Abstract
C28H31ClN2O, monoclinic, P1̄ (no. 2), a = 21.9309(11) Å, b = 9.9648(5) Å, c = 11.0049(7) Å, β = 93.403(6)°, V = 2400.7(2) Å3, Z = 4, Rgt(F) = 0.0566, wRref(F2) = 0.1355, T = 293 K.
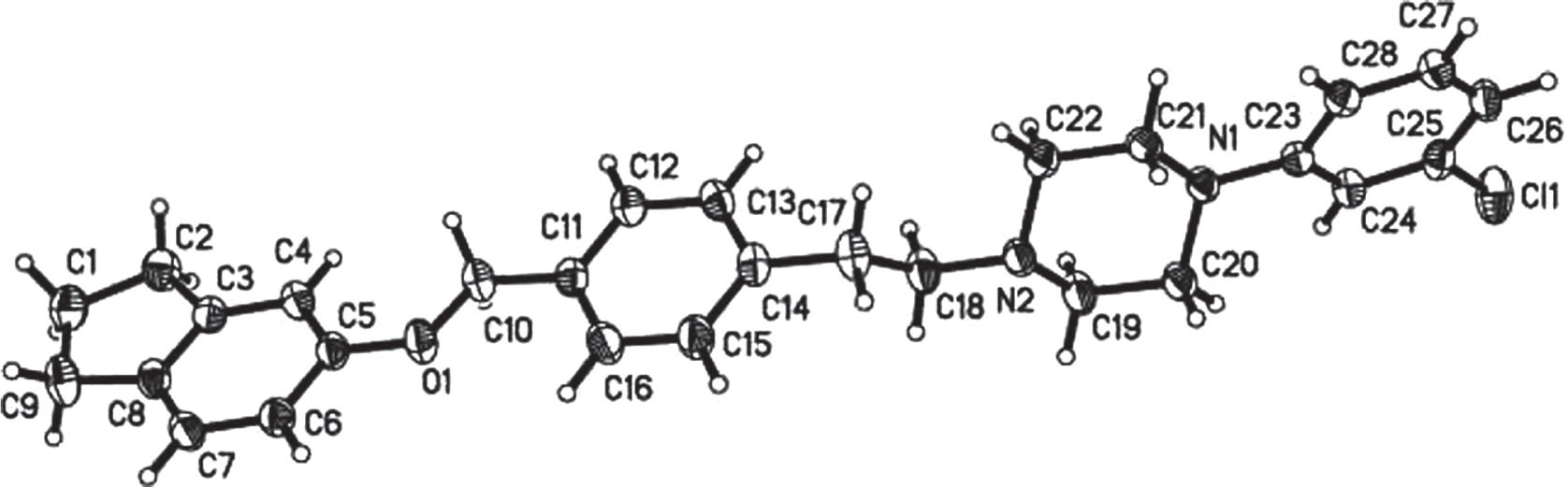
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Yellow block |
| Size: | 0.18 × 0.15 × 0.13 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.18 mm−1 |
| Diffractometer, scan mode: | SuperNova, ω |
| θmax, completeness: | 25.0°, >99% |
| N(hkl)measured, N(hkl)unique, Rint: | 12758, 4213, 0.028 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 2973 |
| N(param)refined: | 289 |
| Programs: | SHELX [1], Olex2 [2], CrysAlisPRO [3] |
Source of material
The title compound was synthesized from 4-(((2,3-dihydro-1H-inden-5-yl)oxy)methyl)phenethyl 4-methylbenzenesulfonate in form of light yellow crystals. To a solution of 4-(((2,3-dihydro-1H-inden-5-yl)oxy)methyl)phenethyl 4-methyl-benzenesulfonate (100 mg, 0.23 mmol) in acetonitrile (CH3CN, 10 mL) was added 1-(3-chlorophenyl)piperazine (54 mg, 0.27 mmol) and potassium carbonate (190 mg, 1.38 mmol). The reaction mixture was stirred at reflux for 16 h. The mixture was filtered before cooling to ambient. The filtrate was concentrated in vacuo and the residue was purified by silica gel column chromatography using ethyl acetate/petroleum ether (1/15, v/v) as eluent to obtain the product as a light yellow solid. Yield: 71%; Mp. 83-84 °C (HCl salt); 1H NMR (500 MHz, CDCl3) δ [ppm] 7.40 (d, J = 7.9 Hz, 2H), 7.28 (d, J = 2.3 Hz, 1H), 7.20 (t, J = 8.1 Hz, 1H), 7.15 (d, J = 8.2 Hz, 1H), 6.92 (t, J = 2.0 Hz, 1H), 6.90 (s, 1H), 6.86–6.79 (m, 3H), 5.04 (s, 2H), 3.35–3.18 (m, 4H), 2.99–2.81 (m, 6H), 2.71 (dd, J = 16.0, 7.0 Hz, 6H), 2.16–2.05 (m, 2H). 13C NMR (126 MHz, CDCl3) δ [ppm] 157.81, 152.29, 145.75, 139.72, 136.48, 135.29, 134.99, 130.05, 128.92, 127.74, 124.76, 119.36, 115.81, 113.91, 112.84, 110.94, 70.10, 60.30, 53.00, 48.62, 33.21, 32.02, 29.73, 25.86; HRMS (ESI) m/z [M + 1]+: calcd. for C28H31ClN2O, 447.2198, found, 447.2196.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | Uiso*/Ueq |
|---|---|---|---|---|
| Cl1 | 0.95801(3) | 0.13164(8) | 0.32362(9) | 0.0820(3) |
| O1 | 0.30527(8) | 0.36018(18) | 0.92112(17) | 0.0575(5) |
| N1 | 0.75550(8) | 0.3461(2) | 0.42215(18) | 0.0448(5) |
| N2 | 0.65779(9) | 0.3809(2) | 0.58260(18) | 0.0482(5) |
| C1 | 0.05755(13) | 0.1458(4) | 1.0455(3) | 0.0779(10) |
| H1A | 0.0207 | 0.1734 | 0.9990 | 0.094* |
| H1B | 0.0481 | 0.0677 | 1.0934 | 0.094* |
| C2 | 0.10706(12) | 0.1124(3) | 0.9606(3) | 0.0633(8) |
| H2A | 0.1223 | 0.0222 | 0.9755 | 0.076* |
| H2B | 0.0916 | 0.1191 | 0.8764 | 0.076* |
| C3 | 0.15654(11) | 0.2140(2) | 0.9881(2) | 0.0471(6) |
| C4 | 0.21119(11) | 0.2312(3) | 0.9320(2) | 0.0497(6) |
| H4 | 0.2211 | 0.1751 | 0.8686 | 0.060* |
| C5 | 0.25040(11) | 0.3321(3) | 0.9713(2) | 0.0475(6) |
| C6 | 0.23540(11) | 0.4163(3) | 1.0657(2) | 0.0502(7) |
| H6 | 0.2619 | 0.4847 | 1.0916 | 0.060* |
| C7 | 0.18104(12) | 0.3982(3) | 1.1210(2) | 0.0529(7) |
| H7 | 0.1711 | 0.4544 | 1.1842 | 0.063* |
| C8 | 0.14149(11) | 0.2969(3) | 1.0826(2) | 0.0498(6) |
| C9 | 0.08040(13) | 0.2580(3) | 1.1277(3) | 0.0719(9) |
| H9A | 0.0848 | 0.2279 | 1.2116 | 0.086* |
| H9B | 0.0524 | 0.3334 | 1.1225 | 0.086* |
| C10 | 0.32434(11) | 0.2709(3) | 0.8299(2) | 0.0533(7) |
| H10A | 0.2936 | 0.2676 | 0.7628 | 0.064* |
| H10B | 0.3292 | 0.1813 | 0.8633 | 0.064* |
| C11 | 0.38385(11) | 0.3184(2) | 0.7847(2) | 0.0463(6) |
| C12 | 0.40245(11) | 0.2679(3) | 0.6751(2) | 0.0508(7) |
| H12 | 0.3778 | 0.2068 | 0.6313 | 0.061* |
| C13 | 0.45739(11) | 0.3079(3) | 0.6307(2) | 0.0525(7) |
| H13 | 0.4690 | 0.2723 | 0.5574 | 0.063* |
| C14 | 0.49538(11) | 0.3990(3) | 0.6920(3) | 0.0507(7) |
| C15 | 0.47636(12) | 0.4489(3) | 0.8006(3) | 0.0602(8) |
| H15 | 0.5008 | 0.5107 | 0.8441 | 0.072* |
| C16 | 0.42141(12) | 0.4086(3) | 0.8465(3) | 0.0591(7) |
| H16 | 0.4100 | 0.4434 | 0.9202 | 0.071* |
| C17 | 0.55386(12) | 0.4451(3) | 0.6399(3) | 0.0621(8) |
| H17A | 0.5448 | 0.4760 | 0.5572 | 0.074* |
| H17B | 0.5699 | 0.5208 | 0.6871 | 0.074* |
| C18 | 0.60224(11) | 0.3379(3) | 0.6388(3) | 0.0596(8) |
| H18A | 0.6128 | 0.3104 | 0.7219 | 0.072* |
| H18B | 0.5855 | 0.2604 | 0.5951 | 0.072* |
| C19 | 0.70695(11) | 0.2854(3) | 0.6097(3) | 0.0619(8) |
| H19A | 0.6950 | 0.1979 | 0.5781 | 0.074* |
| H19B | 0.7142 | 0.2776 | 0.6972 | 0.074* |
| C20 | 0.76509(11) | 0.3295(3) | 0.5540(2) | 0.0571(7) |
| H20A | 0.7787 | 0.4139 | 0.5903 | 0.069* |
| H20B | 0.7968 | 0.2632 | 0.5715 | 0.069* |
| C21 | 0.70578(10) | 0.4401(3) | 0.3939(2) | 0.0470(6) |
| H21A | 0.6983 | 0.4467 | 0.3064 | 0.056* |
| H21B | 0.7171 | 0.5284 | 0.4248 | 0.056* |
| C22 | 0.64837(11) | 0.3936(3) | 0.4506(2) | 0.0495(6) |
| H22A | 0.6158 | 0.4574 | 0.4315 | 0.059* |
| H22B | 0.6360 | 0.3076 | 0.4162 | 0.059* |
| C23 | 0.80744(10) | 0.3531(2) | 0.3537(2) | 0.0426(6) |
| C24 | 0.85340(11) | 0.2564(2) | 0.3714(2) | 0.0469(6) |
| H24 | 0.8505 | 0.1916 | 0.4316 | 0.056* |
| C25 | 0.90288(11) | 0.2568(3) | 0.3000(2) | 0.0501(7) |
| C26 | 0.90951(12) | 0.3506(3) | 0.2096(3) | 0.0559(7) |
| H26 | 0.9431 | 0.3495 | 0.1619 | 0.067* |
| C27 | 0.86455(12) | 0.4456(3) | 0.1928(2) | 0.0555(7) |
| H27 | 0.8680 | 0.5102 | 0.1325 | 0.067* |
| C28 | 0.81439(11) | 0.4480(3) | 0.2628(2) | 0.0479(6) |
| H28 | 0.7848 | 0.5140 | 0.2490 | 0.057* |
Experimental details
The hydrogen atoms were placed in calculated positions and refined using a riding model on attached atoms with isotropic thermal parameters 1.2 times those of their carrier atoms.
Comment
Arylpiperazines are great important kinds of drug molecules, and have various bioactivties such as antiarrhythmic, diuretic, antiallergic, antidepressant, anxiolytic, antipsychotic, antimalarial, and antiplasmodial [4], [5], [6], [7], [8], [9], [10], [11], [12]. Some of these compounds have good receptor-blocking properties [13]. According to experiments of the testprostate cancer cells, arylpiperazine derivatives have displayed significant cytotoxic [14]. Although some crystal structures of arylpiperazine derivatives have been reported recently [15], [16], the different functional groups and organic ligands would lead to different properties. Therefore, to further study and explore the new arylpiperazine, we report the crystal structure of the title compound.
The molecule of the title compound displays the 2,3-dihydro-1H-indene, piperazine ring, and two benzene ring groups. One nitrogen atom of piperazine connects the phenylethyl moiety via C—N bond, and the other N links to the carbon atom of chlorobenzene. Furthermore, the above mentioned phenylethyl extends connection of 2,3-dihydro-1H-indene group a C—O bond (see the figure). The 2,3-dihydro-1H-indene group, piperazine unit, and two aryl moieties are not in the same plane. The whole molecule displays a twisty conformation. In the molecule, the Cl1—C25, O1—C10, O1—C5, N1—C20, N1—C23, N1—C21, N2—C22, N2—C19, and N2—C18 bond lengths are found to be 1.746(3) Å, 1.423(3) Å, 1.382(3) Å, 1.464(3) Å, 1.404(3) Å, 1.457(3) Å, 1.461(3) Å, 1.455(3) Å, and 1.463(3) Å, respectively, which are nearly equal to other typical single bonds. The bond angles C5—O1—C10, C23—N1—C20, C23—N1—C21, C21—N1—C20, C22—N2—C18, C19—N2—C22, C19—N2—C18, N2—C22—C21, N1—C20—C19, C24—C23—N1, C28—C23—N1, N1—C21—C22, C24—C25—Cl1, C26—C25—Cl1, N2—C19—C20, N2—C18—C17, O1—C10—C11, O1—C5—C6, 115.3(2)°, and C4—C5—O1 are 117.05(19)°, 117.67(19)°, 118.31(19)°, 110.22(19)°, 112.0(2)°, 108.7(2)°, 110.32(19)°, 111.4(2)°, 111.1(2)°, 119.4(2)°, 123.1(2)°, 110.3(2)°, 118.4(2)°, 119.32(19)°, 111.0(2)°, 113.6(2)°, 109.6(2)°, and 115.3(2)°, respectively.
Acknowledgements
The work was supported by Science and Technology Planning Project of Henan Province of China (172102110105) for financial assistances.
References
1. Sheldrick, G. M.: Crystal structure refinement with SHELXL. Acta Crystallogr. C71 (2015) 3–8.10.1107/S2053229614024218Suche in Google Scholar
2. Dolomanov, O. V.; Bourhis, L. J.; Gildea, R. J.; Howard, J. A. K.; Puschmann, H.: OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 42 (2009) 339–341.10.1107/S0021889808042726Suche in Google Scholar
3. Agilent Technologies: CrysAlisPRO Software system. Agilent Technologies UK Ltd, Oxford, UK (2011).Suche in Google Scholar
4. Szkaradek, N.; Rapacz, A.; Pytka, K.; Filipek, B.; Siwek, A.; Cegla, M.; Marona, H.: Synthesis and preliminary evaluation of pharmacological properties of some piperazine derivatives of xanthone. Bioorg. Med. Chem. 21 (2013) 514–522.10.1016/j.bmc.2012.11.014Suche in Google Scholar
5. Cecchetti, V.; Fravolini, A.; Schiaffella, F.; Tabarrini, O.; Bruni, G.; Segret, G.: o-Chlorobenzenesulfonamidic derivatives of (aryloxy) propanolamines as beta-blocking/diuretic agents. J. Med. Chem. 36 (1993) 157–161.10.1021/jm00053a020Suche in Google Scholar
6. Walsh, D. A.; Chen, Y. H.; Green, J. B.; Nolan, J. C.; Yannit, J. M.: The synthesis and antiallergy activity of 1-(aryloxy)-4-(4-arylpiperazinyl)-2-butanol derivatives. J. Med. Chem. 33 (1990) 1823–1827.10.1021/jm00168a044Suche in Google Scholar
7. Seo, H. J.; Park, E. J.; Kim, M. J.; Kang, S. Y.; Lee, S. H.; Kim, H. J.; Lee, K. N.; Jung, M. E.; Lee, M.; Kim, M. S.; Son, E. J.; Park, W. K.; Kim, J.; Lee, J.: Design and synthesis of novel arylpiperazine derivatives containing the imidazole core targeting 5-HT(2A) receptor and 5-HT transporter. J. Med. Chem. 54 (2011) 6305–6318.10.1021/jm200682bSuche in Google Scholar
8. Kikumoto, R.; Tobe, A.; Fukami, H.; Egawa, M.: Synthesis and antianxiety activity of (omega-piperazinylalkoxy)indan derivatives. J. Med. Chem. 26 (1983) 246–250.10.1021/jm00356a024Suche in Google Scholar
9. Jaen, J. C.; Wise, L. D.; Heffner, T. G.; Pugsley, T. A.; Meltzed, L. T.: Dopamine autoreceptor agonists as potential antipsychotics. 1. (aminoalkoxy)anilines. J. Med. Chem. 31 (1988) 1621–1625.10.1021/jm00403a022Suche in Google Scholar
10. Cross, R. M.; Namelikonda, N. K.; Mutka, T. S.; Luong, L.; Kyle, D. E.; Manetsch, R.: Synthesis, antimalarial activity, and structure-activity relationship of 7-(2-phenoxyethoxy)-4(1H)-quinolones. J. Med. Chem. 54 (2011) 8321–8327.10.1021/jm200718mSuche in Google Scholar
11. Clarkson, C.; Musonda, C. C.; Chibale, K.; Campbella, W. E.; Smitha, P.: Synthesis of totarol amino alcohol derivatives and their antiplasmodial activity and cytotoxicity. Bioorg. Med. Chem. 11 (2003) 4417–4422.10.1016/S0968-0896(03)00491-7Suche in Google Scholar
12. Berardi, F.; Abate, C.; Ferorelli, S.; De Robertis, A. F.; Leopoldo, M.; Colabufo, N. A.; Niso, M.; Perrone, R.: Novel 4-(4-aryl)cyclohexyl-1-(2-pyridyl)piperazines as Δ8-Δ7 sterol isomerase (emopamil binding protein) selective ligands with antiproliferative activity. J. Med. Chem. 51 (2008) 7523–7531.10.1021/jm800965bSuche in Google Scholar PubMed
13. Leopoldo, M.; Lacivita, E.; Passafiume, E.; Contino, M.; Colabufo, N. A.; Berardi, F.; Perrone, R.: 4-[ω-[4-Arylpiperazin-1-yl]alkoxy]phenyl) imidazo[1,2-a]pyridine derivatives: fluorescent high-affinity dopamine D3 receptor ligands as potential probes for receptor visualization. J. Med. Chem. 50 (2007) 5043–5047.10.1021/jm070721+Suche in Google Scholar PubMed
14. Chen, H.; Yu, Y.-Z.; Tian, X.-M.; Wang, C.-L.; Qian, Y.-N.; Deng, Z.-A.; Zhang, J.-X.; Lv, D.-J.; Zhang, H.-B.; Shen, J.-L.; Yuan, M.; Zhao, S.-C.: Synthesis and biological evaluation of arylpiperazine derivatives as potential anti-prostate cancer agents. Bioorg. Med. Chem. 27 (2019) 133–143.10.1016/j.bmc.2018.11.029Suche in Google Scholar PubMed
15. Chen, H.; Jia, H.-X.: Crystal structure of 2-(4-(2-fluorophenyl) piperazin-1-yl) ehthyl) benzyl)benzoisothiazol-3(2H)-one1,1-dioxide, C26H26FN3O3S – a saccharin derivative. Z. Kristalllogr. NCS 233 (2018) 111–113.10.1515/ncrs-2017-0197Suche in Google Scholar
16. Chen, H.; Jia, H.-X.; Xu, Q.-T.: Crystal structure of ure of 1-(4-((benzo(1,3)dioxol-5-yloxy)methyl)phenethyl)-4-(3-chlorophenyl)piperazin-1-ium chloride, C26H28Cl2N2O3. Z. Kristalllogr. NCS 233 (2018) 107–109.10.1515/ncrs-2017-0196Suche in Google Scholar
©2020 Aiqing Feng et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 Public License.
Artikel in diesem Heft
- Frontmatter
- Crystal structure of bis [1-(phenylsulfonyl)-2-(1-(pyrazin-2-yl)ethylidene)hydrazin-1-ido-κ3N,N′,O]cobalt(II), C24H22N8O4S2Co
- The crystal structure of 1,3-bis(4-(methoxycarbonyl)benzyl)-2-methyl-1H-benzo[d]imidazol-3-ium bromide, C26H25BrN2O4
- Crystal structure of {tris((1H-benzo[d]imidazol-2-yl)methyl)amine-κ4N,N′,N′′,N′′′}-(nitrito-κ2O,O′)nickel(II) perchlorate – ethanol (1/1), C26H27ClN8NiO7
- Crystal structure of catena-poly[aqua[(μ2-4,5-dicarboxylato-2-(2-carboxylatophenyl)imidazol-1-ido-κ4N,O,O′:N′)](μ2-4,4′-bipyridine-κ2N:N′)dicopper(II)], C22H14Cu2N4O7
- Crystal structure of chlorido-tris(4-methylbenzyl-κC)-(triphenylarsine oxide-κO)tin(IV), C42H42AsClOSn
- The crystal structure of 4,4′-bipyridinium bis(3-carboxy-2-nitrobenzoate) tetrahydrate, C13H13N2O8
- Crystal structure of 1-(3-chlorophenyl)-4-(4-(((2,3-dihydro-1H-inden-5-yl)oxy)methyl)phenethyl)piperazine, C28H31ClN2O
- Crystal structure of catena-poly[diaqua-bis(μ2-5,5′-(1H-imidazole-4,5-diyl)bis(tetrazol-2-ido)-κ4N,N′:N′′,N′′′)magnesium], C10H8N20O2Mg
- The crystal structure of (E)-2-((2-hydroxy-4-ethoxybenzylidene)amino)-2-methylpropane-1,3-diol monohydrate, C13H21NO5
- Crystal structure of catena-poly[diaqua-(μ2-bipyridine-κ2N:N′)-bis(3,5-dichloroisonicotinato-κO)cadmium(II)] dihydrate, C22H20CdCl4N4O8
- The crystal structure of 4-(4-chlorophenyl)cyclohexane-1-carboxylic acid, C13H15ClO2
- Redetermination of the crystal structure of yttrium(III) trinitrate(V) pentahydrate, Y(NO3)3 ⋅ 5 H2O, H10N3O14Y
- Crystal structure of catena-poly[di-μ2-chlorido-1,10-phenanthroline-κ2N,N′-cadmium(II)], C12H8Cl2CdN2
- Crystal structure of 4-((2-methyl-6-(trifluoromethyl)pyrimidin-4-yl)oxy)benzoic acid, C13H9F3N2O3
- Crystal structure of 3-acetyl-4-hydroxybenzoic acid, C18H16O8
- Crystal structure of bis(N,2-bis(4-ethoxybenzylidene)hydrazine-1-carbohydrazonothioato-κ2N,S)nickel(II) — N,N-dimethylformamide (1/2), C44H56N10S2O6Ni
- The crystal structure of 5-chloro-4,6-dimethoxypyrimidin-2-amine, C6H8ClN3O2
- Crystal structure of poly[aqua-(μ4-benzene-1,2,4,5-tetracarboxylato-κ4O,O′,O′′,O′′′)bis(μ2-1-(4-(1H-imidazol-1-yl)benzyl)-1H-1,2,4-triazole-κ2N:N)dinickel(II)], NiC17H14N5O5
- Crystal structure of poly[aqua(5-dimethylamino)naphthalene-1-sulfonato-κ2N:O)(μ2-4,4′-bipyridyl -κ2N:N′)silver(I)], C44H44Ag2N6O8S2
- Crystal structure of 1-[3-(trifluoromethyl)cinnamoyl]-3-(pyridin-2-yl-κN)pyrazole-κ2N-bis(2-phenylpyridinato-k2C,N)iridium(III) hexafluorophosphate complex, [C40H28F3IrN5O]PF6
- Crystal structure of catena-poly[aqua(μ6-piperazine-1,4-bisethanesulfonato-κ6N:N′:O:O′:O′′:O′′′)(μ2-pyrazinyl-κ2N:N′)disilver(I)sesquihydrate], C12H30Ag2N4O11S2
- Crystal structure of (E)-1-(2-nitrophenyl)-N-(o-tolyl)methanimine, C14H12N2O2
- Crystal structure of 4′-amino-3′,5′-diisopropyl-(1,1′-biphenyl)-4-carbonitrile, C19H22N2
- The crystal structure of poly[bis(N,N-dimethylformamide-κ1O)-tetrakis(μ2-cyanido-κ2C:N)dinickel(II)], C10H14N6O2Ni2
- Crystal structure of rac-trans-N,N′-bis(3-bromo-5-chlorosalicylidene)-1,2-cyclohexanediamine, C20H18Br2Cl2N2O2
- Crystal structure of rac-trans-N,N′-bis(3,5-dibromosalicylidene)-1,2-cyclohexanediamine, C20H18Br4N2O2
- The crystal structure of (dichromato-κ2O,O′)bis(1,10-phenanthroline-κ2N,N′)nickel(II), C12H16N4O7Cr2Ni
- The crystal structure of 3-((1R,2S)-1-methylpyrrolidin-1-ium-2-yl)pyridin-1-ium tetrachloridozincate(II) monohydrate, C10H18Cl4ZnN2O
- Crystal structure of bis(μ2-azido-k2N,N)-bis(2-amino-1-(N-(3-bromosalicylaldiminato))ethane)-dicopper(II), C20H18Br4N2O2
- Crystal structure of (η6-1-methyl-4-isopropylbenzene)-[5-bromo-2-(2-pyridyl)phenyl-κ2C,N]-chloro-ruthenium(II), C21H21BrClNRu
- Crystal structure of N-(methyl(oxo)(1-(6-(trifluoromethyl)pyridin-3-yl)ethyl)-λ6-sulfanylidene)cyanamide, C10H10F3N3OS
- Crystal structure of 6,6′-((cyclohexane-1,2-diylbis(azanylylidene))bis(methanylylidene))bis(2-bromo-4-chlorophenolato-κ4N,N′,O,O′)nickel(II), C20H16Br2Cl2NiN2O2
- Redetermination of the crystal structure of catena-poly[aqua-(1,10-phenanthroline-κ2N,N′)-(μ2-tetraoxidomolybdato(VI)-κ2O:O′)manganese(II) monohydrate, C12H12N2O6MoMn
- The crystal structure tetrakis(μ2-o-chlorobenzoato-κ2O:O′)-bis(methanol-κ1O)dirhodium(II), C30H24Cl4O10Rh2
- Crystal structure of bis(2,3-diphenyltetrazolidine-5-thione-κ1S)-(nitrato-κ1O)-(nitrato-κ2O,O′)lead(II), C26H20N10O6S2Pb
- Crystal structure of bis(3-bromo-N-(1-(3-methylpyrazin-2-yl)ethylidene)benzohydrazonato-κ3O,N,N′)cadmium(II) hemihydrate, C28H25N8O2.5Br2Cd
- Crystal structure of catena-poly[tetrakis(μ2-trifluoroacetato-κ2O:O′)(μ2-2,5-dimethylpyrazine-κ2N,N′)dicopper(II)], C7H4CuF6NO4
- The crystal structure of catena-poly[bis[3-azoniapentane-1,5-diammonium][bis(μ4-oxo)-tetrakis(μ3-oxo)-heptakis(μ2-oxo)-tetradecaoxo-octa-molybdenum] dihydrate], (C8H36N6O29Mo8)n
- Crystal structure of tetraaqua-bis(2-((3,5,6-trichloropyridin-2-yl)oxy)acetato-κO)-nickel(II)—diaqua-bis(2-((3,5,6-trichloropyridin-2-yl)oxy)acetato-nickel(II), C28H24Cl12N4Ni2O18
- The crystal structure of bis(2-hydroxypyrimidinium) pentachloridobismuthate(III), (C4N2H5O)2BiCl5
- The crystal structure of catena-poly[(μ2-4,4′-dipyridine-κ2N,N′)-bis(3,5,6-trichloropyridine-2-oxyacetato-κO)-bis(ethanol-κO)nickel(II)], C28H26Cl6N4NiO8
- Crystal structure and anti-inflammatory activity of (3E,5E)-1-((4-chlorophenyl)sulfonyl)-3,5-bis(4-fluorobenzylidene)piperidin-4-one-dichloromethane (1/1), C26H20Cl3F2NO3S
- The crystal structure of 5-bromopicolinic acid monohydrate, C6H6BrNO3
- The crystal structure of 2-(3-(4-bromophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)-8H-indeno[1,2-d]thiazole, C25H17BrFN3S
- The crystal structure of catena-poly[(μ2-2-((3-bromo-2-oxidobenzylidene)amino)acetato-κ4O,N,O′:O′′)-(dimethylformamide-κ1O)]zinc(II), C12H13N2O4BrZn
- Crystal structure of aqua-azido-κ1N-(6,6′-((propane-1,3-diylbis(azanylylidene))bis(methanylylidene))bis(3-bromophenolato)-κ4N,N′,O,O′iron(III), C17H16Br2FeN5O3
- The crystal structure of tris(1-ethylimidazole-κ1N)-(sulfato-κ2O,O′)vanadium(IV), C15H24N6O5SV
- Crystal structure of (E)-3-methoxy-N′-(1-(pyridin-2-yl)ethylidene)benzohydrazide, C15H15N3O2
- Crystal structure of dichloro-bis-(1-butyl-1H-benzo[d]imidazole)-nickel(II), C22H28Cl2N4Ni
- The crystal structure of 2-(2,3-dimethoxyphenyl)-3-hydroxy-4H-chromen-4-one, C17H14O5
- The crystal structure of 5-(2-(4-fluorophenyl)hydrazono)-4-methyl-2-((3-(5-methyl-1-(4-methylphenyl)-1H-1,2,3-triazol-4-yl)-1-phenyl-1H-pyrazol-4-yl)methylene) hydrazono)-2,5-dihydrothiazole dimethylformamide monosolvate, C30H25FN10S⋅C3H7NO
- The crystal structure of 1,8-bis(pyridin-4-ylethynyl)anthracene-1,2,4,5-tetrafluoro-3,6-diiodobenzene (2/1), C62H32F4I2N4
- The crystal structure of 3,6-di-tert-butyl-1,8-diiodo-9-methyl-9H-carbazole, C21H25I2N
- The crystal structure of 8-((4-chlorophenylamino)methylene)-6,10-dioxaspiro[4.5]decane-7,9-dione, C15H14ClNO4
- The crystal structure of catena-poly[oktaaqua-bis(μ2-4,4′-ethene-1,2-diyldipyridine-κ2N:N′)-(μ2-3,3′-(1-oxidodiazene-1,2-diyl)diphthalato-κ2O:O′)dicobalt(II)] dihydrate, C28H36N4O19Co2
- Crystal structure of (E)-1-(2-cyano-3-oxo-1-phenylprop-1-en-1-yl)-3,7-diphenylindolizine-6-carbonitrile, C31H19N3O
- Crystal structure of 1,1′-bis(diphenylphosphino)ferrocene-(1,1′-bis(diphenylphosphino)ferrocene-κ2P,P′)-(O-isobutyl sulfurodithioito-κ2S,S′)copper(I), C39H37CuFeOP2S2
- Crystal structure of poly[(5-bimethylamino-1-naphthalenesulfonato-κO)-(μ3-hexamethylenetetramino-κ3N:N′:N′′)silver(I)] dihydrate, C36H52Ag2N10O8S2
- Crystal structure of poly[μ2-diaqua-(μ2-2-amino-4,5-dicyano-κ2N:N′-imidazol-1-ide)sodium(I)], C5H6N5O2Na
- Crystal structure of (1,3-propanediamine-κ2N,N′)(N-(3-aminopropyl)-α-methyl aspartato-κ4N,N′,O,O′)cobalt(III) chloride, C11H24ClCoN4O4
- Crystal structure and anti-inflammatory activity of (3E,5E)-3,5-bis(4-fluorobenzylidene)-1-((4-fluorophenyl)sulfonyl)piperidin-4-one-dichloromethane (1/1), C26H20Cl2F3NO3S
- Crystal structure of (S)-(+)-1-cyclohexylethylaminium chloride, C8H18NCl
- The crystal structure of tris(nitrato-κ2O,O′)-bis(4,4,5,5-tetramethyl-2-(o-pyridyl)imidazoline-1-oxyl 3-oxide-κ2N,O)yttrium(III), C24H32N9O13Y
- Hydrogen bonding versus packing effects in the crystal structure of 3-((1R,2S)-1-methylpyrrolidin-1-ium-2-yl)pyridin-1-ium tetraiodidozincate(II), C10H16I4ZnN2
- Dimerization of 2-[(2-((2-aminophenyl)thio)phenyl)amino]-cyclohepta-2,4,6-trien-1-one through hydrogen bonding, C19H16N2OS
- Crystal structure of 1-(4-chloro-phenyl)-7-ethoxyl-6,8-difluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid, C18H12ClF2NO4
- Crystal structure of 7-ethoxy-6,8-difluoro-4-oxo-1-pyridin-2-ylmethyl-1,4-dihydro-quinoline-3-carboxylic acid, C18H14F2N2O4
- Crystal structure of octahydro-7aR,8′R-dimethylspiro[isobenzofuran-4(1H), 4′ (3′H)-[1H-7,9a]methanocyclohepta[c]pyran]-1′,3, 9′ (3aH,4′aH)-trione, C20H26O5
- Crystal structure of bis(5-ethoxy-2-(((1-hydroxy-2-methyl-3-oxidopropan-2-yl)imino)methyl)phenolato-κ3N,O,O’)manganese(IV) – methanol (1/1), C27H38MnN2O9
- Crystal structure of 8a,8a′′-oxybis(8aH-8,9-dioxa-3a1λ4-aza-8aλ4-borabenzo[fg]tetracene), C34H22B2N2O5
- Crystal structure of bromido-triphenyl-(triphenylarsine oxide-κO)tin(IV), C36H30AsBrOSn
- Crystal structure of catena-poly[chlorido-(μ2-formato-κ2O:O′)-(1,10-phenathroline-κ2N,N′)copper(II)], C26H18Cl2Cu2N4O4
- The crystal structure of poly[(μ10-5-carboxyisophthalato-κ10O)disodium], C9H4Na2O6
- The crystal structure of 3,5-difluoroisonicotinic acid, C6H3F2NO2
- The crystal structure of ethyl-1-(N-(adamantan-1-yl)-carbamothioyl)piperidine-4-carboxylate, C19H30N2O2S
- Crystal structure of 5-methyl-3-phenyl-1-tosyl-1,2,3,4-tetrahydropyridine, C19H21NO2S
- Crystal structure of bis((3-chlorosalicylidene)-ethylenediaminato-κ4N,N′,O,O′)nickel (II), C16H12Cl2NiN2O2
- Crystal structure of (E)-N′-(2-chloro-6-hydroxybenzylidene)-4-hydroxybenzohydrazide — dihydrofuran-2(3H)-one (1/1), C18H17ClN2O5
- Crystal structure of bis((3-bromosalicylidene)-ethylenediaminato-κ4N,N′,O,O′) nickel (II), C16H12Br2NiN2O2
- Crystal structure of trimethylsulfoxonium tetrachloridocobaltate(II) [(CH3)3SO]2CoCl4
Artikel in diesem Heft
- Frontmatter
- Crystal structure of bis [1-(phenylsulfonyl)-2-(1-(pyrazin-2-yl)ethylidene)hydrazin-1-ido-κ3N,N′,O]cobalt(II), C24H22N8O4S2Co
- The crystal structure of 1,3-bis(4-(methoxycarbonyl)benzyl)-2-methyl-1H-benzo[d]imidazol-3-ium bromide, C26H25BrN2O4
- Crystal structure of {tris((1H-benzo[d]imidazol-2-yl)methyl)amine-κ4N,N′,N′′,N′′′}-(nitrito-κ2O,O′)nickel(II) perchlorate – ethanol (1/1), C26H27ClN8NiO7
- Crystal structure of catena-poly[aqua[(μ2-4,5-dicarboxylato-2-(2-carboxylatophenyl)imidazol-1-ido-κ4N,O,O′:N′)](μ2-4,4′-bipyridine-κ2N:N′)dicopper(II)], C22H14Cu2N4O7
- Crystal structure of chlorido-tris(4-methylbenzyl-κC)-(triphenylarsine oxide-κO)tin(IV), C42H42AsClOSn
- The crystal structure of 4,4′-bipyridinium bis(3-carboxy-2-nitrobenzoate) tetrahydrate, C13H13N2O8
- Crystal structure of 1-(3-chlorophenyl)-4-(4-(((2,3-dihydro-1H-inden-5-yl)oxy)methyl)phenethyl)piperazine, C28H31ClN2O
- Crystal structure of catena-poly[diaqua-bis(μ2-5,5′-(1H-imidazole-4,5-diyl)bis(tetrazol-2-ido)-κ4N,N′:N′′,N′′′)magnesium], C10H8N20O2Mg
- The crystal structure of (E)-2-((2-hydroxy-4-ethoxybenzylidene)amino)-2-methylpropane-1,3-diol monohydrate, C13H21NO5
- Crystal structure of catena-poly[diaqua-(μ2-bipyridine-κ2N:N′)-bis(3,5-dichloroisonicotinato-κO)cadmium(II)] dihydrate, C22H20CdCl4N4O8
- The crystal structure of 4-(4-chlorophenyl)cyclohexane-1-carboxylic acid, C13H15ClO2
- Redetermination of the crystal structure of yttrium(III) trinitrate(V) pentahydrate, Y(NO3)3 ⋅ 5 H2O, H10N3O14Y
- Crystal structure of catena-poly[di-μ2-chlorido-1,10-phenanthroline-κ2N,N′-cadmium(II)], C12H8Cl2CdN2
- Crystal structure of 4-((2-methyl-6-(trifluoromethyl)pyrimidin-4-yl)oxy)benzoic acid, C13H9F3N2O3
- Crystal structure of 3-acetyl-4-hydroxybenzoic acid, C18H16O8
- Crystal structure of bis(N,2-bis(4-ethoxybenzylidene)hydrazine-1-carbohydrazonothioato-κ2N,S)nickel(II) — N,N-dimethylformamide (1/2), C44H56N10S2O6Ni
- The crystal structure of 5-chloro-4,6-dimethoxypyrimidin-2-amine, C6H8ClN3O2
- Crystal structure of poly[aqua-(μ4-benzene-1,2,4,5-tetracarboxylato-κ4O,O′,O′′,O′′′)bis(μ2-1-(4-(1H-imidazol-1-yl)benzyl)-1H-1,2,4-triazole-κ2N:N)dinickel(II)], NiC17H14N5O5
- Crystal structure of poly[aqua(5-dimethylamino)naphthalene-1-sulfonato-κ2N:O)(μ2-4,4′-bipyridyl -κ2N:N′)silver(I)], C44H44Ag2N6O8S2
- Crystal structure of 1-[3-(trifluoromethyl)cinnamoyl]-3-(pyridin-2-yl-κN)pyrazole-κ2N-bis(2-phenylpyridinato-k2C,N)iridium(III) hexafluorophosphate complex, [C40H28F3IrN5O]PF6
- Crystal structure of catena-poly[aqua(μ6-piperazine-1,4-bisethanesulfonato-κ6N:N′:O:O′:O′′:O′′′)(μ2-pyrazinyl-κ2N:N′)disilver(I)sesquihydrate], C12H30Ag2N4O11S2
- Crystal structure of (E)-1-(2-nitrophenyl)-N-(o-tolyl)methanimine, C14H12N2O2
- Crystal structure of 4′-amino-3′,5′-diisopropyl-(1,1′-biphenyl)-4-carbonitrile, C19H22N2
- The crystal structure of poly[bis(N,N-dimethylformamide-κ1O)-tetrakis(μ2-cyanido-κ2C:N)dinickel(II)], C10H14N6O2Ni2
- Crystal structure of rac-trans-N,N′-bis(3-bromo-5-chlorosalicylidene)-1,2-cyclohexanediamine, C20H18Br2Cl2N2O2
- Crystal structure of rac-trans-N,N′-bis(3,5-dibromosalicylidene)-1,2-cyclohexanediamine, C20H18Br4N2O2
- The crystal structure of (dichromato-κ2O,O′)bis(1,10-phenanthroline-κ2N,N′)nickel(II), C12H16N4O7Cr2Ni
- The crystal structure of 3-((1R,2S)-1-methylpyrrolidin-1-ium-2-yl)pyridin-1-ium tetrachloridozincate(II) monohydrate, C10H18Cl4ZnN2O
- Crystal structure of bis(μ2-azido-k2N,N)-bis(2-amino-1-(N-(3-bromosalicylaldiminato))ethane)-dicopper(II), C20H18Br4N2O2
- Crystal structure of (η6-1-methyl-4-isopropylbenzene)-[5-bromo-2-(2-pyridyl)phenyl-κ2C,N]-chloro-ruthenium(II), C21H21BrClNRu
- Crystal structure of N-(methyl(oxo)(1-(6-(trifluoromethyl)pyridin-3-yl)ethyl)-λ6-sulfanylidene)cyanamide, C10H10F3N3OS
- Crystal structure of 6,6′-((cyclohexane-1,2-diylbis(azanylylidene))bis(methanylylidene))bis(2-bromo-4-chlorophenolato-κ4N,N′,O,O′)nickel(II), C20H16Br2Cl2NiN2O2
- Redetermination of the crystal structure of catena-poly[aqua-(1,10-phenanthroline-κ2N,N′)-(μ2-tetraoxidomolybdato(VI)-κ2O:O′)manganese(II) monohydrate, C12H12N2O6MoMn
- The crystal structure tetrakis(μ2-o-chlorobenzoato-κ2O:O′)-bis(methanol-κ1O)dirhodium(II), C30H24Cl4O10Rh2
- Crystal structure of bis(2,3-diphenyltetrazolidine-5-thione-κ1S)-(nitrato-κ1O)-(nitrato-κ2O,O′)lead(II), C26H20N10O6S2Pb
- Crystal structure of bis(3-bromo-N-(1-(3-methylpyrazin-2-yl)ethylidene)benzohydrazonato-κ3O,N,N′)cadmium(II) hemihydrate, C28H25N8O2.5Br2Cd
- Crystal structure of catena-poly[tetrakis(μ2-trifluoroacetato-κ2O:O′)(μ2-2,5-dimethylpyrazine-κ2N,N′)dicopper(II)], C7H4CuF6NO4
- The crystal structure of catena-poly[bis[3-azoniapentane-1,5-diammonium][bis(μ4-oxo)-tetrakis(μ3-oxo)-heptakis(μ2-oxo)-tetradecaoxo-octa-molybdenum] dihydrate], (C8H36N6O29Mo8)n
- Crystal structure of tetraaqua-bis(2-((3,5,6-trichloropyridin-2-yl)oxy)acetato-κO)-nickel(II)—diaqua-bis(2-((3,5,6-trichloropyridin-2-yl)oxy)acetato-nickel(II), C28H24Cl12N4Ni2O18
- The crystal structure of bis(2-hydroxypyrimidinium) pentachloridobismuthate(III), (C4N2H5O)2BiCl5
- The crystal structure of catena-poly[(μ2-4,4′-dipyridine-κ2N,N′)-bis(3,5,6-trichloropyridine-2-oxyacetato-κO)-bis(ethanol-κO)nickel(II)], C28H26Cl6N4NiO8
- Crystal structure and anti-inflammatory activity of (3E,5E)-1-((4-chlorophenyl)sulfonyl)-3,5-bis(4-fluorobenzylidene)piperidin-4-one-dichloromethane (1/1), C26H20Cl3F2NO3S
- The crystal structure of 5-bromopicolinic acid monohydrate, C6H6BrNO3
- The crystal structure of 2-(3-(4-bromophenyl)-5-(4-fluorophenyl)-4,5-dihydro-1H-pyrazol-1-yl)-8H-indeno[1,2-d]thiazole, C25H17BrFN3S
- The crystal structure of catena-poly[(μ2-2-((3-bromo-2-oxidobenzylidene)amino)acetato-κ4O,N,O′:O′′)-(dimethylformamide-κ1O)]zinc(II), C12H13N2O4BrZn
- Crystal structure of aqua-azido-κ1N-(6,6′-((propane-1,3-diylbis(azanylylidene))bis(methanylylidene))bis(3-bromophenolato)-κ4N,N′,O,O′iron(III), C17H16Br2FeN5O3
- The crystal structure of tris(1-ethylimidazole-κ1N)-(sulfato-κ2O,O′)vanadium(IV), C15H24N6O5SV
- Crystal structure of (E)-3-methoxy-N′-(1-(pyridin-2-yl)ethylidene)benzohydrazide, C15H15N3O2
- Crystal structure of dichloro-bis-(1-butyl-1H-benzo[d]imidazole)-nickel(II), C22H28Cl2N4Ni
- The crystal structure of 2-(2,3-dimethoxyphenyl)-3-hydroxy-4H-chromen-4-one, C17H14O5
- The crystal structure of 5-(2-(4-fluorophenyl)hydrazono)-4-methyl-2-((3-(5-methyl-1-(4-methylphenyl)-1H-1,2,3-triazol-4-yl)-1-phenyl-1H-pyrazol-4-yl)methylene) hydrazono)-2,5-dihydrothiazole dimethylformamide monosolvate, C30H25FN10S⋅C3H7NO
- The crystal structure of 1,8-bis(pyridin-4-ylethynyl)anthracene-1,2,4,5-tetrafluoro-3,6-diiodobenzene (2/1), C62H32F4I2N4
- The crystal structure of 3,6-di-tert-butyl-1,8-diiodo-9-methyl-9H-carbazole, C21H25I2N
- The crystal structure of 8-((4-chlorophenylamino)methylene)-6,10-dioxaspiro[4.5]decane-7,9-dione, C15H14ClNO4
- The crystal structure of catena-poly[oktaaqua-bis(μ2-4,4′-ethene-1,2-diyldipyridine-κ2N:N′)-(μ2-3,3′-(1-oxidodiazene-1,2-diyl)diphthalato-κ2O:O′)dicobalt(II)] dihydrate, C28H36N4O19Co2
- Crystal structure of (E)-1-(2-cyano-3-oxo-1-phenylprop-1-en-1-yl)-3,7-diphenylindolizine-6-carbonitrile, C31H19N3O
- Crystal structure of 1,1′-bis(diphenylphosphino)ferrocene-(1,1′-bis(diphenylphosphino)ferrocene-κ2P,P′)-(O-isobutyl sulfurodithioito-κ2S,S′)copper(I), C39H37CuFeOP2S2
- Crystal structure of poly[(5-bimethylamino-1-naphthalenesulfonato-κO)-(μ3-hexamethylenetetramino-κ3N:N′:N′′)silver(I)] dihydrate, C36H52Ag2N10O8S2
- Crystal structure of poly[μ2-diaqua-(μ2-2-amino-4,5-dicyano-κ2N:N′-imidazol-1-ide)sodium(I)], C5H6N5O2Na
- Crystal structure of (1,3-propanediamine-κ2N,N′)(N-(3-aminopropyl)-α-methyl aspartato-κ4N,N′,O,O′)cobalt(III) chloride, C11H24ClCoN4O4
- Crystal structure and anti-inflammatory activity of (3E,5E)-3,5-bis(4-fluorobenzylidene)-1-((4-fluorophenyl)sulfonyl)piperidin-4-one-dichloromethane (1/1), C26H20Cl2F3NO3S
- Crystal structure of (S)-(+)-1-cyclohexylethylaminium chloride, C8H18NCl
- The crystal structure of tris(nitrato-κ2O,O′)-bis(4,4,5,5-tetramethyl-2-(o-pyridyl)imidazoline-1-oxyl 3-oxide-κ2N,O)yttrium(III), C24H32N9O13Y
- Hydrogen bonding versus packing effects in the crystal structure of 3-((1R,2S)-1-methylpyrrolidin-1-ium-2-yl)pyridin-1-ium tetraiodidozincate(II), C10H16I4ZnN2
- Dimerization of 2-[(2-((2-aminophenyl)thio)phenyl)amino]-cyclohepta-2,4,6-trien-1-one through hydrogen bonding, C19H16N2OS
- Crystal structure of 1-(4-chloro-phenyl)-7-ethoxyl-6,8-difluoro-4-oxo-1,4-dihydro-quinoline-3-carboxylic acid, C18H12ClF2NO4
- Crystal structure of 7-ethoxy-6,8-difluoro-4-oxo-1-pyridin-2-ylmethyl-1,4-dihydro-quinoline-3-carboxylic acid, C18H14F2N2O4
- Crystal structure of octahydro-7aR,8′R-dimethylspiro[isobenzofuran-4(1H), 4′ (3′H)-[1H-7,9a]methanocyclohepta[c]pyran]-1′,3, 9′ (3aH,4′aH)-trione, C20H26O5
- Crystal structure of bis(5-ethoxy-2-(((1-hydroxy-2-methyl-3-oxidopropan-2-yl)imino)methyl)phenolato-κ3N,O,O’)manganese(IV) – methanol (1/1), C27H38MnN2O9
- Crystal structure of 8a,8a′′-oxybis(8aH-8,9-dioxa-3a1λ4-aza-8aλ4-borabenzo[fg]tetracene), C34H22B2N2O5
- Crystal structure of bromido-triphenyl-(triphenylarsine oxide-κO)tin(IV), C36H30AsBrOSn
- Crystal structure of catena-poly[chlorido-(μ2-formato-κ2O:O′)-(1,10-phenathroline-κ2N,N′)copper(II)], C26H18Cl2Cu2N4O4
- The crystal structure of poly[(μ10-5-carboxyisophthalato-κ10O)disodium], C9H4Na2O6
- The crystal structure of 3,5-difluoroisonicotinic acid, C6H3F2NO2
- The crystal structure of ethyl-1-(N-(adamantan-1-yl)-carbamothioyl)piperidine-4-carboxylate, C19H30N2O2S
- Crystal structure of 5-methyl-3-phenyl-1-tosyl-1,2,3,4-tetrahydropyridine, C19H21NO2S
- Crystal structure of bis((3-chlorosalicylidene)-ethylenediaminato-κ4N,N′,O,O′)nickel (II), C16H12Cl2NiN2O2
- Crystal structure of (E)-N′-(2-chloro-6-hydroxybenzylidene)-4-hydroxybenzohydrazide — dihydrofuran-2(3H)-one (1/1), C18H17ClN2O5
- Crystal structure of bis((3-bromosalicylidene)-ethylenediaminato-κ4N,N′,O,O′) nickel (II), C16H12Br2NiN2O2
- Crystal structure of trimethylsulfoxonium tetrachloridocobaltate(II) [(CH3)3SO]2CoCl4

