Abstract
This study investigated the role of N6-methyladenosine (m6A) and alternative splicing (AS) in bladder cancer (BLCA). The BLCA-related RNA expression profiles and AS events were downloaded from the UCSC Xena and SpliceSeq databases, respectively. Differentially expressed AS (DEAS) was screened, and prognostic-related DEAS events were used to construct prognostic risk models based on Cox proportional hazards regression analysis. Receiver operating characteristic curves and multivariate Cox analysis were used to evaluate the predictive efficiency and independence of these models. We also constructed a protein-to-protein interaction (PPI) network and a regulation network of splicing factors (SFs) and DEAS events. In total, 225 m6A-related prognostic-related DEAS events were identified. The predictive ability of each prognostic model was good, and the alternate terminator model showed the best performance when the area under the curve was 0.793. The risk score of the model was an independent prognostic factor for BLCA. The PPI network revealed that AKT serine/threonine kinase 1, serine- and arginine-rich SF6, and serine- and arginine-rich SF2 had higher-node degrees. A complex regulator correlation was shown in the SF and DEAS networks. This study provides insights for the subsequent understanding of the role of AS events in BLCA.
1 Introduction
Bladder cancer (BLCA) is a disease where a malignant tumor develops in the mucous membrane of the bladder and is a common malignant tumor in the urinary system. The etiology of BLCA is complicated and includes both external environmental and internal genetic factors [1]. The Bacillus Calmette–Guérin vaccine is mainly used to treat and prevent recurrence after transurethral resection of bladder tumors. Patients who do not respond to the drug require additional treatments. Advances in genetic understanding of BLCA and immunotherapy have led to new treatments [2,3].
Like many diseases involving tumors, BLCA can inactivate cytotoxic T cells by downregulating tumor antigen expression. Similarly, the immune system can be avoided by updating immune checkpoints and maintaining the immune environment [4]. Many immune-based biomarkers are being studied for the development of BLCA biomarkers. These studies include predictive biomarkers for patient selection, pharmacodynamic markers of target engagement, and identification and standardization of surrogate markers/endpoints to develop immunotherapy [5].
N6-Methyladenosine (m6A) is the most common form of RNA modification, and its regulatory factors are involved in the development of many cancers [6]. The level of modification of transcript m6A is regulated by “writers,” “readers,” and “erasers” [7]. Methyltransferases are known as “writers” and include KIAA1429, methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit (METTL3), methyltransferase 14, N6-adenosine-methyltransferase subunit (METTL14), RNA-binding motif protein 15 (RBM15), WT1-associated protein (WTAP), and zinc finger CCCH-type containing 13 (ZC3H13). Its main function is to add a methyl group to the nitrogen of the sixth carbon of the aromatic ring of an adenosine residue [8]. “Reader” m6A-binding proteins mainly include heterogeneous nuclear ribonucleoprotein C (HNRNPC), YTH domain containing 1 (YTHDC1), YTH domain containing 2 (YTHDC2), YTH m6A RNA-binding protein 1 (YTHDF1), and YTH m6A RNA-binding protein 2 (YTHDF2). It preferentially binds to RNA and regulates downstream functions [9]. Demethylase, an “eraser,” specifically targets RNA m6A, mainly AlkB homolog 5, RNA demethylase (ALKBH5), and FTO alpha-ketoglutarate-dependent dioxygenase (FTO) [10]. Dysregulation of m6A regulatory factors leads to reduced cell proliferation, loss of self-renewal ability, developmental defects, and cell death [11]. Recently, some scholars have started studying the relationship between m6A RNA methylation regulators and BLCA. Han et al. reported that METTL3 promotes bladder tumor proliferation in an m6A-dependent manner by accelerating pri-miR221/222 [12]. Ying et al. reported that BLAC development could be promoted by m6A modification of CUB domain containing protein 1 mRNA [13].
Alternative splicing (AS) refers to the production of multiple types of mRNA from a single gene and is an important mechanism of post-transcriptional regulation. AS affects the normal growth and development of the human body and plays an important role in the occurrence and development of many diseases, including cancer. Studies have found that the loss of wild-type periostin by downregulation or AS, which produces Variant I, is closely correlated with the development of BLCA [14]. A study also found that splicing mutations were more common in both BLCA and normal bladder tissues [15].
However, it is still unclear how m6A methylation-related AS affects BLCA. In this study, we investigated how AS events related to BLCA prognosis and m6A methylation play a role in BLCA to provide new possible targets for future treatment and intervention of BLCA. A flow chart of the overall study design is shown in Figure 1.
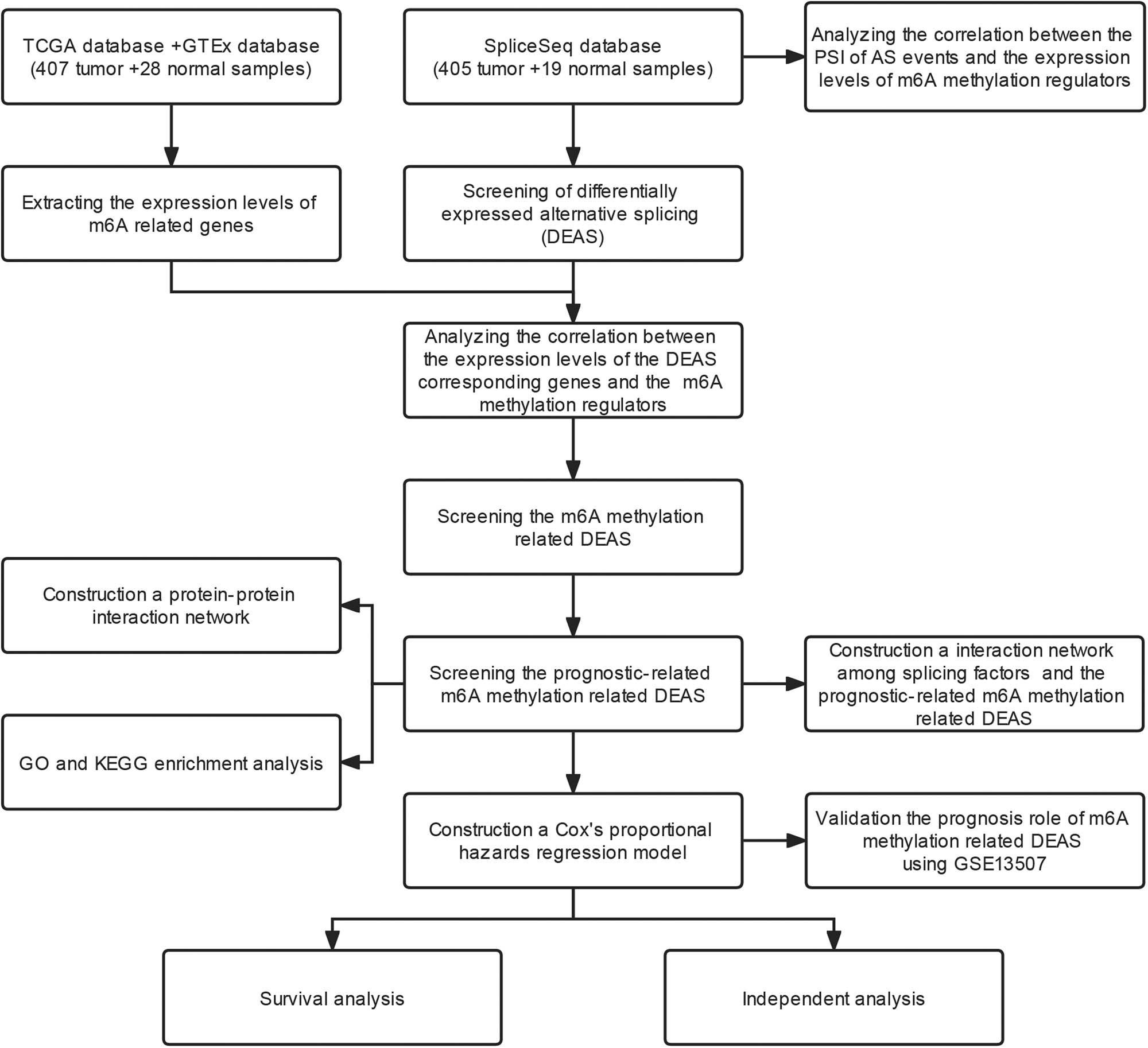
Research flow chart.
2 Materials and methods
2.1 Data collection
The cohort gene expression RNAseq-RSEM transcripts per million of BLCA was downloaded from The Cancer Genome Atlas (TCGA) database, and 407 BLCA samples were obtained (https://portal.gdc.cancer.gov/projects/TCGA-BLCA). The Genotype-Tissue Expression database was used to download the normal bladder samples’ information, and 28 normal bladder samples were obtained (https://www.gtexportal.org).
The specific percentage spliced in (PSI) value for the AS event of TCGA-BLCA was downloaded from the SpliceSeq database (http://projects.insilico.us.com/TCGASpliceSeq/) [16]. To generate as reliable a set of AS events as possible, we implemented a series of stringent filters (percentage of samples with PSI value ≥75, average PSI value ≥0.05). A total of 418 samples were obtained, including 19 normal and 405 tumor samples.
2.2 Differential expression analysis
The seven AS events were alternate acceptors (AA), alternate donors (AD), exon skipping (ES), retained intron (RI), alternate promoters (AP), alternate terminators (AT), and mutually exclusive exons (ME). Each AS event detected by the PSI value in BLCA vs. normal tissue was identified and counted using the software R-3.4.3. The corresponding p values were obtained using the t-test in the generalized linear model, and the difference multiplication relations were calculated to screen the AS with significant differences. Finally, an adjusted p value of <0.05 was set as a criterion to screen the significantly differentially expressed AS (DEAS).
2.3 Screening of m6A methylation-related DEAS
The expression of m6A methylation regulators [METTL3, METTL14, METTL15, WTAP, RBM15, RNA-binding motif protein 15B, KIAA1429, ZC3H13, FTO, ALKBH5, RNA-binding motif protein X-linked, YTHDC1, YTHDC2, insulin-like growth factor 2 mRNA-binding protein 1, insulin-like growth factor 2 mRNA-binding protein 2, insulin-like growth factor 2 mRNA-binding protein 3, YTHDF1, YTHDF2, YTH m6A RNA-binding protein 3, heterogeneous nuclear ribonucleoprotein a2/b1 (HNRNPA2B1), and HNRNPC] was extracted from the BLCA samples from TCGA. The Pearson correlation coefficient between the expression levels of the DEAS-corresponding genes and the expression levels of the m6A methylation regulators was calculated, and p < 0.05 and ǀrǀ > 0.5 were set as the threshold to identify the m6A methylation-related DEAS.
In addition, we analyzed the correlation between PSI of AS events and the expression levels of m6A methylation regulators.
2.4 Prognostic analysis of m6A methylation-related DEAS
The prognosis information of BLCA, including overall survival (OS) and OS status, was downloaded from TCGA database. Prognostic-related m6A methylation-related DEAS was screened using univariate Cox hazard analysis in the survival package (version 2.41-1, http://bioconductor.org/packages/survivalr/) of R3.6.1. Statistical significance was set at p < 0.05.
2.5 Functional enrichment and pathway analysis of differentially expressed genes
The prognostic-related m6A methylation-related DEAS in the same AS type was used to perform Gene Ontology biological process (GO BP) [17] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [18] enrichment analysis using clusterProfiler [19]. The significance threshold of the hypergeometric test p < 0.01 was set as the significant enrichment result.
2.6 Protein-to-protein interaction (PPI) network and module mining analysis
The interaction between proteins was analyzed using the STRING database (Version: 11.0, http://www.string-db.org/) [20]. The PPI score was set to 0.4. The network was then constructed using Cytoscape (version 3.6.1) software [21] after the PPI edges were obtained.
2.7 Relationship between splicing factors (SFs) and the prognostic-related m6A methylation-related DEAS
To study the relationship between SFs and prognostic-related m6A methylation-related DEAS, we obtained 71 SFs through a literature search [22]. Subsequently, survival analysis was performed on the genes corresponding to the SFs to determine prognosis-related SFs. Pearson’s test was used to assess the correlation between the expression levels of SFs and PSI of AS. p < 0.05 and ǀrǀ > 0.3 were set as the significant threshold.
2.8 Construction of Cox proportional hazards regression model
Based on the different AS types, prognostic-related m6A methylation-related DEAS was used to conduct multivariate Cox proportional hazards regression survival analysis and construct risk models. The Cox regression coefficients and PSI for each DEAS were then obtained.
2.9 Survival analysis
The patients were divided into high-risk and low-risk groups based on their risk scores. Kaplan–Meier survival curves were constructed to compare whether there was a significant difference in prognosis between patients in the high- and low-risk groups. The survival receiver operating characteristic curve (ROC) package was used to perform an ROC analysis of these risk models.
2.10 Validation of the role of m6A methylation-related DEAS in BLCA prognosis
The GSE13507 BLCA dataset obtained from the GPL6102 Illumina human-6 v2.0 expression BeadChip sequencing platform was downloaded from the NCBI GEO (https://www.ncbi.nlm.nih.gov/) database on May 23, 2022. After excluding samples with missing survival information, 165 samples were retained for validation analysis.
2.11 Independent analysis
Univariate and multivariate Cox regression analyses were used to assess whether clinical characteristics (TNM stage, age, and sex) and risk score were independent prognostic factors for BLCA. Log-rank p < 0.05 was set as the threshold for screening significantly independent prognostic factors.
3 Results
3.1 AS event statistics and differential analysis
The seven types of AS patterns are shown in Figure 2a. Furthermore, we counted the number of occurrences of each AS type and the number of corresponding genes. As shown in Figure 2b, there were 2,237, 2,278, 1,900, 2,769, 3,558, 9,547, and 39 events for AA, RI, AD, AP, AT, ES, and ME, respectively. There were 1,840, 1,616, 1,549, 2,769, 3,558, 5,332, and 39 genes for AA, RI, AD, AP, AT, ES, and ME events, respectively. ES was the most frequent event among the seven AS types, whereas ME was the least frequent. We also identified 3,715 DEAS, including 1,226 downregulated and 2,489 upregulated DEAS. These 3,715 DEAS corresponded to 2,766 genes (Table S1).
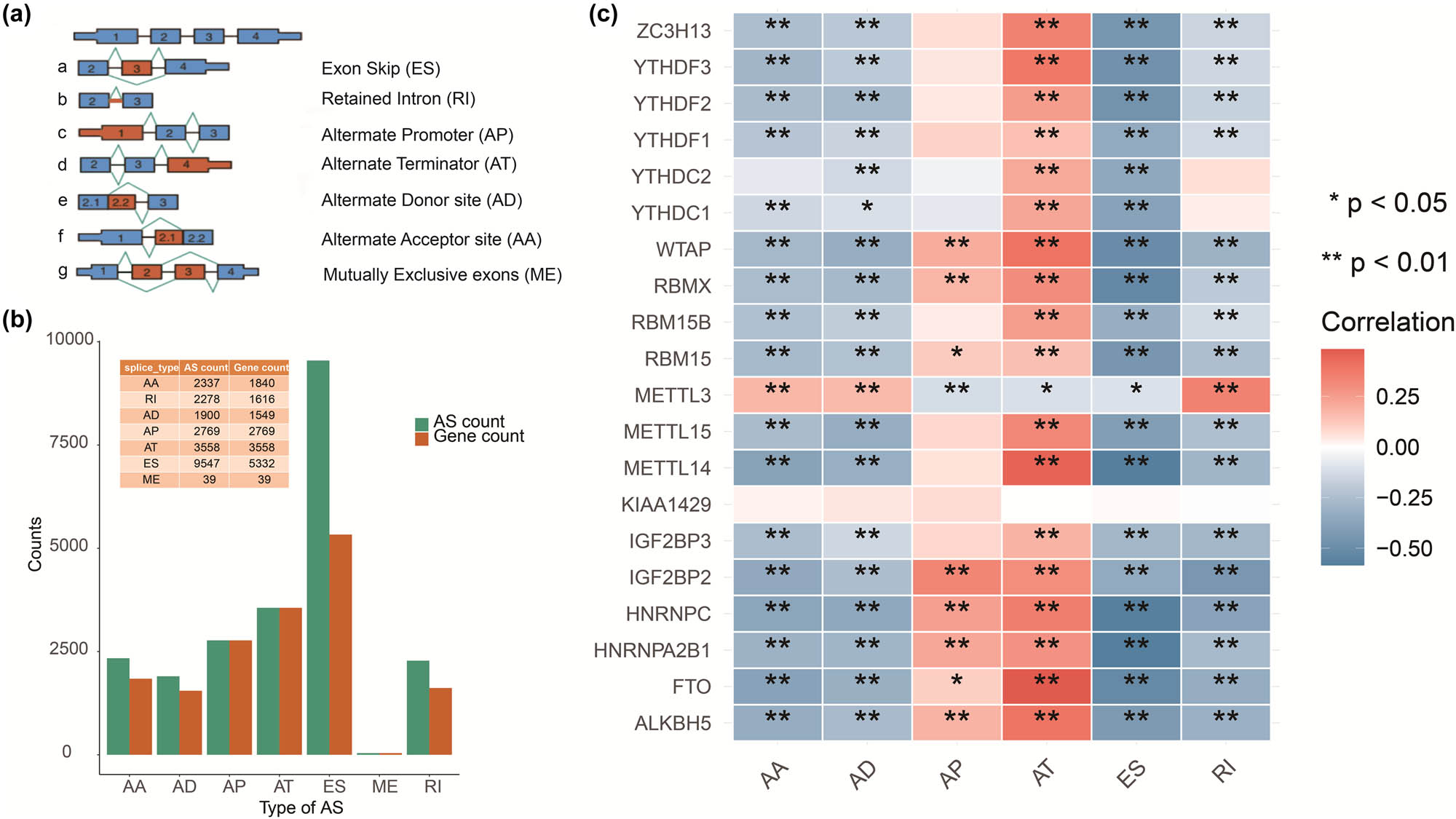
Different AS. (a) Schematic representation of AS events, AA and AD sites, AP, AT, ES, RI, and ME. (b) Numbers of AS events and corresponding genes in enrolled cases. (c) Correlation between m6A methylation regulators and AS events.
3.2 Screening of prognostic-related DEAS
A total of 1,310 m6A methylation-related DEAS were identified, corresponding to 1,015 genes (Table S2). After the univariate Cox hazard analysis, six AS types were significantly associated with OS (p < 0.05). There were 13, 6, 38, 52, 57, and 59 AS events in AA, AD, AP, AT, ES, and RI significantly related to OS, respectively (Table S3, p < 0.05).We then calculated the Pearson correlation coefficients between m6A methylation regulators and AS events. As shown in Figure 2c, there was a strong correlation between m6A methylation regulators and AS events (Table S4).
3.3 Function and pathway enrichment analysis of prognostic-related DEAS
The genes corresponding to the RI event were enriched in 4 KEGG and 210 GO BP pathways. These terms included spliceosome, regulation of RNA splicing, Golgi to plasma membrane protein transport, sister chromatid segregation, regulation of mRNA splicing via spliceosome, and mitochondrial calcium ion transmembrane transport (Figure S1a and b). A total of 3 KEGG pathways and 152 terms were obtained from the AT enrichment analysis. These terms were mainly involved in base excision repair and the regulation of the G2/M cell cycle phase (Figure S2a and b). The genes corresponding to AA were mainly involved in the MAPK signaling pathway, VEGF signaling pathway, and histone acetylation (Figure S3a and b). The genes corresponding to AP were mainly involved in endocrine resistance and responses to prostaglandins (Figure S4a and b). A total of 6 KEGG pathways and 174 terms were obtained from the ES enrichment analysis. These terms were mainly involved in mitotic centrosome separation, cytokinesis, and centrosome separation (Figure S5a and b). As shown in Figure S6a and b, genes in AD were mainly involved in the phospholipase D signaling pathway, antigen processing, and antigen presentation by MHC II.
3.4 The correlation network among genes in different AS events
The constructed PPI network included 110 edges and 81 nodes (Table S5 and Figure 3). The ten genes with the highest degrees in the network were Akt serine/threonine kinase 1 (AKT1), serine- and arginine-rich splicing factor 6 (SRSF6), serine- and arginine-rich splicing factor 2 (SRSF2), heterogeneous nuclear ribonucleoprotein a1 (HNRNPA1), nucleoporin 62 (NUP62), HNRNPA2B1, serine- and arginine-rich splicing factor 7 (SRSF7), minichromosome maintenance complex component 7 (MCM7), heterogeneous nuclear ribonucleoprotein l (HNRNPL), and heterogeneous nuclear ribonucleoprotein D like (HNRNPDL). Their respective cutting events were AP, ES, RI, ES, RI, AA, RI, AP, ES, and ES (Table 1).
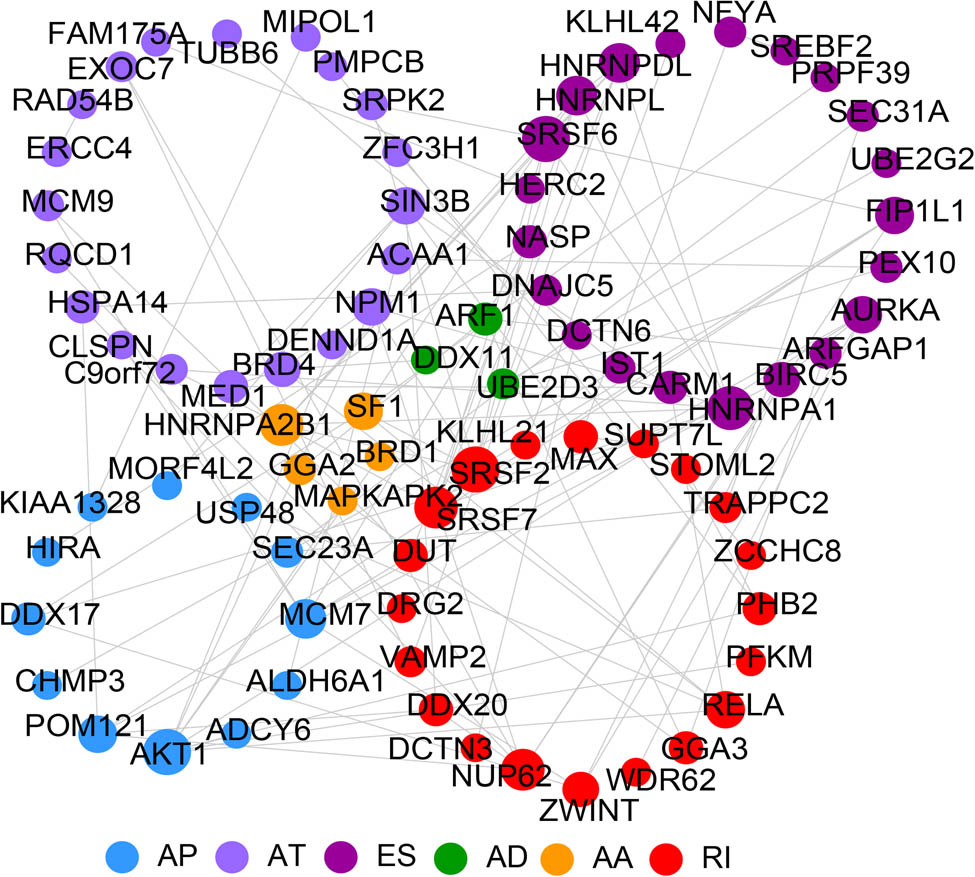
PPI network of prognostic-related m6A methylation-related DEAS. The blue, lavender, purple, green, yellow, and red nodes represent genes where AP, AT, ES, AD, AA, and RI have occurred, respectively. AA and AD sites, AP, AT, ES, RI. The larger the nodes, the more proteins interact with them.
TOP10 genes and corresponding alternative splicing events in protein–protein interaction network
| Name | as_id | Betweenness | Closeness | Degree | Splice_type |
|---|---|---|---|---|---|
| AKT1 | 29559 | 935.6667 | 0.032219 | 9 | AP |
| SRSF6 | 59434 | 102.6048 | 0.031733 | 9 | ES |
| SRSF2 | 43660 | 228.1071 | 0.031911 | 9 | RI |
| HNRNPA1 | 22149 | 264.0103 | 0.031923 | 8 | ES |
| NUP62 | 51123 | 535.3992 | 0.032116 | 7 | RI |
| HNRNPA2B1 | 79038 | 13.77143 | 0.031484 | 7 | AA |
| SRSF7 | 53279 | 23.4 | 0.031658 | 7 | RI |
| MCM7 | 80881 | 470 | 0.031583 | 6 | AP |
| HNRNPL | 49699 | 0.333333 | 0.031471 | 6 | ES |
| HNRNPDL | 69705 | 0.333333 | 0.031471 | 6 | ES |
3.5 Network of SFs and the prognostic-related DEAS
A total of eight SFs related to prognosis were obtained. These were NOVA AS regulator 1 (NOVA1), heterogeneous nuclear ribonucleoprotein H1 (HNRNPH1), QKI, KH domain containing RNA binding (QKI), splicing factor 3b subunit 1 (SF3B1), NOVA AS regulator 2 (NOVA2), TIA1 cytotoxic granule-associated RNA-binding protein (TIA1), ELAV-like RNA-binding protein 3 (ELAVL3), and RNA-binding Fox-1 homolog 2 (RBFOX2). Furthermore, we divided the patients into high- and low-expression groups according to the expression values of these eight SFs. A Kaplan–Meier (KM) curve was used to measure the difference in OS between the groups (Figure 4). Patients with low expression levels of NOVA1, NOVA2, and QKI had a better prognosis than those with high expression levels (p < 0.05) (Figure 4a–c). Low expression of ITA1, ELAV3, HNRNPH1, and SF3B1 was correlated with a significantly worse prognosis (p < 0.05) (Figure 4d–g). There was no significant difference between the prognosis of patients in the RBFOX2 high- and low-expression groups (p = 0.05) (Figure 4h).
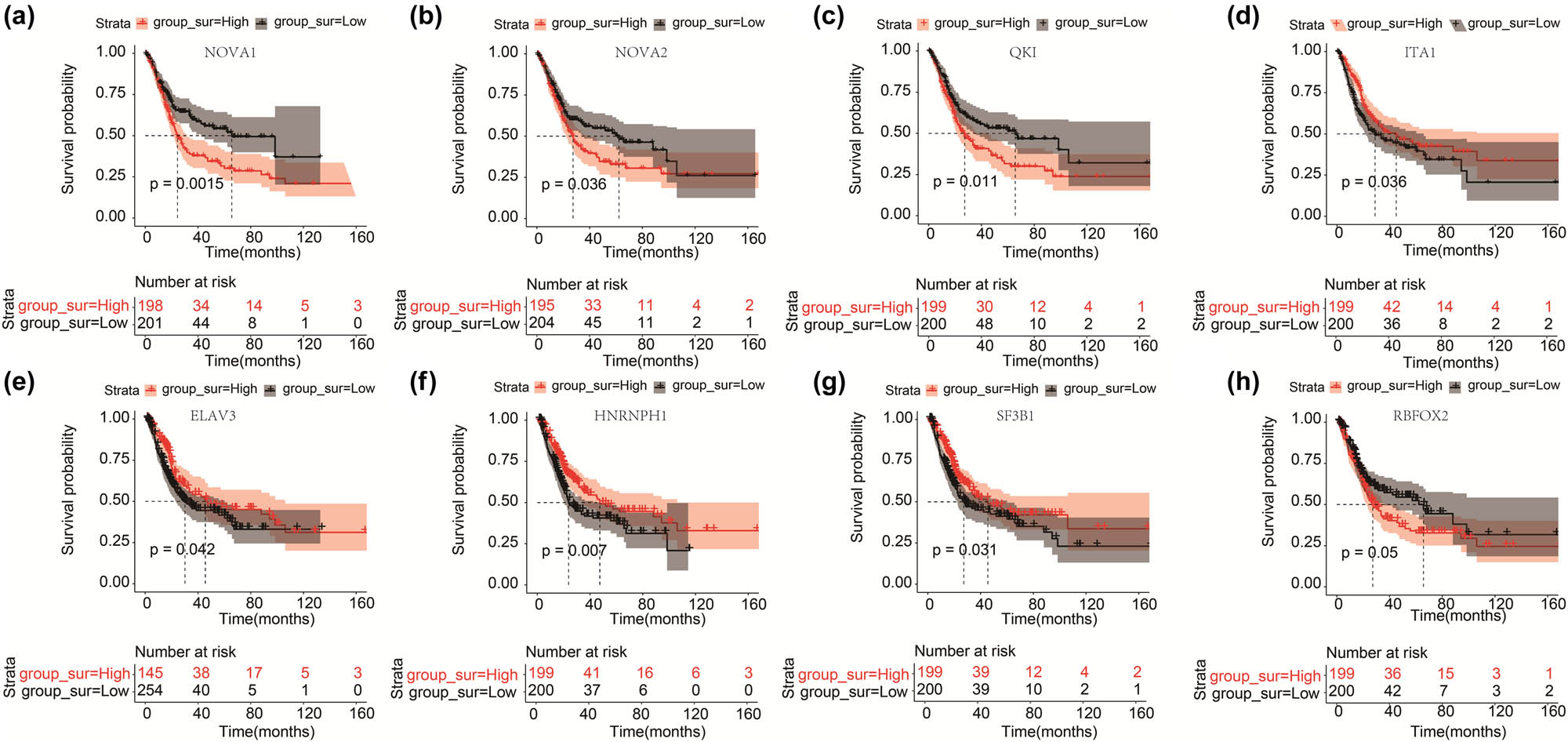
KM curves of the OS difference between high and low expression of SF. (a–h) Represent the KM curves of the OS difference between the high and low expression of NOVA1, NOVA2, QKI, ITA1, ELVA3, HNRNPH1, SF3B1, and RBFOX2, respectively.
The relationship between SFs expression and the PSI of AS events was analyzed. A regulatory network was constructed based on this relationship (Figure 5). This regulatory network contained 32 genes that corresponded to 34 AS events and 4 SFs (QK1, RBFOX2, TIA1, and HNRNPH1) (Table S6).
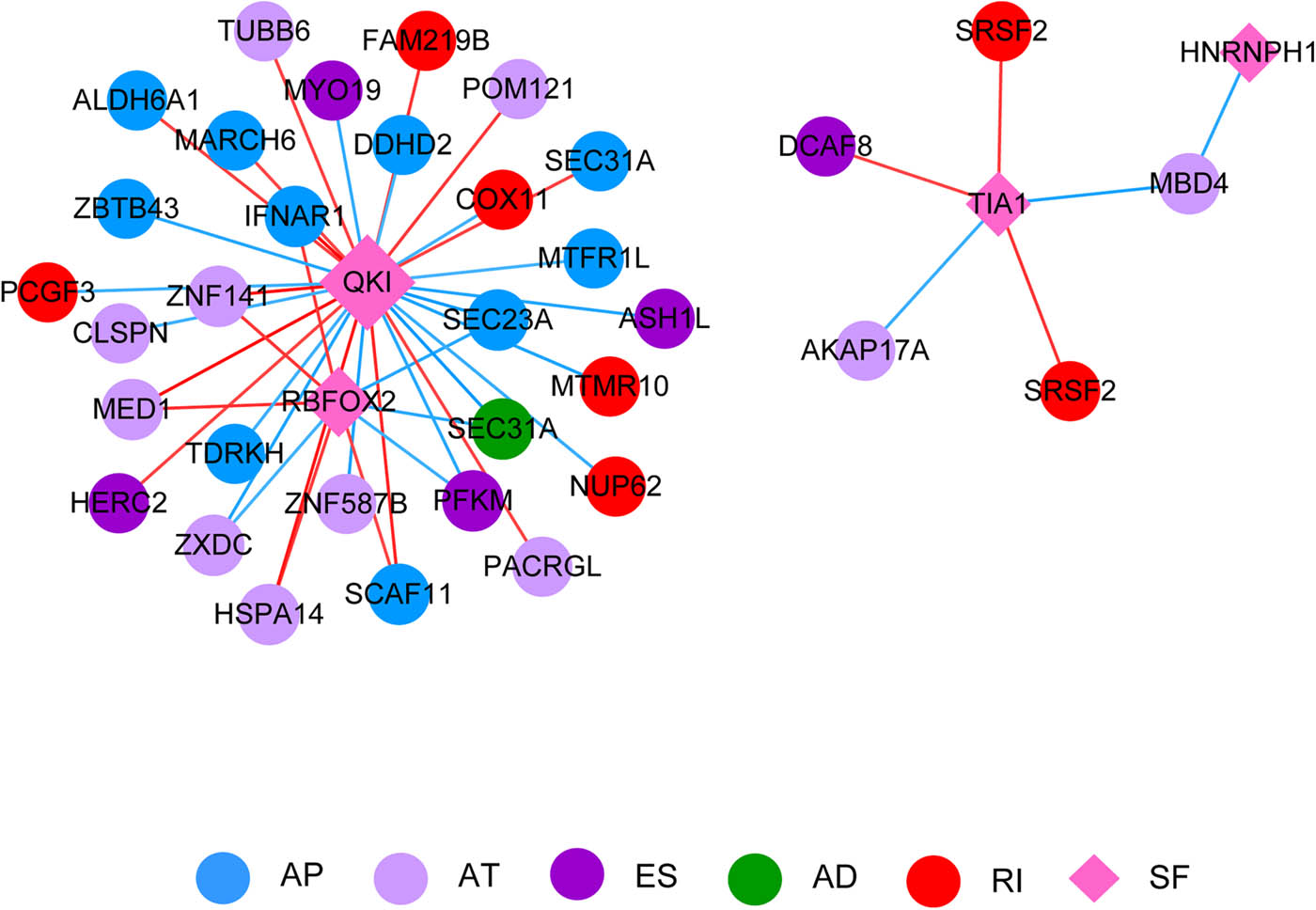
Correlation network between SF and the genes corresponding to AS events. Blue, lavender, purple, green, and red nodes represent AP, AT, ES, AD, and RI events, respectively. The pink diamond represents SF. AD, AP, AT, ES, RI. The red line indicates a positive correlation, and the blue line indicates a negative correlation.
3.6 Cox proportional hazards regression model
Six Cox proportional hazards regression models were constructed using the above six OS-related AS types (AA, AD, AP, AT, ES, and RI). Patients were divided into two groups based on the risk scores calculated in each model. In either models, patients with low-risk scores had a better OS than those with high-risk scores (p < 0.05) (Figure 6a–f). Furthermore, we constructed an ROC curve to measure the performance of each model. The AUC of AT (0.793) was the highest among all models, while that of AD (0.624) was the lowest. The AUCs for RI, AP, ES, and AA were 0.729, 0.703, 0.786, and 0.652, respectively (Figure 6g).
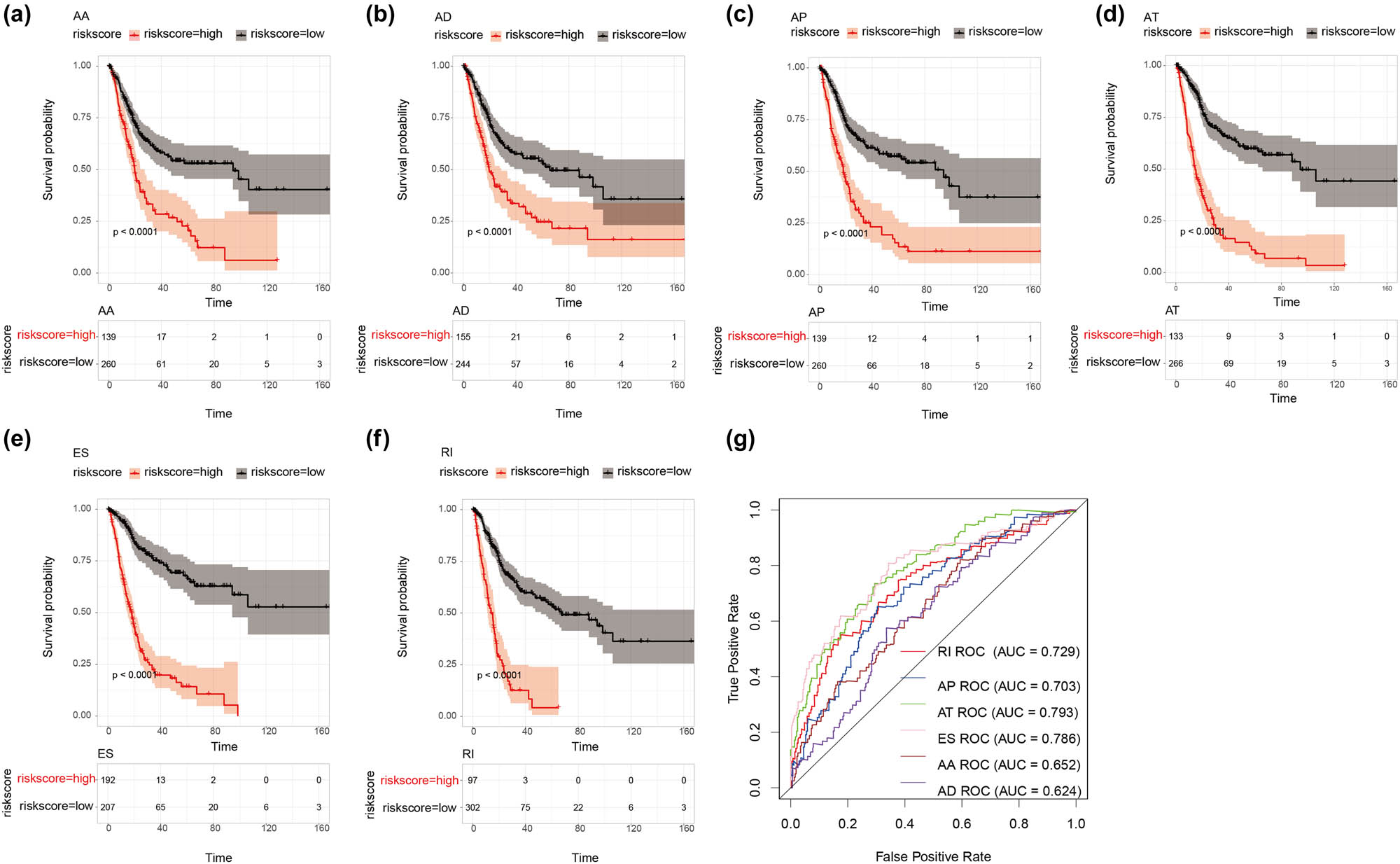
KM and ROC of prognostic predictors in the TCGA. (a–f) Represent the KM plot depicting the survival probability over time for the prognostic predictor of six types of AS events with the high- (red) and low- (blue) risk groups, respectively. (g) ROC analysis for all prognostic predictors.
3.7 Correlation between m6A methylation-related DEAS and BLCA prognosis in the validation dataset
Based on GSE13507, we validated that the prognosis of patients in the high-risk score group was significantly worse than that in the low-risk score group in all six models (AA, AD, AP, AT, ES, and RI) (Figure 7a–f). Further ROC analysis showed that all six models had good predictive performances. The AUCs for RI, AP, AT, AA, ES, and AD were 0.845, 0.809, 0.759, 0.738, 0.677, and 0.672, respectively (Figure 7g).
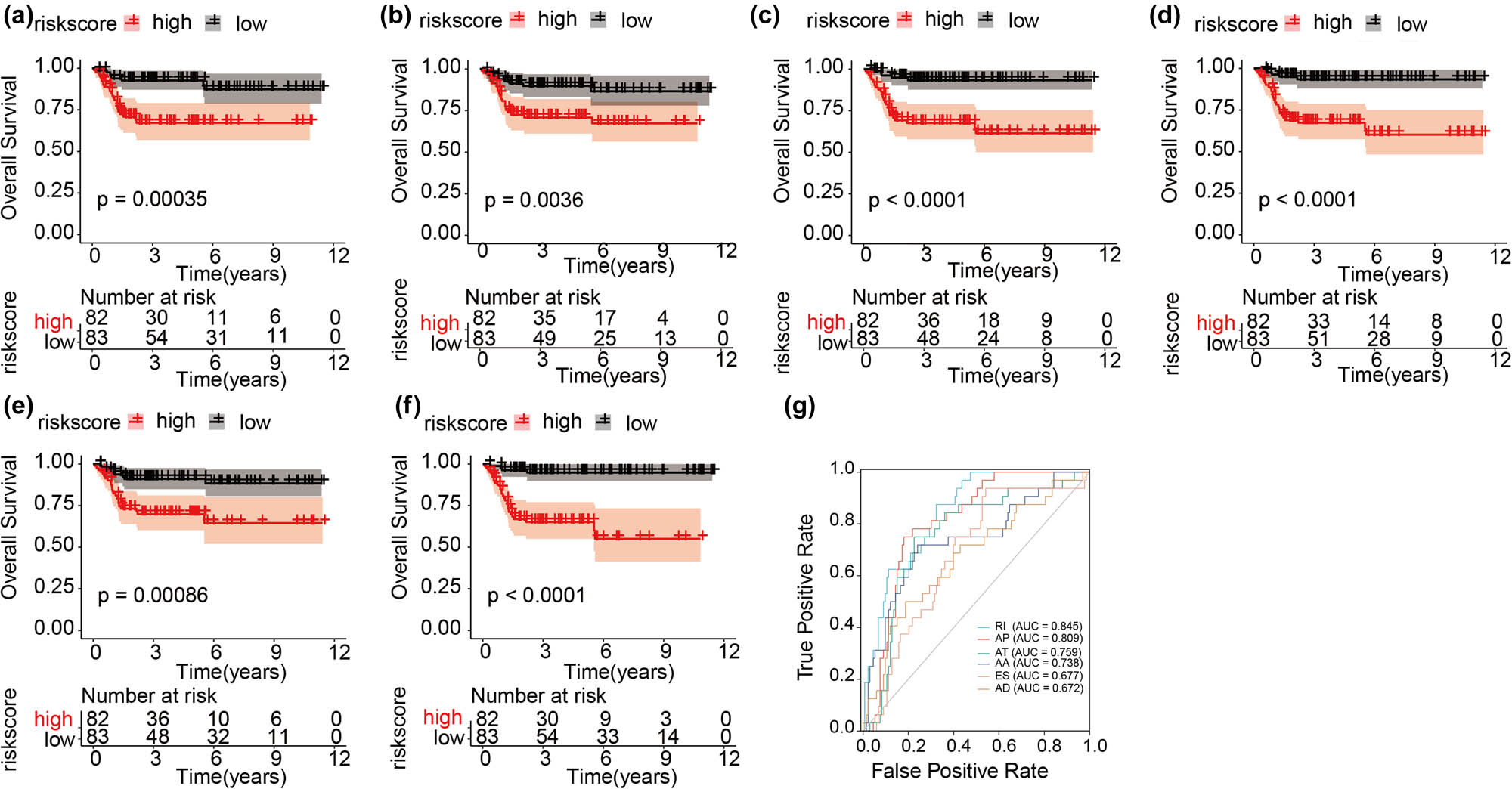
KM and ROC of prognostic predictors in GEO. (a–f) Represent the KM plot depicting the survival probability over time for the prognostic predictor of six types of AS events with the high- (red) and low- (blue) risk groups, respectively. (g) ROC analysis for all prognostic predictors.
3.8 Screening of independent prognostic clinical factors
After performing a univariate Cox analysis, we found that TNM stage, age, and risk score were significantly associated with prognosis (data not shown, p < 0.05). Further multivariate Cox analysis showed that in the AS groups, except for AP (p = 0.148), the risk score was an independent prognostic factor of BLCA (p < 0.05) (Figure 8).
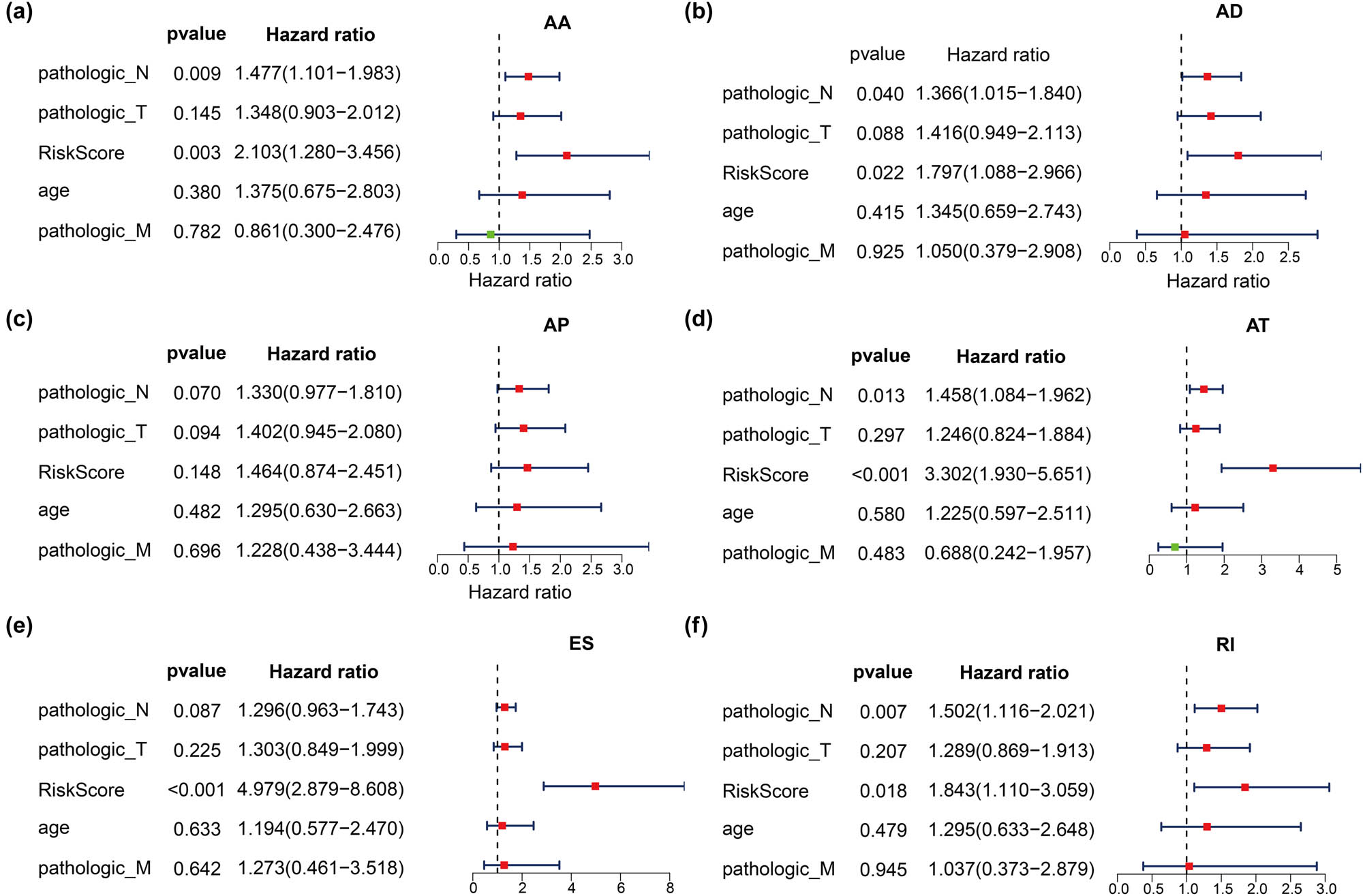
Forest plots for independent prognostic clinical factors based on multivariate Cox regression analysis. (a–g) Represent Forest plots for independent prognostic clinical factors in AA, AD, AP, AT, ES, and RI, respectively. AA and AD sites, AP, AT, ES, RI.
4 Discussion
BLCA is a common disease of the urinary system involving malignant tumors with a 60–70% recurrence rate after therapy [23]. Genetic factors, including AS, have been proved by many studies to play a role in the metastasis and development of BLCA [24]. m6A methylation modification is a potential target for cancer therapy and has been extensively studied in recent years [25]. However, there are few reports on combining m6A methylation modification and AS to analyze its role in BLCA species. The main purpose of this study was to explore whether the combined analysis of m6A methylation modification and AS will provide new insights into understanding the mechanism of BLCA.
We found that six AS types were significantly associated with OS. A significant correlation was also observed between m6A methylation regulators and AS events. In another study, m6A methylation regulators were found to play an important regulatory role in gene AS events [26]. Future studies will investigate the possible function of prognostic-related DEAS. Enrichment analysis revealed that they were mainly involved in RNA splicing regulation, cell cycle G2/M regulation, the MAPK signaling pathway, and prostaglandin response. These functions have been associated with various cancers [27]. Prostaglandins are lipids derived from polyunsaturated fatty acids. Recent studies suggest that it also plays a role in BLCA. Prostaglandin E2 (PGE2) is a highly expressed prostaglandin that controls a variety of normal, tissue-dependent physiological processes [28]. PGE2 has a tumor-promoting effect. It blocks cell death, enhances angiogenesis, promotes tumor inflammation, and prevents proper immune surveillance of tumors [29]. PGE2 shifts the phenotype of macrophages from antitumor M1 macrophages to pro-tumor M2 macrophages. PGE2/EP4 signaling can regulate macrophage plasticity [30]. Cyclooxygenase (COX)-2, one of the key proteins responsible for angiogenesis and tumorigenesis, is highly expressed in BLCA. Prostaglandin levels were positively correlated with COX2 expression and BLCA progression [31]. Inhibition of COX enzymes by non-vehicle anti-inflammatory drugs results in decreased prostaglandin D2 levels, ultimately reducing the occurrence of BLCA [32].
There was an interaction relationship between the genes corresponding to the prognosis-related DEAS, and a PPI network consisting of 81 points was formed. The top three nodes in the network are AKT1, SRSF6, and SRSF2. TIA1 is an important tumor suppressor gene involved in many aspects of cancer occurrence and development and is considered a new tumor suppressor [33]. It regulates the expression of various mRNAs involved in proliferation, apoptosis, and angiogenesis of cancer cells [34]. TIA1 regulates RAB40B to inhibit cell proliferation [35]. In addition, TIA1 can promote apoptosis by regulating Fas AS [36]. SRSF6 and SRSF2 are serine/arginine-rich SFs. SRSF6 is essential for normal growth and development. SRSF6 also protects the genome from the formation of DNA–RNA hybrids (R-loops) following transcription [37]. Mutations in SRSF2 are associated with many diseases [38].
In addition, these prognosis-related DEAS interacted with SFs. Most poor prognostic AS events were inversely associated with SF expression, consistent with previous findings [39]. Upregulated expression of oncogenic SFs increases the oncogenic potential of protein variants and promotes cancer development [40]. These regulatory axes provide important guidance for subsequent exploration of the specific functional mechanisms of DEAS events.
We constructed six Cox proportional hazards regression models for the six OS-related AS types. An ROC curve was developed to evaluate the predictive efficiency of these models. The results revealed that the AUC of the AT model (0.793) was the highest, followed by ES (0.786), RI (0.729), AA (0.652), and AD (0.624). The above results show that our model has good predictive efficiency. Similarly, we also demonstrated the good predictive performance of the model in the GEO validation cohort. There has been no research on constructing a BLCA prognostic risk model using different AS types. Interestingly, AT showed the best predictive performance in the different types of AS-based prognostic risk models constructed for other cancers, such as glioma (AUC = 0.907) [41]. In another study on head and neck cancer, AT events were the most powerful prognostic factor [42].
Independence analysis revealed that the prognostic risk model based on multiple AS types was an independent prognostic factor for BLCA. A recent study demonstrated that high-throughput sequencing analysis is a key technique for characterizing the most common genetic aberrations in spliceosomes and splice sites [43]. In the future, clinical factors independent of age and TNM stage will be used to predict the prognosis of BLCA, and a combination of multiple clinical factors may yield better results.
This study had certain limitations. First, it was a retrospective study based on public databases. In the future, multiple centers, large samples, and more clinical features are needed to verify the results of this study. Second, BLCA is also affected by external environmental factors, such as smoking, in addition to genetic factors. Therefore, the constructed model needs further stratified verification. Third, whether the AS events obtained in our research actually exist still requires further experimental verification.
In conclusion, this study screened for prognostic-related AS events based on m6A methylation modification and constructed a reliable prognostic risk model. Further research on upstream SF predicted a possible regulatory relationship between SF and DEAS. This study provides insights for the subsequent understanding of the role of AS events in BLCA.
-
Funding information: This study was supported by the 12th Five-Year Social Science Research Project of the Jilin Provincial Education Department (No. 104115023).
-
Author contributions: Yuan Chang and Shukun Yu conceived and designed the study, and Miao Zhang and Tianshu Jiang participated in acquiring the data. Yuan Chang performed the analysis and interpretation of the data. Yuan Liu and Xiuyun Zhu designed the study and performed statistical analysis. Shukun Yu participated in obtaining funding. Yuan Chang and Shukun Yu conceived the study, participated in its design and coordination, helped draft the article, and revised the manuscript for important intellectual content. All authors have read and approved the final manuscript.
-
Conflict of interest: Authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Xu Y, Wu G, Li J, Li J, Ruan N, Ma L, et al. Screening and identification of key biomarkers for bladder cancer: A study based on TCGA and GEO data. BioMed Res Int. 2020;2020:1–20.10.1155/2020/8283401Search in Google Scholar PubMed PubMed Central
[2] Ramirez D, Gupta A, Canter D, Harrow B, Dobbs RW, Kucherov V, et al. Microscopic haematuria at time of diagnosis is associated with lower disease stage in patients with newly diagnosed bladder cancer. BJU Int. 2016;117(5):783–6.10.1111/bju.13345Search in Google Scholar PubMed
[3] Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, et al. Bladder cancer. Nat Rev Dis Primers. 2017;3(1):1–19.10.1038/nrdp.2017.22Search in Google Scholar PubMed
[4] Pelly VS, Moeini A, Roelofsen LM, Bonavita E, Bell CR, Hutton C, et al. Anti-inflammatory drugs remodel the tumor immune environment to enhance immune checkpoint blockade efficacy. Cancer Discovery. 2021;11(10):2602–19.10.1158/2159-8290.CD-20-1815Search in Google Scholar PubMed PubMed Central
[5] Lopez-Beltran A, Cimadamore A, Blanca A, Massari F, Vau N, Scarpelli M, et al. Immune checkpoint inhibitors for the treatment of bladder cancer. Cancers. 2021;13(1):131.10.3390/cancers13010131Search in Google Scholar PubMed PubMed Central
[6] He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):1–15.10.1186/s12943-019-1109-9Search in Google Scholar PubMed PubMed Central
[7] Chen M, Nie Z-Y, Wen X-H, Gao Y-H, Cao H, Zhang S-F. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep. 2019;39(12):BSR20192892.10.1042/BSR20192892Search in Google Scholar PubMed PubMed Central
[8] Tuncel G, Kalkan R. Importance of m N(6)-methyladenosine (m(6)A) RNA modification in cancer. Med Oncol. 2019;36(4):36.10.1007/s12032-019-1260-6Search in Google Scholar PubMed
[9] Zhang C, Fu J, Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front Immunol. 2019;10:922.10.3389/fimmu.2019.00922Search in Google Scholar PubMed PubMed Central
[10] Song Y, Xu Q, Wei Z, Zhen D, Su J, Chen K. Predict epitranscriptome targets and regulatory functions of N (6)-methyladenosine (m(6)A) writers and erasers. Evolut Bioinforma. 2019;15:1176934319871290.10.1177/1176934319871290Search in Google Scholar
[11] Li J, Rao B, Yang J, Liu L, Huang M, Liu X, et al. Dysregulated m6A-related regulators are associated with tumor metastasis and poor prognosis in osteosarcoma. Front Oncol. 2020;10:769.10.3389/fonc.2020.00769Search in Google Scholar PubMed PubMed Central
[12] Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110.10.1186/s12943-019-1036-9Search in Google Scholar PubMed PubMed Central
[13] Ying X, Jiang X, Zhang H, Liu B, Huang Y, Zhu X, et al. Programmable N6-methyladenosine modification of CDCP1 mRNA by RCas9-methyltransferase like 3 conjugates promotes bladder cancer development. Mol Cancer. 2020;19(1):169.10.1186/s12943-020-01289-0Search in Google Scholar PubMed PubMed Central
[14] Kim CJ, Isono T, Tambe Y, Chano T, Okabe H, Okada Y, et al. Role of alternative splicing of periostin in human bladder carcinogenesis. Int J Oncol. 2008;32(1):161–9.10.3892/ijo.32.1.161Search in Google Scholar
[15] Xie R, Chen X, Cheng L, Huang M, Zhou Q, Zhang J, et al. NONO inhibits lymphatic metastasis of bladder cancer via alternative splicing of SETMAR. Mol Ther. 2021;29(1):291–307.10.1016/j.ymthe.2020.08.018Search in Google Scholar PubMed PubMed Central
[16] Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44(D1):D1018–D22.10.1093/nar/gkv1288Search in Google Scholar PubMed PubMed Central
[17] Hayles J. The gene ontology resource: 20 years and still GOing strong. England: Nucleic Acid Research; 2019.Search in Google Scholar
[18] Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.10.1093/nar/28.1.27Search in Google Scholar PubMed PubMed Central
[19] Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J Integr Biol. 2012;16(5):284–7.10.1089/omi.2011.0118Search in Google Scholar PubMed PubMed Central
[20] Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids Res. 2015;43(D1):D447–52.10.1093/nar/gku1003Search in Google Scholar PubMed PubMed Central
[21] Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.10.1101/gr.1239303Search in Google Scholar PubMed PubMed Central
[22] Xiong Y, Deng Y, Wang K, Zhou H, Zheng X, Si L, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: A study based on large-scale sequencing data. EBioMedicine. 2018;36:183–95.10.1016/j.ebiom.2018.09.021Search in Google Scholar PubMed PubMed Central
[23] Shariat SF, Sfakianos JP, Droller MJ, Karakiewicz PI, Meryn S, Bochner BH. The effect of age and gender on bladder cancer: A critical review of the literature. BJU Int. 2010;105(3):300.10.1111/j.1464-410X.2009.09076.xSearch in Google Scholar PubMed PubMed Central
[24] Guo Z, Zhu H, Xu W, Wang X, Liu H, Wu Y, et al. Alternative splicing related genetic variants contribute to bladder cancer risk. Mol Carcinog. 2020;59(8):923–9.10.1002/mc.23207Search in Google Scholar PubMed
[25] Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):1–16.10.1038/s41392-020-00450-xSearch in Google Scholar PubMed PubMed Central
[26] Zhao Z, Cai Q, Zhang P, He B, Peng X, Tu G, et al. N6-Methyladenosine RNA methylation regulator-related alternative splicing (AS) gene signature predicts non–small cell lung cancer prognosis. Front Mol Biosci. 2021;8:486.10.3389/fmolb.2021.657087Search in Google Scholar PubMed PubMed Central
[27] Park C, Cha H-J, Lee H, Hwang-Bo H, Ji SY, Kim MY, et al. Induction of G2/M cell cycle arrest and apoptosis by genistein in human bladder cancer T24 cells through inhibition of the ROS-dependent PI3k/Akt signal transduction pathway. Antioxidants. 2019;8(9):327.10.3390/antiox8090327Search in Google Scholar PubMed PubMed Central
[28] Finetti F, Travelli C, Ercoli J, Colombo G, Buoso E, Trabalzini L. Prostaglandin E2 and cancer: Insight into tumor progression and immunity. Biology. 2020;9(12):434.10.3390/biology9120434Search in Google Scholar PubMed PubMed Central
[29] Woolbright BL, Pilbeam CC, Taylor JA. Prostaglandin E2 as a therapeutic target in bladder cancer: From basic science to clinical trials. Prostaglandins Other Lipid Mediators. 2020;148:106409.10.1016/j.prostaglandins.2020.106409Search in Google Scholar PubMed
[30] Mizuno R, Kawada K, Sakai Y. Prostaglandin E2/EP signaling in the tumor microenvironment of colorectal cancer. Int J Mol Sci. 2019;20(24):6254.10.3390/ijms20246254Search in Google Scholar PubMed PubMed Central
[31] Bourn J, Cekanova M. Cyclooxygenase inhibitors potentiate receptor tyrosine kinase therapies in bladder cancer cells in vitro. Drug Des Dev Ther. 2018;12:1727–42.10.2147/DDDT.S158518Search in Google Scholar PubMed PubMed Central
[32] Jara-Gutiérrez Á, Baladrón V. The role of prostaglandins in different types of cancer. Cells. 2021;10(6):1487.10.3390/cells10061487Search in Google Scholar PubMed PubMed Central
[33] Sánchez-Jiménez C, Izquierdo JM. T-cell intracellular antigens in health and disease. Cell Cycle. 2015;14(13):2033–43.10.1080/15384101.2015.1053668Search in Google Scholar PubMed PubMed Central
[34] Hamdollah Zadeh MA, Amin EM, Hoareau-Aveilla C, Domingo E, Symonds KE, Ye X, et al. Alternative splicing of TIA−1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. Mol Oncol. 2015;9(1):167–78.10.1016/j.molonc.2014.07.017Search in Google Scholar PubMed PubMed Central
[35] Reyes R, Alcalde J, Izquierdo JM. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009;10(8):R87.10.1186/gb-2009-10-8-r87Search in Google Scholar PubMed PubMed Central
[36] Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19(4):475–84.10.1016/j.molcel.2005.06.015Search in Google Scholar PubMed
[37] Juge F, Fernando C, Fic W, Tazi J. The SR protein B52/SRp55 is required for DNA topoisomerase I recruitment to chromatin, mRNA release and transcription shutdown. PLoS Genet. 2010;6(9):e1001124.10.1371/journal.pgen.1001124Search in Google Scholar PubMed PubMed Central
[38] Rahman MA, Lin KT, Bradley RK, Abdel-Wahab O, Krainer AR. Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes Dev. 2020;34(5–6):413–27.10.1101/gad.332270.119Search in Google Scholar PubMed PubMed Central
[39] Zong Z, Li H, Yi C, Ying H, Zhu Z, Wang H. Genome-wide profiling of prognostic alternative splicing signature in colorectal cancer. Front Oncol. 2018;8:1–9.10.3389/fonc.2018.00537Search in Google Scholar PubMed PubMed Central
[40] Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2019;68(1):118–29.10.1136/gutjnl-2017-314983Search in Google Scholar PubMed
[41] Xie ZC, Wu HY, Dang YW, Chen G. Role of alternative splicing signatures in the prognosis of glioblastoma. Cancer Med. 2019;8(18):7623–36.10.1002/cam4.2666Search in Google Scholar PubMed PubMed Central
[42] Liang Y, Song J, He D, Xia Y, Wu Y, Yin X, et al. Systematic analysis of survival-associated alternative splicing signatures uncovers prognostic predictors for head and neck cancer. J Cell Physiol. 2019;234(9):15836–46.10.1002/jcp.28241Search in Google Scholar PubMed PubMed Central
[43] Casamassimi A, Federico A, Rienzo M, Esposito S, Ciccodicola A. Transcriptome profiling in human diseases: New advances and perspectives. Int J Mol Sci. 2017;18(8):1652.10.3390/ijms18081652Search in Google Scholar PubMed PubMed Central
© 2022 Yuan Chang et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Biomedical Sciences
- Effects of direct oral anticoagulants dabigatran and rivaroxaban on the blood coagulation function in rabbits
- The mother of all battles: Viruses vs humans. Can humans avoid extinction in 50–100 years?
- Knockdown of G1P3 inhibits cell proliferation and enhances the cytotoxicity of dexamethasone in acute lymphoblastic leukemia
- LINC00665 regulates hepatocellular carcinoma by modulating mRNA via the m6A enzyme
- Association study of CLDN14 variations in patients with kidney stones
- Concanavalin A-induced autoimmune hepatitis model in mice: Mechanisms and future outlook
- Regulation of miR-30b in cancer development, apoptosis, and drug resistance
- Informatic analysis of the pulmonary microecology in non-cystic fibrosis bronchiectasis at three different stages
- Swimming attenuates tumor growth in CT-26 tumor-bearing mice and suppresses angiogenesis by mediating the HIF-1α/VEGFA pathway
- Characterization of intestinal microbiota and serum metabolites in patients with mild hepatic encephalopathy
- Functional conservation and divergence in plant-specific GRF gene family revealed by sequences and expression analysis
- Application of the FLP/LoxP-FRT recombination system to switch the eGFP expression in a model prokaryote
- Biomedical evaluation of antioxidant properties of lamb meat enriched with iodine and selenium
- Intravenous infusion of the exosomes derived from human umbilical cord mesenchymal stem cells enhance neurological recovery after traumatic brain injury via suppressing the NF-κB pathway
- Effect of dietary pattern on pregnant women with gestational diabetes mellitus and its clinical significance
- Potential regulatory mechanism of TNF-α/TNFR1/ANXA1 in glioma cells and its role in glioma cell proliferation
- Effect of the genetic mutant G71R in uridine diphosphate-glucuronosyltransferase 1A1 on the conjugation of bilirubin
- Quercetin inhibits cytotoxicity of PC12 cells induced by amyloid-beta 25–35 via stimulating estrogen receptor α, activating ERK1/2, and inhibiting apoptosis
- Nutrition intervention in the management of novel coronavirus pneumonia patients
- circ-CFH promotes the development of HCC by regulating cell proliferation, apoptosis, migration, invasion, and glycolysis through the miR-377-3p/RNF38 axis
- Bmi-1 directly upregulates glucose transporter 1 in human gastric adenocarcinoma
- Lacunar infarction aggravates the cognitive deficit in the elderly with white matter lesion
- Hydroxysafflor yellow A improved retinopathy via Nrf2/HO-1 pathway in rats
- Comparison of axon extension: PTFE versus PLA formed by a 3D printer
- Elevated IL-35 level and iTr35 subset increase the bacterial burden and lung lesions in Mycobacterium tuberculosis-infected mice
- A case report of CAT gene and HNF1β gene variations in a patient with early-onset diabetes
- Study on the mechanism of inhibiting patulin production by fengycin
- SOX4 promotes high-glucose-induced inflammation and angiogenesis of retinal endothelial cells by activating NF-κB signaling pathway
- Relationship between blood clots and COVID-19 vaccines: A literature review
- Analysis of genetic characteristics of 436 children with dysplasia and detailed analysis of rare karyotype
- Bioinformatics network analyses of growth differentiation factor 11
- NR4A1 inhibits the epithelial–mesenchymal transition of hepatic stellate cells: Involvement of TGF-β–Smad2/3/4–ZEB signaling
- Expression of Zeb1 in the differentiation of mouse embryonic stem cell
- Study on the genetic damage caused by cadmium sulfide quantum dots in human lymphocytes
- Association between single-nucleotide polymorphisms of NKX2.5 and congenital heart disease in Chinese population: A meta-analysis
- Assessment of the anesthetic effect of modified pentothal sodium solution on Sprague-Dawley rats
- Genetic susceptibility to high myopia in Han Chinese population
- Potential biomarkers and molecular mechanisms in preeclampsia progression
- Silencing circular RNA-friend leukemia virus integration 1 restrained malignancy of CC cells and oxaliplatin resistance by disturbing dyskeratosis congenita 1
- Endostar plus pembrolizumab combined with a platinum-based dual chemotherapy regime for advanced pulmonary large-cell neuroendocrine carcinoma as a first-line treatment: A case report
- The significance of PAK4 in signaling and clinicopathology: A review
- Sorafenib inhibits ovarian cancer cell proliferation and mobility and induces radiosensitivity by targeting the tumor cell epithelial–mesenchymal transition
- Characterization of rabbit polyclonal antibody against camel recombinant nanobodies
- Active legumain promotes invasion and migration of neuroblastoma by regulating epithelial-mesenchymal transition
- Effect of cell receptors in the pathogenesis of osteoarthritis: Current insights
- MT-12 inhibits the proliferation of bladder cells in vitro and in vivo by enhancing autophagy through mitochondrial dysfunction
- Study of hsa_circRNA_000121 and hsa_circRNA_004183 in papillary thyroid microcarcinoma
- BuyangHuanwu Decoction attenuates cerebral vasospasm caused by subarachnoid hemorrhage in rats via PI3K/AKT/eNOS axis
- Effects of the interaction of Notch and TLR4 pathways on inflammation and heart function in septic heart
- Monosodium iodoacetate-induced subchondral bone microstructure and inflammatory changes in an animal model of osteoarthritis
- A rare presentation of type II Abernethy malformation and nephrotic syndrome: Case report and review
- Rapid death due to pulmonary epithelioid haemangioendothelioma in several weeks: A case report
- Hepatoprotective role of peroxisome proliferator-activated receptor-α in non-cancerous hepatic tissues following transcatheter arterial embolization
- Correlation between peripheral blood lymphocyte subpopulations and primary systemic lupus erythematosus
- A novel SLC8A1-ALK fusion in lung adenocarcinoma confers sensitivity to alectinib: A case report
- β-Hydroxybutyrate upregulates FGF21 expression through inhibition of histone deacetylases in hepatocytes
- Identification of metabolic genes for the prediction of prognosis and tumor microenvironment infiltration in early-stage non-small cell lung cancer
- BTBD10 inhibits glioma tumorigenesis by downregulating cyclin D1 and p-Akt
- Mucormycosis co-infection in COVID-19 patients: An update
- Metagenomic next-generation sequencing in diagnosing Pneumocystis jirovecii pneumonia: A case report
- Long non-coding RNA HOXB-AS1 is a prognostic marker and promotes hepatocellular carcinoma cells’ proliferation and invasion
- Preparation and evaluation of LA-PEG-SPION, a targeted MRI contrast agent for liver cancer
- Proteomic analysis of the liver regulating lipid metabolism in Chaohu ducks using two-dimensional electrophoresis
- Nasopharyngeal tuberculosis: A case report
- Characterization and evaluation of anti-Salmonella enteritidis activity of indigenous probiotic lactobacilli in mice
- Aberrant pulmonary immune response of obese mice to periodontal infection
- Bacteriospermia – A formidable player in male subfertility
- In silico and in vivo analysis of TIPE1 expression in diffuse large B cell lymphoma
- Effects of KCa channels on biological behavior of trophoblasts
- Interleukin-17A influences the vulnerability rather than the size of established atherosclerotic plaques in apolipoprotein E-deficient mice
- Multiple organ failure and death caused by Staphylococcus aureus hip infection: A case report
- Prognostic signature related to the immune environment of oral squamous cell carcinoma
- Primary and metastatic squamous cell carcinoma of the thyroid gland: Two case reports
- Neuroprotective effects of crocin and crocin-loaded niosomes against the paraquat-induced oxidative brain damage in rats
- Role of MMP-2 and CD147 in kidney fibrosis
- Geometric basis of action potential of skeletal muscle cells and neurons
- Babesia microti-induced fulminant sepsis in an immunocompromised host: A case report and the case-specific literature review
- Role of cerebellar cortex in associative learning and memory in guinea pigs
- Application of metagenomic next-generation sequencing technique for diagnosing a specific case of necrotizing meningoencephalitis caused by human herpesvirus 2
- Case report: Quadruple primary malignant neoplasms including esophageal, ureteral, and lung in an elderly male
- Long non-coding RNA NEAT1 promotes angiogenesis in hepatoma carcinoma via the miR-125a-5p/VEGF pathway
- Osteogenic differentiation of periodontal membrane stem cells in inflammatory environments
- Knockdown of SHMT2 enhances the sensitivity of gastric cancer cells to radiotherapy through the Wnt/β-catenin pathway
- Continuous renal replacement therapy combined with double filtration plasmapheresis in the treatment of severe lupus complicated by serious bacterial infections in children: A case report
- Simultaneous triple primary malignancies, including bladder cancer, lymphoma, and lung cancer, in an elderly male: A case report
- Preclinical immunogenicity assessment of a cell-based inactivated whole-virion H5N1 influenza vaccine
- One case of iodine-125 therapy – A new minimally invasive treatment of intrahepatic cholangiocarcinoma
- S1P promotes corneal trigeminal neuron differentiation and corneal nerve repair via upregulating nerve growth factor expression in a mouse model
- Early cancer detection by a targeted methylation assay of circulating tumor DNA in plasma
- Calcifying nanoparticles initiate the calcification process of mesenchymal stem cells in vitro through the activation of the TGF-β1/Smad signaling pathway and promote the decay of echinococcosis
- Evaluation of prognostic markers in patients infected with SARS-CoV-2
- N6-Methyladenosine-related alternative splicing events play a role in bladder cancer
- Characterization of the structural, oxidative, and immunological features of testis tissue from Zucker diabetic fatty rats
- Effects of glucose and osmotic pressure on the proliferation and cell cycle of human chorionic trophoblast cells
- Investigation of genotype diversity of 7,804 norovirus sequences in humans and animals of China
- Characteristics and karyotype analysis of a patient with turner syndrome complicated with multiple-site tumors: A case report
- Aggravated renal fibrosis is positively associated with the activation of HMGB1-TLR2/4 signaling in STZ-induced diabetic mice
- Distribution characteristics of SARS-CoV-2 IgM/IgG in false-positive results detected by chemiluminescent immunoassay
- SRPX2 attenuated oxygen–glucose deprivation and reperfusion-induced injury in cardiomyocytes via alleviating endoplasmic reticulum stress-induced apoptosis through targeting PI3K/Akt/mTOR axis
- Aquaporin-8 overexpression is involved in vascular structure and function changes in placentas of gestational diabetes mellitus patients
- Relationship between CRP gene polymorphisms and ischemic stroke risk: A systematic review and meta-analysis
- Effects of growth hormone on lipid metabolism and sexual development in pubertal obese male rats
- Cloning and identification of the CTLA-4IgV gene and functional application of vaccine in Xinjiang sheep
- Antitumor activity of RUNX3: Upregulation of E-cadherin and downregulation of the epithelial–mesenchymal transition in clear-cell renal cell carcinoma
- PHF8 promotes osteogenic differentiation of BMSCs in old rat with osteoporosis by regulating Wnt/β-catenin pathway
- A review of the current state of the computer-aided diagnosis (CAD) systems for breast cancer diagnosis
- Bilateral dacryoadenitis in adult-onset Still’s disease: A case report
- A novel association between Bmi-1 protein expression and the SUVmax obtained by 18F-FDG PET/CT in patients with gastric adenocarcinoma
- The role of erythrocytes and erythroid progenitor cells in tumors
- Relationship between platelet activation markers and spontaneous abortion: A meta-analysis
- Abnormal methylation caused by folic acid deficiency in neural tube defects
- Silencing TLR4 using an ultrasound-targeted microbubble destruction-based shRNA system reduces ischemia-induced seizures in hyperglycemic rats
- Plant Sciences
- Seasonal succession of bacterial communities in cultured Caulerpa lentillifera detected by high-throughput sequencing
- Cloning and prokaryotic expression of WRKY48 from Caragana intermedia
- Novel Brassica hybrids with different resistance to Leptosphaeria maculans reveal unbalanced rDNA signal patterns
- Application of exogenous auxin and gibberellin regulates the bolting of lettuce (Lactuca sativa L.)
- Phytoremediation of pollutants from wastewater: A concise review
- Genome-wide identification and characterization of NBS-encoding genes in the sweet potato wild ancestor Ipomoea trifida (H.B.K.)
- Alleviative effects of magnetic Fe3O4 nanoparticles on the physiological toxicity of 3-nitrophenol to rice (Oryza sativa L.) seedlings
- Selection and functional identification of Dof genes expressed in response to nitrogen in Populus simonii × Populus nigra
- Study on pecan seed germination influenced by seed endocarp
- Identification of active compounds in Ophiopogonis Radix from different geographical origins by UPLC-Q/TOF-MS combined with GC-MS approaches
- The entire chloroplast genome sequence of Asparagus cochinchinensis and genetic comparison to Asparagus species
- Genome-wide identification of MAPK family genes and their response to abiotic stresses in tea plant (Camellia sinensis)
- Selection and validation of reference genes for RT-qPCR analysis of different organs at various development stages in Caragana intermedia
- Cloning and expression analysis of SERK1 gene in Diospyros lotus
- Integrated metabolomic and transcriptomic profiling revealed coping mechanisms of the edible and medicinal homologous plant Plantago asiatica L. cadmium resistance
- A missense variant in NCF1 is associated with susceptibility to unexplained recurrent spontaneous abortion
- Assessment of drought tolerance indices in faba bean genotypes under different irrigation regimes
- The entire chloroplast genome sequence of Asparagus setaceus (Kunth) Jessop: Genome structure, gene composition, and phylogenetic analysis in Asparagaceae
- Food Science
- Dietary food additive monosodium glutamate with or without high-lipid diet induces spleen anomaly: A mechanistic approach on rat model
- Binge eating disorder during COVID-19
- Potential of honey against the onset of autoimmune diabetes and its associated nephropathy, pancreatitis, and retinopathy in type 1 diabetic animal model
- FTO gene expression in diet-induced obesity is downregulated by Solanum fruit supplementation
- Physical activity enhances fecal lactobacilli in rats chronically drinking sweetened cola beverage
- Supercritical CO2 extraction, chemical composition, and antioxidant effects of Coreopsis tinctoria Nutt. oleoresin
- Functional constituents of plant-based foods boost immunity against acute and chronic disorders
- Effect of selenium and methods of protein extraction on the proteomic profile of Saccharomyces yeast
- Microbial diversity of milk ghee in southern Gansu and its effect on the formation of ghee flavor compounds
- Ecology and Environmental Sciences
- Effects of heavy metals on bacterial community surrounding Bijiashan mining area located in northwest China
- Microorganism community composition analysis coupling with 15N tracer experiments reveals the nitrification rate and N2O emissions in low pH soils in Southern China
- Genetic diversity and population structure of Cinnamomum balansae Lecomte inferred by microsatellites
- Preliminary screening of microplastic contamination in different marine fish species of Taif market, Saudi Arabia
- Plant volatile organic compounds attractive to Lygus pratensis
- Effects of organic materials on soil bacterial community structure in long-term continuous cropping of tomato in greenhouse
- Effects of soil treated fungicide fluopimomide on tomato (Solanum lycopersicum L.) disease control and plant growth
- Prevalence of Yersinia pestis among rodents captured in a semi-arid tropical ecosystem of south-western Zimbabwe
- Effects of irrigation and nitrogen fertilization on mitigating salt-induced Na+ toxicity and sustaining sea rice growth
- Bioengineering and Biotechnology
- Poly-l-lysine-caused cell adhesion induces pyroptosis in THP-1 monocytes
- Development of alkaline phosphatase-scFv and its use for one-step enzyme-linked immunosorbent assay for His-tagged protein detection
- Development and validation of a predictive model for immune-related genes in patients with tongue squamous cell carcinoma
- Agriculture
- Effects of chemical-based fertilizer replacement with biochar-based fertilizer on albic soil nutrient content and maize yield
- Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions
- Agronomic and economic performance of mung bean (Vigna radiata L.) varieties in response to rates of blended NPS fertilizer in Kindo Koysha district, Southern Ethiopia
- Influence of furrow irrigation regime on the yield and water consumption indicators of winter wheat based on a multi-level fuzzy comprehensive evaluation
- Discovery of exercise-related genes and pathway analysis based on comparative genomes of Mongolian originated Abaga and Wushen horse
- Lessons from integrated seasonal forecast-crop modelling in Africa: A systematic review
- Evolution trend of soil fertility in tobacco-planting area of Chenzhou, Hunan Province, China
- Animal Sciences
- Morphological and molecular characterization of Tatera indica Hardwicke 1807 (Rodentia: Muridae) from Pothwar, Pakistan
- Research on meat quality of Qianhua Mutton Merino sheep and Small-tail Han sheep
- SI: A Scientific Memoir
- Suggestions on leading an academic research laboratory group
- My scientific genealogy and the Toronto ACDC Laboratory, 1988–2022
- Erratum
- Erratum to “Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study”
- Erratum to “A two-microRNA signature predicts the progression of male thyroid cancer”
- Retraction
- Retraction of “Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis”
Articles in the same Issue
- Biomedical Sciences
- Effects of direct oral anticoagulants dabigatran and rivaroxaban on the blood coagulation function in rabbits
- The mother of all battles: Viruses vs humans. Can humans avoid extinction in 50–100 years?
- Knockdown of G1P3 inhibits cell proliferation and enhances the cytotoxicity of dexamethasone in acute lymphoblastic leukemia
- LINC00665 regulates hepatocellular carcinoma by modulating mRNA via the m6A enzyme
- Association study of CLDN14 variations in patients with kidney stones
- Concanavalin A-induced autoimmune hepatitis model in mice: Mechanisms and future outlook
- Regulation of miR-30b in cancer development, apoptosis, and drug resistance
- Informatic analysis of the pulmonary microecology in non-cystic fibrosis bronchiectasis at three different stages
- Swimming attenuates tumor growth in CT-26 tumor-bearing mice and suppresses angiogenesis by mediating the HIF-1α/VEGFA pathway
- Characterization of intestinal microbiota and serum metabolites in patients with mild hepatic encephalopathy
- Functional conservation and divergence in plant-specific GRF gene family revealed by sequences and expression analysis
- Application of the FLP/LoxP-FRT recombination system to switch the eGFP expression in a model prokaryote
- Biomedical evaluation of antioxidant properties of lamb meat enriched with iodine and selenium
- Intravenous infusion of the exosomes derived from human umbilical cord mesenchymal stem cells enhance neurological recovery after traumatic brain injury via suppressing the NF-κB pathway
- Effect of dietary pattern on pregnant women with gestational diabetes mellitus and its clinical significance
- Potential regulatory mechanism of TNF-α/TNFR1/ANXA1 in glioma cells and its role in glioma cell proliferation
- Effect of the genetic mutant G71R in uridine diphosphate-glucuronosyltransferase 1A1 on the conjugation of bilirubin
- Quercetin inhibits cytotoxicity of PC12 cells induced by amyloid-beta 25–35 via stimulating estrogen receptor α, activating ERK1/2, and inhibiting apoptosis
- Nutrition intervention in the management of novel coronavirus pneumonia patients
- circ-CFH promotes the development of HCC by regulating cell proliferation, apoptosis, migration, invasion, and glycolysis through the miR-377-3p/RNF38 axis
- Bmi-1 directly upregulates glucose transporter 1 in human gastric adenocarcinoma
- Lacunar infarction aggravates the cognitive deficit in the elderly with white matter lesion
- Hydroxysafflor yellow A improved retinopathy via Nrf2/HO-1 pathway in rats
- Comparison of axon extension: PTFE versus PLA formed by a 3D printer
- Elevated IL-35 level and iTr35 subset increase the bacterial burden and lung lesions in Mycobacterium tuberculosis-infected mice
- A case report of CAT gene and HNF1β gene variations in a patient with early-onset diabetes
- Study on the mechanism of inhibiting patulin production by fengycin
- SOX4 promotes high-glucose-induced inflammation and angiogenesis of retinal endothelial cells by activating NF-κB signaling pathway
- Relationship between blood clots and COVID-19 vaccines: A literature review
- Analysis of genetic characteristics of 436 children with dysplasia and detailed analysis of rare karyotype
- Bioinformatics network analyses of growth differentiation factor 11
- NR4A1 inhibits the epithelial–mesenchymal transition of hepatic stellate cells: Involvement of TGF-β–Smad2/3/4–ZEB signaling
- Expression of Zeb1 in the differentiation of mouse embryonic stem cell
- Study on the genetic damage caused by cadmium sulfide quantum dots in human lymphocytes
- Association between single-nucleotide polymorphisms of NKX2.5 and congenital heart disease in Chinese population: A meta-analysis
- Assessment of the anesthetic effect of modified pentothal sodium solution on Sprague-Dawley rats
- Genetic susceptibility to high myopia in Han Chinese population
- Potential biomarkers and molecular mechanisms in preeclampsia progression
- Silencing circular RNA-friend leukemia virus integration 1 restrained malignancy of CC cells and oxaliplatin resistance by disturbing dyskeratosis congenita 1
- Endostar plus pembrolizumab combined with a platinum-based dual chemotherapy regime for advanced pulmonary large-cell neuroendocrine carcinoma as a first-line treatment: A case report
- The significance of PAK4 in signaling and clinicopathology: A review
- Sorafenib inhibits ovarian cancer cell proliferation and mobility and induces radiosensitivity by targeting the tumor cell epithelial–mesenchymal transition
- Characterization of rabbit polyclonal antibody against camel recombinant nanobodies
- Active legumain promotes invasion and migration of neuroblastoma by regulating epithelial-mesenchymal transition
- Effect of cell receptors in the pathogenesis of osteoarthritis: Current insights
- MT-12 inhibits the proliferation of bladder cells in vitro and in vivo by enhancing autophagy through mitochondrial dysfunction
- Study of hsa_circRNA_000121 and hsa_circRNA_004183 in papillary thyroid microcarcinoma
- BuyangHuanwu Decoction attenuates cerebral vasospasm caused by subarachnoid hemorrhage in rats via PI3K/AKT/eNOS axis
- Effects of the interaction of Notch and TLR4 pathways on inflammation and heart function in septic heart
- Monosodium iodoacetate-induced subchondral bone microstructure and inflammatory changes in an animal model of osteoarthritis
- A rare presentation of type II Abernethy malformation and nephrotic syndrome: Case report and review
- Rapid death due to pulmonary epithelioid haemangioendothelioma in several weeks: A case report
- Hepatoprotective role of peroxisome proliferator-activated receptor-α in non-cancerous hepatic tissues following transcatheter arterial embolization
- Correlation between peripheral blood lymphocyte subpopulations and primary systemic lupus erythematosus
- A novel SLC8A1-ALK fusion in lung adenocarcinoma confers sensitivity to alectinib: A case report
- β-Hydroxybutyrate upregulates FGF21 expression through inhibition of histone deacetylases in hepatocytes
- Identification of metabolic genes for the prediction of prognosis and tumor microenvironment infiltration in early-stage non-small cell lung cancer
- BTBD10 inhibits glioma tumorigenesis by downregulating cyclin D1 and p-Akt
- Mucormycosis co-infection in COVID-19 patients: An update
- Metagenomic next-generation sequencing in diagnosing Pneumocystis jirovecii pneumonia: A case report
- Long non-coding RNA HOXB-AS1 is a prognostic marker and promotes hepatocellular carcinoma cells’ proliferation and invasion
- Preparation and evaluation of LA-PEG-SPION, a targeted MRI contrast agent for liver cancer
- Proteomic analysis of the liver regulating lipid metabolism in Chaohu ducks using two-dimensional electrophoresis
- Nasopharyngeal tuberculosis: A case report
- Characterization and evaluation of anti-Salmonella enteritidis activity of indigenous probiotic lactobacilli in mice
- Aberrant pulmonary immune response of obese mice to periodontal infection
- Bacteriospermia – A formidable player in male subfertility
- In silico and in vivo analysis of TIPE1 expression in diffuse large B cell lymphoma
- Effects of KCa channels on biological behavior of trophoblasts
- Interleukin-17A influences the vulnerability rather than the size of established atherosclerotic plaques in apolipoprotein E-deficient mice
- Multiple organ failure and death caused by Staphylococcus aureus hip infection: A case report
- Prognostic signature related to the immune environment of oral squamous cell carcinoma
- Primary and metastatic squamous cell carcinoma of the thyroid gland: Two case reports
- Neuroprotective effects of crocin and crocin-loaded niosomes against the paraquat-induced oxidative brain damage in rats
- Role of MMP-2 and CD147 in kidney fibrosis
- Geometric basis of action potential of skeletal muscle cells and neurons
- Babesia microti-induced fulminant sepsis in an immunocompromised host: A case report and the case-specific literature review
- Role of cerebellar cortex in associative learning and memory in guinea pigs
- Application of metagenomic next-generation sequencing technique for diagnosing a specific case of necrotizing meningoencephalitis caused by human herpesvirus 2
- Case report: Quadruple primary malignant neoplasms including esophageal, ureteral, and lung in an elderly male
- Long non-coding RNA NEAT1 promotes angiogenesis in hepatoma carcinoma via the miR-125a-5p/VEGF pathway
- Osteogenic differentiation of periodontal membrane stem cells in inflammatory environments
- Knockdown of SHMT2 enhances the sensitivity of gastric cancer cells to radiotherapy through the Wnt/β-catenin pathway
- Continuous renal replacement therapy combined with double filtration plasmapheresis in the treatment of severe lupus complicated by serious bacterial infections in children: A case report
- Simultaneous triple primary malignancies, including bladder cancer, lymphoma, and lung cancer, in an elderly male: A case report
- Preclinical immunogenicity assessment of a cell-based inactivated whole-virion H5N1 influenza vaccine
- One case of iodine-125 therapy – A new minimally invasive treatment of intrahepatic cholangiocarcinoma
- S1P promotes corneal trigeminal neuron differentiation and corneal nerve repair via upregulating nerve growth factor expression in a mouse model
- Early cancer detection by a targeted methylation assay of circulating tumor DNA in plasma
- Calcifying nanoparticles initiate the calcification process of mesenchymal stem cells in vitro through the activation of the TGF-β1/Smad signaling pathway and promote the decay of echinococcosis
- Evaluation of prognostic markers in patients infected with SARS-CoV-2
- N6-Methyladenosine-related alternative splicing events play a role in bladder cancer
- Characterization of the structural, oxidative, and immunological features of testis tissue from Zucker diabetic fatty rats
- Effects of glucose and osmotic pressure on the proliferation and cell cycle of human chorionic trophoblast cells
- Investigation of genotype diversity of 7,804 norovirus sequences in humans and animals of China
- Characteristics and karyotype analysis of a patient with turner syndrome complicated with multiple-site tumors: A case report
- Aggravated renal fibrosis is positively associated with the activation of HMGB1-TLR2/4 signaling in STZ-induced diabetic mice
- Distribution characteristics of SARS-CoV-2 IgM/IgG in false-positive results detected by chemiluminescent immunoassay
- SRPX2 attenuated oxygen–glucose deprivation and reperfusion-induced injury in cardiomyocytes via alleviating endoplasmic reticulum stress-induced apoptosis through targeting PI3K/Akt/mTOR axis
- Aquaporin-8 overexpression is involved in vascular structure and function changes in placentas of gestational diabetes mellitus patients
- Relationship between CRP gene polymorphisms and ischemic stroke risk: A systematic review and meta-analysis
- Effects of growth hormone on lipid metabolism and sexual development in pubertal obese male rats
- Cloning and identification of the CTLA-4IgV gene and functional application of vaccine in Xinjiang sheep
- Antitumor activity of RUNX3: Upregulation of E-cadherin and downregulation of the epithelial–mesenchymal transition in clear-cell renal cell carcinoma
- PHF8 promotes osteogenic differentiation of BMSCs in old rat with osteoporosis by regulating Wnt/β-catenin pathway
- A review of the current state of the computer-aided diagnosis (CAD) systems for breast cancer diagnosis
- Bilateral dacryoadenitis in adult-onset Still’s disease: A case report
- A novel association between Bmi-1 protein expression and the SUVmax obtained by 18F-FDG PET/CT in patients with gastric adenocarcinoma
- The role of erythrocytes and erythroid progenitor cells in tumors
- Relationship between platelet activation markers and spontaneous abortion: A meta-analysis
- Abnormal methylation caused by folic acid deficiency in neural tube defects
- Silencing TLR4 using an ultrasound-targeted microbubble destruction-based shRNA system reduces ischemia-induced seizures in hyperglycemic rats
- Plant Sciences
- Seasonal succession of bacterial communities in cultured Caulerpa lentillifera detected by high-throughput sequencing
- Cloning and prokaryotic expression of WRKY48 from Caragana intermedia
- Novel Brassica hybrids with different resistance to Leptosphaeria maculans reveal unbalanced rDNA signal patterns
- Application of exogenous auxin and gibberellin regulates the bolting of lettuce (Lactuca sativa L.)
- Phytoremediation of pollutants from wastewater: A concise review
- Genome-wide identification and characterization of NBS-encoding genes in the sweet potato wild ancestor Ipomoea trifida (H.B.K.)
- Alleviative effects of magnetic Fe3O4 nanoparticles on the physiological toxicity of 3-nitrophenol to rice (Oryza sativa L.) seedlings
- Selection and functional identification of Dof genes expressed in response to nitrogen in Populus simonii × Populus nigra
- Study on pecan seed germination influenced by seed endocarp
- Identification of active compounds in Ophiopogonis Radix from different geographical origins by UPLC-Q/TOF-MS combined with GC-MS approaches
- The entire chloroplast genome sequence of Asparagus cochinchinensis and genetic comparison to Asparagus species
- Genome-wide identification of MAPK family genes and their response to abiotic stresses in tea plant (Camellia sinensis)
- Selection and validation of reference genes for RT-qPCR analysis of different organs at various development stages in Caragana intermedia
- Cloning and expression analysis of SERK1 gene in Diospyros lotus
- Integrated metabolomic and transcriptomic profiling revealed coping mechanisms of the edible and medicinal homologous plant Plantago asiatica L. cadmium resistance
- A missense variant in NCF1 is associated with susceptibility to unexplained recurrent spontaneous abortion
- Assessment of drought tolerance indices in faba bean genotypes under different irrigation regimes
- The entire chloroplast genome sequence of Asparagus setaceus (Kunth) Jessop: Genome structure, gene composition, and phylogenetic analysis in Asparagaceae
- Food Science
- Dietary food additive monosodium glutamate with or without high-lipid diet induces spleen anomaly: A mechanistic approach on rat model
- Binge eating disorder during COVID-19
- Potential of honey against the onset of autoimmune diabetes and its associated nephropathy, pancreatitis, and retinopathy in type 1 diabetic animal model
- FTO gene expression in diet-induced obesity is downregulated by Solanum fruit supplementation
- Physical activity enhances fecal lactobacilli in rats chronically drinking sweetened cola beverage
- Supercritical CO2 extraction, chemical composition, and antioxidant effects of Coreopsis tinctoria Nutt. oleoresin
- Functional constituents of plant-based foods boost immunity against acute and chronic disorders
- Effect of selenium and methods of protein extraction on the proteomic profile of Saccharomyces yeast
- Microbial diversity of milk ghee in southern Gansu and its effect on the formation of ghee flavor compounds
- Ecology and Environmental Sciences
- Effects of heavy metals on bacterial community surrounding Bijiashan mining area located in northwest China
- Microorganism community composition analysis coupling with 15N tracer experiments reveals the nitrification rate and N2O emissions in low pH soils in Southern China
- Genetic diversity and population structure of Cinnamomum balansae Lecomte inferred by microsatellites
- Preliminary screening of microplastic contamination in different marine fish species of Taif market, Saudi Arabia
- Plant volatile organic compounds attractive to Lygus pratensis
- Effects of organic materials on soil bacterial community structure in long-term continuous cropping of tomato in greenhouse
- Effects of soil treated fungicide fluopimomide on tomato (Solanum lycopersicum L.) disease control and plant growth
- Prevalence of Yersinia pestis among rodents captured in a semi-arid tropical ecosystem of south-western Zimbabwe
- Effects of irrigation and nitrogen fertilization on mitigating salt-induced Na+ toxicity and sustaining sea rice growth
- Bioengineering and Biotechnology
- Poly-l-lysine-caused cell adhesion induces pyroptosis in THP-1 monocytes
- Development of alkaline phosphatase-scFv and its use for one-step enzyme-linked immunosorbent assay for His-tagged protein detection
- Development and validation of a predictive model for immune-related genes in patients with tongue squamous cell carcinoma
- Agriculture
- Effects of chemical-based fertilizer replacement with biochar-based fertilizer on albic soil nutrient content and maize yield
- Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions
- Agronomic and economic performance of mung bean (Vigna radiata L.) varieties in response to rates of blended NPS fertilizer in Kindo Koysha district, Southern Ethiopia
- Influence of furrow irrigation regime on the yield and water consumption indicators of winter wheat based on a multi-level fuzzy comprehensive evaluation
- Discovery of exercise-related genes and pathway analysis based on comparative genomes of Mongolian originated Abaga and Wushen horse
- Lessons from integrated seasonal forecast-crop modelling in Africa: A systematic review
- Evolution trend of soil fertility in tobacco-planting area of Chenzhou, Hunan Province, China
- Animal Sciences
- Morphological and molecular characterization of Tatera indica Hardwicke 1807 (Rodentia: Muridae) from Pothwar, Pakistan
- Research on meat quality of Qianhua Mutton Merino sheep and Small-tail Han sheep
- SI: A Scientific Memoir
- Suggestions on leading an academic research laboratory group
- My scientific genealogy and the Toronto ACDC Laboratory, 1988–2022
- Erratum
- Erratum to “Changes of immune cells in patients with hepatocellular carcinoma treated by radiofrequency ablation and hepatectomy, a pilot study”
- Erratum to “A two-microRNA signature predicts the progression of male thyroid cancer”
- Retraction
- Retraction of “Lidocaine has antitumor effect on hepatocellular carcinoma via the circ_DYNC1H1/miR-520a-3p/USP14 axis”

