Abstract
Background
Gastric cancer, one of the most common cancers in the world, is a multifactorial disease in which environmental and genetic factors play a role. In our study, we aimed to determine the expression levels of four miRNAs (miR127-5p, miR-544a, miR-369-3p and miR-655-3p) on chromosome 14q32 in gastric cancer.
Materials and methods
Total RNA was isolated from blood samples taken from 66 gastric cancer and 66 healthy individuals. The gene expression levels determined by cDNA and quantitative real-time polymerase chain reaction were analyzed according to the 2−∆∆Ct method. SPSS 22 were used for statistical analysis and p < 0.05 was considered as statistically significant.
Results and discussion
miR-655-3p (fold change: 100, p = 0.026), miR-127-5p (fold change: 48, p < 0.001) and miR-369-3p (fold change: 1.6, p > 0.05) was less expressed in the gastric cancer group than control group. miR-544a was found 15.5-fold more expressed in the patient group than control group (fold change: 15.47, p < 0.001).
Conclusion
miR127-5p, miR-544a, and miR-655-3p may be evaluated as biomarkers in gastric cancer.
Öz
Amaç
Dünyadaki en yaygın kanserlerden biri olan mide kanseri, çevresel ve genetik faktörlerin rol oynadığı multifaktöriyel bir hastalıktır. Çalışmamızda mide kanserinde 14q32 kromozomunda dört miRNA’nın (miR127-5p, miR-544a, miR-369-3p ve miR-655-3p) ekspresyon seviyelerini belirlemeyi amaçladık.
Gereç ve Yöntemler
Total RNA 66 mide kanserinden ve 66 sağlıklı bireyden alınan kan örneklerinden izole edildi. cDNA ve kantitatif gerçek zamanlı polimeraz zincir reaksiyonu ile belirlenen gen ekspresyon seviyeleri 2−∆∆Ct metoduna göre analiz edildi. İstatistiksel analiz için SPSS22 kullanıldı ve p < 0,05 istatistiksel olarak anlamlı kabul edildi.
Bulgular ve Tartışma
miR-655-3p (kat değişimi: 100, p = 0,026), miR-127-5p (kat değişimi: 48, p < 0,001) ve miR-369-3p (kat değişikliği: 1,6, p > 0,05) mide kanseri grubunda kontrol grubundan daha az ifade edildi. miR-544a hasta grubunda kontrol grubundan 15,5 kat daha fazla ifade edildi (kat değişimi: 15,47, p < 0,001).
Sonuç
miR127-5p, miR-544a ve miR-655-3p, mide kanserinde biyobelirteçler olarak değerlendirilebilir.
Introduction
Gastric cancer is the fourth frequently seen among malignant tumors in the world, while the fifth in Turkey and one of the most common causes of cancer-related deaths worldwide [1]. Gastric cancer has a low survival rate because of the lack of early diagnosis techniques is usually noticed late [2]. Gastric cancer is a heterogeneous disease; genetic factors, Helicobacter pylori infection, heavy alcohol consumption, smoking, high salt consumption and various environmental factors contribute to carcinogenesis [3]. The anomalies of various oncogene, tumor suppressor genes, transcription factors and growth factors might have crucial roles in tumor pathogenesis [4], [5], [6]. miRNAs play crucial role in the regulation of numerous metabolic and cellular pathways, particularly in the control of apoptosis and cell proliferation [7]. MicroRNAs (miRNAs) regulate gene expression targeting mRNA by causing translation inhibition or mRNA degradation and consist of 19–25 nucleotides [8], [9]. It is predicted that more than 60% of all protein-coding genes is controlling by miRNAs [10]. Several miRNAs such as miR-655-3p, miR-127-5p, miR-369-3p, miR-544a encoded in the 14q32 locus was found associated with metastatic phenotype in cancer [11], [12]. We aimed to determine the expressions of these miRNAs in our study and to evaluate their potential for biomarker and treatment of gastric cancer.
Materials and methods
Groups selection
The Ethics Committee of the Istanbul University approved our study (2015/1291) and all participants involved have provided written informed consent. The 5 mL fresh blood samples collected from newly diagnosed gastric cancer patients with no anticancer therapy at Istanbul University, Department of General Surgery formed gastric cancer group. Patients who received anticancer therapy and underwent surgery for treatment purposes were excluded from the study. The control group consisted of 5 mL fresh blood collected from healthy individuals who are not diagnosed with any type of cancer. The serum were collected from the fresh blood samples and stored at −80°C until further analyses.
MicroRNA isolation
The 200 μL of the serum samples was spiked with 2 μL of 25 fmol synthetic ce-miR-39 (miRNA mimic 10 pmol) as the external reference. Total RNA enriched for small RNAs was isolated simultaneously from the serum with the miRNeasy serum/plasma Isolation Kit (Qiagen, Germany, Product Number: 217184). The purities and concentrations were measured at 260 and 280 nm with NanoDrop spectrophotometer (NanoDrop 2000, Wilmington, DE, USA).
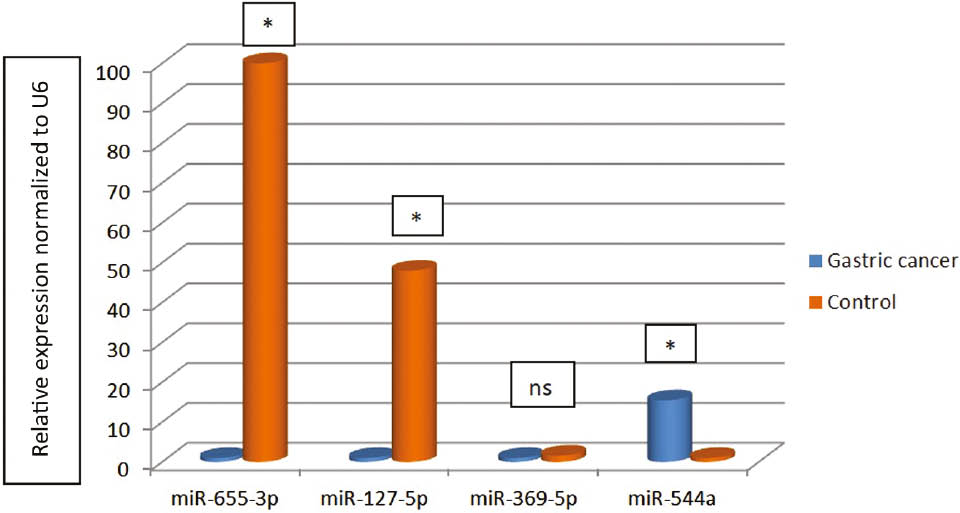
Relative expression normalized to U6 for miR-127-5p, miR-544a, miR-369-3p and miR-655-3p.
*p<0.05; ns, Non-statistical.
Reverse transcription and quantitative real-time polimerase chain reaction
The extracted microRNA was reverse transcribed to cDNA in 20 μL using miscript II RT cDNA Synthesis Kit (Qiagen, Germany, Product Number: 218161) and total volume was placed in thermal cycler with 60 min at 37°C followed by 5 min at 95°C. Reverse transcription components and volumes were given in Table 1 and run in duplicate. The polimerase chain (PCR) reaction was performed for amplification using miscript SYBR Green PCR kit (Qiagen, Germany, Product Number: 218073) and miScript Primer Assay kit (for each microRNA: Qiagen, Germany, Product Number: 218300) on ABI PRISM™ 7000 Sequence Detection System. U6 was used as housekeeping gene. Each quantitative PCR reaction solution contained diluted cDNA, 2×Quanti Tect (with SYBR), the microRNA-specific forward primer and a universal reverse primer to a total volume of 25 μL and the components were given in Table 2. The qPCR reaction parameters were predenaturation at 94°C for 15 min, 40 cycles of 94°C for 15 s, 55°C for 30 s and extension at 72°C for 30 s.
The components of reverse transcriptase reaction.
| Component | Volume |
|---|---|
| 5× miScript Hispec buffer | 4 μL |
| 10× miScript Nucleics Mix | 2 μL |
| miScript Reverse Transcriptase mix | 2 μL |
| Template RNA | 2 μL |
| RNase-Free Water | 10 μL |
| Total volume | 20 μL |
The components for real-time PCR.
| Component | Volume |
|---|---|
| 2× Quanti Tect SYBR Green PCR master Mix. | 12.5 μL |
| Universal Primer | 2.5 μL |
| 10× miScript Primer Assay | 2.5 μL |
| Template cDNA | 2 μL |
| RNase-Free water | 5.5 μL |
| Total volume | 25 μL |
Statistical analysis
The 2−ΔΔCT method was used for the determining expression levels of the genes between the groups. The distribution of gene expressions in the patient and control groups was analyzed by Kolmogorov Smirnov test and Mann Whitney U test was used for not normally distributed values in SPSS software (Version 22; SPSS Inc., Chicago, IL, USA). Spearman ρ test was used for correlation analysis and p value <0.05 was considered as statistically significant.
Results
Gastric cancer group consisted of 26 women and 40 men, while the number of women was 35 and the number of men was 31 in the control group. Five people reported that they consumed alcohol and 23 people said they were smoking in the gastric cancer group. The mean age was 57.53±19.87 in the gastric cancer group and 57.42±16.98 in the females and 57.6±21.76 in the males. The mean age was 39.2±9.74 in the control group, 36.58±8 in the females and 42.25±10.77 in the males. The age difference in the patient and control groups was statistically significant (p<0.001).
The expression levels of four microRNAs (miR-127-5p, miR-544a, miR-369-3p and miR-655-3p) of gastric cancer group compared to the control group with 2−ΔΔCT method and relative fold change was determined. miR-655-3p was found 100-fold less expressed in the gastric cancer group compared to the control group (fold change: 0.01, p=0.026). The expression levels of miR-127-5p were found about 48-fold less expressed in the gastric cancer group than the control group (fold change: 0.021). This fold change in the groups was found statistically significantly (p<0.001). miR-369-3p was found approximately 1.6-fold less expressed in the gastric cancer group to the control group (fold change: 0.63, p=0.054). It was found that miR-544a was expressed about 15.5-fold more in gastric cancer group to the control group (fold change: 15.47, p<0.001) (Table 3) (Figure 1).
The gene expression values for microRNAs according to 2−ΔΔCT.
| Gastric cancer | Control | Gastric cancer | Control | ||||
|---|---|---|---|---|---|---|---|
| miR-655-3p | miR-127-5p | ||||||
| ΔCT | 9.31±1.586 | 2.71±4.65 | ΔCT | 9.67±2.27 | 4.13±3.79 | ||
| ΔΔCT | 6.59 | ΔΔCT | 5.55 | ||||
| 2−ΔΔCT | 0.01 | 2−ΔΔCT | 0.021 | ||||
| p-Value | 0.026 | p-Value | <0.001 | ||||
| miR-369-5p | miR-544a | ||||||
| ΔCT | 9.68±2.27 | 9.02±2.26 | ΔCT | 4.77±2.21 | 8.72±2.62 | ||
| ΔΔCT | 0.66 | ΔΔCT | −3.951 | ||||
| 2−ΔΔCT | 0.63 | 2−ΔΔCT | 15.47 | ||||
| p-Value | 0.054 | p-Value | <0.001 | ||||
Metastasis was detected in 23 patients in gastric cancer group. When the correlation between the gene expression levels of microRNAs and metastasis is examined; correlation was determined with miR-127-5p and miR-369-5p (p<0.001). It was observed that miR-127-5p and miR-369-5p expression levels were lower in the presence of metastases (data not shown).
Discussion
A case control study was conducted in gastric cancer and we found miR-655-3p (fold change: 100, p=0.026), miR-127-5p (fold change: 48, p<0.001) and miR-369-3p (fold change: 1.6, p>0.05) downregulated in the gastric cancer. However, miR-544a was found 15.5-fold more expressed in the patient group than control group (fold change: 15.47, p<0.001).
The overexpression of miR-655 targeted pituitary tumor-transforming gene-1 (PTTG1) acting as a tumor suppressor [13]. Han et al. found that expression levels of miR-655-3p significantly decrease in cervical cancer [14]. Downregulated miR-655-3p levels were found correlated with advanced tumor stage and lymph node metastasis in hepatocellular carcinoma [15]. miR-655-3p also directly targets ADAM10 and inhibits β-catenin pathway. Wu et al. showed decreased expression levels of miR-655-3p in in hepatocellular carcinoma tissues and cell lines. Tumor stage was negatively related to low miR-655-3p expression [16]. However, tumor growth was suppressed using miR-655-3p with conventional cytotoxic agents such as oxaliplatin via tumor-specific nanoscale coordination polymers on colorectal liver metastases [17]. In our study, we found miR-655-3p was found 100-fold less expressed in the patient group (fold change: 0.01, p=0.026). Our result shows similarity with other cancer types and miR-655-3p may be downregulated via methylation. However, we did not evaluate methylation status.
The miR-127 is known to target the proto-oncogene B-cell CLL/lymphoma 6 (BCL6) [18]. It was shown that downregulation of miR-127 in rat liver cells and overexpression of miR-127 directly inhibits BCL6 expression and suppresses cell growth [19]. miR-127-5p was found downregulated in more than half of hepatocellular carcinoma samples. Moreover, researchers showed that miR-127-5p suppresses nuclear factor-κB activity by directly targeting biliverdin reductase B [20]. Downregulation of miR-127-5p was also found associated with colon cancer [21]. The expression levels of miR-127-5p were found about 48-fold less expressed in the gastric cancer in our study (fold change: 0.021, p<0.001). Pronina et al. found an association between hypermethylation of miR-127, breast cancer progression and metastasis [22]. We also found miR-127-5p expression levels were lower in the presence of metastases (p<0.001).
Low expressions of miR-369-3p were found associated with shorter overall survival in thyroid cancer patients and overexpression of miR-369-3p promoted apoptosis and suppressed proliferation in papillary thyroid cancer [23]. Thayanithy et al. reported that miR-369-3p in 14q32 locus may contributes to osteosarcoma pathogenesis [12]. The reprogramming of miR-369-3p or -5p inhibited cell proliferation and invasion in colon cancer cells [24]. Another study showed that glycolysis and tumor growth is suppressed by miR-369-3p through inhibition of lactate dehydrogenase A. The expression levels of miR-369-3p were found lower in primary colorectal cancer specimens [25]. We observed miR-369-3p approximately 1.6-fold less expressed in the gastric cancer group to the control group (fold change: 0.63, p=0.54). Moreover, the correlation was found between metastasis and miR-369-5p expression (p<0.001) and miR-369-5p expression levels were lower in the presence of metastases.
Liu et al. reported that upregulation of miR-544a in lung cancer tissues and cells and proliferation and invasion is facilitated by miR-544a [26]. Upregulated miR-544a expression was found in metastatic tumor samples and colorectal cancer cell lines. Furthermore, researchers suggested that metastatic and invasive properties of colorectal cancer are regulated by miR-544a via modulating Homeobox A10 [27]. The expression levels of cadherin 1 were found negatively correlate with miR-544a and miR-544a expression was higher in breast cancer MDA-MB-231 cells compared to the normal breast Hs578Bst cells [28]. It was demonstrated that the expression of e-cadherin is reduced via overexpression of miR-544a and resulting epithelial-mesenchymal transition (EMT) in gastric cancer [29]. It was found that miR-544a was expressed about 15.5-fold more in gastric cancer group (fold change: 15.47, p<0.001).
Conclusion
Our study demonstrated that miR-655-3p, miR-127-5p and miR-544a might be assessed as biomarker in gastric cell. However, these microRNAs expressions should be evaluated at methylation levels in both of cell lines and human tissues. Furthermore, miR-655-3p and miR-127-5p might be introduced to the gastric cancer cell lines to show their therapeutic effects because of the downregulated in gastric cancer. On the contrary, miR-544a can be silenced in gastric cancer cell lines to observe the effects on carcinogenic profile.
Acknowledgements
This study was supported by Istanbul University Scientific Research Project Unit (Project No: 20515).
Conflict of interest: The authors declare that there is no conflict of interests regarding the publication of this article.
References
1. Polat FR, Duran Y. Gastric cancer and the importance of early diagnosis. Namık Kemal Med J 2018;6:32–5.Search in Google Scholar
2. Zhang ZZ, Wang CJ, Niu L, Xu J, Wang M, Cao H, et al. Analysis of plasma MicroRNAs to identifying early diagnostic molecule for gastric cancer. Int J Clin Exp Med 2015;8:3700–6.Search in Google Scholar
3. Massarrat S, Stolte M. Development of gastric cancer and its prevention. Arch Iran Med 2014;17:514–20.Search in Google Scholar
4. Xie K, Huang S. Regulation of cancer metastasis by stress pathways. Clin Exp Metastasis 2003;20:31–43.10.1023/A:1022590402748Search in Google Scholar
5. Vlasova-St Louis I, Bohjanen PR. Post-transcriptional regulation of cytokine and growth factor signaling in cancer. Cytokine Growth Factor Rev 2017;33:83–93.10.1016/j.cytogfr.2016.11.004Search in Google Scholar
6. Zhang Y, Liu J, Lin J, Zhou L, Song Y, Wei B, et al. The transcription factor GATA1 and the histone methyltransferase SET7 interact to promote VEGF-mediated angiogenesis and tumor growth and predict clinical outcome of breast cancer. Oncotarget 2016;7:9859–75.10.18632/oncotarget.7126Search in Google Scholar
7. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522–31.10.1038/nrg1379Search in Google Scholar
8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97.10.1016/S0092-8674(04)00045-5Search in Google Scholar
9. Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008;27:5959–74.10.1038/onc.2008.274Search in Google Scholar PubMed
10. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105.10.1101/gr.082701.108Search in Google Scholar PubMed PubMed Central
11. Uppal A, Wightman SC, Mallon S, Oshima G, Pitroda SP, Zhang Q, et al. 14q32-encoded microRNAs mediate an oligometastatic phenotype. Oncotarget 2015;6:3540–52.10.18632/oncotarget.2920Search in Google Scholar PubMed PubMed Central
12. Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, et al. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone 2012;50:171–81.10.1016/j.bone.2011.10.012Search in Google Scholar PubMed PubMed Central
13. Wang Y, Zang W, Du Y, Ma Y, Li M, Li P, et al. Mir-655 up-regulation suppresses cell invasion by targeting pituitary tumortransforming gene-1 in esophageal squamous cell carcinoma. J Transl Med 2013;11:301.10.1186/1479-5876-11-301Search in Google Scholar PubMed PubMed Central
14. Han MS, Lee JM, Kim SN, Kim JH, Kim HS. Human papillomavirus 16 oncoproteins downregulate the expression of miR-148a-3p, miR-190a-5p, and miR-199b-5p in cervical cancer. Biomed Res Int 2018;2018:1942867.10.1155/2018/1942867Search in Google Scholar PubMed PubMed Central
15. Zhao XQ, Liang B, Jiang K, Zhang HY. Down-regulation of miR-655-3p predicts worse clinical outcome in patients suffering from hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2017;21:748–52.Search in Google Scholar
16. Wu G, Zheng K, Xia S, Wang Y, Meng X, Qin X, et al. MicroRNA-655-3p functions as a tumor suppressor by regulating ADAM10 and β-catenin pathway in hepatocellular carcinoma. J Exp Clin Cancer Res 2016;35:89.10.1186/s13046-016-0368-1Search in Google Scholar PubMed PubMed Central
17. Oshima G, Guo N, He C, Stack ME, Poon C, Uppal A, et al. In vivo delivery and therapeutic effects of a microRNA on colorectal liver metastases. Mol Ther 2017;25:1588–95.10.1016/j.ymthe.2017.04.005Search in Google Scholar PubMed PubMed Central
18. Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006;9:435–43.10.1016/j.ccr.2006.04.020Search in Google Scholar PubMed
19. Pan C, Chen H, Wang L, Yang S, Fu H, Zheng Y, et al. Down-regulation of MiR-127 facilitates hepatocyte proliferation during rat liver regeneration. PLoS One 2012;7:e39151.10.1371/journal.pone.0039151Search in Google Scholar PubMed PubMed Central
20. Huan L, Bao C, Chen D, Li Y, Lian J, Ding J, et al. MicroRNA-127-5p targets the biliverdin reductase B/nuclear factor-κB pathway to suppress cell growth in hepatocellular carcinoma cells. Cancer Sci 2016;107:258–66.10.1111/cas.12869Search in Google Scholar PubMed PubMed Central
21. Pathak S, Meng WJ, Nandy SK, Ping J, Bisgin A, Helmfors L, et al. Radiation and SN38 treatments modulate the expression of microRNAs, cytokines and chemokines in colon cancer cells in a p53-directed manner. Oncotarget 2015;6: 44758–80.10.18632/oncotarget.5815Search in Google Scholar PubMed PubMed Central
22. Pronina IV, Loginov VI, Burdennyy AM, Fridman MV, Senchenko VN, Kazubskaya TP, et al. DNA methylation contributes to deregulation of 12 cancer-associated microRNAs and breast cancer progression. Gene 2017;604:1–8.10.1016/j.gene.2016.12.018Search in Google Scholar PubMed
23. Li P, Dong M, Wang Z. Downregulation of TSPAN13 by miR-369-3p inhibits cell proliferation in papillary thyroid cancer (PTC). Bosn J Basic Med Sci 2019;19:146–54.10.17305/bjbms.2018.2865Search in Google Scholar PubMed PubMed Central
24. Ogawa H, Wu X, Kawamoto K, Nishida N, Konno M, Koseki J, et al. MicroRNAs induce epigenetic reprogramming and suppress malignant phenotypes of human colon cancer cells. PLoS One 2015;10:e0127119.10.1371/journal.pone.0127119Search in Google Scholar PubMed PubMed Central
25. Wang J, Wang H, Liu A, Fang C, Hao J, Wang Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 2015;6:19456–68.10.18632/oncotarget.3318Search in Google Scholar PubMed PubMed Central
26. Liu X, Ma J, Xu F, Li L. TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. Biomed Pharmacother 2018;99:9–17.10.1016/j.biopha.2018.01.049Search in Google Scholar PubMed
27. Sun S, Su C, Zhu Y, Li H, Liu N, Xu T, et al. MicroRNA-544a regulates migration and invasion in colorectal cancer cells via regulation of Homeobox A10. Dig Dis Sci 2016;61: 2535–44.10.1007/s10620-016-4186-2Search in Google Scholar PubMed
28. Lu P, Gu Y, Li L, Wang F, Qiu X. miR-544a promotes breast cancer cell migration and invasion reducing cadherin 1 expression. Oncol Res 2016;23:165–70.10.3727/096504016X14519157902726Search in Google Scholar PubMed PubMed Central
29. Yanaka Y, Muramatsu T, Uetake H, Kozaki K, Inazawa J. miR-544a induces epithelial-mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis 2015;36:1363–71.10.1093/carcin/bgv106Search in Google Scholar PubMed
©2019 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Research Articles
- Investigating the impact of polysomy 17 in breast cancer patients with HER2 amplification through meta-analysis
- Diagnostic performance of microRNAs in the circulation in differential diagnosis of BPH, chronic prostatitis and prostate cancer
- Enhanced anticancer effect of cetuximab combined with stabilized silver ion solution in EGFR-positive lung cancer cells
- CA125, YKL-40, HE-4 and Mesothelin: a new serum biomarker combination in discrimination of benign and malign epithelial ovarian tumor
- Paricalcitol pretreatment attenuates renal ischemia/reperfusion injury by inhibiting p38 MAPK and activating PI3K/Akt signaling pathways
- Identification of cytoplasmic sialidase NEU2-associated proteins by LC-MS/MS
- Investigation of tyrosinase inhibition by some 1,2,4 triazole derivative compounds: in vitro and in silico mechanisms
- Investigation of alanine, propionylcarnitine (C3) and 3-hydroxyisovalerylcarnitine (C5-OH) levels in patients with partial biotinidase deficiency
- The expression levels of miR-655-3p, miR127-5p, miR-369-3p, miR-544a in gastric cancer
- Evaluation of the JAK2 V617F gene mutation in myeloproliferative neoplasms cases: a one-center study from Eastern Anatolia
- Effects of Rituximab on JAK-STAT and NF-κB signaling pathways in acute lymphoblastic leukemia and chronic lymphocytic leukemia
- Analysis of the effect of DEK overexpression on the survival and proliferation of bone marrow stromal cells
- Serum fetuin-A levels and association with hematological parameters in chronic kidney disease and hemodialysis patients
- Investigation of relaxation times in 5-fluorouracil and human serum albumin mixtures
- Oxydative stress markers and cytokine levels in rosuvastatin-medicated hypercholesterolemia patients
- The protective effects of urapidil on lung tissue after intestinal ischemia-reperfusion injury
- Effects of SR-BI rs5888 and rs4238001 variations on hypertension
- Antioxidant and cytotoxic activity of three Turkish marine-derived fungi
- Is spectrophotometric enzymatic method a cost-effective alternative to indirect Ion Selective Electrode based method to measure electrolytes in small clinical laboratories?
- Plasma presepsin in determining gastric leaks following bariatric surgery
Articles in the same Issue
- Frontmatter
- Research Articles
- Investigating the impact of polysomy 17 in breast cancer patients with HER2 amplification through meta-analysis
- Diagnostic performance of microRNAs in the circulation in differential diagnosis of BPH, chronic prostatitis and prostate cancer
- Enhanced anticancer effect of cetuximab combined with stabilized silver ion solution in EGFR-positive lung cancer cells
- CA125, YKL-40, HE-4 and Mesothelin: a new serum biomarker combination in discrimination of benign and malign epithelial ovarian tumor
- Paricalcitol pretreatment attenuates renal ischemia/reperfusion injury by inhibiting p38 MAPK and activating PI3K/Akt signaling pathways
- Identification of cytoplasmic sialidase NEU2-associated proteins by LC-MS/MS
- Investigation of tyrosinase inhibition by some 1,2,4 triazole derivative compounds: in vitro and in silico mechanisms
- Investigation of alanine, propionylcarnitine (C3) and 3-hydroxyisovalerylcarnitine (C5-OH) levels in patients with partial biotinidase deficiency
- The expression levels of miR-655-3p, miR127-5p, miR-369-3p, miR-544a in gastric cancer
- Evaluation of the JAK2 V617F gene mutation in myeloproliferative neoplasms cases: a one-center study from Eastern Anatolia
- Effects of Rituximab on JAK-STAT and NF-κB signaling pathways in acute lymphoblastic leukemia and chronic lymphocytic leukemia
- Analysis of the effect of DEK overexpression on the survival and proliferation of bone marrow stromal cells
- Serum fetuin-A levels and association with hematological parameters in chronic kidney disease and hemodialysis patients
- Investigation of relaxation times in 5-fluorouracil and human serum albumin mixtures
- Oxydative stress markers and cytokine levels in rosuvastatin-medicated hypercholesterolemia patients
- The protective effects of urapidil on lung tissue after intestinal ischemia-reperfusion injury
- Effects of SR-BI rs5888 and rs4238001 variations on hypertension
- Antioxidant and cytotoxic activity of three Turkish marine-derived fungi
- Is spectrophotometric enzymatic method a cost-effective alternative to indirect Ion Selective Electrode based method to measure electrolytes in small clinical laboratories?
- Plasma presepsin in determining gastric leaks following bariatric surgery

