Abstract
C10H5NO4, triclinic,
Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Yellow block |
| Size: | 0.10 × 0.09 × 0.06 mm |
| Wavelength: | Mo kα radiation (0.71073 Å) |
| μ: | 0.12 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, |
| θ max, completeness: | 26.4°, >99 % |
| N(hkl)measured, N(hkl)unique, R int: | 33,183, 5487, 0.055 |
| Criterion for I obs, N(hkl)gt: | I obs > 2σ(I obs), 3217 |
| N(param)refined: | 406 |
| Programs: | Bruker [1], Olex2 [2, 3], SHELX [4], PLATON [5] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| C1C | 0.8189 (3) | −0.0060 (3) | 1.0218 (2) | 0.0624 (7) |
| C1B | 0.8013 (3) | 0.4327 (3) | 0.94615 (18) | 0.0602 (7) |
| C1A | 0.8264 (3) | 0.5575 (3) | 0.6313 (2) | 0.0656 (7) |
| C2B | 0.7121 (4) | 0.5346 (3) | 0.9383 (2) | 0.0789 (9) |
| H2B | 0.7202 (4) | 0.6053 (3) | 0.9975 (2) | 0.0946 (11)* |
| C2A | 0.7252 (3) | 0.5477 (3) | 0.5317 (2) | 0.0739 (8) |
| H2A | 0.6624 (3) | 0.6088 (3) | 0.5294 (2) | 0.0887 (10)* |
| C2C | 0.8341 (4) | 0.1051 (3) | 1.1149 (2) | 0.0784 (9) |
| H2C | 0.9111 (4) | 0.1161 (3) | 1.1746 (2) | 0.0941 (11)* |
| C3B | 0.6215 (4) | 0.5295 (3) | 0.8510 (2) | 0.0754 (9) |
| H3B | 0.5681 (4) | 0.5971 (3) | 0.8504 (2) | 0.0905 (10)* |
| C3A | 0.7208 (3) | 0.4552 (3) | 0.4456 (2) | 0.0684 (8) |
| H3A | 0.6536 (3) | 0.4525 (3) | 0.3848 (2) | 0.0821 (9)* |
| C3C | 0.7426 (4) | 0.1904 (3) | 1.1173 (2) | 0.0736 (8) |
| H3C | 0.7567 (4) | 0.2594 (3) | 1.1787 (2) | 0.0883 (10)* |
| C4B | 0.6010 (3) | 0.4217 (2) | 0.75449 (19) | 0.0553 (6) |
| C4A | 0.8172 (3) | 0.3567 (2) | 0.44194 (19) | 0.0529 (6) |
| C4C | 0.6198 (3) | 0.1803 (3) | 1.0269 (2) | 0.0604 (7) |
| C5C | 0.5001 (3) | 0.0622 (2) | 0.83660 (18) | 0.0482 (6) |
| C5B | 0.6817 (3) | 0.2112 (2) | 0.67086 (16) | 0.0457 (5) |
| C5A | 1.0114 (3) | 0.2682 (2) | 0.54634 (18) | 0.0512 (6) |
| C6B | 0.7745 (3) | 0.1204 (2) | 0.6723 (2) | 0.0588 (7) |
| H6B | 0.7668 (3) | 0.0510 (2) | 0.6126 (2) | 0.0706 (8)* |
| C6A | 1.1124 (3) | 0.2755 (3) | 0.6368 (2) | 0.0655 (7) |
| H6A | 1.1747 (3) | 0.2138 (3) | 0.6380 (2) | 0.0787 (9)* |
| C6C | 0.4883 (3) | −0.0350 (3) | 0.7459 (2) | 0.0652 (7) |
| H6C | 0.4162 (3) | −0.0413 (3) | 0.6856 (2) | 0.0783 (9)* |
| C7B | 0.8786 (3) | 0.1342 (3) | 0.7635 (2) | 0.0626 (7) |
| H7B | 0.9438 (3) | 0.0747 (3) | 0.7652 (2) | 0.0751 (9)* |
| C7A | 1.1198 (3) | 0.3758 (3) | 0.7256 (2) | 0.0714 (8) |
| H7A | 1.1880 (3) | 0.3823 (3) | 0.7873 (2) | 0.0857 (10)* |
| C7C | 0.5853 (4) | −0.1236 (3) | 0.7454 (2) | 0.0753 (8) |
| H7C | 0.5790 (4) | −0.1899 (3) | 0.6843 (2) | 0.0903 (10)* |
| C8B | 0.8878 (3) | 0.2347 (3) | 0.85228 (19) | 0.0559 (6) |
| H8B | 0.9558 (3) | 0.2410 (3) | 0.91400 (19) | 0.0671 (8)* |
| C8C | 0.6910 (3) | −0.1143 (3) | 0.8346 (2) | 0.0651 (7) |
| H8C | 0.7549 (3) | −0.1750 (3) | 0.8336 (2) | 0.0781 (9)* |
| C8A | 1.0273 (3) | 0.4664 (3) | 0.72374 (19) | 0.0628 (7) |
| H8A | 1.0316 (3) | 0.5326 (3) | 0.78432 (19) | 0.0753 (9)* |
| C9B | 0.7952 (3) | 0.3270 (2) | 0.84968 (16) | 0.0454 (5) |
| C9C | 0.7031 (3) | −0.0155 (2) | 0.92578 (18) | 0.0484 (6) |
| C9A | 0.9275 (3) | 0.4596 (2) | 0.63195 (17) | 0.0488 (6) |
| C10B | 0.6922 (2) | 0.3181 (2) | 0.75754 (16) | 0.0410 (5) |
| C10C | 0.6068 (3) | 0.0761 (2) | 0.92869 (16) | 0.0432 (5) |
| C10A | 0.9188 (3) | 0.3599 (2) | 0.54060 (16) | 0.0443 (5) |
| N1B | 0.5625 (3) | 0.1846 (2) | 0.57292 (16) | 0.0589 (6) |
| N1C | 0.3932 (3) | 0.1528 (2) | 0.83158 (19) | 0.0638 (6) |
| N1A | 1.0014 (4) | 0.1545 (3) | 0.45486 (19) | 0.0751 (7) |
| O1C | 0.8964 (3) | −0.0899 (2) | 1.02191 (19) | 0.0946 (8) |
| O1B | 0.8725 (3) | 0.4325 (2) | 1.02835 (14) | 0.0947 (8) |
| O1A | 0.8271 (3) | 0.6412 (2) | 0.70996 (18) | 0.1060 (9) |
| O2A | 0.8171 (3) | 0.2777 (2) | 0.36167 (14) | 0.0792 (6) |
| O2B | 0.5156 (3) | 0.4185 (2) | 0.67494 (16) | 0.0938 (8) |
| O2C | 0.5290 (4) | 0.2516 (3) | 1.0325 (2) | 0.1314 (12) |
| O3A | 0.8975 (4) | 0.0609 (2) | 0.42725 (19) | 0.1095 (9) |
| O3B | 0.5972 (3) | 0.2115 (3) | 0.50857 (16) | 0.0967 (8) |
| O3C | 0.4323 (3) | 0.2530 (3) | 0.8264 (3) | 0.1441 (14) |
| O4B | 0.4352 (2) | 0.1297 (2) | 0.56118 (16) | 0.0890 (7) |
| O4A | 1.0981 (3) | 0.1581 (3) | 0.4153 (2) | 0.1204 (10) |
| O4C | 0.2674 (3) | 0.1210 (3) | 0.8296 (3) | 0.1398 (13) |
1 Source of materials
The 5-nitronaphtoquinone was prepared following the reported methodology [6] starting from naphthoquinone in H2SO4 in presence of NaNO3, the crude was precipitated in crushed ice to then vacuum filtered and washed with saturated NaHCO3 dissolution. The solid obtained was recrystallized in acetone and yellow block-like crystals were obtained. The structure was confirmed by melting point [7] and, 1 H NMR (400 MHz, Acetone-d6) d 8.32 (dd, J = 7.8, 1.2 Hz, 1H), 8.13 (t, J = 7.8 Hz, 1H), 8.05 (dd, J = 8.0, 1.2 Hz, 1H), 7.18 (d, J = 10.4 Hz, 1H), 7.12 (d, J = 10.4 Hz, 1H) 13 C NMR (101 MHz, Acetone) d 182.76, 181.78, 138.97, 138.41, 135.25, 133.04, 128.46, 127.56, 122.65.
2 Experimental details
Using Olex2 [2], with the olex2.solve [3] using Charge Flipping and refined with the SHELXL [4] the structure was solved. SADABS-2016/2 (Bruker, 2016/2) was used for absorption correction. Interactions were calculated using Platon [5]. Reflections were merged by SHELXL according to the crystal class for the calculation of statistics and refinement. H atoms were finally included in their calculated positions and treated as riding on their parent atom with constrained thermal parameters as U iso(H) = 1.2 U eq(C), the constraint distances of C–H was 0.93 Å.
3 Comment
Quinones are organic compounds that consist in two-carbonyl keto with two carbon–carbon double bonds in a six membered ring and depending on the aromatic ring fused to the quinone we can find three types of quinones, benzoquinones, naphthoquinones and anthraquinones [6]. Quinones are natural occurring compounds which are found in plants, bacteria or insects [7, 8]. Quinones are interesting compounds due to multiple biological properties such as antibacterial, antifungal, antioxidant and anti-cancer [9], [10], [11], [12], [13]. Quinones are vastly studied due to redox properties which can be used for energy storage applications [14, 15].
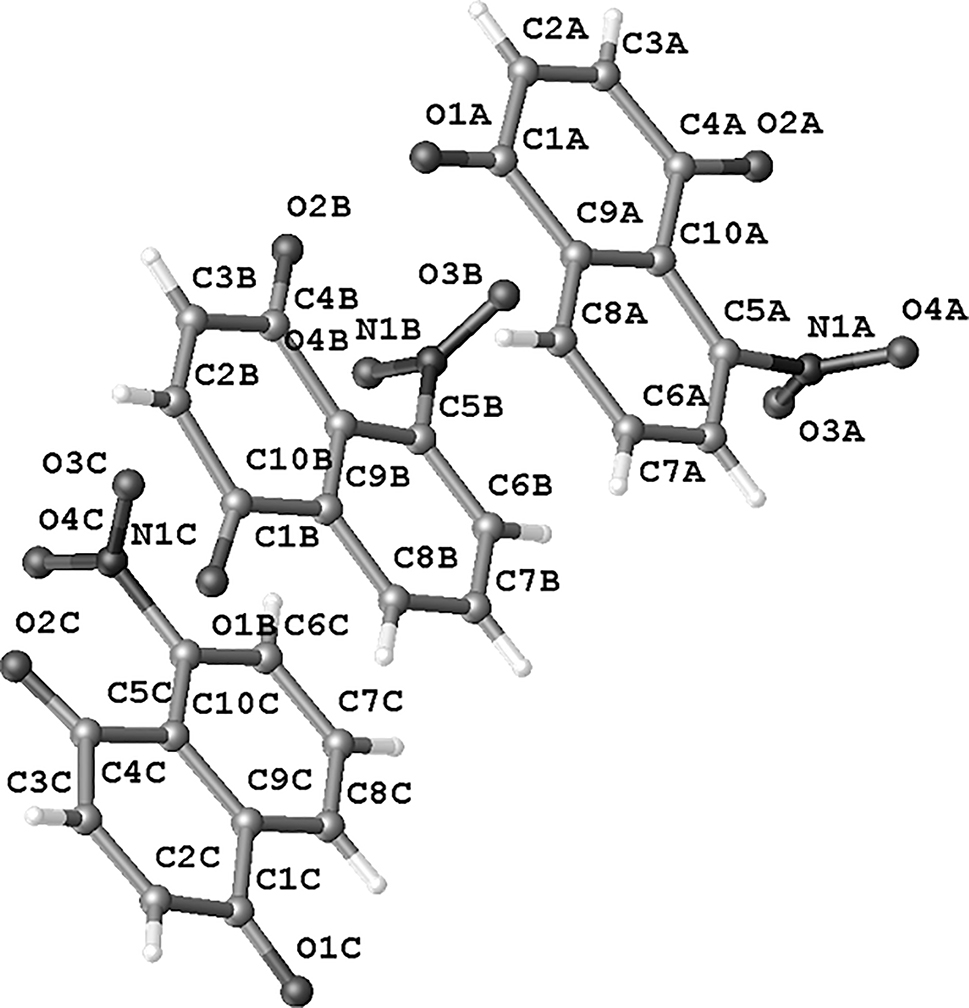
In this structure, there are three crystallographically independent molecules in the asymmetric unit. (see the figure) Half-normal probability plot analysis was used to identify systematic geometrical differences. A comparison of the bond distances and angles of the fitted residues reveals that the molecules do not show any significant geometrical differences. The main difference lies in the conformations of the nitro-group, which are involved in intermolecular interactions and crystal-packing effects. The largest difference (0.0460 Å) is between the N1C–O3C bond in the first molecule and N1B–O4B in the second molecule (9.200 Δ/σ). The N–O bond lengths in the nitro group range from 1.170(3) to 1.220(4) Å. The angle between O–N–O in these structures range from 121.0(3) to 125.0(3)°, O–N–C range between 116.4(2) and 119.9(2)°, similar value to reported by us in Barrientos et al. [16, 17]. There are no classical hydrogen bonds in the crystal [5], only intermolecular C–H⋯O interactions with distances of 3.129(5)–3.273(4) Å between the donor and acceptor atoms are found [18]. The crystal structure exhibits C–O⋯p and N–O⋯p interactions [C(4A)–O(2A)⋯Cg(1) = 3.817(3) Å, C(4C)–O(2C)⋯Cg(7) = 3.939(4) Å, C(4C)–O(2C)⋯Cg(8) = 3.937(4) Å, N(1B)–O(3B)⋯Cg(1) = 3.101(3) Å, N(1C)–O(3C)⋯Cg(4) = 2.899(4) Å. In crystal-packing the interaction between the quinone ring and the quinone oxygen of the next unit is observed (C(4A)–O(2A)⋯Cg(1)) [19], [20], [21], simultaneously in the next unit the interaction between the oxygen of the nitro group with the quinone ring is observed (N(1B)–O(3B)⋯Cg(1) and N(1C)–O(3C)⋯Cg(4)) generating a turn between the molecules that form the asymmetric unit.
Acknowledgements
We gratefully acknowledge support by FONDEQUIP EQM200138 for D8 Venture diffractometer.
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: FONDEQUIP EQM200138.
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
References
1. BRUKER. SAINT, APEX2 and SADABS; Bruker AXS Inc.: Madison, Wisconsin, USA, 2009.Search in Google Scholar
2. Bourhis, L. J., Dolomanov, O. V., Gildea, R. J., Howard, J. A. K., Puschmann, H. The anatomy of a comprehensive constrained, restrained refinement program for the modern computing environment-Olex2 dissected. Acta Crystallogr. 2015, A71, 59–75.10.1107/S2053273314022207Search in Google Scholar PubMed PubMed Central
3. Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341.10.1107/S0021889808042726Search in Google Scholar
4. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8.10.1107/S2053229614024218Search in Google Scholar PubMed PubMed Central
5. Spek, A. L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13.10.1107/S0021889802022112Search in Google Scholar
6. Shvartsberg, M. S., Kolodina, E. A., Lebedeva, N. I., Fedenok, L. G. Vicinal acetylenic derivatives of 2-amino-1,4-naphthoquinone as a key precursors of heterocyclic quinones. Russ. Chem. Bull. 2012, 61, 582–588.10.1007/s11172-012-0084-8Search in Google Scholar
7. Ivashkina, N. V., Romanov, V. S., Moroz, A. A., Shvartsberg, M. S. 5-Arylethynyl-1,4-naphthoquinones. Russ. Chem. Bull. 1984, 33, 2345–2348.10.1007/BF00948851Search in Google Scholar
8. Dulo, B., Phan, K., Githaiga, J., Raes, K., De Meester, S. Natural quinone dyes: a review on structure, extraction techniques, analysis and application potential. Waste Biomass. Valorization 2021, 12, 6339–6374.10.1007/s12649-021-01443-9Search in Google Scholar
9. Abaham, I., Joshi, R., Pardasani, P., Pardasani, R. T. Recent advances in 1,4-benzoquinone chemistry. J. Braz. Chem. Soc. 2011, 22, 385–421.10.1590/S0103-50532011000300002Search in Google Scholar
10. Borges, M. E., Tejera, R. L., Diaz, L., Esparza, P., Ibanez, E. Natural dyes extraction from cochineal (Dactylopius coccus). New extraction methods. Food Chem. 2012, 132, 1855–1860.10.1016/j.foodchem.2011.12.018Search in Google Scholar
11. Srivastava, S., Chowdhury, A. R., Maurya, S. Antimicrobial efficacy of methylated lac dye, an anthraquinone derivative. Indian J. Microbiol. 2017, 57, 470–476.10.1007/s12088-017-0682-0Search in Google Scholar PubMed PubMed Central
12. Khan, S. A., Ahmad, A., Khan, M. I., Yusuf, M., Shahid, M., Manzoor, N., Mohammad, F. Antimicrobial activity of wool yarn dyed with Rheum emodi L. (Indian Rhubarb). Dyes Pigments 2012, 95, 206–214.10.1016/j.dyepig.2012.04.010Search in Google Scholar
13. Tsao, Y.-C., Chang, Y.-J., Wang, C.-H., Chen, L. Discovery of isoplumbagin as a novel NQO1 substrate and anti-cancer quinone. Int. J. Mol. Sci. 2020, 21, 4378.10.3390/ijms21124378Search in Google Scholar PubMed PubMed Central
14. Jones, A., Ejigu, A., Wang, B., Adams, R., Bissett, M., Dryfe, R. Quinone voltammetry for redox-flow battery applications. J. Electroanal. Chem. 2022, 920, 116572.10.1016/j.jelechem.2022.116572Search in Google Scholar
15. Sedenho, G., De Porcellinis, D., Jing, Y., Kerr, E., Mejia-Mendoza, L. M., Vazquez-Mayagoitia, A., Aspuru-Guzik, A., Gordon, R. G., Crespilho, F., Aziz, M. J. Effect of molecular structure of quinones and carbon electrode surfaces on the interfacial electron transfer process. ACS Appl. Energy Mater. 2020, 3, 1933–1943.10.1021/acsaem.9b02357Search in Google Scholar
16. Barrientos, C., Barahona, P., Guevara, J. L., Squella, J. A., Moris, S. The crystal structure of 4-(pyren-1-yl)butyl-3-nitrobenzoate, C27H21NO4. Z. Kristallogr. N. Cryst. Struct. 2019, 234, 1213–1214.10.1515/ncrs-2019-0340Search in Google Scholar
17. Barrientos, C., Squella, J. A., Moris, S. The crystal structure of 4-(pyren-1-yl)butyl-4-nitrobenzoate, C27H21NO4. Z. Kristallogr. N. Cryst. Struct. 2023, 238, 459–461.10.1515/ncrs-2023-0034Search in Google Scholar
18. Moris, S., Galdamez, A., Jara, P., Saitz-Barria, C. Synthesis of novel p-tert-butylcalix[4]arene derivative: structural characterization of a methanol inclusion compound. Crystals 2016, 6, 114.10.3390/cryst6090114Search in Google Scholar
19. Steed, K. M., Steed, J. W. Packing problems: high Z′ crystal structures and their relationship to cocrystals, inclusion compounds, and polymorphism. Chem. Rev. 2015, 115, 2895–2933.10.1021/cr500564zSearch in Google Scholar PubMed
20. Desiraju, G. R. On the presence of multiple molecules in the crystal asymmetric unit (Z′ > 1). Cryst. Eng. Comm. 2007, 9, 91–92.10.1039/B614933BSearch in Google Scholar
21. Nichol, G. S., Clegg, W. The importance of weak C–H⋯O bonds and π⋯π stacking interactions in the formation of organic 1,8-bis(dimethylamino)naphthalene complexes with Z′ > 1. Cryst. Growth Des. 2006, 6, 451–460.10.1021/cg0503806Search in Google Scholar
© 2023 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of a polymorph of potassium picrate, C6H2KN3O7
- The crystal structure of (1E,2E)-1,2-bis(quinolin-2-ylmethylene)hydrazine, C20H14N4
- 5-Amino-2-chloro-4-fluoro-N-(N-isopropyl-N-methylsulfamoyl) benzamide, C11H15O3ClFN3S
- Crystal structure of trans-N 1,N 8-bis(2-cyanoethyl)-5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane, C22H42N6
- The crystal structure of [N-{[2-(oxy)-4-methoxyphenyl](phenyl)methylidene}alaninato]-diphenyl-silicon(IV) – chloroform (1/1), C29H25NO4Si·CHCl3
- Crystal structure of tetracarbonyl-{μ-[N-(diphenylphosphanyl)-N,P,P-triphenylphosphinous amide]}-bis[μ-(phenylmethanethiolato)]diiron (Fe–Fe), C48H39Fe2NO4P2S2
- Crystal structure of baryte from Mine du Pradet (France)
- The crystal structure of [(2,2′-bipyridine-6-carboxylato-κ3 N,N,O)-(6-phenylpyridine-2-carboxylate-κ2 N,O)copper(II)] monohydrate, C23H17N3O5Cu
- Crystal structure of bis(μ-benzeneselenolato)-(μ-[N-benzyl-N-(diphenylphosphanyl)-P,P-diphenylphosphinous amide])-tetracarbonyl diiron (Fe–Fe), C47H37Fe2NO4P2Se2
- The crystal structure of diaqua-methanol-κ1 O- (3-thiophenecarboxylato-κO)-(2,2′-dipyridyl-κ2 N,N′)manganese(II) 3-thiophenecarboxylate, C21H22N2O7S2Mn
- Crystal structure of catena-poly[tetrakis(butyl)-μ2-2-((oxido(phenyl)methylene)hydrazineylidene)propanoato-κ4 O:O,O′,N-μ2-2-((oxido(phenyl)methylene)hydrazineylidene)propanoato-κ4 O,N,O′:N′-ditin(IV)], C34H50N6O6Sn2
- Crystal structure of 4-chloro-N′-[(1E)-(2-nitrophenyl)methylidene]benzohydrazide, C14H10ClN3O3
- The crystal structure of 3-(1′-deoxy-3′,5′-O-dibenzy-β-d-ribosyl)adenine dichloromethane solvate, C49H52Cl2N10O6
- The crystal structure of (Z)-4-amino-N′-(1-(o-tolyl)ethylidene)benzohydrazide, C16H17N3O
- The co-crystal structure of etoricoxib–phthalic acid (1/1), C18H15ClN2O2S·C8H6O4
- Crystal structure of (glycinto-κ 2 O,N ′)-[5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane-κ 4 N,N ′,N ″,N ‴]nickel(II) perchlorate monohydrate C18H42ClN5NiO7
- The crystal structure of catena-poly[bis(1-ethylimidazole-k1 N)-(μ 2-benzene-1-carboxyl-3,5-dicarboxylato-κ 2 O, O′)zinc(II)], C19H20N4O6Zn
- Crystal structure of 3-(thiazol-2-ylcarbamoyl)-7-oxabicyclo[2.2.1]heptane-2-carboxylic acid, C11H12N2O4S
- Rietveld structure analysis of keatite, a rare, metastable SiO2 polymorph
- Crystal structure of catena-poly[(μ2-isophthalato-k3 O,O′:O″)(4-(4-pyridyl)-2,5-dipyrazylpyridine-k3 N,N′,N″)cobalt(II)] trihydrate C26H22N6O7Co1
- Crystal structure of 3,5–di-O-benzoyl-1,2-O-isopropylidene-α–D-ribose, C22H22O7
- The crystal structure of fac-tricarbonyl(6-bromo-2,2-bipyridine-κ2 N,N)-(nitrato-κO)rhenium(I), C13H7BrN3O6Re
- The crystal structure of (E)-N′-(4-hydroxy-3-methoxybenzylidene)-2-naphthohydrazide monohydrate, C19H18N2O4
- The crystal structure of 5,5′-diselanediyl-bis(2-hydroxybenzaldehyde), C14H10O4Se2
- The crystal structure of catena-poly[diaqua-m2-dicyanido-κ2 C:N-dicyanido-κ1 C-bis(4-(pyridin-4-yl)benzaldehyde-κ1N)iron(II)-platinum(II), C28H22N6O4PtFe
- Redetermination of the crystal structure of 5,14-dihydro-6,17-dimethyl-8,15-diphenyldibenzo(b,i)(1,4,8,11)tetra-azacyclotetradecine, C32H28N4
- Crystal structure of poly[(μ3-2-(3,5-dicarboxyphenyl) benzimidazole-6-carboxylato-κ4O:O:O′:O″)lead(II)] monohydrate, C16H10N2O7Pb
- The crystal structure of fac-tricarbonyl(2-pyridin-2-yl-quinoline-κ2 N,N′)-(pyrazole-κN)rhenium(I)nitrate, C20H14N4O3ReNO3
- Crystal structure of dibromo-dicarbonyl-bis(tricyclohexylphosphine)-osmium(II) dichloromethane solvate, C38H66Br2O2OsP2
- Crystal structure of poly[bis(μ 2-2,6-bis(1-imidazoly)pyridine-κ 2 N:N′)copper(II)] diperchlorate dihydrate, C22H22Cl2CuN10O10
- The crystal structure of fac-tricarbonyl(N-benzoyl-N,N-cyclohexylmethylcarbamimidothioato-κ2 S,O)-(pyridine-κN)rhenium(I), C23H24N3O4ReS
- Crystal structure of (E)-7-fluoro-2-(4-fluorobenzylidene)-3,4-dihydronaphthalen-1(2H)-one, C17H12F2O
- Synthesis and crystal structure of 1-((3R,10S,13S, 17S)-3-((2-methoxyphenyl)amino)-10,13-dimethylhexadecahydro-1H-cyclopenta[α]phenanthren-17-yl)ethan-1-one, C28H41NO2
- The crystal structure of fac-tricarbonyl((pyridin-2-yl)methanamino-κ2 N,N′)-((pyridin-2-yl)methanamino-κN)rhenium(I) nitrate, C15H16O3N4Re
- The crystal structure of (1-(pyridin-2-yl)-N-(pyridin-2-ylmethyl)-N-((1-(4-vinylbenzyl)-1H-benzo[d]imidazol-2-yl)methyl)methanamine-κ 4 N,N′,N″,N‴)tris(nitrato-kO,O′)-erbium(III), C29H27ErN8O9
- Crystal structure of tetracene-5,12-dione, C18H10O2
- Crystal structure of (3R,3aS,6R,6aR)-6-hexyl-3-methyltetrahydrofuro[3,4-b]furan-2,4-dione, C13H20O4
- The crystal structure of N′1,N′3-bis((E)-thiophen-2-ylmethylene)isophthalohydrazide monohydrate, C18H16N4O3S2
- Crystal structure of methyl ((4-aminobenzyl)sulfonyl)-L-prolinate, C13H18N2O4S
- Crystal structure of (E)-3-(3-methoxybenzylidene)benzofuran-2(3H)-one, C16H12O3
- Synthesis and crystal structure (E)-1-(4-bromo-2-hydroxyphenyl)-3-(dimethylamino)prop-2-en-1-one, C11H12BrNO2
- Synthesis and crystal structure of (S,E)-4-hydroxy-3-(2-((4aR,6aS,7R,10aS,10bR)-3,3,6a,10b-tetramethyl-8-methylenedecahydro-1H-naphtho[2,1-d][1,3]dioxin-7-yl)ethylidene)dihydrofuran-2(3H)-one, C23H34O5
- The crystal structure of N,N′-(1,2-phenylene)bis (2-((2-oxopropyl)selanyl)benzamide), C26H24N2O4Se2
- The crystal structure of 1-ethyl-2-nitro-imidazole oxide, C5H7N3O3
- The crystal structure of 2-(2-fluorophenyl)naphtho[2,1-d]thiazole, C17H10FNS
- Crystal structure of (E)-2,4-di-tert-butyl-6-(((2-fluorophenyl)imino) methyl)phenol, C21H26FNO
- Synthesis and crystal structure of 3-methyl-2-(methylthio)-4H-chromen-4-one, C12H12O2S
- Crystal structure of dithieno[2,3-d:2′,3′-d′]benzo[1,2-b:4,5-b′]dithiophene-5,10-dione, C14H4O2S4
- The crystal structure of dimethyl 2,2ʹ-((adamantane-1,3-diylbis(4,1-phenylene)) bis(oxy))diacetate, C28H32O6
- The crystal structure of N-(6-chloro-2-methyl-2H-indazol-5-yl)acetamide, C10H10ClN3O
- Crystal structure of triaqua-(5-bromoisophthalato-κ1 O)-(2,2′-bipyridine-κ2 N:N′)nickel(II) hydrate, C18H19BrN2NiO8
- The crystal structure of 2-amino-4-carboxypyridin-1-ium perchlorate, C6H7ClN2O6
- The crystal structure of catena-poly[5-aminonicotinic acid-k1 N-m2-bromido-copper(I)], Cu(C6N2H6O2)Br
- The crystal structure of 2,2-bis(3-methoxyphenyl)-1-tosyl-1,2-dihydro- 2λ4,3λ4 -[1,3,2]diazaborolo[4,5,1-ij]quinoline - dichloromethane (1/1)
- The crystal structure of catena-poly[bis(6-phenylpyridine-2-carboxylato-κ2 N,O)-(μ2-4,4′-bipyridne-κ2 N:N)cadmium(II)], C34H24N4O4Cd
- The crystal structure of 5,7-dinitropyrazolo[5,1-b]quinazolin-9(4H)-one, C10H5N5O5
- Crystal structure of rac-1,8-bis(2-carbamoylethyl)-5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane, C22H46N6O2
- The crystal structure of (E)-N ′-(2-bromobenzylidene)-2-naphthohydrazide, C36H26Br2N4O2
- The crystal structure of 5-nitronaphthoquinone, C10H5NO4
- The crystal structure of (S, R p )-4–benzhydrylideneamino-12-(4-tert-butyl oxazolin-2-yl)[2.2]paracyclophane, C36H36N2O
- Synthesis and crystal structure of 2-(2-oxo-2-(o-tolyl)ethyl)-4H-chromen-4-one, C18H14O3
- Crystal structure of 2-(thiazol-2-yl)hexahydro-1H-4,7-epoxyisoindole-1,3(2H)-dione, C11H10N2O3S
- Crystal structure of N-(diaminomethylene)-1-(dimethylamino)-1-iminiomethanaminium dichloride, C4H13Cl2N5
- Crystal structure of poly[(μ3-3, 5-dichloro-2-hydroxy-benzoato-κ4 Cl,O:O′:O″) silver(I)], C7H3AgCl2O3
- The crystal structure of tetrakis(1-isopropylimidazole-κ1 N)-[μ2- imidazole-4,5-dicarboxylato-κ4 O,N,O′,N′)]- trioxido-divanadium, C29H41N10O7V2
- Crystal structure of catena-[(μ3-bromido)-(1H-1,2,4-triazol-1-yl)benzoato-κ1 N)copper(I)], C9H7BrCuN3O2
- The crystal structure of (E)-4-fluoro-N′-(1-(2-hydroxyphenyl)propylidene)benzohydrazide, C16H15FN2O2
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of a polymorph of potassium picrate, C6H2KN3O7
- The crystal structure of (1E,2E)-1,2-bis(quinolin-2-ylmethylene)hydrazine, C20H14N4
- 5-Amino-2-chloro-4-fluoro-N-(N-isopropyl-N-methylsulfamoyl) benzamide, C11H15O3ClFN3S
- Crystal structure of trans-N 1,N 8-bis(2-cyanoethyl)-5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane, C22H42N6
- The crystal structure of [N-{[2-(oxy)-4-methoxyphenyl](phenyl)methylidene}alaninato]-diphenyl-silicon(IV) – chloroform (1/1), C29H25NO4Si·CHCl3
- Crystal structure of tetracarbonyl-{μ-[N-(diphenylphosphanyl)-N,P,P-triphenylphosphinous amide]}-bis[μ-(phenylmethanethiolato)]diiron (Fe–Fe), C48H39Fe2NO4P2S2
- Crystal structure of baryte from Mine du Pradet (France)
- The crystal structure of [(2,2′-bipyridine-6-carboxylato-κ3 N,N,O)-(6-phenylpyridine-2-carboxylate-κ2 N,O)copper(II)] monohydrate, C23H17N3O5Cu
- Crystal structure of bis(μ-benzeneselenolato)-(μ-[N-benzyl-N-(diphenylphosphanyl)-P,P-diphenylphosphinous amide])-tetracarbonyl diiron (Fe–Fe), C47H37Fe2NO4P2Se2
- The crystal structure of diaqua-methanol-κ1 O- (3-thiophenecarboxylato-κO)-(2,2′-dipyridyl-κ2 N,N′)manganese(II) 3-thiophenecarboxylate, C21H22N2O7S2Mn
- Crystal structure of catena-poly[tetrakis(butyl)-μ2-2-((oxido(phenyl)methylene)hydrazineylidene)propanoato-κ4 O:O,O′,N-μ2-2-((oxido(phenyl)methylene)hydrazineylidene)propanoato-κ4 O,N,O′:N′-ditin(IV)], C34H50N6O6Sn2
- Crystal structure of 4-chloro-N′-[(1E)-(2-nitrophenyl)methylidene]benzohydrazide, C14H10ClN3O3
- The crystal structure of 3-(1′-deoxy-3′,5′-O-dibenzy-β-d-ribosyl)adenine dichloromethane solvate, C49H52Cl2N10O6
- The crystal structure of (Z)-4-amino-N′-(1-(o-tolyl)ethylidene)benzohydrazide, C16H17N3O
- The co-crystal structure of etoricoxib–phthalic acid (1/1), C18H15ClN2O2S·C8H6O4
- Crystal structure of (glycinto-κ 2 O,N ′)-[5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane-κ 4 N,N ′,N ″,N ‴]nickel(II) perchlorate monohydrate C18H42ClN5NiO7
- The crystal structure of catena-poly[bis(1-ethylimidazole-k1 N)-(μ 2-benzene-1-carboxyl-3,5-dicarboxylato-κ 2 O, O′)zinc(II)], C19H20N4O6Zn
- Crystal structure of 3-(thiazol-2-ylcarbamoyl)-7-oxabicyclo[2.2.1]heptane-2-carboxylic acid, C11H12N2O4S
- Rietveld structure analysis of keatite, a rare, metastable SiO2 polymorph
- Crystal structure of catena-poly[(μ2-isophthalato-k3 O,O′:O″)(4-(4-pyridyl)-2,5-dipyrazylpyridine-k3 N,N′,N″)cobalt(II)] trihydrate C26H22N6O7Co1
- Crystal structure of 3,5–di-O-benzoyl-1,2-O-isopropylidene-α–D-ribose, C22H22O7
- The crystal structure of fac-tricarbonyl(6-bromo-2,2-bipyridine-κ2 N,N)-(nitrato-κO)rhenium(I), C13H7BrN3O6Re
- The crystal structure of (E)-N′-(4-hydroxy-3-methoxybenzylidene)-2-naphthohydrazide monohydrate, C19H18N2O4
- The crystal structure of 5,5′-diselanediyl-bis(2-hydroxybenzaldehyde), C14H10O4Se2
- The crystal structure of catena-poly[diaqua-m2-dicyanido-κ2 C:N-dicyanido-κ1 C-bis(4-(pyridin-4-yl)benzaldehyde-κ1N)iron(II)-platinum(II), C28H22N6O4PtFe
- Redetermination of the crystal structure of 5,14-dihydro-6,17-dimethyl-8,15-diphenyldibenzo(b,i)(1,4,8,11)tetra-azacyclotetradecine, C32H28N4
- Crystal structure of poly[(μ3-2-(3,5-dicarboxyphenyl) benzimidazole-6-carboxylato-κ4O:O:O′:O″)lead(II)] monohydrate, C16H10N2O7Pb
- The crystal structure of fac-tricarbonyl(2-pyridin-2-yl-quinoline-κ2 N,N′)-(pyrazole-κN)rhenium(I)nitrate, C20H14N4O3ReNO3
- Crystal structure of dibromo-dicarbonyl-bis(tricyclohexylphosphine)-osmium(II) dichloromethane solvate, C38H66Br2O2OsP2
- Crystal structure of poly[bis(μ 2-2,6-bis(1-imidazoly)pyridine-κ 2 N:N′)copper(II)] diperchlorate dihydrate, C22H22Cl2CuN10O10
- The crystal structure of fac-tricarbonyl(N-benzoyl-N,N-cyclohexylmethylcarbamimidothioato-κ2 S,O)-(pyridine-κN)rhenium(I), C23H24N3O4ReS
- Crystal structure of (E)-7-fluoro-2-(4-fluorobenzylidene)-3,4-dihydronaphthalen-1(2H)-one, C17H12F2O
- Synthesis and crystal structure of 1-((3R,10S,13S, 17S)-3-((2-methoxyphenyl)amino)-10,13-dimethylhexadecahydro-1H-cyclopenta[α]phenanthren-17-yl)ethan-1-one, C28H41NO2
- The crystal structure of fac-tricarbonyl((pyridin-2-yl)methanamino-κ2 N,N′)-((pyridin-2-yl)methanamino-κN)rhenium(I) nitrate, C15H16O3N4Re
- The crystal structure of (1-(pyridin-2-yl)-N-(pyridin-2-ylmethyl)-N-((1-(4-vinylbenzyl)-1H-benzo[d]imidazol-2-yl)methyl)methanamine-κ 4 N,N′,N″,N‴)tris(nitrato-kO,O′)-erbium(III), C29H27ErN8O9
- Crystal structure of tetracene-5,12-dione, C18H10O2
- Crystal structure of (3R,3aS,6R,6aR)-6-hexyl-3-methyltetrahydrofuro[3,4-b]furan-2,4-dione, C13H20O4
- The crystal structure of N′1,N′3-bis((E)-thiophen-2-ylmethylene)isophthalohydrazide monohydrate, C18H16N4O3S2
- Crystal structure of methyl ((4-aminobenzyl)sulfonyl)-L-prolinate, C13H18N2O4S
- Crystal structure of (E)-3-(3-methoxybenzylidene)benzofuran-2(3H)-one, C16H12O3
- Synthesis and crystal structure (E)-1-(4-bromo-2-hydroxyphenyl)-3-(dimethylamino)prop-2-en-1-one, C11H12BrNO2
- Synthesis and crystal structure of (S,E)-4-hydroxy-3-(2-((4aR,6aS,7R,10aS,10bR)-3,3,6a,10b-tetramethyl-8-methylenedecahydro-1H-naphtho[2,1-d][1,3]dioxin-7-yl)ethylidene)dihydrofuran-2(3H)-one, C23H34O5
- The crystal structure of N,N′-(1,2-phenylene)bis (2-((2-oxopropyl)selanyl)benzamide), C26H24N2O4Se2
- The crystal structure of 1-ethyl-2-nitro-imidazole oxide, C5H7N3O3
- The crystal structure of 2-(2-fluorophenyl)naphtho[2,1-d]thiazole, C17H10FNS
- Crystal structure of (E)-2,4-di-tert-butyl-6-(((2-fluorophenyl)imino) methyl)phenol, C21H26FNO
- Synthesis and crystal structure of 3-methyl-2-(methylthio)-4H-chromen-4-one, C12H12O2S
- Crystal structure of dithieno[2,3-d:2′,3′-d′]benzo[1,2-b:4,5-b′]dithiophene-5,10-dione, C14H4O2S4
- The crystal structure of dimethyl 2,2ʹ-((adamantane-1,3-diylbis(4,1-phenylene)) bis(oxy))diacetate, C28H32O6
- The crystal structure of N-(6-chloro-2-methyl-2H-indazol-5-yl)acetamide, C10H10ClN3O
- Crystal structure of triaqua-(5-bromoisophthalato-κ1 O)-(2,2′-bipyridine-κ2 N:N′)nickel(II) hydrate, C18H19BrN2NiO8
- The crystal structure of 2-amino-4-carboxypyridin-1-ium perchlorate, C6H7ClN2O6
- The crystal structure of catena-poly[5-aminonicotinic acid-k1 N-m2-bromido-copper(I)], Cu(C6N2H6O2)Br
- The crystal structure of 2,2-bis(3-methoxyphenyl)-1-tosyl-1,2-dihydro- 2λ4,3λ4 -[1,3,2]diazaborolo[4,5,1-ij]quinoline - dichloromethane (1/1)
- The crystal structure of catena-poly[bis(6-phenylpyridine-2-carboxylato-κ2 N,O)-(μ2-4,4′-bipyridne-κ2 N:N)cadmium(II)], C34H24N4O4Cd
- The crystal structure of 5,7-dinitropyrazolo[5,1-b]quinazolin-9(4H)-one, C10H5N5O5
- Crystal structure of rac-1,8-bis(2-carbamoylethyl)-5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradecane, C22H46N6O2
- The crystal structure of (E)-N ′-(2-bromobenzylidene)-2-naphthohydrazide, C36H26Br2N4O2
- The crystal structure of 5-nitronaphthoquinone, C10H5NO4
- The crystal structure of (S, R p )-4–benzhydrylideneamino-12-(4-tert-butyl oxazolin-2-yl)[2.2]paracyclophane, C36H36N2O
- Synthesis and crystal structure of 2-(2-oxo-2-(o-tolyl)ethyl)-4H-chromen-4-one, C18H14O3
- Crystal structure of 2-(thiazol-2-yl)hexahydro-1H-4,7-epoxyisoindole-1,3(2H)-dione, C11H10N2O3S
- Crystal structure of N-(diaminomethylene)-1-(dimethylamino)-1-iminiomethanaminium dichloride, C4H13Cl2N5
- Crystal structure of poly[(μ3-3, 5-dichloro-2-hydroxy-benzoato-κ4 Cl,O:O′:O″) silver(I)], C7H3AgCl2O3
- The crystal structure of tetrakis(1-isopropylimidazole-κ1 N)-[μ2- imidazole-4,5-dicarboxylato-κ4 O,N,O′,N′)]- trioxido-divanadium, C29H41N10O7V2
- Crystal structure of catena-[(μ3-bromido)-(1H-1,2,4-triazol-1-yl)benzoato-κ1 N)copper(I)], C9H7BrCuN3O2
- The crystal structure of (E)-4-fluoro-N′-(1-(2-hydroxyphenyl)propylidene)benzohydrazide, C16H15FN2O2

