Abstract
2[C5H10N3][C8F4O4], monoclinic, P21/n (no. 14), a = 6.1490(2) Å, b = 11.0063(3) Å, c = 14.0214(3) Å, β = 94.386(5)∘, V = 946.16(5) Å3, Z = 2, R gt (F) = 0.0371, wR ref (F2) = 0.1127, T = 178 K.
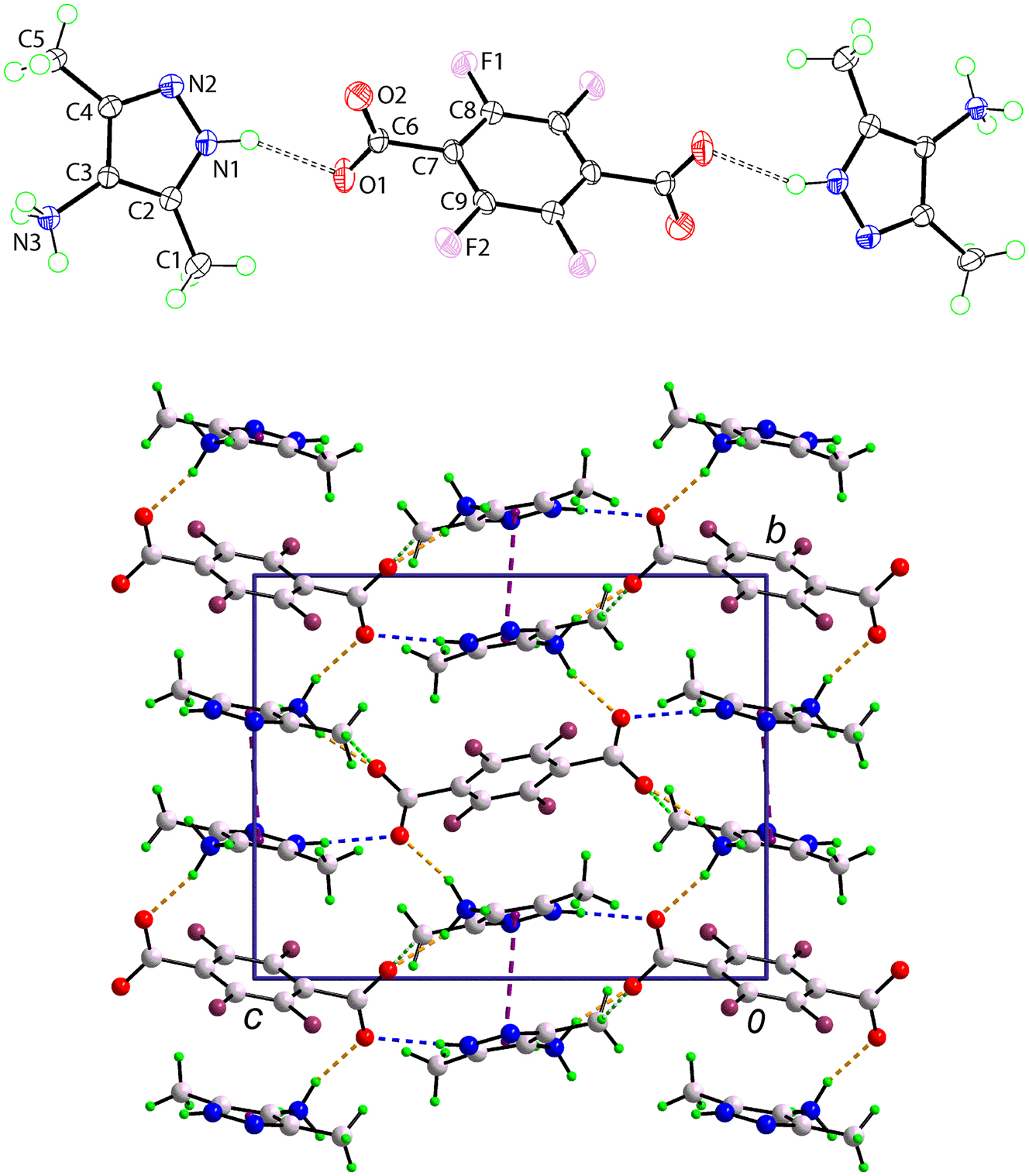
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colourless block |
| Size: | 0.16 × 0.14 × 0.10 mm |
| Wavelength: | MoKα radiation (0.71073 Å) |
| μ: | 0.14 mm−1 |
| Diffractometer, scan mode: | Rigaku XtaLAB P200, ω |
| θmax, completeness: | 29.6°, >99 % |
| N(hkl)measured, N(hkl)unique, Rint: | 6985, 2414, 0.021 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 2259 |
| N(param)refined: | 159 |
| Programs: | CrysAlisPRO [1], IL MILIONE [2], SHELX [3], WinGX/ORTEP [4] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| N1 | 0.36750 (15) | 0.33299 (9) | −0.08110 (7) | 0.0178 (2) |
| H1N | 0.291 (2) | 0.3370 (14) | −0.1373 (8) | 0.021* |
| N2 | 0.26894 (15) | 0.36280 (9) | −0.00029 (7) | 0.0182 (2) |
| N3 | 0.83825 (14) | 0.32853 (9) | 0.08590 (6) | 0.0156 (2) |
| H2N | 0.841 (3) | 0.2570 (10) | 0.1197 (10) | 0.023* |
| H3N | 0.853 (3) | 0.3915 (12) | 0.1283 (10) | 0.023* |
| H4N | 0.955 (2) | 0.3320 (15) | 0.0496 (11) | 0.023* |
| C1 | 0.72698 (19) | 0.28321 (12) | −0.14236 (8) | 0.0230 (2) |
| H1A | 0.733071 | 0.194500 | −0.147514 | 0.034* |
| H1B | 0.874302 | 0.315120 | −0.127015 | 0.034* |
| H1C | 0.667020 | 0.317498 | −0.203310 | 0.034* |
| C2 | 0.58520 (17) | 0.31739 (10) | −0.06537 (7) | 0.0155 (2) |
| C3 | 0.62920 (16) | 0.33752 (9) | 0.03093 (7) | 0.0139 (2) |
| C4 | 0.43077 (17) | 0.36511 (9) | 0.06909 (7) | 0.0155 (2) |
| C5 | 0.39585 (18) | 0.39193 (11) | 0.17112 (8) | 0.0211 (2) |
| H5A | 0.239157 | 0.398388 | 0.178724 | 0.032* |
| H5B | 0.467426 | 0.468767 | 0.189879 | 0.032* |
| H5C | 0.458021 | 0.326241 | 0.211774 | 0.032* |
| F1 | −0.27376 (11) | 0.38486 (7) | −0.38662 (5) | 0.02198 (18) |
| F2 | 0.40163 (11) | 0.57251 (7) | −0.42807 (5) | 0.02257 (18) |
| O1 | 0.25345 (15) | 0.35392 (8) | −0.28398 (6) | 0.0240 (2) |
| O2 | 0.08167 (14) | 0.52050 (9) | −0.23902 (6) | 0.0265 (2) |
| C6 | 0.14165 (17) | 0.44777 (10) | −0.29906 (7) | 0.0157 (2) |
| C7 | 0.06848 (17) | 0.47559 (9) | −0.40327 (7) | 0.0145 (2) |
| C8 | −0.13667 (17) | 0.44180 (9) | −0.44222 (7) | 0.0152 (2) |
| C9 | 0.20297 (17) | 0.53508 (9) | −0.46292 (7) | 0.0153 (2) |
1 Source of material
Under an argon atmosphere, the reaction of 4-amino-3,5-dimethyl-1-pyrazole (0.050 g, 0.45 mmol) with tetrafluoroterephthalic acid (0.049 g, 0.21 mmol) in anhydrous methanol (10 mL) was conducted at room temperature overnight. The solvent was removed in vacuo. The colourless crystals of (I), suitable for X-ray analysis, were obtained by slow evaporation at room temperature from an anhydrous methanol and toluene solution (7:3 v/v) (0.042 g, 0.091 mmol, 43 % yield). Anal. Calcd. for C18H20F4N6O4: C 46.96, H 4.38, N 18.25 %. Found: C 46.85, H 4.41, N 18.27 %. 1H NMR (CD3OD, 500 MHz): δ 2.17 (s, 12H, CH3); N–H protons were not observed. IR (KBr, cm−1): 3197 s ν(N–H), 2894 s ν(C–H), 1639 s ν(C=O), 1607 s ν(C=O), 1466 s, 1382 s, 978 s, 728 s.
2 Experimental details
The C-bound H atoms were geometrically placed (C–H = 0.98 Å) and refined as riding with Uiso(H) = 1.5Ueq(C). The N-bound H atoms were located in a difference Fourier map and refined with pyrazolyl–N–H = 0.88 ± 0.01 Å and ammonium–N–H = 0.91 ± 0.01 Å with Uiso(H) = 1.2 and 1.5Ueq(N), respectively.
3 Comment
Recently, covalent organic frameworks (COFs) became recognised as a new class of materials featuring linkages supported by strong covalent bonds to form two-or three-dimensional, porous and stable crystalline materials [5]. In COFs the covalent links can arise from, for example, B–O, imine–C=N and amide–C–N bond formation [6]. The title salt (I, systematic name: bis(3,5-dimethyl-1H-pyrazol-4-aminium) 2,3,5,6-tetrafluorobenzene-1,4-dicarboxylate) was generated in an on-going study into imine–C=N bond formation, employing an aliphatic or aromatic amine reacting with an aldehyde followed by dehydration. Previously, the structure of the result of C=N bond formation by the direct reaction of 4-amino-3,5-diisopropyl-1-pyrazole (L1HpzNH2) [7] with terephthalaldehyde (benzene-1,4-dicarboxaldehyde) was described [8]. Moreover, the structure obtained by the reaction of the less hindered methyl-substituted pyrazole viz. 4-amino-3,5-dimethyl-1-pyrazole (L0HpzNH2) with terephthalaldehyde in anhydrous methanol solution has also been described recently [9]. In this article, the results of the exploration of amide bond formation by the reaction of L0HpzNH2 with a carboxylic acid, i.e. tetrafluoroterephthalic acid, are outlined. While initially terephthalic acid was employed as the carboxylic acid source, this acid has very poor solubility in normal organic solvents such as methanol, ethanol and chloroform. Therefore, tetrafluoroterephthalic acid was evaluated as the carboxylic acid precursor, since the presence of fluoride improves the solubility in regular solvents due to the polarized Cδ+ and Fδ− atoms [10, 11]. In the IR spectrum, salt (I) has a characteristic absorption band at 3197 cm−1, which is assigned to N–H stretching. It is also noted that the sharp N–H2 stretching band evident at 3345 cm−1 in the IR spectrum of L0HpzNH2 disappeared. In addition, new characteristic C=O stretching bands, split at 1639 and 1607 cm−1, which are clearly shifted from those of the other starting material, tetrafluoroterephthalic acid (1700 cm−1).
The molecular structures of the constituents of salt (I) are shown in the upper view of the figure (70 % displacement ellipsoids, dashed lines indicate N–H⋯O hydrogen bonds and unlabelled atoms are related by the symmetry operation – x, 1 – y, 1 – z). The asymmetric-unit comprises a 3,5-dimethyl-1H-pyrazol-4-ammonium cation, in a general position, and half a 2,3,5,6-tetrafluorobenzene-1,4-dicarboxylate dianion, being situated about a centre of inversion. The confirmation of proton transfer during co-crystallisation is evident in the near equivalence of the C6–O1,O2 bond lengths [1.2498(13) & 1.2381(13) Å] and the wider angle at the N1 atom [C2–N1–N2 = 113.09(9)∘] compared with that at the N2 atom [C4–N2–N1 = 104.80(9)∘].
The pyrazolyl ring is planar to ±0.002(1) Å and an evaluation of the bond lengths within the five-membered ring is consistent with the delocalisation of π-electron density over the constituent atoms. Thus, the formally double-bonded C4–N2 [1.3376(14) Å] and C2–C3 [1.3745(13) Å] bonds are elongated and the formally single-bonded N1–N2 [1.3653(13) Å], C2–N1 [1.3509(14) Å] and C3–C4 [1.4026(14) Å] bonds are shortened; the exocyclic C3–N3 bond length is 1.4508(13) Å. The dianion is not planar with the carboxylate residues being close to orthogonal to the phenyl ring to which they are connected to, with the dihedral angle between the respective least-squares planes being 83.14(6)∘. The dihedral angle between the carboxylate residue and the hydrogen bonded pyrazolyl ring is 41.84(8)∘.
The crystal structure of the neutral 3,5-dimethyl-4-aminopyrazole molecule has been described [12]. There is a high degree of concordance in the geometric parameters in the pyrazolyl rings although the N1–N2 bond length of 1.353(2)° is shorter than that in (I) [1.3653(13) Å]. Further, two molecules of 3,5-dimethyl-4-aminopyrazole coordinate the cobalt(II) atom of CoCl2 via the imine–N atoms to generate a tetrahedral complex [13]. Finally, a cation of 3,5-dimethyl-4-aminopyrazole with a 3,4-dicarboxy-3-hydroxybutanoate counter-anion, as a monohydrate, has been reported [14]. Here, protonation has occurred at the ring rather than at the amino–N atom in (I).
As anticipated from the compositon of (I), extensive hydrogen-bonding is evident in the crystal; all specified hydrogen bonds are charge-assisted. The connection between the pyrazolyl–N–H and the carboxylate–O1 atoms [N1–H1n⋯O1: H1n⋯O1 = 2.060(11) Å, N1⋯O1 = 2.8865(13) Å with angle at H1n = 154.5(11)∘] is highlighted as blue-dashed bonds in the unit-cell diagram shown in the lower view of the figure. When viewed down the a-axis direction, the crystal comprises columns of cations and anions such that each column comprising cations is surrounded by six columns of alternatively charged species, whereas each column of anions is surrounded by six columns of cations. Hydrogen bonds linking cations within a column are of the type ammonium–N–H⋯N(pyrazolyl) [N3–H4n⋯N2 i : H4n⋯N2 i = 2.130(13) Å, N3⋯N2 i = 3.0176(13) Å with angle at H4n = 164.1(13)∘ for symmetry operation (i) 1 + x, y, z]. The remaining ammonium–N–H⋯O(carboxylate) hydrogen bonds involve two different carboxylate residues within the same column [N3–H2n⋯O1 ii : H2n⋯O1 ii = 1.928(14) Å, N3⋯O1 ii = 2.7890(13) Å with angle at H2n = 155.4(15)∘ and N3–H3n⋯O2 iii = H3n⋯O2 iii = 1.848(14) Å, N3⋯O2 iii = 2.7297(13) Å with angle at H3n = 161.7(13)∘ for (ii) – x, 1 – y, – z and (iii) 1 – x, 1 – y, – z]. All specified contacts occur within a three-dimensional assembly. Complementing the hydrogen-bonding interactions are close methyl–C–H⋯O(carboxylate) [C5–H5a⋯O2 iv : H5a⋯O2 iv = 2.38 Å, C5⋯O2 iv = 3.2992(14) Å with angle at H5a = 156° for (iv) 1/2 + x, 1/2 – y, 1/2 + z] interactions, shown as bright-green dashed bonds in the unit-cell diagram, and π(pyrazolyl)⋯π(pyrazolyl) [Cg(pyrazolyl)⋯Cg(pyrazolyl) iii = 3.5008(7) Å] contacts between centrosymmetrically related molecules occupying different columns (purple dashed lines). Longer [3.7880(6) Å] π(pyrazolyl)⋯π(phenyl) contacts are also noted.
This analysis was augmented by the calculation of the Hirshfeld surfaces and of the full and delineated two-dimensional fingerprint plots with the aid of Crystal Explorer 21 [15] and standard protocols [16]. The calculations were based on the three-molecule aggregate shown in the upper view of the figure, and highlight the prominent role hydrogen plays in the molecular packing by participating in 86.0 % of all surface contacts. While H⋯H contacts contribute 23.5 % to the calculated surface, the greatest contribution comes from O⋯H/H⋯O contacts, reflecting, in part, the prominent role of hydrogen-bonding involving oxygen. After these, the next most dominant contacts involve fluoride with F⋯H/H⋯F contacts contributing 19.8 % to the surface but at separations greater than the van der Waals criterion. At 9.2 and 7.7 %, significant contributions are made by N⋯H/H⋯N and C⋯H/H⋯C contacts, respectively. Smaller contributions are made by F⋯F [3.8 %], C⋯C [3.7 %], N⋯C/C⋯N [3.6 %] and F⋯C. C⋯F [2.1 %] surface contacts. Naturally, when the individual ions were evaluated for their specific surface contacts, vastly different distributions of surface contacts are apparent. Thus, for the cation, the most signficant contributions to the surface about this species were H⋯H [35.0 %], H⋯O [21.3 %], H⋯N/N⋯H [13.9 %], H⋯F [13.5 %] and H⋯C/C⋯H [6.2 %]. For the di-anion, O⋯H [42.6 %], F⋯H [26.3 %], F⋯F [9.3 %] and C⋯H [8.8 %] surface contacts predominate.
Acknowledgments
KF is grateful for support from the joint usage/research programme “Artificial Photosynthesis” based at Osaka City University.
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: This study was financially supported by the Joint Usage/Research Center for Catalysis (Proposals 22DS0143 and 23DS0198).
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. Rigaku Oxford Diffraction. CrysAlisPRO; Rigaku Corporation: Oxford, UK, 2021.Search in Google Scholar
2. Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G., Siliqi, D., Spagna, R. IL MILIONE: a suite of computer programs for crystal structure solution of proteins. J. Appl. Cryst. 2007, 40, 609–613; https://doi.org/10.1107/S0021889807010941.Search in Google Scholar
3. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, C71, 3–8; https://doi.org/10.1107/S2053229614024218.Search in Google Scholar PubMed PubMed Central
4. Farrugia, L. J. WinGX and ORTEP for windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854; https://doi.org/10.1107/S0021889812029111.Search in Google Scholar
5. Côté, A. P., Benin, A. I., Ockwig, N. W., O′Keeffe, M., Matzger, A. J., Yaghi, O. M. Porous, crystalline, covalent organic frameworks. Science 2005, 310, 1166–1170; https://www.science.org/doi/10.1126/science.1120411.10.1126/science.1120411Search in Google Scholar PubMed
6. Diercks, C. S., Yaghi, O. M. The atom, the molecule, and the covalent organic framework. Science 2017, 355, eaa1585; https://www.science.org/doi/10.1126/science.aal1585.10.1126/science.aal1585Search in Google Scholar PubMed
7. Fujisawa, K., Ageishi, K., Okano, M., Tiekink, E. R. T. The crystal structure of 3,5-bis(propan-2-yl)-1H-pyrazol-4-amine, C9H17N3. Z. Kristallogr. N. Cryst. Struct. 2022, 237, 1055–1057; https://doi.org/10.1515/ncrs-2022–0362.10.1515/ncrs-2022-0362Search in Google Scholar
8. Fujisawa, K., Ageishi, K., Tiekink, E. R. T. Crystal structure of [N(E),N′(E)]-N,N′-(1,4-phenylenedimethylidyne)bis-3,5-bis(propan-2-yl)-1H-pyrazol-4-amine, C26H36N6. Z. Kristallogr. N. Cryst. Struct. 2023, 238, 973–976. https://doi.org/10.1515/ncrs-2023–0305.10.1515/ncrs-2023-0305Search in Google Scholar
9. Fujisawa, K., Ageishi, K., Harakuni, S., Tiekink, E. R. T. Crystal structure of [N(E),N′(E)]-N,N′-(1,4-phenylenedimethylidyne)bis-3,5-dimethyl-1H-pyrazol-4-amine di-methanol solvate, C18H20N6⋅2(CH3OH). Z. Kristallogr. N. Cryst. Struct. 2023, 238, 1005–1008. https://doi.org/10.1515/ncrs-2023–0322.10.1515/ncrs-2023-0322Search in Google Scholar
10. O′Hagan, D. Understanding organofluorine chemistry. An introduction to the C–F bond. Chem. Soc. Rev. 2008, 37, 308–319; https://doi.org/10.1039/b711844a.Search in Google Scholar PubMed
11. Purser, S., Moore, P. R., Swallow, S., Gouverneur, V. Fluorine in medicinal chemistry. Chem. Soc. Rev. 2008, 37, 320–330; https://doi.org/10.1039/b610213c.Search in Google Scholar PubMed
12. Infantes, L., Foces-Foces, C., Claramunt, R. M., López, C., Elguero, J. Aminopyrazoles and their conjugated acids. An X-ray study of 3,5-dimethyl-4-aminopyrazole and the picrate of 3(5)-aminopyrazole. J. Heterocycl. Chem. 1999, 36, 595–600; https://doi.org/10.1002/jhet.5570360303.Search in Google Scholar
13. Cai, X.-W., Zhao, Y.-Y., Han, G.-F. Dichloridobis(3,5-dimethyl-1H-pyrazol-4-amine-κN2)cobalt(II). Acta Crystallogr. 2008, E64, m1012; https://doi.org/10.1107/S1600536808020461.Search in Google Scholar PubMed PubMed Central
14. Radhika, B., Prashanth, J., Basavoju, S., Jyothi, S., Venkatram Reddy, B. Synthesis, single-crystal X-ray diffraction, NLO and DFT studies of centrosymmetric 4-amino-3,5-dimethyl-1H-pyrazolium citrate monohydrate salt. Mol. Phys. 2022, 120, e2022797; https://doi.org/10.1080/00268976.2021.2022797.Search in Google Scholar
15. Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D., Spackman, M. A. CrystalExplorer: a program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011; https://doi.org/10.1107/S1600576721002910.Search in Google Scholar PubMed PubMed Central
16. Tan, S. L., Jotani, M. M., Tiekink, E. R. T. Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. 2019, E75, 308–318; https://doi.org/10.1107/S2056989019001129.Search in Google Scholar PubMed PubMed Central
© 2023 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of (N-([1,1′:4′,1″-terphenyl]-4,4′-diethyl)-2-(bis(pyridin-2-ylmethyl)amino)acetamide-κ4N,N,N″, O)tri(nitrato-kO, O′) samarium(III) - methanol - acetonitrile (1/1/1), C40H39SmN8O14
- The crystal structure of 6,6′-(((2-(dimethylamino)ethyl)azanediyl)bis(methylene))bis(2-chloro-4-methyl phenolate-κ4N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)-titanium(IV), C27H27Cl2N3O6Ti
- N′-[(1E)-(4–Fluorophenyl)methylidene]adamantane-1-carbohydrazide, C18H21FN2O
- Crystal structure of 4-bromo-3-nitro-1H-pyrazole-5-carboxylic acid monohydrate, C4H2N3BrO4·H2O
- Crystal structure of dipyridine-k1N-tris(2,2,6,6-tetramethyl-5-oxohept-3-en-3-olato-k2O,O′)dysprosium(III), DyC43H67O6N2
- Crystal structure of cyclo[tetraiodido-bis{μ2-1-[(benzotriazol-1-yl)methyl]-1-H-1,3-(2-isopropyl-imidazol)-k2N:N}dicadmiun(II)], C26H30N10Cd2I4
- The crystal structure of tert-butyl (E)-3-(2-(benzylideneamino)phenyl)-1H-indole-1-carboxylate, C26H24N2O2
- The crystal structure of 4-(3-carboxy-1-cyclopropyl-6-fluoro-8-methoxy-4-oxo-1,4- dihydroquinolin-7-yl)-2-methylpiperazin-1-ium 2,5-dihydroxybenzoate methanol solvate, C27H32FN3O9
- Crystal structure of (μ2-1-(4,4′-bipyridine-κ2N:N′)-bis[diaqua-(4-iodopyridine-2,6-dicarboxylato-κ3O,N,O′)–cobalt(II)], C24H20Co2I2N4O12
- The crystal structure of dimethyl 4,4′-(10,20-diphenylporphyrin-5,15-diyl)dibenzoate dichloromethane solvate, C49H36N4O4Cl2
- (E)-2-((E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-ylidene)hydrazine-1-carbothioamide C14H23N3S1
- The crystal structure of [1-(4-(trifluoromethyl)phenyl)-3,4-dihydroquinolin-2(1H)-one], C16H12F3NO
- Crystal structure of (E)-2-amino-N′-((3-hydroxy-5-(hydroxymethyl)-2-methylpyridin-4-yl)methylene)benzohydrazide – dimethylformamide – water (1/1/2), C15H16N4O3·C3H7NO·2H2O
- Crystal structure of 3-(4-bromophenyl)-5-methyl-1H-pyrazole, C10H9BrN2
- Crystal structure of 1,10-phenanthrolinium bromide dihydrate, C12H9N2Br
- Crystal structure of N-(4′-chloro-[1,1′-biphenyl]-2-yl)formamide, C13H10ClNO
- The crystal structure of nitroterephthalic acid, C8H5NO6
- Crystal structure of (2-((4-bromo-2,6-dichlorophenyl)amino)phenyl) (morpholino)methanone, C17H15BrCl2N2O2
- Crystal structure of tetraaqua-bis(ethanol-κO)-tetrakis(μ2-trifluoroacetate-κ2O:O′)-bis(trifluoroacetate-κ2O)digadolinium(III) Gd2C16H20O18F18
- The crystal structure of dimethyl 4,4′-[10,20-bis(2,6-difluorophenyl)porphyrin-5,15-diyl]dibenzoate chloroform solvate, C50H32Cl6F4N4O4
- The crystal structure of N,N′-((nitroazanediyl)bis(methylene))diacetamide, C6H12O4N4
- The crystal structure of [bis(2,2′-bipyridine-6-carboxylato-κ3N,N,O)magnesium(II)]dihydrate, C22H18N4O6Mg
- Crystal structure of poly[diaqua-(bis(μ2-1,4-bis(imidazol-1-ylmethyl)benzene)-κ2N,N′] cobalt(II)-tetraqua-bis(1,4-bis(imidazol-1-ylmethyl)benzene)-κ1N)-cobalt(II) di(2,5-thiophenedicarboxylate) dihydrate, C68H76Co2N16O16S2
- Crystal structure of poly[chlorido-μ2-chlorido-(μ2-1-[(2-ethyl-4-methyl-1H-imidazol-1-yl)methyl]-1H-benzotriazole-κN:N’)cadmium(II)], C13H15CdN5Cl2
- The crystal structure of (4-hydroxybenzenesulfonate)-k1O-6,6′-((1E,1′E)- (ethane-1,2-diylbis(azaneylylidene))bis(methaneylylidene)) bis(2-methoxyphenol)-κ2N,N,μ2O,O,κ2O, O)-(methanol)-cobalt(II) sodium(I), C25H27CoN2NaO9S
- Crystal structure of (1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl)(4-((2-methyl-6-(trifluoromethyl)pyrimidin-4-yl)amino)piperidin-1-yl)methanone, C17H18F6N6O
- Crystal structure of bis{[(cyclohexylimino)(phenylimino)-l5-(methyl)diethylazane-κ2N:N′]-(ethyl)-zinc(II)]}, C38H62N6Zn2
- Crystal structure of 2-[(4-bromobenzyl)thio]-5-(5-bromothiophen-2-yl)-1,3,4-oxadiazole, C13H8Br2N2OS2
- Crystal structure of 10-methoxy-7,11b,12,13-tetrahydro-6H-pyrazino [2′,3′:5,6]pyrazino[2,1-a]isoquinoline, C15H16N4O
- The crystal structure of 1-propyl-2-nitro-imidazole oxide, C6H9N3O3
- The crystal structure of 3-nitrobenzene-1,2-dicarboxylic acid–2-ethoxybenzamide (1/1), C17H16N2O8
- The structure of RUB-1, (C8H16N)6[B6Si48O108], a boron containing levyne-type zeolite, occluding N-methyl-quinuclidinium in the cage-like pores
- The crystal structure of diaqua-(naphthalene-4,5-dicarboxylate-1,8-dicarboxylic anhydride-κ1O)-(4′-(4-(1H-benzimidazolyl-1-yl)phenyl)-2,2′:6′,2″-terpyridine-κ3N,N′,N″)–manganese(II) dihydrate, C42H27MnN5O9·2H2O
- Crystal structure of 6,6′-((1E,1′E)-hydrazine-1,2-diylidenebis(methanylylidene))bis (3-(3-bromopropoxy)phenol), C20H22Br2N2O4
- The crystal structure of 3-(2-hydroxyphenyl)-4-phenyl-6-(p-tolyl)-2H-pyran-2-one, C24H18O3
- Crystal structure of bis(μ2-2-(1,5-dimethyl–3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)imino)methyl)phenolato-κ4O:O,N,O′)-(nitrato-κ2O,O′)dicobalt(II), C36H32Co2N8O4
- Synthesis and crystal structure of (3E,5S,10S,13S,14S,17Z)-17-ethylidene-10,13-dimethylhexadecahydro-3H-cyclopenta[α] phenanthren-3-one O-(4-fluorobenzoyl) oxime, C28H36FNO2
- The crystal structure of 4-aminiumbiphenyl benzenesulfonate, C18H17NO3S
- Synthesis and crystal structure of 1-(7-hydroxy-3-(4-hydroxy-3-nitrophenyl)-4-oxo-4H-chromen-8-yl)-N,N-dimethylmethanaminiumnitrate, C18H17N3O9
- Crystal structure of N-(Ar)-N′-(Ar′)-formamidine, C14H12Br2N2O
- The crystal structure of 4-(2,4-dichlorophenyl)-2-(4-fluorophenyl)-5-methyl-1H-imidazole, C16H11Cl2FN2
- Crystal structure of 1-(4–chlorophenyl)-4-benzoyl-3-methyl-1H-pyrazol-5-ol, C17H13ClN2O2
- The crystal structure of 5-amino-1-methyl-4-nitroimidazole, C4H6O2N4
- Crystal structure of 1,3-diisopropyl-4,5-dimethylimidazol-2-ylidene-N,N′-bis(1,3-bis(2,6-diisopropylphenyl)-1,3-dihydro-2H-1,3,2-diazaborol-2-yl)-l2-germenediamine, C63H94B2GeN8
- The crystal structure of (bromido, chlorido)-tricarbonyl-(5,5′-dimethyl-2,2′-bipyridine)-rhenium(I), C15H12Br0.2Cl0.8N2O3Re1
- Crystal structure of [N(E),N′(E)]-N,N′-(1,4-phenylenedimethylidyne)bis-3,5-bis(propan-2-yl)-1H-pyrazol-4-amine, C26H36N6
- The crystal structure of poly[2-(4-carboxypyridin-3-yl)terephthalpoly[diaqua-(μ4-2-(6-carboxylatopyridin-3-yl)terephthalato-κ5O,N:O′:O″,O‴)]) cadmium(II)] dihydrate, C28H20Cd3N2O16
- Crystal structure of [tetraaqua-bis((3-carboxy-5-(pyridin-4-yl)benzoate-κ1N)cobalt(II)] tetrahydrate, C26H32CoN2O16
- Crystal structure of bis(μ2-azido-κ2N:N)-tetrakis(azido-κ1N)-tetrakis(1,10-phenanthroline-κ2N,N′)dibismuth(III), C48H32N26Bi2
- Crystal structure of (Z)-N-(4-(4-(4-((4,5,6-trimethoxy-3-oxobenzofuran-2(3H)-ylidene)methyl)phenoxy)butoxy)phenyl)acetamide, C30H31NO8
- Crystal structure of poly[diaqua-(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2N:N′)-bis(μ2-5-carboxybenzene-1,3-dicarboxylato-O,O′:O″)-aqua-di-zinc dihydrate solvate], C27H28N4O16Zn2
- Crystal structure of 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile, C12H14N2
- Crystal structure of chlorido-(5-nitro-2-phenylpyridine-κ2N,C)-[(methylsulfinyl)methane-κ1S]platinum(II), C13H13ClN2O3PtS
- The crystal structure of the co-crystal 1,4-dioxane–4,6-bis(nitroimino)-1,3,5-triazinan-2-one(2/1), C11H19N7O9
- Crystal structure of [N(E),N′(E)]-N,N′-(1,4-phenylenedimethylidyne)bis-3,5-dimethyl-1H-pyrazol-4-amine di-methanol solvate, C18H20N6·2(CH3OH)
- Crystal structure of catena-poly[bis(μ2-azido-k2N:N′)-(nitrato-K2N:N′)-bis(1,10-phenanthroline-K2N:N′)samarium(III)], C24H16N11O3Sm
- Crystal structure of (Z)-2-(4-((5-bromopentyl)oxy)benzylidene)-4,5,6-trimethoxybenzofuran-3(2H)-one, C23H25BrO6
- Crystal structure of bis(3,5-dimethyl-1H-pyrazol-4-ammonium) tetrafluoroterephthate, 2[C5H10N3][C8F4O4]
- Crystal structure of 2-amino-4-(2-fluoro-4-(trifluoromethyl)phenyl)-9-methoxy-1,4,5,6-tetrahydrobenzo[h]quinazolin-3-ium chloride, C20H18ClF4N3O
- Crystal structure of 6-(pyridin-3-yl)-1,3,5-triazine-2,4-diamine-sebacic acid (2/1), C13H17N6O2
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of (N-([1,1′:4′,1″-terphenyl]-4,4′-diethyl)-2-(bis(pyridin-2-ylmethyl)amino)acetamide-κ4N,N,N″, O)tri(nitrato-kO, O′) samarium(III) - methanol - acetonitrile (1/1/1), C40H39SmN8O14
- The crystal structure of 6,6′-(((2-(dimethylamino)ethyl)azanediyl)bis(methylene))bis(2-chloro-4-methyl phenolate-κ4N,N′,O,O′)-(pyridine-2,6-dicarboxylato-N,O,O′)-titanium(IV), C27H27Cl2N3O6Ti
- N′-[(1E)-(4–Fluorophenyl)methylidene]adamantane-1-carbohydrazide, C18H21FN2O
- Crystal structure of 4-bromo-3-nitro-1H-pyrazole-5-carboxylic acid monohydrate, C4H2N3BrO4·H2O
- Crystal structure of dipyridine-k1N-tris(2,2,6,6-tetramethyl-5-oxohept-3-en-3-olato-k2O,O′)dysprosium(III), DyC43H67O6N2
- Crystal structure of cyclo[tetraiodido-bis{μ2-1-[(benzotriazol-1-yl)methyl]-1-H-1,3-(2-isopropyl-imidazol)-k2N:N}dicadmiun(II)], C26H30N10Cd2I4
- The crystal structure of tert-butyl (E)-3-(2-(benzylideneamino)phenyl)-1H-indole-1-carboxylate, C26H24N2O2
- The crystal structure of 4-(3-carboxy-1-cyclopropyl-6-fluoro-8-methoxy-4-oxo-1,4- dihydroquinolin-7-yl)-2-methylpiperazin-1-ium 2,5-dihydroxybenzoate methanol solvate, C27H32FN3O9
- Crystal structure of (μ2-1-(4,4′-bipyridine-κ2N:N′)-bis[diaqua-(4-iodopyridine-2,6-dicarboxylato-κ3O,N,O′)–cobalt(II)], C24H20Co2I2N4O12
- The crystal structure of dimethyl 4,4′-(10,20-diphenylporphyrin-5,15-diyl)dibenzoate dichloromethane solvate, C49H36N4O4Cl2
- (E)-2-((E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-ylidene)hydrazine-1-carbothioamide C14H23N3S1
- The crystal structure of [1-(4-(trifluoromethyl)phenyl)-3,4-dihydroquinolin-2(1H)-one], C16H12F3NO
- Crystal structure of (E)-2-amino-N′-((3-hydroxy-5-(hydroxymethyl)-2-methylpyridin-4-yl)methylene)benzohydrazide – dimethylformamide – water (1/1/2), C15H16N4O3·C3H7NO·2H2O
- Crystal structure of 3-(4-bromophenyl)-5-methyl-1H-pyrazole, C10H9BrN2
- Crystal structure of 1,10-phenanthrolinium bromide dihydrate, C12H9N2Br
- Crystal structure of N-(4′-chloro-[1,1′-biphenyl]-2-yl)formamide, C13H10ClNO
- The crystal structure of nitroterephthalic acid, C8H5NO6
- Crystal structure of (2-((4-bromo-2,6-dichlorophenyl)amino)phenyl) (morpholino)methanone, C17H15BrCl2N2O2
- Crystal structure of tetraaqua-bis(ethanol-κO)-tetrakis(μ2-trifluoroacetate-κ2O:O′)-bis(trifluoroacetate-κ2O)digadolinium(III) Gd2C16H20O18F18
- The crystal structure of dimethyl 4,4′-[10,20-bis(2,6-difluorophenyl)porphyrin-5,15-diyl]dibenzoate chloroform solvate, C50H32Cl6F4N4O4
- The crystal structure of N,N′-((nitroazanediyl)bis(methylene))diacetamide, C6H12O4N4
- The crystal structure of [bis(2,2′-bipyridine-6-carboxylato-κ3N,N,O)magnesium(II)]dihydrate, C22H18N4O6Mg
- Crystal structure of poly[diaqua-(bis(μ2-1,4-bis(imidazol-1-ylmethyl)benzene)-κ2N,N′] cobalt(II)-tetraqua-bis(1,4-bis(imidazol-1-ylmethyl)benzene)-κ1N)-cobalt(II) di(2,5-thiophenedicarboxylate) dihydrate, C68H76Co2N16O16S2
- Crystal structure of poly[chlorido-μ2-chlorido-(μ2-1-[(2-ethyl-4-methyl-1H-imidazol-1-yl)methyl]-1H-benzotriazole-κN:N’)cadmium(II)], C13H15CdN5Cl2
- The crystal structure of (4-hydroxybenzenesulfonate)-k1O-6,6′-((1E,1′E)- (ethane-1,2-diylbis(azaneylylidene))bis(methaneylylidene)) bis(2-methoxyphenol)-κ2N,N,μ2O,O,κ2O, O)-(methanol)-cobalt(II) sodium(I), C25H27CoN2NaO9S
- Crystal structure of (1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl)(4-((2-methyl-6-(trifluoromethyl)pyrimidin-4-yl)amino)piperidin-1-yl)methanone, C17H18F6N6O
- Crystal structure of bis{[(cyclohexylimino)(phenylimino)-l5-(methyl)diethylazane-κ2N:N′]-(ethyl)-zinc(II)]}, C38H62N6Zn2
- Crystal structure of 2-[(4-bromobenzyl)thio]-5-(5-bromothiophen-2-yl)-1,3,4-oxadiazole, C13H8Br2N2OS2
- Crystal structure of 10-methoxy-7,11b,12,13-tetrahydro-6H-pyrazino [2′,3′:5,6]pyrazino[2,1-a]isoquinoline, C15H16N4O
- The crystal structure of 1-propyl-2-nitro-imidazole oxide, C6H9N3O3
- The crystal structure of 3-nitrobenzene-1,2-dicarboxylic acid–2-ethoxybenzamide (1/1), C17H16N2O8
- The structure of RUB-1, (C8H16N)6[B6Si48O108], a boron containing levyne-type zeolite, occluding N-methyl-quinuclidinium in the cage-like pores
- The crystal structure of diaqua-(naphthalene-4,5-dicarboxylate-1,8-dicarboxylic anhydride-κ1O)-(4′-(4-(1H-benzimidazolyl-1-yl)phenyl)-2,2′:6′,2″-terpyridine-κ3N,N′,N″)–manganese(II) dihydrate, C42H27MnN5O9·2H2O
- Crystal structure of 6,6′-((1E,1′E)-hydrazine-1,2-diylidenebis(methanylylidene))bis (3-(3-bromopropoxy)phenol), C20H22Br2N2O4
- The crystal structure of 3-(2-hydroxyphenyl)-4-phenyl-6-(p-tolyl)-2H-pyran-2-one, C24H18O3
- Crystal structure of bis(μ2-2-(1,5-dimethyl–3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)imino)methyl)phenolato-κ4O:O,N,O′)-(nitrato-κ2O,O′)dicobalt(II), C36H32Co2N8O4
- Synthesis and crystal structure of (3E,5S,10S,13S,14S,17Z)-17-ethylidene-10,13-dimethylhexadecahydro-3H-cyclopenta[α] phenanthren-3-one O-(4-fluorobenzoyl) oxime, C28H36FNO2
- The crystal structure of 4-aminiumbiphenyl benzenesulfonate, C18H17NO3S
- Synthesis and crystal structure of 1-(7-hydroxy-3-(4-hydroxy-3-nitrophenyl)-4-oxo-4H-chromen-8-yl)-N,N-dimethylmethanaminiumnitrate, C18H17N3O9
- Crystal structure of N-(Ar)-N′-(Ar′)-formamidine, C14H12Br2N2O
- The crystal structure of 4-(2,4-dichlorophenyl)-2-(4-fluorophenyl)-5-methyl-1H-imidazole, C16H11Cl2FN2
- Crystal structure of 1-(4–chlorophenyl)-4-benzoyl-3-methyl-1H-pyrazol-5-ol, C17H13ClN2O2
- The crystal structure of 5-amino-1-methyl-4-nitroimidazole, C4H6O2N4
- Crystal structure of 1,3-diisopropyl-4,5-dimethylimidazol-2-ylidene-N,N′-bis(1,3-bis(2,6-diisopropylphenyl)-1,3-dihydro-2H-1,3,2-diazaborol-2-yl)-l2-germenediamine, C63H94B2GeN8
- The crystal structure of (bromido, chlorido)-tricarbonyl-(5,5′-dimethyl-2,2′-bipyridine)-rhenium(I), C15H12Br0.2Cl0.8N2O3Re1
- Crystal structure of [N(E),N′(E)]-N,N′-(1,4-phenylenedimethylidyne)bis-3,5-bis(propan-2-yl)-1H-pyrazol-4-amine, C26H36N6
- The crystal structure of poly[2-(4-carboxypyridin-3-yl)terephthalpoly[diaqua-(μ4-2-(6-carboxylatopyridin-3-yl)terephthalato-κ5O,N:O′:O″,O‴)]) cadmium(II)] dihydrate, C28H20Cd3N2O16
- Crystal structure of [tetraaqua-bis((3-carboxy-5-(pyridin-4-yl)benzoate-κ1N)cobalt(II)] tetrahydrate, C26H32CoN2O16
- Crystal structure of bis(μ2-azido-κ2N:N)-tetrakis(azido-κ1N)-tetrakis(1,10-phenanthroline-κ2N,N′)dibismuth(III), C48H32N26Bi2
- Crystal structure of (Z)-N-(4-(4-(4-((4,5,6-trimethoxy-3-oxobenzofuran-2(3H)-ylidene)methyl)phenoxy)butoxy)phenyl)acetamide, C30H31NO8
- Crystal structure of poly[diaqua-(μ2-1,3-di(1H-imidazol-1-yl)propane-κ2N:N′)-bis(μ2-5-carboxybenzene-1,3-dicarboxylato-O,O′:O″)-aqua-di-zinc dihydrate solvate], C27H28N4O16Zn2
- Crystal structure of 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile, C12H14N2
- Crystal structure of chlorido-(5-nitro-2-phenylpyridine-κ2N,C)-[(methylsulfinyl)methane-κ1S]platinum(II), C13H13ClN2O3PtS
- The crystal structure of the co-crystal 1,4-dioxane–4,6-bis(nitroimino)-1,3,5-triazinan-2-one(2/1), C11H19N7O9
- Crystal structure of [N(E),N′(E)]-N,N′-(1,4-phenylenedimethylidyne)bis-3,5-dimethyl-1H-pyrazol-4-amine di-methanol solvate, C18H20N6·2(CH3OH)
- Crystal structure of catena-poly[bis(μ2-azido-k2N:N′)-(nitrato-K2N:N′)-bis(1,10-phenanthroline-K2N:N′)samarium(III)], C24H16N11O3Sm
- Crystal structure of (Z)-2-(4-((5-bromopentyl)oxy)benzylidene)-4,5,6-trimethoxybenzofuran-3(2H)-one, C23H25BrO6
- Crystal structure of bis(3,5-dimethyl-1H-pyrazol-4-ammonium) tetrafluoroterephthate, 2[C5H10N3][C8F4O4]
- Crystal structure of 2-amino-4-(2-fluoro-4-(trifluoromethyl)phenyl)-9-methoxy-1,4,5,6-tetrahydrobenzo[h]quinazolin-3-ium chloride, C20H18ClF4N3O
- Crystal structure of 6-(pyridin-3-yl)-1,3,5-triazine-2,4-diamine-sebacic acid (2/1), C13H17N6O2


