Abstract
C4H11NO2, monoclinic, P21/n (no. 14), a = 6.0864(4) Å, b = 10.9696(8) Å, c = 8.5928(5) Å, β = 93.360(3)°, V = 572.72(7) Å3, Z = 4, Rgt(F) = 0.0307, wRref(F2) = 0.0891, T = 200 K.
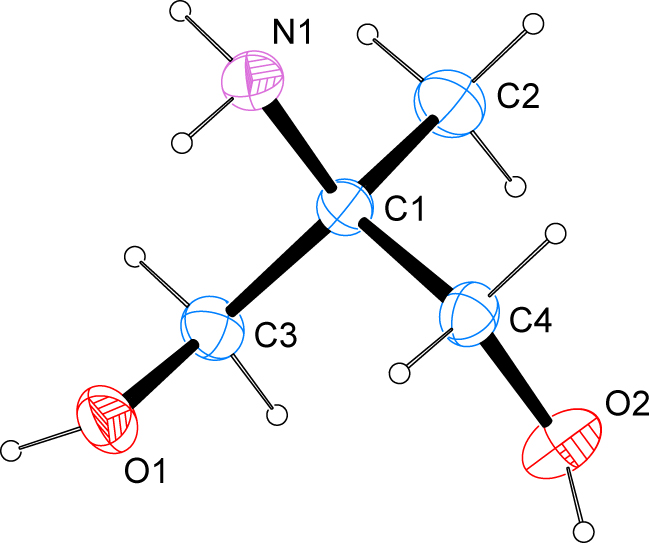
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colourless blocks |
| Size: | 0.59 × 0.53 × 0.35 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.10 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θmax, completeness: | 28.3°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 6559, 1422, 0.016 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 1299 |
| N(param)refined: | 75 |
| Programs: | Bruker [1], [2], SHELX [3], WinGX/ORTEP [4], Mercury [5], PLATON [6] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | Z | Uiso*/Ueq |
|---|---|---|---|---|
| O1 | 0.75720 (9) | 0.32652 (6) | 0.52692 (7) | 0.02653 (17) |
| H1 | 0.861363 | 0.306999 | 0.590497 | 0.040* |
| O2 | 0.52222 (11) | 0.55170 (6) | 0.29563 (8) | 0.03188 (18) |
| H2 | 0.436659 | 0.585810 | 0.355318 | 0.048* |
| N1 | 0.59156 (12) | 0.22262 (6) | 0.23927 (8) | 0.02410 (18) |
| C1 | 0.66847 (12) | 0.35027 (7) | 0.25099 (9) | 0.01995 (18) |
| C2 | 0.76768 (15) | 0.38609 (9) | 0.09854 (10) | 0.0298 (2) |
| H2A | 0.657587 | 0.374985 | 0.011835 | 0.045* |
| H2B | 0.895809 | 0.334561 | 0.082129 | 0.045* |
| H2C | 0.813381 | 0.471728 | 0.103728 | 0.045* |
| C3 | 0.84325 (13) | 0.36530 (8) | 0.38456 (9) | 0.02338 (19) |
| H3A | 0.974613 | 0.316158 | 0.363340 | 0.028* |
| H3B | 0.888401 | 0.451857 | 0.393101 | 0.028* |
| C4 | 0.46442 (13) | 0.42694 (7) | 0.27876 (9) | 0.02309 (19) |
| H4A | 0.355016 | 0.417145 | 0.189757 | 0.028* |
| H4B | 0.396708 | 0.398216 | 0.374115 | 0.028* |
| H1A | 0.548 (2) | 0.2002 (12) | 0.3331 (16) | 0.037 (3)* |
| H1B | 0.704 (2) | 0.1733 (13) | 0.2192 (15) | 0.039 (3)* |
Source of materials
The compound was obtained commercially (Merck). Crystals were taken directly from the provided product.
Carbon-bound H atoms were placed in calculated positions (C–H 0.99 Å for methylene groups) and were included in the refinement in the riding model approximation, with U(H) set to 1.2Ueq(C). The H atoms of the methyl group were allowed to rotate with a fixed angle around the C–C bond to best fit the experimental electron density (HFIX 137 in the SHELX program [2]), with U(H) set to 1.5Ueq(C). The H atoms of the hydroxyl groups were allowed to rotate with a fixed angle around the C–O bond to best fit the experimental electron density (HFIX 147 in the SHELX program [2]), with U(H) set to 1.5Ueq(O). Both nitrogen-bound H atoms were refined freely.
Comment
Chelate ligands have found widespread use in coordination chemistry due to the increased stability of coordination compounds they can form in comparison to monodentate ligands [7]. Aminols are particularly interesting in this aspect as they offer different donor sites of markedly diverging acidity as potential bonding partners. Upon variation of the substitution pattern on the hydrocarbon backbone, the acidity of the donor sites can be varied over a wide range and they may serve as probes for establishing the rules in which pKa range coordination to various central atoms of variable Lewis acidity can be observed. Furthermore, the spatial pretense of the substitution pattern can also be exploited to enable unusual coordination numbers. The title compound was chosen as it features one amino group next to two alcohol groups and can give rise to either five-membered heteroleptic chelate rings or six-membered homoleptic chelate rings. While the unit cell dimensions of the aminole are apparent in the literature no 3D coordinates have been deposited [8]. The crystal and molecular structures of similar aminoles have been reported earlier [9], [10], [11], [12], [13], [14], [15], [16].
The structure shows the expected connectivity. The C–O bond lengths of 1.4184(10) and 1.4237(10) Å as well as the C–N bond length of 1.4780(10) Å are in agreement with values reported for similar compounds deposited with the Cambridge Structural Database [17].
In the crystal, classical hydrogen bonds of the O–H⃛O, O–H⃛N and N–H⃛O type are apparent that employ all hydrogen atoms bonded to heteroatoms as donors as well as all heteroatoms as acceptors. One of the hydrogen atoms of the amino group establishes an intramolecular hydrogen bond to one of the hydroxyl groups. In terms of graph-set analysis [18], the descriptor for these hydrogen bonds is S(5)
Funding source: National Science Foundation
Author contribution: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: The corresponding author thanks the National Research Foundation for financial support.
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Bruker. APEX2; Bruker AXS Inc.: Madison, Wisconsin, USA, 2012.Search in Google Scholar
2. Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122; https://doi.org/10.1107/s0108767307043930.Search in Google Scholar
3. Farrugia, L. J. WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854; https://doi.org/10.1107/s0021889812029111.Search in Google Scholar
4. Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez–Monge, L., Taylor, R., van de Streek, J., Wood, P. A. Mercury CSD 2.0 – new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470; https://doi.org/10.1107/s0021889807067908.Search in Google Scholar
5. Spek, A. L. Structure validation in chemical crystallography. Acta Crystallogr. 2009, D65, 148–155; https://doi.org/10.1107/s090744490804362x.Search in Google Scholar
6. Bruker. SADABS; Bruker AXS Inc.: Madison, Wisconsin, USA, 2008.Search in Google Scholar
7. Gade, L. H. Koordinationschemie, 1. Auflage; Wiley-VCH: Weinheim, 1998.10.1002/9783527663927Search in Google Scholar
8. Rose, H. A., Van Camp, A. Crystallographic data. 139. 2-amino-2-methyl-1,3-propanediol. Anal. Chem. 1956, 28, 1790–1791; https://doi.org/10.1021/ac60119a048.Search in Google Scholar
9. Wikete, C., Wu, P., Zampella, G., De Gioia, L., Licini, G., Rehder, D. Glycine- and sarcosine–based models of vanadate–dependent haloperoxidases in sulfoxygenation reactions. Inorg. Chem. 2007, 46, 196–207; https://doi.org/10.1021/ic061534p.Search in Google Scholar
10. Hossain, M. K., Haukka, M., Sillanpaa, R., Hrovat, D. A., Richmond, M. G., Nordlander, E., Lehtonen, A. Syntheses and catalytic oxotransfer activities of oxo molybdenum(VI) complexes of a new aminoalcohol phenolate ligand. Dalton Trans. 2017, 46, 7051–7060; https://doi.org/10.1039/c7dt00846e.Search in Google Scholar
11. Wu, P., Santoni, G., Froba, M., Rehder, D. Modelling the sulfoxygenation activity of vanadate–dependent peroxidases. Chem. Biodivers. 2008, 5, 1913–1926; https://doi.org/10.1002/cbdv.200890179.Search in Google Scholar
12. Kocakaya, S. O., Karakaplan, M., Scopelliti, R. Synthesis and crystal structure of a chiral lactam and three amino alcohols as potential protein tyrosine phosphates 1B inhibitors. Tetrahedron Asymmetry 2017, 28, 1342–1349. https://doi.org/10.1016/j.tetasy.2017.09.014.Search in Google Scholar
13. Deshpande, S. H., Kelkar, A. A., Gonnade, R. G., Shingote, S. K., Chaudhari, R. V. Catalytic asymmetric transfer hydrogenation of ketones using [Ru(p-cymene) Cl2]2 with chiral amino alcohol ligands. Catal. Lett. 2010, 138, 231–238; https://doi.org/10.1007/s10562-010-0408-y.Search in Google Scholar
14. Wikete, C., Wu, P., Zampella, G., De Gioia, L., Lincini, G., Rehder, D. Glycine- and sarcosine-based models of vanadate-dependent haloperoxidases in sulfoxygenation reactions. Inorg. Chem. 2007, 46, 196–207; https://doi.org/10.1021/ic061534p.Search in Google Scholar
15. Betz, R., Gerber, T., Schalekamp, H. 5-Aminopentan-1-ol. Acta Crystallogr. 2011, E67, o1841; https://doi.org/10.1107/s1600536811024731.Search in Google Scholar
16. Betz, R., Gerber, T., Hosten, E. (3-Aminophenyl)methanol. Acta Crystallogr. 2011, E67, o2118; https://doi.org/10.1107/s1600536811029163.Search in Google Scholar
17. Allen, F. H. The Cambridge structural database: a quarter of a million crystal structures and rising. Acta Crystallogr. 2002, B58, 380–388; https://doi.org/10.1107/s0108768102003890.Search in Google Scholar
18. Bernstein, J., Davis, R. E., Shimoni, L., Chang, N.-L. Patterns in hydrogen bonding: functionality and graph set analysis in crystals. Angew Chem. Int. Ed. Engl. 1995, 34, 1555–1573; https://doi.org/10.1002/anie.199515551.Search in Google Scholar
© 2020 Eric C. Hosten and Richard Betz, published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of 4-hydroxybenzene-1,3-diaminium dichloride, C6H10Cl2N2O
- The crystal structure of 3-chloropropylammonium chloride, C3H9Cl2N
- The crystal structure of 1-chloro-2-(dimethylamino)ethane hydrochloride, C4H11Cl2N
- Crystal structure of N-(2-(trifluoromethyl)phenyl)hexanamide, C13H16F3NO
- Redetermination of the crystal structure of para-toluidine, C7H9N
- The crystal structure of bis(1,3-dihydroxy-2-methylpropan-2-aminium) carbonate, C9H24N2O7
- The crystal structure of 4-chloro-1-methylpiperidin-1-ium chloride, C6H13Cl2N
- Crystal structure of (Z)-3-(6-bromo-1H-indol-3-yl)-1,3-diphenylprop-2-en-1-one, C23H16BrNO
- The crystal structure of ethyl 2-amino-4-(3,5-difluorophenyl)-7,7-dimethyl-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carboxylate, C20H21F2NO4
- Crystal structure of 6,6'‐((1E,1'E)‐(propane‐1,3‐diylbis(azaneylylidene))bis(methaneylylidene))bis(3‐bromophenol), C34H32Br4N4O4
- The crystal structure of (E)-2-(2-((2-picolinoylhydrazono)methyl)phenoxy)acetic acid dihydrate, C15H17N3O6
- Crystal structure of (E)-4-bromo-N′-(3-chloro-2-hydroxybenzylidene)benzohydrazide, C14H10BrClN2O2
- Crystal structure of N,N′-bis(4-bromosalicylidene) ethylene-1,2-diaminopropan, C34H32Br4N4O4
- Crystal structure of 4-bromo-N′-[(3-bromo-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H14Br2N2O3
- The crystal structure of 1,2-bis(1H-benzo[d]imidazol-2-yl)ethane-1,2-diol — N-(2-aminophenyl)-3-(1H-benzo[d]imidazol-2-yl)-2,3-dihydroxypropanamide (1/1), C32H30N8O5
- The crystal structure of para-trifluoromethyl-aniline hemihydrate, C14H14F6N2O
- Redetermination of the crystal structure of 2-amino-2-methyl-propane-1,3-diole, C4H11NO2
- The crystal structure of methacholine chloride, C8H18ClNO2
- Crystal structure of 5,7,7-trimethyl-4,6,7,8-tetrahydrocyclopenta[g]isochromen-1(3H)-one, C15H18O2
- Crystal structure of poly[diammine-bis(μ4-4-hydroxypyridine-3-sulfonato-κ5N:O, O′:O′′:O′′)(μ2-pyrazinyl-κ2N:N′)tetrasilver(I)], C7H8Ag2N3O4S
- Crystal structure of ethyl (E)-5-(((3′,6′-bis(ethylamino)-3-oxospiro[isoindoline-1,9′-xanthen]-2-yl)imino)methyl)-2,4-dimethyl-1H-pyrrole-3-carboxylate — ethanol (1/1), C38H45N5O5
- Crystal structure of 4-bromo-N′-[(3-chloro-2-hydroxyphenyl)methylidene]benzohydrazide, C14H7Br2N2O2
- Redetermination of the crystal structure of 3,3,3-triphenylpropanoic acid, C21H18O2 – Deposition of hydrogen atomic coordinates
- Structure redetermination of dextromethorphan hydrobromide monohydrate, C18H28BrNO2 – localization of hydrogen atoms
- Crystal structure of tris(azido-κ1N)-(N-(2-aminoethyl)-N-methyl-1,3-propanediamine-κ3N,N′,N′′)cobalt(III), C7H19CoN12
- Crystal structure of tetraaqua-bis(1H-indazole-6-carboxylate-κN)cadmium (II), C16H18CdN4O8
- Crystal structure of dichloride-bis(1-propylimidazole-κ1N)zinc(II), C12H20Cl2N4Zn
- Crystal structure of (E)-resveratrol 3-O-β-D-xylopyranoside, C19H22O8
- Crystal structure of 3,3′-(1,2-phenylene-bis(methylene))bis(1-vinyl- 1H-imidazol-3-ium) bis(hexafluoro phosphate)(V), C18H20F12N4P2
- Crystal structure of diaqua[bis(benzimidazol-2-yl-methyl)amine-κ3N,N′,N″]-phthalato-κ1O-nickel(II)-methanol (1/2), C26H31N5NiO8
- Crystal structure of 6,7-difluoro-1-methyl-3-(trifluoromethyl)quinoxalin-2(1H)-one, C10H5F5N2O
- Crystal structure of dichlorido-bis(1-hexyl-1H-benzotriazole-k1N)zinc(II), C24H34N6Cl2Zn
- The crystal structre of 2-(4-bromophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H12BBrN2
- Crystal structure of diethyl 3,9-bis(4-fluorophenyl)-6,12-diphenyl-3,9-diazapentacyclo[6.4.0.02,7.04,11]dodecane-1,5-dicarboxylate, C40H36F2N2O4
- Crystal structure of (E)-7-methoxy-2-((5-methoxypyridin-3-yl)methylene)-3,4- dihydronaphthalen-1(2H)-one, C18H17NO3
- Crystal structure of (E)-2-chloro-6-(((1,3-dihydroxy-2-(oxidomethyl)propan-2-yl)imino)methyl)phenolate-κ3N,O,O’)manganese(IV), C22H24Cl2MnN2O8
- The crystal structure of α-(meta-methoxyphenoxy)-ortho-tolylic acid, C15H14O4
- The crystal structure of N-(2-chloroethyl)-N,N-diethylammonium chloride, C6H15Cl2N
- The crystal structure of tris(2,3,4,6,7,8,9,10-octahydro-1H-pyrimido[1,2-a]azepin-5-ium) trihydrodecavanadate(V), C27H54N6O28V10
- Crystal structure of 1,3-bis(octyl)benzimidazolium perchlorate C23H39ClN2O4
- Crystal structure of tetrakis[(Z)-(2-(1-(furan-2-yl)-2-methylpropylidene)-1-phenylhydrazin-1-ido-κ2N,N′)] zirconium(IV), C56H60N8O4Zr
- The crystal structure of 2-(naphthalen-2-yloxy)-4-phenyl-6-(prop-2-yn-1-yloxy)-1,3,5-triazine, C22H15N3O2
- The crystal structure of trimethylsulfonium tris(trifluoromethylsulfonyl)methanide, C7H9F9O6S4
- Crystal structure of 4-bromo-N′-[3,5-dichloro-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H13BrCl2N2O3
- The crystal structure of 4-(4-bromophenyl)-2-(3-(4-bromophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)thiazole, C24H16Br2FN3S
- The crystal structure of N-(adamantan-1-yl)-piperidine-1-carbothioamide, C16H26N2S
- The crystal structure of 1-phenyl-N-(4,5,6,7-tetrabromo-1,3-dioxoisoindolin-2-yl)-5-(thiophen-2-yl)-1H-pyrazole-3-carboxamide-dimethylformamide (1/1) C22H10Br4N4O3S
- The crystal structure of benzeneseleninic acid anhydride, C12H10O3Se2
- The crystal structure of diphenyalmine hydrochloride antimony trichloride co-crystallizate, C12H12Cl4NSb – Localization of hydrogen atoms
- The crystal structure of para-nitrobenzylbromide, C7H6BrNO2 – A second polymorph and correction of 3D coordinates
- Crystal structure of catena-poly[(5H-pyrrolo[3,2-b:4,5-b′]dipyridine-κ2N,N′)-(μ4-hexaoxidodivanadato)dizinc(II)],C10H9N3O6V2Zn
- Crystal structure of N,N′-(2-hydroxypropane-1,3-diyl)bis(pyridine-2-aldimine)-κ5N,N′,N′′,N′′′,O]-tris(nitrato-κ2O,O′) cerium(III), C15H16CeN7O10
- Synthesis and crystal structure of oktakis(dimethylsulphoxide-κ1O)gadolinium(III) [tetrabromido-μ2-bromido-μ2-sulfido-di-μ3-sulfido-μ4-sulfido-tetracopper(I)-tungsten(VI)], C16H48O8S12Br5Cu4GdW
- Crystal structure of {tris((1H-benzo[d]imidazol-2- yl)methyl)amine-κ4N,N′,N′′,N′′′}-(succinato-κ2O,O′)nickel(II) – methanol (1/4), C32H41N7NiO8
- Crystal structure of catena-poly[trans-tetraaqua(μ2-1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol)-k2N:N′)cobalt(II)] dinitrate – 1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol) – water (1/3/2), C72H68CoN18O12
- Crystal structure of bis(μ2-2-oxido-2-phenylacetate-κ3O:O,O′)-bis(1-isopropoxy-2-oxo-2-phenylethan-1-olato-κ2O,O′)-bis(propan-2-olato-κ1O)dititanium(IV), C44H52O14Ti2
- The crystal structure of 5-carboxy-2-(hydroxymethyl)-1H-imidazol-3-ium-4-carboxylate, C6H8N2O6
- The crystal structure of 2,6-dibromo-4-fluoroaniline, C6H4Br2FN
- The crystal structure of 4-chloro-N-(2-phenoxyphenyl)benzamide, C19H14ClNO2
- The crystal structure of 2-methyl-β-naphthothiazole, C12H9NS
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of 4-hydroxybenzene-1,3-diaminium dichloride, C6H10Cl2N2O
- The crystal structure of 3-chloropropylammonium chloride, C3H9Cl2N
- The crystal structure of 1-chloro-2-(dimethylamino)ethane hydrochloride, C4H11Cl2N
- Crystal structure of N-(2-(trifluoromethyl)phenyl)hexanamide, C13H16F3NO
- Redetermination of the crystal structure of para-toluidine, C7H9N
- The crystal structure of bis(1,3-dihydroxy-2-methylpropan-2-aminium) carbonate, C9H24N2O7
- The crystal structure of 4-chloro-1-methylpiperidin-1-ium chloride, C6H13Cl2N
- Crystal structure of (Z)-3-(6-bromo-1H-indol-3-yl)-1,3-diphenylprop-2-en-1-one, C23H16BrNO
- The crystal structure of ethyl 2-amino-4-(3,5-difluorophenyl)-7,7-dimethyl-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carboxylate, C20H21F2NO4
- Crystal structure of 6,6'‐((1E,1'E)‐(propane‐1,3‐diylbis(azaneylylidene))bis(methaneylylidene))bis(3‐bromophenol), C34H32Br4N4O4
- The crystal structure of (E)-2-(2-((2-picolinoylhydrazono)methyl)phenoxy)acetic acid dihydrate, C15H17N3O6
- Crystal structure of (E)-4-bromo-N′-(3-chloro-2-hydroxybenzylidene)benzohydrazide, C14H10BrClN2O2
- Crystal structure of N,N′-bis(4-bromosalicylidene) ethylene-1,2-diaminopropan, C34H32Br4N4O4
- Crystal structure of 4-bromo-N′-[(3-bromo-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H14Br2N2O3
- The crystal structure of 1,2-bis(1H-benzo[d]imidazol-2-yl)ethane-1,2-diol — N-(2-aminophenyl)-3-(1H-benzo[d]imidazol-2-yl)-2,3-dihydroxypropanamide (1/1), C32H30N8O5
- The crystal structure of para-trifluoromethyl-aniline hemihydrate, C14H14F6N2O
- Redetermination of the crystal structure of 2-amino-2-methyl-propane-1,3-diole, C4H11NO2
- The crystal structure of methacholine chloride, C8H18ClNO2
- Crystal structure of 5,7,7-trimethyl-4,6,7,8-tetrahydrocyclopenta[g]isochromen-1(3H)-one, C15H18O2
- Crystal structure of poly[diammine-bis(μ4-4-hydroxypyridine-3-sulfonato-κ5N:O, O′:O′′:O′′)(μ2-pyrazinyl-κ2N:N′)tetrasilver(I)], C7H8Ag2N3O4S
- Crystal structure of ethyl (E)-5-(((3′,6′-bis(ethylamino)-3-oxospiro[isoindoline-1,9′-xanthen]-2-yl)imino)methyl)-2,4-dimethyl-1H-pyrrole-3-carboxylate — ethanol (1/1), C38H45N5O5
- Crystal structure of 4-bromo-N′-[(3-chloro-2-hydroxyphenyl)methylidene]benzohydrazide, C14H7Br2N2O2
- Redetermination of the crystal structure of 3,3,3-triphenylpropanoic acid, C21H18O2 – Deposition of hydrogen atomic coordinates
- Structure redetermination of dextromethorphan hydrobromide monohydrate, C18H28BrNO2 – localization of hydrogen atoms
- Crystal structure of tris(azido-κ1N)-(N-(2-aminoethyl)-N-methyl-1,3-propanediamine-κ3N,N′,N′′)cobalt(III), C7H19CoN12
- Crystal structure of tetraaqua-bis(1H-indazole-6-carboxylate-κN)cadmium (II), C16H18CdN4O8
- Crystal structure of dichloride-bis(1-propylimidazole-κ1N)zinc(II), C12H20Cl2N4Zn
- Crystal structure of (E)-resveratrol 3-O-β-D-xylopyranoside, C19H22O8
- Crystal structure of 3,3′-(1,2-phenylene-bis(methylene))bis(1-vinyl- 1H-imidazol-3-ium) bis(hexafluoro phosphate)(V), C18H20F12N4P2
- Crystal structure of diaqua[bis(benzimidazol-2-yl-methyl)amine-κ3N,N′,N″]-phthalato-κ1O-nickel(II)-methanol (1/2), C26H31N5NiO8
- Crystal structure of 6,7-difluoro-1-methyl-3-(trifluoromethyl)quinoxalin-2(1H)-one, C10H5F5N2O
- Crystal structure of dichlorido-bis(1-hexyl-1H-benzotriazole-k1N)zinc(II), C24H34N6Cl2Zn
- The crystal structre of 2-(4-bromophenyl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborinine, C16H12BBrN2
- Crystal structure of diethyl 3,9-bis(4-fluorophenyl)-6,12-diphenyl-3,9-diazapentacyclo[6.4.0.02,7.04,11]dodecane-1,5-dicarboxylate, C40H36F2N2O4
- Crystal structure of (E)-7-methoxy-2-((5-methoxypyridin-3-yl)methylene)-3,4- dihydronaphthalen-1(2H)-one, C18H17NO3
- Crystal structure of (E)-2-chloro-6-(((1,3-dihydroxy-2-(oxidomethyl)propan-2-yl)imino)methyl)phenolate-κ3N,O,O’)manganese(IV), C22H24Cl2MnN2O8
- The crystal structure of α-(meta-methoxyphenoxy)-ortho-tolylic acid, C15H14O4
- The crystal structure of N-(2-chloroethyl)-N,N-diethylammonium chloride, C6H15Cl2N
- The crystal structure of tris(2,3,4,6,7,8,9,10-octahydro-1H-pyrimido[1,2-a]azepin-5-ium) trihydrodecavanadate(V), C27H54N6O28V10
- Crystal structure of 1,3-bis(octyl)benzimidazolium perchlorate C23H39ClN2O4
- Crystal structure of tetrakis[(Z)-(2-(1-(furan-2-yl)-2-methylpropylidene)-1-phenylhydrazin-1-ido-κ2N,N′)] zirconium(IV), C56H60N8O4Zr
- The crystal structure of 2-(naphthalen-2-yloxy)-4-phenyl-6-(prop-2-yn-1-yloxy)-1,3,5-triazine, C22H15N3O2
- The crystal structure of trimethylsulfonium tris(trifluoromethylsulfonyl)methanide, C7H9F9O6S4
- Crystal structure of 4-bromo-N′-[3,5-dichloro-2-hydroxyphenyl)methylidene]benzohydrazide methanol solvate, C15H13BrCl2N2O3
- The crystal structure of 4-(4-bromophenyl)-2-(3-(4-bromophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)thiazole, C24H16Br2FN3S
- The crystal structure of N-(adamantan-1-yl)-piperidine-1-carbothioamide, C16H26N2S
- The crystal structure of 1-phenyl-N-(4,5,6,7-tetrabromo-1,3-dioxoisoindolin-2-yl)-5-(thiophen-2-yl)-1H-pyrazole-3-carboxamide-dimethylformamide (1/1) C22H10Br4N4O3S
- The crystal structure of benzeneseleninic acid anhydride, C12H10O3Se2
- The crystal structure of diphenyalmine hydrochloride antimony trichloride co-crystallizate, C12H12Cl4NSb – Localization of hydrogen atoms
- The crystal structure of para-nitrobenzylbromide, C7H6BrNO2 – A second polymorph and correction of 3D coordinates
- Crystal structure of catena-poly[(5H-pyrrolo[3,2-b:4,5-b′]dipyridine-κ2N,N′)-(μ4-hexaoxidodivanadato)dizinc(II)],C10H9N3O6V2Zn
- Crystal structure of N,N′-(2-hydroxypropane-1,3-diyl)bis(pyridine-2-aldimine)-κ5N,N′,N′′,N′′′,O]-tris(nitrato-κ2O,O′) cerium(III), C15H16CeN7O10
- Synthesis and crystal structure of oktakis(dimethylsulphoxide-κ1O)gadolinium(III) [tetrabromido-μ2-bromido-μ2-sulfido-di-μ3-sulfido-μ4-sulfido-tetracopper(I)-tungsten(VI)], C16H48O8S12Br5Cu4GdW
- Crystal structure of {tris((1H-benzo[d]imidazol-2- yl)methyl)amine-κ4N,N′,N′′,N′′′}-(succinato-κ2O,O′)nickel(II) – methanol (1/4), C32H41N7NiO8
- Crystal structure of catena-poly[trans-tetraaqua(μ2-1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol)-k2N:N′)cobalt(II)] dinitrate – 1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol) – water (1/3/2), C72H68CoN18O12
- Crystal structure of bis(μ2-2-oxido-2-phenylacetate-κ3O:O,O′)-bis(1-isopropoxy-2-oxo-2-phenylethan-1-olato-κ2O,O′)-bis(propan-2-olato-κ1O)dititanium(IV), C44H52O14Ti2
- The crystal structure of 5-carboxy-2-(hydroxymethyl)-1H-imidazol-3-ium-4-carboxylate, C6H8N2O6
- The crystal structure of 2,6-dibromo-4-fluoroaniline, C6H4Br2FN
- The crystal structure of 4-chloro-N-(2-phenoxyphenyl)benzamide, C19H14ClNO2
- The crystal structure of 2-methyl-β-naphthothiazole, C12H9NS

