Abstract
C32H30N8O5, orthorhombic, P212121 (no. 19), a = 9.1258(4) Å, b = 10.6225(5) Å, c = 30.1870(14) Å, V = 2926.3(2) Å3, Z = 4, Rgt(F) = 0.0367, wRref(F2) = 0.0837, T = 200 K.
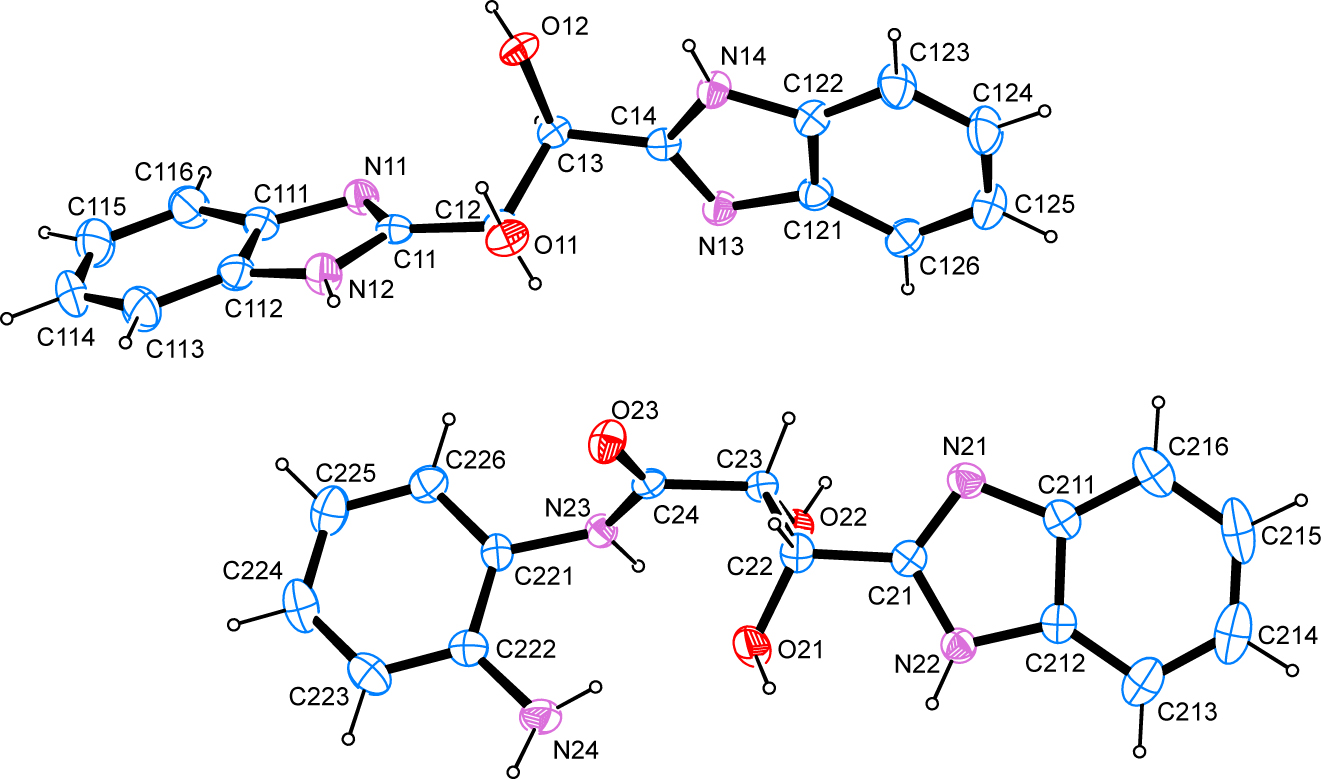
Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colourless rod |
| Size: | 0.55 × 0.15 × 0.06 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.10 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θmax, completeness: | 28.3°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 39064, 7280, 0.034 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 6175 |
| N(param)refined: | 446 |
| Programs: | Bruker [1], [2], SHELX [3], WinGX/ORTEP [4], Mercury [5], PLATON [6] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| O11 | 0.14620 (17) | 0.52437 (13) | 0.55431 (5) | 0.0254 (3) |
| O12 | −0.04913 (15) | 0.33833 (14) | 0.52593 (5) | 0.0221 (3) |
| O21 | 0.7834 (2) | 0.71613 (16) | 0.48936 (5) | 0.0356 (4) |
| O22 | 0.73500 (15) | 0.47932 (13) | 0.45153 (5) | 0.0201 (3) |
| O23 | 0.47180 (16) | 0.58894 (14) | 0.53054 (5) | 0.0278 (3) |
| N11 | 0.18880 (19) | 0.21491 (16) | 0.60449 (6) | 0.0222 (4) |
| N12 | 0.1950 (2) | 0.40536 (18) | 0.63450 (6) | 0.0273 (4) |
| N13 | 0.26775 (18) | 0.35960 (16) | 0.45259 (6) | 0.0216 (4) |
| N14 | 0.03971 (19) | 0.43346 (17) | 0.44705 (6) | 0.0221 (4) |
| N21 | 0.6082 (2) | 0.70821 (17) | 0.38000 (6) | 0.0254 (4) |
| N22 | 0.8329 (2) | 0.76985 (17) | 0.40019 (6) | 0.0228 (4) |
| N23 | 0.67444 (18) | 0.46668 (16) | 0.53706 (5) | 0.0198 (3) |
| N24 | 0.8680 (2) | 0.61172 (19) | 0.58518 (7) | 0.0300 (4) |
| C11 | 0.1880 (2) | 0.33647 (18) | 0.59689 (7) | 0.0202 (4) |
| C12 | 0.1913 (2) | 0.39755 (17) | 0.55193 (7) | 0.0198 (4) |
| H12 | 0.295697 | 0.397524 | 0.541840 | 0.024* |
| C13 | 0.1030 (2) | 0.32362 (18) | 0.51742 (6) | 0.0182 (4) |
| H13 | 0.129238 | 0.232391 | 0.519356 | 0.022* |
| C14 | 0.1389 (2) | 0.37158 (18) | 0.47207 (7) | 0.0189 (4) |
| C21 | 0.7007 (2) | 0.72366 (18) | 0.41284 (7) | 0.0200 (4) |
| C22 | 0.6637 (2) | 0.69591 (19) | 0.46038 (7) | 0.0227 (4) |
| H22 | 0.580261 | 0.750863 | 0.469710 | 0.027* |
| C23 | 0.6184 (2) | 0.55756 (17) | 0.46556 (6) | 0.0175 (4) |
| H23 | 0.529794 | 0.540544 | 0.446994 | 0.021* |
| C24 | 0.5809 (2) | 0.53774 (17) | 0.51421 (6) | 0.0178 (4) |
| C111 | 0.1984 (2) | 0.2035 (2) | 0.65061 (7) | 0.0231 (4) |
| C112 | 0.2029 (2) | 0.3229 (2) | 0.66990 (7) | 0.0263 (5) |
| C113 | 0.2149 (3) | 0.3402 (2) | 0.71540 (8) | 0.0379 (6) |
| H113 | 0.218860 | 0.421892 | 0.728172 | 0.045* |
| C114 | 0.2207 (3) | 0.2331 (3) | 0.74101 (8) | 0.0409 (6) |
| H114 | 0.230735 | 0.240989 | 0.772212 | 0.049* |
| C115 | 0.2123 (3) | 0.1138 (2) | 0.72231 (8) | 0.0404 (6) |
| H115 | 0.214259 | 0.042204 | 0.741147 | 0.049* |
| C116 | 0.2012 (3) | 0.0964 (2) | 0.67706 (8) | 0.0324 (5) |
| H116 | 0.195752 | 0.014530 | 0.664536 | 0.039* |
| C121 | 0.2502 (2) | 0.41740 (19) | 0.41123 (7) | 0.0221 (4) |
| C122 | 0.1071 (2) | 0.4637 (2) | 0.40741 (7) | 0.0233 (4) |
| C123 | 0.0568 (2) | 0.5225 (2) | 0.36916 (7) | 0.0317 (5) |
| H123 | −0.040979 | 0.552381 | 0.366800 | 0.038* |
| C124 | 0.1552 (3) | 0.5354 (2) | 0.33483 (8) | 0.0372 (6) |
| H124 | 0.124946 | 0.575539 | 0.308245 | 0.045* |
| C125 | 0.2991 (3) | 0.4905 (2) | 0.33839 (8) | 0.0360 (6) |
| H125 | 0.364626 | 0.501291 | 0.314226 | 0.043* |
| C126 | 0.3478 (2) | 0.4308 (2) | 0.37630 (7) | 0.0303 (5) |
| H126 | 0.445218 | 0.399878 | 0.378392 | 0.036* |
| C211 | 0.6842 (2) | 0.75012 (19) | 0.34284 (7) | 0.0243 (4) |
| C212 | 0.8252 (2) | 0.7899 (2) | 0.35496 (7) | 0.0239 (4) |
| C213 | 0.9257 (3) | 0.8358 (2) | 0.32441 (9) | 0.0387 (6) |
| H213 | 1.020769 | 0.862466 | 0.333108 | 0.046* |
| C214 | 0.8807 (4) | 0.8407 (3) | 0.28080 (9) | 0.0468 (7) |
| H214 | 0.946478 | 0.871479 | 0.258947 | 0.056* |
| C215 | 0.7421 (4) | 0.8020 (2) | 0.26809 (8) | 0.0460 (7) |
| H215 | 0.715366 | 0.806900 | 0.237728 | 0.055* |
| C216 | 0.6415 (3) | 0.7563 (2) | 0.29834 (8) | 0.0372 (6) |
| H216 | 0.546641 | 0.729935 | 0.289245 | 0.045* |
| C221 | 0.6754 (2) | 0.45380 (19) | 0.58432 (7) | 0.0207 (4) |
| C222 | 0.7771 (2) | 0.5266 (2) | 0.60795 (7) | 0.0247 (4) |
| C223 | 0.7911 (3) | 0.5056 (2) | 0.65315 (8) | 0.0339 (5) |
| H223 | 0.860700 | 0.552393 | 0.669779 | 0.041* |
| C224 | 0.7040 (3) | 0.4169 (2) | 0.67414 (8) | 0.0380 (6) |
| H224 | 0.715266 | 0.402953 | 0.705039 | 0.046* |
| C225 | 0.6012 (3) | 0.3484 (2) | 0.65080 (8) | 0.0350 (5) |
| H225 | 0.540528 | 0.289258 | 0.665631 | 0.042* |
| C226 | 0.5873 (2) | 0.3668 (2) | 0.60562 (7) | 0.0263 (5) |
| H226 | 0.517305 | 0.319681 | 0.589239 | 0.032* |
| H1A | 0.826 (3) | 0.642 (2) | 0.5593 (9) | 0.035 (7)* |
| H1B | 0.912 (3) | 0.675 (3) | 0.6043 (9) | 0.049 (8)* |
| H11A | 0.056 (3) | 0.526 (3) | 0.5610 (9) | 0.040 (8)* |
| H12A | 0.200 (3) | 0.485 (3) | 0.6350 (9) | 0.043 (8)* |
| H12B | −0.091 (3) | 0.270 (3) | 0.5320 (9) | 0.036 (7)* |
| H14A | −0.048 (3) | 0.449 (2) | 0.4546 (8) | 0.028 (6)* |
| H21A | 0.818 (4) | 0.788 (3) | 0.4879 (10) | 0.059 (10)* |
| H22A | 0.708 (3) | 0.422 (3) | 0.4327 (9) | 0.042 (8)* |
| H22B | 0.898 (3) | 0.801 (3) | 0.4180 (9) | 0.041 (8)* |
| H23A | 0.758 (3) | 0.440 (2) | 0.5230 (9) | 0.035 (7)* |
Source of material
Approximately 0.1 mol o-phenylene diamine and 0.1 mol tartaric acid were refluxed in 100 mL 4 M HCl in a round bottomed flask for 1 h. The flask was allowed to cool to room temperature and then the purple solution was neutralized with conc. NH3 solution. The brown precipitate was filtered off and recrystallised from a mixture of water and ethanol. Brown crystals were formed.
Experimental details
Carbon-bound H atoms were placed in calculated positions (C–H 0.95 Å for aromatic carbon atoms) and were included in the refinement in the riding model approximation, with U(H) set to 1.2Ueq(C). All oxygen- and nitrogen-bound H atoms were located on a difference Fourier map and refined freely.
Comment
Chelate ligands have found widespread use in coordination chemistry due to the increased stability of coordination compounds they can form in comparison to monodentate ligands. The stability of these compounds is enhanced further if the denticity of the ligand is increased upon incorporation of more and more potential donor sites [7]. Especially dicarboxylic acids are interesting in this aspect as chemical factors such as pH values could influence on the protonation/deprotonation of acidic functional groups and fine-tune the denticity of the resulting ligand species. Examples in this aspect are imidazole-dicarboxylic acids that can be synthesized according to a published procedure [8]. The structure of piperazine-1,4-diium bis(hydrogen 2-propyl-1H-imidazole-4,5-dicarboxylate) monohydrate has been reported earlier and confirms the validity of the general synthetic protocol [9]. In continuation of our studies of the structures of polyfunctional carboxylic acids [10], [11], [12], [13], we attempted the synthesis of a novel imidazole-dicarboxylic acid.
The structure solution shows a surprising result confirming that the synthesis reaction was interrupted too early for the complete conversion of the starting materials. The asymmetric unit contains two complete, neutral molecular species 1,2-bis(1H-benzo[d]imidazol-2-yl)ethane-1,2-diol and N-(2-aminophenyl)-3-(1H-benzo[d]imidazol-2-yl)-2,3-dihydroxypropanamide. The latter can be expected as a first step on the conversion pathway of the educts to the intended product. C–N bond lengths in the symmetric molecular entity clearly show the distinction between the imino and the amino-type nitrogen atom with values of 1.312(3) and 1.320(3) Å for the former and 1.352(3) and 1.350(3) Å for the latter with regards to the bond towards the carbon atom connected to both nitrogen atoms within each heterocyclic moiety. These values also tally with the corresponding bond lengths in the benzimidazole motif of the asymmetric second molecular entity present in the asymmetric unit of the crystal structures where values of 1.312(3) and 1.358(3) Å are apparent. In each case, these values are in good agreement with other benzimidazole derivatives whose molecular and crystal structures were determined by means of diffraction studies performed on single crystals and whose metrical parameters have been deposited with the Cambridge Structural Database. The C=O bond in the asymmetric benzimidazol moiety present in the crystal structure is found at 1.237(2) Å. The least-squares planes as defined by the respective non-hydrogen atoms of the two benzimidazole systems present in the 1,2-bis(2-benzimidazyl)-1,2,-dihydroxyethane molecule intersect at an angle of 75.04(7)° while the least-squares planes as defined by the respective non-hydrogen atoms of the benzimidazole moiety on the one hand and the non-hydrogen atoms of the aminophenyl moiety on the other hand in the asymmetric molecule present in the crystal structure intersect at an angle of 23.86(7)° only.
In the crystal, classical hydrogen bonds of the O–H⋯O, O–H⋯N, N–H⋯O and N–H⋯N type are present next to C–H⋯N contacts whose range falls by more than 0.1 Å below the sum of van-der-Waals radii of the atoms participating in them. All hydrogen atoms bonded to heteroatoms act as donors while all imino-type nitrogen atoms as well as all oxygen atoms act as acceptors. These classical hydrogen bonds are established between the two molecular entities of the title compound as well as symmetry-generated equivalents thereof. One intramolecular N–H⋯O hydrogen bond is formed by the amino group of the aminophenyl moiety. The C–H⋯N contact is supported by one of the hydrogen atoms in ortho position to the heterocyclic five-membered ring in the 1,1,2-bis(1H-benzo[d]imidazol-2-yl)ethane-1,2-diol molecule. In terms of graph-set analysis [14], the descriptor for these contacts is S(9)DDDDC11(5)C11(6) C11(7)C11(9)C11(10). In total, the entities present in the asymmetric unit are connected to sheets perpendicular to the crystallographic c axis. Furthermore, a number of N–H⋯π as well as C–H⋯π interactions could be discussed whose metrical details have been tabulated.
Funding source: National Research Foundation
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Researchfunding: CRC thanks the National Research Foundation for financial support in connection with the Thuthuka funding initiative.
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Bruker. APEX2; Bruker AXS Inc.: Madison, Wisconsin, USA, 2012.Search in Google Scholar
2. Bruker. SADABS; Bruker AXS Inc.: Madison, Wisconsin, USA, 2008.Search in Google Scholar
3. Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122; https://doi.org/10.1107/s0108767307043930.Search in Google Scholar PubMed
4. Farrugia, L. J. WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854; https://doi.org/10.1107/s0021889812029111.Search in Google Scholar
5. Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J., Wood, P. A. Mercury CSD 2.0 – new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470; https://doi.org/10.1107/s0021889807067908.Search in Google Scholar
6. Spek, A. L. Structure validation in chemical crystallography. Acta Crystallogr. 2009, D65, 148–155; https://doi.org/10.1107/s090744490804362x.Search in Google Scholar
7. Gade, L. H. Koordinationschemie, 1. Auflage; Wiley-VCH: Weinheim, 1998.10.1002/9783527663927Search in Google Scholar
8. Zheng, S. R., Cai, S. L., Pan, M., Fan, J., Xiao, T. T., Zhang, W. G. The construction of coordination networks based on imidazole-based dicarboxylate ligand containing hydroxymethyl group. CrystEngComm 2011, 13, 883–888; https://doi.org/10.1039/c0ce00369g.Search in Google Scholar
9. Gao, Z. Q., Gu, J. Z. Piperazine-1,4-diium bis(hydrogen 2-propyl-1H-imidazole-4,5-dicarboxylate) monohydrate. Acta Crystallogr. 2011, E67, o31; https://doi.org/10.1107/s1600536810049822.Search in Google Scholar
10. Betz, R., Klüfers, P. 1-Hydroxycyclopropane-1-carboxylic acid. Acta Crystallogr. 2007, E63, o3891; https://doi.org/10.1107/s1600536807041256.Search in Google Scholar
11. Betz, R., Klüfers, P. 1-Hydroxycyclobutane-1-carboxylic acid. Acta Crystallogr. 2007, E63, o4032; https://doi.org/10.1107/s1600536807043747.Search in Google Scholar
12. Betz, R., Klüfers, P. 1-Hydroxycyclopentane-1-carboxylic acid. Acta Crystallogr. 2007, E63, o3932; https://doi.org/10.1107/s1600536807042183.Search in Google Scholar
13. Betz, R., Gerber, T. I. A., Schalekamp, H. 2-Hydroxy-6-isopropyl-3-methylbenzoic acid. Acta Crystallogr. 2011, E67, o1063; https://doi.org/10.1107/s1600536811011998.Search in Google Scholar
14. Bernstein, J., Davis, R. E., Shimoni, L., Chang, N.-L. Patterns in hydrogen bonding: functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573; https://doi.org/10.1002/anie.199515551.Search in Google Scholar
© 2020 Candyce R. Clark et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of 4-hydroxybenzene-1,3-diaminium dichloride, C6H10Cl2N2O
- The crystal structure of 3-chloropropylammonium chloride, C3H9Cl2N
- The crystal structure of 1-chloro-2-(dimethylamino)ethane hydrochloride, C4H11Cl2N
- Crystal structure of N-(2-(trifluoromethyl)phenyl)hexanamide, C13H16F3NO
- Redetermination of the crystal structure of para-toluidine, C7H9N
- The crystal structure of bis(1,3-dihydroxy-2-methylpropan-2-aminium) carbonate, C9H24N2O7
- The crystal structure of 4-chloro-1-methylpiperidin-1-ium chloride, C6H13Cl2N
- Crystal structure of (Z)-3-(6-bromo-1H-indol-3-yl)-1,3-diphenylprop-2-en-1-one, C23H16BrNO
- The crystal structure of ethyl 2-amino-4-(3,5-difluorophenyl)-7,7-dimethyl-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carboxylate, C20H21F2NO4
- Crystal structure of 6,6'‐((1E,1'E)‐(propane‐1,3‐diylbis(azaneylylidene))bis(methaneylylidene))bis(3‐bromophenol), C34H32Br4N4O4
- The crystal structure of (E)-2-(2-((2-picolinoylhydrazono)methyl)phenoxy)acetic acid dihydrate, C15H17N3O6
- Crystal structure of (E)-4-bromo-N′-(3-chloro-2-hydroxybenzylidene)benzohydrazide, C14H10BrClN2O2
- Crystal structure of N,N′-bis(4-bromosalicylidene) ethylene-1,2-diaminopropan, C34H32Br4N4O4
- Crystal structure of 4-bromo-N′-[(3-bromo-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H14Br2N2O3
- The crystal structure of 1,2-bis(1H-benzo[d]imidazol-2-yl)ethane-1,2-diol — N-(2-aminophenyl)-3-(1H-benzo[d]imidazol-2-yl)-2,3-dihydroxypropanamide (1/1), C32H30N8O5
- The crystal structure of para-trifluoromethyl-aniline hemihydrate, C14H14F6N2O
- Redetermination of the crystal structure of 2-amino-2-methyl-propane-1,3-diole, C4H11NO2
- The crystal structure of methacholine chloride, C8H18ClNO2
- Crystal structure of 5,7,7-trimethyl-4,6,7,8-tetrahydrocyclopenta[g]isochromen-1(3H)-one, C15H18O2
- Crystal structure of poly[diammine-bis(μ4-4-hydroxypyridine-3-sulfonato-κ5N:O, O′:O′′:O′′)(μ2-pyrazinyl-κ2N:N′)tetrasilver(I)], C7H8Ag2N3O4S
- Crystal structure of ethyl (E)-5-(((3′,6′-bis(ethylamino)-3-oxospiro[isoindoline-1,9′-xanthen]-2-yl)imino)methyl)-2,4-dimethyl-1H-pyrrole-3-carboxylate — ethanol (1/1), C38H45N5O5
- Crystal structure of 4-bromo-N′-[(3-chloro-2-hydroxyphenyl)methylidene]benzohydrazide, C14H7Br2N2O2
- Redetermination of the crystal structure of 3,3,3-triphenylpropanoic acid, C21H18O2 – Deposition of hydrogen atomic coordinates
- Structure redetermination of dextromethorphan hydrobromide monohydrate, C18H28BrNO2 – localization of hydrogen atoms
- Crystal structure of tris(azido-κ1N)-(N-(2-aminoethyl)-N-methyl-1,3-propanediamine-κ3N,N′,N′′)cobalt(III), C7H19CoN12
- Crystal structure of tetraaqua-bis(1H-indazole-6-carboxylate-κN)cadmium (II), C16H18CdN4O8
- Crystal structure of dichloride-bis(1-propylimidazole-κ1N)zinc(II), C12H20Cl2N4Zn
- Crystal structure of (E)-resveratrol 3-O-β-D-xylopyranoside, C19H22O8
- Crystal structure of 3,3′-(1,2-phenylene-bis(methylene))bis(1-vinyl- 1H-imidazol-3-ium) bis(hexafluoro phosphate)(V), C18H20F12N4P2
- Crystal structure of diaqua[bis(benzimidazol-2-yl-methyl)amine-κ3N,N′,N″]-phthalato-κ1O-nickel(II)-methanol (1/2), C26H31N5NiO8
- Crystal structure of 6,7-difluoro-1-methyl-3-(trifluoromethyl)quinoxalin-2(1H)-one, C10H5F5N2O
- Crystal structure of dichlorido-bis(1-hexyl-1H-benzotriazole-k1N)zinc(II), C24H34N6Cl2Zn
- The crystal structre of 2-(4-bromophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H12BBrN2
- Crystal structure of diethyl 3,9-bis(4-fluorophenyl)-6,12-diphenyl-3,9-diazapentacyclo[6.4.0.02,7.04,11]dodecane-1,5-dicarboxylate, C40H36F2N2O4
- Crystal structure of (E)-7-methoxy-2-((5-methoxypyridin-3-yl)methylene)-3,4- dihydronaphthalen-1(2H)-one, C18H17NO3
- Crystal structure of (E)-2-chloro-6-(((1,3-dihydroxy-2-(oxidomethyl)propan-2-yl)imino)methyl)phenolate-κ3N,O,O’)manganese(IV), C22H24Cl2MnN2O8
- The crystal structure of α-(meta-methoxyphenoxy)-ortho-tolylic acid, C15H14O4
- The crystal structure of N-(2-chloroethyl)-N,N-diethylammonium chloride, C6H15Cl2N
- The crystal structure of tris(2,3,4,6,7,8,9,10-octahydro-1H-pyrimido[1,2-a]azepin-5-ium) trihydrodecavanadate(V), C27H54N6O28V10
- Crystal structure of 1,3-bis(octyl)benzimidazolium perchlorate C23H39ClN2O4
- Crystal structure of tetrakis[(Z)-(2-(1-(furan-2-yl)-2-methylpropylidene)-1-phenylhydrazin-1-ido-κ2N,N′)] zirconium(IV), C56H60N8O4Zr
- The crystal structure of 2-(naphthalen-2-yloxy)-4-phenyl-6-(prop-2-yn-1-yloxy)-1,3,5-triazine, C22H15N3O2
- The crystal structure of trimethylsulfonium tris(trifluoromethylsulfonyl)methanide, C7H9F9O6S4
- Crystal structure of 4-bromo-N′-[3,5-dichloro-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H13BrCl2N2O3
- The crystal structure of 4-(4-bromophenyl)-2-(3-(4-bromophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)thiazole, C24H16Br2FN3S
- The crystal structure of N-(adamantan-1-yl)-piperidine-1-carbothioamide, C16H26N2S
- The crystal structure of 1-phenyl-N-(4,5,6,7-tetrabromo-1,3-dioxoisoindolin-2-yl)-5-(thiophen-2-yl)-1H-pyrazole-3-carboxamide-dimethylformamide (1/1) C22H10Br4N4O3S
- The crystal structure of benzeneseleninic acid anhydride, C12H10O3Se2
- The crystal structure of diphenyalmine hydrochloride antimony trichloride co-crystallizate, C12H12Cl4NSb – Localization of hydrogen atoms
- The crystal structure of para-nitrobenzylbromide, C7H6BrNO2 – A second polymorph and correction of 3D coordinates
- Crystal structure of catena-poly[(5H-pyrrolo[3,2-b:4,5-b′]dipyridine-κ2N,N′)-(μ4-hexaoxidodivanadato)dizinc(II)],C10H9N3O6V2Zn
- Crystal structure of N,N′-(2-hydroxypropane-1,3-diyl)bis(pyridine-2-aldimine)-κ5N,N′,N′′,N′′′,O]-tris(nitrato-κ2O,O′) cerium(III), C15H16CeN7O10
- Synthesis and crystal structure of oktakis(dimethylsulphoxide-κ1O)gadolinium(III) [tetrabromido-μ2-bromido-μ2-sulfido-di-μ3-sulfido-μ4-sulfido-tetracopper(I)-tungsten(VI)], C16H48O8S12Br5Cu4GdW
- Crystal structure of {tris((1H-benzo[d]imidazol-2- yl)methyl)amine-κ4N,N′,N′′,N′′′}-(succinato-κ2O,O′)nickel(II) – methanol (1/4), C32H41N7NiO8
- Crystal structure of catena-poly[trans-tetraaqua(μ2-1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol)-k2N:N′)cobalt(II)] dinitrate – 1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol) – water (1/3/2), C72H68CoN18O12
- Crystal structure of bis(μ2-2-oxido-2-phenylacetate-κ3O:O,O′)-bis(1-isopropoxy-2-oxo-2-phenylethan-1-olato-κ2O,O′)-bis(propan-2-olato-κ1O)dititanium(IV), C44H52O14Ti2
- The crystal structure of 5-carboxy-2-(hydroxymethyl)-1H-imidazol-3-ium-4-carboxylate, C6H8N2O6
- The crystal structure of 2,6-dibromo-4-fluoroaniline, C6H4Br2FN
- The crystal structure of 4-chloro-N-(2-phenoxyphenyl)benzamide, C19H14ClNO2
- The crystal structure of 2-methyl-β-naphthothiazole, C12H9NS
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of 4-hydroxybenzene-1,3-diaminium dichloride, C6H10Cl2N2O
- The crystal structure of 3-chloropropylammonium chloride, C3H9Cl2N
- The crystal structure of 1-chloro-2-(dimethylamino)ethane hydrochloride, C4H11Cl2N
- Crystal structure of N-(2-(trifluoromethyl)phenyl)hexanamide, C13H16F3NO
- Redetermination of the crystal structure of para-toluidine, C7H9N
- The crystal structure of bis(1,3-dihydroxy-2-methylpropan-2-aminium) carbonate, C9H24N2O7
- The crystal structure of 4-chloro-1-methylpiperidin-1-ium chloride, C6H13Cl2N
- Crystal structure of (Z)-3-(6-bromo-1H-indol-3-yl)-1,3-diphenylprop-2-en-1-one, C23H16BrNO
- The crystal structure of ethyl 2-amino-4-(3,5-difluorophenyl)-7,7-dimethyl-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carboxylate, C20H21F2NO4
- Crystal structure of 6,6'‐((1E,1'E)‐(propane‐1,3‐diylbis(azaneylylidene))bis(methaneylylidene))bis(3‐bromophenol), C34H32Br4N4O4
- The crystal structure of (E)-2-(2-((2-picolinoylhydrazono)methyl)phenoxy)acetic acid dihydrate, C15H17N3O6
- Crystal structure of (E)-4-bromo-N′-(3-chloro-2-hydroxybenzylidene)benzohydrazide, C14H10BrClN2O2
- Crystal structure of N,N′-bis(4-bromosalicylidene) ethylene-1,2-diaminopropan, C34H32Br4N4O4
- Crystal structure of 4-bromo-N′-[(3-bromo-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H14Br2N2O3
- The crystal structure of 1,2-bis(1H-benzo[d]imidazol-2-yl)ethane-1,2-diol — N-(2-aminophenyl)-3-(1H-benzo[d]imidazol-2-yl)-2,3-dihydroxypropanamide (1/1), C32H30N8O5
- The crystal structure of para-trifluoromethyl-aniline hemihydrate, C14H14F6N2O
- Redetermination of the crystal structure of 2-amino-2-methyl-propane-1,3-diole, C4H11NO2
- The crystal structure of methacholine chloride, C8H18ClNO2
- Crystal structure of 5,7,7-trimethyl-4,6,7,8-tetrahydrocyclopenta[g]isochromen-1(3H)-one, C15H18O2
- Crystal structure of poly[diammine-bis(μ4-4-hydroxypyridine-3-sulfonato-κ5N:O, O′:O′′:O′′)(μ2-pyrazinyl-κ2N:N′)tetrasilver(I)], C7H8Ag2N3O4S
- Crystal structure of ethyl (E)-5-(((3′,6′-bis(ethylamino)-3-oxospiro[isoindoline-1,9′-xanthen]-2-yl)imino)methyl)-2,4-dimethyl-1H-pyrrole-3-carboxylate — ethanol (1/1), C38H45N5O5
- Crystal structure of 4-bromo-N′-[(3-chloro-2-hydroxyphenyl)methylidene]benzohydrazide, C14H7Br2N2O2
- Redetermination of the crystal structure of 3,3,3-triphenylpropanoic acid, C21H18O2 – Deposition of hydrogen atomic coordinates
- Structure redetermination of dextromethorphan hydrobromide monohydrate, C18H28BrNO2 – localization of hydrogen atoms
- Crystal structure of tris(azido-κ1N)-(N-(2-aminoethyl)-N-methyl-1,3-propanediamine-κ3N,N′,N′′)cobalt(III), C7H19CoN12
- Crystal structure of tetraaqua-bis(1H-indazole-6-carboxylate-κN)cadmium (II), C16H18CdN4O8
- Crystal structure of dichloride-bis(1-propylimidazole-κ1N)zinc(II), C12H20Cl2N4Zn
- Crystal structure of (E)-resveratrol 3-O-β-D-xylopyranoside, C19H22O8
- Crystal structure of 3,3′-(1,2-phenylene-bis(methylene))bis(1-vinyl- 1H-imidazol-3-ium) bis(hexafluoro phosphate)(V), C18H20F12N4P2
- Crystal structure of diaqua[bis(benzimidazol-2-yl-methyl)amine-κ3N,N′,N″]-phthalato-κ1O-nickel(II)-methanol (1/2), C26H31N5NiO8
- Crystal structure of 6,7-difluoro-1-methyl-3-(trifluoromethyl)quinoxalin-2(1H)-one, C10H5F5N2O
- Crystal structure of dichlorido-bis(1-hexyl-1H-benzotriazole-k1N)zinc(II), C24H34N6Cl2Zn
- The crystal structre of 2-(4-bromophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H12BBrN2
- Crystal structure of diethyl 3,9-bis(4-fluorophenyl)-6,12-diphenyl-3,9-diazapentacyclo[6.4.0.02,7.04,11]dodecane-1,5-dicarboxylate, C40H36F2N2O4
- Crystal structure of (E)-7-methoxy-2-((5-methoxypyridin-3-yl)methylene)-3,4- dihydronaphthalen-1(2H)-one, C18H17NO3
- Crystal structure of (E)-2-chloro-6-(((1,3-dihydroxy-2-(oxidomethyl)propan-2-yl)imino)methyl)phenolate-κ3N,O,O’)manganese(IV), C22H24Cl2MnN2O8
- The crystal structure of α-(meta-methoxyphenoxy)-ortho-tolylic acid, C15H14O4
- The crystal structure of N-(2-chloroethyl)-N,N-diethylammonium chloride, C6H15Cl2N
- The crystal structure of tris(2,3,4,6,7,8,9,10-octahydro-1H-pyrimido[1,2-a]azepin-5-ium) trihydrodecavanadate(V), C27H54N6O28V10
- Crystal structure of 1,3-bis(octyl)benzimidazolium perchlorate C23H39ClN2O4
- Crystal structure of tetrakis[(Z)-(2-(1-(furan-2-yl)-2-methylpropylidene)-1-phenylhydrazin-1-ido-κ2N,N′)] zirconium(IV), C56H60N8O4Zr
- The crystal structure of 2-(naphthalen-2-yloxy)-4-phenyl-6-(prop-2-yn-1-yloxy)-1,3,5-triazine, C22H15N3O2
- The crystal structure of trimethylsulfonium tris(trifluoromethylsulfonyl)methanide, C7H9F9O6S4
- Crystal structure of 4-bromo-N′-[3,5-dichloro-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H13BrCl2N2O3
- The crystal structure of 4-(4-bromophenyl)-2-(3-(4-bromophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)thiazole, C24H16Br2FN3S
- The crystal structure of N-(adamantan-1-yl)-piperidine-1-carbothioamide, C16H26N2S
- The crystal structure of 1-phenyl-N-(4,5,6,7-tetrabromo-1,3-dioxoisoindolin-2-yl)-5-(thiophen-2-yl)-1H-pyrazole-3-carboxamide-dimethylformamide (1/1) C22H10Br4N4O3S
- The crystal structure of benzeneseleninic acid anhydride, C12H10O3Se2
- The crystal structure of diphenyalmine hydrochloride antimony trichloride co-crystallizate, C12H12Cl4NSb – Localization of hydrogen atoms
- The crystal structure of para-nitrobenzylbromide, C7H6BrNO2 – A second polymorph and correction of 3D coordinates
- Crystal structure of catena-poly[(5H-pyrrolo[3,2-b:4,5-b′]dipyridine-κ2N,N′)-(μ4-hexaoxidodivanadato)dizinc(II)],C10H9N3O6V2Zn
- Crystal structure of N,N′-(2-hydroxypropane-1,3-diyl)bis(pyridine-2-aldimine)-κ5N,N′,N′′,N′′′,O]-tris(nitrato-κ2O,O′) cerium(III), C15H16CeN7O10
- Synthesis and crystal structure of oktakis(dimethylsulphoxide-κ1O)gadolinium(III) [tetrabromido-μ2-bromido-μ2-sulfido-di-μ3-sulfido-μ4-sulfido-tetracopper(I)-tungsten(VI)], C16H48O8S12Br5Cu4GdW
- Crystal structure of {tris((1H-benzo[d]imidazol-2- yl)methyl)amine-κ4N,N′,N′′,N′′′}-(succinato-κ2O,O′)nickel(II) – methanol (1/4), C32H41N7NiO8
- Crystal structure of catena-poly[trans-tetraaqua(μ2-1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol)-k2N:N′)cobalt(II)] dinitrate – 1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol) – water (1/3/2), C72H68CoN18O12
- Crystal structure of bis(μ2-2-oxido-2-phenylacetate-κ3O:O,O′)-bis(1-isopropoxy-2-oxo-2-phenylethan-1-olato-κ2O,O′)-bis(propan-2-olato-κ1O)dititanium(IV), C44H52O14Ti2
- The crystal structure of 5-carboxy-2-(hydroxymethyl)-1H-imidazol-3-ium-4-carboxylate, C6H8N2O6
- The crystal structure of 2,6-dibromo-4-fluoroaniline, C6H4Br2FN
- The crystal structure of 4-chloro-N-(2-phenoxyphenyl)benzamide, C19H14ClNO2
- The crystal structure of 2-methyl-β-naphthothiazole, C12H9NS

