Abstract
Increased concerns for sustainable agriculture have led to increased use of beneficial rhizobacteria as biofertilizers. Soil bacteria play a significant role in the nutrient cycle of soil, but their presence can be affected by abiotic stress, such as salinity. This study aimed to compare the chemical characteristics of slightly saline and non-saline rice soil and examine the bacterial community structure in both rhizosphere and bulk soil. We utilized 16SrRNA gene sequencing and performed arithmetic means clustering, a type of hierarchical clustering, on the samples collected from the rice fields of Cimrutu and Rawaapu Village in Cilacap Regency, Indonesia. Although the nutrient content was similar in both soils, there was a noticeable difference in their electrical conductivity (EC) despite the two locations being less than 4 km apart. The EC value in the Cimrutu soil suggests that it is non-saline, while the Rawaapu soil exhibits a low salinity level. The study found that Proteobacteria was the most prevalent phylum in saline rhizospheric soil. In contrast, Firmicutes was the most abundant group in saline bulk soil and non-saline rhizospheric and bulk soil. Additionally, Halothiobacillus, Thioalkalispira-Slvurivermis, and Acidothermus genera dominated the saline rhizospheric soil, suggesting that halotolerant microbes play a significant role as plant growth-promoting rhizobacteria in saline soil. The study provides valuable insights into cultured or uncultured bacterial populations and structure in saline and non-saline soil to develop future strategies related to salinity by introducing beneficial microbes.
1 Introduction
The richness of beneficial microbes in agricultural soil plays a crucial role in determining food crop productivity. The bacterial communities in soil dictate various pathways of the nutrient cycle, including nitrogen (N), phosphorous (P), potassium (K), and even micronutrients such as iron (Fe), zinc (Zn), and manganese (Mn). Moreover, certain soil bacteria play a significant role in providing phytohormones. The rhizosphere is a dynamic area around the plant roots which is the most active area of soil as it is rich in nutrients released by roots. Moreover, the community structure of microbes in the rhizosphere is more varied resulting in a higher microbial population than in bulk soil.
The composition of bacteria in soil is influenced by biotic and abiotic factors. Bacterial composition depends mostly on soil salinity which reduces the water availability rather then temperature and pH [1]. Some lowland rice farms in tropical areas located in coastal and tidal-influenced regions are the most affected by increased salinity. Rice farming in the coastal areas of the Cilacap district of Central Java, Indonesia, often experiences floods due to climate change. Moreover, the major challenge of growing rice in the tidal area is low productivity due to salinity; during the rainy season, the salinity will reduce and vice versa in the dry season. Due to inundation, the salinity of soil in Cilacap increased from 1.47 to 7.36 dS m−1 [2].
Farmers grow rice varieties that are not tolerant enough to submerge conditions. Frequent floods over a longer period cause abiotic stress which adversely affects the growth of rice plants. Rice fields in the coastal area of Cilacap Regency had low rice productivity of 3.78–4.97 t ha−1 [3]. Farmers rely on inorganic fertilizers to promote plant productivity, but integrated plant nutrition by using chemical, organic, and biofertilizer is recommended for maintaining soil health and saving the environment. The biofertilizer formulation required plant growth-promoting rhizobacteria to supply nutrition, mainly nitrogen and phosphor, and stimulate plant growth. In general, the bacteria for biofertilizer formulation are isolated from plant rhizosphere.
The diversity of bacterial communities in bulk soil is usually lower than that in the rhizosphere due to the richest nutrients from root exudates [4,5]. Based on the 16sRNA gene, the abundance and diversity of Bacillus, Pseudomonas, Staphylococcus, nitrogen-fixing bacteria, and denitrifying bacteria are reported in the rice rhizosphere [6,7]. The rhizobacteria Bacillus sp., Bulkholderia sp. Pseudomonas sp., Streptococcus sp., and Staphylococcus sp. in rice produce indole acetic acid [8,9], while Pseudomonas aeruginosa and a different strain of Bacillus subtilis provide available P for plant growth [10]. Moreover, indigenous Bacillus spp. control Rice blast (Pyricularia oryzae) diseases and increase rice yield [11]. For decades, the role of rhizospheric microbes to withstand abiotic stress conditions is reported [12]. Despite the abundance of data about the bacterial community in bulk soil in the rice field, the studies of bacterial groups in the rice rhizosphere grown in tropical slightly saline soil by 16SrRNA gene sequencing are limited.
The composition and community structure of bacteria in soil largely depend on organic matter and salinity. The effect of organic matter on the abundance of bacteria in soil has been reported for heterotrophic microbes [13,14]. High level of dissolved inorganic salts in the soil can increase the osmotic pressure inside the microbial cells and induce a rapid flow of cell water out of the cell along the osmotic gradient, resulting in decreased turgor and cytoplasmic dehydration [15]. Certain soil microbes accumulate the osmolytes as a mechanism to withstand the high salinity environment; the osmo-protectant produced by halotolerant microbes includes glycine, betaine, carnitine, and proline [16,17].
According to a meta-analysis of global studies, salinity, rather than pH, temperature, or other physicochemical environmental factors, is the most important environmental determinant of microbial community composition [1]. The reduced microbial diversity and richness due to saline conditions are reported elsewhere [18,19]. The objectives of this research were to compare the chemical characteristics of saline and non-saline rice soil, to study the biodiversity in saline and non-saline rice soil in Cilacap, Central Java, and to identify the bacterial community structure in the rice rhizosphere and bulk soil of both soils.
2 Materials and methods
2.1 Study area
The soil samples were collected from a rice field in Rawaapu and Cimrutu Village in Patimuan District, Cilacap Regency, Central Java, Indonesia (Figure 1). Patimuan District is located in the tropic with an equatorial climate. According to the Indonesian Central Bureau of Statistics [20], the average annual temperature in Patimuan was 32–36°C, the average relative humidity was 55–70%, and the rainfall distribution of 21–50 mm per month. Most of this area is lowland with an average slope of 4% and is located at an average altitude of 25–100 m above sea level; the study area has slightly wavy to hilly topography. The Patimuan area has a high clay Vertisol configuration; the soil expands during the rainy season and cracks in the dry season.
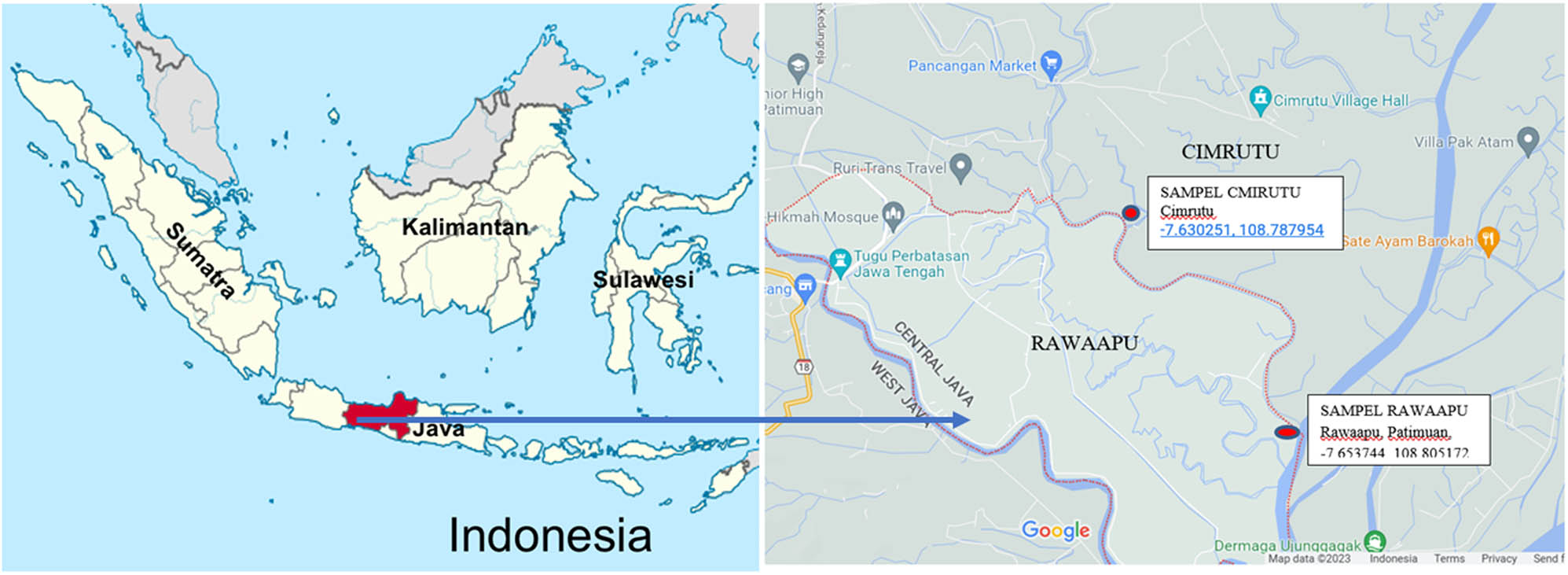
Location map of study sites in Cimrutu (A soil) and Rawaapu (B soil) of Cilacap Regency, Central Java, Indonesia. Source: Google map.
2.2 Site description and soil sampling
Soil samples were collected from two rice fields: non-saline (A) and saline (B) areas at Cimrutu and Rawaapu Village, respectively (Table 1). Both rice fields were grown with different rice varieties. The soil of Cimrutu was non-saline while that of Rawaapu was slightly saline.
Locations of sampling sites in Patimuan District, Cilacap Regency, Central Java Province
| Location code | Geographical coordinates | Descriptions |
|---|---|---|
| A | −7.630251, 108.787954 |
|
| B | −7.653744, 108.805172 |
|
The rhizosphere and bulk soil samples were collected by composite method from five sampling points of each location. Rhizospheric soil samples were collected from the remaining soil attached to the roots of rice plants using a clean brush. Soil samples taken from the five plant roots in each location were mixed well, put in sealed plastic bags, and stored in a cold box. Meanwhile, bulk soil samples were taken from the 20 cm-deep top soil around the roots of five rice plants in each location. Soil samples were put in a plastic bag and stored in a cold box. Double sampling was performed for either rhizosphere or bulk soil samples of both locations. Composite soil samples were transported in the cold box to the laboratory for analysis of physicochemical parameters and 5 g of each sample was stored at −20°C for metagenomic analysis.
2.3 Physico-chemical soil analysis
Already established procedures of soil analysis [21,22] were used to evaluate the physical and chemical properties of the soil. The pH of the soil was measured from the soil suspended in deionizer water of 1:2.5 soil–water ratio by using a glass electrode. The Walkley and Black wet oxidation techniques were used to detect organic carbon. The total N was calculated by using the Kjeldahl method. The total P2O5 and K2O contents were measured after soil extraction with using 25% HCl. Exchangeable bases such as K+, Ca2+, Na+, and Mg2+ were measured of soil extraction using 1 M NH4OAc, and their atomic absorption spectra were then used to measure the cationic concentration. The soil texture was determined by using the pipet approach [22]. The cation exchange capacity (CEC) and base saturation (BS) of soil were determined by calculation. The electrical conductivity (EC) of soil was measured by a conductivity meter on the soil–water suspension (1:5).
2.4 Molecular analysis of microbial community
Total soil DNA was extracted from a 0.25 g soil sample using a MoBio PowerSoil DNA Isolation Kit (Mobio Laboratory, Carlsbad, CA, USA) following the manufacturer’s instructions. DNA concentration and purity were monitored on 1% (w/v) agarose gels. According to the concentration, DNA was diluted to 1 ng/μL using sterile water. Bacterial DNA was amplified from each soil sample for Illumina sequencing analysis. The V4 region of the bacterial 16S rRNA gene was amplified using the forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [23].
Polymerase chain reactions (PCRs) were carried out using Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA). The PCR amplification program consisted of an initial heating to 98°C for 1 min, 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and extension at 72°C for 60 s, followed by a 5 min extension at 72°C. The PCR products were mixed with the same volume of 1× loading buffer (containing SYBR green) and were detected through electrophoresis on 2% agarose gel. A Qiagen Gel Extraction Kit (Qiagen, Hilden, DE) was used to purify PCR amplification products. Sequencing libraries were generated with NEBNext® UltraTM DNA Library Prep Kit for Illumina and quantified via Qubit and qPCR. The libraries were sequenced on an Illumina platform (Illumina, Inc., San Diego, CA, US), generating 250 bp paired-end reads, by the Novogene Bioinformatics Technology Company (Beijing, China).
2.5 Sequencing data processing
Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequences. The 250 bp paired-end reads were merged using Fast Length Adjustment of short reads of FLASH version 1.2.7 [24]. Quality filtering on the raw tags was performed under specific filtering conditions to obtain high-quality clean tags [25], according to the QIIME (version 1.7.0) quality-controlled process [26]. The tags were compared with the reference database (GOLD database) using the UCHIME algorithm to detect chimera sequences [27]. The chimera sequences were removed [28]. Subsequently, the effective tags were finally obtained.
Sequence analysis was performed by UPARSE software (Uparse version 7.0.1001) using all the effective tags [29]. Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs). The representative sequence for each OTU was screened for further annotation. For each representative sequence, Mothur software analysis was performed against the SSUrRNA database of SILVA Database available at http://www.arb-silva.de/ [30] for species annotation at each taxonomic rank (Threshold:0.8–1), including kingdom, phylum, class, order, family, genus, and species [31]. In order to obtain the phylogenetic relationship of all OTU representative sequences, MUSCLE (version 3.8.31) was used to compare multiple sequences rapidly [32]. The OTU abundance information was normalized using a standard sequence number corresponding to the sample with the least sequences. Subsequent analyses of alpha diversity and beta diversity were all performed based on these output normalized data.
The Alpha diversity (Observed-species, Chao1, Shannon, Simpson, ACE, and Good-coverage) was calculated with QIIME (version 1.7.0) and displayed with R software (version 2.15.3). Beta diversity was calculated by QIIME software (version 1.7.0). Unweighted pair-group method with arithmetic means (UPGMA) clustering was performed as a type of hierarchical clustering method to interpret the distance matrix using average linkage and was conducted by QIIME software (Version 1.7.0).
3 Results
3.1 Soil properties
The texture of location A and B soil was clay loam and clay, respectively (Table 2), which agrees with vertisol texture in general. The chemical properties of the soil samples from locations A and B were slightly different (Table 2). The pH of soil sample A was highly acidic, while the pH of soil sample B was approximately neutral. At both sites, organic-C and total-N were very high, but the C/N ratio was low, indicating an advanced decomposition. The available-P content in locations A and B was very low, only 1.44 and 2.37 mg kg−1, respectively, relating to a high level of P fixation, and so P became unavailable. Soil salinity at location A was low with an EC of only 0.21 dS m−1, indicating that it is non-saline soil. Meanwhile, the soil at location B has low salinity with an EC of 2.30 dS m−1.
Texture and chemical properties of soil samples taken from Cimrutu and Rawaapu Village
| Properties | Location A (Cimrutu) | Criteria1 | Location B (Rawaapu) | Criteria1 |
|---|---|---|---|---|
| Texture | ||||
| Sand (%) | 3.97 | Clay loam | 6.57 | Clay |
| Silt (%) | 36.08 | 38.32 | ||
| Clay (%) | 59.96 | 55.12 | ||
| pH (in H2O) | 4.99 | Acid | 6.65 | Neutral |
| C-organic (%) | 5.46 | Very high | 7.00 | Very high |
| Total N (%) | 0.99 | Very high | 0.99 | Very high |
| C/N ratio | 5.45 | Low | 7.07 | Low |
| P (mg kg−1) | 1.44 | Very low | 2.37 | Very low |
| K (cmol kg−1) | 2.34 | Very high | 2.03 | Very high |
| Caex (cmol kg−1) | 12.67 | High | 26.59 | Very high |
| Mgex (cmol kg−1) | 10.18 | High | 10.34 | High |
| Naex (cmol kg−1) | 2.54 | High | 9.11 | High |
| CEC3 (cmol kg−1) | 29.44 | High | 41.17 | Very high |
| Alex (cmol kg−1) | 0.87 | Low | 0.77 | Low |
| Hex (cmol kg−1) | 0.30 | Low | 0.25 | Low |
| BS4 (%) | 95.89 | Very high | 97.93 | Very high |
| EC5 (dS m−1) | 0.21 | Non-saline2 | 2.30 | Low salinity2 |
3.2 Bacterial diversity and composition
A total of 429,729 high-quality sequences were obtained after quality filtering. The high-quality sequences in all samples ranged between 96,488 and 118,592 sequences with an average length of 420 bp. Table 3 shows the alpha diversity of the bacterial community in the bulk soil and rhizospheric soil of rice plants. Good coverage (Table 3) and rarefaction curves (Figure 2) indicated that the DNA libraries were sufficient to estimate most bacterial diversity in the samples. The results showed that non-saline soil (AR and AB) had more diverse bacteria than low-saline soil (BR and BB) according to observed species, Shannon and Simpson indices.
Alpha diversity of the bulk soil and rhizospheric soil of rice plants grown in flooding areas
| Sample name | Observed species | Shannon | Simpson | Chao1 | ACE | Good’s coverage | PD whole tree |
|---|---|---|---|---|---|---|---|
| AR | 2,383 ± 102 | 7.81 ± 0.63 | 0.96 ± 0.03 | 2,541 ± 85 | 2,570 ± 77 | 0.996 | 162.46 ± 6.24 |
| AB | 1,902 ± 193 | 8.01 ± 0.47 | 0.99 ± 0.00 | 1,931 ± 183 | 1,968 ± 197 | 0.998 | 186.05 ± 1.87 |
| BR | 1,842 ± 82 | 6.06 ± 1.29 | 0.85 ± 0.13 | 2,025 ± 78 | 2,051 ± 76 | 0.996 | 133.59 ± 6.60 |
| BB | 1,441 ± 240 | 3.85 ± 1.12 | 0.64 ± 0.14 | 1,635 ± 199 | 1,682 ± 204 | 0.996 | 115.53 ± 15.76 |
Data were calculated at 3% genetic distance level with standard deviation based on the same number of sequences from each replicate in Mothur.
Values are the mean values of two replicates ± SE. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
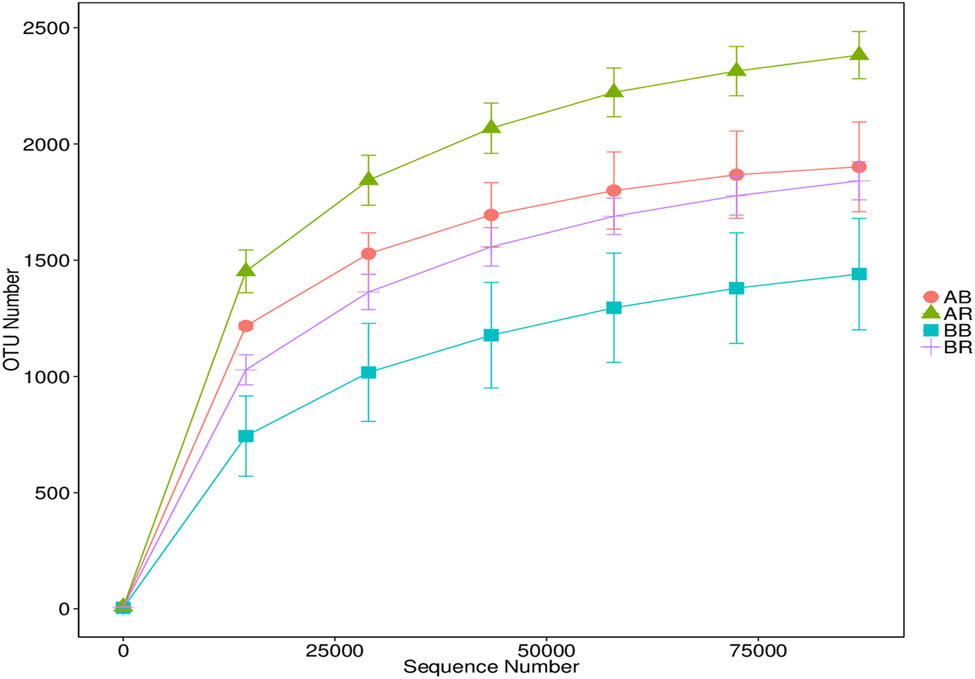
Rarefaction curves based on the V4 region of the 16S rRNA gene obtained from bulk soil and rhizospheric rice soil samples. Error bars represent the standard error of two replicates. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
The Venn diagram shows that a total of 4,075 OTUs were successfully detected across all samples, in which 1,305 OTUs were common to all samples. The shared OTUs mainly belonged to Firmicutes (266 OTUs), Proteobacteria (258 OTUs), Chloroflexi (135 OTUs), and Actinobacteria (98 OTUs) at the phylum level (Table 4). The AB sample had more specific OTUs (688 OTUs) in comparison with other samples (Figure 3).
Top ten bacterial phyla shared among the samples
| Bacterial phylum | OTU number |
|---|---|
| Firmicutes | 266 |
| Proteobacteria | 258 |
| Chloroflexi | 135 |
| Actinobacteria | 98 |
| Patescibacteria | 66 |
| Bacteroidetes | 58 |
| Myxomycota | 49 |
| Acidobacteria | 41 |
| Cyanobacteria | 15 |
| Others | 302 |
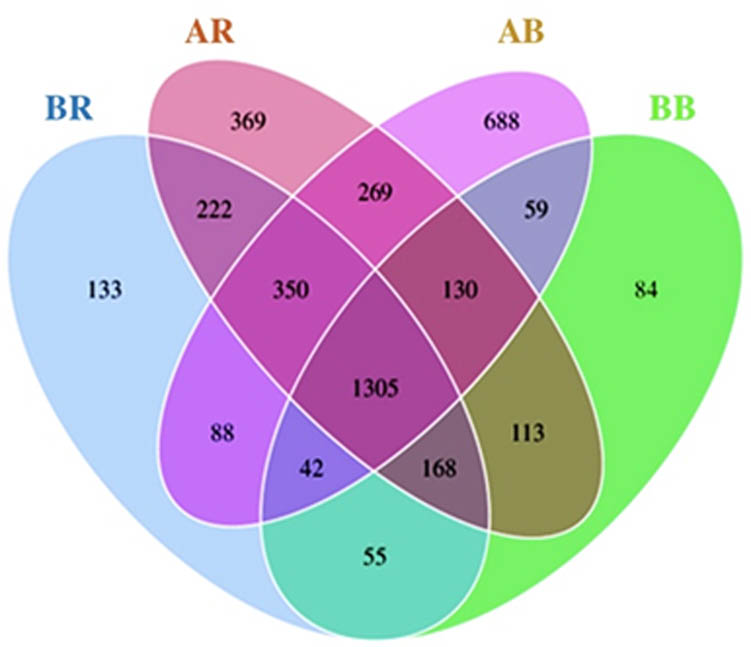
Venn diagram of bacterial OTUs shared among different samples. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
The diversity of bacterial communities in different samples was revealed by Illumina-based high-throughput sequencing at different taxonomic levels. The top ten bacterial phyla with each relative abundance >1% are presented in Figure 4. Proteobacteria was the most abundant phylum in BR (54.07%), followed by Actinobacteria (14.38%) and Firmicutes (13.68%). On the other hand, Firmicutes became the most dominant phylum in BB, AR, and AB, accounting for 48.53, 32.73, and 29.39%, respectively. Proteobacteria (BB: 37.91%, AR: 27.93%, and AB: 20.43%) and Actinobacteria (BB: 4.63%, AR: 9.72%, and AB: 16.45%) were the second and third most abundant phyla in these three samples. We observed Acidobacteriota (0.9%) in BB, which were less abundant compared to other samples (BR: 2.46%, AR: 6.97%, and AB: 3.95%). In addition, Halobacterota was observed as less dominant in BB (0.0075%) than in other samples (BR: 0.12%, AR: 0.37%, and AB: 1.51%).
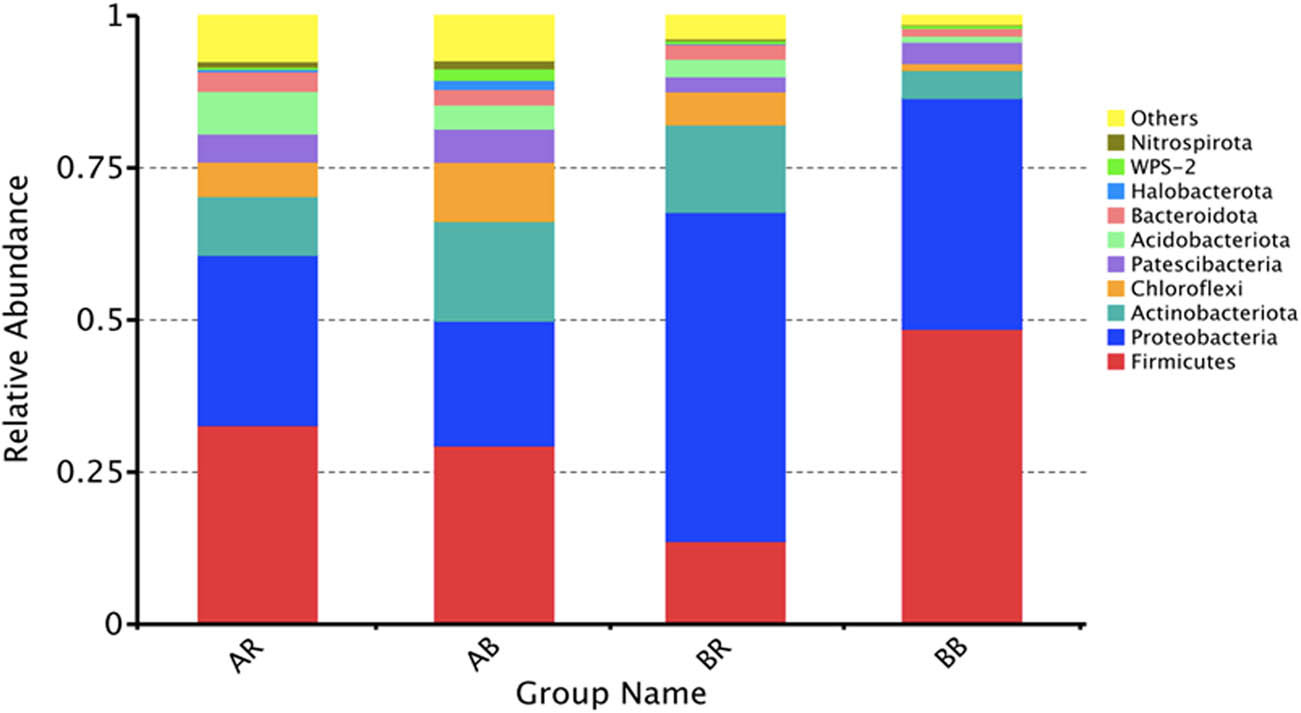
The relative abundances of bacteria at the phylum level in different samples. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
Figure 5 shows the relative abundance of bacterial order in each sample. At the order level of taxonomy, Bacillales had the most abundance in AR (19.67%) and BB (38.69%), followed by Burkholderiales as the second most prevalent group in AR (10.92%) and BB (31.96%). Meanwhile, Burkholderiales was the most abundant group in AB (8.29%) and BR (35.74%). Bacillales was the second most dominant group in AB (7.20%). In contrast, Frankiales was observed as the second most common order in BR, accounting for 7.74%.
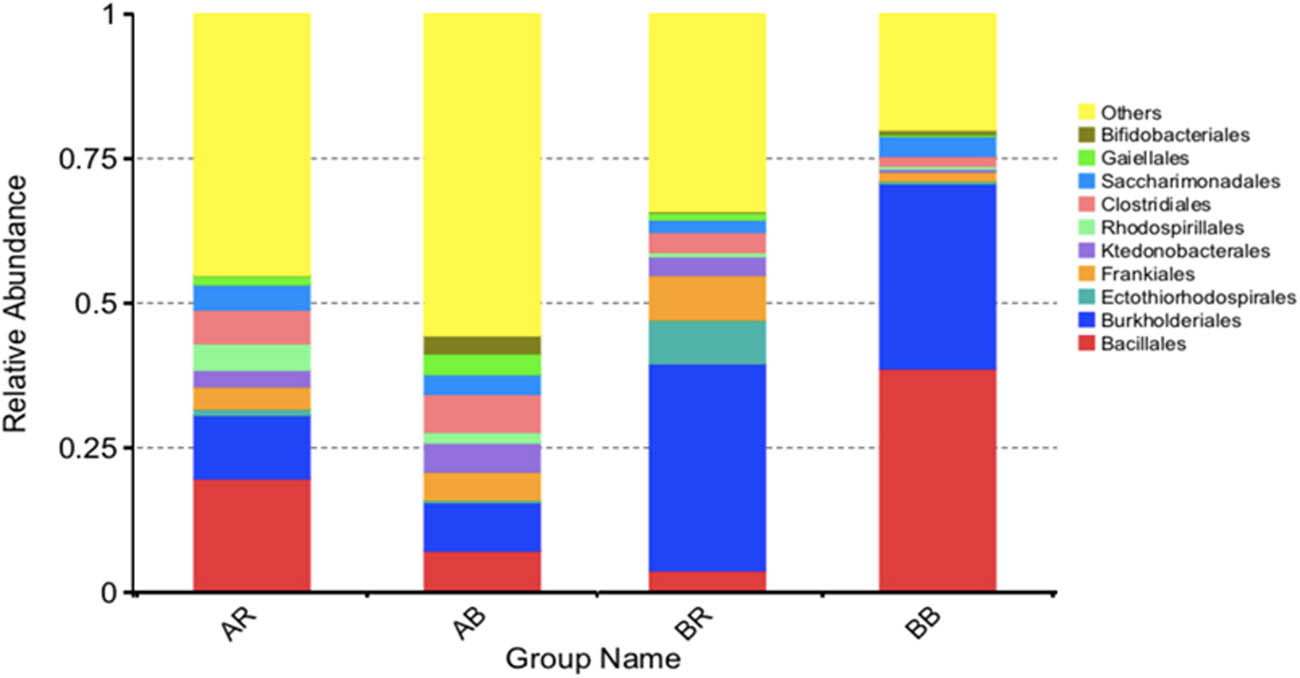
The relative abundances of bacteria at the order level in different samples. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
The most abundant 35 bacterial genera were clustered as shown in Figure 6. Bifidobacterium (3.12%), Clostridioides (1.15%), Paenibacillus (1.03%), WPS-2 (1.86%), Candidatus_Levybacteria (1.84%), Streptomyces (0.76%), Enterobacter (0.86%), and Clostridium_sensu_stricto_1 (2.53%) were predominantly distributed in the AB. Meanwhile, AR had more species belong to Sideroxydans (2.51%), RCP2-54 (1.30%), Lysinibacillus (12.70%), Thiobacillus (0.99%), Magnetospirillaceae (2.85%), TM7a (1.11%), Streptococcus (1.13%), Subgroup_2 (2.25%), and Subgroup_13 (0.84%). Staphylococcus (9.11%), Bacillus (37.19%), and Veillonella (8.26%) were mainly distributed in the BB, while Thioalkalispira-Sulfurivermis (7.58%), Halothiobacillus (2.27%), and Acidothermus (7.03%) were the most dominant group in the BR.
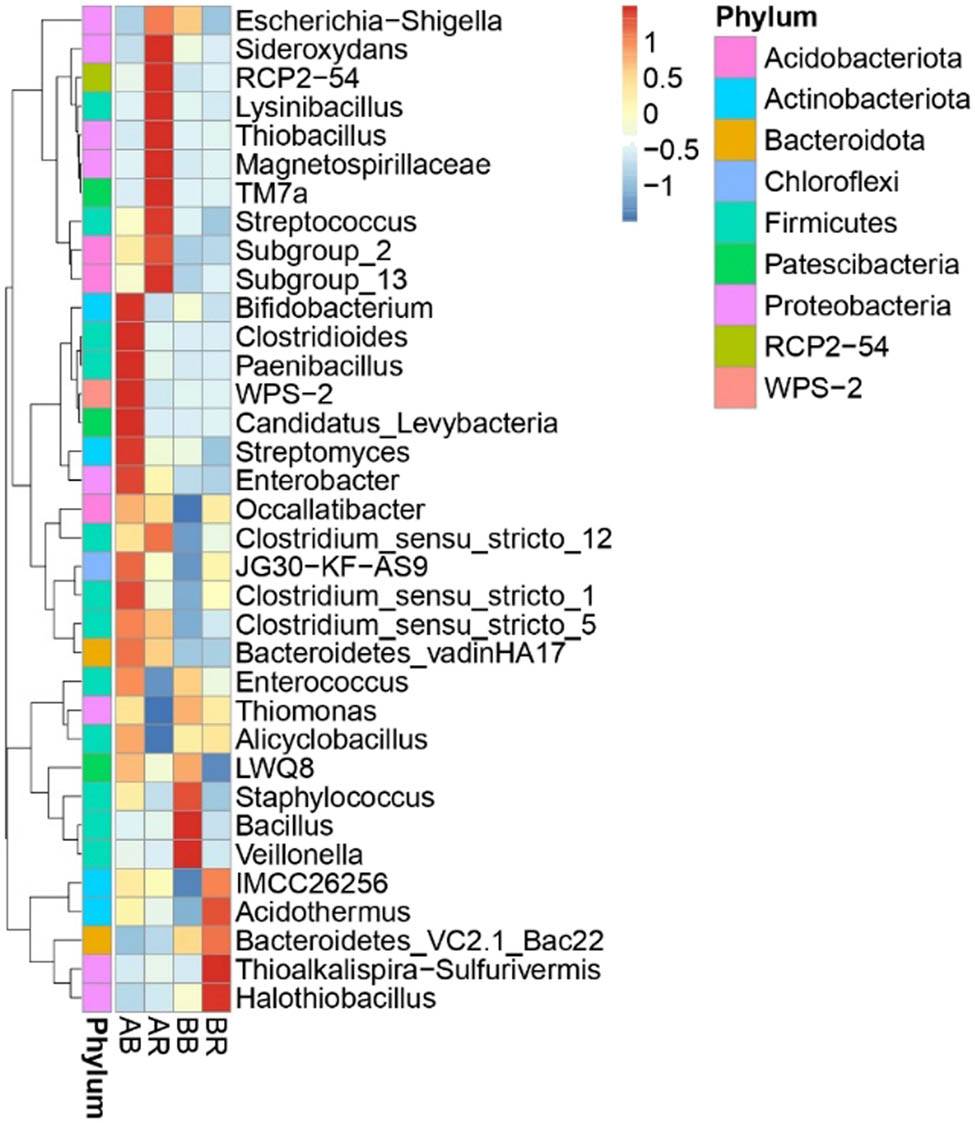
Heatmap showing the relative abundance of the most abundant bacterial genera in different samples. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
The results showed two distinct clusters at the phylum level in the UPGMA tree, i.e., cluster 1 (AR and AB, non-saline soil) and cluster 2 (BR and BB, slightly-saline soil), as shown in Figure 7. This finding revealed that the bacterial community in AR was more similar to those of the AB. In addition, BB and BR had more similar bacterial diversity and composition. It is observed that the clustering was based on soil salinity (non-saline and saline soil) rather than soil type (rhizospheric and bulk soil).
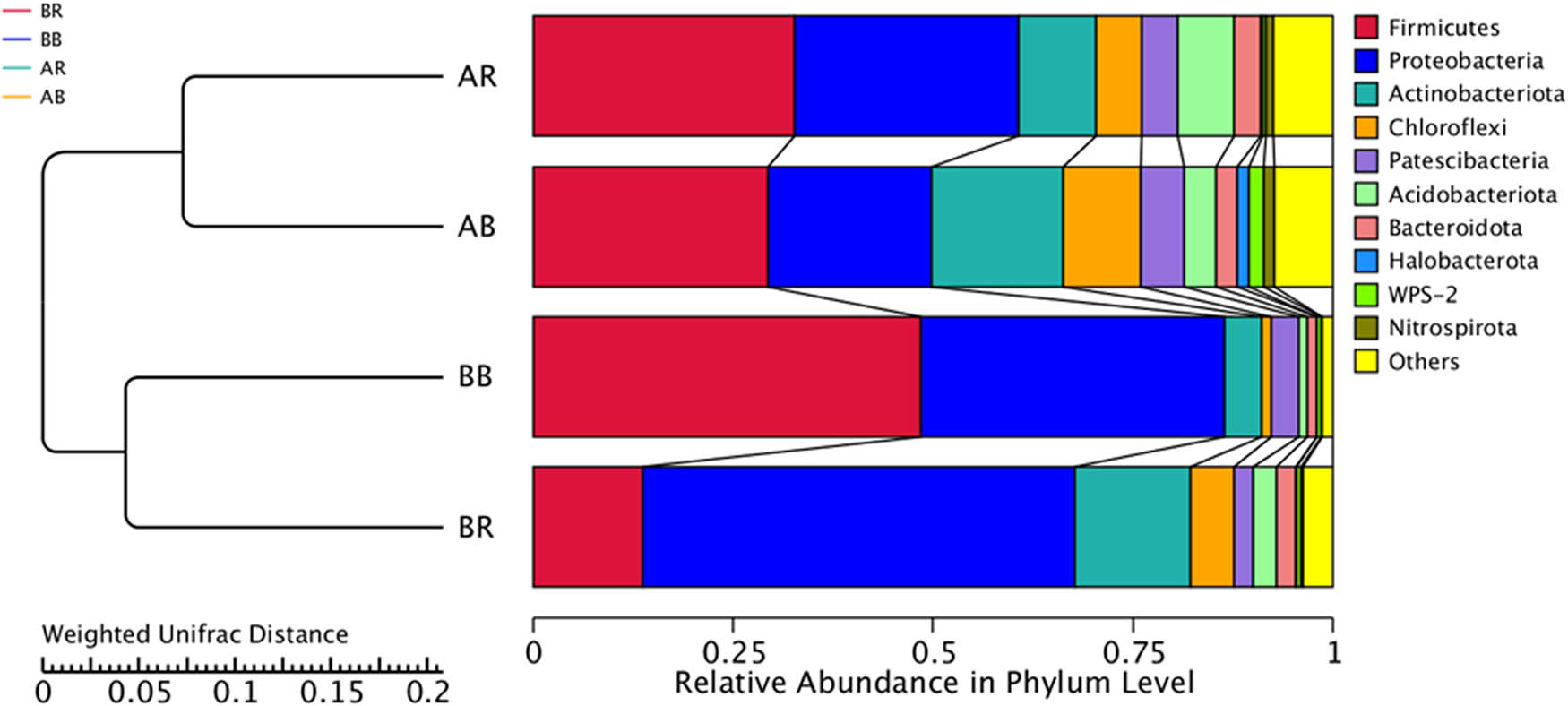
UPGMA clustering tree of the bacterial community structure based on Weighted UniFrac Distance at the phylum level. AR – non-saline rhizospheric soil; AB – non-saline bulk soil; BR – saline rhizospheric soil; BB – saline bulk soil.
4 Discussion
The impact of salinity on bacterial diversity in rice field can be complex and depends on several factors, including the severity and duration of salinity, the specific bacterial taxa present, and soil environmental conditions. This complexity was also observed in a study conducted in Rawaapu and Cimrutu Village, Cilacap Regency, where the impact of salinity on the bacterial community in rice fields was investigated.
In general, the nutrient composition of the two soils is quite similar. Both soils exhibit a significantly high level of organic carbon, total nitrogen, and potassium but a markedly low level of phosphorus content. However, there was a noticeable difference in the EC of the soils in Cimrutu and Rawaapu, despite the locations being less than 4 km apart. The EC in Cimrutu soil is inadequate, indicating that the soil is non-saline, whereas the Rawaapu soil has a low level of salinity according to the salinity classification proposed by food and agriculture organization (FAO) [34]. The cause of the difference in salinity between Cimrutu and Rawaapu is unclear. There are several possible causes for the difference in salinity between the two adjacent locations, including differences in water source, irrigation practices, and drainage system.
The non-saline soil exhibited an acidic reaction, whereas the low-salinity soil had a neutral pH. Furthermore, the sodium content in the Rawaapu soil was higher than that in the non-saline Cimrutu soil, which is consistent with the previous findings [35], which reported an increase in soil pH and exchangeable Na in saline soils. In addition, the neutral pH in the Rawaapu soil may be attributed to a higher ex-Ca concentration compared to the Cimrutu soil. This is in line with the observation that soils with a high Ca concentration tend to have a higher pH [36].
The high nitrogen content in both soils can be attributed to the frequent use of urea and NPK chemical fertilizers in rice cultivation. Such high nutrient levels can have an impact on the abundance and diversity of bacteria in the rhizosphere. Specifically, excessive nitrogen levels can inhibit the activity of N2-fixing bacteria, while low levels of available phosphorus can stimulate the growth of phosphate-solubilizing microbes [37]. Additionally, the high level of organic carbon in the soil supports the growth of heterotrophic bacterial communities that utilize organic carbon for their oxidative metabolism [38].
The results of this study indicated that the EC values of A and B soils were 0.21 and 2.30 dS m−1, respectively (Table 2), which fell within the tolerable salinity range of less than 4 dS m−1 recommended by FAO [34]. Rice is sensitive to the high salt content in the soil: more than 50% loss was recorded in soil with EC up to 6 dS m−1 [39]. Outside of this range, rice yield may be significantly reduced, and the plants may exhibit symptoms of salt stress, such as stunted growth, reduced tillering, and decreased photosynthetic activity.
The findings of this study indicated that the diversity of bacteria in slightly saline soils in Rawaapu (BR and BB) was lower than that in non-saline soils in Cimrutu (AR and AB), as evidenced by the Observed species, Shannon, and Simpson indices (Table 3). Furthermore, non-saline soils (AR and AB) exhibited a greater number of specific OTUs than slightly saline soils (BR and BB) depicted in Figure 3. These results are in agreement with findings that salinity is a significant factor influencing the community structure of soil microbes [1,40]. Saline conditions generally reduce microbial diversity and richness. Elevated salt levels can alter microbial physiology and activity directly by modifying microbial communities or indirectly via changes in pH and nutrient availability [18]. High levels of ambient salt can cause osmotic stress in microbial cells, thereby reducing their physiological fitness [41]. Microbes that fail to adjust their intracellular osmolarity will eventually perish or become inactive. As a result, salinization may alter the microbial community directly by favoring halophilic or halotolerant microbes [40].
The diversity of bacteria in soil is influenced by several factors, including soil characteristics, environmental conditions, and plant–microbe interactions [42]. When it comes to slightly saline soil, the presence of salt can limit bacterial diversity. As a result, the diversity of bacteria in slightly saline soil is typically less than in non-saline soil. Salt can alter the physical and chemical properties of soil, reducing water availability, which in turn limits the growth and survival of some bacterial species. Additionally, salt can change the soil’s ionic composition, impacting nutrient availability and altering microbial communities. This can create a selective pressure in favor of salt-tolerant bacteria, which can outcompete other bacterial species and reduce bacterial diversity.
Our results revealed an enriched population of salt-tolerant species, indicating that microbial communities were abundant and diverse even in high-salinity soils. Non-saline bulk soil Cimrutu (AB) is dominated by Bifidobacterium, Clostridioides, Paenibacillus, WPS-2, Candidatus_Levybacteria, Streptomyces, Enterobacteria, and Clostridium_sensu_stricto_1 (Figure 6). On the other hand, non-saline rhizospheric soil Cimrutu (AR) had more bacterial species belonging to Sideroxydans, RCP2-54, Lysinibacillus, Thiobacillus, Magnetospirillaceae, TM7a Streptococcus, Subgroup_2, and Subgroup_23 (Figure 6), which was identified as having a function as a biofertilizer. Sideroxydans are microaerobic, neutrophilic iron-oxidizing Beta-proteobacteria in rice soils [43]. Lysinibacillus are naturally colonizing rice roots and can potentially be highly resistant biofertilizers due to their spore production [44]; moreover, Thiobacillus is a nitrate-dependent neutrophilic Fe-oxidizer in rice soils. The members of the family Magnetospirillaceae, Magnetospirillum, are also found as diazotrophs in rice roots [45,46]. The diversity of bacteria that can serve as biofertilizers in high-salinity rice fields is extensive, and different bacterial species can offer various advantages to rice plants. The saline rice soil provides a valuable opportunity to explore and discover new beneficial indigenous microbes that have a main function as biofertilizers and enable to withstand high-salinity stress, offering potential benefits to rice cultivation.
In our study, rice varieties under different salinity affected the abundance and diversity of bacteria around the roots. Rice plants release exudates rich in organic acids, such as malate and citrate, which are known to chelate and solubilize toxic ions in the rhizosphere under salinity stress [47]. Some rice varieties may produce specific exudates in response to salinity stress, which can modify the rhizosphere’s physicochemical properties, such as pH and ion concentrations [48]. Salinity stress affects the composition and quantity of exudates released by rice plants, which, in turn, influences the selection of specific microbial communities in the rhizosphere [47].
The difference in bacterial community structure in the rhizosphere among the two locations might also be due to the root exudate differences between the rice cultivar Mendawak and Ciherang. Plant genotype may influence root exudation, and hence the rhizosphere microbiota. Root exudate composition depends on the expression level of specific genes [49]. The rice genotype is one of the factors affecting the microbial communities in the endosphere, rhizoplane, and rhizosphere [50]. Rice variety significantly altered the active fungal and Gram-negative bacterial groups [51]. Moreover, the rice plant with the seedling salt-tolerant gene under salt stress influence the rhizosphere bacterial community, proving that the key genes of plants play a significant role in shaping the rhizosphere microbiome [52].
5 Conclusions
The soil nutrient content was similar in both Cimrutu (location A) and Rawaapu (location B) rice fields, but the soil reaction in Cimurutu was acidic while the pH in Rawaapu was near neutral. There was a noticeable difference in their EC despite the two locations being less than 4 km apart. The EC value in the Cimrutu soil suggests that it is non-saline, while the Rawaapu soil exhibits a low level of salinity. The soil in both locations had high C-organic and total-N but a low ratio of C to N. The soils were very low in P and very high in K; the CEC and BS of soils were high. Those soil properties indicated that the soils were fertile.
The predominant phylum in saline rhizospheric soil Rawaapu (BR) was observed to be Proteobacteria, while Firmicutes was the most abundant group in non-saline rhizospheric soil Cimrutu (AR) and non-saline bulk soil Cimrutu (AB) as well as saline bulk soil Rawaapu (BB). Halothiobacillus, Thioalkalispira-Slvurivermis, and Acidothermus were the predominant genera in saline rhizospheric soil Rawaapu (BR), indicating the significant roles played by halotolerant microbes as plant growth-promoting rhizobacteria in saline soil. The current study showed that the salinity instead of chemical fertility was the main driving force for the different bacterial community diversity in rice field. The difference was possibly related to the root exudates in the rhizosphere of rice grown in saline and non-saline soil, as well as metabolic characteristics of predominant species. The study provided an important understanding of cultured or uncultured bacterial populations and structure in saline and non-saline soil to develop future strategies related to salinity by introducing beneficial microbes. Further, a more comprehensive study to determine the role and contribution of saline-resistance microbes found in this study is needed for better plant nutrient management in rice fields near the coastal area.
Acknowledgments
This research was funded by the Academic Leadership Grant of Universitas Padjadjaran year 2021.
-
Funding information: This study was funded by Universitas Padjadjaran, Indonesia.
-
Author contributions: RH and EP composed the research proposal and methods; RH and TS performed the sample collection and soil analysis; EP and YSF carried out the metagenomic analysis and its data analysis; RH, EP, and YSF wrote the manuscript. The authors have reviewed the manuscript and agreed to submit the manuscript.
-
Conflict of interest: The authors state no conflict of interest.
-
Data availability statement: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
[1] Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104(27):11436–40. 10.1073/pnas.0611525104.Suche in Google Scholar PubMed PubMed Central
[2] Rose ME, Huerbin MB, Melick J, Marion DW, Palmer AM, Schiding JK, et al. Regulation of interstitial excitatory amino acid concentrations after cortical contusion injury. Brain Res. 2002;935(1–2):40–6. 10.1016/S0006-8993(02)02445-9Suche in Google Scholar PubMed
[3] Simarmata T, Prayoga MK, Setiawati MR, Adinata K, Stöber S. Improving the climate resilience of rice farming in flood-prone areas through Azolla biofertilizer and saline-tolerant varieties. Sustainability. 2021;13(21):12308. 10.3390/su132112308.Suche in Google Scholar
[4] Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, et al. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: Plant-dependent enrichment and seasonal shifts revealed. Appl Env Microbiol. 2001;67(10):4742–51. 10.1128/AEM.67.10.4742-4751.2001.Suche in Google Scholar PubMed PubMed Central
[5] Cui H, Yang X, Lu D, Jin H, Yan Z, Chen J, et al. Isolation and characterization of bacteria from the rhizosphere and bulk soil of Stellera chamaejasme L. Can J Microbiol. 2015;61(3):171–81. 10.1139/cjm-2014-0543.Suche in Google Scholar PubMed
[6] Shu W, Pablo GP, Jun Y, Danfeng H. Abundance and diversity of nitrogen-fixing bacteria in rhizosphere and bulk paddy soil under different duration of organic management. World J Microbiol Biotechnol. 2012l;2(2):493–503. 10.1007/s11274-011-0840-1.Suche in Google Scholar PubMed
[7] Wang N, Zhao Y-H, Yu J-G, Xue L-H, Li H-B, Yang L-Z. Roles of bulk and rhizosphere denitrifying bacteria in denitrification from paddy soils under straw return condition. J Soils Sediment. 2021;21:2179–91. 10.1007/s11368-021-02942-x.Suche in Google Scholar
[8] Wu Y, Sun J, Yu P, Zhang W, Lin Y, Ma D. The rhizosphere bacterial community contributes to the nutritional competitive advantage of weedy rice over cultivated rice in paddy soil. BMC Microbiol. 2022;22:232. 10.1186/s12866-022-02648-1.Suche in Google Scholar PubMed PubMed Central
[9] Purwanto P, Agustono T, Widjonarko BR, Widiatmoko T. Indole acetic acid production of indigenous plant growth promotion rhizobacteria from paddy soil. Planta Tropika: J Agrosains (J Agro Sci). 2019;7(1):1–7. 10.18196/pt.2019.087.1-7.Suche in Google Scholar
[10] Gupta R, Kumari A, Sharma S, Alzahrani OM, Noureldeen A, Darwish H. Identification, characterization and optimization of phosphate solubilizing rhizobacteria (PSRB) from rice rhizosphere. Saudi J Biol Sci. 2022;29(1):35–42. 10.1016/j.sjbs.2021.09.075.Suche in Google Scholar PubMed PubMed Central
[11] Rais A, Shakeel M, Malik K, Hafeez FY, Yasmin H, Mumtaz S, et al. Antagonistic Bacillus spp. reduce blast incidence on rice and increase grain yield under field conditions. Microbiol Res. 2018;208:54–62. 10.1016/j.micres.2018.01.009.Suche in Google Scholar PubMed
[12] Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009;14(1):1–4. 10.1016/j.tplants.2008.10.004.Suche in Google Scholar PubMed
[13] Smith AP, Marín-Spiott E, de Graaff MA, Balser TC. Microbial community structure varies across soil organic matter aggregate pools during tropical land cover change. Soil Biol Biochem. 2014;77:292–303. 10.1016/j.soilbio.2014.05.030.Suche in Google Scholar
[14] Whitman T, Pepe-Ranney C, Enders A, Koechli C, Campbell A, Buckley DH, et al. Dynamics of microbial community composition and soil organic carbon mineralization in soil following addition of pyrogenic and fresh organic matter. ISME J. 2016;10(12):2918–30. 10.1038/ismej.2016.68.Suche in Google Scholar PubMed PubMed Central
[15] Wood JM. Bacterial responses to osmotic challenges. J Gen Physiol. 2015;145(5):381–8. 10.1085/jgp.201411296.Suche in Google Scholar PubMed PubMed Central
[16] Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 2002;26(1):49–71. 10.1111/j.1574-6976.2002.tb00598.x.Suche in Google Scholar PubMed
[17] Takagi H. Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol. 2008;81(2):211–23. 10.1007/s00253-008-1698-5.Suche in Google Scholar PubMed
[18] Dang C, Morrissey EM, Neubaue SC, Franklin RB. Novel microbial community composition and carbon biogeochemistry emerge over time following saltwater intrusion in wetlands. Glob Change Biol. 2019;25(2):549–61. 10.1111/gcb.14486.Suche in Google Scholar PubMed
[19] Chen H, Ma K, Huang Y, Fu Q, Qiu Y, Yao Z. Significant response of microbial community to increased salinity across wetland ecosystems. Geoderma. 2022;415:115778. 10.1016/j.geoderma.2022.115778.Suche in Google Scholar
[20] Badan Pusat Statistik. Statistik Kabupaten Cilacap (Statistics of Cilacap Regency); 2020. https://cilacapkab.bps.go.id/.Suche in Google Scholar
[21] Van Reuuwijk LP. Procedures for soil analysis. Wageningen, The Netherlands: International Soil References and Information Centre; 2002.Suche in Google Scholar
[22] AOAC. Official methods of analysis. 19th edn. Arlington, VA: Association of Official Analytical Chemists; 2012.Suche in Google Scholar
[23] Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–22. 10.1073/pnas.1000080107.Suche in Google Scholar PubMed PubMed Central
[24] Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. 10.1093/bioinformatics/btr507.Suche in Google Scholar PubMed PubMed Central
[25] Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from ollumina amplicon sequencing. Nat Methods. 2013;10(1):57–9. 10.1038/nmeth.2276.Suche in Google Scholar PubMed PubMed Central
[26] Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. 10.1038/nmeth.f.303.Suche in Google Scholar PubMed PubMed Central
[27] Edgar RC, Haas BI, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. 10.1093/bioinformatics/btr381.Suche in Google Scholar PubMed PubMed Central
[28] Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. 10.1101/gr.112730.110.Suche in Google Scholar PubMed PubMed Central
[29] Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. 10.1038/nmeth.2604.Suche in Google Scholar PubMed
[30] Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Env Microbiol. 2007;73(16):5261–7. 10.1128/AEM.00062-07.Suche in Google Scholar PubMed PubMed Central
[31] Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl Acids Res. 2013;41:D590–6. 10.1093/nar/gks1219.Suche in Google Scholar PubMed PubMed Central
[32] Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. 10.1093/nar/gkh340.Suche in Google Scholar PubMed PubMed Central
[33] Eviati S. Analisis Kimia tanah, tanaman, air dan pupuk (Chemical analysis of soil, plant, water, and fertilizer) In Indonesian. Bogor, Indonesia: Balai Penelitian Tanah (Indonesia Soil Research Institute); 2009.Suche in Google Scholar
[34] FAO. Saline soils and their management. Rome: FAO; 2005. https://www.fao.org/3/x5871e/x5871e04.htm.Suche in Google Scholar
[35] Zhang WW, Wang C, Xue R, Wang LJ. Effects of salinity on the soil microbial community and soil fertility. J Integr Agric. 2019;18(6):1360–8. 10.1016/S2095-3119(18)62077-5.Suche in Google Scholar
[36] Chong IQ, Azman EA, Ng JF, Ismail R, Awang A, Hasbullah NA, et al. Improving selected chemical properties of a paddy soil in Sabah amended with calcium silicate: A Laboratory incubation study. Sustainability. 2022;14:13214. 10.3390/su142013214.Suche in Google Scholar
[37] Long XE, Yao H, Huang Y, Wei W, Zhu YG. Phosphate levels influence the utilisation of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Biol Biochem. 2018;118:103–14. 10.1016/j.soilbio.2017.12.014.Suche in Google Scholar
[38] Six J, Frey SD, Thiet RK, Batten KM. Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci Soc Am J. 2006;70(2):555–69. 10.2136/sssaj2004.0347.Suche in Google Scholar
[39] Al-Tamimi N, Oakey H, Tester M, Negrão S. Assessing rice salinity tolerance: From phenomics to association mapping. In: Bandyopadhyay A, Thilmony R, editors. Rice genome engineering and gene editing. New York: Humana Press; vol. 2238, 2021. p. 339–75. 10.1007/978-1-0716-1068-8_23.Suche in Google Scholar PubMed
[40] Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4(1):2. 10.1186/1746-1448-4-2.Suche in Google Scholar PubMed PubMed Central
[41] Galinski EA, Trüper HG. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol. Rev. 1994;15(2–3):95–108. https://www.sciencedirect.com/science/article/abs/pii/0168644594901066.10.1111/j.1574-6976.1994.tb00128.xSuche in Google Scholar
[42] Wu Z, Liu Q, Li Z, Li Z, Cheng W, Sun J, et al. Environmental factors shaping the diversity of bacterial communities that promote rice production. BMC Microbiol. 2018;18(1):51. 10.1186/s12866-018-1174-z.Suche in Google Scholar PubMed PubMed Central
[43] Watanabe T, Sumida H, Nhut Minh DO, Yano K, Asakawa S, Kimura M. Bacterial consortia in iron-deposited colonies formed on paddy soil surface under microaerobic conditions. Soil Sci Plant Nutr. 2013;59(3):337–46. 10.1080/00380768.2013.791807.Suche in Google Scholar
[44] Ramírez AMD, Agake S, Maeda M, Kojima K, Ohkama-Ohtsu N, Yokoyama T. Diversity of fast-growth spore-forming microbes and their activity as plant partners. Microorganisms. 2023;11(2):232. 10.3390/microorganisms11020232.Suche in Google Scholar PubMed PubMed Central
[45] Das S, Jeong ST, Das S, Kim PJ. Composted cattle manure increases microbial activity and soil fertility more than composted swine manure in a submerged rice paddy. Front Microbiol. 2017;8:1702. 10.3389/fmicb.2017.01702.Suche in Google Scholar PubMed PubMed Central
[46] Meyer-Cifuentes I, Martinez-Lavanchy PM, Marin-Cevada V, Böhnke S, Harms H, Műller JA, et al. Isolation and characterization of Magnetospirillum sp. strain 15-1 as a representative anaerobic toluene-degrader from a constructed wetland model. PLoS One. 2017;12(4):e0174750. 10.1371/journal.pone.0174750.Suche in Google Scholar PubMed PubMed Central
[47] Chen Y, Yao Z, Sun Y, Wang E, Tian C, Sun Y, et al. Current studies of the effects of drought stress on root exudates and rhizosphere microbiomes of crop plant species. Int J Mol Sci. 2022;23(4):2374. 10.3390/ijms23042374.Suche in Google Scholar PubMed PubMed Central
[48] Naveed M, Brown LK, Raffan AC, George TS, Bengough AG, Roose T, et al. Plant exudates may stabilize or weaken soil depending on species, origin and time. Eur J Soil Sci. 2017;68:806–16. 10.1111/ejss.12487.Suche in Google Scholar PubMed PubMed Central
[49] Rolfe SA, Griffiths J, Ton J. Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble healthpromoting soil microbiomes. Curr Opin Microbiol. 2019;49:73–82. 10.1016/j.mib.2019.10.003.Suche in Google Scholar PubMed
[50] Edwards J, Johnson C, Santos-Medellína C, Luriea E, Podishetty NK, Bhatnagar S, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2020;112(8):E911–20. 10.1073/pnas.1414592112.Suche in Google Scholar PubMed PubMed Central
[51] Balasooriya WK, Huygens D, Rajapaksha RMCP, Boeckx P. Effect of rice variety and fertilizer type on the active microbial community structure in tropical paddy fields in Sri Lanka. Geoderma. 2016;265:87–95. 10.1016/j.geoderma.2015.11.Suche in Google Scholar
[52] Lian T, Huang Y, Xie X, Huo X, Shahid MQ, Tian L, et al. Variation shapes the rhizosphere bacterial community, conferring tolerance to salt stress through regulating soil metabolites. Systems. 2020;5:e00721-20. 10.1128/mSystems.00721-20.Suche in Google Scholar PubMed PubMed Central
© 2023 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Artikel in diesem Heft
- Regular Articles
- The impact of COVID-19 pandemic on business risks and potato commercial model
- Effects of potato (Solanum tuberosum L.)–Mucuna pruriens intercropping pattern on the agronomic performances of potato and the soil physicochemical properties of the western highlands of Cameroon
- Machine learning-based prediction of total phenolic and flavonoid in horticultural products
- Revamping agricultural sector and its implications on output and employment generation: Evidence from Nigeria
- Does product certification matter? A review of mechanism to influence customer loyalty in the poultry feed industry
- Farmer regeneration and knowledge co-creation in the sustainability of coconut agribusiness in Gorontalo, Indonesia
- Lablab purpureus: Analysis of landraces cultivation and distribution, farming systems, and some climatic trends in production areas in Tanzania
- The effects of carrot (Daucus carota L.) waste juice on the performances of native chicken in North Sulawesi, Indonesia
- Properties of potassium dihydrogen phosphate and its effects on plants and soil
- Factors influencing the role and performance of independent agricultural extension workers in supporting agricultural extension
- The fate of probiotic species applied in intensive grow-out ponds in rearing water and intestinal tracts of white shrimp, Litopenaeus vannamei
- Yield stability and agronomic performances of provitamin A maize (Zea mays L.) genotypes in South-East of DR Congo
- Diallel analysis of length and shape of rice using Hayman and Griffing method
- Physicochemical and microbiological characteristics of various stem bark extracts of Hopea beccariana Burck potential as natural preservatives of coconut sap
- Correlation between descriptive and group type traits in the system of cow’s linear classification of Ukrainian Brown dairy breed
- Meta-analysis of the influence of the substitution of maize with cassava on performance indices of broiler chickens
- Bacteriocin-like inhibitory substance (BLIS) produced by Enterococcus faecium MA115 and its potential use as a seafood biopreservative
- Meta-analysis of the benefits of dietary Saccharomyces cerevisiae intervention on milk yield and component characteristics in lactating small ruminants
- Growth promotion potential of Bacillus spp. isolates on two tomato (Solanum lycopersicum L.) varieties in the West region of Cameroon
- Prioritizing IoT adoption strategies in millennial farming: An analytical network process approach
- Soil fertility and pomelo yield influenced by soil conservation practices
- Soil macrofauna under laying hens’ grazed fields in two different agroecosystems in Portugal
- Factors affecting household carbohydrate food consumption in Central Java: Before and during the COVID-19 pandemic
- Properties of paper coated with Prunus serotina (Ehrh.) extract formulation
- Fertiliser cost prediction in European Union farms: Machine-learning approaches through artificial neural networks
- Molecular and phenotypic markers for pyramiding multiple traits in rice
- Natural product nanofibers derived from Trichoderma hamatum K01 to control citrus anthracnose caused by Colletotrichum gloeosporioides
- Role of actors in promoting sustainable peatland management in Kubu Raya Regency, West Kalimantan, Indonesia
- Small-scale coffee farmers’ perception of climate-adapted attributes in participatory coffee breeding: A case study of Gayo Highland, Aceh, Indonesia
- Optimization of extraction using surface response methodology and quantification of cannabinoids in female inflorescences of marijuana (Cannabis sativa L.) at three altitudinal floors of Peru
- Production factors, technical, and economic efficiency of soybean (Glycine max L. Merr.) farming in Indonesia
- Economic performance of smallholder soya bean production in Kwara State, Nigeria
- Indonesian rice farmers’ perceptions of different sources of information and their effect on farmer capability
- Feed preference, body condition scoring, and growth performance of Dohne Merino ram fed varying levels of fossil shell flour
- Assessing the determinant factors of risk strategy adoption to mitigate various risks: An experience from smallholder rubber farmers in West Kalimantan Province, Indonesia
- Analysis of trade potential and factors influencing chili export in Indonesia
- Grade-C kenaf fiber (poor quality) as an alternative material for textile crafts
- Technical efficiency changes of rice farming in the favorable irrigated areas of Indonesia
- Palm oil cluster resilience to enhance indigenous welfare by innovative ability to address land conflicts: Evidence of disaster hierarchy
- Factors determining cassava farmers’ accessibility to loan sources: Evidence from Lampung, Indonesia
- Tailoring business models for small-medium food enterprises in Eastern Africa can drive the commercialization and utilization of vitamin A rich orange-fleshed sweet potato puree
- Revitalizing sub-optimal drylands: Exploring the role of biofertilizers
- Effects of salt stress on growth of Quercus ilex L. seedlings
- Design and fabrication of a fish feed mixing cum pelleting machine for small-medium scale aquaculture industry
- Indicators of swamp buffalo business sustainability using partial least squares structural equation modelling
- Effect of arbuscular mycorrhizal fungi on early growth, root colonization, and chlorophyll content of North Maluku nutmeg cultivars
- How intergenerational farmers negotiate their identity in the era of Agriculture 4.0: A multiple-case study in Indonesia
- Responses of broiler chickens to incremental levels of water deprivation: Growth performance, carcass characteristics, and relative organ weights
- The improvement of horticultural villages sustainability in Central Java Province, Indonesia
- Effect of short-term grazing exclusion on herbage species composition, dry matter productivity, and chemical composition of subtropical grasslands
- Analysis of beef market integration between consumer and producer regions in Indonesia
- Analysing the sustainability of swamp buffalo (Bubalus bubalis carabauesis) farming as a protein source and germplasm
- Toxicity of Calophyllum soulattri, Piper aduncum, Sesamum indicum and their potential mixture for control Spodoptera frugiperda
- Consumption profile of organic fruits and vegetables by a Portuguese consumer’s sample
- Phenotypic characterisation of indigenous chicken in the central zone of Tanzania
- Diversity and structure of bacterial communities in saline and non-saline rice fields in Cilacap Regency, Indonesia
- Isolation and screening of lactic acid bacteria producing anti-Edwardsiella from the gastrointestinal tract of wild catfish (Clarias gariepinus) for probiotic candidates
- Effects of land use and slope position on selected soil physicochemical properties in Tekorsh Sub-Watershed, East Gojjam Zone, Ethiopia
- Design of smart farming communication and web interface using MQTT and Node.js
- Assessment of bread wheat (Triticum aestivum L.) seed quality accessed through different seed sources in northwest Ethiopia
- Estimation of water consumption and productivity for wheat using remote sensing and SEBAL model: A case study from central clay plain Ecosystem in Sudan
- Agronomic performance, seed chemical composition, and bioactive components of selected Indonesian soybean genotypes (Glycine max [L.] Merr.)
- The role of halal requirements, health-environmental factors, and domestic interest in food miles of apple fruit
- Subsidized fertilizer management in the rice production centers of South Sulawesi, Indonesia: Bridging the gap between policy and practice
- Factors affecting consumers’ loyalty and purchase decisions on honey products: An emerging market perspective
- Inclusive rice seed business: Performance and sustainability
- Design guidelines for sustainable utilization of agricultural appropriate technology: Enhancing human factors and user experience
- Effect of integrate water shortage and soil conditioners on water productivity, growth, and yield of Red Globe grapevines grown in sandy soil
- Synergic effect of Arbuscular mycorrhizal fungi and potassium fertilizer improves biomass-related characteristics of cocoa seedlings to enhance their drought resilience and field survival
- Control measure of sweet potato weevil (Cylas formicarius Fab.) (Coleoptera: Curculionidae) in endemic land of entisol type using mulch and entomopathogenic fungus Beauveria bassiana
- In vitro and in silico study for plant growth promotion potential of indigenous Ochrobactrum ciceri and Bacillus australimaris
- Effects of repeated replanting on yield, dry matter, starch, and protein content in different potato (Solanum tuberosum L.) genotypes
- Review Articles
- Nutritional and chemical composition of black velvet tamarind (Dialium guineense Willd) and its influence on animal production: A review
- Black pepper (Piper nigrum Lam) as a natural feed additive and source of beneficial nutrients and phytochemicals in chicken nutrition
- The long-crowing chickens in Indonesia: A review
- A transformative poultry feed system: The impact of insects as an alternative and transformative poultry-based diet in sub-Saharan Africa
- Short Communication
- Profiling of carbonyl compounds in fresh cabbage with chemometric analysis for the development of freshness assessment method
- Special Issue of The 4th International Conference on Food Science and Engineering (ICFSE) 2022 - Part I
- Non-destructive evaluation of soluble solid content in fruits with various skin thicknesses using visible–shortwave near-infrared spectroscopy
- Special Issue on FCEM - International Web Conference on Food Choice & Eating Motivation - Part I
- Traditional agri-food products and sustainability – A fruitful relationship for the development of rural areas in Portugal
- Consumers’ attitudes toward refrigerated ready-to-eat meat and dairy foods
- Breakfast habits and knowledge: Study involving participants from Brazil and Portugal
- Food determinants and motivation factors impact on consumer behavior in Lebanon
- Comparison of three wine routes’ realities in Central Portugal
- Special Issue on Agriculture, Climate Change, Information Technology, Food and Animal (ACIFAS 2020)
- Environmentally friendly bioameliorant to increase soil fertility and rice (Oryza sativa) production
- Enhancing the ability of rice to adapt and grow under saline stress using selected halotolerant rhizobacterial nitrogen fixer
Artikel in diesem Heft
- Regular Articles
- The impact of COVID-19 pandemic on business risks and potato commercial model
- Effects of potato (Solanum tuberosum L.)–Mucuna pruriens intercropping pattern on the agronomic performances of potato and the soil physicochemical properties of the western highlands of Cameroon
- Machine learning-based prediction of total phenolic and flavonoid in horticultural products
- Revamping agricultural sector and its implications on output and employment generation: Evidence from Nigeria
- Does product certification matter? A review of mechanism to influence customer loyalty in the poultry feed industry
- Farmer regeneration and knowledge co-creation in the sustainability of coconut agribusiness in Gorontalo, Indonesia
- Lablab purpureus: Analysis of landraces cultivation and distribution, farming systems, and some climatic trends in production areas in Tanzania
- The effects of carrot (Daucus carota L.) waste juice on the performances of native chicken in North Sulawesi, Indonesia
- Properties of potassium dihydrogen phosphate and its effects on plants and soil
- Factors influencing the role and performance of independent agricultural extension workers in supporting agricultural extension
- The fate of probiotic species applied in intensive grow-out ponds in rearing water and intestinal tracts of white shrimp, Litopenaeus vannamei
- Yield stability and agronomic performances of provitamin A maize (Zea mays L.) genotypes in South-East of DR Congo
- Diallel analysis of length and shape of rice using Hayman and Griffing method
- Physicochemical and microbiological characteristics of various stem bark extracts of Hopea beccariana Burck potential as natural preservatives of coconut sap
- Correlation between descriptive and group type traits in the system of cow’s linear classification of Ukrainian Brown dairy breed
- Meta-analysis of the influence of the substitution of maize with cassava on performance indices of broiler chickens
- Bacteriocin-like inhibitory substance (BLIS) produced by Enterococcus faecium MA115 and its potential use as a seafood biopreservative
- Meta-analysis of the benefits of dietary Saccharomyces cerevisiae intervention on milk yield and component characteristics in lactating small ruminants
- Growth promotion potential of Bacillus spp. isolates on two tomato (Solanum lycopersicum L.) varieties in the West region of Cameroon
- Prioritizing IoT adoption strategies in millennial farming: An analytical network process approach
- Soil fertility and pomelo yield influenced by soil conservation practices
- Soil macrofauna under laying hens’ grazed fields in two different agroecosystems in Portugal
- Factors affecting household carbohydrate food consumption in Central Java: Before and during the COVID-19 pandemic
- Properties of paper coated with Prunus serotina (Ehrh.) extract formulation
- Fertiliser cost prediction in European Union farms: Machine-learning approaches through artificial neural networks
- Molecular and phenotypic markers for pyramiding multiple traits in rice
- Natural product nanofibers derived from Trichoderma hamatum K01 to control citrus anthracnose caused by Colletotrichum gloeosporioides
- Role of actors in promoting sustainable peatland management in Kubu Raya Regency, West Kalimantan, Indonesia
- Small-scale coffee farmers’ perception of climate-adapted attributes in participatory coffee breeding: A case study of Gayo Highland, Aceh, Indonesia
- Optimization of extraction using surface response methodology and quantification of cannabinoids in female inflorescences of marijuana (Cannabis sativa L.) at three altitudinal floors of Peru
- Production factors, technical, and economic efficiency of soybean (Glycine max L. Merr.) farming in Indonesia
- Economic performance of smallholder soya bean production in Kwara State, Nigeria
- Indonesian rice farmers’ perceptions of different sources of information and their effect on farmer capability
- Feed preference, body condition scoring, and growth performance of Dohne Merino ram fed varying levels of fossil shell flour
- Assessing the determinant factors of risk strategy adoption to mitigate various risks: An experience from smallholder rubber farmers in West Kalimantan Province, Indonesia
- Analysis of trade potential and factors influencing chili export in Indonesia
- Grade-C kenaf fiber (poor quality) as an alternative material for textile crafts
- Technical efficiency changes of rice farming in the favorable irrigated areas of Indonesia
- Palm oil cluster resilience to enhance indigenous welfare by innovative ability to address land conflicts: Evidence of disaster hierarchy
- Factors determining cassava farmers’ accessibility to loan sources: Evidence from Lampung, Indonesia
- Tailoring business models for small-medium food enterprises in Eastern Africa can drive the commercialization and utilization of vitamin A rich orange-fleshed sweet potato puree
- Revitalizing sub-optimal drylands: Exploring the role of biofertilizers
- Effects of salt stress on growth of Quercus ilex L. seedlings
- Design and fabrication of a fish feed mixing cum pelleting machine for small-medium scale aquaculture industry
- Indicators of swamp buffalo business sustainability using partial least squares structural equation modelling
- Effect of arbuscular mycorrhizal fungi on early growth, root colonization, and chlorophyll content of North Maluku nutmeg cultivars
- How intergenerational farmers negotiate their identity in the era of Agriculture 4.0: A multiple-case study in Indonesia
- Responses of broiler chickens to incremental levels of water deprivation: Growth performance, carcass characteristics, and relative organ weights
- The improvement of horticultural villages sustainability in Central Java Province, Indonesia
- Effect of short-term grazing exclusion on herbage species composition, dry matter productivity, and chemical composition of subtropical grasslands
- Analysis of beef market integration between consumer and producer regions in Indonesia
- Analysing the sustainability of swamp buffalo (Bubalus bubalis carabauesis) farming as a protein source and germplasm
- Toxicity of Calophyllum soulattri, Piper aduncum, Sesamum indicum and their potential mixture for control Spodoptera frugiperda
- Consumption profile of organic fruits and vegetables by a Portuguese consumer’s sample
- Phenotypic characterisation of indigenous chicken in the central zone of Tanzania
- Diversity and structure of bacterial communities in saline and non-saline rice fields in Cilacap Regency, Indonesia
- Isolation and screening of lactic acid bacteria producing anti-Edwardsiella from the gastrointestinal tract of wild catfish (Clarias gariepinus) for probiotic candidates
- Effects of land use and slope position on selected soil physicochemical properties in Tekorsh Sub-Watershed, East Gojjam Zone, Ethiopia
- Design of smart farming communication and web interface using MQTT and Node.js
- Assessment of bread wheat (Triticum aestivum L.) seed quality accessed through different seed sources in northwest Ethiopia
- Estimation of water consumption and productivity for wheat using remote sensing and SEBAL model: A case study from central clay plain Ecosystem in Sudan
- Agronomic performance, seed chemical composition, and bioactive components of selected Indonesian soybean genotypes (Glycine max [L.] Merr.)
- The role of halal requirements, health-environmental factors, and domestic interest in food miles of apple fruit
- Subsidized fertilizer management in the rice production centers of South Sulawesi, Indonesia: Bridging the gap between policy and practice
- Factors affecting consumers’ loyalty and purchase decisions on honey products: An emerging market perspective
- Inclusive rice seed business: Performance and sustainability
- Design guidelines for sustainable utilization of agricultural appropriate technology: Enhancing human factors and user experience
- Effect of integrate water shortage and soil conditioners on water productivity, growth, and yield of Red Globe grapevines grown in sandy soil
- Synergic effect of Arbuscular mycorrhizal fungi and potassium fertilizer improves biomass-related characteristics of cocoa seedlings to enhance their drought resilience and field survival
- Control measure of sweet potato weevil (Cylas formicarius Fab.) (Coleoptera: Curculionidae) in endemic land of entisol type using mulch and entomopathogenic fungus Beauveria bassiana
- In vitro and in silico study for plant growth promotion potential of indigenous Ochrobactrum ciceri and Bacillus australimaris
- Effects of repeated replanting on yield, dry matter, starch, and protein content in different potato (Solanum tuberosum L.) genotypes
- Review Articles
- Nutritional and chemical composition of black velvet tamarind (Dialium guineense Willd) and its influence on animal production: A review
- Black pepper (Piper nigrum Lam) as a natural feed additive and source of beneficial nutrients and phytochemicals in chicken nutrition
- The long-crowing chickens in Indonesia: A review
- A transformative poultry feed system: The impact of insects as an alternative and transformative poultry-based diet in sub-Saharan Africa
- Short Communication
- Profiling of carbonyl compounds in fresh cabbage with chemometric analysis for the development of freshness assessment method
- Special Issue of The 4th International Conference on Food Science and Engineering (ICFSE) 2022 - Part I
- Non-destructive evaluation of soluble solid content in fruits with various skin thicknesses using visible–shortwave near-infrared spectroscopy
- Special Issue on FCEM - International Web Conference on Food Choice & Eating Motivation - Part I
- Traditional agri-food products and sustainability – A fruitful relationship for the development of rural areas in Portugal
- Consumers’ attitudes toward refrigerated ready-to-eat meat and dairy foods
- Breakfast habits and knowledge: Study involving participants from Brazil and Portugal
- Food determinants and motivation factors impact on consumer behavior in Lebanon
- Comparison of three wine routes’ realities in Central Portugal
- Special Issue on Agriculture, Climate Change, Information Technology, Food and Animal (ACIFAS 2020)
- Environmentally friendly bioameliorant to increase soil fertility and rice (Oryza sativa) production
- Enhancing the ability of rice to adapt and grow under saline stress using selected halotolerant rhizobacterial nitrogen fixer

