Abstract
In this study, we aimed to characterize the liver protein profile of Chaohu ducks using two-dimensional electrophoresis and proteomics. The livers were quickly collected from 120 healthy, 84-day-old Chaohu ducks. The intramuscular fat (IMF) content of the left pectoralis muscle was determined using the Soxhlet extraction method. The total protein of liver tissues from the high and low IMF groups was extracted for proteomics. Functional enrichment analysis of the differentially expressed proteins (DEPs) was conducted using gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG). In total, 43 DEPs were identified. Functional enrichment analysis indicated that these DEPs were significantly related to four lipid metabolic processes: carboxylic acid metabolic process, ATP metabolic process, oxoacid metabolic process, and organic acid metabolic process. Three pathways correlated with lipid metabolism were identified using KEGG analysis: glycolysis/gluconeogenesis, pentose phosphate pathway, fructose, and mannose metabolism. Eight key proteins associated with lipid metabolism were identified: ALDOB, GAPDH, ENO1, RGN, TPI1, HSPA9, PRDX1, and GPX1. Protein–protein interaction analysis revealed that the glycolysis/gluconeogenesis pathway mediated the interaction relationship. Key proteins and metabolic pathways were closely related to lipid metabolism and showed a strong interaction in Chaohu ducks.
1 Introduction
Poultry meat is one of the most common animal food sources, accounting for approximately 30% of global meat consumption; duck products (second only to chicken products) are increasingly gaining popularity [1]. The body fat of livestock is affected by many factors, and the genetic characteristics of poultry determine the rate of fat deposition [2,3]. Therefore, selecting and breeding high-quality and lean poultry varieties is essential to reduce excessive body fat deposition [4]. Chaohu duck is a meat–egg type duck variety, originally bred around the Chaohu Lake basin in Anhui Province; it is an excellent local waterfowl variety in the nation. Chaohu duck is favored by consumers owing to its tender meat, low subcutaneous fat content, and delicious taste. However, the current feeding method for Chaohu ducks causes excessive fat deposition in their bodies owing to improved growth speed and shorter breeding cycles [5,6]. Lipid metabolism affects the growth and development of poultry and meat quality traits. Therefore, exploring the molecular mechanism of body fat deposition in Chinese native ducks and understanding how to cultivate appropriate body fat content is essential in poultry breeding.
The fat deposited in the muscles primarily affects the muscle quality, and an appropriate increase in intramuscular fat (IMF) can improve the meat quality [7] and shorten the sexual maturation time [8]. The deposition of IMF is comprehensively regulated by multiple signaling pathways [9]. The liver is the main site for the biosynthesis of sugar, fat, and protein, and these three nutrients are interconvertible through key metabolites (such as acetyl-CoA) [10]. Using two-dimensional gel electrophoresis (2-DE) proteomics, the livers from obese and lean Peking ducks were compared, and 76 proteins were found to be differentially expressed in the two types of ducks [11]. We conducted a proteomic analysis of the breast muscle of Beijing You-chicken from the embryonic stage to the early postnatal stage. There were 77 proteins in the slow type and 68 in the fast type of You-chicken [12]. Lipid production in domestic birds differs from that in mammals. Adipose tissue may play a minor role, whereas the liver may play a major role in de novo fatty acid synthesis. Approximately 96% of body fat is derived from liver synthesis and transport. Few in-depth studies are conducted on the correlation between the changes in IMF of Chaohu ducks and the regulatory proteins related to lipogenesis in the liver.
In this study, the liver of Chaohu ducks was selected as the study material; the proteins involved in the key regulation of fat generation, transport, and metabolism were screened through 2-DE, which provided a reference for studying the molecular mechanism of fat metabolism of Chaohu ducks and poultry and identifying candidate proteins for the genetic improvement of the quality traits of Chaohu duck meat.
2 Methods
2.1 Animals and feeding
In total, 500 healthy male ducks that were 1-day old were randomly selected from the original population of Chaohu ducks. They were bred under the same environmental conditions at Anhui Yongqiang Agricultural Science and Technology Co., Ltd (Anqing City, China). These ducks freely consumed basal pellet feed and water. At 21 days of age, 180 drakes were selected according to their body weight; they were fed for 84 days in separate duck sheds (60 ducks per shed). The indoor temperature of the duck house was 25 ± 2°C and the relative humidity was 70.5 ± 8.0%. During the entire feeding period, the basic feed of Chaohu ducks was formulated according to the nutritional standard of meat ducks of the China Agricultural Standard (Standard number: NY/T 2122-2012). All methods were carried out in accordance with the relevant guidelines and regulations and were reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org) for reporting animal experiments.
-
Ethical approval: The research related to animal use has been complied with all the relevant national regulations and institutional policies for the care and use of animals. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the Anhui Agricultural University, Hefei, China, under permit No. ZXD-P20140809.
2.2 Group design and sample collection
Chaohu ducks aged 84 days were fasted for 12 h and weighed. Based on the average body weight, 40 ducks were selected from each cage, and 120 ducks were selected for sample collection. The ducks were slaughtered using electric shock. Twenty grams of the left pectoral muscle was collected and stored at −20°C for IMF determination. Five grams of the liver was collected, stored in liquid nitrogen, and transferred to −80°C for protein extraction. The results of IMF content and statistical analysis of Chaohu ducks have been reported in our previous study [2]. The Chaohu ducts were divided into two groups according to the distribution rule of IMF content in breast muscle: extremely high group (CH) and extremely low group (CL). Three liver samples were collected from the CH and CL groups.
2.3 Extraction and concentration determination of total liver protein
Total protein was extracted from the liver samples using a mammalian proteome kit (Focus™ CAT# 786-246, G-Biosciences, Inc., USA) according to the manufacturer’s instructions. Briefly, 200 mg of the liver sample was added to 500 μL of protein solubilization buffer and homogenized for 4 min at 4°C at intervals of 10 s. The homogenate was centrifuged at 20,000×g for 30 min at 20°C. Subsequently, the supernatant was transferred to a clean tube and stored at −80°C until use. The total protein concentration was determined by the Bradford method (P0006C, Beyotime Biotechnology Co., Ltd, China) using an automatic enzyme label analyzer (iMark 168-1002XC, Bio-Rad, Inc., USA).
The proteins were pretreated for desalination using the acetone precipitation method before conducting 2-DE. Briefly, the protein solution was mixed with acetone at a volume ratio of 1:4 and incubated at −20°C for 12 h. Next, the mixture was centrifuged at 12,000×g for 10 min. After discarding the supernatant, the precipitant was air-dried at 20°C and was dissolved in a 10 mL buffer (same as the 2-DE loading buffer I) containing 4.805 g urea, 0.4 g CHAPS, 0.098 g DTT, 50 μL Bio-Lyte, 10 μL bromophenol blue, and ultrapure water. Desalted protein was used in the 2D test.
2.4 Separation of total protein from samples
The main reagents for 2-DE test were prepared, including immobilized pH gradient (IPG, pH 3–10, 7 cm, 163–2,000, Bio-Rad Inc., USA), hydrated loading buffer (I), tape balance buffer (I), tape balance buffer (II), 12% SDS-PAGE polyacrylamide gel, and Coomassie Bright Blue R-250 staining solution.
Isoelectric focusing electrophoresis (IEF): First, the sample protein (300 μg) was added to the hydration loading buffer (I) to prepare a protein-loading liquid system with a concentration of 3 mg/mL. The protein-loading liquid was added to the focusing tray, and the IPG tape was placed. The focusing plate was placed in an IEF apparatus (Protean IEF, Bio-Rad Inc., USA), and the isoelectric focusing program was set (Table 1). After electrophoresis, the tape was removed, and a tape-balancing process was performed.
SDS-PAGE electrophoresis: The 1× buffer solution was added to a vertical electrophoresis tank (Mini-PROTEAN Tetra cell, Bio-Rad Inc., USA). The balanced IPG strips were carefully transferred to the long glass plate in the interlayer to prepare the gel electrophoresis device. Electrophoresis was performed at 70 V. After the bromophenol blue indicator was completely separated from the IPG tape and concentrated in a line (approximately 20 min), the voltage was increased to 120 V, and the electrophoresis was continued for approximately 1 h.
Dyeing and decolorization of gels: The SDS-PAGE gel was transferred to a decolorization tray, stained with a Coomassie Bright Blue R-250 stain solution, and decolorized using a decolorization solution.
Procedure setting by IEF
| Step | Voltage (V) | Gradient | Time (h/Vhr) | Function |
|---|---|---|---|---|
| Hydrating | 50 | Gradually | 14 | Active hydration |
| S1 | 100 | Linear | 0.5 | Desalination |
| S2 | 250 | Linear | 1 | Desalination |
| S3 | 500 | Linear | 1 | Desalination |
| S4 | 1,000 | Linear | 1 | Desalination |
| S5 | 2,000 | Linear | 1 | Desalination |
| S6 | 3,000 | Linear | 1 | Desalination |
| S7 | 4,000 | Linear | 3 | Desalination |
| S8 | 4,000 | Rapid | 20,000 | Focusing |
| S9 | 500 | Rapid | Any time | Protection |
2.5 Gel scanning and image analysis
According to the operating procedures of a calibrated optical densimeter (GS-900™, Bio-Rad Inc., USA), the gel stained with Coomassie Bright Blue was scanned, and the image information regarding three samples in the CH group and three samples in the CL group was obtained. According to the operating procedure of PDQuest 2-DE analysis software (V8.0.1) from Bio-Rad, the abundance of protein spots was analyzed on the scanning maps of each sample in the CH group and CL group, and the difference in protein spots between the two groups was screened. The analysis parameters were as follows: low quantitative value, 20; protein spot sensitivity, 15; scale size, 3; minimum peak value, 4,000; t-test, 95% confidence interval. The gel spots with a difference ratio of optical density (OD) ≥1.5 were identified as differentially expressed protein (DEP) spots.
2.6 Enzymatic hydrolysis and mass spectrometry analysis of differential protein gels
According to the image analysis data, gel points at the same position in the two groups of samples were cut and placed in a new centrifuge tube.
The gel point was rinsed thrice with ultrapure water and decolorized. Each centrifuge tube was treated with 5 μL trypsin (0.01 μg/μL) and placed on a 384-well stainless-steel plate. These samples were analyzed using mass spectrometry (MALDI-TOF-MS Analyzer 5800, AB Sciex Inc., USA). The original MS files collected by mass spectrometer were processed, retrieved, analyzed, and identified by Mascot software (V2.5.1), with reference to the NCBI-Anas platyrhynchos protein database (https://www.uniprot.org/).
2.7 Bioinformatics analysis of DEPs
The DEPs identified through mass spectrometry (P ≤ 0.05) were analyzed using bioinformatics, including gene ontology (GO) enrichment analysis, kyoto encyclopedia of genes and genomes (KEGG) pathway analysis, and protein–protein interaction (PPI) analysis. Based on the QuickGO database (http://www.geneontology.org) and the KEGG database (https://www.kegg.jp/), the significantly enriched GO terms and KEGG pathways were screened at P ≤ 0.05. A PPI pathway map was developed by mapping the DEPs and their related pathways into STRING online database (http://string.embl.de).
3 Results
3.1 Total protein concentration of the sample
According to the standard protein curve of Bradford’s method (Figure 1), protein concentrations of six samples in the CL and CH groups were calculated, which ranged from 21.64 to 27.24 μg/μL, and the total protein content of each sample was more than 6,492 μg (Table 2).
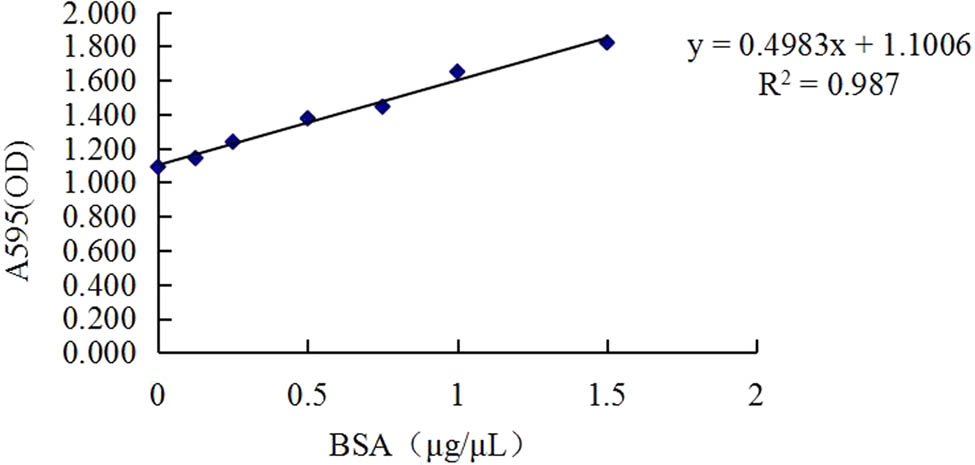
Standard Bradford curve for protein contents. Note: The x-axis denotes BSA, bovine serum albumin concentration (μg/μL); y-axis denotes OD value at A595, absorbance of the microplate reader at a wavelength of 595 nm.
Concentration of liver protein sample from Chaohu ducks
| Sample | OD | Concentration (μg/μL) | Volume (μL) | Protein weight (μg) |
|---|---|---|---|---|
| CL-1 | 2.30 | 24.13 | 300 | 7,239 |
| CL-2 | 2.25 | 23.15 | 300 | 6,945 |
| CL-3 | 2.18 | 21.64 | 300 | 6,492 |
| CH-1 | 2.46 | 27.24 | 300 | 8,172 |
| CH-2 | 2.35 | 25.03 | 300 | 7,509 |
| CH-3 | 2.29 | 23.99 | 300 | 7,197 |
3.2 Gel analysis of the total protein in CH and CL groups
The total liver proteins of the CH and CL groups were separated using 2-DE, and the protein separation maps in each group showed repetitions, with clear protein spots and few background impurities (Figure 2). The PDQuest analysis software was used for image comparison and analysis. There were 68 differential protein spots in the total protein map of liver samples between the CH and CL groups. The screened differential protein spots were labeled on the gel map of the CH and CL groups (Figure 3).
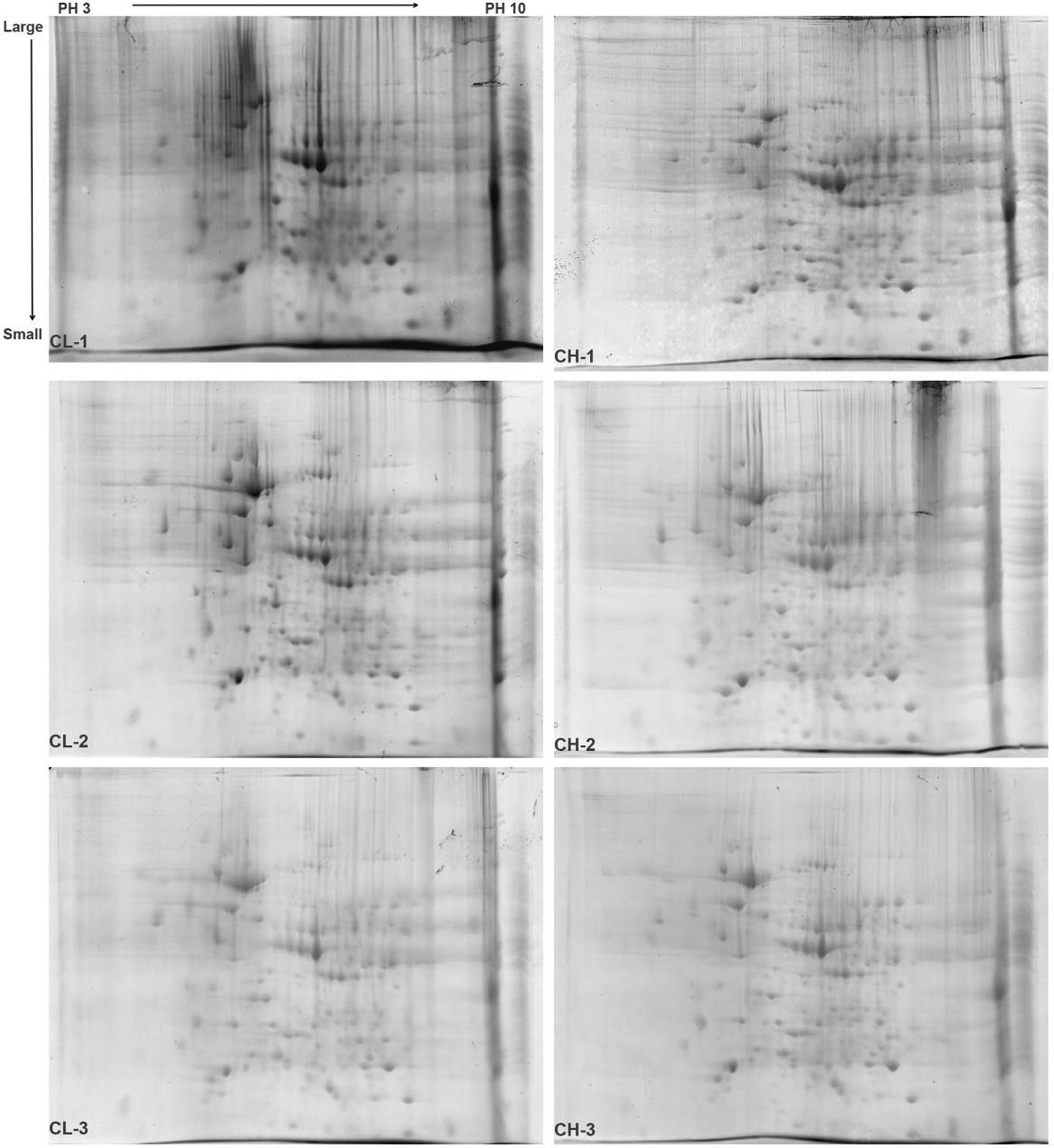
2-DE gel diagram of liver protein samples in Chaohu ducks. Note: PH 3 → 10 is the isoelectric point of the protein from the IPG strip, which gradually increases from left to right. Large → Small is the molecular weight of the protein that gradually decreases from top to bottom.
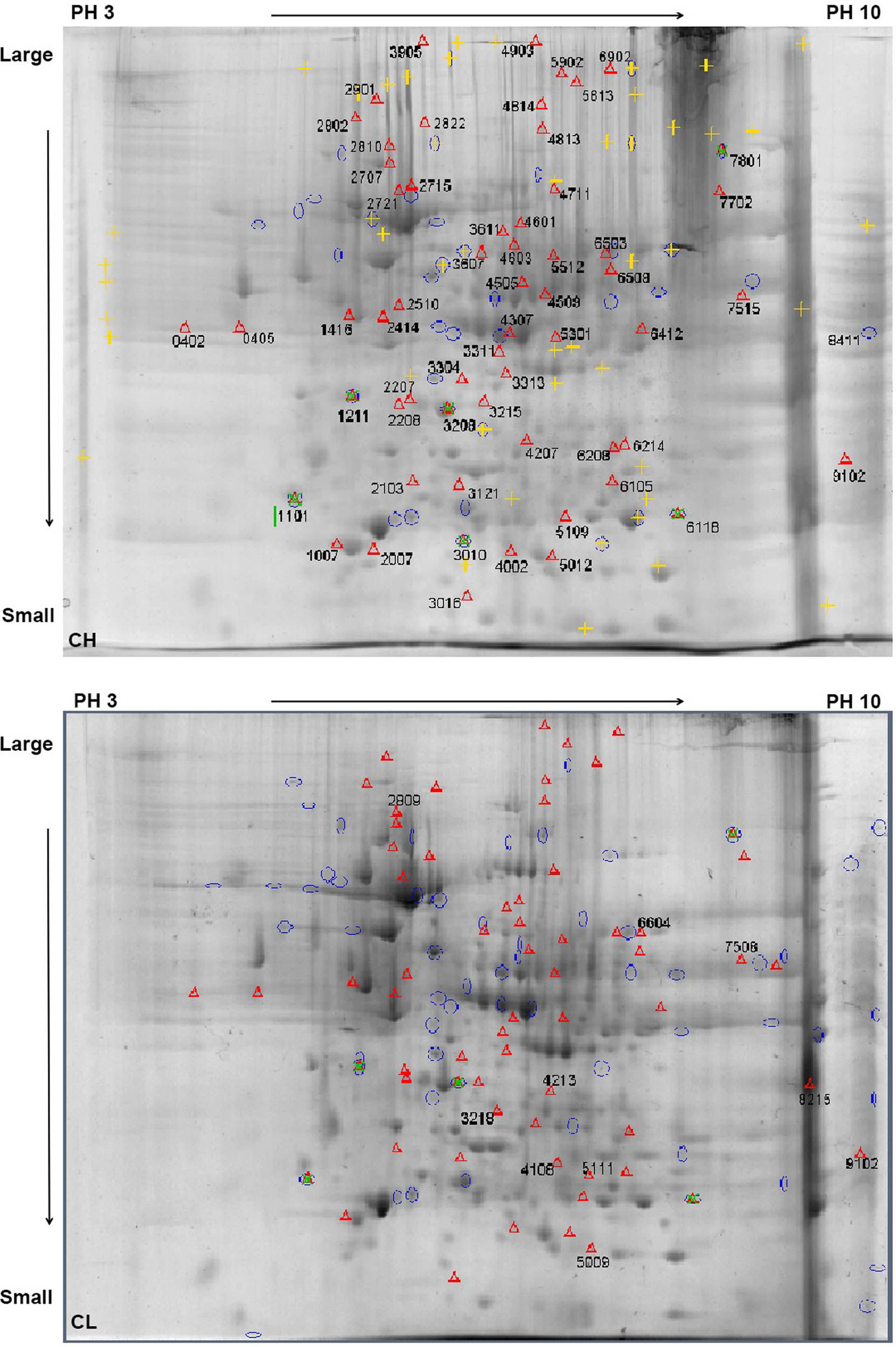
Alignment of differential protein points from Chaohu duck protein samples in 2-DE gel. Note: Red triangle mark and serial number “Δ0402” on the gel are the different protein points of the CH group compared with the CL group. pH 3 → 10 is the isoelectric point of the protein from the IPG strip, which gradually increases from left to right. Large → Small is the molecular weight of the protein gradually decreasing from top to bottom.
3.3 Identification of differential protein spots using mass spectrometry
The 68 DEPs were digested with trypsin and identified using mass spectrometry. According to the NCBI-Anas platyrhynchos protein database, 43 DEPs, including fructose bisphosphate aldolase B (ALDOB), were screened. All the DEPs were upregulated proteins (Table A1).
3.4 GO enrichment analysis of DEPs
The 43 DEPs were mapped to the GO database for enrichment analysis. The significantly enriched GO functional items included 598 biological processes (1,448 in total), 77 cell components (214 in total), and 118 molecular functions (256 in total).
Considering the top ten biological processes, the small molecule biosynthetic process (P = 2.22 × 10−12) had the highest statistically significant difference. Four metabolic processes were significantly related to lipid metabolism, including carboxylic acid, ATP, oxoacid, and organic acid metabolic processes (Figure 4). Additionally, eight DEPs were found to be significantly enriched: ALDOB, triosephosphate isomerase (TPI1), alpha-enolase (ENO1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), regucalcin (RGN), peroxiredoxin-1 (PRDX1), glutathione peroxidase (GPX1), and stress-70 protein family mitochondria (HSPA9). These DEPs were simultaneously involved in multiple GO functional processes related to carbohydrate and lipid metabolism, suggesting that these proteins are closely related to fat deposition in Chaohu ducks.
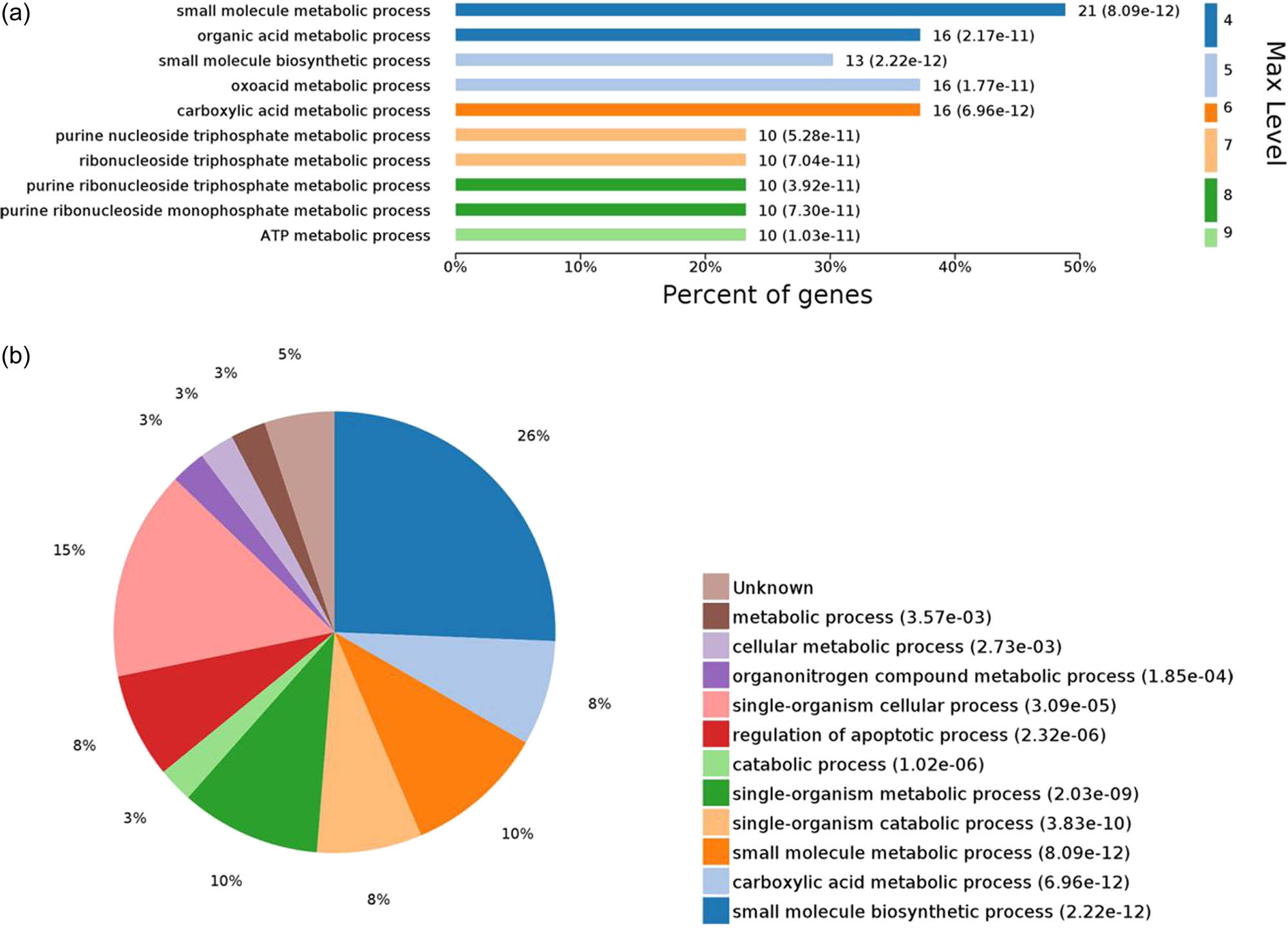
GO enriched biological process terms with statistically significant differences. Note: Map (a) shows the top ten biological processes for statistically significant differences; y-axis denotes the difference order at the maximum level terms; x-axis denotes the percentage of enriched proteins in each term. Map (b) shows the proportion distribution of all biological processes, with P ≤ 0.05.
3.5 KEGG pathway enrichment analysis of DEPs
The above-mentioned 43 DEPs were significantly enriched in nine pathways (P-value ≤ 0.05), including the biosynthesis of amino acids, glycolysis/gluconeogenesis, and pentose phosphate pathway. Among these, the biosynthesis of amino acids showed the highest statistically significant difference (P = 1.65 × 10−09). The following three pathways were related to lipid metabolism: glycolysis/gluconeogenesis (P = 2.60 × 10−8), pentose phosphate pathway (P = 7.85 × 10−5), and fructose and mannose metabolism (P = 2.47 × 10−4) (Figure 5). Six DEPs were significantly enriched: ALDOB, ENO1, GAPDH, TPI1, RGN, and HSPA9. In addition to lipid metabolism pathways, these DEPs were also involved in the biosynthesis of amino acids, carbon metabolism, RNA degradation, ascorbate, and aldarate metabolism. Thus, metabolic pathways of carbohydrates, lipids, and proteins are interrelated and responsible for fat deposition in the body.
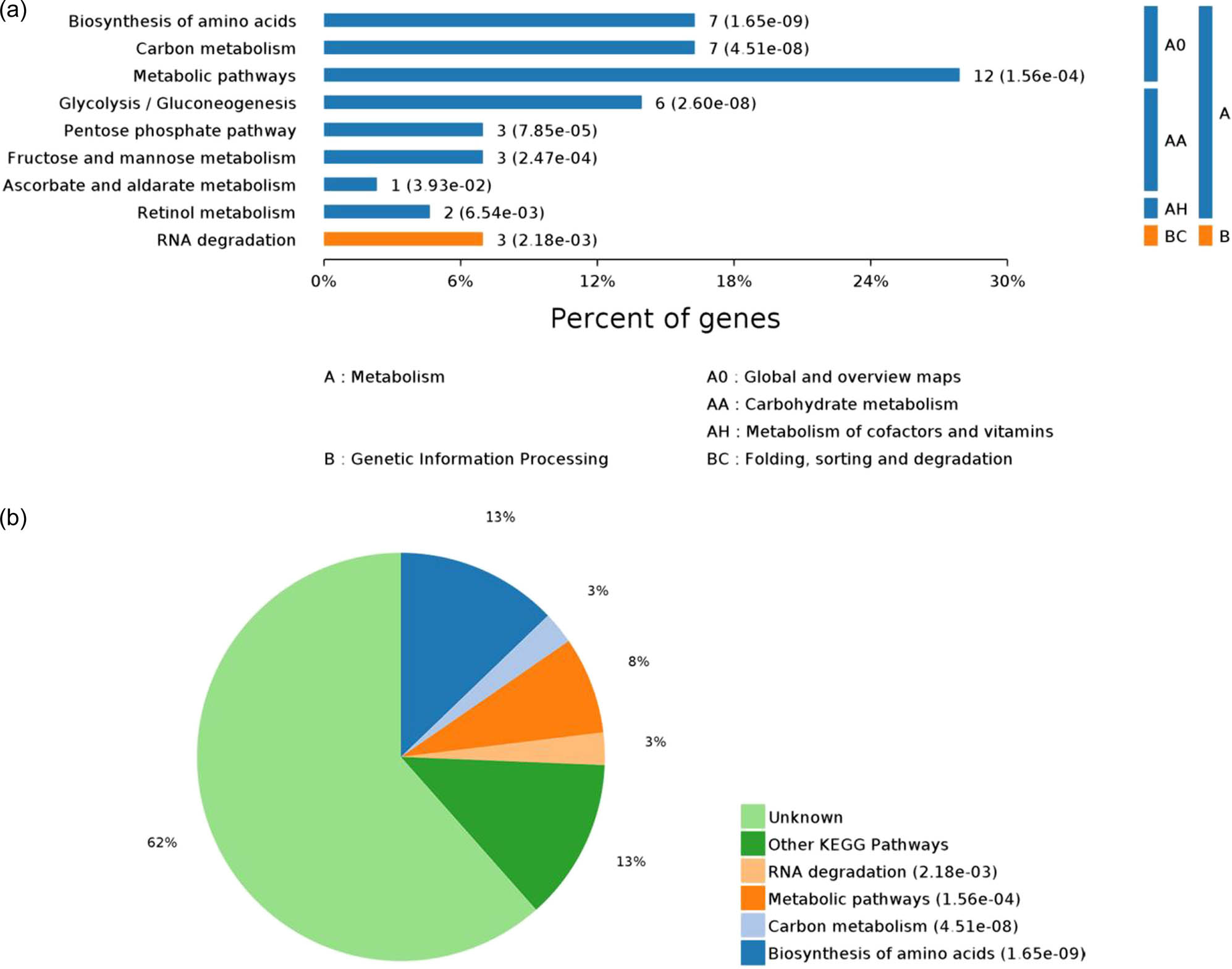
Pathways with significant differences in KEGG enrichment. Note: Map (a) shows all KEGG enriched pathways with P ≤ 0.05; y-axis denotes the pathway classification and name; x-axis denotes the percentage of enriched proteins in each pathway. Map (b) shows the proportion distribution of pathways with P ≤ 0.05.
3.6 PPI of DEPs related to lipid metabolism
Through the bioinformatic statistical analysis, three key metabolic pathways and eight key DEPs were found to play an important role in the regulation of lipid metabolism. The three key metabolic pathways included glycolysis/gluconeogenesis, pentose phosphate pathway, and fructose and mannose metabolism. The eight key proteins were ALDOB, GAPDH, ENO1, RGN, TPI1, HSPA9, PRDX1, and GPX1 (Table 3).
Lipid metabolism related regulatory proteins information
| Gene name | Uniprot-ID | Log2 fold change | Identity (%) | E-value | InterPro description |
|---|---|---|---|---|---|
| PRDX1 | P0CB50 | 3.063502942 | 98.99 | 5.00 × 10−145 | Peroxiredoxin-1 |
| HSPA9 | Q5ZM98 | 1.555816155 | 98.69 | 0 | Stress-70 protein mitochondrial |
| TPI1 | P00940 | 1.469885976 | 99.6 | 0 | Triosephosphate isomerase |
| ENO1 | P51913 | 1.189033824 | 97.24 | 0 | Alpha-enolase |
| GAPDH | P00356 | 1.163498732 | 99.1 | 0 | Glyceraldehyde-3-phosphate dehydrogenase |
| ALDOB | P07341 | 0.831877241 | 94.15 | 7.00 × 10−117 | Fructose-bisphosphate aldolase B |
| GPX1 | R4GH86 | 0.799087306 | 100 | 4.00 × 10−114 | Glutathione peroxidase |
| RGN | Q9I923 | 0.782408565 | 88.76 | 7.00 × 10−179 | Regucalcin |
The regulatory networks of these key metabolic pathways and DEPs were analyzed. The glycolysis and gluconeogenesis pathways showed the highest degree of interaction. TPI1, ENO1, GAPDH, ALDOB, and HSPA9 proteins were of high degree. The fold changes in the expression of ALDOB and PRDX1 were the highest (Figure 6).
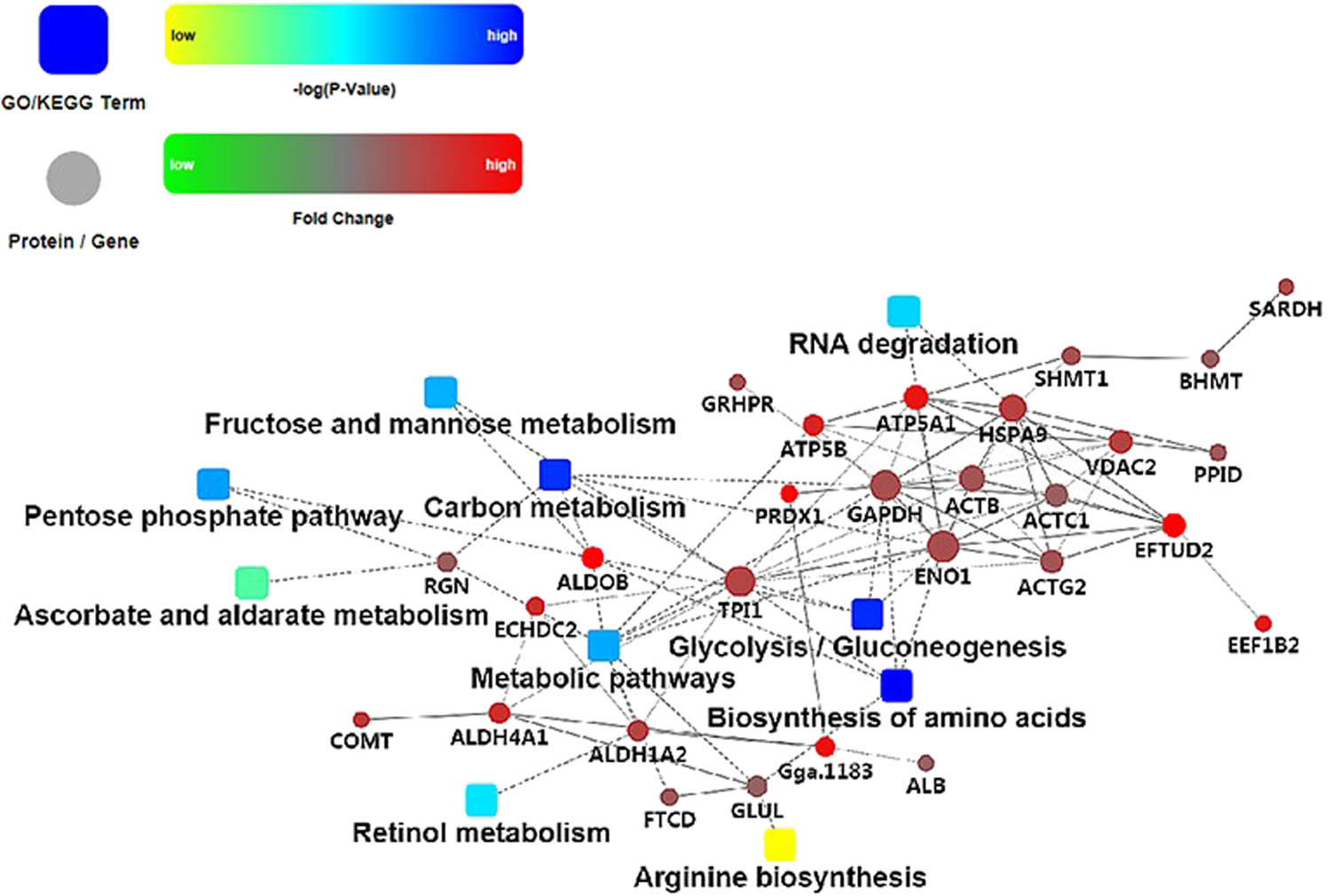
PPI from key regulatory proteins and metabolic pathways. Note: Dot represents proteins or genes, and the dot size indicates the intensity of interaction. Box represents GO or KEGG processes. Red and green are different fold changes; blue and yellow are the different log P-values. The solid line indicates interaction score >0.5 (dotted line indicates interaction score <0.5).
4 Discussion
Meat quality is reportedly affected by the IMF content [13]. In recent years, the meat quality of Chaohu ducks has deteriorated due to faster growth speed and shorter breeding cycles. Therefore, genetic improvements are needed to breed new varieties as per the needs of duck meat consumption. In this study, we used 2-DE combined with mass spectrometry to screen the key candidate proteins and elucidate the molecular mechanisms of lipid metabolism and fat deposition. We identified 43 DEPs between Chaohu ducks with high and low IMF, including ALDOB, GAPDH, ENO1, RGN, TPI1, HSPA9, PRDX1, and GPX1. Functional enrichment analysis revealed that DEPs were closely related to carbohydrate, lipid, and protein metabolism, including glycolysis/gluconeogenesis, pentose phosphate pathway, fructose, and mannose metabolism.
ALDOB (expressed in the liver) is one of the three isozymes of fructose-1,6-diphosphate aldolase in mammalian tissues, with the exception of ALDOA (expressed in muscle) and ALDOC (expressed in nervous tissue) [14]. Fructose-1,6-bisphosphate and aldolase mediate glucose metabolism by regulating the AMP-activated protein kinase pathway [15]. It plays a role in fructose catabolism, gluconeogenesis, and lipogenesis [16]. Enolase (ENO1 gene code), also known as 2-phospho-d-glycerol hydrolase, is normally present in the cytoplasm and contributes to glycolysis by catalyzing the conversion of 2-phosphoglycerate to phosphoenolpyruvate in the glycolytic pathway [17]. Enolases are abundant in the cytoplasm but are also present in the cell membrane and nucleus [18]. Previous studies speculated that ENO1 promotes the accumulation of tRNA transport-binding proteins on the surface of mitochondria and is involved in the transport of aminoacyl-tRNA synthase into mitochondria, playing a regulatory role in gene transcription [19]. Enolase also participates in pyruvate synthesis from d-glyceraldehyde-3-phosphate. The expression of these two enzymes in the liver increased significantly in this study and was associated with the decomposition and transformation of carbohydrates. These enzymes also provide raw materials for fat synthesis, causing excessive fat accumulation.
Glyceraldehyde-3-phosphate dehydrogenase (encoded by GAPDH) plays a role in glycolysis and nuclear function. As a key enzyme in glycolysis, GAPDH catalyzes the first step of the pathway by converting d-glyceraldehyde-3-phosphate to 3-phosphate-d-glyceryl phosphate [20]. In 1957, it was demonstrated that acyl phosphatase (GAPDH) isolated from the muscle catalyzes the hydrolysis of 1,3-bisphosphoglycerate to 3-phosphoglycerate. GAPDH is also involved in cellular and cytoplasmic vesicle transport and oxidative stress [21]. Propanone phosphate isomerase (TPI1) is a highly efficient metabolic enzyme that catalyzes the conversion of dihydroxyacetone phosphate (DHAP) to phosphoglyceraldehyde during glycolysis and gluconeogenesis [22]. Studies have shown that deficiency of TPI1 is related to abnormally high levels of DHAP accumulation and oxidative stress [23]. Propanone phosphoisomerase is responsible for ATP production from glycolysis; it also produces a pyruvate molecule for conversion to glucose molecules under aerobic and anaerobic conditions, allowing rapid equilibrium of the propanone phosphoaldase produced by glycolysis. This is associated with lipid metabolism through the glycerol-3-phosphate and pentose cycles [24]. We found that these two enzymes were also significantly enriched in the glycolysis/gluconeogenesis metabolic pathway of IMF deposition, with an upstream and downstream relationship in Chaohu ducks, which confirmed the results of previous studies.
Additionally, several proteins related to lipid metabolism and oxidative stress were identified in this study. RGN encoding regucalcin is a calcium-binding protein that regulates intracellular Ca2+ homeostasis, oxidative stress, cell survival, and apoptosis [25]. Yamaguchi et al. reported that RGN is distributed in rat hepatocellular plasma and has reversible effects on the activation and inhibition of various enzymes bound to Ca2+ [26]. A previous study also reported that overexpression of RGN enhanced lipid accumulation in adipocyte cells, suggesting that RGN may be a novel regulator of adipocyte differentiation [27]. In this study, RGN was significantly upregulated in the CH group, which is consistent with a previous study [27].
HSPA9 is a member of the heat shock protein 70 (HSP70) family. Zhang et al. performed iTRAQ-based proteomic analysis to identify proteins in duck muscles related to lipid oxidation; they found that heat shock protein were correlated with lipid oxidation [28]. 2-DE proteomic studies showed that HSPA9 is highly expressed in the proliferation of preadipocytes of the omentum and is closely related to adipogenesis [29]. A growing number of studies have linked chaperone molecules to adipogenesis, obesity, and diabetes [30,31]. The chaperone protein HSPA9 forms a complex with the iron–sulfur cluster and uses the energy released by ATP hydrolysis to drive the conformational change and refolding of the target protein [32]. Heat shock proteins could protect lipids against reactive oxygen species (ROS)-induced lipid oxidation [33]. The overexpression of HSPA9 reduces the accumulation of ROS in glucose-deficient PC-12 cells [30]. In this study, it was found that HSPA9 was significantly upregulated in the CH group, was enriched in the process of cellular heat stress, and played an important role in the formation of mitochondrial iron–sulfur clusters.
GPX1 is a cellular and mitochondrial enzyme that catalyzes the reduction of organic hydroperoxides and hydrogen peroxide by glutathione, thereby protecting the cells against oxidative damage. Inactivation of the GPX1 gene leads to growth retardation in mice, possibly because of decreased mitochondrial energy from increased oxidative stress [34]. As a selenium-dependent enzyme that reduces the concentration of intracellular hydrogen peroxide and lipid peroxides, GPX1 plays an important role in regulating glucose homeostasis, lipogenesis, and liposis [35]. GPX1 activity is elevated in the liver and adipose tissues of pigs; it mediates lipid accumulation and fatty acid profile changes [36]. Additionally, GPX1 overexpression in mice induces elevated lipid concentrations in the plasma and tissues [37]. In this study, GPX1 was significantly enriched in several biological processes, including adipocyte differentiation and internal apoptotic signaling pathway in response to oxidative stress. Simultaneously, it was found that GPX1 was significantly upregulated, indicating the expression and activity of this enzyme in meat ducks, which is consistent with previous studies.
Peroxiredoxin 1 (encoded by PRDX1), a mercaptan-specific peroxidase, catalyzes the reduction of hydrogen peroxide and organic peroxides to water and alcohol, respectively. PRDX1 is a scavenger of ROS, which may be involved in signaling cascades of growth factor and tumor necrosis factor-α by regulating intracellular H2O2 concentration [38]. The expression of PRDX1 is necessary to control the response of corneal endothelial cells to agents that cause lipid peroxidation [39]. PRDX1 has a crucial role in the maintenance of lipophagic flux in macrophages [40]. Future research should investigate the implementation of protective functions, such as antioxidant stress and immunity, in the process of fat deposition and metabolism.
5 Conclusion
Three important pathways were identified in this study: glycolysis, gluconeogenesis, pentose phosphate pathway, and metabolism of fructose and mannose. Eight key proteins were identified, namely ALDOB, ENO1, RGN, GAPDH, TPI1, HSPA9, PRDX1, and GPX1. These proteins interact strongly with metabolic pathways and play an important role in the regulation of liver fat metabolism in Chaohu ducks. These results provide a good reference regarding the molecular mechanism of IMF deposition and fat metabolism in waterfowl for the regulation of protein expression.
-
Funding information: This study was supported by the Natural Science Research Project of West Anhui University (No. WXZR201931) and the High-level Talents Scientific Research Start-up Fund Project of West Anhui University (No. WGKQ2021028).
-
Author contributions: K.G. and Z.G. designed the research, and K.G. carried them out. K.G. and Z.G. analyzed and interpreted the data. K.G. drafted the manuscript with contributions from all co-authors. All authors read and approved the final manuscript. The authors applied the SDC approach for the sequence of authors.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Appendix
Forty-three DEPs identified by MOLDI-TOF-MS
| Protein ID | Log2 fold change | Identity (%) | E-value | Uniprot-ID | Gene name |
|---|---|---|---|---|---|
| U3IHG8 | 3.321928095 | 94.78 | 0 | P07341 | ALDOB |
| A0A1D5PS29 | 3.317593505 | 38.95 | 0 | Q5F3X4 | EFTUD2 |
| R0LJ39 | 3.063502942 | 98.99 | 5.00 × 10−145 | P0CB50 | PRDX1 |
| U3ISH9 | 2.941106311 | 95.07 | 0 | F1NGJ7 | Gga.1183 |
| A0A1D5PN54 | 2.776103988 | 100 | 0 | F1NI22 | ATP5A1 |
| R0LNI3 | 2.627606838 | 98.48 | 1.00 × 10−95 | F1NYA9 | EEF1B2 |
| A0A226NPQ5 | 2.375734539 | 99.19 | 0 | Q5ZLC5 | ATP5B |
| F1NR44 | 2.13093087 | 37 | 5.00 × 10−43 | F1NSS6 | ECHDC2 |
| R0K4R7 | 2.084064265 | 92.41 | 0 | E1C8Z8 | ALDH4A1 |
| R0KX92 | 1.799087306 | 67.79 | 0 | O93344 | ALDH1A2 |
| R0JKM2 | 1.790772038 | 90.25 | 2.00 × 10−160 | E1BTA0 | COMT |
| R0L7N0 | 1.555816155 | 98.69 | 0 | Q5ZM98 | HSPA9 |
| A0A1L1RLH6 | 1.531069493 | 100 | 0 | Q9I9D1 | VDAC2 |
| A0A226MZ03 | 1.469885976 | 99.6 | 0 | P00940 | TPI1 |
| A0A226PNR7 | 1.459431619 | 75 | 0 | O93344 | ALDH1A2 |
| R0LLL3 | 1.207892852 | 94.22 | 0 | E1BRE5 | EEA1 |
| F1NBJ7 | 1.207892852 | 100 | 0 | F1NBJ7 | SARDH |
| P19140 | 1.189033824 | 97.24 | 0 | P51913 | ENO1 |
| A0A346M329 | 1.163498732 | 99.1 | 0 | P00356 | GAPDH |
| U3IR52 | 1.097610797 | 97.7 | 0 | P51913 | ENO1 |
| G1N0B8 | 1.097610797 | 97.33 | 0 | E1BS67 | SHMT1 |
| F1NX57 | 1.084064265 | 100 | 0 | F1NX57 | GRHPR |
| R0LHA7 | 1.042644337 | 100 | 0 | P60706 | ACTB |
| R0LBX0 | 1.028569152 | 97 | 0 | P63270 | ACTG2 |
| U3ITD3 | 0.963474124 | 19.12 | 0.0009 | F1NZZ2 | ENSGALG00000005321 |
| R0K8X5 | 0.933572638 | 90.24 | 0 | F1NY09 | C1H11ORF54 |
| R0JKJ9 | 0.910732662 | 89.31 | 0 | Q9YH58 | FTCD |
| R0K9P5 | 0.910732662 | 80.46 | 0 | Q8JG30 | SULT1B1 |
| R0K2N7 | 0.90303827 | 78.96 | 0 | E1C6Z5 | Gga.55096 |
| R0LIW0 | 0.879705766 | 85.13 | 0 | E1BV78 | FGG |
| E1BXG9 | 0.879705766 | 100 | 0 | E1BXG9 | PPID |
| B4Z854 | 0.831877241 | 94.15 | 7.00 × 10−117 | P07341 | ALDOB |
| R0JHQ1 | 0.831877241 | 95.34 | 0 | E1BZS9 | APC |
| D2J267 | 0.799087306 | 100 | 4.00 × 10−114 | R4GH86 | GPX1 |
| R0M714 | 0.790772038 | 95.29 | 0 | E1BSH9 | BHMT |
| R0L3A1 | 0.782408565 | 88.76 | 7.00 × 10−179 | Q9I923 | RGN |
| U3I8T6 | 0.722466024 | 99.72 | 0 | P68034 | ACTC1 |
| R0K525 | 0.713695815 | 81.85 | 8.00 × 10−151 | F1N9C1 | LOC415661 |
| T2HNS8 | 0.704871964 | 100 | 1.00 × 10−112 | F1NI22 | ATP5A1 |
| R0JL78 | 0.678071905 | 92.07 | 0 | H9KYX6 | SELENBP1 |
| R0M0W6 | 0.641546029 | 85.27 | 0 | P19121 | ALB |
| R0JE50 | 0.622930351 | 98.65 | 0 | P16580 | GLUL |
| U3J7S2 | 0.613531653 | 96.52 | 0 | E1BYD4 | NDRG1 |
References
[1] Fan W, Liu W, Liu H, Meng Q, Xu Y, Guo Y, et al. Dynamic accumulation of fatty acids in duck (Anas platyrhynchos) breast muscle and its correlations with gene expression. BMC Genomics. 2020;21(1):58.10.1186/s12864-020-6482-7Search in Google Scholar PubMed PubMed Central
[2] Ge K, Chen X, Kuang J, Yang L, Geng Z. Comparison of liver transcriptome from high- and low-intramuscular fat Chaohu ducks provided additional candidate genes for lipid selection. 3 Biotech. 2019;9(7):251.10.1007/s13205-019-1780-ySearch in Google Scholar PubMed PubMed Central
[3] Liu L, Cui H, Fu R, Zheng M, Liu R, Zhao G, et al. The regulation of IMF deposition in pectoralis major of fast- and slow-growing chickens at hatching. J Anim Sci Biotechnol. 2017;8:77.10.1186/s40104-017-0207-zSearch in Google Scholar PubMed PubMed Central
[4] Kokoszyński D, Wilkanowska A, Arpášová H, Hrnčár C. Comparison of some meat quality and liver characteristics in Muscovy and mule ducks. Arch Anim Breed. 2020;63(1):137–44.10.5194/aab-63-137-2020Search in Google Scholar PubMed PubMed Central
[5] He J, Zheng H, Pan D, Liu T, Sun Y, Cao J, et al. Effects of aging on fat deposition and meat quality in Sheldrake duck. Poult Sci. 2018;97(6):2005–10.10.3382/ps/pey077Search in Google Scholar PubMed
[6] Zhang C, Ah Kan Razafindrabe RH, Chen K, Zhao X, Yang L, Wang L, et al. Effects of different rearing systems on growth performance, carcass traits, meat quality and serum biochemical parameters of Chaohu ducks. Anim Sci J = Nihon Chikusan Gakkaiho. 2018;89(4):672–8.10.1111/asj.12976Search in Google Scholar PubMed
[7] Hugo A, Roodt E. Significance of porcine fat quality in meat technology: a review. Food Rev Int. 2007;23(2):175–98.10.1080/87559120701225037Search in Google Scholar
[8] Yang N, Jiang RS. Recent advances in breeding for quality chickens. World Poult Sci J. 2005;61(3):373–81.10.1079/WPS200563Search in Google Scholar
[9] Mir NA, Rafiq A, Kumar F, Singh V, Shukla V. Determinants of broiler chicken meat quality and factors affecting them: a review. J Food Sci Technol. 2017;54(10):2997–3009.10.1007/s13197-017-2789-zSearch in Google Scholar PubMed PubMed Central
[10] Rao S, Huang J, Shen Z, Xiang C, Zhang M, Lu X. Inhibition of TREM-1 attenuates inflammation and lipid accumulation in diet-induced nonalcoholic fatty liver disease. J Cell Biochem. 2019;120(7):11867–77.10.1002/jcb.28468Search in Google Scholar PubMed PubMed Central
[11] Zheng A, Chang W, Hou S, Zhang S, Cai H, Chen G, et al. Unraveling molecular mechanistic differences in liver metabolism between lean and fat lines of Pekin duck (Anas platyrhynchos domestica): a proteomic study. J Proteom. 2014;98:271–88.10.1016/j.jprot.2013.12.021Search in Google Scholar PubMed
[12] Liu R, Wang H, Liu J, Wang J, Zheng M, Tan X, et al. Uncovering the embryonic development-related proteome and metabolome signatures in breast muscle and intramuscular fat of fast- and slow-growing chickens. BMC Genomics. 2017;18(1):816.10.1186/s12864-017-4150-3Search in Google Scholar PubMed PubMed Central
[13] Wen Y, Liu H, Liu K, Cao H, Mao H, Dong X, et al. Analysis of the physical meat quality in partridge (Alectoris chukar) and its relationship with intramuscular fat. Poult Sci. 2020;99(2):1225–31.10.1016/j.psj.2019.09.009Search in Google Scholar PubMed PubMed Central
[14] Chang YC, Yang YC, Tien CP, Yang CJ, Hsiao M. Roles of aldolase family genes in human cancers and diseases. Trends Endocrinol Metab: TEM. 2018;29(8):549–59.10.1016/j.tem.2018.05.003Search in Google Scholar PubMed
[15] Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548(7665):112–6.10.1038/nature23275Search in Google Scholar PubMed PubMed Central
[16] Niu L, Geyer PE, Wewer Albrechtsen NJ, Gluud LL, Santos A, Doll S, et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol Syst Biol. 2019;15(3):e8793.10.15252/msb.20188793Search in Google Scholar PubMed PubMed Central
[17] Ji H, Wang J, Guo J, Li Y, Lian S, Guo W, et al. Progress in the biological function of alpha-enolase. Anim Nutr (Zhongguo Xu Mu Shou Yi Xue Hui). 2016;2(1):12–7.10.1016/j.aninu.2016.02.005Search in Google Scholar PubMed PubMed Central
[18] Díaz-Ramos A, Roig-Borrellas A, García-Melero A, López-Alemany R. α-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol. 2012;2012:156795.10.1155/2012/156795Search in Google Scholar PubMed PubMed Central
[19] Avilán L, Gualdrón-López M, Quiñones W, González-González L, Hannaert V, Michels PA, et al. Enolase: a key player in the metabolism and a probable virulence factor of trypanosomatid parasites-perspectives for its use as a therapeutic target. Enzyme Res. 2011;2011:932549.10.4061/2011/932549Search in Google Scholar PubMed PubMed Central
[20] Yang JS, Hsu JW, Park SY, Li J, Oldham WM, Beznoussenko GV, et al. GAPDH inhibits intracellular pathways during starvation for cellular energy homeostasis. Nature. 2018;561(7722):263–7.10.1038/s41586-018-0475-6Search in Google Scholar PubMed PubMed Central
[21] Nakajima H, Itakura M, Kubo T, Kaneshige A, Harada N, Izawa T, et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) aggregation causes mitochondrial dysfunction during oxidative stress-induced cell death. J Biol Chem. 2017;292(11):4727–42.10.1074/jbc.M116.759084Search in Google Scholar
[22] Dua A. Enzyme kinetics at the molecular level. Resonance. 2019;24(3):297–311.10.1007/s12045-019-0781-9Search in Google Scholar
[23] Ahmed N, Battah S, Karachalias N, Babaei-Jadidi R, Horányi M, Baróti K, et al. Increased formation of methylglyoxal and protein glycation, oxidation and nitrosation in triosephosphate isomerase deficiency. Biochim Biophys Acta. 2003;1639(2):121–32.10.1016/j.bbadis.2003.08.002Search in Google Scholar
[24] Olivares-Illana V, Riveros-Rosas H, Cabrera N, Tuena de Gómez-Puyou M, Pérez-Montfort R, Costas M, et al. A guide to the effects of a large portion of the residues of triosephosphate isomerase on catalysis, stability, druggability, and human disease. Proteins. 2017;85(7):1190–211.10.1002/prot.25299Search in Google Scholar
[25] Marques R, Maia CJ, Vaz C, Correia S, Socorro S. The diverse roles of calcium-binding protein regucalcin in cell biology: from tissue expression and signalling to disease. Cell Mol Life Sci. 2014;71(1):93–111.10.1007/s00018-013-1323-3Search in Google Scholar
[26] Yamaguchi M, Hamano T, Misawa H. Expression of Ca(2+)-binding protein regucalcin in rat brain neurons: inhibitory effect on protein phosphatase activity. Brain Res Bull. 2000;52(5):343–8.10.1016/S0361-9230(00)00270-7Search in Google Scholar
[27] Murata T, Yamaguchi M, Kohno S, Takahashi C, Risa W, Hatori K, et al. Regucalcin enhances adipocyte differentiation and attenuates inflammation in 3T3-L1 cells. FEBS Open Bio. 2020;10(10):1967–84.10.1002/2211-5463.12947Search in Google Scholar PubMed PubMed Central
[28] Zhang M, Wang D, Xu X, Xu W, Zhou G. iTRAQ-based proteomic analysis of duck muscle related to lipid oxidation. Poult Sci. 2021;100(4):101029.10.1016/j.psj.2021.101029Search in Google Scholar PubMed PubMed Central
[29] Picard B, Gagaoua M. Meta-proteomics for the discovery of protein biomarkers of beef tenderness: an overview of integrated studies. Food Res Int (Ottawa, Ont). 2020;127:108739.10.1016/j.foodres.2019.108739Search in Google Scholar PubMed
[30] Kimura T, Jennings W, Epand RM. Roles of specific lipid species in the cell and their molecular mechanism. Prog Lipid Res. 2016;62:75–92.10.1016/j.plipres.2016.02.001Search in Google Scholar PubMed
[31] Ullery JC, Marnett LJ. Protein modification by oxidized phospholipids and hydrolytically released lipid electrophiles: investigating cellular responses. Biochim Biophys Acta. 2012;1818(10):2424–35.10.1016/j.bbamem.2012.04.014Search in Google Scholar PubMed PubMed Central
[32] Fernández-Fernández MR, Valpuesta JM. Hsp70 chaperone: a master player in protein homeostasis. F1000Research. 2018;7:7.10.12688/f1000research.15528.1Search in Google Scholar PubMed PubMed Central
[33] Zhang M, Wang D, Geng Z, Li P, Sun Z, Xu W. Effect of heat shock protein 90 against ROS-induced phospholipid oxidation. Food Chem. 2018;240:642–7.10.1016/j.foodchem.2017.08.005Search in Google Scholar PubMed
[34] Mai HN, Chung YH, Shin EJ, Kim DJ, Sharma N, Lee YJ, et al. Glutathione peroxidase-1 overexpressing transgenic mice are protected from cocaine-induced drug dependence. Neurochem Int. 2019;124:264–73.10.1016/j.neuint.2019.01.018Search in Google Scholar PubMed
[35] Huang JQ, Zhou JC, Wu YY, Ren FZ, Lei XG. Role of glutathione peroxidase 1 in glucose and lipid metabolism-related diseases. Free Radic Biol Med. 2018;127:108–15.10.1016/j.freeradbiomed.2018.05.077Search in Google Scholar PubMed PubMed Central
[36] Zhao Z, Barcus M, Kim J, Lum KL, Mills C, Lei XG. High dietary selenium intake alters lipid metabolism and protein synthesis in liver and muscle of pigs. J Nutr. 2016;146(9):1625–33.10.3945/jn.116.229955Search in Google Scholar PubMed PubMed Central
[37] McClung JP, Roneker CA, Mu W, Lisk DJ, Langlais P, Liu F, et al. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc Natl Acad Sci U S A. 2004;101(24):8852–7.10.1073/pnas.0308096101Search in Google Scholar PubMed PubMed Central
[38] Ahmed W, Lingner J. PRDX1 and MTH1 cooperate to prevent ROS-mediated inhibition of telomerase. Genes Dev. 2018;32(9-10):658–69.10.1101/gad.313460.118Search in Google Scholar PubMed PubMed Central
[39] Lovatt M, Adnan K, Kocaba V, Dirisamer M, Peh GSL, Mehta JS. Peroxiredoxin-1 regulates lipid peroxidation in corneal endothelial cells. Redox Biol. 2020;30:101417.10.1016/j.redox.2019.101417Search in Google Scholar PubMed PubMed Central
[40] Jeong SJ, Kim S, Park JG, Jung IH, Lee MN, Jeon S, et al. Prdx1 (peroxiredoxin 1) deficiency reduces cholesterol efflux via impaired macrophage lipophagic flux. Autophagy. 2018;14(1):120–33.10.1080/15548627.2017.1327942Search in Google Scholar PubMed PubMed Central
© 2022 Kai Ge and Zhaoyu Geng, published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Effects of direct oral anticoagulants dabigatran and rivaroxaban on the blood coagulation function in rabbits
- The mother of all battles: Viruses vs humans. Can humans avoid extinction in 50–100 years?
- Knockdown of G1P3 inhibits cell proliferation and enhances the cytotoxicity of dexamethasone in acute lymphoblastic leukemia
- LINC00665 regulates hepatocellular carcinoma by modulating mRNA via the m6A enzyme
- Association study of CLDN14 variations in patients with kidney stones
- Concanavalin A-induced autoimmune hepatitis model in mice: Mechanisms and future outlook
- Regulation of miR-30b in cancer development, apoptosis, and drug resistance
- Informatic analysis of the pulmonary microecology in non-cystic fibrosis bronchiectasis at three different stages
- Swimming attenuates tumor growth in CT-26 tumor-bearing mice and suppresses angiogenesis by mediating the HIF-1α/VEGFA pathway
- Characterization of intestinal microbiota and serum metabolites in patients with mild hepatic encephalopathy
- Functional conservation and divergence in plant-specific GRF gene family revealed by sequences and expression analysis
- Application of the FLP/LoxP-FRT recombination system to switch the eGFP expression in a model prokaryote
- Biomedical evaluation of antioxidant properties of lamb meat enriched with iodine and selenium
- Intravenous infusion of the exosomes derived from human umbilical cord mesenchymal stem cells enhance neurological recovery after traumatic brain injury via suppressing the NF-κB pathway
- Effect of dietary pattern on pregnant women with gestational diabetes mellitus and its clinical significance
- Potential regulatory mechanism of TNF-α/TNFR1/ANXA1 in glioma cells and its role in glioma cell proliferation
- Effect of the genetic mutant G71R in uridine diphosphate-glucuronosyltransferase 1A1 on the conjugation of bilirubin
- Quercetin inhibits cytotoxicity of PC12 cells induced by amyloid-beta 25–35 via stimulating estrogen receptor α, activating ERK1/2, and inhibiting apoptosis
- Nutrition intervention in the management of novel coronavirus pneumonia patients
- circ-CFH promotes the development of HCC by regulating cell proliferation, apoptosis, migration, invasion, and glycolysis through the miR-377-3p/RNF38 axis
- Bmi-1 directly upregulates glucose transporter 1 in human gastric adenocarcinoma
- Lacunar infarction aggravates the cognitive deficit in the elderly with white matter lesion
- Hydroxysafflor yellow A improved retinopathy via Nrf2/HO-1 pathway in rats
- Comparison of axon extension: PTFE versus PLA formed by a 3D printer
- Elevated IL-35 level and iTr35 subset increase the bacterial burden and lung lesions in Mycobacterium tuberculosis-infected mice
- A case report of CAT gene and HNF1β gene variations in a patient with early-onset diabetes
- Study on the mechanism of inhibiting patulin production by fengycin
- SOX4 promotes high-glucose-induced inflammation and angiogenesis of retinal endothelial cells by activating NF-κB signaling pathway
- Relationship between blood clots and COVID-19 vaccines: A literature review
- Analysis of genetic characteristics of 436 children with dysplasia and detailed analysis of rare karyotype
- Bioinformatics network analyses of growth differentiation factor 11
- NR4A1 inhibits the epithelial–mesenchymal transition of hepatic stellate cells: Involvement of TGF-β–Smad2/3/4–ZEB signaling
- Expression of Zeb1 in the differentiation of mouse embryonic stem cell
- Study on the genetic damage caused by cadmium sulfide quantum dots in human lymphocytes
- Association between single-nucleotide polymorphisms of NKX2.5 and congenital heart disease in Chinese population: A meta-analysis
- Assessment of the anesthetic effect of modified pentothal sodium solution on Sprague-Dawley rats
- Genetic susceptibility to high myopia in Han Chinese population
- Potential biomarkers and molecular mechanisms in preeclampsia progression
- Silencing circular RNA-friend leukemia virus integration 1 restrained malignancy of CC cells and oxaliplatin resistance by disturbing dyskeratosis congenita 1
- Endostar plus pembrolizumab combined with a platinum-based dual chemotherapy regime for advanced pulmonary large-cell neuroendocrine carcinoma as a first-line treatment: A case report
- The significance of PAK4 in signaling and clinicopathology: A review
- Sorafenib inhibits ovarian cancer cell proliferation and mobility and induces radiosensitivity by targeting the tumor cell epithelial–mesenchymal transition
- Characterization of rabbit polyclonal antibody against camel recombinant nanobodies
- Active legumain promotes invasion and migration of neuroblastoma by regulating epithelial-mesenchymal transition
- Effect of cell receptors in the pathogenesis of osteoarthritis: Current insights
- MT-12 inhibits the proliferation of bladder cells in vitro and in vivo by enhancing autophagy through mitochondrial dysfunction
- Study of hsa_circRNA_000121 and hsa_circRNA_004183 in papillary thyroid microcarcinoma
- BuyangHuanwu Decoction attenuates cerebral vasospasm caused by subarachnoid hemorrhage in rats via PI3K/AKT/eNOS axis
- Effects of the interaction of Notch and TLR4 pathways on inflammation and heart function in septic heart
- Monosodium iodoacetate-induced subchondral bone microstructure and inflammatory changes in an animal model of osteoarthritis
- A rare presentation of type II Abernethy malformation and nephrotic syndrome: Case report and review
- Rapid death due to pulmonary epithelioid haemangioendothelioma in several weeks: A case report
- Hepatoprotective role of peroxisome proliferator-activated receptor-α in non-cancerous hepatic tissues following transcatheter arterial embolization
- Correlation between peripheral blood lymphocyte subpopulations and primary systemic lupus erythematosus
- A novel SLC8A1-ALK fusion in lung adenocarcinoma confers sensitivity to alectinib: A case report
- β-Hydroxybutyrate upregulates FGF21 expression through inhibition of histone deacetylases in hepatocytes
- Identification of metabolic genes for the prediction of prognosis and tumor microenvironment infiltration in early-stage non-small cell lung cancer
- BTBD10 inhibits glioma tumorigenesis by downregulating cyclin D1 and p-Akt
- Mucormycosis co-infection in COVID-19 patients: An update
- Metagenomic next-generation sequencing in diagnosing Pneumocystis jirovecii pneumonia: A case report
- Long non-coding RNA HOXB-AS1 is a prognostic marker and promotes hepatocellular carcinoma cells’ proliferation and invasion
- Preparation and evaluation of LA-PEG-SPION, a targeted MRI contrast agent for liver cancer
- Proteomic analysis of the liver regulating lipid metabolism in Chaohu ducks using two-dimensional electrophoresis
- Nasopharyngeal tuberculosis: A case report
- Characterization and evaluation of anti-Salmonella enteritidis activity of indigenous probiotic lactobacilli in mice
- Aberrant pulmonary immune response of obese mice to periodontal infection
- Bacteriospermia – A formidable player in male subfertility
- In silico and in vivo analysis of TIPE1 expression in diffuse large B cell lymphoma
- Effects of KCa channels on biological behavior of trophoblasts
- Interleukin-17A influences the vulnerability rather than the size of established atherosclerotic plaques in apolipoprotein E-deficient mice
- Multiple organ failure and death caused by Staphylococcus aureus hip infection: A case report
- Prognostic signature related to the immune environment of oral squamous cell carcinoma
- Primary and metastatic squamous cell carcinoma of the thyroid gland: Two case reports
- Neuroprotective effects of crocin and crocin-loaded niosomes against the paraquat-induced oxidative brain damage in rats
- Role of MMP-2 and CD147 in kidney fibrosis
- Geometric basis of action potential of skeletal muscle cells and neurons
- Babesia microti-induced fulminant sepsis in an immunocompromised host: A case report and the case-specific literature review
- Role of cerebellar cortex in associative learning and memory in guinea pigs
- Application of metagenomic next-generation sequencing technique for diagnosing a specific case of necrotizing meningoencephalitis caused by human herpesvirus 2
- Case report: Quadruple primary malignant neoplasms including esophageal, ureteral, and lung in an elderly male
- Long non-coding RNA NEAT1 promotes angiogenesis in hepatoma carcinoma via the miR-125a-5p/VEGF pathway
- Osteogenic differentiation of periodontal membrane stem cells in inflammatory environments
- Knockdown of SHMT2 enhances the sensitivity of gastric cancer cells to radiotherapy through the Wnt/β-catenin pathway
- Continuous renal replacement therapy combined with double filtration plasmapheresis in the treatment of severe lupus complicated by serious bacterial infections in children: A case report
- Simultaneous triple primary malignancies, including bladder cancer, lymphoma, and lung cancer, in an elderly male: A case report
- Preclinical immunogenicity assessment of a cell-based inactivated whole-virion H5N1 influenza vaccine
- One case of iodine-125 therapy – A new minimally invasive treatment of intrahepatic cholangiocarcinoma
- S1P promotes corneal trigeminal neuron differentiation and corneal nerve repair via upregulating nerve growth factor expression in a mouse model
- Early cancer detection by a targeted methylation assay of circulating tumor DNA in plasma
- Calcifying nanoparticles initiate the calcification process of mesenchymal stem cells in vitro through the activation of the TGF-β1/Smad signaling pathway and promote the decay of echinococcosis
- Evaluation of prognostic markers in patients infected with SARS-CoV-2
- N6-Methyladenosine-related alternative splicing events play a role in bladder cancer
- Characterization of the structural, oxidative, and immunological features of testis tissue from Zucker diabetic fatty rats
- Effects of glucose and osmotic pressure on the proliferation and cell cycle of human chorionic trophoblast cells
- Investigation of genotype diversity of 7,804 norovirus sequences in humans and animals of China
- Characteristics and karyotype analysis of a patient with turner syndrome complicated with multiple-site tumors: A case report
- Aggravated renal fibrosis is positively associated with the activation of HMGB1-TLR2/4 signaling in STZ-induced diabetic mice
- Distribution characteristics of SARS-CoV-2 IgM/IgG in false-positive results detected by chemiluminescent immunoassay
- SRPX2 attenuated oxygen–glucose deprivation and reperfusion-induced injury in cardiomyocytes via alleviating endoplasmic reticulum stress-induced apoptosis through targeting PI3K/Akt/mTOR axis
- Aquaporin-8 overexpression is involved in vascular structure and function changes in placentas of gestational diabetes mellitus patients
- Relationship between CRP gene polymorphisms and ischemic stroke risk: A systematic review and meta-analysis
- Effects of growth hormone on lipid metabolism and sexual development in pubertal obese male rats
- Cloning and identification of the CTLA-4IgV gene and functional application of vaccine in Xinjiang sheep
- Antitumor activity of RUNX3: Upregulation of E-cadherin and downregulation of the epithelial–mesenchymal transition in clear-cell renal cell carcinoma
- PHF8 promotes osteogenic differentiation of BMSCs in old rat with osteoporosis by regulating Wnt/β-catenin pathway
- A review of the current state of the computer-aided diagnosis (CAD) systems for breast cancer diagnosis
- Bilateral dacryoadenitis in adult-onset Still’s disease: A case report
- A novel association between Bmi-1 protein expression and the SUVmax obtained by 18F-FDG PET/CT in patients with gastric adenocarcinoma
- The role of erythrocytes and erythroid progenitor cells in tumors
- Relationship between platelet activation markers and spontaneous abortion: A meta-analysis
- Abnormal methylation caused by folic acid deficiency in neural tube defects
- Silencing TLR4 using an ultrasound-targeted microbubble destruction-based shRNA system reduces ischemia-induced seizures in hyperglycemic rats
- Plant Sciences
- Seasonal succession of bacterial communities in cultured Caulerpa lentillifera detected by high-throughput sequencing
- Cloning and prokaryotic expression of WRKY48 from Caragana intermedia
- Novel Brassica hybrids with different resistance to Leptosphaeria maculans reveal unbalanced rDNA signal patterns
- Application of exogenous auxin and gibberellin regulates the bolting of lettuce (Lactuca sativa L.)
- Phytoremediation of pollutants from wastewater: A concise review
- Genome-wide identification and characterization of NBS-encoding genes in the sweet potato wild ancestor Ipomoea trifida (H.B.K.)
- Alleviative effects of magnetic Fe3O4 nanoparticles on the physiological toxicity of 3-nitrophenol to rice (Oryza sativa L.) seedlings
- Selection and functional identification of Dof genes expressed in response to nitrogen in Populus simonii × Populus nigra
- Study on pecan seed germination influenced by seed endocarp
- Identification of active compounds in Ophiopogonis Radix from different geographical origins by UPLC-Q/TOF-MS combined with GC-MS approaches
- The entire chloroplast genome sequence of Asparagus cochinchinensis and genetic comparison to Asparagus species
- Genome-wide identification of MAPK family genes and their response to abiotic stresses in tea plant (Camellia sinensis)
- Selection and validation of reference genes for RT-qPCR analysis of different organs at various development stages in Caragana intermedia
- Cloning and expression analysis of SERK1 gene in Diospyros lotus
- Integrated metabolomic and transcriptomic profiling revealed coping mechanisms of the edible and medicinal homologous plant Plantago asiatica L. cadmium resistance
- A missense variant in NCF1 is associated with susceptibility to unexplained recurrent spontaneous abortion
- Assessment of drought tolerance indices in faba bean genotypes under different irrigation regimes
- The entire chloroplast genome sequence of Asparagus setaceus (Kunth) Jessop: Genome structure, gene composition, and phylogenetic analysis in Asparagaceae
- Food Science
- Dietary food additive monosodium glutamate with or without high-lipid diet induces spleen anomaly: A mechanistic approach on rat model
- Binge eating disorder during COVID-19
- Potential of honey against the onset of autoimmune diabetes and its associated nephropathy, pancreatitis, and retinopathy in type 1 diabetic animal model
- FTO gene expression in diet-induced obesity is downregulated by Solanum fruit supplementation
- Physical activity enhances fecal lactobacilli in rats chronically drinking sweetened cola beverage
- Supercritical CO2 extraction, chemical composition, and antioxidant effects of Coreopsis tinctoria Nutt. oleoresin
- Functional constituents of plant-based foods boost immunity against acute and chronic disorders
- Effect of selenium and methods of protein extraction on the proteomic profile of Saccharomyces yeast
- Microbial diversity of milk ghee in southern Gansu and its effect on the formation of ghee flavor compounds
- Ecology and Environmental Sciences
- Effects of heavy metals on bacterial community surrounding Bijiashan mining area located in northwest China
- Microorganism community composition analysis coupling with 15N tracer experiments reveals the nitrification rate and N2O emissions in low pH soils in Southern China
- Genetic diversity and population structure of Cinnamomum balansae Lecomte inferred by microsatellites
- Preliminary screening of microplastic contamination in different marine fish species of Taif market, Saudi Arabia
- Plant volatile organic compounds attractive to Lygus pratensis
- Effects of organic materials on soil bacterial community structure in long-term continuous cropping of tomato in greenhouse
- Effects of soil treated fungicide fluopimomide on tomato (Solanum lycopersicum L.) disease control and plant growth
- Prevalence of Yersinia pestis among rodents captured in a semi-arid tropical ecosystem of south-western Zimbabwe
- Effects of irrigation and nitrogen fertilization on mitigating salt-induced Na+ toxicity and sustaining sea rice growth
- Bioengineering and Biotechnology
- Poly-l-lysine-caused cell adhesion induces pyroptosis in THP-1 monocytes
- Development of alkaline phosphatase-scFv and its use for one-step enzyme-linked immunosorbent assay for His-tagged protein detection
- Development and validation of a predictive model for immune-related genes in patients with tongue squamous cell carcinoma
- Agriculture
- Effects of chemical-based fertilizer replacement with biochar-based fertilizer on albic soil nutrient content and maize yield
- Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions
- Agronomic and economic performance of mung bean (Vigna radiata L.) varieties in response to rates of blended NPS fertilizer in Kindo Koysha district, Southern Ethiopia
- Influence of furrow irrigation regime on the yield and water consumption indicators of winter wheat based on a multi-level fuzzy comprehensive evaluation
- Discovery of exercise-related genes and pathway analysis based on comparative genomes of Mongolian originated Abaga and Wushen horse
- Lessons from integrated seasonal forecast-crop modelling in Africa: A systematic review
- Evolution trend of soil fertility in tobacco-planting area of Chenzhou, Hunan Province, China
- Animal Sciences
- Morphological and molecular characterization of Tatera indica Hardwicke 1807 (Rodentia: Muridae) from Pothwar, Pakistan
- Research on meat quality of Qianhua Mutton Merino sheep and Small-tail Han sheep
- SI: A Scientific Memoir
- Suggestions on leading an academic research laboratory group
- My scientific genealogy and the Toronto ACDC Laboratory, 1988–2022
- Erratum
- Erratum to “Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study”
- Erratum to “A two-microRNA signature predicts the progression of male thyroid cancer”
- Retraction
- Retraction of “Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis”
Articles in the same Issue
- Biomedical Sciences
- Effects of direct oral anticoagulants dabigatran and rivaroxaban on the blood coagulation function in rabbits
- The mother of all battles: Viruses vs humans. Can humans avoid extinction in 50–100 years?
- Knockdown of G1P3 inhibits cell proliferation and enhances the cytotoxicity of dexamethasone in acute lymphoblastic leukemia
- LINC00665 regulates hepatocellular carcinoma by modulating mRNA via the m6A enzyme
- Association study of CLDN14 variations in patients with kidney stones
- Concanavalin A-induced autoimmune hepatitis model in mice: Mechanisms and future outlook
- Regulation of miR-30b in cancer development, apoptosis, and drug resistance
- Informatic analysis of the pulmonary microecology in non-cystic fibrosis bronchiectasis at three different stages
- Swimming attenuates tumor growth in CT-26 tumor-bearing mice and suppresses angiogenesis by mediating the HIF-1α/VEGFA pathway
- Characterization of intestinal microbiota and serum metabolites in patients with mild hepatic encephalopathy
- Functional conservation and divergence in plant-specific GRF gene family revealed by sequences and expression analysis
- Application of the FLP/LoxP-FRT recombination system to switch the eGFP expression in a model prokaryote
- Biomedical evaluation of antioxidant properties of lamb meat enriched with iodine and selenium
- Intravenous infusion of the exosomes derived from human umbilical cord mesenchymal stem cells enhance neurological recovery after traumatic brain injury via suppressing the NF-κB pathway
- Effect of dietary pattern on pregnant women with gestational diabetes mellitus and its clinical significance
- Potential regulatory mechanism of TNF-α/TNFR1/ANXA1 in glioma cells and its role in glioma cell proliferation
- Effect of the genetic mutant G71R in uridine diphosphate-glucuronosyltransferase 1A1 on the conjugation of bilirubin
- Quercetin inhibits cytotoxicity of PC12 cells induced by amyloid-beta 25–35 via stimulating estrogen receptor α, activating ERK1/2, and inhibiting apoptosis
- Nutrition intervention in the management of novel coronavirus pneumonia patients
- circ-CFH promotes the development of HCC by regulating cell proliferation, apoptosis, migration, invasion, and glycolysis through the miR-377-3p/RNF38 axis
- Bmi-1 directly upregulates glucose transporter 1 in human gastric adenocarcinoma
- Lacunar infarction aggravates the cognitive deficit in the elderly with white matter lesion
- Hydroxysafflor yellow A improved retinopathy via Nrf2/HO-1 pathway in rats
- Comparison of axon extension: PTFE versus PLA formed by a 3D printer
- Elevated IL-35 level and iTr35 subset increase the bacterial burden and lung lesions in Mycobacterium tuberculosis-infected mice
- A case report of CAT gene and HNF1β gene variations in a patient with early-onset diabetes
- Study on the mechanism of inhibiting patulin production by fengycin
- SOX4 promotes high-glucose-induced inflammation and angiogenesis of retinal endothelial cells by activating NF-κB signaling pathway
- Relationship between blood clots and COVID-19 vaccines: A literature review
- Analysis of genetic characteristics of 436 children with dysplasia and detailed analysis of rare karyotype
- Bioinformatics network analyses of growth differentiation factor 11
- NR4A1 inhibits the epithelial–mesenchymal transition of hepatic stellate cells: Involvement of TGF-β–Smad2/3/4–ZEB signaling
- Expression of Zeb1 in the differentiation of mouse embryonic stem cell
- Study on the genetic damage caused by cadmium sulfide quantum dots in human lymphocytes
- Association between single-nucleotide polymorphisms of NKX2.5 and congenital heart disease in Chinese population: A meta-analysis
- Assessment of the anesthetic effect of modified pentothal sodium solution on Sprague-Dawley rats
- Genetic susceptibility to high myopia in Han Chinese population
- Potential biomarkers and molecular mechanisms in preeclampsia progression
- Silencing circular RNA-friend leukemia virus integration 1 restrained malignancy of CC cells and oxaliplatin resistance by disturbing dyskeratosis congenita 1
- Endostar plus pembrolizumab combined with a platinum-based dual chemotherapy regime for advanced pulmonary large-cell neuroendocrine carcinoma as a first-line treatment: A case report
- The significance of PAK4 in signaling and clinicopathology: A review
- Sorafenib inhibits ovarian cancer cell proliferation and mobility and induces radiosensitivity by targeting the tumor cell epithelial–mesenchymal transition
- Characterization of rabbit polyclonal antibody against camel recombinant nanobodies
- Active legumain promotes invasion and migration of neuroblastoma by regulating epithelial-mesenchymal transition
- Effect of cell receptors in the pathogenesis of osteoarthritis: Current insights
- MT-12 inhibits the proliferation of bladder cells in vitro and in vivo by enhancing autophagy through mitochondrial dysfunction
- Study of hsa_circRNA_000121 and hsa_circRNA_004183 in papillary thyroid microcarcinoma
- BuyangHuanwu Decoction attenuates cerebral vasospasm caused by subarachnoid hemorrhage in rats via PI3K/AKT/eNOS axis
- Effects of the interaction of Notch and TLR4 pathways on inflammation and heart function in septic heart
- Monosodium iodoacetate-induced subchondral bone microstructure and inflammatory changes in an animal model of osteoarthritis
- A rare presentation of type II Abernethy malformation and nephrotic syndrome: Case report and review
- Rapid death due to pulmonary epithelioid haemangioendothelioma in several weeks: A case report
- Hepatoprotective role of peroxisome proliferator-activated receptor-α in non-cancerous hepatic tissues following transcatheter arterial embolization
- Correlation between peripheral blood lymphocyte subpopulations and primary systemic lupus erythematosus
- A novel SLC8A1-ALK fusion in lung adenocarcinoma confers sensitivity to alectinib: A case report
- β-Hydroxybutyrate upregulates FGF21 expression through inhibition of histone deacetylases in hepatocytes
- Identification of metabolic genes for the prediction of prognosis and tumor microenvironment infiltration in early-stage non-small cell lung cancer
- BTBD10 inhibits glioma tumorigenesis by downregulating cyclin D1 and p-Akt
- Mucormycosis co-infection in COVID-19 patients: An update
- Metagenomic next-generation sequencing in diagnosing Pneumocystis jirovecii pneumonia: A case report
- Long non-coding RNA HOXB-AS1 is a prognostic marker and promotes hepatocellular carcinoma cells’ proliferation and invasion
- Preparation and evaluation of LA-PEG-SPION, a targeted MRI contrast agent for liver cancer
- Proteomic analysis of the liver regulating lipid metabolism in Chaohu ducks using two-dimensional electrophoresis
- Nasopharyngeal tuberculosis: A case report
- Characterization and evaluation of anti-Salmonella enteritidis activity of indigenous probiotic lactobacilli in mice
- Aberrant pulmonary immune response of obese mice to periodontal infection
- Bacteriospermia – A formidable player in male subfertility
- In silico and in vivo analysis of TIPE1 expression in diffuse large B cell lymphoma
- Effects of KCa channels on biological behavior of trophoblasts
- Interleukin-17A influences the vulnerability rather than the size of established atherosclerotic plaques in apolipoprotein E-deficient mice
- Multiple organ failure and death caused by Staphylococcus aureus hip infection: A case report
- Prognostic signature related to the immune environment of oral squamous cell carcinoma
- Primary and metastatic squamous cell carcinoma of the thyroid gland: Two case reports
- Neuroprotective effects of crocin and crocin-loaded niosomes against the paraquat-induced oxidative brain damage in rats
- Role of MMP-2 and CD147 in kidney fibrosis
- Geometric basis of action potential of skeletal muscle cells and neurons
- Babesia microti-induced fulminant sepsis in an immunocompromised host: A case report and the case-specific literature review
- Role of cerebellar cortex in associative learning and memory in guinea pigs
- Application of metagenomic next-generation sequencing technique for diagnosing a specific case of necrotizing meningoencephalitis caused by human herpesvirus 2
- Case report: Quadruple primary malignant neoplasms including esophageal, ureteral, and lung in an elderly male
- Long non-coding RNA NEAT1 promotes angiogenesis in hepatoma carcinoma via the miR-125a-5p/VEGF pathway
- Osteogenic differentiation of periodontal membrane stem cells in inflammatory environments
- Knockdown of SHMT2 enhances the sensitivity of gastric cancer cells to radiotherapy through the Wnt/β-catenin pathway
- Continuous renal replacement therapy combined with double filtration plasmapheresis in the treatment of severe lupus complicated by serious bacterial infections in children: A case report
- Simultaneous triple primary malignancies, including bladder cancer, lymphoma, and lung cancer, in an elderly male: A case report
- Preclinical immunogenicity assessment of a cell-based inactivated whole-virion H5N1 influenza vaccine
- One case of iodine-125 therapy – A new minimally invasive treatment of intrahepatic cholangiocarcinoma
- S1P promotes corneal trigeminal neuron differentiation and corneal nerve repair via upregulating nerve growth factor expression in a mouse model
- Early cancer detection by a targeted methylation assay of circulating tumor DNA in plasma
- Calcifying nanoparticles initiate the calcification process of mesenchymal stem cells in vitro through the activation of the TGF-β1/Smad signaling pathway and promote the decay of echinococcosis
- Evaluation of prognostic markers in patients infected with SARS-CoV-2
- N6-Methyladenosine-related alternative splicing events play a role in bladder cancer
- Characterization of the structural, oxidative, and immunological features of testis tissue from Zucker diabetic fatty rats
- Effects of glucose and osmotic pressure on the proliferation and cell cycle of human chorionic trophoblast cells
- Investigation of genotype diversity of 7,804 norovirus sequences in humans and animals of China
- Characteristics and karyotype analysis of a patient with turner syndrome complicated with multiple-site tumors: A case report
- Aggravated renal fibrosis is positively associated with the activation of HMGB1-TLR2/4 signaling in STZ-induced diabetic mice
- Distribution characteristics of SARS-CoV-2 IgM/IgG in false-positive results detected by chemiluminescent immunoassay
- SRPX2 attenuated oxygen–glucose deprivation and reperfusion-induced injury in cardiomyocytes via alleviating endoplasmic reticulum stress-induced apoptosis through targeting PI3K/Akt/mTOR axis
- Aquaporin-8 overexpression is involved in vascular structure and function changes in placentas of gestational diabetes mellitus patients
- Relationship between CRP gene polymorphisms and ischemic stroke risk: A systematic review and meta-analysis
- Effects of growth hormone on lipid metabolism and sexual development in pubertal obese male rats
- Cloning and identification of the CTLA-4IgV gene and functional application of vaccine in Xinjiang sheep
- Antitumor activity of RUNX3: Upregulation of E-cadherin and downregulation of the epithelial–mesenchymal transition in clear-cell renal cell carcinoma
- PHF8 promotes osteogenic differentiation of BMSCs in old rat with osteoporosis by regulating Wnt/β-catenin pathway
- A review of the current state of the computer-aided diagnosis (CAD) systems for breast cancer diagnosis
- Bilateral dacryoadenitis in adult-onset Still’s disease: A case report
- A novel association between Bmi-1 protein expression and the SUVmax obtained by 18F-FDG PET/CT in patients with gastric adenocarcinoma
- The role of erythrocytes and erythroid progenitor cells in tumors
- Relationship between platelet activation markers and spontaneous abortion: A meta-analysis
- Abnormal methylation caused by folic acid deficiency in neural tube defects
- Silencing TLR4 using an ultrasound-targeted microbubble destruction-based shRNA system reduces ischemia-induced seizures in hyperglycemic rats
- Plant Sciences
- Seasonal succession of bacterial communities in cultured Caulerpa lentillifera detected by high-throughput sequencing
- Cloning and prokaryotic expression of WRKY48 from Caragana intermedia
- Novel Brassica hybrids with different resistance to Leptosphaeria maculans reveal unbalanced rDNA signal patterns
- Application of exogenous auxin and gibberellin regulates the bolting of lettuce (Lactuca sativa L.)
- Phytoremediation of pollutants from wastewater: A concise review
- Genome-wide identification and characterization of NBS-encoding genes in the sweet potato wild ancestor Ipomoea trifida (H.B.K.)
- Alleviative effects of magnetic Fe3O4 nanoparticles on the physiological toxicity of 3-nitrophenol to rice (Oryza sativa L.) seedlings
- Selection and functional identification of Dof genes expressed in response to nitrogen in Populus simonii × Populus nigra
- Study on pecan seed germination influenced by seed endocarp
- Identification of active compounds in Ophiopogonis Radix from different geographical origins by UPLC-Q/TOF-MS combined with GC-MS approaches
- The entire chloroplast genome sequence of Asparagus cochinchinensis and genetic comparison to Asparagus species
- Genome-wide identification of MAPK family genes and their response to abiotic stresses in tea plant (Camellia sinensis)
- Selection and validation of reference genes for RT-qPCR analysis of different organs at various development stages in Caragana intermedia
- Cloning and expression analysis of SERK1 gene in Diospyros lotus
- Integrated metabolomic and transcriptomic profiling revealed coping mechanisms of the edible and medicinal homologous plant Plantago asiatica L. cadmium resistance
- A missense variant in NCF1 is associated with susceptibility to unexplained recurrent spontaneous abortion
- Assessment of drought tolerance indices in faba bean genotypes under different irrigation regimes
- The entire chloroplast genome sequence of Asparagus setaceus (Kunth) Jessop: Genome structure, gene composition, and phylogenetic analysis in Asparagaceae
- Food Science
- Dietary food additive monosodium glutamate with or without high-lipid diet induces spleen anomaly: A mechanistic approach on rat model
- Binge eating disorder during COVID-19
- Potential of honey against the onset of autoimmune diabetes and its associated nephropathy, pancreatitis, and retinopathy in type 1 diabetic animal model
- FTO gene expression in diet-induced obesity is downregulated by Solanum fruit supplementation
- Physical activity enhances fecal lactobacilli in rats chronically drinking sweetened cola beverage
- Supercritical CO2 extraction, chemical composition, and antioxidant effects of Coreopsis tinctoria Nutt. oleoresin
- Functional constituents of plant-based foods boost immunity against acute and chronic disorders
- Effect of selenium and methods of protein extraction on the proteomic profile of Saccharomyces yeast
- Microbial diversity of milk ghee in southern Gansu and its effect on the formation of ghee flavor compounds
- Ecology and Environmental Sciences
- Effects of heavy metals on bacterial community surrounding Bijiashan mining area located in northwest China
- Microorganism community composition analysis coupling with 15N tracer experiments reveals the nitrification rate and N2O emissions in low pH soils in Southern China
- Genetic diversity and population structure of Cinnamomum balansae Lecomte inferred by microsatellites
- Preliminary screening of microplastic contamination in different marine fish species of Taif market, Saudi Arabia
- Plant volatile organic compounds attractive to Lygus pratensis
- Effects of organic materials on soil bacterial community structure in long-term continuous cropping of tomato in greenhouse
- Effects of soil treated fungicide fluopimomide on tomato (Solanum lycopersicum L.) disease control and plant growth
- Prevalence of Yersinia pestis among rodents captured in a semi-arid tropical ecosystem of south-western Zimbabwe
- Effects of irrigation and nitrogen fertilization on mitigating salt-induced Na+ toxicity and sustaining sea rice growth
- Bioengineering and Biotechnology
- Poly-l-lysine-caused cell adhesion induces pyroptosis in THP-1 monocytes
- Development of alkaline phosphatase-scFv and its use for one-step enzyme-linked immunosorbent assay for His-tagged protein detection
- Development and validation of a predictive model for immune-related genes in patients with tongue squamous cell carcinoma
- Agriculture
- Effects of chemical-based fertilizer replacement with biochar-based fertilizer on albic soil nutrient content and maize yield
- Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions
- Agronomic and economic performance of mung bean (Vigna radiata L.) varieties in response to rates of blended NPS fertilizer in Kindo Koysha district, Southern Ethiopia
- Influence of furrow irrigation regime on the yield and water consumption indicators of winter wheat based on a multi-level fuzzy comprehensive evaluation
- Discovery of exercise-related genes and pathway analysis based on comparative genomes of Mongolian originated Abaga and Wushen horse
- Lessons from integrated seasonal forecast-crop modelling in Africa: A systematic review
- Evolution trend of soil fertility in tobacco-planting area of Chenzhou, Hunan Province, China
- Animal Sciences
- Morphological and molecular characterization of Tatera indica Hardwicke 1807 (Rodentia: Muridae) from Pothwar, Pakistan
- Research on meat quality of Qianhua Mutton Merino sheep and Small-tail Han sheep
- SI: A Scientific Memoir
- Suggestions on leading an academic research laboratory group
- My scientific genealogy and the Toronto ACDC Laboratory, 1988–2022
- Erratum
- Erratum to “Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study”
- Erratum to “A two-microRNA signature predicts the progression of male thyroid cancer”
- Retraction
- Retraction of “Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis”


