Discovery of the bacterial HslV protease activators as lead molecules with novel mode of action
-
Sana Aurangzeb
, Khalid Mohammed Khan
, Metab Alharbi
Abstract
The HslVU enzyme complex, a proteasomal analog found in bacteria, consists of two components, i.e., the HslV protease and the HslU ATPase. These proteins come together to form a functional enzyme complex, where the C-terminal helix of each HslU subunit is inserted into the binding pocket of each HslV dimer. This interaction leads to the activation of the HslV protease through allosteric mechanisms, enabling its enzymatic function. This bacterial complex is reflected as an attractive target for drug development due to its presence in disease-causing microorganisms and concurrent absence in humans. The objective of this research was to identify certain promising drug candidates that could excessively stimulate the HslV protease, leading to uncontrolled protein breakdown in the pathogens. Four dihydropyrimidone derivatives have been identified as potential activators of HslV protease exhibiting high docking scores, favorable binding patterns, and significant in vitro activation capabilities. These compounds have demonstrated effective dose 50 values within the sub-micromolar range, i.e., 0.4–0.58 µM. Normal mode analysis investigations provided additional confirmation regarding the stability of the conformational interactions between the HslV protease and the active compounds. In addition, the predicted absorption, distribution, metabolism, excretion, and toxicity properties of these lead compounds remarkably demonstrated their considerable drug-like and non-toxic qualities. This study not only presents more potent small non-peptide activators of the HslV protease but also enhances the understanding regarding the mechanism of HslVU complex activation via small non-peptidic molecules.
1 Introduction
The HslVU complex is a prokaryotic complex that shares significant structural conservation with the eukaryotic proteasome [1,2,3,4]. Like the proteasome, the HslVU complex is responsible for maintaining protein hemostasis in the cell by removing damaged/unnecessary proteins in an adenosine triphosphate (ATP)-dependent manner [1]. The HslVU protease/chaperone complex comprises two multimeric enzymes, the HslV protease and the HslU ATPase [1,4]. Structurally, the HslV protease is a dodecamer of two stacked rings, each consisting of six subunits with active sites located in the central proteolytic chamber [3,5,6]. The HslV dodecamer is flanked by the hexameric HslU-ATPase, which shows structural homology to the base components of the proteasome, i.e., 19S complex, and is grouped in the AAA-ATPase superfamily [5,7,8]. The HslU-ATPase typically recognizes its proteinic substrate followed by unfolding and delivering into the HslV catalytic chamber [5,9]. HslV alone is a weak protease and can only degrade some small hydrophobic peptides [9]; its protease activity increases remarkably when associated with HslU-ATPase [10,11,12]. The carboxy-terminal peptides of HslU (C-tail) are considered as molecular devices crucial for a functional HslVU complex formation [5,13]. These C-tail helices stretch significantly during complex formation and dock via multiple hydrophilic and hydrophobic interactions between the neighboring HslV subunits (Figure 1a). This binding causes marked conformational changes in the active site of the HslV protease, leading to its allosteric activation [5]. It has been proved that HslV protease can be activated using only HslU C-tail instead of whole HslU subunits to hydrolyze the unfolded polypeptide (α-casein) and other smaller peptides [13,14]. This allosteric activation strategy is unique to the HslVU complex compared to the eukaryotic proteasome.
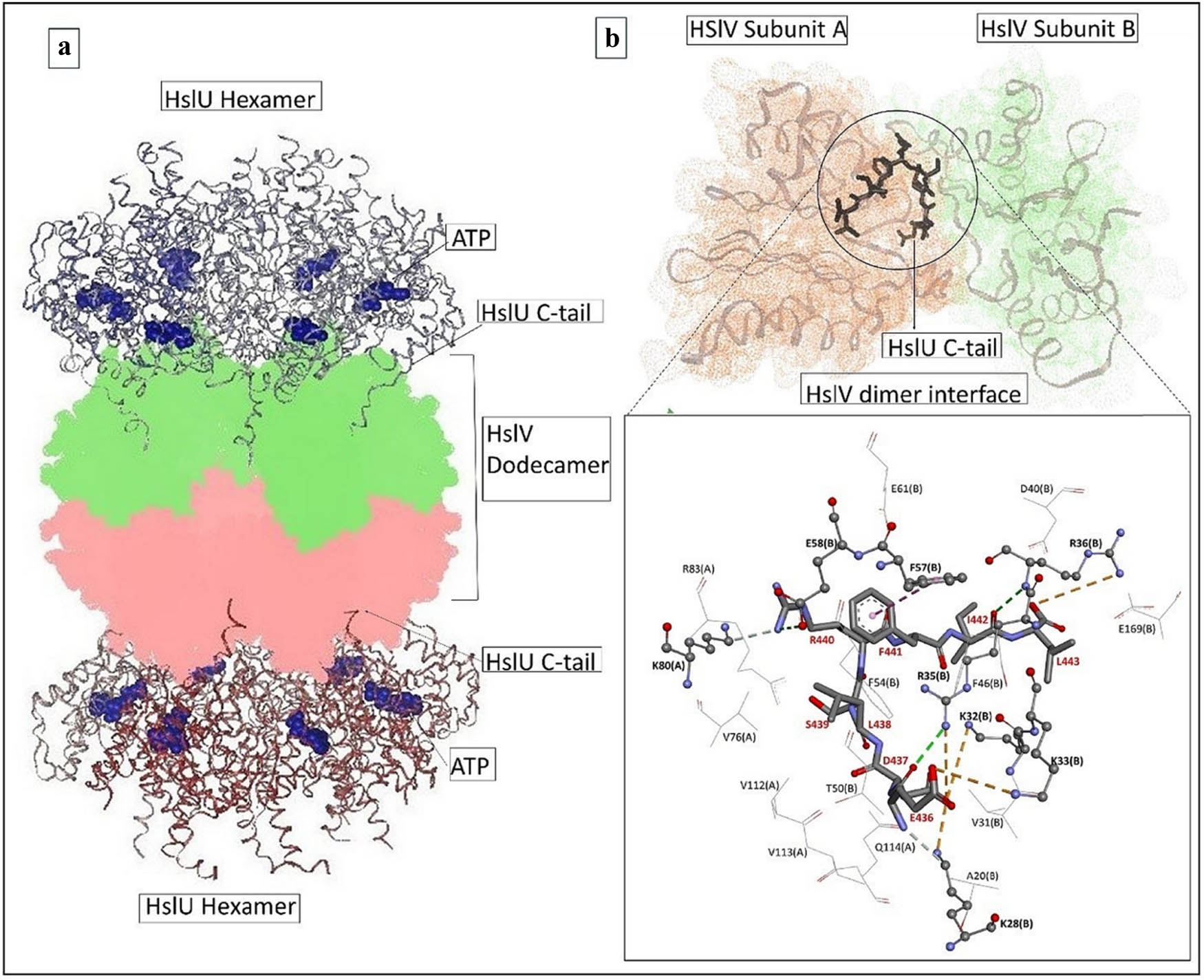
(a) Functional HslVU complex illustrating the C-tail insertions and (b) the docked HslU C-tail (stick) within the HslV-dimeric cleft. The key bonding residues are depicted in ball-and-stick representations, whereas the residues formed van der Waals contacts, which are shown in line representations.
The existence of HslVU complex in disease-causing bacteria and its simultaneous absence in humans make this complex an effective therapeutic target [5,15,16,17,18,19,20,21,22,23]. Our group has reported certain non-peptidic small molecules belonging to different scaffolds as activators of the HslV protease, which strongly mimic the C-tail in binding with the HslV dimeric cleft and in activating the HslV protease many folds [24,25]. In particular, the bacterial HslV protease activation strategy is a novel and less targeted approach in antimicrobial drug discovery [21,22,23,24]. Pyrimidine rings are important pharmacophores, and compounds containing pyrimidine rings have shown a wide range of therapeutic applications [26]. Dihydropyrimidones have become highly significant in the area of medicinal chemistry following the identification of monastrol, a derivative of dihydropyrimidone, as a promising compound capable of inhibiting the formation of bipolar spindles [26,27,28]. Extensive research has been dedicated to dihydropyrimidones due to their significant potential in various pharmacological applications [28,29,30]. During the present studies, the HslV activation potential of dihydropyrimidones was assessed using both in silico and in vitro studies.
2 Methodology
2.1 Molecular docking investigations
The three-dimensional model of the E. coli HslV-dimer complexed with C-tail [24] was utilized as the receptor for molecular docking studies using the Autodock Vina software (version 1.1.2) [31]. The ligand files were prepared by assigning the Gasteiger charges and computing the torsion angles using Marvin Sketch (version 16.2.8) [32] and Autodock tools (version 1.5.6) [33]; the files were then saved in pdbqt format. For docking tests, the HslU C-tail octapeptide was set as the reference ligand. The receptor input file was prepared for docking procedures by the addition of polar hydrogen atoms, assigning Kollman charges, and retaining and fixing the missing atoms in the structure. Grid box dimensions were set to 20 × 18 × 28; x, y, and z coordinates were 182.388, 60.41, and 87.683, respectively, with grid spacing of 1 and exhaustiveness of 8. The two-dimensional images of the docked complexes were generated using DS Visualizer (version 4.5) [34] and VMD (version 1.9.2) software [35].
2.2 HslV protease activation assay
Successful transformation of HslV gene containing PET12b+ was carried out into BL21 codon plus (DE3) RIL cells. The HslV protease was expressed and purified as reported earlier [3,9]. Four compounds with the best docking scores were used for the HslV protease assay using the same reaction parameters described earlier [24,25]. Dihydropyrimidone derivatives considered for in vitro examination were acquired from the ICCBS Compound Bank (University of Karachi), offering a variety of isolated/synthetic chemical compounds in pure forms. The procedure of the synthesis of these derivatives has already been documented [36].
During the in vitro HslV activation analysis, the synthetic HslU C-tail (ADEDLSRFIL) (AnaSpec. Inc. USA) was used as a reference, whereas the four selected hit compounds were examined for their activation effects at five distinct micro-molar concentrations, i.e., 0.1, 0.25, 0.5, 1.0, and 1.5. The HslV protease activation activity was determined by constantly observing the fluorescence of 7-amido-4-methyl coumarin (AMC) at excitation = 355 nm and emission = 460 nm following its dissociation from the fluorogenic peptide substrate (Z-GGL-AMC). The graphs between Vi/Vo (where Vi and Vo are the velocities in the presence and absence of compound) and compound concentrations were utilized to determine the effective concentration of each compound at which HslV exhibited half-maximal activity (i.e., the effective dose 50 or effective dose 50 [ED50]). The 96-well plate reader Varioskan LUX (Thermo Scientific) using SkanIt™ software version 6.0 was employed to measure the fluorescence in relative fluorescence unit tail [24,25].
2.3 Procedure of normal mode analysis (NMA)
Comparing the dynamics of significant protein complexes with their normal modes can indeed demonstrate an alternative and more efficient practice to the expensive atomistic simulations [37,38,39]. To better understand the functional motions of the proteins, this approach combines the related concepts from protein dynamics and structural biology. The iMODS server [40], which is quick, convenient, and potent, was used to run the molecular dynamics simulation to study the conformational stabilities of the interactions between the HslV-dimer and lead compounds via NMA. The docked complexes of the four leads in PDB format were uploaded to the server, and as a reference, the docked HslV-dimer with C-tail complex was used. The iterative Krylov-subspace method is implemented by the iMODS server to perform the faster calculations after reading the input PDB file and calculates the lowest frequency normal modes, which depicts the biological motions in internal coordinates [41]. The simulation tests were executed by setting all of the parameters default. By examining the normal modes of the internal coordinates, this tool reveals details regarding the collective motions via deformability, mobility profiles, eigenvalues, variance, covariance map, and elastic network of the docked complexes.
2.4 ADME profiling of the lead compounds
Many potential drugs fail clinical trials because they have an unfavorable ADME (Absorption, Distribution, Metabolism, and Excretion) profile. Here, ADME profiling was performed to examine drug similarity, medicinal chemistry, and friendliness of lead compounds employing a web server Swiss ADME (http://www.swissadme.ch/) [42]. The hit compounds’ two-dimensional structures were served as input. Recognition of ADME characteristics of the compound is of utmost importance in drug discovery and development. This information aids in the optimization of drug formulations and dosage techniques as well as the evaluation of possible drug interactions.
A free web server, i.e., Protox-II (http://tox.charite.de/protox II), predicts in silico toxicity of the chemical compounds. This server is a great selection for predicting the detrimental risk associated by the chemical compounds. The design and development of specific experimental follow-up investigations, hit selection, and lead optimization processes are made easier by the in silico toxicity prediction [43]. Like Swiss ADME server, this tool also utilized two-dimensional structures of chemical compounds as input. The chemical compound toxicity profiles of the selected model are shown, along with a confidence scores. The toxicity prediction model used in this study takes into account the following variables: oral toxicity, organ toxicity (hepatotoxicity), toxicity endpoints (cytotoxicity), toxicological route (PPAR Gamma), and toxicological target (phosphoprotein tumor suppressor p53).
2.5 Antibacterial activities of lead compounds
The antibacterial activity of SHS-1-61 was evaluated using the microtiter broth dilution method [44]. E. coli ATCC 25922 was incubated with SHS-1-61 at concentrations of 0.0, 0.625, 1.25, 2.5, 5, 10, 20, 40, 80, 160, and 320 μM in Luria-Bertani broth at 37°C for 24 h. Bacterial growth was measured by optical density at 600 nm (OD600). The percentage inhibition was calculated relative to the control. The inhibitory concentration 50 (IC50) value was determined using graph by fitting the data to a sigmoidal dose–response curve. The experiments were performed in triplicate to calculate IC50 value.
3 Results
3.1 Molecular docking interpretations
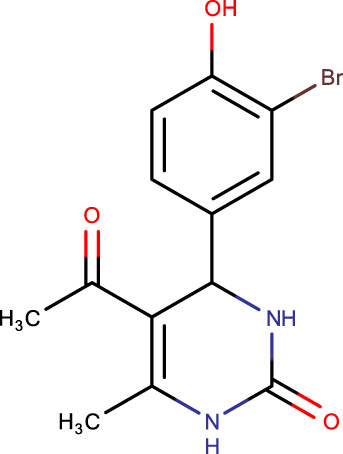
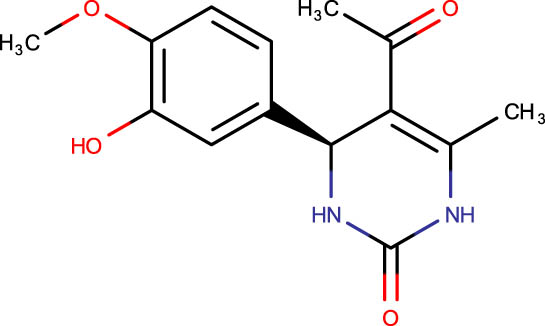
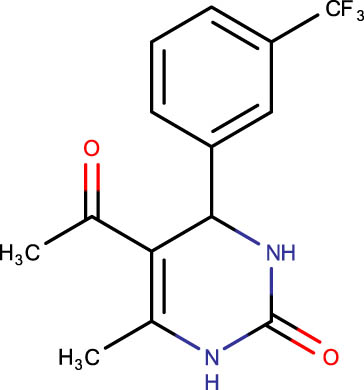
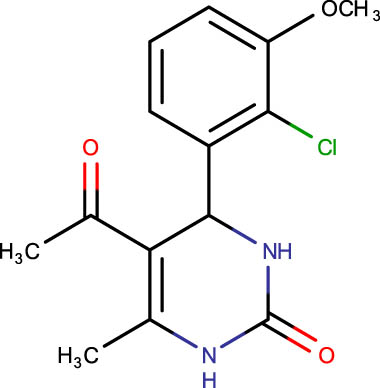
The crystal structure of E. coli HslVU complex available at Protein Data Bank (PDB ID: 1E94) displays a wrong association between the HslV dodecamer and HslU hexamers (Figure 1). The HslU C-tails of HslU hexamer are on the incorrect side of the complex, i.e., far from HslV, failing to illustrate the insertion into the HslV dimeric cleft and allosteric activation mechanism. Because of this, molecular docking studies could not be performed on that structure; instead, the homology model of E. coli HslV-dimeric structure complexed with the HslU C-tail was used as described previously [24,25]. Along with 43 distinct dihydropyrimidone derivatives, the HslU-C tail was docked into the E. coli HslV-dimer’s C tail binding pocket (Figure 1b and Table 1). The docking score for HslU C-tail was lower. The four derivatives, SHS-1-61, SHS-1-30, SHS-1-60, and SHS-1-6, were chosen for the in vitro HslV activation assay because they exhibited considerably higher docking scores (Table 1).
Description of codes, structures, and the docking scores of dihydropyrimidone derivatives
| Compound code | Structures | Docking score | Compound code | Structures | Docking score |
|---|---|---|---|---|---|
| SHS-1-61 |
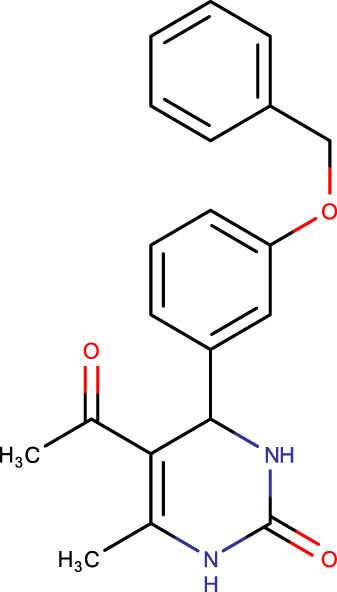 |
−8.5 | SHS-1-75 |
 |
−6.4 |
| SHS-1-30 |
 |
−8.0 | SHS-1-29 |
 |
−6.3 |
| SHS-1-60 |
 |
−7.3 | SHS-1-67 |
 |
−6.3 |
| SHS-1-6 |
 |
−7.3 | SHS-1-33 |
 |
−6.3 |
| SHS-1-49 |
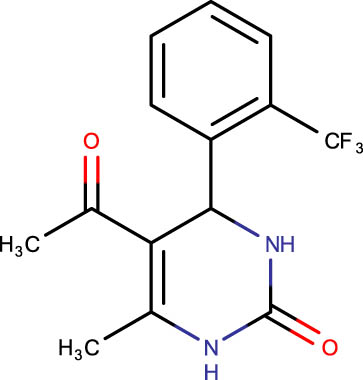 |
−7.2 | SHS-1-3 |
 |
−6.3 |
| SHS-1-83 |
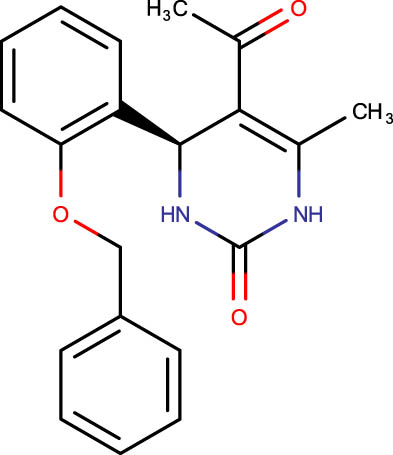 |
−7.1 | SHS-1-31 |
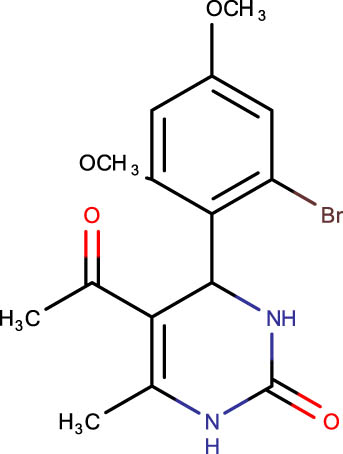 |
−6.2 |
| SHS-1-85 |
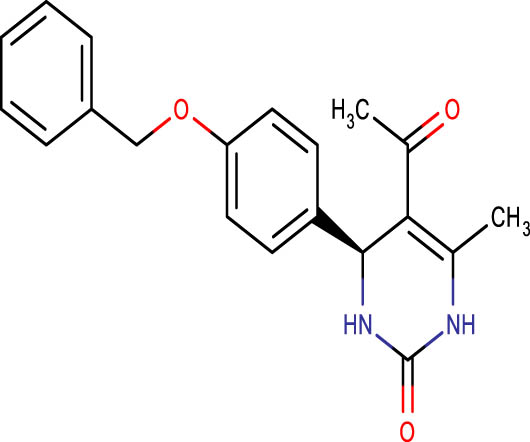 |
−7.1 | SHS-1-70 |
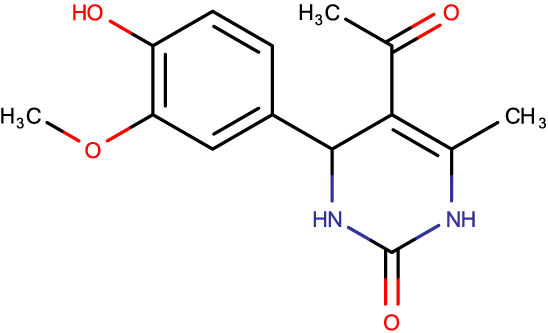 |
−6.2 |
| SHS-1-45 |
 |
−7.0 | SHS-1-63 |
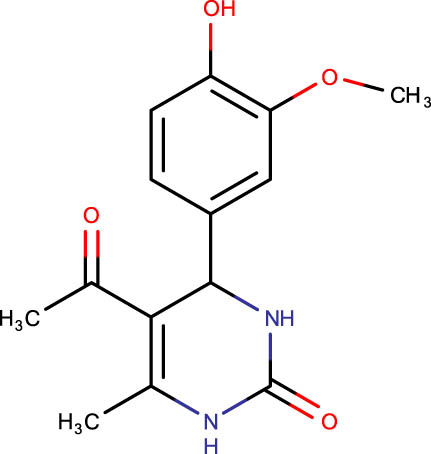 |
−6.2 |
| SHS-1-81 |
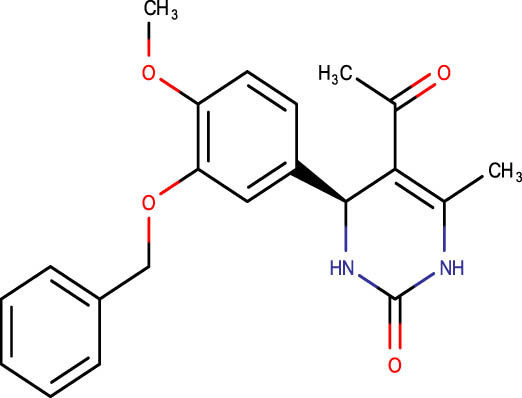 |
−7.0 | SHS-1-65 |
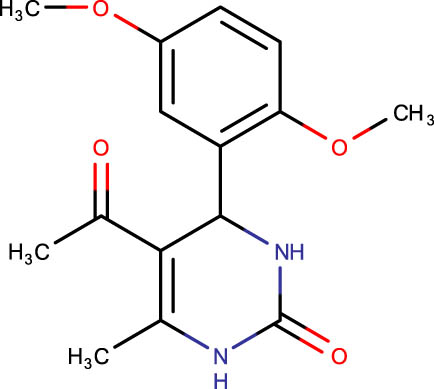 |
−6.2 |
| SHS-1-87 |
 |
−7.0 | SHS-1-15 |
 |
−6.1 |
| SHS-1-89 |
 |
−7.0 | SHS-1-36 |
 |
−6.1 |
| SHS-1-91 |
 |
−7.0 | SHS-1-8 |
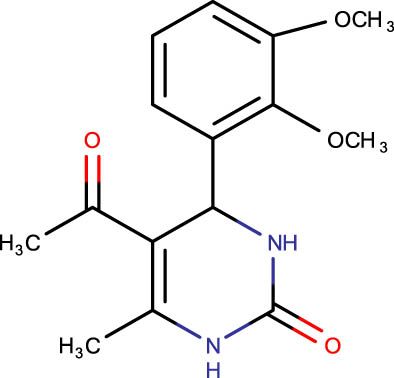 |
−6.0 |
| SHS-1-93 |
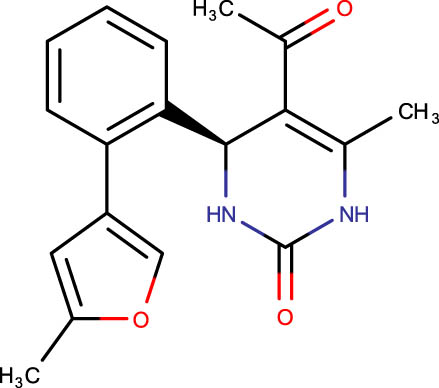 |
−7.0 | SHS-1-16 |
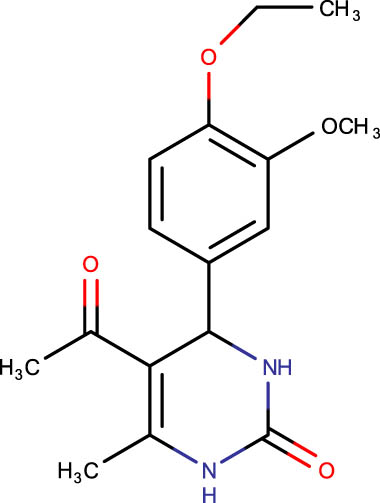 |
−6.0 |
| SHS-1-35 |
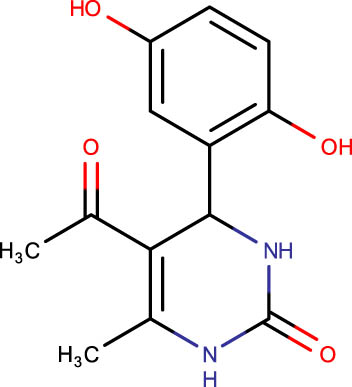 |
−6.8 | SHS-1-73 |
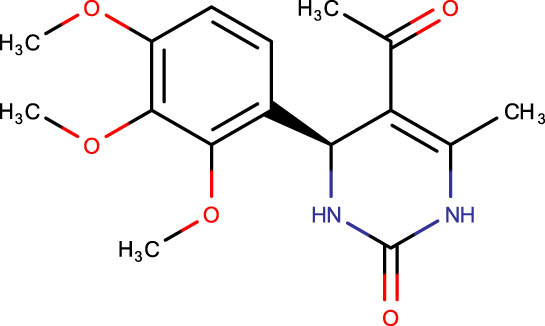 |
−6.0 |
| SHS-1-37 |
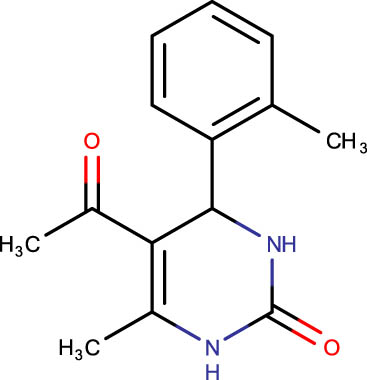 |
−6.7 | SHS-1-22 |
 |
−6.0 |
| SHS-1-80 |
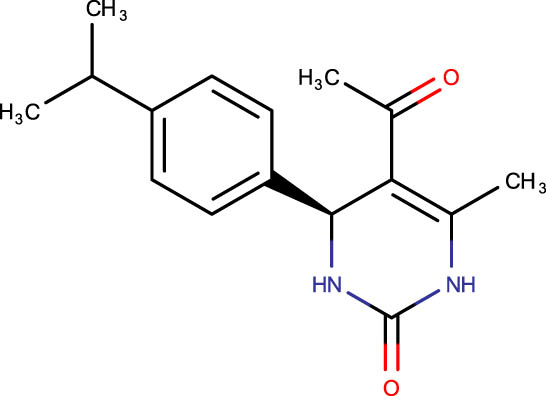 |
6.7 | SHS-1-55 |
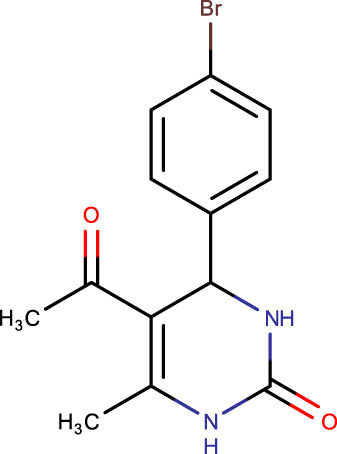 |
−6.0 |
| SHS-1-25 |
 |
−6.6 | SHS-1-21 |
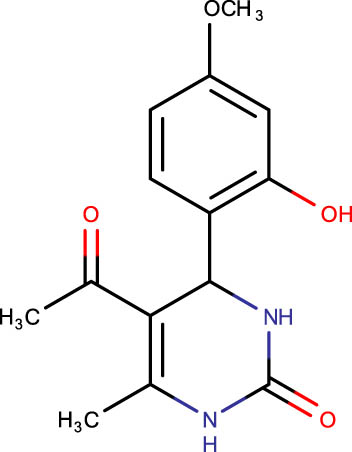 |
−5.9 |
| SHS-1-5 |
 |
−6.5 | SHS-1-7 |
 |
−5.8 |
| SHS-1-26 |
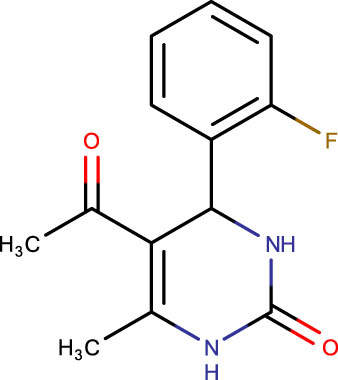 |
−6.5 | SHS-1-77 |
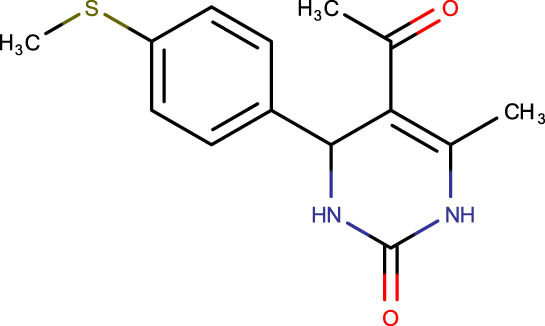 |
−5.8 |
| SHS-1-28 |
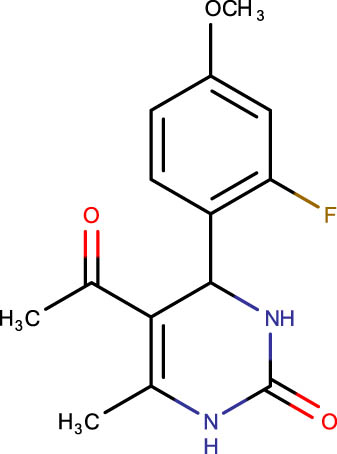 |
−6.4 | SHS-1-46 |
 |
−5.5 |
| SHS-1-34 |
 |
−6.4 | SHS-1-66 |
 |
−5.4 |
| SHS-1-53 |
 |
−6.4 |
3.2 HslV protease activation assay
Purified E. coli HslV protease was obtained after successful transformation, expression, and purification experiments (data not shown). The HslV activation potential of five different doses of each lead compound, ranging from 0.1 to 1.5 µM, was examined without adding its natural activator HslU or its C-tail (Figure 2). The HslU C-tail was not applicable to the same concentration range because there was not any apparent HslV activation at these concentrations. Nevertheless, the concentration range of 1.0–5.0 µM was used to analyze the HslU C-tail activation potential. The C-tail’s ED50 value was observed to be 2.6 µM. However, the ED50 values for the hit compounds were substantially lower than that of the C-tail, i.e., 0.4–0.58 µM (Table 2).
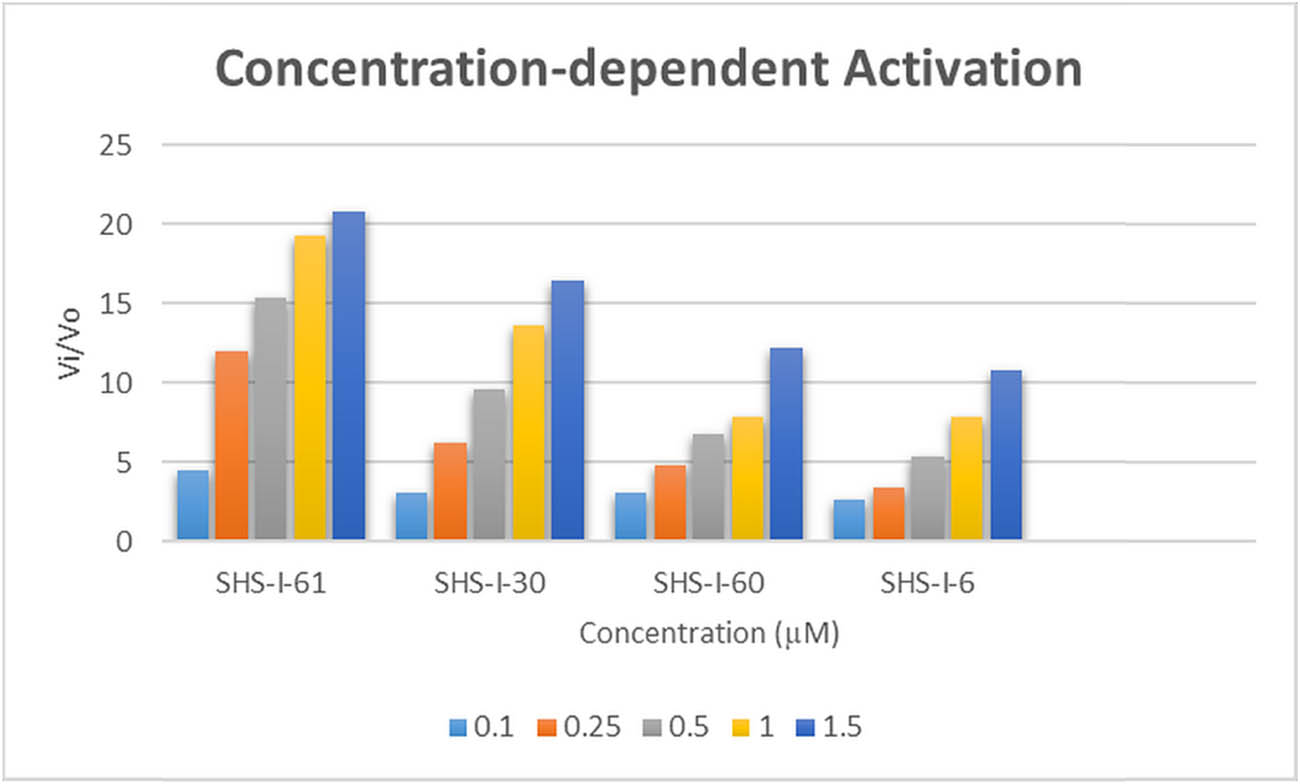
Graphical illustration of concentration-dependent activation of E. coli HslV protease by dihydropyrimidone derivatives.
Molecular docking scores and calculated ED50 values relating the HslU C-tail and lead compounds
| Codes | Docking scores (kcal/mol) | ED50 (µM) | |
|---|---|---|---|
| 1 | HslU C-tail (reference) | −6.0 | 2.6 ± 0.34 |
| 2 | SHS-1-61 | −8.5 | 0.4 ± 0.03 |
| 3 | SHS-1-30 | −8.0 | 0.52 ± 0.005 |
| 4 | SHS-1-60 | −7.3 | 0.58 ± 0.01 |
| 5 | SHS-1-6 | −7.3 | 0.52 ± 0.01 |
3.3 Analysis of the docked complexes
The main interaction pattern observed between the C-tail and the HslV-dimer involved three conventional hydrogen bonds with the residues Glu58 (B), Arg35 (B), and Arg36 (B) and five salt bridges involving the residues Lys28 (B), Lys32 (B), Lys33 (B), Arg35 (B), and Arg36 (B) of the HslV-dimer. Twelve HslV residues, including four residues from chain-A (Val112, Val113, Gln114, and Val76) and eight residues from chain-B (Ala20, Val31, Val34, Phe46, Asp40, Phe54, Glu61, and Glu169), formed hydrophobic (T-shaped and van der Waals) contacts with the HslU C-tail (Figure 1b).
The interaction analysis between the HslV-dimer and 5-acetyl-4-[3-(benzyloxy) phenyl]-6-methyl-1,2,3,4-tetrahydropyrimidin-2-one (SHS-1-61) docked complex is depicted in Figure 3a and b. The residues Lys28 and Gln114 made conventional hydrogen bonds with the aryl ring and the pyrimidone ring of the compound. Arg35(B), the key interacting polar residue, showed a π-cation bond with the aryl ring of the compound. Different hydrophobic bonds (π-sigma, π-alkyl, and van der Waals) between the compound and the HslV dimeric residues, i.e., four residues of chain-A, Arg83, Val72, Val76, Val113, and the eight residues of chain-B Ala20, Val31, Thr50, Phe54, Lys32, Arg83, Val72, and Val76, have been detected. The other lead compound, i.e., 5-acetyl-6-methyl-4-(naphthalene-1-yl)-1,2,3,4-tetrahydropyrimidin-2-one (SHS-1-30), also made significant bonds with the HslV-dimer C-tail pocket residues (Figure 3c and d). A hydrogen bond was established between the Arg35(B) and pyrimidone ring, and two π–π stacked bonds formed between the Phe54(B) and the naphthalene of the compound. Different hydrophobic bonds were also found, i.e., Val112(A) formed two alkyls and π-alkyl bonds with the methyl group of this compound. The six residues from chain-A, i.e., Arg83, Val112, Val76, Asp111, Val113, Gln114, and the three residues from chain-B, i.e., Val31, Arg28, and Thr50, formed hydrophobic (van der Waals) contacts with this ligand. The lead compound, i.e., 5-acetyl-6-methyl-4-(naphthalen-2-yl)-1,2,3,4-tetrahydropyrimidin-2-one, (SHS-1-60) stabilized by forming important interactions with the HslV-dimer residues (Figure 3e and f). Arg35(B) made a hydrogen bond with the pyrimidone ring, and Phe54(B) formed π–π stacked bond with the naphthalene of the compound. Like other leads, this compound also showed various hydrophobic bonds (π-sigma, π-alkyl, and van der Waals) with the three residues of chain-A. i.e., Val113, Asp111, and Val112, and the four residues of chain-B Ala20, Val31, Lys28, and Thr50. The compound, i.e., 5-acetyl-6-methyl-4-[3-(trifluoromethyl) phenyl]-1,2,3,4-tetrahydropyrimidin-2-one (SHS-1-6), made three hydrogen bonds with the key residues of chain-B of HslV-dimer, i.e., Lys28, Lys32, Arg35. Lys28 and Arg35 interact with fluorine atoms and Lys32 with the pyrimidone ring of the compound. This lead compound also showed varied hydrophobic contacts (π-alkyl and van der Waals) with the five residues of chain-A, i.e., Arg83, Val76, Val112, Val113, and Gln114, and the two residues of chain-B, i.e., Val31 and Phe54 (Figure 3g and h).
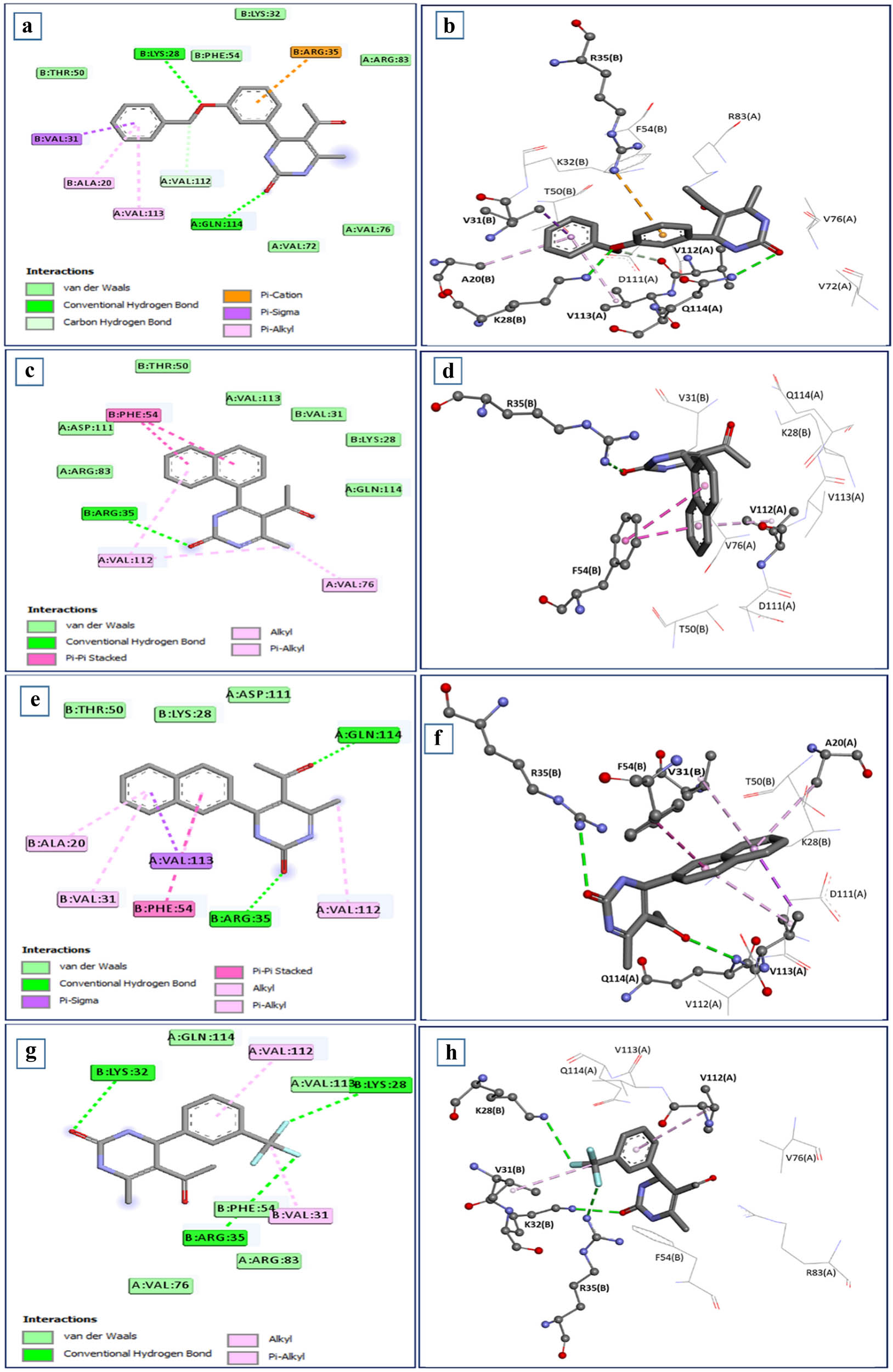
Intermolecular interactions between the HslV dimer and dihydropyrimidone derivatives: (a) and (b) SHS-1-61, (c) and (d) SHS-1-30, (e) and (f) SHS-1-60 and (g) and (h) SHS-1-6 are depicted in two-dimensional and three-dimensional representations. The pyrimidine rings of the leads show hydrogen bonds with the important residues.
3.4 NMA outcomes
NMA examines molecular dynamics and vibrations, aiding in understanding drug interactions in protein and molecular modeling [45]. NMA, conducted via the iMODS server, revealed the dynamic behavior and adaptability of docked HslV protease complexes. Deformability graphs indicated that docked complexes (Figure 4a–d) exhibited increased flexibility in specific regions compared to the reference docked complex (HslV dimer with C-tail), which displayed greater structural stability (Figure 4e). This suggests that docked compounds are better suited for dynamic roles, while the C-tail peptide is optimized for stable structural functions. B-factor graphs (Figure 4f–i) validated these findings, showing higher atomic fluctuations in docked complexes, which is consistent with experimental data, whereas the reference complex exhibited uniform fluctuations (Figure 4j).
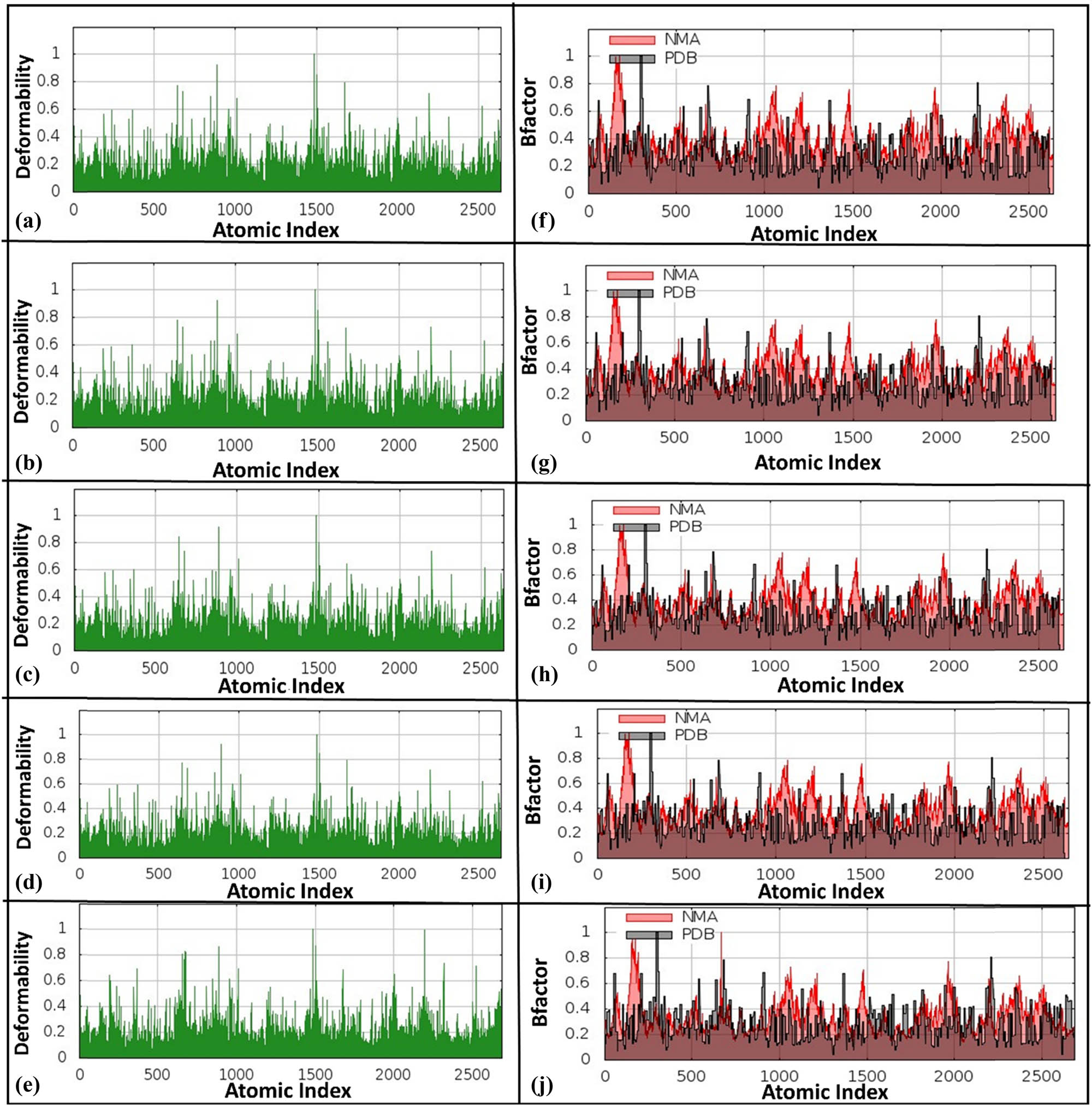
Deformability graphs and B-factor graphs for different HslV compound-docked complexes. The deformability graphs are shown for (a) HslV-SHS-1-61, (b) HslV-SHS-1-30, (c) HslV-SHS-1-60, (d) HslV-SHS-1-6, and (e) HslV-C-tail. Likewise, the B-factor graphs are presented for (f) HslV-SHS-1-61, (g) HslV-SHS-1-30, (h) HslV-SHS-1-60, (i) HslV-SHS-1-6, and (j) HslV-C-tail.
The generated eigenvalues of the four docked complexes, i.e., HslV-dimer with SHS-1-61, HslV-dimer with SHS-1-30, HslV-dimer with SHS-1-60, and HslV-dimer with SHS-1-6, were 5.564078 × 10−4, 5.546997 × 10−4, 5.355598 × 10−4, and 5.732494 × 10−4, respectively. However, the determined eigenvalue of the reference docked complex (HslV-dimer with C-tail) was 6.434125 × 10−4 (Figure 5a–e). Variance analysis revealed dominant contributions from specific normal modes in docked complexes (Figure 5f–i), while the reference maintained even distribution, reflecting stable motion (Figure 5j).
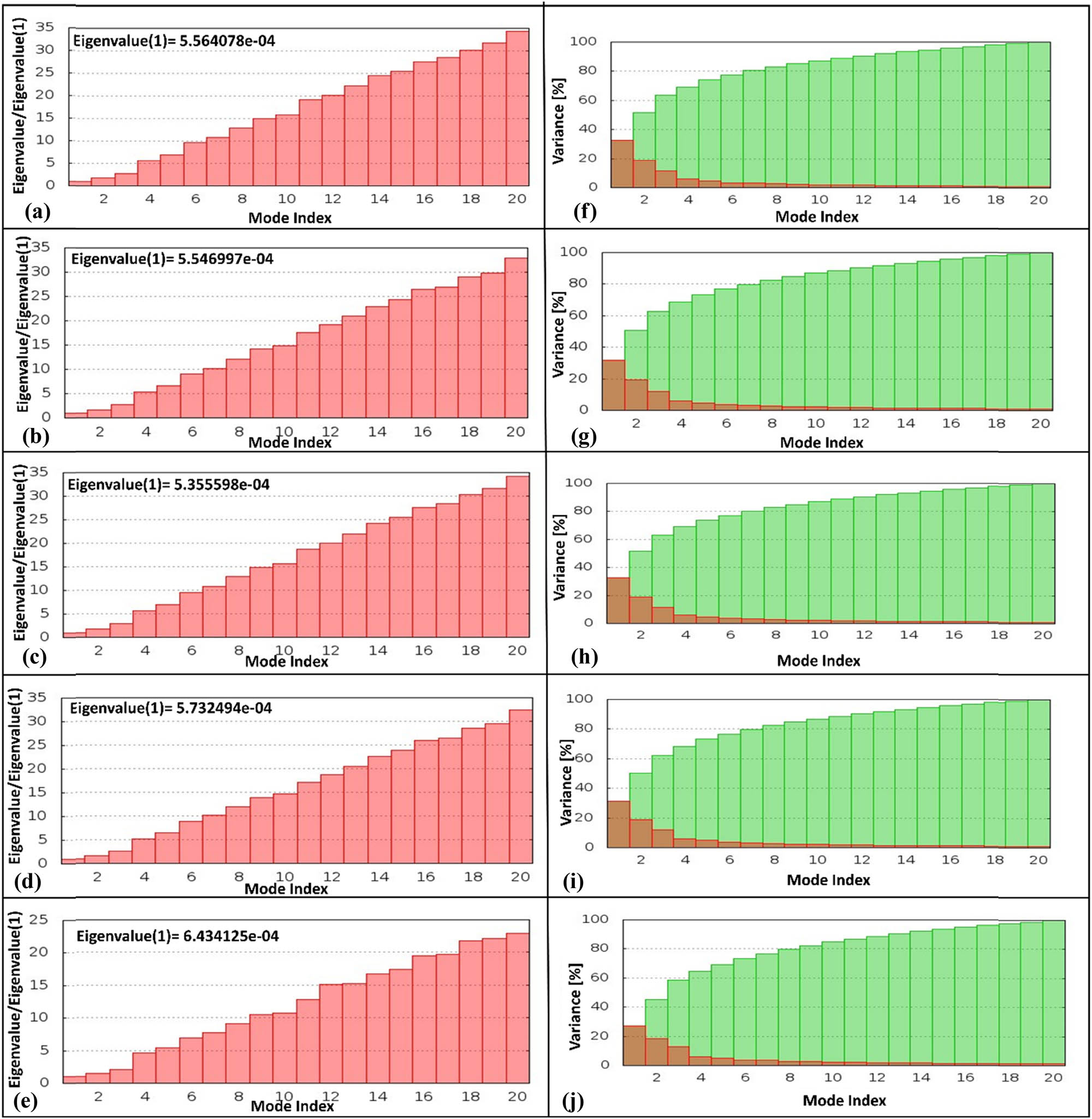
Eigenvalue graphs and variance graphs for different HslV compound-docked complexes. The eigenvalue graphs are depicted for (a) HslV-SHS-1-61, (b) HslV-SHS-1-30, (c) HslV-SHS-1-60, (d) HslV-SHS-1-6, and (e) HslV-C-tail. Likewise, the variance graphs are shown for (f) HslV-SHS-1-61, (g) HslV-SHS-1-30, (h) HslV-SHS-1-60, (i) HslV-SHS-1-6, and (j) HslV-C-tail.
Covariance maps (Figure 6a–d) highlighted altered inter-residue dynamics in docked complexes, particularly in binding regions, suggesting enhanced modulation and new functional pathways, while the reference maintained stable dynamics (Figure 6e). Elastic network maps are the simplified presentations of the complex system, where the docked complex is modeled as a network of interconnected atoms or residues via springs. Each atom in the complex is represented as a spot in the network, with springs indicating interactions between atoms. The darkness of the spots reflects the strength of the springs, with darker spots representing stiffer springs and lighter dots indicating weaker springs. Elastic network maps (Figure 6f–i) showed reduced stiffness in docked complexes, enabling flexibility critical for binding and conformational transitions, compared to the uniformly high stiffness of the reference complex (Figure 6j).
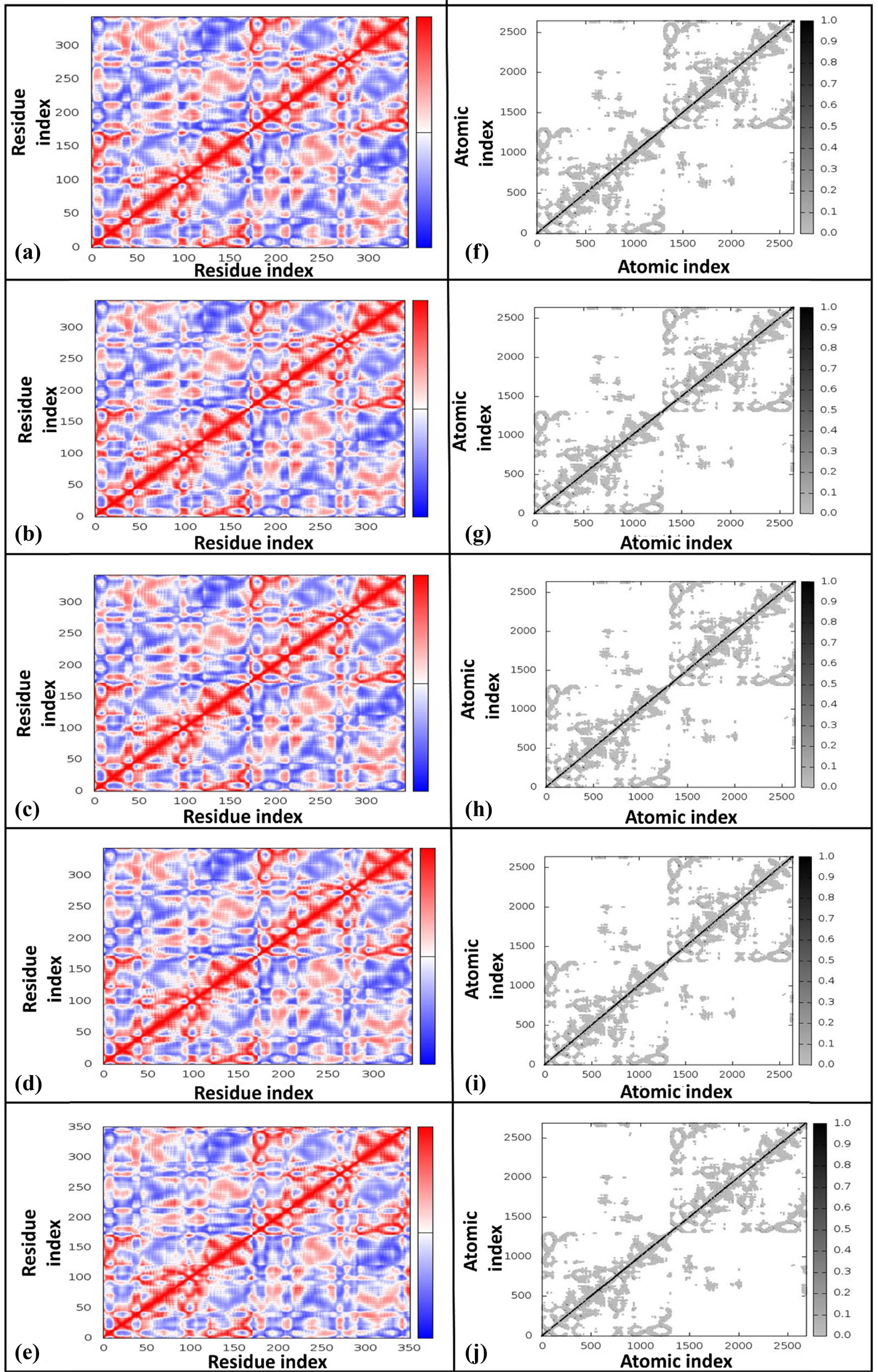
Covariance maps and elastic maps for different HslV compound-docked complexes. The covariance maps are presented for (a) HslV-SHS-1-61, (b) HslV-SHS-1-30, (c) HslV-SHS-1-60, (d) HslV-SHS-1-6, and (e) HslV-C-tail. Likewise, the corresponding elastic maps are illustrated for (f) HslV-SHS-1-61, (g) HslV-SHS-1-30, (h) HslV-SHS-1-60, (i) HslV-SHS-1-6, and (j) HslV-C-tail. The colors in co-variance maps, i.e., red, white, and blue, correspond to the positive (correlated), zero (un-correlated), and negative (anti-correlated) correlations, respectively.
3.5 In silico evaluations of drug-likeness and toxicity of lead compounds
Using the Swiss ADME server, the lead compounds found during this research were assessed for their drug-like qualities [42] (Table 3). The analysis revealed that all the leads have fewer than 5 hydrogen bond donors and fewer than 10 hydrogen bond acceptors. The topological surface area (TPSA) is a measure of surface area of the compound, i.e., available for the interaction with the other molecules (enzymes or receptors). A TPSA of less than 140 Ų is a best predictor of oral bioavailability. The leads of this study displayed that TPSA values ranged from 58.20 to 67.43 Ų, meaning that the leads below the threshold are more likely to be absorbed into the bloodstream and possess a higher chance to being effective as a drug. The synthetic accessibility (SA) is a measure of how easy or difficult to synthesize a specific compound or drugs. Those compounds with a score of 1 are easy to synthesize, while those with a score of 10 are challenging. The leads of this study showed that score ranged from 3.51 to 3.75, which showed easiness of their synthesis and showed a significant impact on the commercial viability of the compound. The lipophilicity is also one of the important factors that is used to analyze the solubility of the compounds in fats or lipids, and how it is absorbed in the body. To optimize the lipophilicity of the leads consensus, the log P o/w value was determined and this value should not exceed than 5. The leads exhibited this value between 2.20 and 2.55, which provided the best outcomes with better efficacy and safety. Furthermore, the leads strictly followed the Lipinski, Veber, Egan, and Muegge guidelines widely used in the drug discovery. The Lipinski rule demonstrated the excellent intestinal and solubility of lead compounds and potential to become a drug [46]. The Ghose rule emphasizes the relevance of improved physiochemical characteristics in the identification of leads with drug-like effects [47]. According to the Veber rule [48], these leads have potential to be effective and safe oral drug candidates. The Egan rule’s prediction showed how effortlessly the human passive gut may absorb lead compounds [49]. The Muegge rule distinguishes between chemical substances that are similar to drugs and those that are not [50]. All of these compounds have a high gastrointestinal (GI) absorption, strong solubility, and a substantial TPSA, which makes them the drug-like molecules. The cytochrome P450 is the enzyme family metabolized by almost most drugs. The two enzymes, i.e., CYP3A4 and CYP2D6, are the most significant [51]. The lead compound SHS-I-61 showed inhibition against CYP1A2, CYP2C9, CYP2D6, and CYP3A4. The compounds, i.e., SHS-1-30 and SHS-1-60, showed inhibition against CYP1A2 and CYP2C19 only. However, the compound SHS-1-6 has not shown inhibition against anyone of the enzyme (Table 3). The metabolism of a drug is subject to diverse levels of modulation by inhibitors, contingent upon both the dosage administered and the inhibitor’s ability to effectively bind to the target enzyme [52]. These predictions aid in reducing the possibilities of risks produced in response to drug reactions and interactions. The toxicity profiles of the leads were evaluated through the assessment of toxicity endpoints such as cytotoxicity (Table 3).
In silico ADMET predictions of the lead compounds
| Properties | SHS-1-61 | SHS-1-30 | SHS-1-60 | SHS-1-6 |
|---|---|---|---|---|
| General properties | ||||
| Molecular weight (g/mol) | 336.38 | 280.32 | 280.32 | 298.26 |
| Number of H-bond acceptor | 3 | 2 | 2 | 5 |
| Number of H-bond donors | 2 | 2 | 2 | 2 |
| Number of rotatable bonds | 5 | 2 | 2 | 3 |
| Molar refractivity | 102.86 | 102.86 | 89.39 | 76.89 |
| TPSA (Å2) | 67.43 | 58.20 | 58.20 | 58.20 |
| Solubility | −3.41 (soluble) | −3.12 (soluble) | −3.12 (soluble) | −2.79 (soluble) |
| GI absorption | High | High | High | High |
| Blood brain barrier permeant | Yes | Yes | Yes | Yes |
| Log K p (cm/s) | −6.71 | −6.53 | −6.53 | −6.89 |
| Consensus log P o/w | 2.55 | 2.20 | 2.22 | 2.36 |
| Bioavailability | 0.55 | 0.55 | 0.55 | 0.55 |
| Drug-likeness | ||||
| Five rules (Lipinski, Ghose, Egan, Muegge, Veber) | Yes | Yes | Yes | Yes |
| Leadlikeness | Yes | Yes | Yes | Yes |
| SA | 3.75 | 3.55 | 3.55 | 3.51 |
| Drug metabolism | ||||
| P-gp substrate | Yes | Yes | Yes | No |
| CYP2C9 inhibitor | Yes | No | No | No |
| CYP2D6 inhibitor | Yes | No | No | No |
| CYP3A4 inhibitor | Yes | No | No | No |
| CYP1A2 inhibitor | Yes | Yes | Yes | No |
| CYP2C19 inhibitor | No | Yes | Yes | No |
| Drug toxicity | ||||
| Oral toxicity (mg/kg) | 6,200 | 500 | 500 | 800 |
| Predicted toxicity class | Class 6 | Class 4 | Class 4 | Class 4 |
| Average similarity (%) | 45.65 | 48.04 | 48.04 | 44.73 |
| Prediction accuracy (%) | 54.26 | 54.26 | 54.26 | 54.26 |
| Organ toxicity (hepatotoxicity) | 0.52 (inactive) | 0.55 (inactive) | 0.52 (inactive) | 0.55 (inactive) |
| Cytotoxicity | 0.84 (inactive) | 0.67 (inactive) | 0.84 (inactive) | 0.67 (inactive) |
| PPAR gamma | 0.93 (inactive) | 0.96 (inactive) | 0.93 (inactive) | 0.96 (inactive) |
| Phosphoprotein tumor suppressor p53 | 0.79 (inactive) | 0.78 (inactive) | 0.78 (inactive) | 0.75 (inactive) |
The prediction of toxicity can be performed in both qualitative and quantitative manners. In qualitative assessment, it can be categorized as binary, indicating whether the compound is active or inactive against specific cell types and assays. On the other hand, quantitative measurements involve determining lethal dose 50 (LD50) values, which represent the lethal dosage required to cause fatality in specific cell types and assays. Additionally, the compound’s effects were examined in relation to various areas, including cytotoxicity, hepatotoxicity, PPAR gamma, and the phosphoprotein tumor suppressor p53 [43]. The results of the toxicity prediction analysis indicated that the LD50 values for compounds SHS-1-61, SHS-1-30, SHS-1-60, and SHS-1-6 were found to be 6,200, 500, 500, and 800 mg/kg, respectively. It was determined SHS-1-61 falls under toxicity class six, signifies it as a non-toxic compound. On the other hand, SHS-1-30, SHS-1-60, and SHS-1-6 were classified to toxicity class four, which suggests that they are considered harmful if swallowed, with LD50 values ranging from 300 to 2,000 mg/kg. The average similarity observed among the compounds falls within a range of 44.73–48.04%. Furthermore, it is worth noting that the prediction accuracy for all the compounds was uniformly measured at 54.26%.
In terms of cytotoxicity, compounds SHS-1-61, SHS-1-30, SHS-1-60, and SHS-1-6 exhibited no cytotoxic effects with values of 0.84, 0.67, 0.84, and 0.97, respectively. Additionally, the activity toward PPAR gamma was observed to be inactive for SHS-1-61 and SHS-1-60, while SHS-1-30 and SHS-1-6 showed the activity values of 0.93 and 0.96, respectively. For the phosphoprotein tumor suppressor p53, all compounds demonstrated inactivity with values of 0.79, 0.78, 0.78, and 0.75, respectively. These findings suggest that none of the compounds exhibit significant cytotoxicity or adverse effects on the tested tumor suppressor proteins, as predicted by in silico absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis, and they demonstrate a promising potential for safe use as medicinal agents.
3.6 Antibacterial activity results
The lead compounds under investigation, i.e., SHS-1-61, SHS-1-30, SHS-1-60, and SHS-1-6, exhibited promising HslV activation potential during in silico and in vitro experiments. However, their antibacterial assay using the microtiter broth dilution method [44] revealed that only SHS-1-61 effectively inhibited the growth of E. coli ATCC 25922 (Table 4). The dose–response analysis for SHS-1-61 against E. coli ATCC 25922 revealed an IC50 value of 13.55 μM (Figure 7).
Antibacterial activity of SHS-1-61 compound
| Concentrations (μM) | Mean OD ± SEM | % Inhibition |
|---|---|---|
| 0.0 | 1.03 ± 0.035 | 0 |
| 0.625 | 0.91 ± 0.120 | 11.65 |
| 1.25 | 0.82 ± 0.009 | 20.39 |
| 2.5 | 0.77 ± 0.021 | 25.24 |
| 5 | 0.62 ± 0.012 | 39.81 |
| 10 | 0.55 ± 0.164 | 46.60 |
| 20 | 0.45 ± 0.021 | 56.31 |
| 40 | 0.37 ± 0.121 | 64.08 |
| 80 | 0.24 ± 0.164 | 76.70 |
| 160 | 0.19 ± 0.140 | 81.55 |
| 320 | 0.11 ± 0.006 | 89.32 |
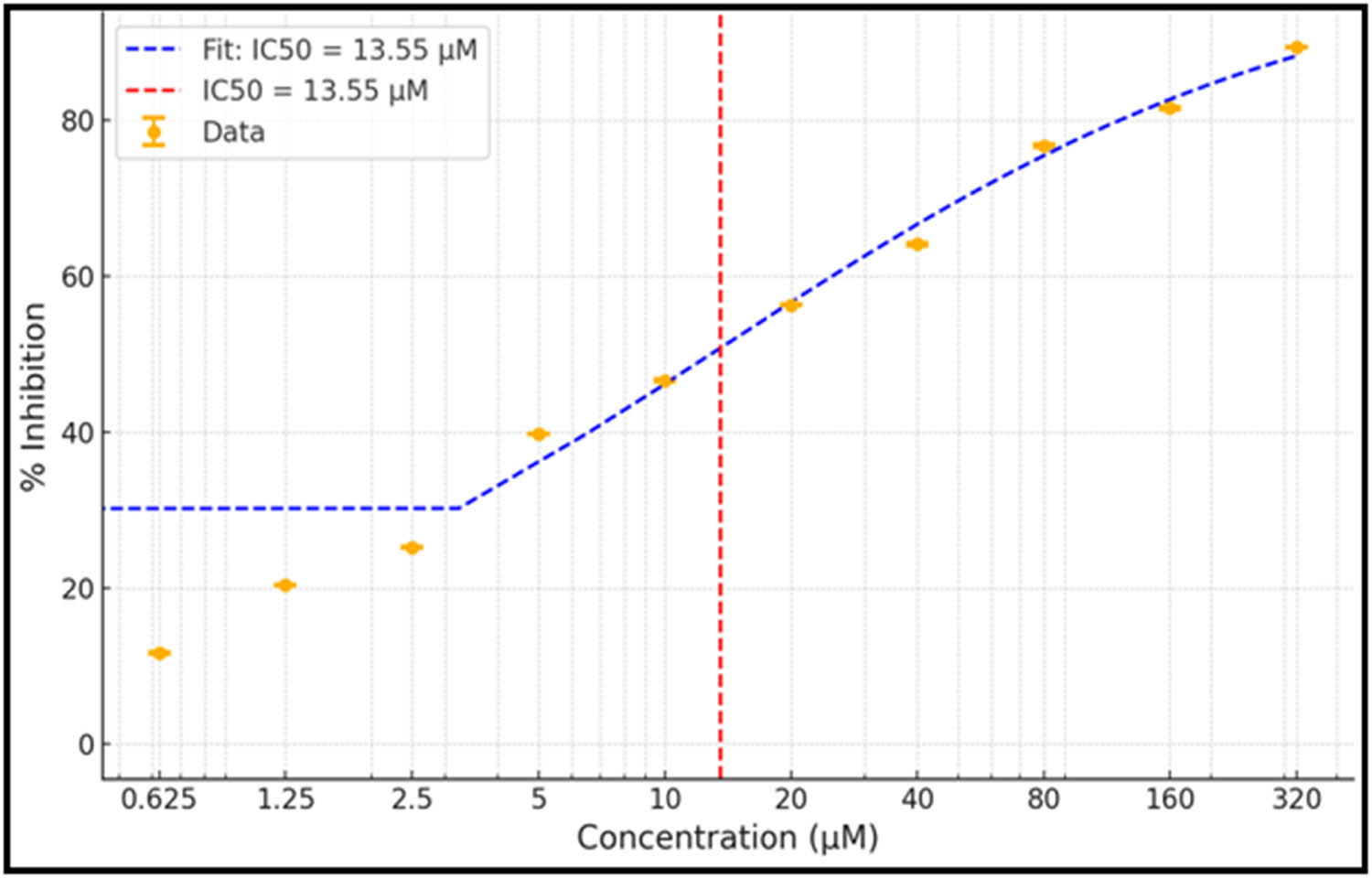
Dose–response curve. The dose–response analysis to calculate IC50 value for SHS-1-61 compound against E. coli ATCC 25922 is depicted.
4 Discussion
In this study, dihydropyrimidone derivatives were identified as potential activators of the E. coli HslV protease, highlighting their capability to circumvent natural regulatory mechanisms and enhance enzymatic activity.
Molecular docking revealed significant interactions between HslV protease and the dihydropyrimidone derivatives, elucidating their binding mechanisms and molecular recognition patterns. The binding of SHS-1-61, SHS-1-30, SHS-1-60, and SHS-1-6 involved hydrogen bonds, π-cation interactions, and extensive hydrophobic contacts. These interactions predominantly engaged critical residues such as Arg35 (B), Phe54 (B), and Lys28, which are pivotal in stabilizing the HslV complex. Notably, Arg35 (B) exhibited a key role in hydrogen bonding and π-cation interactions, thereby facilitating the compounds’ allosteric activation potential. Similar interaction mechanisms were observed in previous studies exploring protease activators like acyldepsipeptides (ADEPs), which destabilized bacterial proteostasis by hyperactivating ClpP proteases [53].
The in vitro activation assays corroborated the computational predictions, with the lead compounds demonstrating significantly lower ED50 values compared to the natural HslU C-tail peptide. This increased potency indicates the efficacy of the compounds in modulating protease activity even at lower concentrations, aligning with the emerging paradigm of using protease activators to induce bacterial cell death. Comparable findings in ClpP protease systems have shown that small molecules can bypass native chaperone regulation, leading to non-specific proteolysis and bacterial cytotoxicity [54].
NMA underscored the dynamic adaptability of the docked complexes, particularly in regions crucial for enzymatic activation. The reduced eigenvalues and enhanced flexibility observed in ligand-bound complexes suggest that the compounds facilitate conformational changes critical for HslV activation. This dynamic adaptability is consistent with the mechanism of activators like ADEPs, which induce allosteric transitions in ClpP, leading to unregulated proteolysis [55]. These insights highlight the compounds’ suitability for dynamic biological roles such as allosteric modulation and enzymatic activation, distinguishing them from the relatively static behavior of the natural C-tail peptide.
The ADMET analysis predicted the drug-like properties of the lead compounds, including compliance with Lipinski’s rule of five, high GI absorption, and favorable SA scores. The compounds exhibited low toxicity profiles, with SHS-1-61 categorized as non-toxic and the others as minimally harmful. Their metabolic stability, characterized by minimal inhibition of cytochrome P450 enzymes, further supports their potential as therapeutic agents. Similar studies on insulin degrading enzyme activators have demonstrated that optimizing physicochemical and pharmacokinetic properties can enhance the clinical applicability of protease activators [56].
The present study has made a remarkable discovery, as the identified HslV protease activators are significantly more potent than the previously identified ones [24]. This signifies a considerable advancement in the understanding of protease activation and opens up new possibilities for further exploration and development in this field. The allosteric activation mechanisms and enhanced protease dynamics observed in NMA align with similar findings in ClpP protease systems, suggesting a broader applicability of these compounds in protease modulation.
5 Conclusion
Our research builds upon previous findings identifying non-peptidic small molecules capable of activating the HslV protease in E. coli at micromolar concentrations. Using molecular docking analysis, we have identified additional promising candidates. Ligand interaction studies and simulations provided robust evidence for the compatibility of these compounds with the HslV-dimer’s C-tail cleft. Experimental assays confirmed their ability to bind and activate HslV via allosteric mechanisms, with significantly lower ED50 values compared to the HslU C-tail. These findings suggest that these compounds could serve as novel antimicrobial agents, leveraging uncontrolled proteolysis to target pathogenic microorganisms.
Acknowledgements
The authors greatly acknowledge and express their gratitude to the Researchers Supporting Project Number (RSP2025R462), King Saud University, Riyadh, Saudi Arabia. Furthermore, we would like to extend our utmost appreciation to Professor Robert T. Sauer from the Department of Biology at the Massachusetts Institute of Technology in the United States for his timely provision HslV gene containing the PET12b+ vector.
-
Funding information: We express our sincere gratitude to the Higher Education Commission of Pakistan for generously providing financial assistance through the National Research Program for Universities NRPU-2014 (Project No. 20-4102).
-
Author contributions: Conceptualization, S.A, M.A, S.S, Y.R, A.A, and T.A; methodology, S.A, M.A, S.S, Y.R, A.A, and T.A; software, S.A and Y.R; validation, A.A.S; formal analysis, A.F.A.; investigation, S.A, M.A, S.S, Y.R, A.A, and T.A; resources, M.A and A.A.S.; data curation, A.F.A.; writing – original draft preparation, T.A and Y.R.; writing – review and editing, S.A, M.A, S.S, Y.R, A.A, and T.A and U.S; visualization, A.F.A, A.S; supervision, T.A and Y.R.; project administration, A.A.S and M.A; funding acquisition, T.A.
-
Conflict of interest: The authors declare no conflict of interest.
-
Ethical approval: The conducted research is not related to either human or animal use.
-
Data availability statement: All data generated or analyzed during this study are included in this published article.
References
[1] Missiakas D, Schwager F, Betton JM, Georgopoulos C, Raina S. Identification and characterization of HsIV HsIU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15(24):6899–909. 10.1002/j.1460-2075.1996.tb01082.x.Search in Google Scholar
[2] Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386(6624):463–71. 10.1038/386463a0.Search in Google Scholar PubMed
[3] Bochtler M, Ditzel L, Groll M, Huber R. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc Natl Acad Sci USA. 1997;94(12):6070–4. 10.1073/pnas.94.12.6070.Search in Google Scholar PubMed PubMed Central
[4] Azim MK. Structure of prokaryotic HslVU protease-chaperone complex. Pak J Biochem Mol Biol. 2021;54(1–2):6–13.Search in Google Scholar
[5] Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease–chaperone complex. Cell. 2000;103(4):633–43. 10.1016/s0092-8674(00)00166-5.Search in Google Scholar PubMed
[6] Song HK, Bochtler M, Azim MK, Hartmann C, Huber R, Ramachandran R. Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV. Biophys Chem. 2002;100(1–3):437–52. 10.1016/s0301-4622(02)00297-1.Search in Google Scholar PubMed
[7] Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. 10.1101/gr.9.1.27.Search in Google Scholar
[8] Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HslU and the ATP-dependent protease HslU–HslV. Nature. 2000;403(6771):800–5. 10.1038/35001629.Search in Google Scholar PubMed
[9] Yoo SJ, Seol JH, Shin DH, Rohrwild M, Kang MS, Tanaka K, et al. Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem. 1996;271(24):14035–40. 10.1074/jbc.271.24.14035.Search in Google Scholar PubMed
[10] Seol JH, Yoo SJ, Shin DH, Shim YK, Kang MS, Goldberg AL, et al. The heat‐shock protein HslVU from Escherichia coli is a protein‐activated ATPase as well as an ATP‐dependent proteinase. Eur J Biochem. 1997;247(3):1143–50. 10.1111/j.1432-1033.1997.01143.x.Search in Google Scholar PubMed
[11] Azim MK, Goehring W, Song HK, Ramachandran R, Bochtler M, Goettig P. Characterization of the HslU chaperone affinity for HslV protease. Protein Sci. 2005;14(5):1357–62. 10.1110/ps.04970405.Search in Google Scholar PubMed PubMed Central
[12] Park E, Lee JW, Eom SH, Seol JH, Chung CH. Binding of MG132 or deletion of the Thr active sites in HslV subunits increases the affinity of HslV protease for HslU ATPase and makes this interaction nucleotide-independent. J Biol Chem. 2008;283(48):33258–66. 10.1074/jbc.M805411200.Search in Google Scholar PubMed PubMed Central
[13] Seong IS, Kang MS, Choi MK, Lee JW, Koh OJ, Wang J, et al. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J Biol Chem. 2002;277(29):25976–82. 10.1074/jbc.M202793200.Search in Google Scholar PubMed
[14] Ramachandran R, Hartmann C, Song HK, Huber R, Bochtler M. Functional interactions of HslV (ClpQ) with the ATPase HslU (ClpY). Proc Natl Acad Sci USA. 2002;99(11):7396–401. 10.1073/pnas.102188799.Search in Google Scholar PubMed PubMed Central
[15] Du RJ, Ho B. Surface localized heat shock protein 20 (HslV) of Helicobacter pylori. Helicobacter. 2003;8(4):257–67. 10.1046/j.1523-5378.2003.00153.x.Search in Google Scholar PubMed
[16] Frees D, Thomsen LE, Ingmer H. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch Microbiol. 2005;183:286–91. 10.1007/s00203-005-0773-x.Search in Google Scholar PubMed
[17] Ramasamy G, Gupta D, Mohmmed A, Chauhan VS. Characterization and localization of Plasmodium falciparum homolog of prokaryotic ClpQ/HslV protease. Mol Biochem Parasitol. 2007;152(2):139–48. 10.1016/j.molbiopara.2007.01.002.Search in Google Scholar PubMed
[18] Subramaniam S, Mohmmed A, Gupta D. Molecular modeling studies of the interaction between Plasmodium falciparum HslU and HslV subunits. J Biomol Struct Dyn. 2009;26(4):473–9. 10.1080/07391102.2009.10507262.Search in Google Scholar PubMed
[19] Tschan S, Kreidenweiss A, Stierhof YD, Sessler N, Fendel R, Mordmüller B. Mitochondrial localization of the threonine peptidase PfHslV, a ClpQ ortholog in Plasmodium falciparum. Int J Parasitol. 2010;40(13):1517–23. 10.1016/j.ijpara.2010.05.006.Search in Google Scholar PubMed
[20] Barboza NR, Cardoso J, de Paula Lima CV, Soares MJ, Gradia DF, Hangai NS, et al. Expression profile and subcellular localization of HslV, the proteasome-related protease from Trypanosoma cruzi. Exp Parasitol. 2012;130(2):171–7. 10.1016/j.exppara.2011.10.011.Search in Google Scholar PubMed
[21] Kebe NM, Samanta K, Singh P, Lai-Kee-Him J, Apicella V, Payrot N, et al. The HslV protease from Leishmania major and its activation by C-terminal HslU peptides. Int J Mol Sci. 2019;20(5):1021. 10.3390/ijms20051021.Search in Google Scholar PubMed PubMed Central
[22] Jeong S, Ahn J, Kwon AR, Ha NC. Cleavage-dependent activation of ATP-dependent protease HslUV from Staphylococcus aureus. Mol Cell. 2020;43(8):694–704. 10.14348/molcells.2020.0074.Search in Google Scholar PubMed PubMed Central
[23] Singh P, Samanta K, Kebe NM, Michel G, Legrand B, Sitnikova VE, et al. The C-terminal segment of Leishmania major HslU: Toward potential inhibitors of LmHslVU activity. Bioorg Chem. 2022;119:105539. 10.1016/j.bioorg.2021.105539.Search in Google Scholar PubMed
[24] Rashid Y, Azim MK, Saify ZS, Khan KM, Khan R. Small molecule activators of proteasome-related HslV peptidase. Bioorg Med Chem Lett. 2012;22(19):6089–94. 10.1016/j.bmcl.2012.08.033.Search in Google Scholar PubMed
[25] Aurangzeb S, Hamid M, Salar U, Rashid Y, Khan KM, Azim MK, et al. 3-Substituted coumarin derivatives over-activate the HslV protease: A potential drug target for antibacterial activity. Pak J Pharm Sci. 2022;35(4):1241–50. org/10.36721/PJPS.2022.35.4.SP.1243-1252.1.Search in Google Scholar
[26] Beena KP, Suresh R, Rajasekaran A, Manna PK. Dihydropyrimidinones – a versatile scaffold with diverse biological activity. J Pharm Sci Res. 2016;8(8):741.Search in Google Scholar
[27] Kapoor TM, Mayer TU, Desai A, Maddox P, Salmon ED, Schreiber SL, et al. Investigating the inhibition of bipolar spindle formation by monastrol, a small molecule kinesin inhibitor. Mol Biol Cell. 1999;10(1):128A.Search in Google Scholar
[28] Matos LH, Masson FT, Simeoni LA, Homem-de-Mello M. Biological activity of dihydropyrimidinone (DHPM) derivatives: A systematic review. Eur J Med Chem. 2018;143:1779–89. 10.1016/j.ejmech.2017.10.073.Search in Google Scholar PubMed
[29] Kaur R, Chaudhary S, Kumar K, Gupta MK, Rawal RK. Recent synthetic and medicinal perspectives of dihydropyrimidinones: A review. Eur J Med Chem. 2017;132:108–34. 10.1016/j.ejmech.2017.03.025.Search in Google Scholar PubMed PubMed Central
[30] Khasimbi S, Ali F, Manda K, Sharma A, Chauhan G, Wakode S. Dihydropyrimidinones scaffold as a promising nucleus for synthetic profile and various therapeutic targets: A review. Curr Org Synth. 2021;18(3):270–93. 10.2174/1570179417666201207215710.Search in Google Scholar PubMed
[31] Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. 10.1002/jcc.21334.Search in Google Scholar PubMed PubMed Central
[32] Csizmadia P. MarvinSketch and MarvinView: Molecule applets for the World Wide Web. In The 3rd International Electronic Conference on Synthetic Organic Chemistry; 1999 Nov; [online]. MDPI. Session: Information and Compound Archives Management and Internet Application. 10.3390/ecsoc-3-01775.Search in Google Scholar
[33] Dallakyan SJ. MGLTools. Reference Source; 2010.Search in Google Scholar
[34] Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–8. 10.1016/0263-7855(96)00018-5.Search in Google Scholar PubMed
[35] Visualizer DS. Accelrys software inc. Discovery Stud Visualizer. 2005;2.Search in Google Scholar
[36] Shamim S, Khan KM, Salar U, Ali F, Lodhi MA, Taha M, et al. 5-Acetyl-6-methyl-4-aryl-3,4-dihydropyrimidin-2(1H)-ones: As potent urease inhibitors; synthesis, in vitro screening, and molecular modeling study. Bioorg Chem. 2018;76:37–52. 10.1016/j.bioorg.2017.10.021.Search in Google Scholar PubMed
[37] Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9(9):646–52. 10.1038/nsb0902-646.Search in Google Scholar PubMed
[38] Meroueh S. Normal mode analysis: Theoretical and applications to biological and chemical systems. Brief Bioinform. 2007;8(5):378–9. 10.1093/bib/bbm010.Search in Google Scholar
[39] López-Blanco JR, Garzón JI, Chacón P. iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27(20):2843–50. 10.1093/bioinformatics/btr497.Search in Google Scholar PubMed
[40] López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):W271–6. 10.1093/nar/gku339.Search in Google Scholar PubMed PubMed Central
[41] López-Blanco JR, Reyes R, Aliaga JI, Badia RM, Chacon P, Quintana-Orti ES. Exploring large macromolecular functional motions on clusters of multicore processors. J Comput Phys. 2013;246:275–88. 10.1016/j.jcp.2013.03.032.Search in Google Scholar
[42] Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717. 10.1038/srep42717.Search in Google Scholar PubMed PubMed Central
[43] Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46(W1):W257–63. 10.1093/nar/gky318.Search in Google Scholar PubMed PubMed Central
[44] Eloff JN. A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Planta Med. 1998;64(8):711–3. 10.1055/s-2006-957563.Search in Google Scholar PubMed
[45] Bauer JA, Bauerová-Hlinková V. Normal mode analysis: A tool for better understanding protein flexibility and dynamics with application to homology models. In: Maia RT, Moraes Filho RM, Campos M, editors. Homology molecular modeling – perspectives and applications. 1st edn. IntechOpen; 2020. p. 1–9. 10.5772/intechopen.94139.Search in Google Scholar
[46] Lipinski CA. Avoiding investment in doomed drugs. Curr Drug Discov. 2001;1:17–9.Search in Google Scholar
[47] Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1(1):55–68. 10.1021/cc9800071.Search in Google Scholar PubMed
[48] Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–23. 10.1021/jm020017n.Search in Google Scholar PubMed
[49] Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Med Chem. 2000;43(21):3867–77. 10.1021/jm000292e.Search in Google Scholar PubMed
[50] Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem. 2001;44(12):1841–6. 10.1021/jm015507e.Search in Google Scholar PubMed
[51] Lynch T, Price AM. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician. 2007;76(3):391–6.Search in Google Scholar
[52] Sproule BA, Otton SV, Cheung SW, Zhong XH, Romach MK, Sellers EM. CYP2D6 inhibition in patients treated with sertraline. J Clin Psychopharmacol. 1997;17(2):102–6. 10.1097/00004714-199704000-00007.Search in Google Scholar PubMed
[53] Gersch M, Famulla K, Dahmen M, Göbl C, Malik I, Richter K, et al. AAA+ chaperones and acyldepsipeptides activate the ClpP protease via conformational control. Nat Commun. 2015;6(1):6320. 10.1038/ncomms7320.Search in Google Scholar PubMed
[54] Leung E, Datti A, Cossette M, Goodreid J, McCaw SE, Mah M, et al. Activators of cylindrical proteases as antimicrobials: Identification and development of small molecule activators of ClpP protease. Chem Biol. 2011;18(9):1167–78. 10.1016/j.chembiol.2011.07.023.Search in Google Scholar PubMed
[55] Kirstein J, Hoffmann A, Lilie H, Schmidt R, Rübsamen-Waigmann H, Brötz-Oesterhelt H, et al. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol Med. 2009;1(1):37–49. 10.1002/emmm.200900002.Search in Google Scholar PubMed PubMed Central
[56] Cabrol C, Huzarska MA, Dinolfo C, Rodriguez MC, Reinstatler L, Ni J, et al. Small-molecule activators of insulin-degrading enzyme discovered through high-throughput compound screening. PLoS ONE. 2009;4(4):e5274. 10.1371/journal.pone.0005274.Search in Google Scholar PubMed PubMed Central
© 2025 the author(s), published by De Gruyter
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Research Articles
- Phytochemical investigation and evaluation of antioxidant and antidiabetic activities in aqueous extracts of Cedrus atlantica
- Influence of B4C addition on the tribological properties of bronze matrix brake pad materials
- Discovery of the bacterial HslV protease activators as lead molecules with novel mode of action
- Characterization of volatile flavor compounds of cigar with different aging conditions by headspace–gas chromatography–ion mobility spectrometry
- Effective remediation of organic pollutant using Musa acuminata peel extract-assisted iron oxide nanoparticles
- Analysis and health risk assessment of toxic elements in traditional herbal tea infusions
- Cadmium exposure in marine crabs from Jiaxing City, China: Insights into health risk assessment
- Green-synthesized silver nanoparticles of Cinnamomum zeylanicum and their biological activities
- Tetraclinis articulata (Vahl) Mast., Mentha pulegium L., and Thymus zygis L. essential oils: Chemical composition, antioxidant and antifungal properties against postharvest fungal diseases of apple, and in vitro, in vivo, and in silico investigation
- Exploration of plant alkaloids as potential inhibitors of HIV–CD4 binding: Insight into comprehensive in silico approaches
- Recovery of phenylethyl alcohol from aqueous solution by batch adsorption
- Electrochemical approach for monitoring the catalytic action of immobilized catalase
- Green synthesis of ZIF-8 for selective adsorption of dyes in water purification
- Optimization of the conditions for the preparation of povidone iodine using the response surface methodology
- A case study on the influence of soil amendment on ginger oil’s physicochemical properties, mineral contents, microbial load, and HPLC determination of its vitamin level
- Removal of antiviral favipiravir from wastewater using biochar produced from hazelnut shells
- Effect of biochar and soil amendment on bacterial community composition in the root soil and fruit of tomato under greenhouse conditions
- Bioremediation of malachite green dye using Sargassum wightii seaweed and its biological and physicochemical characterization
- Evaluation of natural compounds as folate biosynthesis inhibitors in Mycobacterium leprae using docking, ADMET analysis, and molecular dynamics simulation
- Novel insecticidal properties of bioactive zoochemicals extracted from sea urchin Salmacis virgulata
- Elevational gradients shape total phenolic content and bioactive potential of sweet marjoram (Origanum majorana L.): A comparative study across altitudinal zones
- Study on the CO2 absorption performance of deep eutectic solvents formed by superbase DBN and weak acid diethylene glycol
- Preparation and wastewater treatment performance of zeolite-modified ecological concrete
- Multifunctional chitosan nanoparticles: Zn2+ adsorption, antimicrobial activity, and promotion of aquatic health
- Comparative analysis of nutritional composition and bioactive properties of Chlorella vulgaris and Arthrospira platensis: Implications for functional foods and dietary supplements
- Growth kinetics and mechanical characterization of boride layers formed on Ti6Al4V
- Enhancement of water absorption properties of potassium polyacrylate-based hydrogels in CaCl2-rich soils using potassium di- and tri-carboxylate salts
- Electrochemical and microbiological effects of dumpsite leachates on soil and air quality
- Modeling benzene physicochemical properties using Zagreb upsilon indices
- Characterization and ecological risk assessment of toxic metals in mangrove sediments near Langen Village in Tieshan Bay of Beibu Gulf, China
- Protective effect of Helicteres isora, an efficient candidate on hepatorenal toxicity and management of diabetes in animal models
- Valorization of Juglans regia L. (Walnut) green husk from Jordan: Analysis of fatty acids, phenolics, antioxidant, and cytotoxic activities
- Molecular docking and dynamics simulations of bioactive terpenes from Catharanthus roseus essential oil targeting breast cancer
- Selection of a dam site by using AHP and VIKOR: The Sakarya Basin
- Characterization and modeling of kidney bean shell biochar as adsorbent for caffeine removal from aquatic environments
- The effects of short-term and long-term 2100 MHz radiofrequency radiation on adult rat auditory brainstem response
- Biochemical insights into the anthelmintic and anti-inflammatory potential of sea cucumber extract: In vitro and in silico approaches
- Resveratrol-derived MDM2 inhibitors: Synthesis, characterization, and biological evaluation against MDM2 and HCT-116 cells
- Phytochemical constituents, in vitro antibacterial activity, and computational studies of Sudanese Musa acuminate Colla fruit peel hydro-ethanol extract
- Chemical composition of essential oils reviewed from the height of Cajuput (Melaleuca leucadendron) plantations in Buru Island and Seram Island, Maluku, Indonesia
- Phytochemical analysis and antioxidant activity of Azadirachta indica A. Juss from the Republic of Chad: in vitro and in silico studies
- Stability studies of titanium–carboxylate complexes: A multi-method computational approach
- Efficient adsorption performance of an alginate-based dental material for uranium(vi) removal
- Synthesis and characterization of the Co(ii), Ni(ii), and Cu(ii) complexes with a 1,2,4-triazine derivative ligand
- Evaluation of the impact of music on antioxidant mechanisms and survival in salt-stressed goldfish
- Optimization and validation of UPLC method for dapagliflozin and candesartan cilexetil in an on-demand formulation: Analytical quality by design approach
- Biomass-based cellulose hydroxyapatite nanocomposites for the efficient sequestration of dyes: Kinetics, response surface methodology optimization, and reusability
- Multifunctional nitrogen and boron co-doped carbon dots: A fluorescent probe for Hg2+ and biothiol detection with bioimaging and antifungal applications
- Separation of sulphonamides on a C12-diol mixed-mode HPLC column and investigation of their retention mechanism
- Characterization and antioxidant activity of pectin from lemon peels
- Fast PFAS determination in honey by direct probe electrospray ionization tandem mass spectrometry: A health risk assessment insight
- Correlation study between GC–MS analysis of cigarette aroma compounds and sensory evaluation
- Synthesis, biological evaluation, and molecular docking studies of substituted chromone-2-carboxamide derivatives as anti-breast cancer agents
- The influence of feed space velocity and pressure on the cold flow properties of diesel fuel
- Acid etching behavior and mechanism in acid solution of iron components in basalt fibers
- Protective effect of green synthesized nanoceria on retinal oxidative stress and inflammation in streptozotocin-induced diabetic rat
- Evaluation of the antianxiety activity of green zinc nanoparticles mediated by Boswellia thurifera in albino mice by following the plus maze and light and dark exploration tests
- Yeast as an efficient and eco-friendly bifunctional porogen for biomass-derived nitrogen-doped carbon catalysts in the oxygen reduction reaction
- Novel descriptors for the prediction of molecular properties
- 10.1515/chem-2025-0198
- Turmeric oil-fortified nutraceutical-SNEDDS: An approach to boost therapeutic effectiveness of dapagliflozin during treatment of diabetic patients
- Special Issue on Advancing Sustainable Chemistry for a Greener Future
- One-pot fabrication of highly porous morphology of ferric oxide-ferric oxychloride/poly-O-chloroaniline nanocomposite seeded on poly-1H pyrrole: Photocathode for green hydrogen generation from natural and artificial seawater
- High-efficiency photocathode for green hydrogen generation from sanitation water using bismuthyl chloride/poly-o-chlorobenzeneamine nanocomposite
- Special Issue on Phytochemicals, Biological and Toxicological Analysis of Plants
- Comparative analysis of fruit quality parameters and volatile compounds in commercially grown citrus cultivars
- Total phenolic, flavonoid, flavonol, and tannin contents as well as antioxidant and antiparasitic activities of aqueous methanol extract of Alhagi graecorum plant used in traditional medicine: Collected in Riyadh, Saudi Arabia
- Study on the pharmacological effects and active compounds of Apocynum venetum L.
- Chemical profile of Senna italica and Senna velutina seed and their pharmacological properties
- Essential oils from Brazilian plants: A literature analysis of anti-inflammatory and antimalarial properties and in silico validation
- Toxicological effects of green tea catechin extract on rat liver: Delineating safe and harmful doses
- Unlocking the potential of Trigonella foenum-graecum L. plant leaf extracts against diabetes-associated hypertension: A proof of concept by in silico studies
Articles in the same Issue
- Research Articles
- Phytochemical investigation and evaluation of antioxidant and antidiabetic activities in aqueous extracts of Cedrus atlantica
- Influence of B4C addition on the tribological properties of bronze matrix brake pad materials
- Discovery of the bacterial HslV protease activators as lead molecules with novel mode of action
- Characterization of volatile flavor compounds of cigar with different aging conditions by headspace–gas chromatography–ion mobility spectrometry
- Effective remediation of organic pollutant using Musa acuminata peel extract-assisted iron oxide nanoparticles
- Analysis and health risk assessment of toxic elements in traditional herbal tea infusions
- Cadmium exposure in marine crabs from Jiaxing City, China: Insights into health risk assessment
- Green-synthesized silver nanoparticles of Cinnamomum zeylanicum and their biological activities
- Tetraclinis articulata (Vahl) Mast., Mentha pulegium L., and Thymus zygis L. essential oils: Chemical composition, antioxidant and antifungal properties against postharvest fungal diseases of apple, and in vitro, in vivo, and in silico investigation
- Exploration of plant alkaloids as potential inhibitors of HIV–CD4 binding: Insight into comprehensive in silico approaches
- Recovery of phenylethyl alcohol from aqueous solution by batch adsorption
- Electrochemical approach for monitoring the catalytic action of immobilized catalase
- Green synthesis of ZIF-8 for selective adsorption of dyes in water purification
- Optimization of the conditions for the preparation of povidone iodine using the response surface methodology
- A case study on the influence of soil amendment on ginger oil’s physicochemical properties, mineral contents, microbial load, and HPLC determination of its vitamin level
- Removal of antiviral favipiravir from wastewater using biochar produced from hazelnut shells
- Effect of biochar and soil amendment on bacterial community composition in the root soil and fruit of tomato under greenhouse conditions
- Bioremediation of malachite green dye using Sargassum wightii seaweed and its biological and physicochemical characterization
- Evaluation of natural compounds as folate biosynthesis inhibitors in Mycobacterium leprae using docking, ADMET analysis, and molecular dynamics simulation
- Novel insecticidal properties of bioactive zoochemicals extracted from sea urchin Salmacis virgulata
- Elevational gradients shape total phenolic content and bioactive potential of sweet marjoram (Origanum majorana L.): A comparative study across altitudinal zones
- Study on the CO2 absorption performance of deep eutectic solvents formed by superbase DBN and weak acid diethylene glycol
- Preparation and wastewater treatment performance of zeolite-modified ecological concrete
- Multifunctional chitosan nanoparticles: Zn2+ adsorption, antimicrobial activity, and promotion of aquatic health
- Comparative analysis of nutritional composition and bioactive properties of Chlorella vulgaris and Arthrospira platensis: Implications for functional foods and dietary supplements
- Growth kinetics and mechanical characterization of boride layers formed on Ti6Al4V
- Enhancement of water absorption properties of potassium polyacrylate-based hydrogels in CaCl2-rich soils using potassium di- and tri-carboxylate salts
- Electrochemical and microbiological effects of dumpsite leachates on soil and air quality
- Modeling benzene physicochemical properties using Zagreb upsilon indices
- Characterization and ecological risk assessment of toxic metals in mangrove sediments near Langen Village in Tieshan Bay of Beibu Gulf, China
- Protective effect of Helicteres isora, an efficient candidate on hepatorenal toxicity and management of diabetes in animal models
- Valorization of Juglans regia L. (Walnut) green husk from Jordan: Analysis of fatty acids, phenolics, antioxidant, and cytotoxic activities
- Molecular docking and dynamics simulations of bioactive terpenes from Catharanthus roseus essential oil targeting breast cancer
- Selection of a dam site by using AHP and VIKOR: The Sakarya Basin
- Characterization and modeling of kidney bean shell biochar as adsorbent for caffeine removal from aquatic environments
- The effects of short-term and long-term 2100 MHz radiofrequency radiation on adult rat auditory brainstem response
- Biochemical insights into the anthelmintic and anti-inflammatory potential of sea cucumber extract: In vitro and in silico approaches
- Resveratrol-derived MDM2 inhibitors: Synthesis, characterization, and biological evaluation against MDM2 and HCT-116 cells
- Phytochemical constituents, in vitro antibacterial activity, and computational studies of Sudanese Musa acuminate Colla fruit peel hydro-ethanol extract
- Chemical composition of essential oils reviewed from the height of Cajuput (Melaleuca leucadendron) plantations in Buru Island and Seram Island, Maluku, Indonesia
- Phytochemical analysis and antioxidant activity of Azadirachta indica A. Juss from the Republic of Chad: in vitro and in silico studies
- Stability studies of titanium–carboxylate complexes: A multi-method computational approach
- Efficient adsorption performance of an alginate-based dental material for uranium(vi) removal
- Synthesis and characterization of the Co(ii), Ni(ii), and Cu(ii) complexes with a 1,2,4-triazine derivative ligand
- Evaluation of the impact of music on antioxidant mechanisms and survival in salt-stressed goldfish
- Optimization and validation of UPLC method for dapagliflozin and candesartan cilexetil in an on-demand formulation: Analytical quality by design approach
- Biomass-based cellulose hydroxyapatite nanocomposites for the efficient sequestration of dyes: Kinetics, response surface methodology optimization, and reusability
- Multifunctional nitrogen and boron co-doped carbon dots: A fluorescent probe for Hg2+ and biothiol detection with bioimaging and antifungal applications
- Separation of sulphonamides on a C12-diol mixed-mode HPLC column and investigation of their retention mechanism
- Characterization and antioxidant activity of pectin from lemon peels
- Fast PFAS determination in honey by direct probe electrospray ionization tandem mass spectrometry: A health risk assessment insight
- Correlation study between GC–MS analysis of cigarette aroma compounds and sensory evaluation
- Synthesis, biological evaluation, and molecular docking studies of substituted chromone-2-carboxamide derivatives as anti-breast cancer agents
- The influence of feed space velocity and pressure on the cold flow properties of diesel fuel
- Acid etching behavior and mechanism in acid solution of iron components in basalt fibers
- Protective effect of green synthesized nanoceria on retinal oxidative stress and inflammation in streptozotocin-induced diabetic rat
- Evaluation of the antianxiety activity of green zinc nanoparticles mediated by Boswellia thurifera in albino mice by following the plus maze and light and dark exploration tests
- Yeast as an efficient and eco-friendly bifunctional porogen for biomass-derived nitrogen-doped carbon catalysts in the oxygen reduction reaction
- Novel descriptors for the prediction of molecular properties
- 10.1515/chem-2025-0198
- Turmeric oil-fortified nutraceutical-SNEDDS: An approach to boost therapeutic effectiveness of dapagliflozin during treatment of diabetic patients
- Special Issue on Advancing Sustainable Chemistry for a Greener Future
- One-pot fabrication of highly porous morphology of ferric oxide-ferric oxychloride/poly-O-chloroaniline nanocomposite seeded on poly-1H pyrrole: Photocathode for green hydrogen generation from natural and artificial seawater
- High-efficiency photocathode for green hydrogen generation from sanitation water using bismuthyl chloride/poly-o-chlorobenzeneamine nanocomposite
- Special Issue on Phytochemicals, Biological and Toxicological Analysis of Plants
- Comparative analysis of fruit quality parameters and volatile compounds in commercially grown citrus cultivars
- Total phenolic, flavonoid, flavonol, and tannin contents as well as antioxidant and antiparasitic activities of aqueous methanol extract of Alhagi graecorum plant used in traditional medicine: Collected in Riyadh, Saudi Arabia
- Study on the pharmacological effects and active compounds of Apocynum venetum L.
- Chemical profile of Senna italica and Senna velutina seed and their pharmacological properties
- Essential oils from Brazilian plants: A literature analysis of anti-inflammatory and antimalarial properties and in silico validation
- Toxicological effects of green tea catechin extract on rat liver: Delineating safe and harmful doses
- Unlocking the potential of Trigonella foenum-graecum L. plant leaf extracts against diabetes-associated hypertension: A proof of concept by in silico studies

