Abstract
C17H17N5, monoclinic, P21/c (no. 14), a = 9.4347(9) Å, b = 15.6392(15) Å, c = 11.0939(11) Å, β =
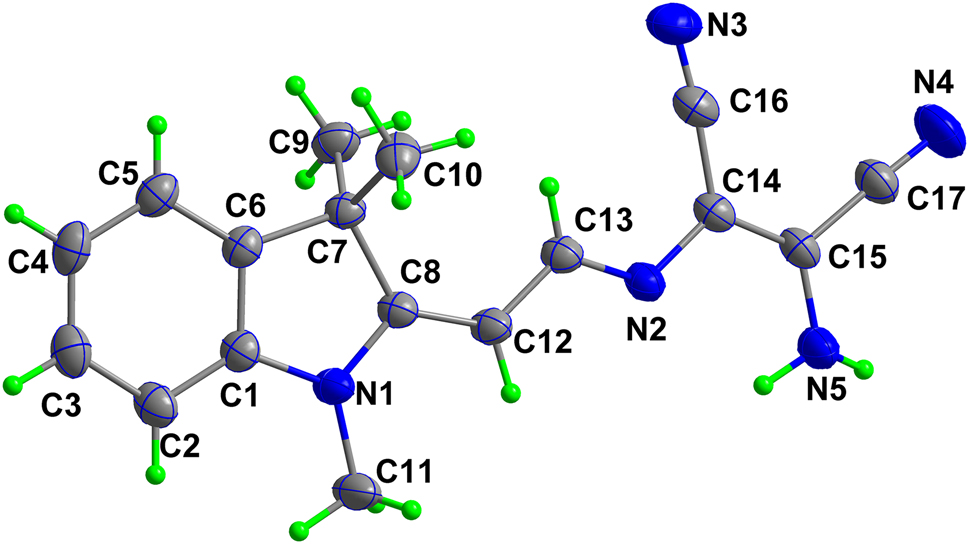
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Red rod |
| Size: | 0.25 × 0.12 × 0.10 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.08 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θ max, completeness: | 25.0°, >99% |
| N(hkl)measured , N(hkl)unique, R int: | 8087, 2837, 0.023 |
| Criterion for I obs, N(hkl)gt: | I obs > 2σ(I obs), 2179 |
| N(param)refined: | 214 |
| Programs: | SHELX [1], Bruker [2] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| N1 | 0.45302 (14) | −0.05812 (7) | 0.22928 (11) | 0.0542 (3) |
| N2 | 0.82583 (13) | 0.02201 (8) | 0.55349 (10) | 0.0508 (3) |
| N3 | 0.93074 (19) | −0.13704 (11) | 0.76901 (16) | 0.0837 (5) |
| N4 | 1.1906 (2) | 0.06644 (13) | 0.90620 (16) | 0.1033 (6) |
| N5 | 0.99322 (17) | 0.15520 (10) | 0.65207 (14) | 0.0648 (4) |
| H5A | 0.915 (2) | 0.1593 (12) | 0.5930 (18) | 0.078 (6)* |
| H5B | 1.024 (2) | 0.2030 (14) | 0.6964(18) | 0.091 (7)* |
| C1 | 0.35602 (16) | −0.12407 (9) | 0.18802 (13) | 0.0505(4) |
| C2 | 0.24188 (18) | −0.12403 (11) | 0.09088 (15) | 0.0617 (4) |
| H2 | 0.2211 | −0.0763 | 0.0408 | 0.074* |
| C3 | 0.15956 (19) | −0.19771 (12) | 0.07087 (16) | 0.0681 (5) |
| H3 | 0.0817 | −0.1995 | 0.0065 | 0.082* |
| C4 | 0.19108 (19) | −0.26816 (12) | 0.14460 (16) | 0.0670(5) |
| H4 | 0.1350 | −0.3172 | 0.1290 | 0.080* |
| C5 | 0.30601(18) | −0.26709(10) | 0.24244(15) | 0.0582(4) |
| H5 | 0.3262 | −0.3148 | 0.2926 | 0.070* |
| C6 | 0.38949 (16) | −0.19447 (9) | 0.26406 (13) | 0.0477 (4) |
| C7 | 0.51933 (16) | −0.17412 (9) | 0.36154(13) | 0.0466 (4) |
| C8 | 0.54813 (16) | −0.08085 (9) | 0.33193 (12) | 0.0458 (4) |
| C9 | 0.4809 (2) | −0.18485 (11) | 0.48919 (14) | 0.0656 (5) |
| H9A | 0.4030 | −0.1471 | 0.4976 | 0.098* |
| H9B | 0.5633 | −0.1713 | 0.5503 | 0.098* |
| H9C | 0.4523 | −0.2429 | 0.4997 | 0.098* |
| C10 | 0.64776 (18) | −0.23082 (11) | 0.34572 (18) | 0.0686 (5) |
| H10A | 0.6249 | −0.2896 | 0.3585 | 0.103* |
| H10B | 0.7308 | −0.2144 | 0.4044 | 0.103* |
| H10C | 0.6678 | −0.2239 | 0.2644 | 0.103* |
| C11 | 0.4462 (2) | 0.02617 (11) | 0.17427 (17) | 0.0765 (6) |
| H11A | 0.3665 | 0.0286 | 0.1072 | 0.111 (7)* |
| H11B | 0.5341 | 0.0374 | 0.1448 | 0.118 (9)* |
| H11C | 0.4334 | 0.0683 | 0.2344 | 0.129 (9)* |
| C12 | 0.64775 (16) | −0.02544 (9) | 0.39179 (13) | 0.0514 (4) |
| H12 | 0.6512 | 0.0282 | 0.3562 | 0.062* |
| C13 | 0.74614 (16) | −0.03963 (9) | 0.50105 (13) | 0.0498 (4) |
| H13 | 0.7540 | −0.0939 | 0.5360 | 0.060* |
| C14 | 0.92156 (16) | 0.00967 (10) | 0.66198 (13) | 0.0494 (4) |
| C15 | 1.00174 (16) | 0.07803 (10) | 0.70943 (13) | 0.0521 (4) |
| C16 | 0.93162 (17) | −0.07085 (11) | 0.72491(15) | 0.0588 (4) |
| C17 | 1.10605 (19) | 0.07038 (12) | 0.81975 (16) | 0.0674 (5) |
1 Source of material
The starting materials 1,3,3-trimethyl-2-(formylmethylene)indoline (2.01 g, 10 mmol) and diaminomaleonitrile (1.08 g, 10 mmol) in an ethanol solution (100 mL) were stirred for 5 h under refluxing. The formed red powder was filtered, and the mother liquid was left in air for a few days, yielding red rod crystals.
2 Experimental details
The structure was solved by Direct Methods and refined with the SHELX crystallographic software package [1]. The hydrogen atoms were placed at calculated positions and refined as riding atoms with isotropic displacement parameters.
3 Comment
Schiff bases have been extensively studied due to their excellent coordinative abilities, biological activities, and fluorescence properties, etc. [3]. Particularly, diaminomaleonitrile-based Schiff bases have been developed as fluorescent probes for the detection of Cu2+ [4] or ClO− [5]. The title compound, derived from 1,3,3-trimethyl-2-(formylmethylene)indoline and diaminomaleonitrile, could act as a fluorescence turn-on probe for detecting Cu2+ [6].
In the title structure, the bond length of C13–N2 is 1.2953(18) Å (see the Figure), indicating the Schiff base formation. Bond lengths and angles are all in the expected ranges [7]. In the solid state, intermolecular N–H···N hydrogen bonds link the molecules into a chain along the b axis, which are further connected with each other by two types of intermolecular C–H···N hydrogen bonds to form a two-dimensional wave-like supermolecular network.
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: None declared.
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122; https://doi.org/10.1107/s0108767307043930.Search in Google Scholar PubMed
2. Bruker. SMART and SAINT; Bruker AXS Inc.: Madison, Wisconsin, (USA), 2007.Search in Google Scholar
3. Dong, W.-K., Li, X.-L., Wang, L., Zhang, Y., Ding, Y.-J. A new application of Salamo-type bisoximes: as a relay–sensor for Zn2+/Cu2+ and its novel complexes for successive sensing of H+/OH−. Sens. Actuators B 2016, 229, 370–378; https://doi.org/10.1016/j.snb.2016.01.139.Search in Google Scholar
4. Xi, W., Gong, Y., Mei, B., Zhang, X., Zhang, Y., Chen, B., Wu, J., Tian, Y., Zhou, H. Schiff base derivatives based on diaminomaleonitrile: colorimetric and fluorescent recognition of Cu(II), cell imaging application, polymorph-dependent fluorescence and aggregation-enhanced emission. Sens. Actuators B 2014, 205, 158–167; https://doi.org/10.1016/j.snb.2014.08.068.Search in Google Scholar
5. Jiang, Y., Wu, S., Jin, C., Wang, B., Shen, J. Novel diaminomaleonitrile-based fluorescent probe for ratiometric detection and bioimaging of hypochlorite. Sens. Actuators B 2018, 265, 365–370; https://doi.org/10.1016/j.snb.2018.01.202.Search in Google Scholar
6. Xue, X.-Y., Jiang, D.-Y., Feng, C., Zhang, H., Wang, Z.-F., Zhao, H. A highly selective and sensitive turn-on fluorescent probe for copper(II) ion with a large Stokes shift. Inorg. Chem. Commun. 2017, 86, 258–261; https://doi.org/10.1016/j.inoche.2017.10.019.Search in Google Scholar
7. Kim, S.-H., Matsumoto, S. Electrochromism and X-ray crystal structure of a new azomethine dye derived from diaminomaleonitrile. Dyes Pigments 2007, 72, 406–408; https://doi.org/10.1016/j.dyepig.2005.09.023.Search in Google Scholar
© 2023 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of phenyl(3,3-dichloro-1,3-dihydro-2H-pyrrolo[2,3-b]pyridin-2-one)methanone, C7H4Cl2N2O
- Crystal structure of poly[diaqua-bis(μ 2-1,4-diaminobutane-N:N′)cobalt(II)] dichloride, C8H28Cl2CoN4O2
- Synthesis and crystal structure of (4aR,7S)-7-hydroxy-7-isopropyl-1,1-dimethyldecahydro-2H,6H-8a,4a-(epoxymethano)phenanthren-12-one, C20H32O3
- The crystal structure of 1-(2-chlorobenzyl)-3-(3,5-dichlorophenyl)urea, C14H11Cl3N2O
- Crystal structure of tetrapropylammonium-1,3,5-thiadiazole-5-amido-2-carbamate – 1,2,4-thiadiazole-3,5-diamine – water (1/1/1), C17H37N9O3S2
- Tetrabutylammonium 1,3,5-thiadiazole-5-amido-2-carbamate—1,2,4-thiadiazole-3,5-diamine— water (1/1/1), C21H45N9O3S2
- The crystal structure of ((E)-2,4-dichloro-6-(((2-hydroxy-5-nitrophenyl)imino)methyl)phenolato-κ 3 N,O,O′)tris(pyridine-κN)manganese(II), C28H21Cl2MnN5O4
- The crystal structure of aqua-bis{2-bromo-6-((2-(2-phenylacetyl)hydrazineylidene)methyl)phenolato-κ3 N,O,O′}-dimethylformamide-κ1 O-erbium(III) chloride – dimethylformamide – water (1/2/1), C39H49N7O9Br2ClEr
- Crystal structure of (diaqua-bis(phenanthroline-K 2 N,N′)-tetrakis(m 2-3,4,5,6-tetrafluorophthalato-K 4 O,O:O′:O″;K 2 O:O′)dierbium (III) phenanthroline (1/2), C80H38Er2F16N8O18
- Crystal structure of (E)-7-methoxy-2-(4-methoxy-2-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C20H17F3O3
- The crystal structure of 4–(4,4,5,5–tetramethyl–1,3,2–dioxaborolan–2–yl)morpholine, C10H20BNO3
- The crystal structure of catena–poly[aqua(1-naphthoato-κ 2 O,O′)-(μ-1-naphthoato-κ 4 O:O,O′:O′)lead(II)], C22H16O5Pb
- The crystal structure of 1-(4-chlorophenyl)-3-cycloheptylurea, C14H19ClN2O
- The crystal structure of 4-(pyren-1-yl)butyl-4-nitrobenzoate, C27H21NO4
- Crystal structure of cyclo-(bis(µ2-3,3′-(1H-imidazole-3-ium-1,3-diyl)dipropionato-κ4 O,O′:O″,O″′)-dinitrato-κ2 O,O′-tetraoxido-diuranium(VI) C18H22N6O18U2
- The crystal structure of catena-[nitrato-κ 2 O,O′-(μ 3-3-iodobenzene-1,2-dicarboxylato-κ 4 O:O′:O″,O‴)-(2,2′:6′,2″-terpyridine-κ 3 N,N′,N″)lanthanum(III)], C23H14IN4O7La
- Redetermination of crystal structure of [bis(pyridin-2-ylmethyl)amine-κ 3 N,Nʹ,Nʺ]chloridopalladium(II) chloride monohydrate
- Crystal structure of catena-poly[triaqua-[bis(m2-4-(1H-1,2,4-triazol-1-yl)benzoato-k2O:O')-(4-(1H-1,2,4-triazol-1-yl)benzoato-k1O)-praseodymium (III) monohydrate], C27H26N9O10Pr
- Crystal structure of trans-diaqua-bis(methyl methylcarbamohydrazonothioato-κ2 N,N′) nickel(II) iodide semihydrate, C6H22N6O2NiS2I2·0.5H2O
- The crystal structure of 2-(2-fluoro-4-methyl-5-((2,2,2-trifluoroethyl)thio)phenyl)isoindolin-1-one, C17H13F4NOS
- The crystal structure of di-μ-1-naphthylacetato-κ 3 O,O′:O;κ 3 O:O,O′-bis[(1-naphthylacetato-κ 2 O,O′)(2,2′-bipyridine-κ 2 N,N′)lead(II)] monohydrate, C68H54N4O9Pb2
- Crystal structure of tetrapropylammonium guanidinium 4,4′-sulfonyldibenzoate monohydrate, C27H44N4O7S
- Crystal structure of bis(tetrapropylammonium) terephthalate – 1-(diaminomethylene)thiourea – water (1/2/4) C18H40N5O4S
- Crystal structure of (E)-7-fluoro-2-(4-morpholinobenzylidene)-3,4-dihydronaphthalen-1(2H)-one, C21H20FNO2
- The crystal structure of poly[diaqua-bis(μ 3-5-bromobenzene-1,3-dicarboxylato-κ 3 O,O,O′)-(μ 2-1,3-bis-(4-pyridyl)-propane-κ 2 N,N′)-dizinc(II))] – 5-bromobenzene-1,3-dicarboxylic acid [2/1], C37H29Br3N2O14Zn2
- The crystal structure of 2-bromo-1,3-phenylene bis(4-methylbenzenesulfonate), C20H17BrO6S2
- Crystal structure of europium dichromium icosaaluminum, EuCr2Al20
- The crystal structure of N′1,N′3-di((E)-benzylidene) isophthalohydrazide dihydrate, C 22 H 22 N 4 O 4
- Crystal structure of 7α,11α-dihydroxy-15-oxo-ent-kauran-16-en-19,6β-olide, C20H26O5
- Crystal structure of 4-chloro-N′-[(1E)-pyridin-3-ylmethylidene]benzohydrazide, C13H10ClN3O
- The crystal structure of (Z)-3-(1-(2-((E)-4-isopropylbenzylidene)hydrazinyl)ethylidene) chroman-2,4-dione, C21H20N2O3
- Crystal structure of E-7-fluoro-2-(2-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C18H12F4O
- Crystal structure of bis(6-aminopyridine-2-carboxylato–k2O,N)-bis(N,N-dimethylformamide-k1 O)zinc(II), C18H24N6O6Zn
- Crystal structure of 5-(adamantan-1-yl)-3-[(4-{[2-(trifluoromethyl)phenyl]-methyl}piperazin-1-yl)methyl]-1,3,4-oxadiazole-2(3H)-thione, C25H31F3N4OS
- Crystal structure of tetrapropylammonium bicarbonate–1-(diaminomethylene)thiourea – water (2/2/1), C30H72N10O7S2
- Crystal structure of tris(2,2′-bipyridine-κ2 N,N′)iron(II) triiodide – dichloromethane (2/1), C61H50Cl2Fe2I12N12
- Crystal structure of 2-amino-3-[2-(1,3,3-trimethyl-1,3-dihydro-indol-2-ylidene)-ethylideneamino]-but-2-enedinitrile, C17H17N5
- The crystal structure of 1-(2-chlorophenyl)-3-cycloheptylurea, C14H19ClN2O
- Crystal structure of potassium bis(pentaselenido-κ 2 Se 1,Se 5)palladate(II), K2[Pd(Se5)2]
- The crystal structure of 5,10-bis(2-methoxyethyl)-5,10-dihydro-[1,2,3,4]tetrathiocino[5,6-b:8, 7-b′]diindole, C22H22N2O2S4
- The crystal structure of 4-(4-iodophenyl)-5H-1,2,3-dithiazole-5-thione, C8H4INS3
- Crystal structure of bis{μ2-(4-acetyl-phenoxy)acetato-κ2 O:O′}-bis{μ2-(4-acetyl-phenoxy)acetato-κ3 O,O′:O)- bis{(4-acetyl-phenoxy)acetato-κ2 O,O′}-bis(phenanthrolin-κ2 N,N′)didysprosium(III) tetrahydrate, C84H78N4O28Dy2
- Crystal structure of Eu2Pd3.37(1)Zn13.63(1)
- Crystal structure of 2-methoxy-4-(methoxy-carbonyl)phenyl 2-chloro-4-fluorobenzoate, C16H12ClFO5
- Crystal structure of catena-poly[bis(μ2-dicyanamide-κ2 N:N′)-bis(4-vinylpyridine-κN)-copper(II)], C18H14CuN8
- The crystal structure of iguratimod-dimethylformamide (1/1), C17H14N2O6S·C3H7NO
- Synthesis and crystal structure of 1-((3R,10S,13S,17S)-10,13-dimethyl-3-(m-tolylamino)hexadecahydro-1H-cyclopenta[α]phenanthren-17-yl)ethan-1-one, C28H41NO
- The crystal structure of diaqua-bis(4-bromo-2-formylphenoxy)zinc(II), C14H12Br2O6Zn
- The crystal structure of tetra(1-ethylimidazole-κ 1 N)-[μ 4-imidazole-4,5-dicarboxylato-κ 4 O, N, O′, N′]-trioxido-divanadium, C25H33N10O7V2
- The crystal structure of (E)-N′-(1-(4-fluorophenyl)propylidene)-2-hydroxybenzohydrazide, C16H15FN2O2
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of phenyl(3,3-dichloro-1,3-dihydro-2H-pyrrolo[2,3-b]pyridin-2-one)methanone, C7H4Cl2N2O
- Crystal structure of poly[diaqua-bis(μ 2-1,4-diaminobutane-N:N′)cobalt(II)] dichloride, C8H28Cl2CoN4O2
- Synthesis and crystal structure of (4aR,7S)-7-hydroxy-7-isopropyl-1,1-dimethyldecahydro-2H,6H-8a,4a-(epoxymethano)phenanthren-12-one, C20H32O3
- The crystal structure of 1-(2-chlorobenzyl)-3-(3,5-dichlorophenyl)urea, C14H11Cl3N2O
- Crystal structure of tetrapropylammonium-1,3,5-thiadiazole-5-amido-2-carbamate – 1,2,4-thiadiazole-3,5-diamine – water (1/1/1), C17H37N9O3S2
- Tetrabutylammonium 1,3,5-thiadiazole-5-amido-2-carbamate—1,2,4-thiadiazole-3,5-diamine— water (1/1/1), C21H45N9O3S2
- The crystal structure of ((E)-2,4-dichloro-6-(((2-hydroxy-5-nitrophenyl)imino)methyl)phenolato-κ 3 N,O,O′)tris(pyridine-κN)manganese(II), C28H21Cl2MnN5O4
- The crystal structure of aqua-bis{2-bromo-6-((2-(2-phenylacetyl)hydrazineylidene)methyl)phenolato-κ3 N,O,O′}-dimethylformamide-κ1 O-erbium(III) chloride – dimethylformamide – water (1/2/1), C39H49N7O9Br2ClEr
- Crystal structure of (diaqua-bis(phenanthroline-K 2 N,N′)-tetrakis(m 2-3,4,5,6-tetrafluorophthalato-K 4 O,O:O′:O″;K 2 O:O′)dierbium (III) phenanthroline (1/2), C80H38Er2F16N8O18
- Crystal structure of (E)-7-methoxy-2-(4-methoxy-2-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C20H17F3O3
- The crystal structure of 4–(4,4,5,5–tetramethyl–1,3,2–dioxaborolan–2–yl)morpholine, C10H20BNO3
- The crystal structure of catena–poly[aqua(1-naphthoato-κ 2 O,O′)-(μ-1-naphthoato-κ 4 O:O,O′:O′)lead(II)], C22H16O5Pb
- The crystal structure of 1-(4-chlorophenyl)-3-cycloheptylurea, C14H19ClN2O
- The crystal structure of 4-(pyren-1-yl)butyl-4-nitrobenzoate, C27H21NO4
- Crystal structure of cyclo-(bis(µ2-3,3′-(1H-imidazole-3-ium-1,3-diyl)dipropionato-κ4 O,O′:O″,O″′)-dinitrato-κ2 O,O′-tetraoxido-diuranium(VI) C18H22N6O18U2
- The crystal structure of catena-[nitrato-κ 2 O,O′-(μ 3-3-iodobenzene-1,2-dicarboxylato-κ 4 O:O′:O″,O‴)-(2,2′:6′,2″-terpyridine-κ 3 N,N′,N″)lanthanum(III)], C23H14IN4O7La
- Redetermination of crystal structure of [bis(pyridin-2-ylmethyl)amine-κ 3 N,Nʹ,Nʺ]chloridopalladium(II) chloride monohydrate
- Crystal structure of catena-poly[triaqua-[bis(m2-4-(1H-1,2,4-triazol-1-yl)benzoato-k2O:O')-(4-(1H-1,2,4-triazol-1-yl)benzoato-k1O)-praseodymium (III) monohydrate], C27H26N9O10Pr
- Crystal structure of trans-diaqua-bis(methyl methylcarbamohydrazonothioato-κ2 N,N′) nickel(II) iodide semihydrate, C6H22N6O2NiS2I2·0.5H2O
- The crystal structure of 2-(2-fluoro-4-methyl-5-((2,2,2-trifluoroethyl)thio)phenyl)isoindolin-1-one, C17H13F4NOS
- The crystal structure of di-μ-1-naphthylacetato-κ 3 O,O′:O;κ 3 O:O,O′-bis[(1-naphthylacetato-κ 2 O,O′)(2,2′-bipyridine-κ 2 N,N′)lead(II)] monohydrate, C68H54N4O9Pb2
- Crystal structure of tetrapropylammonium guanidinium 4,4′-sulfonyldibenzoate monohydrate, C27H44N4O7S
- Crystal structure of bis(tetrapropylammonium) terephthalate – 1-(diaminomethylene)thiourea – water (1/2/4) C18H40N5O4S
- Crystal structure of (E)-7-fluoro-2-(4-morpholinobenzylidene)-3,4-dihydronaphthalen-1(2H)-one, C21H20FNO2
- The crystal structure of poly[diaqua-bis(μ 3-5-bromobenzene-1,3-dicarboxylato-κ 3 O,O,O′)-(μ 2-1,3-bis-(4-pyridyl)-propane-κ 2 N,N′)-dizinc(II))] – 5-bromobenzene-1,3-dicarboxylic acid [2/1], C37H29Br3N2O14Zn2
- The crystal structure of 2-bromo-1,3-phenylene bis(4-methylbenzenesulfonate), C20H17BrO6S2
- Crystal structure of europium dichromium icosaaluminum, EuCr2Al20
- The crystal structure of N′1,N′3-di((E)-benzylidene) isophthalohydrazide dihydrate, C 22 H 22 N 4 O 4
- Crystal structure of 7α,11α-dihydroxy-15-oxo-ent-kauran-16-en-19,6β-olide, C20H26O5
- Crystal structure of 4-chloro-N′-[(1E)-pyridin-3-ylmethylidene]benzohydrazide, C13H10ClN3O
- The crystal structure of (Z)-3-(1-(2-((E)-4-isopropylbenzylidene)hydrazinyl)ethylidene) chroman-2,4-dione, C21H20N2O3
- Crystal structure of E-7-fluoro-2-(2-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C18H12F4O
- Crystal structure of bis(6-aminopyridine-2-carboxylato–k2O,N)-bis(N,N-dimethylformamide-k1 O)zinc(II), C18H24N6O6Zn
- Crystal structure of 5-(adamantan-1-yl)-3-[(4-{[2-(trifluoromethyl)phenyl]-methyl}piperazin-1-yl)methyl]-1,3,4-oxadiazole-2(3H)-thione, C25H31F3N4OS
- Crystal structure of tetrapropylammonium bicarbonate–1-(diaminomethylene)thiourea – water (2/2/1), C30H72N10O7S2
- Crystal structure of tris(2,2′-bipyridine-κ2 N,N′)iron(II) triiodide – dichloromethane (2/1), C61H50Cl2Fe2I12N12
- Crystal structure of 2-amino-3-[2-(1,3,3-trimethyl-1,3-dihydro-indol-2-ylidene)-ethylideneamino]-but-2-enedinitrile, C17H17N5
- The crystal structure of 1-(2-chlorophenyl)-3-cycloheptylurea, C14H19ClN2O
- Crystal structure of potassium bis(pentaselenido-κ 2 Se 1,Se 5)palladate(II), K2[Pd(Se5)2]
- The crystal structure of 5,10-bis(2-methoxyethyl)-5,10-dihydro-[1,2,3,4]tetrathiocino[5,6-b:8, 7-b′]diindole, C22H22N2O2S4
- The crystal structure of 4-(4-iodophenyl)-5H-1,2,3-dithiazole-5-thione, C8H4INS3
- Crystal structure of bis{μ2-(4-acetyl-phenoxy)acetato-κ2 O:O′}-bis{μ2-(4-acetyl-phenoxy)acetato-κ3 O,O′:O)- bis{(4-acetyl-phenoxy)acetato-κ2 O,O′}-bis(phenanthrolin-κ2 N,N′)didysprosium(III) tetrahydrate, C84H78N4O28Dy2
- Crystal structure of Eu2Pd3.37(1)Zn13.63(1)
- Crystal structure of 2-methoxy-4-(methoxy-carbonyl)phenyl 2-chloro-4-fluorobenzoate, C16H12ClFO5
- Crystal structure of catena-poly[bis(μ2-dicyanamide-κ2 N:N′)-bis(4-vinylpyridine-κN)-copper(II)], C18H14CuN8
- The crystal structure of iguratimod-dimethylformamide (1/1), C17H14N2O6S·C3H7NO
- Synthesis and crystal structure of 1-((3R,10S,13S,17S)-10,13-dimethyl-3-(m-tolylamino)hexadecahydro-1H-cyclopenta[α]phenanthren-17-yl)ethan-1-one, C28H41NO
- The crystal structure of diaqua-bis(4-bromo-2-formylphenoxy)zinc(II), C14H12Br2O6Zn
- The crystal structure of tetra(1-ethylimidazole-κ 1 N)-[μ 4-imidazole-4,5-dicarboxylato-κ 4 O, N, O′, N′]-trioxido-divanadium, C25H33N10O7V2
- The crystal structure of (E)-N′-(1-(4-fluorophenyl)propylidene)-2-hydroxybenzohydrazide, C16H15FN2O2

