Crystal structure of 72,73,75,76-tetrafluoro-25,44-dimethyl-31,33,36,38-tetraoxo-31,32,33,36,37,38-hexahydro-3(2,7)-benzo[lmn][3,8]phenanthrolina-1,5(4,1)-dipyridin-1-iuma-2,4(1,2),7(1,4)-tribenzenacyclooctaphane-11,51-diium hexafluoridophosphate, [C46H28F4N4O4][PF6]2, a dicationic cyclophane
Abstract
C46H28F16N4O4P2, monoclinic, P21/n (no. 14), a = 9.3041(2) Å, b = 22.4679(5) Å, c = 19.9446(3) Å, β = 98.154(2)°, V = 4,127.14(14) Å3, Z = 4, Rgt (F) = 0.0478, wRref (F 2) = 0.1411, T = 173(2) K.
Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colorless prism |
| Size: | 0.26 × 0.21 × 0.20 mm |
| Wavelength: | Cu Kα radiation (1.54184 Å) |
| μ: | 2.11 mm−1 |
| Diffractometer, scan mode: | RIGAKU XtaLAB P200, φ and ω |
| θ max, completeness: | 73.9°, >99 % |
| N(hkl)measured, N(hkl)unique, R int: | 24,672, 8,169, 0.030 |
| Criterion for I obs, N(hkl)gt: | I obs > 2 σ(I obs), 6,826 |
| N(param)refined: | 651 |
| Programs: | Bruker, 1 SHELX, 2 , 3 Olex2 4 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| P001 | 0.69407 (7) | 0.51807 (3) | 0.38952 (3) | 0.03046 (15) |
| F1 | 0.49150 (17) | 0.68605 (6) | 0.43832 (7) | 0.0375 (3) |
| O1 | 0.48231 (18) | 0.54505 (7) | 0.07785 (9) | 0.0340 (4) |
| N1 | 0.5653 (2) | 0.68551 (8) | 0.24132 (10) | 0.0285 (4) |
| C1 | 0.5183 (3) | 0.70573 (9) | 0.17805 (12) | 0.0290 (5) |
| H1 | 0.4431 | 0.7346 | 0.1711 | 0.035* |
| P002 | 0.51806 (8) | 0.22304 (3) | 0.14149 (4) | 0.03846 (17) |
| F2 | 0.31869 (18) | 0.60855 (7) | 0.48862 (7) | 0.0408 (4) |
| O2 | 0.94359 (18) | 0.48221 (7) | 0.15257 (8) | 0.0297 (3) |
| N2 | 0.7131 (2) | 0.51324 (8) | 0.11432 (9) | 0.0250 (4) |
| C2 | 0.5789 (3) | 0.68465 (9) | 0.12392 (12) | 0.0284 (5) |
| H2 | 0.5466 | 0.6994 | 0.0797 | 0.034* |
| F3 | 0.31345 (18) | 0.62016 (7) | 0.21714 (7) | 0.0424 (4) |
| O3 | 0.20501 (17) | 0.32481 (7) | 0.27488 (8) | 0.0287 (3) |
| N3 | 0.4352 (2) | 0.30320 (8) | 0.32486 (9) | 0.0258 (4) |
| C3 | 0.6885 (2) | 0.64132 (9) | 0.13371 (11) | 0.0267 (5) |
| F4 | 0.14714 (18) | 0.54069 (7) | 0.26835 (8) | 0.0456 (4) |
| O4 | 0.66646 (19) | 0.27980 (7) | 0.37181 (9) | 0.0332 (4) |
| N4 | 0.1738 (2) | 0.46717 (8) | 0.41146 (10) | 0.0319 (4) |
| C4 | 0.7369 (3) | 0.62307 (10) | 0.19998 (12) | 0.0300 (5) |
| H4 | 0.8134 | 0.5949 | 0.2085 | 0.036* |
| F5 | 0.6962 (2) | 0.58933 (7) | 0.39752 (8) | 0.0524 (4) |
| C5 | 0.6742 (3) | 0.64571 (10) | 0.25282 (12) | 0.0306 (5) |
| H5 | 0.7079 | 0.6332 | 0.2978 | 0.037* |
| F6 | 0.59622 (19) | 0.51433 (8) | 0.44920 (8) | 0.0502 (4) |
| C6 | 0.7491 (2) | 0.61629 (10) | 0.07547 (11) | 0.0259 (4) |
| F7 | 0.55357 (18) | 0.52266 (8) | 0.33482 (8) | 0.0493 (4) |
| C7 | 0.7873 (3) | 0.65436 (10) | 0.02536 (11) | 0.0279 (5) |
| H7 | 0.7712 | 0.6959 | 0.0290 | 0.034* |
| F8 | 0.6942 (2) | 0.44807 (7) | 0.38362 (9) | 0.0564 (5) |
| C8 | 0.8482 (3) | 0.63282 (10) | −0.02959 (11) | 0.0287 (5) |
| F9 | 0.79308 (18) | 0.52315 (8) | 0.33052 (8) | 0.0468 (4) |
| C9 | 0.8902 (3) | 0.67474 (11) | −0.08247 (12) | 0.0347 (5) |
| H9A | 0.8041 | 0.6965 | −0.1035 | 0.052* |
| H9B | 0.9310 | 0.6520 | −0.1172 | 0.052* |
| H9C | 0.9629 | 0.7030 | −0.0611 | 0.052* |
| F10 | 0.83624 (18) | 0.51497 (7) | 0.44573 (8) | 0.0459 (4) |
| C10 | 0.8703 (3) | 0.57155 (10) | −0.03398 (11) | 0.0293 (5) |
| H10 | 0.9155 | 0.5561 | −0.0701 | 0.035* |
| F11 | 0.51311 (19) | 0.29313 (7) | 0.12781 (9) | 0.0504 (4) |
| C11 | 0.8271 (3) | 0.53295 (10) | 0.01367 (11) | 0.0281 (5) |
| H11 | 0.8390 | 0.4913 | 0.0088 | 0.034* |
| F12 | 0.5986 (2) | 0.23415 (7) | 0.21726 (8) | 0.0533 (4) |
| C12 | 0.7669 (2) | 0.55476 (9) | 0.06795 (11) | 0.0253 (4) |
| F13 | 0.6737 (2) | 0.21884 (8) | 0.11739 (9) | 0.0540 (4) |
| C13 | 0.5612 (3) | 0.51196 (9) | 0.11421 (11) | 0.0270 (5) |
| F14 | 0.5245 (2) | 0.15311 (7) | 0.15714 (11) | 0.0583 (5) |
| C14 | 0.8136 (2) | 0.47653 (9) | 0.15412 (11) | 0.0250 (4) |
| F15 | 0.4402 (3) | 0.21142 (10) | 0.06742 (12) | 0.0871 (8) |
| C15 | 0.5059 (2) | 0.46879 (9) | 0.16051 (11) | 0.0248 (4) |
| F16 | 0.3675 (2) | 0.22751 (9) | 0.16927 (14) | 0.0770 (7) |
| C16 | 0.7540 (2) | 0.43153 (9) | 0.19700 (11) | 0.0247 (4) |
| C17 | 0.3600 (3) | 0.46748 (10) | 0.16541 (12) | 0.0283 (5) |
| H17 | 0.2969 | 0.4963 | 0.1421 | 0.034* |
| C18 | 0.6026 (2) | 0.42906 (9) | 0.19868 (11) | 0.0242 (4) |
| C19 | 0.8461 (3) | 0.39324 (10) | 0.23634 (12) | 0.0283 (5) |
| H19 | 0.9475 | 0.3947 | 0.2345 | 0.034* |
| C20 | 0.3039 (3) | 0.42366 (10) | 0.20470 (12) | 0.0282 (5) |
| H20 | 0.2025 | 0.4220 | 0.2065 | 0.034* |
| C21 | 0.5469 (2) | 0.38675 (9) | 0.24040 (11) | 0.0243 (4) |
| C22 | 0.7911 (3) | 0.35226 (10) | 0.27901 (12) | 0.0282 (5) |
| H22 | 0.8555 | 0.3265 | 0.3065 | 0.034* |
| C23 | 0.3952 (2) | 0.38313 (9) | 0.24066 (11) | 0.0250 (4) |
| C24 | 0.6437 (2) | 0.34906 (9) | 0.28139 (11) | 0.0252 (4) |
| C25 | 0.3353 (2) | 0.33521 (9) | 0.27957 (11) | 0.0252 (4) |
| C26 | 0.5878 (3) | 0.30767 (9) | 0.32944 (11) | 0.0279 (5) |
| C27 | 0.3787 (3) | 0.26335 (9) | 0.37213 (11) | 0.0266 (4) |
| C28 | 0.4056 (3) | 0.20286 (10) | 0.36826 (12) | 0.0290 (5) |
| H28 | 0.4575 | 0.1878 | 0.3342 | 0.035* |
| C29 | 0.3562 (3) | 0.16432 (10) | 0.41463 (12) | 0.0301 (5) |
| H29 | 0.3757 | 0.1229 | 0.4121 | 0.036* |
| C30 | 0.2788 (3) | 0.18515 (10) | 0.46448 (11) | 0.0292 (5) |
| C31 | 0.2286 (3) | 0.14348 (11) | 0.51588 (12) | 0.0351 (5) |
| H31A | 0.1629 | 0.1136 | 0.4925 | 0.053* |
| H31B | 0.1774 | 0.1662 | 0.5471 | 0.053* |
| H31C | 0.3128 | 0.1236 | 0.5414 | 0.053* |
| C32 | 0.2480 (3) | 0.24585 (10) | 0.46598 (11) | 0.0288 (5) |
| H32 | 0.1918 | 0.2606 | 0.4986 | 0.035* |
| C33 | 0.2982 (3) | 0.28547 (9) | 0.42051 (11) | 0.0272 (5) |
| C34 | 0.2578 (3) | 0.34945 (10) | 0.42200 (11) | 0.0282 (5) |
| C35 | 0.1137 (3) | 0.36595 (10) | 0.42211 (12) | 0.0319 (5) |
| H35 | 0.0426 | 0.3363 | 0.4262 | 0.038* |
| C36 | 0.0736 (3) | 0.42476 (11) | 0.41634 (13) | 0.0344 (5) |
| H36 | −0.0253 | 0.4355 | 0.4158 | 0.041* |
| C37 | 0.3140 (3) | 0.45294 (10) | 0.41483 (13) | 0.0331 (5) |
| H37 | 0.3837 | 0.4836 | 0.4133 | 0.040* |
| C38 | 0.3595 (3) | 0.39452 (10) | 0.42043 (12) | 0.0313 (5) |
| H38 | 0.4597 | 0.3851 | 0.4232 | 0.038* |
| C39 | 0.1246 (3) | 0.53109 (10) | 0.40713 (15) | 0.0377 (6) |
| H39A | 0.0298 | 0.5335 | 0.3776 | 0.045* |
| H39B | 0.1100 | 0.5448 | 0.4529 | 0.045* |
| C40 | 0.2295 (3) | 0.57166 (10) | 0.38012 (13) | 0.0329 (5) |
| C41 | 0.3190 (3) | 0.61012 (10) | 0.42123 (11) | 0.0306 (5) |
| C42 | 0.4063 (3) | 0.65059 (9) | 0.39512 (11) | 0.0300 (5) |
| C43 | 0.4077 (3) | 0.65662 (9) | 0.32627 (12) | 0.0285 (5) |
| C44 | 0.4927 (3) | 0.70561 (10) | 0.29904 (12) | 0.0320 (5) |
| H44A | 0.4265 | 0.7390 | 0.2841 | 0.038* |
| H44B | 0.5670 | 0.7204 | 0.3357 | 0.038* |
| C45 | 0.3177 (3) | 0.61765 (10) | 0.28505 (12) | 0.0315 (5) |
| C46 | 0.2329 (3) | 0.57637 (10) | 0.31106 (13) | 0.0337 (5) |
1 Source of materials
Bis(4-methyl-2-(4-pyridyl)phenyl)-naphthalenediimide (156 mg, 0.26 mmol) and 1,4-bis(bromomethyl)tetrafluorobenzene (87 mg, 0.26 mmol) were stirred in acetonitrile at 60 °C for 4 h under nitrogen protection. The yellow precipitate was collected by vacuum filtration and dissolved in water. When a saturated aqueous solution of ammonium hexafluorophosphate was added to the reaction mixture, the resulting precipitate was filtered off and then washed with deionized water. The precipitate was dried to afford yellowish solids. Single crystals were grown by slow diffusion of diethyl ether into an acetonitrile solution over 7 days of 72,73,75,76-tetrafluoro-25,44-dimethyl-31,33,36,38-tetraoxo-31,32,33,36,37,38-hexahydro-3(2,7)-benzo[lmn][3,8]phenanthrolina-1,5(4,1)-dipyridin-1-iuma-2,4(1,2),7(1,4)-tribenzenacyclooctaphane-11,51-diium hexafluoridophosphate.
2 Experimental details
The crystal structure was determined using the Shelxt program 2 and subsequently underwent refinement via the Shelxl tools 3 available in the Olex2 suite. 4 The hydrogen atoms were placed in idealized positions and allowed to ride on the relevant carbon atoms.
3 Comment
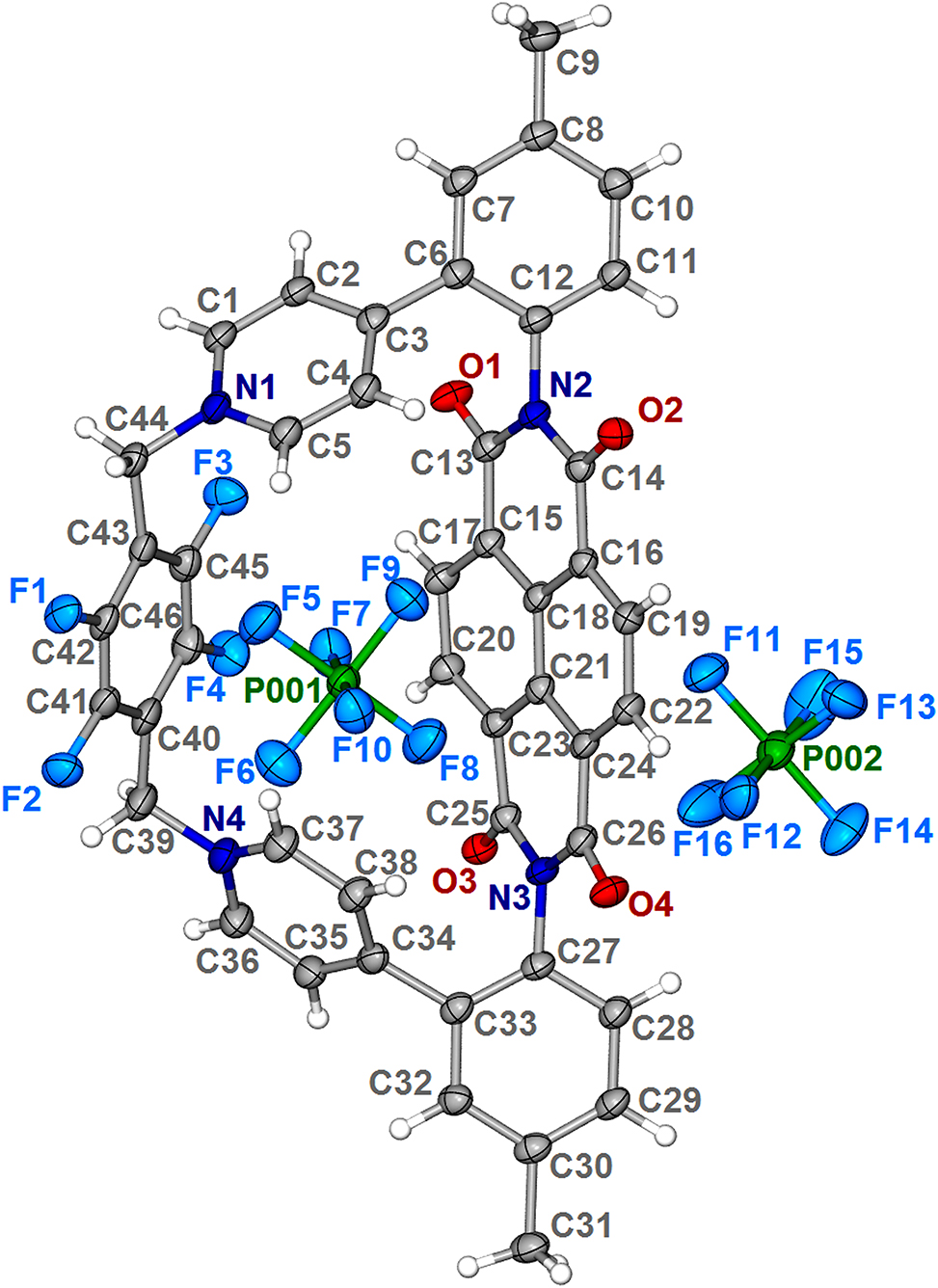
Macrocyclic hosts with cavities are the major workhorses in supramolecular chemistry, 5 and they can play a central role in molecular recognition 6 and be used to construct complex systems such as molecular machines 7 and other ‘smart’ materials. 8 Despite many recent advances in the development of new macrocyclic hosts, 9 their rational design remains a great challenge, due to low yields and tedious purifications. It is known that structural preorganization of building blocks facilitates the ring-closure process for constructing macrocyclic structures. 10 Herein, an syn-atropisomer based on naphthalene diimide (NDI) can be employed as a preorganized precursor to rationally construct a dicationic organic macrocycle, and the crystallographic structure of this macrocycle in the form of hexafluorophosphate salt has been elucidated.
The title compound crystallizes in the monoclinic space group P21/n with one whole macrocycle molecule in the asymmetric unit (see the figure). As depicted in the graphical representation, this macrocycle is composed of an NDI skeleton, a tetrafluorobenzene linker and two lateral pyridinium groups, exhibiting an anticipated trapezoid-shape box-like structure. The centroid–centroid separation of the parallel NDI and phenyl units is ~6.1 Å, which is sufficient to accommodate planar molecules via face-to-face π–π stacking. 11 , 12 , 13 The crystal lattice also contains two counter anions hexafluorophosphate. Bond lengths and angles are all in the normal range, and are very similar to those given in the literature. 12 , 13 , 14 In addition, the hexafluorophosphate ions interact with the electron-deficient NDI and tetrafluorobenzene rings via anion-p interactions, wherein the distances between F and centroid are 3.0322(1), 3.0537(1) and 3.0799(1) Å, which are all typical for anion-π interactions. 15
-
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Conflict of interest: The authors declare no conflicts of interest regarding this article.
-
Research funding: The Beijing Natural Science Foundation (2232027 and 2242004) and the National Natural Science Foundation of China (22171021).
References
1. Bruker. SAINT, APEX2 and SADABS; Bruker AXS Inc.: Madison, WI, USA, 2012.Search in Google Scholar
2. Sheldrick, G. M. SHELXTL – Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. 2015, A71, 3–8.10.1107/S2053273314026370Search in Google Scholar PubMed PubMed Central
3. Sheldrick, G. M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
4. Dolomanov, O. V.; Bourhis, L. J.; Gildea, R. J.; Howard, J. A. K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
5. Davis, F.; Higson, S. Macrocycles: Construction, Chemistry and Nanotechnology Applications; Wiley VCH: Chichester, 2011.10.1002/9780470980200Search in Google Scholar
6. Steed, J. W.; Atwood, J. L. Supramolecular Chemistry, 2nd ed.; Wiley: Chichester, 2009.10.1002/9780470740880Search in Google Scholar
7. Stoddart, J. F. Mechanically Interlocked Molecules (MIMs)—Molecular Shuttles, Switches, and Machines (Nobel Lecture). Angew. Chem., Int. Ed. 2017, 56, 11094–11125; https://doi.org/10.1002/anie.201703216.Search in Google Scholar PubMed
8. Dong, S.; Zheng, B.; Wang, F.; Huang, F. Supramolecular Polymers Constructed from Macrocycle-Based Host-Guest Molecular Recognition Motifs. Acc. Chem. Res. 2014, 47, 1982–1994; https://doi.org/10.1021/ar5000456.Search in Google Scholar PubMed
9. Liu, Z.; Nalluri, S. K. M.; Stoddart, J. F. Surveying Macrocyclic Chemistry: From Flexible Crown Ethers to Rigid Cyclophanes. Chem. Soc. Rev. 2017, 46, 2459–2478; https://doi.org/10.1039/c7cs00185a.Search in Google Scholar PubMed
10. Martι-Centelles, V.; Pandey, M. D.; Burguete, M. I.; Luis, S. V. Macrocyclization Reactions: The Importance of Conformational, Configurational, and Template-Induced Preorganization. Chem. Rev. 2015, 115, 8736–8834; https://doi.org/10.1021/acs.chemrev.5b00056.Search in Google Scholar PubMed
11. Yang, F.; Li, R.; Wei, W.; Ding, X.; Xu, Z.; Wang, P.; Wang, G.; Xu, Y.; Fu, H.; Zhao, Y. Water-Soluble Doubly-Strapped Isolated Perylene Diimide Chromophore. Angew. Chem., Int. Ed. 2022, 61, e202202491; https://doi.org/10.1002/anie.202202491.Search in Google Scholar PubMed
12. Yang, F.; Liu, C.; Yin, D.; Xu, Y.; Wu, M.; Wei, W. Atropisomer-based Construction of Macrocyclic Hosts That Selectively Recognize Tryptophan from Standard Amino Acids. Chem. Commun. 2019, 55, 14335; https://doi.org/10.1039/c9cc07646h.Search in Google Scholar PubMed
13. Yang, F.; Li, Y.; Li, R.; Wang, X.; Cui, X.; Wei, W.; Xu, Y. Fine-Tuning Macrocycle Cavity to Selectively Bind Guests in Water for Near-Infrared Photothermal Conversion. Org. Chem. Front. 2022, 9, 2902–2909; https://doi.org/10.1039/d2qo00443g.Search in Google Scholar
14. Li, R.; Yang, F.; Zhang, L.; Li, M.; Wang, G.; Wang, W.; Xu, Y.; Wei, W. Manipulating Host-Guest Charge Transfer of a Water-Soluble Double-Cavity Cyclophane for NIR–II Photothermal Therapy. Angew. Chem., Int. Ed. 2023, 62, e202301267; https://doi.org/10.1002/ange.202301267.Search in Google Scholar
15. Wang, D.-X.; Wang, M.-X. Exploring Anion-Πi Interactions and Their Applications in Supramolecular Chemistry. Acc. Chem. Res. 2020, 53, 1364–1380; https://doi.org/10.1021/acs.accounts.0c00243.Search in Google Scholar PubMed
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of the co-crystal 2,4,6-triamino-1,3,5-triazine-1,3-dioxide — acetic acid (1/2) C7H14N6O6
- Crystal structure of the dinuclear mercury(II) complex bis(μ2-bromido)-dibromido-bis{1-[(benzotriazol-1-yl)methyl]-1-H-1,3-(2-ethyl-5-methyl-imidazol)-κ1 N} dimercury(II), C26H30N10Hg2Br4
- Crystal structure of poly[hexaqua-pentakis(μ4-2,2′-bipyridine-4,4′-dicarboxylato-κ4 O:O′:O″:O‴)-(μ2-2,2′-bipyridine-4,4′-dicarboxylato-κ2 O:O)tetraytterbium(III)] hydrate, C36H26N6O16Yb2
- Hydrothermal synthesis and crystal structure of catena-poly[(1,10-phenanthroline-κ 2 N,N′)-bis(μ 2-nitroisophthalato-κ 3 O,O′:O″)nickel(II)], C20H13NiN3O7
- Crystal structure of 72,73,75,76-tetrafluoro-25,44-dimethyl-31,33,36,38-tetraoxo-31,32,33,36,37,38-hexahydro-3(2,7)-benzo[lmn][3,8]phenanthrolina-1,5(4,1)-dipyridin-1-iuma-2,4(1,2),7(1,4)-tribenzenacyclooctaphane-11,51-diium hexafluoridophosphate, [C46H28F4N4O4][PF6]2, a dicationic cyclophane
- Crystal structure of (E)-2-(4-(1H-imidazol-1-yl)benzylidene)-7-fluoro-3,4-dihydronaphthalen-1(2H)-one, C20H15FN2O
- The salt crystal structure of etoricoxib hydrochloride, C18H16Cl2N2O2S
- The structure of t-butyl 7-[2-(4-fluorophenyl)-3-phenyl-4-(phenylcarbamoyl)-5-(propan-2-yl)-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoate, C37H43FN2O5
- The crystal structure of (μ4-oxo)-tri(μ4-2,2′-bipyridine-6,6′-bis(olato)-κ5 O,O′:N:N′:O″)tetrazinc(II) – methylformamide (1/1), C33H25N7O8Zn4
- The co-crystal structure of 4-chlorobenzophenone–salicylhydrazide(1/1), C20H17ClN2O3
- Crystal structure of 9-fluoro-4-(6-methoxypyridin-2-yl)-5,6-dihydrobenzo[h]quinazolin-2-amine, C18H15FN4O
- The crystal structure of the co-crystal composed of benzhydrazide and 5-aminoisophthalic acid, C8H7NO4⋅C7H8N2O
- The cocrystal structure of praziquantel-hesperetin (1/1), C35H38N2O8
- Crystal structure of new barium manganese fluorides dihydrates, Ba10Mn2F25·2H2O
- The crystal structure of bis[μ2-(3-(3-(pyridin-2-yl)-1H-pyrazol-1-yl)propanoate-κ2O:N)-bis(2,2′-bipyridine-κ2 N, N′)dicopper(II)]dinitrate, C42H36Cu2N12O10
- Crystal structure of (3,6-di(2-pyridyl)-4-phenylaminopyridazine-κ2N,N′)-bis(2-(p-toluene)pyridinyl-κ2C,N)-iridium(III) hexafluorophosphate –dichloromethane (1/1), C45H37Cl2F6IrN7P
- The crystal structure of 2-(2′-carboxybenzyl)benzoic acid, C15H12O5
- The crystal structure of dichlorido-[(E)-N′,N″-bis((2E,3E)-3-(hydroxyimino)butan-2-ylidene)-2-((E)-3-(hydroxyimino)butan-2-ylidene)hydrazine-1-carbohydrazonhydrazide-κ 4 N 4]cobalt(II), C13H22N9O3Cl2Co
- Crystal structure of (−)-flavesine H, C15H22N2O2
- Crystal structure of 3-methoxybenzyl 2-(6-methoxynaphthalen-2-yl)propanoate, C22H22O4
- Crystal structure of dicarbonyl(2-oxopyridin-1(2H)-olato-κ 2 O,O)iridium(I), C7H4IrNO4
- The crystal structure of 4-(3-(triphenylphosphonio)propyl)piperazin-1-ium dibromide trihydrate, C25H37Br2N2O3P
- The crystal structure of ethyl 5,6-dihydroxybenzofuran-3-carboxylate, C11H10O5
- Crystal structure of 14-(R)-(2′-cyano-phenoxy)-3,19-diacetyl andrographolide, C31H37NO7
- The twinned crystal structure of 10-(4-methyl benzoate)-2,8-diethyl-5,5-difluoro-1,3,7,9-tetramethyl-5H-di-pyrrolo[1,2-c:2′,1′-f] [1,3,2]diazaborinin-4-ium-5-uide, C25H29BF2N2O2
- The crystal structure of (9H-thioxanthen-9- ylidene)hydrazine monohydrate, C13H11N2SO0.5
- The crystal structure of pyridinium diaqua-{1,2-phenylenebis((carboxylatocarbonyl)amido-κ4 N,N′,O,O′)manganese(III), C15H14MnN3O8
- Crystal structure of the hydrogen storage active high entropy phase Tb0.82Sm0.18Ni0.83Co0.17Mg
- Crystal structure of diaqua-bis[5-methyl-1-(1H-pyrazol-3-yl)-1H-1,2,3-triazole-4-carboxylato-κ 2 N,O)]manganese(II), C14H16MnN10O6
- Crystal structures of diiodido-3-((pyridin-2-ylmethylene)amino)-2-(pyridin-3-yl)-2,3-dihydroquinazolin-4(1H)-one-cadmium(II)
- Synthesis and crystal structure of methyl 4-(2-ethoxy-2-oxoethoxy)-3,5-dimethoxybenzoate, C14H18O7
- Crystal structure of isoxazolo[4,5-b]pyridin-3-amine, C6H5N3O
- Crystal structure of 4-chloro-1-isobutyl-1H-imidazo, C14H14ClN3
- The crystal structure of 1,1,1,2,2,2-hexakis(2-methyl-2-phenylpropyl)distannane,C60H78Sn2
- The crystal structure of (2,7-dimethoxynaphthalene-1,8-diyl)bis((3-nitrophenyl)methanone), C26H18N2O8
- Crystal structure of diaqua-tetra((E)-(RS)-1-(2,4-dichlorophenyl)-4,4-dimethyl-2-(1,2,4-triazol-1-yl)pent-1-en-3-ol-κ 1 N)zinc(II) dinitrate dihydrate, C60H76Cl8N14O14Zn
- The crystal structure of diphenyl bis(2-((diphenoxyphosphoryl)amino)ethyl)phosphoramidate monohydrate C40H42N3O10P3
- Crystal structure of 4,4′-bis(dibromomethyl)-1,1′-biphenyl, C14H10Br4
- Crystal structure of CaPtZn
- Crystal structure of 3-chloro-5-(trifluoromethyl)pyridine-2-carboxylic acid, C7H3ClF3NO2
- The crystal structure of (3′-(2-bromophenyl)-2-phenyl-[2,2′-bioxiran]-3-yl)(phenyl)methanone, C92H68O12Br4
- Crystal structure of ethyl 4-(4-benzylpiperazin-1-yl)benzoate, C20H24N2O2
- The crystal structure of bis(selenocyanato-κ1 N)-bis(methanol)-bis((1E,2E)-1,2-bis (1-(pyridin-4-yl)ethylidene)-hydrazine)iron(II) methanol solvate, C34H44FeN10O4Se2
- Crystal structure of (E)-1-(5-bromo-2-hydroxyphenyl)-3-(5-(4-methoxyphenoxy)-3-methyl-1-phenyl-1H-pyrazol-4-yl)prop-2-en-1-one, C26H21BrN2O4
- The crystal structure of methyl 4-(4-(methylsulfonyl)phenyl)-2-methyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate, C19H21NO5S
- Crystal structure of 1′,3′-dihydro-2,2′-spirobi[indene]-1,3-dione, C17H12O2
- Crystal structure of (E)-2,2′,3,3′-tetrahydro-[1,1′-biindenylidene]-4,4′-diol, C18H16O2
- Crystal structure of di-glycylglycinium squarate dihydrate, C12H22N4O12, at 105 K
- Crystal structure of {[(4-fluorophenyl)methyl]triphenylphosphonium}dibromocopper(I), [C25H21FP]+[CuBr2]−
- Crystal structure of poly[diaqua-bis(μ2-5-((pyridin-4-yl-methyl)amino)benzene-1,3-dicarboxylato-κ 2 N:O)cadmium(II)], C28H26CdN4O10
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of the co-crystal 2,4,6-triamino-1,3,5-triazine-1,3-dioxide — acetic acid (1/2) C7H14N6O6
- Crystal structure of the dinuclear mercury(II) complex bis(μ2-bromido)-dibromido-bis{1-[(benzotriazol-1-yl)methyl]-1-H-1,3-(2-ethyl-5-methyl-imidazol)-κ1 N} dimercury(II), C26H30N10Hg2Br4
- Crystal structure of poly[hexaqua-pentakis(μ4-2,2′-bipyridine-4,4′-dicarboxylato-κ4 O:O′:O″:O‴)-(μ2-2,2′-bipyridine-4,4′-dicarboxylato-κ2 O:O)tetraytterbium(III)] hydrate, C36H26N6O16Yb2
- Hydrothermal synthesis and crystal structure of catena-poly[(1,10-phenanthroline-κ 2 N,N′)-bis(μ 2-nitroisophthalato-κ 3 O,O′:O″)nickel(II)], C20H13NiN3O7
- Crystal structure of 72,73,75,76-tetrafluoro-25,44-dimethyl-31,33,36,38-tetraoxo-31,32,33,36,37,38-hexahydro-3(2,7)-benzo[lmn][3,8]phenanthrolina-1,5(4,1)-dipyridin-1-iuma-2,4(1,2),7(1,4)-tribenzenacyclooctaphane-11,51-diium hexafluoridophosphate, [C46H28F4N4O4][PF6]2, a dicationic cyclophane
- Crystal structure of (E)-2-(4-(1H-imidazol-1-yl)benzylidene)-7-fluoro-3,4-dihydronaphthalen-1(2H)-one, C20H15FN2O
- The salt crystal structure of etoricoxib hydrochloride, C18H16Cl2N2O2S
- The structure of t-butyl 7-[2-(4-fluorophenyl)-3-phenyl-4-(phenylcarbamoyl)-5-(propan-2-yl)-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoate, C37H43FN2O5
- The crystal structure of (μ4-oxo)-tri(μ4-2,2′-bipyridine-6,6′-bis(olato)-κ5 O,O′:N:N′:O″)tetrazinc(II) – methylformamide (1/1), C33H25N7O8Zn4
- The co-crystal structure of 4-chlorobenzophenone–salicylhydrazide(1/1), C20H17ClN2O3
- Crystal structure of 9-fluoro-4-(6-methoxypyridin-2-yl)-5,6-dihydrobenzo[h]quinazolin-2-amine, C18H15FN4O
- The crystal structure of the co-crystal composed of benzhydrazide and 5-aminoisophthalic acid, C8H7NO4⋅C7H8N2O
- The cocrystal structure of praziquantel-hesperetin (1/1), C35H38N2O8
- Crystal structure of new barium manganese fluorides dihydrates, Ba10Mn2F25·2H2O
- The crystal structure of bis[μ2-(3-(3-(pyridin-2-yl)-1H-pyrazol-1-yl)propanoate-κ2O:N)-bis(2,2′-bipyridine-κ2 N, N′)dicopper(II)]dinitrate, C42H36Cu2N12O10
- Crystal structure of (3,6-di(2-pyridyl)-4-phenylaminopyridazine-κ2N,N′)-bis(2-(p-toluene)pyridinyl-κ2C,N)-iridium(III) hexafluorophosphate –dichloromethane (1/1), C45H37Cl2F6IrN7P
- The crystal structure of 2-(2′-carboxybenzyl)benzoic acid, C15H12O5
- The crystal structure of dichlorido-[(E)-N′,N″-bis((2E,3E)-3-(hydroxyimino)butan-2-ylidene)-2-((E)-3-(hydroxyimino)butan-2-ylidene)hydrazine-1-carbohydrazonhydrazide-κ 4 N 4]cobalt(II), C13H22N9O3Cl2Co
- Crystal structure of (−)-flavesine H, C15H22N2O2
- Crystal structure of 3-methoxybenzyl 2-(6-methoxynaphthalen-2-yl)propanoate, C22H22O4
- Crystal structure of dicarbonyl(2-oxopyridin-1(2H)-olato-κ 2 O,O)iridium(I), C7H4IrNO4
- The crystal structure of 4-(3-(triphenylphosphonio)propyl)piperazin-1-ium dibromide trihydrate, C25H37Br2N2O3P
- The crystal structure of ethyl 5,6-dihydroxybenzofuran-3-carboxylate, C11H10O5
- Crystal structure of 14-(R)-(2′-cyano-phenoxy)-3,19-diacetyl andrographolide, C31H37NO7
- The twinned crystal structure of 10-(4-methyl benzoate)-2,8-diethyl-5,5-difluoro-1,3,7,9-tetramethyl-5H-di-pyrrolo[1,2-c:2′,1′-f] [1,3,2]diazaborinin-4-ium-5-uide, C25H29BF2N2O2
- The crystal structure of (9H-thioxanthen-9- ylidene)hydrazine monohydrate, C13H11N2SO0.5
- The crystal structure of pyridinium diaqua-{1,2-phenylenebis((carboxylatocarbonyl)amido-κ4 N,N′,O,O′)manganese(III), C15H14MnN3O8
- Crystal structure of the hydrogen storage active high entropy phase Tb0.82Sm0.18Ni0.83Co0.17Mg
- Crystal structure of diaqua-bis[5-methyl-1-(1H-pyrazol-3-yl)-1H-1,2,3-triazole-4-carboxylato-κ 2 N,O)]manganese(II), C14H16MnN10O6
- Crystal structures of diiodido-3-((pyridin-2-ylmethylene)amino)-2-(pyridin-3-yl)-2,3-dihydroquinazolin-4(1H)-one-cadmium(II)
- Synthesis and crystal structure of methyl 4-(2-ethoxy-2-oxoethoxy)-3,5-dimethoxybenzoate, C14H18O7
- Crystal structure of isoxazolo[4,5-b]pyridin-3-amine, C6H5N3O
- Crystal structure of 4-chloro-1-isobutyl-1H-imidazo, C14H14ClN3
- The crystal structure of 1,1,1,2,2,2-hexakis(2-methyl-2-phenylpropyl)distannane,C60H78Sn2
- The crystal structure of (2,7-dimethoxynaphthalene-1,8-diyl)bis((3-nitrophenyl)methanone), C26H18N2O8
- Crystal structure of diaqua-tetra((E)-(RS)-1-(2,4-dichlorophenyl)-4,4-dimethyl-2-(1,2,4-triazol-1-yl)pent-1-en-3-ol-κ 1 N)zinc(II) dinitrate dihydrate, C60H76Cl8N14O14Zn
- The crystal structure of diphenyl bis(2-((diphenoxyphosphoryl)amino)ethyl)phosphoramidate monohydrate C40H42N3O10P3
- Crystal structure of 4,4′-bis(dibromomethyl)-1,1′-biphenyl, C14H10Br4
- Crystal structure of CaPtZn
- Crystal structure of 3-chloro-5-(trifluoromethyl)pyridine-2-carboxylic acid, C7H3ClF3NO2
- The crystal structure of (3′-(2-bromophenyl)-2-phenyl-[2,2′-bioxiran]-3-yl)(phenyl)methanone, C92H68O12Br4
- Crystal structure of ethyl 4-(4-benzylpiperazin-1-yl)benzoate, C20H24N2O2
- The crystal structure of bis(selenocyanato-κ1 N)-bis(methanol)-bis((1E,2E)-1,2-bis (1-(pyridin-4-yl)ethylidene)-hydrazine)iron(II) methanol solvate, C34H44FeN10O4Se2
- Crystal structure of (E)-1-(5-bromo-2-hydroxyphenyl)-3-(5-(4-methoxyphenoxy)-3-methyl-1-phenyl-1H-pyrazol-4-yl)prop-2-en-1-one, C26H21BrN2O4
- The crystal structure of methyl 4-(4-(methylsulfonyl)phenyl)-2-methyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate, C19H21NO5S
- Crystal structure of 1′,3′-dihydro-2,2′-spirobi[indene]-1,3-dione, C17H12O2
- Crystal structure of (E)-2,2′,3,3′-tetrahydro-[1,1′-biindenylidene]-4,4′-diol, C18H16O2
- Crystal structure of di-glycylglycinium squarate dihydrate, C12H22N4O12, at 105 K
- Crystal structure of {[(4-fluorophenyl)methyl]triphenylphosphonium}dibromocopper(I), [C25H21FP]+[CuBr2]−
- Crystal structure of poly[diaqua-bis(μ2-5-((pyridin-4-yl-methyl)amino)benzene-1,3-dicarboxylato-κ 2 N:O)cadmium(II)], C28H26CdN4O10

