Abstract
C12H8N2O5, triclinic, P1¯ (no. 2), a = 6.9215(2) Å, b = 8.0572(2) Å, c = 19.8378(6) Å, α = 98.657(1)°, β = 94.076(1)°, γ = 90.495(1)°, V = 1090.73(5) Å3, Z = 2, Rgt(F) = 0.0602, wRref(F2) = 0.1329, T = 293(2) K.
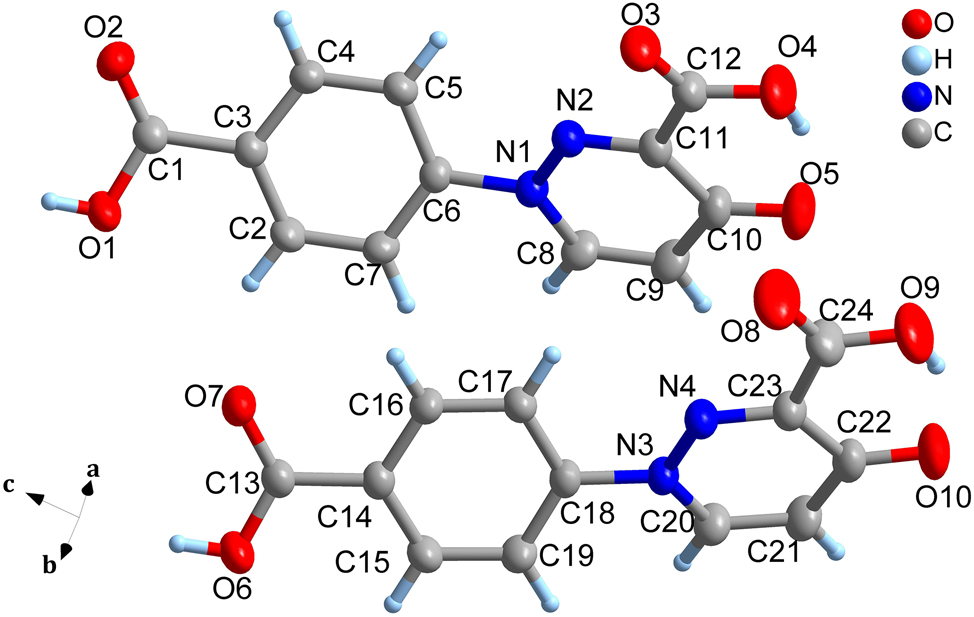
The molecular structure is shown in the figure. Table 1 contains the crystallographic data. The list of the atoms including atomic coordinates and displacement parameters can be found in the cif-file attached to this article.
Data collection and handling.
| Crystal: | Plate, clear light colourless |
| Size: | 0.8 × 0.8 × 0.3 mm |
| Wavelength: | MoKα radiation (0.71073 Å) |
| μ: | 0.13 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω-scans |
| θmax, completeness: | 28.3°, >99 % |
| N(hkl)measured, N(hkl)unique, Rint: | 30883, 5392, 0.063 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2σ(Iobs), 4,107 |
| N(param)refined: | 350 |
| Programs: | Bruker programs, 1 OLEX2, 2 SHELX 3 , 4 |
1 Source of material
The ethyl 1-(4-(ethoxycarbonyl)phenyl)-5-methyl-4-oxo-1,4-dihydropyridazine-3-carboxylate (0.4 mmol) and sodium hydroxide (0.5 mmol) were dissolved in 100 mL N,N-dimethylformamide, then added to a 250 mL flask and heated at 323 K for 2 h and cooled down to room temperature acidified by addition of 15 % HCl solution until pH = 2. The residue was thoroughly washed, carefully filtered, and subsequently dried, ultimately yielding the desired compound in the form of colorless block crystals.
2 Experimental details
Using Olex2, 2 the structure was solved with the ShelXT 3 structure solution program and refined with the ShelXL 4 refinement package.
3 Comment
Pyridazine derivatives constitute a particularly significant class of biologically active heterocycles. These compounds have garnered considerable attention as ligands due to their remarkable structural and synthetic versatility, precise tunability, and selectivity towards transition metal atoms. 5 Their diverse therapeutic potential, encompassing anticancer, 6 , 7 antidiabetic, 8 , 9 and antibiotic activities, 10 has been extensively documented. Leveraging this understanding, we have achieved the successful synthesis of a novel pyridazine carboxyl derivative, achieved by incorporating a benzene ring adorned with carboxyl groups onto the pyridazine ring. Simultaneously, numerous studies have reported the syntheses of transition metal complexes utilizing this derivative, specifically 1-(4-carboxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxylic acid, as a ligand and explored their properties. 11 , 12 , 13 , 14 , 15 , 16 However, despite these reports on complex formation and properties, the crystal structure of this pyridazine carboxylic acid ligand itself remains unreported to date.
Single-crystal X-ray diffraction analysis reveals that the asymmetric unit of the title compound contains two crystallographically independent molecules of 1-(4-carboxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxylic acid. These two molecules exhibit similar spatial configurations, differing primarily in the dihedral angles between their respective benzene and pyridazine rings, measured as 34.74° and 23.90°. The nitrogen-nitrogen bond lengths (N1–N2: 1.3306(16) Å; N3–N4: 1.3342(17) Å), as well as pertinent carbon–nitrogen bond lengths, exhibit intermediate values between those typical for N–N single bonds (1.45 Å) and N=N double bonds (1.25 Å). This observation indicates significant delocalization of p-electrons within the molecular framework. Finally, adjacent molecules are interconnected via intermolecular hydrogen bonding and π–π stacking interactions, collectively forming a three-dimensional supramolecular architecture.
-
Conflict of interest: The authors declare no conflicts of interest regarding this article.
-
Research funding: The work was supported by science and technology innovation project of colleges and universities in Shanxi Province (No. 2023L435).
-
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
References
1. BRUKER. SAINT, APEX2 and SADABS; Bruker AXS Inc.: Madison, Wisconsin, USA, 2009.Search in Google Scholar
2. Dolomanov, O. V.; Bourhis, L. J.; Gildea, R. J.; Howard, J. A. K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis program. J. Appl. Crystallogr. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
3. Sheldrick, G. M. SHELXTL – Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. 2015, A71, 3–8.10.1107/S2053273314026370Search in Google Scholar PubMed PubMed Central
4. Sheldrick, G. M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
5. Chen, S.; Zhang, M. H.; Feng, S.; Gong, C. Y.; Zhou, Y. X.; Xing, L.; He, B. C.; Wu, Y. J.; Xue, W. Design, Synthesis and Biological Activity of Chalcone Derivatives Containing Pyridazine. Arab. J. Chem. 2023, 16, 104852; https://doi.org/10.1016/j.arabjc.2023.104852.Search in Google Scholar
6. Jaballah, M. Y.; Serya, R. T.; Abouzid, K. Pyridazine Based Scaffolds as Privileged Structures in Anti-cancer Therapy. Drug Resist. Updat. 2017, 67, 138–148; https://doi.org/10.1055/s-0042-119992.Search in Google Scholar PubMed
7. Kota, T. V. R.; Gandham, H.; Sanasi, P. D. Synthesis, Characterization, and Antidiabetic Activity of 6-Methoxyimidazo[1,2-b] Pyridazine Derivatives. J. Chin. Chem. Soc. 2019, 66, 630–637; https://doi.org/10.1002/jccs.201800332.Search in Google Scholar
8. Tsuji, T.; Yamaguchi, M.; Kuroyanagi, J.; Furuzono, S.; Konishi, M.; Terayama, K.; Tanaka, J.; Saito, M.; Kobayashi, Y. Discovery of Novel Pyridazine Derivatives as Glucose Transporter Type 4 (GLUT4) Translocation Activators. Bioorg. Med. Chem. Lett. 2019, 29, 1785–1790; https://doi.org/10.1016/j.bmcl.2019.05.013.Search in Google Scholar PubMed
9. Singh, B.; Bhatia, R.; Pani, B.; Gupta, D. Synthesis, Crystal Structures and Biological Evaluation of New Pyridazine Derivatives. J. Mol. Struct. 2020, 1200, 127084; https://doi.org/10.1016/j.molstruc.2019.127084.Search in Google Scholar
10. Huang, P. P.; Wu, T. T.; Tuo, M. Q.; Ge, J.; Huang, P.; Wang, W. Q.; Yang, J. P.; Pan, H. B.; Lu, J. F. Supramolecular Complexes of Co(II), Zn(II) and Mn(II) Based on a Pyridazine Dicarboxylic Derivative: Synthesis, Crystal Structures and Properties. J. Mol. Struct. 2024, 1307, 138061; https://doi.org/10.1016/j.molstruc.2024.138061.Search in Google Scholar
11. Gao, J. H.; Wang, J. X.; Huang, P. P.; Liu, J.; Zheng, N.; Shi, J.; Xu, H. T.; Yue, S. Y.; Lu, J. F. A New Pyrazine Carboxyl Derivative and Its Two d10 Metal Coordination Polymers: Syntheses, Characterization, DFT and Property. J. Mol. Struct. 2023, 1290, 135935–135946; https://doi.org/10.1016/j.molstruc.2023.135935.Search in Google Scholar
12. Gao, J. H.; Huang, P. P.; Liu, J.; Zheng, N.; Wang, D.; Yue, S. Y.; Liu, E. N.; Liu, Q.; Liu, B.; Lu, J. F. Construction of Three Novel Co/Zn/Cd(II) Coordination Polymers Based on the Same Ligands: Different Spatial Structures, Electrocatalysis and Photoluminescence Properties. Arab. J. Chem. 2023, 16, 105314–105330; https://doi.org/10.1016/j.arabjc.2023.105314.Search in Google Scholar
13. Gao, J. H.; Huang, P. P.; Zhang, Z. J.; Tian, F. W.; Ge, J.; Cao, X. Y.; Liu, J.; Wang, D.; Zheng, N.; Lu, J. F.; Liu, B. A New 3D Cd-MOF with 2fold Interpenetrated as “Turn-On/Turn-Off” Fluorescent Sensor for Selective and Sensitive Detection of Cu2+, Al3+ and Fe3+ Ions. J. Mol. Struct. 2024, 1299, 137162–137171; https://doi.org/10.1016/j.molstruc.2023.137162.Search in Google Scholar
14. Pan, H. B.; Gao, J. H.; Huang, P. P.; Wang, J. X.; Wu, T. T.; Lu, J. F. Crystal Structure of Hexaaquazinc(II) catena-poly[Bis(1-(3-carboxyphenyl)-5-Methyl-4-Oxo-1,4-Dihydropyridazine-3-Carboxylato-κ2O,O′)-Bis(μ2-1-(3-carboxyphenyl)-5-methyl-4-oxo-1,4-dihydropyridazine-3-carboxylato-κ2O:O′)trizinc(II)] hexahydrate. Z. Kristallogr. – N. Cryst. Struct. 2024, 239, 159–161. https://doi.org/10.1515/ncrs-2023–039810.1515/ncrs-2023-0398Search in Google Scholar
15. Zhao, J.; Liu, M. L.; Guo, S. B.; Sun, M.; Zheng, J. L.; Ma, S. T.; Lu, J. F.; Ge, H. G. Two Cu(II) Complexes Constructed by Pyridazine Carboxyl Derivatives: Synthesis, Crystal Structure and Property. Inorg. Nano-Metal Chem. 2021, 239, 1952247; https://doi.org/10.1080/24701556.2021.1952247.Search in Google Scholar
16. Liu, W. J.; Wang, D. F.; Cao, X.; Wang, Q. W.; Shen, L. G. Crystal Structure of catena-poly[Triaqua-(μ2-1-(4-carboxylatophenyl)-4-oxo-1,4-dihydropyridazine-3-carboxylato-O,O′:O″)cobalt(II)], C12H12N2O8Co. Z. Kristallogr. – N. Cryst. Struct. 2024, 239, 121–123. https://doi.org/10.1515/ncrs-2023–0465.10.1515/ncrs-2023-0465Search in Google Scholar
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of (1Z, 2Z)-3-phenyl-2-propenal 2-(4-bromo-2-fluorophenyl)hydrazone, C15H12BrFN2
- Refinement of crystal structure of 2-(2,3-dihydro-3-oxo-1 H -inden-1-ylidene)-1 H -indene-1,3(2 H )-dione C18H10O3
- The crystal structure of 3-(1-fluoro-2-(naphthalen-2-yl)-2-oxoethyl)-2-methoxy-3,4-dihydroisoquinolin-1(2H)-one, C22H18FNO3
- Crystal structure of the dinuclear copper(II) complex bis(μ2-2,2′ -{[1,3-phenylenebis-(methylene)]bis(oxy)}dibenzoaot-κ4O,O′:O′′,O′′′)-bis(dimethylformamide-κ1O)dicopper(II), C50H44Cu2N2O14
- Crystal structure of poly[triaqua-(μ9-biphenyl-3,3′,5,5′-tetracarboxylic-κ8 O,O:O,O′: O,O″:O,O‴)samarium(III)sodium(I)], C16H12NaSmO11
- The crystal structure of 5-benzyl-1-(4-fluorobenzyl)-4-((4-fluorobenzyl)oxy)-1,5-dihydro-2H-pyrrol-2-one, C25H21F2NO2
- The crystal structure of diammonium 2,5-dihydroxyterephthalate, C8H12N2O6
- Crystal structure of (E)-4-(4-(1H-1,2,4-triazol-1-yl)benzylidene)-6,8-dimethoxy-3,4-dihydrobenzo[b]oxepin-5(2H)-one, C21H19N3O4
- Crystal structure of poly[oktakis(μ2-oxido-κ2O:O)-tetrakis(oxido-κ1O)-bis(μ2-1,1′-[1,4-phenylenebis(methylene)]di(1H-imidazole-κ2N:N′))-tetravanadium(V)-dizinc(II)] monohydrate, C28H30Zn2N8O13V4
- Crystal structure of acotiamide hydrochloride dimethylacetamide solvate (1/1), C25H40ClN5O6S
- Crystal structure of catena-poly[monoaqua (u2-(3-(3,5-dicarboxyphenyl)pyridine-2,6-dicarboxylic-k4O:O′:O″:N)zinc(II))] monohydrate, C15H11NO10Zn
- Crystal structure of dichlorido{2,6-bis(3,5-diisopropyl-N-pyrazolyl)pyridine}zinc(II), C23H33Cl2N5Zn
- Crystal structure of nitrato-κ2O,O′-[hydridotris(3,5-diethylpyrazol-1-yl)borato-κ3N,N′,N″]copper(II), C21H34BCuN7O3
- Crystal structure of 2,7-bis(3,5-diethyl-1H-pyrazol-4-yl)-benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, C28H26N6O4
- Crystal structure of 2-(4-chlorophenyl)benzothiazole, C13H8ClNS
- Synthesis and crystal structure (3R,4′S)-4′-(3,5-dibromophenyl)-1′-methyl-2H-dispiro [benzofuran-3,3′-pyrrolidine-2′,2″-indene]-1″,2,3″-trione, C26H17Br2NO4
- Crystal structure of bis(((3a,7a-dihydro-1H-benzo[d][1,2,3]triazol-1-yl)methyl) triphenylphosphonium) tetrachloridomanganate(II), C50H42Cl4MnN6P2
- The crystal structure of 4,9-bis(4-chlorophenyl)-1,6-bis(2-cyanobenzyl)-2,4a,5,6,7,7a-hexahydro-1H-2,7,5-(epiprop[2]ene[1,1,3]triyl)pyrrolo[3,4-b]pyridine-3,10-dicarbonitrile, C40H26Cl2N6
- The crystal structure of poly((μ2-3-(3-nitro-4-carboxylphenyl)benzoate-κ3O, O′:O″)-μ2-1,4-bis(1-imidazolyl)benzene-κ2N:N′-cadmium(II)), C26H17N5O6Cd
- The crystal structure of 6-hydroxy-5H-pyrrolo[3,4-b]pyridine-5,7(6H)-dione monohydrate, C7H6N2O4
- Crystal structure of 4-((cyclohexylsulfonyl)methyl)-1,2,3,4-tetrahydrobenzo [4,5]imidazo[1,2-a]pyridine, C18H24N2O2S
- Crystal structure of 4,7-diphenyl-1,10-phenanthroline-κ2N,N′)-bis(2,4-di(fluorine)-1-phenylpyridine-κ2C,N)-iridium(III) hexafluorophosphate–dichloromethane (1/1), C47H30Cl2F10IrN4P
- Crystal structure of (4-(1-phenyl-1H-benzo[d]imidazol-2-yl)phenyl)boronic acid, C19H15BN2O2
- The crystal structure of (E)-(2-((pyridin-2-ylmethylene)amino)phenyl)arsonic acid, C12H11AsN2O3
- The crystal structure of N(benzyl(phenyl)carbomothioyl)benzamide, C21H18N2OS
- The crystal structure of bis(2-picolinium) hexachlorostannate dichloromethane monosolvate, C13H18Cl8N2Sn
- Crystal structure of poly[tetraaqua-bis(μ4-3–1-(carboxylatomethyl)-1H-1,2,4-triazole-3-carboxylato)-κ4O:O′,O″,N)zinc(II)], C5H7N3O6Zn
- The crystal structure of the co-crystal isonicotinamide – 2-(nitrophenyl)methanol (1/1), C6H6N2O·C7H7NO3
- The crystal structure of 4-(3-carboxy-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinolin-7-yl)piperazin-1-ium 2-fluorobenzoate hydrate, C23H25F2N3O6
- Crystal structure of [diaqua-{1H-benzo[d]imidazol-3-ium-5,6-dicarboxylato-κ2O,O′}magnesium(II)] C18H14MgN4O10
- Crystal structure of (3-(dimethoxymethyl)-5-methoxy-1H-indol-1-yl) (2-iodo-5-methoxyphenyl)methanone, C20H20INO5
- The crystal structure of 3,7,11-trimethylbenzo[5,6][1,4]thiazino[2,3,4-kl]phenothiazine 5,5,9,9-tetraoxide, C21H17NO4S2
- Crystal structure of tris(piperazine-1,4-diium)bis(2-hydroxy-1,2,3-propane-tricarboxylate) pentahydrate, C24H56N6O19
- Crystal structure of 2-chloro-5-((5-isopropyl-2-methylphenoxy)methyl)pyridine, C16H18ClNO
- Crystal structure of (E)-4-(4-(1H-pyrrol-1-yl)benzylidene)-6,8-dimethoxy-3,4-dihydrobenzo[(b)]oxepin-5(2H)-one, C23H21NO4
- Crystal structure of (E)-N′-(3,4-dichlorobenzylidene)-3,4,5-trimethoxybenzohydrazide, C17H16Cl2N2O4
- The crystal structure of 2-(2-hydroxyphenyl)-3-(pyridin-2-yl)-2,3- dihydroquinazolin-4(1H)-one, C19H15N3O2
- Crystal structure of 5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-8-methylchroman-4-one, C17H16O5
- Crystal structure of bis[(3,4-dimethoxybenzyl)triphenylphosphonium]di-μ2-bromido-dibromidodicopper(I)
- Crystal structure of bis [(1,3-dioxolan-2-ylmethyl)triphenylphosphonium] tetrabromidodicopper(I), C22H22Br2CuO2P
- Crystal structure of [1-(4-carboxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxylic acid], C12H8N2O5
- The crystal structure of one-dimensional cooridnation polymer bis(thiocyanato)-bis((1E,2E)-1,2-bis(1-(pyridin-3-yl)ethylidene)-hydrazine κ2N:N)iron(II), (C30H28N10S2Fe)n
- Crystal structure of ((4-acetamidophenyl)sulfonyl)-l-alanine, C11H14N2O5S
- Crystal structure of [(1-naphthalen-1-yl-methyl)triphenylphosphonium] dichloridocopper(I), [C29H24P]+[CuCl2]−
- RbTm3S5: the first rubidium lanthanoid(III) sulfide with CsEr3Se5-type crystal structure
- Crystal structure of 2,2′-((ethane-1,2-diylbis(methylammoniumdiyl))bis(methylene))bis(pyridin-1-ium) diiodido-tris(μ2-iodido-κ2I:I)dicopper(II) chloride dihydrate, C16H30Cu2I6N4O2
- The crystal structure of 4-(trifluoromethyl)pyridine-2-carboxylic acid, C7H4F3NO2
- The crystal structure of (E)-2-ethoxy-1-methoxy-4-(2-(methylsulfonyl)vinyl)benzene, C12H16O4S
- Crystal structure of potassium 1H,1H,2H,2H-perfluorooctanesulfonate, C8H4O3F13SK
- Crystal structure of 4-(4-(quinolin-8-yloxy)-1,2,5-thiadiazol-3-yl)morpholine, C15H14O2N4S
- The crystal structure of 1,4-bis(bromomethyl)-2,5-dimethylbenzene, C10H12Br2
- The crystal structure of imidazo[4,5-e][1,3]diazepine-4,6,8-triamine methanol solvate, C7H11N7O
- The crystal structure of chlorido-bis(1,10-phenantroline-κ2N,N′)-(2-formylphenoxyacetato-κ2O,O) lead(II), C33H23N4O4ClPb
- Crystal structure of pyridinium tetrakis(1,1,1-trifluoro-2,4-pentadionato-κ2O,O′)yttrium(III) C20F12H16YO8C5H6N
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of (1Z, 2Z)-3-phenyl-2-propenal 2-(4-bromo-2-fluorophenyl)hydrazone, C15H12BrFN2
- Refinement of crystal structure of 2-(2,3-dihydro-3-oxo-1 H -inden-1-ylidene)-1 H -indene-1,3(2 H )-dione C18H10O3
- The crystal structure of 3-(1-fluoro-2-(naphthalen-2-yl)-2-oxoethyl)-2-methoxy-3,4-dihydroisoquinolin-1(2H)-one, C22H18FNO3
- Crystal structure of the dinuclear copper(II) complex bis(μ2-2,2′ -{[1,3-phenylenebis-(methylene)]bis(oxy)}dibenzoaot-κ4O,O′:O′′,O′′′)-bis(dimethylformamide-κ1O)dicopper(II), C50H44Cu2N2O14
- Crystal structure of poly[triaqua-(μ9-biphenyl-3,3′,5,5′-tetracarboxylic-κ8 O,O:O,O′: O,O″:O,O‴)samarium(III)sodium(I)], C16H12NaSmO11
- The crystal structure of 5-benzyl-1-(4-fluorobenzyl)-4-((4-fluorobenzyl)oxy)-1,5-dihydro-2H-pyrrol-2-one, C25H21F2NO2
- The crystal structure of diammonium 2,5-dihydroxyterephthalate, C8H12N2O6
- Crystal structure of (E)-4-(4-(1H-1,2,4-triazol-1-yl)benzylidene)-6,8-dimethoxy-3,4-dihydrobenzo[b]oxepin-5(2H)-one, C21H19N3O4
- Crystal structure of poly[oktakis(μ2-oxido-κ2O:O)-tetrakis(oxido-κ1O)-bis(μ2-1,1′-[1,4-phenylenebis(methylene)]di(1H-imidazole-κ2N:N′))-tetravanadium(V)-dizinc(II)] monohydrate, C28H30Zn2N8O13V4
- Crystal structure of acotiamide hydrochloride dimethylacetamide solvate (1/1), C25H40ClN5O6S
- Crystal structure of catena-poly[monoaqua (u2-(3-(3,5-dicarboxyphenyl)pyridine-2,6-dicarboxylic-k4O:O′:O″:N)zinc(II))] monohydrate, C15H11NO10Zn
- Crystal structure of dichlorido{2,6-bis(3,5-diisopropyl-N-pyrazolyl)pyridine}zinc(II), C23H33Cl2N5Zn
- Crystal structure of nitrato-κ2O,O′-[hydridotris(3,5-diethylpyrazol-1-yl)borato-κ3N,N′,N″]copper(II), C21H34BCuN7O3
- Crystal structure of 2,7-bis(3,5-diethyl-1H-pyrazol-4-yl)-benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, C28H26N6O4
- Crystal structure of 2-(4-chlorophenyl)benzothiazole, C13H8ClNS
- Synthesis and crystal structure (3R,4′S)-4′-(3,5-dibromophenyl)-1′-methyl-2H-dispiro [benzofuran-3,3′-pyrrolidine-2′,2″-indene]-1″,2,3″-trione, C26H17Br2NO4
- Crystal structure of bis(((3a,7a-dihydro-1H-benzo[d][1,2,3]triazol-1-yl)methyl) triphenylphosphonium) tetrachloridomanganate(II), C50H42Cl4MnN6P2
- The crystal structure of 4,9-bis(4-chlorophenyl)-1,6-bis(2-cyanobenzyl)-2,4a,5,6,7,7a-hexahydro-1H-2,7,5-(epiprop[2]ene[1,1,3]triyl)pyrrolo[3,4-b]pyridine-3,10-dicarbonitrile, C40H26Cl2N6
- The crystal structure of poly((μ2-3-(3-nitro-4-carboxylphenyl)benzoate-κ3O, O′:O″)-μ2-1,4-bis(1-imidazolyl)benzene-κ2N:N′-cadmium(II)), C26H17N5O6Cd
- The crystal structure of 6-hydroxy-5H-pyrrolo[3,4-b]pyridine-5,7(6H)-dione monohydrate, C7H6N2O4
- Crystal structure of 4-((cyclohexylsulfonyl)methyl)-1,2,3,4-tetrahydrobenzo [4,5]imidazo[1,2-a]pyridine, C18H24N2O2S
- Crystal structure of 4,7-diphenyl-1,10-phenanthroline-κ2N,N′)-bis(2,4-di(fluorine)-1-phenylpyridine-κ2C,N)-iridium(III) hexafluorophosphate–dichloromethane (1/1), C47H30Cl2F10IrN4P
- Crystal structure of (4-(1-phenyl-1H-benzo[d]imidazol-2-yl)phenyl)boronic acid, C19H15BN2O2
- The crystal structure of (E)-(2-((pyridin-2-ylmethylene)amino)phenyl)arsonic acid, C12H11AsN2O3
- The crystal structure of N(benzyl(phenyl)carbomothioyl)benzamide, C21H18N2OS
- The crystal structure of bis(2-picolinium) hexachlorostannate dichloromethane monosolvate, C13H18Cl8N2Sn
- Crystal structure of poly[tetraaqua-bis(μ4-3–1-(carboxylatomethyl)-1H-1,2,4-triazole-3-carboxylato)-κ4O:O′,O″,N)zinc(II)], C5H7N3O6Zn
- The crystal structure of the co-crystal isonicotinamide – 2-(nitrophenyl)methanol (1/1), C6H6N2O·C7H7NO3
- The crystal structure of 4-(3-carboxy-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinolin-7-yl)piperazin-1-ium 2-fluorobenzoate hydrate, C23H25F2N3O6
- Crystal structure of [diaqua-{1H-benzo[d]imidazol-3-ium-5,6-dicarboxylato-κ2O,O′}magnesium(II)] C18H14MgN4O10
- Crystal structure of (3-(dimethoxymethyl)-5-methoxy-1H-indol-1-yl) (2-iodo-5-methoxyphenyl)methanone, C20H20INO5
- The crystal structure of 3,7,11-trimethylbenzo[5,6][1,4]thiazino[2,3,4-kl]phenothiazine 5,5,9,9-tetraoxide, C21H17NO4S2
- Crystal structure of tris(piperazine-1,4-diium)bis(2-hydroxy-1,2,3-propane-tricarboxylate) pentahydrate, C24H56N6O19
- Crystal structure of 2-chloro-5-((5-isopropyl-2-methylphenoxy)methyl)pyridine, C16H18ClNO
- Crystal structure of (E)-4-(4-(1H-pyrrol-1-yl)benzylidene)-6,8-dimethoxy-3,4-dihydrobenzo[(b)]oxepin-5(2H)-one, C23H21NO4
- Crystal structure of (E)-N′-(3,4-dichlorobenzylidene)-3,4,5-trimethoxybenzohydrazide, C17H16Cl2N2O4
- The crystal structure of 2-(2-hydroxyphenyl)-3-(pyridin-2-yl)-2,3- dihydroquinazolin-4(1H)-one, C19H15N3O2
- Crystal structure of 5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-8-methylchroman-4-one, C17H16O5
- Crystal structure of bis[(3,4-dimethoxybenzyl)triphenylphosphonium]di-μ2-bromido-dibromidodicopper(I)
- Crystal structure of bis [(1,3-dioxolan-2-ylmethyl)triphenylphosphonium] tetrabromidodicopper(I), C22H22Br2CuO2P
- Crystal structure of [1-(4-carboxyphenyl)-4-oxo-1,4-dihydropyridazine-3-carboxylic acid], C12H8N2O5
- The crystal structure of one-dimensional cooridnation polymer bis(thiocyanato)-bis((1E,2E)-1,2-bis(1-(pyridin-3-yl)ethylidene)-hydrazine κ2N:N)iron(II), (C30H28N10S2Fe)n
- Crystal structure of ((4-acetamidophenyl)sulfonyl)-l-alanine, C11H14N2O5S
- Crystal structure of [(1-naphthalen-1-yl-methyl)triphenylphosphonium] dichloridocopper(I), [C29H24P]+[CuCl2]−
- RbTm3S5: the first rubidium lanthanoid(III) sulfide with CsEr3Se5-type crystal structure
- Crystal structure of 2,2′-((ethane-1,2-diylbis(methylammoniumdiyl))bis(methylene))bis(pyridin-1-ium) diiodido-tris(μ2-iodido-κ2I:I)dicopper(II) chloride dihydrate, C16H30Cu2I6N4O2
- The crystal structure of 4-(trifluoromethyl)pyridine-2-carboxylic acid, C7H4F3NO2
- The crystal structure of (E)-2-ethoxy-1-methoxy-4-(2-(methylsulfonyl)vinyl)benzene, C12H16O4S
- Crystal structure of potassium 1H,1H,2H,2H-perfluorooctanesulfonate, C8H4O3F13SK
- Crystal structure of 4-(4-(quinolin-8-yloxy)-1,2,5-thiadiazol-3-yl)morpholine, C15H14O2N4S
- The crystal structure of 1,4-bis(bromomethyl)-2,5-dimethylbenzene, C10H12Br2
- The crystal structure of imidazo[4,5-e][1,3]diazepine-4,6,8-triamine methanol solvate, C7H11N7O
- The crystal structure of chlorido-bis(1,10-phenantroline-κ2N,N′)-(2-formylphenoxyacetato-κ2O,O) lead(II), C33H23N4O4ClPb
- Crystal structure of pyridinium tetrakis(1,1,1-trifluoro-2,4-pentadionato-κ2O,O′)yttrium(III) C20F12H16YO8C5H6N


