2O6
- The crystal structure of poly[aqua-(μ2-4,4′- bis(imidazolyl)biphenyl-κ2N:N′)-(μ2-3-nitrobenzene-1,2-dicarboxylato-κ2O:O′)]copper (II) hydrate, C26H21N5O8Cu
- The crystal structure of bis(4-(6-carboxy-8-ethyl-3-fluoro-5-oxo-5,8-dihydro-1,8-naphthyridin-2- yl)piperazin-1-ium) adipate tetrahydrate, C36H52F2N8O14
- Synthesis and crystal structure of poly[aqua(μ4-(1R,2S,4R)-4-hydroxy-1-((7-hydroxy-3-(4-hydroxy-3-sulfonatophenyl)-4-oxo-4H-chromen-8-yl)methyl)pyrrolidin-1-ium-2-carboxylate-κ4O:O′:O″:O‴)sodium(I)] monohydrate, C21H22NNaO12S
- Crystal structure of chlorido-(η6-toluene)(2,2′-bipyridine-κ2N,N′)ruthenium(II) hexafluorophosphate, C17H16ClN2RuPF6
- The crystal structure of (R)-6-hydroxy-8-methoxy-3-methylisochroman-1-one, C11H12O4
- Crystal structure of catena-poly[(5,5,7,12,12,14-hexamethyl -1,4,8,11-tetraazacyclotetradecane- κ4N,N′,Nʺ,N‴)nickel(II)-(μ2-perchlorato-κ2O:O′)] 3,5-dicarboxybenzoate – methanol (1/2), C27H49ClN4NiO12
- The crystal structure of 4-(chloromethyl)benzonitrile, C8H6ClN
- The crystal structure of dimethylammonium 8-[(7,9-dioxo-6,10-dioxaspiro[4.5]decan-8-ylidene)methyl]-9-oxo-6,10-dioxaspiro[4.5]dec-7-en-7-olate, C19H25NO8
- Crystal structure of (2R,3S,4S,5R,6S)-2-(acetoxymethyl)-6-((1-acetyl-5-bromo-4-chloro-1H-indol-3-yl)oxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate hemihydrate C24H25BrClNO11
- The crystal structure of the co-crystal tetrakis[2-(tris(4-methoxyphenyl)stannyl)ethyl]silane – tetrahydrofuran – toluene – tetrahydrofurane (1/1/1), C103H116O13SiSn4
- Crystal structure of methyl 3-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)propanoate, C16H13NO4
- Crystal structure of ethyl (Z)-3-amino-2-cyano-3-(2-oxo-2H-chromen-3-yl)acrylate, C15H12N2O4
- Crystal structure of methyl 2-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)acetate, C15H11NO4
- Crystal structure of catena-poly[diaqua-bis(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2N:N′)cobalt(II)] tetrafluoroterephthalate, C26H28N8O6F4Co

