Abstract
C18H20N6O4, monoclinic, Pc, a = 4.4412(8) Å, b = 14.100(3) Å, c = 15.206(3) Å, β = 95.920(4)°, V = 947.1(3) Å3, Z = 2, Rgt(F) = 0.0352, wRref(F2) = 0.0770, T = 296.15 K.
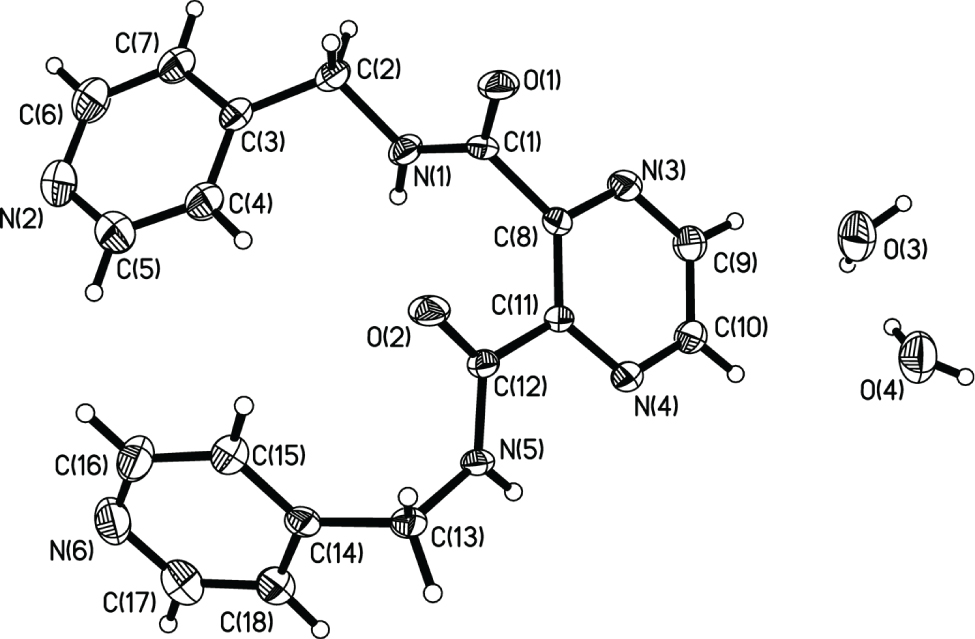
The crystal structure is shown in the figure. Tables 1 and 2 contain details on crystal structure and measurement conditions and a list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Yellow block |
| Size: | 0.28 × 0.22 × 0.16 |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 1.0 cm−1 |
| Diffractometer, scan mode: | Bruker, φ and ω |
| 2θmax, completeness: | 55.6°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 5693, 2793, 0.022 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 2326 |
| N(param)refined: | 253 |
| Programs: | Bruker programs [8], SHELX [9, 10] , OLEX2 [11] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| O1 | 0.0764(4) | 0.50400(13) | 0.69686(11) | 0.0456(4) |
| O2 | −0.1529(4) | 0.58860(11) | 0.50554(11) | 0.0450(5) |
| N1 | 0.4514(5) | 0.58862(14) | 0.64393(13) | 0.0382(5) |
| H1 | 0.5572 | 0.5968 | 0.6003 | 0.046* |
| N2 | 0.6937(7) | 0.93853(17) | 0.6340(2) | 0.0700(8) |
| N3 | 0.4427(6) | 0.36879(15) | 0.59609(14) | 0.0465(6) |
| N4 | 0.1944(5) | 0.38785(14) | 0.42142(14) | 0.0430(5) |
| N5 | −0.0888(5) | 0.55002(13) | 0.36457(12) | 0.0337(5) |
| H5 | 0.0044 | 0.5142 | 0.3304 | 0.040* |
| N6 | 0.1070(7) | 0.90226(17) | 0.3298(2) | 0.0646(8) |
| C1 | 0.2571(5) | 0.51721(15) | 0.64218(14) | 0.0308(5) |
| C2 | 0.4845(6) | 0.65575(17) | 0.71719(17) | 0.0432(6) |
| H2A | 0.6452 | 0.6343 | 0.7607 | 0.052* |
| H2B | 0.2983 | 0.6573 | 0.7453 | 0.052* |
| C3 | 0.5563(6) | 0.75401(17) | 0.68745(16) | 0.0388(6) |
| C4 | 0.4248(7) | 0.7919(2) | 0.6094(2) | 0.0530(7) |
| H4 | 0.2870 | 0.7565 | 0.5727 | 0.064* |
| C5 | 0.4986(9) | 0.8829(2) | 0.5858(2) | 0.0677(9) |
| H5A | 0.4059 | 0.9069 | 0.5328 | 0.081* |
| C6 | 0.8168(9) | 0.9011(2) | 0.7106(3) | 0.0740(11) |
| H6 | 0.9500 | 0.9384 | 0.7470 | 0.089* |
| C7 | 0.7571(7) | 0.8111(2) | 0.7383(2) | 0.0555(8) |
| H7 | 0.8523 | 0.7885 | 0.7916 | 0.067* |
| C8 | 0.2830(6) | 0.44608(16) | 0.56901(15) | 0.0316(5) |
| C9 | 0.4760(8) | 0.30252(18) | 0.53578(19) | 0.0535(8) |
| H9 | 0.5829 | 0.2476 | 0.5527 | 0.064* |
| C10 | 0.3573(7) | 0.31265(18) | 0.44917(18) | 0.0531(8) |
| H10 | 0.3921 | 0.2654 | 0.4088 | 0.064* |
| C11 | 0.1544(5) | 0.45461(15) | 0.48162(14) | 0.0305(5) |
| C12 | −0.0427(5) | 0.53776(16) | 0.45160(14) | 0.0310(5) |
| C13 | −0.2905(6) | 0.62335(16) | 0.32515(17) | 0.0390(6) |
| H13A | −0.4698 | 0.6257 | 0.3565 | 0.047* |
| H13B | −0.3542 | 0.6063 | 0.2643 | 0.047* |
| C14 | −0.1485(5) | 0.72047(17) | 0.32697(16) | 0.0375(6) |
| C15 | −0.1882(7) | 0.78357(19) | 0.39381(19) | 0.0520(7) |
| H15 | −0.3005 | 0.7665 | 0.4397 | 0.062* |
| C16 | −0.0596(9) | 0.8724(2) | 0.3919(2) | 0.0642(9) |
| H16 | −0.0918 | 0.9142 | 0.4372 | 0.077* |
| C17 | 0.1435(8) | 0.8410(2) | 0.2651(2) | 0.0640(9) |
| H17 | 0.2573 | 0.8599 | 0.2202 | 0.077* |
| C18 | 0.0205(7) | 0.7505(2) | 0.2614(2) | 0.0517(7) |
| H18 | 0.0523 | 0.7102 | 0.2148 | 0.062* |
| O4 | 0.2917(6) | 0.08204(14) | 0.41438(19) | 0.0796(7) |
| H4A | 0.3102 | 0.0307 | 0.3909 | 0.119* |
| H4B | 0.4755 | 0.0921 | 0.4399 | 0.119* |
| O3 | 0.8244(6) | 0.09840(15) | 0.52465(18) | 0.0794(7) |
| H3A | 0.8494 | 0.0438 | 0.5539 | 0.119* |
| H3B | 0.9698 | 0.0981 | 0.4933 | 0.119* |
Source of material
All reagents and solvents for syntheses were purchased from commercial sources and used as received without further purification. The title compound was prepared by the condensation reaction of dimethyl pyrazine-2,3-dicarboxylate (3.92 g, 20 mmol) with 4-aminomethylpyridine (2.16 g, 20 mmol) under the reflux condition in 15 mL of methanol for 12 h. Followed by rotary evaporation to remove the solvent, a yellow solid was obtained. Recrystallization in CH3OH gave light yellow crystals.
Experimental details
The Uiso values of hydrogen atoms of the water molecules were set to 1.5Ueq(O) and the Uiso values of all other hydrogen atoms were set to 1.2Ueq(C).
Discussion
Pyrazine-based amide ligands have played a very important role in coordination chemistry [1, 2] . Some transition metal complexes of pyrazine-2,3-bisamide ligands were explored to construct supramolecular architectures such as molecular grids. For example, a symmetrical diamide ligand containing a central bridging pyrazinyl unit connected to two tetradentate binding pockets that forms bi- and tetranuclear copper(II) complexes have been reported [3], [4], [5], [6], [7]. The two amide hydrogens of these compounds can be deprotonated, allowing the anionic amidate N donor atoms to coordinate to a metal center. The neutral molecule forms a centrosymmetric dimer. Whereas in the presence of triethylamine, a tetranuclear copper(II) complex is formed, where the pyrazine unit bridges between metal centers [3]. In order to expand the research scope of pyrazine-2,3-bisamide ligand system, we prepared the title compound to investigate its transition metal chemistry.
The title structure consists of one central pyrazine and two pyridin-4-ylmethyl group connected by two amide group and two water molecules. The coordination modes of H2L could be more flexible due to two outside N atom from the pyridine. So, it may be used as a bridging ligand to construct metal complexes with 3D networks or metal organic frameworks.
H2L molecules are connected by hydrogen bonds. The water molecules are connected by hydrogen bonds to form a chain along [100].
Acknowledgement
The author would like to thank the talent introduction project of Chongqing University of Arts and Sciences(No⋅R2012CH12), and Science and technology research project of Chongqing Municipal Education Committee (No⋅KJ131215) for financial support.
References
1 Cati, D. S.; Ribas, J.; Ribas-Ariño, J.; Stoeckli-Evans,H.: Self-assembly of CuII and NiII [2×2] grid complexes and a binuclear CuII complex with a new semiflexible substituted pyrazine ligand: multiple anion encapsulation and magnetic properties. Inorg. Chem. 43 (2004) 1021–1030.10.1021/ic030174sSearch in Google Scholar PubMed
2 Klingele, J.; Boas, J. F.; Pilbrow, J. R.; Moubaraki, B.; Murray, K. S.; Berry, K. J.; Hunter, K. A.; Jameson, G. B.; Boyd, P. D. W.; Brooker, S.: A [2×2] nickel(II) grid and a copper(II) square result from differing binding modes of a pyrazine-based diamide ligand. Dalton. Trans. (2007) 633–645.10.1039/B614796HSearch in Google Scholar PubMed
3 Hausmann, J.; Jamesonb, G. B.; Brooker, S.: Control of molecular architecture by the degree of deprotonation: self-assembled di- and tetranuclear copper(II) complexes of N,N′-bis(2-pyridylmethyl)pyrazine-2,3-dicarboxamide. Chem. Commun. (2003) 2992–2993.10.1039/B308263FSearch in Google Scholar PubMed
4 Hausmann, J.; Brooker, S.: Control of molecular architecture by use of the appropriate ligand isomer: a mononuclear “corner-type” versus a tetranuclear [2×2]grid-type cobalt(III) complex. Chem. Commun. (2004) 1531–1531.10.1039/B403905JSearch in Google Scholar PubMed
5 Cati, D. S.; Stoeckli-Evans, H.: A mononuclear cobalt(III) complex of the ligand N,N′-bis(pyridin-2-ylmethyl)pyrazine-2,3-dicarboxamide. Acta Crystallogr. E60 (2004) m174–m176.10.1107/S1600536804000431Search in Google Scholar
6 Cati, D. S.; Stoeckli-Evans, H.: A macrocyclic binuclear copper(II) complex of the ligand N,N′-bis(pyridin-2-ylmethyl)pyrazine-2,3-dicarboxamide. Acta Crystallogr. E60 (2004) m177–m179.10.1107/S1600536804000522Search in Google Scholar
7 Cati, D. S.; Stoeckli-Evans, H.: A neutral macrocyclic binuclear copper(II) complex of the deprotonated form of the ligand N,N′-bis(pyridin-2-ylmethyl)pyrazine-2,3- dicarboxamide. Acta Crystallogr. E60 (2004) m180–m182.10.1107/S1600536804000534Search in Google Scholar
8 Bruker. SAINT . Brucker AXS Inc., Madison, WI, USA (2013).Search in Google Scholar
9 Sheldrick, G. M.: Crystal structure refinement with SHELXL. Acta Crystallogr. A71 (2015) 3–8.10.1107/S2053273314026370Search in Google Scholar PubMed PubMed Central
10 Sheldrick, G. M.: SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. C71 (2015) 3–8.10.1107/S2053273314026370Search in Google Scholar
11 Dolomanov, O. V.; Bourhis, L. J.; Gildea, R. J.; Howard, J. A. K.; Puschmann, H.: OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 42 (2009) 339–341.10.1107/S0021889808042726Search in Google Scholar
©2017 Zhaodong Wang et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 3.0 License.
Articles in the same Issue
- Cover and Frontmatter
- Crystal structure of 2,5-diiodo-4-nitro-1H-imidazole hemihydrate, C6H4I4N6O5
- Crystal structure of catena-poly[μ2-2,2′-(1,3-phenylene)diacetato-κ4O,O′:O′′,O′′′)-(μ2-1,6-bis(2-methyl-1H-benzo[d]imidazol-1-yl)hexane-κ2N:N′)cadmium(II)], C32H34CdN4O4
- Crystal structure of poly[aqua-(2,2′-bipyridine-κN,N′)-(μ4-5,5′-(hexane-1,6-diyl)-bis(oxy)diisophthalato κ8O1,O2:O3,O4:O5,O6:O7,O8)manganese(II)], C21H21MnN2O7
- Crystal structure of poly-[(μ2-((1,3-bis(benzimidazol-1-yl)propane-κ2N:N′)(μ2-4-tert-butyl-phthalato-κ2O:O′)cobalt(II)] monohydrate, C29H30CoN4O5
- Crystal structure of 2-amino-4-(3,4,5-trimethoxy-phenyl)-5-(oxo-5,6,7,8-tetrahydro-4H-chromene)-3-carbonitrile – ethanol (1/1), C21H26N2O6
- Crystal structure of ethyl 1-benzyl-5-phenyl-1H-pyrazole-3-carboxylate, C19H18N2O2
- Crystal structure of 2-(1-benzyl-3-phenyl-1H-pyrazol-5-yl)-5-(4-nitrobenzylthio)-1,3,4-oxadiazole, C25H19N5O3S
- Structure and photochromism of 1,2-bis[2-methyl-5-(2-chlorophenyl)-3-thienyl]-3,3,4,4,5,5-hexafluorocyclopent-1-ene, C27H16Cl2F6S2
- The crystal structure of the Schiff base (E)-2,6-diisopropyl-N-(pyridin-4-ylmethylene)aniline, C18H22N2
- Crystal structure of (E)-2,4-dibromo-6-(((4-methyl-2-nitrophenyl)imino) methyl)phenol, C10H14Br2N2O3
- Crystal structure of (bis(2,2′-bipyridine-κ2N,N′))-(3,5-dinitrosalicylato-κ2O,O′)nickel(II), C27H18N6NiO7
- Crystal structure of 1-(diethoxy phosphonomethyl) 2-benzoyl-3-chloro-2-cyclohexen-1-ol, C18H24ClO5P
- Crystal structure of tetraaqua-bis(1,3-benzimidazol-3-ium-1,3-diacetato-κO)copper(II) hemihydrate, C22H27CuN4O12.50
- Crystal structure of 1α,11-dihydroxyeremophil-9-en-8-one, C15H24O3
- Crystal structure of 1-ferrocenylsulfonyl-1H-imidazo[4,5-b]pyridine, C16H13FeN3O2S
- Crystal structure of bis(μ2-azido-κ2N:N)-dichlorido-bis(μ2-2-(pyridin-2-yl)ethan-1-ol-κ2O,N)dicopper(II), C14H18Cl2Cu2N8O2
- Crystal structure of (5,15-cis-bis(2-hydroxy-1-naphthyl)-10-phenyl-20-(4-hydroxyphenyl)-porphyrinato)-(pyridine)-zinc(ii) pyridine solvate, C67H47N7O3Zn
- Crystal structure of (μ2-[2,2′-bis(diphenylphosphino)-1,1′-binaphthalene oxide-κ2O,P])-iodido copper(I), C44H32CuIOP2
- Crystal structure of 6,8-diphenyl-2-(4-fluorophenyl)-2,3-dihydroquinolin-4(3H)-one, C27H20FNO
- Crystal structure of 5,11,17,23-tetra(tert-butyl)-25,26,27,28-tetrahexoxycalix[4]arene, C68H104O4
- Crystal structure of N,N′–bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide dihydrate, C18H20N6O4
- Crystal structure of a diaqua-bis(3,5-di(1H-imidazol-1-yl)pyridine-κN)-bis(2-(4-carboxy-phenyl)acetato-κO]manganese(II), C40H36MnN10O10
- Crystal structure of 4-(4-hydroxy-3-methoxy-phenyl)-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydro-quinoline-3-carboxylic acid methyl ester, C21H25NO5
- Crystal structure of (E)-4,4′-(ethene-1,2-diyl)bis(3-nitrobenzoic acid) 1.5 hydrate, C16H13N2O9.5
- Crystal structure of (E)-2-(5-(4-fluorophenyl)-3-(furan-2-yl)-4,5-dihydro-1H-pyrazol-1-yl)-5-((4-fluorophenyl)diazenyl)-4-methylthiazole, C23H17F2N5OS
- Crystal structure of the co-crystalline adduct 4-((4,4-dimethyl-2,6-dioxocyclohexylidene)methylamino)-N-(4,6-dimethylpyrimidin-2-yl)benzenesulfonamide - acetic acid (1/1), C21H24N4O4S ⋅ C2H4O2
- Synthesis and crystal structure of 2-((5-chlorobenzo[c][1,2,5]thiadiazol-4-yl)amino)-4,5-dihydro-1H-imidazol-3-ium tetraphenylborate, C33H29BClN5S
- Crystal structure of (4-chlorophenyl)(3-ferrocenyl-5-(trifluoromethyl)-1H-pyrazol-1-yl)methanone, C21H14ClF3FeN2O
- Crystal structure of (S)-benzyl 3-(benzylcarba-moyl)-3,4-dihydroisoquinoline-2(1H)-carboxylate, C25H24N2O3
- Crystal structure of 5-acetyl-3-(3-fluoro-4-morpholinophenyl)oxazolidin-2-one, C15H17FN2O4
- Crystal structure of 2-(4-fluorophenyl)-1,3-dimethyl-1H-perimidin-3-ium iodide, C19H16FIN2
- Crystal structure of methyl 1H-indole-2-carboxylate, C10H9NO2
- Crystal structure of 2,3-diphenyl-1-[(dipropylamino)acetyl]-1,3-diazaspiro[4.5]decan-4-one, C28H37N3O2
- Crystal structure of 1-(2H-1,3-benzodioxol-5-yl)-3-(1H-imidazol-1-yl)propan-1-one, C13H12N2O3
- Crystal structure of 2,9-dibromo-1,10-phenanthroline, C12H6Br2N2
- Crystal structure of 3-(adamantan-1-yl)-4-(4-fluorophenyl)-1H-1,2,4-triazole-5(4H)-thione, C18H20FN3S
- Crystal structure of trans-bis((E)-7-oxo-4-(phenyldiazenyl)cyclohepta-1,3,5-trien-1-olato)-κ2O,O′)-bis(pyridine-κN)cobalt(II), C36H28CoN6O4
- Crystal structure of 2-(4-methyl-3-phenylthiazol-2(3H)-ylidene)malononitrile, C13H9N3S
- Crystal structure of (Z)-3-(adamantan-1-yl)-1-(3-chlorophenyl)-S-benzylisothiourea, C24H27ClN2S
- Crystal structure of chlorido{[3-(η5-cyclopenta-dienyl)-2,2,3-trimethyl-1-phenylbutylidene] azanido-κN}[η2(N,O)-N,N-dimethylhydroxylaminato]titanium(IV), C20H27ClN2OTi
- Crystal Structure of 1,1′-dimethyl-[4,4′-bipyridine]-1,1′-diium tetrachloridozincate(II), C12H14Cl4N2Zn
- Crystal structure of 5-nitro-2-(pyrrolidin-1-yl)benzaldehyde, C11H12N2O3
- Crystal structure of 2,3-diphenyl-1-(morpholin-4-ylacetyl)-1,3-diazaspiro[4.5]decan-4-one, C26H31N3O3
- Crystal structure of 3,3-dimethyl-3,4-dihydro-1H-benzo[c]chromene-1,6(2H)-dione, C15H14O3
- Crystal structure of bis(2-(2-hydroxymethyl)pyridine-κ2N,O)-bis(pivalato-κO)nickel(II), C22H32N2NiO6
- Crystal structure of (1,10-phenanthroline-κ2N,N′)-bis(1H-pyrazole-3-carboxylato-κ2N,O)manganese(II) trihydrate, C20H20N6O7Mn
- The crystal structure of 3-aminopropan-1-aminium iodide, C3H11N2I
- Crystal structure of ethyl 1-(4-chlorophenyl)-5-methyl-1H-1,2,3-triazole-4 carboxylate, C12H12ClN3O2
- Crystal structure of 4,4′-((1Z,1′Z)-2,2′-(2,5-diethoxy-1,4-phenylene)bis(ethene-2,1-diyl))dipyridine, C24H24N2O2
- Crystal structure of (16S)-12,16-epoxy-11,14-dihydroxy-17(15/16)-abeo-3a,18-cyclo-8,11,13-abietatrien-7-one, C20H24O4
- Crystal structure of aquadichlorido(2,4,6-tri-2-pyridyl-1,3,5-triazine-κ3N,N′,N′′)nickel(II) monohydrate, C18H16Cl2N6NiO2
- Crystal structure of catena-poly[dichlorido-(μ-ethane-1,2-diyl-bis-(pyridyl-4-carboxylate-κN:N′)mercury(II)], C15H14Cl2HgN2O4
- Crystal structure of methyl 2-acetamido-5-chlorbenzoate, C10H10ClNO3
- Crystal structure of tetrakis(μ2-3,3-dimethylacrylato-κ2O,O′)-bis(2-aminopyrimidine-κN) dicopper(II), C28H38Cu2N6O8
- Crystal structure of 3-amino-8-methoxy-1-phenyl-1H-benzo[f]chromene-2-carbonitrile, C21H16N2O2
- Crystal structure of 4-(2-ammonioethyl)morpholin-4-ium dichloride monohydrate, C6H18Cl2N2O2
- Crystal structure of 1-(3-((5-bromo-2-hydroxybenzylidene)amino)phenyl)ethanone O-benzyl oxime, C22H19BrN2O2
- Crystal structure of 2-(4-(dimethylamino)-2-fluorophenyl)-1,3-bis(1H-1,2,4-triazol-1-yl)propan-2-ol monohydrate, C15H20FN7O2
- Crystal structure of 4-bromo-2-(1H-pyrazol-3-yl)phenol, C9H7BrN2O
- Crystal structure of 1,2,3,4,5-pentamethyl-1,3-cyclopentadiene, C10H16
Articles in the same Issue
- Cover and Frontmatter
- Crystal structure of 2,5-diiodo-4-nitro-1H-imidazole hemihydrate, C6H4I4N6O5
- Crystal structure of catena-poly[μ2-2,2′-(1,3-phenylene)diacetato-κ4O,O′:O′′,O′′′)-(μ2-1,6-bis(2-methyl-1H-benzo[d]imidazol-1-yl)hexane-κ2N:N′)cadmium(II)], C32H34CdN4O4
- Crystal structure of poly[aqua-(2,2′-bipyridine-κN,N′)-(μ4-5,5′-(hexane-1,6-diyl)-bis(oxy)diisophthalato κ8O1,O2:O3,O4:O5,O6:O7,O8)manganese(II)], C21H21MnN2O7
- Crystal structure of poly-[(μ2-((1,3-bis(benzimidazol-1-yl)propane-κ2N:N′)(μ2-4-tert-butyl-phthalato-κ2O:O′)cobalt(II)] monohydrate, C29H30CoN4O5
- Crystal structure of 2-amino-4-(3,4,5-trimethoxy-phenyl)-5-(oxo-5,6,7,8-tetrahydro-4H-chromene)-3-carbonitrile – ethanol (1/1), C21H26N2O6
- Crystal structure of ethyl 1-benzyl-5-phenyl-1H-pyrazole-3-carboxylate, C19H18N2O2
- Crystal structure of 2-(1-benzyl-3-phenyl-1H-pyrazol-5-yl)-5-(4-nitrobenzylthio)-1,3,4-oxadiazole, C25H19N5O3S
- Structure and photochromism of 1,2-bis[2-methyl-5-(2-chlorophenyl)-3-thienyl]-3,3,4,4,5,5-hexafluorocyclopent-1-ene, C27H16Cl2F6S2
- The crystal structure of the Schiff base (E)-2,6-diisopropyl-N-(pyridin-4-ylmethylene)aniline, C18H22N2
- Crystal structure of (E)-2,4-dibromo-6-(((4-methyl-2-nitrophenyl)imino) methyl)phenol, C10H14Br2N2O3
- Crystal structure of (bis(2,2′-bipyridine-κ2N,N′))-(3,5-dinitrosalicylato-κ2O,O′)nickel(II), C27H18N6NiO7
- Crystal structure of 1-(diethoxy phosphonomethyl) 2-benzoyl-3-chloro-2-cyclohexen-1-ol, C18H24ClO5P
- Crystal structure of tetraaqua-bis(1,3-benzimidazol-3-ium-1,3-diacetato-κO)copper(II) hemihydrate, C22H27CuN4O12.50
- Crystal structure of 1α,11-dihydroxyeremophil-9-en-8-one, C15H24O3
- Crystal structure of 1-ferrocenylsulfonyl-1H-imidazo[4,5-b]pyridine, C16H13FeN3O2S
- Crystal structure of bis(μ2-azido-κ2N:N)-dichlorido-bis(μ2-2-(pyridin-2-yl)ethan-1-ol-κ2O,N)dicopper(II), C14H18Cl2Cu2N8O2
- Crystal structure of (5,15-cis-bis(2-hydroxy-1-naphthyl)-10-phenyl-20-(4-hydroxyphenyl)-porphyrinato)-(pyridine)-zinc(ii) pyridine solvate, C67H47N7O3Zn
- Crystal structure of (μ2-[2,2′-bis(diphenylphosphino)-1,1′-binaphthalene oxide-κ2O,P])-iodido copper(I), C44H32CuIOP2
- Crystal structure of 6,8-diphenyl-2-(4-fluorophenyl)-2,3-dihydroquinolin-4(3H)-one, C27H20FNO
- Crystal structure of 5,11,17,23-tetra(tert-butyl)-25,26,27,28-tetrahexoxycalix[4]arene, C68H104O4
- Crystal structure of N,N′–bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide dihydrate, C18H20N6O4
- Crystal structure of a diaqua-bis(3,5-di(1H-imidazol-1-yl)pyridine-κN)-bis(2-(4-carboxy-phenyl)acetato-κO]manganese(II), C40H36MnN10O10
- Crystal structure of 4-(4-hydroxy-3-methoxy-phenyl)-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydro-quinoline-3-carboxylic acid methyl ester, C21H25NO5
- Crystal structure of (E)-4,4′-(ethene-1,2-diyl)bis(3-nitrobenzoic acid) 1.5 hydrate, C16H13N2O9.5
- Crystal structure of (E)-2-(5-(4-fluorophenyl)-3-(furan-2-yl)-4,5-dihydro-1H-pyrazol-1-yl)-5-((4-fluorophenyl)diazenyl)-4-methylthiazole, C23H17F2N5OS
- Crystal structure of the co-crystalline adduct 4-((4,4-dimethyl-2,6-dioxocyclohexylidene)methylamino)-N-(4,6-dimethylpyrimidin-2-yl)benzenesulfonamide - acetic acid (1/1), C21H24N4O4S ⋅ C2H4O2
- Synthesis and crystal structure of 2-((5-chlorobenzo[c][1,2,5]thiadiazol-4-yl)amino)-4,5-dihydro-1H-imidazol-3-ium tetraphenylborate, C33H29BClN5S
- Crystal structure of (4-chlorophenyl)(3-ferrocenyl-5-(trifluoromethyl)-1H-pyrazol-1-yl)methanone, C21H14ClF3FeN2O
- Crystal structure of (S)-benzyl 3-(benzylcarba-moyl)-3,4-dihydroisoquinoline-2(1H)-carboxylate, C25H24N2O3
- Crystal structure of 5-acetyl-3-(3-fluoro-4-morpholinophenyl)oxazolidin-2-one, C15H17FN2O4
- Crystal structure of 2-(4-fluorophenyl)-1,3-dimethyl-1H-perimidin-3-ium iodide, C19H16FIN2
- Crystal structure of methyl 1H-indole-2-carboxylate, C10H9NO2
- Crystal structure of 2,3-diphenyl-1-[(dipropylamino)acetyl]-1,3-diazaspiro[4.5]decan-4-one, C28H37N3O2
- Crystal structure of 1-(2H-1,3-benzodioxol-5-yl)-3-(1H-imidazol-1-yl)propan-1-one, C13H12N2O3
- Crystal structure of 2,9-dibromo-1,10-phenanthroline, C12H6Br2N2
- Crystal structure of 3-(adamantan-1-yl)-4-(4-fluorophenyl)-1H-1,2,4-triazole-5(4H)-thione, C18H20FN3S
- Crystal structure of trans-bis((E)-7-oxo-4-(phenyldiazenyl)cyclohepta-1,3,5-trien-1-olato)-κ2O,O′)-bis(pyridine-κN)cobalt(II), C36H28CoN6O4
- Crystal structure of 2-(4-methyl-3-phenylthiazol-2(3H)-ylidene)malononitrile, C13H9N3S
- Crystal structure of (Z)-3-(adamantan-1-yl)-1-(3-chlorophenyl)-S-benzylisothiourea, C24H27ClN2S
- Crystal structure of chlorido{[3-(η5-cyclopenta-dienyl)-2,2,3-trimethyl-1-phenylbutylidene] azanido-κN}[η2(N,O)-N,N-dimethylhydroxylaminato]titanium(IV), C20H27ClN2OTi
- Crystal Structure of 1,1′-dimethyl-[4,4′-bipyridine]-1,1′-diium tetrachloridozincate(II), C12H14Cl4N2Zn
- Crystal structure of 5-nitro-2-(pyrrolidin-1-yl)benzaldehyde, C11H12N2O3
- Crystal structure of 2,3-diphenyl-1-(morpholin-4-ylacetyl)-1,3-diazaspiro[4.5]decan-4-one, C26H31N3O3
- Crystal structure of 3,3-dimethyl-3,4-dihydro-1H-benzo[c]chromene-1,6(2H)-dione, C15H14O3
- Crystal structure of bis(2-(2-hydroxymethyl)pyridine-κ2N,O)-bis(pivalato-κO)nickel(II), C22H32N2NiO6
- Crystal structure of (1,10-phenanthroline-κ2N,N′)-bis(1H-pyrazole-3-carboxylato-κ2N,O)manganese(II) trihydrate, C20H20N6O7Mn
- The crystal structure of 3-aminopropan-1-aminium iodide, C3H11N2I
- Crystal structure of ethyl 1-(4-chlorophenyl)-5-methyl-1H-1,2,3-triazole-4 carboxylate, C12H12ClN3O2
- Crystal structure of 4,4′-((1Z,1′Z)-2,2′-(2,5-diethoxy-1,4-phenylene)bis(ethene-2,1-diyl))dipyridine, C24H24N2O2
- Crystal structure of (16S)-12,16-epoxy-11,14-dihydroxy-17(15/16)-abeo-3a,18-cyclo-8,11,13-abietatrien-7-one, C20H24O4
- Crystal structure of aquadichlorido(2,4,6-tri-2-pyridyl-1,3,5-triazine-κ3N,N′,N′′)nickel(II) monohydrate, C18H16Cl2N6NiO2
- Crystal structure of catena-poly[dichlorido-(μ-ethane-1,2-diyl-bis-(pyridyl-4-carboxylate-κN:N′)mercury(II)], C15H14Cl2HgN2O4
- Crystal structure of methyl 2-acetamido-5-chlorbenzoate, C10H10ClNO3
- Crystal structure of tetrakis(μ2-3,3-dimethylacrylato-κ2O,O′)-bis(2-aminopyrimidine-κN) dicopper(II), C28H38Cu2N6O8
- Crystal structure of 3-amino-8-methoxy-1-phenyl-1H-benzo[f]chromene-2-carbonitrile, C21H16N2O2
- Crystal structure of 4-(2-ammonioethyl)morpholin-4-ium dichloride monohydrate, C6H18Cl2N2O2
- Crystal structure of 1-(3-((5-bromo-2-hydroxybenzylidene)amino)phenyl)ethanone O-benzyl oxime, C22H19BrN2O2
- Crystal structure of 2-(4-(dimethylamino)-2-fluorophenyl)-1,3-bis(1H-1,2,4-triazol-1-yl)propan-2-ol monohydrate, C15H20FN7O2
- Crystal structure of 4-bromo-2-(1H-pyrazol-3-yl)phenol, C9H7BrN2O
- Crystal structure of 1,2,3,4,5-pentamethyl-1,3-cyclopentadiene, C10H16

