Abstract
C9H11NS2, monoclinic, P21/n (no. 14), a = 5.6183(1) Å, b = 18.2426(3) Å, c = 9.5185(2) Å, β = 96.835(2)°, V = 968.64(3) Å3, Z = 4, Rgt(F) = 0.0303, wRref(F2) = 0.0826, T = 100(2) K.
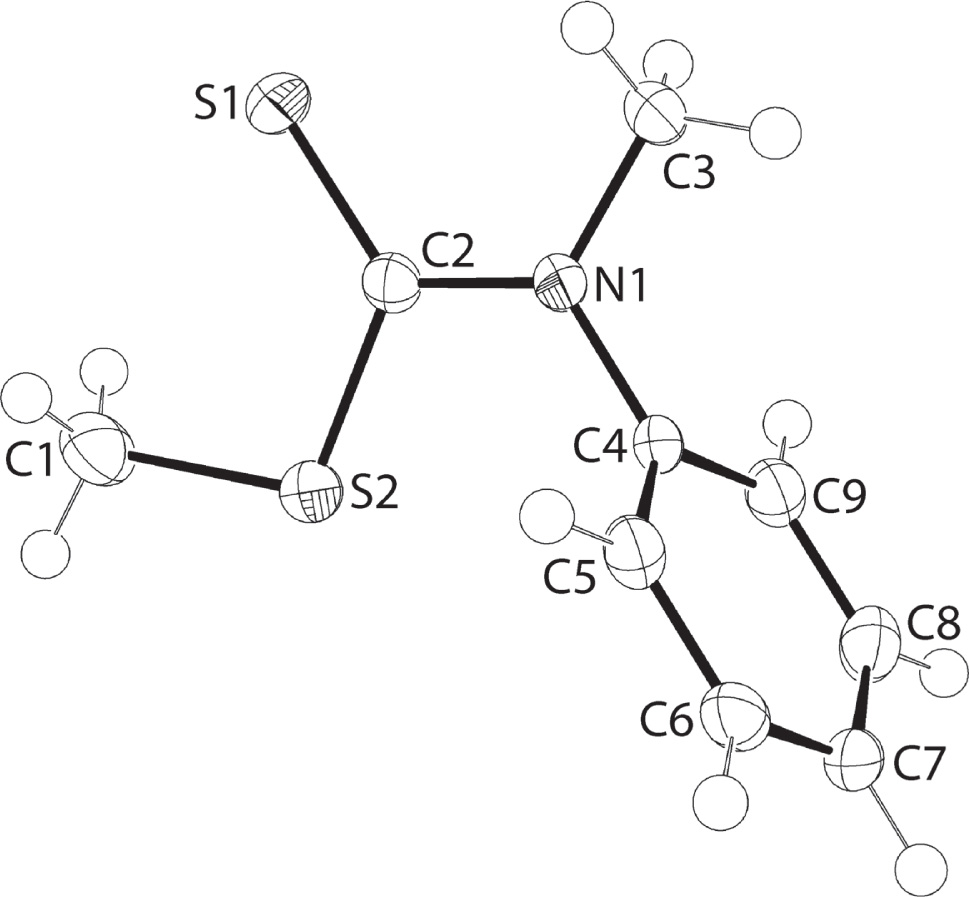
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colourless prism |
| Size: | 0.17 × 0.09 × 0.04 mm |
| Wavelength: | Cu Kα radiation (1.54184 Å) |
| μ: | 4.51 mm−1 |
| Diffractometer, scan mode: | XtaLAB Synergy, ω |
| θmax, completeness: | 67.1°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 11698, 1735, 0.050 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 1558 |
| N(param)refined: | 111 |
| Programs: | CrysAlisPRO [1], SHELX [2], [3], WinGX/ORTEP [4] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| S1 | 0.52898(8) | 0.54031(2) | 0.22540(5) | 0.02023(15) |
| S2 | 0.31034(8) | 0.65491(2) | 0.39930(5) | 0.01934(15) |
| N1 | 0.6011(3) | 0.68373(8) | 0.21179(16) | 0.0153(3) |
| C1 | 0.1812(4) | 0.57046(11) | 0.4487(2) | 0.0257(4) |
| H1A | 0.083225 | 0.549454 | 0.366476 | 0.039* |
| H1B | 0.080440 | 0.579626 | 0.524144 | 0.039* |
| H1C | 0.309249 | 0.536056 | 0.482660 | 0.039* |
| C2 | 0.4920(3) | 0.62716(10) | 0.26972(18) | 0.0145(4) |
| C3 | 0.7748(3) | 0.66948(10) | 0.11115(19) | 0.0181(4) |
| H3A | 0.917919 | 0.646158 | 0.160744 | 0.027* |
| H3B | 0.819704 | 0.715878 | 0.069587 | 0.027* |
| H3C | 0.702498 | 0.636909 | 0.036002 | 0.027* |
| C4 | 0.5614(3) | 0.75900(9) | 0.24878(18) | 0.0146(4) |
| C5 | 0.3513(3) | 0.79439(10) | 0.19358(19) | 0.0176(4) |
| H5 | 0.233802 | 0.769028 | 0.131899 | 0.021* |
| C6 | 0.3147(3) | 0.86695(10) | 0.22923(19) | 0.0190(4) |
| H6 | 0.170928 | 0.891202 | 0.192572 | 0.023* |
| C7 | 0.4874(3) | 0.90418(10) | 0.31828(19) | 0.0186(4) |
| H7 | 0.461426 | 0.953743 | 0.343003 | 0.022* |
| C8 | 0.6988(3) | 0.86889(11) | 0.37137(19) | 0.0206(4) |
| H8 | 0.817922 | 0.894582 | 0.431345 | 0.025* |
| C9 | 0.7357(3) | 0.79614(10) | 0.33673(19) | 0.0180(4) |
| H9 | 0.879708 | 0.771934 | 0.373098 | 0.022* |
Source of material
All chemicals and solvents were used as purchased without purification. The melting point was determined using a Mel-temp II digital melting point apparatus and was uncorrected. The solid-state IR spectrum was obtained on a Bruker Vertex 70v FTIR Spectrometer from 4000 to 400 cm−1. The 1H and 13C{1H} NMR spectra were recorded at room temperature in DMSO-d6 solution on a Bruker Ascend 400 MHz NMR spectrometer with chemical shifts relative to tetramethylsilane.
The dithiocarbamate ligand was prepared in situ (acetone) from the reaction of CS2 (Merck, 0.25 mmol) with N-methylaniline (Merck, 0.25 mmol) and NaOH (0.02 mL; 50% w/v); CS2 was added dropwise into the acetone solution (15 ml). The resulting mixture solution was kept at 273 K for 1 h. Next, dimethyltin dichloride (Merck, 0.25 mmol, 0.055 g) in acetone (10 ml) was added into the solution and the resulting mixture was stirred for 2.5 h. The filtrate was evaporated until a white precipitate was obtained. The precipitate was washed with n-hexane and recrystallized from a methanol-acetone solution. The title ester was a side-product obtained from the slow evaporation of the solvent. Yield: 0.005 g (10.2%). M.pt: 481–483 K. IR (cm−1) 1490 (m) ν(C—N), 1112 (s) ν(C—S), 1082 (s) ν(C—S). 1H NMR (DMSO-d6, p.p.m.): δ 2.85 (s, 3H, SCH3), 3.50 (s, 3H, NCH3), 6.91–7.30 (m, 5H, Ph—H). 13C{1H} NMR (DMSO-d6, p.p.m.): δ 23.5 (SCH3), 43.1 (NCH3), 121.1, 124.5, 129.1, 138.9 (Ph—C), 197.8 (CS2).
Experimental details
The C-bound H atoms were geometrically placed (C—H = 0.95–0.98 Å) and refined as riding with Uiso(H) = 1.2–1.5Ueq(C).
Comment
Dithiocarbamate ligands (−S2CNRR′) are very well known as important ligands for metals/main group elements/lanthanides owing to their strong chelating ability. Recent interest in our laboratories has focused upon the structural chemistry of the homoleptic zinc-triad dithiocarbamates [5] along with their adducts with bipyridyl-type ligands [6]. It is salient to note that even with such a substantial body of structural data [5], in recent years two unprecedented structural motifs for cadmium dithiocarbamates have been discovered [7], [8], underscoring the value of systematic and thorough structural studies. An allied focus is upon the evaluation of the potential activity and biological modes of action of metal compounds of this important class of ligands [9], such as anti-cancer activity of bismuth(III) [10], [11] and zinc(II) [12] species, and anti-bacterial activity of gold(I) derivatives [13], [14]. In this context, it is noteworthy that all-organic, dithiocarbamate esters, R′′S2CNRR′, also attract on-going interest in the context of developing pharmaceuticals [15], [16]. In view of the above, herein, the crystal and molecular structures of a simple example of a dithiocarbamate ester, namely, MeSC(= S)N(Me)Ph, are described, which was obtained as a side-product while developing some organotin chemistry (see Source of material).
The title molecule is shown in the figure (70% displacement ellipsoids). The central CNS2 residue is strictly planar (r.m.s. deviation = 0.0034 Å) with the appended C1 [0.096(3) Å] and C4 [0.043(3) Å] atoms lying to one side of the plane, and the C3 [0.1145 (0.0027) Å] atom to the other side. The formal C2= S1 double bond of 1.6590(18) Å is significantly shorter that either of the C2—S2 [1.7662(17) Å] and C1—S1 [1.789(2) Å] bonds. The C2—N1 bond length is 1.351(2) Å, and is considerably shorter than either of the C3—N1 [1.470(2) Å] and C4—N1 [1.442(2) Å] bond lengths. These observations suggest some delocalization of π-electron density over the central CNS2 chromophore. Nevertheless, the bond angles subtended at the C2 atom, with those involving the S1 atom being systematically wider by approximately 10° [S1—C2—S2 = 123.47(11)° and S1—C2—N1 = 123.32(13)°] than the third angle [S2—C2—N1 = 113.21(13)°], suggest appreciable double-bond character in the C2=S1 bond. The phenyl ring is inclined with respect to the central plane, with the dihedral angle of 77.74(4)° indicative of an almost orthogonal relationship. This occurs to minimize steric repulsion between the terminal N1-bound residues.
The most prominent supramolecular contact in the molecular packing is a nitrogen-bound-methyl-C—H⋯S(ester) interaction [C3—H3b⋯S2i: C3—H3b⋯S2i =2.86 Å, C3⋯S2i = 3.8033(19) Å with angle at H3b = 162° for symmetry operation (i) 1/2 + x, 3/2 – y, −1/2 + z]. These interactions lead to a zigzag supramolecular chain along the c-axis, being propagated by glide symmetry. The only other contacts of note are sulfur-bound-methyl-C—H⋯π(phenyl) [C1—H1b⋯Cg(C4—C9)ii: = H1b⋯Cg(C4—C9)ii = 3.00 Å, C1⋯Cg(C4—C9)ii = 3.841(2) Å with angle at H1b = 145° for (ii) −1/2 + x, 3/2 – y, 1/2 + z]. These serve to connect the aforementioned chains into a supramolecular layer in the ac-plane. Layers stack along the b-axis without directional interactions between them.
To analyse the molecular packing in more detail, Crystal Explorer 17 [17] was utilized, following established procedures [18], to calculate the Hirshfeld surfaces along with the overall and decomposed two-dimensional fingerprint plots in order to ascertain the dominant contacts in the packing. There are three predominant surface contacts, amounting to 99.5% of the entire surface, i.e. H⋯H [53.7%], S⋯H/H⋯S [27.4%] and C⋯H/H⋯C [18.4%].
The crystal structure of a closely related compound, i.e. MeSC(= S)NMe2, is available [19] but, only as a communication to the Cambridge Structural Database. The key geometric parameters, i.e. C2=S1 [1.665(5) Å], C2—S2 [1.772(6) Å], C1—S1 [1.789(7) Å] and C2—N1 [1.335(6) Å] bear a close similarity to those for the title compound. In terms of molecular packing, no directional interactions are apparent [20]. Accordingly, an analysis of the Hirshfeld surface indicates a greater dominance of H⋯H [59.0%] contacts and significant surface contacts of the type S⋯H/H⋯S [32.1%], both enhanced compared with the title structure, at the expense of C⋯H/H⋯C [5.9%] contacts. Other contacts are evident, e.g. S⋯N/N⋯S [1.3%] but, at separations greater than the sum of the van der Waals radii.
Acknowledgements
Sunway University Sdn Bhd is thanked for financial support of this work through Grant No. STR-RCTR-RCCM-001–2019.
References
1. Rigaku Oxford Diffraction: CrysAlisPRO. Rigaku Corporation, Oxford, UK (2018).Suche in Google Scholar
2. Sheldrick, G. M.: A short history of SHELX. Acta Crystallogr. A64 (2008) 112–122.10.1107/S0108767307043930Suche in Google Scholar PubMed
3. Sheldrick, G. M.: Crystal structure refinement with SHELXL. Acta Crystallogr. C71 (2015) 3–8.10.1107/S2053229614024218Suche in Google Scholar PubMed PubMed Central
4. Farrugia, L. J.: WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 45 (2012) 849–854.10.1107/S0021889812029111Suche in Google Scholar
5. Tiekink, E. R. T.: Exploring the topological landscape exhibited by binary zinc-triad 1,1-dithiolates. Crystals 8 (2018) 292.10.3390/cryst8070292Suche in Google Scholar
6. Tiekink, E. R. T.: Perplexing coordination behaviour of potentially bridging bipyridyl-type ligands in the coordination chemistry of zinc and cadmium 1,1-dithiolate compounds. Crystals 8 (2018) 18.10.3390/cryst8010018Suche in Google Scholar
7. Tan, Y. S.; Halim, S. N. A.; Tiekink, E. R. T.: Exploring the crystallization landscape of cadmium bis(N-hydroxyethyl, N-isopropyl-dithiocarbamate), Cd[S2CN(iPr)CH2CH2OH]2. Z. Kristallogr. – Cryst. Mater. 231 (2016) 113–126.10.1515/zkri-2015-1889Suche in Google Scholar
8. Ahmad, J.; How, F. N.-F.; Halim, S. N. A.; Jotani, M. M.; Lee, S. M.; Tiekink, E. R. T.: A new structural motif for cadmium dithiocarbamates: crystal structures and Hirshfeld surface analyses of homoleptic zinc and cadmium morpholine dithiocarbamates. Z. Kristallogr. – Cryst. Mater. 234 (2019) 341–349.10.1515/zkri-2018-2141Suche in Google Scholar
9. Hogarth, G.: Metal-dithiocarbamate complexes: chemistry and biological activity. Mini Rev. Med. Chem. 12 (2012) 1202–1215.10.2174/138955712802762095Suche in Google Scholar PubMed
10. Li, H.; Lai, C. S.; Wu, J.; Ho, P. C.; de Vos, D.; Tiekink, E. R. T.: Cytotoxicity, Qualitative Structure-Activity Relationship (QSAR), and anti-tumor activity of bismuth dithiocarbamate complexes. J. Inorg. Biochem. 101 (2007) 809–816.10.1016/j.jinorgbio.2007.01.010Suche in Google Scholar PubMed
11. Ishak, D. H. A.; Ooi, K. K.; Ang, K. P.; Akim, A. M.; Cheah, Y. K.; Nordin, N.; Halim, S. N. B. A.; Seng, H.-L.; Tiekink, E. R. T.: A bismuth diethyldithiocarbamate compound promotes apoptosis in HepG2 carcinoma, cell cycle arrest and inhibits cell invasion through modulation of the NF-κB activation pathway. J. Inorg. Biochem. 130 (2014) 38–51.10.1016/j.jinorgbio.2013.09.018Suche in Google Scholar PubMed
12. Tan, Y. S.; Ooi, K. K.; Ang, K. P.; Akim, A. M.; Cheah, Y.-K.; Halim, S. N. A.; Seng, H.-L.; Tiekink, E. R. T.: Molecular mechanisms of apoptosis and cell selectivity of zinc dithiocarbamates functionalized with hydroxyethyl substituents. J. Inorg. Biochem. 150 (2015) 48–62.10.1016/j.jinorgbio.2015.06.009Suche in Google Scholar PubMed
13. Chen, B.-J.; Jamaludin, N. S.; Khoo, C.-H.; See, T.-H.; Sim, J.-H.; Cheah, Y.-K.; Halim, S. N. A.; Seng, H.-L.; Tiekink, E. R. T.: In vitro antibacterial and time kill evaluation of mononuclear phosphanegold(I) dithiocarbamates. J. Inorg. Biochem. 163 (2016) 68–80.10.1016/j.jinorgbio.2016.08.002Suche in Google Scholar PubMed
14. Tan, Y. J.; Tan, Y. S.; Yeo, C. I.; Chew, J.; Tiekink, E. R. T.: In vitro anti-bacterial and time kill evaluation of binuclear tricyclohexylphosphanesilver(I) dithiocarbamates, {Cy3PAg(S2CNRR′)}2. J. Inorg. Biochem. 192 (2019) 107–118.10.1016/j.jinorgbio.2018.12.017Suche in Google Scholar PubMed
15. Liu, B.; Yuan, X.; Xu, B.; Zhang, H.; Li, R.; Wang, X.; Ge, Z.: Synthesis of novel 7-azaindole derivatives containing pyridin-3-ylmethyl dithiocarbamate moiety as potent PKM2 activators and PKM2 nucleus. translocation inhibitors. Eur. J. Med. Chem. 170 (2019) 1–15.10.1016/j.ejmech.2019.03.003Suche in Google Scholar PubMed
16. Su, Y.; Li, R.; Ning, X.; Lin, Z.; Zhao, X.; Zhou, J.; Liu, J.; Jin, Y.; Yin, Y.: Discovery of 2,4-diarylaminopyrimidine derivatives bearing dithiocarbamate moiety as novel FAK inhibitors with antitumor and anti-angiogenesis activities. Eur. J. Med. Chem. 177 (2019) 32–46.10.1016/j.ejmech.2019.05.048Suche in Google Scholar PubMed
17. Turner, M. J.; McKinnon, J. J.; Wolff, S. K.; Grimwood, D. J.; Spackman, P. R.; Jayatilaka, D.; Spackman, M. A.: Crystal Explorer v17. The University of Western Australia, Australia (2017).Suche in Google Scholar
18. Tan, S. L.; Jotani, M. M.; Tiekink, E. R. T.: Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. E75 (2019) 308–318.10.1107/S2056989019001129Suche in Google Scholar PubMed PubMed Central
19. Koley, S.; Singh, M. S.: Private communication to the Cambridge Structural Database. Refode: PAMPAX (2016).Suche in Google Scholar
20. Spek, A. L.: Structure validation in chemical crystallography. Acta Crystallogr. D65 (2009) 148–155.10.1107/S090744490804362XSuche in Google Scholar PubMed PubMed Central
© 2019 Kong Mun Lo et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 Public License.
Artikel in diesem Heft
- Frontmatter
- Crystal structure of [aqua[2,2′-(1,2-phenylene)bis(1H-imidazole-4-carboxylato-5-carboxy)-κ4N3,N3′,O4,O4′] zinc(II)] monohydrate, C16H10N4O9Zn⋅H2O
- Crystal structure of ethyl 3-(4-methoxyphenyl)-5-methylcarbamoyl-1H-pyrazole-4-carboxylate, C15H17N3O4
- 7-(4-Fluorobenzylidene)-3-(4-fluorophenyl)-N-phenyl-3,3a,4,5,6,7-hexahydro-2H-indazole-2-carbothioamide–dimethylformamide (2/1), C27H23F2N3S, 0.5(C3H7NO)
- Crystal structure of 4,4′-(hydrazonomethylene)diphenol dihydrate, C13H16N2O4
- Crystal structure of 4-methoxyphenyl-3-phenylpropiolate, C16H12O3
- Crystal Structure of tris(tetrakis{1-vinyl-1H-imidazole-κN}copper(II)) bis[tri-μ2-bromido-tetrabromido-bis(1-vinyl-1H-imidazole-κN)tetracopper(I)], C80H96N32Cu11Br14
- Crystal structure of (E)-2-(3,6-bis(diethylamino)-9H-xanthen-9-yl)-N′-(quinoxalin-2-ylmethylene)benzohydrazide, C37H36N6O2
- Crystal structure of 4-(1-phenylimidazo[1,5-a]pyridin-3-yl)benzoic acid (C20H14N2O2)
- Crystal structure of 3-fluoro-3-methyl-1-((2-nitrophenyl)sulfonyl)-5,5-diphenylpiperidine, C24H23FN2O4S
- Crystal structure of dimethyl 3,12-dibenzyl-6,10-diphenyl-3,12-diazapentacyclo [6.3.1.02.7.04.11.05.9]-dodecane-7,11-dicarboxylate — acetone (2/1), C40H38N2O2 ⋅ 0.5C3H6O
- Crystal structure of poly[(μ2-2-(1H-1,2,4-triazol-1-yl)benzoato-κ4O:O′:N:N′)silver(I)] monohydrate, C9H8AgO3N3
- Crystal structure of poly[(μ2-9H-carbazole-3,6-dicarboxylate-κ4O1,O2:O3,O4)(μ2-1,3-di(pyridin-4-yl)propane-κ2N:N′)cadmium(II)]monohydrate, C27H23N3O5Cd
- The synthesis and crystal structure of bis(2-(benzo[d]thiazol-2-yl)-5-methylbenzen-1-ido-κ2C,N)-(N,N′-diethyldithiocarbamato-κ2S,S′)iridium(III), C33H30N3S4Ir
- The crystal structure of 5-amino-1-(2,6-dichloro-4-(trifluoromethyl)phenyl)-4-(trifluoromethylsulfonyl)-1H-pyrazole-3-carboxamide, C12H6N4Cl2F6O3S
- Synthesis and crystal structure of poly[(μ2-nitrato-κ4O,O′:O′,O′′)-nitrato-κO-(μ2-1,4-bis((1H-imidazol-1-yl)methyl)benzene-κ2N:N′)cadmium(II)], C14H14N6O6Cd
- Crystal structure of ethyl (Z)-(4-oxo-4-phenylbut-2-en-2-yl)glycinate, C14H17NO3
- Halogen bonds in the crystal structure of 5-bromo-3,4′-bipyridine – 1,4-diiodotetrafluorobenzene (2/1), C26H14Br2F4I2N4
- Crystal structure of bis(2,2′-bipyridine-κ2N,N′)-tetrakis(μ2-3-(phenylsulfonamido)propanoato-κ2O:O′)-bis(3-(phenylsulfonamido)propanoato-κ2O,O′)digadolinium(III) – 2,2′-bipyridine (1/1), C84H84Gd2N12O24S6
- Crystal structure of poly[aqua(μ2-2-amino-1,4-benzenedisulfonato-κ2O:O′)bis(μ2-pyrazin-κ2N:N′)silver(I)], C14H16Ag2N5O8S2
- The crystal structure of 1,6-di-tert-butyl-1,1,3,3,4,4,6,6-octamethyl-2,2,5,5-tetrakis (trimethylsilyl)hexasilane, C28H78Si10
- Crystal structure of discandium triruthenium tetrasilicide, Sc2Ru3Si4
- Crystal structure of poly[(μ2-4-amino-1,5-naphthalenedisulfonato-κ4O,N:O′, N′)bis(μ2-hexamethylenetetramino-κ2N;N′)silver(I)], {C22H30Ag2N9O6S2}n
- Crystal structure of diaqua[5,5′-dicarboxy-2,2′-(propane-1,3-diyl)bis(1H-imidazole-4-carboxylato-κ4O,O′,N,N′)]zinc(II) dihydrate, C13H18N4O12Zn
- The crystal structure of poly [(μ3-N1,N4-bis(pyridin-3-yl)cyclohexane-1,4-dicarboxamide-κ3-O:N:N′)-(p-toluenesulfonato-κ2O,O′)silver(I)], C25H27Ag1N4O5S
- The crystal structure of 1,2-bis(3-bromophenoxy) ethane, C14H12Br2O2
- The crystal structure of 4-(pyren-1-yl)butyl-3-nitrobenzoate, C27H21NO4
- Crystal structure of bis[(2-(4-chlorophenyl)-5-methyl-1,3-dioxane-5-carboxylato-κ1O) (5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane-κ4N,N′,N′′,N′′′)]nickel(II), C40H60Cl2N4NiO8
- The crystal structure of 1,5-dinitro-2,3,4-trichlorobenzene, C6H1Cl3N2O4
- The crystal structure of the solid solution of 3,5-dinitropyrazole and 4-chlorine-3,5-dinitropyrazole, C3H1.24Cl0.76N4O4
- The cocrystal structure of 4-nitropyrazole — acetic acid (1/1), C5H7N3O4
- The crystal structure of propan-2-one O-(2,4,6-trinitrophenyl) oxime, C9H8N4O7
- The crystal structure of ethyl 2-(3-(2-ethoxy-2-oxoethyl)benzo[d] thiazol-2(3H)-ylidene)acetate, C15H17NO4S
- Crystal structure of (acetic acid-κ1O)-bis(μ2-2-chlorobenzoato-κ2O:O′)-(2-chlorobenzoato-κ1O)-(μ2-hydroxy-κ2O:O)-bis(1,10-phenanthroline-κ2N,N′)dimanganese(II) — methanol (1/1), C48H37Cl3Mn2N4O10
- Crystal structure of 3-methyl-2-phenyl-1,8-naphthyridine, C15H12N2
- Crystal structure of chlorido-(5-acetyl-2-(5-methylpyridin-2-yl)benzen-1-ido-κ2C,N)-pyridine-κN-palladium(II), C19H17ClN2OPd
- Crystal structure of (4-methyl-benzoato-κ2O,O′)-(5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane-κ4N,N′,N′′,N′′′)nickel(II) perchlorate monohydrate, C24H45ClN4NiO7
- Crystal structure of (1,4,7,10,13,16-hexaoxacyclooctadecane-κ6O6) 1,2,3,4,5-pentamethyl-cyclopenta-2,4-dien-1-yl(potassium, rubidium) — ammonia (1/2), [K0.3Rb0.7(18-crown-6)]Cp*⋅2 NH3, C22H45K0.3N2O6Rb0.7
- Crystal structure of (3E,5E)-1-((4-fluorophenyl)sulfonyl)-3,5-bis(3-nitrobenzylidene)piperidin-4-one — dichloromethane (2/1), C51H38Cl2F2N6O14S2
- Crystal structure of (E)-N′-((1,6-dihydropyren-1-yl)methylene)isonicotinohydrazide — methanol (1/1), C24H19N3O2
- Crystal structure of poly[aqua(μ2-2-amino-1,4-benzenedisulfonato-κ3N,O:O′)-(μ4-hexamethylenetetramino-κ4N:N′:N′′:N′′′)disilver(I)] monohydrate, C12H21Ag2N5O8S2
- Crystal structure of bis(acridin-10-ium) 2,5-dihydroxyterephthalate — 2,5-dihydroxyterephthalic acid (1/1), C21H15NO6
- The crystal structure of 1,12-diazaperylene, C18H10N2
- Crystal structure of 1-(5-(4-chlorophenyl)-3-(2-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)ethan-1-one, C17H14N2OFCl
- Crystal structure of (4aR,6aR,6bR,10S,12aR)-10-acetoxy-1,2,3,4, 4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a, 12b,13,14b-icosahydro-2,2,4a,6b,9,9,12a-heptamethylpicene-6a-carboxylic acid, C32H50O4
- The crystal structure of tetrachlorido-bis{1,3-bis(2,6-diisopropylphenyl)-1H-3λ4-imidazol-2-yl}-(μ2-pyrimidine-κ2N:N′)dipalladium(IV) — dichloromethane (1/2), C60H80Cl8N6Pd2
- The crystal structure of (E)-4-(7-methoxy-2-oxo-2H-chromen-8-yl)-2-methylbut-2-en-1-yl 4-nitrobenzoate, C22H19NO7
- Crystal structure of 3-methyl-N-(pyrimidin-5-ylmethyl)pyridin-2-amine, C11H12N4
- The crystal structure of 2,5-dichloroterephthalic acid dihydrate, C8H8Cl2O6
- The crystal structure of 2,4,6-tris[4-(1H-imidazol-1-yl)phenyl]-1,3,5-triazine — dimethylformamide (1/1), C33H28N10O
- Crystal structure of N-(adamantan-1-yl)-5-(dimethylamino)naphthalene-1-sulfonamide, C22H28N2O2S
- Crystal structure of poly[diaqua-(μ4-4-(3,5-dicarboxy-κ1O-phenoxy)phthalato-κ3O:O′:O′)cadmium(II)], C16H12CdO11
- Crystal structure of poly[diaqua-bis(μ2-3-((1H-imidazol-1-yl)methyl)benzoato-κ2N:O)manganese(II)], C22H22MnN4O6
- Crystal structure of 9-(3-phenoxyphenyl)-3,4,6,7,9,10-hexahydroacridine-1,8(2H,5H)-dione, C25H23NO3
- The crystal structure of poly[(μ3-2,4,6-tris[4-(1H-imidazol-1-yl)phenyl]-1,3,5-triazine-k3N:N′:N′′)-(nitrato-k2O,O)-(nitrato-k1O)zinc(II)] - N,N-dimethylacetamide (1/2), C38H39N13O8Zn
- Crystal structure of poly[(μ7-4-(3,5-dicarboxylatophenoxy)phthalato)-(1,10-phenanthroline-κ2N,N′)dizinc(II)], C28H14N2O9Zn2
- The crystal structure of methyl 2-(benzylamino)-5-(benzyloxy)benzoate, C22H21NO3
- Crystal structure of (1,4,8,11-tetraazacyclotetradecane)palladium(II) tetracyanoplatinate(II), C14H24N8PdPt
- Crystal structure of (pyridine-2-carboxylato-κ2N,O)-[2-(2-pyridyl)phenyl-κ2N,C1]palladium(II), C17H12N2O2Pd
- Crystal structure of (cyclohexane-1,4-diammonium) 4-[(4-carboxylatophenyl)disulfanyl]benzoate dimethylsulphoxide hydrate (1/1/1/1), [C6H16N2]2+[C14H8O4S2]2−⋅C2H6OS⋅H2O
- Crystal structure of the 2:1 co-crystal 2-[(2-carboxyphenyl)disulfanyl]benzoic acid – 3-bromobenzoic acid, 2(C14H10O4S2)⋅C7H5BrO2
- Crystal structure of chlorido-dimethyl-(phenylpiperazine-1-carbodithioato-κ2S,S′)tin(IV), C13H19ClN2S2Sn
- Crystal structure of (N-n-butyl, N-methyl-dithiocarbamato-κ2 S,S′)-chlorido-dimethyl-tin(IV), C8H18ClNS2Sn
- Crystal structure of (2,2′-bipyridyl)bis(4-bromobenzyl)dibromidotin(IV), C24H20Br4N2Sn
- Crystal structure of (2,2′-bipyridyl)bis(4-chlorobenzyl)dichloridotin(IV), C24H20Cl4N2Sn
- Crystal structure of N-methyl-N-phenyl(methylsulfanyl)carbothioamide, C9H11NS2
- Crystal structure of 4-phenylpiperazin-1-ium (4-phenylpiperazin-1-yl)carbothioylsulfanide, [C10H15N2][C11H13N2S2]
- Crystal structure of catena-{di-aqua-sodium [n-butyl(methyl)carbamothioyl]sulfanide}n, [C6H16NNaO2S2]n
- Crystal structure of (2-([1,1-bis(hydroxymethyl)-2-oxyethyl]iminomethyl)-5-(n-decyl)phenolato)-dimethyl-tin(IV), C23H39NO5Sn
- Crystal structure of 4-chloro-N′-[(1E)-(3-ethoxy-2-hydroxyphenyl)methylidene]benzohydrazide – a Z′ = 3 structure, C16H15ClN2O3
Artikel in diesem Heft
- Frontmatter
- Crystal structure of [aqua[2,2′-(1,2-phenylene)bis(1H-imidazole-4-carboxylato-5-carboxy)-κ4N3,N3′,O4,O4′] zinc(II)] monohydrate, C16H10N4O9Zn⋅H2O
- Crystal structure of ethyl 3-(4-methoxyphenyl)-5-methylcarbamoyl-1H-pyrazole-4-carboxylate, C15H17N3O4
- 7-(4-Fluorobenzylidene)-3-(4-fluorophenyl)-N-phenyl-3,3a,4,5,6,7-hexahydro-2H-indazole-2-carbothioamide–dimethylformamide (2/1), C27H23F2N3S, 0.5(C3H7NO)
- Crystal structure of 4,4′-(hydrazonomethylene)diphenol dihydrate, C13H16N2O4
- Crystal structure of 4-methoxyphenyl-3-phenylpropiolate, C16H12O3
- Crystal Structure of tris(tetrakis{1-vinyl-1H-imidazole-κN}copper(II)) bis[tri-μ2-bromido-tetrabromido-bis(1-vinyl-1H-imidazole-κN)tetracopper(I)], C80H96N32Cu11Br14
- Crystal structure of (E)-2-(3,6-bis(diethylamino)-9H-xanthen-9-yl)-N′-(quinoxalin-2-ylmethylene)benzohydrazide, C37H36N6O2
- Crystal structure of 4-(1-phenylimidazo[1,5-a]pyridin-3-yl)benzoic acid (C20H14N2O2)
- Crystal structure of 3-fluoro-3-methyl-1-((2-nitrophenyl)sulfonyl)-5,5-diphenylpiperidine, C24H23FN2O4S
- Crystal structure of dimethyl 3,12-dibenzyl-6,10-diphenyl-3,12-diazapentacyclo [6.3.1.02.7.04.11.05.9]-dodecane-7,11-dicarboxylate — acetone (2/1), C40H38N2O2 ⋅ 0.5C3H6O
- Crystal structure of poly[(μ2-2-(1H-1,2,4-triazol-1-yl)benzoato-κ4O:O′:N:N′)silver(I)] monohydrate, C9H8AgO3N3
- Crystal structure of poly[(μ2-9H-carbazole-3,6-dicarboxylate-κ4O1,O2:O3,O4)(μ2-1,3-di(pyridin-4-yl)propane-κ2N:N′)cadmium(II)]monohydrate, C27H23N3O5Cd
- The synthesis and crystal structure of bis(2-(benzo[d]thiazol-2-yl)-5-methylbenzen-1-ido-κ2C,N)-(N,N′-diethyldithiocarbamato-κ2S,S′)iridium(III), C33H30N3S4Ir
- The crystal structure of 5-amino-1-(2,6-dichloro-4-(trifluoromethyl)phenyl)-4-(trifluoromethylsulfonyl)-1H-pyrazole-3-carboxamide, C12H6N4Cl2F6O3S
- Synthesis and crystal structure of poly[(μ2-nitrato-κ4O,O′:O′,O′′)-nitrato-κO-(μ2-1,4-bis((1H-imidazol-1-yl)methyl)benzene-κ2N:N′)cadmium(II)], C14H14N6O6Cd
- Crystal structure of ethyl (Z)-(4-oxo-4-phenylbut-2-en-2-yl)glycinate, C14H17NO3
- Halogen bonds in the crystal structure of 5-bromo-3,4′-bipyridine – 1,4-diiodotetrafluorobenzene (2/1), C26H14Br2F4I2N4
- Crystal structure of bis(2,2′-bipyridine-κ2N,N′)-tetrakis(μ2-3-(phenylsulfonamido)propanoato-κ2O:O′)-bis(3-(phenylsulfonamido)propanoato-κ2O,O′)digadolinium(III) – 2,2′-bipyridine (1/1), C84H84Gd2N12O24S6
- Crystal structure of poly[aqua(μ2-2-amino-1,4-benzenedisulfonato-κ2O:O′)bis(μ2-pyrazin-κ2N:N′)silver(I)], C14H16Ag2N5O8S2
- The crystal structure of 1,6-di-tert-butyl-1,1,3,3,4,4,6,6-octamethyl-2,2,5,5-tetrakis (trimethylsilyl)hexasilane, C28H78Si10
- Crystal structure of discandium triruthenium tetrasilicide, Sc2Ru3Si4
- Crystal structure of poly[(μ2-4-amino-1,5-naphthalenedisulfonato-κ4O,N:O′, N′)bis(μ2-hexamethylenetetramino-κ2N;N′)silver(I)], {C22H30Ag2N9O6S2}n
- Crystal structure of diaqua[5,5′-dicarboxy-2,2′-(propane-1,3-diyl)bis(1H-imidazole-4-carboxylato-κ4O,O′,N,N′)]zinc(II) dihydrate, C13H18N4O12Zn
- The crystal structure of poly [(μ3-N1,N4-bis(pyridin-3-yl)cyclohexane-1,4-dicarboxamide-κ3-O:N:N′)-(p-toluenesulfonato-κ2O,O′)silver(I)], C25H27Ag1N4O5S
- The crystal structure of 1,2-bis(3-bromophenoxy) ethane, C14H12Br2O2
- The crystal structure of 4-(pyren-1-yl)butyl-3-nitrobenzoate, C27H21NO4
- Crystal structure of bis[(2-(4-chlorophenyl)-5-methyl-1,3-dioxane-5-carboxylato-κ1O) (5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane-κ4N,N′,N′′,N′′′)]nickel(II), C40H60Cl2N4NiO8
- The crystal structure of 1,5-dinitro-2,3,4-trichlorobenzene, C6H1Cl3N2O4
- The crystal structure of the solid solution of 3,5-dinitropyrazole and 4-chlorine-3,5-dinitropyrazole, C3H1.24Cl0.76N4O4
- The cocrystal structure of 4-nitropyrazole — acetic acid (1/1), C5H7N3O4
- The crystal structure of propan-2-one O-(2,4,6-trinitrophenyl) oxime, C9H8N4O7
- The crystal structure of ethyl 2-(3-(2-ethoxy-2-oxoethyl)benzo[d] thiazol-2(3H)-ylidene)acetate, C15H17NO4S
- Crystal structure of (acetic acid-κ1O)-bis(μ2-2-chlorobenzoato-κ2O:O′)-(2-chlorobenzoato-κ1O)-(μ2-hydroxy-κ2O:O)-bis(1,10-phenanthroline-κ2N,N′)dimanganese(II) — methanol (1/1), C48H37Cl3Mn2N4O10
- Crystal structure of 3-methyl-2-phenyl-1,8-naphthyridine, C15H12N2
- Crystal structure of chlorido-(5-acetyl-2-(5-methylpyridin-2-yl)benzen-1-ido-κ2C,N)-pyridine-κN-palladium(II), C19H17ClN2OPd
- Crystal structure of (4-methyl-benzoato-κ2O,O′)-(5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane-κ4N,N′,N′′,N′′′)nickel(II) perchlorate monohydrate, C24H45ClN4NiO7
- Crystal structure of (1,4,7,10,13,16-hexaoxacyclooctadecane-κ6O6) 1,2,3,4,5-pentamethyl-cyclopenta-2,4-dien-1-yl(potassium, rubidium) — ammonia (1/2), [K0.3Rb0.7(18-crown-6)]Cp*⋅2 NH3, C22H45K0.3N2O6Rb0.7
- Crystal structure of (3E,5E)-1-((4-fluorophenyl)sulfonyl)-3,5-bis(3-nitrobenzylidene)piperidin-4-one — dichloromethane (2/1), C51H38Cl2F2N6O14S2
- Crystal structure of (E)-N′-((1,6-dihydropyren-1-yl)methylene)isonicotinohydrazide — methanol (1/1), C24H19N3O2
- Crystal structure of poly[aqua(μ2-2-amino-1,4-benzenedisulfonato-κ3N,O:O′)-(μ4-hexamethylenetetramino-κ4N:N′:N′′:N′′′)disilver(I)] monohydrate, C12H21Ag2N5O8S2
- Crystal structure of bis(acridin-10-ium) 2,5-dihydroxyterephthalate — 2,5-dihydroxyterephthalic acid (1/1), C21H15NO6
- The crystal structure of 1,12-diazaperylene, C18H10N2
- Crystal structure of 1-(5-(4-chlorophenyl)-3-(2-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)ethan-1-one, C17H14N2OFCl
- Crystal structure of (4aR,6aR,6bR,10S,12aR)-10-acetoxy-1,2,3,4, 4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a, 12b,13,14b-icosahydro-2,2,4a,6b,9,9,12a-heptamethylpicene-6a-carboxylic acid, C32H50O4
- The crystal structure of tetrachlorido-bis{1,3-bis(2,6-diisopropylphenyl)-1H-3λ4-imidazol-2-yl}-(μ2-pyrimidine-κ2N:N′)dipalladium(IV) — dichloromethane (1/2), C60H80Cl8N6Pd2
- The crystal structure of (E)-4-(7-methoxy-2-oxo-2H-chromen-8-yl)-2-methylbut-2-en-1-yl 4-nitrobenzoate, C22H19NO7
- Crystal structure of 3-methyl-N-(pyrimidin-5-ylmethyl)pyridin-2-amine, C11H12N4
- The crystal structure of 2,5-dichloroterephthalic acid dihydrate, C8H8Cl2O6
- The crystal structure of 2,4,6-tris[4-(1H-imidazol-1-yl)phenyl]-1,3,5-triazine — dimethylformamide (1/1), C33H28N10O
- Crystal structure of N-(adamantan-1-yl)-5-(dimethylamino)naphthalene-1-sulfonamide, C22H28N2O2S
- Crystal structure of poly[diaqua-(μ4-4-(3,5-dicarboxy-κ1O-phenoxy)phthalato-κ3O:O′:O′)cadmium(II)], C16H12CdO11
- Crystal structure of poly[diaqua-bis(μ2-3-((1H-imidazol-1-yl)methyl)benzoato-κ2N:O)manganese(II)], C22H22MnN4O6
- Crystal structure of 9-(3-phenoxyphenyl)-3,4,6,7,9,10-hexahydroacridine-1,8(2H,5H)-dione, C25H23NO3
- The crystal structure of poly[(μ3-2,4,6-tris[4-(1H-imidazol-1-yl)phenyl]-1,3,5-triazine-k3N:N′:N′′)-(nitrato-k2O,O)-(nitrato-k1O)zinc(II)] - N,N-dimethylacetamide (1/2), C38H39N13O8Zn
- Crystal structure of poly[(μ7-4-(3,5-dicarboxylatophenoxy)phthalato)-(1,10-phenanthroline-κ2N,N′)dizinc(II)], C28H14N2O9Zn2
- The crystal structure of methyl 2-(benzylamino)-5-(benzyloxy)benzoate, C22H21NO3
- Crystal structure of (1,4,8,11-tetraazacyclotetradecane)palladium(II) tetracyanoplatinate(II), C14H24N8PdPt
- Crystal structure of (pyridine-2-carboxylato-κ2N,O)-[2-(2-pyridyl)phenyl-κ2N,C1]palladium(II), C17H12N2O2Pd
- Crystal structure of (cyclohexane-1,4-diammonium) 4-[(4-carboxylatophenyl)disulfanyl]benzoate dimethylsulphoxide hydrate (1/1/1/1), [C6H16N2]2+[C14H8O4S2]2−⋅C2H6OS⋅H2O
- Crystal structure of the 2:1 co-crystal 2-[(2-carboxyphenyl)disulfanyl]benzoic acid – 3-bromobenzoic acid, 2(C14H10O4S2)⋅C7H5BrO2
- Crystal structure of chlorido-dimethyl-(phenylpiperazine-1-carbodithioato-κ2S,S′)tin(IV), C13H19ClN2S2Sn
- Crystal structure of (N-n-butyl, N-methyl-dithiocarbamato-κ2 S,S′)-chlorido-dimethyl-tin(IV), C8H18ClNS2Sn
- Crystal structure of (2,2′-bipyridyl)bis(4-bromobenzyl)dibromidotin(IV), C24H20Br4N2Sn
- Crystal structure of (2,2′-bipyridyl)bis(4-chlorobenzyl)dichloridotin(IV), C24H20Cl4N2Sn
- Crystal structure of N-methyl-N-phenyl(methylsulfanyl)carbothioamide, C9H11NS2
- Crystal structure of 4-phenylpiperazin-1-ium (4-phenylpiperazin-1-yl)carbothioylsulfanide, [C10H15N2][C11H13N2S2]
- Crystal structure of catena-{di-aqua-sodium [n-butyl(methyl)carbamothioyl]sulfanide}n, [C6H16NNaO2S2]n
- Crystal structure of (2-([1,1-bis(hydroxymethyl)-2-oxyethyl]iminomethyl)-5-(n-decyl)phenolato)-dimethyl-tin(IV), C23H39NO5Sn
- Crystal structure of 4-chloro-N′-[(1E)-(3-ethoxy-2-hydroxyphenyl)methylidene]benzohydrazide – a Z′ = 3 structure, C16H15ClN2O3


