Abstract
C6H11NO4, monoclinic, P21/c (no. 14), a = 19.1903(7) Å, b = 18.4525(6) Å, c = 9.2197(3) Å, β = 95.879(1)°, V = 3247.61(19) Å3, Z = 16, Rgt(F) = 0.0495, wRref (F2) = 0.1194, T = 173 K.
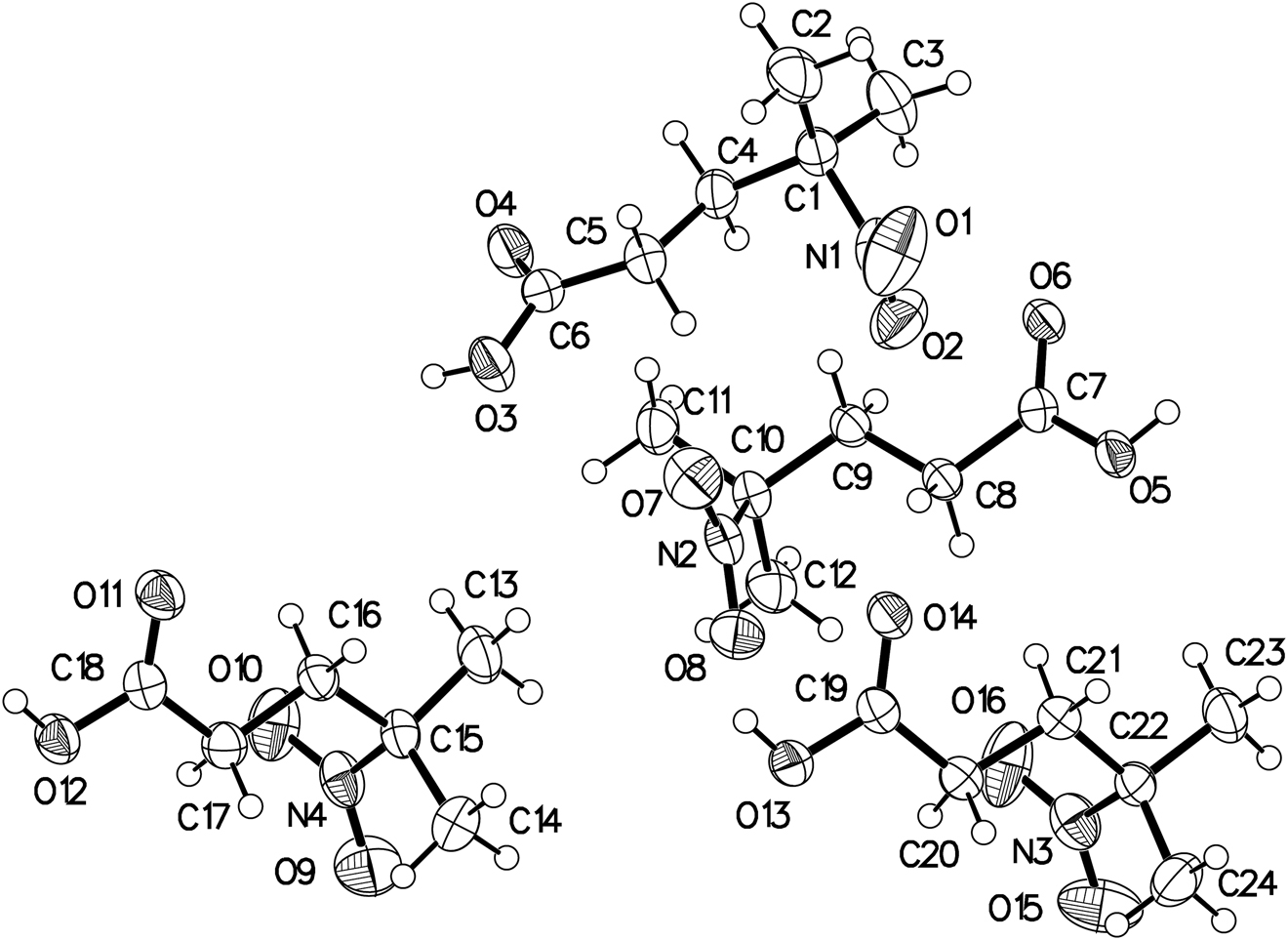
The asymmetric unit (four molecules) of the title crystal structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colourless block |
| Size: | 0.19 × 0.12 × 0.08 mm |
| Wavelength: | MoKα radiation (0.71073 Å) |
| μ: | 0.11 mm−1 |
| Diffractometer, scan mode: | D8 VENTURE, φ and ω |
| θmax, completeness: | 25.7°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 34982, 6176, 0.065 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ (Iobs), 4192 |
| N(param)refined: | 409 |
| Programs: | Bruker [1], Olex2 [2], SHELX [3, 4] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| C1 | 0.75608 (11) | 0.64871 (12) | −0.0723 (2) | 0.0367 (5) |
| C2 | 0.73326 (14) | 0.72754 (13) | −0.0825 (3) | 0.0585 (7) |
| H2A | 0.706299 | 0.739142 | −0.000821 | 0.088* |
| H2B | 0.704067 | 0.735546 | −0.174664 | 0.088* |
| H2C | 0.774710 | 0.758787 | −0.078538 | 0.088* |
| C3 | 0.80611 (13) | 0.63013 (15) | −0.1853 (3) | 0.0565 (7) |
| H3A | 0.847565 | 0.661336 | −0.170800 | 0.085* |
| H3B | 0.782473 | 0.637847 | −0.283447 | 0.085* |
| H3C | 0.820428 | 0.579287 | −0.174277 | 0.085* |
| C4 | 0.69429 (11) | 0.59658 (12) | −0.0813 (2) | 0.0359 (5) |
| H4A | 0.665347 | 0.604531 | −0.175119 | 0.043* |
| H4B | 0.712503 | 0.546397 | −0.081792 | 0.043* |
| C5 | 0.64757 (11) | 0.60351 (11) | 0.0415 (2) | 0.0350 (5) |
| H5A | 0.675130 | 0.590960 | 0.134848 | 0.042* |
| H5B | 0.632397 | 0.654583 | 0.047987 | 0.042* |
| C6 | 0.58415 (11) | 0.55602 (11) | 0.0218 (2) | 0.0337 (5) |
| C7 | 0.92257 (10) | 0.44876 (11) | 0.0671 (2) | 0.0296 (5) |
| C8 | 0.86350 (10) | 0.40871 (11) | 0.1245 (2) | 0.0293 (5) |
| H8A | 0.846285 | 0.437086 | 0.204732 | 0.035* |
| H8B | 0.881073 | 0.361752 | 0.165345 | 0.035* |
| C9 | 0.80290 (10) | 0.39479 (11) | 0.0083 (2) | 0.0306 (5) |
| H9A | 0.821137 | 0.368898 | −0.073968 | 0.037* |
| H9B | 0.784470 | 0.442072 | −0.029089 | 0.037* |
| C10 | 0.74218 (10) | 0.35086 (11) | 0.0585 (2) | 0.0293 (5) |
| C11 | 0.67860 (11) | 0.35406 (13) | −0.0543 (3) | 0.0433 (6) |
| H11A | 0.638989 | 0.329650 | −0.015980 | 0.065* |
| H11B | 0.689465 | 0.329817 | −0.143807 | 0.065* |
| H11C | 0.666394 | 0.404785 | −0.075686 | 0.065* |
| C12 | 0.76261 (12) | 0.27347 (11) | 0.0974 (3) | 0.0439 (6) |
| H12A | 0.802466 | 0.273427 | 0.172937 | 0.066* |
| H12B | 0.775717 | 0.248413 | 0.010533 | 0.066* |
| H12C | 0.722874 | 0.248497 | 0.134020 | 0.066* |
| C13 | 0.56382 (12) | 0.35839 (13) | 0.3456 (3) | 0.0472 (6) |
| H13A | 0.600837 | 0.321590 | 0.353621 | 0.071* |
| H13B | 0.584818 | 0.406546 | 0.361079 | 0.071* |
| H13C | 0.538046 | 0.356125 | 0.248310 | 0.071* |
| C14 | 0.55212 (12) | 0.34131 (13) | 0.6122 (3) | 0.0478 (6) |
| H14A | 0.519089 | 0.328427 | 0.682318 | 0.072* |
| H14B | 0.572771 | 0.388805 | 0.637438 | 0.072* |
| H14C | 0.589296 | 0.304768 | 0.615088 | 0.072* |
| C15 | 0.51379 (10) | 0.34429 (11) | 0.4602 (2) | 0.0329 (5) |
| C16 | 0.45390 (10) | 0.39909 (11) | 0.4473 (2) | 0.0325 (5) |
| H16A | 0.473254 | 0.447322 | 0.476092 | 0.039* |
| H16B | 0.434680 | 0.402127 | 0.343691 | 0.039* |
| C17 | 0.39417 (11) | 0.38264 (11) | 0.5381 (2) | 0.0354 (5) |
| H17A | 0.375231 | 0.333948 | 0.511865 | 0.043* |
| H17B | 0.412538 | 0.381622 | 0.642437 | 0.043* |
| C18 | 0.33636 (11) | 0.43665 (11) | 0.5171 (2) | 0.0323 (5) |
| C19 | 0.82245 (11) | 0.43839 (11) | 0.5703 (2) | 0.0315 (5) |
| C20 | 0.88257 (10) | 0.38917 (11) | 0.6127 (2) | 0.0331 (5) |
| H20A | 0.866486 | 0.338432 | 0.598889 | 0.040* |
| H20B | 0.897645 | 0.396017 | 0.717734 | 0.040* |
| C21 | 0.94537 (10) | 0.40079 (11) | 0.5274 (2) | 0.0333 (5) |
| H21A | 0.966544 | 0.448418 | 0.554780 | 0.040* |
| H21B | 0.928839 | 0.402546 | 0.422200 | 0.040* |
| C22 | 1.00167 (11) | 0.34277 (11) | 0.5520 (2) | 0.0331 (5) |
| C23 | 1.05905 (12) | 0.35373 (13) | 0.4508 (3) | 0.0503 (6) |
| H23A | 1.092088 | 0.313142 | 0.461992 | 0.075* |
| H23B | 1.083956 | 0.399105 | 0.475913 | 0.075* |
| H23C | 1.037897 | 0.355992 | 0.349520 | 0.075* |
| C24 | 1.03104 (13) | 0.33656 (13) | 0.7097 (3) | 0.0503 (6) |
| H24A | 0.993059 | 0.325579 | 0.769665 | 0.075* |
| H24B | 1.053214 | 0.382466 | 0.741832 | 0.075* |
| H24C | 1.065913 | 0.297617 | 0.720061 | 0.075* |
| N1 | 0.79737 (9) | 0.63679 (11) | 0.0767 (2) | 0.0426 (5) |
| N2 | 0.72169 (9) | 0.38875 (10) | 0.1960 (2) | 0.0354 (4) |
| N3 | 0.96799 (10) | 0.26962 (10) | 0.5037 (2) | 0.0436 (5) |
| N4 | 0.48169 (10) | 0.26949 (10) | 0.4223 (2) | 0.0402 (5) |
| O1 | 0.80612 (10) | 0.68733 (11) | 0.1613 (2) | 0.0805 (7) |
| O2 | 0.81854 (9) | 0.57622 (10) | 0.1064 (2) | 0.0614 (5) |
| O3 | 0.55162 (8) | 0.54987 (9) | 0.13922 (16) | 0.0439 (4) |
| H3 | 0.515449 | 0.524521 | 0.120596 | 0.066* |
| O4 | 0.56416 (8) | 0.52655 (9) | −0.09352 (16) | 0.0455 (4) |
| O5 | 0.97344 (7) | 0.46448 (9) | 0.16907 (15) | 0.0397 (4) |
| H5 | 1.005219 | 0.486990 | 0.131798 | 0.059* |
| O6 | 0.92375 (7) | 0.46551 (8) | −0.06038 (15) | 0.0372 (4) |
| O7 | 0.69537 (9) | 0.44870 (9) | 0.18189 (19) | 0.0539 (5) |
| O8 | 0.73406 (9) | 0.35905 (9) | 0.31411 (17) | 0.0511 (4) |
| O9 | 0.50447 (10) | 0.21668 (9) | 0.4906 (2) | 0.0674 (5) |
| O10 | 0.43602 (9) | 0.26558 (9) | 0.3203 (2) | 0.0580 (5) |
| O11 | 0.34033 (8) | 0.49239 (8) | 0.44751 (17) | 0.0440 (4) |
| O12 | 0.28062 (8) | 0.41778 (8) | 0.57958 (17) | 0.0411 (4) |
| H12 | 0.249494 | 0.449598 | 0.563435 | 0.062* |
| O13 | 0.76674 (7) | 0.42231 (8) | 0.63628 (16) | 0.0407 (4) |
| H13 | 0.733639 | 0.450048 | 0.606388 | 0.061* |
| O14 | 0.82438 (7) | 0.48858 (8) | 0.48409 (17) | 0.0411 (4) |
| O15 | 0.99058 (12) | 0.21474 (9) | 0.5639 (2) | 0.0753 (6) |
| O16 | 0.92169 (9) | 0.26922 (10) | 0.4027 (2) | 0.0699 (6) |
Source of material
A mixture of methyl 4-methyl-4-nitropentanoate (13.6 g, 77.7 mmol), H2O (20 mL), and sodium hydroxide (6.2 g, 155.0 mmol) was stirred in methanol (200 mL) at 25 °C for 4 h. The reaction was monitored by thin-layer chromatography. The solution was concentrated and diluted with water, and the pH was adjusted to two by adding hydrochloric acid solution (4 mol/L). Then the mixture was extracted with ethyl acetate and washed with brine. The organic layer was concentrated under reduced pressure, and the residue was purified by column chromatography using petroleum ether and ethyl acetate (5:1, v/v) as eluent. Crystals were obtained by slow evaporation of the aforementioned mixed solution at room temperature after a week.
Experimental details
All hydrogen atoms were placed in theoretical positions and refined in riding models with the U iso values set to 1.2 or 1.5 times of those of the attached atoms. The distances of C–H bonds were constrained from 0.98 to 0.99 Å. Oxygen-bound H atoms were also included in the refinement in the riding model with O–H = 0.84 Å and their U iso = 1.5 U eq (O).
Comment
The γ-aminobutyric acid (GABA) is a kind of non-protein amino acid widely distributed in nature, such as in plants, animals, and microorganisms. It can be produced by plant extraction, isolation, microbial fermentation, and chemical synthesis [5, 6]. Some γ-aminobutyric acids are the primary inhibitory transmitters of the mammalian central nervous system. They have significant physiological functions such as lowering blood pressure, sedation, anti-anxiety, anti-epilepsy, enhancing memory, preventing diabetes, and regulating insomnia [7], [8], [9], [10], [11]. Therefore, γ-aminobutyric acid and its derivatives have broad application prospects in pharmaceutical research [12, 13]. The 4-methyl-4-nitro-pentanoic acid is an essential intermediate for the synthesis of γ- aminobutyric acid derivatives. In addition, it has been widely used in the preparation of lactam, nitro acid, and nitro amide. As a precursor of γ-amino acid, the crystal structure of the title compound would provide a crucial pharmacodynamic fragment for drug molecular design and synthesis.
The asymmetric unit contains four crystallographically independent molecules, as shown in the figure. The bond lengths of C6–O4, C7–O6, C18–O11, and C19–O14 in the title molecule are 1.221(3), 1.218(3), 1.219(3), and 1.223(3) Å, respectively. It illustrated characteristic C=O double bonds. In addition, the bond length of N1–O1, N1–O2, N2–O7, N2–O8, N3–O15, N3–O16, N4–O9, N4–O10 in the title molecule are in the range of 1.211(3)–1.220(3) Å. The bond lengths of C–C, C–N are in the range of 1.489–1.529 Å and 1.531–1.542 Å, respectively. These bond distances are all in their normal ranges. The molecules are connected to each other through hydrogen bonds between carboxyl groups and carboxyl groups, including O3…H3…O4, O13…H13…O11, O12…H12…O14, and O5…H5…O6. The lengths of hydrogen bonds are 1.7906 (16), 1.8015 (15), 1.8374 (14), and 1.8010 (15) Å. It can be seen that the hydrogen bonds are stable and strong.
Funding source: Natural Science Foundation of Shannxi Province http://dx.doi.org/10.13039/501100007128
Award Identifier / Grant number: 2019JQ-924, 2021JQ-883
Funding source: Collaborative Innovation Center of Green Manufacturing Technology for Traditional Chinese Medicine in Shaanxi Province http://dx.doi.org/10.13039/501100009103
Award Identifier / Grant number: 2019XT-1–03
Funding source: Shaanxi University Association for Science and Technology Young Talent Support Program Project http://dx.doi.org/10.13039/501100017550
Award Identifier / Grant number: 20210313
Funding source: Key Laboratory of Molecular Imaging and Drug Synthesis of Xianyang City http://dx.doi.org/10.13039/501100011710
Award Identifier / Grant number: 2021QXNL–PT-0008
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: This work was financially supported by the projects of Natural Science Foundation of Shannxi Province (2019JQ-924, 2021JQ-883), Key breeding program by Collaborative Innovation Center of Green Manufacturing Technology for Traditional Chinese Medicine in Shaanxi Province (2019XT-1–03), Shaanxi University Association for Science and Technology Young Talent Support Program Project (20210313) and Key Laboratory of Molecular Imaging and Drug Synthesis of Xianyang City (2021QXNL–PT-0008).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Bruker. SMART APEX-II CCD; Bruker AXS Inc.: Madison, WI, USA, 2006.Search in Google Scholar
2. Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
3. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar
4. Sheldrick, G. M. SHELXTL – integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8; https://doi.org/10.1107/s2053273314026370.Search in Google Scholar
5. Morse, D. E., Hooker, N., Duncan, H., Jensen, L. γ–aminobutyric acid, a neurotransmitter, induces planktonic abalone larvae to settle and begin metamorphosis. Science 1979, 204, 407–410; https://doi.org/10.1126/science.204.4391.407.Search in Google Scholar
6. Komatsuzaki, N. Production of γ-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 2005, 22, 497–504; https://doi.org/10.1016/j.fm.2005.01.002.Search in Google Scholar
7. Braun, M., Ramracheya, R., Bengtsson, M., Clark, A., Walker, J. N., Johnson, P. R., Rorsman, P. γ–aminobutyric acid (GABA) is an autocrine excitatory transmitter in human pancreatic β-cells. Diabetes 2010, 59, 1694–1701; https://doi.org/10.2337/db09-0797.Search in Google Scholar
8. Hasegawa, T. Anti-hypertensive effect of γ-aminobutyric acid (GABA)-rich on high-normal blood pressure and borderline hypertension in placebo-controlled double blind study. Clin. Exp. Hypertens. 2009, 31, 342–354.10.1080/10641960902977908Search in Google Scholar PubMed
9. Caraiscos, V. B., Elliott, E. M., You-Ten, K. E., Cheng, V. Y., Belelli, D., Newell, J. G., Jackson, M. F., Lambert, J. J., Rosahl, T. W., Wafford, K. A., MacDonald, J. F., Orser, B. A. Tonic inhibition in mouse hippocampal CA1 pyramidal neurons is mediated by α5 subunit-containing γ-aminobutyric acid type A receptors. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 3662–3667; https://doi.org/10.1073/pnas.0307231101.Search in Google Scholar
10. Takeshima, K., Yamatsu, A., Yamashita, Y., Watabe, K., Horie, N., Masuda, K., Kim, M. Subchronic toxicity evaluation of γ-aminobutyric acid (GABA) in rats. Food Chem. Toxicol. 2014, 68, 128–134; https://doi.org/10.1016/j.fct.2014.02.005.Search in Google Scholar
11. Patel, A. B., de Graaf, R. A., Rothman, D. L., Behar, K. L. Effects of γ–aminobutyric acid transporter 1 inhibition by tiagabine on brain glutamate and γ–aminobutyric acid metabolism in the anesthetized rat in vivo. J. Neurosci. Res. 2015, 93, 1101–1108; https://doi.org/10.1002/jnr.23548.Search in Google Scholar
12. Voronina, T. A., Glozman, O. M., Orlova, É. K., Meshcheryakova, L. M., Zauer, V., Ékkard, R., Garibova, T. L., Rakhmankulova, I. K., Rostok, A., Zigemund, K. Synthesis and pharmacological properties of amine analogs of piracetam. Pharm. Chem. J. 1990, 24, 798–802; https://doi.org/10.1007/bf00768378.Search in Google Scholar
13. Chatterjie, N., Alexander, G., Wang, H. Synthesis of valproic acid amides of a melatonin derivative, a piracetam and amantadine for biological tests. Neurochem. Res. 2001, 26, 1171–1176; https://doi.org/10.1023/a:1012383125480.10.1023/A:1012383125480Search in Google Scholar
© 2022 Hongjuan Tong and Wenqiang Tang, published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of (E)-(4-imidazol-1-yl-phenyl)-(2-methoxy-benzylidene)-amine monohydrate, C17H17N3O2
- Crystal structure of 6-methyl-3-(pyrrolidine-1-carbonyl)-2H-chromen-2-one, C15H15N1O3
- Crystal structure of 4-methyl-4-nitropentanoic acid, C6H11NO4
- The crystal structure of (E)-3-(furan-2-yl)acrylonitrile, C7H5NO
- Crystal structure of 3-(difluoromethyl)-1-methyl-N-(4,11,11-trimethyl-1,2,3,4-tetrahydro-1,4-methanoacridin-9-yl)-1H-pyrazole-4-carboxamide monohydrate, C23H26F2N4O3
- Crystal structure of 2-(4-bromobenzyloxy)-6-chloropyridine, C12H9BrClNO
- Crystal structure of N-(4-bromo-2,6-dichloro-phenyl)pyrazin-2-amine, C10H6BrCl2N3
- Crystal structure of (E)-1-(2–nitrophenyl)-3-phenylprop-2-en-1-one, C15H11NO3
- The crystal structure of (E)-3-chloro-2-(2-(4-fluorobenzylidene)hydrazinyl)pyridine, C12H9ClFN3
- Crystal structure of (E)-amino(2-(thiazol-2-ylmethylene)hydrazineyl)methaniminium nitrate, C10H16N12O6S2
- Crystal structure of 9-methoxy-2,3,4,4a,5,6-hexahydro-1H-pyrido [1′,2′:1,6]pyrazino[2,3-b]quinoxaline, C15H18N4O
- The crystal structure bis(dimethylsulfoxide-κ1O)-dipyridine-κ1 N-bis(m2-(Z)-3-methyl-2-oxido-N-((Z)-oxido(phenyl)methylene)benzohydrazonato-κ5)trinickel(II) - dimethylsulfoxide (1/2), C48H56N6Ni3O10S4
- Crystal structure of bis(bis(triphenylphosphine)iminium) tetradecacarbonyltetratelluridopentaferrate(2-), (PPN)2[Fe5Te4(CO)14]
- Crystal structure of 4-Hydroxy-3-(naphthalen-2-ylthio)pent-3-en-2-one, C15H14O2S
- The crystal structure of [(1,10-phenanthroline-κ2 N,N)-bis(6-phenylpyridine-2-carboxylate-κ2 N,O)nickel(II)] monohydrate, C36H26N4O5Ni
- Crystal structure of 3,3′-(pyridine-2,6-diylbis(methylene))bis(1-propyl-1H-imidazol-3-ium) ditetrafluoroborate, C19H27B2F8N5
- The crystal structure of (E)-1-(4-aminophenyl)-3-(p-tolyl)prop-2-en-1-one, C16H15NO
- The crystal structure of poly[(μ2-terephthalato-κ4O,O′: O″,O‴)-(μ4-terephthalato-κ4O:O′:O″:O‴)-{μ4-(1,2,4,5-tetrakis(1,2,4-triazol-1-ylmethyl)-benzene-κ4O:O′:O″,O‴)}dicadmium(II)] – water – acetronitrile (1/2/2), C38H36N14O10Cd2
- The crystal structure of diaqua-bis(6-phenylpyridine-2-carboxylato-κ2 N,O)cobalt(II)–water–N,N-dimethylformamide(1/2/1), C27H31N3O9Co
- The co-crystal structure of 4-hydroxy-3-methoxybenzoic acid – 4,4′-bipyridine, C8H8O4·C10H8N2
- Crystal structure of catena-poly[(μ2-1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol)-κ2N:N′)-bis(4-bromobenzoate-κ1O)cobalt(II)], C32H22Br2CoN4O4
- Crystal structure of (E)-5-propyl-4-((pyridin-2-ylmethylene)amino)-2,4-dihydro-3H-1,2,4-triazole-3-thione – methanol (1/1), C11H13N5S
- The crystal structure of (Z)-4-bromo-6-(((1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl)amino)methylene)cyclohexa-2,4-dien-1-one monohydrate, C11H16BrNO5
- Crystal structure of bis(tetrapropylammonium) nonaselenidotetrastannate(IV), (Pr4N)2[Sn4Se9]
- Crystal structure of 2,6-di-tert-butyl-4-(4-chlorobenzylidene)cyclohexa-2,5-dien-1-one, C21H25ClO
- Crystal structure of (2,2′-((naphthalen-1-ylmethyl)azanediyl)diacetato-κ3 N,O,O′)-(1,10-phenanthroline-κ2 N,N′)-copper(II) trihydrate, CuC27H27N3O7
- The crystal structure of tetrakis(6-phenylpyridine-2-carboxylato-κ2N,O)-bis(1H-pyrazol-3-ylamine-κ2 N:N)dicobalt(II) dihydrate, C27H23N5O5Co
- The crystal structure of bis((E)-2-((tert-butylimino)methyl)-4-chlorophenolato-κ2N,O)zinc(II), C22H26Cl2N2O2Zn
- The crystal structure of poly[diaqua-(μ3-5-nitrobenzene-1,2,3-tricarboxylato-κ3O:O′:O′)-(μ2-4,4′-dipyridylamine-κ2N:N′)copper(II)], C38H30Cu3N8O20
- The crystal structure of (E)-1-ferrocenyl-3-(naphthalen-1-yl)prop-2-en-1-one, C23H18FeO
- The crystal structure of (E)-1-ferrocenyl-3-(4-isopropylphenyl)prop-2-en-1-one, C22H22FeO
- Crystal structure of 6-hydroxy-2,2-dimethyl-4Hbenzo[d][1,3]dioxin-4-one, C10H10O4
- The crystal structure of (2E,4E)-1-ferrocenyl-5-phenylpenta-2,4-dien-1-one, C21H18FeO
- Crystal structure of alaninato-κ2N,O-bis(hydroxylamido-κ2N,O)-oxido-vanadium(V), C3H10N3O5V
- Crystal structure of catena-poly[aqua-bis[μ2-6-(1H-imidazol-1-yl)nicotinato-κ2 N,O]copper(II)], C18H14N6O5Cu
- Crystal structure of diethyl 4,6-diphenyl-1,9-di-p-tolylhexahydro-3H-2,7,3,5-(epimethanetriyliminomethanetriyl)cyclopenta[b]pyridine-3,5(2H)-dicarboxylate, C42H42N2O4
- The crystal structure of cobalt cadmium bis(hydrogenphosphate) bis(phosphate(V)) tetrahydrate, H10O20P4Co3.14Cd1.86
- Crystal structure of dimethyl 1,4,6,9-tetraphenylhexahydro-3H-2,7,3,5-(epimethanetriyliminomethanetriyl)cyclopenta[b]pyridine-3,7(2H)-dicarboxylate, C38H34N2O4
- Crystal structure of (Z)-4-(furan-2-yl((4-iodophenyl)amino)methylene)-5-methyl-2(p-tolyl)-2,4-dihydro-3H-pyrazol-3-one, C21H16I N3O2
- Crystal structure of (E)-1-(4-(3,5-dimethoxystyryl)phenyl)-7-ethylheptanedioate, C25H30O6
- Crystal structure of 6-bromo-2-(4-chlorophenyl)chroman-4-one (6-bromo-4′-chloroflavanone), C15H10BrClO2
- The crystal structure of 2-(benzhydryloxy)-3-nitropyridine, C18H14N2O3
- The crystal structure of 1,3(4,1)-dipyridin-1-iuma-2(1,8)-diethynylanthracena-5(1,3)-benzenacyclohexaphane-11,31-diium bis(hexafluoridophosphate), C36H24F12N2P2
- Crystal structure of 3,6-di-tert-butyl-1-iodo-9-methyl-8-(pyren-1-ylethynyl)-9H-carbazole, C39H34IN
- The cocrystal 2-(dimethylammonio)-5-nitrobenzoate – 2-(dimethylamino)-5-nitrobenzoic acid, C9H10N2O4
- Crystal structure of 5-nitroquinazolin-4(3H)-one, C8H5N3O3
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of (E)-(4-imidazol-1-yl-phenyl)-(2-methoxy-benzylidene)-amine monohydrate, C17H17N3O2
- Crystal structure of 6-methyl-3-(pyrrolidine-1-carbonyl)-2H-chromen-2-one, C15H15N1O3
- Crystal structure of 4-methyl-4-nitropentanoic acid, C6H11NO4
- The crystal structure of (E)-3-(furan-2-yl)acrylonitrile, C7H5NO
- Crystal structure of 3-(difluoromethyl)-1-methyl-N-(4,11,11-trimethyl-1,2,3,4-tetrahydro-1,4-methanoacridin-9-yl)-1H-pyrazole-4-carboxamide monohydrate, C23H26F2N4O3
- Crystal structure of 2-(4-bromobenzyloxy)-6-chloropyridine, C12H9BrClNO
- Crystal structure of N-(4-bromo-2,6-dichloro-phenyl)pyrazin-2-amine, C10H6BrCl2N3
- Crystal structure of (E)-1-(2–nitrophenyl)-3-phenylprop-2-en-1-one, C15H11NO3
- The crystal structure of (E)-3-chloro-2-(2-(4-fluorobenzylidene)hydrazinyl)pyridine, C12H9ClFN3
- Crystal structure of (E)-amino(2-(thiazol-2-ylmethylene)hydrazineyl)methaniminium nitrate, C10H16N12O6S2
- Crystal structure of 9-methoxy-2,3,4,4a,5,6-hexahydro-1H-pyrido [1′,2′:1,6]pyrazino[2,3-b]quinoxaline, C15H18N4O
- The crystal structure bis(dimethylsulfoxide-κ1O)-dipyridine-κ1 N-bis(m2-(Z)-3-methyl-2-oxido-N-((Z)-oxido(phenyl)methylene)benzohydrazonato-κ5)trinickel(II) - dimethylsulfoxide (1/2), C48H56N6Ni3O10S4
- Crystal structure of bis(bis(triphenylphosphine)iminium) tetradecacarbonyltetratelluridopentaferrate(2-), (PPN)2[Fe5Te4(CO)14]
- Crystal structure of 4-Hydroxy-3-(naphthalen-2-ylthio)pent-3-en-2-one, C15H14O2S
- The crystal structure of [(1,10-phenanthroline-κ2 N,N)-bis(6-phenylpyridine-2-carboxylate-κ2 N,O)nickel(II)] monohydrate, C36H26N4O5Ni
- Crystal structure of 3,3′-(pyridine-2,6-diylbis(methylene))bis(1-propyl-1H-imidazol-3-ium) ditetrafluoroborate, C19H27B2F8N5
- The crystal structure of (E)-1-(4-aminophenyl)-3-(p-tolyl)prop-2-en-1-one, C16H15NO
- The crystal structure of poly[(μ2-terephthalato-κ4O,O′: O″,O‴)-(μ4-terephthalato-κ4O:O′:O″:O‴)-{μ4-(1,2,4,5-tetrakis(1,2,4-triazol-1-ylmethyl)-benzene-κ4O:O′:O″,O‴)}dicadmium(II)] – water – acetronitrile (1/2/2), C38H36N14O10Cd2
- The crystal structure of diaqua-bis(6-phenylpyridine-2-carboxylato-κ2 N,O)cobalt(II)–water–N,N-dimethylformamide(1/2/1), C27H31N3O9Co
- The co-crystal structure of 4-hydroxy-3-methoxybenzoic acid – 4,4′-bipyridine, C8H8O4·C10H8N2
- Crystal structure of catena-poly[(μ2-1,1′-(biphenyl-4,4′-diyl)bis(1H-imidazol)-κ2N:N′)-bis(4-bromobenzoate-κ1O)cobalt(II)], C32H22Br2CoN4O4
- Crystal structure of (E)-5-propyl-4-((pyridin-2-ylmethylene)amino)-2,4-dihydro-3H-1,2,4-triazole-3-thione – methanol (1/1), C11H13N5S
- The crystal structure of (Z)-4-bromo-6-(((1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl)amino)methylene)cyclohexa-2,4-dien-1-one monohydrate, C11H16BrNO5
- Crystal structure of bis(tetrapropylammonium) nonaselenidotetrastannate(IV), (Pr4N)2[Sn4Se9]
- Crystal structure of 2,6-di-tert-butyl-4-(4-chlorobenzylidene)cyclohexa-2,5-dien-1-one, C21H25ClO
- Crystal structure of (2,2′-((naphthalen-1-ylmethyl)azanediyl)diacetato-κ3 N,O,O′)-(1,10-phenanthroline-κ2 N,N′)-copper(II) trihydrate, CuC27H27N3O7
- The crystal structure of tetrakis(6-phenylpyridine-2-carboxylato-κ2N,O)-bis(1H-pyrazol-3-ylamine-κ2 N:N)dicobalt(II) dihydrate, C27H23N5O5Co
- The crystal structure of bis((E)-2-((tert-butylimino)methyl)-4-chlorophenolato-κ2N,O)zinc(II), C22H26Cl2N2O2Zn
- The crystal structure of poly[diaqua-(μ3-5-nitrobenzene-1,2,3-tricarboxylato-κ3O:O′:O′)-(μ2-4,4′-dipyridylamine-κ2N:N′)copper(II)], C38H30Cu3N8O20
- The crystal structure of (E)-1-ferrocenyl-3-(naphthalen-1-yl)prop-2-en-1-one, C23H18FeO
- The crystal structure of (E)-1-ferrocenyl-3-(4-isopropylphenyl)prop-2-en-1-one, C22H22FeO
- Crystal structure of 6-hydroxy-2,2-dimethyl-4Hbenzo[d][1,3]dioxin-4-one, C10H10O4
- The crystal structure of (2E,4E)-1-ferrocenyl-5-phenylpenta-2,4-dien-1-one, C21H18FeO
- Crystal structure of alaninato-κ2N,O-bis(hydroxylamido-κ2N,O)-oxido-vanadium(V), C3H10N3O5V
- Crystal structure of catena-poly[aqua-bis[μ2-6-(1H-imidazol-1-yl)nicotinato-κ2 N,O]copper(II)], C18H14N6O5Cu
- Crystal structure of diethyl 4,6-diphenyl-1,9-di-p-tolylhexahydro-3H-2,7,3,5-(epimethanetriyliminomethanetriyl)cyclopenta[b]pyridine-3,5(2H)-dicarboxylate, C42H42N2O4
- The crystal structure of cobalt cadmium bis(hydrogenphosphate) bis(phosphate(V)) tetrahydrate, H10O20P4Co3.14Cd1.86
- Crystal structure of dimethyl 1,4,6,9-tetraphenylhexahydro-3H-2,7,3,5-(epimethanetriyliminomethanetriyl)cyclopenta[b]pyridine-3,7(2H)-dicarboxylate, C38H34N2O4
- Crystal structure of (Z)-4-(furan-2-yl((4-iodophenyl)amino)methylene)-5-methyl-2(p-tolyl)-2,4-dihydro-3H-pyrazol-3-one, C21H16I N3O2
- Crystal structure of (E)-1-(4-(3,5-dimethoxystyryl)phenyl)-7-ethylheptanedioate, C25H30O6
- Crystal structure of 6-bromo-2-(4-chlorophenyl)chroman-4-one (6-bromo-4′-chloroflavanone), C15H10BrClO2
- The crystal structure of 2-(benzhydryloxy)-3-nitropyridine, C18H14N2O3
- The crystal structure of 1,3(4,1)-dipyridin-1-iuma-2(1,8)-diethynylanthracena-5(1,3)-benzenacyclohexaphane-11,31-diium bis(hexafluoridophosphate), C36H24F12N2P2
- Crystal structure of 3,6-di-tert-butyl-1-iodo-9-methyl-8-(pyren-1-ylethynyl)-9H-carbazole, C39H34IN
- The cocrystal 2-(dimethylammonio)-5-nitrobenzoate – 2-(dimethylamino)-5-nitrobenzoic acid, C9H10N2O4
- Crystal structure of 5-nitroquinazolin-4(3H)-one, C8H5N3O3

