Abstract
C39H46BClCuN6·CH2Cl2, monoclinic, C2/c (no. 15), a = 18.1441(7) Å, b = 15.6735(8) Å, c = 28.1341(13) Å, β = 90.247(4)°, V = 8000.7(6) Å3, Z = 8, Rgt(F) = 0.0563, wRref(F2) = 0.1306, T = 168 K.
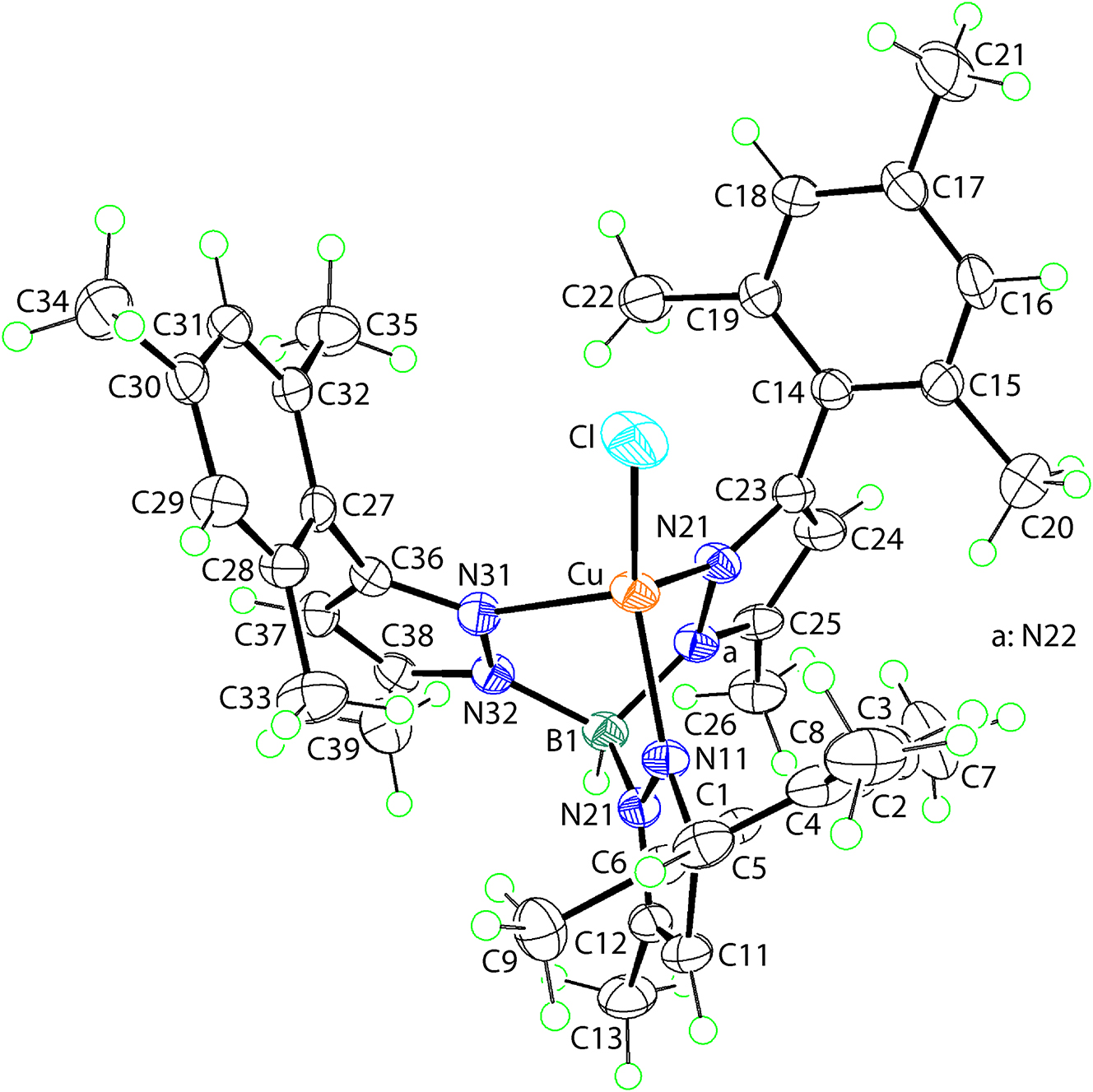
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
1 Source of material
A solution of {hydridotris[3-mesityl-5-methyl-1H-pyrazol-1-yl-κN3]borato}thallium(I) 5 ([Tl{HB(3–Mes-5–Mepz)3}], 144.36 mg, 0.178 mmol) in dichloromethane (15 mL) was added slowly to a solution of CuCl2⋅2H2O (34.7 mg, 0.204 mmol) in methanol (5 mL). After the mixture was stirred for 60 min, the solvent was evaporated under vacuum and the resulting solid was extracted with dichloromethane (25 mL) to separate the resulting salts. A red powder was obtained after evaporation of the solvent. Red crystals for X-ray crystallography were obtained by the slow evaporation of a saturated dichloromethane/n-heptane (1:1 v/v) solution held at room temperature which were characterised as [Cu(Cl){HB(3–Mes-5–Mepz)3}]·CH2Cl2 (72.2 mg, 0.102 mmol, 58 %). Anal. calcd. for C39H46BClCuN6: C, 66.10; H, 6.54; N, 11.86. Found: C; 65.94, H; 6.58, N; 11.90 %. IR (KBr, cm−1): 3002 m ν(C–H), 2954 s ν(C–H), 2920 s ν(C–H), 2858 m ν(C–H), 2527 s ν(B–H), 1615 s ν(C=N). Far–IR (CsI, cm−1): 383 ν(Cu–Cl). UV–Vis (CH2Cl2, λmax, nm (ε, M−1 cm−1)) 298 (1100), 376 (2400), 505 (480), 1020 (180).
Data collection and handling.
| Crystal: | Red prism |
| Size: | 0.25 × 0.13 × 0.05 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 0.78 mm−1 |
| Diffractometer, scan mode: | Rigaku XtaLAB P200, ω scan |
| θmax, completeness: | 27.5°, 100 % |
| N(hkl)measured, N(hkl)unique, Rint: | 31658, 9164, 0.050 |
| Criterion for Iobs, N(hkl)gt: | Iobs > 2 σ(Iobs), 5,911 |
| N(param)refined: | 472 |
| Programs: | CrysAlisPRO, 1 IL MILIONE, 2 SHELX, 3 WinGx 4 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| x | y | z | U iso */U eq | |
|---|---|---|---|---|
| Cu | 0.26421 (2) | 0.01644 (3) | 0.10440 (2) | 0.02686 (11) |
| Cl1 | 0.21368 (5) | 0.01407 (7) | 0.17240 (3) | 0.0519 (3) |
| N11 | 0.33615 (12) | 0.07677 (16) | 0.06479 (9) | 0.0251 (6) |
| N12 | 0.34979 (12) | 0.04100 (16) | 0.02147 (9) | 0.0243 (6) |
| N21 | 0.18930 (13) | 0.01001 (17) | 0.05018 (9) | 0.0264 (6) |
| N22 | 0.22011 (12) | −0.00805 (17) | 0.00684 (9) | 0.0250 (6) |
| N31 | 0.31034 (13) | −0.10051 (17) | 0.08635 (9) | 0.0265 (6) |
| N32 | 0.31682 (13) | −0.11085 (16) | 0.03788 (9) | 0.0247 (6) |
| C1 | 0.38309 (15) | 0.1962 (2) | 0.11361 (11) | 0.0255 (7) |
| C2 | 0.32592 (16) | 0.2548 (2) | 0.12103 (12) | 0.0334 (8) |
| C3 | 0.32716 (17) | 0.3056 (2) | 0.16155 (13) | 0.0366 (8) |
| H3 | 0.288139 | 0.344936 | 0.166534 | 0.044* |
| C4 | 0.38347 (17) | 0.3006 (2) | 0.19495 (12) | 0.0336 (8) |
| C5 | 0.44039 (17) | 0.2445 (2) | 0.18596 (12) | 0.0322 (8) |
| H5 | 0.480152 | 0.241483 | 0.207975 | 0.039* |
| C6 | 0.44190 (15) | 0.1925 (2) | 0.14617 (11) | 0.0271 (7) |
| C7 | 0.2642 (2) | 0.2635 (3) | 0.08477 (15) | 0.0543 (11) |
| H7A | 0.233766 | 0.313138 | 0.092666 | 0.081* |
| H7B | 0.233653 | 0.211913 | 0.085213 | 0.081* |
| H7C | 0.285253 | 0.271000 | 0.053035 | 0.081* |
| C8 | 0.3820 (2) | 0.3539 (3) | 0.23946 (14) | 0.0487 (10) |
| H8A | 0.347434 | 0.401405 | 0.235217 | 0.073* |
| H8B | 0.431441 | 0.376510 | 0.245849 | 0.073* |
| H8C | 0.366198 | 0.318628 | 0.266294 | 0.073* |
| C9 | 0.50672 (17) | 0.1334 (2) | 0.13858 (13) | 0.0401 (9) |
| H9A | 0.488956 | 0.078247 | 0.126763 | 0.060* |
| H9B | 0.532875 | 0.125025 | 0.168768 | 0.060* |
| H9C | 0.540311 | 0.158595 | 0.115284 | 0.060* |
| C10 | 0.38347 (15) | 0.1416 (2) | 0.07084 (11) | 0.0245 (7) |
| C11 | 0.42890 (16) | 0.1467 (2) | 0.03106 (11) | 0.0302 (7) |
| H11 | 0.467577 | 0.186425 | 0.025914 | 0.036* |
| C12 | 0.40658 (15) | 0.0826 (2) | 0.00072 (11) | 0.0278 (7) |
| C13 | 0.43632 (19) | 0.0585 (2) | −0.04693 (13) | 0.0416 (9) |
| H13A | 0.481968 | 0.090127 | −0.052852 | 0.062* |
| H13B | 0.399958 | 0.072516 | −0.071576 | 0.062* |
| H13C | 0.446520 | −0.002869 | −0.047569 | 0.062* |
| C14 | 0.06661 (15) | 0.0413 (2) | 0.08312 (11) | 0.0250 (7) |
| C15 | 0.03953 (16) | 0.1231 (2) | 0.09150 (12) | 0.0296 (7) |
| C16 | −0.00651 (16) | 0.1361 (2) | 0.12996 (12) | 0.0327 (8) |
| H16 | −0.024367 | 0.192021 | 0.135901 | 0.039* |
| C17 | −0.02744 (16) | 0.0711 (2) | 0.15995 (12) | 0.0323 (8) |
| C18 | −0.00135 (17) | −0.0098 (2) | 0.15020 (12) | 0.0354 (8) |
| H18 | −0.015699 | −0.055775 | 0.170163 | 0.043* |
| C19 | 0.04493 (16) | −0.0262 (2) | 0.11237 (12) | 0.0310 (7) |
| C20 | 0.0588 (2) | 0.1974 (3) | 0.05985 (16) | 0.0550 (11) |
| H20A | 0.106613 | 0.187025 | 0.044891 | 0.083* |
| H20B | 0.061344 | 0.249672 | 0.078900 | 0.083* |
| H20C | 0.020852 | 0.203788 | 0.035196 | 0.083* |
| C21 | −0.0785 (2) | 0.0877 (3) | 0.20142 (15) | 0.0558 (11) |
| H21A | −0.061264 | 0.055835 | 0.229273 | 0.084* |
| H21B | −0.128534 | 0.069300 | 0.193070 | 0.084* |
| H21C | −0.078786 | 0.148886 | 0.208715 | 0.084* |
| C22 | 0.0706 (2) | −0.1160 (2) | 0.10319 (16) | 0.0578 (12) |
| H22A | 0.058360 | −0.132137 | 0.070426 | 0.087* |
| H22B | 0.046011 | −0.155075 | 0.125245 | 0.087* |
| H22C | 0.124041 | −0.119398 | 0.107905 | 0.087* |
| C23 | 0.11748 (15) | 0.0245 (2) | 0.04264 (11) | 0.0266 (7) |
| C24 | 0.10205 (16) | 0.0173 (2) | −0.00548 (12) | 0.0344 (8) |
| H24 | 0.055528 | 0.025339 | −0.020441 | 0.041* |
| C25 | 0.16784 (16) | −0.0038 (2) | −0.02754 (11) | 0.0280 (7) |
| C26 | 0.18343 (18) | −0.0172 (3) | −0.07905 (12) | 0.0383 (8) |
| H26A | 0.137231 | −0.014500 | −0.097160 | 0.057* |
| H26B | 0.206244 | −0.073332 | −0.083533 | 0.057* |
| H26C | 0.217048 | 0.027273 | −0.090273 | 0.057* |
| C27 | 0.33158 (15) | −0.18728 (19) | 0.15870 (11) | 0.0254 (7) |
| C28 | 0.39049 (16) | −0.1535 (2) | 0.18510 (12) | 0.0297 (7) |
| C29 | 0.39646 (17) | −0.1730 (2) | 0.23303 (12) | 0.0340 (8) |
| H29 | 0.436063 | −0.149279 | 0.250846 | 0.041* |
| C30 | 0.34633 (17) | −0.2262 (2) | 0.25598 (12) | 0.0309 (7) |
| C31 | 0.28692 (16) | −0.2560 (2) | 0.22951 (12) | 0.0292 (7) |
| H31 | 0.250578 | −0.289676 | 0.244835 | 0.035* |
| C32 | 0.27878 (15) | −0.2382 (2) | 0.18164 (11) | 0.0270 (7) |
| C33 | 0.44760 (19) | −0.0978 (3) | 0.16150 (13) | 0.0465 (10) |
| H33A | 0.423220 | −0.049098 | 0.146229 | 0.070* |
| H33B | 0.474004 | −0.131044 | 0.137492 | 0.070* |
| H33C | 0.482624 | −0.077013 | 0.185460 | 0.070* |
| C34 | 0.35854 (19) | −0.2526 (3) | 0.30682 (12) | 0.0435 (9) |
| H34A | 0.373630 | −0.202849 | 0.325587 | 0.065* |
| H34B | 0.397232 | −0.296145 | 0.308260 | 0.065* |
| H34C | 0.312695 | −0.275959 | 0.319726 | 0.065* |
| C35 | 0.21411 (18) | −0.2749 (3) | 0.15440 (13) | 0.0449 (9) |
| H35A | 0.196330 | −0.232814 | 0.131277 | 0.067* |
| H35B | 0.174424 | −0.288952 | 0.176580 | 0.067* |
| H35C | 0.229669 | −0.326662 | 0.137684 | 0.067* |
| C36 | 0.32879 (15) | −0.1749 (2) | 0.10646 (11) | 0.0254 (7) |
| C37 | 0.34756 (16) | −0.2330 (2) | 0.07111 (12) | 0.0292 (7) |
| H37 | 0.363071 | −0.290358 | 0.075600 | 0.035* |
| C38 | 0.33931 (15) | −0.1914 (2) | 0.02856 (11) | 0.0265 (7) |
| C39 | 0.35084 (19) | −0.2245 (2) | −0.02078 (12) | 0.0390 (9) |
| H39A | 0.366304 | −0.284312 | −0.019326 | 0.058* |
| H39B | 0.389093 | −0.190651 | −0.036495 | 0.058* |
| H39C | 0.304683 | −0.220029 | −0.038832 | 0.058* |
| B1 | 0.30227 (18) | −0.0346 (2) | 0.00414 (13) | 0.0269 (8) |
| H1 | 0.317126 | −0.051942 | −0.033230 | 0.032* |
| C40 | 0.31133 (19) | 0.0121 (3) | 0.27321 (13) | 0.0437 (9) |
| H40A | 0.303123 | −0.040660 | 0.254541 | 0.052* |
| H40B | 0.264487 | 0.044430 | 0.273681 | 0.052* |
| Cl2 | 0.37910 (6) | 0.07350 (8) | 0.24567 (4) | 0.0652 (3) |
| Cl3 | 0.33624 (6) | −0.01559 (7) | 0.33200 (4) | 0.0537 (3) |
2 Experimental details
The B- and C-bound H atoms were geometrically placed (B–H = 1.12 Å and C–H = 0.95–1.00 Å) and refined as riding with Uiso(H) = 1.2–1.5 Ueq(B, C).
3 Discussion
Hyridotris(pyrazolyl)borate ligands are still recognised as one of the most well-studied classes of ligands in contemporary coordination chemistry. The reason for their popularity relates their easy derivatisation to introduce substituents of varying steric and electronic profiles into the pyrazolyl rings. This key feature of this ligands flexibility for the fine-tuning of the coordination environments for a wide range of metal centres. In 2016, the structure and properties of a thallium(I) salt ligated by a relatively sterically hindered mesityl-substituted pyrazolyl ring, i.e. [Tl{HB(3–Mes-5–Mepz)3}], were described. 5 Herein, in continuation of these studies, a related chlorido copper(II) complex, [Cu(Cl){HB(3–Mes-5–Mepz)3}], isolated as its dichloromethane mono-solvate, (I), is described.
The complex, [Cu(Cl){HB(3–Mes-5–Mepz)3}], was obtained by the reaction of [Tl{HB(3–Mes-5–Mepz)3}] with CuCl2⋅2H2O in 58 % yield. The IR spectrum of [Cu(Cl){HB(3–Mes-5–Mepz)3}] shows the B–H stretching band at 2527 cm−1 is slightly shifted from the absorption band at 2522 cm−1 for [Tl{HB(3–Mes-5–Mepz)3}]. 5 In the far–IR spectrum, [Cu(Cl){HB(3–Mes-5–Mepz)3}] shows a characteristic band at 383 cm−1, which is assignable to Cu–Cl stretching. Similar bands were also observed at 375 cm−1 for the phenyl analogue [Cu(Cl){HB(3–Ph-5–Mepz)3}] 6 and at 359 cm−1 for the t-butyl/i-propyl analogue, [Cu(Cl){HB(3-tBu-5-iPrpz)3}]. 7 The d-d transition band was observed at 1020 nm (180 M−1 cm−1), which compares to 960 nm (180 M−1 cm−1) reported for [Cu(Cl){HB(3–Ph-5–Mepz)3}]. 6 This low-energy shift of the d-d transition arises from the ground state change from d(x2 − y2) to d(z2). 6 , 7
The molecular structure of (I) is shown in figure (50 % probability ellipsoids; the solvent dichloromethane molecule is not shown). The copper(II) centre exists within a ClN3 donor set defined by a chlorido ligand and three pyrazolyl-nitrogen atoms derived from a tridentate {HB(3–Mes-5–Me-pz)3}− anion. The angles subtended at the copper(II) atom range from 90.96(10)°, for N11–Cu–N31, to 144.09(8)°, for N11–Cu–Cl1. Using the τ4 parameter as a guide, 8 which is calculated from the equation, τ4 = [360 − (α + β)/141], the α and β angles being the two widest angles subtended at the copper(II) centre, an indication of the nature of the coordination geometry is given. In (I), τ4 computes to 0.73, a value intermediate between 0.64, for a see-saw geometry, and 0.85, for a trigonal-pyramidal geometry. 8 The angles substended at the copper(II) atom by the tridentate ligand span a narrow range, i.e. 90.96(10)° [N11–Cu–N31] to 92.40(10)° [N11–Cu–N21] and are systematically narrower than the N–Cu–Cl angles. The latter fall in two distinct values, i.e. 112.54(7) and 112.34(8)° for N21, N31–Cu–Cl1, respectively, and a wide 144.09(8) for N11–Cu–Cl1. The pattern of bond lengths indicates a significant deviation from the putative three-fold axis incorporating the Cu, Cl and B atom. The reason for the wide N11–Cu–Cl1 angle could relate the presence of a weak Cl⃛H contact between the complex and solvent dichloromethane molecule [C40–H40b⃛Cl1: H40b⃛Cl1 = 3.03 Å with angle at H40b = 100°] as well as a relatively close intramolecular contact [C22–H22c⃛Cl1: H22c⃛Cl1 = 3.21 Å with angle at H22c = 123°]. It is also noted the Cu–N11 bond length of 1.963(2) Å is significantly shorter than the Cu–N21, N31 bond lengths of 2.041(2) and 2.079(3) Å, respectively.
The Cu–Cl bond length in (I) is 2.1253(9) Å, which can be compared with other Cu–Cl bonds in related tetrahedral chlorido copper(II) complexes having tridentate hydridotris(pyrazolyl)borate ligands. Thus, the Cu–Cl bond in (I) is experimentally equivalent to the Cu–Cl bond length of 2.125(6) Å in [Cu(Cl){HB(3,5-iPr2pz)3}], 9 but shorter than 2.167(1) Å, in [Cu(Cl){HB(3-tBu-5-iPrpz)3}], 7 2.1706(9) Å, in [Cu(Cl){HB(3–Ad-5-iPrpz)3}], 7 and 2.1738(14) and 2.1760(13) Å for the two independent molecules in [Cu(Cl){HB(3-tBu-5–Mepz)3}]. 10
In the crystal of (I), several interactions are noted. First and foremost are the interactions between the complex molecule and the dichloromethane molecule of crystallisation. Here, the solvent molecule bridges two mesityl substituents via C–Cl⃛π [C40–Cl2⃛Cg(C1–C6): Cl2⃛Cg(C1–C6) = 3.7745(18) Å with angle at Cl2 = 136.31(14)°] and C–H⃛π [C40–H40a⃛Cg(C27–C32): H40a⃛Cg(C27–C32) = 2.98 Å, C40⃛Cg(C27–C32) = 3.916(5) Å with angle at H40a = 158°] interactions to form a two-molecule aggregate. These form a linear chain along [1 −1 0] featuring C–H⃛π [C21–H21c⃛Cg(C27–C32) i : H21c⃛Cg(C27–C32) i = 2.73 Å, C21⃛Cg(C27–C32) i = 3.576(5) Å with angle at H21c = 145° for symmetry operation (i) −1/2 + x, 1/2 + y, z] interactions. Centrosymmetrically related chains assemble into double-chains which feature additional C–H⃛π [C26–H26a⃛Cg(C14–C19) ii : H26a⃛Cg(C14–C19) ii = 2.99 Å, C26–H26a⃛Cg(C14–C19) ii = 3.907(4) Å with angle at H26a = 157° for (ii) −x, −y, −z] interactions. The chains assemble in the crystal without directional interactions between them.
Acknowledgments
This research was supported by the Joint Usage/Research Centre for Catalysis and the Koyanagi Foundation.
-
Author contributions: All authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Conflict of interest: The authors declare no conflicts of interest.
-
Research funding: This study was financially supported by the Joint Usage/Research Centre for Catalysis (Proposals 22DS0143, 23DS0198 and 24ES0584).
References
1. Rigaku Oxford Diffraction. CrysAlis PRO; Rigaku Corporation: Oxford, UK, 2021.Search in Google Scholar
2. Burla, M. C.; Caliandro, R.; Camalli, M.; Carrozzini, B.; Cascarano, G. L.; De Caro, L.; Giacovazzo, C.; Polidori, G.; Siliqi, D.; Spagna, R. IL MILIONE: A Suite of Computer Programs for Crystal Structure Solution of Proteins. J. Appl. Cryst. 2007, 40, 609–613. https://doi.org/10.1107/S0021889807010941.Search in Google Scholar
3. Sheldrick, G. M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. https://doi.org/10.1107/S2053229614024218.Search in Google Scholar PubMed PubMed Central
4. Farrugia, L. J. WinGX and ORTEP for Windows: An Update. J. Appl. Cryst. 2012, 45, 849–854. https://doi.org/10.1107/S0021889812029111.Search in Google Scholar
5. Fujisawa, K.; Shimizu, D.; Szilagyi, R. K. Comparison of Thallium(I) Complexes with Mesityl-Substituted Tris(pyrazolyl)hydroborate Ligands, [Tl{HB(3–Ms-5–Mepz)3}] and [Tl{HB(3–Ms-5–Mepz)2(3–Me-5–Mspz)}]. Acta Crystallogr. 2016, C72, 786–790. https://doi.org/10.1107/S2053229615023797.Search in Google Scholar PubMed
6. Fujisawa, K.; Shimizu, M.; Tiekink, E. R. T. Crystal Structure of Chlorido {hydridotris[3-phenyl-5-methylpyrazol-1-yl-N3]borato}copper(II), C30H28BClCuN6. Z. Kristallogr. New Cryst. Struct. 2021, 236, 135–138. https://doi.org/10.1515/ncrs-2020–0410.10.1515/ncrs-2020-0410Search in Google Scholar
7. Fujisawa, K.; Tada, N.; Ishikawa, Y.; Higashimura, H.; Miyashita, Y.; Okamoto, K. The Most Hindered Hydrotris(pyrazolyl)borate Ligand, X-Ray Structure of Chlorocopper(II) Complex: [Cu(Cl){HB(3–Ad-5–Pripz)3}] as Compared with [Cu(Cl){HB(3–But-5–Pripz)3}]. Inorg. Chem. Commun. 2004, 7, 209–212. https://doi.org/10.1016/j.inoche.2003.10.030.Search in Google Scholar
8. Yang, L.; Powell, D. R.; Houser, R. P. Structural Variation in Copper(I) Complexes with Pyridylmethylamide Ligands: Structural Analysis with a New Four-Coordinate Geometry Index, τ4. Dalton Trans. 2007, 955–964. https://doi.org/10.1039/B617136B.Search in Google Scholar PubMed
9. Kitajima, N.; Fujisawa, K.; Moro-oka, Y. Tetrahedral Copper(II) Complexes Supported by a Hindered Pyrazolylborate. Formation of the Thiolato Complex, Which Closely Mimics the Spectroscopic Characteristics of Blue Copper Proteins. J. Am. Chem. Soc. 1990, 112, 3210–3212. https://doi.org/10.1021/ja00164a052.Search in Google Scholar
10. Fujisawa, K.; Iwamoto, H.; Tobita, K.; Miyashita, Y.; Okamoto, K. Copper(II) Nitrato and Chloro Complexes with Sterically Hindered Tridentate Ligands: Influence of Ligand Framework and Charge on Their Structure and Physicochemical Properties. Inorg. Chim. Acta 2009, 362, 4500–4509. https://doi.org/10.1016/j.ica.2009.05.016.Search in Google Scholar
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of 5,5′-bis(2,4,6-trinitrophenyl)-2,2′-bi(1,3,4-oxadiazole), C16H4N10O14
- Crystal structure of catena-poly[(μ3-4,4′-oxydibenzoato- κ5 O,O′: O″,O‴:O‴)-bis(2,4,6-tri(3-pyridine)-1,3,5-triazine-κ1 N)cadmium(II)], C50H32CdN12O5
- The crystal structure of 1,4-diazepane-1,4-diium potassium trinitrate, C5H14KN5O9
- The crystal structure of benzyl 2,2,5,5-tetramethylthiazolidine-4-carboxylate, C15H21NO2S
- Crystal structure of 2-hydroxyethyl-triphenylphosphonium tetracyanidoborate, C24H20BN4OP
- The crystal structure of 1-methyl-3-(N-methylnitrous amide–N-methylene) imidazolidine-2,4,5-trione
- Crystal structure of N-((3-cyano-1-(2,6-dichloro-4-(trifluoromethyl)phenyl)-4-(2,2,2-trifluoroacetyl)-1H-pyrazol-5-yl)carbamoyl)-2,6-difluorobenzamide, C20H7Cl2F8N5O3S
- Crystal structure of 5-(2,2-difluoropropyl)-5-methylbenzo[4,5]imidazo[2,1-a] isoquinolin-6(5H)-one, C20H18F2N2O
- The crystal structure of N′,N″-[1,2-bis(4-chlorophenyl)ethane-1,2-diylidene]bis(furan-2- carbohydrazide), C24H16Cl2N4O4
- Crystal structure of [(4-bromobenzyl)triphenylphosphonium] tetrabromoantimony(III), [C25H21BrP]+[SbBr4]−
- Crystal structure of [(4-bromobenzyl)triphenylphosphonium] tetrabromidoindium(III), [C25H21BrP]+[InBr4]−
- The crystal structure of 4-carboxy-2-oxobutan-1-aminium chloride, C5H10ClNO3
- Crystal structure of (4-(4-chlorophenyl)-1H-pyrrole-3-carbonyl)ferrocene, C21H16ClFeNO
- The crystal structure of dichlorido(η6-p-cymene)(triphenylarsine)ruthenium(II), C28H29AsCl2Ru
- Crystal structure of (Z)-2-hydroxy-N′-(1-(o-tolyl)ethylidene)benzohydrazide, C16H16N2O2
- The crystal structure of 10-(1-bromoethyl)-14-(bromomethyl)dibenzo[a, c]acridine, C24H17NBr2
- Synthesis and crystal structure of 6-methoxy-7-[(4-methoxyphenyl)methoxy]-2H-1-benzopyran-2-one, C18H16O5
- Synthesis and crystal structure of ethyl 4-((4-trifluoromethylbenzyl)amino)benzo, C17H16F3NO2
- The crystal structure of (Z)-2-(tert-butyl)-6-(7-(tert-butyl)-5-methylbenzo[d][1,3]oxathiol-2-ylidene)-4-methylcyclohexa-2,4-dien-1-one, C23H28O2S
- The crystal structure of (R)-2-aminobutanamide hydrochloride, C4H11ClN2O
- Crystal structure of bromido[hydridotris(3-tert-butyl-5-isopropylpyrazolyl)borato-κ3 N,N′,N″]copper(II), C30H52BBrCuN6
- Crystal structure of chlorido{hydridotris[3-mesityl-5-methyl-1H-pyrazol-1-yl-κN3]borato}-copper(II) dichloromethane monosolvate
- Crystal structure of 4-[3,5-bis(propan-2-yl)-1H-pyrazol-4-yl]pyridine, C14H19N3
- Crystal structure of ((4-(4-bromophenyl)-1H-pyrrol-3-yl)methyl)ferrocene, C21H16BrFeNO
- Crystal structure of [(4-chlorobenzyl)triphenylphosphonium] dichloridocopper(I), {[C25H21ClP]+[CuCl2]−}n
- The crystal structure of {Cu(2,9-diisopropyl-4,7-diphenyl-1,10-phenanthroline)[4,5-bis(diphenylphosphino)-9,9-dimethylxanthene]}+ PF6−·1.5(EtOAC)
- Crystal structure of 3,5-bis(t-butyl)-1H-pyrazol-4-amine, C11H21N3
- Crystal structure of [(2,4-dichlorobenzyl)triphenylphosphonium] trichloridocopper(II), [C25H20Cl2P]+[CuCl3]−
- The crystal structure of dipotassium sulfide, K2S
- Crystal structure of (4-(4-methoxyphenyl)-1H-pyrrole-3-carbonyl)ferrocene, C22H19FeNO2
- Crystal structure of (E)-6-(4-methylpiperazin-1-yl)-2-(4-(trifluoromethyl)benzylidene)-3, 4-dihydronaphthalen-1(2H)-one, C23H23F3N2O
- Crystal structure of (E)-6-morpholino-2-(4-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C22H20F3NO2
- Crystal structure of Ce9Ir37Ge25
- The crystal structure of ethyl 6-(2-nitrophenyl)imidazo[2,1-b]thiazole-3-carboxylate, C14H11N3O4S
- Crystal structure of (4-(4-isopropylphenyl)-1H-pyrrol-3-yl)(ferrocenyl)methanone, C24H23FeNO
- Crystal structure of bis(methylammonium) tetrathiotungstate(VI), (CH3NH3)2[WS4]
- Crystal structure of 6,11-dihydro-12H-benzo[e]indeno[1,2-b]oxepin-12-one, C17H12O2
- Crystal structure of 3-[(4-phenylpiperidin-1-yl)methyl]-5-(thiophen-2-yl)-2,3-dihydro-1,3,4- oxadiazole-2-thione, C18H19N3OS2
- Crystal structure of N-isopropyl-1,8-naphthalimide C15H13NO2
- TiNiSi-type EuPdBi
- Crystal structure of 1-(p-tolylphenyl)-4-(2-thienoyl)-3-methyl-1H-pyrazol-5-ol, C16H14N2O2S
- The crystal structure of 3-(3-carboxypropyl)-2-nitro-1H-pyrrole 1-oxide, C7H9N3O5
- The crystal structure of tetraaqua-bis(2-(2-methyl-5-nitro-1H-imidazol-1-yl)acetato-k2O:N)-tetrakis(2-(2-methyl-5-nitro-1H-imidazol-1-yl)acetato-k1N)trizinc(II) hexahydrate C36H52N18O32Zn3
- The crystal structure of 4-(3-carboxy-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinolin-7-yl)piperazin-1-ium 4-hydroxy-3,5-dimethoxybenzoate monohydrate, C25H30FN3O9
- Crystal structure of bis(DL-1-carboxy-2-(1H-indol-3-yl)ethan-1-aminium) oxalate — acetic acid (1/2)
- Crystal structure of methyl (E)-4-((4-methylphenyl)sulfonamido)but-2-enoate, C12H15NO4S
- The crystal structure of actarit, C10H11NO3
- The crystal structure of bicyclol, C19H18O9
- The crystal structure of topiroxostat, C13H8N6
- Crystal structure of 2,2-dichloro-N-methyl-N-(4-p-tolylthiazol-2-yl)acetamide, C13H12Cl2N2OS
- Crystal structure of 4-(trifluoromethyl)-7-coumarinyl trifluoromethanesulfonate C11H4F6O5S
- Crystal structure of (1,4,7,10,13,16-hexaoxacyclooctadecane-κ6O6)-((Z)-N,N′-bis(2-(dimethylamino)phenyl)carbamimidato-κ1N)potassium(I)
- Crystal structure of (Z)-2-(5-((4-(dimethylamino)naphthalen-1-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid, C18H16N2O3S2
- Crystal structure of (4-fluorobenzyl)triphenylphosphonium bromide, C25H21BrFP
- The crystal structure of dichlorido-[6-(pyridin-2-yl)phenanthridine-κ2N, N′]zinc(II)-chloroform (1/1), C19H13N2ZnCl5
- Crystal structure of (E)-(3-(2,4-dichlorophenyl)acryloyl)ferrocene, C19H14Cl2FeO
- The crystal structure of (E)-7-chloro-1-cyclopropyl-6-fluoro-3-((2-hydroxybenzylidene)amino)quinolin-4(1H)-one, C19H14ClFN2O2
- Crystal structure of 2-bromo-11-(((fluoromethyl)sulfonyl)methyl)-6-methyl-6,11-dihydrodibenzo[c,f][1,2]thiazepine 5,5-dioxide, C16H13BrFNO4S2
- Crystal structure of 2-chloro-11-(((fluoromethyl)sulfonyl)methyl)-6-methyl-6,11-dihydrodibenzo[c,f][1,2]thiazepine 5,5-dioxide, C16H13ClFNO4S2
- Crystal structure of 5-(2,2-difluoropropyl)-5-methyl-6-oxo-5,6-dihydrobenzo[4,5]imidazo[2,1-a]isoquinoline-3-carbonitrile, C20H15F2N3O
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- Crystal structure of 5,5′-bis(2,4,6-trinitrophenyl)-2,2′-bi(1,3,4-oxadiazole), C16H4N10O14
- Crystal structure of catena-poly[(μ3-4,4′-oxydibenzoato- κ5 O,O′: O″,O‴:O‴)-bis(2,4,6-tri(3-pyridine)-1,3,5-triazine-κ1 N)cadmium(II)], C50H32CdN12O5
- The crystal structure of 1,4-diazepane-1,4-diium potassium trinitrate, C5H14KN5O9
- The crystal structure of benzyl 2,2,5,5-tetramethylthiazolidine-4-carboxylate, C15H21NO2S
- Crystal structure of 2-hydroxyethyl-triphenylphosphonium tetracyanidoborate, C24H20BN4OP
- The crystal structure of 1-methyl-3-(N-methylnitrous amide–N-methylene) imidazolidine-2,4,5-trione
- Crystal structure of N-((3-cyano-1-(2,6-dichloro-4-(trifluoromethyl)phenyl)-4-(2,2,2-trifluoroacetyl)-1H-pyrazol-5-yl)carbamoyl)-2,6-difluorobenzamide, C20H7Cl2F8N5O3S
- Crystal structure of 5-(2,2-difluoropropyl)-5-methylbenzo[4,5]imidazo[2,1-a] isoquinolin-6(5H)-one, C20H18F2N2O
- The crystal structure of N′,N″-[1,2-bis(4-chlorophenyl)ethane-1,2-diylidene]bis(furan-2- carbohydrazide), C24H16Cl2N4O4
- Crystal structure of [(4-bromobenzyl)triphenylphosphonium] tetrabromoantimony(III), [C25H21BrP]+[SbBr4]−
- Crystal structure of [(4-bromobenzyl)triphenylphosphonium] tetrabromidoindium(III), [C25H21BrP]+[InBr4]−
- The crystal structure of 4-carboxy-2-oxobutan-1-aminium chloride, C5H10ClNO3
- Crystal structure of (4-(4-chlorophenyl)-1H-pyrrole-3-carbonyl)ferrocene, C21H16ClFeNO
- The crystal structure of dichlorido(η6-p-cymene)(triphenylarsine)ruthenium(II), C28H29AsCl2Ru
- Crystal structure of (Z)-2-hydroxy-N′-(1-(o-tolyl)ethylidene)benzohydrazide, C16H16N2O2
- The crystal structure of 10-(1-bromoethyl)-14-(bromomethyl)dibenzo[a, c]acridine, C24H17NBr2
- Synthesis and crystal structure of 6-methoxy-7-[(4-methoxyphenyl)methoxy]-2H-1-benzopyran-2-one, C18H16O5
- Synthesis and crystal structure of ethyl 4-((4-trifluoromethylbenzyl)amino)benzo, C17H16F3NO2
- The crystal structure of (Z)-2-(tert-butyl)-6-(7-(tert-butyl)-5-methylbenzo[d][1,3]oxathiol-2-ylidene)-4-methylcyclohexa-2,4-dien-1-one, C23H28O2S
- The crystal structure of (R)-2-aminobutanamide hydrochloride, C4H11ClN2O
- Crystal structure of bromido[hydridotris(3-tert-butyl-5-isopropylpyrazolyl)borato-κ3 N,N′,N″]copper(II), C30H52BBrCuN6
- Crystal structure of chlorido{hydridotris[3-mesityl-5-methyl-1H-pyrazol-1-yl-κN3]borato}-copper(II) dichloromethane monosolvate
- Crystal structure of 4-[3,5-bis(propan-2-yl)-1H-pyrazol-4-yl]pyridine, C14H19N3
- Crystal structure of ((4-(4-bromophenyl)-1H-pyrrol-3-yl)methyl)ferrocene, C21H16BrFeNO
- Crystal structure of [(4-chlorobenzyl)triphenylphosphonium] dichloridocopper(I), {[C25H21ClP]+[CuCl2]−}n
- The crystal structure of {Cu(2,9-diisopropyl-4,7-diphenyl-1,10-phenanthroline)[4,5-bis(diphenylphosphino)-9,9-dimethylxanthene]}+ PF6−·1.5(EtOAC)
- Crystal structure of 3,5-bis(t-butyl)-1H-pyrazol-4-amine, C11H21N3
- Crystal structure of [(2,4-dichlorobenzyl)triphenylphosphonium] trichloridocopper(II), [C25H20Cl2P]+[CuCl3]−
- The crystal structure of dipotassium sulfide, K2S
- Crystal structure of (4-(4-methoxyphenyl)-1H-pyrrole-3-carbonyl)ferrocene, C22H19FeNO2
- Crystal structure of (E)-6-(4-methylpiperazin-1-yl)-2-(4-(trifluoromethyl)benzylidene)-3, 4-dihydronaphthalen-1(2H)-one, C23H23F3N2O
- Crystal structure of (E)-6-morpholino-2-(4-(trifluoromethyl)benzylidene)-3,4-dihydronaphthalen-1(2H)-one, C22H20F3NO2
- Crystal structure of Ce9Ir37Ge25
- The crystal structure of ethyl 6-(2-nitrophenyl)imidazo[2,1-b]thiazole-3-carboxylate, C14H11N3O4S
- Crystal structure of (4-(4-isopropylphenyl)-1H-pyrrol-3-yl)(ferrocenyl)methanone, C24H23FeNO
- Crystal structure of bis(methylammonium) tetrathiotungstate(VI), (CH3NH3)2[WS4]
- Crystal structure of 6,11-dihydro-12H-benzo[e]indeno[1,2-b]oxepin-12-one, C17H12O2
- Crystal structure of 3-[(4-phenylpiperidin-1-yl)methyl]-5-(thiophen-2-yl)-2,3-dihydro-1,3,4- oxadiazole-2-thione, C18H19N3OS2
- Crystal structure of N-isopropyl-1,8-naphthalimide C15H13NO2
- TiNiSi-type EuPdBi
- Crystal structure of 1-(p-tolylphenyl)-4-(2-thienoyl)-3-methyl-1H-pyrazol-5-ol, C16H14N2O2S
- The crystal structure of 3-(3-carboxypropyl)-2-nitro-1H-pyrrole 1-oxide, C7H9N3O5
- The crystal structure of tetraaqua-bis(2-(2-methyl-5-nitro-1H-imidazol-1-yl)acetato-k2O:N)-tetrakis(2-(2-methyl-5-nitro-1H-imidazol-1-yl)acetato-k1N)trizinc(II) hexahydrate C36H52N18O32Zn3
- The crystal structure of 4-(3-carboxy-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinolin-7-yl)piperazin-1-ium 4-hydroxy-3,5-dimethoxybenzoate monohydrate, C25H30FN3O9
- Crystal structure of bis(DL-1-carboxy-2-(1H-indol-3-yl)ethan-1-aminium) oxalate — acetic acid (1/2)
- Crystal structure of methyl (E)-4-((4-methylphenyl)sulfonamido)but-2-enoate, C12H15NO4S
- The crystal structure of actarit, C10H11NO3
- The crystal structure of bicyclol, C19H18O9
- The crystal structure of topiroxostat, C13H8N6
- Crystal structure of 2,2-dichloro-N-methyl-N-(4-p-tolylthiazol-2-yl)acetamide, C13H12Cl2N2OS
- Crystal structure of 4-(trifluoromethyl)-7-coumarinyl trifluoromethanesulfonate C11H4F6O5S
- Crystal structure of (1,4,7,10,13,16-hexaoxacyclooctadecane-κ6O6)-((Z)-N,N′-bis(2-(dimethylamino)phenyl)carbamimidato-κ1N)potassium(I)
- Crystal structure of (Z)-2-(5-((4-(dimethylamino)naphthalen-1-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid, C18H16N2O3S2
- Crystal structure of (4-fluorobenzyl)triphenylphosphonium bromide, C25H21BrFP
- The crystal structure of dichlorido-[6-(pyridin-2-yl)phenanthridine-κ2N, N′]zinc(II)-chloroform (1/1), C19H13N2ZnCl5
- Crystal structure of (E)-(3-(2,4-dichlorophenyl)acryloyl)ferrocene, C19H14Cl2FeO
- The crystal structure of (E)-7-chloro-1-cyclopropyl-6-fluoro-3-((2-hydroxybenzylidene)amino)quinolin-4(1H)-one, C19H14ClFN2O2
- Crystal structure of 2-bromo-11-(((fluoromethyl)sulfonyl)methyl)-6-methyl-6,11-dihydrodibenzo[c,f][1,2]thiazepine 5,5-dioxide, C16H13BrFNO4S2
- Crystal structure of 2-chloro-11-(((fluoromethyl)sulfonyl)methyl)-6-methyl-6,11-dihydrodibenzo[c,f][1,2]thiazepine 5,5-dioxide, C16H13ClFNO4S2
- Crystal structure of 5-(2,2-difluoropropyl)-5-methyl-6-oxo-5,6-dihydrobenzo[4,5]imidazo[2,1-a]isoquinoline-3-carbonitrile, C20H15F2N3O

