Abstract
Melanoma is an aggressive malignant tumor. The crucial role of circular RNAs has been documented in many types of cancer, including melanoma. The objective of this study was to uncover the function of circ_0084043 in the biological process of melanoma and associated mechanism of action. The expression of circ_0084043, miR-31, and Krüppel-like factor 3 (KLF3) was determined by qRT-PCR. Cell proliferation and apoptosis were monitored by the MTT assay and flow cytometry assay, respectively. The progression of glycolysis was evaluated according to the levels of glucose consumption, lactate production, and ATP concentration using appropriate detection kits. The relationship between miR-31 and circ_0084043 or KLF3 was predicted by the bioinformatics tool and ascertained by the dual-luciferase reporter assay. The protein levels of KLF3 and glucose transporter 1 (Glut1) were quantified by western blot. A xenograft model was established to ascertain the role of circ_0084043 in vivo. As a result, circ_0084043 expression was reinforced in melanoma tissues and cells. Circ_0084043 knockdown inhibited cell proliferation, induced cell apoptosis, and restrained glycolysis. MiR-31 was a target of circ_0084043, and miR-31 deficiency reversed the role of circ_0084043 knockdown. KLF3 was targeted by miR-31, and KLF3 upregulation abolished the effects of miR-31 enrichment. Moreover, circ_0084043 knockdown impeded tumor growth in vivo and suppressed the level of Glut1 by modulating miR-31 and KLF3. Circ_0084043 promoted cell proliferation and glycolysis, and blocked apoptosis through the circ_0084043–miR-31–KLF3 regulatory axis in melanoma.
1 Introduction
Melanoma, originating from the malignant transformation of melanocytes, is the most massive malignant tumor in a number of skin cancer–related deaths [1,2]. As an extremely aggressive skin cancer, the mortality rate of melanoma is invariably high. Unfortunately, the incidence with melanoma has increased in recent years [3,4]. Despite significant advances in the diagnosis and treatment with molecular-targeted therapies and immunotherapy in recent years [5,6], the prognosis of melanoma patients is less than ideal and the 5-year survival rate is tragic [3,7]. Therefore, there is an urgent need to discover the molecular mechanisms of melanoma tumorigenesis and development and to identify biomarkers for the diagnosis and treatment of melanoma.
Recently, the crucial role of circular RNAs (circRNAs) in numerous human diseases, including cancers, has attracted wide attention [8]. CircRNAs are a class of noncoding RNAs that have more stable and conservative properties than linear RNAs because there are no 3′-poly (A) tail and 5′-ends [9,10]. CircRNAs have many biological functions, including adjustment of alternative splicing, regulation of parental gene expression, combination with RNA-binding proteins, and their role as microRNA (miRNA) sponges [11]. Several circRNAs have been documented to be associated with melanoma. For example, Yang et al. identified a series of dysregulated circRNAs through the microarray analysis of uveal melanoma tissues and normal tissues, including hsa_circ_0119873, hsa_circ_0128533, and hsa_circ_0047924 [12]. Also, Wang et al. obtained several differently expressed circRNAs from melanoma cells and normal melanocytes by microarray analysis [13]. These data indicated that circRNAs are implicated in the development of melanoma. A previous study identified that circ_0084043 was significantly upregulated in melanoma [14], which attracted our attention. However, the exact role of circ_0084043 in melanoma was not fully elucidated, and more associated mechanisms of action of circ_0084043 need to be explored.
CircRNAs are located mainly in the cytoplasm, which lays a foundation for circRNAs to act as competing endogenous RNAs of miRNAs [15,16]. MiRNAs are a kind of noncoding RNAs with 18–22 nucleotides in length. The crucial role of miR-31 is documented in various types of cancers, such as breast cancer, triple negative breast cancer, and colon cancer [17,18,19]. Unfortunately, the role of miR-31 in melanoma was rarely reported, and the associated mechanism of action is worthy of investigation. Generally, miRNAs function by combining with the 3′-UTR of targeted mRNAs to suppress their expression [20]. Krüppel-like factor 3 (KLF3), which belongs to the Krüppel-like factor family of zinc finger transcription factors, is located at chromosome 4q14 [21]. Research has implicated KLF3 in the regulation of adipogenesis, hematopoiesis, and muscle cell biology [22,23,24]. Recently, the involvement of KLF3 in the progression of cancers, particularly lung cancer, has been partly identified [25]. KLF3 was usually identified as a target of miRNAs. However, the potential role of KLF3 in cancers is not clear and needs to be explored.
Here, the abundance of circ_0084043 was determined in melanoma tissues and cells, and the function of circ_0084043 was investigated both in vitro and in vivo. Besides, the underlying mechanism of action was explored by analyzing the downstream miRNA and mRNA. The objective of our research was to determine the role of circ_0084043 in melanoma and provide a novel mechanism of action so as to further understand the progression of melanoma.
2 Materials and methods
2.1 Specimen collection
A total of 32 melanoma tissues and matched non-tumor tissues were obtained from the First Affiliated Hospital of University of South China. All tissues were frozen in liquid nitrogen and preserved at −80°C.
Informed consent: Informed consent has been obtained from all individuals included in this study.
Ethical approval: The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance with the tenets of the Helsinki Declaration, and has been approved by the Ethics Committee of the First Affiliated Hospital of University of South China.
2.2 Cell lines and cell culture
Melanoma cell lines (A375 and A875) were purchased from BeNa Culture Collection (Suzhou, China). Human epidermal melanocytes (HEMn-LP) were obtained from Gibco (Carlsbad, CA, USA). All cells were maintained in DMEM culture medium (Gibco) with 10% fetal bovine serum (Gibco) at 37°C under 5% CO2.
2.3 Cell transfection
The oligonucleotides and plasmids, including small interfering RNA (siRNA) targeting circ_0084043 (si-circ_0084043; 5′-UAAGCUUACAGGUACUUCCUU-3′), short hairpin RNA (shRNA) targeting circ_0084043 (sh-circ_0084043; 5′-TTTCATTGTATGTAGGTCC-3′), overexpression vector pcDNA-KLF3, and corresponding negative controls (si-NC, sh-NC, and pcDNA-Control), were assembled by GenePharma (Shanghai, China). MiR-31 mimic, miR-31 inhibitor, and relative negative controls (miRNA NC and inhibitor NC) were customized by RiboBio (Guangzhou, China). These oligonucleotides or plasmids were inserted into cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The transfected cells were cultivated for 48 h and used for subsequent tests.
2.4 qRT-PCR analysis
Trizol reagent (Invitrogen) was utilized to obtain total RNA from melanoma tissues and cells. Then a HiScript III First Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) or a miRNA First Strand Synthesis Kit (Vazyme) was used to conduct the reverse transcription reaction. Next, the amplification reaction was performed using SYBR Mix (Vazyme) and a CFX96 Touch Deep Well Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The relative expression was analyzed using the 2−ΔΔCt method and standardized by GAPDH or U6. The primer sequences were as follows: circ_0084043, F: 5′-TTCTAGACAGCCGGGGAGTG-3′ and R: 5′-CCAAAACCTTTCTTTCTTGATGGGA-3′; miR-31, F: 5′-GCCGCAGGCAAGATGCTGGC-3′ and R: 5′-CAGTGCAGGGTCCGAGGT-3′; KLF3, F: 5′-TGTCTCAGTGTCATACCCATCT-3′ and R: 5′-CCTTCTGGGGTCTGAAAGAACTT-3′; GAPDH, F: 5′-ACCACAGTCCATGCCATCAC-3′ and R: 5′-TCCACCACCCTGTTGCTGTA-3′; U6, F: 5′-GCGCGTCGTGAAGCGTTC-3′ and R: 5′-GTGCAGGGTCCGAGGT-3′.
2.5 MTT assay
A375 and A875 cells with different transfections were placed into 96-well plates (5 × 103 cells/well). Then 10 µL of MTT solution (Beyotime, Shanghai, China) was pipetted into each well at different time points (0, 24, 48, and 72 h) to incubate cells for another 4 h at 37°C. Afterward, dimethyl sulfoxide (DMSO; Beyotime) was pipetted into each well to dissolve the formazan. The absorbance at 490 nm was ascertained using an iMark microplate reader (Bio-Rad).
2.6 Flow cytometry assay
A375 and A875 cells with different transfections in 6-well plates were trypsinized and washed using PBS. Then, the cells (5 × 104) were stained with an Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme) following the manufacturer’s instructions. Cells at different time points were analyzed using a flow cytometer (BD Biosciences, San Jose, CA, USA).
2.7 Detection of glucose uptake, lactate production, and ATP level
Glucose uptake, lactate production, and ATP level were detected to determine the progression of glycolysis. Glucose consumption, lactate production, and ATP level were evaluated using a Glucose Assay Kit (BioVision, Milpitas, CA, USA), Lactate Assay Kit (BioVision), and ATP Assay Kit (Beyotime), respectively.
2.8 Target prediction and verification
Bioinformatics tools, including TargetScan (http://www.targetscan.org/vert_72/) and CircInteractome (https://circinteractome.nia.nih.gov/miRNA_Target_Sites/mirna_target_sites.html), were applied to predict the potential targets.
The relationship between miR-31 and circ_0084043 or KLF3 was further validated by the dual-luciferase reporter assay. To be specific, partial wild-type (WT, harboring the specific binding site for miR-31) and mutant-type (MUT, harboring the mutational binding site for miR-31) circ_0084043 sequences were amplified and inserted into the pGL4 vector (Promega, Madison, WI, USA), namely, WT-circ_0084043 and MUT-circ_0084043. Likewise, the 3′-UTR of KLF3 (WT and MUT) were also cloned into the pGL4 vector to generate WT-KLF3 3′-UTR and MUT-KLF3 3′-UTR. These fusion plasmids were transfected into A375 and A875 cells along with miR-31 mimic or miRNA NC. After 48 h, the firefly luminescence intensity was measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized by Renilla luminescence activity.
2.9 Western blot
Western blot was carried out as previously described elsewhere [26]. Briefly, the proteins were separated and transferred to PVDF membranes. Subsequently, the membranes were blocked with 5% skim milk and probed with the primary antibodies against KLF3 (1:1,000; ab154531; Abcam, Cambridge, MA, USA), glucose transporter 1 (Glut1) (1:100,000; ab115730; Abcam) and GAPDH (1:2500; ab9485; Abcam) and with the corresponding secondary antibody (1:5,000; ab205718; Abcam).
2.10 Tumor formation assay in vivo
Experimental mice (BALB/c, 6-week-old), from Shanghai SIPPR-BK Laboratory Animal Co. Ltd (Shanghai, China), were divided into two groups (n = 6). A375 cells with sh-circ_0084043 or sh-NC transfection were subcutaneously inoculated into the left flank of mice back. From the seventh day after inoculation, the tumor volume (length × width2 × 0.5) in the sh-circ_0084043 group and in the sh-NC group was measured every 4 days. All mice were sacrificed after 27 days. Tumor tissues were excised for weighing and expression analysis.
Ethical approval: The research related to animal use has been complied with all the relevant national regulations and institutional policies for the care and use of animals and has been approved by the Animal Ethics Committee of the First Affiliated Hospital of University of South China.
2.11 Data processing
All experiments consisted of at least three independent repetitions. Statistical analysis was performed using SPSS software (SPSS Inc., Chicago, IL, USA), and the data were expressed as mean ± standard deviation. One-way analysis of variance or Student’s t-test was employed to conduct differential analysis. The difference was considered statistically significant at P < 0.05.
3 Results
3.1 Circ_0084043 was aberrantly overexpressed in melanoma tissues and cells
qRT-PCR analysis indicated the abundance of circ_0084043 in cancer tissues and cells. Noticeably, the expression of circ_0084043 was higher in melanoma tissues (n = 32) than in normal nontumor tissues (n = 32) (Figure 1a). Also, the expression of circ_0084043 was significantly elevated in melanoma cell lines (A375 and A875) compared with that in HEMn-LP cells (Figure 1b). The data suggested that the dysregulation of circ_0084043 might be associated with the development of melanoma.

Circ_0084043 was highly regulated in melanoma tissues and cell lines. (a) The abundance of circ_0084043 in melanoma tissues (n = 32) and normal nontumor tissues (n = 32) was detected by qRT-PCR. (b) The abundance of circ_0084043 in melanoma cell lines (A375 and A875) and HEMn-LP cells was detected by qRT-PCR. *P < 0.05.
3.2 Circ_0084043 knockdown impaired cell proliferation, induced cell apoptosis, and inhibited glycolysis in melanoma cells
The endogenous content of circ_0084043 was knocked down in A375 and A875 cells by transfecting with si-circ_0084043 to determine the role of circ_0084043. First, we noticed that the expression of circ_0084043 was clearly decreased in cells with si-circ_0084043 transfection, while the expression of its pre-mRNA (ADAM9) differed little in cells with si-circ_0084043 transfection relative to si-NC (Figure 2a and b). Then, the MTT assay indicated that cell proliferation was notably reduced in cells transfected with si-circ_0084043 relative to si-NC (Figure 2c and d). Next, flow cytometry assay manifested that circ_0084043 knockdown increased the rate of apoptosis (Figure 2e). Moreover, the levels of glucose consumption, lactate production, and ATP concentration were markedly declined in A375 and A875 cells transfected with si-circ_0084043 compared with si-NC (Figure 2f–h). These analyses suggested that circ_0084043 knockdown impaired proliferation and glycolysis but promoted apoptosis in melanoma cells.
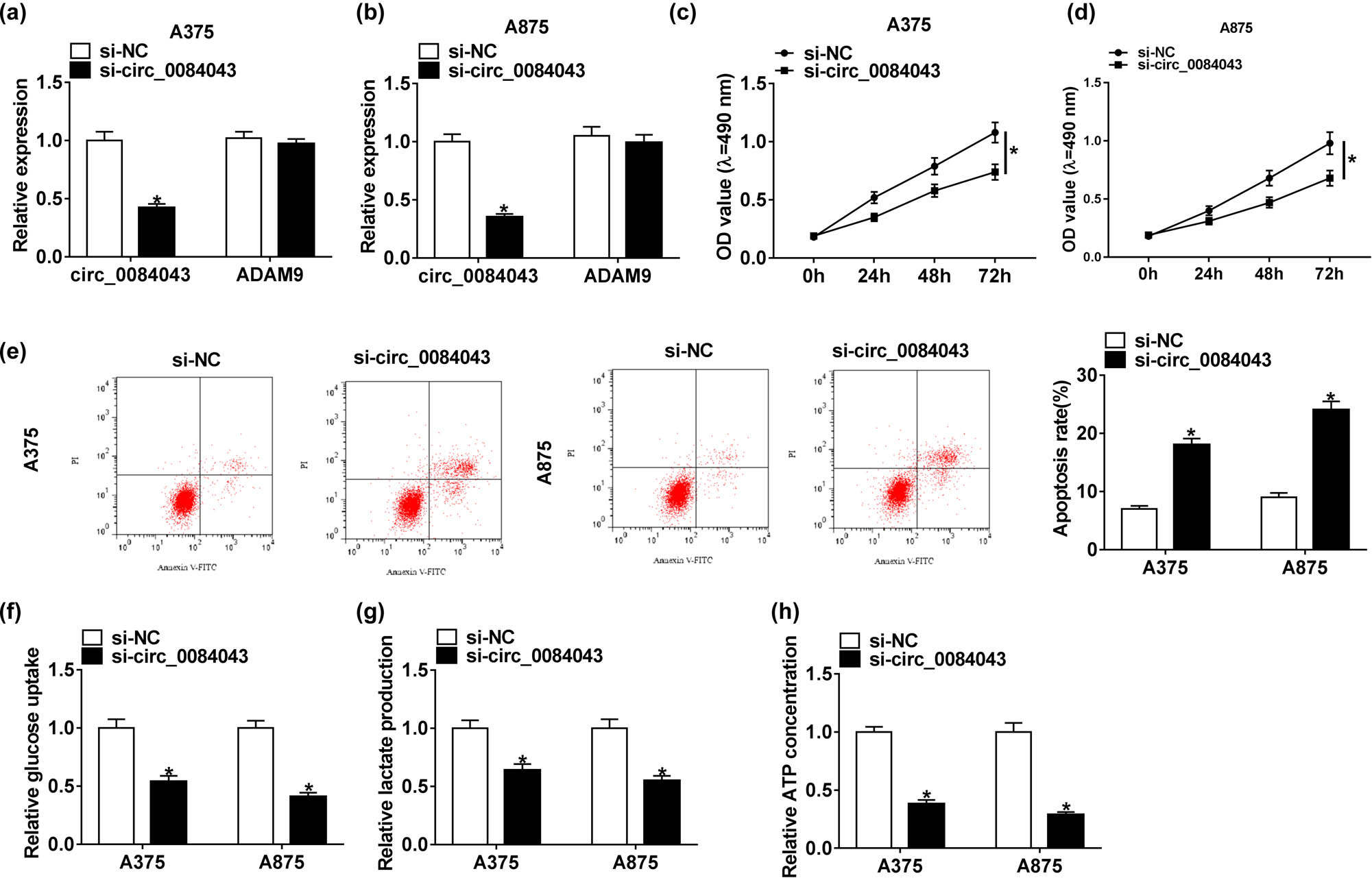
Circ_0084043 knockdown inhibited proliferation and glycolysis but induced apoptosis in melanoma cells. A375 and A875 cells were transfected with si-circ_0084043 or si-NC. (a and b) The expression of circ_0084043 was measured by qRT-PCR. (c and d) Cell proliferation was assessed by the MTT assay. (e) Cell apoptosis was examined by the flow cytometry assay. (f–h) The progression of glycolysis was evaluated, according to glucose consumption, lactate production, and ATP concentration. *P < 0.05.
3.3 MiR-31 was a target of circ_0084043, and miR-31 inhibition reversed the impact of circ_0084043 knockdown in melanoma cells
To detect whether circ_0084043 functioned by targeting its downstream miRNAs, the putative target miRNAs of circ_0084043 were predicted by the online database CircInteractome (Table A1). The partial sequences of circ_0084043 harboring the binding site for miR-31 were amplified and cloned into the pGL4 vector, named WT-circ_0084043. Meanwhile, the mutated sequences were also cloned into the pGL4 vector, named MUT-circ_0084043 (Figure 3a). As shown in Figure 3b and c, the miR-31 mimic prominently diminished the luciferase activity in A375 and A875 cells transfected with WT-circ_0084043 rather than with MUT-circ_0084043. In addition, the expression of miR-31 was obviously enhanced by circ_0084043 knockdown (Figure 3e). Next, the miR-31 expression was inhibited in A375 and A875 cells to observe its role. The transfection efficiency showed that the expression of miR-31 pronouncedly declined in A375 and A875 cells with miR-31 inhibitor transfection (Figure 3d). Furthermore, the cell proliferation inhibited by circ_0084043 knockdown was recovered by miR-31 inhibition (Figure 3f and 3g). However, the apoptosis rate increased in cells with si-circ_0084043 transfection but decreased in cells with si-circ_0084043 + miR-31 inhibitor transfection (Figure 3h). Unsurprisingly, si-circ_0084043 + miR-31 inhibitor transfection restored the glucose consumption, lactate production, and ATP concentration inhibited by si-circ_0084043 transfection (Figure 3i–k). These data indicated that circ_0084043 knockdown inhibited melanoma progression by modulating miR-31 in vitro.

MiR-31 was targeted by circ_0084043, and its inhibition reversed the effects of circ_0084043 knockdown. (a) The binding site between miR-31 and circ_0084043 was analyzed by the online tool CircInteractome. (b and c) The relationship between miR-31 and circ_0084043 was verified by the dual-luciferase reporter assay. (d and e) The expression of miR-31 in the cells transfected with si-circ_0084043 or miR-31 inhibitor. (f and g) Cell proliferation, (h) cell apoptosis, and (i–k) the progression of glycolysis were detected by the MTT assay, flow cytometry assay, and the levels of glucose consumption, lactate production, and ATP concentration, respectively. *P < 0.05.
3.4 KLF3 was targeted by miR-31, and KLF3 upregulation overturned the role of miR-31 mimic in melanoma cells
Generally, miRNAs function by uniting with the 3′-UTR of targeted mRNAs. To determine whether miR-31 exerted its role by targeting downstream mRNAs, the potential mRNAs targeted by miR-31 were screened by the online tool TargetScan (Table A2). Subsequently, the sequences of KLF3 3′-UTR containing the binding site for miR-31 and the corresponding mutant sequences were cloned into the pGL4 vector to generate WT-KLF3 3′-UTR and MUT-KLF3 3′-UTR fusion plasmids (Figure 4a). We found that miR-31 mimic significantly decreased the luciferase activity in A375 and A875 cells introduced with WT-KLF3 3′-UTR but not with MUT-KLF3 3′-UTR (Figure 4b and c). In A375 and A875 cells transfected with miR-31 mimic, the expression of miR-31 was clearly increased (Figure 4d), indicating that the transfection efficiency was competent. Then, we also detected that the KLF3 expression in A375 and A875 cells transfected with miR-31 mimic was obviously impaired at both mRNA and protein levels (Figure 4e and f), suggesting that miR-31 overexpression suppressed the KLF3 expression. The detection of transfection efficiency showed that the expression of KLF3 was substantially elevated in A375 and A875 cells transfected with pcDNA-KLF3 (Figure 4g and i), suggesting that these transfected cells were available. Functionally, the MTT assay showed that the cell proliferation was inhibited in cells introduced with miR-31 mimic but reinforced in cells introduced with miR-31 mimic + pcDNA-KLF3 (Figure 4h and j). Inversely, the apoptosis rate was notably stimulated in cells transfected with the miR-31 mimic but repressed in cells transfected with miR-31 mimic + pcDNA-KLF3 (Figure 4k). Moreover, glucose consumption, lactate production, and ATP concentration were reduced by miR-31 enrichment but elevated by KLF3 overexpression (Figure 4l–n). These data indicated that miR-31 participated in proliferation, apoptosis, and glycolysis by regulating the expression of KLF3 in melanoma cells.
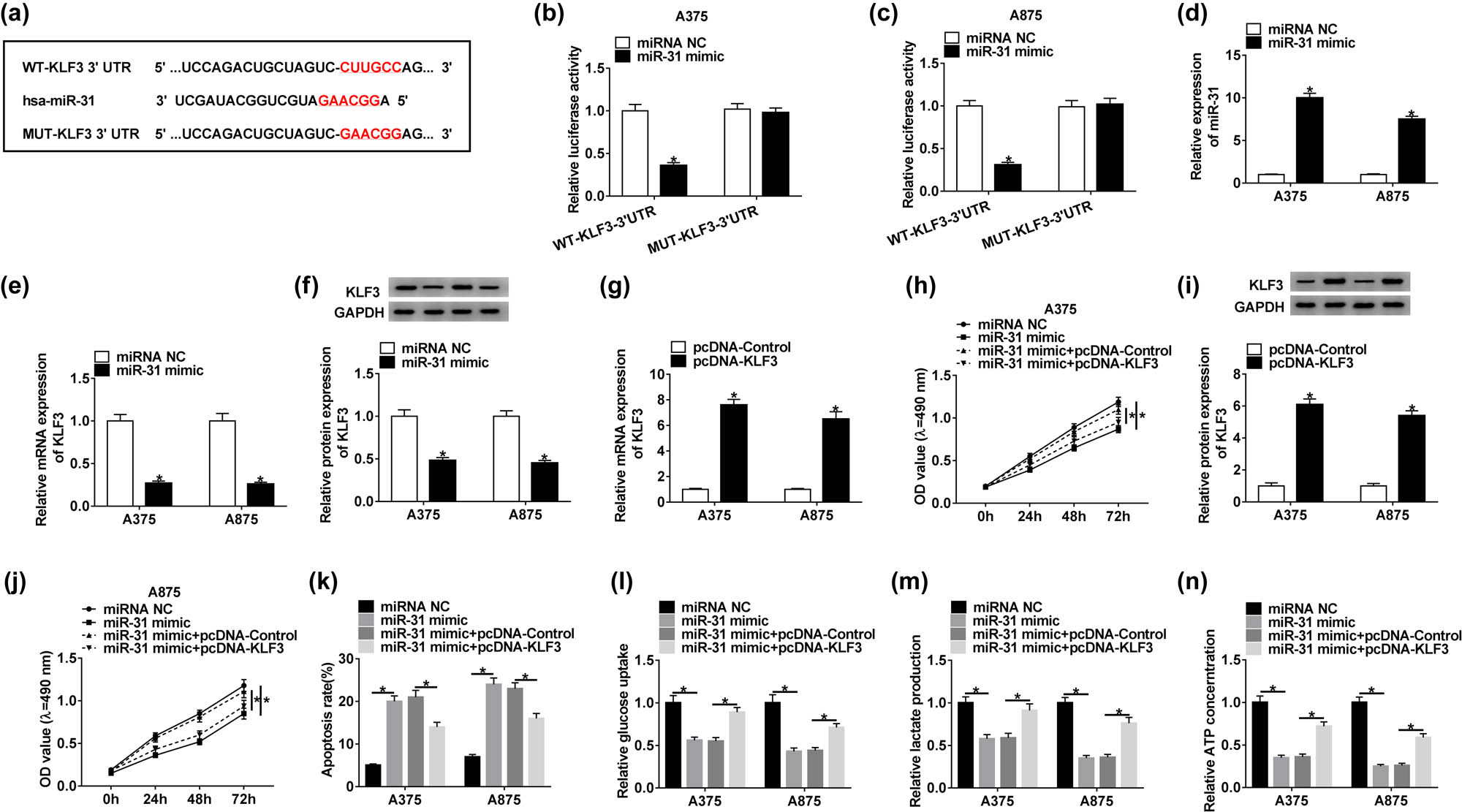
KLF3 was targeted by miR-31, and its overexpression abolished the role of miR-31 mimic. (a) The binding site between miR-31 and KLF3 3′-UTR was forecasted by the online tool TargetScan. (b and c) The interaction between miR-31 and KLF3 was confirmed by the dual-luciferase reporter assay. (d–f) The expression of miR-31 and KLF3 was checked in the cells transfected with miR-31 mimic. (g and i) The expression of KLF3 at mRNA and protein levels in the cells transfected with pcDNA-KLF3 was examined by qRT-PCR and western blot. (h and j) Cell proliferation was assessed by the MTT assay. (k) Cell apoptosis was examined by the flow cytometry assay. (l–n) The progression of glycolysis was evaluated, according to glucose consumption, lactate production, and ATP concentration. *P < 0.05.
3.5 KLF3 overexpression rescued the effects of circ_0084043 knockdown in melanoma cells
To explore the interaction between circ_0084043 and KLF3, rescue experiments were performed. A375 and A875 cells were introduced with si-circ_0084043 or si-circ_0084043 + pcDNA-KLF3, with si-NC or si-circ_0084043 + pcDNA-Control as the control. The transfection efficiency was examined. We found that the expression of KLF3 at both mRNA and protein levels was decreased in cells with si-circ_0084043 transfection but regained in cells with si-circ_0084043 + pcDNA-KLF3 transfection (Figure 5a–c). Cell proliferation inhibited by si-circ_0084043 was elevated by si-circ_0084043 + pcDNA-KLF3 (Figure 5d and e). The apoptosis rate was induced by si-circ_0084043 transfection but suppressed by si-circ_0084043 + pcDNA-KLF3 transfection (Figure 5f). Additionally, KLF3 overexpression restored the glucose consumption, lactate production, and ATP concentration blocked by circ_0084043 knockdown (Figure 5g–i). The data indicated that circ_0084043 regulated cell proliferation, apoptosis, and glycolysis by promoting the KLF3 expression in melanoma cells.

KLF3 overexpression rescued the role of circ_0084043 knockdown. A375 and A875 cells were transfected with si-circ_0084043 or si-circ_0084043 + pcDNA-KLF3, si-NC or si-circ_0084043 + pcDNA-Control as the control. (a–c) The expression of KLF3 at mRNA and protein levels was determined by qRT-PCR and western blot. (d and e) Cell proliferation was assessed by the MTT assay. (f) Cell apoptosis was examined by the flow cytometry assay. (g–i) The progression of glycolysis was evaluated, according to glucose consumption, lactate production, and ATP concentration. *P < 0.05.
3.6 Circ_0084043 regulated the expression of KLF3 by sponging miR-31
A375 and A875 cells were transfected with si-circ_0084043, si-NC, si-circ_0084043 + miR-31 inhibitor, and si-circ_0084043 + inhibitor NC. Apparently, the expression of KLF3 was inhibited in cells with si-circ_0084043 transfection relative to si-NC, while the expression of KLF3 was recovered in cells with si-circ_0084043 + miR-31 inhibitor transfection relative to si-circ_0084043 + inhibitor NC at both mRNA and protein levels (Figure 6a and b). The data suggested that KLF3 was regulated by circ_0084043 through miR-31.

Circ_0084043 regulated the expression of KLF3 by targeting miR-31. A375 and A875 cells were transfected with si-circ_0084043 or si-circ_0084043 + miR-31 inhibitor, si-NC or si-circ_0084043 + inhibitor NC as the control. (a and b) The expression of KLF3 at mRNA and protein levels was quantified by qRT-PCR and western blot. *P < 0.05.
3.7 Circ_0084043 knockdown inhibited tumorigenesis in vivo
To ascertain the role of circ_0084043 in vivo, the xenograft model was established. A375 cells with sh-circ_0084043 transfection were inoculated in the right flank of mice groin, with sh-NC as the control. The tumor volume and tumor weight in the sh-circ_0084043 group were obviously lower than those in the sh-NC group (Figure 7a and b). Subsequently, the expression levels of circ_0084043, miR-31, and KLF3 in the removed tumor tissues were examined. The results displayed that the expression of circ_0084043 was lower in the sh-circ_0084043 group than in the sh-NC group, while the expression of miR-31 was enhanced in the sh-circ_0084043 group (Figure 7c and d). Moreover, the expression of KLF3 was declined in the sh-circ_0084043 group relative to that in the sh-NC group (Figure 7e and f). The data indicated that circ_0084043 knockdown impeded tumor growth in vivo.

Circ_0084043 knockdown inhibited tumor growth in vivo. (a and b) Circ_0084043 knockdown significantly reduced the tumor volume and tumor weight. (c and d) The expression of circ_0084043 and miR-31 in removed tumor tissues was detected by qRT-PCR. (e and f) The expression of KLF3 in removed tumor tissues at both mRNA and protein levels was determined by qRT-PCR and western blot. *P < 0.05.
3.8 Circ_0084043 modulated the expression of Glut1 through the miR-31/KLF3 axis
Glut1 is a key regulator of glycolysis and is closely related to the occurrence and progression of malignant tumors. The expression of Glut1 was reduced in A375 and A378 cells with si-circ_0084043 transfection but recovered in cells with si-circ_0084043 + miR-31 inhibitor transfection at the protein level (Figure 8a). Besides, the expression of Glut1 was reduced by the miR-31 mimic but enhanced by KLF3 overexpression (Figure 8b). The data pointed out that circ_0084043 facilitated the expression of Glut1 via the modulation of the miR-31/KLF3 axis.

The expression of Glut1 was modulated by the circ_0084043/miR-31/KLF3 regulatory axis. (a) The protein level of Glut1 in A375 and A875 cells transfected with si-circ_0084043, si-NC, si-circ_0084043 + miR-31 inhibitor, or si-circ_0084043 + inhibitor NC was measured by western blot. (b) The protein level of Glut1 in A375 and A875 cells transfected with miR-31 mimic, miRNA NC, miR-31 mimic + pcDNA-KLF3, or miR-31 mimic + pcDNA-Control was measured by western blot. *P < 0.05.
4 Discussion
Increasing evidence shows that melanoma is a severe threat to people’s health. It is essential to find novel therapeutic strategies against melanoma. In our study, we discovered that circ_0084043 was abundantly expressed in melanoma tissues and cells, and its downregulation inhibited proliferation and glycolysis but triggered apoptosis in melanoma cells. We also screened and identified the target miRNA (miR-31) of circ_0084043 and the target mRNA (KLF3) of miR-31, revealing a potential role of circ_0084043 in the progression of melanoma.
CircRNAs are involved in the progression of many types of cancers, whereas knowledge of their specific role is limited and inadequate. Using circRNA microarray analysis, a previous study claimed that circ_0084043 was aberrantly upregulated in melanoma tissues and that reintroduction of circ_0084043 stimulated cell proliferation and metastasis in melanoma [14]. Consistent with this research, we also discovered that circ_0084043 was highly expressed in melanoma tissues and cells. Functional analysis revealed that circ_0084043 knockdown restrained cell proliferation, induced cell apoptosis, and impeded glycolysis. This is the first study to explore the regulation of circ_0084043 in glycolysis, and we proposed that circ_0084043 promoted glycolysis metabolism to promote melanoma development. Moreover, circ_0084043 knockdown suppressing tumor growth was also confirmed in vivo. Accumulating evidence has shown that circRNAs participate in the progression of cancers through various mechanisms, such as interaction with miRNAs, translation into proteins, and generation of fusion circRNAs [10,11,27]. The above-mentioned study also found that circ_0084043 served as an oncogene in melanoma by functioning as a molecular sponge of miR-153-3p [14]. To determine the novel mechanisms of circ_0084043 function in melanoma, the downstream miRNAs of circ_0084043 were predicted and screened.
MiR-31 was a target of circ_0084043 with a specific binding site. Besides, the expression of miR-31 was strikingly reduced in melanoma cell lines. Given that the role of miR-31 in melanoma was partly mentioned in preceding studies, miR-31 was thus chosen as a research object in this study. Throughout the former studies, the endogenous expression of miR-31 was consistently reduced in melanoma tissues and cells [28,29,30]. Moreover, miR-31 overexpression blocked tumor growth and chemoresistance of melanoma [30]. In agreement with these studies, we observed that miR-31 enrichment attenuated cell proliferation and glycolysis but enhanced cell apoptosis, and miR-31 inhibition reversed the regulatory effects of circ_0084043 knockdown. These findings emphasized that miR-31 was a tumor suppressor in melanoma.
The circRNA–miRNA–mRNA axis is an indispensable regulatory way that modulates cancer-related biological processes [31]. In our study, KLF3 was verified to be targeted by miR-31 through bioinformatics prediction and dual-luciferase reporter detection. Interestingly, the role of KLF3 varied in different types of cancers. For instance, KLF3 downregulation inhibited tumorigenesis and the development of lung cancer [24]. Inversely, loss of KLF3 led to malignant phenotypes and poor prognosis of colorectal cancer [32]. This might be due to the different expression patterns of KLF3 in different cancers. A previous study elucidated that KLF3 was highly expressed in melanoma cells and that KLF3 knockdown inhibited cell viability, migration, and invasion [33]. Similarly, we also found that KLF3 was upregulated in melanoma cell lines and that KLF3 overexpression abolished the effect of miR-31 enrichment on the activities of melanoma cells, suggesting that KLF3 was indeed an oncogene in melanoma.
Taken together, circ_0084043 expression was abnormally enhanced in melanoma tissues and cells. Circ_0084043 contributed to cell proliferation and glycolysis but inhibited cell apoptosis through the regulation of the circ_0084043–miR-31–KLF3 axis. Our study adds to the understanding of the progression of melanoma and provides a promising biomarker for the treatment of melanoma. The limitation of this study was that we mainly investigated the function of circ_0084043 by its knockdown, and the effects of circ_0084043 overexpression should be monitored in the future work.
Appendix
The partial target miRNAs of circ_0084043 predicted by CircInteractome
| MiRNA name | Targeting sites | Score |
|---|---|---|
| hsa_circ_0084043 (5′…3′) | GAAGUGUGCCACUGGGAAUGCUU | 77 |
| hsa-miR-1179 (3′ …… 5′) | ||||||| | |
| GGUUGGUUACUUUCUUACGAA | ||
| hsa_circ_0084043 (5′…3′) | AAUGGCAUUUGUGGGAACAGUGU | 87 |
| hsa-miR-1208 (3′…5′) | ||||||| | |
| AGGCGGACAGACUUGUCACU | ||
| hsa_circ_0084043 (5′…3′) | UCUCCAGUGACACCUCCCAGAGA | 99 |
| hsa-miR-326 (3′…5′) | ||||||| | |
| GACCUCCUUCCCGGGUCUCC | ||
| hsa_circ_0084043 (5′…3′) | UUCAGAAUGGAUAUCCUUGCCAG | 82 |
| hsa-miR-31 (3′…5′) | |||||| | |
| UCGAUACGGUCGUAGAACGGA | ||
| hsa_circ_0084043 (5′…3′) | GGAGUGUUCCUCGACAUGUUUCU | 89 |
| hsa-miR-494 (3′…5′) | |||| ||||||| | |
| CUCCAAAGGGCACAUACAAAGU | ||
| hsa_circ_0084043 (5′…3′) | UCUCUGGCAAUGAAUACAAGAAG | 73 |
| hsa-miR-578 (3′…5′) | ||||||| | |
| UGUUAGGAUCUCGUGUUCUUC |
The partial target mRNAs of miR-31 predicted by TargetScan
| Target gene | Representative transcript | Gene name |
|---|---|---|
| KLF3 | ENST00000261438.5 | Krüppel-like factor 3 (basic) |
| CEP85 | ENST00000252992.4 | Centrosomal protein 85 kDa |
| UBN1 | ENST00000262376.6 | Ubinuclein 1 |
| AMOTL1 | ENST00000317837.9 | Angiomotin like 1 |
| EIF5 | ENST00000216554.3 | Eukaryotic translation initiation factor 5 |
| SLC30A6 | ENST00000282587.5 | Solute carrier family 30 (zinc transporter), member 6 |
| RAB31 | ENST00000578921.1 | RAB31, member RAS oncogene family |
| NDRG3 | ENST00000373803.2 | NDRG family member 3 |
Conflict of interest: The authors state no conflict of interest.
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA Dermatol. 2015;151:1081–6 [PubMed: 25928283].10.1001/jamadermatol.2015.1187Suche in Google Scholar
[2] Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108 [PubMed: 25651787].10.3322/caac.21262Suche in Google Scholar
[3] Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet. 2014;383:816–27 [PubMed: 24054424].10.1016/S0140-6736(13)60802-8Suche in Google Scholar
[4] Strouse JJ, Fears TR, Tucker MA, Wayne AS. Pediatric melanoma: risk factor and survival analysis of the surveillance, epidemiology and end results database. J Clin Oncol. 2005;23:4735–41 [PubMed: 16034049].10.1200/JCO.2005.02.899Suche in Google Scholar PubMed
[5] Nikolaou VA, Stratigos AJ, Flaherty KT, Tsao H. Melanoma: new insights and new therapies. J Invest Dermatol. 2012;132:854–63 [PubMed: 22217739].10.1038/jid.2011.421Suche in Google Scholar PubMed PubMed Central
[6] Roukos DH. PLX4032 and melanoma: resistance, expectations and uncertainty. Expert Rev Anticancer Ther. 2011;11:325–8 [PubMed: 21417847].10.1586/era.11.3Suche in Google Scholar PubMed
[7] Little EG, Eide MJ. Update on the current state of melanoma incidence. Dermatol Clin. 2012;30:355–61 [PubMed: 22800543].10.1016/j.det.2012.04.001Suche in Google Scholar PubMed
[8] Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25 [PubMed: 28143578].10.1186/s12943-017-0598-7Suche in Google Scholar PubMed PubMed Central
[9] Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–1 [PubMed: 23620042].10.1126/science.1238522Suche in Google Scholar PubMed PubMed Central
[10] Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8 [PubMed: 23446348].10.1038/nature11928Suche in Google Scholar PubMed
[11] Xu S, Zhou L, Ponnusamy M, Zhang L, Dong Y, Zhang Y, et al. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. 2018;6:e5503 [PubMed: 30155370].10.7717/peerj.5503Suche in Google Scholar PubMed PubMed Central
[12] Yang X, Li Y, Liu Y, Xu X, Wang Y, Yan Y, et al. Novel circular RNA expression profile of uveal melanoma revealed by microarray. Chin J Cancer Res. 2018;30:656–68 [PubMed: 30700934].10.21147/j.issn.1000-9604.2018.06.10Suche in Google Scholar PubMed PubMed Central
[13] Wang Q, Chen J, Wang A, Sun L, Qian L, Zhou X, et al. Differentially expressed circRNAs in melanocytes and melanoma cells and their effect on cell proliferation and invasion. Oncol Rep. 2018;39:1813–24 [PubMed: 29436673].10.3892/or.2018.6263Suche in Google Scholar PubMed
[14] Luan W, Shi Y, Zhou Z, Xia Y, Wang J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502:22–9 [PubMed: 29777697].10.1016/j.bbrc.2018.05.114Suche in Google Scholar PubMed
[15] Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–42 [PubMed: 22140119].10.1093/nar/gkr1009Suche in Google Scholar PubMed PubMed Central
[16] Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75:1071–98 [PubMed: 29116363].10.1007/s00018-017-2688-5Suche in Google Scholar PubMed PubMed Central
[17] Soheilyfar S, Velashjerdi Z, Sayed Hajizadeh Y, Fathi Maroufi N, Amini Z, Khorrami A, et al. In vivo and in vitro impact of miR-31 and miR-143 on the suppression of metastasis and invasion in breast cancer. J BUON. 2018; 23:1290–6 [PubMed: 30570849].Suche in Google Scholar
[18] Luo LJ, Yang F, Ding JJ, Yan DL, Wang DD, Yang SJ, et al. MiR-31 inhibits migration and invasion by targeting SATB2 in triple negative breast cancer. Gene. 2016;594:47–58 [PubMed: 27593563].10.1016/j.gene.2016.08.057Suche in Google Scholar PubMed
[19] Zhong L, Huot J, Simard MJ. p38 activation induces production of miR-146a and miR-31 to repress E-selectin expression and inhibit transendothelial migration of colon cancer cells. Sci Rep. 2018;8:2334 [PubMed: 29402939].10.1038/s41598-018-20837-9Suche in Google Scholar PubMed PubMed Central
[20] Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–50 [PubMed: 19182800].10.1038/nsmb.1552Suche in Google Scholar PubMed PubMed Central
[21] Pearson RC, Funnell AP, Crossley M. The mammalian zinc finger transcription factor Kruppel-like factor 3 (KLF3/BKLF). IUBMB Life. 2011;63:86–93 [PubMed: 21360637].10.1002/iub.422Suche in Google Scholar PubMed
[22] Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, et al. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol Cell Biol. 2008;28:3967–78 [PubMed: 18391014].10.1128/MCB.01942-07Suche in Google Scholar PubMed PubMed Central
[23] Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–70 [PubMed: 16380451].10.1182/blood-2005-07-2888Suche in Google Scholar PubMed PubMed Central
[24] Himeda CL, Ranish JA, Pearson RC, Crossley M, Hauschka SD. KLF3 regulates muscle-specific gene expression and synergizes with serum response factor on KLF binding sites. Mol Cell Biol. 2010;30:3430–43 [PubMed: 20404088].10.1128/MCB.00302-10Suche in Google Scholar PubMed PubMed Central
[25] Wang R, Xu J, Xu J, Zhu W, Qiu T, Li J, et al. MiR-326/Sp1/KLF3: a novel regulatory axis in lung cancer progression. Cell Prolif. 2019;52:e12551 [PubMed: 30485570].10.1111/cpr.12551Suche in Google Scholar PubMed PubMed Central
[26] Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19 [PubMed: 29386015].10.1186/s12943-018-0771-7Suche in Google Scholar PubMed PubMed Central
[27] Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64 [PubMed: 28520103].10.1002/hep.29270Suche in Google Scholar PubMed
[28] Greenberg E, Hershkovitz L, Itzhaki O, Hajdu S, Nemlich Y, Ortenberg R, et al. Regulation of cancer aggressive features in melanoma cells by microRNAs. PLoS One. 2011;6:e18936 [PubMed: 21541354].10.1371/journal.pone.0018936Suche in Google Scholar PubMed PubMed Central
[29] Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011–25 [PubMed: 22948084].10.18632/oncotarget.622Suche in Google Scholar PubMed PubMed Central
[30] Zheng Y, Sun Y, Liu Y, Zhang X, Li F, Li L, et al. The miR-31-SOX10 axis regulates tumor growth and chemotherapy resistance of melanoma via PI3K/AKT pathway. Biochem Biophys Res Commun. 2018;503:2451–8 [PubMed: 29969627].10.1016/j.bbrc.2018.06.175Suche in Google Scholar PubMed
[31] Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804–14 [PubMed: 28184940].10.3892/or.2017.5415Suche in Google Scholar PubMed
[32] Wang X, Jiang Z, Zhang Y, Wang X, Liu L, Fan Z. RNA sequencing analysis reveals protective role of kruppel-like factor 3 in colorectal cancer. Oncotarget. 2017;8:21984–93 [PubMed: 28423541].10.18632/oncotarget.15766Suche in Google Scholar PubMed PubMed Central
[33] Ding F, Lai J, Gao Y, Wang G, Shang J, Zhang D, et al. NEAT1/miR-23a-3p/KLF3: a novel regulatory axis in melanoma cancer progression. Cancer Cell Int. 2019;19:217 [PubMed: 31462890].10.1186/s12935-019-0927-6Suche in Google Scholar PubMed PubMed Central
© 2020 Songjiang Wu et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Plant Sciences
- Dependence of the heterosis effect on genetic distance, determined using various molecular markers
- Plant Growth Promoting Rhizobacteria (PGPR) Regulated Phyto and Microbial Beneficial Protein Interactions
- Role of strigolactones: Signalling and crosstalk with other phytohormones
- An efficient protocol for regenerating shoots from paper mulberry (Broussonetia papyrifera) leaf explants
- Functional divergence and adaptive selection of KNOX gene family in plants
- In silico identification of Capsicum type III polyketide synthase genes and expression patterns in Capsicum annuum
- In vitro induction and characterisation of tetraploid drumstick tree (Moringa oleifera Lam.)
- CRISPR/Cas9 or prime editing? – It depends on…
- Study on the optimal antagonistic effect of a bacterial complex against Monilinia fructicola in peach
- Natural variation in stress response induced by low CO2 in Arabidopsis thaliana
- The complete mitogenome sequence of the coral lily (Lilium pumilum) and the Lanzhou lily (Lilium davidii) in China
- Ecology and Environmental Sciences
- Use of phosphatase and dehydrogenase activities in the assessment of calcium peroxide and citric acid effects in soil contaminated with petrol
- Analysis of ethanol dehydration using membrane separation processes
- Activity of Vip3Aa1 against Periplaneta americana
- Thermostable cellulase biosynthesis from Paenibacillus alvei and its utilization in lactic acid production by simultaneous saccharification and fermentation
- Spatiotemporal dynamics of terrestrial invertebrate assemblages in the riparian zone of the Wewe river, Ashanti region, Ghana
- Antifungal activity of selected volatile essential oils against Penicillium sp.
- Toxic effect of three imidazole ionic liquids on two terrestrial plants
- Biosurfactant production by a Bacillus megaterium strain
- Distribution and density of Lutraria rhynchaena Jonas, 1844 relate to sediment while reproduction shows multiple peaks per year in Cat Ba-Ha Long Bay, Vietnam
- Biomedical Sciences
- Treatment of Epilepsy Associated with Common Chromosomal Developmental Diseases
- A Mouse Model for Studying Stem Cell Effects on Regeneration of Hair Follicle Outer Root Sheaths
- Morphine modulates hippocampal neurogenesis and contextual memory extinction via miR-34c/Notch1 pathway in male ICR mice
- Composition, Anticholinesterase and Antipedicular Activities of Satureja capitata L. Volatile Oil
- Weight loss may be unrelated to dietary intake in the imiquimod-induced plaque psoriasis mice model
- Construction of recombinant lentiviral vector containing human stem cell leukemia gene and its expression in interstitial cells of cajal
- Knockdown of lncRNA KCNQ1OT1 inhibits glioma progression by regulating miR-338-3p/RRM2
- Protective effect of asiaticoside on radiation-induced proliferation inhibition and DNA damage of fibroblasts and mice death
- Prevalence of dyslipidemia in Tibetan monks from Gansu Province, Northwest China
- Sevoflurane inhibits proliferation, invasion, but enhances apoptosis of lung cancer cells by Wnt/β-catenin signaling via regulating lncRNA PCAT6/ miR-326 axis
- MiR-542-3p suppresses neuroblastoma cell proliferation and invasion by downregulation of KDM1A and ZNF346
- Calcium Phosphate Cement Causes Nucleus Pulposus Cell Degeneration Through the ERK Signaling Pathway
- Human Dental Pulp Stem Cells Exhibit Osteogenic Differentiation Potential
- MiR-489-3p inhibits cell proliferation, migration, and invasion, and induces apoptosis, by targeting the BDNF-mediated PI3K/AKT pathway in glioblastoma
- Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating the microRNA-34a-5p/NOTCH1 signaling pathway
- Large Brunner’s gland adenoma of the duodenum for almost 10 years
- Neurotrophin-3 accelerates reendothelialization through inducing EPC mobilization and homing
- Hepatoprotective effects of chamazulene against alcohol-induced liver damage by alleviation of oxidative stress in rat models
- FXYD6 overexpression in HBV-related hepatocellular carcinoma with cirrhosis
- Risk factors for elevated serum colorectal cancer markers in patients with type 2 diabetes mellitus
- Effect of hepatic sympathetic nerve removal on energy metabolism in an animal model of cognitive impairment and its relationship to Glut2 expression
- Progress in research on the role of fibrinogen in lung cancer
- Advanced glycation end product levels were correlated with inflammation and carotid atherosclerosis in type 2 diabetes patients
- MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB
- Knockdown of DDX46 inhibits trophoblast cell proliferation and migration through the PI3K/Akt/mTOR signaling pathway in preeclampsia
- Buformin suppresses osteosarcoma via targeting AMPK signaling pathway
- Effect of FibroScan test in antiviral therapy for HBV-infected patients with ALT <2 upper limit of normal
- LncRNA SNHG15 regulates osteosarcoma progression in vitro and in vivo via sponging miR-346 and regulating TRAF4 expression
- LINC00202 promotes retinoblastoma progression by regulating cell proliferation, apoptosis, and aerobic glycolysis through miR-204-5p/HMGCR axis
- Coexisting flavonoids and administration route effect on pharmacokinetics of Puerarin in MCAO rats
- GeneXpert Technology for the diagnosis of HIV-associated tuberculosis: Is scale-up worth it?
- Circ_001569 regulates FLOT2 expression to promote the proliferation, migration, invasion and EMT of osteosarcoma cells through sponging miR-185-5p
- Lnc-PICSAR contributes to cisplatin resistance by miR-485-5p/REV3L axis in cutaneous squamous cell carcinoma
- BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells
- MYL6B drives the capabilities of proliferation, invasion, and migration in rectal adenocarcinoma through the EMT process
- Inhibition of lncRNA LINC00461/miR-216a/aquaporin 4 pathway suppresses cell proliferation, migration, invasion, and chemoresistance in glioma
- Upregulation of miR-150-5p alleviates LPS-induced inflammatory response and apoptosis of RAW264.7 macrophages by targeting Notch1
- Long non-coding RNA LINC00704 promotes cell proliferation, migration, and invasion in papillary thyroid carcinoma via miR-204-5p/HMGB1 axis
- Neuroanatomy of melanocortin-4 receptor pathway in the mouse brain
- Lipopolysaccharides promote pulmonary fibrosis in silicosis through the aggravation of apoptosis and inflammation in alveolar macrophages
- Influences of advanced glycosylation end products on the inner blood–retinal barrier in a co-culture cell model in vitro
- MiR-4328 inhibits proliferation, metastasis and induces apoptosis in keloid fibroblasts by targeting BCL2 expression
- Aberrant expression of microRNA-132-3p and microRNA-146a-5p in Parkinson’s disease patients
- Long non-coding RNA SNHG3 accelerates progression in glioma by modulating miR-384/HDGF axis
- Long non-coding RNA NEAT1 mediates MPTP/MPP+-induced apoptosis via regulating the miR-124/KLF4 axis in Parkinson’s disease
- PCR-detectable Candida DNA exists a short period in the blood of systemic candidiasis murine model
- CircHIPK3/miR-381-3p axis modulates proliferation, migration, and glycolysis of lung cancer cells by regulating the AKT/mTOR signaling pathway
- Reversine and herbal Xiang–Sha–Liu–Jun–Zi decoction ameliorate thioacetamide-induced hepatic injury by regulating the RelA/NF-κB/caspase signaling pathway
- Therapeutic effects of coronary granulocyte colony-stimulating factor on rats with chronic ischemic heart disease
- The effects of yam gruel on lowering fasted blood glucose in T2DM rats
- Circ_0084043 promotes cell proliferation and glycolysis but blocks cell apoptosis in melanoma via circ_0084043-miR-31-KLF3 axis
- CircSAMD4A contributes to cell doxorubicin resistance in osteosarcoma by regulating the miR-218-5p/KLF8 axis
- Relationship of FTO gene variations with NAFLD risk in Chinese men
- The prognostic and predictive value of platelet parameters in diabetic and nondiabetic patients with sudden sensorineural hearing loss
- LncRNA SNHG15 contributes to doxorubicin resistance of osteosarcoma cells through targeting the miR-381-3p/GFRA1 axis
- miR-339-3p regulated acute pancreatitis induced by caerulein through targeting TNF receptor-associated factor 3 in AR42J cells
- LncRNA RP1-85F18.6 affects osteoblast cells by regulating the cell cycle
- MiR-203-3p inhibits the oxidative stress, inflammatory responses and apoptosis of mice podocytes induced by high glucose through regulating Sema3A expression
- MiR-30c-5p/ROCK2 axis regulates cell proliferation, apoptosis and EMT via the PI3K/AKT signaling pathway in HG-induced HK-2 cells
- CTRP9 protects against MIA-induced inflammation and knee cartilage damage by deactivating the MAPK/NF-κB pathway in rats with osteoarthritis
- Relationship between hemodynamic parameters and portal venous pressure in cirrhosis patients with portal hypertension
- Long noncoding RNA FTX ameliorates hydrogen peroxide-induced cardiomyocyte injury by regulating the miR-150/KLF13 axis
- Ropivacaine inhibits proliferation, migration, and invasion while inducing apoptosis of glioma cells by regulating the SNHG16/miR-424-5p axis
- CD11b is involved in coxsackievirus B3-induced viral myocarditis in mice by inducing Th17 cells
- Decitabine shows anti-acute myeloid leukemia potential via regulating the miR-212-5p/CCNT2 axis
- Testosterone aggravates cerebral vascular injury by reducing plasma HDL levels
- Bioengineering and Biotechnology
- PL/Vancomycin/Nano-hydroxyapatite Sustained-release Material to Treat Infectious Bone Defect
- The thickness of surface grafting layer on bio-materials directly mediates the immuno-reacitivity of macrophages in vitro
- Silver nanoparticles: synthesis, characterisation and biomedical applications
- Food Science
- Bread making potential of Triticum aestivum and Triticum spelta species
- Modeling the effect of heat treatment on fatty acid composition in home-made olive oil preparations
- Effect of addition of dried potato pulp on selected quality characteristics of shortcrust pastry cookies
- Preparation of konjac oligoglucomannans with different molecular weights and their in vitro and in vivo antioxidant activities
- Animal Sciences
- Changes in the fecal microbiome of the Yangtze finless porpoise during a short-term therapeutic treatment
- Agriculture
- Influence of inoculation with Lactobacillus on fermentation, production of 1,2-propanediol and 1-propanol as well as Maize silage aerobic stability
- Application of extrusion-cooking technology in hatchery waste management
- In-field screening for host plant resistance to Delia radicum and Brevicoryne brassicae within selected rapeseed cultivars and new interspecific hybrids
- Studying of the promotion mechanism of Bacillus subtilis QM3 on wheat seed germination based on β-amylase
- Rapid visual detection of FecB gene expression in sheep
- Effects of Bacillus megaterium on growth performance, serum biochemical parameters, antioxidant capacity, and immune function in suckling calves
- Effects of center pivot sprinkler fertigation on the yield of continuously cropped soybean
- Special Issue On New Approach To Obtain Bioactive Compounds And New Metabolites From Agro-Industrial By-Products
- Technological and antioxidant properties of proteins obtained from waste potato juice
- The aspects of microbial biomass use in the utilization of selected waste from the agro-food industry
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part I
- Automatic detection and segmentation of adenomatous colorectal polyps during colonoscopy using Mask R-CNN
- The impedance analysis of small intestine fusion by pulse source
- Errata
- Erratum to “Diagnostic performance of serum CK-MB, TNF-α and hs-CRP in children with viral myocarditis”
- Erratum to “MYL6B drives the capabilities of proliferation, invasion, and migration in rectal adenocarcinoma through the EMT process”
- Erratum to “Thermostable cellulase biosynthesis from Paenibacillus alvei and its utilization in lactic acid production by simultaneous saccharification and fermentation”
Artikel in diesem Heft
- Plant Sciences
- Dependence of the heterosis effect on genetic distance, determined using various molecular markers
- Plant Growth Promoting Rhizobacteria (PGPR) Regulated Phyto and Microbial Beneficial Protein Interactions
- Role of strigolactones: Signalling and crosstalk with other phytohormones
- An efficient protocol for regenerating shoots from paper mulberry (Broussonetia papyrifera) leaf explants
- Functional divergence and adaptive selection of KNOX gene family in plants
- In silico identification of Capsicum type III polyketide synthase genes and expression patterns in Capsicum annuum
- In vitro induction and characterisation of tetraploid drumstick tree (Moringa oleifera Lam.)
- CRISPR/Cas9 or prime editing? – It depends on…
- Study on the optimal antagonistic effect of a bacterial complex against Monilinia fructicola in peach
- Natural variation in stress response induced by low CO2 in Arabidopsis thaliana
- The complete mitogenome sequence of the coral lily (Lilium pumilum) and the Lanzhou lily (Lilium davidii) in China
- Ecology and Environmental Sciences
- Use of phosphatase and dehydrogenase activities in the assessment of calcium peroxide and citric acid effects in soil contaminated with petrol
- Analysis of ethanol dehydration using membrane separation processes
- Activity of Vip3Aa1 against Periplaneta americana
- Thermostable cellulase biosynthesis from Paenibacillus alvei and its utilization in lactic acid production by simultaneous saccharification and fermentation
- Spatiotemporal dynamics of terrestrial invertebrate assemblages in the riparian zone of the Wewe river, Ashanti region, Ghana
- Antifungal activity of selected volatile essential oils against Penicillium sp.
- Toxic effect of three imidazole ionic liquids on two terrestrial plants
- Biosurfactant production by a Bacillus megaterium strain
- Distribution and density of Lutraria rhynchaena Jonas, 1844 relate to sediment while reproduction shows multiple peaks per year in Cat Ba-Ha Long Bay, Vietnam
- Biomedical Sciences
- Treatment of Epilepsy Associated with Common Chromosomal Developmental Diseases
- A Mouse Model for Studying Stem Cell Effects on Regeneration of Hair Follicle Outer Root Sheaths
- Morphine modulates hippocampal neurogenesis and contextual memory extinction via miR-34c/Notch1 pathway in male ICR mice
- Composition, Anticholinesterase and Antipedicular Activities of Satureja capitata L. Volatile Oil
- Weight loss may be unrelated to dietary intake in the imiquimod-induced plaque psoriasis mice model
- Construction of recombinant lentiviral vector containing human stem cell leukemia gene and its expression in interstitial cells of cajal
- Knockdown of lncRNA KCNQ1OT1 inhibits glioma progression by regulating miR-338-3p/RRM2
- Protective effect of asiaticoside on radiation-induced proliferation inhibition and DNA damage of fibroblasts and mice death
- Prevalence of dyslipidemia in Tibetan monks from Gansu Province, Northwest China
- Sevoflurane inhibits proliferation, invasion, but enhances apoptosis of lung cancer cells by Wnt/β-catenin signaling via regulating lncRNA PCAT6/ miR-326 axis
- MiR-542-3p suppresses neuroblastoma cell proliferation and invasion by downregulation of KDM1A and ZNF346
- Calcium Phosphate Cement Causes Nucleus Pulposus Cell Degeneration Through the ERK Signaling Pathway
- Human Dental Pulp Stem Cells Exhibit Osteogenic Differentiation Potential
- MiR-489-3p inhibits cell proliferation, migration, and invasion, and induces apoptosis, by targeting the BDNF-mediated PI3K/AKT pathway in glioblastoma
- Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating the microRNA-34a-5p/NOTCH1 signaling pathway
- Large Brunner’s gland adenoma of the duodenum for almost 10 years
- Neurotrophin-3 accelerates reendothelialization through inducing EPC mobilization and homing
- Hepatoprotective effects of chamazulene against alcohol-induced liver damage by alleviation of oxidative stress in rat models
- FXYD6 overexpression in HBV-related hepatocellular carcinoma with cirrhosis
- Risk factors for elevated serum colorectal cancer markers in patients with type 2 diabetes mellitus
- Effect of hepatic sympathetic nerve removal on energy metabolism in an animal model of cognitive impairment and its relationship to Glut2 expression
- Progress in research on the role of fibrinogen in lung cancer
- Advanced glycation end product levels were correlated with inflammation and carotid atherosclerosis in type 2 diabetes patients
- MiR-223-3p regulates cell viability, migration, invasion, and apoptosis of non-small cell lung cancer cells by targeting RHOB
- Knockdown of DDX46 inhibits trophoblast cell proliferation and migration through the PI3K/Akt/mTOR signaling pathway in preeclampsia
- Buformin suppresses osteosarcoma via targeting AMPK signaling pathway
- Effect of FibroScan test in antiviral therapy for HBV-infected patients with ALT <2 upper limit of normal
- LncRNA SNHG15 regulates osteosarcoma progression in vitro and in vivo via sponging miR-346 and regulating TRAF4 expression
- LINC00202 promotes retinoblastoma progression by regulating cell proliferation, apoptosis, and aerobic glycolysis through miR-204-5p/HMGCR axis
- Coexisting flavonoids and administration route effect on pharmacokinetics of Puerarin in MCAO rats
- GeneXpert Technology for the diagnosis of HIV-associated tuberculosis: Is scale-up worth it?
- Circ_001569 regulates FLOT2 expression to promote the proliferation, migration, invasion and EMT of osteosarcoma cells through sponging miR-185-5p
- Lnc-PICSAR contributes to cisplatin resistance by miR-485-5p/REV3L axis in cutaneous squamous cell carcinoma
- BRCA1 subcellular localization regulated by PI3K signaling pathway in triple-negative breast cancer MDA-MB-231 cells and hormone-sensitive T47D cells
- MYL6B drives the capabilities of proliferation, invasion, and migration in rectal adenocarcinoma through the EMT process
- Inhibition of lncRNA LINC00461/miR-216a/aquaporin 4 pathway suppresses cell proliferation, migration, invasion, and chemoresistance in glioma
- Upregulation of miR-150-5p alleviates LPS-induced inflammatory response and apoptosis of RAW264.7 macrophages by targeting Notch1
- Long non-coding RNA LINC00704 promotes cell proliferation, migration, and invasion in papillary thyroid carcinoma via miR-204-5p/HMGB1 axis
- Neuroanatomy of melanocortin-4 receptor pathway in the mouse brain
- Lipopolysaccharides promote pulmonary fibrosis in silicosis through the aggravation of apoptosis and inflammation in alveolar macrophages
- Influences of advanced glycosylation end products on the inner blood–retinal barrier in a co-culture cell model in vitro
- MiR-4328 inhibits proliferation, metastasis and induces apoptosis in keloid fibroblasts by targeting BCL2 expression
- Aberrant expression of microRNA-132-3p and microRNA-146a-5p in Parkinson’s disease patients
- Long non-coding RNA SNHG3 accelerates progression in glioma by modulating miR-384/HDGF axis
- Long non-coding RNA NEAT1 mediates MPTP/MPP+-induced apoptosis via regulating the miR-124/KLF4 axis in Parkinson’s disease
- PCR-detectable Candida DNA exists a short period in the blood of systemic candidiasis murine model
- CircHIPK3/miR-381-3p axis modulates proliferation, migration, and glycolysis of lung cancer cells by regulating the AKT/mTOR signaling pathway
- Reversine and herbal Xiang–Sha–Liu–Jun–Zi decoction ameliorate thioacetamide-induced hepatic injury by regulating the RelA/NF-κB/caspase signaling pathway
- Therapeutic effects of coronary granulocyte colony-stimulating factor on rats with chronic ischemic heart disease
- The effects of yam gruel on lowering fasted blood glucose in T2DM rats
- Circ_0084043 promotes cell proliferation and glycolysis but blocks cell apoptosis in melanoma via circ_0084043-miR-31-KLF3 axis
- CircSAMD4A contributes to cell doxorubicin resistance in osteosarcoma by regulating the miR-218-5p/KLF8 axis
- Relationship of FTO gene variations with NAFLD risk in Chinese men
- The prognostic and predictive value of platelet parameters in diabetic and nondiabetic patients with sudden sensorineural hearing loss
- LncRNA SNHG15 contributes to doxorubicin resistance of osteosarcoma cells through targeting the miR-381-3p/GFRA1 axis
- miR-339-3p regulated acute pancreatitis induced by caerulein through targeting TNF receptor-associated factor 3 in AR42J cells
- LncRNA RP1-85F18.6 affects osteoblast cells by regulating the cell cycle
- MiR-203-3p inhibits the oxidative stress, inflammatory responses and apoptosis of mice podocytes induced by high glucose through regulating Sema3A expression
- MiR-30c-5p/ROCK2 axis regulates cell proliferation, apoptosis and EMT via the PI3K/AKT signaling pathway in HG-induced HK-2 cells
- CTRP9 protects against MIA-induced inflammation and knee cartilage damage by deactivating the MAPK/NF-κB pathway in rats with osteoarthritis
- Relationship between hemodynamic parameters and portal venous pressure in cirrhosis patients with portal hypertension
- Long noncoding RNA FTX ameliorates hydrogen peroxide-induced cardiomyocyte injury by regulating the miR-150/KLF13 axis
- Ropivacaine inhibits proliferation, migration, and invasion while inducing apoptosis of glioma cells by regulating the SNHG16/miR-424-5p axis
- CD11b is involved in coxsackievirus B3-induced viral myocarditis in mice by inducing Th17 cells
- Decitabine shows anti-acute myeloid leukemia potential via regulating the miR-212-5p/CCNT2 axis
- Testosterone aggravates cerebral vascular injury by reducing plasma HDL levels
- Bioengineering and Biotechnology
- PL/Vancomycin/Nano-hydroxyapatite Sustained-release Material to Treat Infectious Bone Defect
- The thickness of surface grafting layer on bio-materials directly mediates the immuno-reacitivity of macrophages in vitro
- Silver nanoparticles: synthesis, characterisation and biomedical applications
- Food Science
- Bread making potential of Triticum aestivum and Triticum spelta species
- Modeling the effect of heat treatment on fatty acid composition in home-made olive oil preparations
- Effect of addition of dried potato pulp on selected quality characteristics of shortcrust pastry cookies
- Preparation of konjac oligoglucomannans with different molecular weights and their in vitro and in vivo antioxidant activities
- Animal Sciences
- Changes in the fecal microbiome of the Yangtze finless porpoise during a short-term therapeutic treatment
- Agriculture
- Influence of inoculation with Lactobacillus on fermentation, production of 1,2-propanediol and 1-propanol as well as Maize silage aerobic stability
- Application of extrusion-cooking technology in hatchery waste management
- In-field screening for host plant resistance to Delia radicum and Brevicoryne brassicae within selected rapeseed cultivars and new interspecific hybrids
- Studying of the promotion mechanism of Bacillus subtilis QM3 on wheat seed germination based on β-amylase
- Rapid visual detection of FecB gene expression in sheep
- Effects of Bacillus megaterium on growth performance, serum biochemical parameters, antioxidant capacity, and immune function in suckling calves
- Effects of center pivot sprinkler fertigation on the yield of continuously cropped soybean
- Special Issue On New Approach To Obtain Bioactive Compounds And New Metabolites From Agro-Industrial By-Products
- Technological and antioxidant properties of proteins obtained from waste potato juice
- The aspects of microbial biomass use in the utilization of selected waste from the agro-food industry
- Special Issue on Computing and Artificial Techniques for Life Science Applications - Part I
- Automatic detection and segmentation of adenomatous colorectal polyps during colonoscopy using Mask R-CNN
- The impedance analysis of small intestine fusion by pulse source
- Errata
- Erratum to “Diagnostic performance of serum CK-MB, TNF-α and hs-CRP in children with viral myocarditis”
- Erratum to “MYL6B drives the capabilities of proliferation, invasion, and migration in rectal adenocarcinoma through the EMT process”
- Erratum to “Thermostable cellulase biosynthesis from Paenibacillus alvei and its utilization in lactic acid production by simultaneous saccharification and fermentation”

